Abstract
The present study aimed to determine the clinical significance and potential molecular mechanisms of C-C motif chemokine receptor (CCR) genes in patients with early-stage pancreatic ductal adenocarcinoma (PDAC). The transcriptomic, survival and clinical data of 112 patients with early-stage PDAC who underwent pancreaticoduodenectomy were obtained from The Cancer Genome Atlas. The prognostic values of the CCR genes involved in early-stage PDAC were evaluated using Kaplan-Meier analysis and the multivariate Cox proportional risk regression model, and the potential molecular mechanisms were determined using bioinformatics tools. The identified CCRs closely interacted with each other at both the gene and protein levels. High expression levels of CCR5 [adjusted P=0.012; adjusted hazard ration (HR)=0.478, 95% confidence interval (CI)=0.269–0.852], CCR6 (adjusted P=0.026; adjusted HR=0.527, 95% CI=0.299–0.927) and CCR9 (adjusted P=0.001; adjusted HR=0.374, 95% CI=0.209–0.670) were significantly associated with longer overall survival times in patients with early-stage PDAC. The contribution of CCR5, CCR6 and CCR9 to the outcome of early-stage PDAC was also demonstrated. Combined survival analysis of CCR5, CCR6 and CCR9 suggested that patients with high expression levels of these CCRs exhibited the most favorable outcomes. A prognostic signature was constructed in terms of the expression level of CC5, CCR6 and CCR9, and time-dependent receiver operating characteristic curves indicated that this signature was able to effectively predict the outcome of patients with early-stage PDAC. The potential molecular mechanisms of CCR5, CC6 and CCR9 in PDAC include its intersection of the P53, nuclear factor (NF)-κB, generic transcription, mitogen-activated protein kinase and STAT signaling pathways. Collectively, this highlights that CCR5, CCR6 and CCR9 are potential prognostic biomarkers for early-stage PDAC.
Keywords: molecular mechanism, C-C motif chemokine receptor, early-stage pancreatic ductal adenocarcinoma, pancreaticoduodenectomy
Introduction
In 2018, ~458 million new cases of pancreatic cancer were diagnosed worldwide, resulting in 432 million mortalities, the seventh highest among all cancer-associated deaths (1). Pancreatic cancer is a lethal malignancy with a <5% five-year survival rate, indicating a mortality rate almost equal to its occurrence (1–4). Pancreatic ductal adenocarcinoma (PDAC), which accounts for ~90% of all pancreatic cancer cases (5) is the third and sixth leading cause of cancer-associated death in the United States and China, respectively (6–8). As 80–90% of patients with PDAC are diagnosed at an advanced stage, when the tumor has usually metastasized, radical resection is not possible (6,9–11). Conventional treatments such as chemotherapy and radiotherapy, as well as targeted therapies, have largely failed to prolong the overall survival (OS) time of patients with PDAC (12), and only a few novel anti-PDAC strategies are currently in use (13,14). Therefore, the identification of early diagnostic markers and therapeutic targets for PDAC is critical. Taking into account the encouraging results of immuno- and gene therapies against PDAC (15–17), the present study aimed to identify novel immunological targets associated with its prognosis.
Chemokines and their receptors are critical mediators of the inflammatory and immune responses (18–20), and have recently been implicated in tumorigenesis (21,22). Chemokine receptors promote tumorigenesis via numerous mechanisms (23), including inflammation, that is known to play a significant role in the pathogenesis and progression of pancreatic cancer (24–26). In addition, the poor prognosis and frequent distant metastasis of pancreatic cancer are also associated with immune surveillance escape (12,27,28). The chemokine receptors are classified into four subfamilies-CCR, CXCR, XCR and CX3CR-based on variations within the cysteine motif. Although several studies have elucidated the potential roles of these CCRs (C-C motif chemokine receptors) in pancreatic cancer, using cell lines or murine models (29–35), the association of CCRs with OS remains ambiguous. Therefore, the aim of the present study was to investigate the association between CCRs and the prognosis of patients with PDAC.
Materials and methods
Data mining and processing
The transcriptome profiles of patients with PDAC were obtained from TCGA database (https://cancergenome.nih.gov/, accessed at April 20, 2017), and normalized using the DESeq Package in R (36,37). The corresponding clinical data were acquired from the University of California, Santa Cruz Xena (UCSC Xena; http://xena.ucsc.edu/, accessed at April 20, 2017). In order to eliminate interference from unrelated factors, the patients were selected based on the following inclusion criteria: i) Histological validation; ii) pathological stage I or II according to the 7th American Joint Committee on Cancer (AJCC); iii) availability of complete survival data; and iv) having undergone pancreaticoduodenectomy.
Bioinformatics and correlation analysis of CCR genes
The CCR genes were functionally annotated using Gene Ontology (GO) terms, and the associated pathways were determined by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using the Database for Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/home.jsp version 6.8, accessed at January 19, 2018) (38). The correlation between CCR genes was analyzed by Pearson's correlation coefficient using the corrplot package in R (version 1.2.1335; www.r-project.org). A protein-protein interaction (PPI) map was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (https://string-db.org/, version 11, accessed at march 30, 2019) (39). All CCR gene symbols (CCR1-CCR10) were entered into the platform for Homo sapiens and an interaction score >0.4 was considered to be significant. Finally, a gene-gene interaction network was constructed using GeneMANIA (http://genemania.org/, accessed at March 30, 2019) (40).
