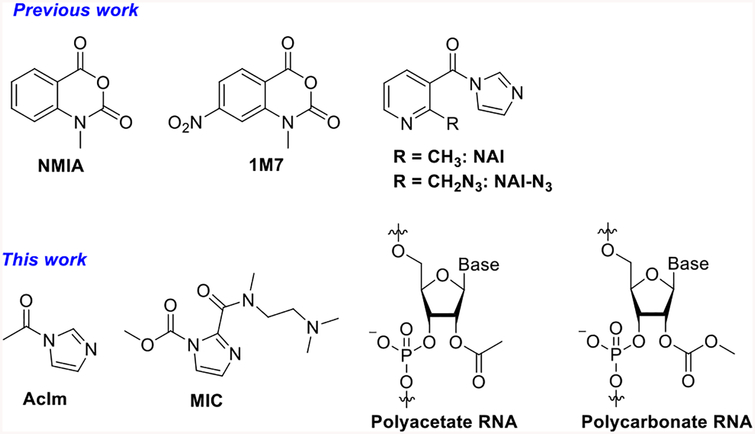
Abstract
Acylation of RNA at 2′-OH groups is widely applied in mapping RNA structure and recently for controlling RNA function. Reactions are described that install the smallest 2-carbon acyl groups on RNA—namely, 2′-O-acetyl and 2′-O-carbonate groups. Hybridization and thermal melting experiments are performed to assess the effects of the acyl groups on duplex formation. Both reagents can be employed at lower concentrations to map RNA secondary structure by reverse transcriptase primer extension (SHAPE) methods.
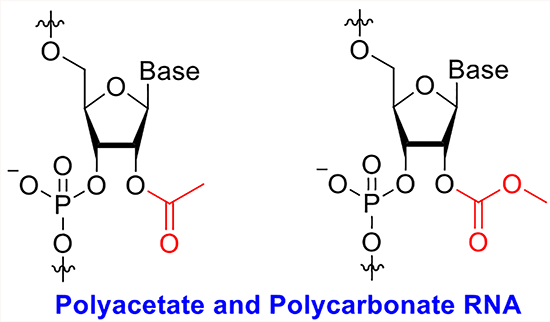
Graphical Abstract
Aqueous bond-forming reactions between small-molecule reagents and biomolecules have become an essential tool for biotechnology and medicine. Simple electrophilic reagents have been employed almost universally in functionalization, labeling, and conjugation of peptides and proteins;1,2 prominent examples include fluorescence labeling of antibodies,3,4 used in imaging and quantification of biomedically important antigens; conjugation of antibodies with toxic drugs for selective targeting of cancer cells;5,6 and activity-based protein profiling, employing small molecule reagents to assess activities in the proteome.7,8 Such reactions typically make use of nucleophilic residues in proteins, such as lysine amines and cysteine thiols, reacting with alkyl and acyl electrophiles.
In the domain of nucleic acids, nucleophilic reactivity is limited considerably relative to that found in proteins. The nucleobases of DNA and RNA are typically weakly reactive due to their aromaticity, and the exocyclic amines found on cytosine, adenine, and guanine are relatively poorly nucleophilic due to lone pair delocalization into the pyrimidine and purine rings. For this reason, reactions with electrophiles have not been widely applied for functionalizing DNA, and for RNA only recently. One functionalization that has received some study is the reaction of diazoketones with phosphates in DNA and RNA;9,10 while this reaction does offer some flexibility in reagent design, it causes instability in longer nucleic acid chains due to lability of the resulting phosphotriester linkages.11 Beyond this, few methods exist for useful internal functionalization of nucleic acids, and so researchers have commonly relied instead on incorporating non-natural reactive residues into the biopolymer during its construction, either via polymerase-mediated synthesis or total chemical synthesis.
