Abstract
TAR DNA-binding protein 43 (TDP-43) has been identified as the major constituent of the proteinaceous inclusions that are characteristic of most forms of amyotrophic lateral sclerosis (ALS) and ubiquitin positive frontotemporal lobar degeneration (FTLD). Wild type TDP-43 inclusions are a pathological hallmark of >95% of patients with sporadic ALS and of the majority of familial ALS cases, and they are also found in a significant proportion of FTLD cases. ALS is the most common form of motor neuron disease, characterized by progressive weakness and muscular wasting, and typically leads to death within a few years of diagnosis. To determine how the translocation and misfolding of TDP-43 contribute to ALS pathogenicity, it is crucial to define the dynamic behavior of this protein within the cellular environment. It is therefore necessary to develop cell models that allow the location of the protein to be defined. We report the use of TDP-43 with a tetracysteine tag for visualization using fluorogenic biarsenical compounds and show that this model displays features of ALS observed in other cell models. We also demonstrate that this labeling procedure enables live-cell imaging of the translocation of the protein from the nucleus into the cytosol.
Amyotrophic lateral sclerosis (ALS) is a progressive, and ultimately fatal, neurodegenerative disorder that primarily affects the upper and lower motor neurons of the central nervous system (CNS). Symptoms of the disease are normally first observed at a focal site of onset and then gradually spread to contiguous regions of the nervous system over time. As is the case with a number of other neurodegenerative diseases, this process is accompanied by the deposition of insoluble inclusions of aggregated protein in the cytoplasm of affected cell types that acts as a pathological signature for the condition. A major component of these inclusions has been identified as hyperphosphorylated and ubiquitinated pathological forms of both full-length and proteolytic cleavage fragments of TDP-43.1−3 Interestingly, scores of additional proteins are found co-deposited in spinal motor neurons with various forms of TDP-43.4
Under physiological conditions, TDP-43 is a predominantly nuclear protein and, among other functions, is involved in mRNA regulation and splicing.1−3,5 In ALS, however, it has been found to mislocalize to the cytoplasm where it forms misfolded aggregates. The formation of preinclusions that do not associate with ubiquitin6 coincides with the movement of TDP-43 from the nucleus, and it is considered to be an early aggregated species that precedes the accumulation of mature, ubiquitin-associated inclusions.6,7 There is great debate surrounding the relative contributions to pathogenesis of the loss of functional TDP-43 from the nucleus [loss of function (LOF)] and of the accumulation of aggregated species with toxic properties in the cytoplasm [gain of function (GOF)], although recent evidence suggests that both mechanisms can contribute to the disease.8 Indeed, cytoplasmic translocation and aggregation of TDP-43 are directly associated with cell death, suggesting that the study of the initial stages of these processes could significantly improve our understanding of how TDP-43 contributes to the molecular pathology of the disease.
Fluorescent labels have been employed in many live-cell models to allow the observation of the intracellular distribution of proteins. The tetracysteine (TC) motif and biarsenical dye system makes use of a derivative of fluorescein, called fluorescein arsenical hairpin binder (FlAsH). This dye binds to a short amino acid sequence with the TC motif, having the general structure Cys-Cys-Xaa-Xaa-Cys-Cys (CCXXCC, in which X denotes any amino acid).9 Both FlAsH and a red-shifted variant, ReAsH, denoting resorufin arsenical hairpin binder, are commercially available and well-characterized fluorescent dyes. These reagents are not fluorescent in their unbound states, but interaction with the TC motif results in a large increase in their fluorescence quantum yields. Previous studies to optimize the flanking regions of the TC peptide sequence resulted in two 12-amino acid motifs, FLNCCPGCCMEP and HRWCCPGCCKTF, with high capacities to accommodate the FlAsH and ReAsH dyes.10 Such optimized TC tags can be genetically inserted into the target protein, allowing the FlAsH or ReAsH dye to bind with high specificity. A significant advantage of the TC tag compared to the use of fluorescent proteins is that its small size reduces the likelihood of it significantly affecting the properties of the protein of interest.11 This system lends itself to the visualization of the target protein inside live cells because of the relatively low cytotoxicity of the dye.12 Furthermore, it has been shown that a variety of proteins in common mammalian cell lines, primary cortical neurons, and also Gram-negative bacteria can be labeled with FlAsH and ReAsH.13 Both reagents have been used successfully to report on the conformation of proteins expressed in cultured cells.14−16 In the context of the study presented here, we note that this labeling strategy has been used in cell models of several neurodegenerative diseases to investigate the conformational states of aberrantly misfolded proteins in a cellular environment.17,18 For example, the incorporation of a TC tag into a variant of huntingtin (Httex1) has been used to probe the aggregation state of the protein in Neuro2A cells, in which the conformational properties of the monomeric form but not the aggregated forms enable the TC tag to bind to ReAsH.17 In addition, transfected SH-SY5Y cells have been used to overexpress TC-labeled α-synuclein, allowing the dynamics and structural properties of the aggregates to be studied using in situ microscopy techniques, including fluorescence recovery after photobleaching (FRAP) and confocal fluorescence anisotropy.18
We discuss in this paper the development of a live-cell model of ALS using transiently transfected SH-SY5Y cells overexpressing TC-tagged wild type (WT) TDP-43 (HA-TDP43-TC) that allows direct visualization of the protein in living cells using the biarsenical dye FlAsH. We demonstrate that the TC tag does not detectably alter the behavior of TDP-43 in this cell model by comparing its behavior to that of cells overexpressing HA-TDP-43 (HA-TDP43), a human influenza hemeagglutinin (HA) epitope-tagged TDP-43 that is a well-characterized system used previously to investigate the role of TDP-43 in ALS.19,20 We show here that the HA-TDP43-TC model recapitulates key biochemical features of TDP-43 proteinopathies, such as association with stress granule markers and phosphorylation.19,21,22 Furthermore, we show it is possible to monitor noninvasively, and with spatiotemporal resolution, the cytoplasmic accumulation of TDP-43 concomitantly with its nuclear clearance over the course of 72 h post-transfection. Finally, we demonstrate that this model is amenable to time-lapse confocal microscopy and observe FlAsH-labeled TDP-43 being transferred from the nucleus into the cytoplasm in real time.
Materials and Methods
Construction of Plasmids
To generate the HA- and TC-tagged WT TDP-43 (HA-TDP43 and HA-TDP43-TC, respectively) sequences, the cDNA encoding TDP-43 was amplified from the pCMV.SPORT6.1_TDP-43 plasmid (Source BioScience, Nottingham, U.K.) by polymerase chain reaction using Phusion High-fidelity DNA polymerase (Thermo Fisher Scientific, Loughborough, U.K.) and primers that either incorporated the DNA sequences of the HA tag and the TC tag [TDP43-TC (forward) and TDP43-TC (reverse) primers] or incorporated only the HA tag [HA-TDP43 (forward) and HA-TDP43 (reverse)]. Sequences of the primers are listed in Table S1, and the DNA sequences encoding HA-TDP43 and HA-TDP43-TC are shown in Figure S1. In both cases, the primers introduced unique BamHI and XbaI sites at the 5′ and 3′ ends of the coding sequences, respectively. These sites were used to clone the genes of interest into the pcDNA3.1(+) vector (Thermo Fisher Scientific). The constructs were transformed using standard heat-shock protocols into chemically competent DH5α Escherichia coli (Thermo Fisher Scientific) and plated on LB-agar plates containing ampicillin (100 μg/mL). DNA was isolated using a Qiaprep Spin mini-prep or maxi-prep kit (Qiagen, Manchester, U.K.) according to the manufacturer’s instructions. Purified DNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific), and constructs were confirmed by DNA sequencing (Department of Biochemistry, University of Cambridge, U.K.).
Cell Culture and Transfection
The human neuroblastoma SH-SY5Y cell line was routinely cultured in complete growth medium [DMEM/F12 supplemented with 10% (v/v) fetal bovine serum (FBS)] in a humidified chamber at 37 °C in 95% air and 5% CO2. Cells were either chemically transfected using Lipofectamine 2000 (Thermo Fisher Scientific) or electroporated using the Neon system (Thermo Fisher Scientific).
