Abstract
Gastric cancer (GC) threatens human health worldwide and we performed this meta-analysis to evaluate the clinical value of Ki-67/MKI67 in patients with GC. The combined hazard ratio (HR), odds ratio (OR) and 95% confidence interval (95% CI) were calculated to assess the relationships of Ki-67/MKI67 expression with prognoses and clinicopathological characteristics. Genes co-expressed with MKI67 were collected for Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and protein-protein interaction (PPI) network analyses. In total, 53 studies with 7078 patients were included in this study. The pooled HRs indicated that an elevated expression of Ki-67/MKI67 predicted an unfavorable overall survival (HR: 1.54, 95% CI: 1.33-1.78, P<0.0001) and disease-free survival (HR: 2.28, 95% CI: 1.43-3.64, P<0.0001) in GC patients. Additionally, in patients with advanced GC, a high Ki-67/MKI67 expression was also significantly connected with OS (HR: 1.37, 95% CI: 1.18-1.60, P<0.0001). The combined ORs showed that Ki-67/MKI67 expression was related to TNM stage (stage III/IV versus stage I/II: OR=1.93, 95% CI=1.34-2.78, P<0.0001), tumor differentiation (poor versus well/moderate: OR=1.94, 95% CI=1.32-2.85, P=0.001), lymph node metastasis (yes versus no: OR=1.67, 95% CI=1.23-2.25, P=0.001), distant metastasis (yes versus no: OR=1.67, 95% CI=1.24-2.26, P=0.001) and tumor invasion depth (T3/T4 versus Tis/T1/T2: OR=1.98, 95% CI=1.60-2.44, P<0.0001). The results of GO, KEGG pathway and PPI network analyses indicated that Ki-67/MKI67 may be involved in the development of GC via influencing P53 signaling pathway. Ki-67/MKI67 could be a potential indicator to predict the prognosis of patients with GC and identify high-risk cases. Detecting Ki-67/MKI67 expression in clinic may be helpful in optimizing individual treatment and further improving the survival expectancy of patients with GC.
Keywords: gastric cancer, Ki-67/MKI67, prognosis, clinical value
Background
Gastric cancer (GC) is one of the most common and aggressive malignancies worldwide according to the World Health Organization 1. In the US, about 28,000 new cases and 10,960 deaths occurred in 2017 2. While in China, GC is the second main cause of cancer-related death with an estimated 498,000 deaths occur annually 3. In spite of the improvements in diagnosis and treatment 4, currently, the long-term survival rate for a large number of GC patients is still dismal, with a 5-year survival rate of less than 20% 5. Most patients have developed tumor metastasis when they are diagnosed with GC, and the high tumor metastasis rate leads to unfavorable survival outcome in patients with GC 6. Identifying a biomarker for early diagnosis and clinical outcome prediction is important to understand the development of GC and further improve the prognosis in patients with GC.
Antigen Ki-67, also known as MKI67 (antigen identified by monoclonal antibody Ki-67), is a cell proliferation-related protein that encoded by gene MKI67. Ki-67/MKI67 exists in G1, S, G2 and mitosis phases of cell cycle 7 and can be put into clinical practice by acting as an index to evaluate cell proliferation by immunohistochemical staining 8. A large number of studies have reported the clinical and prognostic value of Ki-67/MKI67 in various malignancies, such as breast cancer 9, cervical cancer 10, glioma 11, colorectal cancer 12 and hepatocellular carcinoma 13. Our research group previously conducted systematic meta-analyses to explore the prognostic role of Ki-67/MKI67 in lung cancer 14, hepatocellular carcinoma 13, cervical cancer 15, glioma 16 and colorectal cancer 17 and found that Ki-67/MIB-1 could be an independent prognostic biomarker in these types of carcinoma.
Although two previous meta-analyses have reported the potential of Ki-67/MKI67 as a predictive biomarker in patients with GC 18, 19, studies based on more cases and stronger evidence are still needed to corroborate the prognostic and clinicopathological role of Ki-67/MKI67 for GC patients. Thus, we conducted a comprehensive literature search and performed a systematic analysis on the prognostic value and clinical significance of Ki-67/MKI67 in GC. Simultaneously, we investigated the underling action mechanism by which Ki-67/MKI67 affects cell proliferation and promotes tumor development by performing bioinformatics analysis on genes co-expressed with MKI67.
Methods
Publication identification and searching strategy
A comprehensive literature search via PubMed, PMC, Science Direct, Web of Science, Wiley Online, Chinese National Knowledge Infrastructure (CNKI), Chinese WanFang database, Chinese Chongqing VIP and Embase was carried out up to March 5, 2019 based on the following searching terms: (Ki-67 OR Ki67 OR MKI67 OR MIB-1 OR proliferative index OR proliferative activity OR mitotic index OR mitotic activity OR mitotic figure OR labeling index OR mitotic count OR proliferative marker) AND (prognos* OR surviv* OR predict* OR follow-up OR followed-up OR mortality OR outcome OR diagnosis OR diagnostic OR detect) AND (gastric OR stomach OR tummy) AND (cancer OR carcinoma OR tumo* OR neoplas* OR malignan*).
Eligibility criteria for inclusion and exclusion
Two investigators evaluated all of the acquired records independently using the criteria listed below: (1) All patients were distinctly diagnosed with gastric cancer by pathology; (2) The studies were written in English or Chinese; (3) The studies investigated the expression level of Ki-67/MKI67 in gastric cancer and adjacent non-cancerous tissues; (3) The studies reported the relationships of Ki-67/MKI67 expression with prognosis or clinicopathological parameters in gastric cancer patients; (4) The studies provided enough information to estimate the hazard ratios (HRs), odds ratios (ORs) and their 95% confidence intervals (CIs); (5) The latest and most complete study was included when the same patients were reported in different publications; (6) The studies must be performed in humans.
The following publications were excluded: conference abstracts, reviews, meta-analyses, case reports, expert opinions, studies not written in English or Chinese, and studies without sufficient data to calculate HRs, ORs and their 95% CIs. Because of the heterogeneous expression of Ki-67 in GC, tissue microarrays (TMAs) carry the risk of sampling error due to an underestimation and non-representative evaluation of Ki-67 in GC 20. Therefore, we excluded studies using TMAs.
Data extraction
Two researchers independently performed the data collection from all of the enrolled studies, with any discrepancies settled by discussion with a third reviewer. The relevant information covered the following details: first author and publication year, region, cut-off value, antibody, quality score, statistical method, and HR with corresponding 95% CI.
Quality assessment
Two investigators independently read each enrolled prognosis-related record and scored it using the Newcastle-Ottawa scale (NOS) 21. Each study was assessed based on the following three perspectives: selection, comparability and outcome. The score ranged from 0 to 9, and high scores mean high quality of the included records. Additionally, the Quality Assessment of Diagnostic Accuracy Studies-2 (QADAS-2) tool was used to assess the quality of diagnostic accuracy studies 22. The risk of bias and applicability of individual studies were judged as “low”, “high” or “unclear” based on the following four critical domains: patient selection, index test, reference standard, and flow and timing. Both reviewers compared their judgments and achieved a consensus by discussing in conference if the results were controversial.
Statistical analysis
To determine the ability of Ki-67/MKI67 in discriminating tumor from non-tumorous gastric tissues, we performed summary receiver operating characteristic curve (SROC) analysis and calculated the area under the curve (AUC) value, the diagnostic OR and the corresponding sensitivity and specificity by using Meta-DISc 23.
The HRs and 95% CIs were combined to measure the effects of Ki-67/MKI67 expression on overall survival (OS) and disease-free survival (DFS). We directly extracted the HRs and 95% CIs from the original studies if the survival data were provided; otherwise, we calculated them by using Engauge Digitizer Version 4.1 on the basis of the Kaplan-Meier survival curves. Three investigators read the survival curves independently to improve the precision in the extraction of HRs and 95% CIs. Then we combined the individual HRs into a pooled HR by using Stata 12.0 (StataCorp, College Station, TX, USA) to investigate the correlation between Ki-67/MKI67 expression and prognosis in patients with GC. Subgroup analyses were conducted to determine whether different regions, antibodies, cut-off values and statistical methods resulted in the differences of the overall results. An observed HR>1 and it's 95% CI not crossing 1 indicated that a high expression of Ki-67/MKI67 predicted a poor outcome in gastric cancer patients.
The ORs and 95% CIs were calculated to determine the relationships between Ki-67/MKI67 expression and clinicopathological characteristics, such as gender (female versus male), TNM stage (stage III/IV versus stage I/II), tumor differentiation (poor differentiation versus well/moderate differentiation), lymph node metastasis (yes versus no), distant metastasis (yes versus no), invasion depth (T3/T4 versus Tis/T1/T2), Lauren's classification (diffuse versus intestinal) and vascular invasion (yes versus no). The impacts of Ki-67/MKI67 on clinicopathological parameters were considered statistically significant if the 95% CI did not cross 1. The data calculations were conducted by using Stata 12.0.
To evaluate the stability of the pooled results, we conducted sensitivity analysis by excluding individual studies successively. The χ2 test (Chi squared test; Chi2) and the I2 test (Higgins I-squared test; I2) were used to analyze the heterogeneity among studies. If an inter-study heterogeneity existed (P<0.05 and/or I2≥50%), a random-effects model was applied; otherwise, a fixed-effects model was chosen. Begg's funnel plot and Egger's test were used to assess the publication bias of the eligible studies. All P<0.05 (tested by two-sided) were considered statistically significant.
Bioinformatics analysis of genes co-expressed with MKI67
Genes co-expressed with MKI67 were collected from cBioPortal (http://www.cbioportal.org/) online website. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed with the DAVID (https://david.ncifcrf.gov/) online tool. Protein-protein interaction (PPI) network wwas constructed with the STRING (http://string-db.org) database.
Results
Studies selection and characteristics
A total of 53 studies with 7078 patients were included in accordance with our strict inclusion and exclusion criteria (Fig. 1). The main details of the selected studies are concluded in Table 1. All of the studies were retrospective. Of all of the studies, 24 were performed in China 24-47, 12 in Japan 48-59, 3 in Greece 60-62 , 2 in Italy 63, 64, 2 in Poland 65, 66, 1 in Korea 67, 1in Germany 68, 2 in Egypt 69, 70, 1 in Finland 71, 1 in Timisoara 72, 1 in Turkey 73, 1 in Sultanate of Oman 74, 1 in Tunisia 75 and 1 in Saudi Arabia 76. A total of 10 publications were written in Chinese and 43 articles were written in English. The number of patients in each study ranged from 40 to 693. Immunohistochemistry (IHC) method was applied to detect the Ki-67 protein expression: MIB-1 was utilized in 24 studies and anti-Ki-67 antibody was used in 29 studies. Of the 53 included records, 3 records compared the expression of Ki-67/MKI67 in gastric cancerous and adjacent non-cancerous tissues, 44 records reported the prognostic value of Ki-67/MKI67 in GC, and 26 records assessed the relationships between Ki-67/MKI67 and clinicopathological parameters. Among the 44 prognosis-related studies, 20 studies provided HRs and corresponding 95% CIs for OS and DFS directly, and 24 studies only provided Kaplan-Meier survival curves. The methodological quality scores assessed by the NOS scale varied from 6 to 9.