For already-existing RNA strands in particular, including those from living systems, few practical chemical methods are available for functionalization. Although labeling at remote 5′ and 3′ ends is feasible,12–14 functionalization of the internal nucleotides of RNA has received little attention until very recently (Scheme 1). Given the complexities and biomedical importance of RNA biology, the elucidation of new reactivities for this biopolymer could provide useful tools for labeling and analysis in a biological setting.
Scheme 1.
Structures of RNA Acylating Reagents and Adducts
Here we address this issue by studying an RNA-selective reaction, the acylation of the 2′-OH group. This reactivity has proven broadly useful for mapping RNA structure in the SHAPE methodology,15 wherein active acyl compounds (traditionally N-methylisatoic anhydride (NMIA) and 1-methyl-7-nitroisatoic anhydride (1M7), Scheme 1) react with 2′-OH groups at exposed and flexible nucleotides. The steric bulk of the acyl adducts causes reverse transcriptase enzymes to stop, allowing researchers to map their locations in folded RNAs. However, these reagents are not ideal as chemical functionalization tools, as they react only in very low yields (less than 3%), likely due to their short half-lives in water and relatively low solubility.16,17 More recently, isatoic anhydride reagents with higher solubility have been developed,17 and biotinylated isatoic anhydride reagents were applied in an effort to separate RNA from DNA.18 Highest-yielding RNA acylation reactions have recently been achieved with a pyridine-based acylimidazole reagent (NAI and NAI-N3, Scheme 1), which can functionalize RNA super-stoichiometrically, reacting with over half of the 2′-OH groups on an RNA strand if desired.19 The steric bulk of the adducts was used to block RNA folding and RNA–enzyme interactions.19
The research to date on RNA acylation leaves open a number of basic chemical and biochemical questions. This acylation has thus far been performed with specialized reagents, the large majority of which are based on aryl structure. Several issues remain unclear: how well do much smaller acylating reagents react with RNA? Do biological acetylating agents react with RNA? How do such small acyl groups affect the properties of RNA? Finally, can these smallest reagents be employed to map RNA-folded structure, similar to the larger aryl reagents used previously? Here, we address these questions by studying reactions that place the smallest stable acyl groups, acetyl and methylcarbonate, on RNA.
Our first experiments addressed whether activated acetyl reagents or methyl carbamate reagents could react with RNA to produce polyacetyl or poly(methylcarbonate)-substituted strands. Although acetylation of RNA was reported five decades ago,20,21 it was carried out before modern analytical methods were developed, and yields and properties of the resulting RNAs were not well characterized. Carbonate substitution of RNA has been completely unknown until 2018, when photoreactive benzylcarbamate reagents were shown to react with RNA.22