Survival analysis
The association between different clinical factors and the prognosis of patients with early-stage PDAC was determined using Kaplan-Meier analysis with the log-rank test; the relevant factors were then included in the multivariate Cox proportional risk regression model to identify the CCR genes significantly associated with OS. Based on the results of the survival analysis of individual genes, combined effect survival analysis was performed and a nomogram was constructed. The CCR genes associated with PDAC prognosis were assessed using combined effect survival analysis (Kaplan-Meier analysis with log-rank test) and the multivariate Cox proportional risk regression model. The nomogram was constructed in R (version 3.5.2; www.r-project.org) using the rms package, based on clinical variables and the expression levels of CCR genes. The scale marked on the line indicates the value range of each variable, and the length of the line segment reflects the contribution of this factor to the outcome event.
Prognostic signature construction
According to the results of the survival analysis, the CCR genes associated with PDAC prognosis were combined to construct a prognostic model based on gene expression level. The risk score formula was as follows: Risk score=expression of gene1 × β1 + expression of gene2 × β2+… expression of Genen × βn (41,42), where βn is the regression coefficient derived from the result of multivariate Cox proportional hazards regression analysis for the corresponding gene. Based on the median risk score value, the patients were divided into a high- and low risk group. To assess the predictive value of the prognostic signature, a time-dependent ROC curve was constructed using the survivalROC package in R (43). Survival analysis was performed to compare prognoses between the high- and low-risk groups.
Gene set enrichment analysis (GSEA)
In order to identify the pathways in which the CCR gene is enriched, and to determine whether the CCR genes in each gene set are enriched in the upper or lower part of the phenotype-related sorted gene list, genome-wide expression profile datasets and corresponding grouping files determined by the expression of CCR genes were uploaded to GSEA (44) for enrichment analysis with database c2 and c5 of the Molecular Signatures Database (MSigDB) (45). A set of genes with both false discovery rate (FDR) <0.25 and P<0.05 was considered to be statistically significant.
Statistical analysis
Statistical analysis was conducted using SPSS 22.0 (IBM Corporation) or R 3.52 (https://www.r-project.org/). Hazard ratios (HRs) and 95% confidence intervals (CI) were used to indicate the relative risk between the high-C and low-CCR expression groups. Pearson's correlation coefficient was used to determine the correlation between CCR genes, where P<0.05 was considered to indicate a statistically significant result. The FDR control in GSEA was achieved using the Benjamini-Hochberg procedure and adjusted for multiple testing (46–48), where an FDR<0.25 was considered to indicate a statistically significant difference.
Results
Data collection and arrangement
The expression profiles of patients with PDAC were acquired from TCGA database. After screening based on the inclusion criteria, patients that fell outside of these parameters were eliminated, and the profiles of the remaining 112 patients were further analyzed.
Functional annotation and correlation analysis of CCR genes
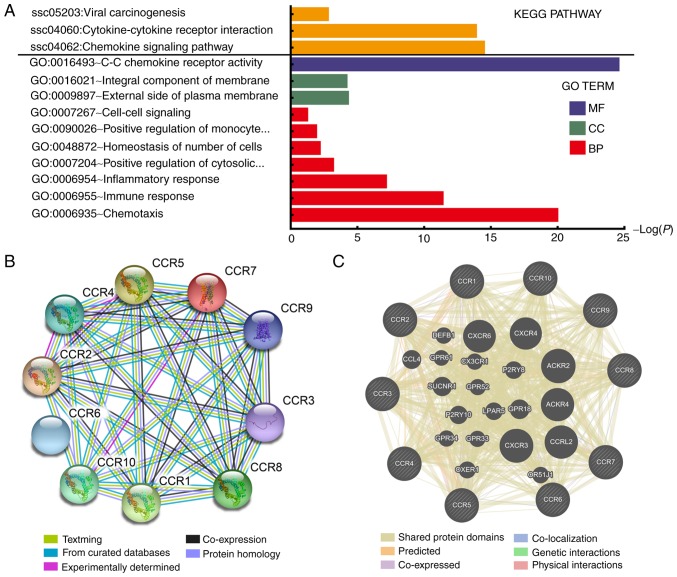
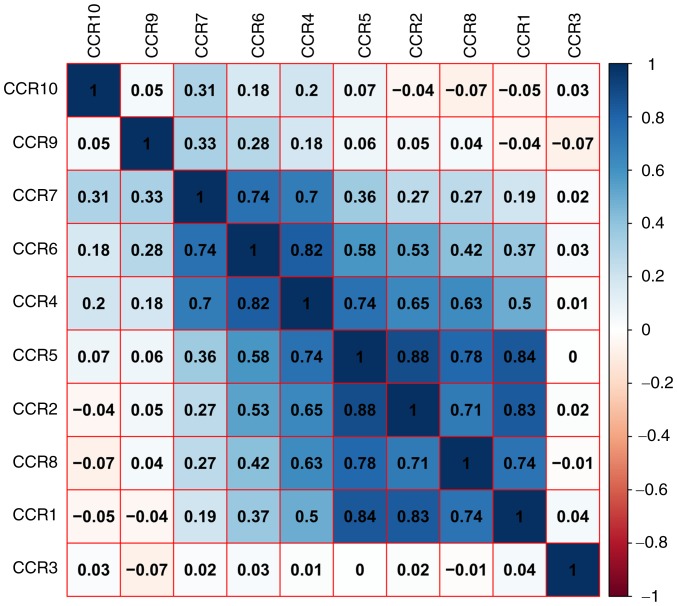
As shown in Fig. 1A and Table SI, the results of GO and KEGG pathway analysis indicated that CCR genes are primarily involved in pathways related to immunity and inflammation, and that the JAK/STAT, mitogen-activated protein kinase (MAPK) and nuclear factor (NF)-κB signaling pathways were associated with chemokine signaling (Fig. S1). PPI and gene-gene interaction network analyses revealed that CCRs interact closely with each other (Fig. 1B and C). A matrix graph of Pearson's correlation analysis indicated that CCR1, CCR2, CCR4, CCR5 and CCR8 are closely related to each other with a correlation coefficient ≥0.5. In addition, a higher degree of correlation was observed among CCR4, CCR6 and CCR7 (correlation coefficient ≥0.7) (Fig. 2). The numbers in each grid represent the correlations between the corresponding genes.