Lipofection
Cells were plated 1 day before transfection in 35 mm glass bottom dishes (Ibidi, ThistleScientific, Glasgow, U.K.) such that the culture was at 70–90% confluence on the day of transfection. Transfections were carried out using Lipofectamine 2000 according to the manufacturer’s instructions in serum free medium (DMEM/F12 without FBS). Briefly, the plasmid/Lipofectamine 2000 complex was prepared at a 1:4 ratio (μg:μL). Lipofectamine 2000 was incubated [room temperature (RT), 5 min] in half the total volume of medium before being mixed with the medium containing the plasmid DNA, followed by further incubation (RT, 20 min). The complete medium was aspirated from the cultured cells and replaced with the DNA/Lipofectamine complex in serum free medium. The cells were incubated in the DNA/Lipofectamine-containing medium (37 °C, 5 h) that was then replaced with complete medium, and the cells were incubated overnight at 37 °C before being used for further experiments.
Electroporation
Two days prior to electroporation, cells were seeded into T-75 flasks with complete growth medium such that the cells were 70–90% confluent on the day of the experiment. The cells were then detached with 0.25% Trypsin-EDTA (3 mL, Thermo Fisher Scientific) and washed in phosphate-buffered saline (pH 7.4) (PBS). The cells were resuspended in the resuspension buffer provided and electroporated (1100 V, 50 ms, once) according to the manufacturer’s protocol (Neon Transfection System, Thermo Fisher Scientific). After electroporation, the cells were seeded in complete growth medium in six-well or 96-well plates (Corning, Appleton Woods Ltd., Birmingham, U.K.) or in 35 mm glass bottom dishes (Ibidi, Thistle Scientific) and left for at least 24 h before being used in experiments.
Immunocytochemistry
SH-SY5Y cells overexpressing TDP-43 were grown in 35 mm glass bottom dishes. To monitor the effects over time, immunocytochemistry was performed with cells 24, 48, and 72 h post-transfection. The cells were washed with chilled PBS and fixed with chilled paraformaldehyde (3.7% in PBS, 15 min). After 15 min, the cells were washed twice with chilled PBS (10 min) and permeabilized by being incubated in chilled PBS-T [0.3% (v/v) Triton-X in PBS, 30 min]. After 30 min, the cells were blocked with BSA [5% (w/v) in PBS-T, 30 min] and washed twice with chilled PBS (5 min). The cells were incubated with the primary antibody (1:1000) (RT for 1 h or 4 °C overnight) while being gently rocked. For co-localization experiments, cells were co-incubated with the primary antibodies anti-HA 3F10 (Sigma-Aldrich UK Ltd., Gillingham U.K.) and either phospho (403/404) TDP-43 (ProteinTech, Manchester, U.K.) or phospo (409/410) TDP43 (ProteinTech) or TIA-1 C-20 (Santa Cruz Biotechnology, Heidelberg, Germany). This was followed by incubation with an appropriate secondary Alexa Fluor antibody (1:1000, Life Technologies, Paisley, U.K.) (RT for 1 h or 4 °C overnight). Wheat germ agglutinin (WGA) Alexa Fluor 647 conjugate (Life Technologies) treatment was performed prior to permeabilization by treating the cells with a 1:500 dilution of WGA conjugate in Hank’s buffered salt solution (HBSS; 10 min, RT). Cells were washed twice in HBSS, and permeabilization was performed with subsequent labeling. Cells were washed twice with chilled PBS after incubation with each of the primary and secondary antibodies. After being washed, the cells were incubated with the nuclear stain Hoechst (1 μg/mL, Life Technologies) for 5 min, and the labeled cells were mounted with Ibidi mounting medium (Ibidi, Thistle Scientific). Imaging of the samples was performed on a Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany) at the Cambridge Advanced Imaging Centre (CAIC) at the University of Cambridge.
In-Cell Biarsenical Dye Labeling
For the labeling of TC-tagged TDP-43 (HA-TDP43-TC) overexpressed in SH-SY5Y cells, the TC-FlAsH II In-Cell Tetracysteine Tag Detection Kit from Molecular Probes (Thermo Fisher Scientific) was used. Transfected SH-SY5Y cells were grown in 35 mm glass bottom dishes. To monitor the effects over time, in-cell FlAsH labeling was performed 24, 48, and 72 h post-transfection. The cells were washed twice with reduced serum Opti-MEM without phenol red (Life Technologies) and incubated with FlAsH (1 μM) in Opti-MEM (37 °C, 30 min, protected from light). After 30 min, the cells were washed twice with 1× BAL (2,3-dimercaptopropanol) buffer in Opti-MEM (37 °C, 1 h, protected from light). For live-cell imaging at each time point, cells were incubated with Hoechst stain (1 μg/mL) in Opti-MEM for 5 min after the BAL buffer wash steps. Finally, the cells were washed with Opti-MEM, cultured in Opti-MEM, and prepared for live-cell imaging.
Quantification of Transfection Efficiency
Cells were transfected with either HA-TDP43 or HA-TDP43-TC using Lipofectamine 2000. Twenty-four hours post-transfection, the cells were fixed and immunostained with an anti-HA antibody and an Alexa Fluor 488 secondary antibody, to detect positively transfected cells, and Hoechst nuclear counterstain, to visualize the total number of cells. The samples were imaged using a Leica TCS SP5 confocal microscope equipped with 40× HPC Fluo Tar and 63× APO oil immersion objectives, and a UV diode and 488 nm argon laser line to visualize the Hoechst and Alexa Fluor 488 fluorescence, respectively (Leica Microsystems, Milton Keynes, U.K.). Three images were obtained at 40× magnification in different regions of the dish for each sample. Subsequent quantification was performed using ImageJ, using the Cell Counter plugin to count manually all cells stained with the anti-HA antibody or Hoechst stain. The number of cells positive for both anti-HA and Hoechst was calculated as a percentage of the total number of Hoechst positive cells.
Time-Lapse Imaging
Cells transfected with HA-TDP43-TC in 35 mm glass dishes were labeled with the Molecular Probes FlAsH-EDT2 dye from Invitrogen Life Technologies, 24–36 h post-transfection. Cells were washed in Opti-MEM (RT, 2×) followed by incubation in FlAsH-EDT2 (1 μM in Opti-MEM) (RT, 30 min, protected from light). The cells were washed twice in 1× BAL buffer (in Opti-MEM) (37 °C, 1 h, protected from light) and imaged in Opti-MEM. The chamber slide was placed in a CO2-UNIT-BL Stage top heated CO2 chamber (37 °C, no CO2 perfusion) 30 min prior to imaging, using confocal microscopy (Leica Microsystems). The built-in software (LAS AF, Leica Microsystems) was used for time-lapse imaging using the “Best Focus” function at different locations within the dish using the “Mark and Find” function (overnight, 15 min intervals).
Quantification of the Depletion of Nuclear Fluorescence from Time-Lapse Images
Each frame of the time-lapse video was analyzed as an individual tiff file in ImageJ, and analysis was performed in the green channel of each image. A region of interest was drawn around the nucleus of the target cell exhibiting fluorescence using the freeform tool. “Set measurements” was selected from the Analyze menu, and “area”, “integrated density”, and “mean gray value” were ticked. “Measure” was selected from the “Analyze” menu to obtain values. The process was repeated for an area of the image without fluorescence to measure the background signal and was performed for all frames up to 11 h of imaging, after which there was no difference between the nuclear fluorescence and the background signal. The results were copied and pasted into Microsoft Excel for further analysis. Equation 1 was used to obtain a value for the corrected fluorescence at each time point.
| 1 |
Comparison of the Cellular Location of TDP-43 in HA-TDP43 and HA-TDP43-TC following Electroporation Treatment
Confocal images acquired from cells expressing HA-TDP43 and HA-TDP43-TC after labeling with an anti-HA antibody were used to determine the location of fluorescence signals by dividing into three categories: (1) predominantly nuclear localization, (2) localization in both cytoplasm and nucleus, and (3) predominantly cytoplasmic localization. Fifty cells were randomly counted from different sample preparations, and the percentage was calculated for each category.
