Abstract
Objective:
To analyze the physical manifestations and genetic features of 2 families segregating X-linked deafness, which is most commonly reported to be caused by mutations of the POU domain gene POU3F4 at the DFN3 locus.
Design:
Computed tomographic study of the temporal bone in probands from each family, followed by mutation screening and deletion mapping of POU3F4 in family members.
Setting:
Two midwestern genetics clinics.
Participants:
Two families with X-linked deafness.
Main Outcome Measures:
Anomalies of the inner ear in the probands; results of gene mapping and severity and effects of hearing loss in the family members.
Results:
In the first family, a large deletion was identified that includes POU3F4 and extends upstream approximately 530 kilobases; in the second family, a novel serine-to-leucine (S228L) amino acid mutation was identified in the POU-specific domain of POU3F4. Both the deletion and the missense mutation segregate with the clinical phenotype and are causally related to the deafness in these families.
Conclusions:
Deafness related to the POU3F4 gene is associated with dilation of the internal auditor canal and a spectrum of other temporal bone anomalies that range in severity from mild to severe dysplasia of the cochlea and semicircular canals. The consequence of these anomalies is a congenital mixed hearing loss, the sensorineural component of which progresses over time. Affected males can also present with vestibular dysfunction that is associated with delayed developmental motor milestones. Intrafamilial variability occurs.
CONGENITAL DEAFNESS IS diagnosed in approximately 1 in every 1000 newborns, half of whom have inherited deafness that is most often autosomal recessive and nonsyndromic. The fractional contribution of X-linked deafness is estimated at 2%, which implies that X-linked deafness affects roughly 1 in every 50 000 newborn males. The Hereditary Hearing Loss Homepage1 lists 6 X-linked nonsyndromic deafness loci (DFN2, DFN3, DFN4, DFN5, DFN6, and DFN8) and provides mapping data for 4 (DFN2, DFN3, DFN4, and DNF6), but in only 1 instance has the deafness-related gene been identified. Deafness at the DFN3 locus is caused by mutations in the coding sequence or deletions upstream of POU3F4.
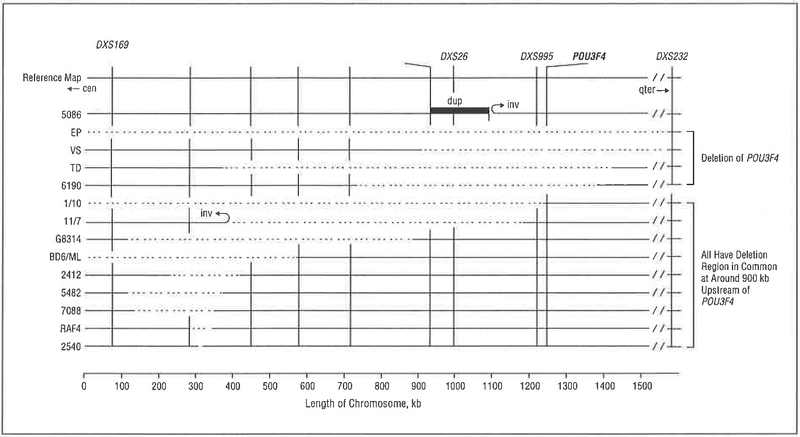
The gene POU3F4 (POU domain, class III, transcriptional factor 4) encodes a transcription factor with bipartite DNA-binding domains, a 75-amino acid POU-specific domain that is linked by 17 amino acids to a 63-amino acid homeobox domain. The latter is common among transcription factors and recognizes specific DNA sequence motifs while the former is unique to the POU gene family and strengthens the binding affinity to DNA. All POU3F4 deafness-causing missense mutations localize to 1 of these 2 DNA-binding domains while deletions affect a putative regulatory element located atleast400 kilobases (kb) upstream of the gene2,3 (Table 1 and Figure 1). In aggregate, these mutations account for about 40% of X-linked deafness.
Table 1.
Mutations of P0U3F4 Associated With DFN3 Deafness
| POU-Specific Domain | PQU Homeodomain | ||
|---|---|---|---|
| Source | Mutation | Source | Mutation |
| Cremers et al,4 2000 | Phe201/Lys202 del | Bitner-Glindzicz et al,5 1995 | 862-866 del |
| De Kok et al,6 1995 | Lys202Stop | Cremers et al,4 2000 | Glu286Stop |
| De Kok et al,6 1995 | Asp215Stop | De Kok et al,6 1995 | Leu298Stop |
| Present report | Ser228Leu | Cremers et al,4 2000 | Pro303Ser |
| Friedman et al,7 1997 | Thr230lle | Bitner-Glindzicz et al,5 1995 | Ala312Val |
| De Kok et al,6 1995 | Leu317Trp | ||
| De Kok et al,8 1997 | Arg323Gly | ||
| Cremers et al,4 2000 | Asn328Thr | ||
| Friedman et al,7 1997 | Arg329Gly | ||
| De Kok et al,2 1995 | Arg330Ser | ||
| De Kok et al,3 1996 | Lys334Glu | ||
Abbreviations: del, deletion; POU3F4, POU domain gene.