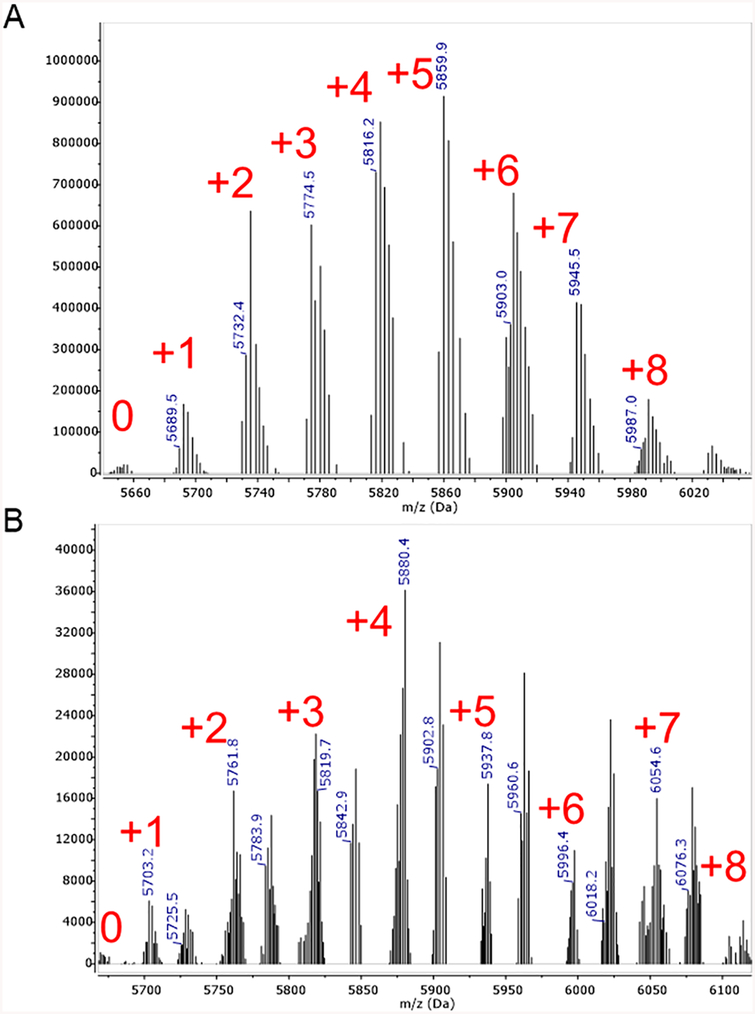
To measure reactivity for esterification of RNA, we reacted acetylimidazole (AcIm) with RNA at varying concentrations and times. We found that fresh preparation of AcIm in dry DMSO was superior to the use of older bottles of AcIm, which are significantly hydrolyzed. The half-life of AcIm in 25 mM aqueous phosphate buffer (pH 7.4) was determined to be ~30 min at room temperature (Figure S1). To test the feasibility of biochemical acetylation, we also tested acetylCoA and acetylphosphate, two common acetylating agents in the cell.23,24 After reaction, RNAs were analyzed by MALDI-MS. The results showed that acetylimidazole does react with RNA in high yields, yielding up to 89% acetylation of nucleotides in a concentration-dependent manner (Figures 1A and S2). AcetylCoA and acetylphosphate did not provide measurable acetylation under these conditions (Figures S3–S5), suggesting that if RNA is biologically acylated in cells it likely requires enzymatic assistance. We similarly tested acylation of RNA by methyl imidazole carbamate compound (MIC) (Scheme 1). Based on previous studies,25 we chose an electron-deficient imidazole as the leaving group to enhance reactivity over unsubstituted imidazole (Scheme 1). The resulting half-life of the electron-deficient MIC was 24 h in 25 mM phosphate buffer (pH 7.4) at room temperature (Figure S1). The results in this case also showed high levels of reactivity with RNA, again yielding adducts up to 89% of nucleotides depending on concentration (Figures 1B and S6), but requiring longer incubation times compared to AcIm. To test the RNA selectivity of acylation reactions, we compared reactions of AcIm with DNA and RNA. The results showed only minor reaction with DNA (Figure S7), confirming the need for 2′-OH groups as nucleophiles in the reactions, and ruling out major reaction with exocyclic amine groups. We also tested reactions of RNAs in duplex with DNA, and acylation yields were reduced markedly (Figure S8), suggesting that the 2′-OH groups in double-stranded helical RNA are not readily accessible even to these small reagents. This suggests their possible use in RNA structure mapping (vide infra).
Figure 1.
MALDI-TOF mass spectral data showing polyacylation of RNA by AcIm and MIC reagents, resulting in (A) polyacetyl and (B) polycarbonate RNA, respectively. An 18 nt RNA strand was reacted as described, and then purified by precipitation prior to analysis. The number of adducts for major peaks is given in red. The spectrum of polycarbonate RNA also shows a sodium adduct for each added carbonate group. Data here are shown for medium-yield acylation; see Figures S2 and S6 for high-level acylation. RNA sequence: 5′-AUC CUG CCG ACU ACG CCA-3′.