Immunoprecipitation Experiments
Transfected cells were rinsed with PBS (10 mL) and detached using a trypsin-EDTA solution followed by the addition of complete growth medium to neutralize the trypsin. The cells were pelleted by centrifugation (4 °C, 5 min, 1500 rpm), and the supernatant was removed. The cell pellet was washed with chilled PBS and further centrifuged (4 °C, 5 min, 1500 rpm). The supernatant was removed, and the cell pellet was lysed with RIPA buffer [1 mL, 50 mM Tris-HCl (pH 8.0), 150 mM sodium chloride, 1.0% Igepal CA-630 (NP-40), 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate] supplemented with EDTA free protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) for 10 min on ice, followed by centrifugation (4 °C, 10 min, 10000 rpm), after which the supernatant was retained for immunoprecipitation. Pierce Anti-HA Magnetic Beads (Thermo Fisher Scientific, 25 μL) were washed twice with TBS-T [Tris-buffered saline, 0.05% (v/v) Tween 20, 300 μL], followed by a final wash with doubly distilled H2O, with brief vortexing between washes. The cell lysate (1 mL) was added to a 1.5 mL microfuge tube followed by addition of the prewashed magnetic beads. The sample was then mixed and incubated (RT, 30 min, 200 rpm) with constant rotation. After incubation, the tube was placed into a DynaMag-Spin Magnet stand (Thermo Fisher Scientific) and the supernatant containing the unbound protein was collected and saved for analysis. To elute the HA-tagged TDP-43 bound to the magnetic beads, Pierce HA Peptide (Thermo Fisher Scientific) (100 μL of a 2 mg/mL solution) was added to the bead slurry, and this was then vortexed and incubated (37 °C, 10 min). The eluted sample was then analyzed by Western blotting.
Western Blotting
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was used to separate protein samples prior to Western blotting. The protein sample (20 μL) was prepared in NuPAGE LDS Sample Buffer (4×) (LifeTechnologies) and NuPAGE Sample Reducing Agent (10×) (Life Technologies) and run on a NuPAGE 4 to 12% gradient Bis-Tris (Life Technologies) gel with MES running buffer (Life Technologies) (200 V, 35 min). The proteins were transferred from the gel to an iBlot Transfer Stack with a polyvinylidene fluoride (PVDF) membrane (0.2 μm pore size) using the iBlot Dry Blotting System (Life Technologies) according to the manufacturer’s protocol. After the transfer, the PVDF membrane was blocked with 5% (w/v) dry skimmed milk powder in PBS with 0.05% Tween 20 (blocking buffer) (RT, 1 h, gentle agitation). The membrane was then incubated with primary antibodies in blocking buffer (1:1000, 4 °C overnight or RT for 1 h) followed by washing four times with PBS-T (RT, 10 min). After being washed, the membrane was incubated with Alexa Fluor 488- or 594-labeled secondary antibody (1:1000, RT, 1 h). After incubation, the membrane was washed four times with PBS-T and imaged using a Typhoon 9400 laser-based scanner (GE Healthcare) at 550 V using a green (532 nm) excitation laser to excite Alexa Fluor 594 or a blue (488 nm) laser to excite Alexa Fluor 488.
Results
Transient Transfection of SH-SY5Y Cells To Overexpress Full-Length TDP-43 Containing a C-Terminal Tetracysteine Tag (TC)
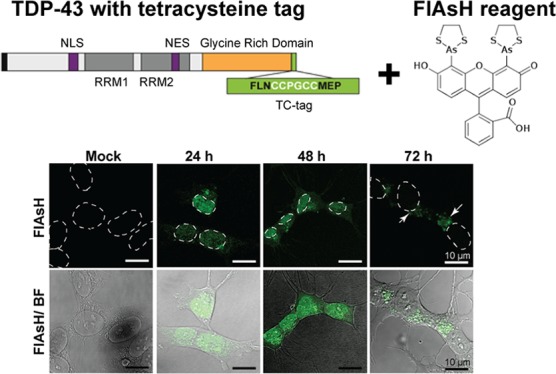
A number of TDP-43 cell models use epitope tags or fluorescent fusion proteins to distinguish between overexpressed TDP-43 and the endogenous protein.7,19,23−25 For our model, we added the 12-amino acid TC tag (FLNCCPGCCMEP)10 to the C-terminus of full-length TDP-43 (Figure 1, i). It has been established that in disease states such as ALS, C-terminally truncated fragments of TDP-43 are found in the inclusions,3,26 so we reasoned that the addition of the TC tag to the C-terminus of our full-length TDP-43 construct will allow us to visualize both the full-length and truncated fragments of TDP-43. We also incorporated an HA epitope tag at the N-terminus of the protein (Figure 1, ii) in the same construct to allow an alternative means of identification using antibody detection.19,23,25,27,28 The HA-TDP43-TC construct was inserted into mammalian expression vector pcDNA3.1(+). Despite the tags introduced being relatively small (∼1 kDa), we checked that the addition of the C-terminal TC tag did not change the cellular behavior of the overexpressed WT TDP-43 by comparing our TC-tagged TDP-43 to an N-terminally HA-tagged full-length TDP-43 [HA-TDP43 (Figure 1, iii)] construct that has been previously shown to have diffuse nuclear localization in transiently transfected cells.19,20
Figure 1.
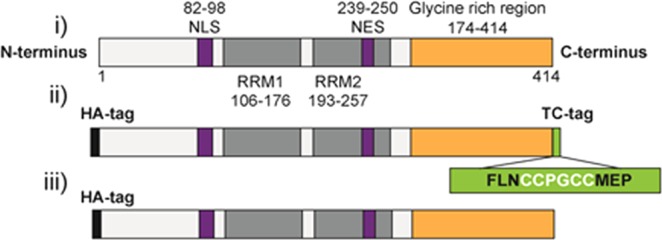
Schematic diagram illustrating (i) full-length wild type TDP-43, (ii) HA-TDP43-TC, and (iii) HA-TDP43 constructs. The HA tag is colored black. The nuclear localization signal (NLS) and nuclear export signal (NES) are colored purple. The RNA recognition motifs are colored dark gray. The glycine rich region is colored orange. The TC tag is colored green.
The HA-TDP43 and HA-TDP43-TC constructs were transiently transfected into SH-SY5Y cells using two methods, lipofection (Lipofectamine 2000) and electroporation (Neon system), to determine which most efficiently delivers the HA-TDP43-TC plasmid into the cells. SH-SY5Y cells are a human neuroblastoma cell line that is well-established and extensively used in neurodegenerative disease models, including for studies involving TDP-43.18,19,25,29,30 We used this cell line for the study presented here as they are consistently amenable to transient transfection protocols and have a favorable nucleus:cytoplasm ratio, which allows clear microscopy imaging. Twenty-four hours post-transfection, the cells were fixed, permeabilized, and probed with an anti-HA primary antibody. For both HA-TDP43- and HA-TDP43-TC-expressing cells, HA-specific labeling was detected as predominantly diffuse nuclear fluorescence (Figure 2A). By analyzing the number of positively stained cells for the two different transfection methods, we observed 10% and 12% transfection efficiencies for the HA-TDP43 and HA-TDP43-TC plasmids, respectively, using the lipofection method and a 14% transfection efficiency for both plasmids with the electroporation method (Table S2). For the HA-TDP43-TC cell line, we could image the live cells 24 h post-transfection using the FlAsH dye (Figure 2B), finding that FlAsH-bound protein was predominantly localized to the nucleus. It is interesting to note that for the lipofected cells, the FlAsH-bound HA-TDP43-TC displayed a weaker signal intensity compared to that of electroporated cells, although this was not statistically significant (n = 1; two-tailed unpaired t test, p = 0.1404) (Table S3). We proceeded with the electroporation method in light of its greater efficiency, using it to characterize the intracellular distribution of HA-TDP43-TC over time. To confirm further the presence of the full-length protein within the cells 24 h post-transfection, the HA-tagged proteins were isolated using immunoprecipitation (anti-HA magnetic beads), and the resulting samples were analyzed by Western blotting (Figure 2C); similar results were observed for the 48 and 72 h post-transfected samples (data not shown).
Figure 2.
