Abstract
Background
Cancer cells reprogram metabolism for proliferation. Phosphoglycerate kinase 1 (PGK1), as a glycolytic enzyme and newly identified protein kinase, coordinates glycolysis and mitochondrial metabolism. However, the clinical significance of PGK1 expression and function in cancer progression is unclear. Here, we investigated the relationship between the progression and prognosis of multiple cancer types and PGK1 expression and its function in the mitochondrial metabolism regulation.
Methods
We performed pan-cancer analyses of PGK1 mRNA level and DNA methylation in 11,908 tumor tissues and 1582 paired normal tissues across 34 cancer types in The Cancer Genome Atlas datasets. Using specific antibodies against PGK1 S203 and PDHK1 T338 phosphorylation, we performed immunohistochemistry with tissue microarray assay in additional 818 cancer cases with 619 paired normal tissues from five cancer types.
Results
The PGK1 mRNA level was significantly elevated with hypomethylation in promotor regions and associated with advanced TNM stage in 15 and four cancer types, respectively. In breast carcinoma, elevated PGK1 mRNA level and promoter hypomethylation were associated with poor prognosis. Positively correlated PGK1 S203 and PDHK1 T338 phosphorylation levels were significantly associated with short overall survival (OS) in cancers of the breast, liver, lung, stomach, and esophagus and with advanced TNM stage in breast and esophageal cancers. PGK1 pS203 and PDHK1 pT338 were also independent predictors of short OS in liver, lung, and stomach cancer.
Conclusions
The elevated expression, promoter hypomethylation, and phosphorylation of PGK1 and PDHK1 were related with disease progression and short OS in diverse types of cancer. PGK1 and PDHK1 phosphorylation may be potential prognostic biomarkers.
Keywords: PGK1, Cancer metabolism, Epigenetics, Phosphorylation, Methylation, The Cancer Genome Atlas, Pan-cancer analysis, Prognosis, Overall survival
Background
Most cancer cells, even in the presence of ample oxygen, predominantly generate adenosine triphosphate (ATP) by a high rate of glycolysis followed by lactate fermentation in the cytosol rather than by oxidation of pyruvate in the mitochondria, as in most normal cells. This phenomenon, known as aerobic glycolysis or the Warburg effect, facilitates tumor cell growth [1–5]. In the glycolytic pathway, phosphoglycerate kinase 1 (PGK1), the first enzyme to generate ATP, catalyzes the transfer of the high-energy phosphate from 1,3-diphosphoglycerate to adenosine diphosphate (ADP), leading to the generation of 3-phosphoglycerate and ATP. In Homo sapiens, PGK has two isozymes, ubiquitously expressed PGK1 and testis-expressed PGK2, both with 87%–88% identical amino acid sequence identity [6].
The reprogramming of metabolism is an emerging hallmark of cancer biology [4, 7, 8]. Recent studies have shown that the protein level of PGK1 was elevated in breast cancer [9], astrocytoma [10], metastatic colon cancer [11], and pancreatic ductal adenocarcinoma [12]; its mRNA levels were increased in gastric cancer [13].
Our previous studies [14] revealed that in tumor cells, PGK1 possesses protein kinase activity in addition to performing its well-established glycolytic function. In response to receptor tyrosine kinase activation, the expression of K-Ras G12V and B-Raf V600E, hypoxia, pyruvate metabolism in mitochondria is suppressed [14, 15]. This is primarily regulated by the mitochondrial translocation of PGK1, which is phosphorylated at S203 by extracellular signal-regulated kinase 1/2 (ERK1/2) and cis–trans isomerized by peptidyl-prolyl cis–trans isomerase NIMA-interacting 1 (PIN1), leading to exposure of the pre-sequence of PGK1 for binding to the translocase of the outer membrane (TOM) complex of mitochondria. In the mitochondria, PGK1 functions as a protein kinase to phosphorylate pyruvate dehydrogenase kinase 1 (PDHK1, also known as PDK1) at T338, which activates PDHK1 to phosphorylate and inhibit the pyruvate dehydrogenase (PDH) complex [14, 15]. Suppression of PDH activity reduces mitochondrial pyruvate utilization and reactive oxygen species production and increases lactate production, thereby promoting tumorigenesis. In addition, PGK1 S203 and PDHK1 T338 phosphorylation levels were found to be positively correlated with each other, and both were correlated with PDH S293 inactivating phosphorylation levels and poor prognosis in patients with glioblastoma (GBM) [14]. However, whether the newly identified protein kinase function of PGK1 applies to other cancer types and the relationship between PGK1 kinase activity and tumor progression remain unknown.
Here, we performed a pan-cancer analysis of clinical relevance of PGK1 using data from 11,908 cases (including 1582 with paired normal tissues) across 34 cancer types from The Cancer Genome Atlas (TCGA) datasets. We also analyzed the clinical relevance of PGK1 S203 and PDHK1 T338 phosphorylation levels by conducting immunohistochemical experiments in an additional 818 independent cancer cases (including 619 with paired normal tissues). We aimed to evaluate the pathological progression value and prognostic values of PGK1 mRNA high expression, PGK1 promoter methylation, and PGK1 mediated-PDHK1 activating phosphorylation in multiple human cancers.
Materials and methods
Data resource
We downloaded clinical records, RNAseqV2 level 3 gene level data, and DNA methylation level 3 data for 11,908 cases across 34 cancer types from TCGA (http://xena.ucsc.edu/welcome-to-ucsc-xena/). Profiling data of the TCGA-retrieved cases were generated using the Illumina HiSeq 2000 RNA Sequencing and Illumina Infinium Human Methylation 450 platforms, as described by the TCGA network [16, 17]. Gene transcription estimates for each gene were presented as in RNA-Seq using the Expectation Maximization (RSEM) software. DNA methylation values are presented as beta values for each CpG probe transformed into M values. The detailed information about data processing is provided in Additional file 1: Methods. A summary of the sample sizes for the PGK1 RNA-Seq and DNA methylation analyses for each cancer type is shown in Additional file 1: Table S1. There are 16 methylation probes that cover the PGK1 gene (Chromosome X; UCSC Gene Accession: NM_000291) (Additional file 1: Table S2).
The histopathologic diagnoses of the TCGA cases are available in the Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/).
PGK gene level data in 16 tissue types were downloaded from the Illumina Body Map Project (https://www.ebi.ac.uk/gxa/home), and the results are presented as transcripts per million (TPM) values.
Patients and tissue samples
We retrospectively collected surgically resected, formalin-fixed, paraffin-embedded tissue samples from the biobank of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital in Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). Tissue samples of 818 treatment-naïve patients who underwent surgery for pathologically diagnosed cancer between 2006 and 2015 were selected as an additional independent cohort, including 145 cases of breast carcinoma (BRCA) (with 69 paired normal specimens), 185 cases of liver hepatocellular carcinoma (LIHC) (with 174 paired normal specimens), 179 cases of lung adenocarcinoma (LUAD) (with 175 paired normal specimens), 95 cases of stomach adenocarcinoma (STAD) (with 55 paired normal specimens), and 214 cases of esophageal carcinoma (ESCA) (with 146 paired normal specimens).
Tissue microarray construction
Rabbit polyclonal antibodies recognizing phospho-PGK1 S203 and phospho-PDHK1 T338 were obtained from Signalway Antibody (College Park, MD, USA). The specificities of these antibodies were previously validated [14]. A rabbit monoclonal antibody recognizing IgG was purchased from Cell Signaling Technology (Danvers, MA, USA). Formalin-fixed, paraffin-embedded tissues were obtained by surgical resection, archived after clinical use for pathological diagnosis, and stained with Mayer’s haematoxylin and eosin (H&E; Biogenex Laboratories, San Ramon, CA, USA).
Tumor samples from the 818 cancer cases with 550 paired normal tissues were subjected to tissue microarray (TMA). Employing an automated tissue array instrument (Minicore® 3, Alphelys, Plaisir, France), cancer tissue (diameter at 2 mm, selected by a pathologist) from each specimen was extracted and fixed into a paraffin block. After quality control, the TMA blocks were sectioned into 3-μm-thick slides for immunohistochemistry analysis.
