Abstract
Hybridization can profoundly affect the genomic composition and phenotypes of closely related species, and provides an opportunity to identify mechanisms that maintain reproductive isolation between species. Recent evidence suggests that hybridization outcomes within a species pair can vary across locations. However, we still don’t know how variable outcomes of hybridization are across geographic replicates, and what mechanisms drive that variation. In this study, we described hybridization outcomes across 27 locations in the North Fork Shoshone River basin (Wyoming, USA) where native Yellowstone cutthroat trout and introduced rainbow trout co-occur. We used genomic data and hierarchical Bayesian models to precisely identify ancestry of hybrid individuals. Hybridization outcomes varied across locations. In some locations, only rainbow trout and advanced backcrossed hybrids towards rainbow trout were present, while trout in other locations had a broader range of ancestry, including both parental species and first-generation hybrids. Later-generation intermediate hybrids were rare relative to backcrossed hybrids and rainbow trout individuals. Using an individual-based simulation, we found that outcomes of hybridization in the North Fork Shoshone River basin deviate substantially from what we would expect under null expectations of random mating and no selection against hybrids. Since this deviation implies that some mechanisms of reproductive isolation function to maintain parental taxa and a diversity of hybrid types, we then modeled hybridization outcomes as a function of environmental variables and stocking history that are likely to affect prezygotic barriers to hybridization. Variables associated with history of fish stocking were the strongest predictors of hybridization outcomes, followed by environmental variables that might affect overlap in spawning time and location.
Keywords: hybridization, reproductive isolation, ancestry, simulation, Oncorhynchus
Introduction
Hybridization has the potential to substantially alter the evolutionary trajectory of species, and outcomes of hybridization also reflect underlying mechanisms of reproductive isolation between species. While hybridization has often been studied to better understand primary divergence and speciation (Barton & Hewitt 1985, Harrison & Larson 2014, Ravinet et al. 2017), hybrid zones reflecting secondary contact between divergent species can reveal how isolation between close relatives is maintained in sympatry (Wagner & Mandeville 2017). When species hybridize following a species introduction or anthropogenic disturbance, hybridization has the potential to threaten biodiversity. Negative effects of hybridization on native species can include genetic or demographic swamping (Rhymer & Simberloff 1996, Wolf et al. 2001, Buerkle et al. 2003, Todesco et al. 2016, Grabenstein & Taylor 2018), potentially leading to extinction or local extirpation of the native species. Hybrids can also compete with parental species, and in some cases have higher fitness (Rieseberg et al. 1999, Fitzpatrick & Shaffer 2007). Taken together, the potential for both ecological and genetic effects on native species has led conservation practitioners to be deeply alarmed about the prospect of interspecific hybridization with non-native species (Allendorf et al. 2001). However, it is far from straightforward to predict outcomes of hybridization, due to the potential for geographic variation in hybridization outcomes (Mandeville et al. 2017), incomplete understanding of the relative fitnesses of hybrids and parental species (Arnold et al. 2012), and the potential for environmental context to influence outcomes of hybridization (Hatfield & Schluter 1999).
It is therefore important to precisely quantify variation in outcomes of hybridization, and to connect variable hybridization outcomes more specifically to mechanisms that cause variation in realized reproductive isolation (Gompert et al. 2017). Geographic variation in outcomes of hybridization remains incompletely characterized, despite recent evidence that hybridization outcomes and mechanisms of reproductive isolation can vary across locations where a pair of species interacts (Buerkle & Rieseberg 2001, Vines et al. 2003, Sweigart et al. 2007, Good et al. 2008, Nolte et al. 2009, Teeter et al. 2010, Haselhorst & Buerkle 2013, Gompert et al. 2014, Mandeville et al. 2017). In some cases incidence of hybridization varies (e.g. Mandeville et al. 2017), while in other cases the genetic ancestry of hybrids varies, reflecting different numbers of generations of hybridization and different degrees of backcrossing to parental species (Haselhorst & Buerkle 2013, Gompert et al. 2014, Mandeville et al. 2017) or different patterns of locus-specific introgression (Nolte et al. 2009, Teeter et al. 2010). Although it is now apparent that hybridization often varies across locations, there are several limitations common to recent studies. Some studies showing geographic variation in hybridization and introgression explore only a few locations (e.g. Vines et al. 2003, Nolte et al. 2009, Mandeville et al. 2015), while others incorporate samples from many locations, but sampling is inconsistent or opportunistic due to the difficulties of sampling across the range of widespread species (e.g. Mandeville et al. 2017). In other cases, studies with excellent geographic sampling have relied on genetic data from few loci due to logistical constraints or desire for compatibility with historical data (Muhlfeld et al. 2014, Young et al. 2016, Muhlfeld et al. 2017). Methods that use thousands of loci sampled across the genome permit fundamentally different kinds of analyses and inferences than studies with only handfuls of loci (Narum et al. 2013, Gompert et al. 2017), and represent a new opportunity to better understand hybridization.
In addition to limitations stemming from sampling, few studies have conclusively identified causes of variation in hybridization. This is a difficult undertaking, but is a necessary next step for truly understanding variation in reproductive isolation between species (Gompert et al. 2017). Hybridization in river-dwelling fishes presents a unique opportunity for understanding variation in hybridization because the structure of fish populations is constrained by the geography of river drainages, and spawning preferences often lead to multiple, distinct spawning zones across relatively small spatial scales. Therefore, it is possible to observe spatially consolidated, replicated outcomes of hybridization in these systems. This geographic constraint and local subdivision of populations in turn makes it more feasible to investigate associations between hybridization outcomes and environmental variables (as in Loxterman et al. 2014, Young et al. 2016, Muhlfeld et al. 2017).
In this study, we used genomic data to describe variation in outcomes of hybridization across 27 tributaries in the North Fork Shoshone river basin (Wyoming, USA) where native Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and non-native rainbow trout (Oncorhynchus mykiss) come into contact. Our goals were to quantify variation in outcomes of hybridization across locations, and to connect patterns of variation explicitly to environmental and ecological attributes of tributaries.
Rainbow trout have been widely introduced in the US (Halverson 2008, 2010), and hybridization with rainbow trout has long been known to pose a threat to cutthroat trout subspecies (Allendorf & Leary 1988, Seiler & Keeley 2007, Muhlfeld et al. 2009, Ostberg et al. 2011, Kovach et al. 2011, Ostberg et al. 2013, Muhlfeld et al. 2014). The North Fork Shoshone River basin has experienced extensive stocking of salmonids over the past century, including rainbow trout and Yellowstone cutthroat trout. Yellowstone cutthroat trout without hybrid ancestry are now rare in this basin and throughout the historic range of the taxon (Kruse et al. 2000, Gresswell 2011, Endicott et al. 2016). Stocking of rainbow trout in the North Fork Shoshone River basin ended in the mid-20th century and was primarily constrained to Buffalo Bill reservoir. Stocking of Yellowstone cutthroat ended in the late 20th century in most locations (Nordberg et al. unpublished data), continuing until 2009 in the headwaters of a single drainage (Trout Creek). Despite extensive hybridization and long-term presence of rainbow trout, some unhybridized Yellowstone cutthroat trout still persist in the North Fork Shoshone river basin, defying predictions of extirpation via hybridization 20 years ago (Kruse et al. 2000).
Although hybridization dynamics in westslope cutthroat trout have been extensively studied (e.g. Allendorf & Leary 1988, 2005, Muhlfeld et al. 2009, Bennett et al. 2010, Hohenlohe et al. 2013, Loxterman et al. 2014, Muhlfeld et al. 2014, Kovach et al. 2015, Young et al. 2016, Muhlfeld et al. 2017), relatively little work has focused on Yellowstone cutthroat trout hybridization (but see Kovach et al. 2011, Ostberg et al. 2013, and others). It is an open question to what extent hybridization dynamics are consistent or variable across hybridizing pairs of species or subspecies within a genus. Unlike with westslope cutthroat systems, upstream refugia for native trout are not known to exist in the North Fork Shoshone River basin (based on extensive longitudinal sampling of streams; Kruse et al. 2000), suggesting that mechanisms of reproductive isolation might differ for these Yellowstone cutthroat trout compared to westslope cutthroat trout. Identifying patterns of hybridization and connecting hybridization outcomes with stocking history and environmental data will yield a more complete understanding of how and to what extent reproductive isolation between Yellowstone cutthroat and rainbow trout is maintained in the North Fork Shoshone river basin.
Methods
Sampling and library preparation
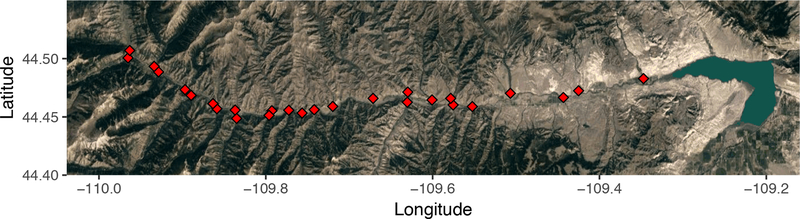
We sampled 26 tributaries of the North Fork Shoshone River and the main stem of the North Fork Shoshone at the upstream limit of our sampling in September 2016 (Fig. 1). We sampled in the fall because of the specifics of the migratory life history of trout in this drainage. Yellowstone cutthroat trout and rainbow trout in the North Fork Shoshone River drainage use the downstream portion of the river and Buffalo Bill Reservoir in the fall and winter. Adult trout migrate into North Fork Shoshone tributaries in the spring (May-June) to spawn. Emergence occurs in mid to late summer with most juvenile trout out-migrating in the fall (Kent 1984). By sampling out-migrating juveniles in the fall, we sampled a single cohort of fish in their natal tributaries.
Figure 1:
Sampling locations for this study. We sampled juvenile trout from 26 tributaries of the North Fork Shoshone River, plus the main stem of the North Fork Shoshone at the upstream limit of our sampling. In each tributary, 2–3 reaches were sampled (summarized by a single point in this map). The North Fork Shoshone River flows down out of the Absaroka Range (left) into Buffalo Bill Reservoir (right). Trout migrate upstream from the reservoir and lower river in the spring to spawn in tributaries. Fish sampled for genetic analysis comprised single cohort of fish, sampled in their natal tributaries prior to outmigration.
We sampled all individuals using backpack electrofishing, and sampled 2–3 reaches in each tributary. Reaches were separated by >200 m. For each reach, we sampled upstream for 10 minutes, capturing all fish observed in each reach. We processed fish in random order, and took fin clips for genetic analysis from the first 20 fish processed in each reach, for a total of 40–60 individuals sampled for each tributary (sampling conducted under IACUC permit 20160803NW00248–02 from University of Wyoming). Sampling was conducted without respect to phenotype (i.e. without attempting to identify juvenile Oncorhynchus to species), and individuals selected for genetic analysis represent a random sample from the reaches surveyed. We stored fin clips in 95% ethanol between sampling and DNA extraction.
