Abstract
The enzyme reverse transcriptase (RT) was discovered in retroviruses almost 50 years ago. The demonstration that other types of viruses, and what are now called retrotransposons, also replicated using an enzyme that could copy RNA into DNA came a few years later. The intensity of the research in both the process of reverse transcription and the enzyme RT was greatly stimulated by the recognition, in the mid-1980s, that human immunodeficiency virus (HIV) was a retrovirus and by the fact that the first successful anti-HIV drug, azidothymidine (AZT), is a substrate for RT. Although AZT monotherapy is a thing of the past, the most commonly prescribed, and most successful, combination therapies still involve one or both of the two major classes of anti-RT drugs. Although the basic mechanics of reverse transcription were worked out many years ago, and the first high-resolution structures of HIV RT are now more than 20 years old, we still have much to learn, particularly about the roles played by the host and viral factors that make the process of reverse transcription much more efficient in the cell than in the test tube. Moreover, we are only now beginning to understand how various host factors that are part of the innate immunity system interact with the process of reverse transcription to protect the host-cell genome, the host cell, and the whole host, from retroviral infection, and from unwanted retrotransposition.
INTRODUCTION
The conversion, well over a billion years ago, of the RNA world into the modern configuration, in which genetic information is maintained primarily in DNA, required reverse transcriptases (RTs), enzymes that were able to copy genetic information from RNA into DNA, a process called reverse transcription. With minor (but important) exceptions, for example telomerases, normal cellular processes no longer require reverse transcription, which is now primarily employed in the replication of hepadnaviruses, retroviruses, and retrotransposons. This chapter will cover the process of reverse transcription, and the RTs that are involved in the replication of retroviruses and the related long terminal repeat (LTR) retrotransposons, which have lifestyles that are similar to a retrovirus that has either lost, or never acquired, the ability to be transmitted horizontally from one cell to another. The RTs of, and reverse transcription by, non-LTR retrotransposons will be considered in the chapters that describe these elements (49–55). A substantial fraction of the work that has been done on reverse transcription and RT has focused on human immunodeficiency virus type 1 (HIV-1); this is entirely appropriate given the extent of the HIV epidemic and the fact that HIV-1 RT is the target of two important classes of anti-HIV drugs. Thus, a substantial portion of this review will describe data and insights obtained in experiments that were done with HIV-1 and HIV-1 RT. However, there are some important differences in the RTs, and the process of reverse transcription, among the different retroviruses and LTR retrotransposons; these differences will also be considered, at least briefly. The literature on RT and reverse transcription is both vast and complex. Any review, including this one, can present no more than a superficial overview of what is known. Much that is important has been omitted, some intentionally, some inadvertently; for these omissions, the author apologizes. For those who are interested, a number of helpful reviews have already been published, most of which are focused on retroviral RTs (1, 2, 3, 4).
THE PROCESS OF REVERSE TRANSCRIPTION
In retroviruses and LTR retrotransposons, reverse transcription is the conversion of a single-stranded RNA (ssRNA) copy of the genome into a double-stranded DNA (dsDNA). To avoid the loss of genetic information, the dsDNA copy is longer, on both ends, than the ssRNA from which it is derived (Fig. 1). Although genomic RNA has both a 5′ cap and a poly(A) tail, the ends of the genomic RNA that are transcribed from DNA have short duplications, called R. After reverse transcription, the DNA copy is flanked by longer duplications called the long terminal repeats, or LTRs (5, 6, 7). The LTRs have in them the sequences (enhancers, a promoter, poly-adenylation signals, etc.) that are needed for the host machinery to generate the genomic RNA from the reverse-transcribed DNA copy. The basic scheme by which retroviruses and LTR retrotransposons synthesize the DNA copies of their genomes is the same. It is theoretically possible to produce the dsDNA version of the genome from a single copy of the ssRNA genome (Fig. 1); in practice, retroviruses and LTR retrotransposons contain two copies of the RNA genome (8). It is likely that the primary reason for the presence of two copies of the RNA genome is that the second copy makes it possible to produce a complete dsDNA copy even if the RNA genome is nicked; RT can use the second RNA strand as an alternate template to bypass the nick. This switching of DNA synthesis between the two RNAs also means, if the two RNAs differ in their sequences, that the resulting dsDNA will be a recombinant; this will be discussed in more detail in a later section. Although retrotransposons and retroviruses encode other proteins that increase the efficiency of reverse transcription, RT has both of the enzymatic activities (DNA polymerase and RNase H) that are needed to carry out all of the steps in the process. The DNA polymerase of RT can copy either an RNA or a DNA template; RNase H cleaves RNA if, and only if, it is part of an RNA/DNA hybrid. Like many other DNA polymerases, the polymerase of RT requires both a template and a primer. Many retroviruses and LTR retrotransposons encapsidate a host tRNA that RT uses as a primer to initiate the synthesis of the first DNA strand; however, there are alternative strategies, including the use of a half tRNA primer (9) and self-priming using a processed form of the RNA template (10). Because the RNA genome is the same sense as the mRNAs, it is a plus strand; the first DNA strand to be synthesized by RT is a minus strand. Retroviruses and retrotransposons use a number of different host tRNAs as minus-strand primers; in such cases, the RNA genome carries, near its 5′ end, the appropriate complementary sequence, called the primer-binding site (PBS) (Fig. 1). This means that only a short piece of DNA (sometimes called the minus-strand strong stop) can be synthesized before the growing minus strand reaches the 5′ end of the RNA template. It would appear, at least for HIV-1 RT, that the initiation of reverse transcription is a particularly difficult step, probably at least in part because HIV RT cannot use an RNA/RNA template/primer efficiently (11, 12). This issue is discussed in more detail in the section on the structure of RT.
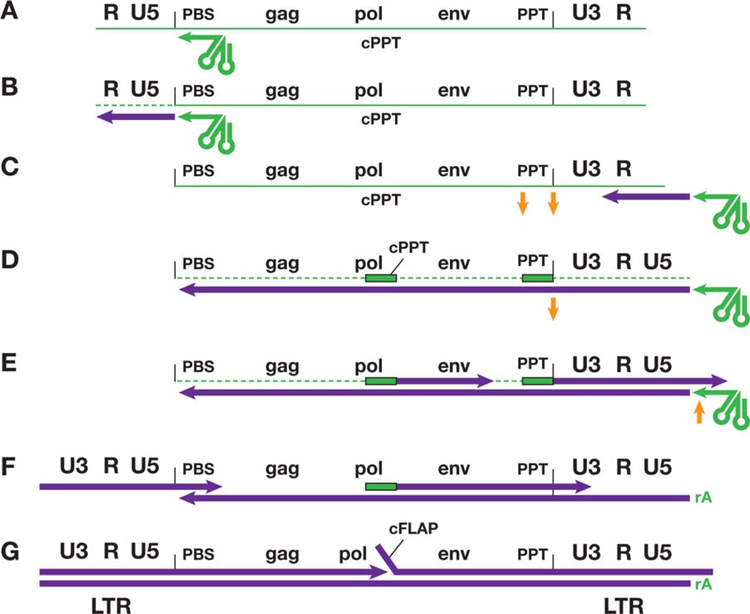
FIGURE 1.
Reverse transcription of the genome of HIV-1. This figure shows, in cartoon form, the steps that are involved in the conversion of the ssRNA genome found in virions into dsDNA. In the figure, RNA is shown in green and DNA in purple. For simplicity, the 5′ cap and the poly(A) tail, which are present on the viral genomic RNA, have been omitted. The various sequence elements in the viral genome, including the genes, are not drawn to scale. (A) The tRNA primer (green arrow on the left) is base paired to the primer-binding site (PBS). (B) RT has initiated minus-strand DNA synthesis from the tRNA primer, and has copied the U5 and R sequences at the 5′ end of the genome. This creates an RNA/DNA duplex, which allows RNase H to degrade the RNA that has been copied (dotted green line). (C) The minus-strand DNA (see text) has been transferred, using the R sequence found at both ends of the viral RNA, to the 3′ end of the viral RNA, and minus-strand DNA synthesis can resume. The HIV-1 genome has two purine-rich sequences (polypurine tracts, or PPTs, one immediately adjacent to U3; the other, the central PPT [cPPT] is in the pol gene). (D–G) The two PPTs are relatively resistant to RNase H (D) and they serve as primers for plus-strand DNA synthesis (E). Once plus-strand DNA synthesis has been initiated, RNase H removes the PPT from the plus-strand DNAs (F, G). The plus-strand that is initiated in U3 is extended until the first 18 bases to the tRNA primer are copied (see text); RNase H then removes the tRNA primer (E). It appears that the entire PPT primers are removed by RNase H; however, RNase H leaves a single riboA on the 5′ end of the minus-strand (E, F). Removing the tRNA primer sets the stage for the second-strand transfer (F). Both the plus- and minus-strand DNAs are then elongated. The plus-strand that was initiated at the U3 junction (on the left in the figure) displaces a segment of the plus-strand that was initiated from the cPPT, creating a small flap, called the central flap (cFLAP). doi:10.1128/microbiolspec.MDNA3–0027-2014.f1
Synthesis of minus-strand DNA creates an RNA/ DNA hybrid that is a substrate for RNase H. There is some controversy about whether there is significant RNase H cleavage by the RT that carries out minus-strand viral DNA synthesis. Cleavage by the polymerizing RT is not required; complete infectious copies of the dsDNA can be made (albeit inefficiently) by a mixture of RTs, some of which have only polymerase activity and some of which have only RNase H activity (13, 14). Moreover, although there is some disagreement in the literature, the bulk of the evidence suggests that there is little, if any, RNase H cleavage by an actively polymerizing RT in in vitro assays (15, 16). This issue is discussed in the section on the structure of RT. The short duplicated sequences at the ends of genomic RNA sequences (R) allow the complementary portion of minus-strand DNA to bind to the R sequence at the 3′ end of the viral RNA after the RNA template has been degraded by RNase H. It appears that minus-strand DNA is equally able to bind to (transfer to) the 3′ end of either of the two viral RNAs; thus, retroviral minus-strand DNA synthesis can either continue on the same RNA template, or switch to the other RNA (17, 18, 19); a similar result was obtained in experiments done with the retrotransposon Ty1 (20). After the minus strand is bound to the 3′ end of one of the two the RNAs (this step is called the first or minus-strand transfer), minus-strand DNA synthesis can continue.
The RNA genomes of retroviruses and retrotransposons all have at least one, and some have two, copies of purine-rich sequences that are relatively resistant to cleavage by RNase H. The essential copy of these polypurine tracts (PPTs) is found near the 3′ end of the genome; the second copy, if one is present, is usually found near the middle of the genome. After minus-strand DNA synthesis has copied the RNA past the PPT, RNase H degrades most of the RNA that has been copied but spares the PPT. The PPT can then act as the primer for plus-strand DNA synthesis (see Fig. 1). The PPT primer is subsequently removed from the plus-strand DNA by RNase H. If a central PPT is present (as it is in HIV-1), it can act as a second site for the initiation of plus-strand DNA synthesis. Based on experiments that were done primarily with HIV vectors, the second PPT does not appear to be essential for virus replication, although it does appear to help the efficiency of plus-strand DNA synthesis (21, 22). Extension of the plus-strand DNA along the minus-strand continues past the end of minus-strand DNA. This second transfer creates the LTRs (U3-R-U5) that will be found at the ends of the DNA genome when it is completed. Once plus-strand DNA synthesis copies the end of U5, synthesis continues until the portion of the tRNA primer that binds the PBS is copied. Experiments done with avian sarcoma leukosis virus (ASLV) showed that the portion of the tRNA that is copied extends to a modified A, which cannot properly base pair with a T; the segment that is copied into plus-strand DNA corresponds exactly to the PBS (23). The minus-strand DNA is also extended and copies the PBS from the RNA genome. It is likely that, in many cases, the RNA 5′ of the PBS has already been copied by RT and degraded by RNase H; in such cases, the 3′ end of the growing minus strand will stop at the 5′ end of the PBS. Copying the PBS and/or the tRNA primer requires RT to displace the tRNA from the RNA genome; RT is capable of this kind of displacement activity in in vitro reactions. After RT copies the end of the tRNA primer, the primer is cleaved from the minus-strand DNA by RNase H, setting the stage for the second (or plus-) strand transfer reaction. In contrast to the first strand transfer, the second strand transfer occurs in cis, presumably because there is only one long minus-strand DNA that can participate in the plus-strand transfer reaction (24). Full extension of the minus and plus strands leads to the synthesis of the dsDNA form of the genome, which is flanked on each end by the LTRs (Fig. 1).