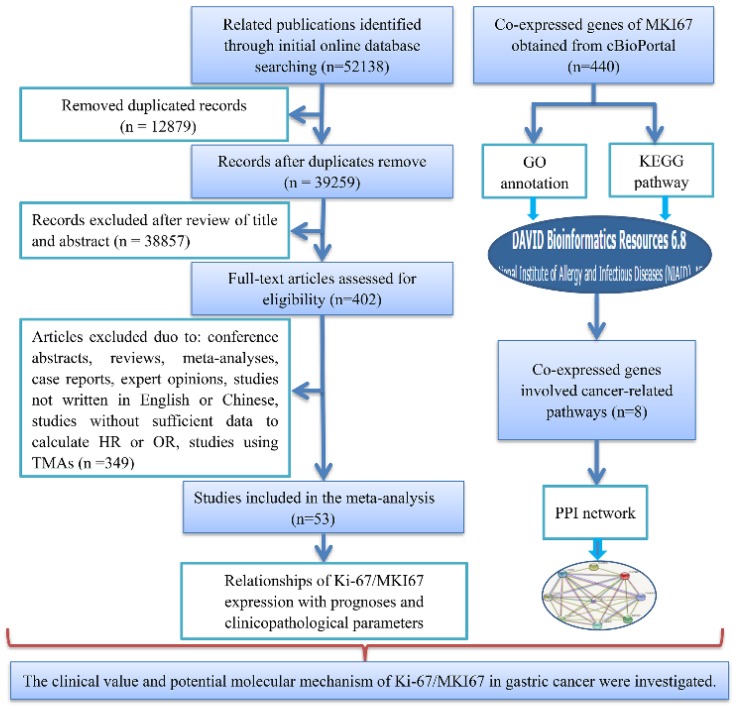
Fig 1.
Flow chart of the overall design of the present study. TMA: tissue microarray; HR: hazard ratio; OR: odds ratio; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: Protein-protein interaction.
Table 1.
Main characteristics of the 53 studies included in the meta-analysis
| ID | First author (publication year) | Regions | Cut-off value | Antibody | Statistical method | HR (95% CI) | Quality score |
|---|---|---|---|---|---|---|---|
| 1 | Ye (2017) | China | 0.25 | Anti-Ki-67 | Multivariate analysis | OS: 1.197 (0.598-2.397) | 7 |
| 2 | Liu (2017) | China | 0.1 | Anti-Ki-67 | Multivariate analysis | OS: 4.290 (2.839-6.483) | 7 |
| 3 | Yu (2017) | China | 0.5 | Anti-Ki-67 | Univariate analysis | OS: 0.807 (0.487-1.336) | 8 |
| 4 | Wang (2016) | China | 0.5 | Anti-Ki-67 | Univariate analysis | OS: 2.974 (1.944-4.550) | 7 |
| 5 | Huang (2016) | China | 0.5 | Anti-Ki-67 | Multivariate analysis | OS: 1.46 (1.215-1.754) | 8 |
| 6 | Calik (2015) | Turkey | 0.1 | MIB-1 | Survival curve | OS: 1.42 (0.54-3.79) | 6 |
| 7 | Li (2015) | China | 0.5 | Anti-Ki-67 | Survival curve Multivariate analysis | OS: 1.01 (0.32-3.22) DFS: 1.358 (0.632-2.917) | 7 |
| 8 | Zhu (2015) | China | 0.5 | Anti-Ki-67 | Multivariate analysis | OS: 2.927 (1.518-5.646) | 8 |
| 9 | Yang (2014) | China | 0.3 | Anti-Ki-67 | Survival curve | OS: 1.64 (1.05-2.56) DFS: 1.61 (1.03-2.52) | 8 |
| 10 | Wu (2014) | China | 0.12 | Anti-Ki-67 | Survival curve | OS: 1.78 (0.69-4.6) | 8 |
| 11 | Sun (2014) | China | 0.05 | Anti-Ki-67 | Survival curve | OS: 1.99 (0.94-4.21) DFS: 2.31 (1.07-4.99) | 8 |
| 12 | Xiao (2013) | Japan | 0.5 | Anti-Ki-67 | Multivariate analysis | OS: 0.98 (0.82-1.17) | 7 |
| 13 | Wang (2013) | China | NA | Anti-Ki-67 | Multivariate analysis | OS: 2.10 (1.40-3.22) | 8 |
| 14 | Su (2013) | China | NA | Anti-Ki-67 | Survival curve | OS: 1.49 (0.55-4.04) | 7 |
| 15 | Huang (2012) | China | 0.25 | Anti-Ki-67 | Survival curve | OS: 1.03 (0.47-2.26) | 8 |
| 16 | Sun (2012) | China | 0.05 | Anti-Ki-67 | Survival curve | OS: 1.21 (0.28-5.16) | 8 |
| 17 | Tang (2012) | China | 0.1 | Anti-Ki-67 | Survival curve | OS: 1.91 (0.91-4) | 7 |
| 18 | Yu (2012) | China | 0.2 | MIB-1 | Multivariate analysis | OS: 1.982 (1.5-2.62) | 8 |
| 19 | Ichinoe (2011) | Japan | 0.4 | MIB-1 | Univariate analysis | OS: 0.907 (0.532-1.547) | 7 |
| 20 | Shomori (2010) | Japan | 0.45 | Anti-Ki-67 | Univariate analysis | OS: 1 (0.54-1.85) | 9 |
| 21 | Tatsuwaki (2010) | Japan | 0.25 | MIB-1 | Univariate analysis | OS: 3.07 (1.13-8.37) | 8 |
| 22 | Lazar (2010) | Timisoara | 0.45 | MIB-1 | Survival curve | OS: 1.06 (0.73-1.52) | 8 |
| 23 | Tzanakis (2009) | Greece | 0.35 | MIB-1 | Univariate analysis | OS: 3.42 (1.27-9.2) | 8 |
| 24 | Tsamandas (2009) | Greece | 0.05 | MIB-1 | Survival curve | OS: 3.86 (2.14-6.95) | 8 |
| 25 | Solcia (2009) | Italy | 0.4 | MIB-1 | Multivariate analysis | OS: 1.54 (1.08-2.19) | 8 |
| 26 | Czyzewska (2009) | Poand | 0.5 | MIB-1 | Survival curve | OS: 1.06 (0.61-1.77) | 9 |
| 27 | Li (2009) | China | 0.1 | Anti-Ki-67 | Survival curve | OS: 1.6 (1.15-2.23) | 7 |
| 28 | Tokuyasu (2008) | Japan | 0.1 | MIB-1 | Survival curve | OS: 1.3 (0.29-5.82) | 9 |
| 29 | Chen (2008) | China | 0.1 | MIB-1 | Survival curve | OS: 1.17 (0.49-2.82) DFS: 6.39 (2.97-13.74) | 7 |
| 30 | Li (2008) | China | 0.5 | Anti-Ki-67 | Survival curve | OS: 3.93 (1.81-8.52) | 8 |
| 31 | Zhang (2007) | China | 0.1 | Anti-Ki-67 | Survival curve | OS: 1.06 (0.35-3.22) | 7 |
| 32 | Joo (2006) | Korea | 0.5 | MIB-1 | Survival curve | OS: 0.99 (0.51-1.92) | 8 |
| 33 | Takahashi (2006) | Japan | 0.5 | MIB-1 | Survival curve | OS: 1.13 (0.45-2.82) | 8 |
| 34 | Wang (2004) | China | 0.53 | Anti-Ki-67 | Survival curve | OS: 2.06 (1.21-3.5) | 7 |
| 35 | Kijima (2003) | Japan | 0.1 | Anti-Ki-67 | Multivariate analysis | OS: 0.5 (0.2-1.3) | 6 |
| 36 | Liu (2001) | Japan | 0.27 | MIB-1 | Multivariate analysis | OS: 1.13 (0.63-2.02) | 7 |
| 37 | Ohtani (1999) | Japan | 0.36 | MIB-1 | Multivariate analysis | OS: 1.31 (0.778-2.2) | 8 |
| 38 | Kikuyama (1998) | Japan | 0.55 | MIB-1 | Multivariate analysis | OS: 3.56 (1.31-9.63) | 7 |
| 39 | de Manzoni (1998) | Italy | 0.1 | Anti-Ki-67 | Multivariate analysis | OS: 1.12 (0.35-3.58) | 7 |
| 40 | Victorzon (1997) | Finland | 0.3 | Anti-Ki-67 | Survival curve | OS: 1.05 (0.78-1.43) | 9 |
| 41 | Muller (1996) | Germany | 0.53 | MIB-1 | Survival curve | OS: 1.05 (0.7-1.32) | 8 |
| 42 | Yonemura (1990) | Japan | 0.25 | MIB-1 | Survival curve | OS: 2.63 (1.09-6.38) | 6 |
| 43 | Han (2013) | China | 0.1 | Anti-Ki-67 | Survival curve | DFS: 6.31 (3.11-12.81) | 8 |
| 44 | Nakashima (2011) | Japan | NA | MIB-1 | Univariate analysis | DFS: 1.41 (0.714-2.835) | 6 |
| 45 | Ahmed (2018) | Saudi Arabia | 0.1 | MIB-1 | NA | NA | 7 |
| 46 | Abdel-Aziz (2017) | Egypt | 0.45 | MIB-1 | NA | NA | 7 |
| 47 | Badary (2017) | Egypt | NA | Anti-Ki-67 | NA | NA | 8 |
| 48 | Zhou (2015) | China | NA | Anti-Ki-67 | NA | NA | 8 |
| 49 | Ayed (2014) | Tunisa | 0.01 | MIB-1 | NA | NA | 7 |
| 50 | Giaginis (2011) | Greece | 0.5 | MIB-1 | NA | NA | 8 |
| 51 | Zhao (2010) | China | 0.5 | Anti-Ki-67 | NA | NA | 7 |
| 52 | Al-Moundhri (2005) | Sultanate of Oman | 0.25 | MIB-1 | NA | NA | 7 |
| 53 | Czyzewska (2004) | Poland | NA | Anti-Ki-67 | NA | NA | 8 |
NA: data not available; OS: overall survival; DFS: disease-free survival; HR: hazard ratio; CI: confidence interval.