In addition to juvenile individuals sampled from wild populations in the North Fork Shoshone River basin, we also included reference individuals from known non-admixed populations. These reference individuals came from three fish hatchery broodstocks maintained by the Wyoming Game and Fish Department, as well as one isolated population of Yellowstone cutthroat trout from outside the study region. Hatchery reference individuals included Yellowstone cutthroat trout from the Tensleep Hatchery (Tensleep, Wyoming, USA), rainbow trout from the Story Hatchery (Story, Wyoming, USA), and Snake River cutthroat trout from the Auburn Hatchery (Auburn, Idaho, USA). Yellowstone cutthroat from an isolated wild population in the Wind River mountains (Wyoming, USA) were also included as reference individuals.
To prepare genomic libraries for high throughput DNA sequencing, we first extracted DNA from fin tissue using Qiagen DNeasy Blood & Tissue kits according to the manufacturer’s instructions, using a QIAcube robot (Qiagen, Inc.). We then prepared reduced-complexity genomic libraries according to protocols in Parchman et al. 2012, similar to previous sequencing projects involving fish with similarly sized genomes (Mandeville et al. 2015, Underwood et al. 2016, Mandeville et al. 2017, Walters et al. 2017). This genotyping-by-sequencing method involves first fragmenting DNA using restriction enzymes (EcoRI and MseI), then ligating unique 8–10 base pair nucleotide barcodes to each individual fish’s DNA. Once barcoded, individual samples were multiplexed and amplified by PCR. For this project, we pooled 192 individuals per library for lanes 1–6, and 153 individuals in the 7th lane. Each library was sequenced on one Illumina HiSeq lane. Prior to sequencing, genomic libraries were size-selected using BluePippin (Sage Science) to retain fragments 350–450 bp in length. DNA sequencing of all 7 libraries (1305 individual fish) on Illumina Hiseq 4000 (1×150) was completed at the University of Texas Genome Sequencing and Analysis Facility, Austin, Texas.
We used the Mount Moran high performance computing cluster at the University of Wyoming (Advanced Research Computing Center 2012) for all bioinformatics processing of data and subsequent analyses. We filtered raw data to remove common contaminants, including E.coli, PhiX (which is used as a control in Illumina sequencing), and leftover primers, barcodes, and adaptors from the library preparation process. We then assigned reads to individual fish by parsing barcodes and removed barcode sequence from the retained sequences using a custom perl script. After parsing barcodes, we aligned reads to the Atlantic salmon genome (ICSASG v2; Lien et al. 2016) using bwa mem (version 0.7.15; Li & Durbin 2009, Li 2013). We chose to use the Atlantic salmon genome because it is a high quality reference genome with chromosome-level assemblies, which makes overassembling paralogous loci together less likely. Additionally, we chose this reference to avoid alignment bias that might result from using the rainbow trout genome - i.e., to avoid a scenario where Yellowstone cutthroat individuals might have lower alignment rates than rainbow trout individuals. Following alignment, we excluded individual fish with less than 10,000 reads aligned to the reference genome (19 individuals). All subsequent analyses were done with the remaining 1,286 individuals with greater than or equal to 10,000 reads aligned.
We then identified variant sites using samtools and bcftools (version 1.3.1; Li 2011). Rather than calling genotypes at these loci, we retained genotypes as genotype likelihoods that reflect differences in information obtained across loci. We filtered these variants using vcftools (version 0.1.14; Danecek et al. 2011) to acquire a set of well-supported SNPs for downstream analyses. To retain a single nucleotide variant, we required that at least 60% of individuals had data at that genomic site. We retained only SNPs where the minor allele frequency was greater than 5%. To avoid paralogous loci, we excluded any sites with >2 alternative alleles. We also plotted per-locus coverage and verified that the distribution is unimodal, suggesting that our retained loci are unlikely to include paralogs. We retained genotype data for 12,666 SNPs in genotype likelihood format, to facilitate analyses carrying through genotype uncertainty in a Bayesian framework, allowing for better quantification of uncertainty in higher-level parameter estimates (Li 2011, Pasaniuc et al. 2012). This probabilistic genotyping approach allowed us to retain more of the data, including loci with relatively low coverage or individuals with more missing data, because we quantified and incorporated uncertainty about genotypes in our estimates of ancestry of individuals. By pooling information across many loci and individuals, we were able to accurately estimate ancestry of individuals despite uncertainty at some loci for some individuals (Buerkle & Gompert 2013).
Although panels of putatively diagnostic loci are frequently used for the assessment of hybrid ancestry in trout populations, we opted to use a genotyping-by-sequencing approach that includes a cross-section of all SNPs identified, rather than focusing on putatively diagnostic loci. We made this choice for several reasons. While putatively diagnostic loci are usually sufficient for simply identifying ancestry of an individual, there are some drawbacks to using diagnostic loci to study hybridization as an evolutionary process, and population genetic processes in wild populations in general. First, diagnostic SNPs are defined relative to reference individuals, so intraspecific variation in parental species might cause misleading inferences if a SNP is not actually fixed in all populations of that species. This is particularly likely to be problematic in species with large and highly structured geographic ranges, as is the case for Yellowstone cutthroat trout. Second, SNPs that are sharply differentiated between parental taxa are more likely to be under selection or associated with reproductive isolation (Gompert & Buerkle 2010, 2011a), and might therefore introgress unusually relative to the overall ancestry of an individual. Indeed, this property of unusual introgression patterns is the concept behind methods which seek to detect loci under selection in hybrid zones, such as introgress, bgc, and other genomic cline methods (Gompert & Buerkle 2010, 2011b). Finally, the use of genotyping-by-sequencing methods allowed us to gather genotype data from a larger number of SNPs across the genome. The number of loci needed to identify a backcrossed individual also scales with the number of generations of hybridization that have occurred (McFarlane & Pemberton 2019), which in this study system might be >25 generations in some locations. For all of these reasons, we chose to sample all SNPs, including those that are potentially variable in both species, for quantifying genome-wide ancestry of hybrid individuals (see also Gompert et al. 2017).
Quantifying genomic outcomes of hybridization
To identify ancestry of individual fish, we ran entropy to estimate proportion of ancestry in each parental species for each individual. entropy is a hierarchical Bayesian model that uses Markov Chain Monte Carlo to converge on estimates of population genetic parameters such as proportion of ancestry in a cluster, and incorporates estimates of uncertainty surrounding these parameters (Gompert et al. 2014). This model requires specification of number of genetic clusters, but does not incorporate a priori assumptions about ancestry of individuals (similar to STRUCTURE; Pritchard et al. 2000, Falush et al. 2003). We ran 3 chains for the k = 2 model for 100,000 steps, discarded the first 80,000 steps as burn-in, and retained every 10th sample from the posterior distribution. To confirm proper mixing and convergence of parameter estimates, we extracted posterior samples using the rhdf5 package in R (Fischer & Pau 2017), then plotted a subset of chains in R and visually assessed convergence and mixing of parameter estimates. From this model output, we generated estimates for q, proportion of ancestry, and Q, interspecific ancestry. Interspecific ancestry (Q) is the proportion of loci in an individual’s genome that are estimated to have ancestry from two parental species.
The bivariate relationship of proportion of ancestry in each cluster (q) and interspecific ancestry (Q) allowed us to assign individuals to hybrid classes representing different histories of hybridization (e.g. F1, F2, backcrossed hybrids, and later-generation recombinant hybrids; as in Gompert et al. 2014, Lindtke et al. 2014, Mandeville et al. 2017). Yellowstone cutthroat individuals were identified as having q greater than 0.9 and Q less than or equal to 0.25. Similarly, rainbow trout individuals were identified as having q less than 0.1 and Q less than or equal to 0.25. Individuals were designated F1 hybrids if they had q estimates 0.4–0.6 and Q estimates greater than 0.8. F2 hybrids were designated as having q 0.4–0.6 (the same as F1 hybrids) but Q 0.4–0.6, consistent with reduced interspecific ancestry due to recombination. We described backcrosses to both parental species using an equation quantifying the proximity of an individual to the expected line between parental species at (0,0) or (1,0) and F1 hybrids at (0.5,1). Backcrosses to rainbow trout were defined as individuals with −0.1 < Q − 2q < 0.1; backcrosses to Yellowstone cutthroat were defined as individuals with 1.9 < Q + 2q < 2.1. Finally, hybrid individuals were classified as “Other” if they did not meet any of these conditions for specific hybrid classes, but were intermediate between parental species.
We chose these criteria to bin individuals into categories based on two factors: q and Q values for reference individuals in this dataset representing parental species, and also on applications of the entropy model to other datasets, including simulated datasets (Gompert et al. 2014, Lindtke et al. 2014, Mandeville et al. 2017). Both of these lines of evidence suggest that it is reasonable to expect among-individual variation in q and Q within a hybrid class. This variation likely stems from several sources. First, there is likely stochastic variation in the exact proportion of the genome coming from each parental species within each individual. This variation might result from variation in outcomes of recombination, and potentially as a result of selection acting on some subset of ancestry combinations. Intraspecific variation in parental species can also affect q and Q estimates; Q estimates in particular are known to be depressed when there is intraspecific variation in parental species (Mandeville et al. 2017) or when parental species are closely related (see simulations in Lindtke et al. 2014). In these trout, it is likely that individuals with <10% ancestry in one cluster are a product of variable intraspecific ancestry in parental species due to historical stocking of both rainbow and cutthroat trout from multiple sources. For some individuals, model uncertainty might also contribute to individuals having <10% ancestry in a cluster, but since credible intervals around ancestry estimates were small (mean 95% credible interval for q was 0.0218; mean 95% credible interval for Q was 0.0421), variable intraspecific ancestry is a more plausible explanation.
We quantified hybridization outcomes for each river using five different statistics, calculated from entropy results. For each river, we quantified 1) proportion Yellowstone cutthroat ancestry, 2) proportion Yellowstone cutthroat individuals, 3) proportion rainbow trout individuals, 4) proportion hybrid individuals, and 5) range of backcrossing. Range of backcrossing was defined as the 95% quantiles q for individuals designated as hybrids in a stream. This summary statistic captures whether backcrossing occurs symmetrically to both parental species or is more constrained (i.e., backcrossing to only one parental species, or hybrids restricted to intermediate proportions of ancestry; as in Mandeville et al. 2017).