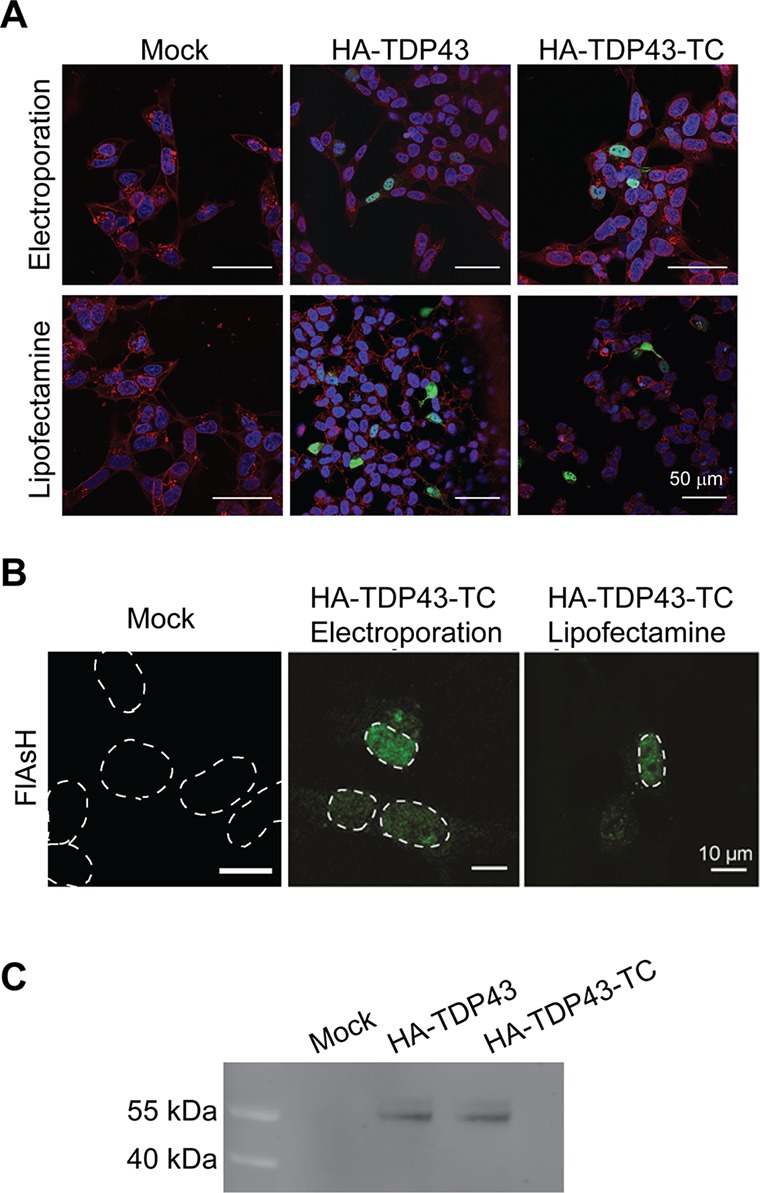
Confocal images of SH-SY5Y cells transfected (by either electroporation or lipofection) to overexpress HA-TDP43 or HA-TDP43-TC, 24 h post-transfection. (A) Immunofluorescence images generated using an anti-HA antibody and an Alexa Fluor 488 secondary antibody (green) and Hoechst nuclear stain (blue). Membranes are stained with wheat germ agglutinin (WGA) Alexa Fluor 647 conjugate (red), and scale bars are 50 μm. (B) Fluorescence images after the addition of the FlAsH dye (24 h post-transfection). For the sake of clarity, the white dotted line denotes the nucleus. Images are representative of multiple independent experiments. (C) Immunoprecipitation followed by Western blot analysis of HA-TDP43 and HA-TDP43-TC isolated from SH-SY5Y cell lysates 24 h post-transfection. Mock transfections are cells transfected with buffer alone.
Monitoring HA-TDP43-TC Expression at Different Time Points Post-Transfection with the FlAsH Dye
In previously reported cell models, WT TDP-43 is overexpressed and remains localized within the nucleus,19,31 whereas disease-related variants show translocation of the protein from the nucleus to the cytoplasm where they form inclusions when monitored up to 72 h post-transfection. When cells expressing WT TDP-43 are exposed to conditions of stress, via the addition of arsenite or through nutrient deprivation, this aberrant behavior is also observed.21,22 To determine whether FlAsH labeling was able to identify HA-TDP43-TC positively more than 72 h post-transfection, we compared live-cell images after the addition of the FlAsH reagent to HA-TDP43-TC-expressing cells 24, 48, and 72 h post-transfection. In Neon-transfected SH-SY5Y cells expressing HA-TDP43-TC, we observed diffuse nuclear staining 24 h post-transfection (Figure 3, 24 h). At 48 h post-transfection, however, FlAsH fluorescence was observed in both the nucleus and the cytoplasm (Figure 3, 48 h), and by 72 h, FlAsH-labeled HA-TDP43-TC was predominantly in the cytoplasm and appeared as inclusions of variable size (Figure 3, 72 h). The cells with strong staining of cytoplasmic inclusions showed a lack of nuclear fluorescence. No FlAsH fluorescence was observed at any time point (24–72 h) when the overexpressed protein lacked the TC tag (Figure S2). Translocation of the HA-TDP43-TC protein had clearly occurred spontaneously in this cell model despite earlier reports that in the absence of exogenous stress, WT TDP-43 remains in the nucleus.32−34 We next examined whether this translocation was due to the C-terminal TC tag perturbing the behavior of WT TDP-43 protein or if the mode of transfection had itself imposed an exogenous stress. To pursue this objective, we used the well-characterized HA-TDP43 cell model19 and compared results from time course experiments in which cells were transfected using either electroporation or lipofection.
Figure 3.
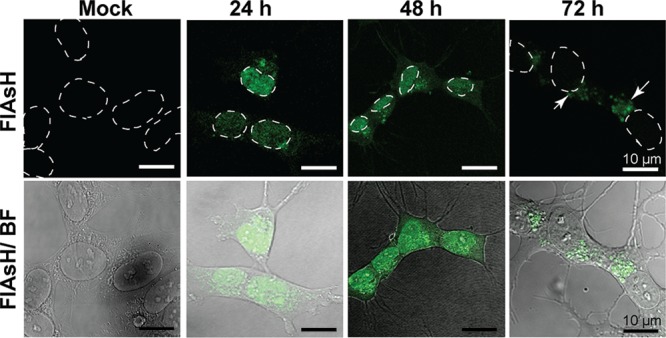
Confocal images of FlAsH-bound HA-TDP43-TC in live SH-SY5Y cells 24, 48, and 72 h post-transfection. FlAsH fluorescence images only (top) or merged with bright field (BF) images (bottom). The images are representative of multiple independent experiments. For the sake of clarity, the outlines of the nuclei are indicated by dashed white lines. The white arrowheads indicate punctate staining of HA-TDP43-TC. Mock transfection indicates cells transfected with buffer alone.
SH-SY5Y cells were transfected to express HA-TDP43 using electroporation, and as seen for HA-TDP43-TC, translocation of the protein was observed 48 and 72 h post-transfection (Figure 4A,B). Comparison of the number of cells containing nuclear or cytoplasmic TDP-43 (detected through HA tag labeling; n = 50 cells per condition) showed that the HA-TDP43 and HA-TDP43-TC models both displayed similar distributions (Figure 4C,D). When the cells underwent lipofection, however, much less translocation was observed in both cell models and 72 h post-transfection the nuclear TDP-43 staining remained prominent (Figure S3), in agreement with the results of previous studies.20,24,25,27,28 Interestingly, when the electroporation method was used to transiently transfect cells to overexpress EGFP-tagged TDP-43, we found that 48 and 72 h post-transfection, the TDP43-EGFP fluorescence was predominantly localized in the nucleus (Figure S4). This predominant nuclear localization by WT TDP43-EGFP 48 and 72 h post-transfection has been reported previously.35,36
Figure 4.