Figure 1.
Diagram of deletion associated with POU3F4-related deafness. The listing along the left side is proband identification; the designations along the top are short tandem repeat polymorphisms, cen Indicates centromere; dup, duplication; inv, inversion; qter, long-arm terminal; and stippled lines, deletions. Modified with permission from De Kok et al.3 Copyright 1996, Oxford University Press.
Although the genes regulated by POU3F4 are unknown, the phenotypic consequence of absence of a functional POU3F4 protein is a range of temporal bone anomalies that make thin-cut computed tomography (CT) of the temporal bones an important diagnostic tool to determine whether mutation screening of POU3F4 is warranted in a male with presumed nonsyndromic deafness. The observed defects of the bony labyrinth are often classified as pseudo–Mondini stage II dysplasia.9 Features include partial hypoplasia of the cochlea and dilation of the internal auditory canal, often with a fistulous communication between its lateral aspect and the basal turn of the cochlea.10 Outward pressure of perilymphatic fluid on the oval window from within the vestibule hinders stapedial vibration,11 and anomalies of the oval window and stapedial footplate also compromise ossicular chain mobility.12 In aggregate, these anomalies result in a conductive component to POU3F4-related deafness. With time, the sensorineural component of the hearing loss progresses as a reflection of ongoing inner ear damage. Stapedectomy is contraindicated because the uncontrolled leak of perilympha (a perilymphatic “gusher”) can severely compromise residual hearing and vestibular function.
In this article, we describe 2 extended families segregating X-linked deafness. Temporal bone CT of the proband from each family showed numerous defects of the bony labyrinth, raising the possibility of mutations in POU3F4. Mutation screening and deletion mapping of this gene were completed, confirming the diagnosis of deafness at the DFN3 locus. The clinical presentation of POU3F4-related deafness is reviewed as an aid to recognizing this type of nonsyndromic hearing loss.
METHODS
SUBJECTS
The 2 families in this study were ascertained through genetics clinics at Southern Illinois University School of Medicine, Springfield (family 6190), and Children’s Mercy Hospitals and Clinics, Kansas City, Mo (family 6270) (Figure 2 and Figure 3, respectively). Blood samples from probands were sent to the Molecular Otolaryngology Research Laboratories at The University of Iowa, Iowa City, for mutation screening. Family histories were obtained by means of a questionnaire and personal interviews. A complete physical examination, including audiometry, was performed on each proband. Audiograms also were obtained from additional family members interested in participating in this study. All persons who wished to participate in the study provided a blood sample. The University of Iowa Hum an Subjects Committee approved all procedures.
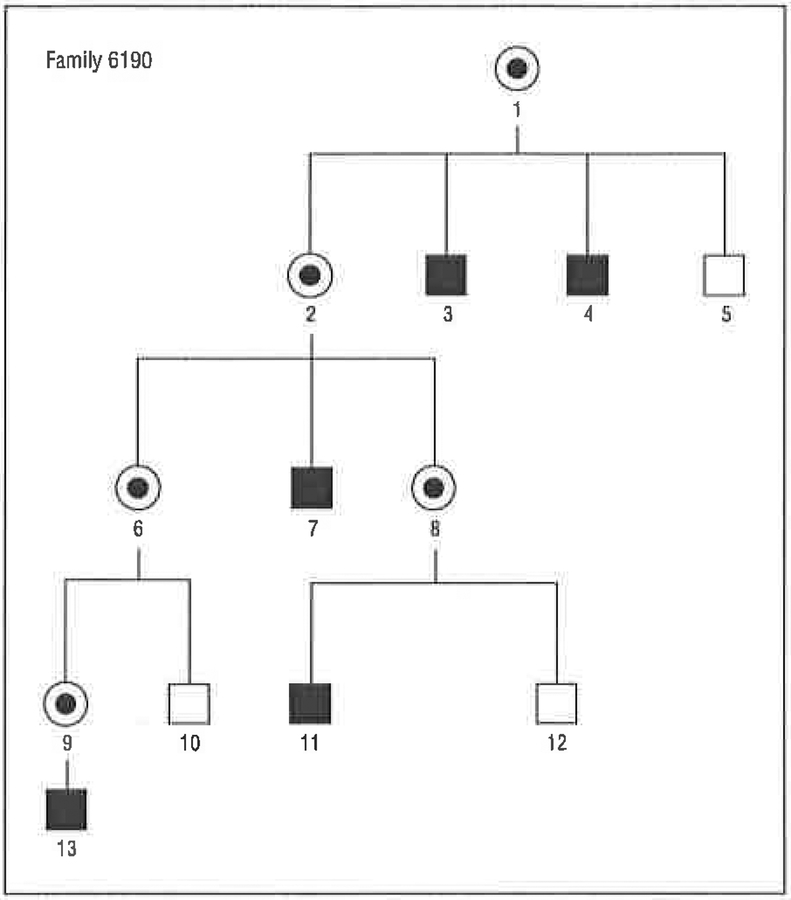
Figure 2.
