Abstract
Defective mismatch repair has been detected in human colorectal and endometrial carcinomas which exhibit microsatellite instability (MIN). The purpose of this study was to search for MIN in melanoma. Paraffin-embedded neoplastic and non-neoplastic control cells were obtained from 20 untreated individuals with cutaneous malignant melanoma. Breslow thickness ranged from 0.2–7.4 mm (mean 1.4). Cells were carefully scraped from glass slides so that tumor and control DNA could be isolated and then amplified by polymerase chain reaction (PCR) at seven separate microsatellites localized to specific chromosome regions: lp22 (D1S187), 5qll.2–13.3 (D5S107), 6q21–23.3 (D6S357), 9p21 (IFNA), llpl5.2 (D11S861), 17pl3.1 (D17S786), and 18qll (D18S34). Heterozygosity indices were ≥0.70. Loci from these chromosome regions were chosen because of cytogenetic abnormalities reported in melanoma (lp, 6q, 9p), location of common oncogenes (lip-HRAS, 17p-TP53), or use in other MIN studies (5q, 18q). Five individuals (25%) demonstrated MIN. There was no correlation with tissue thickness. One individual demonstrated MIN at two loci and one individual demonstrated loss of heterozygosity. The results indicate that MIN occurs in melanoma, albeit less frequently than reported in carcinomas.
INTRODUCTION
Approximately 34,000 new cases of cutaneous malignant melanoma (MM) are diagnosed annually in the United States, with incidence rates increasing more rapidly than that of any other malignancy. This year alone, there will be approximately 7,200 melanoma-related deaths [1]. Approximately 10% of melanoma cases are familial; however, the majority of cases are sporadic [2]. Numerous chromosomal abnormalities in MM have been identified [3–8].
Molecular genetic studies of several malignancies have detected alterations in tumor DNA, relative to normal DNA, which reflect amplification or deletion of interspersed repetitive sequences of DNA [9–15]. These DNA sequences, termed microsatellites, consist of polymorphic tandem repeated elements of the form (CA)n, with the range of n being 15–30, which are widely distributed throughout the human genome [16–17]. Microsatellite instability (MIN) has been described in several tumors, including colon cancer [11], hereditary nonpolyposis colorectal cancer (HNPCC) [10], non-small cell lung carcinoma [9], endometrial carcinoma [12], gastric cancer [13], pancreatic cancer [14], and keratoacanthoma [16], MIN has been proposed as an indirect marker of globally defective DNA mismatch repair in the cells of cancer patients [18]. In this study, we searched for MIN in MM utilizing polymerase chain reaction (PCR) primers for microsatellites on chromosome regions 1p, 6q, 9p, lip, 17p, 5q, and 18q. These particular cytogenetic regions were selected because of: (1) previous reports of abnormalities at these sites in melanoma patients (1p, 6q, 9p) [3, 6, 7]; (2) prior identification of common oncogene sites (11p-HRAS, 17p-TP53) [19, 20]; or (3) their use in other MIN studies (5q, 18q) [11],
MATERIALS AND METHODS
Twenty patients (14 males and six females; average age 54.3 years with a range of 30 to 92 years) with previously untreated histologically confirmed sporadic cutaneous (n = 16) or metastatic (n = 4) MM treated at Vanderbilt University Medical Center were randomly chosen and analyzed. Clinical and genetic data for these patients are listed in Table 1. Hematoxylin- and eosin-stained biopsy slides were reviewed and corresponding paraffin-embedded un-stained slides obtained. Tumor and normal tissues were carefully scraped from the slides using disposable scalpels. The tissue was then deparaffinized and DNA isolated. Forward PCR primers were end-labeled with [γ32P-dATP] (Amersham Co., Arlington Heights, IL). PCR amplification was performed, following established protocols, at seven separate DNA microsatellites localized to chromosome regions: lp22(DlSl87), 5qll.2–13.3(D5Sl07), 6q21–23.3(D6-S357), 9p21(IFNA), Hpl5.2(DllS861), 17pl3.1(Dl7S-786), and 18qll(Dl8S34). Heterozygosity indices for the primers were ≥0.70 [17, 21–26]. Tumor tissue was then compared to normal tissue for each case.
Table 1.