Although the general outline of the process of reverse transcription for retroviruses and LTR retrotransposons has been known for about 35 years, there are some additional points that should be considered. There is the question of whether, at least in some cases, plus-strand DNA synthesis is initiated at positions other than the PPT, making use of residual fragments of the RNA genome that were not completely degraded by RNase H as plus-strand primers. There is quite good evidence that, in ASLVs, plus-strand DNA synthesis is initiated at many sites (25, 26). Conversely, murine leukemia viruses (MLVs) appear to have only one site at which plus-strand DNA synthesis is initiated, the PPT (5). HIV is somewhat controversial; however, there are published data that suggest that, like the ASLVs, HIV plus-strand DNA synthesis has multiple initiation sites (27, 28, 29). However, this raises an interesting question. ASLVs do not have a second PPT (nor, for that matter, do MLVs). If HIV can initiate plus-strand DNA synthesis at multiple points, why does it need a second central PPT?
It also appears, at least in the case of HIV, that plus-strand viral DNA synthesis displaces a portion of 5′ end of the plus-strand viral DNA that was initiated from the central PPT, creating a structure called the central flap. Various functions have been attributed to this flap in the literature. In particular, there are claims from several laboratories that the central flap is involved in nuclear import; however, recent experiments have called this idea into question (30). Although vectors that lack the central PPT (and, by extension, the central flap) replicate reasonably well, several groups have reported that the presence of the central PPT increases the titer of the virus (22, 31, 32). It does appear that the presence of the central PPT allows the virus to complete second-strand DNA synthesis more quickly, making it less susceptible to the host restriction by members of the APOBEC gene family (33, 34) (the APOBECs and their effects are discussed, briefly, in a later section).
In addition, it should be noted that the ends of the viral DNA, which need to be precise because they are the substrates for the integrase (IN) that inserts the dsDNA into the host genome, are defined by the specificity of RNase H cleavages. The U5 end of the genome is defined by the cleavage that removes the tRNA primer (Fig. 1); the U3 end is defined both by the cleavage that creates the 3′ end of the PPT and also by the subsequent cleavage that removes the PPT primer. What is remarkable is that these cleavages are usually made with single-nucleotide precision by an enzyme that has no obvious ability, based on both extensive structural and biochemical analysis, to bind to or otherwise recognize specific sequences. At first glance, it would appear that removing the tRNA primer would be a simple task; all RNase H needs to do is to find the RNA/DNA junction at the 3′ end of the tRNA and remove the entire primer. In most retroviruses, that is exactly what happens; however, HIV-1 is an exception. The RNase H of HIV-1 actually cleaves its tRNA primer one nucleotide from the 3′ end, and the last nucleotide at the 5′ end of the minus-strand of the completed linear viral DNA is a riboA (Fig. 1) (35, 36, 37). This riboA is lost during integration, but its presence, at the very 5′ end of the minus strand in the linear form of HIV-1 viral DNA, shows that RNase H, in removing an RNA primer, does not always simply look for the junction between RNA and DNA. As far as we know (this is a problem that is difficult to investigate directly), the entire PPT primer is always removed; here the problem is how RNase H manages to cleave the RNA genome with single-nucleotide precision. As has already been mentioned, the single-nucleotide precision of these cleavages does not appear to involve sequence recognition in any direct sense; rather the specificity appears to be the result of the structure of the RNA/DNA duplex and its interactions with RT. Altering the sequence of either the PPT (which would alter its structure) or of the portion of the RNase H that contacts the nucleic acid near the PPT can cause a loss of cleavage specificity. Thus, it appears to be the interactions between the RNA/DNA duplex and the polymerase domain that determines the specificity of RNase H cleavage (38, 39, 40, 41, 42); this issue will be discussed in more detail in the next section.
REVERSE TRANSCRIPTASES OF RETROVIRUSES AND LTR RETROTRANSPOSONS
The RTs of retroviruses and LTR retrotransposons are synthesized as polyproteins that are processed by the corresponding viral/retrotransposon proteases (43). This processing takes place in the context of an assembled particle and, depending on which retrovirus/ retrotransposon is being considered, can happen either within the producer cell or outside of it. The details of assembly of the polyproteins, their processing, and subsequent rearrangement to form mature particles are fascinating, but are issues that are beyond the scope of this review. It is important, for the reverse transcription reaction, that RT and its nucleic acid substrates are contained within a particle; why this is important will be considered later. Replication-competent retroviruses/ retrotransposons encode three enzymes, RT, the protease needed to process the polyproteins, and the IN that inserts the dsDNA form of the viral genome into the host genome. These three proteins are most commonly expressed as part of a Gag-Pro-Pol polyprotein, with gag being the gene that encodes the structural components of the particle. Gag is encoded 5′ of Pro and Pol; in most but not all cases, the genomes of retroviruses and retrotransposons have special features that allow protein synthesis to occasionally bypass the stop codon at the end of gag, leading to the synthesis of Gag-Pro-Pol. Gag self-assembles, and Gag-Pro-Pol co-assembles with Gag. The protease processes both Gag and Gag-Pro-Pol to produce the mature forms of the structural proteins and the enzymes. There are exceptions; for example, the foamy viruses, despite being retroviruses, synthesize Pro-Pol separately from Gag, and Pro-Pol co-assembles with Gag by some other mechanism (44, 45). The mature form of RT also differs among various retroviruses and retrotransposons. In its simplest form, RT is a monomer (at least in solution) that contains both the polymerase and RNase H (46). There are also more complex forms of RT; the RT of prototype foamy virus, which also appears to be a monomer, is composed of both protease and RT (47). There are forms of RT that appear to be homodimers of a complete copy of RT (polymerase+ RNase H); however, there is recent structural evidence to suggest, based on the structure of Ty3 RT, that, in at least some homodimeric RTs, the two subunits assume different structures, and it has been suggested that the polymerase activity and the RNaseH activities of Ty3 RT are contributed by different subunits (48). There are also heterodimeric forms of RT. ASLV RT appears to exist as a heterodimer that is composed of a larger subunit (RT+IN) and a smaller subunit (RT alone), and as two types of homodimer composed of the larger and smaller subunits of the heterodimer. All three forms are active in in vitro assays (49, 50, 51); whether all three forms are also active in infected cells is not clear. The RT of HIV-1, which is by far the best characterized, is a heterodimer in which the larger subunit is a complete copy of RT and the smaller subunit is a protease cleavage product that has lost almost all of the RNase H domain (52, 53, 54).
The polymerase domain of viral and retrotransposon RTs are related, both at the structural and sequence levels, to DNA polymerases, including Escherichia coli DNA polymerase I (55). Although the polymerase of RT and DNA polymerase I (Pol I) are only distantly related, the similarity in both structure and function suggests that they had a common ancestor. The RNase H domain of RT has considerable similarity to a cellular RNase H; however, it has been suggested, based in part on comparing RNase H sequences, that retroviruses and LTR retrotransposons were derived from a non-LTR retrotransposon, perhaps by fusion with a DNA transposon (56). This idea is supported by the observation that in many RTs there is a subdomain (the connection; see Fig. 2) that links polymerase and RNase H. It would appear that the connection subdomain was derived from RNase H. Although the two domains of the RTs that are now found in retroviruses and retrotransposons can be linked to cellular enzymes, whether they are related in any direct way to the original RTs that participated in the conversion of the RNA world into a DNA world is not clear. If retroviruses and LTR retrotransposons were derived, at least in part, from non-LTR retrotransposons, then the path back would have to go through the RTs of non-LTR retrotransposons.
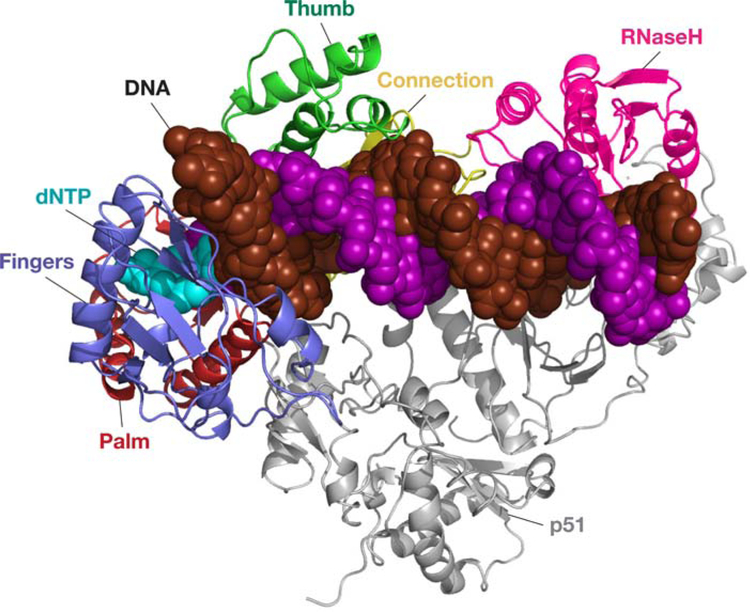
FIGURE 2.
HIV-1 RT in a complex with dsDNA and an incoming dNTP. RT is shown as a ribbon diagram; the DNA and the incoming dNTP are shown as space filling models. The p51 subunit (at the bottom) is shown in gray. The RNase H domain is shown in pink, and the four subdomains to the polymerase domain are shown in different colors: fingers, blue; thumb, green; palm, red; and connection, yellow. The DNA template strand (the strand that is being copied) is dark red and the primer strand (the strand that is being extended) is purple. The incoming dNTP is light blue. doi:10.1128/microbiolspec.MDNA3–0027-2014.f2
HIV-1 RT
As already noted, the RT of HIV-1 is by far the best-studied RT, and it has been extensively analyzed biochemically, genetically, and structurally. It will not be possible, in this review, to do justice to the wealth of biochemical and genetic data that is available, and the description of the structural data will, of necessity, be superficial. The two subunits of HIV-1 RT are, respectively, 560 and 440 amino acids (52). The larger subunit (often called p66) is composed of two domains, the polymerase and RNase H. The polymerase domain, which includes approximately the first 430 amino acids of the p66 subunit, is folded into four subdomains: the fingers, palm, thumb, and connection (53, 54). The smaller subunit (p51) corresponds approximately, but not exactly, to the polymerase domain of the larger subunit and is folded into the same four subdomains. However, the relationship of the four subdomains to each other is quite different in the two subunits (Fig. 2). The active site of the polymerase is composed of a triad of aspartic acids (D110, D185, and D186) in the p66 subunit. The subdomains of p51 are arranged quite differently from their relative positions in p66; one consequence of this difference is that D110, D185, and D186 in the smaller subunit are not involved in any enzymatic function. In the p66 subunit, these three aspartates chelate the two Mg2+ ions that carry out the chemical step of adding nucleotides to the growing DNA chain. There are a number of other amino acids near the catalytic triad that play important roles in the polymerase reaction. For example Y115, and the F at the corresponding position in MLV RT, are situated immediately beneath the 2′ position of incoming dNTPs. These aromatic amino acids act as a steric gate that allows the incorporation of dNTPs, but, because of a clash with a 2′-OH, greatly reduce the ability of the polymerases of these RTs to incorporate NTPs (57, 58).
The RNase H domain is present only in the larger subunit of HIV-1 RT; the RNase H active site is composed of four amino acids that chelate the two Mg2+ ions (D443, E478, D498, and D549). Thus, both of the enzymatic functions of RT are in p66; p51 appears to play a structural role (53, 54). We are fortunate not only to have structures of HIV-1 RT as an unliganded protein (59, 60, 61), but also structures in which RT has bound dsDNA (54, 62) or RNA/DNA hybrids (63, 64). We also have structures in which RT has bound dsDNA and an incoming dNTP or a dNTP analog (65, 66, 67, 68), and numerous structures of RT bound to nonnucleoside RT inhibitors (NNRTIs), only some of which are cited here (53, 69, 70, 71, 72, 73, 74, 75). Although there are still some modest holes in the overall structural picture (it would, for example, be nice to have a structure of HIV-1 RT bound to an RNA/RNA duplex, particularly one that mimicked the initiation of minus-strand DNA synthesis), there are enough different structures available that we have a good idea of the structural changes that take place during the binding of a nucleic acid substrate and the incorporation of a dNTP.
One of the important themes of reverse transcription is that some of the structural elements of HIV-1 RT undergo considerable movement during the polymerase reaction. These movements are an integral part of polymerization: RT really is a small molecular machine. In the unliganded state, the thumb of p66 is closed and reaches over to touch the fingers (Fig. 3). Binding of a template/primer is accompanied by a major movement of the thumb; this allows the double-stranded nucleic acid access to a long groove in the protein where it binds. The double-stranded nucleic acid to which RT binds is relatively long, 17 to 18 bp (54, 63, 65). Binding bends the double-stranded nucleic acid substrate by approximately 40°. This bend involves a segment of the dsDNA several base pairs long and occurs near the base of the thumb of p66. In contrast to most DNA polymerases, which only have to copy DNA, RTs must be able to copy both a DNA and an RNA template. dsDNA can exist as either an A-form or a B-form duplex, although the B form is usually the preferred structure in solution. Because of the 2′-OH, dsRNA cannot exist as a B-form duplex. RNA/DNA duplexes often form a sort of hybrid structure, intermediate between A and B, called H form. RT appears to have resolved the problem of dealing with more than one type of double-stranded nucleic acid substrate by constraining DNA/DNA duplexes in an A-form configuration near the polymerase active site. The transition from primarily A form to B form occurs at the bend in the DNA near the base of the p66 thumb.
