In twelve 4-(4-methoxyphenyl)piperazin-1-ium salts containing organic anions, the hydrogen-bonded supramolecular assembly ranges from simple chains via chains of rings and sheets to three-dimensional framework structures.
Keywords: synthesis, piperazines, crystal structure, disorder, twinning, hydrogen bonding, supramolecular assembly
Abstract
Twelve 4-(4-methoxyphenyl)piperazin-1-ium salts containing organic anions have been prepared and structurally characterized. The monohydrated benzoate, 4-fluorobenzoate, 4-chlorobenzoate and 4-bromobenzoate salts, C11H17N2O+·C7H5O2 −·H2O (I), C11H17N2O+·C7H4FO2 −·H2O (II), C11H17N2O+·C7H4ClO2 −·H2O (III), and C11H17N2O+·C7H4BrO2 −·H2O (IV), respectively, are isomorphous and all exhibit disorder in the 4-methoxyphenyl unit: the components are linked by N—H⋯O and O—H⋯O hydrogen bond to form chains of rings. The unsolvated 2-hydroxybenzoate, pyridine-3-carboxylate and 2-hydroxy-3,5-dinitrobenzoate salts, C11H17N2O+·C7H5O3 − (V), C11H17N2O+·C6H4NO2 − (VI) and C11H17N2O+·C7H3N2O7 − (VII), respectively, are all fully ordered: the components of (V) are linked by multiple N—H⋯O hydrogen bonds to form a chain of rings; those of (VI) are linked into a three-dimensional framework by a combination of N—H⋯O, C—H⋯O and C—H⋯N hydrogen bonds and those of (VII), where the anion has a structure reminiscent of the picrate anion, are linked into a three-dimensional array by N—H⋯O and C—H⋯O hydrogen bonds. The hydrogensuccinate and hydrogenfumarate salts, C11H17N2O+·C4H5O4 − (VIII) and C11H17N2O+·C4H3O3 − (IX), respectively, are isomorphous, and both exhibit disorder in the anionic component: N—H⋯O and O—H⋯O hydrogen bonds link the ions into sheets, which are further linked by C—H⋯π(arene) interactions. The anion of the hydrogenmaleate salt, C11H17N2O+·C4H3O3 − (X), contains a very short and nearly symmetrical O⋯H⋯O hydrogen bond, and N—H⋯O hydrogen bonds link the anions into chains of rings. The ions in the trichloroacetate salt, C11H17N2O+·C2Cl3O2 − (XI), are linked into simple chains by N—H⋯O hydrogen bonds. In the hydrated chloranilate salt, 2C11H17N2O+·C6Cl2O4 2−·2H2O (XII), which crystallizes as a non-merohedral twin, the anion lies across a centre of inversion in space group P21/n, and a combination of N—H⋯O and O—H⋯O hydrogen bonds generates complex sheets. Comparisons are made with the structures of some related compounds.
Chemical context
In recent years, N-(4-methoxyphenyl)piperazine (MeOPP) has emerged as a new addition to the range of designer recreational drugs, and considerable effort has been invested in the development of methods for the detection both of MeOPP itself and of its metabolites N-(4-hydroxyphenyl)piperazine and 4-hydroxyaniline (Arbo et al., 2012 ▸) in human fluids (Staack & Maurer, 2003 ▸; Staack et al., 2004 ▸). MeOPP has euphoric stimulant properties and its action on human physiology is similar to that of amphetamines (Staack & Maurer, 2005 ▸; Wohlfarth et al., 2010 ▸), but it has a significantly lower potential for abuse (Nagai et al., 2007 ▸). However, no therapeutic applications of MeOPP have been reported to date. In view of the reported properties of MeOPP, coupled with the broad range of biological activities exhibited by piperazine derivatives (Asif, 2015 ▸; Brito et al., 2019 ▸), we have recently initiated a programme of study centred on N-(4-methoxyphenyl)piperazine derivatives, and we have recently reported the synthesis and structures of a series of 1-aroyl-4-(4-methoxyphenyl)piperazines (Kiran Kumar et al., 2019 ▸). In a continuation of that work, we have now prepared a series of 4-methoxyphenyl)piperazin-1-ium salts of simple organic acids, (I)–(XII), in order to study the various patterns of hydrogen-bonding interactions present in these salts, which may eventually be of value in pharmacological and pharmaceutical applications (Kavitha et al., 2014 ▸; Kaur et al., 2015 ▸; Shaibah, Yathirajan et al., 2017 ▸; Shaibah, Sagar et al., 2017 ▸; Shaibah et al., 2019 ▸). Salts of this type are readily prepared by co-crystallizations of the piperazine and the acids in methanol solution and, in total, 28 different acids representing a wide range of chemical types were investigated (see Section 5): however, only twelve of these provided crystals suitable for single-crystal X-ray diffraction, and thus we report here the molecular and supramolecular structures of (I)–(XII) (Figs. 1 ▸–12 ▸
▸
▸
▸
▸
▸
▸
▸
▸
▸
▸).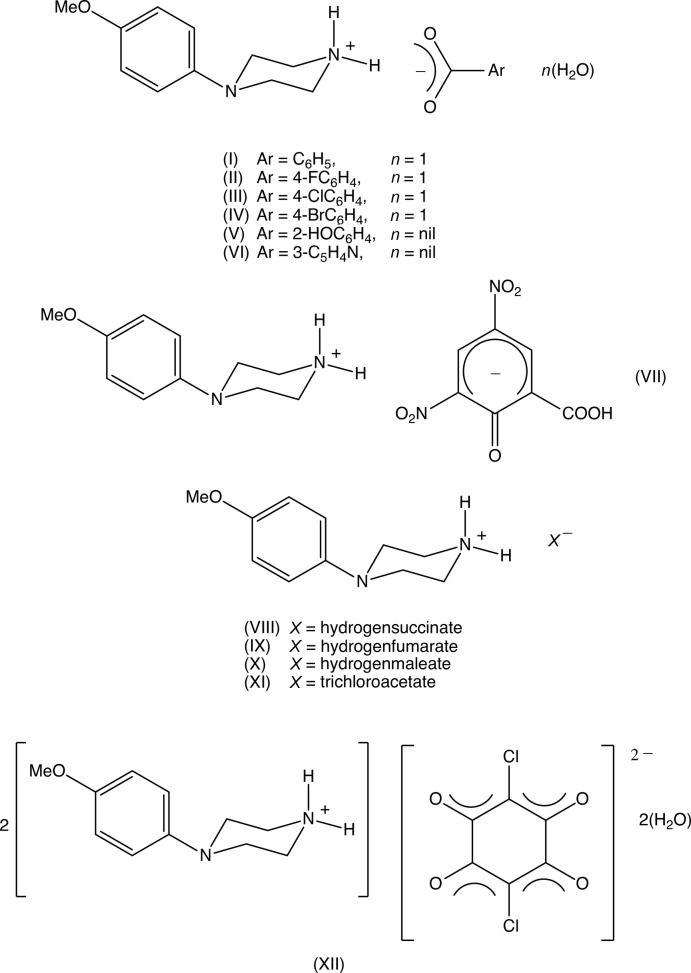
Figure 1.
The independent components of compound (I), showing the atom-labelling scheme, the disorder of the 4-methoxyphenyl group, and the hydrogen bonds within the selected asymmetric unit. The major disorder component is drawn using full lines and the minor disorder component is drawn using dashed lines. Displacement ellipsoids are drawn at the 30% probability level and, for the sake of clarity, a few of the atom labels have been omitted.
Figure 2.
The independent components of compound (II), showing the atom-labelling scheme, the disorder of the 4-methoxyphenyl group, and the hydrogen bonds within the selected asymmetric unit. The major disorder component is drawn using full lines and the minor disorder component is drawn using dashed lines. Displacement ellipsoids are drawn at the 30% probability level and, for the sake of clarity, a few of the atom labels have been omitted.
Figure 3.
The independent components of compound (III), showing the atom-labelling scheme, the disorder of the 4-methoxyphenyl group, and the hydrogen bonds within the selected asymmetric unit. The major disorder component is drawn using full lines and the minor disorder component is drawn using dashed lines. Displacement ellipsoids are drawn at the 30% probability level and, for the sake of clarity, a few of the atom labels have been omitted.
Figure 4.
The independent components of compound (IV), showing the atom-labelling scheme, the disorder of the 4-methoxyphenyl group, and the hydrogen bonds within the selected asymmetric unit. The major disorder component is drawn using full lines and the minor disorder component is drawn dashed broken lines. Displacement ellipsoids are drawn at the 30% probability level and, for the sake of clarity, a few of the atom labels have been omitted.
Figure 5.
The independent components of compound (V), showing the atom-labelling scheme and the hydrogen bonds within the selected asymmetric unit. Displacement ellipsoids are drawn at the 30% probability level.
Figure 6.
The independent components of compound (VI), showing the atom-labelling scheme and the hydrogen bonds within the selected asymmetric unit. Displacement ellipsoids are drawn at the 30% probability level.
Figure 7.
The independent components of compound (VII), showing the atom-labelling scheme and the hydrogen bond within the selected asymmetric unit. Displacement ellipsoids are drawn at the 30% probability level.
Figure 8.
The independent components of compound (VIII), showing the atom-labelling scheme, the disorder of anion, and the hydrogen bonds within the selected asymmetric unit. The major disorder component is drawn using full lines and the minor disorder component is drawn using dashed lines. Displacement ellipsoids are drawn at the 30% probability level.
Figure 9.
The independent components of compound (IX), showing the atom-labelling scheme, the disorder of anion, and the hydrogen bonds within the selected asymmetric unit. The major disorder component is drawn using full lines and the minor disorder component is drawn using dashed lines. Displacement ellipsoids are drawn at the 30% probability level.
Figure 10.
The independent components of compound (X), showing the atom-labelling scheme and the hydrogen bonds within the selected asymmetric unit. Displacement ellipsoids are drawn at the 30% probability level.
Figure 11.
The independent components of compound (XI), showing the atom-labelling scheme and the hydrogen bond within the selected asymmetric unit. Displacement ellipsoids are drawn at the 30% probability level.
Figure 12.
The independent components of compound (XII), showing the atom-labelling scheme and the hydrogen bonds within the selected asymmetric unit. Displacement ellipsoids are drawn at the 30% probability level, and the atoms marked with the suffix ‘a′ are at the symmetry position (1 − x, 1 − y, −z)
Structural commentary
Compounds (I)–(XI) are all 1:1 salts, but in (XII), where the dianion lies across a centre of inversion while the cation lies in a general position, the cation:anion ratio is 2:1. Compounds (I)–(IV) and (XII) all crystallize as hydrates, but compounds (V)–(XI) all crystallize in solvent-free form. Compounds (I)–(IV) are isomorphous (Table 2 ▸), in each of which the 4-methoxyphenyl groups is disordered over two sets of atomic sites (Figs. 1 ▸–4 ▸ ▸ ▸), having occupancies 0.66 (2) and 0.34 (2) in (I), 0.81 (3) and 0.19 (3) in (II), 0.73 (2) and 0.27 (2) in (III) and 0.80 (2) and 0.20 (2) in (IV). Similarly, compounds (VIII) and (IX) are isomorphous, and in both of them the anion exhibits disorder, with occupancies of 0.660 (15) and 0.340 (15) in (VIII), and 0.906 (9) and 0.094 (9) in (IX) (Figs. 8 ▸ and 9 ▸). While compounds (I)–(IV) are isostructural, compounds (VIII) and (IX) are not, because of both the different configurations of their anions and the different degrees of disorder. Examples have been previously reported of compounds that are isomorphous but not strictly isostructural in terms of their intermolecular interactions (Acosta et al., 2009 ▸).
Table 2. Experimental details.
| (I) | (II) | (III) | (IV) | |
|---|---|---|---|---|
| Crystal data | ||||
| Chemical formula | C11H17N2O+·C7H5O2 −·H2O | C11H17N2O+·C7H4FO2 −·H2O | C11H17N2O+·C7H4ClO2 −·H2O | C11H17N2O+·C7H4BrO2 −·H2O |
| M r | 332.39 | 350.38 | 366.83 | 411.28 |
| Crystal system, space group | Triclinic, P

|
Triclinic, P

|
Triclinic, P

|
Triclinic, P

|
| Temperature (K) | 296 | 293 | 293 | 293 |
| a, b, c (Å) | 6.215 (1), 7.547 (1), 18.716 (4) | 6.256 (1), 7.489 (1), 19.097 (2) | 6.211 (1), 7.481 (1), 20.144 (4) | 6.2004 (8), 7.4957 (9), 20.440 (2) |
| α, β, γ (°) | 84.34 (2), 87.14 (2), 84.69 (2) | 84.19 (1), 86.98 (2), 84.62 (2) | 84.90 (2), 87.48 (2), 85.19 (2) | 85.08 (1), 87.37 (1), 85.00 (1) |
| V (Å3) | 869.1 (3) | 885.4 (2) | 928.4 (3) | 942.17 (19) |
| Z | 2 | 2 | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 0.09 | 0.10 | 0.23 | 2.21 |
| Crystal size (mm) | 0.40 × 0.24 × 0.04 | 0.40 × 0.24 × 0.04 | 0.20 × 0.16 × 0.02 | 0.48 × 0.44 × 0.16 |
| Data collection | ||||
| Diffractometer | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) |
| T min, T max | 0.834, 0.996 | 0.973, 0.996 | 0.951, 0.995 | 0.536, 0.719 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 5751, 3442, 1839 | 5760, 3477, 1355 | 5883, 3454, 1343 | 6176, 3818, 2063 |
| R int | 0.029 | 0.046 | 0.041 | 0.018 |
| (sin θ/λ)max (Å−1) | 0.618 | 0.618 | 0.607 | 0.629 |
| Refinement | ||||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.054, 0.134, 1.02 | 0.066, 0.128, 1.01 | 0.065, 0.135, 0.94 | 0.068, 0.197, 1.06 |
| No. of reflections | 3442 | 3477 | 3454 | 3818 |
| No. of parameters | 256 | 265 | 265 | 265 |
| No. of restraints | 17 | 17 | 17 | 17 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.17, −0.19 | 0.13, −0.14 | 0.24, −0.23 | 0.94, −0.64 |
| (V) | (VI) | (VII) | (VIII) | |
|---|---|---|---|---|
| Crystal data | ||||
| Chemical formula | C11H17N2O+·C7H5O3 − | C11H17N2O+·C6H4NO2 − | C7H3N2O7 +·C11H17N2O− | C11H17N2O+·C4H5O4 − |
| M r | 330.38 | 315.37 | 420.38 | 310.35 |
| Crystal system, space group | Orthorhombic, P212121 | Orthorhombic, P b c a | Monoclinic, P21/c | Orthorhombic, P n a21 |
| Temperature (K) | 296 | 296 | 296 | 296 |
| a, b, c (Å) | 6.5009 (8), 7.9735 (9), 32.155 (4) | 9.2817 (7), 11.2905 (7), 30.309 (2) | 7.5500 (9), 7.6489 (9), 32.719 (6) | 9.3225 (9), 28.261 (3), 5.8228 (8) |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 91.30 (1), 90 | 90, 90, 90 |
| V (Å3) | 1666.8 (3) | 3176.2 (4) | 1889.0 (5) | 1534.1 (3) |
| Z | 4 | 8 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 0.09 | 0.09 | 0.12 | 0.10 |
| Crystal size (mm) | 0.42 × 0.42 × 0.34 | 0.46 × 0.42 × 0.36 | 0.18 × 0.12 × 0.06 | 0.44 × 0.42 × 0.24 |
| Data collection | ||||
| Diffractometer | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) |
| T min, T max | 0.899, 0.969 | 0.879, 0.968 | 0.916, 0.993 | 0.816, 0.976 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6249, 3564, 2875 | 22154, 3593, 2616 | 8215, 4074, 2003 | 5828, 2419, 2053 |
| R int | 0.014 | 0.028 | 0.038 | 0.018 |
| (sin θ/λ)max (Å−1) | 0.656 | 0.658 | 0.660 | 0.649 |
| Refinement | ||||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.041, 0.089, 1.05 | 0.048, 0.119, 1.04 | 0.066, 0.128, 1.03 | 0.043, 0.104, 1.14 |
| No. of reflections | 3564 | 3593 | 4074 | 2419 |
| No. of parameters | 228 | 215 | 281 | 233 |
| No. of restraints | 0 | 0 | 0 | 16 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.14, −0.13 | 0.19, −0.16 | 0.22, −0.23 | 0.16, −0.24 |
| Absolute structure | Flack x determined using 1011 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) | – | – | Flack x determined using 460 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | – | – | – | – |
| (IX) | (X) | (XI) | (XII) | |
|---|---|---|---|---|
| Crystal data | ||||
| Chemical formula | C11H17N2O+·C4H3O4 − | C11H17N2O+·C4H3O4 − | C11H17N2O+·C2Cl3O2 − | C11H17N2O+·0.5C6Cl2O4 2−·H2O |
| M r | 308.33 | 308.33 | 355.64 | 314.76 |
| Crystal system, space group | Orthorhombic, P n a21 | Monoclinic, P21/c | Orthorhombic, P c a21 | Monoclinic, P21/n |
| Temperature (K) | 296 | 296 | 296 | 296 |
| a, b, c (Å) | 9.069 (1), 28.528 (3), 5.8375 (9) | 9.063 (1), 6.4956 (9), 26.093 (3) | 10.6117 (11), 13.808 (1), 10.9137 (8) | 9.1597 (5), 15.1434 (8), 10.8742 (6) |
| α, β, γ (°) | 90, 90, 90 | 90, 93.18 (1), 90 | 90, 90, 90 | 90, 102.067 (5), 90 |
| V (Å3) | 1510.3 (3) | 1533.7 (3) | 1599.1 (2) | 1475.02 (14) |
| Z | 4 | 4 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 0.10 | 0.10 | 0.58 | 0.28 |
| Crystal size (mm) | 0.48 × 0.48 × 0.08 | 0.48 × 0.44 × 0.32 | 0.48 × 0.48 × 0.20 | 0.44 × 0.24 × 0.20 |
| Data collection | ||||
| Diffractometer | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD | Oxford Diffraction Xcalibur with Sapphire CCD |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) |
| T min, T max | 0.888, 0.992 | 0.871, 0.968 | 0.476, 0.892 | 0.892, 0.947 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 5834, 2827, 2316 | 6112, 3311, 2459 | 6173, 2428, 2278 | 9650, 9650, 7444 |
| R int | 0.015 | 0.014 | 0.027 | ? |
| (sin θ/λ)max (Å−1) | 0.650 | 0.651 | 0.654 | 0.651 |
| Refinement | ||||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.041, 0.101, 1.05 | 0.040, 0.111, 1.05 | 0.032, 0.086, 1.08 | 0.039, 0.105, 1.02 |
| No. of reflections | 2827 | 3311 | 2428 | 9650 |
| No. of parameters | 221 | 210 | 198 | 204 |
| No. of restraints | 11 | 0 | 1 | 0 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.15, −0.14 | 0.21, −0.13 | 0.25, −0.31 | 0.23, −0.32 |
| Absolute structure | Flack x determined using 769 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) | – | Classical Flack method preferred over Parsons because s.u. lower | – |
| Absolute structure parameter | – | – | 0.11 (7) | – |
In the anion of compound (VII), the carboxyl group is unionized, with C—O distances of 1.220 (3) and 1.309 (3) Å and it is the phenolic H atom which has been lost (Fig. 7 ▸). The C32—O33 distance, 1.280 (3) Å, is closer to that normally found in ketones than to that typical of phenols or phenolates (Allen et al., 1987 ▸): in addition, the C31—C32 and C32—C33 distances, 1.437 (4) and 1.430 (4) Å, respectively, are significantly larger than the other C—C distances in this ring, which lie in the rather narrow range 1.370 (3)–1.385 (4) Å, but the C—N and N—O distances are all typical of their types (Allen et al., 1987 ▸). These observations indicate that the negative charge in this anion is delocalized over the five atoms C31, C33, C34, C35 and C36, but without any significant delocalization onto the nitro groups, as has been observed in trinitrophenolate (picrate) anions (Kavitha et al., 2006 ▸; Sagar et al., 2017 ▸; Shaibah et al., 2017a ▸,b ▸).
The anion of compound (X) contains an almost linear and very short (Emsley, 1980 ▸; Herschlag & Pinney, 2018 ▸) O⋯H⋯O hydrogen bond, in which the H atom is almost, but not exactly, centred between the two O atoms (Table 1 ▸). In the centrosymmetric anion of compound (XII) (Fig. 12 ▸), the two independent C—O distances are identical within experimental uncertainty, 1.244 (2) and 1.246 (2) Å, as are the distances C31—C32 and C32—C33, 1.398 (3) and 1.392 (2) Å. However, the remaining C—C distance in this ring, 1.539 (3) Å is typical of a single C—C bond (Allen et al., 1987 ▸). These observations indicates the delocalization of a negative charge across each of the O–C–C–C–O units, and that these two units are effectively isolated from each other electronically. Despite the apparent simplicity of this dianion, with its high intrinsic symmetry, it is not possible adequately to describe its electronic structure in a single diagrammatic form, and four forms (A)–(D) (Fig. 13 ▸) are required.
Table 1. Hydrogen-bond parameters and short intermolecular contacts (Å, °).
Cg1 andCg2 are the centroids of the C31–C36 and C21–C26 rings, respectively.
| Compound | D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|---|
| (I) | N1—H11⋯O31 | 0.90 (2) | 1.88 (2) | 2.777 (3) | 174.1 (19) |
| N1—H12⋯O41 | 0.97 (2) | 1.85 (2) | 2.808 (3) | 169.7 (18) | |
| O41—H41⋯O32i | 0.88 (3) | 1.75 (3) | 2.631 (3) | 177 (3) | |
| O41—H42⋯O31ii | 0.91 (3) | 1.87 (3) | 2.763 (3) | 169 (3) | |
| C2—H2B⋯O31iii | 0.97 | 2.54 | 3.485 (3) | 165 | |
| C22—H22⋯Cg1ii | 0.93 | 2.85 | 3.603 (3) | 139 | |
| C26—H26⋯Cg1iv | 0.93 | 2.90 | 3.62 (2) | 135 | |
| C56—H56⋯Cg1iv | 0.93 | 2.64 | 3.41 (5) | 141 | |
| (II) | N1—H11⋯O31 | 1.09 (3) | 1.67 (3) | 2.758 (4) | 174.1 (19) |
| N1—H12⋯O41 | 0.86 (3) | 1.96 (3) | 2.818 (4) | 170 (3) | |
| O41—H41⋯O32i | 0.86 (4) | 1.75 (4) | 2.627 (4) | 174 (4) | |
| O41—H42⋯O31ii | 0.91 (4) | 1.88 (4) | 2.768 (3) | 163 (3) | |
| C2—H2B⋯O31iii | 0.97 | 2.58 | 3.529 (4) | 166 | |
| C6—H6B⋯O41i | 0.97 | 2.57 | 3.386 (4) | 142 | |
| C26—H26⋯Cg1iv | 0.93 | 2.81 | 3.56 (2) | 138 | |
| C56—H56⋯Cg1iv | 0.93 | 2.96 | 3.55 (9) | 123 | |
| (III) | N1—H11⋯O31 | 1.09 (3) | 1.71 (3) | 2.790 (4) | 176 (3) |
| N1—H12⋯O41 | 0.83 (3) | 1.98 (3) | 2.811 (4) | 174 (3) | |
| O41—H41⋯O32i | 0.91 (4) | 1.73 (4) | 2.624 (4) | 172 (4) | |
| O41—H42⋯O31ii | 0.94 (4) | 1.84 (4) | 2.775 (4) | 170 (4) | |
| C2—H2B⋯O31iii | 0.97 | 2.52 | 3.467 (4) | 165 | |
| C6—H6B⋯O41i | 0.97 | 2.60 | 3.408 (4) | 141 | |
| C22—H22⋯Cg1iv | 0.93 | 2.89 | 3.631 (13) | 137 | |
| C26—H26⋯Cg1iv | 0.93 | 2.81 | 3.58 (2) | 141 | |
| (IV) | N1—H11⋯O31 | 0.78 (4) | 2.03 (4) | 2.805 (5) | 174 (5) |
| N1—H12⋯O41 | 0.95 (5) | 1.86 (5) | 2.802 (5) | 172 (4) | |
| O41—H41⋯O32i | 0.79 (6) | 1.84 (6) | 2.623 (6) | 170 (6) | |
| O41—H42⋯O31ii | 0.79 (7) | 2.00 (7) | 2.772 (5) | 169 (6) | |
| C2—H2B⋯O31iii | 0.97 | 2.52 | 3.471 (5) | 166 | |
| C22—H22⋯Cg1ii | 0.93 | 2.52 | 3.471 (5) | 166 | |
| C26—H26⋯Cg1iv | 0.93 | 2.84 | 3.58 (2) | 137 | |
| (V) | N1—H11⋯O31 | 0.96 (3) | 1.85 (3) | 2.759 (3) | 156 (3) |
| N1—H11⋯O32 | 0.96 (3) | 2.47 (3) | 3.283 (3) | 142 (2) | |
| N1—H12⋯O32v | 0.95 (3) | 1.87 (3) | 2.806 (3) | 166 (2) | |
| O33—H33A⋯O31 | 0.97 (3) | 1.60 (3) | 2.516 (3) | 156 (3) | |
| C6—H6A⋯O33vi | 0.97 | 2.58 | 3.444 (3) | 148 | |
| C2—H2A⋯Cg1vii | 0.97 | 2.88 | 3.711 (3) | 144 | |
| C26—H26⋯Cg1viii | 0.93 | 2.87 | 3.642 (3) | 141 | |
| (VI) | N1—H11⋯O31 | 0.976 (19) | 1.714 (19) | 2.677 (2) | 168.2 (18) |
| N1—H12⋯O32ix | 0.94 (2) | 1.82 (2) | 2.749 (2) | 168.3 (17) | |
| C2—H2B⋯N31iv | 0.97 | 2.56 | 3.518 (2) | 169 | |
| C36—H36⋯O24x | 0.93 | 2.51 | 3.432 (2) | 172 | |
| C3—H3A⋯Cg1xi | 0.97 | 2.97 | 3.775 (2) | 156 | |
| (VII) | O32—H32⋯O33 | 1.04 (4) | 1.47 (4) | 2.472 (3) | 158 (3) |
| N1—H11⋯O33 | 0.93 (3) | 1.98 (3) | 2.020 (3) | 150 (3) | |
| N1—H11⋯O34 | 0.93 (3) | 2.27 (3) | 2.910 (3) | 126 (2) | |
| N1—H12⋯O31i | 0.93 (3) | 2.04 (3) | 2.931 (3) | 160 (3) | |
| N1—H12⋯O32i | 0.93 (3) | 2.58 (3) | 3.250 (3) | 129 (2) | |
| C34—H34⋯O36xii | 0.93 | 2.53 | 3.449 (3) | 171 | |
| C5—H5B⋯Cg2xiii | 0.97 | 2.84 | 3.639 (3) | 140 | |
| (VIII) | N1—H11⋯O31 | 0.86 (3) | 1.90 (3) | 2.750 (15) | 167 (4) |
| N1—H12⋯O32xiv | 0.98 (3) | 1.77 (4) | 2.741 (19) | 171 (3) | |
| O34—H34⋯O31xv | 0.82 | 1.79 | 2.60 (2) | 168 | |
| N1—H11⋯O41 | 0.86 (3) | 2.18 (4) | 3.03 (3) | 165 (4) | |
| N1—H12⋯O42xiv | 0.98 (3) | 1.82 (5) | 2.77 (4) | 163 (3) | |
| O44—H44⋯O41xv | 0.82 | 1.56 | 2.35 (2) | 161 | |
| C3—H3A⋯Cg2xvi | 0.97 | 2.76 | 3.652 (3) | 154 | |
| (IX) | N1—H11⋯O31 | 0.81 (4) | 2.18 (3) | 2.940 (4) | 155 (3) |
| N1—H12⋯O32xiv | 0.96 (4) | 1.77 (4) | 2.714 (4) | 169 (3) | |
| O34—H34⋯O31xv | 0.82 | 1.71 | 2.522 (5) | 170 | |
| O43—H34⋯O31xv | 0.82 | 1.62 | 2.44 (2) | 175 | |
| C3—H3A⋯Cg2xvi | 0.97 | 2.76 | 3.650 (3) | 153 | |
| (X) | O33—H33A⋯O32 | 1.167 (18) | 1.247 (18) | 2.4121 (16) | 175 (2) |
| N1—H11⋯O31 | 0.915 (17) | 2.126 (16) | 2.9309 (19) | 146.2 (15) | |
| N1—H11⋯O32 | 0.915 (17) | 2.296 (17) | 3.0798 (18) | 143.5 (14) | |
| N1—H12⋯O34xvii | 0.919 (18) | 1.881 (18) | 2.7563 (17) | 158.5 (17) | |
| C2—H2A⋯O34ii | 0.97 | 2.56 | 3.363 (2) | 140 | |
| (XI) | N1—H11⋯O31 | 0.92 (4) | 1.86 (4) | 2.775 (4) | 172 (3) |
| N1—H11⋯O32xviii | 0.97 (3) | 1.80 (3) | 2.724 (3) | 158 (3) | |
| (XII) | N1—H11⋯O31 | 0.89 (3) | 1.96 (3) | 2.802 (3) | 157 (2) |
| N1—H11⋯O33 | 0.89 (3) | 2.29 (2) | 2.838 (3) | 119 (2) | |
| N1—H12⋯O41 | 0.90 (2) | 1.92 (2) | 2.798 (3) | 168 (3) | |
| O41—H41⋯O33i | 0.84 (4) | 1.92 (4) | 2.738 (3) | 166 (3) | |
| O41—H42⋯O24xix | 0.82 (3) | 2.49 (3) | 3.269 (3) | 160 (3) |
Symmetry codes: (i) 1 − x, 1 − y, 1 − z; (ii) 2 − x, 1 − y, 1 − z; (iii) 2 − x, 2 − y, 1 − z; (iv) 1 − x, 2 − y, 1 − z; (v) 1 − x, − + y,
+ y,  − z; (vi) 2 − x, −
− z; (vi) 2 − x, − + y,
+ y,  − z; (vii) 2 − x,
− z; (vii) 2 − x,  + y,
+ y,  − z; (viii) 1 − x,
− z; (viii) 1 − x,  + y,
+ y,  − z; (ix)
− z; (ix)  − x, −
− x, − + y, z; (x) x,
+ y, z; (x) x,  − y,
− y,  + z; (xi) −
+ z; (xi) − + x,
+ x,  − y, 1 − z; (xii) 3 − x, 2 − y, 1 − z; (xiii) 1 − x, −
− y, 1 − z; (xii) 3 − x, 2 − y, 1 − z; (xiii) 1 − x, − + y,
+ y,  − z; (xiv) −
− z; (xiv) − + x,
+ x,  − y, z; (xv) −
− y, z; (xv) − + x,
+ x,  − y, 1 + z; (xvi) 1 − x, 1 − y,
− y, 1 + z; (xvi) 1 − x, 1 − y,  + z; (xvii) −1 + x, y, z; (xviii) −
+ z; (xvii) −1 + x, y, z; (xviii) − + x, 1 − y, z; (xix) −
+ x, 1 − y, z; (xix) − + x,
+ x,  − y, −
− y, − + z.
+ z.
Figure 13.
The canonical forms of the anion in compound (XII).
Supramolecular features
In each of the four isomorphous salts (I)–(IV), the ions are linked by a combination of N—H⋯O and O—H⋯O hydrogen bonds (Table 1 ▸) to form a chain of edge-fused centrosymmetric rings running parallel to the [100] direction, in which  (12) (Etter, 1990 ▸; Etter et al., 1990 ▸; Bernstein et al., 1995 ▸) rings centred at (n,
(12) (Etter, 1990 ▸; Etter et al., 1990 ▸; Bernstein et al., 1995 ▸) rings centred at (n,  ,
,  ) alternate with
) alternate with  (16) rings centred at (n +
(16) rings centred at (n +  ,
,  ,
,  ) , where n represents an integer in each case (Fig. 14 ▸). In each of these four salts, a combination of C—H⋯O and C—H⋯π(arene) hydrogen bonds links the [100] chain into complex sheets lying parallel to (001).
) , where n represents an integer in each case (Fig. 14 ▸). In each of these four salts, a combination of C—H⋯O and C—H⋯π(arene) hydrogen bonds links the [100] chain into complex sheets lying parallel to (001).
Figure 14.
Part of the crystal structure of compound (I) showing the formation of a chain of rings parallel to the [100] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the minor disorder component and the H atoms bonded to C atoms have been omitted.
There is an intermolecular O—H⋯O hydrogen bond in the anion of the unsolvated salt (V). The two anions in the selected asymmetric unit (Fig. 5 ▸) are linked by an asymmetric three-centre N—H⋯(O)2 hydrogen bond, and the resulting ion pairs, which are related by 21 screw axis along ( , y,
, y,  ), are linked by a two-centre N—H⋯O hydrogen bond to form chain of rings running parallel to the [010] direction (Fig. 15 ▸). Chains of this type are weakly linked into sheets lying parallel to (001) by a combination of C—H⋯O and C—H⋯π(arene) hydrogen bonds.
), are linked by a two-centre N—H⋯O hydrogen bond to form chain of rings running parallel to the [010] direction (Fig. 15 ▸). Chains of this type are weakly linked into sheets lying parallel to (001) by a combination of C—H⋯O and C—H⋯π(arene) hydrogen bonds.
Figure 15.
Part of the crystal structure of compound (V) showing the formation of a chain of rings parallel to the [010] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
The component ions in compound (VI) (Fig. 6 ▸) are linked by a two-centre N—H⋯O hydrogen bond and the resulting ion pairs are further linked by a combination of N—H⋯O, C—H⋯O and C—H⋯N hydrogen bonds to form a three-dimensional framework structure, whose formation can readily be analysed in terms of three simple sub-structures (Ferguson et al., 1998a
▸,b
▸; Gregson et al., 2000 ▸). Ion pairs which are related by the b-glide plane at x =  are linked by a second N—H⋯O hydrogen bond to form a
are linked by a second N—H⋯O hydrogen bond to form a  (6) chain running parallel to the [010] direction, and in the second sub-structure, ion pairs which are related by the c-glide plane at y =
(6) chain running parallel to the [010] direction, and in the second sub-structure, ion pairs which are related by the c-glide plane at y =  are linked by a C—H⋯O hydrogen bond (Table 1 ▸) to form a
are linked by a C—H⋯O hydrogen bond (Table 1 ▸) to form a  (17) chain running parallel to the [001] direction. The combination of these two simple chain motifs generates a sheet of
(17) chain running parallel to the [001] direction. The combination of these two simple chain motifs generates a sheet of  (40) rings lying parallel to (100) in the domain
(40) rings lying parallel to (100) in the domain  < x < 1.0 (Fig. 16 ▸). A second sheet of this type, related to the first by inversion lies in the domain 0 < x <
< x < 1.0 (Fig. 16 ▸). A second sheet of this type, related to the first by inversion lies in the domain 0 < x <  , and adjacent sheets are linked by the third sub-structure in which inversion-related ion pairs are linked by C—H⋯N hydrogen bonds into a centrosymmetric
, and adjacent sheets are linked by the third sub-structure in which inversion-related ion pairs are linked by C—H⋯N hydrogen bonds into a centrosymmetric  (18) ring (Fig. 17 ▸): the action of this interaction is to link all of the (100) sheets into a continuous three-dimensional array.
(18) ring (Fig. 17 ▸): the action of this interaction is to link all of the (100) sheets into a continuous three-dimensional array.
Figure 16.
Part of the crystal structure of compound (VI) showing the formation of a sheet of  (40) rings lying parallel to (100). Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms not involved in the motifs shown have been omitted.
(40) rings lying parallel to (100). Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms not involved in the motifs shown have been omitted.
Figure 17.

Part of the crystal structure of compound (VI) showing the formation of the  (18) ring which links the (100) sheets. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the unit-cell outline and the H atoms which are bonded to the C atoms not involved in the motif shown have been omitted. The atoms marked with an asterisk (*) are at the symmetry position (1 − x, 2 − y, 1 − z).
(18) ring which links the (100) sheets. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the unit-cell outline and the H atoms which are bonded to the C atoms not involved in the motif shown have been omitted. The atoms marked with an asterisk (*) are at the symmetry position (1 − x, 2 − y, 1 − z).
There is an intermolecular O—H⋯O hydrogen bond in the anion of compound (VII) (Fig. 7 ▸), but the carboxyl H atom plays no part in the supramolecular assembly. The ions are linked by a combination of N—H⋯O and C—H⋯O hydrogen bonds to form a chain of centrosymmetric rings running parallel to the [210] direction, in which  (10) rings centred at (2n −
(10) rings centred at (2n −  , n,
, n,  ) alternate with
) alternate with  (16) rings centred at (2n +
(16) rings centred at (2n +  , n +
, n +  ,
,  ), where n represents an integer in each case (Fig. 18 ▸). Two chains of this type, related to one another by the translational symmetry operations, pass through each unit cell, and a weak C—H⋯π(arene) hydrogen bond links the chains into a three-dimensional framework structure.
), where n represents an integer in each case (Fig. 18 ▸). Two chains of this type, related to one another by the translational symmetry operations, pass through each unit cell, and a weak C—H⋯π(arene) hydrogen bond links the chains into a three-dimensional framework structure.
Figure 18.
Part of the crystal structure of compound (VII) showing the formation of a chain of  (10) and
(10) and  (16) rings parallel to the [210] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms which are bonded to the C atoms not involved in the motif shown have been omitted.
(16) rings parallel to the [210] direction. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms which are bonded to the C atoms not involved in the motif shown have been omitted.