Simulations
To better understand how our results relate to specific evolutionary scenarios for past hybridization in the North Fork Shoshone River basin, we constructed an individual-based simulation in R to model hybridization outcomes in terms of the summary statistics (proportion of ancestry and interspecific ancestry) that we generated for empirical data using entropy. (Simulation code to be made available in Dryad repository upon acceptance of this MS; available to reviewers on request.) For our null model, we assumed random mating, no immigration, and no selection against any genetic category, including hybrids. For each generation, parents were randomly selected for the next generation, and proportion of ancestry (q) and interspecific ancestry (Q) were calculated for each o spring based on parental q and Q. This makes simulated outcomes of hybridization directly comparable to empirical estimates of ancestry from entropy.
We started with a population of 1000 individuals split between two parental species, and varied the starting proportions of parental species, including starting populations with 50%, 75%, and 90% individuals from Species 1. Since all stocking of both parental species in the North Fork Shoshone ended by 2000 in most tributaries (with the exception of stocking of Yellowstone cutthroat in the headwaters of Trout Creek until 2009; Nordberg et al. in prep.), and fish were sampled for genetic analysis in 2016, we present simulation results for 5 generations and 10 generations of hybridization without stocking of additional individuals from parental species, which is comparable to the known history of stocking in the North Fork Shoshone River basin (assuming generation time of 3 years, which is a conservative estimate; Behnke 1992). We also relaxed the assumption of no immigration of parental species in a subset of simulations; see Supplement for additional details.
Environmental and ecological correlates of hybridization
To identify potential environmental dependence in genomic outcomes of hybridization, we modeled hybridization outcomes as a function of environmental and historical stocking variables. We considered several response variables, including proportion Yellowstone cutthroat trout ancestry, proportion Yellowstone cutthroat individuals, proportion rainbow trout individuals, proportion hybrid individuals, and extent of backcrossing. Each of these variables captures a different aspect of hybridization dynamics, and they are not necessarily correlated in a predictable way in all systems (Mandeville et al. 2017). In the North Fork Shoshone River tributaries, proportion Yellowstone cutthroat (both ancestry and individuals) was strongly positively correlated with both proportion hybrid individuals and extent of backcrossing. These four variables were strongly negatively correlated with proportion rainbow trout individuals. We used three of these response variables for modeling the effects of environmental and ecological predictor variables on hybridization dynamics, because different factors might predict the persistence of Yellowstone cutthroat, the propensity to hybridize, and to what extent hybrids backcross to parental species. Our response variables were therefore proportion Yellowstone cutthroat ancestry, proportion hybrid individuals, and extent of backcrossing.
We selected predictor variables according to our hypotheses about what mechanisms might influence hybridization outcomes. Predictor variables fell into three categories: environmental variables, stocking history variables, and disturbance metrics (Table 1; similar to Yau & Taylor 2013). Environmental variables are likely to affect hybridization outcomes by causing variation in the temporal or spatial overlap of spawning for parental species. For example, in some systems, Yellowstone cutthroat trout are known to spawn several weeks later than rainbow trout, suggesting that temporal asynchrony in spawning could be an isolating mechanism that might allow local persistence of native Yellowstone cutthroat (Henderson et al. 2000, DeRito et al. 2010). Temperature has been previously shown to be important in mediating hybridization between westslope cutthroat and rainbow trout (Yau & Taylor 2013, Muhlfeld et al. 2014, Young et al. 2016, Muhlfeld et al. 2017). Environmental variables included were mean modeled August stream temperature, precipitation, basin size, slope, canopy cover, elevation, and aspect. We acquired data for all of these variables except aspect from the NorWeST Stream Temperature Regional Database (Isaak et al. 2017), which uses historical temperature data and climate models to project current and future water temperatures. We selected the modeled point closest to our fish sampling location, and checked these points visually by plotting both sampling locations and points for modeled data on a shapefile of streams in the North Fork Shoshone River basin.
Table 1:
Predictor variables for outcomes of hybridization fall into major categories according to our hypotheses about what mechanisms drive variable outcomes of hybridization in the North Fork Shoshone River Basin.
| Category | Predictor variables | Hypothesized mechanism |
|---|---|---|
| Environmental variables | Temperature, Basin size, Elevation, Aspect, Canopy, Slope, Precipitation | Environmental variables influence temporal or spatial overlap in spawning, affecting opportunity for hybridization |
| Stocking history | Number of Yellowstone cutthroat stocked, Number of rainbow trout stocked, Distance from core rainbow stocking location (Buffalo Bill Reservoir), Duration of stocking | Relative abundance of parental species could affect probability of hybridization vs. probability of conspecific reproduction |
| Disturbance metrics | Proximity to road (binary, indicating same side of river as road), Total number of fish stocked from all species | Disturbance can promote hybridization by bringing species into contact or changing relative fitnesses |
We also included predictor variables related to stocking history. Since stocking history is likely to have directly affected ratios of Yellowstone cutthroat to rainbow trout in the North Fork Shoshone River basin, and therefore the probability of interspecific hybridization, we used historical stocking data to generate predictor variables for our analysis of hybridization. Prior to this study, stocking records earlier than 1980 had not been digitized. We compiled all available fish stocking data for North Fork Shoshone River between 1905 and the present (Table S2; this dataset is described in more detail in Nordberg et al. in prep). From these data, we calculated the total number of Yellowstone cutthroat and rainbow trout stocked in each tributary, and the duration of stocking at each location. Because far more rainbow trout were stocked in Buffalo Bill Reservoir than into the North Fork Shoshone River and its tributaries directly, we also calculated distance from each sampling site to the confluence of the North Fork Shoshone River with Buffalo Bill Reservoir, using NHDPlusV2 river shapefiles for this basin (National Hydrography Dataset; McKay et al. 2012) and the riverdist package in R (Tyers 2017), and used this distance as a predictor variable in our models. Previous studies of introduced rainbow trout suggest that many individuals remain close to their stocking location, but a subset disperse (Cresswell & Williams 1981, Bennett et al. 2010) and several studies suggest that distance from stocking locations of introduced rainbow trout can influence hybridization outcomes with cutthroat trout, or use indices of propagule pressure that include distance to quantify rainbow trout stocking (Gunnell et al. 2008, Bennett et al. 2010, Muhlfeld et al. 2017).
Finally, predictor variables also included two disturbance metrics. Ecological disturbances can promote hybridization (reviewed in Grabenstein & Taylor 2018), and variation in disturbance intensity could generate variation in hybridization outcomes. The North Fork Shoshone River basin is relatively undisturbed, as it lies almost entirely in Shoshone National Forest or Yellowstone National Park, and headwater portions of many tributary streams are in Wilderness areas. Therefore, we only considered two metrics of disturbance: proximity to the road that parallels the North Fork Shoshone main stem (quantified as a binary variable indicating whether a tributary is on the same side of the river as the road), and total number of fish stocked into a drainage (all species stocked, not just cutthroat and rainbow trout).
We used two methods to account for multicollinearity in predictors. First, we used a stepwise variance inflation factor calculation (vif in the car package in R; Fox & Weisberg 2011) to remove predictors with severe multicollinearity that might negatively affect model inference. We sequentially removed the predictor with highest variance inflation factor until all remaining predictors had variance inflation factors of less than or equal to 10 (as suggested by Marquardt 1970), removing first elevation and then total number of fish stocked. One predictor, distance from Buffalo Bill Reservoir, had variance inflation factor equal to 10. We also used partial correlations (ppcor in R; Kim 2015) to examine which predictors were correlated after removing the effect of all other predictor variables. This analysis also supported removing elevation and total number of fish stocked. Since distance from Buffalo Bill Reservoir did not have a high partial correlation with remaining predictors following removal of elevation and total number of fish stocked, we retained this variable in our final set of predictor variables.
We then conducted AICc-based model-averaging in a generalized linear modeling framework to rank relative importance of predictor variables associated with genomic outcomes of contact between Yellowstone cutthroat and rainbow trout in tributaries of the North Fork Shoshone River (Burnham & Anderson 2002). Specifically, we used the MuMIn package in R to calculate AICc for generalized linear models using all possible additive combinations of our 11 predictor variables. We then calculated the relative importance (RI) of each predictor variable as the sum of AICc weights of models in which that predictor variable was included. Relative importance (RI) scales from 0 to 1, with 0 indicating the variable had little influence on improving model fit and 1 indicating that the variable had very strong influence on improving model fit. In addition to calculating relative importance for each predictor variable, we also identified whether there was a positive or negative relationship between that predictor and the response variable. We also examined the top 5 models as ranked by AICc, and identified which predictors were significant across multiple models within the best-supported subset.
Results
DNA sequencing resulted in 2,047,579,520 reads 150 base pairs in length, representing 7 lanes of sequencing on the Illumina Hiseq 4000 platform (1×150). 1,219,638,920 of the reads matched to barcoded individual fish. Following alignment to the Atlantic salmon genome (Salmo salar; Lien et al. 2016) we retained 1,106,351,121 reads (90.7% of barcoded reads). We identified variable sites in the genome for 1,286 individuals, and retained 12,666 well-supported SNPs, which we then used in all subsequent analyses. Mean coverage at these retained SNPs was approximately 6 reads per locus per individual.
Quantifying genomic outcomes of hybridization
We quantified outcomes of hybridization in each tributary using genome-wide analyses. We used a hierarchical Bayesian model, entropy, to quantify ancestry of each individual fish (Gompert et al. 2014) by estimating both proportion of ancestry from each parental source and interspecific ancestry. These two metrics allowed us to distinguish among histories of hybridization that produced an individual fish, and distinguish first generation, backcrossed, and later generation recombinant hybrids (Fig. 2, 3). Estimates for q and Q for each individual fish were quite precise (mean 95% credible interval for q was 0.0218; mean 95% credible interval for Q was 0.0421).
