Abstract
Reverse transcription quantitative PCR (RT-qPCR) is the most commonly used method to evaluate gene expression. Reliable qPCR results are highly dependent on accurate normalization using suitable reference genes. We investigated expression of commonly used reference genes during murine Cytomegalovirus (mCMV) infection and latency to determine those genes least perturbed by infection. Following mCMV infection in BALB/c mice, lung, salivary gland, liver, spleen and kidney were evaluated. Liver sinusoidal endothelial cells and NIH-3T3 cells were also evaluated. RT-qPCR was performed during acute and latent mCMV infection for 11 commonly used reference genes with comparisons made to uninfected samples. Normfinder, BestKeeper, GeNorm and the comparative delta CT method produced comparable analyses that were combined in RefFinder to generate an overall ranking. Ppia, B2m and Gapdh are the most stable reference genes for in vitro infection studies. For in vivo studies the most suitable reference genes were highly tissue and cell type dependent. Comparing infected and uninfected groups revealed viral influence on transcription of some genes. We provide reference gene guidelines for investigations of gene expression for mCMV Smith strain infection of Balb/cJ mice or NIH-3T3 cells. These results also suggest careful consideration of reference genes for different host tissues evaluated.
Keywords: murine Cytomegalovirus, RT-qPCR, reference gene
1. Introduction
Reverse transcription quantitative PCR (RT-qPCR) is still the most commonly used method to measure levels of gene expression in various biological samples, not only in basic research but also in diagnostic laboratories. The technique’s advantages are high sensitivity, reproducibility, cost effectiveness as well as speed and simplicity of performance.
One major obstacle to RT-qPCR is reproducibility of results. Consideration and disclosure of experimental design, such as nucleic acid extraction and sample information, details of reverse transcription and qPCR performance are all essential to prevent assay variation and ensure result reproducibility (Bustin et al., 2009; Derveaux, Vandesompele, and Hellemans, 2010). Reliable qPCR experiments depend highly on selection of appropriate reference genes (Hellemans and Vandesompele, 2014), but the importance of accurate normalization of results is often underestimated. Depending on experimental conditions, commonly used reference genes may not always represent the best fit (Glare et al., 2002). Several computer based tools are available to help choosing the most suitable control genes, including Normfinder (Andersen, Jensen, and Orntoft, 2004), BestKeeper (Pfaffl et al., 2004), GeNorm (Vandesompele et al., 2002) and the comparative delta CT method (Livak and Schmittgen, 2001; Silver et al., 2006).
The term housekeeping gene has been used to describe genes whose expression is not altered by changes in experimental circumstance. As any gene’s transcription might be influenced depending on experimental circumstances, the idea of universal housekeeping genes is probably erroneous (Glare et al., 2002; Selvey et al., 2001). Host gene expression can also vary depending on the tissue and cell type that is analyzed sometimes making it necessary to use different genes (Barber et al., 2005; Chapman and Waldenstrom, 2015; Suzuki, Higgins, and Crawford, 2000). Different experimental conditions, in particular investigation of cellular transcription after virus infections, may significantly alter expression of commonly used control genes. It is therefore widely accepted that different experimental conditions require specific evaluations to determine the most suitable reference gene. Accurate normalization is a fundamental requirement when studying the significance of gene expression differences. Several studies have been conducted describing the ideal reference gene for many virus infections, including HIV, HSV, VZV, SARS and human CMV to name a few (Neerukonda et al., 2016; Radonic et al., 2005; Watson et al., 2007).
Surprisingly, for the most commonly used animal model of cytomegalovirus infection - the mouse, no general recommendations about reference genes of choice have been published. We describe results from 11 of the most frequently used reference genes in the context of mCMV infection, both in cell culture and in different murine tissues after infection.
2. Materials and Methods
2.1 Animals
Female BALB/cJ mice 6–8 weeks of age were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were euthanized by cervical dislocation under isoflurane inhalation anesthesia. Mouse tissues were dissected aseptically, snap frozen in liquid nitrogen, and stored at −80°C. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines of Beth Israel Deaconess Medical Center.
2.2 Viral infections
For in vitro experiments 70% confluent NIH-3T3 cells (ATCC®, CRL-1658™) were infected with murine CMV strain Smith (ATCC®, VR-194/1981™) at MOI of 0.4. Cells were harvested at 0h, 6h, 24h, 48h and 72h after infection.
For in vivo experiments female Balb/cJ mice were infected intra peritoneal (i.p.) with 1×106 plaque forming units (pfu) of murine CMV strain Smith. All virus stocks were stored at −80°C and before use diluted in Dulbecco’s phosphate buffered saline (DPBS) to reach an injection volume of 100μl. Mock animals were injected with 100μl of sterile DPBS. As previously published, mice were allowed to become latent over the course of at least 4 months (Cook et al., 2002). It has been previously shown that susceptible mice have replicating virus detectable in salivary gland, lungs and liver 2 weeks after infection so this time point was chosen for the acute tissue infection experiments. (Matsuzawa et al., 1995; Selgrade et al., 1984; Shanley and Pesanti, 1985; Yuhasz et al., 1994)
2.3 Isolation of Liver sinusoidal endothelial cells (LSEC)
Non-parenchymal liver cells were isolated as described previously (Seckert et al., 2009). LSEC were isolated from non-parenchymal liver cells by magnetic cell separation using CD146 (LSEC) MicroBeads (Milteny Biotec, cat. no. 130-092-007). Positive selection of CD146 expressing cells was done according to manufacturer’s instructions using LS columns. After cell enumeration, RNA were directly isolated from cell pellets.