For the disordered structure of compound (VIII), the hydrogen bonds formed by the major and minor disorder components are very similar (Table 1 ▸) so that only the major disorder form need be considered in detail. Within the selected asymmetric unit (Fig. 8 ▸), the component ions are linked by a two-centre N—H⋯O hydrogen bond: the ion pairs are linked by a combination of N—H⋯O and O—H⋯O hydrogen bonds to form sheets, whose formation can readily be analysed in terms of two simple sub-structures. In the simpler of these, anions which are related by the a-glide plane at y =  are linked by O—H⋯O hydrogen bonds into C(7) chains running parallel to the [10
are linked by O—H⋯O hydrogen bonds into C(7) chains running parallel to the [10 ] direction (Fig. 19 ▸); in the second sub-structure, ion pairs which are related by the same glide plane are linked by N—H⋯O hydrogen bonds to form a
] direction (Fig. 19 ▸); in the second sub-structure, ion pairs which are related by the same glide plane are linked by N—H⋯O hydrogen bonds to form a  (6) chain running parallel to the [100] direction (Fig. 20 ▸). The combination of these two chain motifs generates a sheet lying parallel to (010), and a single C—H⋯π(arene) hydrogen bond links these sheets into a three-dimensional framework structure. The supramolecular aggregation in the isomorphous compound (IX) is similar to that in (VIII). As noted in Section 2 above, the anion in compound (X) contains a very short and nearly symmetrical O⋯H⋯O hydrogen bond. Within the selected asymmetric unit, the component ions are linked by the three-centre N—H⋯(O)2 hydrogen bond and ion pairs which are related by translation are linked by a two-centre N—H⋯O hydrogen bond to form a C(9)C(9)[
(6) chain running parallel to the [100] direction (Fig. 20 ▸). The combination of these two chain motifs generates a sheet lying parallel to (010), and a single C—H⋯π(arene) hydrogen bond links these sheets into a three-dimensional framework structure. The supramolecular aggregation in the isomorphous compound (IX) is similar to that in (VIII). As noted in Section 2 above, the anion in compound (X) contains a very short and nearly symmetrical O⋯H⋯O hydrogen bond. Within the selected asymmetric unit, the component ions are linked by the three-centre N—H⋯(O)2 hydrogen bond and ion pairs which are related by translation are linked by a two-centre N—H⋯O hydrogen bond to form a C(9)C(9)[ (4)] chain of rings running parallel to the [100] direction (Fig. 21 ▸). The C—H⋯O contact is at the margin of significance (Wood et al., 2009 ▸), but it involves chains related by inversion.
(4)] chain of rings running parallel to the [100] direction (Fig. 21 ▸). The C—H⋯O contact is at the margin of significance (Wood et al., 2009 ▸), but it involves chains related by inversion.
Figure 19.
Part of the crystal structure of compound (VIII) showing the formation of a C(7) chain of anions, parallel to [10 ]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted. The atoms marked with an asterisk (*) or a hash (#) are at the symmetry positions (
]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted. The atoms marked with an asterisk (*) or a hash (#) are at the symmetry positions ( + x,
+ x,  − y, −1 + z) and (−
− y, −1 + z) and (− + x,
+ x,  − y, 1 + z), respectively.
− y, 1 + z), respectively.
Figure 20.
Part of the crystal structure of compound (VIII) showing the formation of a  (6) chain parallel to [100]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
(6) chain parallel to [100]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
Figure 21.
Part of the crystal structure of compound (X) showing the formation of a chain of rings parallel to [100]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
The supramolecular assembly of compound (XI) is extremely simple: two N—H⋯O hydrogen bonds link the ions into a  (6) chain running parallel to the [100] direction (Fig. 22 ▸). In compound (XII), a combination of N—H⋯O and O—H⋯O hydrogen bonds links all three components into a chain of
(6) chain running parallel to the [100] direction (Fig. 22 ▸). In compound (XII), a combination of N—H⋯O and O—H⋯O hydrogen bonds links all three components into a chain of  (18) rings running parallel to the [001] direction (Fig. 23 ▸), while a second O—H⋯O hydrogen bond links a combination of cations and water molecules into a simple
(18) rings running parallel to the [001] direction (Fig. 23 ▸), while a second O—H⋯O hydrogen bond links a combination of cations and water molecules into a simple  (12) chain running parallel to the [101] direction (Fig. 24 ▸) and the combination of these two chain motifs generates a complex sheet lying parallel to (010).
(12) chain running parallel to the [101] direction (Fig. 24 ▸) and the combination of these two chain motifs generates a complex sheet lying parallel to (010).
Figure 22.

Part of the crystal structure of compound (XI) showing the formation of a  (6) chain parallel to [100]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
(6) chain parallel to [100]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
Figure 23.

Part of the crystal structure of compound (XII) showing the formation of an  (18) chain of rings parallel to [001]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
(18) chain of rings parallel to [001]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
Figure 24.
Part of the crystal structure of compound (XII) showing the formation of a  (12) chain of cations and water molecules parallel to [101]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
(12) chain of cations and water molecules parallel to [101]. Hydrogen bonds are drawn as dashed lines and, for the sake of clarity, the H atoms bonded to C atoms have been omitted.
Overall, therefore, the hydrogen-bonded assembly is one-dimensional in each of compounds (X) and (XI), two-dimensional in compounds (I)–(V) and (XII), and three-dimensional in compounds (VI)–(IX). Sub-structures in the form of chains of rings can be identified in compounds (I)–(IV) and in (VII), although (I)–(IV) are all monohydrates, while (VII) is solvent free: within the chain of rings formed by (I)–(IV) it is possible to identify a  (6) motif formed by water molecules and anions only (Fig. 14 ▸), and a
(6) motif formed by water molecules and anions only (Fig. 14 ▸), and a  (6) motif built from alternating cations and anions can, in fact, be identified in each of compounds (V), (VI), (VIII), (IX) and (XI) (Figs. 15 ▸, 16 ▸, 20 ▸, 22 ▸). By contrast, a
(6) motif built from alternating cations and anions can, in fact, be identified in each of compounds (V), (VI), (VIII), (IX) and (XI) (Figs. 15 ▸, 16 ▸, 20 ▸, 22 ▸). By contrast, a  (12) motif, built from water molecules and cations can be identified in the structure of compound (XII) (Fig. 24 ▸), but sub-structural motifs in the form of simple chains are uncommon in this series (Fig. 19 ▸).
(12) motif, built from water molecules and cations can be identified in the structure of compound (XII) (Fig. 24 ▸), but sub-structural motifs in the form of simple chains are uncommon in this series (Fig. 19 ▸).
Database survey
Compounds (I)–(IV), reported here, are isomorphous across the series of anions 4-XC6H4COO−, where X = H, F, Cl or Br, despite the rather disparate sizes of the substituents X. A similar, but more extreme, series of isomorphous salts was found in the substituted anilinium 5-nitro(hydrogenphthalate) salts (4-XC6H4NH3)+·(C8H4NO6)−, which are isomorphous for X = H, Cl, Br and I (Glidewell et al., 2005 ▸). The structures of a number of salts containing the chloranilate dianion have been reported (Ishida, 2004a ▸,b ▸,c ▸,d ▸; Sovago et al., 2016 ▸), and the geometric features previously observed in this anion are fully consistent with the geometry found here in (XII): the nature of the electronic delocalization has been confirmed in several such salts using a combination of deformation density plots and net atomic charge calculations (Sovago et al., 2016 ▸).
The structures of very few salts containing the 4-(methoxyphenyl)piperazin-1-ium cations have been reported. In 4-(4-methoxyphenyl)piperazin-1-ium chloride, two N—H⋯Cl hydrogen bonds link the ions into  (4) chains (Zia-ur-Rehman et al., 2009 ▸), and in the closely related 4-(4-nitrophenyl)piperazin-1-ium chloride monohydrate, a combination of N—H⋯O, O—H⋯Cl and N—H⋯Cl hydrogen bonds links the components into complex ribbons in which each anion accepts three hydrogen bonds (Lu, 2007 ▸). The structure of 4-(3-methoxyphenyl)piperazin-1-ium maleate has been reported (Verdonk et al., 1997 ▸), as have those of the picrate (Verdonk et al., 1997 ▸) and 6-chloro-5-isopropyl-2,4-dioxopyrimidin-1-ide (Al-Omary et al., 2014 ▸) salts of the 4-(2-methoxyphenyl)piperazin-1-ium cation. Finally we note, in addition to the 1-aroyl-4-(4-methoxyphenyl)piperazines referred to in Section 1 above (Kiran Kumar et al., 2019 ▸), the structure of 1-acetyl-(4-hydroxyphenyl)piperazine (Kavitha et al., 2013 ▸), which is an N-acetylated derivative of 4-(4-hydroxyphenyl)piperazines, a metabolite of 4-(4-methoxyphenyl)piperazine.
(4) chains (Zia-ur-Rehman et al., 2009 ▸), and in the closely related 4-(4-nitrophenyl)piperazin-1-ium chloride monohydrate, a combination of N—H⋯O, O—H⋯Cl and N—H⋯Cl hydrogen bonds links the components into complex ribbons in which each anion accepts three hydrogen bonds (Lu, 2007 ▸). The structure of 4-(3-methoxyphenyl)piperazin-1-ium maleate has been reported (Verdonk et al., 1997 ▸), as have those of the picrate (Verdonk et al., 1997 ▸) and 6-chloro-5-isopropyl-2,4-dioxopyrimidin-1-ide (Al-Omary et al., 2014 ▸) salts of the 4-(2-methoxyphenyl)piperazin-1-ium cation. Finally we note, in addition to the 1-aroyl-4-(4-methoxyphenyl)piperazines referred to in Section 1 above (Kiran Kumar et al., 2019 ▸), the structure of 1-acetyl-(4-hydroxyphenyl)piperazine (Kavitha et al., 2013 ▸), which is an N-acetylated derivative of 4-(4-hydroxyphenyl)piperazines, a metabolite of 4-(4-methoxyphenyl)piperazine.
Synthesis and crystallization
All reagents were obtained commercially and were used as received. For the synthesis of each of compounds (I)–(XII), equimolar quantities (0.52 mmol of each component) of N-(4-methoxyphenyl)piperazine and the appropriate acid were separately dissolved in methanol (10 ml) and the two solutions were then mixed, stirred briefly, and then set aside to crystallize, giving the solid products (I)–(XII) after a few days. The products were all collected by filtration and then dried in air. Yields (I) 81%, (II) 83%, (III) 83%, (IV) 81%, (V) 83%, (VI) 78%, (VII) 80%, (VIII) 82%, (IX) 82%, (X) 84%, (XI) 79%, (XII) 82%: melting ranges (I) 513–515 K, (II) 405–407 K, (III) 449–451 K, (IV) 447–449 K, (V) 471–473 K, (VI) 441–443 K, (VII) 475–477 K, (VIII) 439–441 K, (IX) 483–485 K, (X) 429–431 K, (XI) 393–395 K, (XII) 575–577 K. Spectroscopic data (IR and 1H NMR) are provided in the supporting information. Crystals of compounds (I), (II), and (VIII)–(XII) suitable for single-crystal X-ray diffraction analysis were selected directly from the prepared samples. Crystals of compounds (III)–(VII) suitable for single-crystal X-ray diffraction analysis were grown by slow evaporation, at ambient temperature and in the presence of air, of solutions in methanol–ethyl acetate (initial composition 1:1, v/v). A number of other acids were used in similar co-crystallization experiments but they did not provide crystal suitable for single-crystal X-ray diffraction, thus: 2- and 3-fluorobenzoic acids [cf. compound (II)], 2- and 3-chlorobenzoic acids [cf. compound (III)], 2- and 3-bromobenzoic acids [cf. compound (IV)], 2- and 3-iodobenzoic acids, phthalic acid, 3-methylbenzoic acid [cf. compound (I)], 2,4-dichlorobenzoic acid, crotonic and adipic acids [cf. compounds (VIII)–(X)], and ascorbic, aspartic and glutamic acids.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. In each of the isomorphous compounds (I)–(IV), the 4-methoxyphenyl group exhibits disorder over two sets of atomic sites, and in each of (VIII) and (IX), the anion exhibits disorder involving two sets of atomic sites having unequal occupancies. In each case, the bonded distances and the 1,3 non-bonded distances in the minor disorder component were restrained to be the same as the equivalent distances in the major disorder component, subject to s.u. values of 0.01 and 0.02 Å, respectively, and the anisotropic displacement parameters for pairs of partial-occupancy atoms occupying essentially the same physical space were constrained to be equal: in addition, it was found necessary to constrain the minor component of the carboxyl group in (IX) to be planar. The ratio of observed-to-unique data was only 39% for compounds (II) and (III): this is probably a consequence of the ambient temperature data collection allied to the disorder: in both (VII) and (IX), the average U 3/U 1 ratio was > 4.0: this may be consequence of the disorder. Apart from those in the minor disorder components of (I)–(IV), (VIII) and (IX), all H atoms were located in difference maps. The H atoms bonded to C atoms were then treated as riding atoms in geometrically idealized positions with C—H distances of 0.93 Å (alkenyl and aromatic), 0.96 Å (CH3) or 0.97 Å (CH2), and with U iso(H) = kU eq(C), where k = 1.5 for the methyl groups which were permitted to rotate but not to tilt, and 1.2 for all other H atoms bonded to C atoms: the H atoms bonded to C atoms in the minor disorder components were included on the same basis. The H atoms bonded to O atoms in the disordered components of (VIII) and (IX) were treated as riding atoms with O—H = 0.82 Å and U iso(H) = 1.5U eq(O), For the H atoms bonded to N atoms, and for the H atoms bonded to O atoms in (I)–(V), (VII), (X) and (XII), the atomic coordinates were refined with U iso(H) = 1.2U eq(N) or 1.5U eq(O), leading to the N—H and O—H distances shown in Table 1 ▸. The refined occupancies for the disorder components were 0.66 (2) and 0.34 (2) in (I), 0.81 (3) and 0.19 (3) in (II), 0.73 (2) and 0.27 (2) in (III), 0.80 (2) and 0.20 (2) in (IV), 0.660 (15) and 0.340 (15) in (VIII), and 0.906 (9) and 0.094 (9) in (IX). For compound (XI), the correct orientation of the structure relative to the polar axis direction was established using the Flack x parameter (Flack, 1983 ▸), x = 0.11 (7). However, for compounds (V), (VIII) and (IX), where there is very little resonant scattering the values of the Flack x parameter were indeterminate (Flack & Bernardinelli, 2000 ▸), with values −0.3 (5), −0.6 (7) and −0.3 (4), respectively: hence in these three cases, the correct orientation of the structure with respect to the polar axis direction cannot be established, although this has no chemical significance. The refinement of (XII) was treated as a non-merohedral twin, with twin matrix (−1, 0, 0/0, −1, 0/0.496, 0, 1) and with refined twin fractions 0.2467 (9) and 0.7533 (9).
Supplementary Material
Crystal structure: contains datablock(s) global, I, II, III, IV, V, VI, VII, VIII, IX, X, XI, XII. DOI: 10.1107/S2056989019012702/ex2024sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019012702/ex2024Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989019012702/ex2024IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989019012702/ex2024IIIsup4.hkl
Structure factors: contains datablock(s) IV. DOI: 10.1107/S2056989019012702/ex2024IVsup5.hkl
Structure factors: contains datablock(s) V. DOI: 10.1107/S2056989019012702/ex2024Vsup6.hkl
Structure factors: contains datablock(s) VI. DOI: 10.1107/S2056989019012702/ex2024VIsup7.hkl
Structure factors: contains datablock(s) VII. DOI: 10.1107/S2056989019012702/ex2024VIIsup8.hkl
Structure factors: contains datablock(s) VIII. DOI: 10.1107/S2056989019012702/ex2024VIIIsup9.hkl
Structure factors: contains datablock(s) IX. DOI: 10.1107/S2056989019012702/ex2024IXsup10.hkl
Structure factors: contains datablock(s) X. DOI: 10.1107/S2056989019012702/ex2024Xsup11.hkl
Structure factors: contains datablock(s) XI. DOI: 10.1107/S2056989019012702/ex2024XIsup12.hkl
Structure factors: contains datablock(s) XII. DOI: 10.1107/S2056989019012702/ex2024XIIsup13.hkl
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024Isup14.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024IIsup15.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024IIIsup16.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024IVsup17.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024Vsup18.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024VIsup19.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024VIIsup20.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024XIsup21.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
HKK thanks University of Mysore for research facilities.
supplementary crystallographic information
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Crystal data
| C11H17N2O+·C7H5O2−·H2O | Z = 2 |
| Mr = 332.39 | F(000) = 356 |
| Triclinic, P1 | Dx = 1.270 Mg m−3 |
| a = 6.215 (1) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.547 (1) Å | Cell parameters from 3742 reflections |
| c = 18.716 (4) Å | θ = 2.8–28.0° |
| α = 84.34 (2)° | µ = 0.09 mm−1 |
| β = 87.14 (2)° | T = 296 K |
| γ = 84.69 (2)° | Plate, colourless |
| V = 869.1 (3) Å3 | 0.40 × 0.24 × 0.04 mm |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3442 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1839 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.029 |
| ω scans | θmax = 26.1°, θmin = 2.8° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −7→7 |
| Tmin = 0.834, Tmax = 0.996 | k = −9→9 |
| 5751 measured reflections | l = −19→23 |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.054 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.134 | w = 1/[σ2(Fo2) + (0.0611P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 3442 reflections | Δρmax = 0.17 e Å−3 |
| 256 parameters | Δρmin = −0.19 e Å−3 |
| 17 restraints |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Special details
| Experimental. Compound (I). IR (KBr , cm-1) 3328 (OH), 3002 (H2) 2841 (OCH3), 1591 (COO). NMR (CDCl3) δ(1H) 3.22 (m, 4H, piperazine), 3.29 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.86 (m, 4H, methoxyphenyl), 7.39 (m, 2H, phenyl), 7.46 (m, 1H, phenyl), 8.05 (m, 2H, phenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.7874 (3) | 0.7435 (3) | 0.44989 (10) | 0.0482 (5) | |
| H11 | 0.834 (3) | 0.766 (3) | 0.4926 (13) | 0.058* | |
| H12 | 0.748 (3) | 0.621 (3) | 0.4555 (11) | 0.058* | |
| C2 | 0.9747 (3) | 0.7530 (3) | 0.39809 (12) | 0.0519 (6) | |
| H2A | 1.0940 | 0.6709 | 0.4157 | 0.062* | |
| H2B | 1.0215 | 0.8729 | 0.3934 | 0.062* | |
| C3 | 0.9144 (3) | 0.7056 (3) | 0.32596 (12) | 0.0497 (6) | |
| H3A | 1.0363 | 0.7191 | 0.2921 | 0.060* | |
| H3B | 0.8837 | 0.5812 | 0.3301 | 0.060* | |
| N4 | 0.7262 (2) | 0.8168 (2) | 0.29814 (9) | 0.0394 (4) | |
| C5 | 0.5425 (3) | 0.8133 (3) | 0.35024 (11) | 0.0463 (5) | |
| H5A | 0.4932 | 0.6943 | 0.3561 | 0.056* | |
| H5B | 0.4244 | 0.8957 | 0.3320 | 0.056* | |
| C6 | 0.6019 (3) | 0.8640 (3) | 0.42214 (12) | 0.0513 (6) | |
| H6A | 0.6397 | 0.9866 | 0.4173 | 0.062* | |
| H6B | 0.4789 | 0.8557 | 0.4559 | 0.062* | |
| C21 | 0.677 (2) | 0.782 (2) | 0.2275 (6) | 0.034 (2) | 0.66 (2) |
| C22 | 0.8134 (13) | 0.6810 (17) | 0.1831 (5) | 0.0450 (18) | 0.66 (2) |
| H22 | 0.9414 | 0.6235 | 0.2007 | 0.054* | 0.66 (2) |
| C23 | 0.7627 (13) | 0.6649 (18) | 0.1132 (5) | 0.053 (2) | 0.66 (2) |
| H23 | 0.8602 | 0.6005 | 0.0841 | 0.064* | 0.66 (2) |
| C24 | 0.5725 (14) | 0.7413 (16) | 0.0858 (5) | 0.0438 (17) | 0.66 (2) |
| C25 | 0.438 (2) | 0.846 (3) | 0.1278 (8) | 0.0599 (10) | 0.66 (2) |
| H25 | 0.3145 | 0.9097 | 0.1088 | 0.072* | 0.66 (2) |
| C26 | 0.486 (3) | 0.858 (3) | 0.1984 (8) | 0.054 (2) | 0.66 (2) |
| H26 | 0.3857 | 0.9191 | 0.2276 | 0.064* | 0.66 (2) |
| O24 | 0.541 (3) | 0.720 (3) | 0.0149 (7) | 0.078 (3) | 0.66 (2) |
| C27 | 0.335 (3) | 0.773 (4) | −0.0122 (11) | 0.090 (2) | 0.66 (2) |
| H27A | 0.2297 | 0.7017 | 0.0128 | 0.135* | 0.66 (2) |
| H27B | 0.3373 | 0.7580 | −0.0626 | 0.135* | 0.66 (2) |
| H27C | 0.2962 | 0.8970 | −0.0053 | 0.135* | 0.66 (2) |
| C51 | 0.669 (5) | 0.815 (5) | 0.2258 (12) | 0.034 (2) | 0.34 (2) |
| C52 | 0.817 (3) | 0.735 (2) | 0.1784 (11) | 0.0450 (18) | 0.34 (2) |
| H52 | 0.9535 | 0.6917 | 0.1937 | 0.054* | 0.34 (2) |
| C53 | 0.764 (3) | 0.718 (3) | 0.1087 (10) | 0.053 (2) | 0.34 (2) |
| H53 | 0.8615 | 0.6555 | 0.0790 | 0.064* | 0.34 (2) |
| C54 | 0.571 (3) | 0.792 (2) | 0.0828 (10) | 0.0438 (17) | 0.34 (2) |
| C55 | 0.414 (4) | 0.854 (7) | 0.1309 (15) | 0.0599 (10) | 0.34 (2) |
| H55 | 0.2702 | 0.8738 | 0.1184 | 0.072* | 0.34 (2) |
| C56 | 0.474 (5) | 0.886 (7) | 0.1983 (15) | 0.054 (2) | 0.34 (2) |
| H56 | 0.3803 | 0.9570 | 0.2260 | 0.064* | 0.34 (2) |
| O54 | 0.528 (6) | 0.756 (7) | 0.0142 (14) | 0.078 (3) | 0.34 (2) |
| C57 | 0.310 (6) | 0.780 (9) | −0.006 (2) | 0.090 (2) | 0.34 (2) |
| H57A | 0.2919 | 0.7103 | −0.0451 | 0.135* | 0.34 (2) |
| H57B | 0.2718 | 0.9042 | −0.0207 | 0.135* | 0.34 (2) |
| H57C | 0.2171 | 0.7422 | 0.0342 | 0.135* | 0.34 (2) |
| C31 | 0.8112 (3) | 0.7289 (3) | 0.70824 (12) | 0.0441 (5) | |
| C32 | 0.6694 (4) | 0.6501 (3) | 0.75905 (17) | 0.0642 (7) | |
| H32 | 0.5518 | 0.5977 | 0.7441 | 0.077* | |
| C33 | 0.7024 (5) | 0.6491 (3) | 0.83138 (17) | 0.0777 (9) | |
| H33 | 0.6070 | 0.5956 | 0.8649 | 0.093* | |
| C34 | 0.8735 (5) | 0.7259 (3) | 0.85422 (15) | 0.0741 (8) | |
| H34 | 0.8935 | 0.7264 | 0.9031 | 0.089* | |
| C35 | 1.0147 (4) | 0.8017 (3) | 0.80535 (14) | 0.0631 (7) | |
| H35 | 1.1322 | 0.8530 | 0.8209 | 0.076* | |
| C36 | 0.9850 (3) | 0.8032 (3) | 0.73275 (13) | 0.0483 (6) | |
| H36 | 1.0835 | 0.8550 | 0.6999 | 0.058* | |
| C37 | 0.7750 (4) | 0.7325 (3) | 0.62925 (15) | 0.0563 (6) | |
| O31 | 0.9017 (3) | 0.8122 (2) | 0.58536 (9) | 0.0664 (5) | |
| O32 | 0.6242 (3) | 0.6520 (3) | 0.61161 (12) | 0.1072 (8) | |
| O41 | 0.7231 (3) | 0.3783 (2) | 0.45956 (10) | 0.0657 (5) | |
| H41 | 0.607 (5) | 0.372 (4) | 0.4354 (16) | 0.099* | |
| H42 | 0.839 (5) | 0.318 (4) | 0.4391 (16) | 0.099* |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0614 (13) | 0.0460 (11) | 0.0394 (12) | −0.0089 (9) | −0.0119 (10) | −0.0069 (9) |
| C2 | 0.0452 (14) | 0.0584 (14) | 0.0529 (16) | −0.0018 (11) | −0.0116 (11) | −0.0075 (12) |
| C3 | 0.0424 (13) | 0.0583 (14) | 0.0483 (15) | 0.0016 (10) | −0.0047 (10) | −0.0084 (11) |
| N4 | 0.0391 (10) | 0.0403 (10) | 0.0386 (11) | 0.0002 (7) | −0.0018 (8) | −0.0064 (8) |
| C5 | 0.0414 (12) | 0.0553 (13) | 0.0413 (14) | 0.0022 (10) | −0.0001 (10) | −0.0072 (10) |
| C6 | 0.0561 (14) | 0.0529 (14) | 0.0438 (15) | 0.0044 (11) | −0.0012 (11) | −0.0083 (11) |
| C21 | 0.0428 (15) | 0.022 (6) | 0.0377 (14) | −0.001 (3) | −0.0009 (10) | −0.002 (2) |
| C22 | 0.0449 (14) | 0.041 (5) | 0.048 (2) | 0.005 (3) | −0.0021 (13) | −0.007 (3) |
| C23 | 0.0575 (16) | 0.053 (6) | 0.048 (2) | 0.008 (3) | 0.0060 (13) | −0.015 (3) |
| C24 | 0.0628 (16) | 0.031 (5) | 0.0378 (17) | −0.004 (3) | −0.0012 (13) | −0.002 (3) |
| C25 | 0.057 (3) | 0.072 (2) | 0.047 (2) | 0.020 (3) | −0.013 (2) | −0.0097 (16) |
| C26 | 0.057 (2) | 0.054 (7) | 0.0477 (16) | 0.022 (2) | −0.0054 (14) | −0.014 (3) |
| O24 | 0.083 (2) | 0.110 (10) | 0.0410 (12) | 0.009 (3) | −0.0107 (11) | −0.023 (3) |
| C27 | 0.081 (5) | 0.140 (3) | 0.052 (3) | −0.008 (4) | −0.018 (4) | −0.020 (4) |
| C51 | 0.0428 (15) | 0.022 (6) | 0.0377 (14) | −0.001 (3) | −0.0009 (10) | −0.002 (2) |
| C52 | 0.0449 (14) | 0.041 (5) | 0.048 (2) | 0.005 (3) | −0.0021 (13) | −0.007 (3) |
| C53 | 0.0575 (16) | 0.053 (6) | 0.048 (2) | 0.008 (3) | 0.0060 (13) | −0.015 (3) |
| C54 | 0.0628 (16) | 0.031 (5) | 0.0378 (17) | −0.004 (3) | −0.0012 (13) | −0.002 (3) |
| C55 | 0.057 (3) | 0.072 (2) | 0.047 (2) | 0.020 (3) | −0.013 (2) | −0.0097 (16) |
| C56 | 0.057 (2) | 0.054 (7) | 0.0477 (16) | 0.022 (2) | −0.0054 (14) | −0.014 (3) |
| O54 | 0.083 (2) | 0.110 (10) | 0.0410 (12) | 0.009 (3) | −0.0107 (11) | −0.023 (3) |
| C57 | 0.081 (5) | 0.140 (3) | 0.052 (3) | −0.008 (4) | −0.018 (4) | −0.020 (4) |
| C31 | 0.0428 (12) | 0.0353 (11) | 0.0550 (15) | 0.0023 (10) | −0.0041 (11) | −0.0117 (10) |
| C32 | 0.0510 (15) | 0.0506 (15) | 0.092 (2) | −0.0061 (11) | 0.0098 (14) | −0.0174 (14) |
| C33 | 0.091 (2) | 0.0614 (17) | 0.073 (2) | 0.0018 (15) | 0.0343 (17) | 0.0038 (15) |
| C34 | 0.099 (2) | 0.0639 (17) | 0.0554 (19) | 0.0101 (16) | −0.0013 (17) | −0.0025 (14) |
| C35 | 0.0746 (18) | 0.0624 (16) | 0.0530 (18) | −0.0031 (13) | −0.0179 (14) | −0.0044 (13) |
| C36 | 0.0509 (13) | 0.0454 (13) | 0.0492 (16) | −0.0057 (10) | −0.0079 (11) | −0.0023 (10) |
| C37 | 0.0540 (15) | 0.0504 (15) | 0.0680 (19) | 0.0062 (12) | −0.0207 (13) | −0.0233 (13) |
| O31 | 0.0856 (13) | 0.0687 (11) | 0.0468 (11) | −0.0072 (10) | −0.0138 (9) | −0.0092 (9) |
| O32 | 0.0869 (14) | 0.1422 (19) | 0.1065 (18) | −0.0380 (13) | −0.0368 (12) | −0.0380 (15) |
| O41 | 0.0653 (12) | 0.0631 (11) | 0.0715 (14) | −0.0142 (9) | −0.0198 (9) | −0.0044 (9) |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Geometric parameters (Å, º)
| N1—C2 | 1.480 (3) | C27—H27C | 0.9600 |
| N1—C6 | 1.483 (3) | C51—C56 | 1.381 (7) |
| N1—H11 | 0.90 (2) | C51—C52 | 1.385 (7) |
| N1—H12 | 0.97 (2) | C52—C53 | 1.380 (7) |
| C2—C3 | 1.504 (3) | C52—H52 | 0.9300 |
| C2—H2A | 0.9700 | C53—C54 | 1.368 (7) |
| C2—H2B | 0.9700 | C53—H53 | 0.9300 |
| C3—N4 | 1.461 (2) | C54—C55 | 1.374 (9) |
| C3—H3A | 0.9700 | C54—O54 | 1.380 (7) |
| C3—H3B | 0.9700 | C55—C56 | 1.383 (9) |
| N4—C51 | 1.42 (2) | C55—H55 | 0.9300 |
| N4—C21 | 1.428 (10) | C56—H56 | 0.9300 |
| N4—C5 | 1.464 (2) | O54—C57 | 1.415 (9) |
| C5—C6 | 1.507 (3) | C57—H57A | 0.9600 |
| C5—H5A | 0.9700 | C57—H57B | 0.9600 |
| C5—H5B | 0.9700 | C57—H57C | 0.9600 |
| C6—H6A | 0.9700 | C31—C36 | 1.380 (3) |
| C6—H6B | 0.9700 | C31—C32 | 1.391 (3) |
| C21—C26 | 1.382 (4) | C31—C37 | 1.504 (3) |
| C21—C22 | 1.386 (4) | C32—C33 | 1.378 (4) |
| C22—C23 | 1.380 (4) | C32—H32 | 0.9300 |
| C22—H22 | 0.9300 | C33—C34 | 1.363 (4) |
| C23—C24 | 1.367 (4) | C33—H33 | 0.9300 |
| C23—H23 | 0.9300 | C34—C35 | 1.358 (3) |
| C24—C25 | 1.372 (7) | C34—H34 | 0.9300 |
| C24—O24 | 1.378 (4) | C35—C36 | 1.379 (3) |
| C25—C26 | 1.382 (5) | C35—H35 | 0.9300 |
| C25—H25 | 0.9300 | C36—H36 | 0.9300 |
| C26—H26 | 0.9300 | C37—O32 | 1.238 (3) |
| O24—C27 | 1.413 (6) | C37—O31 | 1.258 (3) |
| C27—H27A | 0.9600 | O41—H41 | 0.88 (3) |
| C27—H27B | 0.9600 | O41—H42 | 0.91 (3) |
| C2—N1—C6 | 109.95 (18) | O24—C27—H27A | 109.5 |
| C2—N1—H11 | 106.8 (14) | O24—C27—H27B | 109.5 |
| C6—N1—H11 | 115.6 (14) | H27A—C27—H27B | 109.5 |
| C2—N1—H12 | 108.2 (12) | O24—C27—H27C | 109.5 |
| C6—N1—H12 | 109.2 (12) | H27A—C27—H27C | 109.5 |
| H11—N1—H12 | 106.8 (18) | H27B—C27—H27C | 109.5 |
| N1—C2—C3 | 110.23 (17) | C56—C51—C52 | 116.4 (9) |
| N1—C2—H2A | 109.6 | C56—C51—N4 | 125.0 (16) |
| C3—C2—H2A | 109.6 | C52—C51—N4 | 118.6 (16) |
| N1—C2—H2B | 109.6 | C53—C52—C51 | 121.0 (9) |
| C3—C2—H2B | 109.6 | C53—C52—H52 | 119.5 |
| H2A—C2—H2B | 108.1 | C51—C52—H52 | 119.5 |
| N4—C3—C2 | 112.64 (17) | C54—C53—C52 | 121.0 (9) |
| N4—C3—H3A | 109.1 | C54—C53—H53 | 119.5 |
| C2—C3—H3A | 109.1 | C52—C53—H53 | 119.5 |
| N4—C3—H3B | 109.1 | C53—C54—C55 | 118.1 (9) |
| C2—C3—H3B | 109.1 | C53—C54—O54 | 116.3 (10) |
| H3A—C3—H3B | 107.8 | C55—C54—O54 | 123.3 (12) |
| C51—N4—C3 | 120.7 (9) | C54—C55—C56 | 118.8 (13) |
| C21—N4—C3 | 113.1 (5) | C54—C55—H55 | 120.6 |
| C51—N4—C5 | 114.2 (13) | C56—C55—H55 | 120.6 |
| C21—N4—C5 | 114.4 (6) | C51—C56—C55 | 122.1 (10) |
| C3—N4—C5 | 111.20 (16) | C51—C56—H56 | 119.0 |
| N4—C5—C6 | 111.63 (17) | C55—C56—H56 | 119.0 |
| N4—C5—H5A | 109.3 | C54—O54—C57 | 117.7 (12) |
| C6—C5—H5A | 109.3 | O54—C57—H57A | 109.5 |
| N4—C5—H5B | 109.3 | O54—C57—H57B | 109.5 |
| C6—C5—H5B | 109.3 | H57A—C57—H57B | 109.5 |
| H5A—C5—H5B | 108.0 | O54—C57—H57C | 109.5 |
| N1—C6—C5 | 110.23 (17) | H57A—C57—H57C | 109.5 |
| N1—C6—H6A | 109.6 | H57B—C57—H57C | 109.5 |
| C5—C6—H6A | 109.6 | C36—C31—C32 | 117.9 (2) |
| N1—C6—H6B | 109.6 | C36—C31—C37 | 121.4 (2) |
| C5—C6—H6B | 109.6 | C32—C31—C37 | 120.8 (2) |
| H6A—C6—H6B | 108.1 | C33—C32—C31 | 120.4 (2) |
| C26—C21—C22 | 116.0 (4) | C33—C32—H32 | 119.8 |
| C26—C21—N4 | 119.5 (8) | C31—C32—H32 | 119.8 |
| C22—C21—N4 | 124.4 (8) | C34—C33—C32 | 120.6 (3) |
| C23—C22—C21 | 121.2 (4) | C34—C33—H33 | 119.7 |
| C23—C22—H22 | 119.4 | C32—C33—H33 | 119.7 |
| C21—C22—H22 | 119.4 | C35—C34—C33 | 119.8 (3) |
| C24—C23—C22 | 121.5 (4) | C35—C34—H34 | 120.1 |
| C24—C23—H23 | 119.3 | C33—C34—H34 | 120.1 |
| C22—C23—H23 | 119.3 | C34—C35—C36 | 120.5 (2) |
| C23—C24—C25 | 118.5 (4) | C34—C35—H35 | 119.7 |
| C23—C24—O24 | 116.6 (5) | C36—C35—H35 | 119.7 |
| C25—C24—O24 | 124.6 (6) | C35—C36—C31 | 120.9 (2) |
| C24—C25—C26 | 119.5 (7) | C35—C36—H36 | 119.6 |
| C24—C25—H25 | 120.3 | C31—C36—H36 | 119.6 |
| C26—C25—H25 | 120.3 | O32—C37—O31 | 124.1 (3) |
| C21—C26—C25 | 122.9 (5) | O32—C37—C31 | 117.3 (3) |
| C21—C26—H26 | 118.5 | O31—C37—C31 | 118.6 (2) |
| C25—C26—H26 | 118.5 | H41—O41—H42 | 111 (3) |
| C24—O24—C27 | 118.1 (6) | ||
| C6—N1—C2—C3 | −57.3 (2) | C3—N4—C51—C56 | 166 (4) |
| N1—C2—C3—N4 | 55.6 (2) | C5—N4—C51—C56 | 30 (5) |
| C2—C3—N4—C51 | 168.3 (19) | C21—N4—C51—C52 | −56 (9) |
| C2—C3—N4—C21 | 175.8 (8) | C3—N4—C51—C52 | −13 (4) |
| C2—C3—N4—C5 | −53.8 (2) | C5—N4—C51—C52 | −149 (2) |
| C51—N4—C5—C6 | −164.9 (15) | C56—C51—C52—C53 | −4 (5) |
| C21—N4—C5—C6 | −176.0 (7) | N4—C51—C52—C53 | 175 (2) |
| C3—N4—C5—C6 | 54.3 (2) | C51—C52—C53—C54 | 5 (3) |
| C2—N1—C6—C5 | 58.3 (2) | C52—C53—C54—C55 | −12 (4) |
| N4—C5—C6—N1 | −57.0 (2) | C52—C53—C54—O54 | −176 (3) |
| C51—N4—C21—C26 | −49 (10) | C53—C54—C55—C56 | 18 (6) |
| C3—N4—C21—C26 | 170.7 (16) | O54—C54—C55—C56 | −180 (5) |
| C5—N4—C21—C26 | 42 (2) | C52—C51—C56—C55 | 10 (7) |
| C51—N4—C21—C22 | 128 (12) | N4—C51—C56—C55 | −169 (4) |
| C3—N4—C21—C22 | −12.5 (17) | C54—C55—C56—C51 | −17 (8) |
| C5—N4—C21—C22 | −141.2 (12) | C53—C54—O54—C57 | 161 (4) |
| C26—C21—C22—C23 | 2 (2) | C55—C54—O54—C57 | −2 (7) |
| N4—C21—C22—C23 | −174.5 (11) | C36—C31—C32—C33 | 0.7 (3) |
| C21—C22—C23—C24 | −2.6 (14) | C37—C31—C32—C33 | −179.3 (2) |
| C22—C23—C24—C25 | 4.6 (17) | C31—C32—C33—C34 | 0.3 (4) |
| C22—C23—C24—O24 | 178.6 (15) | C32—C33—C34—C35 | −1.0 (4) |
| C23—C24—C25—C26 | −6 (3) | C33—C34—C35—C36 | 0.7 (4) |
| O24—C24—C25—C26 | −180 (2) | C34—C35—C36—C31 | 0.4 (3) |
| C22—C21—C26—C25 | −4 (3) | C32—C31—C36—C35 | −1.0 (3) |
| N4—C21—C26—C25 | 173 (2) | C37—C31—C36—C35 | 178.9 (2) |
| C24—C25—C26—C21 | 7 (4) | C36—C31—C37—O32 | 175.1 (2) |
| C23—C24—O24—C27 | 171 (2) | C32—C31—C37—O32 | −5.0 (3) |
| C25—C24—O24—C27 | −16 (3) | C36—C31—C37—O31 | −3.1 (3) |
| C21—N4—C51—C56 | 123 (14) | C32—C31—C37—O31 | 176.9 (2) |
4-(4-Methoxyphenyl)piperazin-1-ium benzoate monohydrate (I). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.90 (2) | 1.88 (2) | 2.777 (3) | 174.1 (19) |
| N1—H12···O41 | 0.97 (2) | 1.85 (2) | 2.808 (3) | 169.7 (18) |
| O41—H41···O32i | 0.88 (3) | 1.75 (3) | 2.631 (3) | 177 (3) |
| O41—H42···O31ii | 0.91 (3) | 1.87 (3) | 2.763 (3) | 169 (3) |
| C2—H2B···O31iii | 0.97 | 2.54 | 3.485 (3) | 165 |
| C22—H22···Cg1ii | 0.93 | 2.85 | 3.603 (3) | 139 |
| C26—H26···Cg1iv | 0.93 | 2.90 | 3.62 (2) | 135 |
| C56—H56···Cg1iv | 0.93 | 2.64 | 3.41 (5) | 141 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1; (iii) −x+2, −y+2, −z+1; (iv) −x+1, −y+2, −z+1.
