The change of substituents viz. a chlorine atom in (I) replaced by a methyl group in (II) has not induced any differences in their respective crystal packing features, confirming the validity of the chlorine–methyl exchange rule.
Keywords: crystal structure, acenaphthene, supramolecular, Hirshfeld surface, chlorine-methyl exchange
Abstract
Instances of crystal structures that remain isomorphous in spite of some minor changes in their respective molecules, such as change in a substituent atom/group, can provide insights into the factors that govern crystal packing. In this context, an accurate description of the crystal structures of an isomorphous pair that differ from each other only by a chlorine–methyl substituent, viz. 5′′-(2-chlorobenzylidene)-4′-(2-chlorophenyl)-1′-methyldispiro[acenaphthene-1,2′-pyrrolidine-3′,3′′-piperidine]-2,4′′-dione, C34H28Cl2N2O2, (I), and its analogue 1′-methyl-5′′-(2-methylbenzylidene)-4′-(2-methylphenyl)dispiro[acenaphthene-1,2′-pyrrolidine-3′,3′′-piperidine]-2,4′′-dione, C36H34N2O2, (II), is presented. While there are two C—H⋯O weak intermolecular interactions present in both (I) and (II), the change of substituent from chlorine to methyl has given rise to an additional weak C—H⋯O intermolecular interaction that is relatively stronger than the other two. However, the presence of the stronger C—H⋯O interaction in (II) has not disrupted the validity of the chloro-methyl exchange rule. Details of the crystal structures and Hirshfeld analyses of the two compounds are presented.
Chemical context
The prediction of crystal structures has emerged as an exciting field involving researchers from diverse fields primarily because of its challenging complexity, which is considered analogous to that of the protein-folding problem. Attempts made in the field of crystal-structure prediction, its present status and the challenges ahead were discussed in detail in a recent article (Oganov, 2018 ▸). In this context, instances of crystal structures that remain isomorphous in spite of some minor changes in their respective molecules, such as a change in a substituent atom/group, are worthy of study as they might provide some insights regarding the subtle factors that govern the crystal packing.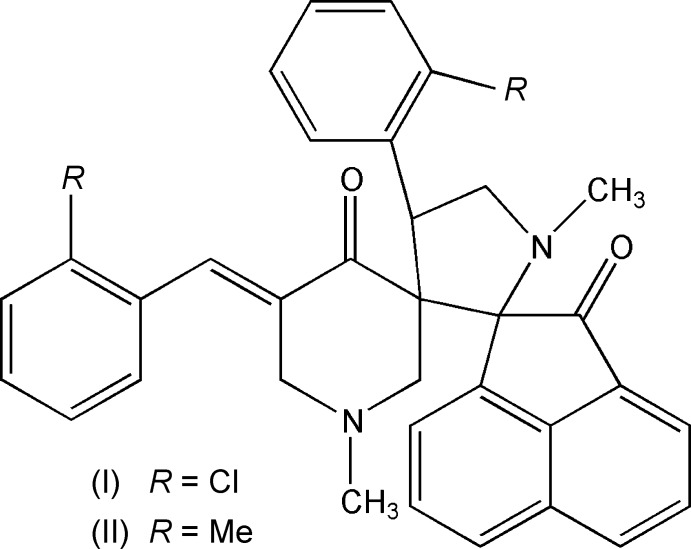
The title compounds (I) and (II) are good examples of crystal structures that obey the Cl–Me exchange rule, complying with the general conclusions arrived at in earlier studies (Jones et al., 1981 ▸; Gnanaguru et al., 1984 ▸; Desiraju & Sarma, 1986 ▸). In some recent studies carried out in our laboratory on molecules that showcase the validity of the Cl–Me exchange rule, it has been observed that factors such as the presence of disorder and minor conformational differences have not disturbed the tendency of molecules to remain as isomorphous pairs (Rajni Swamy, et al., 2013 ▸; Sribala et al., 2018 ▸). Interestingly, the validity of the Cl–Me exchange rule has also been observed in some regularly shaped planar molecules (Nath & Nangia, 2012 ▸).
From a pharmacological view point, the title compounds (I) and (II) are spiro compounds that consist of a methylpyrrole moiety with its 2- and 3- positions as spiro carbons linked, respectively, to acenapthene and methyl pyridinone ring systems. Each of these ring systems has a variety of associated biological properties. Studies on some 4-pyridone derivatives have shown them to be potent antimalarial agents (Bueno et al., 2011 ▸) and effective in the treatment and prophylaxis of the hepatitis B virus infection (Cheng et al., 2018 ▸). Acenaphthene is a pollutant known for its cytotoxicity (Jiang et al., 2019 ▸) but is also useful as a dye intermediate. Derivatives of acenaphthene are found to exhibit antitumor (El-Ayaan et al., 2007 ▸; Zhu et al., 2008 ▸) and fungistatic properties (McDavids & Daniels, 1951 ▸). Pyrrole derivatives belong to an important class of heterocycles owing to their potential applications as antimicrobial, antiviral, antimalarial, antitubercular, anti-inflammatory and anticancer agents (Gholap, 2016 ▸).
Structural commentary
The molecular structures of (I) and (II) (Figs. 1 ▸ and 2 ▸, respectively) differ from each other only by a chlorine atom in (I) being replaced by a methyl group in (II). This replacement has not induced any significant change in their unit-cell parameters, lattice type or space group. Similarly, there are no substantial changes in the torsion angles of the title compounds (see Tables 1 ▸ and 2 ▸), as (I) and (II) are isomorphous.
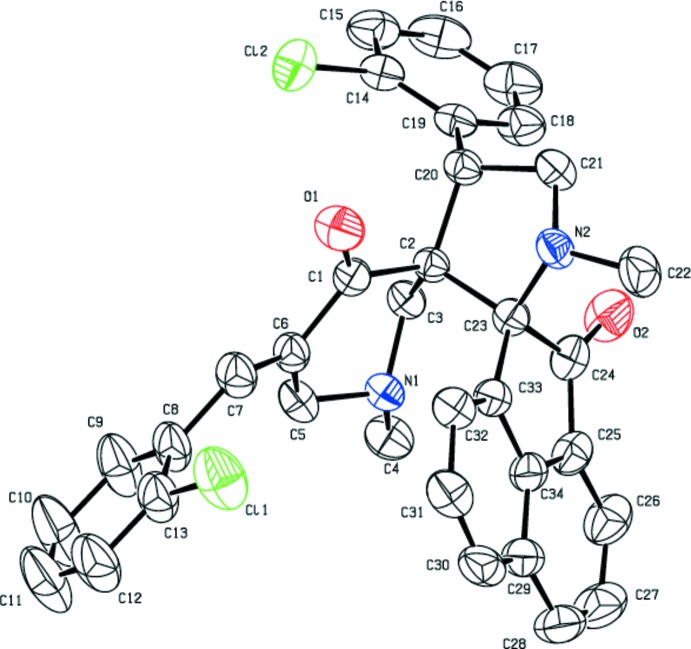
Figure 1.
Displacement ellipsoid plot drawn at 50% probability level for (I) showing the atom-labelling scheme. H atoms have been omitted for clarity.
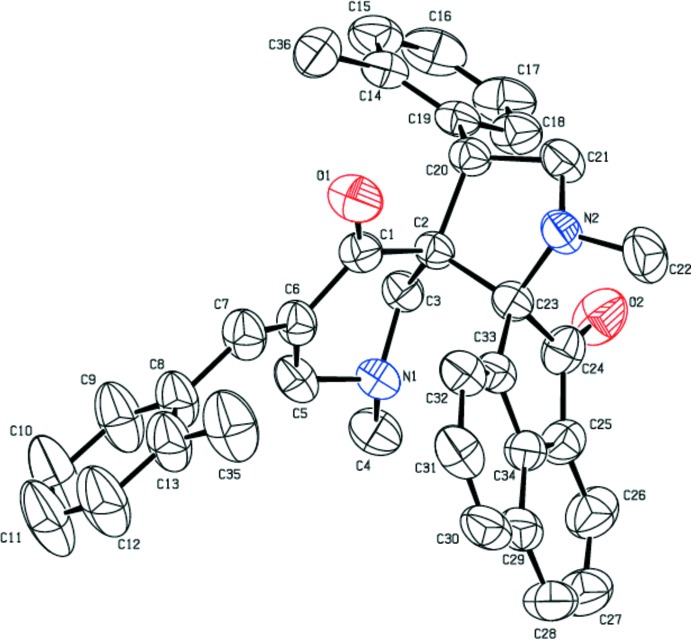
Figure 2.
Displacement ellipsoid plot drawn at 50% probability level for (II), showing the atom-labelling scheme. H atoms have been omitted for clarity.
Table 1. Selected torsion angles (°) for (I) .
| O1—C1—C6—C7 | −14.3 (2) | Cl2—C14—C19—C20 | −0.9 (2) |
| C7—C8—C13—Cl1 | −2.7 (3) | N2—C23—C24—O2 | −51.3 (2) |
Table 2. Selected torsion angles (°) for (II) .
| O1—C1—C6—C7 | −12.8 (3) | C36—C14—C19—C20 | −0.1 (3) |
| C7—C8—C13—C35 | −3.6 (4) | N2—C23—C24—O2 | −51.5 (2) |
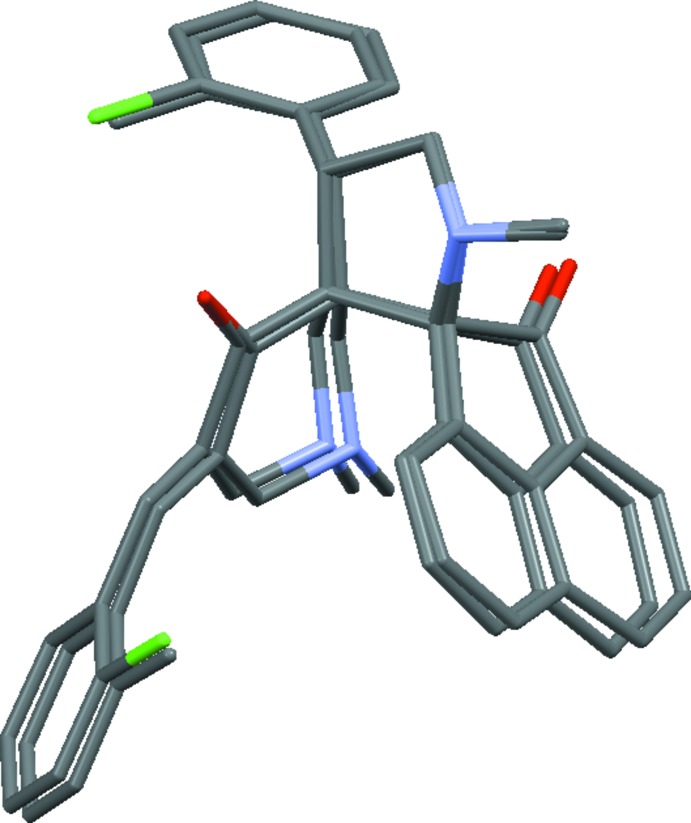
As expected, the conformational features of both compounds are nearly identical, as shown in a overlay diagram (Fig. 3 ▸). The five-membered pyrrolo ring (N2/C21/C20/C2/C23) adopts an envelope conformation on N2 with puckering parameters Q(2) = 0.4011 (2) Å and φ = 180.3733 (3)° for (I), which are comparable with the values of Q(2) = 0.4047 (2) Å and φ = 180.3444 (3)° for (II). In both of the structures, the six-membered pyridinone ring (N1/C3/C2/C1/C6/C5) adopts a screw-boat conformation with puckering parameters Q = 0.5572 (16) Å, θ = 138.9 (2)°, φ = 219.8 (3)° in (I) and Q = 0.5603 (17) Å, θ = 137.7 (2)°, φ = 219.6 (3)° in (II). The acenaphthene ring system is planar in both (I) and (II). However, the O2 atom deviates from the mean plane of the acenaphthene ring system by 0.289 (2) Å in (I) and 0.311 (2) Å in (II), with the r.m.s. deviation of the fitted atoms being 0.043 and 0.044, respectively. This deviation is presumably due to the fact that the O2 atom is involved in two weak C—H⋯O interactions that are characteristic of the molecular interaction patterns of both (I) and (II).
Figure 3.
An overlay diagram depicting the superimposition of molecule (I) and (II) showing no differences in the conformations.