Immunohistochemistry
After deparaffinization, rehydration, and antigen-retrieval, TMA slides were incubated with primary rabbit anti-human phospho-PGK1 S203 (dilution 1:200; Signalway Antibody; SAB487P), primary rabbit anti-human phospho-PDHK1 T338 (dilution 1:500; Signalway Antibody; #11596), or nonspecific IgG (as a negative control) overnight at 4 °C. The slides were then incubated with anti-rabbit secondary antibody (ready-to-use solution; Cell Signaling Technology; #8114), followed by chromogen diaminobenzidine (DAB) staining (Cell Signaling Technology) and hematoxylin counter staining and mounted with xylene-based medium. We quantitatively scored the tissue slides under a microscope according to the percentage of positive cells and staining intensity. We assigned the following proportion scores: 0, 0% of cells being positive; 1, 0% to 1%; 2, 2% to 10%; 3, 11% to 30%; 4, 31% to 70%; and 5, 71% to 100%. We also rated the staining intensity on a scale of 0 to 3: 0, negative; 1, weak; 2, moderate; and 3, strong. The proportion and intensity scores were then combined by addition to obtain a total score (range 0–8), as described previously [18]. Two pathologists (X.F. and S.S.), who were blinded to the clinical information, independently validated the reproducibility of the scoring system.
Statistical analysis
SPSS version 20.0 software (SPSS Inc., Chicago, IL, USA) was used for data analysis. PGK1 and PGK2 mRNA levels in tumor and normal tissues were compared using the independent variable t test. The associations between PGK1 mRNA levels, PGK1 pS203 and PDHK1 pT338 levels and clinicopathologic characteristics of patients were analyzed using one-way analysis of variance (ANOVA) with the post hoc Bonferroni test for multiple comparisons and least significant difference test. The methylation value (M value) method is regarded as more statistically valid than the beta value method [19]. The correlation between the M values of the 11 probes covering the PGK1 gene and PGK1 mRNA levels was analyzed using Spearman’s correlation coefficient. The correlation between PGK1 pS203 and PDHK1 pT338 levels was analyzed using the Pearson correlation coefficient. Overall survival (OS) was defined as the duration from the date of diagnosis to death or the last known date of follow-up. The survival analyses were performed using the K-means cluster analysis to stratify the expression levels of related markers, the Kaplan–Meier method to plot survival curves, the log-rank test to compare survival rate, and a Cox regression model with two-sided Wald tests to calculate hazard ratios (HR) and 95% confidence intervals (CIs). Censored data were used for patients who were alive at last follow-up or lost to follow-up. Variables in univariate analysis with P values less than 0.05 were included in multivariate analysis. P < 0.05 was considered statistically significant. All statistical tests were two-sided.
Results
PGK1 and PGK2 expression in human cancers
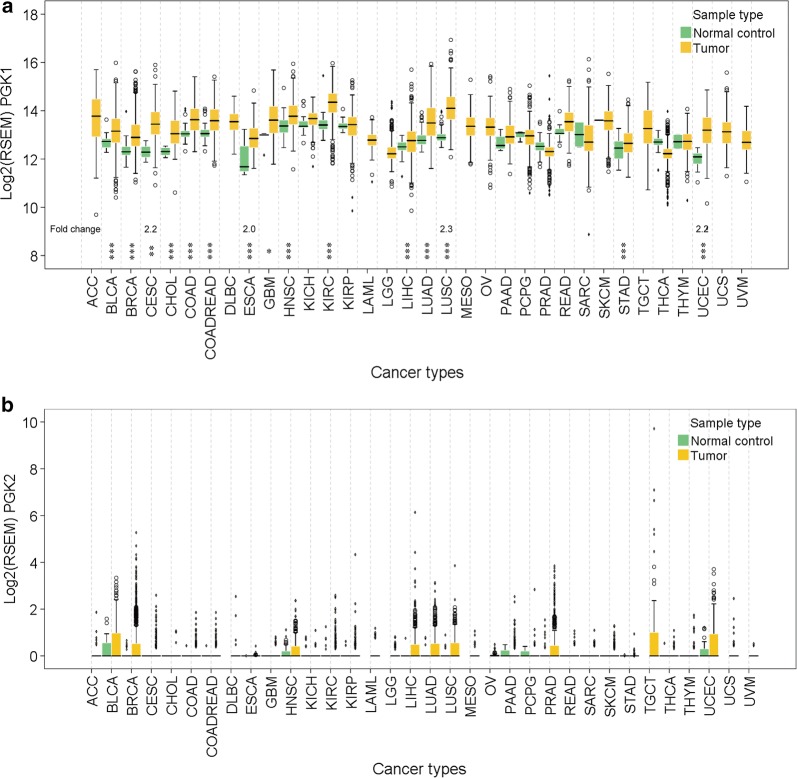
To compare PGK1 and PGK2 mRNA levels between tumor and normal tissues, we analyzed the RNA-Seq data of 11,908 tumor cases with 1582 paired normal tissues across 34 cancer types from TCGA datasets. In all types of cancer, the PGK1 mRNA levels in tumor and matched normal tissues were approximately 212 to 214 times higher than those of PGK2, which could hardly be detected (Fig. 1). In addition, the data from the Illumina Body Map Project further confirmed that PGK1, which had the highest mRNA level in leukocytes, was the major isozyme in all tissues except in the testes (Additional file 1: Fig. S1).
Fig. 1.
PGK1 and PGK2 mRNA levels in human cancer tissues and matched normal tissues. The data of PGK1 (a) and PGK2 (b) mRNA levels in human tumors and paired normal tissues were obtained from TCGA RNA-Seq datasets as log RSEM values (see “Materials and methods” section) and plotted. The fold changes of median PGK1 mRNA level in tumors compared to that in normal controls in four cancer types are provided. Significant changes in median PGK1 mRNA level between tumors and normal controls are marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, independent variable t test). PGK1/2 phosphoglycerate kinase 1/2, TCGA The Cancer Genome Atlas, ACC adrenocortical cancer, BLCA bladder urothelial carcinoma, BRCA breast carcinoma, CESC cervical and endocervical cancer, CHOL cholangiocarcinoma, COAD colon adenocarcinoma, COADREAD colon and rectum adenocarcinoma, DLBC diffuse large B-cell lymphoma, ESCA esophageal carcinoma, GBM glioblastoma multiforme, HNSC head and neck squamous cell carcinoma, KICH kidney chromophobe, KIRC kidney clear cell carcinoma, KIRP kidney papillary cell carcinoma, LAML lymphoblastic acute myeloid leukaemia, LGG brain lower grade glioma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, MESO mesothelioma, OV ovarian serous cystadenocarcinoma, PAAD pancreatic adenocarcinoma, PCPG pheochromocytoma and paraganglioma, PRAD prostate adenocarcinoma, READ rectum adenocarcinoma, SARC sarcoma, SKCM skin cutaneous melanoma, STAD stomach adenocarcinoma, TGCT testicular germ cell tumor, THCA thyroid carcinoma, THYM thymoma, UCEC uterine corpus endometrioid carcinoma, UCS uterine carcinosarcoma, UVM uveal melanoma
Since nine cancer types did not have mRNA level data available for matched normal tissues, we analyzed 25 cancer types and found that 15 had significantly higher PGK1 mRNA level in tumor tissues than in normal tissues and that PGK1 mRNA levels were increased by more than twofolds in 4 of the 15 cancer types, namely, esophageal carcinoma (ESCA), uterine corpus endometrioid carcinoma (UCEC), cervical and endocervical cancer (CESC), and lung squamous cell carcinoma (LUSC) (Fig. 1a). In addition, the 15 cancer types account for 55.0% of cancer incidence and 63.1% of cancer mortality each year worldwide (data from GLOBOCAN 2018) [20], indicating that PGK1 overexpression is prevalent among the most deadly human cancers.