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Crystal data
| C11H17N2O+·C7H4FO2−·H2O | Z = 2 |
| Mr = 350.38 | F(000) = 372 |
| Triclinic, P1 | Dx = 1.314 Mg m−3 |
| a = 6.256 (1) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.489 (1) Å | Cell parameters from 3771 reflections |
| c = 19.097 (2) Å | θ = 2.9–27.9° |
| α = 84.19 (1)° | µ = 0.10 mm−1 |
| β = 86.98 (2)° | T = 293 K |
| γ = 84.62 (2)° | Plate, colourless |
| V = 885.4 (2) Å3 | 0.40 × 0.24 × 0.04 mm |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3477 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1355 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.046 |
| ω scans | θmax = 26.1°, θmin = 2.9° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −7→6 |
| Tmin = 0.973, Tmax = 0.996 | k = −9→9 |
| 5760 measured reflections | l = −23→22 |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.066 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.128 | w = 1/[σ2(Fo2) + (0.0404P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 3477 reflections | Δρmax = 0.13 e Å−3 |
| 265 parameters | Δρmin = −0.14 e Å−3 |
| 17 restraints |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Special details
| Experimental. Compound (II). IR (KBr , cm-1) 3317 (OH), 3011 (NH2), 2838 (OCH3), 1588 (COO), 1365 (CF) NMR (CDCl3) δ(1H) 3.23 (m, 4H, piperazine), 3.29 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.86 (m, 4H, methoxyphenyl), 7.05 (m, 2H, fluorophenyl), 8.05 (m, 2H, fluorophenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.7813 (5) | 0.7442 (3) | 0.45140 (15) | 0.0536 (8) | |
| H11 | 0.823 (4) | 0.778 (3) | 0.5031 (15) | 0.064* | |
| H12 | 0.749 (4) | 0.634 (4) | 0.4565 (14) | 0.064* | |
| C2 | 0.9661 (5) | 0.7540 (4) | 0.39966 (16) | 0.0599 (9) | |
| H2A | 1.0848 | 0.6706 | 0.4164 | 0.072* | |
| H2B | 1.0131 | 0.8746 | 0.3952 | 0.072* | |
| C3 | 0.9056 (5) | 0.7083 (4) | 0.32889 (15) | 0.0574 (9) | |
| H3A | 1.0264 | 0.7229 | 0.2955 | 0.069* | |
| H3B | 0.8755 | 0.5829 | 0.3326 | 0.069* | |
| N4 | 0.7185 (4) | 0.8206 (3) | 0.30227 (12) | 0.0444 (6) | |
| C5 | 0.5377 (5) | 0.8151 (4) | 0.35391 (14) | 0.0527 (8) | |
| H5A | 0.4900 | 0.6947 | 0.3596 | 0.063* | |
| H5B | 0.4192 | 0.8977 | 0.3365 | 0.063* | |
| C6 | 0.5960 (5) | 0.8649 (4) | 0.42410 (14) | 0.0567 (9) | |
| H6A | 0.6324 | 0.9887 | 0.4194 | 0.068* | |
| H6B | 0.4736 | 0.8556 | 0.4572 | 0.068* | |
| C21 | 0.670 (3) | 0.795 (6) | 0.2319 (6) | 0.0442 (18) | 0.81 (3) |
| C22 | 0.8062 (12) | 0.6932 (18) | 0.1883 (4) | 0.064 (2) | 0.81 (3) |
| H22 | 0.9351 | 0.6378 | 0.2053 | 0.077* | 0.81 (3) |
| C23 | 0.7544 (9) | 0.673 (2) | 0.1207 (3) | 0.069 (3) | 0.81 (3) |
| H23 | 0.8520 | 0.6083 | 0.0923 | 0.083* | 0.81 (3) |
| C24 | 0.5644 (10) | 0.7455 (18) | 0.0942 (3) | 0.058 (2) | 0.81 (3) |
| C25 | 0.4255 (15) | 0.846 (3) | 0.1357 (5) | 0.0680 (18) | 0.81 (3) |
| H25 | 0.2967 | 0.9002 | 0.1182 | 0.082* | 0.81 (3) |
| C26 | 0.4782 (18) | 0.866 (3) | 0.2037 (5) | 0.062 (2) | 0.81 (3) |
| H26 | 0.3798 | 0.9313 | 0.2317 | 0.074* | 0.81 (3) |
| O24 | 0.5299 (13) | 0.7159 (19) | 0.0261 (3) | 0.095 (2) | 0.81 (3) |
| C27 | 0.3282 (18) | 0.772 (3) | −0.0015 (6) | 0.107 (4) | 0.81 (3) |
| H27A | 0.2181 | 0.7178 | 0.0279 | 0.160* | 0.81 (3) |
| H27B | 0.3249 | 0.7366 | −0.0483 | 0.160* | 0.81 (3) |
| H27C | 0.3035 | 0.9011 | −0.0029 | 0.160* | 0.81 (3) |
| C51 | 0.655 (11) | 0.79 (3) | 0.236 (2) | 0.0442 (18) | 0.19 (3) |
| C52 | 0.810 (6) | 0.754 (6) | 0.1833 (17) | 0.064 (2) | 0.19 (3) |
| H52 | 0.9531 | 0.7290 | 0.1949 | 0.077* | 0.19 (3) |
| C53 | 0.756 (4) | 0.752 (6) | 0.1146 (15) | 0.069 (3) | 0.19 (3) |
| H53 | 0.8593 | 0.7106 | 0.0818 | 0.083* | 0.19 (3) |
| C54 | 0.554 (4) | 0.808 (5) | 0.0936 (13) | 0.058 (2) | 0.19 (3) |
| C55 | 0.396 (6) | 0.842 (13) | 0.144 (2) | 0.0680 (18) | 0.19 (3) |
| H55 | 0.2524 | 0.8581 | 0.1327 | 0.082* | 0.19 (3) |
| C56 | 0.453 (7) | 0.851 (13) | 0.213 (2) | 0.062 (2) | 0.19 (3) |
| H56 | 0.3509 | 0.8987 | 0.2444 | 0.074* | 0.19 (3) |
| O54 | 0.512 (5) | 0.786 (6) | 0.0251 (14) | 0.095 (2) | 0.19 (3) |
| C57 | 0.300 (7) | 0.827 (12) | 0.004 (3) | 0.107 (4) | 0.19 (3) |
| H57A | 0.2472 | 0.9444 | 0.0166 | 0.160* | 0.19 (3) |
| H57B | 0.2101 | 0.7391 | 0.0266 | 0.160* | 0.19 (3) |
| H57C | 0.2965 | 0.8256 | −0.0463 | 0.160* | 0.19 (3) |
| C31 | 0.8132 (5) | 0.7284 (4) | 0.70495 (17) | 0.0485 (8) | |
| C32 | 0.6742 (5) | 0.6533 (4) | 0.7564 (2) | 0.0676 (10) | |
| H32 | 0.5542 | 0.6028 | 0.7431 | 0.081* | |
| C33 | 0.7114 (6) | 0.6527 (5) | 0.8267 (2) | 0.0798 (11) | |
| H33 | 0.6181 | 0.6022 | 0.8611 | 0.096* | |
| C34 | 0.8883 (7) | 0.7278 (5) | 0.8448 (2) | 0.0752 (11) | |
| F34 | 0.9236 (3) | 0.7291 (3) | 0.91437 (11) | 0.1208 (9) | |
| C35 | 1.0294 (5) | 0.8010 (4) | 0.7961 (2) | 0.0658 (10) | |
| H35 | 1.1495 | 0.8505 | 0.8100 | 0.079* | |
| C36 | 0.9911 (5) | 0.8003 (4) | 0.72606 (17) | 0.0542 (9) | |
| H36 | 1.0870 | 0.8494 | 0.6922 | 0.065* | |
| C37 | 0.7690 (6) | 0.7297 (5) | 0.6289 (2) | 0.0614 (10) | |
| O31 | 0.8943 (4) | 0.8069 (3) | 0.58380 (12) | 0.0715 (7) | |
| O32 | 0.6112 (4) | 0.6566 (4) | 0.61332 (13) | 0.1091 (10) | |
| O41 | 0.7239 (4) | 0.3737 (3) | 0.46049 (12) | 0.0700 (8) | |
| H41 | 0.615 (6) | 0.356 (5) | 0.4356 (17) | 0.105* | |
| H42 | 0.835 (6) | 0.312 (5) | 0.4379 (18) | 0.105* |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.065 (2) | 0.0429 (18) | 0.0550 (18) | −0.0089 (16) | −0.0145 (16) | −0.0061 (15) |
| C2 | 0.050 (2) | 0.060 (3) | 0.070 (2) | −0.0047 (18) | −0.0107 (19) | −0.0069 (18) |
| C3 | 0.049 (2) | 0.065 (2) | 0.059 (2) | −0.0019 (18) | −0.0035 (17) | −0.0091 (18) |
| N4 | 0.0405 (16) | 0.0437 (17) | 0.0483 (16) | 0.0016 (13) | −0.0044 (13) | −0.0054 (12) |
| C5 | 0.049 (2) | 0.057 (2) | 0.052 (2) | −0.0005 (16) | −0.0015 (17) | −0.0083 (16) |
| C6 | 0.059 (2) | 0.057 (2) | 0.053 (2) | 0.0029 (18) | −0.0011 (17) | −0.0049 (17) |
| C21 | 0.044 (3) | 0.042 (3) | 0.047 (2) | −0.004 (4) | 0.001 (2) | −0.008 (3) |
| C22 | 0.055 (2) | 0.070 (7) | 0.068 (3) | 0.009 (3) | −0.005 (2) | −0.020 (3) |
| C23 | 0.063 (3) | 0.080 (8) | 0.067 (3) | 0.009 (3) | 0.004 (2) | −0.030 (4) |
| C24 | 0.066 (3) | 0.058 (6) | 0.053 (3) | −0.007 (3) | −0.003 (2) | −0.017 (3) |
| C25 | 0.061 (3) | 0.087 (3) | 0.054 (4) | 0.012 (4) | −0.009 (3) | −0.009 (4) |
| C26 | 0.056 (3) | 0.070 (5) | 0.056 (3) | 0.016 (4) | −0.001 (2) | −0.013 (4) |
| O24 | 0.094 (3) | 0.129 (7) | 0.0660 (19) | 0.008 (4) | −0.0132 (16) | −0.039 (3) |
| C27 | 0.102 (5) | 0.156 (14) | 0.067 (4) | −0.004 (5) | −0.027 (4) | −0.023 (4) |
| C51 | 0.044 (3) | 0.042 (3) | 0.047 (2) | −0.004 (4) | 0.001 (2) | −0.008 (3) |
| C52 | 0.055 (2) | 0.070 (7) | 0.068 (3) | 0.009 (3) | −0.005 (2) | −0.020 (3) |
| C53 | 0.063 (3) | 0.080 (8) | 0.067 (3) | 0.009 (3) | 0.004 (2) | −0.030 (4) |
| C54 | 0.066 (3) | 0.058 (6) | 0.053 (3) | −0.007 (3) | −0.003 (2) | −0.017 (3) |
| C55 | 0.061 (3) | 0.087 (3) | 0.054 (4) | 0.012 (4) | −0.009 (3) | −0.009 (4) |
| C56 | 0.056 (3) | 0.070 (5) | 0.056 (3) | 0.016 (4) | −0.001 (2) | −0.013 (4) |
| O54 | 0.094 (3) | 0.129 (7) | 0.0660 (19) | 0.008 (4) | −0.0132 (16) | −0.039 (3) |
| C57 | 0.102 (5) | 0.156 (14) | 0.067 (4) | −0.004 (5) | −0.027 (4) | −0.023 (4) |
| C31 | 0.045 (2) | 0.039 (2) | 0.063 (2) | 0.0008 (16) | −0.0084 (18) | −0.0119 (16) |
| C32 | 0.051 (2) | 0.055 (3) | 0.099 (3) | −0.0062 (18) | −0.002 (2) | −0.017 (2) |
| C33 | 0.072 (3) | 0.073 (3) | 0.089 (3) | −0.004 (2) | 0.020 (2) | −0.001 (2) |
| C34 | 0.083 (3) | 0.084 (3) | 0.057 (3) | 0.003 (2) | −0.009 (2) | −0.004 (2) |
| F34 | 0.131 (2) | 0.164 (2) | 0.0633 (15) | 0.0008 (17) | −0.0068 (13) | −0.0036 (14) |
| C35 | 0.059 (2) | 0.068 (3) | 0.072 (3) | −0.0106 (19) | −0.011 (2) | −0.006 (2) |
| C36 | 0.056 (2) | 0.052 (2) | 0.056 (2) | −0.0059 (18) | −0.0122 (17) | −0.0070 (16) |
| C37 | 0.057 (3) | 0.042 (2) | 0.087 (3) | 0.0070 (19) | −0.022 (2) | −0.019 (2) |
| O31 | 0.0848 (18) | 0.0686 (18) | 0.0638 (17) | −0.0057 (15) | −0.0208 (14) | −0.0123 (13) |
| O32 | 0.0871 (19) | 0.129 (3) | 0.124 (2) | −0.0362 (18) | −0.0427 (16) | −0.0343 (17) |
| O41 | 0.0693 (17) | 0.0628 (18) | 0.0816 (18) | −0.0137 (14) | −0.0227 (13) | −0.0073 (13) |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Geometric parameters (Å, º)
| N1—C2 | 1.483 (4) | C27—H27C | 0.9600 |
| N1—C6 | 1.485 (4) | C51—C56 | 1.380 (9) |
| N1—H11 | 1.09 (3) | C51—C52 | 1.390 (19) |
| N1—H12 | 0.87 (3) | C52—C53 | 1.374 (9) |
| C2—C3 | 1.503 (3) | C52—H52 | 0.9300 |
| C2—H2A | 0.9700 | C53—C54 | 1.362 (9) |
| C2—H2B | 0.9700 | C53—H53 | 0.9300 |
| C3—N4 | 1.458 (3) | C54—C55 | 1.370 (13) |
| C3—H3A | 0.9700 | C54—O54 | 1.377 (9) |
| C3—H3B | 0.9700 | C55—C56 | 1.383 (9) |
| N4—C51 | 1.39 (5) | C55—H55 | 0.9300 |
| N4—C21 | 1.429 (10) | C56—H56 | 0.9300 |
| N4—C5 | 1.462 (3) | O54—C57 | 1.408 (10) |
| C5—C6 | 1.499 (3) | C57—H57A | 0.9600 |
| C5—H5A | 0.9700 | C57—H57B | 0.9600 |
| C5—H5B | 0.9700 | C57—H57C | 0.9600 |
| C6—H6A | 0.9700 | C31—C36 | 1.377 (3) |
| C6—H6B | 0.9700 | C31—C32 | 1.388 (4) |
| C21—C26 | 1.379 (6) | C31—C37 | 1.491 (4) |
| C21—C22 | 1.389 (15) | C32—C33 | 1.374 (4) |
| C22—C23 | 1.374 (4) | C32—H32 | 0.9300 |
| C22—H22 | 0.9300 | C33—C34 | 1.363 (4) |
| C23—C24 | 1.361 (5) | C33—H33 | 0.9300 |
| C23—H23 | 0.9300 | C34—C35 | 1.358 (4) |
| C24—C25 | 1.369 (9) | C34—F34 | 1.360 (4) |
| C24—O24 | 1.373 (4) | C35—C36 | 1.373 (4) |
| C25—C26 | 1.383 (5) | C35—H35 | 0.9300 |
| C25—H25 | 0.9300 | C36—H36 | 0.9300 |
| C26—H26 | 0.9300 | C37—O32 | 1.236 (3) |
| O24—C27 | 1.405 (4) | C37—O31 | 1.266 (4) |
| C27—H27A | 0.9600 | O41—H41 | 0.88 (3) |
| C27—H27B | 0.9600 | O41—H42 | 0.91 (4) |
| C2—N1—C6 | 109.5 (2) | O24—C27—H27A | 109.5 |
| C2—N1—H11 | 111.6 (13) | O24—C27—H27B | 109.5 |
| C6—N1—H11 | 111.1 (13) | H27A—C27—H27B | 109.5 |
| C2—N1—H12 | 107 (2) | O24—C27—H27C | 109.5 |
| C6—N1—H12 | 110.0 (19) | H27A—C27—H27C | 109.5 |
| H11—N1—H12 | 107 (2) | H27B—C27—H27C | 109.5 |
| N1—C2—C3 | 110.9 (2) | C56—C51—C52 | 115.3 (18) |
| N1—C2—H2A | 109.5 | C56—C51—N4 | 122 (4) |
| C3—C2—H2A | 109.5 | C52—C51—N4 | 120 (5) |
| N1—C2—H2B | 109.5 | C53—C52—C51 | 121.2 (14) |
| C3—C2—H2B | 109.5 | C53—C52—H52 | 119.4 |
| H2A—C2—H2B | 108.1 | C51—C52—H52 | 119.4 |
| N4—C3—C2 | 112.7 (2) | C54—C53—C52 | 121.5 (11) |
| N4—C3—H3A | 109.0 | C54—C53—H53 | 119.3 |
| C2—C3—H3A | 109.0 | C52—C53—H53 | 119.3 |
| N4—C3—H3B | 109.0 | C53—C54—C55 | 118.3 (12) |
| C2—C3—H3B | 109.0 | C53—C54—O54 | 116.6 (13) |
| H3A—C3—H3B | 107.8 | C55—C54—O54 | 123.2 (15) |
| C51—N4—C3 | 117 (4) | C54—C55—C56 | 119.0 (16) |
| C21—N4—C3 | 114.4 (9) | C54—C55—H55 | 120.5 |
| C51—N4—C5 | 111 (4) | C56—C55—H55 | 120.5 |
| C21—N4—C5 | 115.5 (10) | C51—C56—C55 | 122.8 (13) |
| C3—N4—C5 | 110.9 (2) | C51—C56—H56 | 118.6 |
| N4—C5—C6 | 112.1 (2) | C55—C56—H56 | 118.6 |
| N4—C5—H5A | 109.2 | C54—O54—C57 | 117.8 (15) |
| C6—C5—H5A | 109.2 | O54—C57—H57A | 109.5 |
| N4—C5—H5B | 109.2 | O54—C57—H57B | 109.5 |
| C6—C5—H5B | 109.2 | H57A—C57—H57B | 109.5 |
| H5A—C5—H5B | 107.9 | O54—C57—H57C | 109.5 |
| N1—C6—C5 | 110.6 (2) | H57A—C57—H57C | 109.5 |
| N1—C6—H6A | 109.5 | H57B—C57—H57C | 109.5 |
| C5—C6—H6A | 109.5 | C36—C31—C32 | 118.3 (3) |
| N1—C6—H6B | 109.5 | C36—C31—C37 | 121.4 (3) |
| C5—C6—H6B | 109.5 | C32—C31—C37 | 120.2 (3) |
| H6A—C6—H6B | 108.1 | C33—C32—C31 | 121.0 (3) |
| C26—C21—C22 | 115.6 (7) | C33—C32—H32 | 119.5 |
| C26—C21—N4 | 121.3 (11) | C31—C32—H32 | 119.5 |
| C22—C21—N4 | 123.1 (9) | C34—C33—C32 | 118.3 (4) |
| C23—C22—C21 | 121.3 (4) | C34—C33—H33 | 120.8 |
| C23—C22—H22 | 119.4 | C32—C33—H33 | 120.8 |
| C21—C22—H22 | 119.4 | C35—C34—F34 | 119.1 (4) |
| C24—C23—C22 | 121.8 (4) | C35—C34—C33 | 122.6 (4) |
| C24—C23—H23 | 119.1 | F34—C34—C33 | 118.3 (4) |
| C22—C23—H23 | 119.1 | C34—C35—C36 | 118.6 (3) |
| C23—C24—C25 | 118.5 (4) | C34—C35—H35 | 120.7 |
| C23—C24—O24 | 116.6 (4) | C36—C35—H35 | 120.7 |
| C25—C24—O24 | 124.8 (4) | C35—C36—C31 | 121.2 (3) |
| C24—C25—C26 | 119.5 (6) | C35—C36—H36 | 119.4 |
| C24—C25—H25 | 120.2 | C31—C36—H36 | 119.4 |
| C26—C25—H25 | 120.2 | O32—C37—O31 | 123.5 (4) |
| C21—C26—C25 | 123.2 (6) | O32—C37—C31 | 118.3 (4) |
| C21—C26—H26 | 118.4 | O31—C37—C31 | 118.2 (3) |
| C25—C26—H26 | 118.4 | H41—O41—H42 | 101 (3) |
| C24—O24—C27 | 118.8 (4) | ||
| C6—N1—C2—C3 | −56.3 (3) | C3—N4—C51—C52 | −38 (19) |
| N1—C2—C3—N4 | 55.3 (3) | C5—N4—C51—C52 | −166 (12) |
| C2—C3—N4—C51 | 178 (9) | C56—C51—C52—C53 | −9 (19) |
| C2—C3—N4—C21 | 174 (2) | N4—C51—C52—C53 | −170 (10) |
| C2—C3—N4—C5 | −53.7 (3) | C51—C52—C53—C54 | 8 (11) |
| C51—N4—C5—C6 | −174 (8) | C52—C53—C54—C55 | −10 (7) |
| C21—N4—C5—C6 | −173 (2) | C52—C53—C54—O54 | −175 (4) |
| C3—N4—C5—C6 | 54.6 (3) | C53—C54—C55—C56 | 12 (11) |
| C2—N1—C6—C5 | 57.4 (3) | O54—C54—C55—C56 | 176 (7) |
| N4—C5—C6—N1 | −57.2 (3) | N4—C51—C56—C55 | 172 (13) |
| C3—N4—C21—C26 | 169 (3) | C54—C55—C56—C51 | −14 (15) |
| C5—N4—C21—C26 | 38 (4) | C53—C54—O54—C57 | 174 (5) |
| C3—N4—C21—C22 | −10 (5) | C55—C54—O54—C57 | 10 (8) |
| C5—N4—C21—C22 | −140 (3) | C36—C31—C32—C33 | 0.8 (4) |
| C26—C21—C22—C23 | 2 (4) | C37—C31—C32—C33 | −179.3 (3) |
| N4—C21—C22—C23 | −179 (2) | C31—C32—C33—C34 | 0.1 (5) |
| C21—C22—C23—C24 | −3 (2) | C32—C33—C34—C35 | −0.7 (5) |
| C22—C23—C24—C25 | 2.3 (13) | C32—C33—C34—F34 | 179.2 (3) |
| C22—C23—C24—O24 | −179.7 (6) | F34—C34—C35—C36 | −179.4 (3) |
| C23—C24—C25—C26 | −2 (2) | C33—C34—C35—C36 | 0.5 (5) |
| O24—C24—C25—C26 | −179.9 (14) | C34—C35—C36—C31 | 0.4 (5) |
| C22—C21—C26—C25 | −2 (4) | C32—C31—C36—C35 | −1.0 (4) |
| N4—C21—C26—C25 | 179 (3) | C37—C31—C36—C35 | 179.1 (3) |
| C24—C25—C26—C21 | 2 (3) | C36—C31—C37—O32 | 177.2 (3) |
| C23—C24—O24—C27 | 173.8 (10) | C32—C31—C37—O32 | −2.7 (4) |
| C25—C24—O24—C27 | −8.4 (16) | C36—C31—C37—O31 | −3.3 (4) |
| C3—N4—C51—C56 | 162 (12) | C32—C31—C37—O31 | 176.8 (3) |
| C5—N4—C51—C56 | 34 (18) |
4-(4-Methoxyphenyl)piperazin-1-ium 4-fluorobenzoate monohydrate (II). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 1.09 (3) | 1.67 (3) | 2.758 (4) | 174.1 (19) |
| N1—H12···O41 | 0.86 (3) | 1.96 (3) | 2.818 (4) | 170 (3) |
| O41—H41···O32i | 0.86 (4) | 1.75 (4) | 2.627 (4) | 174 (4) |
| O41—H42···O31ii | 0.91 (4) | 1.88 (4) | 2.768 (3) | 163 (3) |
| C2—H2B···O31iii | 0.97 | 2.58 | 3.529 (4) | 166 |
| C6—H6B···O41i | 0.97 | 2.57 | 3.386 (4) | 142 |
| C22—H22···Cg1ii | 0.93 | 2.93 | 3.664 (12) | 137 |
| C26—H26···Cg1iv | 0.93 | 2.81 | 3.56 (2) | 138 |
| C56—H56···Cg1iv | 0.93 | 2.96 | 3.55 (9) | 123 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1; (iii) −x+2, −y+2, −z+1; (iv) −x+1, −y+2, −z+1.