Expression level of Ki-67/MKI67 in GC
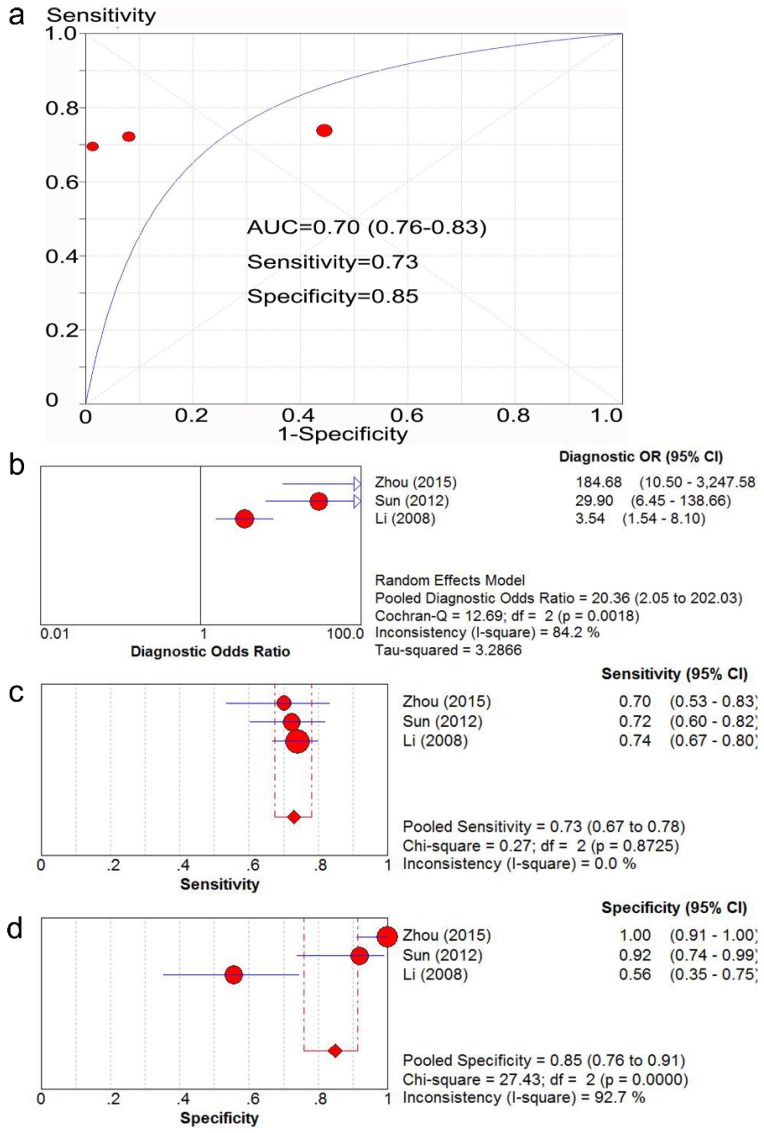
Three studies 35, 40, 44 with 384 patients provided sufficient data to compare the expression of Ki-67/MKI67 in gastric cancerous and adjacent non-cancerous tissues. The potential risk of bias and applicability of the three studies are shown in Table 2. The AUC value of SROC analysis was 0.80 (95% CI: 0.76-0.83; Fig. 2a), the diagnostic OR was 20.36 (95% CI: 2.05-202.03; Fig. 2b), the corresponding sensitivity and specificity were 0.73 (95% CI: 0.67-0.78; Fig. 2c) and 0.85 (95% CI: 0.76-0.91; Fig. 2d), respectively.
Table 2.
Potential risk of bias and applicability of the three diagnostic accuracy studies
| Risk of bias | Applicability | ||||||
|---|---|---|---|---|---|---|---|
| Study | Patient selection | Index test | References standard | Flow and timing | Patient selection | Index test | References standard |
| Li (2008) | J | ? | ? | L | J | J | J |
| Sun (2012) | J | J | ? | L | J | J | J |
| Zhou (2015) | J | L | ? | L | J | L | J |
J: low risk; L: high risk; ?: unclear risk.
Fig 2.
SROC analysis of the ability of Ki-67/MKI67 to distinguish gastric cancer patients from normal controls. The AUC value (a), the diagnostic OR (b), the corresponding sensitivity (c) and specificity (d) were calculated using Meta-DISc software. SROC: summary receiver operating characteristic curve. AUC: area under the value; OR: odds ratio; CI: confidence interval.
Prognostic value of Ki-67/MKI67 in GC
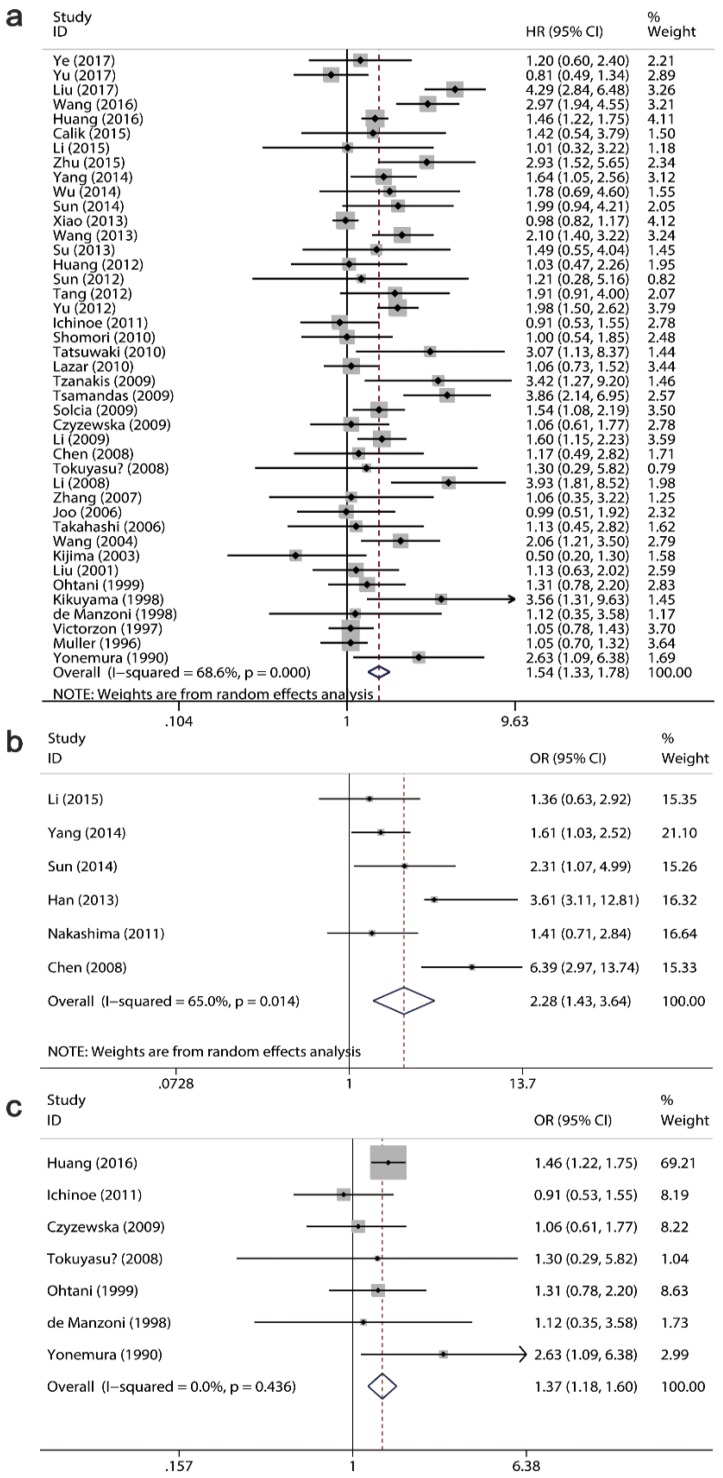
A total 42 studies with 6337 patients assessed the relationship of Ki-67/MKI67 expression with OS, while 6 studies with 564 cases evaluated the association between Ki-67/MKI67 expression and DFS. Among the 42 OS-related studies, 7 studies reported the prognostic role of Ki-67/MKI67 in advanced GC. The pooled HRs from a random-effects model indicated that an elevated expression of Ki-67/MKI67 predicted a poor OS (HR: 1.54, 95% CI: 1.33-1.78, P<0.0001; I2=69.6, P<0.0001; Fig. 3a) and DFS (HR: 2.28, 95% CI: 1.43-3.64, P<0.0001; I2=65, P=0.014; Fig. 3b) in patients with gastric cancer. Additionally, the combined HR from a fixed-effects model showed that a high Ki-67/MKI67 expression was also significantly connected with OS in patients with advanced GC (HR: 1.37, 95% CI: 1.18-1.60, P<0.0001; I2=0, P=0.436; Fig. 3c).
Fig 3.
Forest plots of the pooled harzard ratio (HR) for overall survival (OS) and disease-free survival (DFS). HR>1 indicates a worse OS for the group with an elevated Ki-67/MKI67 expression. (a) A high expression of Ki-67/MKI67 indicated a poor OS in patients with gastric cancer (GC). (b) A high expression of Ki-67/MKI67 indicated a poor DFS in patients with GC. (c) A high expression of Ki-67/MKI67 indicated a poor OS in patients with advanced GC.