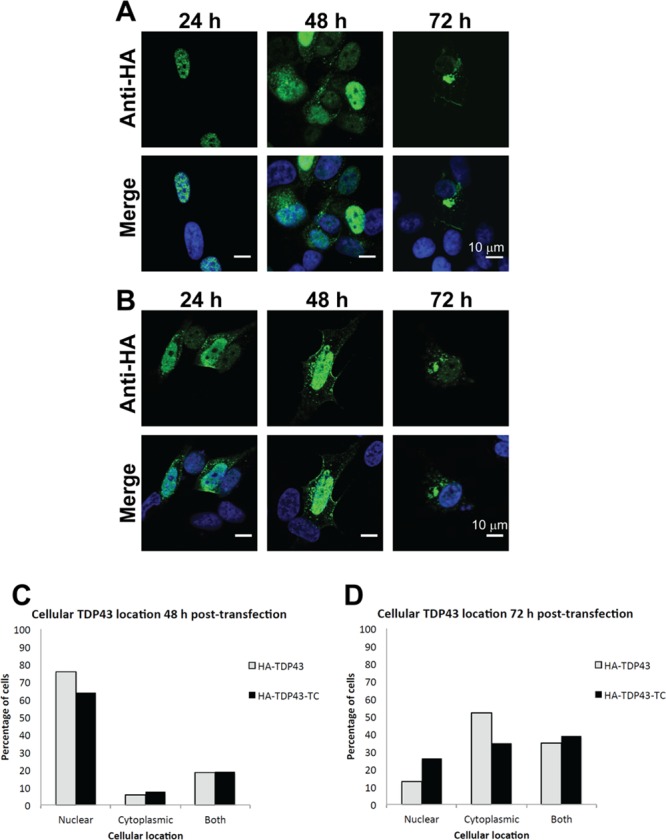
HA-TDP43 and HA-TDP43-TC expression and distribution in SH-SY5Y cells at 48 and 72 h following transfection by electroporation. Representative images of the cellular TDP-43 distribution for (A) HA-TDP43 and (B) HA-TDP43-TC. Scale bars are 10 μm. Merged immunofluorescence images generated using an anti-HA antibody (green) and Hoechst nuclear counterstain (blue). Histograms showing cellular location of HA-TDP43 and HA-TDP43-TC (C) 48 h and (D) 72 h post-transfection. Percentages were calculated by examining cells with positive HA tag labeling from eight independent imaging experiments until 50 cells for each condition (A and B) were identified.
HA-TDP43-TC Shows Co-Localization with the Stress Granule Marker TIA-1
To determine if the cytoplasmic inclusions observed in cells expressing HA-TDP43-TC provide a useful model of ALS pathogenesis, we next examined whether the protein had co-localized with a marker of stress granules (SGs), dynamic RNA-containing complexes that are formed in the cytoplasm when cells are subjected to stress.37 SGs direct translation toward proteins required for cell survival and repair but may under pathological conditions stabilize and act as scaffolds to promote recruitment and aggregation of a variety of proteins, including TDP-43.38,39 SGs in cell culture are formed within minutes of stress induction, normally degrade within 3 h following the cessation of stress, but persist in response to chronic stress.37 It has been reported that in a number of cell models, cytoplasmic TDP-43 aggregates are found to co-localize with stress granule markers such as TIA-1 in both cell culture models22,32,40 and immunocytochemical analyses of ALS and FTLD-TDP tissue samples.21,22,41 Chronic cellular stress may initiate pathological TDP-43 aggregation via its promotion of the assembly of SGs, thereby bringing together high concentrations of aggregation prone proteins and leading to cytoplasmic inclusion.42 At 48 and 72 h post-transfection, we observed that cytoplasmic inclusions containing HA-TDP43-TC co-localized with TIA-1 (Figure 5); similar co-localization was also observed in cells overexpressing HA-TDP43 (Figure 5).
Figure 5.
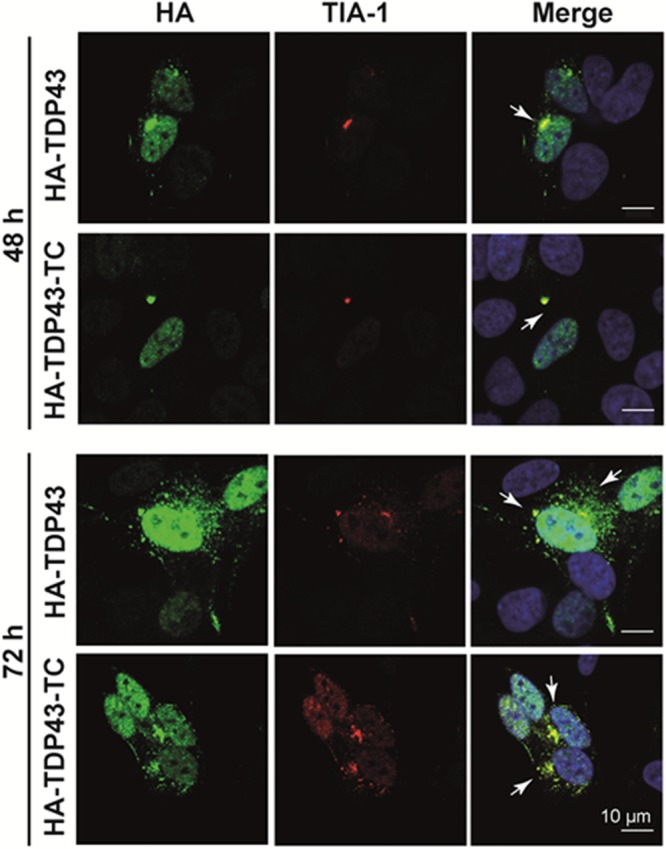
Comparison of the co-localization of TDP-43 and the SG marker TIA-1 between the HA-TDP43 and HA-TDP43-TC SH-SY5Y cell models 48 and 72 h post-transfection using electroporation. Merged immunofluorescence images generated using an anti-HA antibody (green) and Hoechst nuclear counterstain (blue) and anti-TIA-1 staining (red). Yellow denotes co-localization between HA-labeled TDP-43 and TIA-1 (white arrows).
In addition to co-localization with SG markers, phosphorylation of TDP-43 is commonly regarded as a hallmark feature of ALS. Using specific antibodies that recognize phosphorylated Ser403/404 or Ser409/410 epitopes, cytoplasmic inclusions containing phosphorylated TDP-43 have been detected in some cell models; this is not seen, however, in many cell lines expressing WT TDP-43.36,43 We probed transfected SH-SY5Y cells expressing HA-TDP43-TC with antibodies specific for TDP-43 phosphorylation at either Ser403/404 or Ser409/410. In SH-SY5Y cells transfected to express HA-TDP43 or HA-TDP43-TC, 48 and 72 h post-transfection, no specific fluorescence was obtained using the TDP-43 pSer409/410 antibody (data not shown), but fluorescent cytoplasmic inclusions were detected using the pTDP-43 Ser403/404 antibody (Figure 6).
Figure 6.
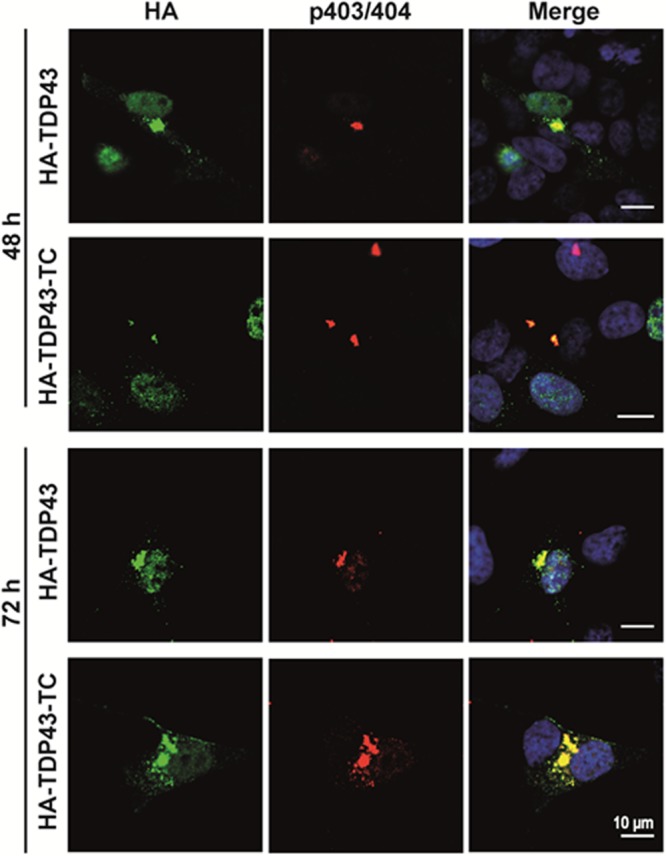
Comparison of co-localization of TDP-43 using an antibody for detection of phosphorylation at serine 403 and 404 residues between the HA-TDP43 and HA-TDP43-TC SH-SY5Y cell models 48 and 72 h post-transfection using electroporation. Merged immunofluorescence images generated using an anti-HA antibody (green), Hoechst nuclear counterstain (blue), and an anti-TDP-43 p403/404 antibody (red). Yellow denotes co-localization between HA-labeled TDP-43 and TDP43-p403/404.