The dihedral angle between the mean planes of the two chlorophenyl groups in (I) is 67.66 (9)°, similarly the corresponding angle between the two methylphenyl groups in (II) is 66.78 (11)°. The dihedral angles between the acenaphthene ring system and the chlorophenyl groups are 69.1 (1) and 49.4 (1)°, respectively. The corresponding angles in the methyl-substituted analogue are 72.3 (1) and 47.8 (1)°, respectively. Thus, it is clear that the minor differences observed in the conformation of the molecules are insufficient to disrupt the tendency of these molecules to remain isomorphous.
Supramolecular features
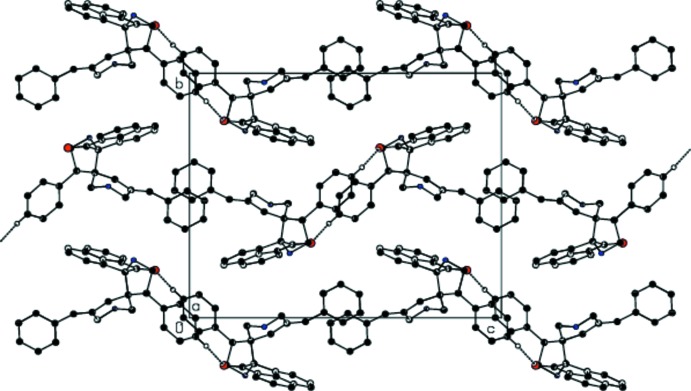
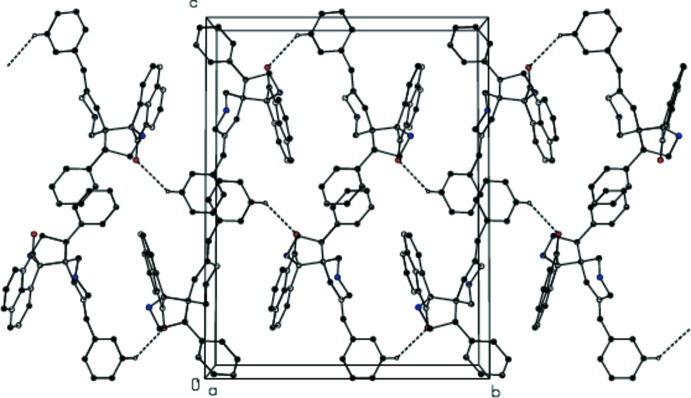
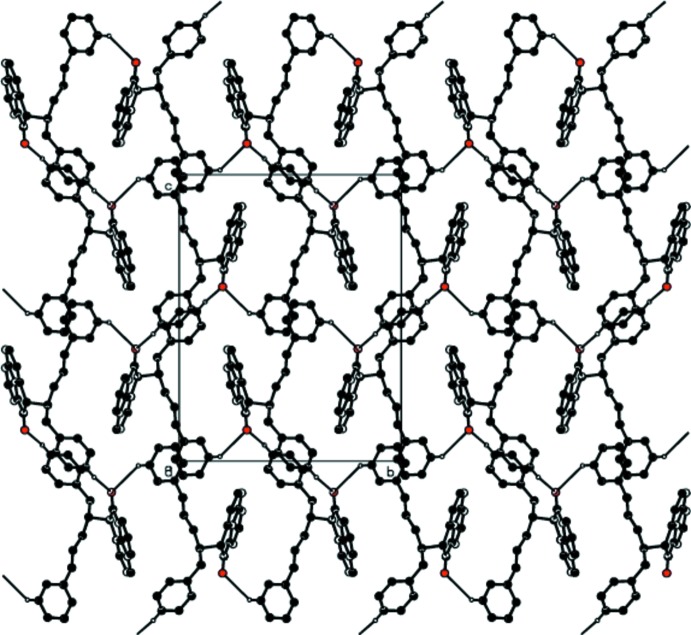
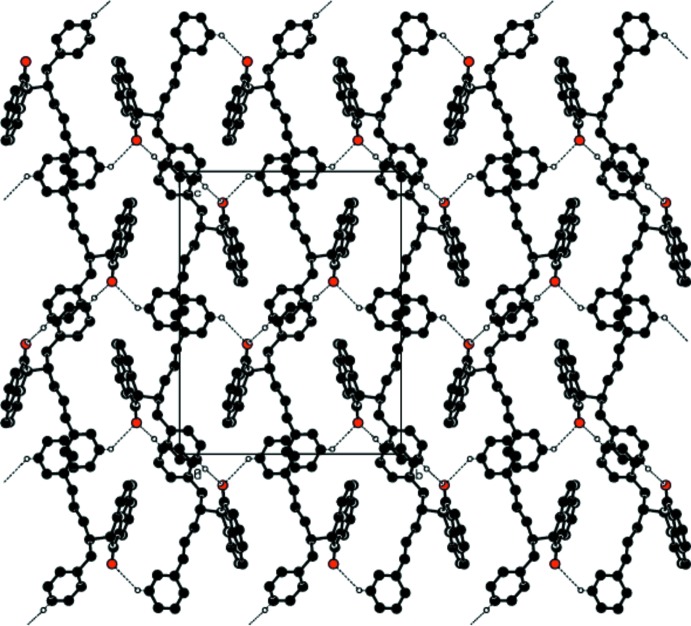
There are no classical hydrogen bonds in the structures of either (I) or (II). However, in both structures two weak C—H⋯O-type intermolecular interactions, viz. C10—H10⋯O2 and C16—H16⋯O2, which are identical in nature and characteristic of similar fundamental molecular interaction patterns are present (Tables 3 ▸ and 4 ▸). The C16—H16⋯O2 interaction occurs between centrosymmetric pairs (Fig. 4 ▸), leading to the formation of 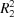 (20) graph-set motifs along the b-axis direction in both (I) and (II). Similarly, in both (I) and (II) the C10—H10⋯O2 interaction links glide-related molecules along the b-axis direction (Fig. 5 ▸). The molecular aggregation pattern may be visualized as being composed of these two characteristic weak interactions in such a manner that centrosymmetric dimeric pairs are linked through glide-related chains of molecules, forming a two-dimensional layer parallel to the bc plane in both structures, as shown in Figs. 6 ▸ and 7 ▸, respectively.
(20) graph-set motifs along the b-axis direction in both (I) and (II). Similarly, in both (I) and (II) the C10—H10⋯O2 interaction links glide-related molecules along the b-axis direction (Fig. 5 ▸). The molecular aggregation pattern may be visualized as being composed of these two characteristic weak interactions in such a manner that centrosymmetric dimeric pairs are linked through glide-related chains of molecules, forming a two-dimensional layer parallel to the bc plane in both structures, as shown in Figs. 6 ▸ and 7 ▸, respectively.
Table 3. Hydrogen-bond geometry (Å, °) for (I) .
Cg1 is the centroid of the C25–C29/C34 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C10—H10⋯O2i | 0.93 | 2.74 | 3.492 (3) | 139 |
| C16—H16⋯O2ii | 0.93 | 2.76 | 3.481 (3) | 135 |
| C5—H5B⋯Cg1i | 0.97 | 2.99 | 3.9466 (19) | 168 |
Symmetry codes: (i) 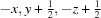 ; (ii)
; (ii) 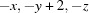 .
.
Table 4. Hydrogen-bond geometry (Å, °) for (II) .
Cg2 is the centroid of the C8–C13 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C36—H36A⋯O1 | 0.96 | 2.66 | 3.586 (3) | 161 |
| C10—H10⋯O2i | 0.93 | 2.77 | 3.529 (3) | 140 |
| C16—H16⋯O2ii | 0.93 | 2.79 | 3.530 (3) | 137 |
| C35—H35F⋯Cg2iii | 0.96 | 2.94 | 3.805 (4) | 151 |
Symmetry codes: (i) 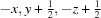 ; (ii)
; (ii) 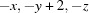 ; (iii)
; (iii) 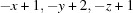 .
.
Figure 4.
Perspective view along the a axis showing the weak C16—H16⋯O2 intermolecular interactions between centrosymmetric pairs of molecules in (I) and (II). Non-participating H atoms, methyl C atoms and Cl atoms have been omitted for clarity.
Figure 5.
Perspective view along the a axis showing the weak C10—H10⋯O2 intermolecular interactions between glide-related molecules in (I) and (II). Non-participating H atoms, methyl C atoms and Cl atoms have been omitted for clarity.
Figure 6.
A view of the crystal structure of (I) showing the formation of 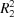 (20) graph-set motifs leading to the formation of layers formed parallel to the bc plane. Dashed lines indicate weak C—H⋯O intermolecular interactions. H atoms not involved in the interactions have been omitted for clarity.
(20) graph-set motifs leading to the formation of layers formed parallel to the bc plane. Dashed lines indicate weak C—H⋯O intermolecular interactions. H atoms not involved in the interactions have been omitted for clarity.
Figure 7.
A view of the crystal structure of (II) showing the 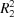 (20) graph-set motifs. Dashed lines indicate weak C—H⋯O intermolecular interactions. H atoms not interactions have been omitted for clarity.
(20) graph-set motifs. Dashed lines indicate weak C—H⋯O intermolecular interactions. H atoms not interactions have been omitted for clarity.
In (II), an additional intermolecular interaction is observed, viz. C36—H36A⋯O1, that is stronger than the two characteristic weak intermolecular interactions and involves the replaced substituent methyl group (C36—H36A) as a donor and the piperidinone O1 atom as an acceptor (see Table 4 ▸). It may be concluded that the presence of this additional C—H⋯O interaction in (II) has not disrupted the validity of the chloro–methyl exchange rule.
In addition, a weak C—H⋯π interaction involving different donor groups and acceptor π-ring systems is present in both (I) and (II). The C5—H5B⋯π interaction observed in (I) is between the C5 atom of the methylpiperidinone ring as a donor and the C25—C29/C34 ring of the acenaphthenone system as an acceptor. Interestingly, a geometrically identical weak π–π interaction about an inversion centre is observed with centroid–centroid Cg3⋯Cg3(1 − x, 2 − y, −z) distances of 3.7459 (2) Å in (I) and 3.8351 (2) Å in (II) with respective slippages of 1.250 and 1.367 Å where Cg3 is the centroid of the C14–C19 ring. The shortest Cl⋯Cl distance observed [Cl1 ⋯Cl1(−x + 1, −y + 2, −z + 1)] is 4.088 (1) Å and bears no structural significance.
Database survey
A thorough search in the Cambridge Structural Database (CSD Version 5.39, update Nov 2017; Groom et al., 2016 ▸) using the main skeleton of the title compounds (having 3D coordinates with no disorder, no ions and no other errors with R factors less than 0.05) gave only three hits: 5′′-(4-chlorobenzylidene)-4′-(4-chlorophenyl)-1′,1′′-dimethyl-2H,4′′H-dispiro[acenaphthylene-1,2′-pyrrolidine-3′,3′′-piperidine]-2,4′′-dione (YIRKUG; Pandiarajan et al., 2008 ▸), 5′′-benzylidene-1′,1′′-dimethyl-4′-phenyl-acenapthene-2-spiro-2′-pyrrolidine-3′-spiro-3′′-piperidine-1,4′′-dione (MAJHEL; Aravindan et al., 2004 ▸) and 1-methyl-4-(4-methylphenyl)pyrrolo-(spiro[2.2′′]acenaphthene-1′′-one)-spiro-[3.3′]-(spiro-[5′.5′′′]-3′′′-(4-chlorophenyl)-4′′′-(4-methylphenyl)-isoxazoline)-1′-methyltetrahydro-4′(1H)-pyridinone (XUQFOF; Kumar, et al., 2009 ▸).