2.4 RNA extraction and cDNA synthesis
RNA were isolated via TRIzol reagent (Ambion, cat. no. 15596-018) following manufacturer’s instructions. Briefly, tissues were homogenized in 1ml TRIzol using tissue lyser II (Qiagen, cat. no. 85300) according to manufacturer protocol ‘purification of RNA from animal tissues’. RNA pellets were resuspended in 35μl of RNase free water and incubated at 55°C for less than 5min. RNA were column purified with RNeasy Mini Kits (Qiagen, cat. no. 74104) with DNase treatment on-column (Qiagen, cat. no. 79254), then eluted in 35μl RNase free water and stored at −80°C until cDNA synthesis was performed. RNA quantifications were done with Nanodrop 1000 (Thermo Scientific). cDNA were produced using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad laboratories, cat. no. 170-8841) according to manufacturer recommendations using RNA input amounts of 800ng, or 200ng when 800ng was not available.
2.5 Quantitative real-time PCR
Eleven reference genes commonly used in qPCR were selected to compare their expression levels in mCMV infected and non-infected cells originating from different tissue types. The following reference genes were chosen for analysis: glyceraldehyde-3-phosphate dehydrogenase (Gapdh); heat shock protein 90 alpha (cytosolic), class B member 1 (Hsp90ab1); beta-2 microglobulin (B2m); glucuronidase, beta (Gusb); TATA box binding protein (Tbp); phosphoglycerate kinase 1 (Pgk1); peptidylprolyl isomerase A (Ppia); actin, beta (Actb); hypoxanthine guanine phosphoribosyl transferase (Hprt); tubulin, beta 4A class IVA (Tubb4a); parathyroid hormone-like peptide (Pthlh). Table 1 lists corresponding primer sequences used, including exon locations, amplicon sizes and IDT catalog numbers. Choosing different exon locations for forward and reverse primers substantially limits the risk of amplifying contaminating DNA. Dilutions of primer pairs were tested in RT-qPCR to optimize primer concentration while yielding the least amount of primer dimers (data not shown).
Table 1.
Reference gene candidates with corresponding primer information
| #a | Refb-gene | Ref-Seqc # | Exon location | Sequence primer-ford | Sequence primer-reve | Size RNA [bp] | Size DNA [bp] | IDTf catalog # |
|---|---|---|---|---|---|---|---|---|
| 1 | Gapdh | NM_008084 | 2–3 | 5′-GTG GAG TCA TAC TGG AAC ATG TAG-3′ | 5′-AAT GGT GAA GGT CGG TGT G-3′ | 150 | 1984 | MmPT 39a1 |
| 2 | Hsp90ab1 | NM_008302 | 1–2 | 5′-CAT GAG CTG GGC AAT TTC TG-3′ | 5′-ACT CGG CTT TCC CGT CA-3′ | 94 | 1349 | MmPT5 841290500 |
| 3 | B2m | NM_009735 | 1–2 | 5′-ACG TAG CAG TTC AGT ATG TTC G-3′ | 5′-GGT CTT TCT GGT GCT TGT CT-3′ | 120 | 3188 | MmPT5 810497647 |
| 4 | Gusb | NM_010368 | 10–11 | 5′-GAA CAG CCT TCT GGT ACT CC-3′ | 5′-GAG AAC TGG TAT AAG ACG CAT CA-3′ | 121 | 3386 | MmPT5 81384361 |
| 5 | Tbp | NM_013684 | 3–4 | 5′-CAA GTT TAC AGC CAA GAT TCA CG-3′ | 5′-TTC ACC AAT GAC TCC TAT GAC C-3′ | 116 | 2811 | MmPT5 810867035 |
| 6 | Pgk1 | NM_008828 | 4–5 | 5′-CTA GTT TGG ACA GTG AGG CT-3′ | 5′-AAC CTC CGC TTT CAT GTA GAG-3′ | 112 | 2178 | MmPT5 813087431 |
| 7 | Ppia | NM_008907 | 4–5 | 5′-TTC ACC TTC CCA AAG ACC AC-3′ | 5′-CAA ACA CAA ACG GTT CCC AG-3′ | 85 | 277 | MmPT3 9a2gs |
| 8 | Actb | NM_007393 | 1–2 | 5′-ATG CCG GAG CCG TTG TC-3′ | 5′-GCG AGC ACA GCT TCT TTG-3′ | 106 | 1065 | MmPT5 833540333 |
| 9 | Hprt | NM_013556 | 2–3 | 5′-AGC AGG TCA GCA AAG AAC T-3′ | 5′-CCT CAT GGA CTG ATT ATG GAC A-3′ | 125 | 3015 | MmPT5 832192191 |
| 10 | Tubb4a | NM_009451 | 1–2 | 5′-GTC GAT GCC GTG CTC AT-3′ | 5′-GAC ACC CGT CCA TCA GAC-3′ | 132 | 1249 | MmPT5 6a9905332 |
| 11 | Pthlh | NM_008970.3 | 3 | 5′-CAA GGG CAA GTC CAT CCA AG-3′ | 5′-GGG ACA CCT CCG AGG TAG CT-3′ | 101 | 101 | custom made |
#-number.