Association between PGK1 mRNA levels and human cancer progression
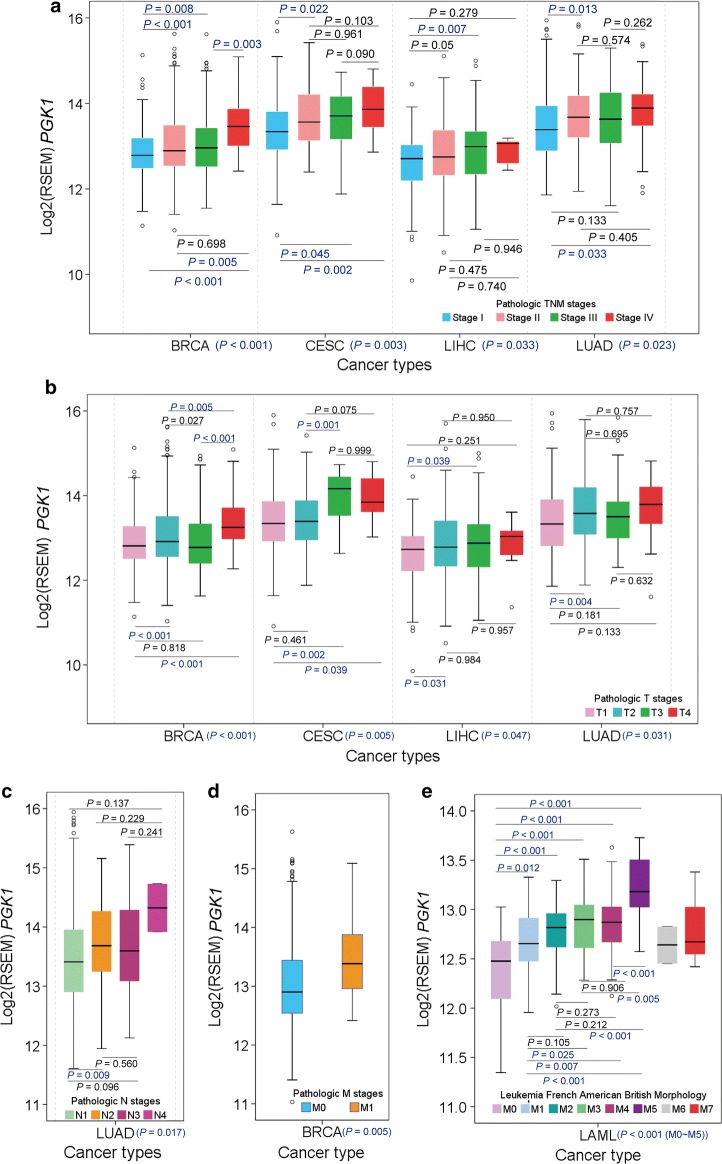
To access whether the PGK1 overexpression in tumor tissues was related to cancer progression, we analyzed the TNM staging data of the 15 cancer types with significantly increased PGK1 mRNA levels. We found that PGK1 mRNA level was significantly associated with progressive pathologic TNM stage in breast carcinoma (BRCA), CESC, liver hepatocellular carcinoma (LIHC), and lung adenocarcinoma (LUAD) (Fig. 2a). Further analyses showed that PGK1 mRNA level was significantly associated with T stage in BRCA, CESC, LIHC, and LUAD (Fig. 2b); N stage in LUAD (Fig. 2c); and M stage in BRCA (Fig. 2d).
Fig. 2.
Association between PGK1 mRNA levels and human cancer progression. a PGK1 mRNA levels are associated with pathologic TNM stage of BRCA, CESC, LIHC, LUAD, and TGCT. b–d PGK1 mRNA levels are associated were with T stage of BRCA, CESC, LIHC, and LUAD (b); N stage of LUAD and TGCT (c); and M stage of BRCA (d). e PGK1 mRNA levels are associated with LAML M0 through M5. BRCA breast carcinoma, CESC cervical and endocervical cancer, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, TGCT testicular germ cell tumor, LAML lymphoblastic acute myeloid leukaemia. The P values of the overall comparison between groups are presented on horizontal axes
Lymphoblastic acute myeloid leukaemia (LAML) has eight subtypes, M0 through M7. Subtypes M0 through M5 have a high percentage of immature myeloblasts, with M0 myeloblasts appearing the least mature and M5 myeloblasts appearing the most mature histologically, whereas subtypes M6 and M7 have a high percentage of immature erythrocytes and megakaryocytes, respectively [21–25]. Among the 16 tissue types, leucocytes had the highest PGK1 mRNA level (Additional file 1: Fig. S1). Notably, PGK1 mRNA levels were associated with LAML M0 through M5 (Fig. 2e), suggesting the association between PGK1 mRNA level and myeloblast maturity.
Association between PGK1 promoter hypomethylation and PGK1 mRNA level elevation
DNA methylation regulates gene expression and is implicated in tumor progression and therapeutic response [26, 27]. We next determined the methylation status of the PGK1 gene in 14 cancer types with significantly elevated PGK1 mRNA levels in the TCGA data. GBM was not included in further comparison and association analysis because there were only two matched normal tissues with DNA methylation data available.
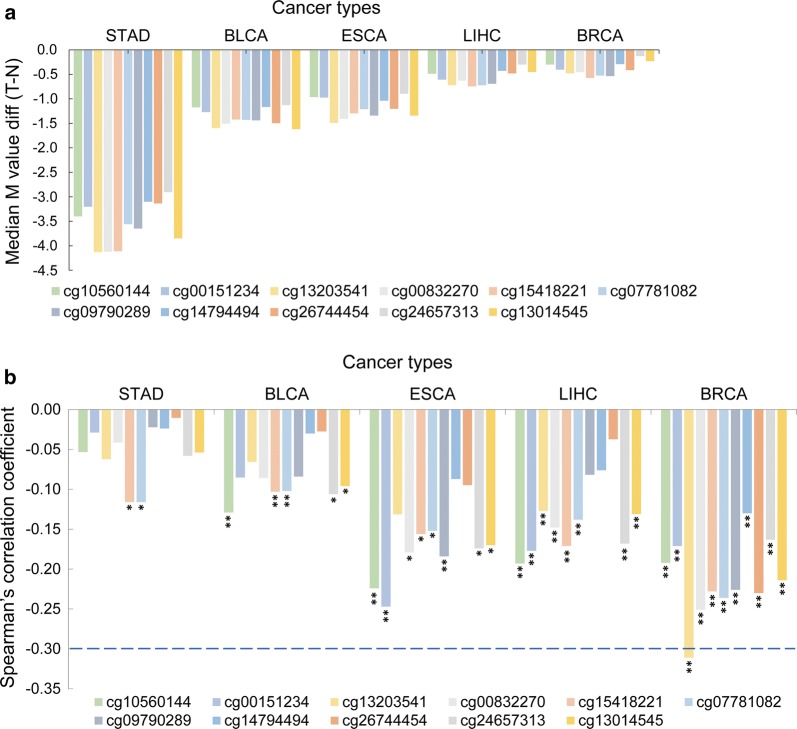
Median differential M values between tumor and normal tissues were plotted in the 14 cancer types (Additional file 1: Fig. S2). We found that 11 methylation probes had unanimous values in five types of cancer [stomach adenocarcinoma (STAD), bladder urothelial carcinoma (BLCA), ESCA, LIHC, and BRCA], and these values were lower in tumor tissues than in normal tissues (Fig. 3a, Additional file 1: Fig. S2). Intriguingly, these 11 probes were all located in the PGK1 promoter regions, ranging from 500 nt upstream of the transcription start site (TSS500) to the 5′-untranslated region (5′-UTR) (Additional file 1: Table S2; Additional file 1: Fig. S3), indicating that the PGK1 promoter regions were hypomethylated in these five types of cancer.
Fig. 3.
Association between PGK1 promoter hypomethylation and PGK1 mRNA level elevation in multiple cancer types. a All 11 methylation probes have unanimous values in five types of cancer, and these values are higher in normal tissues than in tumor tissues. M value the methylation level, diff difference, T − N, the methylation levels of probes in tumor tissues minus that in normal tissues. b PGK1 promoter methylation is significantly associated with PGK1 mRNA levels in STAD, BLCA, ESCA, LIHC, and BRCA (*P < 0.05, **P < 0.01, independent variable t test). All statistical tests were two-sided. BRCA breast carcinoma, LIHC liver hepatocellular carcinoma, ESCA esophageal carcinoma, BLCA bladder urothelial carcinoma, STAD stomach adenocarcinoma
The methylation data for these probes did not follow a normal distribution (1-sample Kolmogorov–Smirnov test, asymptotic P < 0.001, two-tailed; Additional file 1: Table S3). We identified a significant inverse correlation between the methylation levels and mRNA levels of PGK1 in STAD, BLCA, ESCA, LIHC, and BRCA (Fig. 3b; Additional file 1: Table S4), suggesting promoter hypomethylation as a mechanism promoting PGK1 expression. Among the five cancer types, BRCA showed the strongest correlation (Fig. 3b).