Clinical and genetic data for cutaneous sporadic melanoma patients
| Patient | Age/sex | Location | Type | Thicknessa | Instabilityb |
|---|---|---|---|---|---|
| 1 | 73/M | Shoulder | SS | 7.4 mm | ND |
| 2 | 54/M | Back | SS | 0.2 mm | D5S107c |
| 3 | 50/M | Metastasis | UND | — | ND |
| 4 | 40/M | Metastasis | UND | — | ND |
| 5 | 39/F | Metastasis | UND | — | ND |
| 6 | 92/M | Scalp | SS | 1.3 mm | D1S187c |
| 7 | 30/F | Arm | SS | 3.4 mm | ND |
| 8 | 44/M | Shoulder | LM | 0.5 mm | ND |
| 9 | 43/F | Vulva | SS | 1.3 mm | ND |
| 10 | 55/M | Back | SS | 1.5 mm | D17S786c |
| 11 | 34/F | Leg | SS | 1.1 mm | ND |
| 12 | 71/M | Metastasis | UND | — | D1S187c |
| 13 | 44/F | Breast | SS | 0.2 mm | ND |
| 14 | 52/M | Back | SS | 0.8 mm | D6S357c + D1S187c |
| 15 | 63/M | Back | SS | 0.8 mm | ND |
| 16 | 35/F | Arm | SS | 0.7 mm | ND |
| 17 | 64/M | Scalp | SS | 0.5 mm | ND |
| 18 | 76/M | Unknown | SS | 0.9 mm | ND |
| 19 | 45/M | Chest | SS | 0.9 mm | D6S357d |
| 20 | 81/M | Back | SS | 1.2 mm | ND |
Abbreviations: ND, no instability detected; SS, superficial spreading; UND, type not determined; LM, lentigo maligna.
Thickness of metastatic specimens.
Patient’s samples show instability at the indicated sites.
Replication error at locus indicated.
Loss of heterozygosity.
PCR conditions varied according to the primer selected. Primers D18S34, D5S107, D1S187, and IFNA underwent 27 step cycles with 30 seconds at 94°C, 75 seconds at 55°C, 15 seconds at 72°C, and then 6 minutes at 72°C. Primers D17S786 and D6S357 underwent 27 step cycles with 1 minute at 94°C, 2 minutes at 55°C, 2 minutes at 72°C, and then 10 minutes at 72°C. Primer D11S861 underwent 25 step cycles with 1 minute at 94°C, 2 minutes at 57°C, 2 minutes 30s at 72°C, and then 10 minutes at 72°C [17, 21–26].
Following PCR, the samples were denatured at 95°C for 6 minutes. The samples were then immediately placed on ice, and 3.0 microliters of each sample was loaded onto a 6% acrylamide gel. Electrophoresis was performed at 90 watts for 90 minutes and, after cooling, the gel was transferred to gel blot paper and dried. The gel blot paper underwent autoradiography with exposure for 1 to 96 hours at –70°C prior to x-ray film developing and examination.
MIN was recorded when an insertion/deletion of repeat units [replication error) was observed in at least one tumor DNA locus compared to its non-neoplastic control DNA. Loss of heterozygosity was observed when the radio-graphic signal of one allele was reduced in tumor DNA compared with control in those patients heterozygous for a specific locus on repeated analyses.