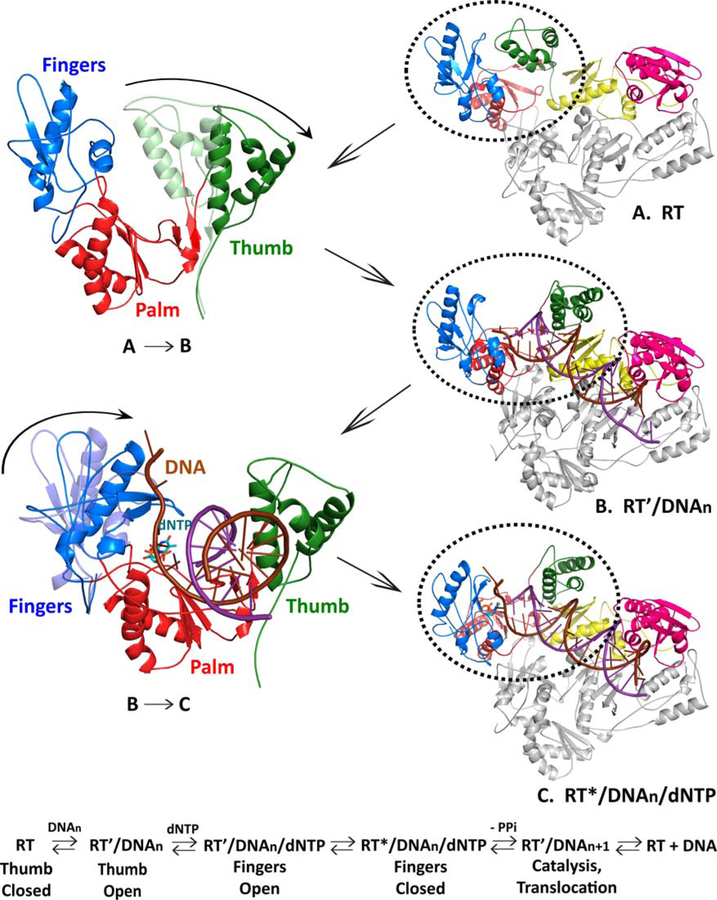
FIGURE 3.
Movement of HIV-1 RT during polymerization. The colors used for the various subunits, domains, and subdomains of RT, and for the two DNA strands, are as in Fig. 2. The p51 subunit is gray. The RNase H domain is shown in pink, and the four subdomains to the polymerase domain are as follows: fingers, blue; thumb, green; palm, red; and connection, yellow. The DNA template is dark red and the primer strand is purple. The incoming dNTP is light blue. The structural changes in RT can be correlated with specific steps in the binding of the substrates and the incorporation of the incoming dNTP. In unliganded RT, the fingers and thumb are closed (A). Before the dsDNA (or other nucleic acid substrate) can be bound, the thumb must move (A → B); this allows the dsDNA to be bound (B). This sets the stage for the binding of the incoming dNTP. When the incoming dNTP binds, the fingers close, which allows the dNTP to be incorporated, with the release of pyrophosphate (PPi). The incorporation of the dNTP temporarily leaves the end of the primer stand in the N or nucleoside triphosphate-binding site (see text). Translocation moves the nucleic acid by 1 bp, which allows the next dNTP to be bound and incorporated. doi:10.1128/microbiolspec.MDNA3–0027-2014.f3
HIV-1 RT prefers to bind a nucleic acid substrate with a 5′ extension that can serve as a template for the addition of dNTPs; this type of substrate is preferentially bound in a fashion that places the 3′ end of the strand that will be extended (the primer stand) immediately adjacent to the polymerase active site, in what is called the P or priming site. In the absence of an incoming dNTP, the fingers of p66 are open (54). Binding an incoming dNTP at the active site (also called the N or nucleoside triphosphate-binding site) induces the fingers to close; this closing of the fingers upon dNTP binding is a feature that HIV-1 RT shares with other DNA polymerases (65, 76). The closing of the fingers sets the stage for the chemical step in which the dNMP portion of the incoming dNTP is incorporated into DNA and pyrophosphate is released. This is essentially an energy-neutral reaction; the total number of high-energy phosphate bonds is unchanged, and the polymerization reaction can be run backwards in vitro in the presence of an appropriate nucleic acid substrate and sufficient pyrophosphate. It is the release and degradation of the pyrophosphate that drives the polymerization reaction. Once pyrophosphate is released, the fingers of p66 open, and the end of the primer, now temporarily in the N site, can move back to the P site, in a process called translocation. This permits the binding and incorporation of the next incoming dNTP, allowing the polymerase of HIV-1 RT to carry out multiple rounds of dNTP incorporation without releasing the nucleic acid substrate. However, in vitro, HIV-1 RT is only modestly processive, usually copying at most a few hundred nucleotides before it falls off the nucleic acid substrate.
It is important to remember that the reactions catalyzed by DNA polymerases depend directly on the sequence of the template that is being copied. As will be discussed in more detail later, RTs carry out this process with considerable precision. Thus, the active site, in the sense that it is the binding site for an incoming dNTP, is composed of both protein and nucleic acid. For the copying reaction to be faithful, the nucleic acid must be held both tightly and precisely by RT. As might be expected, both the polymerase domain and the RNase H domain make extensive contacts with their nucleic acid substrates. Elements have been identified in the polymerase domain (for example, the primer grip and the template grip) that play important roles in positioning the nucleic acid substrate precisely and appropriately (54). However, it is also important to point out that elements of both the RNase H domain and the polymerase domain that are involved in binding the nucleic acid substrates help to position these substrates accurately at the polymerase active site. Conversely, as will be discussed briefly in the next section, the nucleic acidbinding elements in both domains are needed to position the nucleic acid substrate appropriately for proper RNase H cleavage (39, 40, 77, 78, 79, 80, 81, 82, 83).
The structural basis of RNase H activity is somewhat less clear, in part because we have only two published structures of HIV-1 RT in a complex with an RNA/DNA duplex (63, 64). When RT binds a dsDNA duplex, although the RNase H active site is near the template strand (Fig. 2), the active site is not close enough to the template strand for cleavage to take place, even if the template strand was RNA. This separation of the DNA template from the RNase H active site may be important because, strictly speaking, RNase H is not an RNase but a nuclease (84, 85), and it could conceivably cleave a DNA template strand if it contacted the RNase H active site. However, in neither of the structures in which RT is bound to RNA/DNA is there close contact between the active site of RNase H and the RNA strand. In the older structure, the RNA sequence in the nucleic acid duplex was derived from the PPT, and the alignment of the sequence with the enzyme places the RNase H active site over a portion of the PPT where RNase H does not cleave (63). The overall trajectory of the RNA/DNA duplex as it passes through RT is remarkably similar to the overall trajectory of a bound dsDNA whose sequence is unrelated. In particular, there is a similar bend in both nucleic acid duplexes near where they pass the thumb of p66. There is an unusual feature of the PPT RNA/DNA RT structure: in the middle of the nucleic acid duplex there is an unpaired base, two mispaired bases, and then a second unpaired base that brings the duplex back into proper register. There has been considerable speculation over whether this unusual mispaired feature is related to the sequence of the PPT, whether (and to what extent) the mispairing is induced by binding to RT, and whether the mispairing is related to the fact that RNase H cannot cleave within the PPT. We do not have clear answers to any of these questions; however, there are experiments that suggest that portions of the PPT are only weakly base paired when it is not bound to RT, so that binding to RT could easily create (or enhance) the tendency of this segment to be mispaired (41, 42).
As mentioned earlier, it is still unclear exactly how the RNase H of RT is able to make the cleavages that generate and remove the PPT primer (or, for that matter, remove the tRNA primer) with single-nucleotide precision. Although the cleavage site that is chosen is sequence dependent in the sense that, if the sequence is changed, the cleavage site(s) also changes, it appears that this is due to the structure of the PPT when it is bound to RT rather than to the sequence per se. Similarly, sites have been identified in RT, particularly a set of amino acids called the RNase H primer grip, that interact with the DNA strand of an RNA/DNA duplex and are needed to position the RNA strand properly for cleavage (39, 40). As has already been discussed, in thinking about the proper positioning of the RNA/ DNA duplex, contacts in both the RNase H domain and the polymerase domain are important. As might be expected, it would appear that the most important contacts are in the RNase H domain. However, it is worth pointing out that if the RNase H domain of HIV-1 RT is expressed separately from the polymerase domain, it is unable to cleave an RNA/DNA heteroduplex, despite the fact that it is properly folded, because, by itself, the RNase H domain is not able to bind an RNA/DNA heteroduplex. In this sense, RT acts as an integrated whole: two domains work together to properly position the various nucleic acid substrates relative to the two active sites.
Although the more recent structure of RT bound to an RNA/DNA duplex did not involve a duplex whose sequence is based on the PPT, the RNA strand of this structure still does not make close contact with the RNase H active site (64). There are some significant differences in the structures of the two RT/RNA/DNA complexes, for example, there is no evidence of mispaired bases in the newer structure. However, the RNA strand is nicked in this structure and questions have been raised about whether the nick and/or crystal contacts may have affected the structure of this RT/RNA/DNA complex. Thinking about the fact that there are no structures in which a nucleic acid makes close contact with the active site of RNase H brings back the question, raised earlier, of whether an RT that is actively polymerizing can simultaneously carry out RNase H cleavage. As was mentioned previously, there is controversy in the literature, and the available data are based entirely on in vitro assays; however, the bulk of the data suggests that there is little, if any, cleavage of an RNA/DNA duplex in an in vitro reaction if RT is actively polymerizing (15, 16). If this really is the correct interpretation, and if RT must pause before RNase H can cleave, it is possible that there needs to be some repositioning of an RNA/DNA duplex to bring the RNA strand into close contact with the RNase H active site and, conversely, the nucleic acid, when it is properly positioned for RNase H cleavage, might not be properly positioned for polymerization.
There is a large and complicated volume of literature describing the effects of mutations in RT and in critical portions of the RNA genome (such as the PBS and the PPT) on both the polymerase and RNase H activities of RT in vitro and on the process of reverse transcription in infected cells. Most, but not all, of this literature is based on experiments done with purified recombinant HIV-1 RT and with HIV-1-based vectors. All of the obvious features of the enzyme have been mutated (for example, the amino acids in both of the active sites, amino acids that are involved in holding and positioning the nucleic acid in both the polymerase domain and the RNase H domain, and amino acids at the interface between p66 and p51). The good news is that, in general, the genetic and related biochemical data nicely match the structural data. Thus, we can conclude that the interpretations that have been made about what various parts of RT are doing based on where they are in the structure and whether they contact either the nucleic acid or the dNTP substrates are largely correct. There is, in general, relatively good agreement between the effects of mutations on the behavior of purified RT in vitro and their impact on reverse transcription in infected cells. However, as might be expected, there are also some mutations that appear to differentially affect the behavior of purified RT and virus replication. In many of the cases where there appears to be a disagreement, the mutations have a more profound effect on virus replication than on the activity of purified RT.
One simple example involves mutations that affect the ability of RT to be degraded by the protease in virions (86, 87, 88, 89, 90, 91). Many of these RT mutants are temperature sensitive, both in the sense that the extent to which RT is degraded is much greater at the temperature at which the virus normally grows, 37.5°C, than at 32°C, and in the sense that, if the mutant RTs are prepared as purified recombinant proteins, they melt at a lower temperature than wild-type RT. Basically, these temperature-sensitive mutations allow the RT to partially unfold at 37.5°C, and the partially unfolded RTs are susceptible to cleavage by the viral protease. Mutations that confer this phenotype are not difficult to generate; however, as expected, they are, for the most part, mutations that are not usually found in viruses isolated from patients. There is one interesting exception: a mutation, G190E, that confers resistance to NNRTIs. In a standard genetic background, the G190E mutation leads to extensive degradation of RT (88). However, G190E mutants have been found in a small number of patients; in these viruses, the G190E mutation is accompanied by several additional mutations that allow the mutant RT to escape degradation. Taken together, the data suggest that the evolutionary space that is available to RT is constrained not only by the need for the enzyme to be able to carry out its appropriate enzymatic functions efficiently but also by a requirement that RT needs to survive in virions in the presence of the viral protease.
REVERSE TRANSCRIPTION IN CELLS
By the standards of host DNA polymerases, the polymerase of HIV RT is relatively slow. Completing the minus-strand DNA in an infected cell, which involves RT extending a single 3′ end, takes several hours, and the rate of nucleotide addition during minus-strand synthesis in cells has been estimated to be approximately 70 nucleotides per minute (92). There is evidence that it takes less time to complete plus-strand DNA synthesis; the most obvious explanation is that the plus-strand DNA is made in segments. If there are multiple 3′ ends, several RTs can collaborate to complete the task of plus-strand DNA synthesis and the job can be completed more quickly.