Figure 1.
KEGG pathway and GO term analysis of CCR genes and gene interactions. (A) KEGG pathway and GO term analysis of CCR genes. (B) STRING and (C) GeneMANIA protein-protein association networks of CCR genes. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; CCR, C-C motif chemokine receptor; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; MF, Molecular Function; BP, Biological Process; CC, Cellular Component.
Figure 2.
Matrix graphs of Pearson's correlation analysis of CCR genes.
Survival analysis of CCR genes
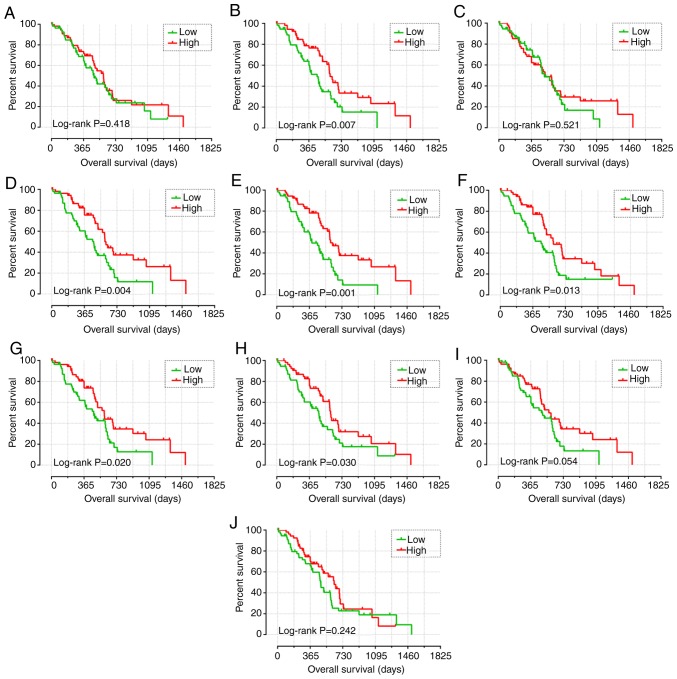
The clinical data of all patients are summarized in Table SII. Histological grade, targeted molecular therapy, radiation therapy and residual resection were all significantly associated with OS. Patients harboring tumors of a higher histological grade, and those who did not receive either targeted or radiation therapies or undergo residual resection were at a higher risk of poor prognosis. In addition, high expression levels of CCR5 (adjusted P=0.012; adjusted HR=0.478, 95% CI=0.269–0.852), CCR6 (adjusted P=0.026; adjusted HR=0.527, 95% CI=0.299–0.927) and CCR9 (adjusted P=0.001; adjusted HR=0.374, 95% CI=0.209–0.670) were significantly associated with lower mortality rates (Table I and Fig. 3). The nomogram also indicated that CCR5, CCR6 and CCR9 may contribute to the prognosis of PDAC, with low expression corresponding to a high point (Fig. 4E).
Table I.
Prognostic value of the expression levels of CCR genes in early-stage pancreatic ductal adenocarcinoma using The Cancer Genome Atlas database.
| Overall survival | |||||||
|---|---|---|---|---|---|---|---|
| Gene expression level | Patients (n=112) | Number of events | Median survival time (days) | Crude HR (95% CI) | Crude Log-rank P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
| CCR1 | |||||||
| Low | 56 | 34 | 485 | 1 | 1 | ||
| High | 56 | 35 | 592 | 0.820 (0.508–1.326) | 0.418 | 0.812 (0.474–1.393) | 0.450 |
| CCR2 | |||||||
| Low | 56 | 38 | 458 | 1 | 1 | ||
| High | 56 | 31 | 603 | 0.518 (0.317–0.846) | 0.007 | 0.668 (0.379–1.178) | 0.163 |
| CCR3 | |||||||
| Low | 56 | 33 | 511 | 1 | 1 | ||
| High | 56 | 36 | 518 | 0.854 (0.527–1.385) | 0.521 | 0.947 (0.556–1.613) | 0.841 |
| CCR4 | |||||||
| Low | 56 | 40 | 470 | 1 | 1 | ||
| High | 56 | 29 | 607 | 0.490 (0.299–0.804) | 0.004 | 0.601 (0.343–1.054) | 0.076 |
| CCR5 | |||||||
| Low | 56 | 39 | 393 | 1 | 1 | ||
| High | 56 | 30 | 603 | 0.433 (0.263–0.714) | 0.001 | 0.478 (0.269–0.852) | 0.012 |
| CCR6 | |||||||
| Low | 56 | 39 | 458 | 1 | 1 | ||
| High | 56 | 30 | 596 | 0.539 (0.329–0.882) | 0.013 | 0.527 (0.299–0.927) | 0.026 |
| CCR7 | |||||||
| Low | 56 | 38 | 473 | 1 | 1 | ||
| High | 56 | 31 | 592 | 0.562 (0.344–0.918) | 0.020 | 0.630 (0.360–1.102) | 0.105 |
| CCR8 | |||||||
| Low | 56 | 38 | 470 | 1 | 1 | ||
| High | 56 | 31 | 603 | 0.588 (0.362–0.955) | 0.030 | 0.642 (0.372–1.107) | 0.111 |
| CCR9 | |||||||
| Low | 56 | 37 | 485 | 1 | 1 | ||
| High | 56 | 32 | 568 | 0.621 (0.381–1.014) | 0.054 | 0.374 (0.209–0.670) | 0.001 |
| CCR10 | |||||||
| Low | 56 | 40 | 481 | 1 | 1 | ||
| High | 56 | 29 | 634 | 0.750 (0.462–1.217) | 0.242 | 0.835 (0.491–1.422) | 0.507 |
Adjusted for histological grade, radiation therapy, radical resection and targeted molecular therapy using the multivariate Cox proportional hazards regression model. CCR, C-C motif chemokine receptor; HR, hazard ratio; CI, confidence interval.