RESULTS
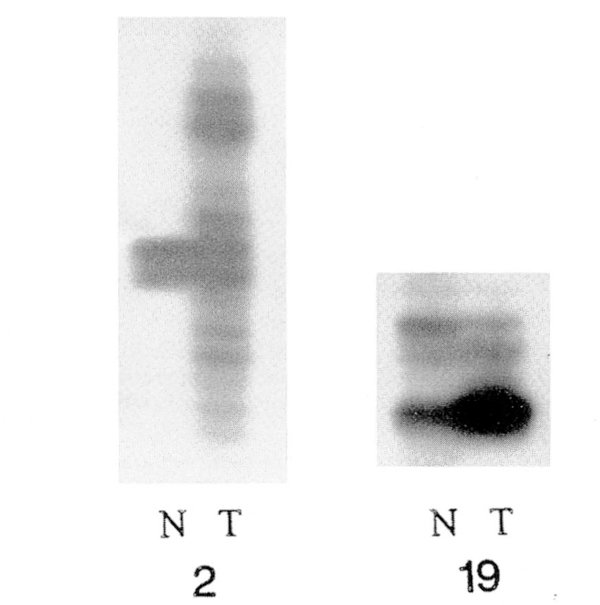
Overall, seven separate DNA microsatellites were studied in our patients. All samples were PCR-amplified with primers representing D1S187, D6S357, IFNA, D11S861, and D17S786 while five patients were analyzed for D18S34 and D5S107. Recording of the Breslow [27] tissue thickness specimens from the microscope slides of the 15 patients with cutaneous MM was undertaken and ranged from 0.2–7.4 mm (mean 1.4 mm). No correlation was found between tissue thickness and the presence of MIN. Instability (MIN or loss of heterozygosity) was noted in six individuals (see Table 1). Five showed an apparent error in replication, i.e., a change in allele size or pattern (see Fig. 1), while one patient repeatedly exhibited loss of heterozygosity in the tumor sample. MIN was detected at two chromosome regions previously displaying cytogenetic abnormalities in reported melanoma studies (i.e., 1p and 6q). Of those samples exhibiting MIN, four were cutaneous lesions and one was metastatic. One patient with a thin cutaneous melanoma had MIN detected at two loci; all other individuals had MIN at a single locus. Loss of heterozygosity was detected in one individual with a thin melanoma of the superficial spreading type.
Figure 1.
Melanoma microsatellite instability studies in two representative patients (2, 19). Patient 2 displays instability at the D5S107 locus, when comparing normal (N) and tumor (T) tissue from each patient.
DISCUSSION
Cutaneous MM lends itself to the study of tumorigenesis and metastasis, due to its natural history [28], The association between dysplastic nevi and melanoma is well-established [29], It is also noted that most types of melanoma exhibit a linear stage of growth, followed by vertical growth and, ultimately, metastasis [28]. Thus, in certain individuals with multiple or giant nevi, identifiable pre-malignant cells become cancerous, and those cells change their behavior as the disease progresses. The triggering and modulating events that dictate this behavior are still unclear. Though our study shows that microsatellite instability exists in melanoma, its role in the malignant process is not well-defined.
Microsatellite instability has recently been identified in melanoma, but with contrasting results. An English study identified a replication error at one locus in only one individual [30], Loss of heterozygosity (LOH) at one locus (8q) was also observed. Clinical parameters were not described. A European study identified a 20% MIN rate in 40 melanoma cases and LOH at 9p21 in 40% and at 9p22 in 22% [31]. Depth of invasion correlated only with LOH at 9p21, when cancer depths were greater than 1.5 mm.
Our results, representing sporadic cutaneous melanoma with primary and distant metastasis from patients from the United States, demonstrated a similar MIN rate (25%) as the European study but a dissimilar rate for LOH. Neither MIN nor LOH correlated with tissue depth in our study.
The microsatellite instability detected in our tumor system is different from those reported in other tumor systems with a relatively lower frequency of MIN at more than one locus. This clearly differs from the genetic instability reported in other tumors (e.g., colon and endome trial cancer), where instability occurs at several loci [10–12, 32]. Four mismatch repair genes (hMSH2, hMLH1, hPMS1, and hPMS2) are known to be mutated in the germ line cells of patients with HNPCC [32]. Mutations in hMSH2 and hMLH1, the human homologs to the Eschri-chia coli mismatch repair genes mutS and mutL, have been clearly identified in colorectal and endometrial cell lines [33]. These findings support the hypothesis that a defect in DNA replication or repair may represent an additional molecular mechanism in human tumorigenesis.
In summary, five of our individuals with MM demonstrated MIN and one additional individual demonstrated LOH, but with no correlation between thickness or type. No correlation was observed between the biological aggressiveness of its tumor, onset of the disease or presence of MIN. Instability was evident at a lower frequency than previously identified in certain tumors displaying specific mismatch repair defects. Malignant melanoma can now be added to the list of malignancies demonstrating somatic alterations in the repeat length of microsatellites.
Acknowledgments
This research was supported in part by a research grant from the Orthopaedic Research and Education Foundation, number 9-005 (HSS). We thank Janie Falkenberg for expert preparation of the manuscript.