Ref, Reference.
Seq, Sequence.
for, forward.
rev, reverse.
IDT, Integrated DNA Technologies.
qPCR were performed on a StepOnePlus real-time PCR system (Applied Biosystems) using microtiter plates in final volumes of 20μl, with the following cycling conditions: 95°C for 5 minutes followed by 45 cycles of 95°C for 10 seconds and a combined annealing/extension step at 60°C for 30 seconds, during which data were collected. Melting curve analyses were performed as follows: 95°C for 15 seconds, 60°C for 1 minute and 95°C for 15 seconds. cDNA were added in a volume of 2μl per reaction equaling 20ng of input RNA. Fluorescent PCR amplicons were detected using QuantiFast SYBR Green PCR master mix (Qiagen, cat. no. 204056) and primers, indicated in Table 1 with the following final concentrations: Pgk1 60nM, Hsp90ab1 and Ppia 100nM, Gusb 150nM, Tbp 200nM, all others 250nM. All primers were predesigned by Integrated DNA Technologies (IDT), except Pthlh specific primers that were designed by the Primer Express 2.0 software (Applied Biosystems). Concomitant “no-RT” reactions, lacking reverse transcriptase, were performed for each sample and run to confirm absence of DNA contamination, as well as no template controls (NTCs) to confirm lack of contamination of all used reagents. RNA integrity was assured by RIN measurement (not shown). A 1:10 dilution series of cDNA was used to construct a standard curve. The PCR efficiency (E) for each reference gene was determined with the following equation using the slope of the standard curve: E= 10 (−1/slope) −1 (Supplementary Table 1). Analyses were done using StepOnePlus Software version 2.3 (Applied Biosystems).
To calculate PCR efficiencies for each reference gene, a 1:10 dilution series of cDNA from uninfected NIH-3T3 cells was evaluated. Resulting standard curves were used to determine efficiency with the following equation E= 10 (−1/slope) −1, as shown in supplementary Table 1. Ideally efficiencies should be 100%, meaning that PCR products double with each cycle. Efficiencies of 90%–110%, corresponding to slopes ranging from −3.6 to −3.1, are generally acceptable (Invitrogen, 2008).
2.6 Analysis of data
Analyses were performed using the Normfinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), GeNorm (Vandesompele et al., 2002) and the comparative delta CT method (Livak and Schmittgen, 2001; Silver et al., 2006). Each of these methods estimates gene stability, with lower values corresponding to more stable gene transcription. These analyses each utilize different statistical approaches to determine stability, and the comparative advantages and disadvantages are beyond the scope of this manuscript. In addition to the current convention of reporting results of all four analyses, it also seems desirable to summarize these results with a final ranking using the RefFinder tool (Xie et al., 2012). RefFinder utilizes geometric means of ranks from Normfinder, BestKeeper, GeNorm and comparative delta CT methods to give an overall stability ranking, thereby allowing direct comparison of all methods (http://150.216.56.64/referencegene.php?type=reference). Three biological replicates were used for each sample type and infection time point.
3. Results
For in vitro experiments NIH-3T3 cells were infected with mCMV Smith strain at MOI 0.4 with RNA evaluated 0, 6, 24, 48 and 72 hours after infection. Age matched non-infected cells were harvested at these time points for controls. For infected cells, there was no significant difference between 6, 24, 48 or 72 hour results so these were all pooled for final analyses. We obtained three biological replicates per time point and analyzed these using Normfinder, BestKeeper, GeNorm and the comparative delta CT method (Andersen et al., 2004; Livak and Schmittgen, 2001; Pfaffl et al., 2004; Silver et al., 2006; Vandesompele et al., 2002). We calculated method-specific stability values for each reference gene and ranked each in order of reliability, where ‘1’ represents the highest reliability. We defined the most stable reference genes as having the least sample to sample variation. Table 2 shows stability values and rankings obtained from the different analysis programs for each gene. An overall ranking was next determined by geometric means based on rankings from each method using the RefFinder tool (Xie et al., 2012). For 3T3 infections, Ppia and B2m were ranked as the two most stable genes by most programs, with only Normfinder placing Gusb as number 2. All programs confirmed Pgk1 and Hprt as the least stable reference genes. In contrast, two of the tested reference genes, Pthlh and Tubb, did not reach acceptable efficiencies in any analyses and are therefore not recommended for use as reference genes following mCMV infection in vitro (not shown). Overall we found that regardless of analysis used the results were similar, and suggest that Ppia/B2m are the best reference genes for mCMV infected NIH-3T3 cells.