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Crystal data
| C11H17N2O+·C7H4ClO2−·H2O | Z = 2 |
| Mr = 366.83 | F(000) = 388 |
| Triclinic, P1 | Dx = 1.312 Mg m−3 |
| a = 6.211 (1) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.481 (1) Å | Cell parameters from 3962 reflections |
| c = 20.144 (4) Å | θ = 2.8–28.8° |
| α = 84.90 (2)° | µ = 0.23 mm−1 |
| β = 87.48 (2)° | T = 293 K |
| γ = 85.19 (2)° | Plate, colourless |
| V = 928.4 (3) Å3 | 0.20 × 0.16 × 0.02 mm |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3454 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1343 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.041 |
| ω scans | θmax = 25.5°, θmin = 2.8° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −7→7 |
| Tmin = 0.951, Tmax = 0.995 | k = −9→8 |
| 5883 measured reflections | l = −24→22 |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.065 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.135 | w = 1/[σ2(Fo2) + (0.0492P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.94 | (Δ/σ)max < 0.001 |
| 3454 reflections | Δρmax = 0.24 e Å−3 |
| 265 parameters | Δρmin = −0.23 e Å−3 |
| 17 restraints |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Special details
| Experimental. Compound (III). IR (KBr , cm-1) 3320 (OH), 3003 (NH2), 2837 (OCH3), 1582 (COO), 772(CCl). NMR (CDCl3) δ(1H) 3.23 (m, 4H, piperazine), 3.28 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.86 (m, 4H, methoxyphenyl), 7.36 (d, J = 8.4 Hz, 2H, chlorophenyl), 7.98 (d, J = 8.4 Hz,2H, chlorophenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.7833 (5) | 0.7442 (4) | 0.45388 (15) | 0.0536 (9) | |
| H11 | 0.827 (5) | 0.777 (4) | 0.5028 (16) | 0.064* | |
| H12 | 0.760 (5) | 0.636 (4) | 0.4593 (16) | 0.064* | |
| C2 | 0.9718 (6) | 0.7498 (5) | 0.40636 (16) | 0.0570 (10) | |
| H2A | 1.0888 | 0.6667 | 0.4234 | 0.068* | |
| H2B | 1.0216 | 0.8699 | 0.4020 | 0.068* | |
| C3 | 0.9136 (6) | 0.6998 (4) | 0.33895 (16) | 0.0545 (10) | |
| H3A | 1.0371 | 0.7116 | 0.3082 | 0.065* | |
| H3B | 0.8803 | 0.5749 | 0.3427 | 0.065* | |
| N4 | 0.7287 (4) | 0.8126 (3) | 0.31215 (12) | 0.0433 (7) | |
| C5 | 0.5425 (5) | 0.8110 (4) | 0.35968 (15) | 0.0516 (9) | |
| H5A | 0.4918 | 0.6912 | 0.3650 | 0.062* | |
| H5B | 0.4260 | 0.8935 | 0.3420 | 0.062* | |
| C6 | 0.5992 (6) | 0.8644 (4) | 0.42667 (15) | 0.0551 (10) | |
| H6A | 0.6377 | 0.9880 | 0.4222 | 0.066* | |
| H6B | 0.4747 | 0.8569 | 0.4573 | 0.066* | |
| C21 | 0.685 (4) | 0.783 (9) | 0.2449 (9) | 0.0446 (14) | 0.73 (2) |
| C22 | 0.8261 (13) | 0.6777 (19) | 0.2052 (4) | 0.060 (3) | 0.73 (2) |
| H22 | 0.9523 | 0.6211 | 0.2230 | 0.072* | 0.73 (2) |
| C23 | 0.7808 (13) | 0.657 (2) | 0.1401 (4) | 0.074 (3) | 0.73 (2) |
| H23 | 0.8807 | 0.5916 | 0.1141 | 0.088* | 0.73 (2) |
| C24 | 0.5923 (18) | 0.730 (3) | 0.1130 (5) | 0.065 (4) | 0.73 (2) |
| C25 | 0.450 (2) | 0.828 (5) | 0.1516 (8) | 0.075 (3) | 0.73 (2) |
| H25 | 0.3196 | 0.8781 | 0.1341 | 0.089* | 0.73 (2) |
| C26 | 0.496 (2) | 0.854 (3) | 0.2162 (6) | 0.060 (3) | 0.73 (2) |
| H26 | 0.3958 | 0.9221 | 0.2413 | 0.072* | 0.73 (2) |
| O24 | 0.564 (2) | 0.697 (2) | 0.0482 (6) | 0.104 (4) | 0.73 (2) |
| C27 | 0.362 (3) | 0.743 (2) | 0.0203 (7) | 0.101 (4) | 0.73 (2) |
| H27A | 0.2517 | 0.6892 | 0.0485 | 0.152* | 0.73 (2) |
| H27B | 0.3623 | 0.6989 | −0.0231 | 0.152* | 0.73 (2) |
| H27C | 0.3331 | 0.8713 | 0.0164 | 0.152* | 0.73 (2) |
| C51 | 0.668 (10) | 0.79 (3) | 0.247 (2) | 0.0446 (14) | 0.27 (2) |
| C52 | 0.833 (4) | 0.755 (4) | 0.1991 (13) | 0.060 (3) | 0.27 (2) |
| H52 | 0.9761 | 0.7413 | 0.2112 | 0.072* | 0.27 (2) |
| C53 | 0.783 (4) | 0.739 (4) | 0.1340 (12) | 0.074 (3) | 0.27 (2) |
| H53 | 0.8924 | 0.7021 | 0.1039 | 0.088* | 0.27 (2) |
| C54 | 0.577 (5) | 0.775 (9) | 0.1126 (15) | 0.065 (4) | 0.27 (2) |
| C55 | 0.418 (6) | 0.825 (15) | 0.158 (2) | 0.075 (3) | 0.27 (2) |
| H55 | 0.2775 | 0.8534 | 0.1437 | 0.089* | 0.27 (2) |
| C56 | 0.464 (5) | 0.834 (8) | 0.2235 (18) | 0.060 (3) | 0.27 (2) |
| H56 | 0.3532 | 0.8701 | 0.2531 | 0.072* | 0.27 (2) |
| O54 | 0.548 (7) | 0.751 (7) | 0.0468 (16) | 0.104 (4) | 0.27 (2) |
| C57 | 0.351 (8) | 0.814 (6) | 0.018 (2) | 0.101 (4) | 0.27 (2) |
| H57A | 0.2346 | 0.7622 | 0.0438 | 0.152* | 0.27 (2) |
| H57B | 0.3518 | 0.7814 | −0.0267 | 0.152* | 0.27 (2) |
| H57C | 0.3326 | 0.9432 | 0.0182 | 0.152* | 0.27 (2) |
| C31 | 0.8074 (6) | 0.7372 (4) | 0.69495 (18) | 0.0470 (9) | |
| C32 | 0.6605 (7) | 0.6665 (5) | 0.7412 (2) | 0.0704 (11) | |
| H32 | 0.5407 | 0.6164 | 0.7267 | 0.084* | |
| C33 | 0.6886 (8) | 0.6688 (5) | 0.8091 (2) | 0.0834 (13) | |
| H33 | 0.5883 | 0.6212 | 0.8400 | 0.100* | |
| C34 | 0.8662 (9) | 0.7424 (6) | 0.8301 (2) | 0.0770 (13) | |
| Cl34 | 0.8970 (3) | 0.74735 (19) | 0.91487 (5) | 0.1383 (7) | |
| C35 | 1.0144 (7) | 0.8114 (5) | 0.7851 (2) | 0.0695 (11) | |
| H35 | 1.1350 | 0.8601 | 0.7997 | 0.083* | |
| C36 | 0.9845 (6) | 0.8084 (4) | 0.71778 (17) | 0.0543 (10) | |
| H36 | 1.0861 | 0.8555 | 0.6872 | 0.065* | |
| C37 | 0.7721 (7) | 0.7351 (5) | 0.6222 (2) | 0.0581 (11) | |
| O31 | 0.9006 (5) | 0.8125 (3) | 0.58079 (12) | 0.0697 (8) | |
| O32 | 0.6183 (5) | 0.6568 (4) | 0.60573 (15) | 0.1085 (11) | |
| O41 | 0.7230 (5) | 0.3744 (3) | 0.46334 (13) | 0.0683 (8) | |
| H41 | 0.614 (7) | 0.363 (5) | 0.4360 (19) | 0.102* | |
| H42 | 0.846 (7) | 0.315 (5) | 0.4434 (19) | 0.102* |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.065 (2) | 0.0467 (18) | 0.0505 (19) | −0.0043 (19) | −0.0122 (18) | −0.0079 (17) |
| C2 | 0.044 (2) | 0.069 (3) | 0.057 (2) | −0.003 (2) | −0.008 (2) | −0.0023 (19) |
| C3 | 0.043 (2) | 0.065 (2) | 0.053 (2) | 0.007 (2) | −0.0072 (19) | −0.0050 (18) |
| N4 | 0.0390 (19) | 0.0473 (17) | 0.0432 (17) | 0.0009 (15) | −0.0023 (15) | −0.0044 (13) |
| C5 | 0.049 (2) | 0.055 (2) | 0.049 (2) | 0.0054 (19) | 0.0007 (19) | −0.0041 (18) |
| C6 | 0.057 (3) | 0.056 (2) | 0.051 (2) | 0.006 (2) | −0.0009 (19) | −0.0052 (18) |
| C21 | 0.038 (4) | 0.046 (7) | 0.049 (3) | 0.001 (5) | 0.001 (3) | −0.005 (2) |
| C22 | 0.054 (3) | 0.064 (8) | 0.059 (3) | 0.021 (4) | −0.007 (2) | −0.011 (4) |
| C23 | 0.073 (4) | 0.084 (9) | 0.059 (3) | 0.035 (5) | −0.002 (3) | −0.020 (5) |
| C24 | 0.074 (4) | 0.077 (12) | 0.043 (2) | 0.008 (3) | −0.004 (2) | −0.016 (4) |
| C25 | 0.064 (5) | 0.104 (4) | 0.053 (4) | 0.021 (7) | −0.013 (4) | −0.013 (5) |
| C26 | 0.048 (4) | 0.081 (6) | 0.049 (4) | 0.012 (5) | 0.003 (3) | −0.012 (4) |
| O24 | 0.104 (3) | 0.148 (12) | 0.059 (2) | 0.039 (5) | −0.0194 (19) | −0.038 (4) |
| C27 | 0.109 (5) | 0.125 (13) | 0.070 (3) | 0.022 (7) | −0.034 (3) | −0.025 (7) |
| C51 | 0.038 (4) | 0.046 (7) | 0.049 (3) | 0.001 (5) | 0.001 (3) | −0.005 (2) |
| C52 | 0.054 (3) | 0.064 (8) | 0.059 (3) | 0.021 (4) | −0.007 (2) | −0.011 (4) |
| C53 | 0.073 (4) | 0.084 (9) | 0.059 (3) | 0.035 (5) | −0.002 (3) | −0.020 (5) |
| C54 | 0.074 (4) | 0.077 (12) | 0.043 (2) | 0.008 (3) | −0.004 (2) | −0.016 (4) |
| C55 | 0.064 (5) | 0.104 (4) | 0.053 (4) | 0.021 (7) | −0.013 (4) | −0.013 (5) |
| C56 | 0.048 (4) | 0.081 (6) | 0.049 (4) | 0.012 (5) | 0.003 (3) | −0.012 (4) |
| O54 | 0.104 (3) | 0.148 (12) | 0.059 (2) | 0.039 (5) | −0.0194 (19) | −0.038 (4) |
| C57 | 0.109 (5) | 0.125 (13) | 0.070 (3) | 0.022 (7) | −0.034 (3) | −0.025 (7) |
| C31 | 0.042 (2) | 0.040 (2) | 0.059 (2) | 0.0033 (19) | −0.006 (2) | −0.0082 (18) |
| C32 | 0.061 (3) | 0.061 (3) | 0.090 (3) | −0.008 (2) | 0.005 (3) | −0.012 (2) |
| C33 | 0.080 (4) | 0.085 (3) | 0.080 (4) | −0.001 (3) | 0.029 (3) | 0.002 (3) |
| C34 | 0.089 (4) | 0.078 (3) | 0.060 (3) | 0.017 (3) | −0.007 (3) | −0.005 (2) |
| Cl34 | 0.1856 (16) | 0.1630 (13) | 0.0572 (8) | 0.0402 (11) | −0.0065 (8) | −0.0083 (8) |
| C35 | 0.070 (3) | 0.077 (3) | 0.062 (3) | −0.002 (2) | −0.015 (3) | −0.010 (2) |
| C36 | 0.054 (3) | 0.054 (2) | 0.055 (3) | −0.007 (2) | −0.007 (2) | −0.0026 (18) |
| C37 | 0.056 (3) | 0.044 (2) | 0.075 (3) | 0.007 (2) | −0.017 (3) | −0.017 (2) |
| O31 | 0.087 (2) | 0.0683 (18) | 0.0553 (17) | −0.0048 (16) | −0.0119 (16) | −0.0086 (14) |
| O32 | 0.095 (2) | 0.132 (3) | 0.110 (2) | −0.033 (2) | −0.040 (2) | −0.0309 (19) |
| O41 | 0.068 (2) | 0.0661 (17) | 0.0737 (19) | −0.0116 (16) | −0.0191 (15) | −0.0057 (14) |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Geometric parameters (Å, º)
| N1—C2 | 1.481 (4) | C27—H27C | 0.9600 |
| N1—C6 | 1.487 (4) | C51—C56 | 1.376 (12) |
| N1—H11 | 1.09 (3) | C51—C52 | 1.40 (2) |
| N1—H12 | 0.83 (3) | C52—C53 | 1.378 (9) |
| C2—C3 | 1.507 (4) | C52—H52 | 0.9300 |
| C2—H2A | 0.9700 | C53—C54 | 1.366 (9) |
| C2—H2B | 0.9700 | C53—H53 | 0.9300 |
| C3—N4 | 1.461 (4) | C54—C55 | 1.363 (14) |
| C3—H3A | 0.9700 | C54—O54 | 1.373 (8) |
| C3—H3B | 0.9700 | C55—C56 | 1.380 (9) |
| N4—C51 | 1.40 (3) | C55—H55 | 0.9300 |
| N4—C21 | 1.434 (12) | C56—H56 | 0.9300 |
| N4—C5 | 1.468 (4) | O54—C57 | 1.401 (9) |
| C5—C6 | 1.505 (4) | C57—H57A | 0.9600 |
| C5—H5A | 0.9700 | C57—H57B | 0.9600 |
| C5—H5B | 0.9700 | C57—H57C | 0.9600 |
| C6—H6A | 0.9700 | C31—C36 | 1.376 (4) |
| C6—H6B | 0.9700 | C31—C32 | 1.378 (5) |
| C21—C26 | 1.376 (12) | C31—C37 | 1.494 (5) |
| C21—C22 | 1.40 (2) | C32—C33 | 1.389 (5) |
| C22—C23 | 1.378 (5) | C32—H32 | 0.9300 |
| C22—H22 | 0.9300 | C33—C34 | 1.372 (5) |
| C23—C24 | 1.366 (6) | C33—H33 | 0.9300 |
| C23—H23 | 0.9300 | C34—C35 | 1.363 (5) |
| C24—C25 | 1.363 (12) | C34—Cl34 | 1.731 (4) |
| C24—O24 | 1.373 (5) | C35—C36 | 1.379 (4) |
| C25—C26 | 1.380 (6) | C35—H35 | 0.9300 |
| C25—H25 | 0.9300 | C36—H36 | 0.9300 |
| C26—H26 | 0.9300 | C37—O32 | 1.233 (4) |
| O24—C27 | 1.400 (6) | C37—O31 | 1.265 (4) |
| C27—H27A | 0.9600 | O41—H41 | 0.91 (4) |
| C27—H27B | 0.9600 | O41—H42 | 0.94 (4) |
| C2—N1—C6 | 110.0 (3) | O24—C27—H27A | 109.5 |
| C2—N1—H11 | 110.7 (16) | O24—C27—H27B | 109.5 |
| C6—N1—H11 | 112.6 (15) | H27A—C27—H27B | 109.5 |
| C2—N1—H12 | 105 (2) | O24—C27—H27C | 109.5 |
| C6—N1—H12 | 114 (2) | H27A—C27—H27C | 109.5 |
| H11—N1—H12 | 105 (3) | H27B—C27—H27C | 109.5 |
| N1—C2—C3 | 110.9 (3) | C56—C51—C52 | 116.1 (16) |
| N1—C2—H2A | 109.5 | C56—C51—N4 | 125 (2) |
| C3—C2—H2A | 109.5 | C52—C51—N4 | 118 (4) |
| N1—C2—H2B | 109.5 | C53—C52—C51 | 120.5 (14) |
| C3—C2—H2B | 109.5 | C53—C52—H52 | 119.7 |
| H2A—C2—H2B | 108.1 | C51—C52—H52 | 119.7 |
| N4—C3—C2 | 112.4 (3) | C54—C53—C52 | 121.5 (11) |
| N4—C3—H3A | 109.1 | C54—C53—H53 | 119.2 |
| C2—C3—H3A | 109.1 | C52—C53—H53 | 119.2 |
| N4—C3—H3B | 109.1 | C55—C54—C53 | 118.5 (10) |
| C2—C3—H3B | 109.1 | C55—C54—O54 | 125.5 (13) |
| H3A—C3—H3B | 107.9 | C53—C54—O54 | 116.0 (12) |
| C51—N4—C3 | 118 (4) | C54—C55—C56 | 120.5 (12) |
| C21—N4—C3 | 113.7 (13) | C54—C55—H55 | 119.8 |
| C51—N4—C5 | 112 (4) | C56—C55—H55 | 119.8 |
| C21—N4—C5 | 115.8 (14) | C51—C56—C55 | 122.4 (12) |
| C3—N4—C5 | 111.1 (2) | C51—C56—H56 | 118.8 |
| N4—C5—C6 | 111.8 (3) | C55—C56—H56 | 118.8 |
| N4—C5—H5A | 109.3 | C54—O54—C57 | 119.2 (14) |
| C6—C5—H5A | 109.3 | O54—C57—H57A | 109.5 |
| N4—C5—H5B | 109.3 | O54—C57—H57B | 109.5 |
| C6—C5—H5B | 109.3 | H57A—C57—H57B | 109.5 |
| H5A—C5—H5B | 107.9 | O54—C57—H57C | 109.5 |
| N1—C6—C5 | 110.5 (3) | H57A—C57—H57C | 109.5 |
| N1—C6—H6A | 109.6 | H57B—C57—H57C | 109.5 |
| C5—C6—H6A | 109.6 | C36—C31—C32 | 118.3 (3) |
| N1—C6—H6B | 109.6 | C36—C31—C37 | 121.7 (4) |
| C5—C6—H6B | 109.6 | C32—C31—C37 | 120.0 (4) |
| H6A—C6—H6B | 108.1 | C31—C32—C33 | 121.0 (4) |
| C26—C21—C22 | 116.2 (8) | C31—C32—H32 | 119.5 |
| C26—C21—N4 | 121.0 (15) | C33—C32—H32 | 119.5 |
| C22—C21—N4 | 122.8 (12) | C34—C33—C32 | 119.1 (4) |
| C23—C22—C21 | 120.9 (6) | C34—C33—H33 | 120.4 |
| C23—C22—H22 | 119.5 | C32—C33—H33 | 120.4 |
| C21—C22—H22 | 119.5 | C35—C34—C33 | 120.7 (4) |
| C24—C23—C22 | 121.4 (5) | C35—C34—Cl34 | 120.7 (4) |
| C24—C23—H23 | 119.3 | C33—C34—Cl34 | 118.6 (4) |
| C22—C23—H23 | 119.3 | C34—C35—C36 | 119.6 (4) |
| C25—C24—C23 | 118.5 (5) | C34—C35—H35 | 120.2 |
| C25—C24—O24 | 125.6 (6) | C36—C35—H35 | 120.2 |
| C23—C24—O24 | 115.9 (6) | C31—C36—C35 | 121.3 (4) |
| C24—C25—C26 | 120.7 (8) | C31—C36—H36 | 119.4 |
| C24—C25—H25 | 119.7 | C35—C36—H36 | 119.4 |
| C26—C25—H25 | 119.7 | O32—C37—O31 | 123.5 (4) |
| C21—C26—C25 | 122.3 (10) | O32—C37—C31 | 117.7 (4) |
| C21—C26—H26 | 118.8 | O31—C37—C31 | 118.7 (4) |
| C25—C26—H26 | 118.8 | H41—O41—H42 | 105 (3) |
| C24—O24—C27 | 119.3 (6) | ||
| C6—N1—C2—C3 | −56.4 (3) | C3—N4—C51—C52 | −37 (18) |
| N1—C2—C3—N4 | 55.4 (4) | C5—N4—C51—C52 | −168 (11) |
| C2—C3—N4—C51 | 175 (9) | C56—C51—C52—C53 | −9 (19) |
| C2—C3—N4—C21 | 173 (3) | N4—C51—C52—C53 | −178 (9) |
| C2—C3—N4—C5 | −54.1 (3) | C51—C52—C53—C54 | 7 (11) |
| C51—N4—C5—C6 | −171 (8) | C52—C53—C54—C55 | −1 (10) |
| C21—N4—C5—C6 | −173 (3) | C52—C53—C54—O54 | −179 (5) |
| C3—N4—C5—C6 | 54.8 (3) | C53—C54—C55—C56 | −2 (14) |
| C2—N1—C6—C5 | 57.2 (3) | O54—C54—C55—C56 | 176 (8) |
| N4—C5—C6—N1 | −56.8 (4) | C52—C51—C56—C55 | 6 (20) |
| C3—N4—C21—C26 | 169 (4) | N4—C51—C56—C55 | 174 (13) |
| C5—N4—C21—C26 | 38 (7) | C54—C55—C56—C51 | −1 (17) |
| C3—N4—C21—C22 | −10 (7) | C55—C54—O54—C57 | 13 (13) |
| C5—N4—C21—C22 | −140 (4) | C53—C54—O54—C57 | −169 (5) |
| C26—C21—C22—C23 | 3 (7) | C36—C31—C32—C33 | 0.8 (5) |
| N4—C21—C22—C23 | −178 (4) | C37—C31—C32—C33 | −179.2 (3) |
| C21—C22—C23—C24 | −3 (4) | C31—C32—C33—C34 | −0.3 (6) |
| C22—C23—C24—C25 | 1 (3) | C32—C33—C34—C35 | −0.4 (6) |
| C22—C23—C24—O24 | −178.6 (14) | C32—C33—C34—Cl34 | 179.1 (3) |
| C23—C24—C25—C26 | 1 (5) | C33—C34—C35—C36 | 0.5 (6) |
| O24—C24—C25—C26 | −180 (3) | Cl34—C34—C35—C36 | −179.0 (3) |
| C22—C21—C26—C25 | −2 (7) | C32—C31—C36—C35 | −0.7 (5) |
| N4—C21—C26—C25 | 180 (4) | C37—C31—C36—C35 | 179.3 (3) |
| C24—C25—C26—C21 | 0 (6) | C34—C35—C36—C31 | 0.1 (5) |
| C25—C24—O24—C27 | −10 (4) | C36—C31—C37—O32 | 174.6 (3) |
| C23—C24—O24—C27 | 169.9 (15) | C32—C31—C37—O32 | −5.3 (5) |
| C3—N4—C51—C56 | 155 (12) | C36—C31—C37—O31 | −5.1 (5) |
| C5—N4—C51—C56 | 24 (19) | C32—C31—C37—O31 | 174.9 (3) |
4-(4-Methoxyphenyl)piperazin-1-ium 4-chlorobenzoate monohydrate (III). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 1.09 (3) | 1.71 (3) | 2.790 (4) | 176 (3) |
| N1—H12···O41 | 0.83 (3) | 1.98 (3) | 2.811 (4) | 174 (3) |
| O41—H41···O32i | 0.91 (4) | 1.73 (4) | 2.624 (4) | 172 (4) |
| O41—H42···O31ii | 0.94 (4) | 1.84 (4) | 2.775 (4) | 170 (4) |
| C2—H2B···O31iii | 0.97 | 2.52 | 3.467 (4) | 165 |
| C6—H6B···O41i | 0.97 | 2.60 | 3.408 (4) | 141 |
| C22—H22···Cg1iv | 0.93 | 2.89 | 3.631 (13) | 137 |
| C26—H26···Cg1iv | 0.93 | 2.81 | 3.58 (2) | 141 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1; (iii) −x+2, −y+2, −z+1; (iv) −x+1, −y+2, −z+1.
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Crystal data
| C11H17N2O+·C7H4BrO2−·H2O | Z = 2 |
| Mr = 411.28 | F(000) = 424 |
| Triclinic, P1 | Dx = 1.450 Mg m−3 |
| a = 6.2004 (8) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.4957 (9) Å | Cell parameters from 3927 reflections |
| c = 20.440 (2) Å | θ = 2.8–27.9° |
| α = 85.08 (1)° | µ = 2.21 mm−1 |
| β = 87.37 (1)° | T = 293 K |
| γ = 85.00 (1)° | Plate, colourless |
| V = 942.17 (19) Å3 | 0.48 × 0.44 × 0.16 mm |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3818 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2063 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.018 |
| ω scans | θmax = 26.6°, θmin = 2.8° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −7→7 |
| Tmin = 0.536, Tmax = 0.719 | k = −8→9 |
| 6176 measured reflections | l = −23→25 |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.068 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.197 | w = 1/[σ2(Fo2) + (0.0819P)2 + 0.9249P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 3818 reflections | Δρmax = 0.94 e Å−3 |
| 265 parameters | Δρmin = −0.64 e Å−3 |
| 17 restraints |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Special details
| Experimental. Compound (IV). IR (KBr , cm-1) 3319 (OH), 3001 (NH2), 2836 (OCH3), 1580 (COO), 600(CBr). . NMR (CDCl3) δ(1H) ) 3.23 (m, 4H, piperazine), 3.30 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.85 (m, 4H, methoxyphenyl), 7.51 (d, J = 8.4 Hz, 2H, bromophenyl), 7.90 (d, J = 8.4 Hz,2H, bromophenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.7803 (6) | 0.7425 (5) | 0.45428 (17) | 0.0475 (9) | |
| H11 | 0.813 (7) | 0.770 (6) | 0.488 (2) | 0.057* | |
| H12 | 0.753 (7) | 0.620 (7) | 0.461 (2) | 0.057* | |
| C2 | 0.9709 (7) | 0.7508 (6) | 0.4079 (2) | 0.0530 (11) | |
| H2A | 1.0899 | 0.6702 | 0.4250 | 0.064* | |
| H2B | 1.0169 | 0.8718 | 0.4034 | 0.064* | |
| C3 | 0.9143 (6) | 0.6978 (6) | 0.3414 (2) | 0.0502 (10) | |
| H3A | 1.0382 | 0.7086 | 0.3110 | 0.060* | |
| H3B | 0.8808 | 0.5732 | 0.3454 | 0.060* | |
| N4 | 0.7286 (5) | 0.8112 (4) | 0.31535 (14) | 0.0389 (7) | |
| C5 | 0.5420 (6) | 0.8089 (6) | 0.36137 (18) | 0.0454 (10) | |
| H5A | 0.4924 | 0.6891 | 0.3665 | 0.055* | |
| H5B | 0.4249 | 0.8909 | 0.3438 | 0.055* | |
| C6 | 0.5976 (7) | 0.8631 (6) | 0.42775 (19) | 0.0485 (10) | |
| H6A | 0.6368 | 0.9862 | 0.4234 | 0.058* | |
| H6B | 0.4723 | 0.8565 | 0.4578 | 0.058* | |
| C21 | 0.6845 (19) | 0.784 (6) | 0.2485 (4) | 0.0421 (11) | 0.80 (2) |
| C22 | 0.8313 (11) | 0.6853 (18) | 0.2086 (3) | 0.066 (3) | 0.80 (2) |
| H22 | 0.9604 | 0.6325 | 0.2256 | 0.079* | 0.80 (2) |
| C23 | 0.7862 (12) | 0.666 (2) | 0.1442 (3) | 0.084 (4) | 0.80 (2) |
| H23 | 0.8895 | 0.6040 | 0.1183 | 0.101* | 0.80 (2) |
| C24 | 0.5980 (13) | 0.7331 (16) | 0.1174 (4) | 0.062 (3) | 0.80 (2) |
| C25 | 0.4512 (15) | 0.826 (5) | 0.1561 (6) | 0.080 (4) | 0.80 (2) |
| H25 | 0.3201 | 0.8733 | 0.1390 | 0.096* | 0.80 (2) |
| C26 | 0.4941 (16) | 0.850 (3) | 0.2209 (4) | 0.064 (4) | 0.80 (2) |
| H26 | 0.3899 | 0.9127 | 0.2462 | 0.077* | 0.80 (2) |
| O24 | 0.5730 (16) | 0.7011 (17) | 0.0530 (3) | 0.101 (3) | 0.80 (2) |
| C27 | 0.373 (2) | 0.743 (3) | 0.0258 (6) | 0.114 (6) | 0.80 (2) |
| H27A | 0.2623 | 0.6966 | 0.0551 | 0.171* | 0.80 (2) |
| H27B | 0.3717 | 0.6905 | −0.0154 | 0.171* | 0.80 (2) |
| H27C | 0.3464 | 0.8712 | 0.0186 | 0.171* | 0.80 (2) |
| C51 | 0.672 (7) | 0.79 (2) | 0.2512 (15) | 0.0421 (11) | 0.20 (2) |
| C52 | 0.838 (5) | 0.786 (6) | 0.2026 (13) | 0.066 (3) | 0.20 (2) |
| H52 | 0.9785 | 0.8027 | 0.2134 | 0.079* | 0.20 (2) |
| C53 | 0.795 (5) | 0.760 (7) | 0.1384 (13) | 0.084 (4) | 0.20 (2) |
| H53 | 0.9078 | 0.7242 | 0.1096 | 0.101* | 0.20 (2) |
| C54 | 0.590 (5) | 0.787 (9) | 0.1169 (14) | 0.062 (3) | 0.20 (2) |
| C55 | 0.426 (6) | 0.81 (2) | 0.163 (2) | 0.080 (4) | 0.20 (2) |
| H55 | 0.2833 | 0.8234 | 0.1492 | 0.096* | 0.20 (2) |
| C56 | 0.466 (6) | 0.818 (13) | 0.2286 (19) | 0.064 (4) | 0.20 (2) |
| H56 | 0.3515 | 0.8441 | 0.2581 | 0.077* | 0.20 (2) |
| O54 | 0.565 (7) | 0.773 (7) | 0.0512 (15) | 0.101 (3) | 0.20 (2) |
| C57 | 0.364 (9) | 0.817 (13) | 0.025 (3) | 0.114 (6) | 0.20 (2) |
| H57A | 0.2541 | 0.7740 | 0.0554 | 0.171* | 0.20 (2) |
| H57B | 0.3582 | 0.7635 | −0.0157 | 0.171* | 0.20 (2) |
| H57C | 0.3404 | 0.9457 | 0.0173 | 0.171* | 0.20 (2) |
| C31 | 0.8064 (6) | 0.7392 (5) | 0.6932 (2) | 0.0436 (9) | |
| C32 | 0.6590 (8) | 0.6690 (7) | 0.7386 (3) | 0.0661 (13) | |
| H32 | 0.5388 | 0.6192 | 0.7245 | 0.079* | |
| C33 | 0.6890 (10) | 0.6724 (8) | 0.8053 (3) | 0.0832 (17) | |
| H33 | 0.5888 | 0.6259 | 0.8360 | 0.100* | |
| C34 | 0.8682 (10) | 0.7450 (8) | 0.8256 (2) | 0.0802 (16) | |
| Br34 | 0.90280 (17) | 0.75220 (15) | 0.91705 (3) | 0.1491 (5) | |
| C35 | 1.0170 (9) | 0.8124 (7) | 0.7814 (2) | 0.0683 (14) | |
| H35 | 1.1383 | 0.8599 | 0.7959 | 0.082* | |
| C36 | 0.9870 (7) | 0.8100 (6) | 0.7146 (2) | 0.0500 (10) | |
| H36 | 1.0884 | 0.8561 | 0.6843 | 0.060* | |
| C37 | 0.7721 (7) | 0.7357 (6) | 0.6201 (2) | 0.0528 (11) | |
| O31 | 0.8989 (6) | 0.8126 (4) | 0.57976 (15) | 0.0631 (9) | |
| O32 | 0.6180 (7) | 0.6578 (6) | 0.6044 (2) | 0.0999 (14) | |
| O41 | 0.7233 (6) | 0.3743 (5) | 0.46351 (17) | 0.0633 (9) | |
| H41 | 0.620 (10) | 0.353 (8) | 0.445 (3) | 0.095* | |
| H42 | 0.820 (11) | 0.313 (9) | 0.449 (3) | 0.095* |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.062 (2) | 0.048 (2) | 0.0349 (17) | −0.0089 (18) | −0.0128 (16) | −0.0048 (16) |
| C2 | 0.044 (2) | 0.067 (3) | 0.049 (2) | −0.004 (2) | −0.0092 (19) | −0.004 (2) |
| C3 | 0.037 (2) | 0.067 (3) | 0.046 (2) | 0.002 (2) | −0.0018 (18) | −0.003 (2) |
| N4 | 0.0365 (17) | 0.047 (2) | 0.0333 (15) | −0.0045 (14) | 0.0000 (13) | −0.0017 (14) |
| C5 | 0.039 (2) | 0.059 (3) | 0.038 (2) | 0.0005 (19) | −0.0004 (17) | −0.0020 (19) |
| C6 | 0.055 (3) | 0.052 (3) | 0.037 (2) | 0.003 (2) | 0.0016 (18) | −0.0034 (18) |
| C21 | 0.044 (3) | 0.048 (3) | 0.034 (2) | −0.006 (3) | 0.0000 (19) | −0.002 (3) |
| C22 | 0.053 (3) | 0.093 (9) | 0.048 (3) | 0.025 (4) | −0.009 (2) | −0.017 (4) |
| C23 | 0.076 (4) | 0.122 (10) | 0.049 (3) | 0.036 (5) | 0.001 (3) | −0.031 (5) |
| C24 | 0.072 (3) | 0.079 (9) | 0.036 (2) | 0.009 (4) | −0.007 (2) | −0.018 (3) |
| C25 | 0.058 (4) | 0.133 (10) | 0.046 (4) | 0.015 (7) | −0.015 (3) | −0.008 (6) |
| C26 | 0.055 (4) | 0.099 (10) | 0.036 (3) | 0.015 (5) | −0.001 (2) | −0.015 (5) |
| O24 | 0.108 (3) | 0.148 (10) | 0.046 (2) | 0.033 (5) | −0.019 (2) | −0.037 (4) |
| C27 | 0.108 (5) | 0.181 (19) | 0.058 (4) | 0.008 (7) | −0.030 (4) | −0.038 (7) |
| C51 | 0.044 (3) | 0.048 (3) | 0.034 (2) | −0.006 (3) | 0.0000 (19) | −0.002 (3) |
| C52 | 0.053 (3) | 0.093 (9) | 0.048 (3) | 0.025 (4) | −0.009 (2) | −0.017 (4) |
| C53 | 0.076 (4) | 0.122 (10) | 0.049 (3) | 0.036 (5) | 0.001 (3) | −0.031 (5) |
| C54 | 0.072 (3) | 0.079 (9) | 0.036 (2) | 0.009 (4) | −0.007 (2) | −0.018 (3) |
| C55 | 0.058 (4) | 0.133 (10) | 0.046 (4) | 0.015 (7) | −0.015 (3) | −0.008 (6) |
| C56 | 0.055 (4) | 0.099 (10) | 0.036 (3) | 0.015 (5) | −0.001 (2) | −0.015 (5) |
| O54 | 0.108 (3) | 0.148 (10) | 0.046 (2) | 0.033 (5) | −0.019 (2) | −0.037 (4) |
| C57 | 0.108 (5) | 0.181 (19) | 0.058 (4) | 0.008 (7) | −0.030 (4) | −0.038 (7) |
| C31 | 0.041 (2) | 0.040 (2) | 0.049 (2) | 0.0019 (18) | −0.0051 (18) | −0.0056 (18) |
| C32 | 0.052 (3) | 0.070 (3) | 0.076 (3) | −0.009 (2) | 0.001 (2) | −0.005 (3) |
| C33 | 0.082 (4) | 0.098 (4) | 0.063 (3) | −0.002 (3) | 0.023 (3) | 0.011 (3) |
| C34 | 0.089 (4) | 0.100 (4) | 0.048 (3) | 0.001 (3) | −0.006 (3) | 0.004 (3) |
| Br34 | 0.1950 (10) | 0.2001 (11) | 0.0444 (4) | 0.0349 (8) | −0.0150 (4) | −0.0114 (4) |
| C35 | 0.073 (3) | 0.082 (4) | 0.053 (3) | −0.006 (3) | −0.021 (3) | −0.010 (3) |
| C36 | 0.049 (2) | 0.056 (3) | 0.046 (2) | −0.010 (2) | −0.0083 (19) | 0.000 (2) |
| C37 | 0.051 (3) | 0.045 (3) | 0.063 (3) | 0.006 (2) | −0.020 (2) | −0.012 (2) |
| O31 | 0.083 (2) | 0.063 (2) | 0.0446 (17) | −0.0054 (18) | −0.0124 (17) | −0.0053 (15) |
| O32 | 0.087 (3) | 0.130 (4) | 0.094 (3) | −0.037 (3) | −0.038 (2) | −0.024 (3) |
| O41 | 0.064 (2) | 0.065 (2) | 0.064 (2) | −0.0133 (17) | −0.0210 (17) | −0.0029 (17) |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Geometric parameters (Å, º)
| N1—C6 | 1.479 (5) | C27—H27C | 0.9600 |
| N1—C2 | 1.483 (6) | C51—C56 | 1.369 (11) |
| N1—H11 | 0.78 (5) | C51—C52 | 1.40 (2) |
| N1—H12 | 0.95 (5) | C52—C53 | 1.383 (11) |
| C2—C3 | 1.513 (6) | C52—H52 | 0.9300 |
| C2—H2A | 0.9700 | C53—C54 | 1.353 (11) |
| C2—H2B | 0.9700 | C53—H53 | 0.9300 |
| C3—N4 | 1.463 (5) | C54—C55 | 1.362 (19) |
| C3—H3A | 0.9700 | C54—O54 | 1.374 (10) |
| C3—H3B | 0.9700 | C55—C56 | 1.392 (11) |
| N4—C51 | 1.40 (4) | C55—H55 | 0.9300 |
| N4—C21 | 1.441 (10) | C56—H56 | 0.9300 |
| N4—C5 | 1.458 (5) | O54—C57 | 1.383 (12) |
| C5—C6 | 1.512 (5) | C57—H57A | 0.9600 |
| C5—H5A | 0.9700 | C57—H57B | 0.9600 |
| C5—H5B | 0.9700 | C57—H57C | 0.9600 |
| C6—H6A | 0.9700 | C31—C32 | 1.377 (6) |
| C6—H6B | 0.9700 | C31—C36 | 1.385 (5) |
| C21—C26 | 1.367 (10) | C31—C37 | 1.521 (6) |
| C21—C22 | 1.399 (19) | C32—C33 | 1.387 (7) |
| C22—C23 | 1.380 (7) | C32—H32 | 0.9300 |
| C22—H22 | 0.9300 | C33—C34 | 1.375 (8) |
| C23—C24 | 1.351 (8) | C33—H33 | 0.9300 |
| C23—H23 | 0.9300 | C34—C35 | 1.360 (8) |
| C24—C25 | 1.362 (16) | C34—Br34 | 1.897 (5) |
| C24—O24 | 1.374 (6) | C35—C36 | 1.388 (6) |
| C25—C26 | 1.392 (8) | C35—H35 | 0.9300 |
| C25—H25 | 0.9300 | C36—H36 | 0.9300 |
| C26—H26 | 0.9300 | C37—O32 | 1.231 (5) |
| O24—C27 | 1.383 (8) | C37—O31 | 1.252 (6) |
| C27—H27A | 0.9600 | O41—H41 | 0.79 (6) |
| C27—H27B | 0.9600 | O41—H42 | 0.79 (7) |
| C6—N1—C2 | 109.7 (3) | O24—C27—H27A | 109.5 |
| C6—N1—H11 | 111 (3) | O24—C27—H27B | 109.5 |
| C2—N1—H11 | 108 (3) | H27A—C27—H27B | 109.5 |
| C6—N1—H12 | 115 (3) | O24—C27—H27C | 109.5 |
| C2—N1—H12 | 106 (3) | H27A—C27—H27C | 109.5 |
| H11—N1—H12 | 106 (4) | H27B—C27—H27C | 109.5 |
| N1—C2—C3 | 110.1 (3) | C56—C51—C52 | 115 (2) |
| N1—C2—H2A | 109.6 | C56—C51—N4 | 125 (3) |
| C3—C2—H2A | 109.6 | C52—C51—N4 | 117 (4) |
| N1—C2—H2B | 109.6 | C53—C52—C51 | 120.4 (19) |
| C3—C2—H2B | 109.6 | C53—C52—H52 | 119.8 |
| H2A—C2—H2B | 108.1 | C51—C52—H52 | 119.8 |
| N4—C3—C2 | 111.3 (3) | C54—C53—C52 | 120.8 (13) |
| N4—C3—H3A | 109.4 | C54—C53—H53 | 119.6 |
| C2—C3—H3A | 109.4 | C52—C53—H53 | 119.6 |
| N4—C3—H3B | 109.4 | C53—C54—C55 | 117.8 (11) |
| C2—C3—H3B | 109.4 | C53—C54—O54 | 116.7 (13) |
| H3A—C3—H3B | 108.0 | C55—C54—O54 | 125.2 (15) |
| C51—N4—C5 | 112 (2) | C54—C55—C56 | 121.2 (16) |
| C21—N4—C5 | 115.3 (7) | C54—C55—H55 | 119.4 |
| C51—N4—C3 | 117 (4) | C56—C55—H55 | 119.4 |
| C21—N4—C3 | 114.0 (11) | C51—C56—C55 | 121.4 (12) |
| C5—N4—C3 | 111.5 (3) | C51—C56—H56 | 119.3 |
| N4—C5—C6 | 111.4 (3) | C55—C56—H56 | 119.3 |
| N4—C5—H5A | 109.4 | C54—O54—C57 | 119.3 (17) |
| C6—C5—H5A | 109.4 | O54—C57—H57A | 109.5 |
| N4—C5—H5B | 109.4 | O54—C57—H57B | 109.5 |
| C6—C5—H5B | 109.4 | H57A—C57—H57B | 109.5 |
| H5A—C5—H5B | 108.0 | O54—C57—H57C | 109.5 |
| N1—C6—C5 | 110.0 (3) | H57A—C57—H57C | 109.5 |
| N1—C6—H6A | 109.7 | H57B—C57—H57C | 109.5 |
| C5—C6—H6A | 109.7 | C32—C31—C36 | 119.5 (4) |
| N1—C6—H6B | 109.7 | C32—C31—C37 | 120.0 (4) |
| C5—C6—H6B | 109.7 | C36—C31—C37 | 120.5 (4) |
| H6A—C6—H6B | 108.2 | C31—C32—C33 | 120.3 (5) |
| C26—C21—C22 | 116.1 (7) | C31—C32—H32 | 119.9 |
| C26—C21—N4 | 121.7 (13) | C33—C32—H32 | 119.9 |
| C22—C21—N4 | 122.2 (10) | C34—C33—C32 | 119.4 (5) |
| C23—C22—C21 | 120.6 (6) | C34—C33—H33 | 120.3 |
| C23—C22—H22 | 119.7 | C32—C33—H33 | 120.3 |
| C21—C22—H22 | 119.7 | C35—C34—C33 | 121.1 (5) |
| C24—C23—C22 | 122.6 (5) | C35—C34—Br34 | 120.3 (5) |
| C24—C23—H23 | 118.7 | C33—C34—Br34 | 118.6 (4) |
| C22—C23—H23 | 118.7 | C34—C35—C36 | 119.7 (5) |
| C23—C24—C25 | 117.4 (5) | C34—C35—H35 | 120.2 |
| C23—C24—O24 | 116.5 (5) | C36—C35—H35 | 120.2 |
| C25—C24—O24 | 126.1 (6) | C31—C36—C35 | 120.1 (4) |
| C24—C25—C26 | 121.1 (10) | C31—C36—H36 | 119.9 |
| C24—C25—H25 | 119.5 | C35—C36—H36 | 119.9 |
| C26—C25—H25 | 119.5 | O32—C37—O31 | 124.0 (5) |
| C21—C26—C25 | 122.1 (10) | O32—C37—C31 | 117.1 (5) |
| C21—C26—H26 | 119.0 | O31—C37—C31 | 118.9 (4) |
| C25—C26—H26 | 119.0 | H41—O41—H42 | 105 (6) |
| C24—O24—C27 | 119.2 (5) | ||
| C6—N1—C2—C3 | −58.3 (5) | C5—N4—C51—C52 | −180 (9) |
| N1—C2—C3—N4 | 56.7 (5) | C3—N4—C51—C52 | −49 (14) |
| C2—C3—N4—C51 | 174 (7) | C56—C51—C52—C53 | −20 (15) |
| C2—C3—N4—C21 | 172.1 (16) | N4—C51—C52—C53 | 179 (8) |
| C2—C3—N4—C5 | −55.2 (4) | C51—C52—C53—C54 | 19 (9) |
| C51—N4—C5—C6 | −171 (8) | C52—C53—C54—C55 | −10 (12) |
| C21—N4—C5—C6 | −172.5 (19) | C52—C53—C54—O54 | 175 (5) |
| C3—N4—C5—C6 | 55.6 (4) | C53—C54—C55—C56 | 3 (19) |
| C2—N1—C6—C5 | 58.6 (4) | O54—C54—C55—C56 | 178 (10) |
| N4—C5—C6—N1 | −57.4 (4) | C52—C51—C56—C55 | 13 (18) |
| C5—N4—C21—C26 | 36 (4) | N4—C51—C56—C55 | 173 (14) |
| C3—N4—C21—C26 | 166 (2) | C54—C55—C56—C51 | −5 (21) |
| C5—N4—C21—C22 | −143 (2) | C53—C54—O54—C57 | −173 (6) |
| C3—N4—C21—C22 | −12 (4) | C55—C54—O54—C57 | 12 (14) |
| C26—C21—C22—C23 | 3 (4) | C36—C31—C32—C33 | 1.2 (7) |
| N4—C21—C22—C23 | −178 (2) | C37—C31—C32—C33 | −179.8 (5) |
| C21—C22—C23—C24 | −3 (2) | C31—C32—C33—C34 | −0.5 (8) |
| C22—C23—C24—C25 | 1 (2) | C32—C33—C34—C35 | −0.4 (9) |
| C22—C23—C24—O24 | −178.9 (9) | C32—C33—C34—Br34 | 178.8 (4) |
| C23—C24—C25—C26 | 0 (4) | C33—C34—C35—C36 | 0.7 (9) |
| O24—C24—C25—C26 | 180 (2) | Br34—C34—C35—C36 | −178.5 (4) |
| C22—C21—C26—C25 | −2 (4) | C32—C31—C36—C35 | −0.9 (6) |
| N4—C21—C26—C25 | 179 (3) | C37—C31—C36—C35 | −179.9 (4) |
| C24—C25—C26—C21 | 1 (4) | C34—C35—C36—C31 | 0.0 (7) |
| C23—C24—O24—C27 | 170.6 (12) | C32—C31—C37—O32 | −4.8 (6) |
| C25—C24—O24—C27 | −9 (3) | C36—C31—C37—O32 | 174.2 (4) |
| C5—N4—C51—C56 | 21 (16) | C32—C31—C37—O31 | 174.5 (4) |
| C3—N4—C51—C56 | 151 (11) | C36—C31—C37—O31 | −6.5 (6) |
4-(4-Methoxyphenyl)piperazin-1-ium 4-bromobenzoate monohydrate (IV). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.78 (4) | 2.03 (4) | 2.805 (5) | 174 (5) |
| N1—H12···O41 | 0.95 (5) | 1.86 (5) | 2.802 (5) | 172 (4) |
| O41—H41···O32i | 0.79 (6) | 1.84 (6) | 2.623 (6) | 170 (6) |
| O41—H42···O31ii | 0.79 (7) | 2.00 (7) | 2.772 (5) | 169 (6) |
| C2—H2B···O31iii | 0.97 | 2.52 | 3.471 (5) | 166 |
| C22—H22···Cg1ii | 0.93 | 2.52 | 3.471 (5) | 166 |
| C26—H26···Cg1iv | 0.93 | 2.84 | 3.58 (2) | 137 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1; (iii) −x+2, −y+2, −z+1; (iv) −x+1, −y+2, −z+1.