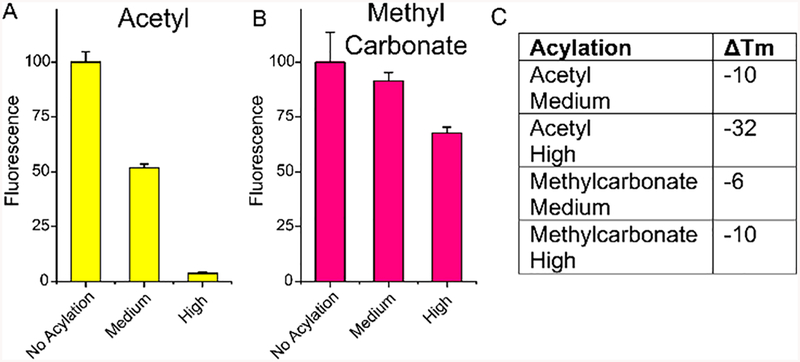
Next, we tested the effects of multisubstituted acetyl and methylcarbonate groups on the ability of RNA to form a duplex with a complementary nucleic acid. The ability to block RNA structure and function has proven useful recently for more complex RNA acyl groups.19,22 For the new 2-carbon groups, it remained to be seen if their lower steric bulk might change their ability to inhibit hybridization. This was tested in two ways, with fluorescence of a complementary molecular beacon (MB), and with thermal melting studies, and was evaluated both for RNA substituted at medium (ca. 40–50% of 2′-OH groups) and high (~70–80%) levels of acylation. The results showed (Figures 2, S9, and S10) that both acetyl and methylcarbonate groups do in fact hinder hybridization, especially at higher levels of substitution, and both lower Tm values of RNAs substantially. Interestingly, the magnitude of disruption was more pronounced for polyacetate modification; the acetyl group lowered Tm significantly more than methylcarbonate did, particularly at high modification levels. The results provide the first data on methylcarbonate-derivatized RNA and confirm one early finding upon acetylation of tRNA, which was reported to disrupt structure.20
Figure 2.
Effects of polyacetate and polycarbonate modification on RNA hybridization. (A, B) Plot of hybridization fluorescence signals with a DNA molecular beacon complement. Melting conditions: 1.0 μM RNA and DNA complement in 5 mM phosphate buffer, 1 mM MgCl2, 140 mM KCl, pH 7.4. Absorbance was measured at 260 nm at 1 °C intervals between 25–90 °C. Beacon hybridization conditions: 100 nM RNA, 80 nM molecular beacon in 5 mM phosphate buffer, 1 mM MgCl2, 140 mM KCl, pH 7.4 at 25 °C, λex = 485 nm, λem = 538 nm. Error bars reflect SD from three experiments. See Figures S8 and S9 for original data. RNA sequence: 5′-AUC CUG CCG ACU ACG CCA-3′; molecular beacon sequence: 5′−6FAM-CGC GGG CGT AGT CGG CAG GAC GCG-Dabcyl-3′. (C) Table of thermal melting values of RNAs with medium and high levels of acylation as compared with unmodified RNA.