Table 2.
Stability values and ranking results for reference genes using NIH-3T3 cells. Lower stability values indicate more stable gene expression.
| Used method and ranking | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt |
|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 3.94 | 3.92 | 4.46 | 4.09 | 4.84 | 5.2 | 4.04 | 10.82 | 10.67 |
| Ranking | 2 | 1 | 5 | 4 | 6 | 7 | 3 | 9 | 8 |
| BestKeeper | 0.57 | 0.88 | 1.56 | 1.09 | 1.33 | 2.13 | 1.02 | 7.27 | 9.50 |
| Ranking | 1 | 2 | 6 | 4 | 5 | 7 | 3 | 8 | 9 |
| Normfinder | 0.21 | 0.12 | 0.49 | 0.28 | 0.16 | 0.55 | 0.18 | 0.77 | 0.56 |
| Ranking | 4 | 1 | 6 | 5 | 2 | 7 | 3 | 9 | 8 |
| GeNorm | 0.56 | 0.56 | 1.12 | 0.92 | 2.07 | 1.52 | 0.74 | 5.78 | 4.34 |
| Ranking | 1 | 1 | 5 | 4 | 7 | 6 | 3 | 9 | 8 |
| Ranking G.M.a | 1 | 2 | 6 | 5 | 4 | 7 | 3 | 9 | 8 |
G.M., geometric mean.
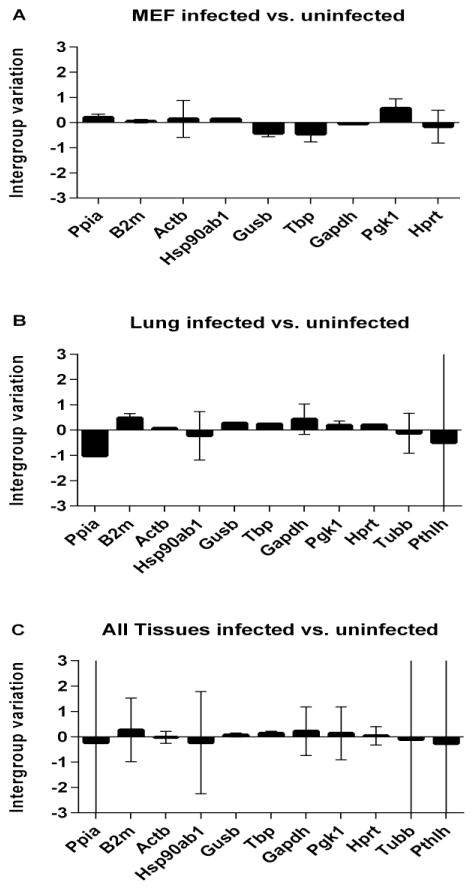
The Normfinder tool allows discrimination between different groups and calculation of inter- and intragroup variations for each gene and we used it to compare uninfected and infected groups. Intergroup variations were evaluated, and are represented by bar size. Intragroup variations within each individual group are represented as error bars for each bar. As shown in Figure 1A, NIH-3T3 cells had very low variations in general, with the largest intergroup variations for Pgk1, Tbp and Gusb. The least intergroup variability occurred for B2m and Gapdh. Intragroup variation was highest for Actb, Hprt, Pgk1 and Tbp. Taken together these results suggest that most of the tested reference genes are not regulated by mCMV infection in NIH-3T3 cells.
Figure 1. Inter- and intragroup variations for the tested reference genes comparing CMV infected with uninfected lung tissues.
Samples were grouped as uninfected or CMV infected and variability of reference gene expression was analyzed for (A) NIH-3T3 cells, (B) lung tissues and (C) all available samples from all tissues taken together. Each bar represents the intergroup variation infected compared to uninfected samples, while the intragroup variation for infected samples is shown as error bars.
We next analyzed commonly studied murine tissues after acute mCMV infection, including lung, salivary gland, spleen and kidney as well as liver and LSEC to determine the most suitable reference genes. Tables 3 and 4 show the stability values and rankings for each tissue comparing the four methods. All tissues were evaluated after acute infection. The lung results however contain latent and acute samples combined, because only subtle differences in reference gene regulation were observed between latent and acute infection (Supplemental Table 2). As shown in Tables 3 and 4, for most tissues there was higher variability in reference gene rankings between methods than for NIH-3T3 cells, except for salivary gland, which showed fairly consistent rankings independent of methodology. More importantly, when investigating different tissues, there were significant differences in reference gene rankings. Unlike in-vitro studies, Pthlh and Tubb showed much better standard curves in tissue derived cDNA so these genes were included for all in vivo analyses. For lung and kidney tissue the most suitable reference genes were Actb, Tbp and Hprt, while for salivary gland Gusb, Tubb and Tbp seemed best. The reference genes of choice for the spleen were Actb, B2m and Hprt.
Table 3.