Figure 3.
Kaplan-Meier survival curve analysis of the association between the high and low expression levels of CCR genes and overall survival in patients with early-stage PDAC, generated using The Cancer Genome Atlas. Overall survival curves for (A) CCR1, (B) CCR2, (C) CCR3, (D) CCR4, (E) CCR5, (F) CCR6, (G) CCR7, (H) CCR8, (I) CCR9 and (J) CCR10. CCR, C-C motif chemokine receptor; PDAC, pancreatic ductal adenocarcinoma.
Figure 4.
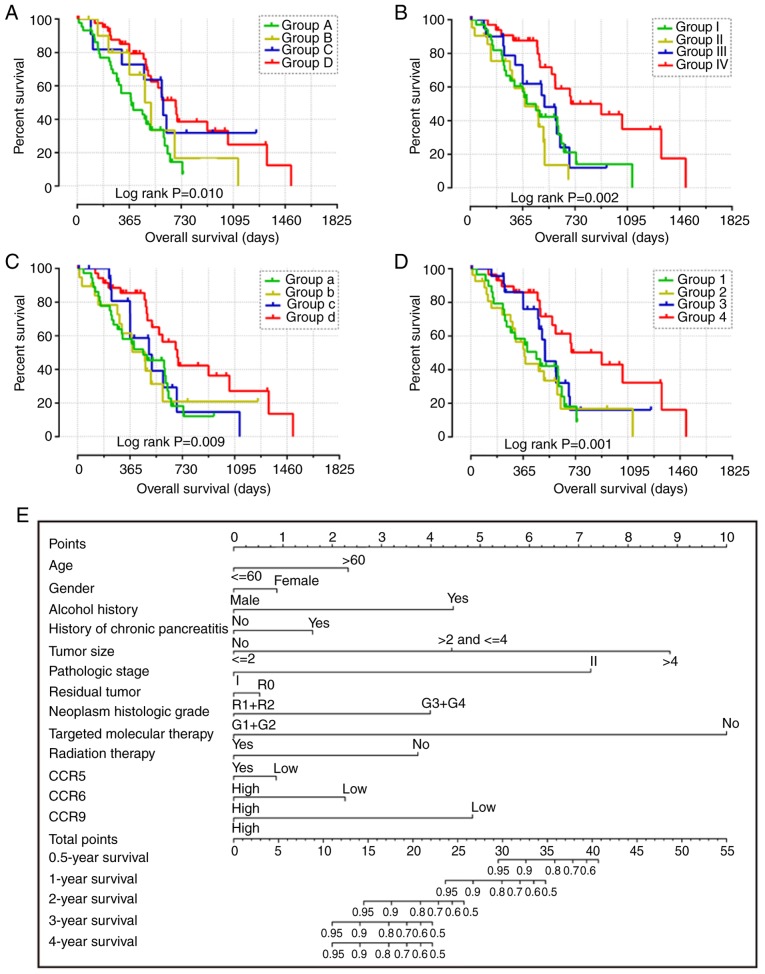
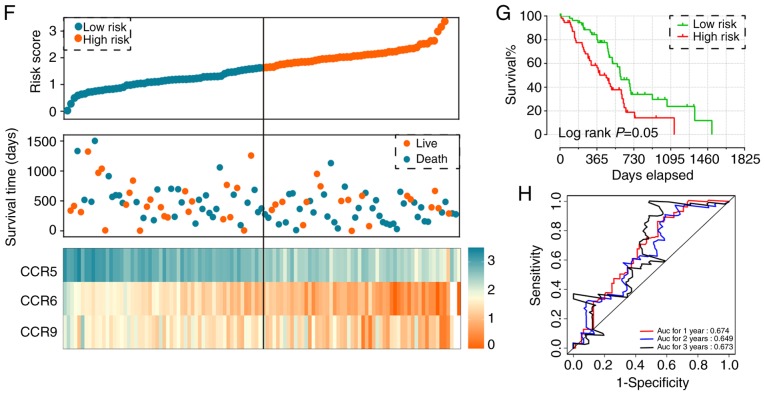
Combined effect of CCR5, CCR6 and CCR9 on the overall survival of patients with early-stage PDAC. Nomogram for predicting 1-, 2- and 3-year events and a prognostic model with risk score, in terms of CCR5, CCR6 and CCR9 expression in early-stage PDAC. (A) Overall survival curves for the combined effect of CCR5 and CCR6. Group A, Low CCR5 + Low CCR6; Group B, Low CCR5 + High CCR6; Group C, High CCR5 + Low CCR6; Group D, High CCR5 + High CCR6. (B) Overall survival curves for the combined effect of CCR5 and CCR9. Group I, Low CCR5 + Low CCR9; Group II, Low CCR5 + High CCR9; Group III, High CCR5 + Low CCR9; Group IV, High CCR5 + High CCR9. (C) Overall survival curves for the combined effect of CCR6 and CCR9. Group a, Low CCR5 + Low CCR9; Group b, Low CCR5 + High CCR9; Group c, High CCR5 + Low CCR9; Group d, High CCR5 + High CCR9. (D) Overall survival curves for the combined effect of CCR5, CCR6 and CCR9. Group 1, Low CCR5 + Low CCR6 + Low CCR9; Group 2, Low CCR5 + Low CCR6 + High CCR9, High CCR5 + Low CCR6 + Low CCR9, and Low CCR5 + High CCR6 + Low CCR9; Group 3, Low CCR5 + High CCR6 + High CCR9, High CCR5 + High CCR6 + Low CCR9, and High CCR5 + Low CCR6 + High CCR9; Group 4, High CCR5 + High CCR6 + High CCR9. (E) Nomogram for predicting 1-, 2- and 3-year events (mortalities) that combine clinical data with CCR5, CCR6 and CCR7 expression. (F) From top to bottom; risk score plot, survival status scatter plot and heat map of the expression levels of CCR5, CCR6 and CCR9 in low- and high-risk groups. (G) Kaplan-Meier curves for low- and high-risk groups. (H) Receiver operating characteristic curve for predicting 1-, 2- and 3-year survival in patients with early-stage PDAC by risk score. CCR, C-C motif chemokine receptor; PDAC, pancreatic ductal adenocarcinoma.
Combined effect survival analysis of CCR genes