Associations between PGK1 promoter hypomethylation and PGK1 mRNA level elevation and poor prognosis in BRCA patients
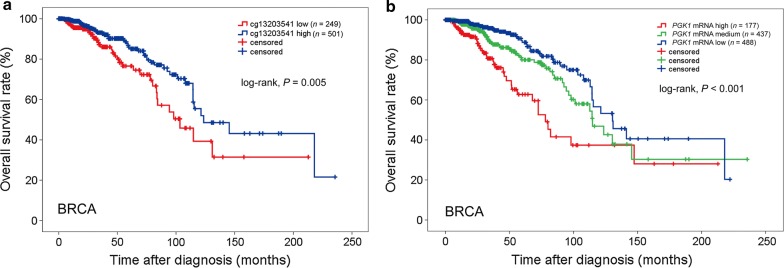
We next analyzed the association between PGK1 promoter hypomethylation and the survival of patients with STAD, BLCA, ESCA, LIHC, and BRCA, and found that only in BRCA, hypomethylation of cg13203541 was associated with short OS (HR = 0.551, 95% CI 0.361–0.841, P = 0.005; Additional file 1: Table S5; Fig. 4a). A multivariate Cox regression model showed that cg13203541 methylation was an independent predictor of prolonged OS in BRCA (HR = 0.599, 95% CI 0.382–0.939, P = 0.026; Additional file 1: Table S5). In line with these results, an inverse correlation between PGK1 mRNA level and the OS of BRCA patients was also identified (HR = 1.966, 95% CI 1.535–2.519, P < 0.001; Fig. 4b). These results suggest that PGK1 promoter methylation and mRNA level may be prognostic markers for BRCA patients.
Fig. 4.
Associations of PGK1 promoter hypomethylation and PGK1 mRNA level with OS of BRCA patients. a High cg13203541 methylation levels are associated with prolonged OS of BRCA patients. b High PGK1 mRNA levels are associated with short OS of BRCA patients. All statistical tests were two-sided. OS overall survival, BRCA breast carcinoma
Correlation between elevated PGK1 pS203 and PDHK1 pT338 levels and their associations with cancer prognosis
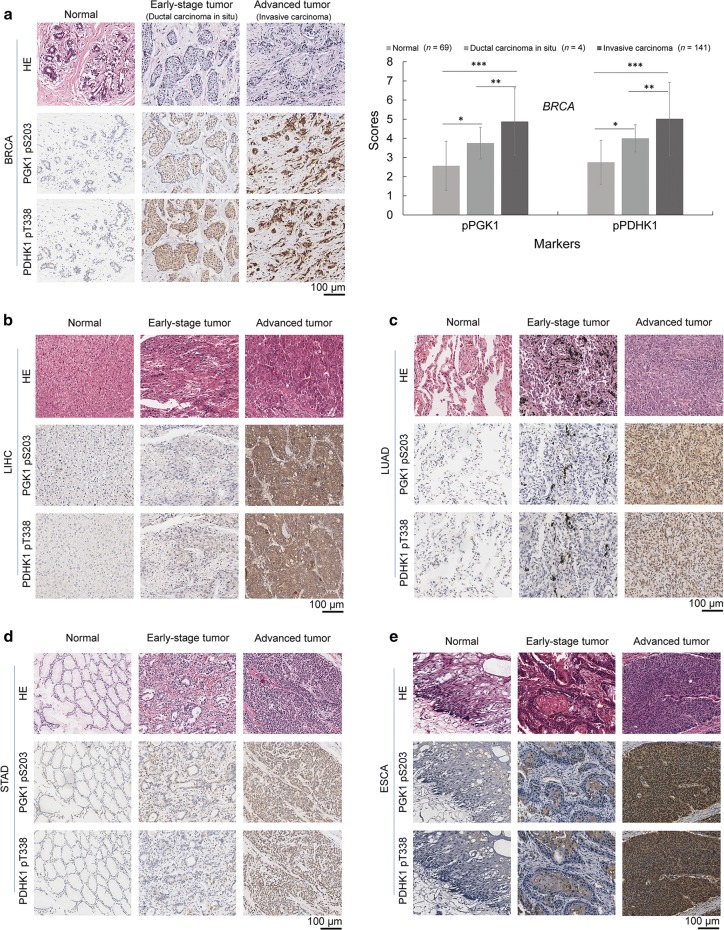
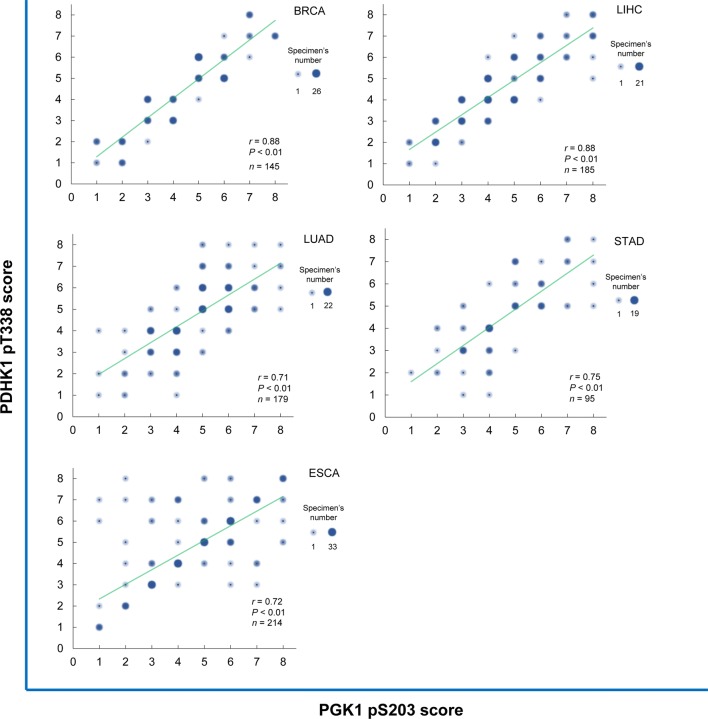
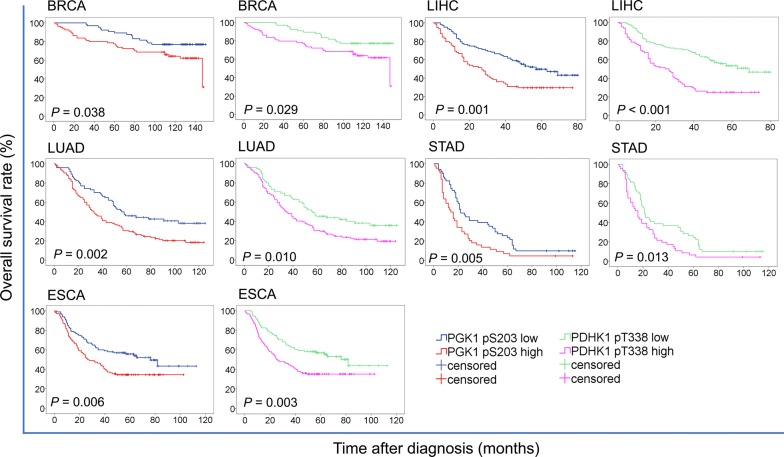
ERK-phosphorylated PGK1 S203 (PGK1 pS203) and PGK1-phosphorylated PDHK1 T338 (PDHK1 pT338) levels were found to be strongly correlated with each other and were both associated with GBM prognosis [14]. Here, we examined the phosphorylation levels of these two proteins in additional 818 independent cancer cases. We found that in all five cancer types, PGK1 pS203 and PDHK1 pT338 levels were higher in most tumor tissues than in their matched normal tissues (Additional file 1: Fig. S4), increasing from normal tissues to early-stage cancer tissues and to advanced carcinoma tissues (Fig. 5a–e), and positively correlating with each other in tumor tissues (Fig. 6). Kaplan–Meier analysis showed that higher levels of both PGK1 pS203 and PDHK1 pT338 were associated with shorter OS in patients with these five cancer types (all P < 0.05) (Fig. 7).