Stability values and ranking results of reference genes using different tissues. Lower stability values indicate more stable gene expression.
| Lung | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt | Tubb | Pthlh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 2.50 | 1.34 | 1.11 | 1.71 | 1.09 | 1.08 | 1.82 | 1.32 | 1.11 | 1.64 | 1.87 |
| Ranking | 11 | 6 | 3 | 8 | 2 | 1 | 9 | 5 | 4 | 7 | 10 |
| BestKeeper | 2.02 | 1.80 | 1.61 | 2.49 | 1.63 | 1.70 | 1.22 | 1.44 | 1.49 | 2.46 | 1.26 |
| Ranking | 9 | 8 | 5 | 11 | 6 | 7 | 1 | 3 | 4 | 10 | 2 |
| Normfinder | 2.34 | 0.84 | 0.16 | 1.37 | 0.20 | 0.17 | 1.62 | 0.74 | 0.24 | 1.25 | 1.67 |
| Ranking | 11 | 6 | 1 | 8 | 3 | 2 | 9 | 5 | 4 | 7 | 10 |
| GeNorm | 1.51 | 0.57 | 0.31 | 1.01 | 0.40 | 0.38 | 1.18 | 0.66 | 0.31 | 0.87 | 1.29 |
| Ranking | 11 | 5 | 1 | 8 | 4 | 3 | 9 | 6 | 1 | 7 | 10 |
| Ranking G.M.a | 11 | 7 | 1 | 10 | 4 | 2 | 6 | 5 | 3 | 9 | 8 |
| Salivary Gland | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt | Tubb | Pthlh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 0.93 | 1.36 | 0.99 | 0.76 | 0.72 | 0.77 | 0.84 | 1.05 | 1 | 0.74 | 1.53 |
| Ranking | 6 | 10 | 7 | 3 | 1 | 4 | 5 | 9 | 8 | 2 | 11 |
| BestKeeper | 0.90 | 1.35 | 0.56 | 0.88 | 0.64 | 0.62 | 0.42 | 1.20 | 1.22 | 0.59 | 1.03 |
| Ranking | 7 | 11 | 2 | 6 | 5 | 4 | 1 | 9 | 10 | 3 | 8 |
| Normfinder | 0.62 | 1.18 | 0.70 | 0.38 | 0.14 | 0.40 | 0.49 | 0.82 | 0.81 | 0.23 | 1.41 |
| Ranking | 6 | 10 | 7 | 3 | 1 | 4 | 5 | 9 | 8 | 2 | 11 |
| GeNorm | 0.54 | 0.85 | 0.72 | 0.44 | 0.30 | 0.30 | 0.40 | 0.66 | 0.59 | 0.36 | 0.97 |
| Ranking | 5 | 9 | 8 | 4 | 1 | 1 | 3 | 7 | 6 | 2 | 11 |
| Ranking G.M.a | 7 | 11 | 6 | 5 | 1 | 3 | 4 | 9 | 8 | 2 | 10 |
| Spleen | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt | Tubb | Pthlh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 0.48 | 0.34 | 0.35 | 0.51 | 0.42 | 0.5 | 0.65 | 0.4 | 0.43 | 0.72 | 0.57 |
| Ranking | 6 | 1 | 2 | 8 | 4 | 7 | 10 | 3 | 5 | 11 | 9 |
| BestKeeper | 0.32 | 0.29 | 0.26 | 0.46 | 0.45 | 0.48 | 0.47 | 0.42 | 0.25 | 0.47 | 0.31 |
| Ranking | 5 | 3 | 2 | 8 | 7 | 11 | 10 | 6 | 1 | 9 | 4 |
| Normfinder | 0.34 | 0.03 | 0.03 | 0.37 | 0.24 | 0.36 | 0.58 | 0.19 | 0.23 | 0.66 | 0.46 |
| Ranking | 6 | 1 | 1 | 8 | 5 | 7 | 10 | 3 | 4 | 11 | 9 |
| GeNorm | 0.24 | 0.06 | 0.06 | 0.36 | 0.20 | 0.33 | 0.44 | 0.18 | 0.28 | 0.49 | 0.39 |
| Ranking | 5 | 1 | 1 | 8 | 4 | 7 | 10 | 3 | 6 | 11 | 9 |
| Ranking G.M.a | 6 | 2 | 1 | 9 | 5 | 8 | 10 | 4 | 3 | 11 | 7 |
G.M., geometric mean.
Table 4.