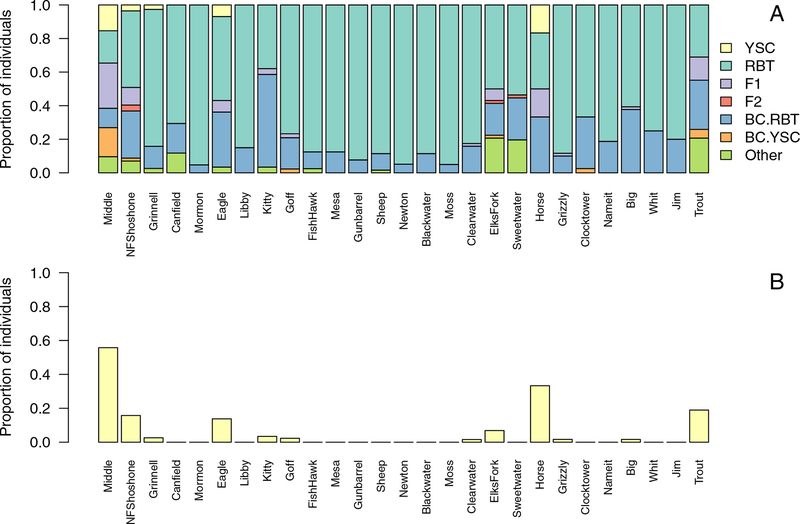
Figure 2:
A) Distribution of parental species and hybrids varied across locations, shown in upstream (left) to downstream (right) order. Individuals sampled included Yellowstone cutthroat trout (YSC), rainbow trout (RBT), first generation hybrids (F1), second generation hybrids (F2), backcrosses towards both parental species (BC.RBT, BC.YSC), and additional unclassified hybrids (Other). B) Proportion of juvenile Oncorhynchus sampled that had at least one Yellowstone cutthroat parent. Although juvenile Yellowstone cutthroat were rare, we were able to infer adult Yellowstone cutthroat spawners from the distribution of hybrid ancestry.
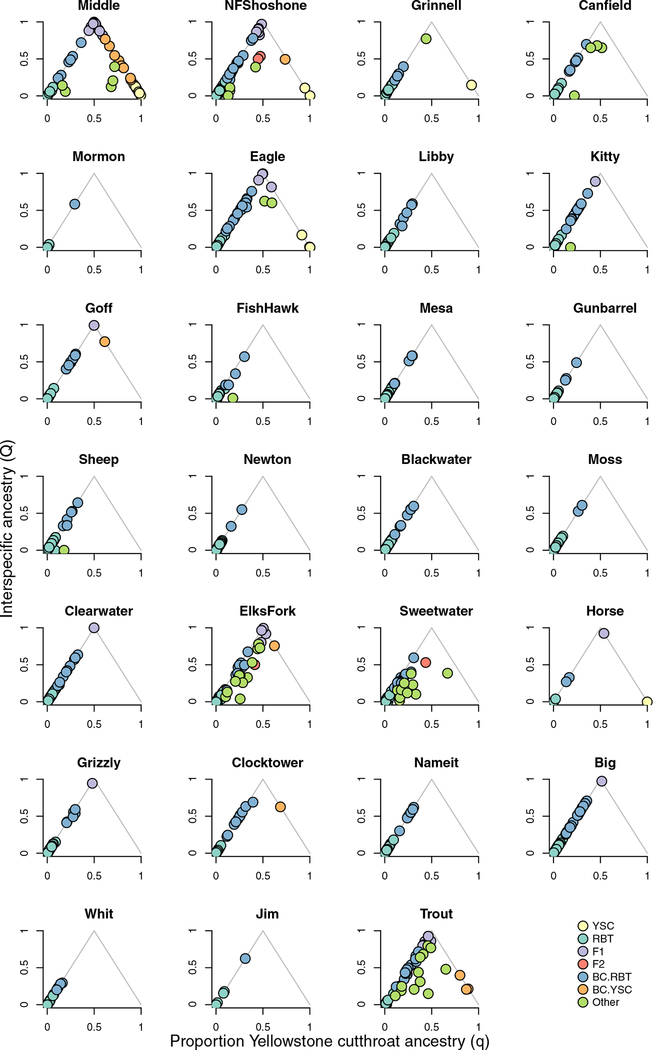
Figure 3:
Hybridization outcomes varied among tributaries (ordered upstream-downstream) where we sampled juvenile trout. In a few locations (e.g. Middle Creek), both parental species, first generation hybrids, and backcrosses were sampled; in other locations (e.g. Blackwater Creek) only rainbow trout and backcrosses to rainbow trout were sampled. Abbreviations are the same as those used in Fig. 2.
Our results indicate that non-hybridized juvenile Yellowstone cutthroat trout were very rare in the areas we sampled in the North Fork Shoshone river drainage, comprising less than 2% of sampled juveniles. We sampled Yellowstone cutthroat juveniles in only five out of 27 locations (Fig. 2A), although we can infer that adult Yellowstone cutthroat used at least 11 of the tributaries due to the presence of hybrids that had at least one Yellowstone cutthroat parent (F1 hybrids and advanced backcrosses towards Yellowstone cutthroat; see Fig. 2B). Most juveniles sampled were either rainbow trout or hybrid individuals. In 13 tributaries, non-native rainbow trout comprised >80% of sampled individuals, and all of the remaining individuals were hybrids.
Hybridization outcomes varied among the 27 tributaries we sampled (Fig. 2, Fig. 3). The majority of hybrids we sampled (65% of hybrids; 19% of sampled juveniles) were backcrosses towards rainbow trout, and we sampled backcrosses towards rainbow trout at all locations. In some tributaries, we also sampled F1 hybrids (11 locations), backcrosses to Yellowstone cutthroat trout (6 locations), and F2 or unidentified later-generation recombinant hybrids (11 locations).
Simulations
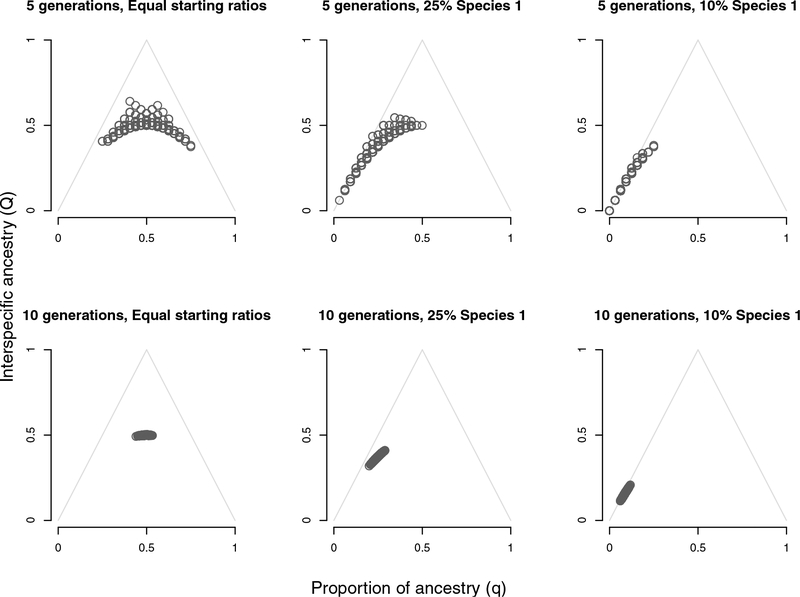
We used individual-based simulations to explore expectations for genomic outcomes of hybridization under a set of null expectations. These simulations assumed random mating, no selection against any genetic category of hybrids or parental species, and no immigration of additional parental individuals after the onset of hybridization. These results were informative for comparisons against empirical data to infer potential explanations for variation in hybridization. Results of simulations (Fig. 4; Fig. S2–S6 in Supplement) suggested that under most scenarios we explored, including scenarios where starting populations of parental species were skewed to be 90% one species, hybridization with random mating and no selection against hybrids led to loss of parental taxa and convergence of hybrid individuals on very homogeneous genetic ancestry within 10 generations with a starting population of 1000 individuals. In contrast, individuals from at least one parental species were present in each location sampled in the North Fork Shoshone River, and both summary statistics of hybrid ancestry (q and Q, proportion of ancestry and interspecific ancestry) were variable among the hybrid individuals at each sampling location. Later-generation intermediate hybrids (F2 and beyond) were also less common in the North Fork Shoshone drainage than we would expect if there was no assortative mating or selection against hybrids. Similarly, individuals with at least one parent from a parental species - those individuals along the sides of the triangle in Fig. 4 - were over-represented in the North Fork Shoshone relative to expectations generated by our simulations. Taken together, these features of our simulation results suggest that in the North Fork Shoshone River drainage, some mechanism or mechanisms of reproductive isolation operate to explain this deviation from neutral expectations.
Figure 4:
Results of simulations after 5 (top row) and 10 generations (bottom row) of hybridization for a population of 1000 individuals, assuming random mating, no selection against hybrids or either parental species, and no immigration of new individuals. These results suggest that the fish we sampled in the North Fork Shoshone (Fig. 3) include more individuals from parental species than we would expect given the number of generations of hybridization we know have occurred post-stocking. Simulations also suggest that we see fewer intermediate later generation hybrids (F2 and later) than we would expect under null expectations of random mating and no selection.
Environmental and ecological correlates of hybridization
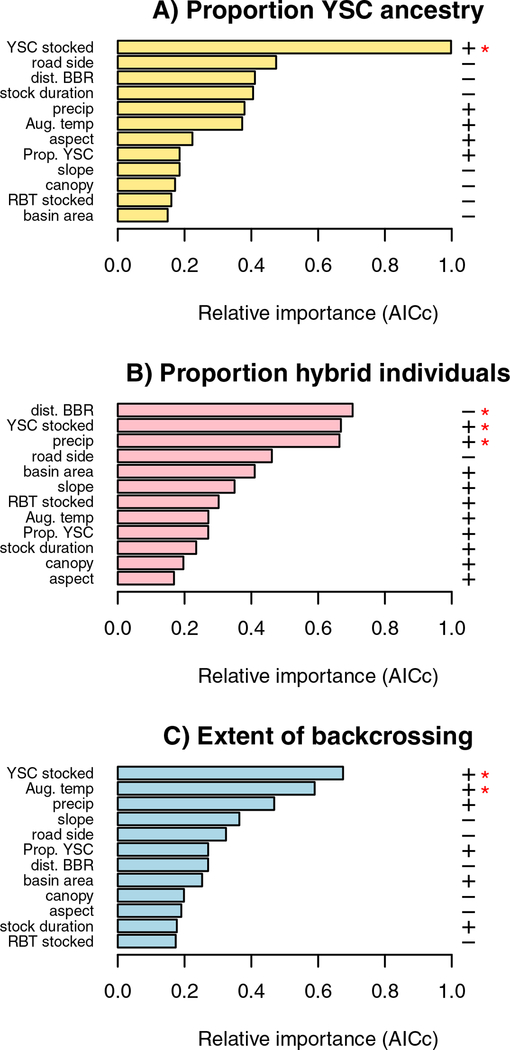
We modeled hybridization outcomes as a function of environmental variables, stocking history, and disturbance metrics (Table 1) to identify factors that contribute to the variation we observed in hybridization outcomes (Fig. 2, 3) and that might influence reproductive isolation between Yellowstone cutthroat and rainbow trout. We used a model averaging approach to rank predictors using relative importance (Burnham & Anderson 2002). For proportion Yellowstone cutthroat ancestry (Fig. 5A), total number of cutthroat trout stocked was the single most important predictor variable, and was the only significant predictor that was included in at least four of the top five models as ranked by AICc. Drainages where more Yellowstone cutthroat were stocked currently have a higher proportion of Yellowstone cutthroat alleles, suggesting that stocking has bolstered Yellowstone cutthroat populations. Only number of Yellowstone cutthroat trout stocked was a significant predictor in all of the five top models ranked by AICc.