Hirshfeld surface analysis
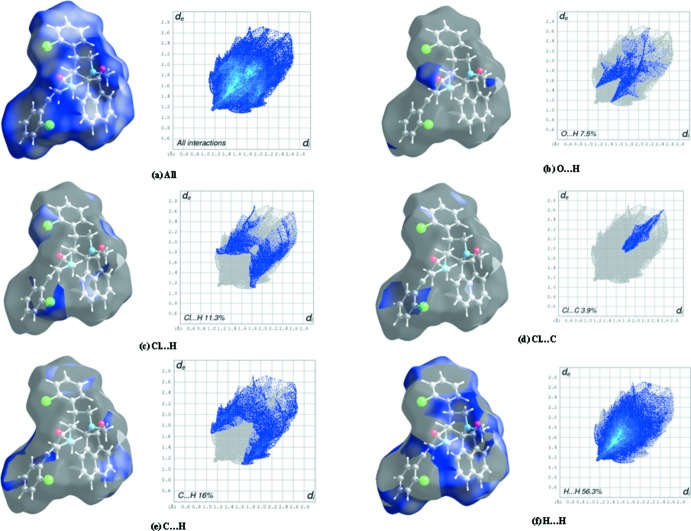
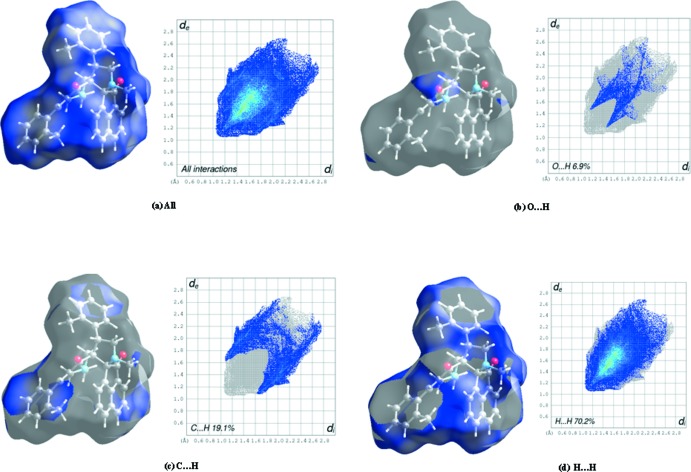
Hirshfeld surface (HS) analysis was used to investigate and visualize the weak intermolecular interactions influential in the packing of the molecules in the crystal. The visual representation of molecular interactions on this isosurface is determined using two parameters, viz. d i and d e , which represent the distances from a given point on the surface to the nearest atom inside and outside the surface, respectively. The normalized contact distance, d norm is based on the values of d i and d e.
In the present work, the Hirshfeld surfaces (Spackman & Jayatilaka, 2009 ▸) and the associated two-dimensional fingerprint plots for title compounds (I) and (II) were generated using CrystalExplorer3.0 (Wolff et al., 2012 ▸). The Hirshfeld surfaces mapped over d norm together with decomposed finger print plots (McKinnon et al., 2007 ▸; Tan et al., 2019 ▸) for (I) and (II) are presented in Figs. 8 ▸ and 9 ▸, respectively. Being isomorphic and isostructural in nature, both (I) and (II) display similar C—H⋯O intermolecular interactions. The combined O⋯H and H⋯O interactions appear symmetrically as distinct spikes at the bottom of the fingerprint plot and contribute 7.5 and 6.9%, respectively, of the total surface in compounds (I) and (II).
Figure 8.
Hirshfeld surface of (I) mapped over shape-index and d norm and decomposed fingerprint plots of the dominant interactions.
Figure 9.
Hirshfeld surface of (II) mapped over shape-index and d norm and decomposed fingerprint plots of the dominant interactions.
The symmetrical internal wing-like projections correspond to C⋯H/H⋯C contacts, which account for 16% of the HS in (I) and 19.1% in (II). The dominant contribution is from the H⋯H contacts [56.3% in (I) and 70.2% in (II)], as shown by the area occupied between the spikes. Such prominent differences may be accounted for by the presence of a Cl⋯H/H⋯Cl contact in (I) (11.3% contribution) and its absence in (II).
Synthesis and crystallization
For (I), a mixture of 1-methyl-3,5-bis[(E)-2-chlorophenylmethylidene] tetrahydro-4(1H)- pyridinone (1 mmol), acenaphthenequinone (1 mmol) and sarcosine (1 mmol) was dissolved in methanol (15 mL) and refluxed for 30 min. After completion of the reaction, as evident from TLC, the mixture was poured into water (50 mL) and the precipitated solid was filtered and washed with water (100 mL) to obtain pure (I) as a yellow solid, (0.31 g, 98%), mp 448–449 K, Rf (petroleum ether/EtOAc, 4:1) 0.40. Suitable crystals for single-crystal X-ray studies were obtained by recrystallization of the product from ethanol.
A similar procedure for (II) was adopted by dissolving a mixture of 1-methyl-3,5-bis[(E)-2-methylphenylmethylidene] tetrahydro-4(1H)-pyridinone (1 mmol), acenaphthenequinone (1 mmol) and sarcosine (1 mmol) in methanol (15 mL) to yield yellow crystals.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 5 ▸. C-bound H atoms were included in calculated positions and treated as riding, with C—H = 0.95–1.00 Å and U iso(H) = 1.5U eq(C) for methyl H atoms or 1.2Ueq(C) otherwise. The H atoms of the methyl atoms C35 and C36 in (II) were refined as idealized and disordered over two positions since significant residual electron densities were noticed between the three hydrogen atoms of the respective methyl C atoms. The introduction of a disordered model for these two methyl groups had appreciable impact on the final structural parameters.
Table 5. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C34H28Cl2N2O2 | C36H34N2O2 |
| M r | 567.48 | 526.65 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 8.6710 (4), 15.6756 (7), 20.2284 (9) | 8.7507 (5), 15.9089 (8), 20.2879 (10) |
| β (°) | 93.036 (2) | 92.935 (2) |
| V (Å3) | 2745.6 (2) | 2820.7 (3) |
| Z | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.27 | 0.08 |
| Crystal size (mm) | 0.31 × 0.22 × 0.19 | 0.32 × 0.24 × 0.18 |
| Data collection | ||
| Diffractometer | Bruker SMART APEXII CCD | Bruker SMART APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Bruker, 2001 ▸) | Multi-scan (SADABS; Bruker, 2001 ▸) |
| T min, T max | 0.771, 1.000 | 0.816, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 33739, 7274, 5025 | 28946, 5339, 3349 |
| R int | 0.026 | 0.040 |
| (sin θ/λ)max (Å−1) | 0.683 | 0.610 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.044, 0.122, 1.02 | 0.045, 0.121, 1.02 |
| No. of reflections | 7274 | 5339 |
| No. of parameters | 361 | 362 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.33, −0.46 | 0.17, −0.15 |
Supplementary Material
Crystal structure: contains datablock(s) I, II. DOI: 10.1107/S2056989019012428/jj2215sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019012428/jj2215Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989019012428/jj2215IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the Sophisticated Analytical Instrumental Facility (SAIF), Indian Institute of Technology, Chennai, for the data collection and the Management of Thiagarajar College, Madurai, for financial support in establishing the Cambridge Structural Database in the Department of Physics.
supplementary crystallographic information
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Crystal data
| C34H28Cl2N2O2 | F(000) = 1184 |
| Mr = 567.48 | Dx = 1.373 Mg m−3Dm = 1.37 Mg m−3Dm measured by floatation method |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.6710 (4) Å | Cell parameters from 7274 reflections |
| b = 15.6756 (7) Å | θ = 4.8–57.2° |
| c = 20.2284 (9) Å | µ = 0.27 mm−1 |
| β = 93.036 (2)° | T = 293 K |
| V = 2745.6 (2) Å3 | Block, yellow |
| Z = 4 | 0.31 × 0.22 × 0.19 mm |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Data collection
| Bruker SMART APEXII CCD diffractometer | 5025 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.026 |
| φ and ω scans | θmax = 29.1°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −11→11 |
| Tmin = 0.771, Tmax = 1.000 | k = −21→21 |
| 33739 measured reflections | l = −27→27 |
| 7274 independent reflections |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.044 | H-atom parameters constrained |
| wR(F2) = 0.122 | w = 1/[σ2(Fo2) + (0.0458P)2 + 1.2817P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max = 0.001 |
| 7274 reflections | Δρmax = 0.33 e Å−3 |
| 361 parameters | Δρmin = −0.46 e Å−3 |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.38200 (9) | 0.88710 (4) | 0.49943 (3) | 0.0749 (2) | |
| Cl2 | 0.53437 (6) | 1.07386 (3) | 0.14315 (3) | 0.05500 (14) | |
| O1 | 0.48210 (14) | 0.92211 (9) | 0.26952 (6) | 0.0509 (3) | |
| O2 | 0.01681 (16) | 0.80480 (10) | 0.10859 (7) | 0.0587 (4) | |
| N1 | 0.02281 (15) | 0.96435 (9) | 0.23726 (6) | 0.0345 (3) | |
| N2 | 0.32804 (16) | 0.76391 (9) | 0.17821 (7) | 0.0379 (3) | |
| C1 | 0.34543 (18) | 0.93585 (10) | 0.26001 (8) | 0.0339 (3) | |
| C2 | 0.25834 (17) | 0.90781 (10) | 0.19571 (7) | 0.0298 (3) | |
| C3 | 0.12354 (18) | 0.96919 (10) | 0.18237 (8) | 0.0330 (3) | |
| H3A | 0.161841 | 1.026946 | 0.177987 | 0.040* | |
| H3B | 0.066923 | 0.953767 | 0.141473 | 0.040* | |
| C4 | −0.1310 (2) | 0.99694 (13) | 0.21990 (10) | 0.0486 (4) | |
| H4A | −0.175090 | 0.965861 | 0.182623 | 0.073* | |
| H4B | −0.194961 | 0.990171 | 0.256864 | 0.073* | |
| H4C | −0.124570 | 1.056321 | 0.208758 | 0.073* | |
| C5 | 0.0931 (2) | 1.00991 (11) | 0.29351 (8) | 0.0412 (4) | |
| H5A | 0.029809 | 1.002742 | 0.331249 | 0.049* | |
| H5B | 0.096552 | 1.070296 | 0.283217 | 0.049* | |
| C6 | 0.2542 (2) | 0.97887 (10) | 0.31150 (8) | 0.0367 (3) | |
| C7 | 0.3209 (2) | 0.98384 (12) | 0.37223 (9) | 0.0453 (4) | |
| H7 | 0.419755 | 0.961011 | 0.377724 | 0.054* | |