The patients were stratified into groups based on the expression levels of different CCR genes. The expression levels of CCR genes in different groups are summarized in Table II. Favorable overall survival was observed in Group D (compared with Groups A, B and C; adjusted P=0.012; adjusted HR=0.434, 95% CI= 0.226–0.833), Group IV (compared with Group I, II and III; adjusted P<0.001; adjusted HR=0.236, 95% CI=0.107–0.520), Group d (compared with Group a, b and c; adjusted P=0.001; adjusted HR=0.284, 95% CI=0.136–0.595) and Group 4 (compared with Group 1, 2 and 3; adjusted P=0.001; adjusted HR=0.253, 95% CI=0.112–0.574) (Table II and Fig. 4A-D).
Table II.
Joint effects survival analysis of CCR gene expression levels with overall survival in patients with early-stage pancreatic ductal adenocarcinoma derived from The Cancer Genome Atlas database.
| Group | CCR5 | CCR6 | CCR9 | Patients (n) | Number of events | Median survival time (days) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|---|---|
| A | Low | Low | 45 | 32 | 381 | 1 | 1 | |||
| B | Low | High | 11 | 7 | 476 | 0.703 (0.307–1.612) | 0.406 | 0.996 (0.374–2.651) | 0.994 | |
| C | High | Low | 11 | 7 | 603 | 0.451 (0.196–1.038) | 0.061 | 0.803 (0.283–2.280) | 0.680 | |
| D | High | High | 45 | 32 | 591 | 0.386 (0.218–0.681) | 0.010 | 0.434 (0.226–0.833) | 0.012 | |
| I | Low | Low | 35 | 24 | 393 | 1 | 1 | |||
| II | Low | High | 21 | 15 | 381 | 1.469 (0.759–2.841) | 0.253 | 0.780 (0.367–1.657) | 0.518 | |
| III | High | Low | 21 | 13 | 517 | 0.846 (0.430–1.667) | 0.630 | 1.158 (0.550–2.437) | 0.699 | |
| IV | High | High | 35 | 17 | 702 | 0.345 (0.178–0.667) | 0.002 | 0.236 (0.107–0.520) | <0.001 | |
| A | Low | Low | 37 | 26 | 458 | 1 | 1 | |||
| B | Low | High | 19 | 13 | 470 | 1.020 (0.521–1.998) | 0.954 | 0.568 (0.256–1.261) | 0.165 | |
| C | High | Low | 19 | 11 | 695 | 0.828 (0.412–1.702) | 0.624 | 0.931 (0.446–1.943) | 0.849 | |
| D | High | High | 37 | 19 | 518 | 0.438 (0.235–0.817) | 0.009 | 0.284 (0.136–0.595) | 0.001 | |
| 1 | Low | Low | Low | 30 | 21 | 458 | 1 | 1 | ||
| 2 | Low | Low | High | 27 | 19 | 375 | 1.094 (0.585–2.047) | 0.778 | 0.874 (0.451–1.692) | 0.689 |
| High | Low | Low | – | – | – | |||||
| Low | High | Low | – | – | – | |||||
| 3 | Low | High | High | 24 | 14 | 518 | 0.667(0.338–1.316) | 0.243 | 0.747(0.347–1.609) | 0.457 |
| High | High | Low | – | – | – | |||||
| High | Low | High | – | – | – | |||||
| 4 | High | High | High | 31 | 15 | 913 | 0.350(0.172–0.711) | 0.004 | 0.253(0.112–0.574) | 0.001 |
Adjusted for histological grade, radiation therapy, radical resection and targeted molecular therapy using the multivariate Cox proportional hazards regression model. CCR, C-C motif chemokine receptor; HR, hazard ratio; CI, confidence interval.