Pedigree of family 6190, showing an X-linked pattern of segregation of hearing loss. Note that only males (squares) are affected and that there is no male-to-male transmission of the affected phenotype. Circles indicate females; fully shaded squares, individuals with hearing loss; and the central dot in the female symbol, obligate carrier of the affected X chromosome.
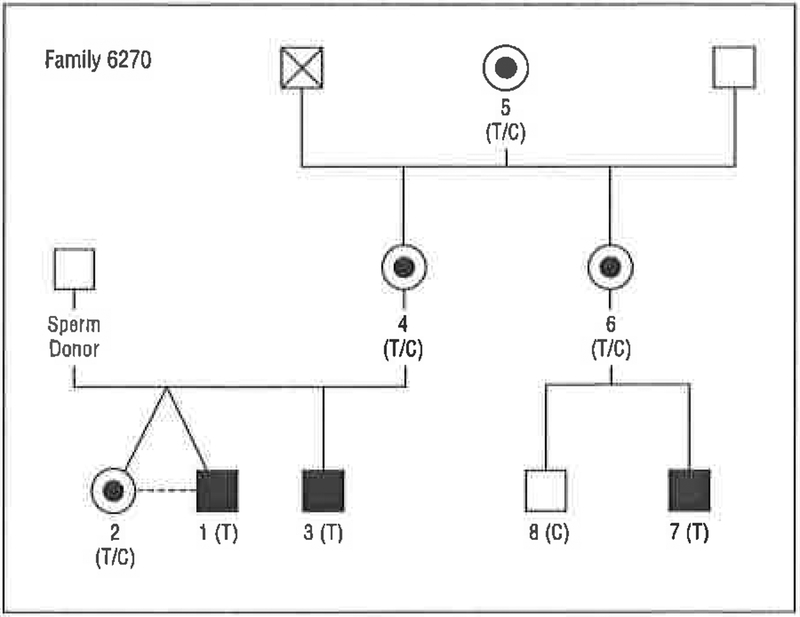
Figure 3.
Pedigree of family 6270, showing an X-linked pattern of segregation of hearing loss. The nucleotide mutation is a transition of cytosine to thymine. The result is a codon change from serine to leucine at position 228 (S228L). T indicates thymine; C, cytosine; squares, males; circles, females; fully shaded squares, individuals with hearing loss; and the central dot in the female symbol, obligate carrier of the affected X chromosome.
PREPARATION OF DNA
Samples of DNA were extracted from peripheral whole blood (approximately 10 mL) following established procedures.13 Each polymerase chain reaction (PCR) was performed in a total volume of 10 μL with 1 μL of 10× buffer, 0.33 μL of magnesium chloride, 0.15 μL of primer A, 0.15 μL of primer B, 0.4 μL of deoxynucleotide triphosphate, 0.04 μL of Taq polymerase, 1.5 μL of 20-ng/μL DNA, and 6.43 μL of water, hot-starting the reaction for 3 minutes at 95°C, followed by 35 cycles of denaturation for 30 seconds at 94°C, annealing for 30 seconds at 66°C, and extension for 30 seconds at 72°C, culminating in a 10-minute extension at 72°C after PCR.
DELETION MAPPING IN FAMILY 6190
Short tandem repeat polymorphisms were used to identify the approximate size of the deletion. Breakpoint mapping was completed using 29 primer pairs upstream of POU3F4, which generated amplimers spaced across 57 kb of genomic DNA at roughly equal intervals (a list of the primer pairs is available from the authors on request). Polymerase chain reaction was carried out as described in the preceding section, and products were resolved by means of gel electrophoresis and scored as present or absent. All PCRs were repeated at least twice.