Then we conducted subgroup analyses to determine whether different regions, antibodies, cut-off values and statistical methods resulted in the alterations of the pooled results (Table 3). Subgroup analysis on regions for OS suggested that a high Ki-67/MKI67 expression predicted a worse survival outcome both in Asian (HR: 1.58, 95% CI: 1.33-1.88, P<0.0001) and non-Asian populations (HR: 1.40, 95% CI: 1.06-1.85, P=0.02). For DFS, the pooled HR in Asian subgroup was 2.52 (95% CI=1.48-4.32, P<0.0001). There was only one study evaluating the relationship between Ki-67/MKI67 expression and DFS in non-Asian subgroup, and the HR was 1.41 (95% CI: 0.71-2.81, P=0.329), respectively. Subgroup analysis on antibodies showed that a highly expressed Ki-67/MKI67 indicated poor OS both in MIB-1 group (HR=1.49, 95% CI=1.21-1.84, P<0.0001) and anti-Ki-67 group (HR=1.56, 95% CI=1.27-1.92, P<0.0001). For DFS, the pooled HRs in MIB-1 and anti-Ki-67 groups were 2.97 (95% CI=0.68-13.07, P=0.149) and 2.00 (95% CI=1.30-3.00, P=0.001). We stratified all of the included studies into high and low cut-off value groups according to the cut-off values of 0.2, 0.25, 0.3, 0.4 and 0.45 to identify a cut-off value of Ki-67/MKI67 expression that could be strongly related to survival expectancy (data not shown). We found that the group with a cut-off value less than 0.25 showed the highest HR, and the group with a cut-off value more than 0.25 showed the lowest HR. For OS, the HRs in high and low cut-off value groups were 1.38 (95% CI: 1.17-1.63, P<0.0001) and 1.77 (95% CI: 1.37-2.28, P<0.0001), respectively. Two studies did not provide the cut-off value of Ki-67/MKI67 expression, and the pooled HR of the two articles was 2.00 (95% CI: 1.36-2.93, P<0.0001). For DFS, the overall results suggested that highly expressed Ki-67/MKI67 was related to worse survival outcome both in high cut-off value group (HR: 1.51, 95% CI: 1.08-2.11, P=0.017) and low cut-off value group (HR: 3.76, 95% CI: 2.14-6.60, P<0.0001).
Table 3.
Results of subgroup analyses for OS and DFS
| Heterogeneity test | Publication bias | ||||||
|---|---|---|---|---|---|---|---|
| Group | Number of studies | HR (95% CI) | P | I2 (%) | P | Begg's P | Egger's P |
| OS | 42 | 1.54 (1.33-1.78) | <0.0001 | 68.6 | <0.0001 | 0.573 | 0.19 |
| Region | |||||||
| Asian | 33 | 1.58 (1.33-1.88) | <0.0001 | 69.3 | <0.0001 | ||
| Non-Asian | 9 | 1.40 (1.06-1.85) | 0.02 | 65.3 | 0.003 | ||
| Antibody | |||||||
| MIB-1 | 18 | 1.49 (1.21-1.84) | <0.0001 | 56.6 | 0.002 | ||
| Anti-Ki-67 | 24 | 1.56 (1.27-1.92) | <0.0001 | 74.8 | <0.0001 | ||
| Cut-off value | |||||||
| Low | 18 | 1.77 (1.37-2.28) | <0.0001 | 56.2 | 0.002 | ||
| High | 22 | 1.38 (1.17-1.63) | <0.0001 | 67.2 | <0.0001 | ||
| NA | 2 | 2.00 (1.36-2.93) | <0.0001 | 0 | 0.534 | ||
| Statistical methods | |||||||
| Multivariate analysis | 13 | 1.62 (1.24-2.12) | <0.0001 | 82.1 | <0.0001 | ||
| Univariate analysis | 6 | 1.59 (0.90-2.80) | 0.111 | 80.4 | <0.0001 | ||
| Survival curve | 23 | 1.46 (1.23-1.74) | <0.0001 | 41.4 | 0.021 | ||
| DFS | 6 | 2.28 (1.43-3.64) | 0.001 | 65 | 0.014 | 0.133 | 0.376 |
| Regions | |||||||
| Asian | 5 | 2.52 (1.48-4.32) | 0.001 | 68.4 | 0.013 | ||
| Non-Asian | 1 | 1.41 (0.71-2.81) | 0.329 | None | None | ||
| Antibody | |||||||
| MIB-1 | 2 | 2.97 (0.68-13.07) | 0.149 | 87.9 | 0.004 | ||
| Anti-Ki-67 | 4 | 2.00 (1.30-3.00) | 0.001 | 35.6 | 0.199 | ||
| Cut-off value | |||||||
| Low | 3 | 3.76 (2.14-6.60) | <0.0001 | 41 | 0.183 | ||
| High | 3 | 1.51 (1.08-2.11) | 0.017 | 0 | 0.909 | ||
| Statistical methods | |||||||
| Multivariate analysis | 1 | 1.36 (0.63-2.92) | 0.433 | None | None | ||
| Univariate analysis | 1 | 1.41(0.71-2.81) | 0.329 | None | None | ||
| Survival curve | 4 | 2.92 (1.41-5.97) | 0.001 | 71.6 | 0.014 | ||
NA: data not available; OS: overall survival; DFS: disease-free survival; HR: hazard ratio; CI: confidence interval.
Subgroup analysis on the type of statistical methods applied to extract HR was also conducted. For OS, the pooled HRs in multivariate analysis, univariate analysis and survival curve subgroups were 1.62 (95% CI=1.24-2.12, P<0.0001), 1.59 (95% CI=0.90-2.80, P=0.111) and 1.46 (95% CI=1.23-1.74, P<0.0001), respectively. For DFS, the combined HR in survival curve subgroup was 2.92 (95% CI=1.41-5.97, P<0.0001). While there was only one study evaluating the relationship between Ki-67/MKI67 expression and DFS in multivariate analysis and univariate analysis subgroups, and the HRs were 1.36 (95% CI=0.63-2.92, P=0.433) and 1.41 (95% CI: 0.71-2.81, P=0.329), respectively.
Clinicopathological significance of Ki-67/MKI67 for GC patients
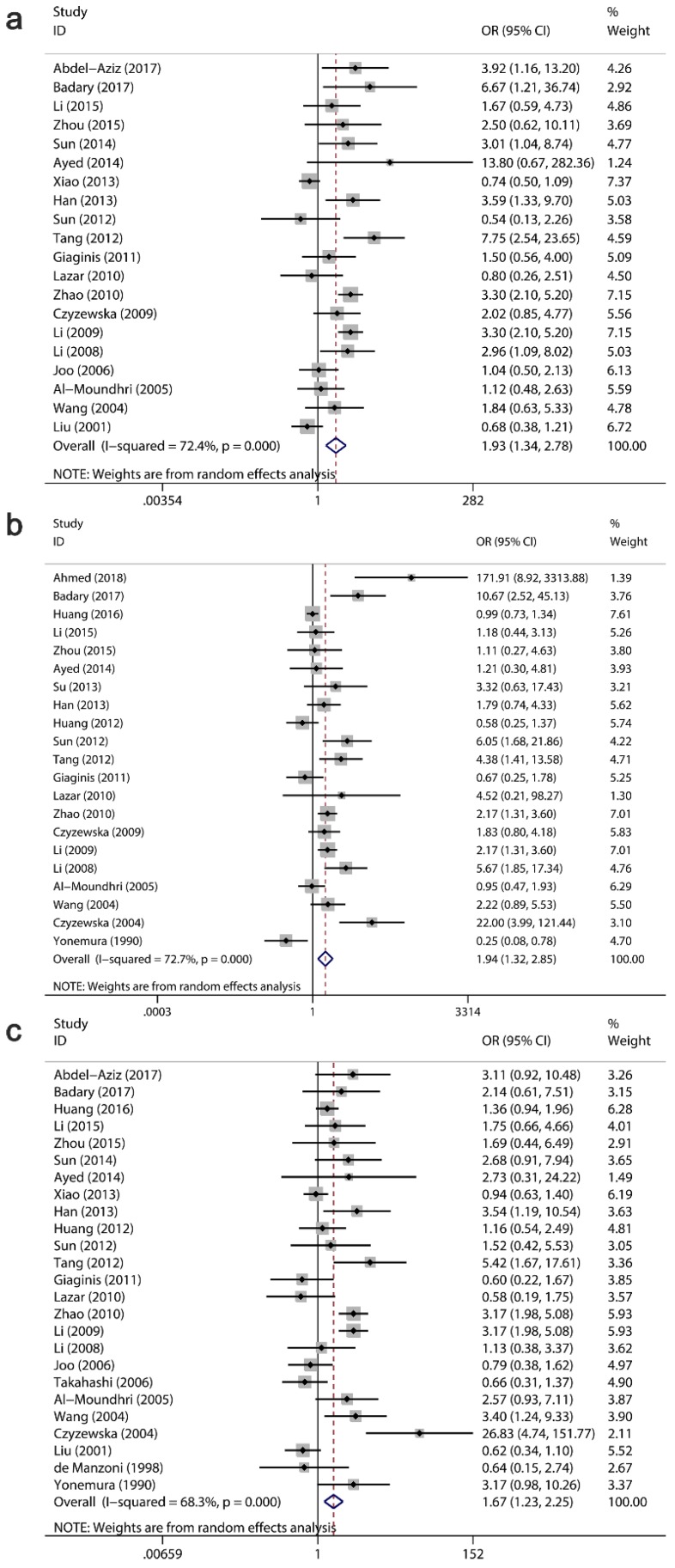
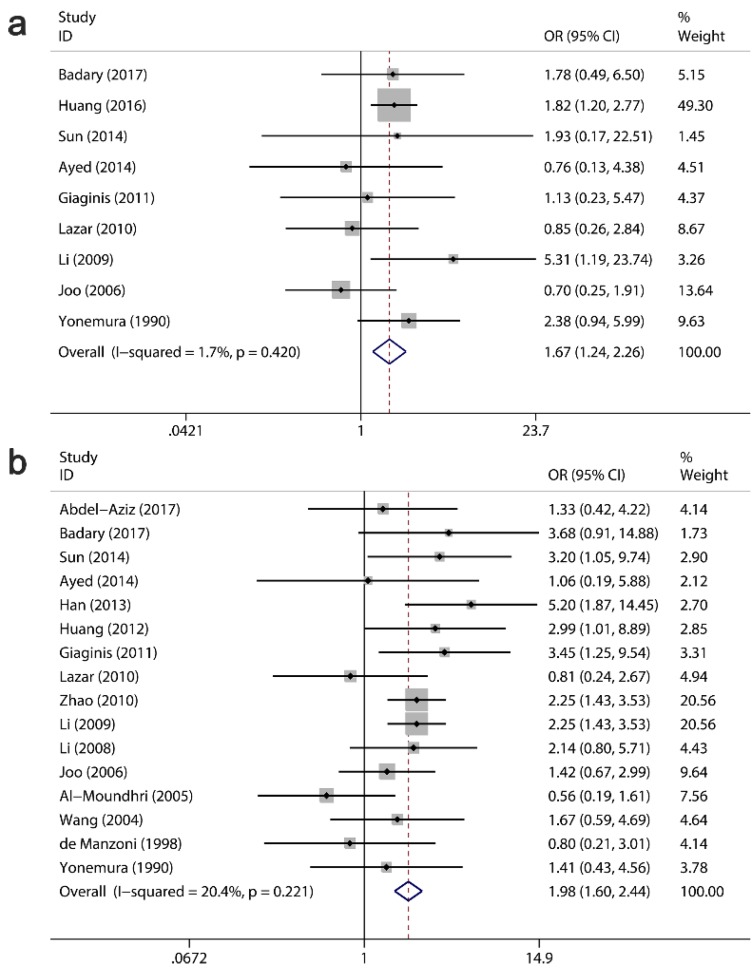
The clinicopathological value of Ki-67/MKI67 expression for GC patients was assessed (Table 4). The combined ORs from a random-effects model suggested that an up-regulation of Ki-67/MKI67 was related to TNM stage (stage III/IV versus stage I/II: OR=1.93, 95% CI=1.34-2.78, P<0.0001; I²=72.4, P<0.0001; Fig. 4a), tumor differentiation (poor versus well/moderate: OR=1.94, 95% CI=1.32-2.85, P =0.001; I²=72.4, P<0.0001; Fig. 4b) and lymph node metastasis (yes versus no: OR=1.67, 95% CI=1.23-2.25, P=0.001; I²=68.3, P<0.0001; Fig. 4c). The pooled ORs from a fixed-effects model showed that an increased expression of Ki-67/MKI67 was correlated with distant metastasis (yes versus no: OR=1.67, 95% CI=1.24-2.26, P=0.001; I²=1.7, P=0.42; Fig. 5a) and tumor invasion depth (T3/T4 versus Tis/T1/T2: OR=1.98, 95% CI=1.60-2.44, P<0.0001; I²=20.4, P=0.221; Fig. 5b). No statistically significant correlations were found of Ki-67/MKI67 expression with gender (female versus male: OR=1.00, 95% CI=0.84-1.19, P=0.986; I²=0, P=0.541), Lauren's classification (intestinal versus diffuse: OR=1.21, 95% CI=0.97-1.53, P=0.096; I²=46.3, P=0.053) and vascular invasion (yes versus no: OR=1.66, 95% CI=0.70-3.95, P=0.253; I²=72.2, P=0.006).