Fig. 5.
PGK1 pS203 and PDHK1 pT338 levels increased from normal to tumor tissues in human cancers. PGK1 pS203 and PDHK1 pT338 levels in normal tissues, early-stage tumor tissues, and advanced tumor tissues were compared using immunohistochemical staining. Representative images are shown. a Breast carcinoma (BRCA). b Liver hepatocellular carcinoma (LIHC). c Lung adenocarcinoma (LUAD). d Stomach adenocarcinoma (STAD). e Esophageal carcinoma (ESCA). *P < 0.05, **P < 0.01, ***P < 0.001 (2-tailed). PGK1 pS203 phosphorylated phosphoglycerate kinase 1 (PGK1) S203, PDHK1 pT338 phosphorylated phosphorylate pyruvate dehydrogenase kinase 1 (PDHK1) T338, HE hematoxylin–eosin staining
Fig. 6.
PGK1 pS203 and PDHK1 pT338 levels are positively correlated with each other in human cancers. Scatter diagrams show the statistical results of the correlation between PGK1 pS203 and PDHK1 pT338 levels (analyzed using the Pearson correlation coefficient). The size of each dot reflects the number of specimens. PGK1 pS203 phosphorylated phosphoglycerate kinase 1 (PGK1) S203, PDHK1 pT338 phosphorylated phosphorylate pyruvate dehydrogenase kinase 1 (PDHK1) T338; BRCA breast carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, STAD stomach adenocarcinoma, ESCA esophageal carcinoma
Fig. 7.
PGK1 pS203 and PDHK1 pT338 levels are associated with poor prognosis in cancer patients. K-Means cluster analysis was used to divide the indicated cancer patients into two groups with high and low levels of PGK1 pS203 and PDHK1 pT338. Kaplan–Meier survival curves were compared using the log-rank test. All statistical tests were two-sided. PGK1 pS203 phosphorylated phosphoglycerate kinase 1 (PGK1) S203, PDHK1 pT338 phosphorylated phosphorylate pyruvate dehydrogenase kinase 1 (PDHK1) T338, BRCA breast carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, STAD stomach adenocarcinoma, ESCA esophageal carcinoma
Prognostic values of PGK1 pS203 and PDHK1 pT338 in cancer
An independent variable t test showed that both PGK1 pS203 and PDHK1 pT338 were associated with advanced TNM stage in patients with BRCA and ESCA (all P < 0.05) (Table 1). Univariate and multivariate Cox regression analyses showed that PGK1 pS203 was an independent predictor of short OS for LIHC (HR = 1.574, 95% CI 1.064–2.327, P = 0.023), LUAD (HR = 1.800, 95% CI 1.238–2.617, P = 0.002), and STAD (HR = 2.603, 95% CI 1.630–4.155, P < 0.001); PDHK1 pT338 was also an independent predictor of short OS for LIHC (HR = 2.060, 95% CI 1.390–3.052, P < 0.001), LUAD (HR = 1.634, 95% CI 1.129–2.364, P = 0.009), and STAD (HR = 2.397, 95% CI 1.501–3.829, P < 0.001) (Tables 2 and 3), suggesting that PGK1 phosphorylation and PGK1 protein kinase activity-mediated phosphorylation and activation of PDHK1 were instrumental for tumor progression and OS in multiple cancer types.
Table 1.
Associations of PGK1 S203 and PDHK1 T338 phosphorylation levels with clinicopathologic characteristics in patients with BRCA, LIHC, LUAD, STAD and ESCA
| Cancer type | Characteristic | Total (cases) | PGK1 pS203 (IHC staining score) | PDHK1 pT338 (IHC staining score) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | 95% CI | P value | Mean | 95% CI | P value | |||
| BRCA | Age (years) | 0.478 | 0.975 | |||||
| ≤ 60 | 96 | 4.70 | 4.34–5.06 | 4.99 | 4.60–5.38 | |||
| > 60 | 49 | 4.92 | 4.41–5.42 | 5.00 | 4.47–5.53 | |||
| TNM stagea | 0.027 | 0.018 | ||||||
| I/II | 95 | 4.54 | 4.18–4.90 | 4.73 | 4.34–5.11 | |||
| III/IV | 48 | 5.23 | 4.73–5.73 | 5.52 | 4.99–6.06 | |||
| LIHC | Age (years) | |||||||
| ≤ 60 | 75 | 4.28 | 3.91–4.65 | 0.615 | 4.31 | 3.96–4.66 | 0.406 | |
| > 60 | 110 | 4.42 | 4.06–4.78 | 4.52 | 4.19–4.85 | |||
| Gender | 0.138 | 0.365 | ||||||
| Male | 159 | 4.44 | 4.17–4.71 | 4.47 | 4.22–4.72 | |||
| Female | 26 | 3.88 | 3.10–4.67 | 4.15 | 3.39–4.92 | |||
| TNM stageb | 0.643 | 0.226 | ||||||
| I/II | 73 | 4.18 | 3.81–4.55 | 4.16 | 3.83–4.50 | |||
| III/IV | 104 | 4.30 | 3.95–4.64 | 4.46 | 4.13–4.79 | |||
| LUAD | Age (years) | 0.763 | 0.385 | |||||
| ≤ 60 | 84 | 4.71 | 4.44–4.99 | 4.85 | 4.54–5.15 | |||
| > 60 | 95 | 4.78 | 4.46–5.10 | 4.65 | 4.34–4.97 | |||
| Gender | 0.722 | 0.700 | ||||||
| Male | 104 | 4.72 | 4.45–4.99 | 4.71 | 4.42–5.00 | |||
| Female | 75 | 4.79 | 4.45–5.14 | 4.79 | 4.45–5.14 | |||
| TNM stage | 0.339 | 0.236 | ||||||
| I/II | 105 | 4.65 | 4.36–4.94 | 4.62 | 4.33–4.91 | |||
| III/IV | 74 | 4.86 | 4.54–5.18 | 4.89 | 4.56–5.22 | |||
| STAD | Age (years) | 0.665 | 0.638 | |||||
| ≤ 60 | 56 | 4.55 | 4.18–4.93 | 4.61 | 4.21–5.01 | |||
| > 60 | 39 | 4.69 | 4.14–5.24 | 4.44 | 3.82–5.05 | |||
| Gender | 0.149 | 0.593 | ||||||
| Male | 60 | 4.78 | 4.39–5.18 | 4.47 | 4.01–4.92 | |||
| Female | 35 | 4.31 | 3.81–4.82 | 4.66 | 4.14–5.18 | |||
| TNM stagec | 0.356 | 0.195 | ||||||
| I/II | 24 | 4.88 | 4.19–5.56 | 4.96 | 4.26–5.66 | |||
| III/IV | 69 | 4.54 | 4.17–4.90 | 4.45 | 4.06–4.84 | |||
| ESCA | Age (years) | 0.781 | 0.089 | |||||
| ≤ 60 | 112 | 4.58 | 4.23–4.93 | 4.63 | 4.30–4.96 | |||
| > 60 | 102 | 4.65 | 4.33–4.97 | 5.03 | 4.71–5.34 | |||
| Gender | 0.559 | 0.216 | ||||||
| Male | 176 | 4.58 | 4.31–4.84 | 4.76 | 4.50–5.01 | |||
| Female | 38 | 4.76 | 4.23–5.29 | 5.13 | 4.56–5.70 | |||
| TNM stage | 0.006 | 0.008 | ||||||
| I/II | 98 | 4.26 | 3.89–4.62 | 4.49 | 4.14–4.84 | |||
| III/IV | 116 | 4.91 | 4.61–5.22 | 5.1 | 4.81–5.40 | |||
BRCA breast carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, STAD stomach adenocarcinoma, ESCA esophageal carcinoma, 95% CI 95% confidence interval, PGK1 pS203 PGK1 S203 phosphorylation level, PDHK1 pT338 PDHK1 T338 phosphorylation level
aThe data of TNM stage in BRCA were available in 143 patients
bThe data of TNM stage in LIHC were available in 177 patients
cThe data of TNM stage in STAD were available in 93 patients
Table 2.