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Crystal data
| C11H17N2O+·C7H5O3− | Dx = 1.317 Mg m−3 |
| Mr = 330.38 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, P212121 | Cell parameters from 3564 reflections |
| a = 6.5009 (8) Å | θ = 2.5–27.8° |
| b = 7.9735 (9) Å | µ = 0.09 mm−1 |
| c = 32.155 (4) Å | T = 296 K |
| V = 1666.8 (3) Å3 | Block, colourless |
| Z = 4 | 0.42 × 0.42 × 0.34 mm |
| F(000) = 704 |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3564 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2875 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.014 |
| ω scans | θmax = 27.8°, θmin = 2.5° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −6→8 |
| Tmin = 0.899, Tmax = 0.969 | k = −10→6 |
| 6249 measured reflections | l = −38→41 |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.041 | w = 1/[σ2(Fo2) + (0.0299P)2 + 0.3737P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.089 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.14 e Å−3 |
| 3564 reflections | Δρmin = −0.13 e Å−3 |
| 228 parameters | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0244 (17) |
| Primary atom site location: difference Fourier map | Absolute structure: Flack x determined using 1011 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Special details
| Experimental. Compound (V). IR (KBr , cm-1) 3650 (OH), 3040 (NH2), 2835 (OCH3), 1571 (COO). NMR (CDCl3) δ(1H) 3.31 (m, 8H, piperazine), 3.77 (s, 3H, OCH3), 6.85 (m, 5H, hydroxyphenyl and methoxyphenyl), 6.92 (m, 1H, hydroxyphenyl), 7.35 (t, 1H, hydroxyphenyl), 7.87 (m, 1H, hydroxyphenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.5261 (3) | 0.3567 (3) | 0.21309 (7) | 0.0468 (5) | |
| H11 | 0.616 (4) | 0.369 (4) | 0.2367 (9) | 0.056* | |
| H12 | 0.452 (4) | 0.254 (4) | 0.2144 (8) | 0.056* | |
| C2 | 0.6568 (4) | 0.3607 (4) | 0.17563 (8) | 0.0488 (6) | |
| H2A | 0.7532 | 0.2680 | 0.1765 | 0.059* | |
| H2B | 0.7353 | 0.4642 | 0.1753 | 0.059* | |
| C3 | 0.5299 (4) | 0.3490 (3) | 0.13672 (8) | 0.0443 (6) | |
| H3A | 0.6189 | 0.3643 | 0.1128 | 0.053* | |
| H3B | 0.4705 | 0.2377 | 0.1349 | 0.053* | |
| N4 | 0.3654 (3) | 0.4727 (3) | 0.13512 (6) | 0.0381 (5) | |
| C5 | 0.2422 (4) | 0.4788 (3) | 0.17339 (7) | 0.0426 (6) | |
| H5A | 0.1606 | 0.3773 | 0.1755 | 0.051* | |
| H5B | 0.1485 | 0.5733 | 0.1720 | 0.051* | |
| C6 | 0.3752 (4) | 0.4953 (3) | 0.21130 (8) | 0.0460 (6) | |
| H6A | 0.4472 | 0.6018 | 0.2106 | 0.055* | |
| H6B | 0.2898 | 0.4933 | 0.2360 | 0.055* | |
| C21 | 0.2490 (4) | 0.4737 (3) | 0.09771 (7) | 0.0373 (5) | |
| C22 | 0.3077 (4) | 0.3820 (4) | 0.06280 (8) | 0.0515 (7) | |
| H22 | 0.4232 | 0.3134 | 0.0642 | 0.062* | |
| C23 | 0.1976 (5) | 0.3911 (4) | 0.02617 (8) | 0.0544 (7) | |
| H23 | 0.2412 | 0.3295 | 0.0033 | 0.065* | |
| C24 | 0.0260 (4) | 0.4886 (3) | 0.02292 (8) | 0.0460 (6) | |
| C25 | −0.0385 (4) | 0.5770 (4) | 0.05738 (8) | 0.0529 (7) | |
| H25 | −0.1573 | 0.6418 | 0.0560 | 0.063* | |
| C26 | 0.0728 (4) | 0.5697 (4) | 0.09394 (8) | 0.0503 (7) | |
| H26 | 0.0277 | 0.6312 | 0.1168 | 0.060* | |
| O24 | −0.0704 (3) | 0.4899 (3) | −0.01523 (5) | 0.0638 (6) | |
| C27 | −0.2374 (6) | 0.6009 (5) | −0.02052 (10) | 0.0833 (11) | |
| H27A | −0.2851 | 0.5953 | −0.0487 | 0.125* | |
| H27B | −0.1938 | 0.7133 | −0.0144 | 0.125* | |
| H27C | −0.3469 | 0.5697 | −0.0020 | 0.125* | |
| C31 | 0.9904 (4) | 0.4542 (3) | 0.32938 (7) | 0.0370 (5) | |
| C32 | 1.1705 (4) | 0.3584 (3) | 0.32627 (7) | 0.0394 (5) | |
| C33 | 1.3127 (4) | 0.3600 (4) | 0.35834 (8) | 0.0501 (7) | |
| H33 | 1.4324 | 0.2967 | 0.3561 | 0.060* | |
| C34 | 1.2782 (5) | 0.4544 (4) | 0.39334 (8) | 0.0557 (7) | |
| H34 | 1.3746 | 0.4546 | 0.4147 | 0.067* | |
| C35 | 1.1023 (5) | 0.5484 (4) | 0.39698 (8) | 0.0584 (8) | |
| H35 | 1.0790 | 0.6119 | 0.4208 | 0.070* | |
| C36 | 0.9613 (4) | 0.5480 (3) | 0.36539 (8) | 0.0501 (7) | |
| H36 | 0.8427 | 0.6123 | 0.3681 | 0.060* | |
| O33 | 1.2074 (3) | 0.2609 (2) | 0.29254 (6) | 0.0548 (5) | |
| H33A | 1.082 (5) | 0.275 (4) | 0.2766 (10) | 0.082* | |
| C37 | 0.8348 (4) | 0.4561 (3) | 0.29516 (8) | 0.0453 (6) | |
| O31 | 0.8614 (3) | 0.3519 (3) | 0.26569 (6) | 0.0601 (6) | |
| O32 | 0.6867 (3) | 0.5531 (3) | 0.29674 (7) | 0.0682 (6) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0464 (13) | 0.0489 (12) | 0.0452 (12) | −0.0102 (11) | −0.0143 (11) | 0.0037 (11) |
| C2 | 0.0363 (13) | 0.0520 (15) | 0.0581 (15) | −0.0003 (12) | −0.0058 (13) | −0.0004 (14) |
| C3 | 0.0364 (13) | 0.0475 (14) | 0.0489 (14) | 0.0030 (12) | 0.0007 (11) | −0.0024 (12) |
| N4 | 0.0353 (10) | 0.0416 (11) | 0.0374 (10) | 0.0037 (9) | −0.0007 (8) | −0.0024 (9) |
| C5 | 0.0388 (12) | 0.0494 (14) | 0.0395 (12) | 0.0027 (12) | 0.0008 (11) | −0.0041 (11) |
| C6 | 0.0484 (14) | 0.0492 (15) | 0.0404 (13) | −0.0030 (13) | 0.0000 (12) | −0.0022 (12) |
| C21 | 0.0396 (12) | 0.0350 (12) | 0.0374 (12) | −0.0018 (11) | 0.0015 (10) | −0.0011 (10) |
| C22 | 0.0559 (16) | 0.0543 (16) | 0.0444 (14) | 0.0172 (14) | 0.0005 (13) | −0.0062 (13) |
| C23 | 0.0699 (19) | 0.0551 (17) | 0.0381 (13) | 0.0109 (15) | 0.0013 (14) | −0.0102 (12) |
| C24 | 0.0555 (15) | 0.0431 (14) | 0.0393 (13) | −0.0032 (13) | −0.0070 (12) | 0.0005 (11) |
| C25 | 0.0501 (15) | 0.0582 (17) | 0.0503 (16) | 0.0139 (14) | −0.0088 (13) | −0.0047 (13) |
| C26 | 0.0523 (15) | 0.0567 (17) | 0.0418 (14) | 0.0153 (14) | −0.0030 (12) | −0.0121 (12) |
| O24 | 0.0793 (14) | 0.0684 (14) | 0.0435 (10) | 0.0051 (12) | −0.0190 (10) | −0.0036 (9) |
| C27 | 0.080 (2) | 0.102 (3) | 0.068 (2) | 0.014 (2) | −0.0331 (19) | 0.000 (2) |
| C31 | 0.0382 (12) | 0.0331 (12) | 0.0396 (13) | −0.0030 (10) | −0.0004 (10) | 0.0076 (10) |
| C32 | 0.0416 (12) | 0.0353 (12) | 0.0413 (13) | −0.0017 (11) | −0.0006 (12) | 0.0047 (11) |
| C33 | 0.0398 (14) | 0.0552 (16) | 0.0554 (16) | 0.0006 (13) | −0.0092 (13) | 0.0093 (14) |
| C34 | 0.0586 (17) | 0.0642 (18) | 0.0442 (15) | −0.0139 (16) | −0.0139 (14) | 0.0084 (14) |
| C35 | 0.076 (2) | 0.0595 (18) | 0.0401 (15) | −0.0090 (17) | 0.0015 (14) | −0.0046 (13) |
| C36 | 0.0524 (15) | 0.0472 (15) | 0.0506 (15) | 0.0064 (14) | 0.0064 (13) | 0.0028 (12) |
| O33 | 0.0547 (12) | 0.0555 (12) | 0.0542 (11) | 0.0139 (10) | −0.0054 (10) | −0.0087 (9) |
| C37 | 0.0431 (14) | 0.0409 (14) | 0.0517 (15) | −0.0037 (13) | −0.0069 (12) | 0.0127 (12) |
| O31 | 0.0639 (13) | 0.0586 (12) | 0.0580 (12) | 0.0036 (11) | −0.0245 (10) | −0.0057 (10) |
| O32 | 0.0524 (11) | 0.0730 (14) | 0.0793 (14) | 0.0200 (11) | −0.0115 (11) | 0.0136 (11) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Geometric parameters (Å, º)
| N1—C2 | 1.474 (3) | C24—C25 | 1.379 (4) |
| N1—C6 | 1.479 (3) | C25—C26 | 1.382 (4) |
| N1—H11 | 0.96 (3) | C25—H25 | 0.9300 |
| N1—H12 | 0.95 (3) | C26—H26 | 0.9300 |
| C2—C3 | 1.502 (3) | O24—C27 | 1.411 (4) |
| C2—H2A | 0.9700 | C27—H27A | 0.9600 |
| C2—H2B | 0.9700 | C27—H27B | 0.9600 |
| C3—N4 | 1.456 (3) | C27—H27C | 0.9600 |
| C3—H3A | 0.9700 | C31—C36 | 1.391 (3) |
| C3—H3B | 0.9700 | C31—C32 | 1.401 (3) |
| N4—C21 | 1.421 (3) | C31—C37 | 1.495 (3) |
| N4—C5 | 1.469 (3) | C32—O33 | 1.356 (3) |
| C5—C6 | 1.500 (3) | C32—C33 | 1.385 (3) |
| C5—H5A | 0.9700 | C33—C34 | 1.372 (4) |
| C5—H5B | 0.9700 | C33—H33 | 0.9300 |
| C6—H6A | 0.9700 | C34—C35 | 1.372 (4) |
| C6—H6B | 0.9700 | C34—H34 | 0.9300 |
| C21—C26 | 1.383 (3) | C35—C36 | 1.368 (4) |
| C21—C22 | 1.393 (3) | C35—H35 | 0.9300 |
| C22—C23 | 1.380 (4) | C36—H36 | 0.9300 |
| C22—H22 | 0.9300 | O33—H33A | 0.97 (3) |
| C23—C24 | 1.364 (4) | C37—O32 | 1.236 (3) |
| C23—H23 | 0.9300 | C37—O31 | 1.272 (3) |
| C24—O24 | 1.378 (3) | ||
| C2—N1—C6 | 109.5 (2) | C24—C23—H23 | 119.3 |
| C2—N1—H11 | 107.1 (16) | C22—C23—H23 | 119.3 |
| C6—N1—H11 | 110.9 (17) | C23—C24—O24 | 116.4 (2) |
| C2—N1—H12 | 110.3 (16) | C23—C24—C25 | 118.6 (2) |
| C6—N1—H12 | 108.2 (16) | O24—C24—C25 | 125.0 (2) |
| H11—N1—H12 | 111 (2) | C24—C25—C26 | 120.2 (2) |
| N1—C2—C3 | 111.3 (2) | C24—C25—H25 | 119.9 |
| N1—C2—H2A | 109.4 | C26—C25—H25 | 119.9 |
| C3—C2—H2A | 109.4 | C25—C26—C21 | 122.1 (2) |
| N1—C2—H2B | 109.4 | C25—C26—H26 | 118.9 |
| C3—C2—H2B | 109.4 | C21—C26—H26 | 118.9 |
| H2A—C2—H2B | 108.0 | C24—O24—C27 | 117.5 (2) |
| N4—C3—C2 | 113.0 (2) | O24—C27—H27A | 109.5 |
| N4—C3—H3A | 109.0 | O24—C27—H27B | 109.5 |
| C2—C3—H3A | 109.0 | H27A—C27—H27B | 109.5 |
| N4—C3—H3B | 109.0 | O24—C27—H27C | 109.5 |
| C2—C3—H3B | 109.0 | H27A—C27—H27C | 109.5 |
| H3A—C3—H3B | 107.8 | H27B—C27—H27C | 109.5 |
| C21—N4—C3 | 115.14 (18) | C36—C31—C32 | 117.8 (2) |
| C21—N4—C5 | 114.77 (18) | C36—C31—C37 | 121.0 (2) |
| C3—N4—C5 | 113.17 (19) | C32—C31—C37 | 121.2 (2) |
| N4—C5—C6 | 111.69 (19) | O33—C32—C33 | 118.8 (2) |
| N4—C5—H5A | 109.3 | O33—C32—C31 | 121.2 (2) |
| C6—C5—H5A | 109.3 | C33—C32—C31 | 120.0 (2) |
| N4—C5—H5B | 109.3 | C34—C33—C32 | 120.4 (3) |
| C6—C5—H5B | 109.3 | C34—C33—H33 | 119.8 |
| H5A—C5—H5B | 107.9 | C32—C33—H33 | 119.8 |
| N1—C6—C5 | 110.4 (2) | C35—C34—C33 | 120.4 (3) |
| N1—C6—H6A | 109.6 | C35—C34—H34 | 119.8 |
| C5—C6—H6A | 109.6 | C33—C34—H34 | 119.8 |
| N1—C6—H6B | 109.6 | C36—C35—C34 | 119.6 (3) |
| C5—C6—H6B | 109.6 | C36—C35—H35 | 120.2 |
| H6A—C6—H6B | 108.1 | C34—C35—H35 | 120.2 |
| C26—C21—C22 | 116.5 (2) | C35—C36—C31 | 121.9 (3) |
| C26—C21—N4 | 121.2 (2) | C35—C36—H36 | 119.1 |
| C22—C21—N4 | 122.2 (2) | C31—C36—H36 | 119.1 |
| C23—C22—C21 | 121.2 (3) | C32—O33—H33A | 102 (2) |
| C23—C22—H22 | 119.4 | O32—C37—O31 | 123.0 (2) |
| C21—C22—H22 | 119.4 | O32—C37—C31 | 120.2 (3) |
| C24—C23—C22 | 121.3 (2) | O31—C37—C31 | 116.7 (2) |
| C6—N1—C2—C3 | −57.5 (3) | C22—C21—C26—C25 | −1.0 (4) |
| N1—C2—C3—N4 | 52.5 (3) | N4—C21—C26—C25 | 177.7 (3) |
| C2—C3—N4—C21 | 176.2 (2) | C23—C24—O24—C27 | −174.6 (3) |
| C2—C3—N4—C5 | −49.1 (3) | C25—C24—O24—C27 | 5.7 (4) |
| C21—N4—C5—C6 | −174.1 (2) | C36—C31—C32—O33 | 178.5 (2) |
| C3—N4—C5—C6 | 51.0 (3) | C37—C31—C32—O33 | −1.6 (3) |
| C2—N1—C6—C5 | 59.7 (3) | C36—C31—C32—C33 | −0.3 (3) |
| N4—C5—C6—N1 | −56.4 (3) | C37—C31—C32—C33 | 179.5 (2) |
| C3—N4—C21—C26 | 171.8 (2) | O33—C32—C33—C34 | −178.6 (2) |
| C5—N4—C21—C26 | 37.8 (3) | C31—C32—C33—C34 | 0.3 (4) |
| C3—N4—C21—C22 | −9.6 (3) | C32—C33—C34—C35 | 0.0 (4) |
| C5—N4—C21—C22 | −143.6 (2) | C33—C34—C35—C36 | −0.2 (4) |
| C26—C21—C22—C23 | 1.7 (4) | C34—C35—C36—C31 | 0.2 (4) |
| N4—C21—C22—C23 | −176.9 (3) | C32—C31—C36—C35 | 0.1 (4) |
| C21—C22—C23—C24 | −0.8 (4) | C37—C31—C36—C35 | −179.8 (2) |
| C22—C23—C24—O24 | 179.2 (3) | C36—C31—C37—O32 | 6.6 (4) |
| C22—C23—C24—C25 | −1.0 (4) | C32—C31—C37—O32 | −173.2 (2) |
| C23—C24—C25—C26 | 1.8 (4) | C36—C31—C37—O31 | −171.9 (2) |
| O24—C24—C25—C26 | −178.5 (3) | C32—C31—C37—O31 | 8.3 (3) |
| C24—C25—C26—C21 | −0.7 (5) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxybenzoate (V). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.96 (3) | 1.85 (3) | 2.759 (3) | 156 (3) |
| N1—H11···O32 | 0.96 (3) | 2.47 (3) | 3.283 (3) | 142 (2) |
| N1—H12···O32i | 0.95 (3) | 1.87 (3) | 2.806 (3) | 166 (2) |
| O33—H33A···O31 | 0.97 (3) | 1.60 (3) | 2.516 (3) | 156 (3) |
| C6—H6A···O33ii | 0.97 | 2.58 | 3.444 (3) | 148 |
| C2—H2A···Cg1iii | 0.97 | 2.88 | 3.711 (3) | 144 |
| C26—H26···Cg1iv | 0.93 | 2.87 | 3.642 (3) | 141 |
Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) −x+2, y+1/2, −z+1/2; (iii) −x+2, y−1/2, −z+1/2; (iv) −x+1, y+1/2, −z+1/2.
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Crystal data
| C11H17N2O+·C6H4NO2− | Dx = 1.319 Mg m−3 |
| Mr = 315.37 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 3593 reflections |
| a = 9.2817 (7) Å | θ = 2.6–27.9° |
| b = 11.2905 (7) Å | µ = 0.09 mm−1 |
| c = 30.309 (2) Å | T = 296 K |
| V = 3176.2 (4) Å3 | Block, colourless |
| Z = 8 | 0.46 × 0.42 × 0.36 mm |
| F(000) = 1344 |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3593 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2616 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.028 |
| ω scans | θmax = 27.9°, θmin = 2.6° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −11→11 |
| Tmin = 0.879, Tmax = 0.968 | k = −14→13 |
| 22154 measured reflections | l = −38→35 |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.048 | w = 1/[σ2(Fo2) + (0.0416P)2 + 1.5726P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.119 | (Δ/σ)max = 0.001 |
| S = 1.03 | Δρmax = 0.19 e Å−3 |
| 3593 reflections | Δρmin = −0.16 e Å−3 |
| 215 parameters | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0074 (6) |
| Primary atom site location: difference Fourier map |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Special details
| Experimental. Compound (VI). IR (KBr , cm-1) 3040 (NH2), 2829 (OCH3), 1584 (COO). NMR (CDCl3) δ(1H) 3.27 (m, 4H, piperazine), 3.34 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.90 (m, 4H, methoxyphenyl), 7.33 (m, 1H, nicotinate), 8.67 (m, 2H, nicotinate), 9.24 (m, 1H, nicotinate). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.66467 (17) | 0.63416 (13) | 0.53807 (5) | 0.0405 (4) | |
| H12 | 0.711 (2) | 0.5701 (18) | 0.5241 (6) | 0.049* | |
| H11 | 0.650 (2) | 0.6962 (17) | 0.5161 (6) | 0.049* | |
| C2 | 0.52299 (18) | 0.59306 (15) | 0.55418 (6) | 0.0401 (4) | |
| H2A | 0.4642 | 0.5684 | 0.5294 | 0.048* | |
| H2B | 0.4739 | 0.6577 | 0.5690 | 0.048* | |
| C3 | 0.54099 (18) | 0.49095 (14) | 0.58566 (5) | 0.0368 (4) | |
| H3A | 0.4473 | 0.4669 | 0.5967 | 0.044* | |
| H3B | 0.5831 | 0.4242 | 0.5702 | 0.044* | |
| N4 | 0.63342 (14) | 0.52403 (11) | 0.62269 (4) | 0.0337 (3) | |
| C5 | 0.77180 (19) | 0.57192 (16) | 0.60810 (6) | 0.0440 (4) | |
| H5A | 0.8273 | 0.5094 | 0.5942 | 0.053* | |
| H5B | 0.8255 | 0.5995 | 0.6336 | 0.053* | |
| C6 | 0.7534 (2) | 0.67296 (16) | 0.57600 (6) | 0.0479 (5) | |
| H6A | 0.7070 | 0.7391 | 0.5907 | 0.058* | |
| H6B | 0.8470 | 0.6990 | 0.5656 | 0.058* | |
| C21 | 0.63956 (17) | 0.44215 (13) | 0.65799 (5) | 0.0317 (3) | |
| C22 | 0.5429 (2) | 0.35039 (16) | 0.66255 (6) | 0.0449 (4) | |
| H22 | 0.4750 | 0.3379 | 0.6405 | 0.054* | |
| C23 | 0.5436 (2) | 0.27603 (15) | 0.69898 (6) | 0.0464 (5) | |
| H23 | 0.4759 | 0.2155 | 0.7010 | 0.056* | |
| C24 | 0.64245 (19) | 0.29057 (14) | 0.73195 (5) | 0.0391 (4) | |
| C25 | 0.7405 (2) | 0.38118 (18) | 0.72806 (6) | 0.0558 (5) | |
| H25 | 0.8087 | 0.3926 | 0.7501 | 0.067* | |
| C26 | 0.7395 (2) | 0.45523 (17) | 0.69203 (6) | 0.0523 (5) | |
| H26 | 0.8073 | 0.5157 | 0.6903 | 0.063* | |
| O24 | 0.65388 (16) | 0.22083 (12) | 0.76910 (4) | 0.0551 (4) | |
| C27 | 0.5378 (3) | 0.14351 (19) | 0.77845 (7) | 0.0632 (6) | |
| H27A | 0.5581 | 0.0999 | 0.8049 | 0.095* | |
| H27B | 0.4512 | 0.1888 | 0.7824 | 0.095* | |
| H27C | 0.5252 | 0.0893 | 0.7544 | 0.095* | |
| N31 | 0.6143 (2) | 1.14730 (14) | 0.39733 (6) | 0.0587 (5) | |
| C32 | 0.6210 (2) | 1.07950 (15) | 0.43309 (6) | 0.0439 (4) | |
| H32 | 0.6092 | 1.1158 | 0.4604 | 0.053* | |
| C33 | 0.64434 (17) | 0.95882 (13) | 0.43243 (5) | 0.0332 (4) | |
| C34 | 0.66384 (19) | 0.90618 (16) | 0.39174 (6) | 0.0419 (4) | |
| H34 | 0.6786 | 0.8249 | 0.3896 | 0.050* | |
| C35 | 0.6611 (2) | 0.97549 (19) | 0.35436 (6) | 0.0517 (5) | |
| H35 | 0.6767 | 0.9422 | 0.3267 | 0.062* | |
| C36 | 0.6353 (2) | 1.09376 (19) | 0.35863 (7) | 0.0588 (6) | |
| H36 | 0.6321 | 1.1397 | 0.3332 | 0.071* | |
| C37 | 0.64844 (18) | 0.88736 (15) | 0.47434 (6) | 0.0378 (4) | |
| O31 | 0.60454 (17) | 0.78319 (11) | 0.47188 (4) | 0.0589 (4) | |
| O32 | 0.69492 (15) | 0.93503 (12) | 0.50849 (4) | 0.0543 (4) |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0484 (9) | 0.0353 (7) | 0.0379 (8) | 0.0052 (7) | 0.0025 (7) | 0.0083 (6) |
| C2 | 0.0387 (9) | 0.0413 (9) | 0.0403 (9) | 0.0052 (7) | −0.0030 (7) | 0.0026 (8) |
| C3 | 0.0337 (9) | 0.0384 (9) | 0.0383 (9) | −0.0016 (7) | −0.0021 (7) | 0.0027 (7) |
| N4 | 0.0313 (7) | 0.0354 (7) | 0.0344 (7) | −0.0020 (6) | 0.0001 (6) | 0.0032 (6) |
| C5 | 0.0382 (9) | 0.0509 (10) | 0.0428 (9) | −0.0112 (8) | −0.0039 (8) | 0.0099 (8) |
| C6 | 0.0515 (11) | 0.0435 (10) | 0.0488 (11) | −0.0119 (8) | −0.0021 (9) | 0.0082 (8) |
| C21 | 0.0315 (8) | 0.0321 (8) | 0.0314 (8) | 0.0024 (6) | 0.0031 (6) | −0.0009 (6) |
| C22 | 0.0483 (11) | 0.0452 (10) | 0.0413 (10) | −0.0135 (8) | −0.0089 (8) | 0.0042 (8) |
| C23 | 0.0536 (11) | 0.0405 (9) | 0.0452 (10) | −0.0153 (8) | 0.0004 (8) | 0.0044 (8) |
| C24 | 0.0465 (10) | 0.0363 (8) | 0.0345 (8) | 0.0008 (8) | 0.0049 (7) | 0.0038 (7) |
| C25 | 0.0547 (12) | 0.0632 (12) | 0.0495 (11) | −0.0151 (10) | −0.0169 (9) | 0.0159 (10) |
| C26 | 0.0487 (11) | 0.0556 (11) | 0.0526 (11) | −0.0201 (9) | −0.0122 (9) | 0.0165 (9) |
| O24 | 0.0657 (9) | 0.0559 (8) | 0.0436 (7) | −0.0092 (7) | −0.0005 (6) | 0.0169 (6) |
| C27 | 0.0701 (14) | 0.0575 (12) | 0.0620 (13) | −0.0050 (11) | 0.0117 (11) | 0.0232 (10) |
| N31 | 0.0794 (13) | 0.0393 (8) | 0.0573 (10) | −0.0007 (9) | −0.0084 (9) | 0.0127 (8) |
| C32 | 0.0537 (11) | 0.0356 (9) | 0.0425 (10) | 0.0018 (8) | −0.0021 (8) | 0.0007 (8) |
| C33 | 0.0283 (8) | 0.0318 (8) | 0.0396 (9) | −0.0024 (6) | −0.0031 (7) | 0.0036 (6) |
| C34 | 0.0434 (10) | 0.0364 (9) | 0.0458 (10) | 0.0001 (8) | −0.0040 (8) | −0.0040 (8) |
| C35 | 0.0523 (12) | 0.0664 (13) | 0.0363 (10) | −0.0086 (10) | −0.0004 (8) | −0.0028 (9) |
| C36 | 0.0712 (14) | 0.0595 (13) | 0.0458 (11) | −0.0115 (11) | −0.0079 (10) | 0.0195 (10) |
| C37 | 0.0343 (9) | 0.0377 (9) | 0.0415 (9) | −0.0008 (7) | −0.0032 (7) | 0.0065 (7) |
| O31 | 0.0833 (11) | 0.0368 (7) | 0.0566 (8) | −0.0120 (7) | −0.0157 (7) | 0.0144 (6) |
| O32 | 0.0670 (9) | 0.0575 (8) | 0.0385 (7) | −0.0166 (7) | −0.0106 (6) | 0.0060 (6) |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Geometric parameters (Å, º)
| N1—C2 | 1.477 (2) | C24—C25 | 1.375 (3) |
| N1—C6 | 1.480 (2) | C24—O24 | 1.3781 (19) |
| N1—H12 | 0.94 (2) | C25—C26 | 1.375 (2) |
| N1—H11 | 0.97 (2) | C25—H25 | 0.9300 |
| C2—C3 | 1.506 (2) | C26—H26 | 0.9300 |
| C2—H2A | 0.9700 | O24—C27 | 1.416 (2) |
| C2—H2B | 0.9700 | C27—H27A | 0.9600 |
| C3—N4 | 1.461 (2) | C27—H27B | 0.9600 |
| C3—H3A | 0.9700 | C27—H27C | 0.9600 |
| C3—H3B | 0.9700 | N31—C32 | 1.328 (2) |
| N4—C21 | 1.4152 (19) | N31—C36 | 1.334 (3) |
| N4—C5 | 1.462 (2) | C32—C33 | 1.380 (2) |
| C5—C6 | 1.509 (2) | C32—H32 | 0.9300 |
| C5—H5A | 0.9700 | C33—C34 | 1.381 (2) |
| C5—H5B | 0.9700 | C33—C37 | 1.505 (2) |
| C6—H6A | 0.9700 | C34—C35 | 1.377 (3) |
| C6—H6B | 0.9700 | C34—H34 | 0.9300 |
| C21—C22 | 1.377 (2) | C35—C36 | 1.363 (3) |
| C21—C26 | 1.395 (2) | C35—H35 | 0.9300 |
| C22—C23 | 1.387 (2) | C36—H36 | 0.9300 |
| C22—H22 | 0.9300 | C37—O32 | 1.244 (2) |
| C23—C24 | 1.366 (3) | C37—O31 | 1.247 (2) |
| C23—H23 | 0.9300 | ||
| C2—N1—C6 | 109.37 (13) | C24—C23—C22 | 120.82 (16) |
| C2—N1—H12 | 108.5 (12) | C24—C23—H23 | 119.6 |
| C6—N1—H12 | 108.6 (12) | C22—C23—H23 | 119.6 |
| C2—N1—H11 | 109.1 (11) | C23—C24—C25 | 118.14 (16) |
| C6—N1—H11 | 113.2 (11) | C23—C24—O24 | 125.48 (16) |
| H12—N1—H11 | 107.9 (15) | C25—C24—O24 | 116.38 (16) |
| N1—C2—C3 | 110.55 (13) | C24—C25—C26 | 121.05 (17) |
| N1—C2—H2A | 109.5 | C24—C25—H25 | 119.5 |
| C3—C2—H2A | 109.5 | C26—C25—H25 | 119.5 |
| N1—C2—H2B | 109.5 | C25—C26—C21 | 121.82 (17) |
| C3—C2—H2B | 109.5 | C25—C26—H26 | 119.1 |
| H2A—C2—H2B | 108.1 | C21—C26—H26 | 119.1 |
| N4—C3—C2 | 110.87 (13) | C24—O24—C27 | 117.22 (15) |
| N4—C3—H3A | 109.5 | O24—C27—H27A | 109.5 |
| C2—C3—H3A | 109.5 | O24—C27—H27B | 109.5 |
| N4—C3—H3B | 109.5 | H27A—C27—H27B | 109.5 |
| C2—C3—H3B | 109.5 | O24—C27—H27C | 109.5 |
| H3A—C3—H3B | 108.1 | H27A—C27—H27C | 109.5 |
| C21—N4—C3 | 115.94 (13) | H27B—C27—H27C | 109.5 |
| C21—N4—C5 | 115.77 (13) | C32—N31—C36 | 116.71 (16) |
| C3—N4—C5 | 112.21 (13) | N31—C32—C33 | 124.38 (17) |
| N4—C5—C6 | 112.02 (15) | N31—C32—H32 | 117.8 |
| N4—C5—H5A | 109.2 | C33—C32—H32 | 117.8 |
| C6—C5—H5A | 109.2 | C32—C33—C34 | 117.29 (16) |
| N4—C5—H5B | 109.2 | C32—C33—C37 | 121.41 (15) |
| C6—C5—H5B | 109.2 | C34—C33—C37 | 121.31 (14) |
| H5A—C5—H5B | 107.9 | C35—C34—C33 | 119.20 (17) |
| N1—C6—C5 | 109.88 (14) | C35—C34—H34 | 120.4 |
| N1—C6—H6A | 109.7 | C33—C34—H34 | 120.4 |
| C5—C6—H6A | 109.7 | C36—C35—C34 | 118.80 (18) |
| N1—C6—H6B | 109.7 | C36—C35—H35 | 120.6 |
| C5—C6—H6B | 109.7 | C34—C35—H35 | 120.6 |
| H6A—C6—H6B | 108.2 | N31—C36—C35 | 123.59 (18) |
| C22—C21—C26 | 116.02 (15) | N31—C36—H36 | 118.2 |
| C22—C21—N4 | 122.74 (15) | C35—C36—H36 | 118.2 |
| C26—C21—N4 | 121.11 (14) | O32—C37—O31 | 124.84 (16) |
| C21—C22—C23 | 122.14 (16) | O32—C37—C33 | 118.62 (15) |
| C21—C22—H22 | 118.9 | O31—C37—C33 | 116.54 (15) |
| C23—C22—H22 | 118.9 | ||
| C6—N1—C2—C3 | −59.85 (18) | O24—C24—C25—C26 | 179.32 (18) |
| N1—C2—C3—N4 | 57.18 (18) | C24—C25—C26—C21 | 0.1 (3) |
| C2—C3—N4—C21 | 170.15 (13) | C22—C21—C26—C25 | −0.4 (3) |
| C2—C3—N4—C5 | −53.72 (18) | N4—C21—C26—C25 | 175.52 (18) |
| C21—N4—C5—C6 | −170.28 (14) | C23—C24—O24—C27 | −13.4 (3) |
| C3—N4—C5—C6 | 53.52 (19) | C25—C24—O24—C27 | 167.31 (18) |
| C2—N1—C6—C5 | 58.59 (19) | C36—N31—C32—C33 | −1.7 (3) |
| N4—C5—C6—N1 | −55.7 (2) | N31—C32—C33—C34 | 0.9 (3) |
| C3—N4—C21—C22 | −13.9 (2) | N31—C32—C33—C37 | −179.08 (17) |
| C5—N4—C21—C22 | −148.45 (17) | C32—C33—C34—C35 | 0.9 (3) |
| C3—N4—C21—C26 | 170.48 (16) | C37—C33—C34—C35 | −179.12 (16) |
| C5—N4—C21—C26 | 35.9 (2) | C33—C34—C35—C36 | −1.8 (3) |
| C26—C21—C22—C23 | 0.7 (3) | C32—N31—C36—C35 | 0.7 (3) |
| N4—C21—C22—C23 | −175.17 (16) | C34—C35—C36—N31 | 1.0 (3) |
| C21—C22—C23—C24 | −0.6 (3) | C32—C33—C37—O32 | −32.1 (2) |
| C22—C23—C24—C25 | 0.3 (3) | C34—C33—C37—O32 | 147.86 (17) |
| C22—C23—C24—O24 | −178.99 (17) | C32—C33—C37—O31 | 148.09 (18) |
| C23—C24—C25—C26 | 0.0 (3) | C34—C33—C37—O31 | −31.9 (2) |
4-(4-Methoxyphenyl)piperazin-1-ium pyridine-3-carboxylate (VI). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.976 (19) | 1.714 (19) | 2.677 (2) | 168.2 (18) |
| N1—H12···O32i | 0.94 (2) | 1.82 (2) | 2.749 (2) | 168.3 (17) |
| C2—H2B···N31ii | 0.97 | 2.56 | 3.518 (2) | 169 |
| C36—H36···O24iii | 0.93 | 2.51 | 3.432 (2) | 172 |
| C3—H3A···Cg1iv | 0.97 | 2.97 | 3.775 (2) | 156 |
Symmetry codes: (i) −x+3/2, y−1/2, z; (ii) −x+1, −y+2, −z+1; (iii) x, −y+3/2, z−1/2; (iv) x−1/2, −y+3/2, −z+1.