MUTATION SCREENING IN FAMILY 6270
Mutation screening of the coding region of POU3F4 was completed using the primers listed in Table 2. The PCR products were resolved by means of gel electrophoresis to ensure product amplification and sequenced using an automated sequencer (Applied Biosystems Model 3700; Applied Biosystems, Inc, Foster City, Calif). Data were analyzed using the Sequencer 4.2 program package (Gene Codes, Inc, Ann Arbor, Mich).
Table 2.
Primers Used to Amplify and Sequence the Coding Regions of P0U3F4
| Primer | Sequence | Annealing Temperature, °C |
Product Size, bp |
|---|---|---|---|
| P0U1 | |||
| Forward | ACTAGTAGGGGATCCTGACCG | 66 | 242 |
| Reverse | GTGGCCAGTGTGGAGGAC | 66 | 242 |
| P0U2 | |||
| Forward | TCACTGGGGGACCAGTCTGA | 66 | 239 |
| Reverse | AGTACACGTTGAGGGGTTGG | 66 | 239 |
| POU3 | |||
| Forward | CACTCACCGCACACTMCCA | 57 | 400 |
| Reverse | GMCCTGCAGATGGTGGTCT | 57 | 400 |
| POU3.5 | |||
| Forward | GATCACTCCGACGAGGAGAC | 57 | 230 |
| Reverse | CCTCCAGCCACTTGTTCAGC | 57 | 230 |
| POU4 | |||
| Forward | GATCACTCCGACGAGGAGAC | 66 | 277 |
| Reverse | CACGATCTTGTCMTGCTG | 66 | 277 |
| POU5 | |||
| Forward | TGAACAAGTGGCTGGAGG | 66 | 263 |
| Reverse | TGGCGGAGTCATTCTTTTCT | 66 | 263 |
| POU6 | |||
| Forward | GAAGGMGTGGTGCGTGTCT | 66 | 265 |
| Reverse | CGAGCGMCGACAGAGAGAG | 66 | 265 |
Abbreviations: bp, base pairs P0U3F4, POU domain gene.
RESULTS
FAMILY 6190
The probandin family 6190 (6190–13) was diagnosed as having congenital severe-to-profound hearing loss after auditory brainstem response testing (ABR) at 1 month of age; delayed latencies also suggested a conductive component to the loss. A temporal bone CT scan obtained 1 month later was consistent with incomplete partitioning of the cochlea and interpreted as pseudo-Mondini stage II dysplasia (Figure 4). Visual reinforcement audiometry at age 7 months gave fairly reliable responses, suggesting a moderate-to-severe hearing loss downsloping from 55 dB in the low frequencies to 80 dB in the high frequencies (Figure 5A).
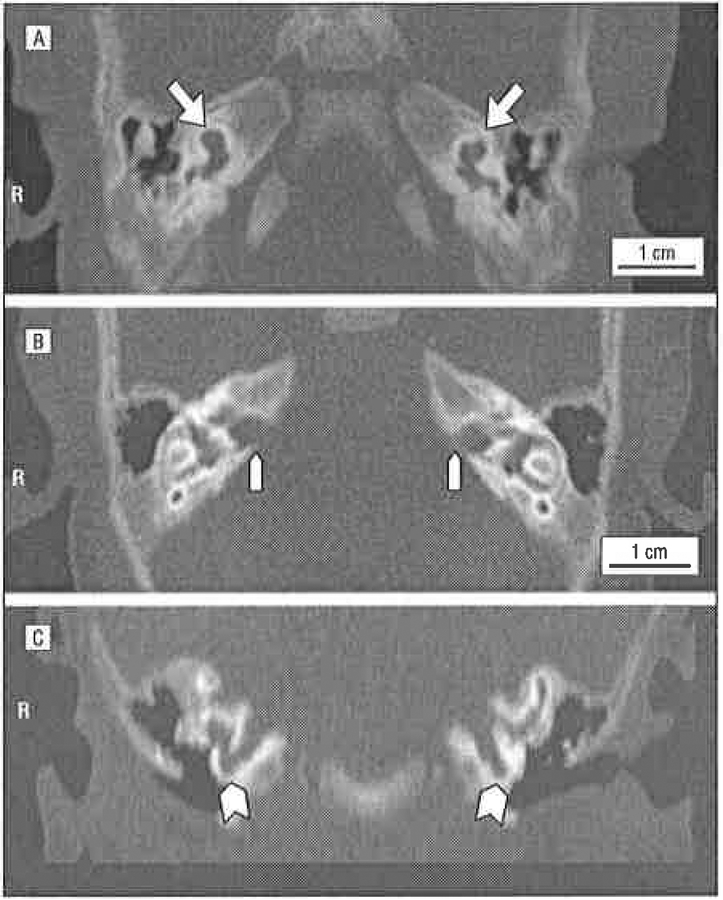
Figura 4.