Stability values and ranking results of reference genes using different tissues-continued from Table 4. Lower stability values indicate more stable gene expression.
| Kidney | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt | Tubb | Pthlh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 1.01 | 1.26 | 0.87 | 1.23 | 0.92 | 0.89 | 0.84 | 1.2 | 0.89 | 2.67 | 1.81 |
| Ranking | 6 | 9 | 2 | 8 | 5 | 3 | 1 | 7 | 3 | 11 | 10 |
| BestKeeper | 0.73 | 0.75 | 0.72 | 1.04 | 0.72 | 0.64 | 0.81 | 1.16 | 0.66 | 1.97 | 0.91 |
| Ranking | 5 | 6 | 3 | 9 | 3 | 1 | 7 | 10 | 2 | 11 | 8 |
| Normfinder | 0.42 | 0.99 | 0.13 | 0.85 | 0.36 | 0.26 | 0.13 | 0.76 | 0.18 | 2.61 | 1.70 |
| Ranking | 6 | 9 | 1 | 8 | 5 | 4 | 2 | 7 | 3 | 11 | 10 |
| GeNorm | 0.47 | 0.75 | 0.38 | 0.66 | 0.24 | 0.26 | 0.32 | 0.58 | 0.24 | 1.24 | 0.92 |
| Ranking | 6 | 9 | 5 | 8 | 1 | 3 | 4 | 7 | 1 | 11 | 10 |
| Ranking G.M.a | 6 | 8 | 2 | 9 | 5 | 3 | 4 | 7 | 1 | 11 | 10 |
| Liver | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt | Tubb | Pthlh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 0.94 | 0.98 | 1.00 | 1.25 | 0.95 | 1.70 | 0.93 | 1.07 | 1.08 | 3.93 | 1.90 |
| Ranking | 2 | 4 | 5 | 8 | 3 | 9 | 1 | 6 | 7 | 11 | 10 |
| BestKeeper | 0.71 | 0.55 | 0.51 | 0.70 | 0.70 | 0.86 | 0.71 | 1.04 | 0.87 | 2.92 | 1.47 |
| Ranking | 5 | 2 | 1 | 3 | 4 | 7 | 6 | 9 | 8 | 11 | 10 |
| Normfinder | 0.19 | 0.15 | 0.37 | 0.86 | 0.10 | 1.28 | 0.10 | 0.42 | 0.51 | 3.86 | 1.58 |
| Ranking | 4 | 3 | 5 | 8 | 2 | 9 | 1 | 6 | 7 | 11 | 10 |
| GeNorm | 0.20 | 0.27 | 0.29 | 0.47 | 0.20 | 0.68 | 0.24 | 0.40 | 0.36 | 1.43 | 0.87 |
| Ranking | 1 | 4 | 5 | 8 | 1 | 9 | 3 | 7 | 6 | 11 | 10 |
| Ranking G.M.a | 3 | 4 | 5 | 6 | 2 | 9 | 1 | 7 | 8 | 11 | 10 |
| LSEC | Ppia | B2m | Actb | Hsp90ab1 | Gusb | Tbp | Gapdh | Pgk1 | Hprt | Tubb | Pthlh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Delta CT | 1.69 | 1.06 | 1.01 | 1.34 | 0.82 | 0.79 | 0.79 | 1.10 | 0.93 | 0.79 | 1.46 |
| Ranking | 11 | 7 | 6 | 9 | 4 | 2 | 3 | 8 | 5 | 1 | 10 |
| BestKeeper | 1.14 | 0.81 | 1.22 | 1.61 | 1.03 | 1.04 | 0.81 | 1.28 | 0.69 | 0.89 | 0.37 |
| Ranking | 8 | 3 | 9 | 11 | 6 | 7 | 4 | 10 | 2 | 5 | 1 |
| Normfinder | 1.59 | 0.72 | 0.72 | 1.22 | 0.13 | 0.15 | 0.15 | 0.89 | 0.45 | 0.22 | 1.28 |
| Ranking | 11 | 7 | 6 | 9 | 1 | 3 | 2 | 8 | 5 | 4 | 10 |
| GeNorm | 1.07 | 0.73 | 0.51 | 0.81 | 0.37 | 0.24 | 0.19 | 0.58 | 0.65 | 0.19 | 0.93 |
| Ranking | 11 | 8 | 5 | 9 | 4 | 3 | 1 | 6 | 7 | 1 | 10 |
| Ranking G.M.a | 11 | 7 | 8 | 10 | 3 | 4 | 2 | 9 | 5 | 1 | 6 |
| G.M. of all tissues | 7.90 | 5.96 | 3.94 | 5.89 | 1.19 | 1.41 | 6.45 | 7.45 | 3.46 | 10.00 | 11.00 |
| Overall ranking G.M. a | 9 | 6 | 4 | 5 | 1 | 2 | 7 | 8 | 3 | 10 | 11 |
G.M., geometric mean.
We evaluated further whether distinct cell subsets within an organ might show differences from isolates from whole organs. For this we compared reference gene expression in LSEC to those from whole liver tissues. LSEC are actively infected during acute infection, and also harbor latent infection (Seckert et al., 2009). Comparison of reference gene stability for LSEC and whole liver tissue revealed different ranking results (Table 4). Although two of the three most stable genes were the same for both sample types, namely Gusb and Gapdh, Tubb seems suitable only for LSEC analysis whereas Ppia seems better for whole liver tissue. For a summary of the rankings for all tested tissues and cells see Table 5.
Table 5.