Univariate analyses of overall survival in additional independent cases of BRCA, LIHC, LUAD, STAD, and ESCA
| Cancer type | Characteristic | Total (cases) | HR (95% CI) | P |
|---|---|---|---|---|
| BRCA | Age (years) | |||
| ≤ 60 | 96 | 1.000 | ||
| > 60 | 49 | 1.467 (0.864–2.752) | 0.323 | |
| TNBCa | ||||
| No | 109 | 1.000 | ||
| Yes | 24 | 2.296 (1.169–4.508) | 0.016 | |
| TNM stageb | ||||
| I/II | 95 | 1.000 | ||
| III/IV | 48 | 2.296 (1.279–4.120) | 0.005 | |
| PGK1 pS203 | ||||
| Low | 65 | 1.000 | ||
| High | 80 | 1.905 (1.024–3.546) | 0.042 | |
| PDHK1 pT338 | ||||
| Low | 66 | 1.000 | ||
| High | 79 | 1.969 (1.058–3.665) | 0.032 | |
| LIHC | Age (years) | |||
| ≤ 60 | 75 | 1.000 | ||
| > 60 | 110 | 1.351 (0.863–1.962) | 0.527 | |
| Gender | ||||
| Male | 159 | 1.000 | ||
| Female | 26 | 0.597 (0.327–1.087) | 0.092 | |
| TNM stagec | ||||
| I/II | 73 | 1.000 | ||
| III/IV | 104 | 2.796 (1.848–4.229) | < 0.001 | |
| PGK1 pS203 | ||||
| Low | 107 | 1.000 | ||
| High | 78 | 1.909 (1.311–2.781) | 0.001 | |
| PDHK1 pT338 | ||||
| Low | 105 | 1.000 | ||
| High | 80 | 2.354 (1.610–3.441) | < 0.001 | |
| LUAD | Age (years) | |||
| ≤ 60 | 84 | 1.000 | ||
| > 60 | 95 | 1.339 (0.944–1.900) | 0.102 | |
| Gender | ||||
| Male | 104 | 1.000 | ||
| Female | 75 | 0.775 (0.542–1.107) | 0.162 | |
| TNM stage | ||||
| I/II | 105 | 1.000 | ||
| III/IV | 74 | 1.861 (1.309–2.647) | 0.001 | |
| PGK1 pS203 | ||||
| Low | 74 | 1.000 | ||
| High | 105 | 1.760 (1.221–2.537) | 0.002 | |
| PDHK1 pT338 | ||||
| Low | 75 | 1.000 | ||
| High | 104 | 1.595 (1.111–2.291) | 0.011 | |
| STAD | Age (years) | |||
| ≤ 60 | 56 | 1.000 | ||
| > 60 | 39 | 1.754 (1.144–2.689) | 0.010 | |
| Gender | ||||
| Male | 60 | 1.000 | ||
| Female | 35 | 1.017 (0.660–1.567) | 0.938 | |
| TNM staged | ||||
| I/II | 24 | 1.000 | ||
| III/IV | 69 | 2.590 (1.536–4.365) | < 0.001 | |
| PGK1 pS203 | ||||
| Low | 51 | 1.000 | ||
| High | 44 | 1.797 (1.173–2.752) | 0.007 | |
| PDHK1 pT338 | ||||
| Low | 49 | 1.000 | ||
| High | 46 | 1.694 (1.105–2.596) | 0.016 | |
| ESCA | Age (years) | |||
| ≤ 60 | 112 | 1.000 | ||
| > 60 | 102 | 1.869 (1.300–2.686) | 0.001 | |
| Gender | ||||
| Male | 176 | 1.000 | ||
| Female | 38 | 1.127 (0.709–1.789) | 0.614 | |
| TNM stage | ||||
| I/II | 98 | 1.000 | ||
| III/IV | 116 | 6.447 (4.139–10.043) | < 0.001 | |
| PGK1 pS203 | ||||
| Low | 103 | 1.000 | ||
| High | 111 | 1.669 (1.157–2.406) | 0.006 | |
| PDHK1 pT338 | ||||
| Low | 94 | 1.000 | ||
| High | 120 | 1.763 (1.213–2.563) | 0.003 | |
BRCA breast carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, STAD stomach adenocarcinoma, ESCA esophageal carcinoma, HR hazard ratio, 95% CI 95% confidence interval, TNBC triple-negative breast cancer, PGK1 pS203 phosphorylated phosphoglycerate kinase 1 (PGK1) S203, PDHK1 pT338 phosphorylated phosphorylate pyruvate dehydrogenase kinase 1 (PDHK1) T338
aThe data of TNBC in BRCA were available in 133 patients
bThe data of TNM stage in BRCA were available in 143 patients
cThe data of TNM stage in LIHC were available in 177 patients
dThe data of TNM stage in STAD were available in 93 patients
Table 3.
Multivariate analyses of overall survival in additional independent cases of BRCA, LIHC, LUAD, STAD, and ESCA
| Cancer type | Characteristic | Total (cases) | PGK1 pS203 | PDHK1 pT338 | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| BRCA | TNBCa | |||||
| No | 109 | 1.000 | 1.000 | |||
| Yes | 24 | 2.561 (1.205–5.444) | 0.014 | 2.532 (1.191 to 5.383) | 0.016 | |
| TNM stageb | ||||||
| I/II | 95 | 1.000 | 1.000 | |||
| III/IV | 48 | 2.241 (1.172–4.286) | 0.015 | 2.238 (1.174 to 4.265) | 0.014 | |
| PGK1 pS203 | ||||||
| Low | 65 | 1.000 | ||||
| High | 80 | 1.596 (0.805–3.168) | 0.181 | |||
| PDHK1 pT338 | ||||||
| Low | 66 | 1.000 | ||||
| High | 79 | 1.661 (0.837 to 3.298) | 0.147 | |||
| LIHC | TNM stagec | |||||
| I/II | 73 | 1.000 | 1.000 | |||
| III/IV | 104 | 2.853 (1.878 to 4.335) | 0.000 | 2.905 (1.910 to 4.419) | 0.000 | |
| PGK1 pS203 | ||||||
| Low | 107 | 1.000 | ||||
| High | 78 | 1.574 (1.064 to 2.327) | 0.023 | |||
| PDHK1 pT338 | ||||||
| Low | 105 | 1.000 | ||||
| High | 80 | 2.060 (1.390 to 3.052) | 0.000 | |||
| LUAD | TNM stage | |||||
| I/II | 105 | 1.000 | 1.000 | |||
| III/IV | 74 | 1.805 (1.267 to 2.570) | 0.001 | 1.828 (1.284 to 2.602) | 0.001 | |
| PGK1 pS203 | ||||||
| Low | 74 | 1.000 | ||||
| High | 105 | 1.800 (1.238 to 2.617) | 0.002 | |||
| PDHK1 pT338 | ||||||
| Low | 75 | 1.000 | ||||
| High | 104 | 1.634 (1.129 to 2.364) | 0.009 | |||
| STAD | Age (years) | |||||
| ≤ 60 | 56 | 1.000 | 1.000 | |||
| > 60 | 39 | 2.258 (1.441 to 3.538) | 0.000 | 2.255 (1.436 to 3.540) | 0.000 | |
| TNM staged | ||||||
| I/II | 24 | 1.000 | 1.000 | |||
| III/IV | 69 | 3.382 (1.957 to 5.843) | 0.000 | 3.314 (1.920 to 5.720) | 0.000 | |
| PGK1 pS203 | ||||||
| Low | 51 | 1.000 | ||||
| High | 44 | 2.603 (1.630 to 4.155) | 0.000 | |||
| PDHK1 pT338 | ||||||
| Low | 49 | 1.000 | ||||
| High | 46 | 2.397 (1.501 to 3.829) | 0.000 | |||
| ESCA | Age (years) | |||||
| ≤ 60 | 112 | 1.000 | 1.000 | |||
| > 60 | 102 | 1.657 (1.146 to 2.397) | 0.007 | 1.614 (1.114 to 2.338) | 0.011 | |
| TNM stage | ||||||
| I/II | 98 | 1.000 | 1.000 | |||
| III/IV | 116 | 6.041 (3.867 to 9.437) | 0.000 | 6.022 (3.852 to 9.415) | 0.000 | |
| PGK1 pS203 | ||||||
| Low | 103 | 1.000 | ||||
| High | 111 | 1.440 (0.997 to 2.079) | 0.052 | |||
| PDHK1 pT338 | ||||||
| Low | 94 | 1.000 | ||||
| High | 120 | 1.453 (0.996 to 2.119) | 0.053 | |||
BRCA breast carcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, STAD stomach adenocarcinoma, ESCA esophageal carcinoma, HR hazard ratio, 95% CI 95% confidence interval, TNBC triple-negative breast cancer, HER2 human epidermal growth factor receptor 2, PGK1 pS203 PGK1 S203 phosphorylation level, PDHK1 pT338 PDHK1 T338 phosphorylation level
aThe data of TNBC in BRCA were available in 133 patients
bThe data of TNM stage in BRCA were available in 143 patients
cThe data of TNM stage in LIHC were available in 177 patients
dThe data of TNM stage in STAD were available in 93 patients
Discussion
In the present study, we identified relationships of high PGK1 mRNA level and promoter hypomethylation with advanced TNM stage and short OS in multiple cancer types in a pan-cancer analysis of TCGA data involving 11,908 cases covering 34 cancer types. Additional analyses of a cohort of 818 cases revealed that the phosphorylation levels of PGK1 S203 and PDHK1 T338 were independent prognostic biomarkers for LIHC, LUAD, and STAD. All these findings suggest that PGK1 gene modification and PGK1-mitochondrial function were significantly associated with clinical behaviors of cancer patients.