Temporal bone computed tomographic scans of proband 6190–13 demonstrating the pseudo-Mondini stage II dysplasia with cochlear hypoplasia (A, arrows, axial section), dilation of the internal auditory canal (IAC) (B, arrowheads, axial section), and a fistula between the IAC and the cochlea (C, arrowheads, coronal section). R indicates right ear.
Figure 5.
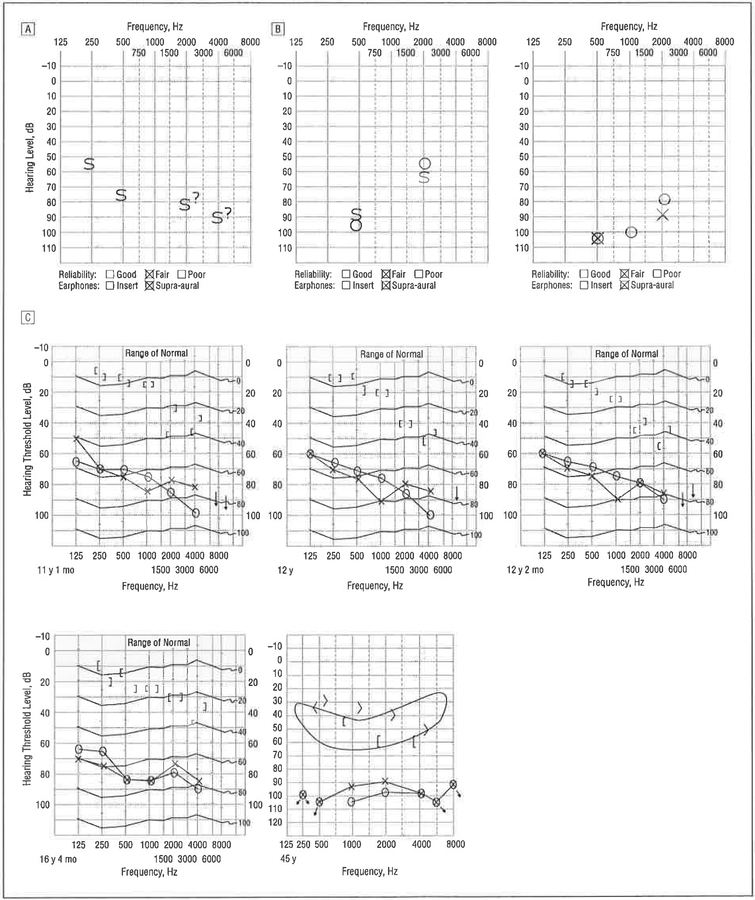
Documentation of hearing loss in family 6190. A, Audiogram from family member 6190–13 obtained at age 7 months using visual reinforcement^ audiometry. Responses were of fair reliability; warble tones and bursts of narrow-band noise revealed threshold responses from 55 dB in the low frequencies to 80 dB in the high frequencies. B, Play audiometry performed in family member 6190–11 illustrates progression of hearing loss and the characteristic upsloping pattern. The audiogram on the left was obtained at 4 years of age; the one on the right, 9 months later. C, Audiograms from family member 6190–7 show progression of hearing loss. S indicates response in sound fields, not ear specific; 0, right ear, X, left ear; </>, right ear/left ear unmasked tone conduction; [/], right ear/left ear masked tone conduction; ?, uncertain response.
The strong family history of deafness was consistent with X-linked inheritance (Figure 2). A review of the medical reports indicated that the proband’s great-great-uncle, 6190–3, had experienced a perilymphatic gusher while undergoing surgery. The proband’s first cousin once removed, 6190–11, was born at full term and failed his newborn hearing screen. Follow-up ABR at 1 month of age showed a bilateral absence of waveforms consistent with at least a severe hearing loss in both ears, and ABR results at 18 months of age were consistent with moderate-to-severe hearing loss. By age 4 years, hearing thresholds were recorded at 95 and 55 dB at 500 and 2000 Hz, respectively (Figure 5B).
The history of the proband’s great-uncle, 6190–7, is particularly instructive. He was born with hearing loss, which was diagnosed as a severe mixed loss. At 6 years of age, he underwent an exploratory tympanotomy to correct the conductive component of his hearing loss. The attempted stapedectomy resulted in a perilymphatic gusher and, although his hearing deteriorated in the operated-on ear, hearing in the contralateral ear remained stable (Figure 5C). Computed tomography was never performed.