Although it is convenient to study reverse transcription in vitro, using purified recombinant RT, this approach is a considerable simplification relative to what happens in cells. Reverse transcription is initiated in the cytoplasm, within a complex structure, usually called the reverse transcription complex (RTC), which is comprised of RT and other viral proteins (93, 94, 95, 96). In addition, there are host proteins that can affect the process of reverse transcription and the outcome. At least initially, the RTC has an outer shell, called the capsid; it is composed of a single protein, also called capsid, or CA, that was derived from Gag by protease cleavage. In the literature, there is much speculation about what happens to the capsid shell during reverse transcription, and there is evidence that the shell is modified during reverse transcription (94, 97, 98). Much of the discussion of what happens to the RTC during reverse transcription involves a process that is called “uncoating.” This name is misleading; there is growing evidence that the capsid shell is largely intact for most or all of the process of reverse transcription, although it appears to be remodeled, either during reverse transcription or perhaps by the process of reverse transcription.
Both the foamy retroviruses and the hepadnaviruses carry out reverse transcription in producer cells, rather than in newly infected cells (45, 99, 100). Thus, for these viruses, it is a dsDNA copy of the viral genome that is inside the virion and inside the capsid shell. This shows that the capsids of these viruses can accommodate a complete, or nearly complete, dsDNA copy of their viral genomes inside their capsid shells, and that, for these viruses reverse transcription is either entirely, or almost entirely, cytoplasmic. The total amount of DNA (number of nucleotides) that is present following reverse transcription is only slightly larger than the total amount of ssRNA in the two strands of genomic RNA that were initially packaged. If, when the plus-strand DNA is synthesized, it is segmented, then the dsDNA could be folded into a relatively compact structure. It has recently been shown that the capsid of some retroviruses protects their viral DNA from host sensors that are part of the innate immune system (101). These sensors are designed to detect the presence of DNA in the cytoplasm; the fact that the capsid protects (or at least partially protects) the viral DNA from these sensors suggests that the capsid remains intact until relatively late in the reverse transcription process. Finally, and probably most importantly, the capsid shell serves to keep the proteins that are needed to carry out both the reverse transcription and integration together with their nucleic acid substrates. Structural data suggests that the mature capsids of retroviruses have holes in them that are large enough to permit dNTPs to enter and pyrophosphate to leave but small enough to prevent the loss of proteins (102, 103). Not surprisingly, there are a number of mutations in the capsid protein that completely block reverse transcription. In particular, mutations in the capsid protein that disrupt the structure of the mature capsid shell block reverse transcription, even if the virions contain a full complement of the other proteins (including active RT) needed to carry out the process (94, 104). Capsid mutants that do not form a normal capsid shell that properly encapsidates the genomic RNA, nucleocapsid (NC), RT, and integrase (IN) fail to initiate reverse transcription, presumably because the necessary components fail to remain associated in the cytoplasm of the newly infected cell. In thinking about the role of the capsid shell, it should be remembered that the RTCs of most retroviruses and retrotransposons contain a fairly large number of copies of RT and IN (estimated to be 50 to 100 for HIV-1). In the case of HIV, reducing the number of enzymatically active copies of RT more than 2- to 3-fold dramatically reduces the ability of the virus to complete the process of reverse transcription (14). This result illustrates the need to retain most or all of the RTs (and, by extension, the other protein components) that were present inside the capsid shell when the particle was formed.
There are other capsid mutants that are able to form what appears to be a reasonably normal capsid shell but a shell that appears to be either too unstable or too stable; both types of mutant can affect the process of reverse transcription (94). Either during the process of reverse transcription or concomitant with the completion of reverse transcription, the RTC is converted into the preintegration complex (PIC) (94, 97, 98, 101, 105). The exact composition of the PIC is not well understood, and whether the PICs of different retroviruses and retrotransposons are or are not different is also unclear. Retroviruses in which reverse transcription is carried out in the infected cell can complete the process in the cytoplasm; fully competent PICs can be isolated from the cytoplasm of infected cells (106, 107). However, it is not clear that reverse transcription must always be completed in the cytoplasm. In the end, the type and the state of the infected cell could play a critical role in determining whether any reverse transcription takes place in the nucleus. In addition, for those retroviruses/ retrotransposons that synthesize their plus-strand DNA in multiple segments (25, 26, 108), it is likely that the final filling in and ligation of a complete, intact plus-strand takes place in the nucleus, and is carried out by host DNA-repair enzymes.
In addition to RT and capsid, the other viral protein that is known to play a major role in reverse transcription is the NC. Like CA, the NC is synthesized as part of the Gag polyprotein; in most cases,the processing of Gag by protease separates the CA and NC. However, the processing of the Gag protein of foamy viruses, while essential for virus replication, removes only a small piece from the C terminus, and, in foamy viruses, the NC equivalent is part of the capsid (109, 110). NC plays the role of a nucleic acid chaperone, facilitating the formation of low-energy RNA and DNA structures. Because NC (in the context of Gag) has an important role in the packaging of genomic RNA, it has not been a simple matter to determine exactly what NC does during reverse transcription in cells. In vitro, NC can, under the right circumstances, help RT get though regions of secondary structure, facilitate transfers between templates, and help prevent “turnaround” DNA synthesis in which the 3′ end of a ssDNA folds back on itself, leading to the synthesis of a DNA hairpin (111, 112, 113, 114, 115, 116, 117, 118).
There are several other viral proteins that have been proposed to have a role in reverse transcription; however, for these other proteins, the data are less compelling. There is no question that a significant fraction of the IN mutants that have been analyzed have defects in reverse transcription, and there are IN mutants that have a profound negative impact on reverse transcription in both HIV and retrotransposons (119, 120, 121, 122, 123, 124, 125). There are claims that an association between RT and IN is needed for reverse transcription. There are, however, no clear data showing that IN helps the process of reverse transcription or the enzymatic activities of RT in vitro. It is likely that the IN mutants have an indirect effect on reverse transcription. At least in the case of HIV-1, the IN mutations that block reverse transcription also affect the structure of the capsid shell. It is unclear why or how IN is involved in forming a proper capsid shell; however, it does appear that the disruption of the capsid shell is the root cause of the failure of these IN mutants to carry out reverse transcription. The mutant virions contain normal amounts of properly processed viral proteins, including RT, and if the virions are broken open in vitro, the RT is fully active. However, as judged by electron microscopy images of the virions that carry these IN mutants, the capsid shell does not enclose the RNA. It is likely that this leads to a dissociation of RT and the RNA early in infection; the IN mutants that have defective capsid shells do not initiate reverse transcription and thus resemble, in their phenotypic behavior, the CA mutants that fail to form a proper capsid shell. This interpretation supports the idea that an intact capsid shell, which surrounds the RNA, helps the RTC retain RT and NC, making it essential for reverse transcription. There have also been claims that other viral proteins, for example Vpr and Tat, can affect the activity of RT in vitro; however, it is not clear that these proteins play an important role in reverse transcription in infected cells (126, 127, 128, 129, 130, 131). These factors are present in only a small subset of retroviruses, and it seems, for that reason, unlikely that they play a fundamental role in reverse transcription.
There are host factors that have been reported to help reverse transcription. The best example is dUTPase. Some retroviruses encode their own dUTPases (132, 133, 134); others appear to use Vpr to interact with host dUTPases (135, 136, 137, 138). However, the use of a dUTPase of either host or viral origin is not universal, and it is unclear why some retroviruses make (or steal) them and others do not. However, the fact that some retroviruses make use of host or viral dUTPases does suggest that these viruses are using these enzymes to increase the fidelity of their replication, and proposals that the replication of the viruses is deliberately sloppy are probably incorrect. This issue will be considered later, in the section devoted to fidelity. It has also been claimed that, for HIV, the host lysyl aminoacyl-tRNA synthetase plays a role in the selective packaging of the correct tRNA primer (139, 140); however, HIV can replicate reasonably efficiently using other tRNA primers (141, 142, 143). It has also been proposed that an RNA debranching enzyme is involved in the first-strand transfer during reverse transcription (144, 145); this claim should also be viewed with some caution, at least until more and better evidence has been presented.
There are host proteins that can negatively affect reverse transcription. The well-characterized negative host factors appear to be involved in host defense: the host would like to protect itself both from retroviral infections and from uncontrolled replication of retrotransposons. Three of the best-known host defense factors, Trim5a, TrimCyp, and FV-1, appear to affect the stability of the capsid shell in the cytoplasm (146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156). This reprises the theme that has been touched on several times already, that the capsid shell needs to pass the Goldilocks test: not too unstable or it will fall apart and the proteins, including RT, that are needed to complete reverse transcription (and subsequently, integration) will be lost; but not too stable or there will be problems, less well defined but possibly related to the completion of reverse transcription and/or the conversion of the RTC into a PIC. Another well-characterized host factor known to have a significant negative impact on the replication of some retrotransposons and retroviruses that acts at the stage of reverse transcription is SAMHD1. SAMHD1, which is found, in an active form, in some non-dividing cells, catalyzes the conversion of dNTPs to their corresponding nucleosides and free triphosphate, and it can reduce the levels of dNTPs to a point where the reverse transcription of retroviruses is blocked (157, 158, 159, 160, 161, 162). Although SAMHD1 was discovered because it can block the replication of HIV-1 in some cultured cells and because HIV-2 makes a protein that counteracts it, based on the phenotype of humans who fail to make SAMHD1, its primary role may be to help control the level of replication of retrotransposons and retroposons (163, 164).
The other well-defined group of host proteins that negatively affects reverse transcription is the APOBECs. These are cytosine deaminases, and the members of the family that affect reverse transcription act on dC residues in a ssDNA template, converting dC to dU (165, 166, 167, 168, 169, 170). There are a number of credible reports that some APOBECs can affect reverse transcription (and apparently integration), even in the absence of an ability to deaminate dC (171, 172, 173, 74). Exactly how this is accomplished is unclear, but the APOBECs bind single-stranded nucleic acids, and it is possible that binding alone is sufficient, at least in some cases, to interfere with reverse transcription and/or integration. However, it is also clear that the conversion of dC to dU is an important part of the antiviral/anti-retrotransposon effects of at least some of the APOBECs. To be active they (usually) need to be packaged into the virion in the producer cell. The antiviral/antiretrotransposon APOBECs are DNA specific: they act primarily on minus-strand DNA after the RNA template has been removed by cleavage by RNase H but before the plus-strand DNA has been synthesized. As was mentioned briefly in a previous section, this could help to explain the role of the central PPT in HIV and perhaps in other retroviruses and retrotransposons. The presence of the central PPT reduces the time that the first DNA strand is single stranded, thus reducing the susceptibility of the virus to APOBECs (33, 34).
The primary signature of the APOBECs is the conversion, in the newly synthesized plus-strand DNA, and subsequently in the RNA, of multiple G residues to As. The various APOBECs have slightly different preferences for the sequences adjacent to the dC residues they deaminate; the preference of the best studied of the antiviral APOBECs, APOBEC3G, causes it to have a tendency to generate termination codons. The fact that, in most cases, APOBECs are only able to modify the genomes of retroviruses if they are expressed in the producer cells and packaged into virions suggests that the capsid shell protects the viral DNA not only from cytoplasmic host-cell DNA sensors in a newly infected cell but also from any APOBEC proteins that might be present. This provides one more piece of evidence that the capsid shell remains largely intact, at least until the RNA genome has been converted into dsDNA. There are a modest number of exceptions to this general rule, and there are some circumstances (or perhaps more accurately, there are some cells) in which APOBEC proteins that are present in a newly infected cell are able to modify the viral genome. As intriguing as these examples are, they are exceptions, and it is not clear how they fit into the broad picture in which most of reverse transcription takes place inside the capsid shell.
FIDELITY
A number of factors contribute to the complex array of mutations that are seen in populations of retroviruses and retrotransposons. Some endogenous elements spend much of their existence hiding in the host genome and, if they are expressed at a high level, they can be deleterious to the host (163, 164). For endogenous viruses and retrotransposons that are primarily transmitted vertically, the position at which the element is integrated can be a major factor in determining whether it will be selected against; in such cases, mutations in the genome can be fixed because the element occupies a fortuitously favorable integration site, rather than because the mutations the element carries help it replicate efficiently. Conversely, for viruses that are transmitted horizontally and replicate rapidly, like HIV, the replicative fitness of the virus is quite important. HIV-1 infections are often initiated by a single virion (175); however, in an untreated patient, within a few years, the virus population has diverged to the point where no two viral RNAs isolated from the blood have the same sequence. This rapid diversification is due to the fact that virus replicates rapidly, and to the relatively large population of replicating virus (176). The viruses that are seen in the blood are those in which mutations have had, in general, a minimal impact on the overall replicative fitness of the virus. In addition, there is selection, in patients, for variants that reduce the immunological susceptibility of the virus. However, all of the variation that is seen in these virus populations derives from errors that are made during replication. Three enzymes participate in the replication of the genomes of retroviruses and retrotransposons: the host RNA Pol II, RT, and the host’s replicative DNA polymerase. The degree to which the host DNA polymerase makes a significant contribution depends on the life-style of the element in question. For actively replicating viruses, like HIV-1, the contribution of the host DNA polymerase, which, in combination with its ancillary proofreading machinery, has a very low error rate, is negligible. However, for some endogenous retroviruses and retrotransposons that are vertically transmitted and are passively replicated for many generations, the host DNA polymerase is the major source of errors. Because the process of reverse transcription creates two LTRs that have identical sequences when they are first synthesized, comparing the sequences of the two LTRs of ancient proviruses/retrotransposons is an important tool in molecular paleontology.