| C8 | 0.2571 (2) | 1.02102 (12) | 0.43141 (9) | 0.0498 (5) | |
| C9 | 0.1760 (3) | 1.09734 (15) | 0.42992 (11) | 0.0700 (7) | |
| H9 | 0.157858 | 1.125378 | 0.389746 | 0.084* | |
| C10 | 0.1219 (4) | 1.13245 (17) | 0.48645 (13) | 0.0900 (9) | |
| H10 | 0.067945 | 1.183728 | 0.484082 | 0.108* | |
| C11 | 0.1467 (4) | 1.09265 (18) | 0.54595 (12) | 0.0925 (10) | |
| H11 | 0.109239 | 1.116503 | 0.584021 | 0.111* | |
| C12 | 0.2269 (3) | 1.01755 (16) | 0.54950 (11) | 0.0761 (7) | |
| H12 | 0.244309 | 0.990238 | 0.590018 | 0.091* | |
| C13 | 0.2817 (3) | 0.98246 (13) | 0.49292 (9) | 0.0546 (5) | |
| C14 | 0.40575 (19) | 1.04221 (11) | 0.07931 (8) | 0.0395 (4) | |
| C15 | 0.3746 (2) | 1.09964 (13) | 0.02837 (10) | 0.0524 (5) | |
| H15 | 0.424605 | 1.152112 | 0.028284 | 0.063* | |
| C16 | 0.2695 (3) | 1.07856 (15) | −0.02190 (10) | 0.0604 (6) | |
| H16 | 0.247308 | 1.116892 | −0.056185 | 0.073* | |
| C17 | 0.1973 (2) | 1.00115 (16) | −0.02157 (10) | 0.0589 (6) | |
| H17 | 0.125775 | 0.986900 | −0.055648 | 0.071* | |
| C18 | 0.2301 (2) | 0.94401 (13) | 0.02908 (9) | 0.0475 (4) | |
| H18 | 0.180273 | 0.891469 | 0.028376 | 0.057* | |
| C19 | 0.33571 (18) | 0.96284 (11) | 0.08114 (8) | 0.0355 (3) | |
| C20 | 0.37056 (18) | 0.90127 (10) | 0.13774 (7) | 0.0333 (3) | |
| H20 | 0.474518 | 0.914578 | 0.156241 | 0.040* | |
| C21 | 0.3698 (2) | 0.80722 (11) | 0.11831 (9) | 0.0418 (4) | |
| H21A | 0.294485 | 0.796255 | 0.082138 | 0.050* | |
| H21B | 0.470866 | 0.789228 | 0.105323 | 0.050* | |
| C22 | 0.3020 (3) | 0.67327 (12) | 0.17056 (11) | 0.0576 (5) | |
| H22A | 0.275049 | 0.649399 | 0.212106 | 0.086* | |
| H22B | 0.219323 | 0.663728 | 0.137946 | 0.086* | |
| H22C | 0.394350 | 0.646490 | 0.156564 | 0.086* | |
| C23 | 0.20239 (17) | 0.81269 (10) | 0.20545 (8) | 0.0322 (3) | |
| C24 | 0.0411 (2) | 0.79856 (11) | 0.16774 (9) | 0.0410 (4) | |
| C25 | −0.06990 (19) | 0.77207 (11) | 0.21615 (10) | 0.0429 (4) | |
| C26 | −0.2263 (2) | 0.75748 (13) | 0.21054 (12) | 0.0572 (5) | |
| H26 | −0.281931 | 0.764906 | 0.170370 | 0.069* | |
| C27 | −0.2990 (2) | 0.73129 (15) | 0.26675 (14) | 0.0684 (7) | |
| H27 | −0.405325 | 0.722892 | 0.264134 | 0.082* | |
| C28 | −0.2189 (3) | 0.71761 (13) | 0.32545 (13) | 0.0628 (6) | |
| H28 | −0.271567 | 0.698729 | 0.361561 | 0.075* | |
| C29 | −0.0578 (2) | 0.73134 (11) | 0.33300 (10) | 0.0461 (4) | |
| C30 | 0.0409 (3) | 0.71831 (12) | 0.38919 (10) | 0.0526 (5) | |
| H30 | 0.001188 | 0.698030 | 0.428006 | 0.063* | |
| C31 | 0.1947 (2) | 0.73521 (12) | 0.38725 (9) | 0.0496 (5) | |
| H31 | 0.258210 | 0.724857 | 0.424875 | 0.060* | |
| C32 | 0.2616 (2) | 0.76802 (11) | 0.32996 (9) | 0.0410 (4) | |
| H32 | 0.366809 | 0.779746 | 0.330281 | 0.049* | |
| C33 | 0.16946 (18) | 0.78193 (10) | 0.27480 (8) | 0.0339 (3) | |
| C34 | 0.01184 (19) | 0.76132 (10) | 0.27645 (9) | 0.0379 (4) |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.1164 (5) | 0.0651 (4) | 0.0426 (3) | 0.0162 (3) | −0.0028 (3) | 0.0052 (2) |
| Cl2 | 0.0520 (3) | 0.0486 (3) | 0.0634 (3) | −0.0065 (2) | −0.0062 (2) | −0.0027 (2) |
| O1 | 0.0375 (6) | 0.0762 (9) | 0.0382 (7) | 0.0023 (6) | −0.0059 (5) | −0.0024 (6) |
| O2 | 0.0606 (8) | 0.0712 (10) | 0.0422 (8) | −0.0164 (7) | −0.0156 (6) | 0.0065 (7) |
| N1 | 0.0344 (7) | 0.0376 (7) | 0.0316 (7) | 0.0049 (5) | 0.0030 (5) | −0.0007 (6) |
| N2 | 0.0460 (8) | 0.0329 (7) | 0.0352 (8) | 0.0044 (6) | 0.0067 (6) | −0.0027 (6) |
| C1 | 0.0384 (8) | 0.0354 (8) | 0.0275 (8) | −0.0033 (6) | −0.0013 (6) | 0.0035 (6) |
| C2 | 0.0309 (7) | 0.0335 (8) | 0.0252 (7) | 0.0007 (6) | 0.0013 (6) | 0.0008 (6) |
| C3 | 0.0365 (8) | 0.0355 (8) | 0.0268 (8) | 0.0039 (6) | −0.0005 (6) | 0.0016 (6) |
| C4 | 0.0411 (9) | 0.0537 (11) | 0.0511 (11) | 0.0131 (8) | 0.0035 (8) | 0.0005 (9) |
| C5 | 0.0517 (10) | 0.0408 (9) | 0.0315 (9) | 0.0077 (7) | 0.0039 (7) | −0.0038 (7) |
| C6 | 0.0465 (9) | 0.0344 (8) | 0.0291 (8) | −0.0030 (7) | 0.0010 (7) | −0.0008 (6) |
| C7 | 0.0571 (11) | 0.0466 (10) | 0.0318 (9) | −0.0019 (8) | −0.0023 (8) | −0.0012 (7) |
| C8 | 0.0732 (13) | 0.0468 (10) | 0.0288 (9) | −0.0039 (9) | −0.0025 (8) | −0.0063 (8) |
| C9 | 0.117 (2) | 0.0542 (13) | 0.0381 (11) | 0.0107 (13) | −0.0047 (12) | −0.0076 (9) |
| C10 | 0.153 (3) | 0.0611 (15) | 0.0555 (15) | 0.0301 (16) | 0.0057 (16) | −0.0159 (12) |
| C11 | 0.163 (3) | 0.0710 (17) | 0.0448 (14) | 0.0170 (18) | 0.0209 (16) | −0.0168 (12) |
| C12 | 0.133 (2) | 0.0629 (14) | 0.0337 (11) | 0.0001 (15) | 0.0128 (13) | −0.0019 (10) |
| C13 | 0.0821 (14) | 0.0460 (11) | 0.0356 (10) | −0.0045 (10) | 0.0017 (9) | −0.0034 (8) |
| C14 | 0.0358 (8) | 0.0465 (9) | 0.0369 (9) | 0.0042 (7) | 0.0078 (7) | 0.0023 (7) |
| C15 | 0.0570 (11) | 0.0509 (11) | 0.0507 (12) | 0.0045 (9) | 0.0165 (9) | 0.0135 (9) |
| C16 | 0.0635 (13) | 0.0777 (15) | 0.0409 (11) | 0.0173 (11) | 0.0104 (9) | 0.0262 (10) |
| C17 | 0.0525 (11) | 0.0899 (17) | 0.0337 (10) | 0.0036 (11) | −0.0023 (8) | 0.0126 (10) |
| C18 | 0.0463 (10) | 0.0642 (12) | 0.0319 (9) | −0.0031 (8) | 0.0010 (7) | 0.0051 (8) |
| C19 | 0.0337 (8) | 0.0459 (9) | 0.0275 (8) | 0.0032 (7) | 0.0065 (6) | 0.0027 (7) |
| C20 | 0.0324 (7) | 0.0403 (8) | 0.0273 (8) | 0.0011 (6) | 0.0013 (6) | 0.0002 (6) |
| C21 | 0.0485 (9) | 0.0432 (9) | 0.0343 (9) | 0.0054 (7) | 0.0075 (7) | −0.0040 (7) |
| C22 | 0.0804 (14) | 0.0358 (10) | 0.0577 (13) | 0.0030 (9) | 0.0133 (11) | −0.0059 (9) |
| C23 | 0.0335 (7) | 0.0336 (8) | 0.0292 (8) | 0.0001 (6) | −0.0006 (6) | 0.0014 (6) |
| C24 | 0.0413 (9) | 0.0364 (9) | 0.0443 (10) | −0.0033 (7) | −0.0059 (7) | 0.0012 (7) |
| C25 | 0.0367 (8) | 0.0342 (9) | 0.0574 (11) | −0.0009 (7) | −0.0002 (8) | 0.0006 (8) |
| C26 | 0.0391 (9) | 0.0519 (11) | 0.0798 (15) | −0.0016 (8) | −0.0034 (10) | −0.0004 (10) |
| C27 | 0.0395 (10) | 0.0635 (14) | 0.104 (2) | −0.0058 (9) | 0.0173 (12) | −0.0037 (13) |
| C28 | 0.0549 (12) | 0.0518 (12) | 0.0851 (17) | −0.0068 (9) | 0.0344 (12) | −0.0017 (11) |
| C29 | 0.0539 (10) | 0.0309 (8) | 0.0555 (12) | 0.0016 (7) | 0.0216 (9) | 0.0003 (8) |
| C30 | 0.0744 (13) | 0.0377 (9) | 0.0482 (11) | 0.0050 (9) | 0.0253 (10) | 0.0054 (8) |
| C31 | 0.0711 (13) | 0.0406 (10) | 0.0374 (10) | 0.0110 (9) | 0.0051 (9) | 0.0074 (8) |
| C32 | 0.0461 (9) | 0.0382 (9) | 0.0389 (9) | 0.0067 (7) | 0.0033 (7) | 0.0048 (7) |
| C33 | 0.0377 (8) | 0.0288 (7) | 0.0355 (9) | 0.0029 (6) | 0.0049 (6) | 0.0026 (6) |
| C34 | 0.0403 (8) | 0.0261 (7) | 0.0479 (10) | 0.0012 (6) | 0.0090 (7) | 0.0000 (7) |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Geometric parameters (Å, º)
| Cl1—C13 | 1.731 (2) | C14—C19 | 1.386 (2) |
| Cl2—C14 | 1.7336 (18) | C15—C16 | 1.369 (3) |
| O1—C1 | 1.2098 (19) | C15—H15 | 0.9300 |
| O2—C24 | 1.208 (2) | C16—C17 | 1.365 (3) |
| N1—C5 | 1.450 (2) | C16—H16 | 0.9300 |
| N1—C3 | 1.4506 (19) | C17—C18 | 1.379 (3) |
| N1—C4 | 1.454 (2) | C17—H17 | 0.9300 |
| N2—C22 | 1.446 (2) | C18—C19 | 1.390 (2) |
| N2—C21 | 1.451 (2) | C18—H18 | 0.9300 |
| N2—C23 | 1.462 (2) | C19—C20 | 1.516 (2) |
| C1—C6 | 1.501 (2) | C20—C21 | 1.526 (2) |
| C1—C2 | 1.533 (2) | C20—H20 | 0.9800 |
| C2—C3 | 1.527 (2) | C21—H21A | 0.9700 |
| C2—C20 | 1.566 (2) | C21—H21B | 0.9700 |
| C2—C23 | 1.584 (2) | C22—H22A | 0.9600 |
| C3—H3A | 0.9700 | C22—H22B | 0.9600 |
| C3—H3B | 0.9700 | C22—H22C | 0.9600 |
| C4—H4A | 0.9600 | C23—C33 | 1.525 (2) |
| C4—H4B | 0.9600 | C23—C24 | 1.573 (2) |
| C4—H4C | 0.9600 | C24—C25 | 1.469 (3) |
| C5—C6 | 1.505 (2) | C25—C26 | 1.374 (2) |
| C5—H5A | 0.9700 | C25—C34 | 1.388 (3) |
| C5—H5B | 0.9700 | C26—C27 | 1.391 (3) |
| C6—C7 | 1.332 (2) | C26—H26 | 0.9300 |
| C7—C8 | 1.466 (3) | C27—C28 | 1.360 (3) |
| C7—H7 | 0.9300 | C27—H27 | 0.9300 |
| C8—C9 | 1.387 (3) | C28—C29 | 1.414 (3) |