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Crystal data
| C7H3N2O7+·C11H17N2O− | F(000) = 880 |
| Mr = 420.38 | Dx = 1.478 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.5500 (9) Å | Cell parameters from 4078 reflections |
| b = 7.6489 (9) Å | θ = 2.7–28.0° |
| c = 32.719 (6) Å | µ = 0.12 mm−1 |
| β = 91.30 (1)° | T = 296 K |
| V = 1889.0 (5) Å3 | Block, colourless |
| Z = 4 | 0.18 × 0.12 × 0.06 mm |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 4074 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2003 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.038 |
| ω scans | θmax = 28.0°, θmin = 2.7° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −9→5 |
| Tmin = 0.916, Tmax = 0.993 | k = −9→9 |
| 8215 measured reflections | l = −42→41 |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.066 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.128 | w = 1/[σ2(Fo2) + (0.0413P)2 + 0.4471P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 4074 reflections | Δρmax = 0.22 e Å−3 |
| 281 parameters | Δρmin = −0.23 e Å−3 |
| 0 restraints |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Special details
| Experimental. Compound (VII). IR (KBr , cm-1) 3084 (NH2), 2834 (OCH3), 1568 (COO), 1499 (NO2). NMR (CDCl3) δ(1H) 3.05 (m, 4H, piperazine), 3.37 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.85 (m, 4H, methoxyphenyl), 7.52 (s, 1H, 3,5-dinitrosalicylate), 8.09 (s, 1H, 3,5-dinitrosalicylate), 8.99 (s, 1H, 3,5-dinitrosalicylate). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.6215 (3) | 0.5723 (4) | 0.61103 (8) | 0.0465 (7) | |
| H11 | 0.703 (4) | 0.587 (4) | 0.5906 (9) | 0.056* | |
| H12 | 0.533 (4) | 0.494 (4) | 0.6028 (9) | 0.056* | |
| C2 | 0.5349 (4) | 0.7433 (4) | 0.61776 (9) | 0.0531 (9) | |
| H2A | 0.4783 | 0.7826 | 0.5925 | 0.064* | |
| H2B | 0.6235 | 0.8292 | 0.6258 | 0.064* | |
| C3 | 0.3982 (4) | 0.7294 (4) | 0.65055 (9) | 0.0489 (8) | |
| H3A | 0.3504 | 0.8446 | 0.6559 | 0.059* | |
| H3B | 0.3016 | 0.6558 | 0.6407 | 0.059* | |
| N4 | 0.4712 (3) | 0.6567 (3) | 0.68854 (7) | 0.0399 (6) | |
| C5 | 0.5669 (4) | 0.4924 (4) | 0.68187 (9) | 0.0483 (8) | |
| H5A | 0.4832 | 0.4024 | 0.6734 | 0.058* | |
| H5B | 0.6233 | 0.4554 | 0.7074 | 0.058* | |
| C6 | 0.7057 (4) | 0.5118 (4) | 0.64972 (8) | 0.0505 (8) | |
| H6A | 0.7947 | 0.5955 | 0.6588 | 0.061* | |
| H6B | 0.7637 | 0.4003 | 0.6454 | 0.061* | |
| C21 | 0.3501 (4) | 0.6515 (4) | 0.72152 (9) | 0.0382 (7) | |
| C22 | 0.1878 (4) | 0.7370 (4) | 0.72002 (10) | 0.0510 (8) | |
| H22 | 0.1538 | 0.7966 | 0.6963 | 0.061* | |
| C23 | 0.0745 (4) | 0.7358 (4) | 0.75303 (10) | 0.0522 (8) | |
| H23 | −0.0327 | 0.7954 | 0.7512 | 0.063* | |
| C24 | 0.1201 (4) | 0.6473 (4) | 0.78814 (9) | 0.0455 (8) | |
| C25 | 0.2807 (4) | 0.5624 (4) | 0.79002 (9) | 0.0547 (9) | |
| H25 | 0.3138 | 0.5028 | 0.8138 | 0.066* | |
| C26 | 0.3934 (4) | 0.5636 (4) | 0.75758 (9) | 0.0499 (8) | |
| H26 | 0.5008 | 0.5044 | 0.7598 | 0.060* | |
| O24 | 0.0184 (3) | 0.6354 (3) | 0.82246 (6) | 0.0640 (7) | |
| C27 | −0.1485 (4) | 0.7203 (5) | 0.82152 (11) | 0.0745 (11) | |
| H27A | −0.2061 | 0.7017 | 0.8470 | 0.112* | |
| H27B | −0.2206 | 0.6733 | 0.7996 | 0.112* | |
| H27C | −0.1318 | 0.8433 | 0.8173 | 0.112* | |
| C31 | 0.8836 (3) | 0.6893 (4) | 0.46865 (8) | 0.0355 (7) | |
| C32 | 0.8864 (3) | 0.7026 (4) | 0.51247 (9) | 0.0374 (7) | |
| C33 | 1.0424 (3) | 0.7812 (4) | 0.52995 (8) | 0.0352 (7) | |
| C34 | 1.1812 (3) | 0.8371 (4) | 0.50706 (8) | 0.0372 (7) | |
| H34 | 1.2797 | 0.8892 | 0.5195 | 0.045* | |
| C35 | 1.1728 (3) | 0.8150 (4) | 0.46526 (9) | 0.0368 (7) | |
| C36 | 1.0252 (3) | 0.7425 (4) | 0.44590 (9) | 0.0383 (7) | |
| H36 | 1.0217 | 0.7298 | 0.4176 | 0.046* | |
| C37 | 0.7243 (4) | 0.6189 (4) | 0.44690 (10) | 0.0446 (8) | |
| O31 | 0.7120 (3) | 0.6063 (3) | 0.40978 (6) | 0.0609 (7) | |
| O32 | 0.5931 (2) | 0.5689 (3) | 0.46961 (7) | 0.0626 (7) | |
| H32 | 0.638 (4) | 0.590 (5) | 0.4996 (12) | 0.094* | |
| O33 | 0.7553 (2) | 0.6499 (3) | 0.53332 (6) | 0.0530 (6) | |
| N33 | 1.0576 (3) | 0.8102 (4) | 0.57403 (8) | 0.0481 (7) | |
| O34 | 0.9695 (3) | 0.7215 (4) | 0.59704 (7) | 0.0868 (9) | |
| O35 | 1.1628 (3) | 0.9203 (3) | 0.58660 (6) | 0.0627 (7) | |
| N35 | 1.3207 (3) | 0.8725 (3) | 0.44092 (9) | 0.0492 (7) | |
| O36 | 1.4399 (3) | 0.9552 (3) | 0.45754 (7) | 0.0715 (7) | |
| O37 | 1.3199 (3) | 0.8357 (3) | 0.40434 (7) | 0.0687 (7) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0429 (15) | 0.061 (2) | 0.0360 (15) | −0.0125 (14) | 0.0016 (11) | −0.0055 (14) |
| C2 | 0.064 (2) | 0.053 (2) | 0.0417 (19) | −0.0007 (17) | 0.0004 (16) | 0.0075 (17) |
| C3 | 0.0565 (19) | 0.053 (2) | 0.0371 (18) | 0.0086 (16) | 0.0021 (15) | 0.0053 (16) |
| N4 | 0.0438 (13) | 0.0416 (16) | 0.0342 (14) | 0.0040 (11) | −0.0003 (11) | 0.0033 (12) |
| C5 | 0.0505 (18) | 0.055 (2) | 0.0397 (19) | 0.0108 (16) | 0.0033 (14) | 0.0063 (16) |
| C6 | 0.0477 (18) | 0.066 (2) | 0.0374 (18) | 0.0033 (16) | −0.0019 (14) | −0.0007 (17) |
| C21 | 0.0430 (16) | 0.0340 (18) | 0.0375 (17) | −0.0001 (13) | −0.0035 (13) | −0.0012 (14) |
| C22 | 0.0511 (19) | 0.057 (2) | 0.045 (2) | 0.0118 (16) | 0.0001 (15) | 0.0135 (17) |
| C23 | 0.0480 (18) | 0.053 (2) | 0.056 (2) | 0.0132 (15) | 0.0034 (16) | 0.0039 (18) |
| C24 | 0.0482 (18) | 0.045 (2) | 0.044 (2) | 0.0021 (15) | 0.0057 (15) | −0.0026 (16) |
| C25 | 0.062 (2) | 0.064 (2) | 0.0382 (19) | 0.0154 (18) | −0.0005 (15) | 0.0126 (18) |
| C26 | 0.0490 (18) | 0.059 (2) | 0.0417 (19) | 0.0167 (16) | −0.0010 (15) | 0.0055 (17) |
| O24 | 0.0614 (15) | 0.0814 (18) | 0.0497 (14) | 0.0129 (12) | 0.0142 (11) | 0.0028 (13) |
| C27 | 0.057 (2) | 0.096 (3) | 0.071 (3) | 0.014 (2) | 0.0207 (19) | −0.007 (2) |
| C31 | 0.0344 (15) | 0.0352 (17) | 0.0366 (17) | −0.0028 (13) | −0.0008 (13) | −0.0002 (14) |
| C32 | 0.0318 (15) | 0.0367 (18) | 0.0437 (18) | 0.0004 (13) | 0.0011 (13) | 0.0046 (15) |
| C33 | 0.0367 (16) | 0.0371 (18) | 0.0315 (16) | 0.0003 (13) | −0.0022 (13) | 0.0003 (14) |
| C34 | 0.0352 (15) | 0.0340 (18) | 0.0422 (18) | −0.0016 (13) | −0.0053 (13) | −0.0020 (15) |
| C35 | 0.0316 (15) | 0.0345 (17) | 0.0443 (19) | −0.0064 (13) | 0.0036 (13) | 0.0017 (15) |
| C36 | 0.0405 (16) | 0.0355 (17) | 0.0388 (17) | −0.0019 (13) | 0.0018 (13) | −0.0015 (14) |
| C37 | 0.0397 (17) | 0.048 (2) | 0.046 (2) | −0.0086 (14) | −0.0020 (15) | 0.0005 (16) |
| O31 | 0.0570 (13) | 0.0868 (19) | 0.0389 (14) | −0.0254 (12) | −0.0012 (10) | −0.0093 (13) |
| O32 | 0.0429 (13) | 0.099 (2) | 0.0455 (13) | −0.0297 (12) | −0.0024 (10) | 0.0073 (14) |
| O33 | 0.0396 (12) | 0.0793 (17) | 0.0404 (13) | −0.0153 (11) | 0.0032 (10) | 0.0039 (12) |
| N33 | 0.0381 (14) | 0.063 (2) | 0.0424 (17) | 0.0001 (13) | −0.0041 (12) | 0.0013 (15) |
| O34 | 0.0712 (16) | 0.146 (3) | 0.0427 (15) | −0.0443 (16) | 0.0011 (12) | 0.0102 (16) |
| O35 | 0.0688 (15) | 0.0690 (17) | 0.0495 (14) | −0.0132 (13) | −0.0138 (11) | −0.0063 (13) |
| N35 | 0.0460 (16) | 0.0512 (18) | 0.0505 (18) | −0.0110 (13) | 0.0022 (13) | 0.0012 (15) |
| O36 | 0.0604 (14) | 0.092 (2) | 0.0616 (16) | −0.0425 (13) | 0.0006 (12) | −0.0031 (14) |
| O37 | 0.0643 (15) | 0.095 (2) | 0.0474 (15) | −0.0258 (13) | 0.0132 (11) | −0.0123 (15) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Geometric parameters (Å, º)
| N1—C6 | 1.478 (4) | C25—H25 | 0.9300 |
| N1—C2 | 1.481 (4) | C26—H26 | 0.9300 |
| N1—H11 | 0.93 (3) | O24—C27 | 1.417 (3) |
| N1—H12 | 0.93 (3) | C27—H27A | 0.9600 |
| C2—C3 | 1.509 (4) | C27—H27B | 0.9600 |
| C2—H2A | 0.9700 | C27—H27C | 0.9600 |
| C2—H2B | 0.9700 | C31—C36 | 1.378 (4) |
| C3—N4 | 1.458 (3) | C31—C32 | 1.437 (4) |
| C3—H3A | 0.9700 | C31—C37 | 1.485 (4) |
| C3—H3B | 0.9700 | C32—O33 | 1.280 (3) |
| N4—C21 | 1.431 (3) | C32—C33 | 1.430 (4) |
| N4—C5 | 1.469 (3) | C33—C34 | 1.370 (3) |
| C5—C6 | 1.509 (4) | C33—N33 | 1.461 (3) |
| C5—H5A | 0.9700 | C34—C35 | 1.378 (4) |
| C5—H5B | 0.9700 | C34—H34 | 0.9300 |
| C6—H6A | 0.9700 | C35—C36 | 1.385 (4) |
| C6—H6B | 0.9700 | C35—N35 | 1.455 (3) |
| C21—C22 | 1.389 (4) | C36—H36 | 0.9300 |
| C21—C26 | 1.391 (4) | C37—O31 | 1.220 (3) |
| C22—C23 | 1.393 (4) | C37—O32 | 1.309 (3) |
| C22—H22 | 0.9300 | O32—H32 | 1.04 (4) |
| C23—C24 | 1.371 (4) | N33—O34 | 1.222 (3) |
| C23—H23 | 0.9300 | N33—O35 | 1.223 (3) |
| C24—C25 | 1.376 (4) | N35—O36 | 1.218 (3) |
| C24—O24 | 1.378 (3) | N35—O37 | 1.229 (3) |
| C25—C26 | 1.375 (4) | ||
| C6—N1—C2 | 109.4 (2) | C25—C24—O24 | 116.1 (3) |
| C6—N1—H11 | 112.1 (18) | C26—C25—C24 | 121.5 (3) |
| C2—N1—H11 | 107.5 (18) | C26—C25—H25 | 119.2 |
| C6—N1—H12 | 109.6 (18) | C24—C25—H25 | 119.2 |
| C2—N1—H12 | 107.2 (18) | C25—C26—C21 | 121.3 (3) |
| H11—N1—H12 | 111 (3) | C25—C26—H26 | 119.3 |
| N1—C2—C3 | 110.8 (2) | C21—C26—H26 | 119.3 |
| N1—C2—H2A | 109.5 | C24—O24—C27 | 117.6 (3) |
| C3—C2—H2A | 109.5 | O24—C27—H27A | 109.5 |
| N1—C2—H2B | 109.5 | O24—C27—H27B | 109.5 |
| C3—C2—H2B | 109.5 | H27A—C27—H27B | 109.5 |
| H2A—C2—H2B | 108.1 | O24—C27—H27C | 109.5 |
| N4—C3—C2 | 112.4 (2) | H27A—C27—H27C | 109.5 |
| N4—C3—H3A | 109.1 | H27B—C27—H27C | 109.5 |
| C2—C3—H3A | 109.1 | C36—C31—C32 | 121.6 (2) |
| N4—C3—H3B | 109.1 | C36—C31—C37 | 118.6 (3) |
| C2—C3—H3B | 109.1 | C32—C31—C37 | 119.8 (2) |
| H3A—C3—H3B | 107.8 | O33—C32—C33 | 124.0 (3) |
| C21—N4—C3 | 114.7 (2) | O33—C32—C31 | 121.0 (2) |
| C21—N4—C5 | 114.4 (2) | C33—C32—C31 | 115.0 (2) |
| C3—N4—C5 | 112.2 (2) | C34—C33—C32 | 123.1 (3) |
| N4—C5—C6 | 111.8 (3) | C34—C33—N33 | 116.7 (2) |
| N4—C5—H5A | 109.3 | C32—C33—N33 | 120.3 (2) |
| C6—C5—H5A | 109.3 | C33—C34—C35 | 119.0 (2) |
| N4—C5—H5B | 109.3 | C33—C34—H34 | 120.5 |
| C6—C5—H5B | 109.3 | C35—C34—H34 | 120.5 |
| H5A—C5—H5B | 107.9 | C34—C35—C36 | 121.5 (2) |
| N1—C6—C5 | 109.6 (2) | C34—C35—N35 | 119.2 (2) |
| N1—C6—H6A | 109.7 | C36—C35—N35 | 119.4 (3) |
| C5—C6—H6A | 109.7 | C31—C36—C35 | 119.8 (3) |
| N1—C6—H6B | 109.7 | C31—C36—H36 | 120.1 |
| C5—C6—H6B | 109.7 | C35—C36—H36 | 120.1 |
| H6A—C6—H6B | 108.2 | O31—C37—O32 | 120.1 (3) |
| C22—C21—C26 | 116.6 (3) | O31—C37—C31 | 123.2 (3) |
| C22—C21—N4 | 122.6 (3) | O32—C37—C31 | 116.7 (3) |
| C26—C21—N4 | 120.8 (2) | C37—O32—H32 | 104.6 (18) |
| C21—C22—C23 | 121.8 (3) | C32—O33—H32 | 99.0 (13) |
| C21—C22—H22 | 119.1 | O34—N33—O35 | 122.3 (3) |
| C23—C22—H22 | 119.1 | O34—N33—C33 | 119.5 (3) |
| C24—C23—C22 | 120.4 (3) | O35—N33—C33 | 118.2 (3) |
| C24—C23—H23 | 119.8 | O36—N35—O37 | 122.8 (2) |
| C22—C23—H23 | 119.8 | O36—N35—C35 | 118.8 (3) |
| C23—C24—C25 | 118.4 (3) | O37—N35—C35 | 118.5 (2) |
| C23—C24—O24 | 125.6 (3) | ||
| C6—N1—C2—C3 | −58.5 (3) | C36—C31—C32—C33 | 2.6 (4) |
| N1—C2—C3—N4 | 54.3 (3) | C37—C31—C32—C33 | −176.6 (2) |
| C2—C3—N4—C21 | 176.0 (2) | O33—C32—C33—C34 | −179.8 (3) |
| C2—C3—N4—C5 | −51.2 (3) | C31—C32—C33—C34 | −1.3 (4) |
| C21—N4—C5—C6 | −174.0 (2) | O33—C32—C33—N33 | −1.6 (4) |
| C3—N4—C5—C6 | 53.0 (3) | C31—C32—C33—N33 | 176.9 (3) |
| C2—N1—C6—C5 | 60.0 (3) | C32—C33—C34—C35 | −0.9 (4) |
| N4—C5—C6—N1 | −57.5 (3) | N33—C33—C34—C35 | −179.2 (2) |
| C3—N4—C21—C22 | −11.2 (4) | C33—C34—C35—C36 | 2.0 (4) |
| C5—N4—C21—C22 | −142.9 (3) | C33—C34—C35—N35 | −179.4 (2) |
| C3—N4—C21—C26 | 170.9 (3) | C32—C31—C36—C35 | −1.7 (4) |
| C5—N4—C21—C26 | 39.2 (4) | C37—C31—C36—C35 | 177.5 (3) |
| C26—C21—C22—C23 | 0.4 (5) | C34—C35—C36—C31 | −0.6 (4) |
| N4—C21—C22—C23 | −177.6 (3) | N35—C35—C36—C31 | −179.3 (2) |
| C21—C22—C23—C24 | −0.8 (5) | C36—C31—C37—O31 | −0.3 (5) |
| C22—C23—C24—C25 | 0.9 (5) | C32—C31—C37—O31 | 179.0 (3) |
| C22—C23—C24—O24 | −179.3 (3) | C36—C31—C37—O32 | 179.6 (3) |
| C23—C24—C25—C26 | −0.6 (5) | C32—C31—C37—O32 | −1.1 (4) |
| O24—C24—C25—C26 | 179.5 (3) | C34—C33—N33—O34 | −158.4 (3) |
| C24—C25—C26—C21 | 0.3 (5) | C32—C33—N33—O34 | 23.3 (4) |
| C22—C21—C26—C25 | −0.2 (5) | C34—C33—N33—O35 | 19.6 (4) |
| N4—C21—C26—C25 | 177.9 (3) | C32—C33—N33—O35 | −158.7 (3) |
| C23—C24—O24—C27 | 0.6 (5) | C34—C35—N35—O36 | −7.6 (4) |
| C25—C24—O24—C27 | −179.6 (3) | C36—C35—N35—O36 | 171.1 (3) |
| C36—C31—C32—O33 | −178.8 (3) | C34—C35—N35—O37 | 172.5 (3) |
| C37—C31—C32—O33 | 2.0 (4) | C36—C35—N35—O37 | −8.8 (4) |
4-(4-Methoxyphenyl)piperazin-1-ium 2-hydroxy-3,5-dinitrobenzoate (VII). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O32—H32···O33 | 1.04 (4) | 1.47 (4) | 2.472 (3) | 158 (3) |
| N1—H11···O33 | 0.93 (3) | 1.98 (3) | 2.820 (3) | 150 (3) |
| N1—H11···O34 | 0.93 (3) | 2.27 (3) | 2.910 (3) | 126 (2) |
| N1—H12···O31i | 0.93 (3) | 2.04 (3) | 2.931 (3) | 160 (3) |
| N1—H12···O32i | 0.93 (3) | 2.58 (3) | 3.250 (3) | 129 (2) |
| C34—H34···O36ii | 0.93 | 2.53 | 3.449 (3) | 171 |
| C5—H5B···Cg2iii | 0.97 | 2.84 | 3.639 (3) | 140 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+3, −y+2, −z+1; (iii) −x+1, y−1/2, −z+3/2.
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Crystal data
| C11H17N2O+·C4H5O4− | Dx = 1.344 Mg m−3 |
| Mr = 310.35 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 2423 reflections |
| a = 9.3225 (9) Å | θ = 2.6–27.8° |
| b = 28.261 (3) Å | µ = 0.10 mm−1 |
| c = 5.8228 (8) Å | T = 296 K |
| V = 1534.1 (3) Å3 | Block, colourless |
| Z = 4 | 0.44 × 0.42 × 0.24 mm |
| F(000) = 664 |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 2419 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2053 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.018 |
| ω scans | θmax = 27.5°, θmin = 2.6° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −6→11 |
| Tmin = 0.816, Tmax = 0.976 | k = −36→27 |
| 5828 measured reflections | l = −7→5 |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.104 | w = 1/[σ2(Fo2) + (0.0459P)2 + 0.356P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.14 | (Δ/σ)max < 0.001 |
| 2419 reflections | Δρmax = 0.16 e Å−3 |
| 233 parameters | Δρmin = −0.24 e Å−3 |
| 16 restraints | Absolute structure: Flack x determined using 460 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Special details
| Experimental. Compound (VIII). IR (KBr , cm-1) 3135 (NH2), 2836 (OCH3), 1562 (COO). NMR (CDCl3) δ(1H) ) 2.66 (s, 4H, succinate), 3.32 (m, 4H, piperazine), 3.35 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.90 (m, 4H, methoxyphenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.5210 (3) | 0.66867 (8) | 0.5428 (5) | 0.0395 (6) | |
| H11 | 0.580 (3) | 0.6921 (11) | 0.556 (7) | 0.047* | |
| H12 | 0.435 (3) | 0.6801 (10) | 0.623 (7) | 0.047* | |
| C2 | 0.5853 (3) | 0.62541 (9) | 0.6443 (7) | 0.0435 (7) | |
| H2A | 0.6135 | 0.6317 | 0.8018 | 0.052* | |
| H2B | 0.6704 | 0.6167 | 0.5586 | 0.052* | |
| C3 | 0.4786 (3) | 0.58515 (9) | 0.6388 (6) | 0.0414 (7) | |
| H3A | 0.5238 | 0.5568 | 0.6992 | 0.050* | |
| H3B | 0.3981 | 0.5929 | 0.7374 | 0.050* | |
| N4 | 0.4259 (2) | 0.57556 (7) | 0.4075 (5) | 0.0347 (5) | |
| C5 | 0.3676 (3) | 0.61875 (9) | 0.3026 (6) | 0.0439 (7) | |
| H5A | 0.2824 | 0.6284 | 0.3859 | 0.053* | |
| H5B | 0.3397 | 0.6120 | 0.1454 | 0.053* | |
| C6 | 0.4745 (3) | 0.65878 (10) | 0.3038 (6) | 0.0479 (8) | |
| H6A | 0.5570 | 0.6503 | 0.2110 | 0.058* | |
| H6B | 0.4312 | 0.6869 | 0.2380 | 0.058* | |
| C21 | 0.3353 (3) | 0.53502 (9) | 0.3862 (5) | 0.0331 (6) | |
| C22 | 0.3336 (3) | 0.49886 (9) | 0.5496 (6) | 0.0394 (7) | |
| H22 | 0.3926 | 0.5011 | 0.6779 | 0.047* | |
| C23 | 0.2451 (3) | 0.45967 (10) | 0.5241 (6) | 0.0440 (7) | |
| H23 | 0.2456 | 0.4361 | 0.6354 | 0.053* | |
| C24 | 0.1567 (3) | 0.45534 (9) | 0.3356 (6) | 0.0407 (7) | |
| C25 | 0.1589 (3) | 0.48988 (10) | 0.1703 (7) | 0.0484 (8) | |
| H25 | 0.1013 | 0.4870 | 0.0406 | 0.058* | |
| C26 | 0.2469 (3) | 0.52924 (10) | 0.1961 (6) | 0.0464 (8) | |
| H26 | 0.2466 | 0.5524 | 0.0826 | 0.056* | |
| O24 | 0.0728 (2) | 0.41532 (8) | 0.3280 (5) | 0.0620 (8) | |
| C27 | −0.0230 (4) | 0.41098 (13) | 0.1399 (9) | 0.0729 (12) | |
| H27A | −0.0790 | 0.3827 | 0.1572 | 0.109* | |
| H27B | 0.0307 | 0.4094 | −0.0005 | 0.109* | |
| H27C | −0.0856 | 0.4379 | 0.1358 | 0.109* | |
| C31 | 0.7061 (15) | 0.7714 (3) | 0.797 (3) | 0.0296 (18) | 0.660 (15) |
| O31 | 0.7173 (15) | 0.7375 (5) | 0.655 (3) | 0.0463 (18) | 0.660 (15) |
| O32 | 0.7811 (18) | 0.8079 (5) | 0.790 (4) | 0.0443 (15) | 0.660 (15) |
| C32 | 0.5909 (9) | 0.7663 (3) | 0.9790 (16) | 0.0375 (16) | 0.660 (15) |
| H32A | 0.6331 | 0.7517 | 1.1138 | 0.045* | 0.660 (15) |
| H32B | 0.5175 | 0.7450 | 0.9216 | 0.045* | 0.660 (15) |
| C33 | 0.5200 (11) | 0.8123 (3) | 1.0509 (19) | 0.056 (3) | 0.660 (15) |
| H33A | 0.5933 | 0.8339 | 1.1050 | 0.067* | 0.660 (15) |
| H33B | 0.4747 | 0.8265 | 0.9178 | 0.067* | 0.660 (15) |
| C34 | 0.4093 (16) | 0.8060 (6) | 1.237 (3) | 0.0468 (19) | 0.660 (15) |
| O33 | 0.3172 (11) | 0.8361 (3) | 1.279 (2) | 0.086 (3) | 0.660 (15) |
| O34 | 0.4253 (19) | 0.7688 (5) | 1.359 (2) | 0.065 (3) | 0.660 (15) |
| H34 | 0.3533 | 0.7647 | 1.4378 | 0.098* | 0.660 (15) |
| C41 | 0.702 (3) | 0.7808 (8) | 0.792 (6) | 0.0296 (18) | 0.340 (15) |
| O41 | 0.736 (3) | 0.7440 (11) | 0.678 (7) | 0.0463 (18) | 0.340 (15) |
| O42 | 0.782 (4) | 0.8158 (10) | 0.816 (8) | 0.0443 (15) | 0.340 (15) |
| C42 | 0.567 (2) | 0.7768 (6) | 0.934 (3) | 0.0375 (16) | 0.340 (15) |
| H42A | 0.4853 | 0.7819 | 0.8332 | 0.045* | 0.340 (15) |
| H42B | 0.5601 | 0.7447 | 0.9916 | 0.045* | 0.340 (15) |
| C43 | 0.555 (2) | 0.8103 (7) | 1.134 (3) | 0.056 (3) | 0.340 (15) |
| H43A | 0.5570 | 0.8425 | 1.0770 | 0.067* | 0.340 (15) |
| H43B | 0.6369 | 0.8060 | 1.2337 | 0.067* | 0.340 (15) |
| C44 | 0.419 (3) | 0.8032 (11) | 1.272 (5) | 0.0468 (19) | 0.340 (15) |
| O43 | 0.304 (2) | 0.8221 (6) | 1.225 (5) | 0.086 (3) | 0.340 (15) |
| O44 | 0.425 (4) | 0.7694 (12) | 1.418 (5) | 0.065 (3) | 0.340 (15) |
| H44 | 0.3490 | 0.7681 | 1.4892 | 0.098* | 0.340 (15) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0367 (12) | 0.0305 (11) | 0.0512 (17) | −0.0052 (10) | 0.0144 (13) | −0.0049 (11) |
| C2 | 0.0389 (14) | 0.0366 (13) | 0.055 (2) | −0.0012 (11) | −0.0002 (16) | −0.0023 (14) |
| C3 | 0.0440 (15) | 0.0361 (13) | 0.0440 (18) | −0.0024 (11) | −0.0032 (15) | 0.0033 (13) |
| N4 | 0.0388 (12) | 0.0296 (10) | 0.0357 (13) | −0.0015 (9) | 0.0050 (11) | −0.0003 (10) |
| C5 | 0.0592 (17) | 0.0331 (12) | 0.0395 (17) | −0.0042 (12) | −0.0014 (16) | 0.0044 (13) |
| C6 | 0.0606 (18) | 0.0386 (14) | 0.0446 (19) | −0.0070 (13) | 0.0126 (18) | 0.0056 (14) |
| C21 | 0.0345 (13) | 0.0314 (12) | 0.0333 (16) | 0.0023 (10) | 0.0055 (12) | −0.0008 (11) |
| C22 | 0.0421 (15) | 0.0354 (13) | 0.0406 (17) | 0.0004 (11) | −0.0064 (14) | 0.0037 (13) |
| C23 | 0.0483 (16) | 0.0366 (14) | 0.0472 (19) | −0.0025 (12) | −0.0064 (16) | 0.0123 (13) |
| C24 | 0.0370 (13) | 0.0332 (12) | 0.052 (2) | −0.0019 (11) | −0.0022 (15) | 0.0026 (13) |
| C25 | 0.0533 (17) | 0.0456 (15) | 0.046 (2) | −0.0059 (13) | −0.0156 (17) | 0.0056 (15) |
| C26 | 0.0587 (18) | 0.0414 (14) | 0.0391 (19) | −0.0073 (13) | −0.0055 (16) | 0.0104 (14) |
| O24 | 0.0604 (13) | 0.0480 (11) | 0.078 (2) | −0.0194 (10) | −0.0266 (15) | 0.0143 (12) |
| C27 | 0.067 (2) | 0.061 (2) | 0.090 (3) | −0.0233 (17) | −0.036 (2) | 0.011 (2) |
| C31 | 0.0307 (14) | 0.020 (4) | 0.0377 (18) | 0.002 (3) | 0.0121 (15) | 0.002 (4) |
| O31 | 0.039 (4) | 0.043 (4) | 0.056 (4) | −0.018 (2) | 0.025 (4) | −0.025 (2) |
| O32 | 0.0467 (11) | 0.029 (4) | 0.057 (5) | −0.011 (3) | 0.027 (2) | −0.014 (3) |
| C32 | 0.043 (3) | 0.025 (3) | 0.045 (4) | 0.002 (2) | 0.019 (3) | 0.004 (2) |
| C33 | 0.069 (5) | 0.0348 (18) | 0.064 (7) | 0.008 (3) | 0.043 (5) | 0.009 (4) |
| C34 | 0.053 (3) | 0.032 (2) | 0.055 (5) | 0.0033 (16) | 0.031 (3) | −0.001 (2) |
| O33 | 0.097 (3) | 0.054 (5) | 0.107 (7) | 0.039 (4) | 0.067 (4) | 0.016 (4) |
| O34 | 0.0654 (15) | 0.0655 (15) | 0.065 (8) | 0.0228 (12) | 0.054 (5) | 0.026 (4) |
| C41 | 0.0307 (14) | 0.020 (4) | 0.0377 (18) | 0.002 (3) | 0.0121 (15) | 0.002 (4) |
| O41 | 0.039 (4) | 0.043 (4) | 0.056 (4) | −0.018 (2) | 0.025 (4) | −0.025 (2) |
| O42 | 0.0467 (11) | 0.029 (4) | 0.057 (5) | −0.011 (3) | 0.027 (2) | −0.014 (3) |
| C42 | 0.043 (3) | 0.025 (3) | 0.045 (4) | 0.002 (2) | 0.019 (3) | 0.004 (2) |
| C43 | 0.069 (5) | 0.0348 (18) | 0.064 (7) | 0.008 (3) | 0.043 (5) | 0.009 (4) |
| C44 | 0.053 (3) | 0.032 (2) | 0.055 (5) | 0.0033 (16) | 0.031 (3) | −0.001 (2) |
| O43 | 0.097 (3) | 0.054 (5) | 0.107 (7) | 0.039 (4) | 0.067 (4) | 0.016 (4) |
| O44 | 0.0654 (15) | 0.0655 (15) | 0.065 (8) | 0.0228 (12) | 0.054 (5) | 0.026 (4) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Geometric parameters (Å, º)
| N1—C6 | 1.484 (5) | O24—C27 | 1.419 (5) |
| N1—C2 | 1.484 (4) | C27—H27A | 0.9600 |
| N1—H11 | 0.86 (3) | C27—H27B | 0.9600 |
| N1—H12 | 0.98 (3) | C27—H27C | 0.9600 |
| C2—C3 | 1.512 (4) | C31—O32 | 1.247 (5) |
| C2—H2A | 0.9700 | C31—O31 | 1.270 (7) |
| C2—H2B | 0.9700 | C31—C32 | 1.513 (6) |
| C3—N4 | 1.459 (4) | C32—C33 | 1.517 (7) |
| C3—H3A | 0.9700 | C32—H32A | 0.9700 |
| C3—H3B | 0.9700 | C32—H32B | 0.9700 |
| N4—C21 | 1.429 (3) | C33—C34 | 1.505 (6) |
| N4—C5 | 1.469 (3) | C33—H33A | 0.9700 |
| C5—C6 | 1.508 (4) | C33—H33B | 0.9700 |
| C5—H5A | 0.9700 | C34—O33 | 1.233 (9) |
| C5—H5B | 0.9700 | C34—O34 | 1.280 (6) |
| C6—H6A | 0.9700 | O34—H34 | 0.8200 |
| C6—H6B | 0.9700 | C41—O42 | 1.247 (9) |
| C21—C26 | 1.390 (4) | C41—O41 | 1.271 (10) |
| C21—C22 | 1.396 (4) | C41—C42 | 1.514 (9) |
| C22—C23 | 1.388 (4) | C42—C43 | 1.507 (11) |
| C22—H22 | 0.9300 | C42—H42A | 0.9700 |
| C23—C24 | 1.378 (5) | C42—H42B | 0.9700 |
| C23—H23 | 0.9300 | C43—C44 | 1.508 (10) |
| C24—C25 | 1.371 (4) | C43—H43A | 0.9700 |
| C24—O24 | 1.376 (3) | C43—H43B | 0.9700 |
| C25—C26 | 1.390 (4) | C44—O43 | 1.235 (13) |
| C25—H25 | 0.9300 | C44—O44 | 1.282 (9) |
| C26—H26 | 0.9300 | O44—H44 | 0.8200 |
| C6—N1—C2 | 109.6 (2) | C21—C26—C25 | 122.0 (3) |
| C6—N1—H11 | 114 (3) | C21—C26—H26 | 119.0 |
| C2—N1—H11 | 110 (2) | C25—C26—H26 | 119.0 |
| C6—N1—H12 | 106 (2) | C24—O24—C27 | 117.0 (3) |
| C2—N1—H12 | 114 (2) | O24—C27—H27A | 109.5 |
| H11—N1—H12 | 103 (3) | O24—C27—H27B | 109.5 |
| N1—C2—C3 | 110.2 (2) | H27A—C27—H27B | 109.5 |
| N1—C2—H2A | 109.6 | O24—C27—H27C | 109.5 |
| C3—C2—H2A | 109.6 | H27A—C27—H27C | 109.5 |
| N1—C2—H2B | 109.6 | H27B—C27—H27C | 109.5 |
| C3—C2—H2B | 109.6 | O32—C31—O31 | 123.7 (8) |
| H2A—C2—H2B | 108.1 | O32—C31—C32 | 120.0 (6) |
| N4—C3—C2 | 112.4 (3) | O31—C31—C32 | 116.3 (6) |
| N4—C3—H3A | 109.1 | C31—C32—C33 | 114.8 (6) |
| C2—C3—H3A | 109.1 | C31—C32—H32A | 108.6 |
| N4—C3—H3B | 109.1 | C33—C32—H32A | 108.6 |
| C2—C3—H3B | 109.1 | C31—C32—H32B | 108.6 |
| H3A—C3—H3B | 107.9 | C33—C32—H32B | 108.6 |
| C21—N4—C3 | 115.3 (2) | H32A—C32—H32B | 107.5 |
| C21—N4—C5 | 114.3 (2) | C34—C33—C32 | 113.3 (6) |
| C3—N4—C5 | 110.7 (2) | C34—C33—H33A | 108.9 |
| N4—C5—C6 | 112.1 (2) | C32—C33—H33A | 108.9 |
| N4—C5—H5A | 109.2 | C34—C33—H33B | 108.9 |
| C6—C5—H5A | 109.2 | C32—C33—H33B | 108.9 |
| N4—C5—H5B | 109.2 | H33A—C33—H33B | 107.7 |
| C6—C5—H5B | 109.2 | O33—C34—O34 | 122.3 (7) |
| H5A—C5—H5B | 107.9 | O33—C34—C33 | 122.8 (7) |
| N1—C6—C5 | 109.8 (3) | O34—C34—C33 | 114.7 (6) |
| N1—C6—H6A | 109.7 | C34—O34—H34 | 109.5 |
| C5—C6—H6A | 109.7 | O42—C41—O41 | 124.3 (15) |
| N1—C6—H6B | 109.7 | O42—C41—C42 | 119.7 (12) |
| C5—C6—H6B | 109.7 | O41—C41—C42 | 115.3 (12) |
| H6A—C6—H6B | 108.2 | C43—C42—C41 | 116.0 (12) |
| C26—C21—C22 | 116.7 (2) | C43—C42—H42A | 108.3 |
| C26—C21—N4 | 120.9 (3) | C41—C42—H42A | 108.3 |
| C22—C21—N4 | 122.3 (3) | C43—C42—H42B | 108.3 |
| C23—C22—C21 | 121.2 (3) | C41—C42—H42B | 108.3 |
| C23—C22—H22 | 119.4 | H42A—C42—H42B | 107.4 |
| C21—C22—H22 | 119.4 | C42—C43—C44 | 113.1 (15) |
| C24—C23—C22 | 120.8 (3) | C42—C43—H43A | 109.0 |
| C24—C23—H23 | 119.6 | C44—C43—H43A | 109.0 |
| C22—C23—H23 | 119.6 | C42—C43—H43B | 109.0 |
| C25—C24—O24 | 124.8 (3) | C44—C43—H43B | 109.0 |
| C25—C24—C23 | 119.1 (3) | H43A—C43—H43B | 107.8 |
| O24—C24—C23 | 116.0 (3) | O43—C44—O44 | 120.3 (16) |
| C24—C25—C26 | 120.2 (3) | O43—C44—C43 | 123.7 (14) |
| C24—C25—H25 | 119.9 | O44—C44—C43 | 114.8 (12) |
| C26—C25—H25 | 119.9 | C44—O44—H44 | 109.5 |
| C6—N1—C2—C3 | −57.9 (3) | O24—C24—C25—C26 | 179.3 (3) |
| N1—C2—C3—N4 | 56.2 (3) | C23—C24—C25—C26 | −1.5 (5) |
| C2—C3—N4—C21 | 174.2 (2) | C22—C21—C26—C25 | 1.0 (4) |
| C2—C3—N4—C5 | −54.0 (3) | N4—C21—C26—C25 | 179.3 (3) |
| C21—N4—C5—C6 | −172.9 (3) | C24—C25—C26—C21 | 0.4 (5) |
| C3—N4—C5—C6 | 54.8 (3) | C25—C24—O24—C27 | −3.1 (5) |
| C2—N1—C6—C5 | 58.5 (3) | C23—C24—O24—C27 | 177.7 (3) |
| N4—C5—C6—N1 | −57.4 (3) | O32—C31—C32—C33 | 32 (2) |
| C3—N4—C21—C26 | 161.9 (3) | O31—C31—C32—C33 | −146 (2) |
| C5—N4—C21—C26 | 31.8 (4) | C31—C32—C33—C34 | −178.3 (12) |
| C3—N4—C21—C22 | −19.9 (4) | C32—C33—C34—O33 | −162 (2) |
| C5—N4—C21—C22 | −150.0 (3) | C32—C33—C34—O34 | 23 (2) |
| C26—C21—C22—C23 | −1.3 (4) | O42—C41—C42—C43 | −13 (5) |
| N4—C21—C22—C23 | −179.6 (3) | O41—C41—C42—C43 | 157 (4) |
| C21—C22—C23—C24 | 0.2 (5) | C41—C42—C43—C44 | −178 (2) |
| C22—C23—C24—C25 | 1.2 (5) | C42—C43—C44—O43 | −86 (4) |
| C22—C23—C24—O24 | −179.5 (3) | C42—C43—C44—O44 | 82 (4) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogensuccinate (VIII). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.86 (3) | 1.90 (3) | 2.750 (15) | 167 (4) |
| N1—H12···O32i | 0.98 (3) | 1.77 (4) | 2.741 (19) | 171 (3) |
| O34—H34···O31ii | 0.82 | 1.79 | 2.60 (2) | 168 |
| N1—H11···O41 | 0.86 (3) | 2.18 (4) | 3.03 (3) | 165 (4) |
| N1—H12···O42i | 0.98 (3) | 1.82 (5) | 2.77 (4) | 163 (3) |
| O44—H44···O41ii | 0.82 | 1.56 | 2.35 (2) | 161 |
| C3—H3A···Cg2iii | 0.97 | 2.76 | 3.652 (3) | 154 |
Symmetry codes: (i) x−1/2, −y+3/2, z; (ii) x−1/2, −y+3/2, z+1; (iii) −x+1, −y+1, z+1/2.