Figure 5:
Model results identified different variables as most important for proportion Yellowstone cutthroat ancestry (A), proportion hybrid individuals in a tributary (B), and extent of backcrossing (C). Signs of relationships between predictors and an individual response variable are shown to the right of the figure. Predictors marked with red asterisks are those that were significant in at least four out of the five top ranked models by AICc. Abbreviations are as follows: RBT = rainbow trout, YSC = Yellowstone cutthroat trout, BBR = Buffalo Bill Reservoir.
For proportion hybrid individuals (Fig. 5B), the three leading predictors ranked by relative importance were distance from Buffalo Bill Reservoir (negative relationship), total number of Yellowstone cutthroat trout stocked (positive relationship), and precipitation (positive relationship), and all three of these predictors were significant in at least four out of five of the top models ranked by AICc. Extent of backcrossing was positively predicted by two variables that were significant in at least four of the top five models, namely number of Yellowstone cutthroat stocked and mean August temperature (Fig. 5C). The subset of highest-ranked models for each response variable predicted a substantial portion of the variance in the response variable (R2 values 0.41–0.63; see supplemental tables S2–S4).
Discussion
When closely related species come into contact and hybridize, outcomes are often variable across replicate zones of contact (Gompert et al. 2017), as we observed in the study of Yellowstone cutthroat × rainbow trout hybridization in the North Fork Shoshone River basin. While recent work has identified variation in genomic outcomes of hybridization in diverse systems (including plants, fish, mammals, and insects; Lepais et al. 2009, Nolte et al. 2009, Teeter et al. 2010, Haselhorst & Buerkle 2013, Gompert et al. 2014, Mandeville et al. 2017), the causes and evolutionary consequences of variable hybridization remain poorly understood in most systems. Identifying why outcomes of hybridization vary across instances of contact is crucial for progress in understanding mechanisms of reproductive isolation between species, and how variable reproductive isolation is across the range of an interaction between a pair of species (Gompert et al. 2017). Better mechanistic understanding of reproductive isolation is also crucial for predicting outcomes of hybridization in study systems where hybridization poses a threat to the persistence of native species (Todesco et al. 2016).
In this study, we observed variable outcomes of hybridization across the 27 locations we sampled where Yellowstone cutthroat trout interact with rainbow trout (Fig. 3). The distribution of hybrids in different classes (i.e., first-generation vs. backcrossed or recombinant hybrids) varied across locations (Fig. 2,3), and in most places backcrosses towards rainbow trout were far more common than either intermediate hybrids or backcrosses towards Yellowstone cutthroat, suggesting asymmetry in backcrossing and introgression (Fig.3). Spatial heterogeneity in hybridization is consistent with other studies of river-dwelling fishes, including hybridization in Catostomus suckers (Mandeville et al. 2015, 2017), between sculpin species (Nolte et al. 2009), and between westslope cutthroat and rainbow trout (Yau & Taylor 2013, Loxterman et al. 2014, Young et al. 2016, Muhlfeld et al. 2017). However the patterns we observed are distinct in several ways, in particular from observations in other cutthroat trout systems. In this study, juvenile Yellowstone cutthroat trout, the native species, were only present in a subset of locations. This contrasts with recent work on westslope cutthroat trout, which finds high relative abundances of native cutthroat trout despite ongoing introgressive hybridization with rainbow trout in some locations (Young et al. 2016, Mckelvey et al. 2016), although westslope cutthroat are rare relative to hybrids and rainbow trout in many locations too. Additionally, Yellowstone cutthroat × rainbow trout hybrids were sampled at all locations, in contrast with studies that have shown variation in presence of hybridization across locations (e.g. Mandeville et al. 2017, Muhlfeld et al. 2017). Spatial ubiquity of hybridization in the North Fork Shoshone River basin confirmed that rainbow and Yellowstone cutthroat trout lack complete reproductive isolation in this part of the introduced range of rainbow trout. However, simulation results also suggested that some mechanisms of reproductive isolation might maintain an over-representation of parental species and an under-representation of later-generation intermediate hybrids relative to null expectations. Over-representation of parental species relative to null expectations is also clear from our estimates of how many individuals have Yellowstone cutthroat parents (Fig.2), potentially suggesting some degree of assortative mating. We therefore explored relationships between potential predictors of prezygotic isolation and hybridization outcomes.
Why do hybridization outcomes vary?
Linking variation in hybridization to specific mechanisms of reproductive isolation is essential for improving our knowledge of the nature of species boundaries (Harrison & Larson 2014, Gompert et al. 2017), but understanding of the causal mechanisms of reproductive isolation is still limited for most species pairs. Many studies of hybridization and reproductive isolation have focused on barrier loci involved in intrinsic genetic components of reproductive isolation (Payseur 2010, Cutter 2012, Ravinet et al. 2017), and a few studies have identified variation in specific genetic mechanisms of reproductive isolation (Sweigart et al. 2007, Good et al. 2008, Vyskočilová et al. 2009, Kozlowska et al. 2012, Larson et al. 2018). However, hybridization is known to be environment-dependent in some systems (Taylor & Donald McPhail 2000, Vines et al. 2003), and it is likely that prezygotic isolation imposed by features of the environment substantially influences outcomes of hybridization in many species pairs. Based on the results of our simulations of hybridization, and the disparity between simulation results and empirical results (Fig. 3, 4), it is likely that in the North Fork Shoshone river drainage either postzygotic selection acts to constrain which categories of hybrids survive, or that prezygotic mechanisms produce some degree of assortative mating.
Another possibility is that there is ongoing immigration from parental species. Given that both Yellowstone cutthroat and rainbow trout in this system have an adfluvial life history (Kent 1984) - moving upstream from Buffalo Bill Reservoir annually to spawn, then returning to the reservoir - this would not be immigration in the typical sense, but rather a shift in migratory dynamics to favor different spawning tributaries. This scenario is possible, but we have no specific evidence regarding whether such a shift is occurring. Since this river system is bounded at its downstream extent by a dam at the outflow of Buffalo Bill Reservoir, individuals cannot easily enter the North Fork Shoshone river and its tributaries from more distant populations. The addition of more individuals by stocking is also unlikely. No rainbow trout have been stocked in this system since the 1960s, although there is precedent for a lag in long distance dispersal following rainbow trout stocking (Bower & Aitken 2008). Yellowstone cutthroat stocking also ended in nearly all locations by the year 2000 (one drainage, Trout Creek, continued to have some stocking in a headwater lake until 2009). Simulations also suggest that immigration alone is unlikely to explain the dynamics we observed in the North Fork Shoshone River.
We modeled hybridization outcomes as a function of features of the environment and history of fish stocking. The rationale for selecting environmental predictor variables was that environmental features are likely to affect spatial and temporal overlap of spawning for rainbow and Yellowstone cutthroat trout. Although spatial separation of spawning locations appears not to be a driving factor in the North Fork Shoshone River basin (Kruse 1998), temporal separation of spawning might be very important. In other river basins Yellowstone cutthroat and rainbow trout spawning times differ by up to a month, with hybrids spawning at a more similar time to rainbow trout (Henderson et al. 2000, DeRito et al. 2010). Stocking history was included in our set of predictor variables because stocking has likely altered ratios of parental species, therefore potentially influencing the probability of hybridization.
For all three hybridization response variables that we used in our models, stocking variables were most strongly predictive of outcomes of contact between Yellowstone cutthroat trout and rainbow trout (Fig. 5). The best predictor of the proportion of cutthroat ancestry at a site was the total number of Yellowstone cutthroat trout stocked in that location over the past century. For proportion hybrid individuals, two out of the three best predictor variables were related to stocking. The farther upstream a sampling location was from Buffalo Bill Reservoir, the primary location that rainbow trout were stocked in this system, the lower the proportion of hybrids in a stream was (Fig. 5B). These results suggest that hybridization dynamics have been influenced by stocking programs of Yellowstone cutthroat trout and rainbow trout in this river system, and that the persistence of Yellowstone cutthroat trout in the face of population increases of rainbow trout (Kruse 1998, Nordberg et al. in prep.) may have been facilitated by the continued stocking of Yellowstone cutthroat trout until approximately the year 2000, long after the cessation of rainbow trout stocking in the 1960s.
The patterns of hybridization we have observed in this study are consistent with studies of westslope cutthroat × rainbow trout hybridization in the sense that both this study and studies of westslope cutthroat identify proximity to rainbow trout introduction sites, temperature, and precipitation as important predictors of hybridization outcomes (Yau & Taylor 2013, Loxterman et al. 2014, Muhlfeld et al. 2014, Young et al. 2016, Muhlfeld et al. 2017). For both cutthroat subspecies, hybridization is predicted by warmer stream temperatures, although our results showed that increased temperature was more strongly predictive of backcrossing than with the proportion of hybrid individuals in a location (Fig. 5). However, studies of westslope cutthroat hybridization identify cold water temperatures as a mechanism that allows westslope cutthroat to persist (Yau & Taylor 2013, Young et al. 2016), although cold water temperatures might not entirely prevent hybridization (Muhlfeld et al. 2017). In contrast, our study suggests that increased Yellowstone cutthroat ancestry in the form of more symmetrical backcrossing to both rainbow trout and Yellowstone cutthroat trout is associated with warmer temperatures (Fig. 5). It also does not appear that Yellowstone cutthroat trout in this system occupy headwater refugia as westslope cutthroat do (Kruse et al. 2000, Paul & Post 2001), so spatial components of reproductive isolation might differ from the westslope cutthroat trout system, although we note that our sampling locations were not located in headwaters. It is unclear how thermal tolerance and thermal drivers of spawning vary among cutthroat subspecies (Bear et al. 2007), although these subspecies have generally been considered similar enough to combine data across cutthroat subspecies (e.g. Mandeville et al.). It is also likely that relationships between hybridization and temperature also depend on relative fitness and growth rates of Yellowstone cutthroat, rainbow, and hybrid trout (Seiler & Keeley 2009, Al-Chokhachy et al. 2013, Ostberg et al. 2013), and that the exact effect of temperature on hybridization will be complex and context-dependent.