| C8—C13 | 1.390 (3) | C28—H28 | 0.9300 |
| C9—C10 | 1.374 (3) | C29—C30 | 1.401 (3) |
| C9—H9 | 0.9300 | C29—C34 | 1.402 (2) |
| C10—C11 | 1.363 (4) | C30—C31 | 1.362 (3) |
| C10—H10 | 0.9300 | C30—H30 | 0.9300 |
| C11—C12 | 1.367 (4) | C31—C32 | 1.420 (2) |
| C11—H11 | 0.9300 | C31—H31 | 0.9300 |
| C12—C13 | 1.377 (3) | C32—C33 | 1.355 (2) |
| C12—H12 | 0.9300 | C32—H32 | 0.9300 |
| C14—C15 | 1.384 (3) | C33—C34 | 1.406 (2) |
| C5—N1—C3 | 109.25 (13) | C16—C17—C18 | 120.3 (2) |
| C5—N1—C4 | 111.01 (13) | C16—C17—H17 | 119.9 |
| C3—N1—C4 | 112.25 (13) | C18—C17—H17 | 119.9 |
| C22—N2—C21 | 114.51 (14) | C17—C18—C19 | 121.78 (19) |
| C22—N2—C23 | 116.03 (14) | C17—C18—H18 | 119.1 |
| C21—N2—C23 | 107.08 (13) | C19—C18—H18 | 119.1 |
| O1—C1—C6 | 121.17 (15) | C14—C19—C18 | 116.24 (16) |
| O1—C1—C2 | 121.21 (15) | C14—C19—C20 | 121.46 (15) |
| C6—C1—C2 | 117.60 (13) | C18—C19—C20 | 122.29 (16) |
| C3—C2—C1 | 107.89 (12) | C19—C20—C21 | 115.01 (13) |
| C3—C2—C20 | 114.15 (12) | C19—C20—C2 | 114.64 (12) |
| C1—C2—C20 | 110.87 (12) | C21—C20—C2 | 105.17 (13) |
| C3—C2—C23 | 112.25 (12) | C19—C20—H20 | 107.2 |
| C1—C2—C23 | 107.74 (12) | C21—C20—H20 | 107.2 |
| C20—C2—C23 | 103.77 (12) | C2—C20—H20 | 107.2 |
| N1—C3—C2 | 108.55 (12) | N2—C21—C20 | 103.58 (13) |
| N1—C3—H3A | 110.0 | N2—C21—H21A | 111.0 |
| C2—C3—H3A | 110.0 | C20—C21—H21A | 111.0 |
| N1—C3—H3B | 110.0 | N2—C21—H21B | 111.0 |
| C2—C3—H3B | 110.0 | C20—C21—H21B | 111.0 |
| H3A—C3—H3B | 108.4 | H21A—C21—H21B | 109.0 |
| N1—C4—H4A | 109.5 | N2—C22—H22A | 109.5 |
| N1—C4—H4B | 109.5 | N2—C22—H22B | 109.5 |
| H4A—C4—H4B | 109.5 | H22A—C22—H22B | 109.5 |
| N1—C4—H4C | 109.5 | N2—C22—H22C | 109.5 |
| H4A—C4—H4C | 109.5 | H22A—C22—H22C | 109.5 |
| H4B—C4—H4C | 109.5 | H22B—C22—H22C | 109.5 |
| N1—C5—C6 | 112.12 (13) | N2—C23—C33 | 111.16 (12) |
| N1—C5—H5A | 109.2 | N2—C23—C24 | 113.88 (13) |
| C6—C5—H5A | 109.2 | C33—C23—C24 | 101.34 (12) |
| N1—C5—H5B | 109.2 | N2—C23—C2 | 101.90 (12) |
| C6—C5—H5B | 109.2 | C33—C23—C2 | 119.06 (13) |
| H5A—C5—H5B | 107.9 | C24—C23—C2 | 110.04 (12) |
| C7—C6—C1 | 116.54 (16) | O2—C24—C25 | 126.70 (16) |
| C7—C6—C5 | 123.80 (16) | O2—C24—C23 | 124.93 (16) |
| C1—C6—C5 | 119.60 (14) | C25—C24—C23 | 108.28 (14) |
| C6—C7—C8 | 127.74 (18) | C26—C25—C34 | 120.53 (18) |
| C6—C7—H7 | 116.1 | C26—C25—C24 | 132.12 (19) |
| C8—C7—H7 | 116.1 | C34—C25—C24 | 107.35 (15) |
| C9—C8—C13 | 116.62 (18) | C25—C26—C27 | 118.0 (2) |
| C9—C8—C7 | 122.54 (18) | C25—C26—H26 | 121.0 |
| C13—C8—C7 | 120.78 (18) | C27—C26—H26 | 121.0 |
| C10—C9—C8 | 121.5 (2) | C28—C27—C26 | 121.8 (2) |
| C10—C9—H9 | 119.2 | C28—C27—H27 | 119.1 |
| C8—C9—H9 | 119.2 | C26—C27—H27 | 119.1 |
| C11—C10—C9 | 120.5 (2) | C27—C28—C29 | 121.7 (2) |
| C11—C10—H10 | 119.8 | C27—C28—H28 | 119.1 |
| C9—C10—H10 | 119.8 | C29—C28—H28 | 119.1 |
| C10—C11—C12 | 119.8 (2) | C30—C29—C34 | 116.10 (17) |
| C10—C11—H11 | 120.1 | C30—C29—C28 | 128.53 (19) |
| C12—C11—H11 | 120.1 | C34—C29—C28 | 115.4 (2) |
| C11—C12—C13 | 119.8 (2) | C31—C30—C29 | 120.33 (17) |
| C11—C12—H12 | 120.1 | C31—C30—H30 | 119.8 |
| C13—C12—H12 | 120.1 | C29—C30—H30 | 119.8 |
| C12—C13—C8 | 121.8 (2) | C30—C31—C32 | 122.50 (18) |
| C12—C13—Cl1 | 118.43 (17) | C30—C31—H31 | 118.8 |
| C8—C13—Cl1 | 119.76 (15) | C32—C31—H31 | 118.8 |
| C15—C14—C19 | 122.37 (17) | C33—C32—C31 | 118.77 (17) |
| C15—C14—Cl2 | 117.45 (15) | C33—C32—H32 | 120.6 |
| C19—C14—Cl2 | 120.17 (13) | C31—C32—H32 | 120.6 |
| C16—C15—C14 | 119.5 (2) | C32—C33—C34 | 118.36 (15) |
| C16—C15—H15 | 120.3 | C32—C33—C23 | 132.64 (15) |
| C14—C15—H15 | 120.3 | C34—C33—C23 | 108.90 (14) |
| C17—C16—C15 | 119.85 (19) | C25—C34—C29 | 122.39 (17) |
| C17—C16—H16 | 120.1 | C25—C34—C33 | 113.76 (15) |
| C15—C16—H16 | 120.1 | C29—C34—C33 | 123.85 (17) |
| O1—C1—C2—C3 | −151.50 (15) | C19—C20—C21—N2 | −152.40 (13) |
| C6—C1—C2—C3 | 30.10 (18) | C2—C20—C21—N2 | −25.30 (16) |
| O1—C1—C2—C20 | −25.8 (2) | C22—N2—C23—C33 | 60.97 (19) |
| C6—C1—C2—C20 | 155.77 (13) | C21—N2—C23—C33 | −169.74 (13) |
| O1—C1—C2—C23 | 87.09 (18) | C22—N2—C23—C24 | −52.7 (2) |
| C6—C1—C2—C23 | −91.31 (15) | C21—N2—C23—C24 | 76.57 (16) |
| C5—N1—C3—C2 | 75.82 (16) | C22—N2—C23—C2 | −171.16 (15) |
| C4—N1—C3—C2 | −160.58 (14) | C21—N2—C23—C2 | −41.87 (15) |
| C1—C2—C3—N1 | −60.62 (16) | C3—C2—C23—N2 | 147.45 (12) |
| C20—C2—C3—N1 | 175.67 (13) | C1—C2—C23—N2 | −93.90 (14) |
| C23—C2—C3—N1 | 57.94 (16) | C20—C2—C23—N2 | 23.71 (14) |
| C3—N1—C5—C6 | −54.27 (18) | C3—C2—C23—C33 | −89.93 (16) |
| C4—N1—C5—C6 | −178.59 (14) | C1—C2—C23—C33 | 28.72 (17) |
| O1—C1—C6—C7 | −14.3 (2) | C20—C2—C23—C33 | 146.34 (13) |
| C2—C1—C6—C7 | 164.07 (15) | C3—C2—C23—C24 | 26.31 (17) |
| O1—C1—C6—C5 | 168.41 (16) | C1—C2—C23—C24 | 144.96 (13) |
| C2—C1—C6—C5 | −13.2 (2) | C20—C2—C23—C24 | −97.43 (14) |
| N1—C5—C6—C7 | −153.17 (16) | N2—C23—C24—O2 | −51.3 (2) |
| N1—C5—C6—C1 | 23.9 (2) | C33—C23—C24—O2 | −170.73 (17) |
| C1—C6—C7—C8 | −179.00 (17) | C2—C23—C24—O2 | 62.4 (2) |
| C5—C6—C7—C8 | −1.9 (3) | N2—C23—C24—C25 | 125.47 (15) |
| C6—C7—C8—C9 | −42.2 (3) | C33—C23—C24—C25 | 6.04 (16) |
| C6—C7—C8—C13 | 140.7 (2) | C2—C23—C24—C25 | −120.86 (14) |
| C13—C8—C9—C10 | −0.5 (4) | O2—C24—C25—C26 | −8.4 (3) |
| C7—C8—C9—C10 | −177.7 (3) | C23—C24—C25—C26 | 174.85 (19) |
| C8—C9—C10—C11 | −0.1 (5) | O2—C24—C25—C34 | 171.10 (18) |
| C9—C10—C11—C12 | 0.4 (5) | C23—C24—C25—C34 | −5.60 (18) |
| C10—C11—C12—C13 | −0.2 (5) | C34—C25—C26—C27 | −0.4 (3) |
| C11—C12—C13—C8 | −0.4 (4) | C24—C25—C26—C27 | 179.1 (2) |
| C11—C12—C13—Cl1 | −179.7 (2) | C25—C26—C27—C28 | −2.0 (3) |
| C9—C8—C13—C12 | 0.8 (3) | C26—C27—C28—C29 | 1.6 (3) |
| C7—C8—C13—C12 | 178.0 (2) | C27—C28—C29—C30 | −178.3 (2) |
| C9—C8—C13—Cl1 | −179.95 (18) | C27—C28—C29—C34 | 1.2 (3) |
| C7—C8—C13—Cl1 | −2.7 (3) | C34—C29—C30—C31 | 0.5 (3) |
| C19—C14—C15—C16 | 0.9 (3) | C28—C29—C30—C31 | 179.99 (19) |
| Cl2—C14—C15—C16 | −177.66 (15) | C29—C30—C31—C32 | 1.4 (3) |
| C14—C15—C16—C17 | −0.4 (3) | C30—C31—C32—C33 | −1.0 (3) |
| C15—C16—C17—C18 | −0.2 (3) | C31—C32—C33—C34 | −1.4 (2) |
| C16—C17—C18—C19 | 0.3 (3) | C31—C32—C33—C23 | −177.36 (16) |
| C15—C14—C19—C18 | −0.7 (2) | N2—C23—C33—C32 | 50.4 (2) |
| Cl2—C14—C19—C18 | 177.79 (13) | C24—C23—C33—C32 | 171.78 (17) |
| C15—C14—C19—C20 | −179.46 (15) | C2—C23—C33—C32 | −67.5 (2) |
| Cl2—C14—C19—C20 | −0.9 (2) | N2—C23—C33—C34 | −125.80 (14) |
| C17—C18—C19—C14 | 0.1 (3) | C24—C23—C33—C34 | −4.45 (16) |
| C17—C18—C19—C20 | 178.84 (17) | C2—C23—C33—C34 | 116.30 (15) |
| C14—C19—C20—C21 | −145.35 (15) | C26—C25—C34—C29 | 3.3 (3) |
| C18—C19—C20—C21 | 36.0 (2) | C24—C25—C34—C29 | −176.29 (15) |
| C14—C19—C20—C2 | 92.53 (18) | C26—C25—C34—C33 | −177.61 (16) |
| C18—C19—C20—C2 | −86.13 (19) | C24—C25—C34—C33 | 2.8 (2) |
| C3—C2—C20—C19 | 5.57 (19) | C30—C29—C34—C25 | 175.93 (16) |
| C1—C2—C20—C19 | −116.52 (15) | C28—C29—C34—C25 | −3.6 (2) |
| C23—C2—C20—C19 | 128.06 (14) | C30—C29—C34—C33 | −3.0 (2) |
| C3—C2—C20—C21 | −121.76 (14) | C28—C29—C34—C33 | 177.43 (16) |
| C1—C2—C20—C21 | 116.15 (14) | C32—C33—C34—C25 | −175.52 (15) |
| C23—C2—C20—C21 | 0.73 (15) | C23—C33—C34—C25 | 1.33 (19) |
| C22—N2—C21—C20 | 173.28 (15) | C32—C33—C34—C29 | 3.5 (2) |
| C23—N2—C21—C20 | 43.13 (17) | C23—C33—C34—C29 | −179.62 (15) |
5''-(2-Chlorobenzylidene)-4'-(2-chlorophenyl)-1'-methyldispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (I). Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C25–C29/C34 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C10—H10···O2i | 0.93 | 2.74 | 3.492 (3) | 139 |
| C16—H16···O2ii | 0.93 | 2.76 | 3.481 (3) | 135 |
| C5—H5B···Cg1i | 0.97 | 2.99 | 3.9466 (19) | 168 |
Symmetry codes: (i) −x, y+1/2, −z+1/2; (ii) −x, −y+2, −z.