Table 4.
Relationships between Ki-67/MKI67 expression and clinicopathological parameters
| Heterogeneity test | Publication bias | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Clinicopathological features | Number of studies | Number of patients | OR (95% CI) | P value | I² (%) | P value | Model | Begg's P | Egger's P |
| Gender | 20 | 2574 | 1.00 (0.84-1.19) | 0.986 | 0 | 0.541 | Fixed-effects model | 0.074 | 0.387 |
| TNM Stage | 20 | 2452 | 1.93 (1.34-2.78) | <0.0001 | 72.4 | <0.0001 | Random-effects model | 0.27 | 0.253 |
| Tumor differentiation | 21 | 2669 | 1.94 (1.32-2.85) | 0.001 | 72.7 | <0.0001 | Random-effects model | 0.08 | 0.487 |
| Lymph node metastasis | 25 | 3439 | 1.67 (1.23-2.25) | 0.001 | 68.3 | <0.0001 | Random-effects model | 0.207 | 0.317 |
| Distant metastasis | 9 | 1557 | 1.67 (1.24-2.26) | 0.001 | 1.7 | 0.42 | Fixed-effects model | 0.99 | 0.6 |
| Invasion depth | 16 | 1695 | 1.98 (1.60-2.44) | <0.0001 | 20.4 | 0.211 | Fixed-effects model | 0.322 | 0.341 |
| Lauren's classification | 10 | 1602 | 1.21 (0.97-1.53) | 0.096 | 46.3 | 0.053 | Fixed-effects model | 0.074 | 0.155 |
| Vascular invasion | 5 | 1329 | 1.66 (0.70-3.95) | 0.253 | 72.2 | 0.006 | Random-effects model | 0.462 | 0.31 |
OR: odds ratio; CI: confidence interval.
Fig 4.
Relationships between Ki-67/MKI67 expression and TNM stage, tumor differentiation and lymph node metastasis. (a) TNM stage (stage III/IV versus stage I/II). (b) Tumor differentiation (poor versus well/moderate). (c) Lymph node metastasis (yes versus no).
Fig 5.
Relationships between Ki-67/MKI67 expression and distant metastasis, invasion depth and Lauren's classification. (a) Distant metastasis (yes versus no). (b) Tumor invasion depth (T3/T4 versus Tis/T1/T2).
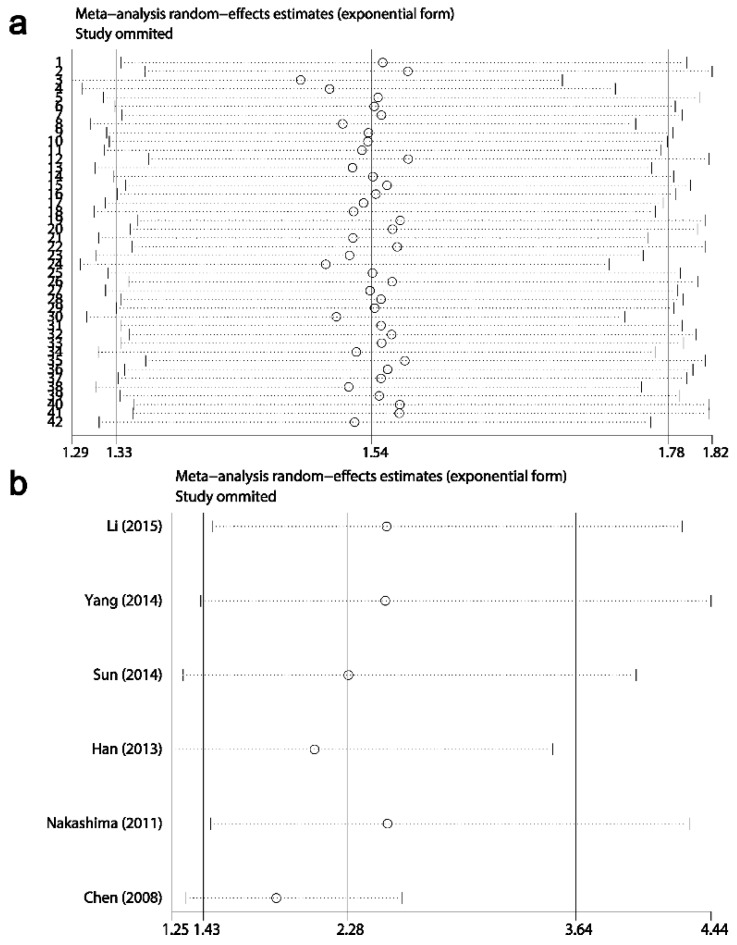
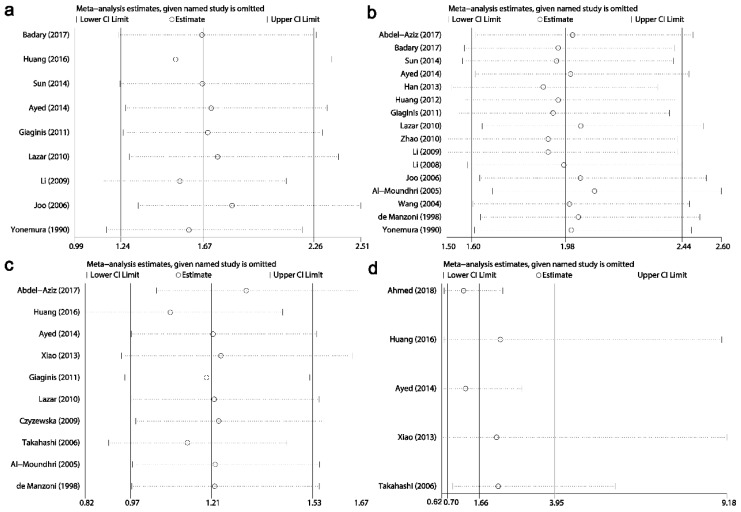
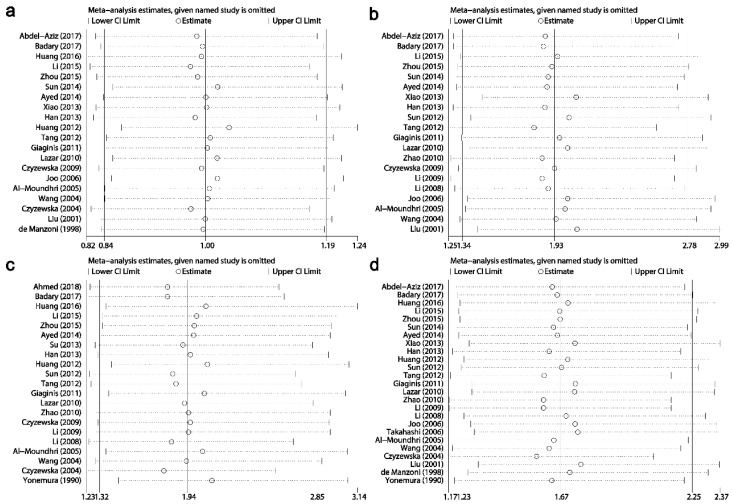
Sensitivity analysis
Results of sensitivity analysis showed that the combined HRs and ORs corresponding to the successive exclusion of individual reports were not significantly altered, indicating that the pooled results of the current meta-analysis were stable (Figs. 6-8).
Fig 6.
Result of sensitivity analysis from a random-effects model for overall survival (a) and disease-free survival (b). Three vertical lines indicated the pooled hazard ratio (HR) with 95% confidence interval (CI) calculated from all included studies. Each dotted horizontal line belongs to an independent meta-analysis. And the middle circle and two sides' short vertical lines represents the pooled HR and its 95% CI corresponding to the successive exclusion of each study.
Fig 8.
Results of sensitivity analysis for relationships between Ki-67/MKI67 expression and clinicopathological characteristics. (a) Distant metastasis. (b) Tumor invasion depth. (c) Lauren's classification. (d) Vascular invasion.