Metabolic reprogramming plays an important role in tumorigenesis [4, 28–32]. It is emerging that the nonmetabolic functions of metabolic enzymes are fundamental to tumorigenesis [33]. We reported that the protein kinase activity of metabolic enzymes, such as PGK1 [14, 15, 34], pyruvate kinase M2 (PKM2) [35, 36], and ketohexokinase isoform A (KHK-A) [37], regulates the Warburg effect, gene expression, cell proliferation, and autophagy [28, 29].We previously found that mitochondrial PGK1 functions as a protein kinase to promote tumor cell proliferation and brain tumorigenesis [14]. In the present study, we investigated the clinical relevance of PGK1 in human cancers and found that elevated PGK1 mRNA level and PGK1 protein kinase activity were associated with advanced TNM stages and poor prognosis in multiple human cancers. Whether other metabolic enzymes with protein kinase activity can act as potential biomarkers for the prediction of progression and prognosis of human cancers should be further analyzed.
The up-regulation of PGK1 involved in the Warburg effect has been detected in several types of human cancer [9–13]. Several other studies also reported the relationship between PGK1 acetylation and its innate enzymatic activity [38, 39]. However, all these studies focused on the metabolic function of PGK1 without elucidating the relationship between cancer progression and gene modification and the mitochondrial function of PGK1. Importantly, recent studies showed that metabolic changes in cancer alter the epigenetic landscape, especially DNA modifications, leading to malignant transformation, adaptation to inadequate nutrition, and tumor development [40]. Therefore, in the present study, we analyzed the DNA methylation data for 14 cancer types from TCGA datasets and identified hypomethylation of the PGK1 promoter (cg13203541) as an independent prognostic biomarker in BRCA patients (Additional file 1: Table S5). We also detected mitochondrial PGK1-dependent PDHK1 T338 phosphorylation in additional cases of five cancer types and demonstrated that mitochondrial function of PGK1 significantly affected the clinical behaviors of patients with these cancers.
Reprogrammed energy metabolism is an emerging hallmark of cancer biology and is an important way to treat cancer [4, 29–31, 41]. One important example is isocitrate dehydrogenase 1 (IDH1) mutation, which has important clinical significance and was found in GBM [42] and myeloid malignancies, such as acute myelocytic leukaemia (AML) [43] and myelodysplastic syndromes (MDS) [44]. A clinical study suggested that IDH1 mutation was an independent, favorable prognostic marker in grade 2–4 glioma [45]. Related clinical trials are ongoing in AML [43, 44]. In the present study, we found another metabolism reprogramming mediated by PGK1 protein kinase activity-dependent phosphorylation, which was associated with clinical behaviors of cancer patients and was an independent prognostic biomarker in multiple types of cancer. In addition, we revealed a high percentage of patients exhibiting elevated protein kinase activity of PGK1 compared to a relatively low IDH1 mutations in cancer patients [42, 43, 45]. Thus, we underscore that PGK1 protein kinase activity is a potential target for cancer treatment.
Our research has several limitations. First, the number of cases of some cancer types were rather limited in TCGA datasets. For example, only 36 tumor samples of cholangiocarcinoma and 66 tumor samples of kidney chromophobe were identified. Second, data of normal samples were not available in several cancer types, therefore, some analyses could not be performed for these cancer types. Third, validation of the association of PGK1 phosphorylation with clinicopathological characteristics could only be made in independent Chinese cohorts covering five cancer types as there were few publicly available datasets regarding the phosphorylation levels of proteins, and we could not validate those associations in Caucasians tumor samples.
Conclusions
We demonstrated a relationship between PGK1 promoter methylation and PGK1 mRNA level and demonstrated the significance of PGK1 mRNA level, PGK1 promoter methylation, and PGK1 pS203 and PDHK1 pT338 levels in tumor progression and cancer patient survival. These findings highlight the potential use of PGK1 mRNA level, PGK1 promoter hypomethylation, and PGK1 pS203 and PDHK1 pT338 levels as biomarkers for cancer progression and prognosis, and the promising significance of PGK1 as a target in cancer treatment.
Supplementary information
Additional file 1. Additional methods, Figures S1–S4 and Tables S1–S5.
Acknowledgements
We gratefully acknowledge contributions from the TCGA Research Network.
Abbreviations
- 5′-UTR
5′-untranslated region
- 95% CI
95% confidence interval
- ACC
adrenocortical cancer
- ADP
adenosine diphosphate
- ANOVA
analysis of variance
- ATP
adenosine triphosphate
- BLCA
bladder urothelial carcinoma
- BRCA
breast carcinoma
- CESC
cervical and endocervical cancer
- CHOL
cholangiocarcinoma
- COAD
colon adenocarcinoma
- COADREAD
colorectal adenocarcinoma
- DLBC
diffuse large B cell lymphoma
- ESCA
esophageal carcinoma
- GBM
glioblastoma multiforme
- GDC
Genomic Data Commons
- H&E
Mayer’s haematoxylin and eosin
- HNSC
head and neck squamous cell carcinoma
- HR
hazard ratio
- KICH
kidney chromophobe
- KIRC
kidney clear cell carcinoma
- KIRP
kidney papillary cell carcinoma
- LAML
lymphoblastic acute myeloid leukaemia
- LGG
brain lower grade glioma
- LIHC
liver hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MESO
mesothelioma
- OV
ovarian serous cystadenocarcinoma
- PAAD
pancreatic adenocarcinoma
- PCPG
pheochromocytoma and paraganglioma
- PDHK1
pyruvate dehydrogenase kinase 1
- PGK1
phosphoglycerate kinase 1
- PIN1
peptidyl-prolyl cis–trans isomerase NIMA-interacting 1
- PRAD
prostate adenocarcinoma
- READ
rectum adenocarcinoma
- RSEM
RNA-Seq by Expectation Maximization
- SARC
sarcoma
- SKCM
skin cutaneous melanoma
- STAD
stomach adenocarcinoma
- TCGA
The Cancer Genome Atlas
- TGCT
testicular germ cell tumor
- THCA
thyroid carcinoma
- THYM
thymoma
- TOM
translocase of the outer membrane
- TPM
transcripts per million
- TSS500
500 nt upstream of the transcription start site
- UCEC
uterine corpus endometrioid carcinoma
- UCS
uterine carcinosarcoma
- UCSC
University of California, Santa Cruz
- UVM
uveal melanoma
Authors’ contributions
JH, ZL and YG conceived the project and supervised all experiments. FS designed the experiments and wrote the manuscript. FS analyzed the data. XY, WW, JW, XF, SS, QX and SG provided support with experimental techniques. FS, XY, WW, JW and WG collected clinical samples and data. The manuscript was revised by FS, YG, ZL, and JH. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by The National Key R&D Program of China (2017YFC1308702, 2017YFC1311000, 2018YFC1312100), the Beijing Municipal Science & Technology Commission (Z181100006218032, Z181100001918002), the CAMS Initiative for Innovative Medicine (2017-I2M-1-005, 2017-I2M-2-003, 2019-I2M-2-002), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2017PT32001, 2017PT32017). The funding sources had no role in the design and conduct of this study, the analysis and interpretation of data, nor the preparation and submission of the manuscript. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Availability of data and materials
The data generated or analyzed during this study are included in this article or are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
This study was approved by the Institute Research Medical Ethics Committee of the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital in Chinese Academy of Medical Sciences and Peking Union Medical College in Beijing. Paired tumor and normal tissue specimens were obtained from the biobank of the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital in Chinese Academy of Medical Sciences and Peking Union Medical College in Beijing. All tissue samples were collected in compliance with an informed consent policy. Written informed consent was obtained from all the patients at the time of admission for the use of their tissue, blood or other samples for scientific research, and patient privacy was protected.