Prognostic signature construction
Using both single gene survival analysis and combined effect survival analysis, CCR5, CCR6 and CCR9 were demonstrated to be associated the prognosis of patients with early-stage PDAC; therefore, these three genes were selected for the construction of the prognostic signature. The regression coefficient of CCR5, CCR6 and CCR9 from the multivariate Cox proportional hazards regression model was −0.836, −0.618 and −0.476 respectively. Because all β-values in this investigation were <0, and to make the result easier to interpret, a constant was added to the end of the following risk score formula: Risk score=expression of CCR5 × −0.836 + expression of CCR × −0.618 + expression of CCR × −0.476 + 4[constant]. The effect of the constant is to ensure that the risk score output is >0. Survival analysis between the high and low risk score groups indicated that a high risk score was significantly associated with the poor outcome of patients with early-stage PDAC (adjusted P=0.018; adjusted HR=1.988, 95% CI=1.125–3.513) (Fig. 4F and G). Time-dependent ROC analysis demonstrated that the prognostic signature effectively predicted the outcome of patients with early-stage PDAC (1-year AUC=0.674; 2-year AUC=0.649; 3-year AUC=0.673; Fig. 4H).
GSEA
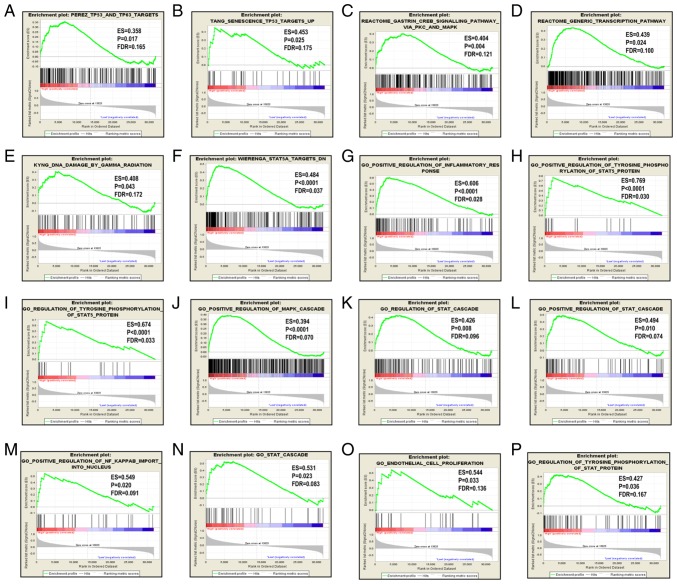
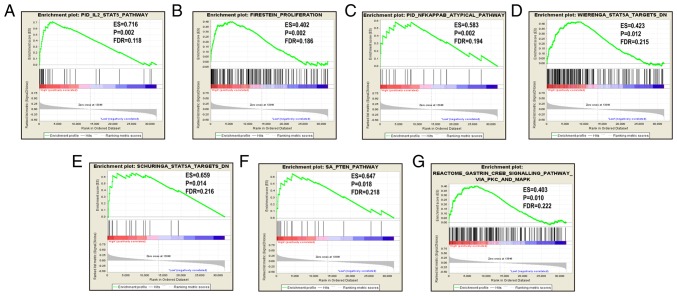
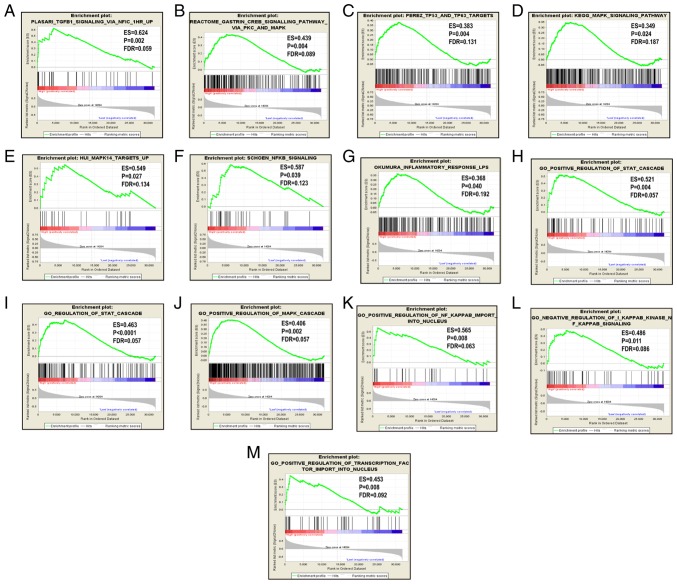
Since CCR5, CCR6 and CCR9 were favorably associated with OS, the patients were stratified according to their respective median expression values. GSEA results are displayed in Figs. 5–7. Analysis of the C2 (curated) gene sets revealed that CCR5 was enriched in TP53 target, TP63 target, MAPK signaling pathway, generic transcription pathway, DNA damage and STAT5A target (Table SIII, Fig. 5A-F); in the C5 (GO) gene sets, CCR5 was enriched in inflammatory response, STAT cascade, MAPK cascade, regulation of NF-κB and endothelial proliferation (Table SIII, Fig. 5G-P); CCR6 was enriched in TGF-β1 signaling pathway, TP53 and TP63 targets, KEGG MAPK signaling pathway, MAPK14 targets and NF-κB signaling in the C2 set (Table SIV and Fig. 6A-G), and in STAT cascade, MAPK cascade, NF-κB import into nucleus, NF-κB signaling and transcription factor importing into nucleus in the C5 set (Table SV, Fig. 6H-M); CCR9 was enriched in the IL-2/STAT5 pathway, proliferation, NF-κB atypical pathway, STAT 5 targets and PTEN pathway in the C2 set (Table SVI and Fig. 7A-G).