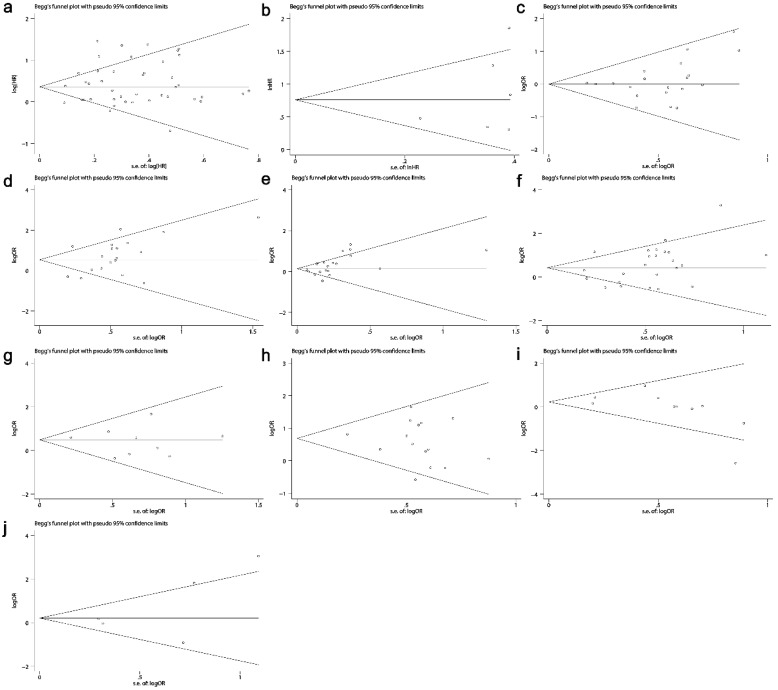
Publication bias
The Begg's funnel plots are presented in Fig. 9. No publication bias existed in all situations (Table 3 and Table 4).
Fig 9.
Funnel plots evaluating potential publication bias among the included studies. (a) Overall survival. (b) Disease-free survival. (c) Gender. (d) TNM stage. (e) Tumor differentiation. (f) Lymph node metastasis. (g) Distant metastasis. (h) Invasion depth. (i) Lauren's classification. (j) Vascular invasion. The circles represent an individual study enrolled in the present meta-analysis.
Potential action mechanism of MKI67 in GC
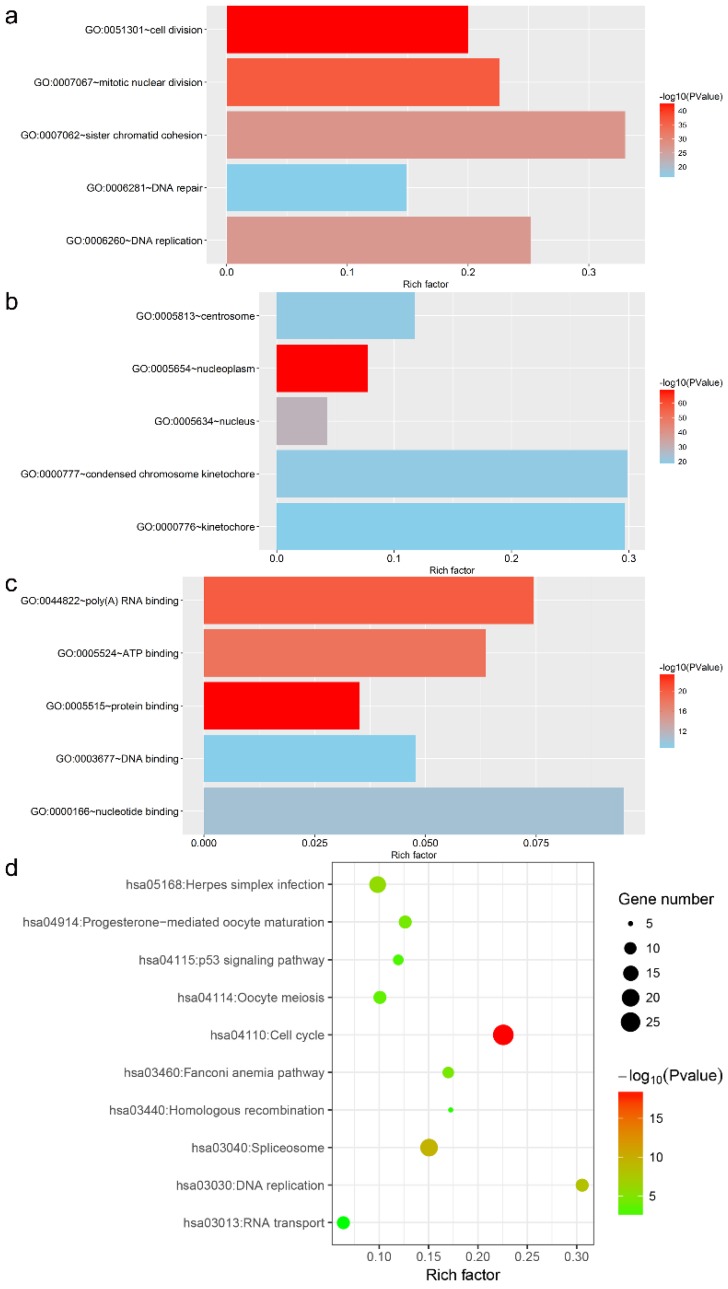
In total, 440 genes co-expressed with MKI67 in GC were collected for GO, KEGG pathway and PPI network analyses. The top five GO functional annotations and top ten KEGG pathways are presented in Fig. 10. The results of GO enrichment analyses indicated that the main biological functions of MKI67 were related to cell division, nucleoplasm and protein binding. The results of KEGG pathway analysis revealed that these co-expressed genes were involved in P53 signaling pathway.
Fig 10.
Top five Gene Ontology functional annotations and top ten Kyoto Encyclopedia of Genes and Genomes pathways of co-expressed genes of MKI67.
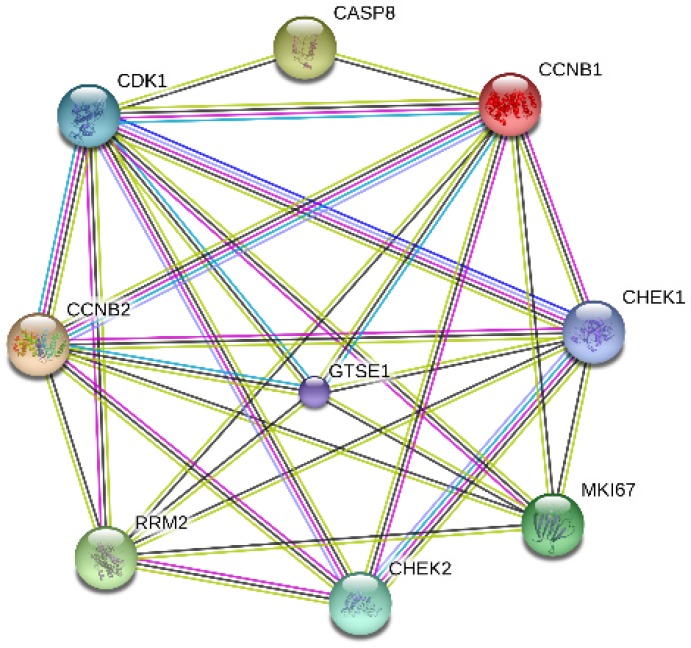
MKI67 and its co-expressed genes that involved in P53 signaling pathway were selected for PPI network construction. The following 8 genes were chosen: cyclin B1 (CCNB1), cyclin-dependent kinase 1 (CDK1), cyclin B1 (CCNB2), ribonucleotide reductase M2 (RRM2), caspase 8 (CASP8), checkpoint kinase 1(CHEK1), checkpoint kinase 2 (CHEK2) and G-2 and S-phase expressed 1 (GTSE1). According to our results, MKI67 directly interacted with genes RRM2, CCNB2, GTSE1, CDK1, CCNB1 and CHEK1 (Fig. 11).
Fig 11.
Interactions between MKI67 and eight genes related to P53 signaling pathway.
Discussion
In the present study, we firstly investigated the ability of Ki-67/MKI67 in discriminating tumor from normal controls. We found that an over-expressed Ki-67/MKI67 could differentiate gastric cancer patients from normal subjects. However, the result should be interpreted with caution because only three relevant studies were included and an inter-study heterogeneity among the three studies was discovered. Further researches with larger sample scales are necessary to validate our result.
A number of studies have indicated that Ki-67/MKI67 is a predictive marker for tumor deterioration and its expression shows an association with unfavorable prognosis and malignant clinical phenotypes in various cancers, such as bladder cancer 77, non-small cell lung cancer 78, hepatocellular carcinoma 13, cervical cancer 10 and glioma 16. The clinical and prognostic value of Ki-67/MKI67 in GC has also been investigated previously. Luo et al. 19 has conducted a meta-analysis based on 29 studies and demonstrated that a high Ki-67/MKI67 expression predicted poor OS and DFS in patients with GC. The authors also discovered that an increased Ki-67/MKI67 expression was related to Lauren's classification and tumor size. Liu et al. 18 also performed a meta-analysis and found that a highly expressed Ki-67/MKI67 predicted an inferior OS in 3825 patients with GC. Compare with Luo's and Liu's studies, our study included much more samples, resulting in more persuasive and stronger conclusions. A total of 53 studies with 7078 patients were enrolled in our meta-analysis. We demonstrated the prognostic and clinicopathological value of Ki-67/MKI67 for GC patients. Especially, we revealed the potential of Ki-67/MKI67 as a prognostic biomarker in advanced gastric cancer patients.
Metastasis is the most important cause of cancer-related death in patients with GC. Identifying a marker to predict the presence of metastasis and stratify patients into different risk groups is helpful in improving the prognosis of GC patients. Here, an up-regulation of Ki-67/MKI67 was discovered in patients with advanced TNM stage, poor tumor differentiation, serosa and neighboring organs invasion, lymph node metastasis and distant metastasis, suggesting that Ki-67/MKI67 could function as an indicator to predict gastric cancer progression and identify high-risk patients, thereby optimizing individual treatment management and improving the prognosis of patients with GC.
Ki-67/MKI67 is a well-known cell cycle-related protein. A recent study has corroborated that specific Ki-67/MKI67 splice variants promote cancer progression by influencing cell cycle 79. It is reported that P53 exert inhibitory effect on Ki-67 promoter by regulating P53- and SP1-dependent pathways 80. Interesting, we also demonstrated the close link between Ki-67/MKI67 and P53 signaling pathway. We found that MKI67 interacted with genes RRM2, CCNB2, GTSE1, CDK1, CCNB1 and CHEK1, which are critical molecules those are involved in P53 signaling network and related to cell cycle progression. Based on previous studies and our evidence, we hypothesize that MKI67 may promote the initiation and progression of GC via interacting with P53 signaling pathway. Further in vitro and in vivo experiments are necessary to confirm our conclusions.