Consent for publication
The study participants provided written consent for the publication of their clinical data.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Fei Shao, Email: shmf2009@live.cn.
Xueying Yang, Email: yangxueying@cicams.ac.cn.
Wei Wang, Email: wangwei_2014@126.com.
Juhong Wang, Email: wjh402826@163.com.
Wei Guo, Email: guowei@cicams.ac.cn.
Xiaoli Feng, Email: fengxl@hotmail.com.
Susheng Shi, Email: shishusheng@sina.com.
Qi Xue, Email: 13801204967@139.com.
Shugeng Gao, Email: gaoshugeng@vip.sina.com.
Yibo Gao, Phone: +86-10-87788798, Email: gaoyibo@cicams.ac.cn.
Zhimin Lu, Phone: +86-571-86971812, Email: zhiminlu@zju.edu.cn.
Jie He, Phone: +86-10-87788207, Email: prof.jiehe@gmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40880-019-0401-9.
References
- 1.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci. 2018;43(4):301–310. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, et al. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol. 1992;154(1):160–168. doi: 10.1016/0012-1606(92)90056-M. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol. 2018;19(9):563–578. doi: 10.1038/s41580-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Xia Y, Lu Z. Metabolic features of cancer cells. Cancer Commun. 2018;38(1):65. doi: 10.1186/s40880-018-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4(11):1686–1696. doi: 10.1074/mcp.M400221-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Yan H, Yang K, Xiao H, Zou YJ, Zhang WB, Liu HY. Over-expression of cofilin-1 and phosphoglycerate kinase 1 in astrocytomas involved in pathogenesis of radioresistance. CNS Neurosci Ther. 2012;18(9):729–736. doi: 10.1111/j.1755-5949.2012.00353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad SS, Glatzle J, Bajaeifer K, Buhler S, Lehmann T, Konigsrainer I, et al. Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer. Int J Oncol. 2013;43(2):586–590. doi: 10.3892/ijo.2013.1971. [DOI] [PubMed] [Google Scholar]
- 12.Hwang TL, Liang Y, Chien KY, Yu JS. Overexpression and elevated serum levels of phosphoglycerate kinase 1 in pancreatic ductal adenocarcinoma. Proteomics. 2006;6(7):2259–2272. doi: 10.1002/pmic.200500345. [DOI] [PubMed] [Google Scholar]
- 13.Zieker D, Konigsrainer I, Tritschler I, Loffler M, Beckert S, Traub F, et al. Phosphoglycerate kinase 1 a promoting enzyme for peritoneal dissemination in gastric cancer. Int J Cancer. 2010;126(6):1513–1520. doi: 10.1002/ijc.24835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell. 2016;61(5):705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian X, Li X, Cai Q, Zhang C, Yu Q, Jiang Y, et al. Phosphoglycerate kinase 1 phosphorylates Beclin1 to induce autophagy. Mol Cell. 2017;65(5):917–931.e6. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Sun F, Yang X, Jin Y, Shi M, Wang L, et al. Correlation between RNA-Seq and microarrays results using TCGA data. Gene. 2017;628:200–204. doi: 10.1016/j.gene.2017.07.056. [DOI] [PubMed] [Google Scholar]
- 17.Zhu XF, Zhu BS, Wu FM, Hu HB. DNA methylation biomarkers for the occurrence of lung adenocarcinoma from TCGA data mining. J Cell Physiol. 2018;233(10):6777–6784. doi: 10.1002/jcp.26531. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Qian X, Jiang H, Xia Y, Zheng Y, Li J, et al. Nuclear PGK1 alleviates ADP-dependent inhibition of CDC7 to promote DNA replication. Mol Cell. 2018;72(4):650–660.e8. doi: 10.1016/j.molcel.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 21.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French–American–British (FAB) co-operative group. Br J Haematol. 1976;33(4):451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 22.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French–American–British Cooperative Group. Ann Intern Med. 1985;103(4):620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 23.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Criteria for the diagnosis of acute leukemia of megakaryocyte lineage (M7). A report of the French–American–British Cooperative Group. Ann Intern Med. 1985;103(3):460–462. doi: 10.7326/0003-4819-103-3-460. [DOI] [PubMed] [Google Scholar]
- 24.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposal for the recognition of minimally differentiated acute myeloid leukaemia (AML-MO) Br J Haematol. 1991;78(3):325–329. doi: 10.1111/j.1365-2141.1991.tb04444.x. [DOI] [PubMed] [Google Scholar]
- 25.Walter RB, Othus M, Burnett AK, Lowenberg B, Kantarjian HM, Ossenkoppele GJ, et al. Significance of FAB subclassification of “acute myeloid leukemia, NOS” in the 2008 WHO classification: analysis of 5848 newly diagnosed patients. Blood. 2013;121(13):2424–2431. doi: 10.1182/blood-2012-10-462440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13(10):679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 28.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem Sci. 2018;43:301–310. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol. 2018;19:563. doi: 10.1038/s41580-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Cao S, Situ B, Zhong J, Hu Y, Li S, et al. Metabolic reprogramming-based characterization of circulating tumor cells in prostate cancer. J Exp Clin Cancer Res. 2018;37(1):127. doi: 10.1186/s13046-018-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017;552(7684):273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, Wang Y. Nonmetabolic functions of metabolic enzymes in cancer development. Cancer Commun. 2018;38(1):63. doi: 10.1186/s40880-018-0336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Zheng Y, Lu Z. PGK1 is a new member of the protein kinome. Cell Cycle. 2016;15(14):1803–1804. doi: 10.1080/15384101.2016.1179037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Li X, Shao F, Lv G, Lv H, Lee JH, et al. The protein kinase activity of fructokinase A specifies the antioxidant responses of tumor cells by phosphorylating p62. Sci Adv. 2019;5(4):eaav4570. doi: 10.1126/sciadv.aav4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology. 2017;65(2):515–528. doi: 10.1002/hep.28887. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Jiang B, Zhang T, Liu L, Wang Y, Wang Y, et al. Insulin and mTOR pathway regulate HDAC3-mediated deacetylation and activation of PGK1. PLoS Biol. 2015;13(9):e1002243. doi: 10.1371/journal.pbio.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang YP, Lei QY. Metabolic recoding of epigenetics in cancer. Cancer Commun. 2018;38(1):25. doi: 10.1186/s40880-018-0302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang L, Wei F, Wu Y, He Y, Shi L, Xiong F, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37(1):87. doi: 10.1186/s13046-018-0758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 44.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27(4):599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 45.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional methods, Figures S1–S4 and Tables S1–S5.
Data Availability Statement
The data generated or analyzed during this study are included in this article or are available from the corresponding author upon reasonable request.