Female members of family 6190 did not have congenital hearing loss, although family members 6190–1 and 6190–2 both developed presbycusis in their seventh decade of life. Patient 6190–1 had interm ittent difficulties with balance and vertigo.
FAMILY 6270
The proband in family 6270 (6270–3) presented with a history of chronic otitis media necessitating an adenotonsillectomy and myringotomies with the placement of pressure-equalizing tubes. Temporal bone CT at age 22 months showed bilateral dilation of the internal auditory canals and cochlear hypoplasia; the vestibule and semicircular canals appeared normal. Auditory brainstem response testing results at 2 years of age were consistent with a symmetrical, moderate low-frequency hearing loss that rose to a mild hearing loss in the mid to high frequencies. The thresholds to click stimuli were 50 and 45 dB in the right and left ears, respectively. Auditory steady-state responses indicated thresholds of 50 and 60 dB, and of 70 and 60 dB at 2000 and 4000 Hz in the right and left ears, respectively. Developmental motor milestones were slightly delayed, with sitting at age 7 to 8 months, crawling at 10 months, and walking at 16 months.
Family member 6270–1, the fraternal twin of 6270–2 and younger brother of 6270–3, was diagnosed as having bilateral mild-to-moderate sensorineural hearing loss at 11 months of age after ABR. Responses to click stimuli were 35 dB in the right ear and 45 dB in the left ear, with responses to 500-Hz tone bursts at 48 dB in both ears. Developmental motor milestones were delayed secondary to presumed vestibular dysfunction, with crawling first noted at age 11 months. By age 20 months, walking was still not possible. Temporal bone CT was not performed.
Family member 6270–7, the 6-year-old affected maternal cousin, had bilateral profound sensorineural hearing loss; developmental motor milestones were reported to be normal. The brother of 6270–7, 6270–8, had normal hearing.
MUTATION SCREENING
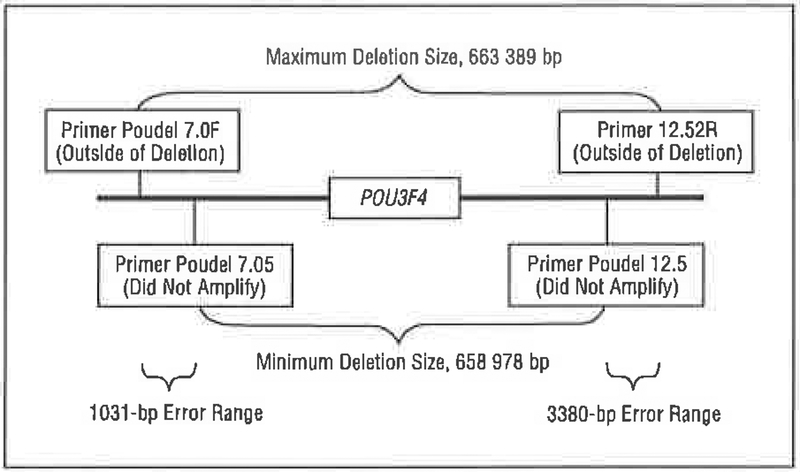
Family 6190 segregates a large deletion that includes POU3F4 and spans a region of approximately 650 kb. The deletion breakpoints are approximately 530 kb up stream and approximately 127 kb downstream of POU3F4 (Figure 6).
Figure 6.
The deletion segregating in family 6190 includes the entire POU3F4 coding interval but does not extend 5’ to the putative regulatory region, bp indicates base pairs. Primer information is available from the authors on request.
Family 6270 segregates a cytosine-to-thymine transition that segregates with the deafness phenotype and predicts a novel missense mutation, S228L (Figure 3). This mutation lies within the POU-specific domain and generates a ConSeq score of 9 (range, 1 [low] to 9 [high]) (http://conseq.bioinfo.tau.ac.il/), consistent with a highly conserved and functionally important amino acid residue.14
COMMENT
Families 6270 and 6190 contribute to our understanding of DFN3 by presenting a novel missense mutation within the POU-specific domain of POU3F4 and a large deletion that includes the entire gene. From the clinical information these cases provide and the available reports of deafness at the DFN3 locus, there appears to be no obvious genotype-phenotype relation associated with POU3F4- related deafness with respect to defects of the bony labyrinth, vestibular dysfunction, the degree of sensorineural hearing loss, and the progression of hearing loss.