Although HIV-1 RT can inefficiently excise the nucleotide analogs that are used to treat HIV infections after they have been incorporated into viral DNA (discussed briefly later in this chapter), and there are RT mutants that are significantly better at excising these analogs than wild-type RT (177, 178), these mutations do not appear to improve the fidelity of HIV replication, and it appears that RT has no editing function that improves its fidelity (179). In contrast, RNA Pol II is able to back up and excise a misincorporated nucleotide (180). This, coupled with the knowledge that HIV-1 RT (and most other retroviral RTs) are relatively error prone in vitro, having an error rate of approximately 10−4, has led to widespread claims that the errors made during virus replication can be attributed to RT. There are several potential problems with this simple and convenient picture. First, the actual error rate of RNA Pol II has not been well defined, and its contribution to the overall errors made during the active replication of retroviruses and retrotransposons is not known (181). Secondly, the overall error rate of retrovirus replication is about 10−5, a much lower error rate than has been measured for HIV-1 RT in vitro (182, 183, 184, 185). This suggests that, in the environment of the RTC and perhaps because of the participation of other viral and/or host proteins, reverse transcription is a higher-fidelity process in an infected cell than it is in vitro. If the overall error rate is about 10−5, then the fidelity of both RNA Pol II and RT cannot be less than 10−5; however, we still do not know which of the two enzymes makes the majority of the errors, or if they make roughly equal contributions. In thinking about the problem, it is important to remember that the flow of information is from RNA Pol II to RT. For this reason, RT cannot be selected to have a higher fidelity, in terms of copying information, than RNA Pol II. If we assume that most retroviruses and retrotransposons are trying to copy their genetic information faithfully, an idea that is supported by the fact that some viruses encode dUTPases, and others make use of host-cell dUTPases, and that retroviruses carry proteins that counteract the APOBEC proteins, then the fidelity of RT would be limited by the fidelity of RNA Pol II, and the fidelity of the two polymerases would be similar.
There are, however, additional layers of complexity. Although it is convenient to think about fidelity in terms of an overall error rate, this does not provide a clear picture of the errors that arise during one round of replication. In the first place, not all substitutions are equally likely. As would be expected, at least for HIV-1, the best-studied system, missense mutations are more frequent than frameshift mutations, and the majority of the missense mutations are transitions rather than transversions (184, 185). In the case of HIV, the most common transition (in the plus-strand) is G to A. If this preference for making G to A mutations is largely due to errors made by RT, which is not clear, it probably reflects a tendency of RT to insert a T opposite a G during first-strand DNA synthesis. (These experiments were done in cells that do not express significant levels of the APOBEC proteins that affect HIV replication, and essentially all of the G-to-A mutations were made in the wrong sequence context to be APOBEC mutations.) However, the mutations were not uniformly distributed but preferentially occurred at specific sites or hotspots. In some cases, exactly the same target sequences were used in the in vitro assays done with purified RT and the virus replication assays; however, the hotspots that were reported in vitro do not match those seen during virus replication (186, 187, 188, 189). To make matters worse, the hotspots seen in experiments done by different laboratories using purified RT to copy the same template do not match. This result, taken together with the fact, already mentioned, that the fidelity of RT, as measured in vitro, is at least 10-fold too low, should serve as a warning to those who are trying to understand fidelity as it applies to the replication of retroviruses/ retrotransposons using in vitro fidelity data that was obtained with purified RT. Not surprisingly, all of the problems that have just been discussed apply with equal or greater force to the very large volume of literature that describes the impact of mutations in RT on fidelity.
As might be expected, mutations in RT affect its fidelity, measured in in vitro assays, and the fidelity of the replication of the parental virus. Here too, most of the experiments have been done with HIV-1, and HIV-1 RT, although there are some data for some other retroviruses and their respective RTs. This is a large and complicated literature, and it is complicated, at least in part, because there have been a large number of assays that have been used to try to measure fidelity in vitro. As was the case with the experiments done with wild-type RT and a vector that replicates using wild-type RT, there is relatively little agreement between the in vitro data obtained with purified RT and the corresponding data, obtained with the same RT mutants, using a viral vector (179, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 202). There are several simple conclusions. Although only a small number of RT mutants have been tested in the context of virus replication, mutations that have been reported to increase the fidelity of RT in vitro caused a small to moderate increase in the overall error rate in virus replication assays (185). Some of the mutations in RT affected the relative proportion of missense and frameshift errors. In addition, mutations in RT that did not cause a substantial increase in the overall error rate still caused substantial increases in the errors made at specific positions in a target gene. This result demonstrates that the overall error rate is not, in many cases, a good measure of the impact of a mutation in RT on the errors made during virus replication. In particular, changes in RT that have little effect on the overall error rate can still increase the chances that mutations will arise that can allow the virus to escape from immunological surveillance and/or develop resistance to particular drugs.
RECOMBINATION
As has already been discussed briefly, there are, during minus-strand DNA synthesis, frequent transfers between the two RNA templates (203, 204, 205, 206, 207, 208, 209, 210, 211, 212). Most or all of these transfers are probably caused by nicks in the genomic RNAs (213). By switching DNA synthesis to the other RNA strand, RT can generate an intact DNA copy of the genome from two nicked RNA templates, so long as both of the RNA strands are not nicked at the same place. The strand transfers that lead to recombination are similar to the first-strand transfer reaction, which has been described in an earlier section of this review. If, as is often the case, the two RNA copies are identical, these multiple strand transfers have no genetic consequences. However, if there are, in the same host cell, DNAs that encode two variants of the same retrovirus or retrotransposon, particles can be produced that contain two related RNA genomes. In such cases, strand transfers that occur during first-strand DNA synthesis will produce a recombinant genome (203). This can have important consequences, if, for example, recombination involves RNAs from two HIV-1s that carry different drug-resistance mutations. Because recombination can only occur if the two parental RNAs are copackaged into a single particle, recombination between different but fairly closely related retroviruses or retrotransposons is rare; however, it does happen. For example, HIVcpz, the chimpanzee retrovirus that gave rise in humans to HIV-1, is a recombinant between two different simian immunodeficiency viruses, one of which is normally found in red-capped mangabeys, and the other in a Cercopithecus monkey (214).
It appears that there are at least three factors that can affect the generation of recombinant retroviruses and retrotransposons: (i) the copackaging of the RNAs; (ii) the similarity of the genomic sequences, which is required for the transfer of DNA synthesis between the templates; and (iii) the viability of any recombinant virus that emerges. The packaging of the retroviral RNA genome is a complex topic, the details of which lie outside of the scope of this review. Briefly, there is a structured segment of the RNA near the 5′ end of the genome, called Ψ, which is important for packaging. The RNA dimer forms before it is packaged, and there is a secondary condensation step that happens after the RNA dimer is packaged (215). In the case of HIV, a kissing loop, called the dimer initiation signal (DIS), is a critical component that has a significant role in the formation of the RNA dimer (216, 217, 218). Even within HIV strains, there is variation in the sequence of the DIS (219, 220), and the compatibility of the DIS (the ability to form a kissing-loop dimer) affects RNA dimerization and, by extension, recombination. However, sequences outside the DIS also affect the ability of two RNA genomes to dimerize and be copackaged (221). Although the compatibility of the DIS affects the ability of two genomic RNAs to be copackaged, the similarity of the sequences of the two RNA genomes will affect the efficiency of the strand transfer. Perhaps even more important, in terms of the ability of any recombinant viruses to grow out, is whether the proteins encoded by the recombinant viruses are fully functional and are able to work together. Studies with artificial HIV/MLV recombinants show that even carefully constructed recombinants of such distantly related viruses replicate very poorly. Even recombinants between different strains of HIV-1 often replicate much less efficiently than the parental viruses.
Not all transfers between templates are precise. Imprecise transfers can create either an insertion or a deletion. It is likely that most of these deletions and duplications are created by nonhomologous strand transfers during first-strand DNA synthesis. In many but not all cases, there are small regions of homology at the site of duplications/deletions. In experiments done with retroviral vectors that carried sizable duplications, the duplications were rapidly lost if the vector was allowed to replicate, and the rate at which duplications were lost was related to both the length of the duplicated sequences and how far they were separated in the genome of the vector (222, 223). There are two RNA targets at which a nonhomologous strand transfer would create a deletion; in contrast, there is only one RNA target at which a strand transfer could create a duplication. In addition, it is likely that many of the erroneous strand transfers are the result of having a break in both RNA strands at the same, or almost exactly the same, site. In such cases, homologous strand transfers are not possible. Thus, any strand transfer that can occur will create either a deletion or a duplication. However, if the breaks are at the same site in the two RNAs, a strand transfer that creates a duplication will not get the growing DNA strand past the breaks in the two RNAs. Conversely, a strand transfer that bypasses the breaks in both RNA strands will, of necessity, create a deletion. These constraints, taken together, nicely account for the large excess of deletions that are seen relative to duplications.
RT DRUGS AND DRUG RESISTANCE
AZT, which inhibits reverse transcription, was the first successful anti-HIV drug; now, some 25 years later, drugs that block reverse transcription are still fundamentally important and widely used components of successful combination anti-HIV therapy. There are, broadly speaking, two major classes of drugs that block reverse transcription, nucleoside analogs (NRTIs) and nonnucleoside RT inhibitors (NNRTIs). NRTIs are, as their name implies, analogs of the normal nucleosides that are used to synthesize DNA; however, all of the NRTIs that have been approved for human use lack a 3′-OH, and, when incorporated into viral DNA by RT, act as chain terminators, blocking viral DNA synthesis. In the strictest sense, NRTIs are not RT inhibitors; their true target is the viral DNA. In contrast, NNRTIs are RT inhibitors; they do not interfere with the ability of RT to bind its substrates but block the chemical step of DNA synthesis (224, 225). With one important exception, NRTIs are given to patients as free nucleosides, and are converted to the triphosphate form in cells in the patient. Tenofovir disoproxil fumarate is the exception; it is given as a monophosphate prodrug. Tenofovir disoproxil fumarate can be taken up by cells because, in the prodrug form, the phosphate is masked by two hydrophobic modifications. In general, NRTIs tend to have toxic effects, particularly after long-term use; a significant part of their toxicity comes from the fact that the triphosphate forms of the NRTIs can, to a greater or lesser extent, be used by host DNA polymerases; the host gamma DNA polymerase found in mitochondria appears to be particularly vulnerable (226, 227).
The other class of approved RT inhibitors, NNRTIs, is a diverse group of hydrophobic compounds that bind in a pocket in the p66 palm of HIV-1 RT, about 10 Å away from the polymerase active site (53, 59, 71, 73). The NNRTI-binding pocket does not exist in unliganded HIV-1 RT, and the binding of an NNRTI alters the structure of RT (60, 61). A bound NNRTI pushes up the palm in a region of the polymerase domain that underlies the cleft where the nucleic acid binds. This, in turn, pushes the end of the nucleic acid substrate upward, moving it away from the polymerase active site (228). In contrast to NRTIs, NNRTIs are quite selective; not only do they not block the activity of host DNA polymerases, they do not in general bind to or block polymerization by other retroviral RTs, including HIV-2 RT. This is both good, in the sense that, for the most part, NNRTIs are relatively non-toxic; and bad, because the first-generation NNRTIs were very susceptible to mutations that made the virus drug resistant. To cite a specific example, attempting to block mother-to-child transmission of HIV at birth by giving the mother a single dose of the first-generation NNRTI nevirapine led to the development of resistance in the majority of treated women (229, 230, 231). Fortunately, it has been possible to develop more advanced NNRTIs that are less susceptible to the development of HIV resistance (232, 233).
Resistance is a major problem in HIV drug therapy. All of the available anti-HIV drugs, including all of the NRTIs and NNRTIs, can, under the proper circumstances, select for resistance mutations that substantially reduce the efficacy of the drugs. Resistance to NRTIs implies that the mutant form of RT, which must still be able to synthesize viral DNA, has an increased ability to discriminate between the NRTI and the corresponding normal nucleoside compared with the wild-type RT. There are, broadly speaking, two mechanisms of NRTI resistance. The first, exclusion, is typified by the M184I/ V mutations that reduce the susceptibility of the virus to 2′,3′-dideoxy-3′-thiacytidine (3TC or lamivudine) and 2′,3′-dideoxy-5-fluoro-3′-thiacytidine (FTC or emtricitabine) (234, 235). As the name implies, exclusion mutations reduce the ability of the mutant RT to bind and/ or incorporate the triphosphate form of the affected NRTI. 3TC and FTC differ from a normal nucleoside both because their pseudosugar ring contains sulfur, and also because the ring is in the L rather than in the normal D configuration. These differences mean that when the triphosphates of these analogs bind at the polymerase active site (N site), a portion of the pseudosugar ring is closer to position 184 than is the case for a normal dNTP. There is room for the analogs to bind and be incorporated if the amino acid at position 184 is M, but there is a steric clash, caused by the β-branch on the side chain of the amino acid, if either I or V is present at this position. Thus, the M184I/V mutations allow RT to selectively discriminate against the incorporation of 3TC triphosphate and FTC triphosphate.