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Crystal data
| C36H34N2O2 | F(000) = 1120 |
| Mr = 526.65 | Dx = 1.240 Mg m−3Dm = 1.24 Mg m−3Dm measured by floatation method |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.7507 (5) Å | Cell parameters from 5358 reflections |
| b = 15.9089 (8) Å | θ = 4.7–42.0° |
| c = 20.2879 (10) Å | µ = 0.08 mm−1 |
| β = 92.935 (2)° | T = 293 K |
| V = 2820.7 (3) Å3 | Block, yellow |
| Z = 4 | 0.32 × 0.24 × 0.18 mm |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Data collection
| Bruker SMART APEXII CCD diffractometer | 3349 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.040 |
| φ and ω scans | θmax = 25.7°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −10→10 |
| Tmin = 0.816, Tmax = 1.000 | k = −19→19 |
| 28946 measured reflections | l = −24→18 |
| 5339 independent reflections |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.045 | w = 1/[σ2(Fo2) + (0.0506P)2 + 0.5169P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.121 | (Δ/σ)max = 0.001 |
| S = 1.02 | Δρmax = 0.17 e Å−3 |
| 5339 reflections | Δρmin = −0.15 e Å−3 |
| 362 parameters | Extinction correction: SHELXL2018 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0040 (6) |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.47706 (15) | 0.91884 (10) | 0.26949 (6) | 0.0632 (4) | |
| O2 | 0.01075 (17) | 0.80552 (10) | 0.11060 (7) | 0.0749 (5) | |
| N1 | 0.02197 (15) | 0.96279 (9) | 0.23966 (7) | 0.0438 (4) | |
| N2 | 0.32038 (17) | 0.76401 (9) | 0.17891 (7) | 0.0488 (4) | |
| C1 | 0.3410 (2) | 0.93366 (11) | 0.26053 (8) | 0.0434 (4) | |
| C2 | 0.25341 (18) | 0.90633 (11) | 0.19667 (8) | 0.0377 (4) | |
| C3 | 0.12046 (19) | 0.96732 (11) | 0.18425 (8) | 0.0420 (4) | |
| H3A | 0.159005 | 1.024095 | 0.179771 | 0.050* | |
| H3B | 0.063116 | 0.952451 | 0.143748 | 0.050* | |
| C4 | −0.1307 (2) | 0.99492 (14) | 0.22319 (10) | 0.0619 (6) | |
| H4A | −0.175538 | 0.964127 | 0.186372 | 0.093* | |
| H4B | −0.192980 | 0.988480 | 0.260484 | 0.093* | |
| H4C | −0.124579 | 1.053358 | 0.211850 | 0.093* | |
| C5 | 0.0937 (2) | 1.00778 (12) | 0.29519 (9) | 0.0529 (5) | |
| H5A | 0.032387 | 1.000780 | 0.333317 | 0.063* | |
| H5B | 0.096585 | 1.067257 | 0.284807 | 0.063* | |
| C6 | 0.2541 (2) | 0.97734 (11) | 0.31209 (8) | 0.0459 (4) | |
| C7 | 0.3224 (2) | 0.98436 (13) | 0.37200 (9) | 0.0566 (5) | |
| H7 | 0.420638 | 0.962182 | 0.376842 | 0.068* | |
| C8 | 0.2630 (3) | 1.02252 (14) | 0.43153 (9) | 0.0637 (6) | |
| C9 | 0.1907 (3) | 1.09994 (16) | 0.42926 (11) | 0.0862 (8) | |
| H9 | 0.176292 | 1.127477 | 0.388985 | 0.103* | |
| C10 | 0.1399 (4) | 1.13691 (19) | 0.48527 (13) | 0.1125 (11) | |
| H10 | 0.092416 | 1.189186 | 0.482807 | 0.135* | |
| C11 | 0.1595 (4) | 1.0966 (2) | 0.54474 (13) | 0.1158 (11) | |
| H11 | 0.124439 | 1.121064 | 0.582782 | 0.139* | |
| C12 | 0.2304 (4) | 1.02073 (18) | 0.54774 (12) | 0.1011 (10) | |
| H12 | 0.242808 | 0.993784 | 0.588346 | 0.121* | |
| C13 | 0.2852 (3) | 0.98180 (14) | 0.49223 (10) | 0.0736 (7) | |
| C14 | 0.4072 (2) | 1.03916 (12) | 0.08230 (9) | 0.0477 (5) | |
| C15 | 0.3741 (2) | 1.09458 (14) | 0.03059 (10) | 0.0627 (6) | |
| H15 | 0.425474 | 1.145727 | 0.029917 | 0.075* | |
| C16 | 0.2684 (3) | 1.07634 (17) | −0.01936 (11) | 0.0716 (7) | |
| H16 | 0.247654 | 1.114850 | −0.053129 | 0.086* | |
| C17 | 0.1933 (2) | 1.00083 (17) | −0.01915 (10) | 0.0693 (6) | |
| H17 | 0.121043 | 0.987782 | −0.052809 | 0.083* | |
| C18 | 0.2252 (2) | 0.94425 (14) | 0.03112 (9) | 0.0563 (5) | |
| H18 | 0.173963 | 0.893017 | 0.030703 | 0.068* | |
| C19 | 0.33151 (19) | 0.96161 (12) | 0.08224 (8) | 0.0429 (4) | |
| C20 | 0.36354 (19) | 0.89945 (11) | 0.13800 (8) | 0.0420 (4) | |
| H20 | 0.466999 | 0.911443 | 0.156377 | 0.050* | |
| C21 | 0.3607 (2) | 0.80656 (12) | 0.11868 (9) | 0.0529 (5) | |
| H21A | 0.284808 | 0.796067 | 0.083082 | 0.063* | |
| H21B | 0.460023 | 0.788322 | 0.105068 | 0.063* | |
| C22 | 0.2938 (3) | 0.67430 (13) | 0.17162 (11) | 0.0731 (7) | |
| H22A | 0.268011 | 0.651006 | 0.213260 | 0.110* | |
| H22B | 0.211144 | 0.664885 | 0.139531 | 0.110* | |
| H22C | 0.384812 | 0.647695 | 0.157250 | 0.110* | |
| C23 | 0.19669 (19) | 0.81255 (11) | 0.20669 (8) | 0.0422 (4) | |
| C24 | 0.0359 (2) | 0.79910 (12) | 0.16969 (10) | 0.0523 (5) | |
| C25 | −0.0735 (2) | 0.77254 (12) | 0.21815 (10) | 0.0540 (5) | |
| C26 | −0.2285 (2) | 0.75824 (14) | 0.21310 (13) | 0.0723 (6) | |
| H26 | −0.284279 | 0.766562 | 0.173375 | 0.087* | |
| C27 | −0.2998 (3) | 0.73101 (16) | 0.26895 (16) | 0.0843 (8) | |
| H27 | −0.405192 | 0.722827 | 0.266586 | 0.101* | |
| C28 | −0.2194 (3) | 0.71598 (14) | 0.32708 (14) | 0.0765 (7) | |
| H28 | −0.270670 | 0.696290 | 0.363029 | 0.092* | |
| C29 | −0.0593 (2) | 0.72969 (12) | 0.33393 (11) | 0.0580 (5) | |
| C30 | 0.0395 (3) | 0.71575 (13) | 0.38964 (11) | 0.0661 (6) | |
| H30 | 0.000748 | 0.694837 | 0.428213 | 0.079* | |
| C31 | 0.1921 (3) | 0.73275 (12) | 0.38742 (10) | 0.0618 (6) | |
| H31 | 0.255708 | 0.721647 | 0.424532 | 0.074* | |
| C32 | 0.2575 (2) | 0.76681 (11) | 0.33047 (9) | 0.0504 (5) | |
| H32 | 0.361543 | 0.778819 | 0.330644 | 0.060* | |
| C33 | 0.1650 (2) | 0.78149 (11) | 0.27577 (8) | 0.0430 (4) | |
| C34 | 0.0085 (2) | 0.76075 (11) | 0.27794 (9) | 0.0473 (5) | |
| C35 | 0.3604 (4) | 0.89722 (16) | 0.49768 (11) | 0.0951 (9) | |
| H35A | 0.391046 | 0.879874 | 0.454998 | 0.143* | 0.5 |
| H35B | 0.448790 | 0.900568 | 0.527609 | 0.143* | 0.5 |
| H35C | 0.289565 | 0.857078 | 0.513916 | 0.143* | 0.5 |
| H35D | 0.361888 | 0.878473 | 0.542684 | 0.143* | 0.5 |
| H35E | 0.304143 | 0.857779 | 0.470073 | 0.143* | 0.5 |
| H35F | 0.463369 | 0.901268 | 0.483766 | 0.143* | 0.5 |
| C36 | 0.5214 (2) | 1.06401 (14) | 0.13658 (11) | 0.0659 (6) | |
| H36A | 0.531595 | 1.019372 | 0.168344 | 0.099* | 0.5 |
| H36B | 0.618648 | 1.074534 | 0.118366 | 0.099* | 0.5 |
| H36C | 0.486814 | 1.114002 | 0.157740 | 0.099* | 0.5 |
| H36D | 0.559776 | 1.119233 | 0.127956 | 0.099* | 0.5 |
| H36E | 0.472723 | 1.064071 | 0.177934 | 0.099* | 0.5 |
| H36F | 0.604557 | 1.024604 | 0.138560 | 0.099* | 0.5 |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0476 (8) | 0.0952 (11) | 0.0459 (8) | 0.0049 (7) | −0.0050 (6) | −0.0030 (7) |
| O2 | 0.0784 (10) | 0.0890 (12) | 0.0551 (10) | −0.0227 (8) | −0.0182 (8) | 0.0099 (8) |
| N1 | 0.0438 (8) | 0.0493 (9) | 0.0386 (9) | 0.0075 (7) | 0.0052 (7) | 0.0005 (7) |
| N2 | 0.0621 (10) | 0.0420 (9) | 0.0434 (9) | 0.0056 (7) | 0.0121 (7) | −0.0019 (7) |
| C1 | 0.0449 (11) | 0.0489 (11) | 0.0361 (10) | −0.0008 (8) | 0.0004 (8) | 0.0042 (8) |
| C2 | 0.0396 (9) | 0.0432 (10) | 0.0303 (9) | 0.0010 (8) | 0.0026 (7) | 0.0004 (8) |
| C3 | 0.0471 (10) | 0.0452 (10) | 0.0336 (10) | 0.0022 (8) | 0.0018 (8) | 0.0018 (8) |
| C4 | 0.0526 (12) | 0.0685 (14) | 0.0650 (14) | 0.0164 (10) | 0.0071 (10) | −0.0003 (11) |
| C5 | 0.0666 (13) | 0.0533 (12) | 0.0391 (11) | 0.0101 (10) | 0.0072 (9) | −0.0033 (9) |
| C6 | 0.0597 (11) | 0.0439 (11) | 0.0340 (10) | −0.0015 (9) | 0.0009 (8) | 0.0001 (8) |
| C7 | 0.0757 (14) | 0.0559 (12) | 0.0377 (11) | −0.0012 (10) | −0.0014 (10) | −0.0005 (9) |
| C8 | 0.0948 (16) | 0.0596 (14) | 0.0362 (12) | 0.0032 (12) | −0.0021 (10) | −0.0079 (10) |
| C9 | 0.143 (2) | 0.0701 (16) | 0.0441 (14) | 0.0211 (16) | −0.0056 (14) | −0.0085 (12) |
| C10 | 0.193 (3) | 0.0801 (19) | 0.0642 (18) | 0.040 (2) | 0.0039 (19) | −0.0204 (15) |
| C11 | 0.202 (3) | 0.093 (2) | 0.0542 (17) | 0.028 (2) | 0.0216 (18) | −0.0222 (16) |