There were still several deficiencies and limitations in our study. First, an obvious inter-study heterogeneity among the included studies was discovered in the present meta-analysis. There were no unified standards for the antibody concentration, IHC staining degree and cut-off value in different studies, which may be part of the reasons for the heterogeneity. Additionally, the differences in clinicopathological characteristics such as age, gender, tumor stage may also lead to the heterogeneity. A random-effects model was applied to eliminate the effect of the heterogeneity on our results. Remarkably, although the heterogeneity existed in our study, the results of sensitivity analyses indicated that no individual researches influenced the pooled HRs and ORs. Second, in 24 studies, HRs with 95% CIs had to be calculated from the Kaplan-Meier survival curves, which may decrease the reliability and accuracy of our meta-analysis. To reduce the deviation, three researchers independently extracted HRs from the Kaplan-Meier curves; the similarity between the copy curves drawn by the extracted data and original curves was the standard to identify which HRs could be used. Third, our meta-analysis depended on fully published studies written in English or Chinese, which may contribute to potential language bias. In addition, studies with positive results are more likely to be published in magazines. Thus, we should not ignore the potential bias even though the results from Begg's and Egger's tests showed no publication bias in the present study.
Conclusions
Our meta-analysis provides evidence that Ki-67/MKI67 not only could be a potential prognostic biomarker in clinic for GC patients but also could be an indicator to predict GC progression and to identify high-risk cases, thereby optimizing individual treatment management and improving the prognosis of GC patients. More interesting, we also found that gene MKI67 probably promoted the occurrence and development of GC by influencing P53 signaling pathway. Further well-designed studies are required to validate our conclusions.
Fig 7.
Results of sensitivity analysis for relationships between Ki-67/MKI67 expression and clinicopathological characteristics. (a) Gender. (b) TNM stage. (c) Tumor differentiation. (d) Lymph node metastasis.
Abbreviations
- GC
gastric cancer
- MKI67
antigen identified by monoclonal antibody Ki-67
- CNKI
Chinese National Knowledge Infrastructure
- HR
hazard ration
- OR
odds ratio
- CI
confidence interval
- TMA
tissue microarray
- NOS
Newcastle-Ottawa scale
- QADAS-2
Quality Assessment of Diagnostic Accuracy Studies-2
- SROC
summary receiver operating characteristic curve
- AUC
area under the value
- OS
overall survival
- DFS
disease-free survival
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein-protein interaction
- IHC
immunohistochemistry
References
- 1.Saricanbaz I, Karahacioglu E, Ekinci O. et al. Prognostic significance of expression of CD133 and Ki-67 in gastric cancer. Asian Pac J Cancer Prev. 2014;15:8215–9. doi: 10.7314/apjcp.2014.15.19.8215. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–15. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- 5.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 6.Song G, Zhang H, Chen C. et al. miR-551b regulates epithelial-mesenchymal transition and metastasis of gastric cancer by inhibiting ERBB4 expression. Oncotarget. 2017;8:45725–35. doi: 10.18632/oncotarget.17392. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Oshima CT, Iriya K, Forones NM. Ki-67 as a prognostic marker in colorectal cancer but not in gastric cancer. Neoplasma. 2005;52:420–4. [PubMed] [Google Scholar]
- 8.Gerdes J, Lemke H, Baisch H. et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5. [PubMed] [Google Scholar]
- 9.Inari H, Suganuma N, Kawachi K. et al. Clinicopathological and prognostic significance of Ki-67 immunohistochemical expression of distant metastatic lesions in patients with metastatic breast cancer. Breast Cancer. 2017;24:748–55. doi: 10.1007/s12282-017-0774-z. [DOI] [PubMed] [Google Scholar]
- 10.Piri R, Ghaffari A, Azami-Aghdash S. et al. Ki-67/MIB-1 as a Prognostic Marker in Cervical Cancer - a Systematic Review with Meta-Analysis. Asian Pac J Cancer Prev. 2015;16:6997–7002. doi: 10.7314/apjcp.2015.16.16.6997. [DOI] [PubMed] [Google Scholar]
- 11.Zeng A, Hu Q, Liu Y. et al. IDH1/2 mutation status combined with Ki-67 labeling index defines distinct prognostic groups in glioma. Oncotarget. 2015;6:30232–8. doi: 10.18632/oncotarget.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Zhang G, Wang HL. et al. Analysis of expression of cyclin E, p27kip1 and Ki67 protein in colorectal cancer tissues and its value for diagnosis, treatment and prognosis of disease. Eur Rev Med Pharmacol Sci. 2016;20:4874–9. [PubMed] [Google Scholar]
- 13.Luo Y, Ren F, Liu Y. et al. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med. 2015;8:10235–47. [PMC free article] [PubMed] [Google Scholar]
- 14.Wei DM, Chen WJ, Meng RM. et al. Augmented expression of Ki-67 is correlated with clinicopathological characteristics and prognosis for lung cancer patients: an up-dated systematic review and meta-analysis with 108 studies and 14,732 patients. Respir Res. 2018;19:150. doi: 10.1186/s12931-018-0843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan D, Wei K, Ling Y. et al. The prognostic role of Ki-67/MIB-1 in cervical cancer: a systematic review with meta-analysis. Med Sci Monit. 2015;21:882–9. doi: 10.12659/MSM.892807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen WJ, He DS, Tang RX. et al. Ki-67 is a valuable prognostic factor in gliomas: evidence from a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2015;16:411–20. doi: 10.7314/apjcp.2015.16.2.411. [DOI] [PubMed] [Google Scholar]
- 17.Xiong DD, Lin XG, He RQ. et al. Ki67/MIB-1 predicts better prognoses in colorectal cancer patients received both surgery and adjuvant radio-chemotherapy: a meta-analysis of 30 studies. Int J Clin Exp Med. 2017;10:1788–804. [Google Scholar]
- 18.Liu G, Xiong D, Zeng J. et al. Clinicopathological and prognostic significance of Ki-67 immunohistochemical expression in gastric cancer: a systematic review and meta-analysis. Onco Targets Ther. 2017;10:4321–8. doi: 10.2147/OTT.S143089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo G, Hu Y, Zhang Z. et al. Clinicopathologic significance and prognostic value of Ki-67 expression in patients with gastric cancer: a meta-analysis. Oncotarget. 2017;8:50273–83. doi: 10.18632/oncotarget.17305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boger C, Behrens HM, Rocken C. Ki67-An unsuitable marker of gastric cancer prognosis unmasks intratumoral heterogeneity. J Surg Oncol. 2016;113:46–54. doi: 10.1002/jso.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 22.Whiting PF, Rutjes AW, Westwood ME. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 23.Zamora J, Abraira V, Muriel A. et al. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, Cao F, Yu X. et al. The expression of HDAC7 in cancerous gastric tissues is positively associated with distant metastasis and poor patient prognosis. Clin Transl Oncol. 2017;19:1045–54. doi: 10.1007/s12094-017-1639-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Li M, Zhang X. et al. Upregulation of CDK7 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Exp Mol Pathol. 2016;100:514–21. doi: 10.1016/j.yexmp.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Huang G, Chen S, Wang D. et al. High Ki67 Expression has Prognostic Value in Surgically-Resected T3 Gastric Adenocarcinoma. Clin Lab. 2016;62:141–53. doi: 10.7754/clin.lab.2015.150610. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Deng W, Ma J. et al. Prognostic evaluation of Nanog, Oct4, Sox2, PCNA, Ki67 and E-cadherin expression in gastric cancer. Med Oncol. 2015;32:433. doi: 10.1007/s12032-014-0433-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Wang Y, Huang H. et al. Upregulation of KPNbeta1 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Tumour Biol. 2015;37:661–72. doi: 10.1007/s13277-015-3839-7. [DOI] [PubMed] [Google Scholar]
- 29.Yang M, Wang X, Zhao Q. et al. Combined evaluation of the expression of NUCKS and Ki-67 proteins as independent prognostic factors for patients with gastric adenocarcinoma. Tumour Biol. 2014;35:7505–12. doi: 10.1007/s13277-014-1880-6. [DOI] [PubMed] [Google Scholar]
- 30.Wu A, Jia Y, Dong B. et al. Apoptosis and KI 67 index correlate with preoperative chemotherapy efficacy and better predict the survival of gastric cancer patients with combined therapy. Cancer Chemother Pharmacol. 2014;73:885–93. doi: 10.1007/s00280-014-2410-3. [DOI] [PubMed] [Google Scholar]
- 31.Sun LM, Zhang J, Zhao JB. et al. Expression and clinical significance of Ki-67 in gastric carcinoma. Chinese Medical Journal. 2014;16:89–92. [Google Scholar]
- 32.Wang Q, Liu X, Zhou J. et al. Ribonucleotide reductase large subunit M1 predicts poor survival due to modulation of proliferative and invasive ability of gastric cancer. PLoS One. 2013;8:e70191. doi: 10.1371/journal.pone.0070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su YF, Wang XJ, Ma JF. Expression and clinical significance of NET-1. FHIT and Ki-67 in gastric cancer tissues. Chinese Journal of surgical oncology. 2013;5:26–9. [Google Scholar]
- 34.Huang L. The expression and significance of p53 protein and Ki-67 antigen in gastric carcinoma. Anhui medical University; 2012. [Google Scholar]
- 35.Sun QQ. Expression and clinical significance of TK1 and Ki-67 in benign and malignant diseases of stomach. Fujian medical University; 2012. [Google Scholar]
- 36.Tang H, Zhao T. Expression and clinical significance of DLC-1 and Ki-67 protein in gastric carcinoma. Anhui Medical University. 2012;33:1473–5. [Google Scholar]
- 37.Yu JR. Expression and prognostic value of Ki-67 protein in young patients with gastric cancer. Nanchang University; 2012. [Google Scholar]
- 38.Li YZ, Zhao P. [Expressions of cyclinB1, FHIT and Ki-67 in 336 gastric carcinoma patients and their clinicopathologic significance] Zhonghua Yi Xue Za Zhi. 2009;89:2337–41. [PubMed] [Google Scholar]