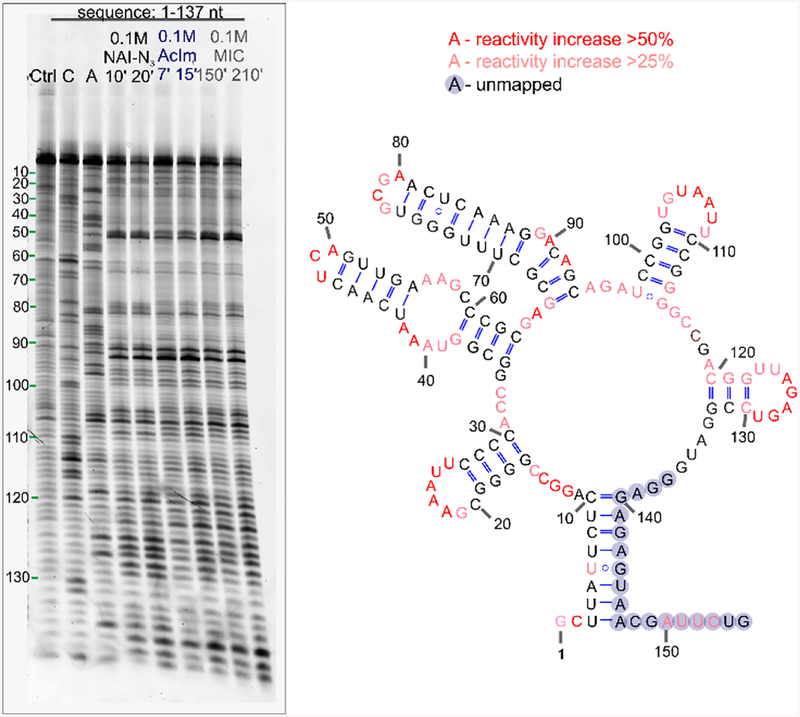
The findings that even these small acyl groups on RNA hinder hybridization and that both reagents prefer single-stranded RNA over double-stranded RNA suggest the possibility that they might be used to probe folded secondary structure by acylation probed by primer extension. These minimalist reagents are more easily prepared than many SHAPE reagents and have aqueous lifetimes that may be more convenient than some traditional reagents as well. However, no data exist on whether these very small substituents might block primer extension by a reverse transcriptase enzyme, which is necessary for the traditional SHAPE methodology.15 As an initial test to explore whether these acylating reagents may be applied for RNA structure mapping, we tested their reaction at standard SHAPE concentrations (100 mM, room temperature, 10–120 min) with FMN riboswitch RNA26 in vitro. The results show (Figure 3) that both reagents give structure-dependent signals, with AcIm requiring even less time than known reagent NAI to give equal distribution of signals (7 min versus 10 min), while the less reactive MIC required longer times (~2.5 h). Both reagents yielded SHAPE mapping signals that are similar to those with NAI-N3, an established larger aryl SHAPE reagent,16,27 as well as those of 1M7 in earlier studies.28 The gel electrophoretic patterns reveal preferential reactivity at known accessible regions of the folded RNA28 and also show similar dynamic ranges of reactivity between the most and least accessible positions in the structure. Interestingly, the new reagents differ slightly in their precise pattern, which may arise from their lower bulk, which may enable reaction at more sterically challenging sites (Figure S11). Overall, we preliminarily conclude that both these reagents, despite their small size and simplicity, can be used to map folded RNA structure by acylation and primer extension. These data represent the first information on the postsynthetic installation and properties of methylcarbonate groups in RNA. For the ester group, 2′-O-acetyl, some previous data have been reported. A recent study aimed at developing synthetic methods for incorporating single acetyl groups into a synthetic oligoribonucleotide revealed a drop in Tm of 3.1 °C for single substitution,29 which is consistent with, and extended by, our current finding of helical destabilization with greater numbers of substitutions. In addition to this, early experiments from 1965 reported that acetylation blocked tRNA structure,20 which is also consistent with our findings. No prior reports have tested either of these acyl groups for their ability to block primer extension; interestingly, a much larger acyl group was recently reported not to block primer extension,30 which is surprising given that much less sterically demanding groups tested here do block a reverse transcriptase.
Figure 3.
Testing reagents AcIm and MIC for their ability to map FMN riboswitch RNA structure at low levels of substitution. Gel electrophoresis SHAPE mapping data reveals stops due to (a) preferential reactivity at accessible residues and (b) pausing of reverse transcriptase at sites of modification. The gel shows FMN structure probing patterns for RNA in buffer and DMSO (Ctrl), next to dideoxy sequencing lanes (C, A) and patterns generated using the acylating compounds (NAI-N3, AcIm, MIC).
Taken together, our data establish the minimal structure needed for reactivity with 2′-OH and for blocking biochemical function. The results document that even very small acylating reagents can efficiently acylate RNA, both at trace levels (for mapping) and at superstoichiometric levels (for blocking hybridization), and can distinguish between single- and double-stranded structure. The resulting 2-carbon acyl groups, acetate and methylcarbonate, are likely the smallest stable acyl groups available for substitution at the 2′-OH position. Our experiments present the first data on the degree by which medium and high levels of acylation destabilize RNA hybridization, and establish that even these very small groups can block primer extension by a polymerase enzyme. This establishes them as the simplest acylating agents available for mapping RNA structure and interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank the U.S. National Institutes of Health for support (GM127295 and GM130704).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.orglett.9b01526.