| C12 | 0.179 (3) | 0.0834 (19) | 0.0420 (14) | 0.013 (2) | 0.0156 (16) | −0.0069 (13) |
| C13 | 0.118 (2) | 0.0631 (15) | 0.0393 (12) | 0.0007 (14) | 0.0031 (12) | −0.0062 (11) |
| C14 | 0.0447 (10) | 0.0549 (12) | 0.0444 (11) | 0.0043 (9) | 0.0126 (8) | 0.0033 (9) |
| C15 | 0.0696 (14) | 0.0620 (14) | 0.0584 (14) | 0.0034 (11) | 0.0222 (11) | 0.0141 (11) |
| C16 | 0.0759 (16) | 0.0903 (18) | 0.0497 (14) | 0.0212 (14) | 0.0140 (12) | 0.0276 (12) |
| C17 | 0.0593 (13) | 0.107 (2) | 0.0415 (12) | 0.0025 (13) | 0.0008 (10) | 0.0154 (12) |
| C18 | 0.0545 (12) | 0.0767 (14) | 0.0377 (11) | −0.0056 (10) | 0.0028 (9) | 0.0068 (10) |
| C19 | 0.0414 (10) | 0.0557 (12) | 0.0322 (10) | 0.0019 (9) | 0.0084 (8) | 0.0030 (8) |
| C20 | 0.0402 (10) | 0.0518 (11) | 0.0342 (10) | −0.0004 (8) | 0.0025 (7) | −0.0005 (8) |
| C21 | 0.0609 (12) | 0.0562 (12) | 0.0425 (11) | 0.0046 (9) | 0.0117 (9) | −0.0047 (9) |
| C22 | 0.1067 (18) | 0.0462 (12) | 0.0682 (15) | 0.0039 (12) | 0.0207 (13) | −0.0071 (11) |
| C23 | 0.0437 (10) | 0.0457 (11) | 0.0372 (10) | 0.0020 (8) | 0.0029 (8) | 0.0007 (8) |
| C24 | 0.0573 (12) | 0.0463 (11) | 0.0524 (13) | −0.0040 (9) | −0.0057 (10) | 0.0040 (9) |
| C25 | 0.0477 (11) | 0.0459 (11) | 0.0684 (14) | −0.0011 (9) | 0.0045 (10) | 0.0018 (10) |
| C26 | 0.0524 (13) | 0.0648 (14) | 0.0994 (19) | −0.0021 (11) | 0.0012 (12) | −0.0008 (13) |
| C27 | 0.0542 (14) | 0.0776 (17) | 0.123 (2) | −0.0061 (13) | 0.0221 (16) | −0.0055 (17) |
| C28 | 0.0690 (16) | 0.0648 (15) | 0.100 (2) | −0.0065 (12) | 0.0418 (15) | −0.0021 (14) |
| C29 | 0.0696 (14) | 0.0404 (11) | 0.0664 (14) | 0.0048 (10) | 0.0249 (12) | 0.0005 (10) |
| C30 | 0.0901 (17) | 0.0501 (13) | 0.0613 (14) | 0.0083 (12) | 0.0364 (13) | 0.0071 (10) |
| C31 | 0.0919 (17) | 0.0501 (12) | 0.0439 (12) | 0.0184 (12) | 0.0088 (11) | 0.0087 (9) |
| C32 | 0.0591 (12) | 0.0465 (11) | 0.0460 (11) | 0.0112 (9) | 0.0069 (9) | 0.0061 (9) |
| C33 | 0.0509 (11) | 0.0366 (10) | 0.0422 (11) | 0.0069 (8) | 0.0082 (8) | 0.0033 (8) |
| C34 | 0.0519 (11) | 0.0349 (10) | 0.0563 (12) | 0.0028 (8) | 0.0149 (10) | 0.0026 (9) |
| C35 | 0.157 (3) | 0.0752 (17) | 0.0521 (14) | 0.0178 (17) | −0.0015 (15) | 0.0049 (12) |
| C36 | 0.0637 (13) | 0.0622 (14) | 0.0717 (15) | −0.0101 (11) | 0.0025 (11) | −0.0023 (11) |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Geometric parameters (Å, º)
| O1—C1 | 1.218 (2) | C17—H17 | 0.9300 |
| O2—C24 | 1.212 (2) | C18—C19 | 1.385 (2) |
| N1—C5 | 1.450 (2) | C18—H18 | 0.9300 |
| N1—C3 | 1.453 (2) | C19—C20 | 1.518 (2) |
| N1—C4 | 1.454 (2) | C20—C21 | 1.529 (3) |
| N2—C22 | 1.452 (2) | C20—H20 | 0.9800 |
| N2—C21 | 1.456 (2) | C21—H21A | 0.9700 |
| N2—C23 | 1.466 (2) | C21—H21B | 0.9700 |
| C1—C6 | 1.496 (2) | C22—H22A | 0.9600 |
| C1—C2 | 1.534 (2) | C22—H22B | 0.9600 |
| C2—C3 | 1.526 (2) | C22—H22C | 0.9600 |
| C2—C20 | 1.573 (2) | C23—C33 | 1.525 (2) |
| C2—C23 | 1.589 (2) | C23—C24 | 1.575 (3) |
| C3—H3A | 0.9700 | C24—C25 | 1.469 (3) |
| C3—H3B | 0.9700 | C25—C26 | 1.374 (3) |
| C4—H4A | 0.9600 | C25—C34 | 1.390 (3) |
| C4—H4B | 0.9600 | C26—C27 | 1.390 (3) |
| C4—H4C | 0.9600 | C26—H26 | 0.9300 |
| C5—C6 | 1.508 (3) | C27—C28 | 1.363 (3) |
| C5—H5A | 0.9700 | C27—H27 | 0.9300 |
| C5—H5B | 0.9700 | C28—C29 | 1.417 (3) |
| C6—C7 | 1.331 (2) | C28—H28 | 0.9300 |
| C7—C8 | 1.470 (3) | C29—C34 | 1.398 (3) |
| C7—H7 | 0.9300 | C29—C30 | 1.405 (3) |
| C8—C9 | 1.384 (3) | C30—C31 | 1.366 (3) |
| C8—C13 | 1.396 (3) | C30—H30 | 0.9300 |
| C9—C10 | 1.374 (3) | C31—C32 | 1.423 (3) |
| C9—H9 | 0.9300 | C31—H31 | 0.9300 |
| C10—C11 | 1.369 (4) | C32—C33 | 1.360 (2) |
| C10—H10 | 0.9300 | C32—H32 | 0.9300 |
| C11—C12 | 1.358 (4) | C33—C34 | 1.412 (2) |
| C11—H11 | 0.9300 | C35—H35A | 0.9600 |
| C12—C13 | 1.392 (3) | C35—H35B | 0.9600 |
| C12—H12 | 0.9300 | C35—H35C | 0.9600 |
| C13—C35 | 1.500 (3) | C35—H35D | 0.9600 |
| C14—C15 | 1.390 (3) | C35—H35E | 0.9600 |
| C14—C19 | 1.400 (3) | C35—H35F | 0.9600 |
| C14—C36 | 1.501 (3) | C36—H36A | 0.9600 |
| C15—C16 | 1.368 (3) | C36—H36B | 0.9600 |
| C15—H15 | 0.9300 | C36—H36C | 0.9600 |
| C16—C17 | 1.369 (3) | C36—H36D | 0.9600 |
| C16—H16 | 0.9300 | C36—H36E | 0.9600 |
| C17—C18 | 1.378 (3) | C36—H36F | 0.9600 |
| C5—N1—C3 | 109.05 (14) | C19—C20—C21 | 116.01 (14) |
| C5—N1—C4 | 111.16 (15) | C19—C20—C2 | 114.95 (14) |
| C3—N1—C4 | 112.30 (14) | C21—C20—C2 | 104.99 (14) |
| C22—N2—C21 | 114.57 (15) | C19—C20—H20 | 106.8 |
| C22—N2—C23 | 116.08 (15) | C21—C20—H20 | 106.8 |
| C21—N2—C23 | 107.05 (13) | C2—C20—H20 | 106.8 |
| O1—C1—C6 | 120.91 (16) | N2—C21—C20 | 103.63 (13) |
| O1—C1—C2 | 120.90 (16) | N2—C21—H21A | 111.0 |
| C6—C1—C2 | 118.18 (15) | C20—C21—H21A | 111.0 |
| C3—C2—C1 | 107.64 (14) | N2—C21—H21B | 111.0 |
| C3—C2—C20 | 114.25 (13) | C20—C21—H21B | 111.0 |
| C1—C2—C20 | 111.07 (13) | H21A—C21—H21B | 109.0 |
| C3—C2—C23 | 112.19 (13) | N2—C22—H22A | 109.5 |
| C1—C2—C23 | 107.70 (13) | N2—C22—H22B | 109.5 |
| C20—C2—C23 | 103.83 (13) | H22A—C22—H22B | 109.5 |
| N1—C3—C2 | 108.55 (13) | N2—C22—H22C | 109.5 |
| N1—C3—H3A | 110.0 | H22A—C22—H22C | 109.5 |
| C2—C3—H3A | 110.0 | H22B—C22—H22C | 109.5 |
| N1—C3—H3B | 110.0 | N2—C23—C33 | 110.81 (14) |
| C2—C3—H3B | 110.0 | N2—C23—C24 | 113.84 (14) |
| H3A—C3—H3B | 108.4 | C33—C23—C24 | 101.21 (14) |
| N1—C4—H4A | 109.5 | N2—C23—C2 | 101.76 (13) |
| N1—C4—H4B | 109.5 | C33—C23—C2 | 119.65 (14) |
| H4A—C4—H4B | 109.5 | C24—C23—C2 | 110.12 (13) |
| N1—C4—H4C | 109.5 | O2—C24—C25 | 126.46 (18) |
| H4A—C4—H4C | 109.5 | O2—C24—C23 | 124.94 (17) |
| H4B—C4—H4C | 109.5 | C25—C24—C23 | 108.48 (16) |
| N1—C5—C6 | 112.26 (15) | C26—C25—C34 | 120.42 (19) |
| N1—C5—H5A | 109.2 | C26—C25—C24 | 132.3 (2) |
| C6—C5—H5A | 109.2 | C34—C25—C24 | 107.31 (16) |
| N1—C5—H5B | 109.2 | C25—C26—C27 | 118.3 (2) |
| C6—C5—H5B | 109.2 | C25—C26—H26 | 120.9 |
| H5A—C5—H5B | 107.9 | C27—C26—H26 | 120.9 |
| C7—C6—C1 | 117.20 (17) | C28—C27—C26 | 121.8 (2) |
| C7—C6—C5 | 123.34 (17) | C28—C27—H27 | 119.1 |
| C1—C6—C5 | 119.41 (15) | C26—C27—H27 | 119.1 |
| C6—C7—C8 | 128.6 (2) | C27—C28—C29 | 121.4 (2) |
| C6—C7—H7 | 115.7 | C27—C28—H28 | 119.3 |
| C8—C7—H7 | 115.7 | C29—C28—H28 | 119.3 |
| C9—C8—C13 | 119.01 (19) | C34—C29—C30 | 116.11 (19) |
| C9—C8—C7 | 121.34 (19) | C34—C29—C28 | 115.7 (2) |
| C13—C8—C7 | 119.6 (2) | C30—C29—C28 | 128.2 (2) |
| C10—C9—C8 | 121.3 (2) | C31—C30—C29 | 120.30 (19) |
| C10—C9—H9 | 119.3 | C31—C30—H30 | 119.8 |
| C8—C9—H9 | 119.3 | C29—C30—H30 | 119.8 |
| C11—C10—C9 | 119.8 (3) | C30—C31—C32 | 122.5 (2) |
| C11—C10—H10 | 120.1 | C30—C31—H31 | 118.8 |
| C9—C10—H10 | 120.1 | C32—C31—H31 | 118.8 |
| C12—C11—C10 | 119.5 (2) | C33—C32—C31 | 118.75 (19) |
| C12—C11—H11 | 120.3 | C33—C32—H32 | 120.6 |
| C10—C11—H11 | 120.3 | C31—C32—H32 | 120.6 |
| C11—C12—C13 | 122.4 (2) | C32—C33—C34 | 118.17 (17) |
| C11—C12—H12 | 118.8 | C32—C33—C23 | 132.64 (16) |
| C13—C12—H12 | 118.8 | C34—C33—C23 | 109.07 (15) |
| C12—C13—C8 | 118.0 (2) | C25—C34—C29 | 122.33 (19) |
| C12—C13—C35 | 120.5 (2) | C25—C34—C33 | 113.55 (16) |
| C8—C13—C35 | 121.46 (19) | C29—C34—C33 | 124.10 (19) |
| C15—C14—C19 | 118.55 (18) | C13—C35—H35A | 109.5 |
| C15—C14—C36 | 119.47 (18) | C13—C35—H35B | 109.5 |
| C19—C14—C36 | 121.98 (17) | H35A—C35—H35B | 109.5 |
| C16—C15—C14 | 122.1 (2) | C13—C35—H35C | 109.5 |
| C16—C15—H15 | 118.9 | H35A—C35—H35C | 109.5 |
| C14—C15—H15 | 118.9 | H35B—C35—H35C | 109.5 |