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Crystal data
| C11H17N2O+·C4H3O4− | Dx = 1.356 Mg m−3 |
| Mr = 308.33 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 2829 reflections |
| a = 9.069 (1) Å | θ = 2.7–27.7° |
| b = 28.528 (3) Å | µ = 0.10 mm−1 |
| c = 5.8375 (9) Å | T = 296 K |
| V = 1510.3 (3) Å3 | Plate, colourless |
| Z = 4 | 0.48 × 0.48 × 0.08 mm |
| F(000) = 656 |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 2827 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2316 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.015 |
| ω scans | θmax = 27.5°, θmin = 2.7° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −8→11 |
| Tmin = 0.888, Tmax = 0.992 | k = −32→35 |
| 5834 measured reflections | l = −7→6 |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.041 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.101 | w = 1/[σ2(Fo2) + (0.0429P)2 + 0.4265P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 2827 reflections | Δρmax = 0.15 e Å−3 |
| 221 parameters | Δρmin = −0.14 e Å−3 |
| 11 restraints | Absolute structure: Flack x determined using 769 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Special details
| Experimental. Compound (IX). IR (KBr , cm-1) 3001 (NH2), 2839 (OCH3), 1562 (COO). NMR (CDCl3) δ(1H) ) 3.09 (m, 4H, piperazine), 3.35 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.26 (s, 2H, fumarate), 6.90 (m, 4H, methoxyphenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.5253 (3) | 0.66688 (8) | 0.5154 (5) | 0.0448 (6) | |
| H11 | 0.588 (4) | 0.6872 (12) | 0.518 (6) | 0.054* | |
| H12 | 0.440 (4) | 0.6776 (11) | 0.596 (6) | 0.054* | |
| C2 | 0.5885 (3) | 0.62425 (10) | 0.6244 (6) | 0.0490 (8) | |
| H2A | 0.6182 | 0.6315 | 0.7801 | 0.059* | |
| H2B | 0.6753 | 0.6144 | 0.5403 | 0.059* | |
| C3 | 0.4771 (3) | 0.58500 (9) | 0.6274 (6) | 0.0449 (7) | |
| H3A | 0.5225 | 0.5572 | 0.6916 | 0.054* | |
| H3B | 0.3952 | 0.5937 | 0.7254 | 0.054* | |
| N4 | 0.4215 (3) | 0.57432 (7) | 0.3988 (4) | 0.0395 (6) | |
| C5 | 0.3637 (4) | 0.61653 (10) | 0.2869 (6) | 0.0505 (8) | |
| H5A | 0.2767 | 0.6272 | 0.3685 | 0.061* | |
| H5B | 0.3344 | 0.6089 | 0.1315 | 0.061* | |
| C6 | 0.4751 (4) | 0.65530 (10) | 0.2808 (6) | 0.0540 (9) | |
| H6A | 0.5589 | 0.6458 | 0.1886 | 0.065* | |
| H6B | 0.4317 | 0.6829 | 0.2106 | 0.065* | |
| C21 | 0.3300 (3) | 0.53374 (9) | 0.3824 (5) | 0.0372 (6) | |
| C22 | 0.3313 (3) | 0.49854 (9) | 0.5482 (6) | 0.0441 (7) | |
| H22 | 0.3919 | 0.5017 | 0.6758 | 0.053* | |
| C23 | 0.2439 (3) | 0.45907 (10) | 0.5261 (7) | 0.0485 (8) | |
| H23 | 0.2468 | 0.4361 | 0.6393 | 0.058* | |
| C24 | 0.1529 (3) | 0.45301 (10) | 0.3401 (6) | 0.0456 (8) | |
| C25 | 0.1521 (4) | 0.48719 (10) | 0.1732 (6) | 0.0518 (8) | |
| H25 | 0.0924 | 0.4836 | 0.0448 | 0.062* | |
| C26 | 0.2391 (4) | 0.52673 (11) | 0.1945 (6) | 0.0509 (8) | |
| H26 | 0.2366 | 0.5493 | 0.0796 | 0.061* | |
| O24 | 0.0698 (3) | 0.41291 (8) | 0.3349 (5) | 0.0668 (8) | |
| C27 | −0.0259 (4) | 0.40672 (14) | 0.1473 (9) | 0.0804 (13) | |
| H27A | −0.0819 | 0.3785 | 0.1680 | 0.121* | |
| H27B | 0.0307 | 0.4045 | 0.0088 | 0.121* | |
| H27C | −0.0918 | 0.4330 | 0.1373 | 0.121* | |
| C31 | 0.7171 (3) | 0.77494 (10) | 0.8023 (6) | 0.0452 (7) | |
| O31 | 0.7323 (3) | 0.73835 (7) | 0.6832 (5) | 0.0673 (8) | |
| O32 | 0.7875 (3) | 0.81180 (7) | 0.7776 (5) | 0.0595 (7) | |
| C32 | 0.6095 (4) | 0.77182 (10) | 0.9941 (6) | 0.0526 (9) | 0.906 (9) |
| H32 | 0.5993 | 0.7427 | 1.0643 | 0.063* | 0.906 (9) |
| C33 | 0.5305 (7) | 0.80489 (15) | 1.0714 (12) | 0.0607 (11) | 0.906 (9) |
| H33 | 0.5456 | 0.8345 | 1.0089 | 0.073* | 0.906 (9) |
| C34 | 0.4163 (5) | 0.80084 (15) | 1.2518 (8) | 0.0518 (9) | 0.906 (9) |
| O33 | 0.3241 (5) | 0.83097 (16) | 1.2845 (8) | 0.0992 (18) | 0.906 (9) |
| O34 | 0.4218 (4) | 0.76290 (13) | 1.3671 (8) | 0.0734 (13) | 0.906 (9) |
| H34 | 0.3526 | 0.7621 | 1.4574 | 0.110* | 0.906 (9) |
| C42 | 0.6095 (4) | 0.77182 (10) | 0.9941 (6) | 0.0526 (9) | 0.094 (9) |
| H42 | 0.5982 | 0.7421 | 1.0566 | 0.063* | 0.094 (9) |
| C43 | 0.531 (6) | 0.8035 (9) | 1.086 (10) | 0.0607 (11) | 0.094 (9) |
| H43 | 0.5533 | 0.8343 | 1.0477 | 0.073* | 0.094 (9) |
| C44 | 0.407 (4) | 0.7961 (12) | 1.247 (6) | 0.0518 (9) | 0.094 (9) |
| O43 | 0.280 (4) | 0.8058 (15) | 1.203 (7) | 0.0992 (18) | 0.094 (9) |
| O44 | 0.444 (4) | 0.7801 (13) | 1.442 (6) | 0.0734 (13) | 0.094 (9) |
| H44 | 0.3697 | 0.7743 | 1.5169 | 0.110* | 0.094 (9) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0436 (14) | 0.0378 (12) | 0.0531 (18) | −0.0049 (10) | 0.0167 (13) | −0.0020 (12) |
| C2 | 0.0444 (16) | 0.0431 (15) | 0.060 (2) | 0.0018 (12) | 0.0054 (16) | −0.0042 (15) |
| C3 | 0.0482 (17) | 0.0398 (14) | 0.047 (2) | −0.0019 (13) | −0.0013 (15) | 0.0052 (13) |
| N4 | 0.0466 (13) | 0.0348 (11) | 0.0371 (14) | 0.0016 (10) | 0.0048 (12) | −0.0005 (10) |
| C5 | 0.072 (2) | 0.0403 (14) | 0.0396 (19) | −0.0043 (14) | −0.0015 (17) | 0.0079 (14) |
| C6 | 0.077 (2) | 0.0423 (14) | 0.043 (2) | −0.0044 (15) | 0.0172 (18) | 0.0048 (15) |
| C21 | 0.0398 (15) | 0.0355 (13) | 0.0363 (17) | 0.0060 (11) | 0.0060 (14) | 0.0009 (12) |
| C22 | 0.0484 (17) | 0.0402 (14) | 0.0437 (19) | 0.0029 (12) | −0.0071 (15) | 0.0040 (14) |
| C23 | 0.0532 (18) | 0.0413 (15) | 0.0510 (19) | 0.0005 (13) | −0.0054 (17) | 0.0123 (15) |
| C24 | 0.0391 (15) | 0.0409 (14) | 0.057 (2) | 0.0015 (12) | −0.0036 (15) | 0.0009 (14) |
| C25 | 0.0560 (19) | 0.0519 (17) | 0.048 (2) | −0.0013 (15) | −0.0160 (17) | 0.0035 (16) |
| C26 | 0.065 (2) | 0.0455 (15) | 0.042 (2) | −0.0041 (14) | −0.0061 (16) | 0.0115 (14) |
| O24 | 0.0633 (15) | 0.0532 (12) | 0.084 (2) | −0.0163 (11) | −0.0224 (15) | 0.0125 (13) |
| C27 | 0.064 (2) | 0.070 (2) | 0.107 (4) | −0.0205 (19) | −0.034 (3) | 0.012 (2) |
| C31 | 0.0431 (16) | 0.0435 (15) | 0.049 (2) | 0.0003 (12) | 0.0227 (15) | −0.0016 (14) |
| O31 | 0.0655 (15) | 0.0577 (13) | 0.079 (2) | −0.0153 (11) | 0.0428 (14) | −0.0234 (13) |
| O32 | 0.0632 (14) | 0.0469 (11) | 0.0684 (17) | −0.0123 (10) | 0.0369 (13) | −0.0050 (11) |
| C32 | 0.0564 (19) | 0.0380 (14) | 0.063 (2) | −0.0018 (14) | 0.0321 (18) | −0.0001 (15) |
| C33 | 0.067 (2) | 0.0474 (16) | 0.067 (3) | 0.0048 (16) | 0.034 (2) | 0.0065 (17) |
| C34 | 0.0558 (19) | 0.0416 (18) | 0.058 (2) | 0.0052 (15) | 0.0280 (18) | −0.0037 (15) |
| O33 | 0.115 (3) | 0.064 (2) | 0.119 (3) | 0.045 (2) | 0.081 (3) | 0.028 (2) |
| O34 | 0.081 (2) | 0.0502 (19) | 0.089 (3) | 0.0215 (17) | 0.061 (2) | 0.0185 (17) |
| C42 | 0.0564 (19) | 0.0380 (14) | 0.063 (2) | −0.0018 (14) | 0.0321 (18) | −0.0001 (15) |
| C43 | 0.067 (2) | 0.0474 (16) | 0.067 (3) | 0.0048 (16) | 0.034 (2) | 0.0065 (17) |
| C44 | 0.0558 (19) | 0.0416 (18) | 0.058 (2) | 0.0052 (15) | 0.0280 (18) | −0.0037 (15) |
| O43 | 0.115 (3) | 0.064 (2) | 0.119 (3) | 0.045 (2) | 0.081 (3) | 0.028 (2) |
| O44 | 0.081 (2) | 0.0502 (19) | 0.089 (3) | 0.0215 (17) | 0.061 (2) | 0.0185 (17) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Geometric parameters (Å, º)
| N1—C6 | 1.481 (4) | C24—C25 | 1.378 (4) |
| N1—C2 | 1.487 (4) | C25—C26 | 1.382 (4) |
| N1—H11 | 0.81 (3) | C25—H25 | 0.9300 |
| N1—H12 | 0.96 (4) | C26—H26 | 0.9300 |
| C2—C3 | 1.508 (4) | O24—C27 | 1.409 (5) |
| C2—H2A | 0.9700 | C27—H27A | 0.9600 |
| C2—H2B | 0.9700 | C27—H27B | 0.9600 |
| C3—N4 | 1.459 (4) | C27—H27C | 0.9600 |
| C3—H3A | 0.9700 | C31—O32 | 1.239 (3) |
| C3—H3B | 0.9700 | C31—O31 | 1.262 (4) |
| N4—C21 | 1.428 (3) | C31—C32 | 1.488 (4) |
| N4—C5 | 1.467 (4) | C32—C33 | 1.268 (4) |
| C5—C6 | 1.499 (4) | C32—H32 | 0.9300 |
| C5—H5A | 0.9700 | C33—C34 | 1.481 (5) |
| C5—H5B | 0.9700 | C33—H33 | 0.9300 |
| C6—H6A | 0.9700 | C34—O33 | 1.214 (4) |
| C6—H6B | 0.9700 | C34—O34 | 1.276 (5) |
| C21—C26 | 1.387 (4) | O34—H34 | 0.8200 |
| C21—C22 | 1.395 (4) | C43—C44 | 1.479 (12) |
| C22—C23 | 1.383 (4) | C43—H43 | 0.9300 |
| C22—H22 | 0.9300 | C44—O43 | 1.212 (12) |
| C23—C24 | 1.375 (5) | C44—O44 | 1.274 (12) |
| C23—H23 | 0.9300 | O44—H44 | 0.8200 |
| C24—O24 | 1.370 (3) | ||
| C6—N1—C2 | 109.4 (2) | C21—C22—H22 | 119.4 |
| C6—N1—H11 | 113 (3) | C24—C23—C22 | 121.3 (3) |
| C2—N1—H11 | 108 (2) | C24—C23—H23 | 119.3 |
| C6—N1—H12 | 106 (2) | C22—C23—H23 | 119.3 |
| C2—N1—H12 | 111 (2) | O24—C24—C23 | 116.9 (3) |
| H11—N1—H12 | 109 (3) | O24—C24—C25 | 124.9 (3) |
| N1—C2—C3 | 110.7 (3) | C23—C24—C25 | 118.2 (3) |
| N1—C2—H2A | 109.5 | C24—C25—C26 | 120.7 (3) |
| C3—C2—H2A | 109.5 | C24—C25—H25 | 119.7 |
| N1—C2—H2B | 109.5 | C26—C25—H25 | 119.7 |
| C3—C2—H2B | 109.5 | C25—C26—C21 | 121.9 (3) |
| H2A—C2—H2B | 108.1 | C25—C26—H26 | 119.0 |
| N4—C3—C2 | 112.1 (3) | C21—C26—H26 | 119.0 |
| N4—C3—H3A | 109.2 | C24—O24—C27 | 117.4 (3) |
| C2—C3—H3A | 109.2 | O24—C27—H27A | 109.5 |
| N4—C3—H3B | 109.2 | O24—C27—H27B | 109.5 |
| C2—C3—H3B | 109.2 | H27A—C27—H27B | 109.5 |
| H3A—C3—H3B | 107.9 | O24—C27—H27C | 109.5 |
| C21—N4—C3 | 115.6 (2) | H27A—C27—H27C | 109.5 |
| C21—N4—C5 | 115.3 (2) | H27B—C27—H27C | 109.5 |
| C3—N4—C5 | 111.1 (2) | O32—C31—O31 | 125.6 (3) |
| N4—C5—C6 | 112.0 (3) | O32—C31—C32 | 118.5 (3) |
| N4—C5—H5A | 109.2 | O31—C31—C32 | 115.9 (2) |
| C6—C5—H5A | 109.2 | C33—C32—C31 | 126.4 (3) |
| N4—C5—H5B | 109.2 | C33—C32—H32 | 116.8 |
| C6—C5—H5B | 109.2 | C31—C32—H32 | 116.8 |
| H5A—C5—H5B | 107.9 | C32—C33—C34 | 126.2 (4) |
| N1—C6—C5 | 110.5 (3) | C32—C33—H33 | 116.9 |
| N1—C6—H6A | 109.6 | C34—C33—H33 | 116.9 |
| C5—C6—H6A | 109.6 | O33—C34—O34 | 123.0 (4) |
| N1—C6—H6B | 109.6 | O33—C34—C33 | 122.5 (4) |
| C5—C6—H6B | 109.6 | O34—C34—C33 | 114.5 (3) |
| H6A—C6—H6B | 108.1 | C34—O34—H34 | 109.5 |
| C26—C21—C22 | 116.7 (3) | C44—C43—H43 | 116.8 |
| C26—C21—N4 | 121.0 (3) | O43—C44—O44 | 121.3 (18) |
| C22—C21—N4 | 122.2 (3) | O43—C44—C43 | 124 (2) |
| C23—C22—C21 | 121.1 (3) | O44—C44—C43 | 114.9 (18) |
| C23—C22—H22 | 119.4 | C44—O44—H44 | 109.5 |
| C6—N1—C2—C3 | −57.2 (3) | C22—C23—C24—O24 | −179.4 (3) |
| N1—C2—C3—N4 | 55.8 (3) | C22—C23—C24—C25 | 0.8 (5) |
| C2—C3—N4—C21 | 172.3 (2) | O24—C24—C25—C26 | 179.4 (3) |
| C2—C3—N4—C5 | −53.8 (3) | C23—C24—C25—C26 | −0.9 (5) |
| C21—N4—C5—C6 | −171.4 (3) | C24—C25—C26—C21 | 0.0 (5) |
| C3—N4—C5—C6 | 54.7 (3) | C22—C21—C26—C25 | 0.9 (5) |
| C2—N1—C6—C5 | 57.9 (3) | N4—C21—C26—C25 | 178.6 (3) |
| N4—C5—C6—N1 | −57.2 (4) | C23—C24—O24—C27 | 178.8 (3) |
| C3—N4—C21—C26 | 162.4 (3) | C25—C24—O24—C27 | −1.4 (5) |
| C5—N4—C21—C26 | 30.5 (4) | O32—C31—C32—C33 | 34.7 (7) |
| C3—N4—C21—C22 | −20.1 (4) | O31—C31—C32—C33 | −148.0 (6) |
| C5—N4—C21—C22 | −151.9 (3) | C31—C32—C33—C34 | 175.5 (5) |
| C26—C21—C22—C23 | −0.9 (4) | C32—C33—C34—O33 | −164.1 (8) |
| N4—C21—C22—C23 | −178.6 (3) | C32—C33—C34—O34 | 14.9 (10) |
| C21—C22—C23—C24 | 0.1 (5) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenfumarate (IX). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.81 (4) | 2.18 (3) | 2.940 (4) | 155 (3) |
| N1—H12···O32i | 0.96 (4) | 1.77 (4) | 2.714 (4) | 169 (3) |
| O34—H34···O31ii | 0.82 | 1.71 | 2.522 (5) | 170 |
| O44—H44···O31ii | 0.82 | 1.62 | 2.44 (2) | 175 |
| C3—H3A···Cg2iii | 0.97 | 2.76 | 3.650 (3) | 153 |
Symmetry codes: (i) x−1/2, −y+3/2, z; (ii) x−1/2, −y+3/2, z+1; (iii) −x+1, −y+1, z+1/2.
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Crystal data
| C11H17N2O+·C4H3O4− | F(000) = 656 |
| Mr = 308.33 | Dx = 1.335 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.063 (1) Å | Cell parameters from 3315 reflections |
| b = 6.4956 (9) Å | θ = 2.7–27.8° |
| c = 26.093 (3) Å | µ = 0.10 mm−1 |
| β = 93.18 (1)° | T = 296 K |
| V = 1533.7 (3) Å3 | Block, colourless |
| Z = 4 | 0.48 × 0.44 × 0.32 mm |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 3311 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2459 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.014 |
| ω scans | θmax = 27.6°, θmin = 2.7° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −11→11 |
| Tmin = 0.871, Tmax = 0.968 | k = −6→8 |
| 6112 measured reflections | l = −26→33 |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.040 | w = 1/[σ2(Fo2) + (0.0521P)2 + 0.3046P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.111 | (Δ/σ)max = 0.001 |
| S = 1.05 | Δρmax = 0.21 e Å−3 |
| 3311 reflections | Δρmin = −0.13 e Å−3 |
| 210 parameters | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0192 (18) |
| Primary atom site location: difference Fourier map |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Special details
| Experimental. Compound (X). IR (KBr , cm-1) 3073 (NH2), 2836 (OCH3), 1565 (COO). NMR (CDCl3) δ(1H) ) 3.34 (m, 4H, piperazine), 3.41 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.29 (s, 2H, maleate), 6.90 (m, 4H, methoxyphenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.54059 (15) | 0.1806 (2) | 0.56144 (5) | 0.0431 (3) | |
| H11 | 0.6147 (19) | 0.166 (3) | 0.5393 (6) | 0.052* | |
| H12 | 0.452 (2) | 0.174 (3) | 0.5426 (7) | 0.052* | |
| C2 | 0.5537 (2) | 0.3928 (3) | 0.58165 (6) | 0.0531 (4) | |
| H2A | 0.5428 | 0.4903 | 0.5535 | 0.064* | |
| H2B | 0.6506 | 0.4121 | 0.5986 | 0.064* | |
| C3 | 0.43600 (18) | 0.4321 (2) | 0.61930 (6) | 0.0468 (4) | |
| H3A | 0.4472 | 0.5706 | 0.6329 | 0.056* | |
| H3B | 0.3392 | 0.4222 | 0.6016 | 0.056* | |
| N4 | 0.44597 (13) | 0.28484 (17) | 0.66144 (4) | 0.0362 (3) | |
| C5 | 0.43395 (19) | 0.0748 (2) | 0.64123 (6) | 0.0481 (4) | |
| H5A | 0.3370 | 0.0559 | 0.6243 | 0.058* | |
| H5B | 0.4438 | −0.0222 | 0.6695 | 0.058* | |
| C6 | 0.55084 (18) | 0.0302 (2) | 0.60367 (6) | 0.0468 (4) | |
| H6A | 0.6480 | 0.0377 | 0.6211 | 0.056* | |
| H6B | 0.5373 | −0.1077 | 0.5900 | 0.056* | |
| C21 | 0.35362 (15) | 0.3268 (2) | 0.70263 (5) | 0.0354 (3) | |
| C22 | 0.26577 (18) | 0.5006 (2) | 0.70426 (6) | 0.0481 (4) | |
| H22 | 0.2632 | 0.5920 | 0.6768 | 0.058* | |
| C23 | 0.18120 (18) | 0.5421 (3) | 0.74593 (6) | 0.0530 (4) | |
| H23 | 0.1247 | 0.6616 | 0.7463 | 0.064* | |
| C24 | 0.18071 (17) | 0.4076 (3) | 0.78649 (5) | 0.0489 (4) | |
| C25 | 0.2673 (2) | 0.2332 (3) | 0.78540 (6) | 0.0537 (4) | |
| H25 | 0.2679 | 0.1410 | 0.8127 | 0.064* | |
| C26 | 0.35276 (18) | 0.1936 (3) | 0.74451 (6) | 0.0464 (4) | |
| H326 | 0.4110 | 0.0757 | 0.7448 | 0.056* | |
| O24 | 0.10103 (14) | 0.4323 (2) | 0.82944 (4) | 0.0706 (4) | |
| C27 | 0.0207 (2) | 0.6185 (4) | 0.83401 (7) | 0.0844 (7) | |
| H27A | −0.0231 | 0.6219 | 0.8667 | 0.127* | |
| H27B | −0.0556 | 0.6260 | 0.8070 | 0.127* | |
| H27C | 0.0864 | 0.7334 | 0.8314 | 0.127* | |
| C31 | 0.81864 (16) | 0.2456 (2) | 0.48041 (7) | 0.0439 (4) | |
| C32 | 0.93279 (17) | 0.2644 (2) | 0.44164 (6) | 0.0460 (4) | |
| H32 | 0.8951 | 0.2815 | 0.4080 | 0.055* | |
| C33 | 1.07926 (16) | 0.2605 (2) | 0.44719 (6) | 0.0429 (4) | |
| H33 | 1.1274 | 0.2743 | 0.4168 | 0.052* | |
| C34 | 1.17846 (15) | 0.2380 (2) | 0.49393 (6) | 0.0370 (3) | |
| O31 | 0.68826 (12) | 0.23449 (19) | 0.46522 (5) | 0.0627 (4) | |
| O32 | 0.85878 (12) | 0.23939 (18) | 0.52832 (4) | 0.0520 (3) | |
| O33 | 1.12495 (12) | 0.23595 (18) | 0.53839 (4) | 0.0488 (3) | |
| H33A | 0.996 (2) | 0.240 (3) | 0.5354 (8) | 0.073* | |
| O34 | 1.31216 (11) | 0.22095 (17) | 0.48789 (4) | 0.0495 (3) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0357 (6) | 0.0568 (8) | 0.0375 (7) | 0.0007 (6) | 0.0084 (5) | −0.0023 (6) |
| C2 | 0.0645 (10) | 0.0474 (9) | 0.0493 (9) | −0.0055 (8) | 0.0214 (8) | 0.0037 (7) |
| C3 | 0.0590 (10) | 0.0388 (8) | 0.0440 (8) | 0.0018 (7) | 0.0146 (7) | 0.0034 (7) |
| N4 | 0.0396 (6) | 0.0349 (6) | 0.0346 (6) | 0.0002 (5) | 0.0054 (5) | 0.0009 (5) |
| C5 | 0.0591 (10) | 0.0385 (8) | 0.0484 (9) | −0.0012 (7) | 0.0186 (7) | 0.0000 (7) |
| C6 | 0.0507 (9) | 0.0450 (9) | 0.0454 (8) | 0.0080 (7) | 0.0087 (7) | −0.0013 (7) |
| C21 | 0.0340 (7) | 0.0391 (7) | 0.0329 (7) | −0.0023 (6) | 0.0007 (5) | −0.0016 (6) |
| C22 | 0.0550 (9) | 0.0482 (9) | 0.0419 (8) | 0.0101 (7) | 0.0106 (7) | 0.0080 (7) |
| C23 | 0.0539 (10) | 0.0584 (10) | 0.0472 (9) | 0.0176 (8) | 0.0085 (7) | 0.0009 (8) |
| C24 | 0.0435 (8) | 0.0715 (11) | 0.0316 (7) | 0.0060 (8) | 0.0023 (6) | −0.0037 (7) |
| C25 | 0.0645 (10) | 0.0662 (11) | 0.0305 (7) | 0.0109 (9) | 0.0042 (7) | 0.0111 (7) |
| C26 | 0.0525 (9) | 0.0508 (9) | 0.0358 (8) | 0.0120 (7) | 0.0001 (6) | 0.0044 (6) |
| O24 | 0.0722 (8) | 0.1024 (11) | 0.0386 (6) | 0.0251 (7) | 0.0175 (6) | 0.0033 (6) |
| C27 | 0.0759 (13) | 0.126 (2) | 0.0528 (11) | 0.0418 (14) | 0.0177 (10) | −0.0042 (12) |
| C31 | 0.0357 (8) | 0.0337 (8) | 0.0631 (10) | 0.0035 (6) | 0.0104 (7) | 0.0045 (7) |
| C32 | 0.0425 (8) | 0.0529 (9) | 0.0427 (8) | 0.0026 (7) | 0.0035 (6) | 0.0069 (7) |
| C33 | 0.0411 (8) | 0.0493 (9) | 0.0395 (8) | 0.0003 (7) | 0.0118 (6) | 0.0033 (7) |
| C34 | 0.0352 (7) | 0.0300 (7) | 0.0463 (8) | −0.0020 (6) | 0.0071 (6) | −0.0005 (6) |
| O31 | 0.0334 (6) | 0.0690 (8) | 0.0858 (9) | 0.0036 (5) | 0.0040 (6) | 0.0110 (7) |
| O32 | 0.0418 (6) | 0.0641 (8) | 0.0517 (7) | 0.0009 (5) | 0.0172 (5) | 0.0003 (5) |
| O33 | 0.0427 (6) | 0.0647 (8) | 0.0393 (6) | 0.0007 (5) | 0.0056 (4) | −0.0020 (5) |
| O34 | 0.0337 (5) | 0.0572 (7) | 0.0580 (7) | −0.0010 (5) | 0.0069 (5) | 0.0012 (5) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Geometric parameters (Å, º)
| N1—C6 | 1.472 (2) | C23—H23 | 0.9300 |
| N1—C2 | 1.478 (2) | C24—O24 | 1.3759 (17) |
| N1—H11 | 0.915 (17) | C24—C25 | 1.379 (2) |
| N1—H12 | 0.917 (18) | C25—C26 | 1.377 (2) |
| C2—C3 | 1.511 (2) | C25—H25 | 0.9300 |
| C2—H2A | 0.9700 | C26—H326 | 0.9300 |
| C2—H2B | 0.9700 | O24—C27 | 1.420 (3) |
| C3—N4 | 1.4567 (18) | C27—H27A | 0.9600 |
| C3—H3A | 0.9700 | C27—H27B | 0.9600 |
| C3—H3B | 0.9700 | C27—H27C | 0.9600 |
| N4—C21 | 1.4248 (17) | C31—O31 | 1.2274 (19) |
| N4—C5 | 1.4645 (19) | C31—O32 | 1.283 (2) |
| C5—C6 | 1.511 (2) | C31—C32 | 1.492 (2) |
| C5—H5A | 0.9700 | C32—C33 | 1.328 (2) |
| C5—H5B | 0.9700 | C32—H32 | 0.9300 |
| C6—H6A | 0.9700 | C33—C34 | 1.482 (2) |
| C6—H6B | 0.9700 | C33—H33 | 0.9300 |
| C21—C22 | 1.383 (2) | C34—O34 | 1.2355 (17) |
| C21—C26 | 1.394 (2) | C34—O33 | 1.2820 (17) |
| C22—C23 | 1.391 (2) | O32—H33A | 1.25 (2) |
| C22—H22 | 0.9300 | O33—H33A | 1.17 (2) |
| C23—C24 | 1.373 (2) | ||
| C6—N1—C2 | 110.57 (12) | C21—C22—C23 | 121.70 (14) |
| C6—N1—H11 | 112.8 (11) | C21—C22—H22 | 119.2 |
| C2—N1—H11 | 105.8 (11) | C23—C22—H22 | 119.2 |
| C6—N1—H12 | 112.6 (11) | C24—C23—C22 | 120.31 (15) |
| C2—N1—H12 | 106.8 (11) | C24—C23—H23 | 119.8 |
| H11—N1—H12 | 107.8 (15) | C22—C23—H23 | 119.8 |
| N1—C2—C3 | 110.13 (13) | C23—C24—O24 | 125.33 (15) |
| N1—C2—H2A | 109.6 | C23—C24—C25 | 118.74 (14) |
| C3—C2—H2A | 109.6 | O24—C24—C25 | 115.93 (14) |
| N1—C2—H2B | 109.6 | C26—C25—C24 | 120.98 (15) |
| C3—C2—H2B | 109.6 | C26—C25—H25 | 119.5 |
| H2A—C2—H2B | 108.1 | C24—C25—H25 | 119.5 |
| N4—C3—C2 | 111.29 (13) | C25—C26—C21 | 121.25 (15) |
| N4—C3—H3A | 109.4 | C25—C26—H326 | 119.4 |
| C2—C3—H3A | 109.4 | C21—C26—H326 | 119.4 |
| N4—C3—H3B | 109.4 | C24—O24—C27 | 117.53 (15) |
| C2—C3—H3B | 109.4 | O24—C27—H27A | 109.5 |
| H3A—C3—H3B | 108.0 | O24—C27—H27B | 109.5 |
| C21—N4—C3 | 115.42 (11) | H27A—C27—H27B | 109.5 |
| C21—N4—C5 | 114.50 (11) | O24—C27—H27C | 109.5 |
| C3—N4—C5 | 109.87 (12) | H27A—C27—H27C | 109.5 |
| N4—C5—C6 | 111.79 (12) | H27B—C27—H27C | 109.5 |
| N4—C5—H5A | 109.3 | O31—C31—O32 | 121.90 (14) |
| C6—C5—H5A | 109.3 | O31—C31—C32 | 118.52 (15) |
| N4—C5—H5B | 109.3 | O32—C31—C32 | 119.57 (14) |
| C6—C5—H5B | 109.3 | C33—C32—C31 | 130.65 (15) |
| H5A—C5—H5B | 107.9 | C33—C32—H32 | 114.7 |
| N1—C6—C5 | 109.95 (13) | C31—C32—H32 | 114.7 |
| N1—C6—H6A | 109.7 | C32—C33—C34 | 130.48 (13) |
| C5—C6—H6A | 109.7 | C32—C33—H33 | 114.8 |
| N1—C6—H6B | 109.7 | C34—C33—H33 | 114.8 |
| C5—C6—H6B | 109.7 | O34—C34—O33 | 122.51 (14) |
| H6A—C6—H6B | 108.2 | O34—C34—C33 | 117.29 (13) |
| C22—C21—C26 | 117.01 (13) | O33—C34—C33 | 120.20 (12) |
| C22—C21—N4 | 122.86 (13) | C31—O32—H33A | 111.7 (9) |
| C26—C21—N4 | 120.09 (13) | C34—O33—H33A | 111.5 (10) |
| C6—N1—C2—C3 | −57.07 (18) | C22—C23—C24—O24 | −179.65 (16) |
| N1—C2—C3—N4 | 57.68 (18) | C22—C23—C24—C25 | 1.0 (3) |
| C2—C3—N4—C21 | 171.43 (13) | C23—C24—C25—C26 | −0.1 (3) |
| C2—C3—N4—C5 | −57.26 (17) | O24—C24—C25—C26 | −179.42 (16) |
| C21—N4—C5—C6 | −171.06 (12) | C24—C25—C26—C21 | −0.7 (3) |
| C3—N4—C5—C6 | 57.15 (17) | C22—C21—C26—C25 | 0.4 (2) |
| C2—N1—C6—C5 | 56.57 (17) | N4—C21—C26—C25 | 178.39 (14) |
| N4—C5—C6—N1 | −57.02 (18) | C23—C24—O24—C27 | −4.6 (3) |
| C3—N4—C21—C22 | −3.3 (2) | C25—C24—O24—C27 | 174.67 (18) |
| C5—N4—C21—C22 | −132.32 (16) | O31—C31—C32—C33 | −173.24 (17) |
| C3—N4—C21—C26 | 178.91 (14) | O32—C31—C32—C33 | 6.0 (2) |
| C5—N4—C21—C26 | 49.85 (18) | C31—C32—C33—C34 | −0.5 (3) |
| C26—C21—C22—C23 | 0.6 (2) | C32—C33—C34—O34 | 173.69 (16) |
| N4—C21—C22—C23 | −177.33 (14) | C32—C33—C34—O33 | −6.1 (2) |
| C21—C22—C23—C24 | −1.3 (3) |
4-(4-Methoxyphenyl)piperazin-1-ium hydrogenmaleate (X). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O33—H33A···O32 | 1.167 (18) | 1.247 (18) | 2.4121 (16) | 175 (2) |
| N1—H11···O31 | 0.915 (17) | 2.126 (16) | 2.9309 (19) | 146.2 (15) |
| N1—H11···O32 | 0.915 (17) | 2.296 (17) | 3.0798 (18) | 143.5 (14) |
| N1—H12···O34i | 0.919 (18) | 1.881 (18) | 2.7563 (17) | 158.5 (17) |
| C2—H2A···O34ii | 0.97 | 2.56 | 3.363 (2) | 140 |
Symmetry codes: (i) x−1, y, z; (ii) −x+2, −y+1, −z+1.