Footnotes
Portions of this work were included in a poster presented at a meeting of the American Association of Cancer Research in Washington, DC in April 1996.
REFERENCES
- 1.Wingo PA, Tony T, Bolden S (1995): Cancer statistics CA. A Cancer Journal for Clinicians. (A Journal of the American Cancer Society) 45:8–30. [DOI] [PubMed] [Google Scholar]
- 2.Greene MH, Fraumei W Jr (1979): The hereditary variant of malignant melanoma In: Human Malignant Melanoma. Clark WH Jr, Goldman U, Mastrangelo MJ, eds. Grune & Stratton, New York, pp. 139–166. [Google Scholar]
- 3.Trent JM, Rosenfeld SB, Meyskens FL (1983): Chromosome 6q involvement in human malignant melanoma. Cancer Genet Cytogenet 9:177–180. [DOI] [PubMed] [Google Scholar]
- 4.Jaspers NGJ, Roza-de-Jongh EJM, Donselaar IG, van Velzen-Tillemans JTM, van Hemel JO, Rümke P, van derKamp AW (1987): Sister chromatid exchange, hyperdiploidy and chromosomal rearrangements studied in cells from melanoma-prone individuals belonging to families with the dysplastic nevus syndrome. Cancer Genet Cytogenet 24:33–43. [DOI] [PubMed] [Google Scholar]
- 5.Greene MH, Clark WH, Tucker MA, Elder DE, Kraemer KH, Guerry D, Witmer WK, Thompson J, Matozzo I, Fraser MC (1985): Acquired precursors of cutaneous malignant melanoma. The familial dysplastic nevus syndrome. New Eng J Med 312:91–97. [DOI] [PubMed] [Google Scholar]
- 6.Bale SJ, Dracopoli NC, Tucker MA (1989): Mapping the gene for hereditary malignant melanoma: dysplastic nevus to chromosome lp. New Eng J Med 320:1367–1372. [DOI] [PubMed] [Google Scholar]
- 7.Cannon-Albright LA, Goldgar DE, Meyer LJ, Lewis CM, Anderson DE, Fountain JW, Hegi ME, Wiseman RW, Petty EM, Boule AE (1992): Assignment of a locus for familial melanoma, MLM, to chromosome 9pl3-p22. Science 258:1148–1152. [DOI] [PubMed] [Google Scholar]
- 8.Milliken D, Meese B, Vogelstein B, Witkowski C, Trent J (1991): Loss of heterozygosity for loci on the long arm of chromosome 6 in human malignant melanoma. Cancer Res 51:5449–5453. [PubMed] [Google Scholar]
- 9.Shridhar V, Siegfried J, Hunt J, del Mar A, Smith DI (1994): Genetic instability of microsatellite sequences in non-small cell lung carcinomas. Cancer Res 54:2084–2087. [PubMed] [Google Scholar]
- 10.Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Meoklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, Petersen GM, Kinzler KW, Vogelstein B, de la Chapelle A (1993): Clues to the pathogenesis of familial colorectal cancer. Science 260:812–816. [DOI] [PubMed] [Google Scholar]
- 11.Thibodeau SN, Bren G, Schaid P (1993): Microsatellite instability in cancer of the proximal colon. Science 260:816–819. [DOI] [PubMed] [Google Scholar]
- 12.Risinger JI, Berchuk A, Kohler MF, Watson P, Lynch HT, Boyd J (1993): Genetic instability of microsatellites in endometrial carcinoma. Cancer Res 53:5100–5103. [PubMed] [Google Scholar]
- 13.Mironov NM, Aguelon MA, Potapova GI, Omori Y, Gorbunov OV, Klimenkov AA, Yamasaki H (1994): Alterations of (CA)n DNA repeats and tumor suppressor genes in human gastric cancer. Cancer Res 54:51–54. [PubMed] [Google Scholar]
- 14.Han H- J, Yanagisawa A, Kato Y, Park J- G, Nakamura Y (1993): Genetic instability in pancreatic cancer and poorly differentiated types of gastric cancer. Cancer Res 53:5087–5089. [PubMed] [Google Scholar]
- 15.Hailing KC, Honchel R, Pittelkow MR, Thibodeau SN (1995): Microsatellite instability in keratoacanthoma. Cancer 76:1765–1771. [DOI] [PubMed] [Google Scholar]