- 39.Chen L, Li X, Wang GL. et al. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 2008;94:531–8. doi: 10.1177/030089160809400415. [DOI] [PubMed] [Google Scholar]
- 40.Li JS. Expression and prognostic significance of Mcm2 protein in gastric cardia carcinogenesis. Shantou University; 2008. [Google Scholar]
- 41.Zhang JN, Liu Q, Lu W. The relationship between Survivin expression and the malignant proliferation of gastric cancer and its prognostic value. Journal of Practical Cancer. 2007;22:350–3. [Google Scholar]
- 42.Wang S. Expression of Tissue Inhibitor of Metalloproteinase 1 and its effect on the biological behavior and prognosis of gastric cancer. Chinese PLA postgraduate medical school; 2004. [Google Scholar]
- 43.Han CC. The relationship between Her-2 and Ki-67 expression and disease-free survival of patients with postoperative adjuvant chemotherapy in gastric cancer. Qinghai University; 2013. [Google Scholar]
- 44.Zhou Y, Li Y, Zheng J. et al. Detecting of gastric cancer by Bcl-2 and Ki67. Int J Clin Exp Pathol. 2015;8:7287–90. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao P, Li Y, Lu Y. Aberrant expression of CD133 protein correlates with Ki-67 expression and is a prognostic marker in gastric adenocarcinoma. BMC Cancer. 2010;10:218. doi: 10.1186/1471-2407-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye F, Jin P, Cai X. et al. High RNA-Binding Motif Protein 3 (RBM3) Expression is Independently Associated with Prolonged Overall Survival in Intestinal-Type Gastric Cancer. Med Sci Monit. 2017;23:6033–41. doi: 10.12659/MSM.905314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D, Sun L, Tong J. et al. Prognostic significance of glutathione peroxidase 2 in gastric carcinoma. Tumour Biol. 2017;39:1010428317701443. doi: 10.1177/1010428317701443. [DOI] [PubMed] [Google Scholar]
- 48.Xiao LJ, Zhao S, Zhao EH. et al. Clinicopathological and prognostic significance of Ki-67, caspase-3 and p53 expression in gastric carcinomas. Oncol Lett. 2013;6:1277–84. doi: 10.3892/ol.2013.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichinoe M, Mikami T, Yoshida T. et al. High expression of L-type amino-acid transporter 1 (LAT1) in gastric carcinomas: comparison with non-cancerous lesions. Pathol Int. 2011;61:281–9. doi: 10.1111/j.1440-1827.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 50.Shomori K, Nishihara K, Tamura T. et al. Geminin, Ki67, and minichromosome maintenance 2 in gastric hyperplastic polyps, adenomas, and intestinal-type carcinomas: pathobiological significance. Gastric Cancer. 2010;13:177–85. doi: 10.1007/s10120-010-0558-z. [DOI] [PubMed] [Google Scholar]
- 51.Tatsuwaki H, Tanigawa T, Watanabe T. et al. Reduction of 15-hydroxyprostaglandin dehydrogenase expression is an independent predictor of poor survival associated with enhanced cell proliferation in gastric adenocarcinoma. Cancer Sci. 2010;101:550–8. doi: 10.1111/j.1349-7006.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tokuyasu N, Shomori K, Nishihara K. et al. Minichromosome maintenance 2 (MCM2) immunoreactivity in stage III human gastric carcinoma: clinicopathological significance. Gastric Cancer. 2008;11:37–46. doi: 10.1007/s10120-008-0451-1. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi H, Murai Y, Tsuneyama K. et al. Overexpression of phosphorylated histone H3 is an indicator of poor prognosis in gastric adenocarcinoma patients. Appl Immunohistochem Mol Morphol. 2006;14:296–302. doi: 10.1097/00129039-200609000-00007. [DOI] [PubMed] [Google Scholar]
- 54.Kijima Y, Ishigami S, Hokita S. et al. The comparison of the prognosis between Epstein-Barr virus (EBV)-positive gastric carcinomas and EBV-negative ones. Cancer Lett. 2003;200:33–40. doi: 10.1016/s0304-3835(03)00410-5. [DOI] [PubMed] [Google Scholar]
- 55.Liu XP, Tsushimi K, Tsushimi M. et al. Expression of p21(WAF1/CIP1) and p53 proteins in gastric carcinoma: its relationships with cell proliferation activity and prognosis. Cancer Lett. 2001;170:183–9. doi: 10.1016/s0304-3835(01)00589-4. [DOI] [PubMed] [Google Scholar]
- 56.Ohtani M, Isozaki H, Fujii K. et al. Impact of the expression of cyclin-dependent kinase inhibitor p27Kip1 and apoptosis in tumor cells on the overall survival of patients with non-early stage gastric carcinoma. Cancer. 1999;85:1711–8. [PubMed] [Google Scholar]
- 57.Kikuyama S, Kubota T, Shimizu K. et al. Ki-67 antigen expression in relation to clinicopathological variables and prognosis in gastric cancer. Oncol Rep. 1998;5:867–70. doi: 10.3892/or.5.4.867. [DOI] [PubMed] [Google Scholar]
- 58.Yonemura Y, Ohoyama S, Kimura H. et al. Assessment of tumor cell kinetics by monoclonal antibody Ki-67. Eur Surg Res. 1990;22:365–70. doi: 10.1159/000129123. [DOI] [PubMed] [Google Scholar]
- 59.Nakashima Y, Yao T, Hirahashi M. et al. Nuclear atypia grading score is a useful prognostic factor in papillary gastric adenocarcinoma. Histopathology. 2011;59:841–9. doi: 10.1111/j.1365-2559.2011.04035.x. [DOI] [PubMed] [Google Scholar]
- 60.Tzanakis NE, Peros G, Karakitsos P. et al. Prognostic significance of p53 and Ki67 proteins expression in Greek gastric cancer patients. Acta Chir Belg. 2009;109:606–11. doi: 10.1080/00015458.2009.11680496. [DOI] [PubMed] [Google Scholar]
- 61.Tsamandas AC, Kardamakis D, Tsiamalos P. et al. The potential role of Bcl-2 expression, apoptosis and cell proliferation (Ki-67 expression) in cases of gastric carcinoma and correlation with classic prognostic factors and patient outcome. Anticancer Res. 2009;29:703–9. [PubMed] [Google Scholar]
- 62.Giaginis C, Giagini A, Tsourouflis G. et al. MCM-2 and MCM-5 expression in gastric adenocarcinoma: clinical significance and comparison with Ki-67 proliferative marker. Dig Dis Sci. 2011;56:777–85. doi: 10.1007/s10620-010-1348-5. [DOI] [PubMed] [Google Scholar]
- 63.Solcia E, Klersy C, Mastracci L. et al. A combined histologic and molecular approach identifies three groups of gastric cancer with different prognosis. Virchows Arch. 2009;455:197–211. doi: 10.1007/s00428-009-0813-z. [DOI] [PubMed] [Google Scholar]
- 64.de Manzoni G, Verlato G, Tomezzoli A. et al. Study on Ki-67 immunoreactivity as a prognostic indicator in patients with advanced gastric cancer. Jpn J Clin Oncol. 1998;28:534–7. doi: 10.1093/jjco/28.9.534. [DOI] [PubMed] [Google Scholar]
- 65.Czyzewska J, Guzinska-Ustymowicz K, Pryczynicz A. et al. Immunohistochemical evaluation of Ki-67, PCNA and MCM2 proteins proliferation index (PI) in advanced gastric cancer. Folia Histochem Cytobiol. 2009;47:289–96. doi: 10.2478/v10042-009-0042-y. [DOI] [PubMed] [Google Scholar]
- 66.Czyzewska J, Guzinska-Ustymowicz K, Lebelt A. et al. Evaluation of proliferating markers Ki-67, PCNA in gastric cancers. Rocz Akad Med Bialymst. 2004;49(Suppl 1):64–6. [PubMed] [Google Scholar]
- 67.Joo YE, Chung IJ, Park YK. et al. Expression of cyclooxygenase-2, p53 and Ki-67 in gastric cancer. J Korean Med Sci. 2006;21:871–6. doi: 10.3346/jkms.2006.21.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muller W, Schneiders A, Meier S. et al. Immunohistochemical study on the prognostic value of MIB-1 in gastric carcinoma. Br J Cancer. 1996;74:759–65. doi: 10.1038/bjc.1996.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Badary DM, Abdel-Wanis ME, Hafez MZ. et al. Immunohistochemical analysis of PTEN, HER2/neu, and ki67 expression in patients with gastric cancer and their association with survival. Pathophysiology. 2017;24:99–106. doi: 10.1016/j.pathophys.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 70.Abdel-Aziz A, Ahmed RA, Ibrahiem AT. Expression of pRb, Ki67 and HER 2/neu in gastric carcinomas: Relation to different histopathological grades and stages. Ann Diagn Pathol. 2017;30:1–7. doi: 10.1016/j.anndiagpath.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 71.Victorzon M, Roberts PJ, Haglund C. et al. Ki-67, ploidy and S-phase fraction as prognostic factors in gastric cancer. Anticancer Res. 1997;17:2923–6. [PubMed] [Google Scholar]
- 72.Lazar D, Taban S, Sporea I. et al. Ki-67 expression in gastric cancer. Results from a prospective study with long-term follow-up. Rom J Morphol Embryol. 2010;51:655–61. [PubMed] [Google Scholar]
- 73.Calik M, Demirci E, Altun E. et al. Clinicopathological importance of Ki-67, p27, and p53 expression in gastric cancer. Turk J Med Sci. 2015;45:118–28. doi: 10.3906/sag-1311-107. [DOI] [PubMed] [Google Scholar]
- 74.Al-Moundhri MS, Nirmala V, Al-Hadabi I. et al. The prognostic significance of p53, p27 kip1, p21 waf1, HER-2/neu, and Ki67 proteins expression in gastric cancer: a clinicopathological and immunohistochemical study of 121 Arab patients. J Surg Oncol. 2005;91:243–52. doi: 10.1002/jso.20324. [DOI] [PubMed] [Google Scholar]
- 75.Ayed DB, Khabir A, Abid M. et al. Clinicopathological and prognostic significance of p53, Ki-67, and Bcl-2 expression in Tunisian gastric adenocarcinomas. Acta Histochem. 2014;116:1244–50. doi: 10.1016/j.acthis.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 76.Ahmed A, Al-Tamimi DM. Incorporation of p-53 mutation status and Ki-67 proliferating index in classifying Her2-neu positive gastric adenocarcinoma. Libyan J Med. 2018;13:1466573. doi: 10.1080/19932820.2018.1466573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian Y, Ma Z, Chen Z. et al. Clinicopathological and Prognostic Value of Ki-67 Expression in Bladder Cancer: A Systematic Review and Meta-Analysis. PLoS One. 2016;11:e0158891. doi: 10.1371/journal.pone.0158891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wen S, Zhou W, Li CM. et al. Ki-67 as a prognostic marker in early-stage non-small cell lung cancer in Asian patients: a meta-analysis of published studies involving 32 studies. BMC Cancer. 2015;15:520. doi: 10.1186/s12885-015-1524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chierico L, Rizzello L, Guan L. et al. The role of the two splice variants and extranuclear pathway on Ki-67 regulation in non-cancer and cancer cells. PLoS One. 2017;12:e0171815. doi: 10.1371/journal.pone.0171815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakano T, Ohno T, Ishikawa H. et al. Current advancement in radiation therapy for uterine cervical cancer. J Radiat Res. 2010;51:1–8. doi: 10.1269/jrr.09132. [DOI] [PubMed] [Google Scholar]