The second mechanism is more interesting, because it depends on the mutant form of HIV-1 RT developing a new ATP-binding site that allows it to excise the AZT monophosphate (and, if appropriate additional mutations are present, other NRTIs) that is blocking the end of the viral DNA after the analog has been incorporated (67, 177, 178, 236). This frees the blocked end of the viral DNA, allowing a complete copy of the genome to be synthesized. Basically, the excision reaction is related to pyrophosphorolysis, which is polymerization run backwards. As described earlier, the incorporation of a dNMP, with the concomitant release of pyrophosphate, is essentially energy neutral. Thus, the polymerization reaction can be run backwards in vitro in the presence of sufficient pyrophosphate. However, it is not possible, because of microscopic reversibility, to speed up an excision reaction that uses pyrophosphate without causing an equivalent increase in the incorporation of the AZT triphosphate. The AZT resistance mutations avoid this problem by creating a novel ATP-binding site: the excision reaction differs from the incorporation reaction because it uses ATP as the pyrophosphate donor. ATP-mediated excision creates a dinucleoside tetraphosphate, AZTppppA. The two primary AZT resistance mutations, T215F/Y and K70R, help create the new ATP-binding site: the F or Y at 215 makes π/π stacking interactions with the adenine ring of ATP, while the R at position 70 interacts with both the ribose ring and the α-phosphate of ATP (67). For this reason, these two mutations (and the other associated AZT resistance mutations) selectively enhance the binding of the ATP that participates in the excision reaction but have little or no effect on the binding or release of the pyrophosphate that is produced during normal polymerization. This means that these mutations selectively enhance the excision of AZT monophosphate without causing a concomitant increase in the incorporation of AZT triphosphate, and thus the mutations cause resistance.
In contrast to NRTI resistance, in which RT must be able to retain the ability to bind and incorporate normal dNTPs, developing resistance to NNRTIs is a simpler problem: almost any mutation that will interfere with NNRTI binding and not distort or disrupt the overall structure and function of RT will be acceptable. As might be expected, the polymerase active site is much more conserved and constrained than the NNRTI-binding site, and the spectrum of mutations that are acceptable to the virus is considerably greater at the NNRTI-binding site. Most of the common NNRTI-resistance mutations affect the binding of NNRTIs, either directly or indirectly. For example, mutations that replace the tyrosine at position 181 or 188 with a non-aromatic amino acid reduce the hydrophobic interactions with a number of NNRTIs (53, 237, 238, 239). There is an interesting exception: the K103N mutation creates a new hydrogen bond (with Y188). This bond helps to keep the entrance to the NNRTI-binding pocket closed; the K103N mutation makes it more difficult for NNRTIs to enter the pocket, and the K103N mutation confers resistance to a number of different NNRTIs (240). The good news is that considerable progress has been made, both in the development of drugs that can effectively block reverse transcription (and other steps in the viral life cycle) and in the development of combination therapies that, if the patients are compliant, can completely suppress the replication of virus and the emergence of resistance. Progress is also being made in the development of new drugs that are effective against many of the extant drug-resistant mutants. There is also now the hope, even in the absence of an effective anti-HIV vaccine that can prevent transmission, that anti-HIV drugs, including RT inhibitors, can help prevent new infections, both because they reduce the viral load in the donor and because, if taken prophylactically, they can block infection in the recipient.
SUMMARY
We have, over the approximately 45 years since RT was discovered, learned a great deal about these fascinating enzymes, their structures, and how they are able to convert a ssRNA genome into a dsDNA copy from which new copies of the genomic RNA can be produced. RTs were essential tools that played a key role in the origins of molecular biology and in the creation of the biotechnology industry, and RTs are widely used in both research laboratories and clinical laboratories today. Although the fundamentals of how reverse transcription works and structures of HIV-1 RT have been available for some time, there are still unsolved puzzles, particularly in relation to understanding how the capsid shell assists the process of reverse transcription in cells and how the shell is remodeled in the process. What is the exact composition and structure of the RTC? How does it help RT carry out DNA synthesis efficiently and effectively? How does the RTC transition into the PIC? Moreover, we are just now beginning to understand how the host’s innate immunity targets the reverse transcription process.
There has been good progress in slowing the spread of HIV not only in the developed world, but also in the developing world; drugs that block reverse transcription have been an essential part of this progress. However, there are still problems with both resistance and toxicity. We will need to supplement our armamentarium of anti-HIV drugs so that we do not fall behind in the battle, particularly in the developing world. We also need to be able to provide less-toxic drugs to those who, at least for the foreseeable future, will need to stay on anti-HIV drug therapy throughout their lives. If the past is any guide, at least some of these new drugs will be RT inhibitors; the more we know about RT and how it works, the more likely we will be to succeed.
ACKNOWLEDGEMENTS
The author would like to thank Teresa Burdette for help with the manuscript, and Karen Kirby and Allen Kane for help with the figures.
REFERENCES
- 1.Telesnitsky A, Goff GP. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121–160. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 2.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol 385:693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu W, Hughes SH. 2012. HIV-1 Reverse Transcription, p. 37–58. In Bushman FD, Nabel GJ, Swanstrom R (ed), HIV from Biology to Prevention and Treatment. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 4.Skalka AM, Goff SP. 1993. Reverse Transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 5.Gilboa E, Mitra SW, Goff S, Baltimore D. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18:93–100. [DOI] [PubMed] [Google Scholar]
- 6.Shank PR, Hughes SH, Kung HJ, Majors JE, Quintrell N, Guntaka RV, Bishop JM, Varmus HE. 1978. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell 15:1383–1395. [DOI] [PubMed] [Google Scholar]
- 7.Hughes SH, Shank PR, Spector DH, Kung HJ, Bishop JM, Varmus HE, Vogt PK, Breitman ML. 1978. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell 15:1397–1410. [DOI] [PubMed] [Google Scholar]
- 8.Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu WS. 2013. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog 9: e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke N, Gao X, Keeney JB, Boeke JD, Voytas DF. 1999. The yeast retrotransposon Ty5 uses the anticodon stem-loop of the initiator methionine tRNA as a primer for reverse transcription. RNA 5:929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JH, Levin HL. 1998. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5–IR stem–loop of retroviruses. Mol Cell Biol 18:6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isel C, Lanchy JM, Le Grice SF, Ehresmann C, Ehresmann B, Marquet R. 1996. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J 15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 12.Lanchy JM, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. 1996. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J 15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 13.Telesnitsky A, Goff SP. 1993. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J 12: 4433–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julias JG, Ferris AL, Boyer PL, Hughes SH. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J Virol 75:6537–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driscoll MD, Golinelli MP, Hughes SH. 2001. In vitro analysis of human immunodeficiency virus type 1 minus-strand strong-stop DNA synthesis and genomic RNA processing. J Virol 75:672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purohit V, Roques BP, Kim B, Bambara RA. 2007. Mechanisms that prevent template inactivation by HIV-1 reverse transcriptase RNase H cleavages. J Biol Chem 282:12598–12609. [DOI] [PubMed] [Google Scholar]
- 17.Panganiban AT, Fiore D. 1988. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science 241:1064–1069. [DOI] [PubMed] [Google Scholar]
- 18.Hu WS, Temin HM. 1990. Retroviral recombination and reverse transcription. Science 250:1227–1233. [DOI] [PubMed] [Google Scholar]
- 19.van Wamel JL, Berkhout B. 1998. The first strand transfer during HIV-1 reverse transcription can occur either intramolecularly or intermolecularly. Virology 244:245–251. [DOI] [PubMed] [Google Scholar]
- 20.Wilhelm M, Boutabout M, Heyman T, Wilhelm FX. 1999. Reverse transcription of the yeast Ty1 retrotransposon: the mode of first strand transfer is either intermolecular or intramolecular. J Mol Biol 288:505–510. [DOI] [PubMed] [Google Scholar]
- 21.Hungnes O, Tjotta E, Grinde B. 1992. Mutations in the central polypurine tract of HIV-1 result in delayed replication. Virology 190:440–442. [DOI] [PubMed] [Google Scholar]
- 22.Charneau P, Alizon M, Clavel F. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol 66:2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanstrom R, Varmus HE, Bishop JM. 1981. The terminal redundancy of the retrovirus genome facilitates chain elongation by reverse transcriptase. J Biol Chem 256:1115–1121. [PubMed] [Google Scholar]
- 24.Yu H, Jetzt AE, Ron Y, Preston BD, Dougherty JP. 1998. The nature of human immunodeficiency virus type 1 strand transfers. J Biol Chem 273:28384–28391. [DOI] [PubMed] [Google Scholar]
- 25.Kung HJ, Fung YK, Majors JE, Bishop JM, Varmus HE. 1981. Synthesis of plus strands of retroviral DNA in cells infected with avian sarcoma virus and mouse mammary tumor virus. J Virol 37:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu TW, Taylor JM. 1982. Single-stranded regions on unintegrated avian retrovirus DNA. J Virol 44:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller MD, Wang B, Bushman FD. 1995. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J Virol 69:3938–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klarmann GJ, Yu H, Chen X, Dougherty JP, Preston BD. 1997. Discontinuous plus-strand DNA synthesis in human immunodeficiency virus type 1-infected cells and in a partially reconstituted cell-free system. J Virol 71:9259–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas JA, Ott DE, Gorelick RJ. 2007. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J Virol 81:4367–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdick RC, Hu WS, Pathak VK. 2013. Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proc Natl Acad Sci U S A 110:E4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ao Z, Yao X, Cohen EA. 2004. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J Virol 78:3170–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Maele B, De Rijck J, De Clercq E, Debyser Z. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J Virol 77:4685–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu C, Saenz DT, Fadel HJ, Walker W, Peretz M, Poeschla EM. 2010. The HIV-1 central polypurine tract functions as a second line of defense against APOBEC3G/F. J Virol 84:11981–11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurtzer S, Goubard A, Mammano F, Saragosti S, Lecossier D, Hance AJ, Clavel F. 2006. Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B. J Virol 80:3679–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitcomb JM, Kumar R, Hughes SH. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol 64:4903–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pullen KA, Ishimoto LK, Champoux JJ. 1992. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J Virol 66:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JS, Roth MJ. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3). J Biol Chem 267:15071–15079. [PubMed] [Google Scholar]
- 38.Pullen KA, Rattray AJ, Champoux JJ. 1993. The sequence features important for plus strand priming by human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem 268:6221–6227. [PubMed] [Google Scholar]
- 39.Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci U S A 99:9515–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice SF. 2002. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 41:4856–4865. [DOI] [PubMed] [Google Scholar]
- 41.Dash C, Rausch JW, Le Grice SF. 2004. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res 32:1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi-Brunozzi HY, Le Grice SF. 2005. Investigating HIV-1 polypurine tract geometry via targeted insertion of abasic lesions in the (–)-DNA template and (+)-RNA primer. J Biol Chem 280:20154–20162. [DOI] [PubMed] [Google Scholar]
- 43.Swanstrom R, Willis JW. 1997. Synthesis, assembly, and processing of viral proteins, p. 263–334. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 44.Linial ML. 1999. Foamy viruses are unconventional retroviruses. J Virol 73:1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579–1582. [DOI] [PubMed] [Google Scholar]
- 46.Roth MJ, Tanese N, Goff SP. 1985. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J Biol Chem 260:9326–9335. [PubMed] [Google Scholar]
- 47.Boyer PL, Stenbak CR, Clark PK, Linial ML, Hughes SH. 2004. Characterization of the polymerase and RNase H activities of human foamy virus reverse transcriptase. J Virol 78:6112–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowak E, Miller JT, Bona MK, Studnicka J, Szczepanowski RH, Jurkowski J, Le Grice SF, Nowotny M. 2014. Ty3 reverse transcriptase complexed with an RNA–DNA hybrid shows structural and functional asymmetry. Nat Struct Mol Biol 21:389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hizi A, Gazit A, Guthmann D, Yaniv A. 1982. DNA-processing activities associated with the purified α, β2, and αβ molecular forms of avian sarcoma virus RNA-dependent DNA polymerase. J Virol 41:974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hizi A, Leis JP, Joklik WK. 1977. RNA-dependent DNA polymerase of avian sarcoma virus B77. II. Comparison of the catalytic properties of the α, β2, and αβ enzyme forms. J Biol Chem 252:2290–2295. [PubMed] [Google Scholar]
- 51.Hizi A, Leis JP, Joklik WK. 1977. The RNA-dependent DNA polymerase of avian sarcoma virus B77. Binding of viral and nonviral ribonucleic acids to the α, β2, and αβ forms of the enzyme. J Biol Chem 252: 6878–6884. [PubMed] [Google Scholar]
- 52.Lightfoote MM, Coligan JE, Folks TM, Fauci AS, Martin MA, Venkatesan S. 1986. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J Virol 60:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783–1790. [DOI] [PubMed] [Google Scholar]