Most prior work with westslope cutthroat has relied on fewer genetic markers than we used here (but see Hohenlohe et al. 2013, Kovach et al. 2016) and has relied more on preselected highly differentiated SNPs. Additionally, previous studies have typically focused on maximizing and quantifying accuracy in admixture proportions (Hohenlohe et al. 2013), but not necessarily estimating the combination of ancestry at specific loci (i.e., our Q parameter), which would allow differentiation between an F1 hybrid and later-generation hybrids with the same admixture proportion. It is therefore somewhat difficult to parse whether the patterns of hybridization we have observed - over-representation of backcrosses, under-representation of later-generation intermediate hybrids - are consistent across cutthroat subspecies when they hybridize with rainbow trout. This would be an interesting avenue for future comparison. We note that the use of smaller sets of diagnostic SNPs for previous studies might have provided narrower estimates of ancestry because smaller amounts of admixture are not detectable with smaller sets of SNPs. A challenge going forward will be distinguishing between small amounts of admixture and variation in parental species, and figuring out what level of admixture is relevant for conservation. Identifying small amounts of admixture correctly is potentially extremely important, given that a study that directly quantified cutthroat trout fitness in the wild suggested that all levels of admixture detectable with a total of ten diagnostic loci were detrimental to fitness in westslope cutthroat trout hybrids (Muhlfeld et al. 2009). However, while this study offered compelling and alarming evidence of the potential fitness consequences of hybridization, we also argue that this number of fixed SNPs might not accurately characterize ancestry of all hybrid individuals, and that there is likely to be significant variation in phenotype, including fitness, among hybrid individuals (Gompert et al. 2017). We believe there is still much work to be done to understand how hybridization affects fitness, both in this system and more generally. Indeed, recent studies of locus-specific hybridization in cutthroat trout provide a more complicated picture - one recent study in Montana suggests that rainbow trout alleles are resistant to introgression and therefore likely to be disadvantageous to fitness (Kovach et al. 2016), while another recent study suggests that in western Canada, some rainbow trout alleles are present at high frequency in hybrids across multiple contact zones, potentially suggesting fitness advantages (Bay et al. 2019).
We emphasize that hybridization may proceed differently across different subspecies of cutthroat trout, and that one contribution of this study is identifying similarities and differences between this system and others. One difference we observed is that numbers of native cutthroat in the North Fork Shoshone appear to be much lower than those in some studied locations within the westslope cutthroat trout range, especially in headwaters (Mckelvey et al. 2016, Young et al. 2016). However, there do not appear to be upstream refuges for Yellowstone cutthroat in the North Fork Shoshone River system, unlike scenarios commonly encountered for westslope cutthroat (Kruse 1998, Paul & Post 2001, Muhlfeld et al. 2014, Young et al. 2016). Additionally, hatchery-derived westslope cutthroat have not generally been stocked in numbers as large as the number of Yellowstone cutthroat stocked into the North Fork Shoshone River (Muhlfeld et al. 2017, Nordberg et al. in prep.). In contrast with studies of westslope cutthroat trout hybridization, the stocking variables that best predict Yellowstone cutthroat × rainbow trout hybridization in the North Fork Shoshone River basin are related to stocking of cutthroat trout, not stocking of rainbow trout. There are also life history differences between Yellowstone cutthroat trout in this system and westslope cutthroat trout. In westslope cutthroats, juveniles often outmigrate as year-old fish the spring after spawning (Behnke 1992), although there is significant variation across locations (Downs et al. 1997). In contrast, outmigration in Yellowstone cutthroat in the North Fork Shoshone happens in the fall of the same year in which they are born in the summer. This life history difference may have important implications for selection in the first 1–2 years of life, which may create differences in the viability and fitness of juveniles, particularly hybrid juveniles, in these systems (Muhlfeld et al. 2009). Indeed, in other salmonids, an age 0 outmigration is associated with low juvenile survival and minimal contribution to the adult population (as in bull trout; Downs et al. 2006). This contrast underscores the need to consider cutthroat trout subspecies individually, and take into account population differences in life history, when considering conservation status and management strategies. It is clear that one cutthroat trout subspecies cannot be equated with all cutthroat trout subspecies because of important life history differences like those highlighted above.
These results suggest some additional factors to bear in mind when considering cutthroat trout conservation. First, it might be important in some studies of fish populations to account for stocking of not just non-native species, but native species as well. It is important to keep in mind that any stocked population will differ genetically from the original wild populations, and the long-term stability of these stocked quasi-native populations is unclear (Létourneau et al. 2017). Replacement of native populations with stocked populations of the same species might have serious implications for the conservation of native fish species if fitness or life history traits in hatchery-derived fish are substantially different from the original native populations. Specifically in this case, it is unclear to what extent the Yellowstone cutthroat trout that persist in the North Fork Shoshone are derived from stocked ancestors, or are derived from a combination of stocked ancestors and the original native populations in the system, and if there are fitness differences among fish with these different ancestries. Preliminary analyses indicate that Yellowstone cutthroat trout in the North Fork Shoshone River are very similar genetically to current Yellowstone cutthroat broodstock maintained in the Tensleep Hatchery in Tensleep, Wyoming. Although this could indicate origins of the Tensleep broodstock from populations closely related to the North Fork Shoshone River populations, we cannot distinguish between this and a predominantly hatchery-derived origin for current North Fork Shoshone Yellowstone cutthroat trout. Range-wide genetic sampling of Yellowstone cutthroat trout is needed to understand whether the genetic variation present in this system likely represents populations that were originally there, or more closely resembles genetic variation in hatchery stocks used during the stocking program of the past 100 years.
Another consideration that our work brings to light is that stocking that occurred outside of the focal area - namely, intensive stocking of rainbow trout in Buffalo Bill reservoir, downstream of the tributaries we sampled - was relevant to outcomes of hybridization in upstream populations. In this case, it was not just stocking within the focal tributaries of this study that mattered, but also stocking downstream and adoption of a migratory life history in the North Fork Shoshone River trout populations, particularly for rainbow trout. Our examination of historical stocking records revealed that very few rainbow trout were stocked into the North Fork Shoshone River basin upstream of Buffalo Bill Reservoir (Nordberg et al. in prep.), which leads us to believe that rainbow trout currently in the tributaries of the North Fork Shoshone are likely descendants of individuals originally stocked into the reservoir. Our finding that distance from Buffalo Bill Reservoir is negatively associated with proportion hybrid individuals is therefore in agreement with previous studies have shown that propagule pressure, a metric of the intensity of stocking that includes distance from introduction location, can drive hybridization outcomes (Bennett et al. 2010, Yau & Taylor 2013, Loxterman et al. 2014, Muhlfeld et al. 2017).
Consequences of variable hybridization
Our observation that hybridization outcomes vary across locations for trout species in the North Fork Shoshone River, and are jointly predicted by both environmental conditions and history of fish stocking, has implications for our broader understanding of the origins and maintenance of reproductive isolation. As we study more replicate instances of interspecific hybridization, it is increasingly clear that the extent of reproductive isolation between a pair of species can vary spatially (Nolte et al. 2009, Teeter et al. 2010, Haselhorst & Buerkle 2013, Gompert et al. 2014, Mandeville et al. 2017). This means that reproductive isolation is itself variable, not uniform across the range of an interaction between a pair of species. Variation in reproductive isolation is potentially a result of complex and polymorphic genetic and phenotypic mechanisms that maintain reproductive isolation between species (Cutter 2012). Intrinsic genetic mechanisms of reproductive isolation like hybrid sterility are known to vary in model organisms (Sweigart et al. 2007, Kozlowska et al. 2012, Larson et al. 2018), but few studies of hybridization in the wild in non-model organisms have included enough replicate locations to robustly identify links to environmental or ecological components of reproductive isolation. Our study of Yellowstone cutthroat × rainbow trout hybridization demonstrates that hybridization dynamics can vary even on a relatively small spatial scale, and are potentially dependent on both historical ecological events (stocking) and abiotic environmental variables, although additional work is required to identify mechanisms of reproductive isolation more precisely.
Variation in outcomes of hybridization also poses a significant challenge for conservation of native species threatened by hybridization (Allendorf et al. 2001, Todesco et al. 2016, Grabenstein & Taylor 2018). Historically, understanding threats to native species by hybridization has been complicated by the uncertain conservation value of hybridized populations (Allendorf et al. 2001, 2004), as well as ambiguity in the legal status of hybrids with ancestry from threatened or endangered species, especially in the U.S. under the Endangered Species Act (Knudsen et al. 2005, Lind-Riehl et al. 2016). While we do not advocate specific conservation actions based on this work at present, especially for individuals with 1%–10% ancestry from rainbow trout, our work suggests that a broader view of hybridization as a conservation challenge might be warranted. Hybridization is often considered as a species-level threat to genetic integrity of native species (Rhymer & Simberloff 1996, Wolf et al. 2001, Todesco et al. 2016, Grabenstein & Taylor 2018), but it is likely that the threat posed by hybridization varies across the range of locations where a pair of species comes into contact. This is intuitive, as we know that when closely related species come into contact where they are both native, they often hybridize infrequently (e.g. Trotter 1989, McDonald et al. 2008), even if the same species hybridize extensively when brought together in locations where they are not historically sympatric. For conservation, heterogeneity in outcomes of hybridization poses a quandary: on the one hand, variation makes managing for persistence of the native species more complicated, as it might be imprudent to apply a single, common strategy across the shared geographic range of hybridizing native and introduced species. However, variation in outcomes of hybridization also presents the opportunity to prioritize conservation efforts where there are strongholds of native species to protect, and also to identify the conditions associated with persistence of native species and attempt to recreate these conditions in other locations.
Conclusions
Describing variation in hybridization outcomes, and understanding the corresponding mechanisms of reproductive isolation, is crucial for better understanding geographic dimensions of reproductive isolation, and how the efficacy of isolating mechanisms can vary across space and potentially through time. In this study, we identified substantial variation in outcomes of hybridization, which is be biologically relevant because it potentially reflects differences in overlap of spawning timing, spawning location, or fitness of hybrids across locations. We modeled this variation as a function of environmental variables and stocking, and were also able to identify a strong influence of stocking history on outcomes of hybridization. This result emphasizes that we need to consider the history of populations and how historical events might shape modern-day distributions of biodiversity. Along with stocking history, our work identified environmental variables as important predictors of hybridization, implying a potential interaction between the history of stocking and effects of current environmental conditions on spawning and hybridization. Additional field studies are needed to identify specific links between these predictors and hybridization outcomes, and quantify relative fitness of parental species and hybrids. In this system, and likely other systems too, multiple mechanisms interacted to determine the efficacy of reproductive isolation between species, with the potential for extensive variation across geographic locations. Beyond the implications of this work for our understanding of the evolution of reproductive isolation, we also identify challenges and opportunities for conservation going forward. While heterogeneity makes managing for persistence of native fish species more challenging from a policy standpoint, it also suggests that some populations might have greater potential for conservation success due to variation in hybridization dynamics.