To date, 6 truncating and 11 nontruncating mutations have been reported in association with POU3F4-related deafness (Table 3). Included in the truncating groups are 5 nonsense mutations and a small deletion of 4 nucleotides that results in a frameshift mutation. The nontruncating mutations include 10 missense mutations and an in-frame deletion of 2 amino acids. With many diseases, including deafness caused by mutations in the genes GJB2 and SLC26A4, phenotypic variability is seen when truncating and nontruncating mutations are compared, reflecting the presence of a translated protein with the latter group of mutations, which may have some, albeit minimal, residual activity. However, from the available clinical descriptions of POU3F4-related deafness, it is clear that no phenotypic-genotypic effect exists (Table 4).
Table 3.
Truncating and Nontruncating Mutations Associated With P0U3F4-Related Deafness
| Truncating | Nontruncating | ||||||
|---|---|---|---|---|---|---|---|
| Nonsense Mutations | Deletions | Mlssense Mutatlons | Deletions | ||||
| Source | Mutation | Source | Deletion | Source | Mutation | Source | Deletion |
| Cremers et al,4 2000 | Pro66Stop | Bitner-Glindzicz et al,5 1995 | 862–866 del | Present report | Ser228Leu | Cremers et al,4 2000 | Phe201/Lys202 |
| De Kok et al,6 1995 | Lys202Stop | Friedman et al,7 1937 | Thr230lle | ||||
| De Kok et al,6 1995 | Asp215Stop | Cremers et al,4 2000 | Pro303Ser | ||||
| Cremers et al,4 2000 | Glu286Stop | Bitner-Glindzicz et al,5 1995 | Ala312Val | ||||
| De Kok et al,6 1995 | Leu298Stop | De Kok et al,6 1995 | Leu317Trp | ||||
| De Kok et al,8 1997 | Arg323Gly | ||||||
| Cremers et al,4 | Asn328Thr | ||||||
| Friedman et al,7 1937 | Arg329Gly | ||||||
| De Kok et al,2 1995 | Arg330Ser | ||||||
| De Kok et al,3 1996 | Lys334Glu | ||||||
Abbreviations: bp, base pairs; P0U3F4, POU domain gene.
Table 4.
Phenotypes Seen With P0U3F4-Related Deafness
| Source | Mutation | Spectrum of Phenotype |
|---|---|---|
| De Kok et al,6 1995 | Asp215Stop | Mixed hearing loss; perilymphatic gusher; bony defect observed on CT |
| De Kok et al,6 1995 | Lys202Stop | Sensorineural hearing loss; bony defect observed on CT; some maternal relatives with cataracts at a young age |
| Friedman et al,7 1997 | Thr230lle | Severe mixed hearing loss; bony defect observed on CT |
| De Kok et at,6 1995 | Leu298Stop | Mixed hearing loss; perilymphatic gusher |
| De Kok et al,6 1995 | Leu317Trp | Mixed hearing loss; bony defect observed on CT |
| De Kok et al,8 1997 | Arg323Gly | Mixed hearing loss; perilymphatic gusher; polytomographic scans of temporal bones interpreted as normal |
| Friedman et al,7 1997 | Arg329Gly | Severe mixed hearing loss; perilymphatic gusher; sensorineural hearing loss in mother and maternal grandmother beginning in sixth decade of life |
| De Kok et al,2 1995 | Arg330Ser | Progressive hearing loss; obesity; growth retardation; intermittent diarrhea; perilymphatic gusher; polytomgraphic scans show dilation of semicircular canals and lateral IAC |
| De Kok et al,3 1996 | Lys334Glu | Progressive mixed hearing loss |
Abbreviations: CT, computed tomography; IAC, internal auditory canal. POU3F4, POU domain gene.
The S228L change we identified in family 6270 is only the second POU-specific domain mutation reported thus far. In this family, intrafamilial variability in temporal bone anomalies and hearing thresholds is clearly evident. Although family members 6270–1 and 6270–3 have yet to experience progression of hearing loss, it is likely that both children will become profoundly deaf. This bleak prognosis reflects the observation made by Cremers and colleagues4 that while the rate of progression of hearing loss can vary among individuals, all persons with POU3F4- related deafness ultimately have profound deafness at varying ages. The 3 affected males in family 6270 also have variable degrees of vestibular dysfunction.
The DFN3-related deafness in family 6190 is due to the deletion of the POU3F4 gene itself. Based on the 5’ breakpoint location, the regulatory region of POU3F4 is unlikely to be affected (Figure 6). Other families have been described that also segregate deletions of POU3F4; however, in most families in which a deletion is found, composite deletion mapping localizes a region approximately 900 kb upstream of POU3F4 as the site of a putative regulatory element.7 In family 6190, the presence of a temporal bone CT scan for only 1 person precludes any comparisons; however, there is a large difference in the audiograms obtained from family members 6190–7, 6190–11, and 6190–13 (Figure 5).