The TC Tag Cell Model Shows Nuclear-to-Cytoplasmic Translocation in Real Time
Having demonstrated that the TC tag cell model reproduces several events relevant to the disease process in ALS, we finally examined whether it is possible to monitor the translocation of TDP-43 from the nucleus into the cytoplasm in real time. Given that the mild stress induced by electroporation appears to stimulate translocation of HA-TDP43-TC, no additional external chemicals or processes are needed to induce this phenomenon. At 24 h post-transfection, the FlAsH reagent was added to these cells and they were imaged using confocal microscopy for a further 20 h (Figure 7).
Figure 7.
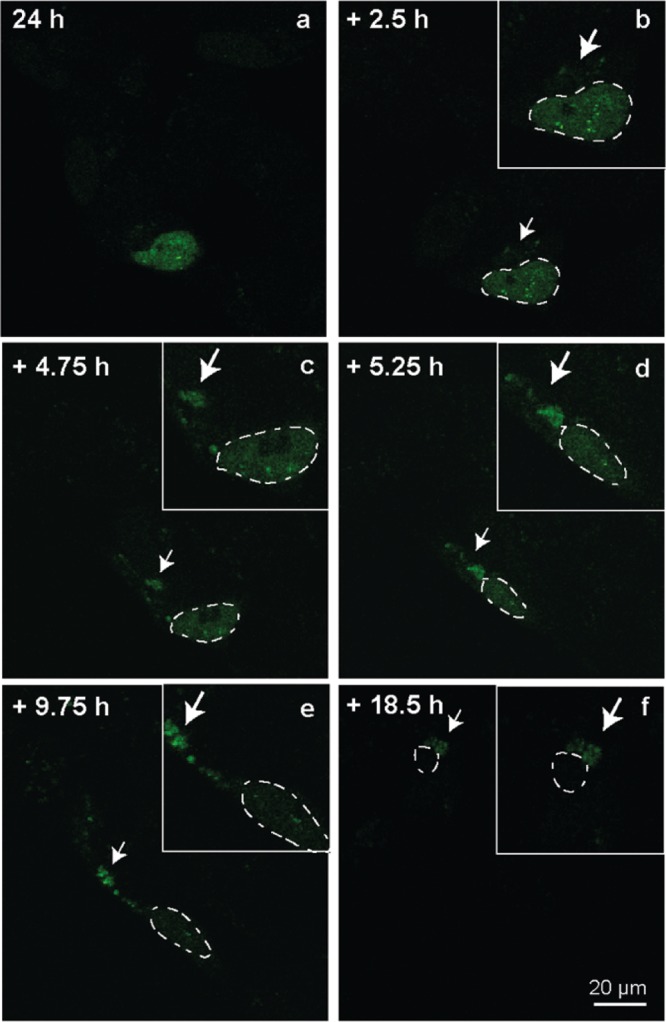
Frames from time-lapse confocal microscopy imaging of HA-TDP43-TC-expressing SH-SY5Y cells labeled with FlAsH dye. Imaging commenced 24 h post-transfection, and the subsequent time points shown are (a) 24:00, (b) 26:30, (c) 28:45, (d) 29:15, (e) 33:45, and (f) 42:30. The inset white boxes contain close-up images of the cell nucleus (2 times larger than the original image). The nuclei of the target cells are denoted by dashed lines, and white arrows denote cytoplasmic puncta. The images are representative of multiple independent experiments.
Initially, individual cells expressing HA-TDP43-TC show predominantly nuclear staining (Figure 7a), but over the time course of the experiment (Figure 7b onward), these cells can be seen to have developed small cytoplasmic inclusions. Interestingly, even at the initial time point when cytoplasmic TDP-43 can be visualized (∼26 h 30 min), the fluorescence signal is not diffuse but appears as small puncta that clearly change position in relation to the cell nucleus over time. They also appear to coalesce during real-time imaging, suggesting that they merge into larger structures or are just in the proximity of one another (Supplementary Video S1). The appearance of HA-TDP43-TC in the cytoplasm coincides with a gradual decrease in nuclear FlAsH signal intensity (Figure 7c onward and Figure S5), indicating nuclear depletion of the labeled TDP-43. Concerns about the high background fluorescence due to endogenous cysteine rich proteins and the potential cytotoxicity of biarsenical dyes have been raised;44 however, with the application of BAL buffer washes, we did not observe high background fluorescence during our live-cell imaging studies, and the cells imaged did not show any signs of morphological change until late into the imaging process [∼44 h (Figure 7f)].
Discussion
Together with fluorescent protein (FP) tags, the HA tag is one of the most extensively used labels in investigations of the role of TDP-43 in ALS.19,24,45 The HA tag, however, has a significant limitation, in that there is a need to fix and immunostain cells to detect the labeled TDP-43, therefore making this model incompatible with live-cell imaging for monitoring intracellular processes. The FP-TDP-43 fusion systems are amenable to live-cell imaging but involve the incorporation of an ∼27 kDa FP. Given the relative size of the FP (∼27 kDa) in relation to TDP-43 (43 kDa), the bulkiness of the fluorescent moiety may perturb the native structure and function of the TDP-43, and therefore, the use of the smaller TC tag in conjunction with biarsenical dyes (∼1 kDa) may prove to be advantageous.
In this paper, we have described a cell model in which the cells are transiently transfected to overexpress HA-tagged TDP-43 with a C-terminal TC tag, which is visualized using the biarsenical dye, FlAsH. This model can be used to monitor the translocation of TDP-43 from the nucleus to the cytoplasm in live cells using time-lapse microscopy. Although our study shows that the HA-TDP43-TC model is very similar to the well-characterized HA-TDP43 cell model, the properties of the TC tag and FlAsH make it a very valuable tool for monitoring the intracellular distribution of TDP-43 in live cells. The TC tag itself is relatively small, while the FlAsH dye is membrane permeable and nonfluorescent until it binds to the TC-tagged protein. In addition, the dye itself is sufficiently photostable, and shows low toxicity, to allow imaging in live cells over several hours.13
Comparisons between the HA-TDP43-TC and HA-TDP43 cell models indicate that the C-terminal TC tag does not detectably alter the properties of HA-TDP43 when overexpressed in SH-SY5Y cells. When transfected by electroporation, 48 and 72 h post-transfection, cells expressing either HA-TDP43 or HA-TDP43-TC show cytoplasmic translocation and accumulation of the overexpressed protein. Interestingly, this observation differs from most previous reports in which WT HA-TDP43 or GFP-TDP43 has been overexpressed in transfected mammalian cell lines.20,46 Nuclear-to-cytoplasmic translocation is more commonly observed in cell models expressing TDP-43 containing familial ALS mutations47,48 or truncated forms of the protein.25,31 For cells expressing WT TDP-43, translocation of the protein to the cytoplasm has generally been reported to occur only following the application of external stress.21,32 For cells expressing HA-TDP43 or HA-TDP43-TC, we observe predominantly nuclear localization when cells were lipofected, with much less cytoplasmic TDP-43 detected at all the time points measured post-transfection than in cells transfected using electroporation. These results suggest that the electroporation method, which slightly increases the transfection efficiency (by 2–4%), may also result in a degree of cell stress;49 we therefore took advantage of this phenomenon to study the translocation of WT TDP-43 in the absence of pharmacological stressors.
Cells transfected by electroporation to express HA-TDP43-TC demonstrate some key biochemical hallmarks of ALS, including the nuclear-to-cytoplasm translocation, an increase in the appearance and size of cytoplasmic inclusions over time, an indication that the TDP-43 is at least partially phosphorylated, and the association of the TDP-43 inclusions with TIA-1, a marker of stress granules.1,3,38 Phosphorylation of TDP-43 at the Ser403/404 or Ser409/410 epitopes has also been associated with ALS pathogenesis.43,50,51 It is not clear whether phosphorylation of TDP-43 leads to aggregate formation and/or neurotoxicity or if this process represents a normal reaction to the presence of an intracellular aggregate, as reports have shown that phosphorylation of WT TDP43 is not always observed.20,24 We did, however, observe positive staining with the antibody specific for p403/404 but not the antibody specific for p409/410 (Figure 6).