Detailed RNA acylation and purification procedure, MS spectra of functionalized RNA, molecular beacon experimental procedure, procedure for melting studies, and description of SHAPE experiments (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Spicer CD; Davis BG Selective Chemical Protein Modification. Nat. Commun 2014, 5, 4740. [DOI] [PubMed] [Google Scholar]
- (2).Boutureira O; Bernardes GJL Advances in Chemical Protein Modification. Chem. Rev 2015, 115 (5), 2174–2195. [DOI] [PubMed] [Google Scholar]
- (3).Yao J; Yang M; Duan Y Chemistry, Biology, and Medicine of Fluorescent Nanomaterials and Related Systems: New Insights into Biosensing, Bioimaging, Genomics, Diagnostics, and Therapy. Chem. Rev 2014, 114 (12), 6130–6178. [DOI] [PubMed] [Google Scholar]
- (4).Coons AH; Kaplan MH Localization o fantigen in tissue cells. J. Exp. Med 1949, 91 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Beck A; Goetsch L; Dumontet C; Corvaïa N Strategies and Challenges for the next Generation of Antibody–drug Conjugates. Nat. Rev. Drug Discovery 2017, 16, 315. [DOI] [PubMed] [Google Scholar]
- (6).Agarwal P; Bertozzi CR Site-Specific Antibody–Drug Conjugates: The Nexus of Bioorthogonal Chemistry, Protein Engineering, and Drug Development. Bioconjugate Chem. 2015, 26 (2), 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cravatt BF; Wright AT; Kozarich JW Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem 2008, 77 (1), 383–414. [DOI] [PubMed] [Google Scholar]
- (8).Nomura DK; Dix MM; Cravatt BF Activity-Based Protein Profiling for Biochemical Pathway Discovery in Cancer. Nat. Rev. Cancer 2010, 10, 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ando H; Furuta T; Tsien RY; Okamoto H Photo-Mediated Gene Activation Using Caged RNA/DNA in Zebrafish Embryos. Nat. Genet 2001, 28 (4), 317–325. [DOI] [PubMed] [Google Scholar]
- (10).Shah S; Rangarajan S; Friedman SH Light-Activated RNA Interference. Angew. Chem 2005, 117 (9), 1352–1356. [DOI] [PubMed] [Google Scholar]
- (11).Blidner RA; Svoboda KR; Hammer RP; Monroe WT Photoinduced RNA Interference Using DMNPE-Caged 2’-Deoxy-2’-Fluoro Substituted Nucleic Acids in Vitro and in Vivo. Mol. BioSyst 2008, 4 (5), 431–440. [DOI] [PubMed] [Google Scholar]
- (12).Huang C; Yu Y-T Synthesis and Labeling of RNA In Vitro. Curr. Protoc. Mol. Biol 2013, 102 (1), 4.15.1–4.15.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rio DC 5′-End Labeling of RNA with [γ−32P]ATP and T4 Polynucleotide Kinase. Cold Spring Harb. Protoc 2014, 2014 (4), pdb.prot080739. [DOI] [PubMed] [Google Scholar]
- (14).Paredes E; Evans M; Das SR RNA Labeling, Conjugation and Ligation. Methods 2011, 54 (2), 251–259. [DOI] [PubMed] [Google Scholar]
- (15).Weeks KM; Mauger DM Exploring RNA Structural Codes with SHAPE Chemistry. Acc. Chem. Res 2011, 44 (12), 1280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee B; Flynn RA; Kadina A; Guo JK; Kool ET; Chang HY Comparison of SHAPE Reagents for Mapping RNA Structures inside Living Cells. RNA 2017, 23 (2), 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fessler AB; Dey A; Garmon CB; Finis DS; Saleh N-A; Fowler AJ; Jones DS; Chakrabarti K; Ogle CA Water-Soluble Isatoic Anhydrides: A Platform for RNA-SHAPE Analysis and Protein Bioconjugation. Bioconjugate Chem. 2018, 29 (9), 3196–3202. [DOI] [PubMed] [Google Scholar]