CONCLUSIONS
Deafness at the DFN3 locus is caused by mutations in POU3F4 or deletions that impact its transcription by eliminating either the coding region or an upsteam regulatory region. There are no clear differences in the phenol-type associated with these different types of mutations. In general, POU3F4-related deafness is mixed and invariably progressive. Vestibular dysfunction associated with delayed developmental motor milestones is not uncommon. Temporal bone CT often shows abnormalities of the bony labyrinth and internal auditory canals. These findings in a male should raise suspicion for POU3F4- related deafness and, if the pedigree is consistent with X-linked inheritance, genetic testing can be considered. If a mutation in POU3F4 is identified, the chance for the couple to have an additional affected child is 25%, and their chance for having another affected male is 50%.
Acknowledgment:
We are grateful to the families who made this research possible.
Funding/Support: This work was supported in part by grant R01 DC03544 from the National Institute on Deafness and Other Communication Disorders, Bethesda, Md (Dr Smith).
Footnotes
Financial Disclosure: None.
REFERENCES
- 1.Camp GV, Smith RJ. Hereditary Hearing Loss Homepage. Available at: http://webhost.ua.ac.be/hhh/. Accessed October 4, 2005.
- 2.De Kok YJM, Merkx GFM, van der Maarel SM, et al. A duplication/paracentric inversion associated with familial X-linked deafness (DFN3) suggests the presence of a regulatory element more than 400 kb upstream of the P0U3F4 gene. Hum Mol Genet. 1995;4:2145–2150. [DOI] [PubMed] [Google Scholar]
- 3.De Kok YJM, Vossenaar ER, Cremers CWRJ, et al. Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene P0U3F4. Hum Mol Genet. 1996;5:1229–1235. [DOI] [PubMed] [Google Scholar]
- 4.Cremers FPM, Cremers CWRJ, Ropers HH. The ins and outs of X-linked deafness type 3. Adv Otorhinolaryngol. 2000;56:184–195. [DOI] [PubMed] [Google Scholar]
- 5.Bitner-Glindzicz M, Turnpenny P, Hoglund P, et al. Further mutations in Brain 4 (P0U3F4) clarify the phenotype in the X-linked deafness, DFN3. Hum Mol Genet. 1995;4:1467–1469. [DOI] [PubMed] [Google Scholar]
- 6.De Kok YJM, van der Maarel SM, Bitner-Glindzicz M, et al. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–683. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RA, Bykhovskaya Y, Tu G, et al. Molecular analysis of the POU3F4 gene in patients with clinical and radiographic evidence of X-linked mixed deafness with perilymphatic gusher. Ann Otol Rhino! Laryngol. 1997;106:320–325. [DOI] [PubMed] [Google Scholar]
- 8.De Kok YJM, Cremers CWRJ, Ropers H-H, Cremers FPM. The molecular basis of X-linked deafness type 3 (DFN3) in two sporadic cases: identification of a somatic mosaicism for a POU3F4 missense mutation. Hum Mutat. 1997;10: 207–211. [DOI] [PubMed] [Google Scholar]
- 9.Phippard D, Heydemann A, Lechner M, et al. Changes in the subcellular localization of the Brn4 gene product precede mesenchymal remodeling of the otic capsule. Hear Res. 1998;120:77–85. [DOI] [PubMed] [Google Scholar]
- 10.Jensen J, Terkildsen K, Thomsen KA. Inner ear malformations with otoliquorrhea: tomographic findings in three cases with a mixed hearing impairment. Arch Otorhinolaryngol. 1977;214:271–282. [DOI] [PubMed] [Google Scholar]
- 11.Cremers CWRJ, Hombergen GCJH, Wentges RTR. Perilymphatic gusher and stapes surgery: a predictable complication? Clin Otolaryngol. 1983;8:235–240. [DOI] [PubMed] [Google Scholar]
- 12.Samadi DS, Sauders JC, Crenshaw EB III. Mutation of the POU-domain gene Brn4/Pou3f4 affects middle-ear sound conduction in the mouse. Hear Res. 2005; 199:11–12. [DOI] [PubMed] [Google Scholar]
- 13.Grimberg J, Nawoschik S, Belluscio L, McKee R.Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berezin C, Glaser F, Rosenberg J, et al. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 2004; 20:1322–1324. [DOI] [PubMed] [Google Scholar]