| C15—C16—C17 | 119.4 (2) | H35D—C35—H35E | 109.5 |
| C15—C16—H16 | 120.3 | H35D—C35—H35F | 109.5 |
| C17—C16—H16 | 120.3 | H35E—C35—H35F | 109.5 |
| C16—C17—C18 | 119.8 (2) | C14—C36—H36A | 109.5 |
| C16—C17—H17 | 120.1 | C14—C36—H36B | 109.5 |
| C18—C17—H17 | 120.1 | H36A—C36—H36B | 109.5 |
| C17—C18—C19 | 121.8 (2) | C14—C36—H36C | 109.5 |
| C17—C18—H18 | 119.1 | H36A—C36—H36C | 109.5 |
| C19—C18—H18 | 119.1 | H36B—C36—H36C | 109.5 |
| C18—C19—C14 | 118.42 (17) | H36D—C36—H36E | 109.5 |
| C18—C19—C20 | 121.30 (17) | H36D—C36—H36F | 109.5 |
| C14—C19—C20 | 120.26 (15) | H36E—C36—H36F | 109.5 |
| O1—C1—C2—C3 | −152.18 (17) | C19—C20—C21—N2 | −153.51 (14) |
| C6—C1—C2—C3 | 29.1 (2) | C2—C20—C21—N2 | −25.45 (17) |
| O1—C1—C2—C20 | −26.4 (2) | C22—N2—C23—C33 | 60.2 (2) |
| C6—C1—C2—C20 | 154.89 (15) | C21—N2—C23—C33 | −170.39 (14) |
| O1—C1—C2—C23 | 86.66 (19) | C22—N2—C23—C24 | −53.0 (2) |
| C6—C1—C2—C23 | −92.04 (17) | C21—N2—C23—C24 | 76.33 (18) |
| C5—N1—C3—C2 | 76.18 (17) | C22—N2—C23—C2 | −171.47 (15) |
| C4—N1—C3—C2 | −160.14 (15) | C21—N2—C23—C2 | −42.10 (16) |
| C1—C2—C3—N1 | −60.49 (17) | C3—C2—C23—N2 | 147.72 (13) |
| C20—C2—C3—N1 | 175.66 (13) | C1—C2—C23—N2 | −94.01 (15) |
| C23—C2—C3—N1 | 57.82 (17) | C20—C2—C23—N2 | 23.85 (15) |
| C3—N1—C5—C6 | −54.01 (19) | C3—C2—C23—C33 | −89.87 (17) |
| C4—N1—C5—C6 | −178.36 (15) | C1—C2—C23—C33 | 28.4 (2) |
| O1—C1—C6—C7 | −12.8 (3) | C20—C2—C23—C33 | 146.26 (14) |
| C2—C1—C6—C7 | 165.91 (16) | C3—C2—C23—C24 | 26.65 (18) |
| O1—C1—C6—C5 | 169.68 (17) | C1—C2—C23—C24 | 144.93 (14) |
| C2—C1—C6—C5 | −11.6 (2) | C20—C2—C23—C24 | −97.21 (15) |
| N1—C5—C6—C7 | −154.62 (18) | N2—C23—C24—O2 | −51.5 (2) |
| N1—C5—C6—C1 | 22.8 (2) | C33—C23—C24—O2 | −170.37 (18) |
| C1—C6—C7—C8 | −178.84 (19) | C2—C23—C24—O2 | 62.1 (2) |
| C5—C6—C7—C8 | −1.4 (3) | N2—C23—C24—C25 | 124.97 (16) |
| C6—C7—C8—C9 | −46.1 (3) | C33—C23—C24—C25 | 6.06 (18) |
| C6—C7—C8—C13 | 136.6 (2) | C2—C23—C24—C25 | −121.50 (15) |
| C13—C8—C9—C10 | −0.4 (4) | O2—C24—C25—C26 | −8.7 (4) |
| C7—C8—C9—C10 | −177.7 (3) | C23—C24—C25—C26 | 174.9 (2) |
| C8—C9—C10—C11 | −0.5 (5) | O2—C24—C25—C34 | 170.5 (2) |
| C9—C10—C11—C12 | 0.6 (6) | C23—C24—C25—C34 | −5.9 (2) |
| C10—C11—C12—C13 | 0.2 (5) | C34—C25—C26—C27 | −0.7 (3) |
| C11—C12—C13—C8 | −1.1 (5) | C24—C25—C26—C27 | 178.4 (2) |
| C11—C12—C13—C35 | −179.0 (3) | C25—C26—C27—C28 | −2.0 (4) |
| C9—C8—C13—C12 | 1.2 (4) | C26—C27—C28—C29 | 1.8 (4) |
| C7—C8—C13—C12 | 178.6 (2) | C27—C28—C29—C34 | 1.1 (3) |
| C9—C8—C13—C35 | 179.0 (3) | C27—C28—C29—C30 | −178.6 (2) |
| C7—C8—C13—C35 | −3.6 (4) | C34—C29—C30—C31 | 0.3 (3) |
| C19—C14—C15—C16 | 1.2 (3) | C28—C29—C30—C31 | −179.9 (2) |
| C36—C14—C15—C16 | −178.30 (19) | C29—C30—C31—C32 | 1.6 (3) |
| C14—C15—C16—C17 | −0.7 (3) | C30—C31—C32—C33 | −1.3 (3) |
| C15—C16—C17—C18 | −0.1 (3) | C31—C32—C33—C34 | −1.0 (3) |
| C16—C17—C18—C19 | 0.3 (3) | C31—C32—C33—C23 | −176.46 (17) |
| C17—C18—C19—C14 | 0.3 (3) | N2—C23—C33—C32 | 50.5 (3) |
| C17—C18—C19—C20 | 178.85 (17) | C24—C23—C33—C32 | 171.57 (19) |
| C15—C14—C19—C18 | −1.0 (3) | C2—C23—C33—C32 | −67.4 (2) |
| C36—C14—C19—C18 | 178.52 (17) | N2—C23—C33—C34 | −125.23 (15) |
| C15—C14—C19—C20 | −179.60 (16) | C24—C23—C33—C34 | −4.16 (17) |
| C36—C14—C19—C20 | −0.1 (3) | C2—C23—C33—C34 | 116.91 (16) |
| C18—C19—C20—C21 | 36.9 (2) | C26—C25—C34—C29 | 3.7 (3) |
| C14—C19—C20—C21 | −144.55 (17) | C24—C25—C34—C29 | −175.53 (17) |
| C18—C19—C20—C2 | −86.1 (2) | C26—C25—C34—C33 | −177.43 (17) |
| C14—C19—C20—C2 | 92.47 (19) | C24—C25—C34—C33 | 3.3 (2) |
| C3—C2—C20—C19 | 6.9 (2) | C30—C29—C34—C25 | 175.91 (18) |
| C1—C2—C20—C19 | −115.06 (16) | C28—C29—C34—C25 | −3.9 (3) |
| C23—C2—C20—C19 | 129.45 (15) | C30—C29—C34—C33 | −2.8 (3) |
| C3—C2—C20—C21 | −121.77 (16) | C28—C29—C34—C33 | 177.44 (17) |
| C1—C2—C20—C21 | 116.24 (16) | C32—C33—C34—C25 | −175.63 (17) |
| C23—C2—C20—C21 | 0.75 (16) | C23—C33—C34—C25 | 0.8 (2) |
| C22—N2—C21—C20 | 173.69 (16) | C32—C33—C34—C29 | 3.2 (3) |
| C23—N2—C21—C20 | 43.47 (18) | C23—C33—C34—C29 | 179.61 (17) |
1'-Methyl-5''-(2-methylbenzylidene)-4'-(2-methylphenyl)dispiro[acenaphthene-1,2'-pyrrolidine-3',3''-piperidine]-2,4''-dione (II). Hydrogen-bond geometry (Å, º)
Cg2 is the centroid of the C8–C13 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C36—H36A···O1 | 0.96 | 2.66 | 3.586 (3) | 161 |
| C10—H10···O2i | 0.93 | 2.77 | 3.529 (3) | 140 |
| C16—H16···O2ii | 0.93 | 2.79 | 3.530 (3) | 137 |
| C35—H35F···Cg2iii | 0.96 | 2.94 | 3.805 (4) | 151 |
Symmetry codes: (i) −x, y+1/2, −z+1/2; (ii) −x, −y+2, −z; (iii) −x+1, −y+2, −z+1.
References
- Aravindan, P. G., Selvanayagam, S., Velmurugan, D., Ravikumar, K., Sridhar, G. & Raghunathan, R. (2004). Acta Cryst. E60, o2149–o2151.
- Bruker (2001). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2012). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bueno, J. M., Manzano, P., García, M. C., Chicharro, J., Puente, M., Lorenzo, M., García, A., Ferrer, S., Gómez, R. M., Fraile, M. T., Lavandera, J. L., Fiandor, J. M., Vidal, J., Herreros, E. & Gargallo-Viola, D. (2011). Bioorg. Med. Chem. Lett. 21, 5214–5218. [DOI] [PubMed]
- Cheng, Z., Han, X., Liang, C. & Yang, S. (2018). US Patent Pub. No. US2018/0134705A1.
- Desiraju, G. R. & Sarma, J. A. R. P. (1986). Proc. - Indian Acad. Sci. Chem. Sci. 96, 599–605.
- El-Ayaan, U., Abdel-Aziz, A. A.-M. & Al-Shihry, S. (2007). Eur. J. Med. Chem. 42, 1325–1333. [DOI] [PubMed]
- Gholap, S. S. (2016). Eur. J. Med. Chem. 110, 13–31. [DOI] [PubMed]
- Gnanaguru, K., Murthy, G. S., Venkatesan, K. & Ramamurthy, V. (1984). Chem. Phys. Lett. 109, 255–258.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jiang, L., Zhang, B., Wang, Y., Sun, J., Ma, X., Wang, G., Fu, S., Lin, C. & Li, Y. (2019). Nat. Prod. Res. https://doi. org/10.1080/14786419.2019.1647422
- Jones, W., Ramdas, S., Theocharis, C. R., Thomas, J. M. & Thomas, N. W. (1981). J. Phys. Chem. 85, 2594–2597.
- Kumar, R. R., Loganayaki, B. & Perumal, S. (2009). Synth. Commun. 39, 3197–3216.
- McDavids, J. E. & Daniels, T. C. (1951). J. Am. Pharm. Assoc. (Sci. ed.), 40, 325–326. [DOI] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Nath, N. K. & Nangia, A. (2012). Cryst. Growth Des. 12, 5411–5425.
- Oganov, A. R. (2018). Faraday Discuss. 211, 643–660. [DOI] [PubMed]
- Pandiarajan, S., Saravanamoorthy, S. N., Kumar, B. R., Kumar, R. R. & Athimoolam, S. (2008). Acta Cryst. E64, o99–o100. [DOI] [PMC free article] [PubMed]
- Rajni Swamy, V., Müller, P., Srinivasan, N., Perumal, S. & Krishnakumar, R. V. (2013). Acta Cryst. C69, 412–415. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sribala, R., Srinivasan, N., Indumathi, S. & Krishnakumar, R. V. (2018). Acta Cryst. E74, 1267–1271. [DOI] [PMC free article] [PubMed]
- Tan, S. L., Jotani, M. M. & Tiekink, E. R. T. (2019). Acta Cryst. E75, 308–318. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wolff, S. K., Grimwood, D. J., McKinnon, J. J., Turner, M. J., Jayatilaka, D. & Spackman, M. A. (2012). Crystal Explorer, University of Western Australia.
- Zhu, W., Dai, M., Xu, Y. & Qian, X. (2008). Bioorg. Med. Chem. 16, 3255–3260. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II. DOI: 10.1107/S2056989019012428/jj2215sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019012428/jj2215Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989019012428/jj2215IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report