Using cells expressing HA-TDP43-TC, we were able to observe translocation of WT TDP-43 in the absence of pharmacological stresses in live cells over time, as indicated by the appearance of fluorescent motile cytoplasmic puncta together with decreased nuclear fluorescence. This process is thought to precede the deposition of cytoplasmic inclusions, which may contribute to the pathogenesis of ALS due to depletion of TDP-43 in the nucleus, preventing the protein from carrying out its normal regulatory functions on mRNA (LOF), and/or to the accumulation of cytoplasmic TDP-43 with inherently toxic properties (GOF).52
The HA-TDP43-TC cell model shares many features of the well-established HA-TDP43 model, which has been successfully utilized to investigate many aspects of TDP-43-mediated disease processes, but offers the considerable advantage of being compatible with live-cell imaging. In particular, we were able to monitor the nuclear-to-cytoplasmic translocation of TDP-43 in live cells using confocal microscopy, which represents an early stage in the deposition of aggregated TDP-43 in the cytoplasm of affected cells.
Having shown that it is possible to image TDP-43 translocation in real time using this model, we find it is clear that this has the potential to provide quantitative information about the translocation process itself and could even be applied to gain further insights into other processes, which occur subsequently to nuclear translocation, such as the aggregation of TDP-43 within the intracellular environment. This in turn may help develop a better understanding of how TDP-43 contributes to neurodegeneration in ALS and related proteinopathies.
Acknowledgments
The authors thank Dr. Kamran Yunus and Dr. Gabi Kaminski-Schierle for providing training and access to the confocal microscope (for live imaging) in the Department of Chemical Engineering and Biotechnology, University of Cambridge. The authors also thank Dr. Leila Luheshi for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00592.
Accession Codes
TDP-43, Q13148-1.
Author Contributions
⊥ J.S.W.N. and M.A.H. contributed equally to this work.
J.S.W.N. was supported by a Yousef Jameel Scholarship and The Cambridge Commonwealth, European & International Trust. M.A.H. was supported by a University of Cambridge CHESS studentship and an Australian Government Endeavour Research Fellowship. The work was supported, in part, by the Wellcome Trust (094425/Z/10/Z; J.R.K. and C.M.D.), the Centre for Misfolding Diseases (J.S.W.N., M.A.H., N.S.M., T.P.B., C.M.D., and J.R.K.), the Wenner-Gren Foundations and the Swedish Research Council (Grant 2016-03902; E.K.E.), and the NHMRC (Grants 1084144 and 1095215; J.J.Y.).
The authors declare no competing financial interest.
Supplementary Material
References
- Arai T.; Hasegawa M.; Akiyama H.; Ikeda K.; Nonaka T.; Mori H.; Mann D.; Tsuchiya K.; Yoshida M.; Hashizume Y.; Oda T. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611. 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Arai T.; Hasegawa M.; Nonoka T.; Kametani F.; Yamashita M.; Hosokawa M.; Niizato K.; Tsuchiya K.; Kobayashi Z.; Ikeda K.; Yoshida M.; Onaya M.; Fujishiro H.; Akiyama H. (2010) Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy. Neuropathology 30, 170–181. 10.1111/j.1440-1789.2009.01089.x. [DOI] [PubMed] [Google Scholar]
- Neumann M.; Sampathu D. M.; Kwong L. K.; Truax A. C.; Micsenyi M. C.; Chou T. T.; Bruce J.; Schuck T.; Grossman M.; Clark C. M.; McCluskey L. F.; Miller B. L.; Masliah E.; Mackenzie I. R.; Feldman H.; Feiden W.; Kretzschmar H. A.; Trojanowski J. Q.; Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Ciryam P.; Lambert-Smith I. A.; Bean D. M.; Freer R.; Cid F.; Tartaglia G. G.; Saunders D. N.; Wilson M. R.; Oliver S. G.; Morimoto R. I.; Dobson C. M.; Vendruscolo M.; Favrin G.; Yerbury J. J. (2017) Spinal motor neuron protein supersaturation patterns are associated with inclusion body formation in ALS. Proc. Natl. Acad. Sci. U. S. A. 114, E3935–E3943. 10.1073/pnas.1613854114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E.; Baralle F. E. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343. 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Farrawell N. E.; Lambert-Smith I. A.; Warraich S. T.; Blair I. P.; Saunders D. N.; Hatters D. M.; Yerbury J. J. (2015) Distinct partitioning of ALS associated TDP-43, FUS and SOD1 mutants into cellular inclusions. Sci. Rep. 5, 13416. 10.1038/srep13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smethurst P.; Newcombe J.; Troakes C.; Simone R.; Chen Y. R.; Patani R.; Sidle K. (2016) In vitro prion-like behaviour of TDP-43 in ALS. Neurobiol. Dis. 96, 236–247. 10.1016/j.nbd.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella R.; Capitini C.; Fani G.; Dobson C. M.; Cecchi C.; Chiti F. (2016) Quantification of the Relative Contributions of Loss-of-function and Gain-of-function Mechanisms in TAR DNA-binding Protein 43 (TDP-43) Proteinopathies. J. Biol. Chem. 291, 19437–19448. 10.1074/jbc.M116.737726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. A.; Adams S. R.; Tsien R. Y. (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272. 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Martin B. R.; Giepmans B. N.; Adams S. R.; Tsien R. Y. (2005) Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 23, 1308–1314. 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- Giepmans B. N.; Adams S. R.; Ellisman M. H.; Tsien R. Y. (2006) The fluorescent toolbox for assessing protein location and function. Science 312, 217–224. 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Roberti M. J.; Bertoncini C. W.; Klement R.; Jares-Erijman E. A.; Jovin T. M. (2007) Fluorescence imaging of amyloid formation in living cells by a functional, tetracysteine-tagged alpha-synuclein. Nat. Methods 4, 345–351. 10.1038/nmeth1026. [DOI] [PubMed] [Google Scholar]
- Hoffmann C.; Gaietta G.; Zurn A.; Adams S. R.; Terrillon S.; Ellisman M. H.; Tsien R. Y.; Lohse M. J. (2010) Fluorescent labeling of tetracysteine-tagged proteins in intact cells. Nat. Protoc. 5, 1666–1677. 10.1038/nprot.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman H.; Wirth A. J.; Gruebele M. (2016) ReAsH as a quantitative probe of in-cell protein dynamics. Biochemistry 55, 1968–1976. 10.1021/acs.biochem.5b01336. [DOI] [PubMed] [Google Scholar]
- Ignatova Z.; Gierasch L. M. (2004) Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc. Natl. Acad. Sci. U. S. A. 101, 523–528. 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irtegun S.; Wood R.; Lackovic K.; Schweiggert J.; Ramdzan Y. M.; Huang D. C.; Mulhern T. D.; Hatters D. M. (2014) A biosensor of SRC family kinase conformation by exposable tetracysteine useful for cell-based screening. ACS Chem. Biol. 9, 1426–1431. 10.1021/cb500242q. [DOI] [PubMed] [Google Scholar]
- Ramdzan Y. M.; Nisbet R. M.; Miller J.; Finkbeiner S.; Hill A. F.; Hatters D. M. (2010) Conformation sensors that distinguish monomeric proteins from oligomers in live cells. Chem. Biol. 17, 371–379. 10.1016/j.chembiol.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M. J.; Jovin T. M.; Jares-Erijman E. (2011) Confocal fluorescence anisotropy and FRAP imaging of alpha-synuclein amyloid aggregates in living cells. PLoS One 6, e23338. 10.1371/journal.pone.0023338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T.; Arai T.; Buratti E.; Baralle F. E.; Akiyama H.; Hasegawa M. (2009) Phosphorylated and ubiquitinated TDP-43 pathological inclusions in ALS and FTLD-U are recapitulated in SH-SY5Y cells. FEBS Lett. 583, 394–400. 10.1016/j.febslet.2008.12.031. [DOI] [PubMed] [Google Scholar]