Figure 5.
GSEA results for CCR5 in patients with pancreatic ductal adenocarcinoma. GSEA results of (A-F) c2-reference and (G-P) c5-reference gene sets for groups with increased CCR5 expression levels. GSEA, gene set enrichment analysis; CCR, C-C motif chemokine receptor; ES, enrichment score; FDR, false discovery rate.
Figure 7.
GSEA results of CCR6 and CCR9 in patients with pancreatic ductal adenocarcinoma. (A-G) GSEA results of c2-reference gene sets for groups with increased CCR9 expression. GSEA, gene set enrichment analysis; CCR, C-C motif chemokine receptor; ES, enrichment score; FDR, false discovery rate.
Figure 6.
GSEA results for CCR5 and CCR6 in patients with pancreatic ductal adenocarcinoma. GSEA results of (A-G) c2-reference and (H-M) c5-reference gene sets for groups with increased CCR6 expression. GSEA, gene set enrichment analysis; CCR, C-C motif chemokine receptor; ES, enrichment score; FDR, false discovery rate.
Discussion
Chemokines and chemokine receptors serve critical roles in oncogenesis and cancer progression via a number of complex mechanisms. It was reported that the chemokines secreted by the tumor, immune and stromal cells were able to initiate the uncontrolled proliferation and metastasis of tumor cells in an autocrine and paracrine manner, by binding to their cognate receptors (49). To date, CXCR4 is the most widely studied and clearly understood chemokine receptor associated with cancer, and was revealed to be involved in the development, growth, invasion, angiogenesis and metastasis of pancreatic cancer in a number of previous studies (50–57). However, a limited number of studies have investigated the role of the CCR gene in PDAC; therefore, the present study primarily focused on the CCR gene family in PDAC.
Chemokine receptors have been reported to impact tumor progression by regulating the MAPK/ERK, JAK/STAT and NF-κB signaling pathways (49,58–60); CCR5 upregulated c-Fos in tumor cells by stimulating the JAK/STAT pathway (60), and CCR5 stimulation by CCL5 restricted the proliferation of breast cancer cells by increasing p53 transcription via the JAK2 and p38-MAPK pathways (61). Further studies have also illustrated the anti-invasive and anti-metastatic roles of CCR5 in mouse models of breast cancer (62,63). However, a contradictory study revealed that the CCR5-Δ53 polymorphism was associated with a greater risk of developing gallbladder cancer (64), highlighting that the role of CCR5 may be cancer type-dependent. In the present study, bioinformatics analysis suggested that CCR genes were primarily involved in immune and inflammatory responses, and also revealed that the JAK/STAT, MAPK and NF-κB signaling pathways are involved in chemokine signaling. These pathways are consistent with the downstream pathways regulated by p53, therefore, it was hypothesized that CCR5 may also serve a role in PDAC by activating p53. Furthermore, CCR3, CCR4, CCR5 and CCR8 were also found to be associated with viral carcinogenesis, a greater number of CCR genes than those identified to be concerned with carcinogenesis in previous studies (23,65–68). Since various studies have shown its close association with inflammation and immunity (69–75), this is highly relevant to PDAC. Also, considering the function of CCR genes in mediating inflammation, and that CCR5, CCR6 and CCR9 were significantly associated with the overall survival of patients with early-stage PDAC (as indicated by the results of survival analysis), it was concluded that CCR5, CCR6 and CCR9 serve important roles in the development and progression of PDAC. Furthermore, the results of the bioinformatics and survival analysis of CCR genes in PDAC also verified previous findings of the role of CCR genes in cancer (23,35,61,76,77).
The present study is believed to be the first to show that high expression levels of CCR5, CCR6 and CCR9 are associated with prolonged overall survival in patients with early-stage PDAC. The role of CCR5 as a protective factor in PDAC is in agreement with previous studies; it was reported that knocking out CCR5 in pancreatic tumor-bearing mice reduced the infiltration and subsequent cytotoxicity of NK cell in tumors (78). Furthermore, smokers carrying a CCR5 mutant allele have a significantly higher risk of developing pancreatic cancer (76). Moreover, the protective role of CCR5 has also been reported in other malignancies. The CCR5 superagonist 1P7 was found to act as an adjuvant to anti-tumor DNA vaccination by inducing specific CD8+ T-cell responses (77), and the CCR5-Δ53 polymorphism was discovered to be associated with susceptibility to breast cancer in the Indian population (35). It has also been noted that in breast cancer, the absence of CCR5 on the tumor cell surface may promote the proliferation of tumor cells which carry wild-type p53, but not those with mutated p53 (61). CCR5 may also serve a role in PDAC via its indirect impact on tumor cells, such as regulating the anti-tumor immune response. CCR5 is also involved in the chemotaxis of activated naive T cells and T-cells homing (79,80).