Overview ranking results of reference genes
| Rank | NIH-3T3 | Lung | Salivary Gland | Spleen | Kidney | Liver | LSEC |
|---|---|---|---|---|---|---|---|
| 1 | Ppia | Actb | Gusb | Actb | Hprt | Gapdh | Gusb |
| 2 | B2m | Tbp | Tubb | B2m | Actb | Gusb | Tbp |
| 3 | Gapdh | Hprt | Tbp | Hprt | Tbp | Ppia | Hprt |
| 4 | Gusb | Gusb | Gapdh | Pgk1 | Gapdh | B2m | Actb |
| 5 | Hsp90ab1 | Pgk1 | Hsp90ab1 | Gusb | Gusb | Actb | Hsp90ab1 |
| 6 | Actb | Gapdh | Actb | Ppia | Ppia | Hsp90ab1 | B2m |
| 7 | Tbp | B2m | Ppia | Pthlh | Pgk1 | Pgk1 | Gapdh |
| 8 | Hprt | Pthlh | Hprt | Tbp | B2m | Hprt | Pgk1 |
| 9 | Pgk1 | Tubb | Pgk1 | Hsp90ab1 | Hsp90ab1 | Tbp | Ppia |
| 10 | nd | Hsp90ab1 | Pthlh | Gapdh | Pthlh | Pthlh | Tubb |
| 11 | nd | Ppia | B2m | Tubb | Tubb | Tubb | Pthlh |
nd-not determined
When compared to an in vitro infection model, investigation of tissues reveals much higher intergroup variation. For example Ppia is downregulated in acutely infected lung (figure 1B) as well as in LSEC (supplementary figure S1). Pthlh is down regulated in all organs except spleen. In general all tested reference genes seemed to be least regulated in the spleen following mCMV infection and most regulated in infected kidneys. The highest intergroup variation for salivary gland was detected with B2m, for liver with Tubb, for LSEC, kidney and lung with Ppia (supplementary figure S1).
When all infected tissue results were grouped and compared to all uninfected tissues (Figure 1C) there was not tremendous intergroup variation. One might conclude from this that CMV infection in general has a low impact on the evaluated reference genes. However when intragroup variations are evaluated, there are large differences, confirming that reference genes are expressed at diverse levels depending on tissue and cell type. Together, these results emphasize the importance of selecting tissue and cell specific reference genes for studies of CMV infection.
4. Discussion
This study describes ideal reference genes for RT-qPCR after Smith strain mCMV infection for several different experimental conditions. We found that there is significant variability in reference gene expression between in vitro cell culture and in vivo tissue infections. Further, we found that within the same host, different tissues vary in reference gene expression stability after viral infection. Finally, our LSEC experiments show that whole organ results are likely a sum of parts, and that what works well for the entire organ might not be transferable to a single cell type of the same organ.
Several analyses have been conducted revealing appropriate reference genes for human virus infections including herpesviruses (Neerukonda et al., 2016; Radonic et al., 2005; Watson et al., 2007). However, for the most commonly used in vivo model, the mouse, there is no study available describing suitable reference genes after mCMV infection. To give a broad overview we investigated the most frequently analyzed tissue types after mCMV infection and used several programs to determine the most suitable reference genes. Tissues studied included lung, salivary gland, liver, spleen and kidney. To discriminate between reference gene expression in whole tissue composed of several cell subsets and an individual cell type, we compared whole livers to LSECs, known to be permissive for mCMV during acute and latent infection. We studied NIH-3T3 cells because they are the most common cell type used for mCMV in vitro studies. Four independent methods were chosen to evaluate the reference genes: Normfinder, BestKeeper, GeNorm and the comparative delta CT method(Andersen et al., 2004; Livak and Schmittgen, 2001; Pfaffl et al., 2004; Silver et al., 2006; Vandesompele et al., 2002). We then applied RefFinder to these results to determine overall rankings and identify ideal reference genes for each condition. Briefly, RefFinder utilizes results from the above mentioned programs, assigning weights for individual reference genes then calculating an overall ranking using geometric means of individual weights (Xie et al., 2012).
Interestingly, stability values obtained by the tools did not differ very much among each other. For in vitro experiments similar ranking hierarches were obtained using the four different methods. We found Ppia, B2m and Gapdh to be the most stable reference genes for in vitro infection studies of NIH-3T3 cells using mCMV strain Smith. This is consistent with data published by Watson et al. from investigations with human CMV strains Towne and Toledo in HFF cell cultures. They described Ppia and Gapdh among their top three most stable genes. Interestingly, they also obtained similar results for VZV, indicating potential similarity in reference gene suitability for herpes viruses in general. Another study in MRC-5 cells infected by human CMV strain AD169 also confirmed Ppia as a recommended reference gene (Radonic et al., 2005).
In contrast to the in vitro results, our in vivo results demonstrate the importance of reevaluating reference genes for specific experimental conditions. We found Actb, Tbp and Hprt to be reference genes of choice for in vivo studies in the lung and kidneys. The most suitable reference genes for spleen tissue are Actb, B2m and Hprt. When analyzing salivary glands, Gusb, Tubb and Tbp appear to be the best options. Finally, for liver and LSEC studies Gapdh and Gusb are the most stable genes, with Tubb suitable for LSEC analysis and Ppia recommended for whole liver. Table 5 summarizes rankings for all tested tissues and cells.