- 54.Jacobo-Molina A, Ding J, Nanni RG, Clark AD Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci U S A 90:6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malik HS, Eickbush TH. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res 11:1187–1197. [DOI] [PubMed] [Google Scholar]
- 57.Gao G, Goff SP. 1998. Replication defect of moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J Virol 72:5905–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2000. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci U S A 97:3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat Struct Biol 2:303–308. [DOI] [PubMed] [Google Scholar]
- 60.Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 92:1222–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsiou Y, Ding J, Das K, Clark AD Jr., Hughes SH, Arnold E. 1996. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4:853–860. [DOI] [PubMed] [Google Scholar]
- 62.Sarafianos SG, Clark AD Jr., Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. 2002. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J 21:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarafianos SG, Das K, Tantillo C, Clark AD Jr., Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J 20:1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lapkouski M, Tian L, Miller JT, Le Grice SF, Yang W. 2013. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat Struct Mol Biol 20: 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang H, Chopra R, Verdine GL, Harrison SC. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669–1675. [DOI] [PubMed] [Google Scholar]
- 66.Lansdon EB, Samuel D, Lagpacan L, Brendza KM, White KL, Hung M, Liu X, Boojamra CG, Mackman RL, Cihlar T, Ray AS, McGrath ME, Swaminathan S. 2010. Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase–DNA complex. J Mol Biol 397:967–978. [DOI] [PubMed] [Google Scholar]
- 67.Tu X, Das K, Han Q, Bauman JD, Clark AD Jr., Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E. 2010. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol 17:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tuske S, Sarafianos SG, Clark AD Jr., Ding J, Naeger LK, White KL, Miller MD, Gibbs CS, Boyer PL, Clark P, Wang G, Gaffney BL, Jones RA, Jerina DM, Hughes SH, Arnold E. 2004. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat Struct Mol Biol 11:469–474. [DOI] [PubMed] [Google Scholar]
- 69.Lansdon EB, Brendza KM, Hung M, Wang R, Mukund S, Jin D, Birkus G, Kutty N, Liu X. 2010. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): implications for drug design. J Med Chem 53:4295–4299. [DOI] [PubMed] [Google Scholar]
- 70.Das K, Bauman JD, Clark AD Jr., Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. 2008. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A 105:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das K, Ding J, Hsiou Y, Clark AD Jr., Moereels H, Koymans L, Andries K, Pauwels R, Janssen PA, Boyer PL, Clark P, Smith RH Jr., Kroeger Smith MB, Michejda CJ, Hughes SH, Arnold E. 1996. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J Mol Biol 264:1085–1100. [DOI] [PubMed] [Google Scholar]
- 72.Esnouf RM, Ren J, Hopkins AL, Ross CK, Jones EY, Stammers DK, Stuart DI. 1997. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc Natl Acad Sci U S A 94:3984–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ren J, Esnouf R, Hopkins A, Ross C, Jones Y, Stammers D, Stuart D. 1995. The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design. Structure 3:915–926. [DOI] [PubMed] [Google Scholar]
- 74.Ren J, Nichols C, Bird LE, Fujiwara T, Sugimoto H, Stuart DI, Stammers DK. 2000. Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding. J Biol Chem 275:14316–14320. [DOI] [PubMed] [Google Scholar]
- 75.Ren J, Esnouf RM, Hopkins AL, Stuart DI, Stammers DK. 1999. Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies. J Med Chem 42:3845–3851. [DOI] [PubMed] [Google Scholar]
- 76.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391:251–258. [DOI] [PubMed] [Google Scholar]
- 77.Palaniappan C, Wisniewski M, Jacques PS, Le Grice SF, Fay PJ, Bambara RA. 1997. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem 272:11157–11164. [DOI] [PubMed] [Google Scholar]
- 78.Gao HQ, Boyer PL, Arnold E, Hughes SH. 1998. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol 277:559–572. [DOI] [PubMed] [Google Scholar]
- 79.Powell MD, Ghosh M, Jacques PS, Howard KJ, Le Grice SF, Levin JG. 1997. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J Biol Chem 272:13262–13269. [DOI] [PubMed] [Google Scholar]
- 80.Sevilya Z, Loya S, Adir N, Hizi A. 2003. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is modulated by residue 294 of the small subunit. Nucleic Acids Res 31:1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sevilya Z, Loya S, Hughes SH, Hizi A. 2001. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is affected by the thumb subdomain of the small protein subunits. J Mol Biol 311:957–971. [DOI] [PubMed] [Google Scholar]
- 82.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix JL, Le Grice SF. 1996. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35: 8553–8562. [DOI] [PubMed] [Google Scholar]
- 83.Powell MD, Beard WA, Bebenek K, Howard KJ, Le Grice SF, Darden TA, Kunkel TA, Wilson SH, Levin JG. 1999. Residues in the αH and αI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J Biol Chem 274:19885–19893. [DOI] [PubMed] [Google Scholar]
- 84.Yang W, Steitz TA. 1995. Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3:131–134. [DOI] [PubMed] [Google Scholar]
- 85.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121:1005–1016. [DOI] [PubMed] [Google Scholar]
- 86.Dunn LL, McWilliams MJ, Das K, Arnold E, Hughes SH. 2009. Mutations in the thumb allow human immunodeficiency virus type 1 reverse transcriptase to be cleaved by protease in virions. J Virol 83:12336–12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dunn LL, Boyer PL, Clark PK, Hughes SH. 2013. Mutations in HIV-1 reverse transcriptase cause misfolding and miscleavage by the viral protease. Virology 444:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang W, Gamarnik A, Limoli K, Petropoulos CJ, Whitcomb JM. 2003. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J Virol 77:1512–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takehisa J, Kraus MH, Decker JM, Li Y, Keele BF, Bibollet-Ruche F, Zammit KP, Weng Z, Santiago ML, Kamenya S, Wilson ML, Pusey AE, Bailes E, Sharp PM, Shaw GM, Hahn BH. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J Virol 81:7463–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang M, Zensen R, Cho M, Martin MA. 1998. Construction and characterization of a temperature-sensitive human immunodeficiency virus type 1 reverse transcriptase mutant. J Virol 72:2047–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Bambara RA, Demeter LM, Dykes C. 2010. Reduced fitness in cell culture of HIV-1 with nonnucleoside reverse transcriptase inhibitor-resistant mutations correlates with relative levels of reverse transcriptase content and RNase H activity in virions. J Virol 84:9377–9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas DC, Voronin YA, Nikolenko GN, Chen J, Hu WS, Pathak VK. 2007. Determination of the ex vivo rates of human immunodeficiency virus type 1 reverse transcription by using novel strand-specific amplification analysis. J Virol 81:4798–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nermut MV, Fassati A. 2003. Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J Virol 77:8196–8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. 2006. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol 80:3712–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A 108:9975–9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403–415. [DOI] [PubMed] [Google Scholar]
- 100.Yu SF, Sullivan MD, Linial ML. 1999. Evidence that the human foamy virus genome is DNA. J Virol 73:1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, Towers GJ. 2013. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469:424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li S, Hill CP, Sundquist WI, Finch JT. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409–413. [DOI] [PubMed] [Google Scholar]
- 104.Tang S, Murakami T, Agresta BE, Campbell S, Freed EO, Levin JG. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J Virol 75:9357–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. 2007. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J 26:3025–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown PO, Bowerman B, Varmus HE, Bishop JM. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347–356. [DOI] [PubMed] [Google Scholar]
- 107.Brown PO, Bowerman B, Varmus HE, Bishop JM. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A 86:2525–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taylor JM, Cywinski A, Smith JK. 1983. Discontinuities in the DNA synthesized by an avian retrovirus. J Virol 48:654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lochelt M, Flugel RM. 1996. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J Virol 70:1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Konvalinka J, Lochelt M, Zentgraf H, Flugel RM, Krausslich HG. 1995. Active foamy virus proteinase is essential for virus infectivity but not for formation of a Pol polyprotein. J Virol 69:7264–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci U S A 93:7577–7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang WH, Hwang CK, Hu WS, Gorelick RJ, Pathak VK. 2002. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J Virol 76:7473–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Driscoll MD, Hughes SH. 2000. Human immunodeficiency virus type 1 nucleocapsid protein can prevent self-priming of minus-strand strong stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J Virol 74:8785–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buckman JS, Bosche WJ, Gorelick RJ. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol 77:1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. 2006. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology 353:41–51. [DOI] [PubMed] [Google Scholar]
- 116.Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. 2008. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J Virol 82: 9318–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thomas JA, Gorelick RJ. 2008. Nucleocapsid protein function in early infection processes. Virus Res 134:39–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol 74:8980–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol 69:2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Masuda T, Planelles V, Krogstad P, Chen IS. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol 69: 6687–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leavitt AD, Robles G, Alesandro N, Varmus HE. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol 70:721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol 73:2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kirchner J, Sandmeyer SB. 1996. Ty3 integrase mutants defective in reverse transcription or 3′-end processing of extrachromosomal Ty3 DNA. J Virol 70:4737–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nymark-McMahon MH, Beliakova-Bethell NS, Darlix JL, Le Grice SF, Sandmeyer SB. 2002. Ty3 integrase is required for initiation of reverse transcription. J Virol 76:2804–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nymark-McMahon MH, Sandmeyer SB. 1999. Mutations in non-conserved domains of Ty3 integrase affect multiple stages of the Ty3 life cycle. J Virol 73:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harrich D, Ulich C, Garcia-Martinez LF, Gaynor RB. 1997. Tat is required for efficient HIV-1 reverse transcription. EMBO J 16:1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kameoka M, Morgan M, Binette M, Russell RS, Rong L, Guo X, Mouland A, Kleiman L, Liang C, Wainberg MA. 2002. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J Virol 76:3637–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liang C, Wainberg MA. 2002. The role of Tat in HIV-1 replication: an activator and/or a suppressor? AIDS Rev 4:41–49. [PubMed] [Google Scholar]
- 129.Apolloni A, Meredith LW, Suhrbier A, Kiernan R, Harrich D. 2007. The HIV-1 Tat protein stimulates reverse transcription in vitro. Curr HIV Res 5:473–483. [DOI] [PubMed] [Google Scholar]
- 130.Henriet S, Sinck L, Bec G, Gorelick RJ, Marquet R, Paillart JC. 2007. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res 35:5141–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carr JM, Coolen C, Davis AJ, Burrell CJ, Li P. 2008. Human immunodeficiency virus 1 (HIV-1) virion infectivity factor (Vif) is part of reverse transcription complexes and acts as an accessory factor for reverse transcription. Virology 372:147–156. [DOI] [PubMed] [Google Scholar]
- 132.Elder JH, Lerner DL, Hasselkus-Light CS, Fontenot DJ, Hunter E, Luciw PA, Montelaro RC, Phillips TR. 1992. Distinct subsets of retroviruses encode dUTPase. J Virol 66:1791–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wagaman PC, Hasselkus-Light CS, Henson M, Lerner DL, Phillips TR, Elder JH. 1993. Molecular cloning and characterization of deoxyuridine triphosphatase from feline immunodeficiency virus (FIV). Virology 196:451–457. [DOI] [PubMed] [Google Scholar]
- 134.Lerner DL, Wagaman PC, Phillips TR, Prospero-Garcia O, Henriksen SJ, Fox HS, Bloom FE, Elder JH. 1995. Increased mutation frequency of feline immunodeficiency virus lacking functional deoxyuridine-triphosphatase. Proc Natl Acad Sci U S A 92:7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Selig L, Benichou S, Rogel ME, Wu LI, Vodicka MA, Sire J, Benarous R, Emerman M. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J Virol 71:4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. 2000. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J Virol 74:7039–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. 2004. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J Biol Chem 279:28419–28425. [DOI] [PubMed] [Google Scholar]
- 138.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol 79:10978–10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cen S, Javanbakht H, Kim S, Shiba K, Craven R, Rein A, Ewalt K, Schimmel P, Musier-Forsyth K, Kleiman L. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J Virol 76:13111–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Javanbakht H, Cen S, Musier-Forsyth K, Kleiman L. 2002. Correlation between tRNALys3 aminoacylation and its incorporation into HIV-1. J Biol Chem 277:17389–17396. [DOI] [PubMed] [Google Scholar]
- 141.Kelly MC, Kosloff BR, Morrow CD. 2007. Forced selection of tRNA (Glu) reveals the importance of two adenosine-rich RNA loops within the U5-PBS for SIV(smmPBj) replication. Virology 366:330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Djekic UV, Morrow CD. 2007. Analysis of the replication of HIV-1 forced to use tRNAMet(i) supports a link between primer selection, translation and encapsidation. Retrovirology 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li M, Eipers PG, Ni N, Morrow CD. 2006. HIV-1 designed to use different tRNAGln isoacceptors prefers to select tRNAThr for replication. Virol J 3:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Galvis AE, Fisher HE, Nitta T, Fan H, Camerini D. 2014. Impairment of HIV-1 cDNA synthesis by DBR1 knockdown. J Virol 88:7054–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ye Y, De Leon J, Yokoyama N, Naidu Y, Camerini D. 2005. DBR1 siRNA inhibition of HIV-1 replication. Retrovirology 2:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Frankel WN, Stoye JP, Taylor BA, Coffin JM. 1989. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J Virol 63: 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. [DOI] [PubMed] [Google Scholar]