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Crystal data
| C11H17N2O+·C2Cl3O2− | Dx = 1.477 Mg m−3 |
| Mr = 355.64 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pca21 | Cell parameters from 2428 reflections |
| a = 10.6117 (11) Å | θ = 3.0–27.7° |
| b = 13.808 (1) Å | µ = 0.58 mm−1 |
| c = 10.9137 (8) Å | T = 296 K |
| V = 1599.1 (2) Å3 | Block, colourless |
| Z = 4 | 0.48 × 0.48 × 0.20 mm |
| F(000) = 736 |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 2428 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2278 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| ω scans | θmax = 27.7°, θmin = 3.0° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −5→13 |
| Tmin = 0.476, Tmax = 0.892 | k = −16→17 |
| 6173 measured reflections | l = −14→5 |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Refinement
| Refinement on F2 | H atoms treated by a mixture of independent and constrained refinement |
| Least-squares matrix: full | w = 1/[σ2(Fo2) + (0.0532P)2 + 0.3843P] where P = (Fo2 + 2Fc2)/3 |
| R[F2 > 2σ(F2)] = 0.032 | (Δ/σ)max < 0.001 |
| wR(F2) = 0.086 | Δρmax = 0.25 e Å−3 |
| S = 1.08 | Δρmin = −0.30 e Å−3 |
| 2428 reflections | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 198 parameters | Extinction coefficient: 0.023 (2) |
| 1 restraint | Absolute structure: Classical Flack method preferred over Parsons because s.u. lower |
| Primary atom site location: difference Fourier map | Absolute structure parameter: 0.11 (7) |
| Hydrogen site location: mixed |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Special details
| Experimental. Compound (XI). IR (KBr , cm-1) 3073 (NH2), 2829 (OCH3), 1561 (COO). NMR (CDCl3) δ(1H) ) 3.07 (m, 4H, piperazine), 3.19 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.89 (m, 4H, methoxyphenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.1394 (2) | 0.44188 (17) | 0.2473 (3) | 0.0301 (5) | |
| H11 | 0.168 (3) | 0.495 (3) | 0.204 (3) | 0.036* | |
| H12 | 0.061 (3) | 0.426 (3) | 0.206 (4) | 0.036* | |
| C2 | 0.1158 (3) | 0.4667 (2) | 0.3775 (3) | 0.0343 (7) | |
| H2A | 0.0542 | 0.5185 | 0.3822 | 0.041* | |
| H2B | 0.1933 | 0.4895 | 0.4148 | 0.041* | |
| C3 | 0.0680 (3) | 0.37961 (19) | 0.4468 (3) | 0.0320 (6) | |
| H3A | 0.0546 | 0.3967 | 0.5320 | 0.038* | |
| H3B | −0.0121 | 0.3591 | 0.4126 | 0.038* | |
| N4 | 0.1586 (2) | 0.30039 (15) | 0.4389 (2) | 0.0261 (5) | |
| C5 | 0.1815 (3) | 0.2750 (2) | 0.3106 (3) | 0.0311 (6) | |
| H5A | 0.1037 | 0.2523 | 0.2738 | 0.037* | |
| H5B | 0.2424 | 0.2227 | 0.3067 | 0.037* | |
| C6 | 0.2304 (2) | 0.3607 (2) | 0.2392 (3) | 0.0324 (6) | |
| H6A | 0.3112 | 0.3808 | 0.2721 | 0.039* | |
| H6B | 0.2423 | 0.3426 | 0.1541 | 0.039* | |
| C21 | 0.1350 (2) | 0.22071 (19) | 0.5194 (3) | 0.0269 (6) | |
| C22 | 0.0345 (3) | 0.2170 (2) | 0.5993 (3) | 0.0347 (6) | |
| H22 | −0.0265 | 0.2653 | 0.5959 | 0.042* | |
| C23 | 0.0216 (3) | 0.1433 (2) | 0.6846 (3) | 0.0396 (7) | |
| H23 | −0.0468 | 0.1430 | 0.7379 | 0.048* | |
| C24 | 0.1103 (3) | 0.0700 (2) | 0.6906 (3) | 0.0398 (7) | |
| C25 | 0.2088 (3) | 0.0707 (2) | 0.6074 (4) | 0.0388 (7) | |
| H25 | 0.2671 | 0.0205 | 0.6083 | 0.047* | |
| C26 | 0.2220 (3) | 0.14432 (19) | 0.5235 (3) | 0.0324 (6) | |
| H26 | 0.2892 | 0.1434 | 0.4689 | 0.039* | |
| O24 | 0.1108 (3) | −0.00368 (18) | 0.7747 (3) | 0.0587 (8) | |
| C27 | 0.0095 (6) | −0.0086 (4) | 0.8579 (5) | 0.0733 (13) | |
| H27A | 0.0234 | −0.0608 | 0.9145 | 0.110* | |
| H27B | −0.0675 | −0.0197 | 0.8138 | 0.110* | |
| H27C | 0.0035 | 0.0513 | 0.9022 | 0.110* | |
| C31 | 0.3163 (2) | 0.61757 (18) | 0.1132 (3) | 0.0266 (5) | |
| O31 | 0.20314 (18) | 0.60081 (16) | 0.1045 (3) | 0.0444 (6) | |
| O32 | 0.39463 (19) | 0.57402 (18) | 0.1749 (3) | 0.0464 (6) | |
| C32 | 0.3653 (3) | 0.7076 (2) | 0.0383 (3) | 0.0304 (6) | |
| Cl1 | 0.52532 (7) | 0.69160 (6) | −0.00662 (9) | 0.0426 (2) | |
| Cl2 | 0.35730 (10) | 0.80958 (5) | 0.13523 (10) | 0.0505 (3) | |
| Cl3 | 0.27618 (8) | 0.72790 (7) | −0.09480 (9) | 0.0540 (3) |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0286 (11) | 0.0228 (11) | 0.0389 (15) | −0.0020 (8) | −0.0003 (11) | 0.0038 (11) |
| C2 | 0.0433 (14) | 0.0199 (13) | 0.0398 (18) | 0.0022 (11) | −0.0004 (14) | −0.0032 (12) |
| C3 | 0.0354 (13) | 0.0242 (13) | 0.0364 (16) | 0.0059 (11) | 0.0060 (12) | −0.0015 (12) |
| N4 | 0.0304 (11) | 0.0178 (10) | 0.0300 (13) | 0.0008 (8) | 0.0049 (9) | −0.0026 (9) |
| C5 | 0.0370 (14) | 0.0234 (13) | 0.0331 (15) | 0.0034 (11) | 0.0027 (13) | −0.0025 (12) |
| C6 | 0.0308 (14) | 0.0307 (14) | 0.0356 (16) | 0.0041 (10) | 0.0064 (13) | 0.0002 (13) |
| C21 | 0.0295 (11) | 0.0196 (11) | 0.0316 (16) | −0.0051 (9) | 0.0001 (11) | −0.0022 (11) |
| C22 | 0.0335 (13) | 0.0295 (13) | 0.0411 (18) | −0.0016 (11) | 0.0036 (13) | −0.0032 (13) |
| C23 | 0.0455 (15) | 0.0342 (15) | 0.0392 (19) | −0.0102 (13) | 0.0117 (14) | −0.0011 (14) |
| C24 | 0.0628 (19) | 0.0206 (12) | 0.0361 (18) | −0.0128 (13) | −0.0005 (15) | −0.0005 (13) |
| C25 | 0.0487 (15) | 0.0198 (12) | 0.048 (2) | 0.0005 (11) | −0.0002 (16) | 0.0004 (13) |
| C26 | 0.0336 (13) | 0.0229 (13) | 0.0408 (18) | −0.0009 (10) | 0.0034 (12) | −0.0015 (13) |
| O24 | 0.0943 (19) | 0.0332 (12) | 0.0485 (17) | −0.0085 (13) | 0.0079 (15) | 0.0129 (13) |
| C27 | 0.095 (3) | 0.064 (3) | 0.061 (3) | −0.028 (2) | 0.008 (3) | 0.023 (2) |
| C31 | 0.0303 (12) | 0.0190 (11) | 0.0305 (15) | −0.0017 (9) | 0.0033 (12) | 0.0005 (11) |
| O31 | 0.0301 (9) | 0.0391 (11) | 0.0640 (17) | −0.0070 (8) | −0.0039 (11) | 0.0214 (12) |
| O32 | 0.0328 (11) | 0.0421 (12) | 0.0643 (18) | −0.0046 (9) | −0.0072 (11) | 0.0244 (12) |
| C32 | 0.0320 (12) | 0.0260 (13) | 0.0332 (15) | −0.0049 (10) | −0.0009 (12) | 0.0029 (12) |
| Cl1 | 0.0343 (3) | 0.0457 (4) | 0.0478 (5) | −0.0125 (3) | 0.0085 (3) | −0.0007 (4) |
| Cl2 | 0.0714 (6) | 0.0236 (3) | 0.0565 (6) | −0.0017 (3) | 0.0089 (5) | −0.0065 (4) |
| Cl3 | 0.0514 (5) | 0.0652 (6) | 0.0455 (5) | −0.0134 (4) | −0.0124 (4) | 0.0233 (5) |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Geometric parameters (Å, º)
| N1—C6 | 1.482 (3) | C22—C23 | 1.386 (5) |
| N1—C2 | 1.484 (4) | C22—H22 | 0.9300 |
| N1—H11 | 0.92 (4) | C23—C24 | 1.383 (5) |
| N1—H12 | 0.97 (3) | C23—H23 | 0.9300 |
| C2—C3 | 1.509 (4) | C24—O24 | 1.370 (4) |
| C2—H2A | 0.9700 | C24—C25 | 1.385 (5) |
| C2—H2B | 0.9700 | C25—C26 | 1.375 (4) |
| C3—N4 | 1.459 (3) | C25—H25 | 0.9300 |
| C3—H3A | 0.9700 | C26—H26 | 0.9300 |
| C3—H3B | 0.9700 | O24—C27 | 1.410 (6) |
| N4—C21 | 1.430 (4) | C27—H27A | 0.9600 |
| N4—C5 | 1.464 (4) | C27—H27B | 0.9600 |
| C5—C6 | 1.509 (4) | C27—H27C | 0.9600 |
| C5—H5A | 0.9700 | C31—O31 | 1.227 (3) |
| C5—H5B | 0.9700 | C31—O32 | 1.227 (4) |
| C6—H6A | 0.9700 | C31—C32 | 1.576 (4) |
| C6—H6B | 0.9700 | C32—Cl3 | 1.756 (3) |
| C21—C22 | 1.379 (4) | C32—Cl2 | 1.763 (3) |
| C21—C26 | 1.402 (4) | C32—Cl1 | 1.781 (3) |
| C6—N1—C2 | 110.0 (2) | C22—C21—N4 | 123.5 (3) |
| C6—N1—H11 | 111 (2) | C26—C21—N4 | 118.9 (2) |
| C2—N1—H11 | 111 (2) | C21—C22—C23 | 121.9 (3) |
| C6—N1—H12 | 111 (2) | C21—C22—H22 | 119.0 |
| C2—N1—H12 | 111 (2) | C23—C22—H22 | 119.0 |
| H11—N1—H12 | 103 (3) | C24—C23—C22 | 120.1 (3) |
| N1—C2—C3 | 110.6 (2) | C24—C23—H23 | 119.9 |
| N1—C2—H2A | 109.5 | C22—C23—H23 | 119.9 |
| C3—C2—H2A | 109.5 | O24—C24—C23 | 125.3 (3) |
| N1—C2—H2B | 109.5 | O24—C24—C25 | 116.2 (3) |
| C3—C2—H2B | 109.5 | C23—C24—C25 | 118.5 (3) |
| H2A—C2—H2B | 108.1 | C26—C25—C24 | 121.3 (3) |
| N4—C3—C2 | 110.3 (2) | C26—C25—H25 | 119.4 |
| N4—C3—H3A | 109.6 | C24—C25—H25 | 119.4 |
| C2—C3—H3A | 109.6 | C25—C26—C21 | 120.7 (3) |
| N4—C3—H3B | 109.6 | C25—C26—H26 | 119.7 |
| C2—C3—H3B | 109.6 | C21—C26—H26 | 119.7 |
| H3A—C3—H3B | 108.1 | C24—O24—C27 | 117.7 (3) |
| C21—N4—C3 | 115.2 (2) | O24—C27—H27A | 109.5 |
| C21—N4—C5 | 115.6 (2) | O24—C27—H27B | 109.5 |
| C3—N4—C5 | 110.2 (2) | H27A—C27—H27B | 109.5 |
| N4—C5—C6 | 111.3 (2) | O24—C27—H27C | 109.5 |
| N4—C5—H5A | 109.4 | H27A—C27—H27C | 109.5 |
| C6—C5—H5A | 109.4 | H27B—C27—H27C | 109.5 |
| N4—C5—H5B | 109.4 | O31—C31—O32 | 127.8 (3) |
| C6—C5—H5B | 109.4 | O31—C31—C32 | 115.6 (2) |
| H5A—C5—H5B | 108.0 | O32—C31—C32 | 116.6 (2) |
| N1—C6—C5 | 109.8 (2) | C31—C32—Cl3 | 112.15 (19) |
| N1—C6—H6A | 109.7 | C31—C32—Cl2 | 107.6 (2) |
| C5—C6—H6A | 109.7 | Cl3—C32—Cl2 | 110.05 (16) |
| N1—C6—H6B | 109.7 | C31—C32—Cl1 | 111.06 (19) |
| C5—C6—H6B | 109.7 | Cl3—C32—Cl1 | 107.82 (18) |
| H6A—C6—H6B | 108.2 | Cl2—C32—Cl1 | 108.08 (15) |
| C22—C21—C26 | 117.4 (3) | ||
| C6—N1—C2—C3 | −57.4 (3) | C22—C23—C24—O24 | 176.6 (3) |
| N1—C2—C3—N4 | 58.2 (3) | C22—C23—C24—C25 | −2.1 (5) |
| C2—C3—N4—C21 | 168.8 (3) | O24—C24—C25—C26 | −176.2 (3) |
| C2—C3—N4—C5 | −58.2 (3) | C23—C24—C25—C26 | 2.6 (5) |
| C21—N4—C5—C6 | −168.7 (2) | C24—C25—C26—C21 | −0.4 (5) |
| C3—N4—C5—C6 | 58.5 (3) | C22—C21—C26—C25 | −2.3 (4) |
| C2—N1—C6—C5 | 56.6 (3) | N4—C21—C26—C25 | 174.1 (3) |
| N4—C5—C6—N1 | −57.6 (3) | C23—C24—O24—C27 | 3.5 (5) |
| C3—N4—C21—C22 | 0.2 (4) | C25—C24—O24—C27 | −177.7 (4) |
| C5—N4—C21—C22 | −130.3 (3) | O31—C31—C32—Cl3 | −29.9 (3) |
| C3—N4—C21—C26 | −175.9 (3) | O32—C31—C32—Cl3 | 151.5 (3) |
| C5—N4—C21—C26 | 53.6 (3) | O31—C31—C32—Cl2 | 91.3 (3) |
| C26—C21—C22—C23 | 2.8 (5) | O32—C31—C32—Cl2 | −87.3 (3) |
| N4—C21—C22—C23 | −173.3 (3) | O31—C31—C32—Cl1 | −150.6 (2) |
| C21—C22—C23—C24 | −0.7 (5) | O32—C31—C32—Cl1 | 30.8 (4) |
4-(4-Methoxyphenyl)piperazin-1-ium trichloroacetate (XI). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.92 (4) | 1.86 (4) | 2.775 (4) | 172 (3) |
| N1—H12···O32i | 0.97 (3) | 1.80 (3) | 2.724 (3) | 158 (3) |
Symmetry code: (i) x−1/2, −y+1, z.
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Crystal data
| C11H17N2O+·0.5C6Cl2O42−·H2O | F(000) = 664 |
| Mr = 314.76 | Dx = 1.417 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.1597 (5) Å | Cell parameters from 3253 reflections |
| b = 15.1434 (8) Å | θ = 2.6–28.0° |
| c = 10.8742 (6) Å | µ = 0.28 mm−1 |
| β = 102.067 (5)° | T = 296 K |
| V = 1475.02 (14) Å3 | Block, colourless |
| Z = 4 | 0.44 × 0.24 × 0.20 mm |
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Data collection
| Oxford Diffraction Xcalibur with Sapphire CCD diffractometer | 9650 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 7444 reflections with I > 2σ(I) |
| Graphite monochromator | θmax = 27.6°, θmin = 2.6° |
| ω scans | h = −11→11 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | k = −19→19 |
| Tmin = 0.892, Tmax = 0.947 | l = −13→14 |
| 9650 measured reflections |
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.039 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.105 | w = 1/[σ2(Fo2) + (0.0573P)2 + 0.263P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 9650 reflections | Δρmax = 0.23 e Å−3 |
| 204 parameters | Δρmin = −0.32 e Å−3 |
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Special details
| Experimental. Compound (XII). IR (KBr , cm-1) 3311 (OH), 3073 (NH2), 2825 (OCH3), 1561 (COO), 793 and 741 (CCl) . NMR (CDCl3) δ(1H) ) 3.11 (m, 4H, piperazine), 3.40 (m, 4H, piperazine), 3.77 (s, 3H, OCH3), 6.88 (m, 4H, methoxyphenyl). |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component twin. |
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.4381 (2) | 0.32824 (14) | 0.35576 (19) | 0.0420 (5) | |
| H11 | 0.437 (2) | 0.3477 (15) | 0.278 (3) | 0.050* | |
| H12 | 0.420 (3) | 0.3725 (16) | 0.405 (2) | 0.050* | |
| C2 | 0.5890 (2) | 0.29024 (17) | 0.4015 (2) | 0.0472 (6) | |
| H2A | 0.6629 | 0.3370 | 0.4125 | 0.057* | |
| H2B | 0.6115 | 0.2490 | 0.3397 | 0.057* | |
| C3 | 0.5965 (2) | 0.24346 (16) | 0.5241 (2) | 0.0427 (6) | |
| H3A | 0.6939 | 0.2164 | 0.5509 | 0.051* | |
| H3B | 0.5827 | 0.2856 | 0.5878 | 0.051* | |
| N4 | 0.48097 (19) | 0.17569 (13) | 0.51047 (18) | 0.0398 (5) | |
| C5 | 0.3334 (2) | 0.21513 (17) | 0.4745 (2) | 0.0455 (6) | |
| H5A | 0.3186 | 0.2572 | 0.5381 | 0.055* | |
| H5B | 0.2576 | 0.1696 | 0.4681 | 0.055* | |
| C6 | 0.3188 (2) | 0.26139 (16) | 0.3494 (2) | 0.0454 (6) | |
| H6A | 0.3256 | 0.2184 | 0.2847 | 0.054* | |
| H6B | 0.2221 | 0.2899 | 0.3271 | 0.054* | |
| C21 | 0.5025 (2) | 0.10929 (15) | 0.6041 (2) | 0.0357 (5) | |
| C22 | 0.6331 (2) | 0.05976 (16) | 0.6228 (2) | 0.0432 (6) | |
| H22 | 0.7033 | 0.0719 | 0.5744 | 0.052* | |
| C23 | 0.6610 (2) | −0.00626 (16) | 0.7103 (2) | 0.0457 (6) | |
| H23 | 0.7503 | −0.0375 | 0.7222 | 0.055* | |
| C24 | 0.5565 (2) | −0.02672 (15) | 0.7812 (2) | 0.0418 (6) | |
| C25 | 0.4243 (2) | 0.01896 (16) | 0.7614 (2) | 0.0452 (6) | |
| H25 | 0.3516 | 0.0040 | 0.8059 | 0.054* | |
| C26 | 0.3991 (2) | 0.08760 (16) | 0.6747 (2) | 0.0419 (6) | |
| H26 | 0.3106 | 0.1195 | 0.6641 | 0.050* | |
| O24 | 0.59700 (19) | −0.09318 (12) | 0.86827 (19) | 0.0615 (5) | |
| C27 | 0.4842 (3) | −0.12833 (17) | 0.9257 (3) | 0.0577 (7) | |
| H27A | 0.5208 | −0.1806 | 0.9720 | 0.086* | |
| H27B | 0.3980 | −0.1429 | 0.8621 | 0.086* | |
| H27C | 0.4573 | −0.0853 | 0.9820 | 0.086* | |
| C31 | 0.4846 (2) | 0.41334 (14) | 0.0515 (2) | 0.0301 (5) | |
| O31 | 0.46952 (17) | 0.34242 (10) | 0.10569 (14) | 0.0429 (4) | |
| C32 | 0.4674 (2) | 0.42353 (14) | −0.0785 (2) | 0.0313 (5) | |
| Cl32 | 0.42518 (7) | 0.33070 (4) | −0.17371 (6) | 0.04725 (19) | |
| O33 | 0.54043 (16) | 0.48217 (10) | 0.25106 (14) | 0.0419 (4) | |
| C33 | 0.5224 (2) | 0.49523 (14) | 0.13592 (19) | 0.0297 (5) | |
| O41 | 0.3458 (2) | 0.45297 (16) | 0.5134 (2) | 0.0698 (6) | |
| H41 | 0.383 (4) | 0.464 (2) | 0.589 (4) | 0.105* | |
| H42 | 0.295 (4) | 0.497 (2) | 0.492 (3) | 0.105* |
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0531 (12) | 0.0465 (13) | 0.0271 (11) | 0.0008 (11) | 0.0101 (9) | 0.0103 (10) |
| C2 | 0.0458 (13) | 0.0526 (15) | 0.0447 (15) | −0.0023 (12) | 0.0132 (11) | 0.0145 (13) |
| C3 | 0.0397 (12) | 0.0497 (14) | 0.0362 (14) | −0.0084 (12) | 0.0026 (10) | 0.0099 (12) |
| N4 | 0.0358 (10) | 0.0461 (11) | 0.0344 (11) | −0.0073 (9) | 0.0003 (8) | 0.0123 (9) |
| C5 | 0.0372 (12) | 0.0559 (16) | 0.0411 (14) | −0.0077 (12) | 0.0028 (10) | 0.0152 (13) |
| C6 | 0.0462 (13) | 0.0509 (15) | 0.0343 (14) | −0.0037 (12) | −0.0026 (10) | 0.0082 (12) |
| C21 | 0.0368 (12) | 0.0404 (13) | 0.0280 (12) | −0.0096 (11) | 0.0027 (9) | 0.0032 (10) |
| C22 | 0.0374 (13) | 0.0471 (15) | 0.0472 (15) | −0.0058 (11) | 0.0139 (11) | 0.0057 (12) |
| C23 | 0.0358 (12) | 0.0451 (14) | 0.0566 (17) | 0.0018 (11) | 0.0103 (11) | 0.0088 (13) |
| C24 | 0.0462 (13) | 0.0399 (14) | 0.0375 (14) | −0.0005 (12) | 0.0043 (10) | 0.0093 (11) |
| C25 | 0.0429 (13) | 0.0540 (16) | 0.0413 (15) | −0.0008 (12) | 0.0146 (10) | 0.0123 (12) |
| C26 | 0.0358 (12) | 0.0498 (15) | 0.0398 (14) | 0.0045 (11) | 0.0070 (10) | 0.0125 (12) |
| O24 | 0.0567 (11) | 0.0620 (12) | 0.0673 (14) | 0.0079 (9) | 0.0164 (9) | 0.0339 (11) |
| C27 | 0.0752 (18) | 0.0480 (16) | 0.0522 (18) | −0.0012 (15) | 0.0190 (14) | 0.0151 (14) |
| C31 | 0.0292 (10) | 0.0331 (12) | 0.0290 (12) | 0.0005 (9) | 0.0083 (8) | 0.0032 (10) |
| O31 | 0.0612 (10) | 0.0355 (9) | 0.0327 (9) | −0.0024 (8) | 0.0115 (7) | 0.0066 (7) |
| C32 | 0.0368 (11) | 0.0315 (11) | 0.0256 (11) | −0.0004 (10) | 0.0060 (8) | −0.0010 (9) |
| Cl32 | 0.0648 (4) | 0.0407 (3) | 0.0347 (3) | −0.0058 (3) | 0.0068 (3) | −0.0065 (3) |
| O33 | 0.0582 (9) | 0.0456 (10) | 0.0215 (8) | −0.0073 (8) | 0.0073 (7) | 0.0036 (7) |
| C33 | 0.0282 (10) | 0.0381 (12) | 0.0232 (11) | 0.0019 (9) | 0.0065 (8) | 0.0014 (9) |
| O41 | 0.0876 (16) | 0.0781 (15) | 0.0408 (12) | 0.0001 (12) | 0.0066 (10) | −0.0146 (12) |
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Geometric parameters (Å, º)
| N1—C6 | 1.481 (3) | C23—C24 | 1.384 (3) |
| N1—C2 | 1.484 (3) | C23—H23 | 0.9300 |
| N1—H11 | 0.89 (3) | C24—C25 | 1.372 (3) |
| N1—H12 | 0.89 (2) | C24—O24 | 1.379 (3) |
| C2—C3 | 1.498 (3) | C25—C26 | 1.390 (3) |
| C2—H2A | 0.9700 | C25—H25 | 0.9300 |
| C2—H2B | 0.9700 | C26—H26 | 0.9300 |
| C3—N4 | 1.459 (3) | O24—C27 | 1.419 (3) |
| C3—H3A | 0.9700 | C27—H27A | 0.9600 |
| C3—H3B | 0.9700 | C27—H27B | 0.9600 |
| N4—C21 | 1.415 (3) | C27—H27C | 0.9600 |
| N4—C5 | 1.455 (3) | C31—O31 | 1.246 (2) |
| C5—C6 | 1.511 (3) | C31—C32 | 1.398 (3) |
| C5—H5A | 0.9700 | C31—C33 | 1.539 (3) |
| C5—H5B | 0.9700 | C32—C33i | 1.392 (3) |
| C6—H6A | 0.9700 | C32—Cl32 | 1.741 (2) |
| C6—H6B | 0.9700 | O33—C33 | 1.244 (2) |
| C21—C26 | 1.378 (3) | C33—C32i | 1.392 (3) |
| C21—C22 | 1.390 (3) | O41—H41 | 0.83 (4) |
| C22—C23 | 1.367 (3) | O41—H42 | 0.82 (3) |
| C22—H22 | 0.9300 | ||
| C6—N1—C2 | 112.16 (18) | C26—C21—N4 | 124.1 (2) |
| C6—N1—H11 | 108.7 (15) | C22—C21—N4 | 118.3 (2) |
| C2—N1—H11 | 105.7 (14) | C23—C22—C21 | 121.7 (2) |
| C6—N1—H12 | 108.2 (15) | C23—C22—H22 | 119.1 |
| C2—N1—H12 | 111.7 (16) | C21—C22—H22 | 119.1 |
| H11—N1—H12 | 110 (2) | C22—C23—C24 | 120.1 (2) |
| N1—C2—C3 | 110.34 (18) | C22—C23—H23 | 120.0 |
| N1—C2—H2A | 109.6 | C24—C23—H23 | 120.0 |
| C3—C2—H2A | 109.6 | C25—C24—O24 | 125.3 (2) |
| N1—C2—H2B | 109.6 | C25—C24—C23 | 119.4 (2) |
| C3—C2—H2B | 109.6 | O24—C24—C23 | 115.3 (2) |
| H2A—C2—H2B | 108.1 | C24—C25—C26 | 120.0 (2) |
| N4—C3—C2 | 110.20 (19) | C24—C25—H25 | 120.0 |
| N4—C3—H3A | 109.6 | C26—C25—H25 | 120.0 |
| C2—C3—H3A | 109.6 | C21—C26—C25 | 121.3 (2) |
| N4—C3—H3B | 109.6 | C21—C26—H26 | 119.4 |
| C2—C3—H3B | 109.6 | C25—C26—H26 | 119.4 |
| H3A—C3—H3B | 108.1 | C24—O24—C27 | 117.40 (19) |
| C21—N4—C5 | 117.85 (18) | O24—C27—H27A | 109.5 |
| C21—N4—C3 | 115.96 (17) | O24—C27—H27B | 109.5 |
| C5—N4—C3 | 110.60 (18) | H27A—C27—H27B | 109.5 |
| N4—C5—C6 | 109.54 (18) | O24—C27—H27C | 109.5 |
| N4—C5—H5A | 109.8 | H27A—C27—H27C | 109.5 |
| C6—C5—H5A | 109.8 | H27B—C27—H27C | 109.5 |
| N4—C5—H5B | 109.8 | O31—C31—C32 | 125.0 (2) |
| C6—C5—H5B | 109.8 | O31—C31—C33 | 116.47 (18) |
| H5A—C5—H5B | 108.2 | C32—C31—C33 | 118.55 (18) |
| N1—C6—C5 | 110.47 (18) | C31—C32—C33i | 123.2 (2) |
| N1—C6—H6A | 109.6 | C31—C32—Cl32 | 118.45 (17) |
| C5—C6—H6A | 109.6 | C33i—C32—Cl32 | 118.31 (16) |
| N1—C6—H6B | 109.6 | O33—C33—C32i | 125.8 (2) |
| C5—C6—H6B | 109.6 | O33—C33—C31 | 115.99 (19) |
| H6A—C6—H6B | 108.1 | C32i—C33—C31 | 118.20 (17) |
| C26—C21—C22 | 117.5 (2) | H41—O41—H42 | 102 (3) |
| C6—N1—C2—C3 | −53.5 (3) | C22—C23—C24—O24 | −178.8 (2) |
| N1—C2—C3—N4 | 56.5 (3) | O24—C24—C25—C26 | 176.9 (2) |
| C2—C3—N4—C21 | 161.1 (2) | C23—C24—C25—C26 | −2.8 (4) |
| C2—C3—N4—C5 | −61.3 (3) | C22—C21—C26—C25 | 0.1 (3) |
| C21—N4—C5—C6 | −162.3 (2) | N4—C21—C26—C25 | 177.1 (2) |
| C3—N4—C5—C6 | 61.0 (3) | C24—C25—C26—C21 | 2.3 (4) |
| C2—N1—C6—C5 | 53.7 (3) | C25—C24—O24—C27 | 12.2 (4) |
| N4—C5—C6—N1 | −56.7 (3) | C23—C24—O24—C27 | −168.1 (2) |
| C5—N4—C21—C26 | −8.7 (3) | O31—C31—C32—C33i | 176.5 (2) |
| C3—N4—C21—C26 | 125.7 (2) | C33—C31—C32—C33i | −2.4 (3) |
| C5—N4—C21—C22 | 168.2 (2) | O31—C31—C32—Cl32 | −1.3 (3) |
| C3—N4—C21—C22 | −57.4 (3) | C33—C31—C32—Cl32 | 179.81 (14) |
| C26—C21—C22—C23 | −2.0 (3) | O31—C31—C33—O33 | 2.8 (3) |
| N4—C21—C22—C23 | −179.1 (2) | C32—C31—C33—O33 | −178.16 (18) |
| C21—C22—C23—C24 | 1.5 (4) | O31—C31—C33—C32i | −176.76 (18) |
| C22—C23—C24—C25 | 0.9 (4) | C32—C31—C33—C32i | 2.2 (3) |
Symmetry code: (i) −x+1, −y+1, −z.
Bis(4-(4-methoxyphenyl)piperazin-1-ium) chloranilate(2-) dihydrate (XII). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O31 | 0.89 (3) | 1.96 (3) | 2.802 (3) | 157 (2) |
| N1—H11···O33 | 0.89 (3) | 2.29 (2) | 2.838 (3) | 119 (2) |
| N1—H12···O41 | 0.90 (2) | 1.92 (2) | 2.798 (3) | 168 (3) |
| O41—H41···O33ii | 0.84 (4) | 1.92 (4) | 2.738 (3) | 166 (3) |
| O41—H42···O24iii | 0.82 (3) | 2.49 (3) | 3.269 (3) | 160 (3) |
Symmetry codes: (ii) −x+1, −y+1, −z+1; (iii) x−1/2, −y+1/2, z−1/2.
Funding Statement
This work was funded by University Grants Commission grant .
References
- Acosta, L. M., Bahsas, A., Palma, A., Cobo, J., Hursthouse, M. B. & Glidewell, C. (2009). Acta Cryst. C65, o92–o96. [DOI] [PubMed]
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–S19.
- Al-Omary, F. A. M., Ghabbour, H. A., El-Emam, A. A., Chidan Kumar, C. S. & Fun, H.-K. (2014). Acta Cryst. E70, o245–o246. [DOI] [PMC free article] [PubMed]
- Arbo, M. D., Bastos, M. L. & Carmo, H. F. (2012). Drug Alcohol Depend. 122, 174–185. [DOI] [PubMed]
- Asif, M. (2015). Int. J. Adv. Sci. Res. 1, 5–11.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Brito, A., Moreira, L. K. S., Menegatti, R. & Costa, E. A. (2019). Fundam. Clin. Pharmacol. 33, 13–24. [DOI] [PubMed]
- Emsley, J. (1980). Chem. Soc. Rev. 9, 91–124.
- Etter, M. C. (1990). Acc. Chem. Res. 23, 120–126.
- Etter, M. C., MacDonald, J. C. & Bernstein, J. (1990). Acta Cryst. B46, 256–262. [DOI] [PubMed]
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998a). Acta Cryst. B54, 129–138.
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998b). Acta Cryst. B54, 139–150.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Flack, H. D. & Bernardinelli, G. (2000). J. Appl. Cryst. 33, 1143–1148.
- Glidewell, C., Low, J. N., Skakle, J. M. S. & Wardell, J. L. (2005). Acta Cryst. C61, o276–o280. [DOI] [PubMed]
- Gregson, R. M., Glidewell, C., Ferguson, G. & Lough, A. J. (2000). Acta Cryst. B56, 39–57. [DOI] [PubMed]
- Herschlag, D. & Pinney, M. M. (2018). Biochemistry, 57, 3338–3352. [DOI] [PubMed]
- Ishida, H. (2004a). Acta Cryst. E60, o974–o976.
- Ishida, H. (2004b). Acta Cryst. E60, o1900–o1901.
- Ishida, H. (2004c). Acta Cryst. E60, o2005–o2006.
- Ishida, H. (2004d). Acta Cryst. E60, o2506–o2508.
- Kaur, M., Jasinski, J. P., Yathirajan, H. S., Kavitha, C. N. & Glidewell, C. (2015). Acta Cryst. E71, 406–413. [DOI] [PMC free article] [PubMed]
- Kavitha, C. N., Jasinski, J. P., Anderson, B. J., Yathirajan, H. S. & Kaur, M. (2013). Acta Cryst. E69, o1671. [DOI] [PMC free article] [PubMed]
- Kavitha, C. N., Yathirajan, H. S., Kaur, M., Hosten, E. C., Betz, R. & Glidewell, C. (2014). Acta Cryst. C70, 805–811. [DOI] [PubMed]
- Kavitha, S. J., Panchanatheswaran, K., Low, J. N., Ferguson, G. & Glidewell, C. (2006). Acta Cryst. C62, o165–o169. [DOI] [PubMed]
- Kiran Kumar, H., Yathirajan, H. S., Sagar, B. K., Foro, S. & Glidewell, C. (2019). Acta Cryst. E75, 1253–1260. [DOI] [PMC free article] [PubMed]
- Lu, Y.-X. (2007). Acta Cryst. E63, o3611.
- Nagai, F., Nonaka, R. & Kamimura, K. S. H. (2007). Eur. J. Pharmacol. 559, 132–137. [DOI] [PubMed]
- Oxford Diffraction (2009). CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd, Abingdon, England.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sagar, B. K., Girisha, M., Yathirajan, H. S., Rathore, R. S. & Glidewell, C. (2017). Acta Cryst. E73, 1320–1325. [DOI] [PMC free article] [PubMed]
- Shaibah, M. A. E., Sagar, B. K., Yathirajan, H. S., Kumar, S. M. & Glidewell, C. (2017a). Acta Cryst. E73, 1513–1516. [DOI] [PMC free article] [PubMed]
- Shaibah, M. A. E., Yathirajan, H. S., Kumar, S. M., Byrappa, K. & Glidewell, C. (2017b). Acta Cryst. E73, 1488–1492. [DOI] [PMC free article] [PubMed]
- Shaibah, M. A. E., Yathirajan, H. S., Rathore, R. S., Furuya, T., Haraguchi, T., Akitsu, T. & Glidewell, C. (2019). Acta Cryst. E75, 292–298. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Sovago, I., Thomas, L. H., Adam, M. S., Capelli, S. C., Wilson, C. C. & Farrugia, L. J. (2016). CrystEngComm, 18, 5697–5709.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Staack, R. F. & Maurer, H. H. (2003). J. Chromatogr. B, 798, 333–342. [DOI] [PubMed]
- Staack, R. F. & Maurer, H. H. (2005). Curr. Drug Metab. 6, 259–274. [DOI] [PubMed]
- Staack, R. F., Theobald, D. S., Paul, D., Springer, D., Kraemer, T. & Maurer, H. H. (2004). Xenobiotica, 34, 179–192. [DOI] [PubMed]
- Verdonk, M. L., Voogd, J. W., Kanters, J. A., Kroon, J., den Besten, R., Brandsma, L., Leysen, D. & Kelder, J. (1997). Acta Cryst. B53, 976–983. [DOI] [PubMed]
- Wohlfarth, A., Weinmann, W. & Dresen, S. (2010). Anal. Bioanal. Chem. 396, 2403–2414. [DOI] [PubMed]
- Wood, P. A., Allen, F. H. & Pidcock, E. (2009). CrystEngComm, 11, 1563–1571.
- Zia-ur-Rehman, Tahir, M. N., Danish, M., Muhammad, N. & Ali, S. (2009). Acta Cryst. E65, o503. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II, III, IV, V, VI, VII, VIII, IX, X, XI, XII. DOI: 10.1107/S2056989019012702/ex2024sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019012702/ex2024Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989019012702/ex2024IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989019012702/ex2024IIIsup4.hkl
Structure factors: contains datablock(s) IV. DOI: 10.1107/S2056989019012702/ex2024IVsup5.hkl
Structure factors: contains datablock(s) V. DOI: 10.1107/S2056989019012702/ex2024Vsup6.hkl
Structure factors: contains datablock(s) VI. DOI: 10.1107/S2056989019012702/ex2024VIsup7.hkl
Structure factors: contains datablock(s) VII. DOI: 10.1107/S2056989019012702/ex2024VIIsup8.hkl
Structure factors: contains datablock(s) VIII. DOI: 10.1107/S2056989019012702/ex2024VIIIsup9.hkl
Structure factors: contains datablock(s) IX. DOI: 10.1107/S2056989019012702/ex2024IXsup10.hkl
Structure factors: contains datablock(s) X. DOI: 10.1107/S2056989019012702/ex2024Xsup11.hkl
Structure factors: contains datablock(s) XI. DOI: 10.1107/S2056989019012702/ex2024XIsup12.hkl
Structure factors: contains datablock(s) XII. DOI: 10.1107/S2056989019012702/ex2024XIIsup13.hkl
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024Isup14.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024IIsup15.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024IIIsup16.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024IVsup17.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024Vsup18.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024VIsup19.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024VIIsup20.cml
Supporting information file. DOI: 10.1107/S2056989019012702/ex2024XIsup21.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report