Supplementary Material
Acknowledgements
We would like to thank the many people who made this work possible. Mark Smith from the Wyoming Game and Fish Department facilitated this work and administered funding. Mike Mazur from the US Fish and Wildlife Service provided reference Yellowstone cutthroat samples from a remote and isolated population outside the study area and provided funding for analysis of those individuals. Shawn Anderson (US Forest Service) assisted in acquiring both samples and funding for this work. We thank Sam Hochhalter, Joe Skorupski, Jessica Dugan, Riley Gallagher, Jake Scoville, Andrew Annear, and Shawn Anderson for field assistance. Personnel at the Story, Auburn, and Tensleep fish hatcheries in Wyoming provided tissue samples from reference individuals. This work was funded by grants from the Wyoming Game and Fish Department (#002433), the US Forest Service (Agreement 17-CS-11021400-012), and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant #2P20GM103432, as well as start-up funding from the University of Wyoming to CEW. CEW was partially supported by NSF Grant DEB-1556963. Computing was accomplished with an allocation from the University of Wyoming’s Advanced Research Computing Center, on its Mount Moran IBM SystemXcluster (http://n2t.net/ark:/85786/m4159c). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The manuscript was improved by comments from Mark Smith, Shawn Anderson, Josh Harrison, members of the Wagner lab at University of Wyoming, associate editor Paul Hohenlohe, and three anonymous reviewers.
Footnotes
Data accessibility
Data and scripts used for analysis are available in the Data Dryad repository (DOI: doi:10.5061/dryad.6s7d02q
References
- Advanced Research Computing Center (2012) Mount Moran: IBM System X cluster. Laramie, WY: University of Wyoming. [Google Scholar]
- Al-Chokhachy R, Alder J, Hostetler S, Gresswell R, Shepard B (2013) Thermal controls of Yellowstone cutthroat trout and invasive fishes under climate change. Global Change Biology, 19, 3069–3081. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Leary RF (1988) Conservation and Distribution of Genetic Variation in a Polytypic Species, the Cutthroat Trout. Conservation Biology, 2, 170–184. [Google Scholar]
- Allendorf FW, Leary RF (2005) Conservation and distribution of genetic variation in a polytypic species, the cutthroat trout. Conservation Biology, 2, 170–184. [Google Scholar]
- Allendorf FW, Leary RF, Hitt NP, Knudsen KL, Lundquist LL, Spruell P (2004) Inter-crosses and the U.S. endangered species act: Should hybridized populations be included as westslope cutthroat trout? Conservation Biology, 18, 1203–1213. [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends in Ecology & Evolution, 16, 613–622. [Google Scholar]
- Arnold ML, Ballerini ES, Brothers AN (2012) Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana Irises. Heredity, 108, 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM (1985) Analysis of hybrid zones. Annual Review of Ecology andSystematics, 16, 113–148. [Google Scholar]
- Bay RA, Taylor EB, Schluter D (2019) Parallel introgression and selection on introduced alleles in a native species. Molecular Ecology, p. mec.15097. [DOI] [PubMed] [Google Scholar]
- Bear EA, McMahon TE, Zale AV (2007) Comparative Thermal Requirements of Westslope Cutthroat Trout and Rainbow Trout: Implications for Species Interactions and Development of Thermal Protection Standards. Transactions of the American Fisheries Society, 136, 1113–1121. [Google Scholar]
- Behnke RJ (1992) Native trout of western North America. American Fisheries Society monograph (USA). no. 6.. [Google Scholar]
- Bennett SN, Olson JR, Kershner JL, Corbett P (2010) Propagule pressure and stream characteristics influence introgression: Cutthroat and rainbow trout in British Columbia. Ecological Applications, 20, 263–277. [DOI] [PubMed] [Google Scholar]
- Bower AD, Aitken SN (2008) Ecological genetics and seed transfer guidelines for \ emph{Pinus albicaulis (Pinaceae)}. American Journal of Botany, 95, 66–76. [DOI] [PubMed] [Google Scholar]
- Buerkle CA, Gompert Z (2013) Population genomics based on low coverage sequencing: how low should we go? Molecular Ecology, 22, 3028–3035. [DOI] [PubMed] [Google Scholar]
- Buerkle CA, Rieseberg LH (2001) Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution, 55, 684–691. [DOI] [PubMed] [Google Scholar]
- Buerkle CA, Wolf DE, Rieseberg LH (2003) The origin and extinction of species through hybridization In: Population Viability in Plants: Conservation, Management, and Modeling of Rare Plants (eds. Brigham CA, Schwartz MW), pp. 117–141, Springer Verlag, New York. [Google Scholar]
- Burnham K, Anderson D (2002) Model Selection and Multimodel Inference. Springer. [Google Scholar]
- Cresswell RC, Williams R (1981) Post-stocking Movements and Recapture of Hatchery-reared Trout Released into Flowing Waters - Effect of a Resident Wild Population. Journal of Fish Biology, 18, 429–442. [Google Scholar]
- Cutter AD (2012) The polymorphic prelude to {Bateson–Dobzhansky–Muller} incompatibilities. Trends in Ecology and Evolution, 27, 209–218. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRito JN, Zale AV, Shepard BB (2010) Temporal Reproductive Separation of Fluvial Yellowstone Cutthroat Trout from Rainbow Trout and Hybrids in the Yellowstone River. North American Journal of Fisheries Management, 30, 866–886. [Google Scholar]
- Downs CC, Horan D, Morgan-Harris E, Jakubowski R (2006) Spawning Demographics and Juvenile Dispersal of an Adfluvial Bull Trout Population in Trestle Creek, Idaho. North American Journal of Fisheries Management, 26, 190–200. [Google Scholar]
- Downs CC, White RG, Shepard BB (1997) Age at Sexual Maturity, Sex Ratio, Fecundity, and Longevity of Isolated Headwater Populations of Westslope Cutthroat Trout. North American Journal of Fisheries Management, 17, 85–92. [Google Scholar]
- Endicott C, Nelson L, Opitz S, et al. (2016) Range-wide status assessment for Yellowstone Cutthroat Trout (Oncorhynchus clarkii bouvieri) Tech. rep, Yellowstone Cutthroat Trout Interagency Coordination Group. [Google Scholar]
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B, Pau G (2017) rhdf5: HDF5 interface to R.
- Fitzpatrick BM, Shaffer HB (2007) Hybrid vigor between native and introduced salamanders raises new challenges for conservation. Proceedings of National Academy of Sciences, 104, 15793–15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S (2011) An R Companion to Applied Regression. 2nd edn, Sage, Thousand Oaks CA. [Google Scholar]
- Gompert Z, Buerkle CA (2010) \emph{introgress}: a software package for mapping components of isolation in hybrids. Molecular Ecology Resources, 10, 378–384. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA (2011a) A Hierarchical {B}ayesian Model for Next-Generation Population Genomics. Genetics, 187, 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA (2011b) Bayesian estimation of genomic clines. Molecular Ecology, 20, 2111–2127. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Lucas LK, Buerkle CA, Forister ML, Fordyce JA, Nice CC (2014) Admixture and the organization of genetic diversity in a butterfly species complex revealed through common and rare genetic variants. Molecular Ecology, 23, 4555–4573. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Mandeville EG, Buerkle CA (2017) Analysis of Population Genomic Data from Hybrid Zones. Annual Review of Ecology Evolution and Systematics, 48, 227–229. [Google Scholar]
- Good JM, Handel MA, Nachman MW (2008) Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution, 62, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenstein KC, Taylor SA (2018) Breaking Barriers: Causes, Consequences, and Experimental Utility of Human-Mediated Hybridization. Trends in Ecology & Evolution, pp. 1–15. [DOI] [PubMed] [Google Scholar]
- Gresswell RE (2011) Biology, status, and management of the yellowstone cutthroat trout. North American Journal of Fisheries Management, 31, 782–812. [Google Scholar]
- Gunnell K, Tada MK, Hawthorne FA, Keeley ER, Ptacek MB (2008) Geographic patterns of introgressive hybridization between native Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and introduced rainbow trout (O. mykiss) in the South Fork of the Snake River watershed, Idaho. Conservation Genetics, 9, 49–64. [Google Scholar]
- Halverson A (2010) An entirely synthetic fish: how rainbow trout beguiled America and over-ran the world. Yale University Press. [Google Scholar]
- Halverson MA (2008) Stocking Trends: A Quantitative Review of Governmental Fish Stocking in the United States , 1931 to 2004 Fisheries, 33, 69–75. [Google Scholar]
- Harrison RG, Larson EL (2014) Hybridization, introgression, and the nature of species boundaries Journal of Heredity, 105, 795–809. [DOI] [PubMed] [Google Scholar]
- Haselhorst MSH, Buerkle CA (2013) Population genetic structure of Picea engelmannii, P. glauca and their previously unrecognized hybrids in the central Rocky Mountains. Tree Genetics & Genomes, 9, 669–681. [Google Scholar]
- Hatfield T, Schluter D (1999) Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution, pp. 866–873. [DOI] [PubMed] [Google Scholar]
- Henderson R, Kershner JL, Toline CA (2000) Timing and Location of Spawning by Nonnative Wild Rainbow Trout and Native Cutthroat Trout in the South Fork Snake River, Idaho, with Implications for Hybridization. North American Journal of Fisheries Management, 20, 584–596. [Google Scholar]
- Hohenlohe PA, Day MD, Amish SJ, et al. (2013) Genomic patterns of introgression in rainbow and westslope cutthroat trout illuminated by overlapping paired-end RAD sequencing. Molecular ecology, 22, 3002–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaak DJ, Wenger SJ, Peterson EE, et al. (2017) The NorWeST Summer Stream Temperature Model and Scenarios for the Western U.S.: A Crowd-Sourced Database and New Geospatial Tools Foster a User Community and Predict Broad Climate Warming of Rivers and Streams. Water Resources Research, pp. 1–25. [Google Scholar]
- Kent R (1984) Fisheries Management Investigations in the Upper Shoshone River Drainage, 1978–1982 Tech. Rep August, Wyoming Game and Fish Department, Fish Division. [Google Scholar]
- Kim S (2015) ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients. Commun Stat Appl Methods, 22, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KL, LEARY RF, SPRUELL P, BOYER MC, HITT NP, ALLENDORF FW (2005) Cutthroat Trout Hybridization and the U.S. Endangered Species Act: One Species, Two Policies. Conservation Biology, 19, 1326–1328. [Google Scholar]
- Kovach RP, Eby LA, Corsi MP (2011) Hybridization between Yellowstone Cutthroat Trout and Rainbow Trout in the Upper Snake River Basin, Wyoming. North American Journal of Fisheries Management, 31, 1077–1087. [Google Scholar]
- Kovach RP, Hand BK, Hohenlohe PA, et al. (2016) Vive la résistance: genome-wide selection against introduced alleles in invasive hybrid zones. Proc. R. Soc. B, 283, 20161380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach RP, Muhlfeld CC, Boyer MC, Lowe WH, Allendorf FW, Luikart G (2015) Dispersal and selection mediate hybridization between a native and invasive species. Proceedings. Biological sciences / The Royal Society, 282, 20142454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowska JL, Ahmad AR, Jahesh E, Cutter AD (2012) Genetic Variation for Postzygotic Reproductive Isolation between \emph{Caenorhabditis briggsae} and \emph{Caenorhabditis} {Sp. 9}. Evolution, 66, 1180–1195. [DOI] [PubMed] [Google Scholar]
- Kruse CG (1998) Influence of Non-native trout and Geomorphology on Distributions of Indigenous trout in the Yellowstone River Drainage of Wyoming Ph.D. thesis, University of Wyoming. [Google Scholar]
- Kruse CG, Hubert WA, Rahel FJ (2000) Status of Yellowstone Cutthroat Trout in Wyoming Waters. North American Journal of Fisheries Management, 20, 693–705. [Google Scholar]
- Larson EL, Vanderpool D, Sarver BAJ, et al. (2018) The evolution of polymorphic hybrid incompatibilities in house mice. Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepais O, Petit R, Guichoux E, et al. (2009) Species relative abundance and direction of introgression in oaks. Molecular Ecology, 18, 2228–2242. [DOI] [PubMed] [Google Scholar]
- Létourneau J, Ferchaud AL, Le Luyer J, Laporte M, Garant D, Bernatchez L (2017) Predicting the genetic impact of stocking in Brook Charr (Salvelinus fontinalis) by combining RAD sequencing and modeling of explanatory variables. Evolutionary Applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011) A statistical framework for {snp} ping calling, mutation discovery, association mapand population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. arXiv preprint arXiv:1303.3997.