- 148.Nisole S, Lynch C, Stoye JP, Yap MW. 2004. A Trim5–cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci U S A 101:13324–13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573. [DOI] [PubMed] [Google Scholar]
- 150.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. [DOI] [PubMed] [Google Scholar]
- 151.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. 2006. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A 103:7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, Wilke CO. 2009. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J Virol 83:8470–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, O’Neil SP, Johnson W. 2008. Evolution of a TRIM5–CypA splice isoform in old world monkeys. PLoS Pathog 4:e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. 2008. Independent genesis of chimeric TRIM5–cyclophilin proteins in two primate species. Proc Natl Acad Sci U S A 105:3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wilson SJ, Webb BL, Ylinen LM, Verschoor E, Heeney JL, Towers GJ. 2008. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci U S A 105:3557–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. 2012. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem 287:12550–12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.St Gelais C, Wu L. 2011. SAMHD1: a new insight into HIV-1 restriction in myeloid cells. Retrovirology 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. 2012. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. 2009. Mutations involved in Aicardi–Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41:829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, Ackroyd S, Allcock R, Bailey KM, Balottin U, Barnerias C, Bernard G, Bodemer C, Botella MP, Cereda C, Chandler KE, Dabydeen L, Dale RC, De Laet C, De Goede CG, Del Toro M, Effat L, Enamorado NN, Fazzi E, Gener B, Haldre M, Lin JP, Livingston JH, Lourenco CM, Marques W Jr., Oades P, Peterson P, Rasmussen M, Roubertie A, Schmidt JL, Shalev SA, Simon R, Spiegel R, Swoboda KJ, Temtamy SA, Vassallo G, Vilain CN, Vogt J, Wermenbol V, Whitehouse WP, Soler D, Olivieri I, Orcesi S, Aglan MS, Zaki MS, Abdel-Salam GM, Vanderver A, Kisand K, Rozenberg F, Lebon P, Crow YJ. 2013. Assessment of interferon-related biomarkers in Aicardi–Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case–control study. Lancet Neurol 12:1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14:1392–1396. [DOI] [PubMed] [Google Scholar]
- 166.Bishop KN, Holmes RK, Sheehy AM, Malim MH. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 167.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol 78:6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. [DOI] [PubMed] [Google Scholar]
- 169.Harris RS, Sheehy AM, Craig HM, Malim MH, Neuberger MS. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat Immunol 4:641–643. [DOI] [PubMed] [Google Scholar]
- 170.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. [DOI] [PubMed] [Google Scholar]
- 171.Bishop KN, Holmes RK, Malim MH. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol 80:8450–8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Holmes RK, Malim MH, Bishop KN. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci 32:118–128. [DOI] [PubMed] [Google Scholar]
- 174.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol 81:7099–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Coffin JM. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489. [DOI] [PubMed] [Google Scholar]
- 177.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell 4:35–43. [DOI] [PubMed] [Google Scholar]
- 178.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol 75:4832–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mansky LM, Bernard LC. 2000. 3′-Azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J Virol 74:9532–9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koyama H, Ito T, Nakanishi T, Sekimizu K. 2007. Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells 12:547–559. [DOI] [PubMed] [Google Scholar]
- 181.O’Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, Preston BD. 2002. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J Biol Chem 277:38053–38061. [DOI] [PubMed] [Google Scholar]
- 182.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 69:5087–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mansky LM. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res Hum Retroviruses 12:307–314. [DOI] [PubMed] [Google Scholar]
- 184.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. 2010. The nature, position and frequency of mutations made in a single-cycle of HIV-1 replication. J Virol 84:9864–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Abram ME, Ferris AL, Das K, Quinones O, Shao W, Tuske S, Alvord WG, Arnold E, Hughes SH. 2014. Mutations in HIV-1 RT affect the errors made in a single cycle of viral replication. J Virol 88:7589–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Roberts JD, Bebenek K, Kunkel TA. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171–1173. [DOI] [PubMed] [Google Scholar]
- 187.Bebenek K, Abbotts J, Roberts JD, Wilson SH, Kunkel TA. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem 264:16948–16956. [PubMed] [Google Scholar]
- 188.Rezende LF, Curr K, Ueno T, Mitsuya H, Prasad VR. 1998. The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J Virol 72:2890–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Boyer PL, Hughes SH. 2000. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J Virol 74:6494–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mansky LM, Pearl DK, Gajary LC. 2002. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J Virol 76: 9253–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Martin-Hernandez AM, Gutierrez-Rivas M, Domingo E, Menendez-Arias L. 1997. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res 25:1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jonckheere H, De Clercq E, Anne J. 2000. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur J Biochem 267: 2658–2665. [DOI] [PubMed] [Google Scholar]
- 193.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad VR. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 271:1282–1285. [DOI] [PubMed] [Google Scholar]
- 194.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. 1996. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry 35:2168–2179. [DOI] [PubMed] [Google Scholar]
- 195.Feng JY, Anderson KS. 1999. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry 38:9440–9448. [DOI] [PubMed] [Google Scholar]
- 196.Rezende LF, Drosopoulos WC, Prasad VR. 1998. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res 26:3066–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Oude Essink BB, Berkhout B. 1999. The fidelity of reverse transcription differs in reactions primed with RNA versus DNA primers. J Biomed Sci 6:121–132. [DOI] [PubMed] [Google Scholar]
- 198.Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad VR, Wainberg MA. 1997. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res 25:4532–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Drosopoulos WC, Prasad VR. 1998. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J Virol 72:4224–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ji J, Loeb LA. 1994. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology 199:323–330. [DOI] [PubMed] [Google Scholar]
- 201.Ji JP, Loeb LA. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954–958. [DOI] [PubMed] [Google Scholar]
- 202.Stuke AW, Ahmad-Omar O, Hoefer K, Hunsmann G, Jentsch KD. 1997. Mutations in the SIV env and the M13 lacZa gene generated in vitro by reverse transcriptases and DNA polymerases. Arch Virol 142:1139–1154. [DOI] [PubMed] [Google Scholar]
- 203.Hu WS, Temin HM. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A 87:1556–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. 1995. Recombination in HIV-1. Nature 374:124–126. [DOI] [PubMed] [Google Scholar]
- 205.Anderson JA, Bowman EH, Hu WS. 1998. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J Virol 72:1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jetzt AE, Yu H, Klarmann GJ, Ron Y, Preston BD, Dougherty JP. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol 74:1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhuang J, Jetzt AE, Sun G, Yu H, Klarmann G, Ron Y, Preston BD, Dougherty JP. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J Virol 76:11273–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dykes C, Balakrishnan M, Planelles V, Zhu Y, Bambara RA, Demeter LM. 2004. Identification of a preferred region for recombination and mutation in HIV-1 gag. Virology 326:262–279. [DOI] [PubMed] [Google Scholar]
- 209.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci U S A 101:4204–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rhodes T, Wargo H, Hu WS. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J Virol 77: 11193–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu WS. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J Virol 79:1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Galli A, Kearney M, Nikolaitchik OA, Yu S, Chin MP, Maldarelli F, Coffin JM, Pathak VK, Hu WS. 2010. Patterns of Human Immunodeficiency Virus type 1 recombination ex vivo provide evidence for coadaptation of distant sites, resulting in purifying selection for intersubtype recombinants during replication. J Virol 84:7651–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Coffin JM. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol 42:1–26. [DOI] [PubMed] [Google Scholar]
- 214.Etienne L, Hahn BH, Sharp PM, Matsen FA, Emerman M. 2013. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 14:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fu W, Rein A. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol 67:5443–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chin MP, Rhodes TD, Chen J, Fu W, Hu WS. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci U S A 102:9002–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen Y, Wu B, Musier-Forsyth K, Mansky LM, Mueller JD. 2009. Fluorescence fluctuation spectroscopy on viral-like particles reveals variable gag stoichiometry. Biophys J 96:1961–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol 81:4002–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hussein IT, Ni N, Galli A, Chen J, Moore MD, Hu WS. 2010. Delineation of the preferences and requirements of the human immunodeficiency virus type 1 dimerization initiation signal by using an in vivo cell-based selection approach. J Virol 84:6866–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.St Louis DC, Gotte D, Sanders-Buell E, Ritchey DW, Salminen MO, Carr JK, McCutchan FE. 1998. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J Virol 72:3991–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chin MP, Chen J, Nikolaitchik OA, Hu WS. 2007. Molecular determinants of HIV-1 intersubtype recombination potential. Virology 363: 437–446. [DOI] [PubMed] [Google Scholar]
- 222.Delviks KA, Pathak VK. 1999. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J Virol 73:7923–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Delviks KA, Pathak VK. 1999. Development of murine leukemia virus-based self-activating vectors that efficiently delete the selectable drug resistance gene during reverse transcription. J Virol 73:8837–8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rittinger K, Divita G, Goody RS. 1995. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc Natl Acad Sci U S A 92:8046–8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Spence RA, Kati WM, Anderson KS, Johnson KA. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Brown JA, Pack LR, Fowler JD, Suo Z. 2011. Pre-steady-state kinetic analysis of the incorporation of anti-HIV nucleotide analogs catalyzed by human X- and Y-family DNA polymerases. Antimicrob Agents Chemother 55:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lim SE, Ponamarev MV, Longley MJ, Copeland WC. 2003. Structural determinants in human DNA polymerase gamma account for mitochondrial toxicity from nucleoside analogs. J Mol Biol 329:45–57. [DOI] [PubMed] [Google Scholar]
- 228.Das K, Martinez SE, Bauman JD, Arnold E. 2012. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol 19:253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Richman DD, Havlir D, Corbeil J, Looney D, Ignacio C, Spector SA, Sullivan J, Cheeseman S, Barringer K, Pauletti D, Shih C-K, Myers M, Griffin J. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol 68:1660–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Johnson JA, Li JF, Morris L, Martinson N, Gray G, McIntyre J, Heneine W. 2005. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis 192:16–23. [DOI] [PubMed] [Google Scholar]
- 231.Palmer S, Boltz V, Martinson N, Maldarelli F, Gray G, McIntyre J, Mellors J, Morris L, Coffin J. 2006. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A 103: 7094–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Das K, Lewi PJ, Hughes SH, Arnold E. 2005. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog Biophys Mol Biol 88:209–231. [DOI] [PubMed] [Google Scholar]
- 233.Das K, Clark AD Jr., Lewi PJ, Heeres J, De Jonge MR, Koymans LM, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, De Bethune MP, Boyer PL, Clark P, Hughes SH, Janssen PA, Arnold E. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem 47:2550–2560. [DOI] [PubMed] [Google Scholar]
- 234.Sarafianos SG, Das K, Clark AD Jr, Ding J, Boyer PL, Hughes SH, Arnold E. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc Natl Acad Sci U S A 96:10027–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gao HQ, Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2000. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J Mol Biol 300:403–418. [DOI] [PubMed] [Google Scholar]
- 236.Meyer PR, Matsuura SE, Tolun AA, Pfeifer I, So AG, Mellors JW, Scott WA. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 46:1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ren J, Nichols C, Bird L, Chamberlain P, Weaver K, Short S, Stuart DI, Stammers DK. 2001. Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors. J Mol Biol 312:795–805. [DOI] [PubMed] [Google Scholar]
- 238.Das K, Sarafianos SG, Clark AD Jr., Boyer PL, Hughes SH, Arnold E. 2007. Crystal structures of clinically relevant Lys103Asn/ Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J Mol Biol 365:77–89. [DOI] [PubMed] [Google Scholar]
- 239.Hsiou Y, Das K, Ding J, Clark AD Jr., Kleim JP, Rosner M, Winkler I, Riess G, Hughes SH, Arnold E. 1998. Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance. J Mol Biol 284:313–323. [DOI] [PubMed] [Google Scholar]
- 240.Hsiou Y, Ding J, Das K, Clark AD Jr., Boyer PL, Lewi P, Janssen PA, Kleim JP, Rosner M, Hughes SH, Arnold E. 2001. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J Mol Biol 309:437–445. [DOI] [PubMed] [Google Scholar]