- Nonaka T.; Masuda-Suzukake M.; Arai T.; Hasegawa Y.; Akatsu H.; Obi T.; Yoshida M.; Murayama S.; Mann D. M.; Akiyama H.; Hasegawa M. (2013) Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 4, 124–134. 10.1016/j.celrep.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Bentmann E.; Neumann M.; Tahirovic S.; Rodde R.; Dormann D.; Haass C. (2012) Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 287, 23079–23094. 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L.; Bilgutay A.; Zhang Y.-J.; Vanderwyde T.; Citro A.; Mehta T.; Zaarur N.; McKee A.; Bowser R.; Sherman M.; Petrucelli L.; Wolozin B. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C.; Bassell G. J.; Rossoll W. (2012) The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet. 21, 3703–3718. 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T.; Kametani F.; Arai T.; Akiyama H.; Hasegawa M. (2009) Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 18, 3353–3364. 10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- Scotter E. L.; Vance C.; Nishimura A. L.; Lee Y. B.; Chen H. J.; Urwin H.; Sardone V.; Mitchell J. C.; Rogelj B.; Rubinsztein D. C.; Shaw C. E. (2014) Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J. Cell Sci. 127, 1263–1278. 10.1242/jcs.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz L. M.; Kwong L. K.; Xu Y.; Truax A. C.; Uryu K.; Neumann M.; Clark C. M.; Elman L. B.; Miller B. L.; Grossman M.; McCluskey L. F.; Trojanowski J. Q.; Lee V. M. (2008) Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 173, 182–194. 10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.; Chen Y.; Zhou X.; Kar A.; Ray P.; Chen X.; Rao E. J.; Yang M.; Ye H.; Zhu L.; Liu J.; Xu M.; Yang Y.; Wang C.; Zhang D.; Bigio E. H.; Mesulam M.; Shen Y.; Xu Q.; Fushimi K.; Wu J. Y. (2011) An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830. 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled S.; Sade D.; Bram Y.; Porat Z.; Kreiser T.; Mimouni M.; Lichtenstein A.; Segal D.; Gazit E. (2017) Single cell imaging and quantification of TDP-43 and alpha-synuclein intercellular propagation. Sci. Rep. 7, 544. 10.1038/s41598-017-00657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevich J.; Langford D. (2013) Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology. Methods Mol. Biol. 1078, 9–21. 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbocker R.; Chia S.; Ruggeri F. S.; Perni M.; Cascella R.; Heller G. T.; Meisl G.; Mannini B.; Habchi J.; Michaels T. C. T.; Challa P. K.; Ahn M.; Casford S. T.; Fernando N.; Xu C. K.; Kloss N. D.; Cohen S. I. A.; Kumita J. R.; Cecchi C.; Zasloff M.; Linse S.; Knowles T. P. J.; Chiti F.; Vendruscolo M.; Dobson C. M. (2019) Trodusquemine enhances A beta(42) aggregation but suppresses its toxicity by displacing oligomers from cell membranes. Nat. Commun. 10, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L.; Lin A. Y.; Ebata A.; Boon J. Y.; Reid W.; Xu Y. F.; Kobrin K.; Murphy G. J.; Petrucelli L.; Wolozin B. (2014) ALS-linked mutations enlarge TDP-43-enriched neuronal RNA granules in the dendritic arbor. J. Neurosci. 34, 4167–4174. 10.1523/JNEUROSCI.2350-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C. M.; Cenik B.; Sephton C. F.; Dries D. R.; Mayer P. 3rd; Good S. K.; Johnson B. A.; Herz J.; Yu G. (2011) TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31, 1098–1108. 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiler M. S.; Strobel B.; Freischmidt A.; Helferich A. M.; Kappel J.; Brewer B. M.; Li D.; Thal D. R.; Walther P.; Ludolph A. C.; Danzer K. M.; Weishaupt J. H. (2015) TDP-43 is intercellularly transmitted across axon terminals. J. Cell Biol. 211, 897–911. 10.1083/jcb.201504057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eersel J.; Ke Y. D.; Gladbach A.; Bi M.; Gotz J.; Kril J. J.; Ittner L. M. (2011) Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS One 6, e22850. 10.1371/journal.pone.0022850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M.; Nonaka T.; Hirai S.; Miwa A.; Okado H.; Arai T.; Hosokawa M.; Akiyama H.; Hasegawa M. (2014) Distinct pathways leading to TDP-43-induced cellular dysfunctions. Hum. Mol. Genet. 23, 4345–4356. 10.1093/hmg/ddu152. [DOI] [PubMed] [Google Scholar]
- Zhang Y. J.; Xu Y. F.; Cook C.; Gendron T. F.; Roettges P.; Link C. D.; Lin W. L.; Tong J.; Castanedes-Casey M.; Ash P.; Gass J.; Rangachari V.; Buratti E.; Baralle F.; Golde T. E.; Dickson D. W.; Petrucelli L. (2009) Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U. S. A. 106, 7607–7612. 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. (2012) Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7, 56. 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C. M.; Cenik B.; Sephton C. F.; Johnson B. A.; Herz J.; Yu G. (2012) TDP-43 aggregation in neurodegeneration: are stress granules the key?. Brain Res. 1462, 16–25. 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S.; Kabuta T.; Nagai Y.; Tsuchiya Y.; Akiyama H.; Wada K. (2013) TDP-43 associates with stalled ribosomes and contributes to cell survival during cellular stress. J. Neurochem. 126, 288–300. 10.1111/jnc.12194. [DOI] [PubMed] [Google Scholar]
- Colombrita C.; Zennaro E.; Fallini C.; Weber M.; Sommacal A.; Buratti E.; Silani V.; Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061. 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- McGurk L.; Lee V. M.; Trojanowksi J. Q.; Van Deerlin V. M.; Lee E. B.; Bonini N. M. (2014) Poly-A binding protein-1 localization to a subset of TDP-43 inclusions in amyotrophic lateral sclerosis occurs more frequently in patients harboring an expansion in C9orf72. J. Neuropathol. Exp. Neurol. 73, 837–845. 10.1097/NEN.0000000000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. R.; King O. D.; Shorter J.; Gitler A. D. (2013) Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372. 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y.; Nonaka T.; Arai T.; Yoshida M.; Hashizume Y.; Beach T. G.; Buratti E.; Baralle F. E.; Akiyama H.; Hisanaga S.; Hasegawa M. (2008) Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Lett. 582, 2899–2904. 10.1016/j.febslet.2008.07.027. [DOI] [PubMed] [Google Scholar]
- Langhorst M. F.; Genisyuerek S.; Stuermer C. A. (2006) Accumulation of FlAsH/Lumio Green in active mitochondria can be reversed by beta-mercaptoethanol for specific staining of tetracysteine-tagged proteins. Histochem. Cell Biol. 125, 743–747. 10.1007/s00418-005-0136-3. [DOI] [PubMed] [Google Scholar]
- Shiina Y.; Toyoda T.; Kawasoe Y.; Tateno S.; Shirai T.; Matsuo K.; Mizuno Y.; Ai T.; Niwa K. (2011) The prevalence and risk factors for cholelithiasis and asymptomatic gallstones in adults with congenital heart disease. Int. J. Cardiol. 152, 171–176. 10.1016/j.ijcard.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Seyfried N. T.; Gozal Y. M.; Dammer E. B.; Xia Q.; Duong D. M.; Cheng D.; Lah J. J.; Levey A. I.; Peng J. (2010) Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol. Cell. Proteomics 9, 705–718. 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budini M.; Romano V.; Avendano-Vazquez S. E.; Bembich S.; Buratti E.; Baralle F. E. (2012) Role of selected mutations in the Q/N rich region of TDP-43 in EGFP-12xQ/N-induced aggregate formation. Brain Res. 1462, 139–150. 10.1016/j.brainres.2012.02.031. [DOI] [PubMed] [Google Scholar]
- Liu R.; Yang G.; Nonaka T.; Arai T.; Jia W.; Cynader M. S. (2013) Reducing TDP-43 aggregation does not prevent its cytotoxicity. Acta Neuropathol. Commun. 1, 49. 10.1186/2051-5960-1-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K.; Soo K. Y.; Sundaramoorthy V.; Parakh S.; Ma Y.; Farg M. A.; Wallace R. H.; Crouch P. J.; Turner B. J.; Horne M. K.; Atkin J. D. (2013) ALS-Associated TDP-43 Induces Endoplasmic Reticulum Stress, Which Drives Cytoplasmic TDP-43 Accumulation and Stress Granule Formation. PLoS One 8, e81170. 10.1371/journal.pone.0081170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M.; Arai T.; Nonaka T.; Kametani F.; Yoshida M.; Hashizume Y.; Beach T. G.; Buratti E.; Baralle F.; Morita M.; Nakano I.; Oda T.; Tsuchiya K.; Akiyama H. (2008) Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70. 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M.; Kwong L. K.; Lee E. B.; Kremmer E.; Flatley A.; Xu Y.; Forman M. S.; Troost D.; Kretzschmar H. A.; Trojanowski J. Q.; Lee V. M. (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149. 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. B.; Lee V. M.; Trojanowski J. Q. (2012) Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 13, 38–50. 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