- (18).Ursuegui S; Chivot N; Moutin S; Burr A; Fossey C; Cailly T; Laayoun A; Fabis F; Laurent A Biotin-Conjugated N-Methylisatoic Anhydride: A Chemical Tool for Nucleic Acid Separation by Selective 2′-Hydroxyl Acylation of RNA. Chem. Commun 2014, 50 (43), 5748–5751. [DOI] [PubMed] [Google Scholar]
- (19).Kadina A; Kietrys AM; Kool ET RNA Cloaking by Reversible Acylation. Angew. Chem., Int. Ed 2018, 57 (12), 3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Knorre DG; Pustoshilova NM; Teplova NM; Shamovskii GG The production of transfer RNA acetylated by 2’-oxy groups. Biokhimiya 1965, 30, 1218–1224. [PubMed] [Google Scholar]
- (21).Knorre DG; Pustoshilova NM; Teplova NM Action of Spleen and Snake Venom Phosphodiesterases on Transfer-RNA Acetylated on the Ribose 2’-Hydroxyl Group. Biokhimiya 1966, 31, 666–669. [PubMed] [Google Scholar]
- (22).Velema WA; Kietrys AM; Kool ET RNA Control by Photoreversible Acylation. J. Am. Chem. Soc 2018, 140 (10), 3491–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pietrocola F; Galluzzi L; Bravo-San Pedro JM; Madeo F; Kroemer G Acetyl Coenzyme A: A Central Metabolite and Second Messenger. Cell Metab. 2015, 21 (6), 805–821. [DOI] [PubMed] [Google Scholar]
- (24).Xu WJ; Wen H; Kim HS; Ko Y-J; Dong S-M; Park I-S; Yook JI; Park S Observation of Acetyl Phosphate Formation in Mammalian Mitochondria Using Real-Time in-Organelle NMR Metabolomics. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (16), 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Velema WA; Kool ET Water-Soluble Leaving Group Enables Hydrophobic Functionalization of RNA. Org. Lett 2018, 20 (20), 6587–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mironov AS; Gusarov I; Rafikov R; Lopez LE; Shatalin K; Kreneva RA; Perumov DA; Nudler E Sensing Small Molecules by Nascent RNA: A Mechanism to Control Transcription in Bacteria. Cell 2002, 111 (5), 747–756. [DOI] [PubMed] [Google Scholar]
- (27).Spitale RC; Flynn RA; Zhang QC; Crisalli P; Lee B; Jung J-W; Kuchelmeister HY; Batista PJ; Torre EA; Kool ET; et al. Structural Imprints in Vivo Decode RNA Regulatory Mechanisms. Nature 2015, 519 (7544), 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Vicens Q; Mondragón E; Batey RT Molecular Sensing by the Aptamer Domain of the FMN Riboswitch: A General Model for Ligand Binding by Conformational Selection. Nucleic Acids Res. 2011, 39 (19), 8586–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Xu J; Duffy CD; Chan CKW; Sutherland JD Solid-Phase Synthesis and Hybrization Behavior of Partially 2′/3′-O-Acetylated RNA Oligonucleotides. J. Org. Chem 2014, 79 (8), 3311–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ursuegui S; Yougnia R; Moutin S; Burr A; Fossey C; Cailly T; Laayoun A; Laurent A; Fabis F A Biotin-Conjugated Pyridine-Based Isatoic Anhydride, a Selective Room Temperature RNA-Acylating Agent for the Nucleic Acid Separation. Org. Biomol. Chem 2015, 13 (12), 3625–3632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.