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Koop BF, Sandve SR, et al. (2016) The Atlantic salmon genome provides insights into rediploidization. Nature, 533, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind-Riehl JF, Mayer AL, Wellstead AM, Gailing O (2016) Hybridization, agency discretion, and implementation of the U.S. Endangered Species Act. Conservation Biology, 30, 1288–1296. [DOI] [PubMed] [Google Scholar]
- Lindtke D, Gompert Z, Lexer C, Buerkle CA (2014) Unexpected ancestry of Populus seedlings from a hybrid zone implies a large role for postzygotic selection in the maintenance of species. Molecular ecology, 23, 4316–4330. [DOI] [PubMed] [Google Scholar]
- Loxterman JL, Keeley ER, Njoroge ZM, Bradford M (2014) Evaluating the influence of stocking history and barriers to movement on the spatial extent of hybridization between westslope cutthroat trout and rainbow trout. Canadian Journal of Fisheries and Aquatic Sciences, 71, 1050–1058. [Google Scholar]
- Mandeville CP, Rahel FJ, Patterson LS, Walters AW () Integrating fish assemblage data, modeled stream temperatures, and thermal tolerance metrics to develop thermal guilds for water temperature regulation: Wyoming case study. Transactions of the American Fisheries Society, 0. [Google Scholar]
- Mandeville E, Parchman T, Song S, et al. (2017) Inconsistent reproductive isolation revealed by interactions between Catostomus fish species. Evolution Letters, 1, 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeville EG, Parchman TL, McDonald DB, Buerkle CA (2015) Highly variable reproductive isolation among pairs of Catostomus species. Molecular ecology, 24, 1856–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt DW (1970) Generalized Inverses, Ridge Regression, Biased Linear Estimation, and Nonlinear Estimation. Technometrics, 12, 591–612. [Google Scholar]
- McDonald DB, Parchman TL, Bower MR, Hubert WA, Rahel FJ (2008) An introduced and a native vertebrate hybridize to form a genetic bridge to a second native species. Proceedings of National Academy of Sciences, 105, 10837–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane SE, Pemberton JM (2019) Detecting the True Extent of Introgression during Anthropogenic Hybridization. Trends in Ecology and Evolution, xx, 1–12. [DOI] [PubMed] [Google Scholar]
- McKay L, Bondelid T, Dewald T, Johnston J, Moore R, Rea A (2012) NHDPlus Version 2: User Guide.
- Mckelvey KS, Young MK, Wilcox TM, Bingham DM, Pilgrim KL, Schwartz MK (2016) Patterns of hybridization among cutthroat trout and rainbow trout in northern Rocky Mountain streams. Ecology and Evolution, 6, 688–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlfeld CC, Kalinowski ST, McMahon TE, et al. (2009) Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters, 5, 328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlfeld CC, Kovach RP, Al-Chokhachy R, et al. (2017) Legacy introductions and climatic variation explain spatiotemporal patterns of invasive hybridization in a native trout. Global Change Biology, pp. 1–12. [DOI] [PubMed] [Google Scholar]
- Muhlfeld CC, Kovach RP, Jones LA, et al. (2014) Invasive hybridization in a threatened species is accelerated by climate change. Nature Climate Change, 4, 620–624. [Google Scholar]
- Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA (2013) Genotyping-by-sequencing in ecological and conservation genomics. Molecular Ecology, 22, 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte AW, Gompert Z, Buerkle CA (2009) Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Molecular Ecology, 18, 2615–2627. [DOI] [PubMed] [Google Scholar]
- Ostberg CO, Duda JJ, Graham JH, et al. (2011) Growth, Morphology, and Developmental Instability of Rainbow Trout, Yellowstone Cutthroat Trout, and Four Hybrid Generations. Transactions of the American Fisheries Society, 140, 334–344. [Google Scholar]
- Ostberg CO, Hauser L, Pritchard VL, Garza JC, Naish Ka (2013) Chromosome rearrangements, recombination suppression, and limited segregation distortion in hybrids between Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and rainbow trout (O. mykiss). BMC genomics, 14, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey F, Benkman CW, Buerkle CA (2012) Genome-wide association genetics of an adaptive trait in lodgepole pine. Molecular Ecology, 21, 2991–3005. [DOI] [PubMed] [Google Scholar]
- Pasaniuc B, Rohland N, McLaren PJ, et al. (2012) Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nature Genetics, 44, 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AJ, Post JR (2001) Spatial Distribution of Native and Nonnative Salmonids in Streams of the Eastern Slopes of the Canadian Rocky Mountains. Transactions of the American Fisheries Society, 130, 417–430. [Google Scholar]
- Payseur BA (2010) Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology Resources, 10, 806–820. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravinet M, Faria R, Butlin R, et al. (2017) Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. Journal of evolutionary biology, 30, 1450–1477. [DOI] [PubMed] [Google Scholar]
- Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annual Review of Ecology and Systematics, 27, 83–109. [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation, and speciation. Heredity, 83, 363–372. [DOI] [PubMed] [Google Scholar]
- Seiler SM, Keeley ER (2007) A comparison of aggressive and foraging behaviour between juvenile cutthroat trout, rainbow trout and F1 hybrids. Animal Behaviour, 74, 1805–1812. [Google Scholar]
- Seiler SM, Keeley ER (2009) Competition between native and introduced salmonid fishes: cutthroat trout have lower growth rate in the presence of cutthroat–rainbow trout hybrids. Canadian Journal of Fisheries and Aquatic Sciences, 66, 133–141. [Google Scholar]
- Sweigart AL, Mason AR, Willis JH (2007) Natural variation for a hybrid incompatibility between two species of \emph{Mimulus}. Evolution, 61, 141–151. [DOI] [PubMed] [Google Scholar]
- Taylor EB, Donald McPhail J (2000) Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proceedings of the Royal Society B: Biological Sciences, 267, 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, Buerkle CA, Nachman MW, Tucker PK (2010) The variable genomic architecture of isolation between hybridizing species of house mouse. Evolution, 64, 472–485. [DOI] [PubMed] [Google Scholar]
- Todesco M, Pascual MA, Owens GL, et al. (2016) Hybridization and extinction. Evolutionary Applications, 9, 892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PC (1989) Coastal cutthroat trout : A life history compendium. Transactions of the American Fisheries Society, 115, 463–473. [Google Scholar]
- Tyers M (2017) riverdist: River Network Distance Computation and Applications.
- Underwood ZE, Mandeville EG, Walters AW (2016) Population connectivity and genetic structure of burbot (Lota lota) populations in the Wind River Basin, Wyoming. Hydrobiologia, 765, 329–342. [Google Scholar]
- Vines TH, Kohler SC, Thiel A, et al. (2003) The maintenance of reproductive isolation in a mosaic hybrid zone between the fire-bellied toads Bombina bombina and B. variegata. Evolution, 57, 1876–1888. [DOI] [PubMed] [Google Scholar]
- Vyskočilová M, Pražanová G, Piálek J (2009) Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mammalian Genome, 20, 83–91. [DOI] [PubMed] [Google Scholar]
- Wagner CE, Mandeville EG (2017) Speciation, species persistence and the goals of studying genomic barriers to gene flow. Journal of Evolutionary Biology, 30, 1512–1515. [DOI] [PubMed] [Google Scholar]
- Walters AW, Mandeville EG, Saunders WC, et al. (2017) Comparison of burbot populations across adjacent native and introduced ranges. Aquatic Invasions, 12, 251–262. [Google Scholar]
- Wolf DE, Takebayashi N, Rieseberg LH (2001) Predicting the risk of extinction through hybridization. Conservation Biology, 15, 1039–1053. [Google Scholar]
- Yau MM, Taylor EB (2013) Environmental and anthropogenic correlates of hybridization between westslope cutthroat trout (Oncorhynchus clarkii lewisi) and introduced rainbow trout (O. mykiss). Conservation Genetics, 14, 885–900. [Google Scholar]
- Young MK, Isaak DJ, McKelvey KS, et al. (2016) Climate, demography, and zoogeography predict introgression thresholds in salmonid hybrid zones in Rocky Mountain streams. PLoS ONE, 11, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.