The Normfinder tool allowed us to group samples and evaluate inter- and intragroup variations of infected versus uninfected specimens. High intergroup variation is indicative of gene regulation caused by virus infection. Despite being the best choice for in vitro studies, Ppia is regulated after infection in lung, kidney and LSEC and is therefore not recommended as a reference gene in these cell/tissue types. Pthlh seems regulated upon virus infection in most tissues. In general reference genes seemed to be more regulated in the kidney, whereas the spleen was least affected by intergroup variations. Given the differences observed between LSEC and whole liver reference genes, we suspect that intergroup variations of reference gene expression is a consequence of the differential regulation of the component cell types composing each tissue.
5. Conclusions
In summary, we present ideal reference genes for varying tissues following mCMV infection. Our results emphasize the importance of experimental condition-dependent fluctuations in reference gene expression after infections. Because our tissue specific data were obtained using Balb/cJ mice and the mCMV strain Smith, any changes in mouse strain, virus strain or tissue of interest may require revalidation of the reference genes of choice.
Supplementary Material
J Virol Methods Highlights.
In this manuscript we evaluate the influence of viral infection on host reference genes commonly used for PCR and RT-PCR studies. We demonstrate variations between ideal reference genes between tissue culture and most of the commonly studied tissues, and make recommendations for ideal reference genes for each of these conditions.
Acknowledgments
This work was funded by NIH grant R01 GM066115 ‘Bacterial Sepsis and Reactivation of Latent Cytomegalovirus’
Footnotes
Conflicts of Interest: The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Author Contributions: MG, MG, and CHC conceived and designed the experiments; MG & MG performed the experiments; MG, MG, & CHC. analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–95. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Waldenstrom J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE. 2015;10:e0141853. doi: 10.1371/journal.pone.0141853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C, Zhang X, McGuinness B, Lahm M, Sedmak D, Ferguson R. Intra-abdominal Bacterial Infection Reactivates Latent Pulmonary Cytomegalovirus in Immunocompetent Mice. J Infect Dis. 2002;185:1395–1400. doi: 10.1086/340508. [DOI] [PubMed] [Google Scholar]
- Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–30. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Glare EM, Divjak M, Bailey MJ, Walters EH. beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–70. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Vandesompele J. Selection of reliable reference genes for RT-qPCR analysis. Springer; New York: 2014. [DOI] [PubMed] [Google Scholar]
- Invitrogen. REAL-TIME PCR: FROM THEORY TO PRACTICE. Invitrogen; 2008. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matsuzawa H, Shimizu K, Okada K, Ando K, Hashimoto K, Koga Y. Analysis of target organs for the latency of murine cytomegalovirus DNA using specific pathogen free and germfree mice. Arch Virol. 1995;140:853–64. doi: 10.1007/BF01314962. [DOI] [PubMed] [Google Scholar]
- Neerukonda SN, Katneni UK, Golovan S, Parcells MS. Evaluation and validation of reference gene stability during Marek’s disease virus (MDV) infection. J Virol Methods. 2016;236:111–6. doi: 10.1016/j.jviromet.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Radonic A, Thulke S, Bae HG, Muller MA, Siegert W, Nitsche A. Reference gene selection for quantitative real-time PCR analysis in virus infected cells: SARS corona virus, Yellow fever virus, Human Herpesvirus-6, Camelpox virus and Cytomegalovirus infections. Virol J. 2005;2:7. doi: 10.1186/1743-422X-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckert CK, Renzaho A, Tervo H-M, Krause C, Deegen P, Kuhnapfel B, Reddehase MJ, Grzimek NKA. Liver Sinusoidal Endothelial Cells Are a Site of Murine Cytomegalovirus Latency and Reactivation. J Virol. 2009;83:8869–8884. doi: 10.1128/JVI.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selgrade MK, Collier AM, Saxton L, Daniels MJ, Graham JA. Comparison of the pathogenesis of murine cytomegalovirus in lung and liver following intraperitoneal or intratracheal infection. J Gen Virol. 1984;65:515–23. doi: 10.1099/0022-1317-65-3-515. [DOI] [PubMed] [Google Scholar]
- Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. Beta-actin--an unsuitable internal control for RT-PCR. Mol Cell Probes. 2001;15:307–11. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- Shanley JD, Pesanti EL. The Relation of Viral Replication to Interstitial Pneumonitis in Murine Cytomegalovirus Lung Infection. J Infect Dis. 1985;151:454–458. doi: 10.1093/infdis/151.3.454. [DOI] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–7. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S, Mercier S, Bye C, Wilkinson J, Cunningham AL, Harman AN. Determination of suitable housekeeping genes for normalisation of quantitative real time PCR analysis of cells infected with human immunodeficiency virus and herpes viruses. Virol J. 2007;4:130. doi: 10.1186/1743-422X-4-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Yuhasz SA, Dissette VB, Cook ML, Stevens JG. Murine cytomegalovirus is present in both chronic active and latent states in persistently infected mice. Virology. 1994;202:272–80. doi: 10.1006/viro.1994.1343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.