- 16.Weber JL, May PE (1989): Abundant class of human DNA polymorphisms which can be typed using polymerase chain reaction. Am J Human Genet 44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 17.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morisette J, Weissenbach J (1996): A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature 380:152–154. [DOI] [PubMed] [Google Scholar]
- 18.Honchel R, Hailing KC, Thibodeau SN (1995): Genomic instability in neoplasia. Semin Cell Biol 6:45–52. [DOI] [PubMed] [Google Scholar]
- 19.Levine AJ, Momand J, Finlay CA (1991): The p53 tumour suppressor gene. Nature 351:453–456. [DOI] [PubMed] [Google Scholar]
- 20.Ramon Y, Cajal S, Suster S, Halabal R, Filvaroff E, Dotto GP (1991): Induction of different morphologic features of malignant melanoma and pigmented lesions after transformation of murine melanocytes with bFGF-cDNA and H-ras, myc, neu, and Ela oncogenes. Am J Pathol 138:349–358. [PMC free article] [PubMed] [Google Scholar]
- 21.Kwiatkowski DJ, Diaz MO (1992): Dinucleotide repeat polymorphism at the IFNA locus (9p22). Human Mol Genet 1:658. [DOI] [PubMed] [Google Scholar]
- 22.Engelstein M, Hudson TJ, Lane MK, Leverone B, Landes GM (1993):A PCR-based linkage map of human chromosome 6. Genomics 15:251–258. [DOI] [PubMed] [Google Scholar]
- 23.Orphanos V, McGown G, Boyle JM, Santibanez-Korf M (1993): Thirteen dinucleotide repeat polymorphisms on chromosome 6. Human Mol Genet 2:2196. [DOI] [PubMed] [Google Scholar]
- 24.Weber JL, Kwitek AE, May PE (1990): Dinucleotide repeat polymorphisms at the D5S107, D5S108, D5S111, D5S117, and D5S118 loci. Nucleic Acids Res 18:4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber JL, May PE (1990): Dinucleotide repeat polymorphism at the D18S34 locus. Nucleic Acids Res 18:3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson TJ, Engelstein M, Lee MK, Ho EC, Rubenfield MJ (1992): Isolation and chromosomal assignment at 100 highly informative human simple sequence repeat polymorphisms. Genomics 13:622–629. [DOI] [PubMed] [Google Scholar]
- 27.Breslow A (1970): Thickness, cross-sectional areas and depth of invasion in the progression of cutaneous melanoma. Annals Surgery 172:902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark WH Jr, Mastrangelo MJ, Ainsworth AM, Berd D, Belief RE, Bernardino EA (1977): Current concepts of the biology of human cutaneous malignant melanoma. Advances Cancer Res 24:267–338. [DOI] [PubMed] [Google Scholar]
- 29.Reimer RR, Clark WH Jr, Greene MH, Ainsworth AM, Frau-meni JF Jr (1978): Precursor lesions in familial malignant melanoma. A new genetic preneoplastic syndrome. JAMA 239:744–746. [PubMed] [Google Scholar]
- 30.Quinn AG, Healy E, Rechman I, Sikkink S, Rees JL (1995): Microsatellite instability in human non-melanoma and melanoma skin cancer. J Invest Dermatol 104:309–312. [DOI] [PubMed] [Google Scholar]
- 31.Peris K, Keller G, Chimenti S, Amantea A, Kerl H, Hofler H (1995): Microsatellite instability and loss of heterozygosity in melanoma. J Invest Dermatol 105:625–628. [DOI] [PubMed] [Google Scholar]
- 32.Nicolaides NC, Papadopoolos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter CJ, Dunlop MG, Hamilton SR, Petersen GM, de la Chapelle A, Vogelstein B, Kinzler KW (1994):Mutations of two PMS homologues in hereditary non-polyposis colon cancer. Nature 371:75–80. [DOI] [PubMed] [Google Scholar]
- 33.Umar A, Boyer JC, Kunkel TA (1994): DNA loop repair by human cell extracts. Science 226:814–816. [DOI] [PubMed] [Google Scholar]