Existing studies of CCR6 expression in pancreatic cancer are ambiguous; while one study reported higher levels of CCR6 expression in the pancreatic tumor relative to the adjacent healthy tissues (81), another showed lower expression levels in pancreatic cancer cell lines than normal pancreatic cells (82). Due to the limited number of studies surrounding CCR6 and CCR9 in pancreatic cancer, it was not possible to support the present findings of these CCRs. Therefore, it was surmised that CCR6 and CCR9 may modulate other tumor suppressor genes to inhibit tumor progression, in the same manner as the CCR5-mediated activation of TP53 (61,83,84). This was supported by the GSEA results of the present study, which suggested that CCR6 was enriched in the p53 and STAT cascade, and that CCR9 was enriched in the STAT cascade and NF-κB signaling pathway. However, further studies are required to elucidate the exact mechanisms involved.
The present study possessed various limitations. Firstly, the sample size was relatively small, which may have led to false negative results. Secondly, since the clinical information of a number of patients was incomplete, the clinical variables used for adjustment were not comprehensive. Thirdly, the relationship between CCR and prognosis was only explored at the transcriptional level. Nevertheless, not only was a novel association between the CCR genes and the prognosis of early-stage PDAC discovered, but also the potential molecular mechanisms. Further studies are required to validate these findings and to establish CCRs as therapeutic targets for PDAC.
Though there were several limitations to this investigation, the present study was the first to reveal the association between the CCR genes and the prognosis of early-stage PDAC. In addition, GSEA was used to identify the potential molecular mechanisms of CCR genes that may impact the prognosis of patients with early-stage PDAC. With subsequent studies to verify these findings, CCR genes may become novel targets for the treatment of PDAC.
In conclusion, CCR5, CCR6 and CCR9 represent potential prognostic biomarkers for patients with early-stage PDAC, and are involved in signaling pathways such as those of p53, NF-κB, generic transcription, MAPK and the STAT cascade.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was partly supported by the National Natural Science Foundation of China (grant no. 81560535, 81802874, 81072321, 30760243, 30460143 and 30560133), the Natural Science Foundation of Guangxi Province of China (grant no. 2018GXNSFBA138013 and 2018GXNSFAA050119), the 2009 Program for New Century Excellent Talents in University, Guangxi Natural Sciences Foundation (grant no. GuiKeGong 1104003A-7) and the Guangxi Health Ministry Medicine Grant (Key-Scientific Research-grant no. Z201018). The present study was also supported by the Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (grant no. Z2016318), the Key laboratory of High-Incidence-Tumor Prevention and Treatment (Guangxi Medical University), Ministry of Education (grant no. GKE2018-01), the Guangxi Key R and D Program (grant no. GKEAB18221019), the Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (grant no. 2018KY0110), the Innovation Project of Guangxi Graduate Education (grant no. JGY2018037) and the 2018 Innovation Project of Guangxi Graduate Education (grant no. YCBZ2018036).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and TP conceived and designed the study; XZ, XL, XW and KH acquired and processed the raw data. CY, TY, JL, CH, GZ, HS, WQ, QH, ZL, JH, YG, XY, ZC and TP performed the data analysis. XZ wrote the manuscript, and Tp guided and supervised the manuscript writing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Quaresma M, Coleman MP, Rachet B. 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971–2011: A population-based study. Lancet. 2015;385:1206–1218. doi: 10.1016/S0140-6736(14)61396-9. [DOI] [PubMed] [Google Scholar]
- 4.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 5.Hidalgo M, Cascinu S, Kleeff J, Labianca R, Lohr JM, Neoptolemos J, Real FX, Van Laethem JL, Heinemann V. Addressing the challenges of pancreatic cancer: Future directions for improving outcomes. Pancreatology. 2015;15:8–18. doi: 10.1016/j.pan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 7.Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroentero. 2015;21:7988–8003. doi: 10.3748/wjg.v21.i26.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 9.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 10.Heinemann V, Boeck S, Hinke A, Labianca R, Louvet C. Meta-analysis of randomized trials: Evaluation of benefit from gemcitabine-based combination chemotherapy applied in advanced pancreatic cancer. BMC Cancer. 2008;8:82. doi: 10.1186/1471-2407-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 12.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardiere C, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 14.Mohammed S, Van Buren G, II, Fisher WE. Pancreatic cancer: Advances in treatment. World J Gastroenterol. 2014;20:9354–9360. doi: 10.3748/wjg.v20.i28.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: The unique properties of PD-1 and their advantages for clinical application. Nat Immunol. 2013;14:1212–1218. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- 16.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 17.Vassaux G, Angelova A, Baril P, Midoux P, Rommelaere J, Cordelier P. The promise of gene therapy for pancreatic cancer. Hum Gene Ther. 2016;27:127–133. doi: 10.1089/hum.2015.141. [DOI] [PubMed] [Google Scholar]
- 18.Mantovani A. The chemokine system: Redundancy for robust outputs. Immunol Today. 1999;20:254–257. doi: 10.1016/S0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 19.Moser B, Willimann K. Chemokines: Role in inflammation and immune surveillance. Ann Rheum Dis. 2004;63(Suppl 2):ii84–ii89. doi: 10.1136/ard.2004.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zabel BA, Zuniga L, Ohyama T, Allen SJ, Cichy J, Handel TM, Butcher EC. Chemoattractants, extracellular proteases, and the integrated host defense response. Exp Hematol. 2006;34:1021–1032. doi: 10.1016/j.exphem.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Van Damme J, Proost P, Lenaerts JP, Opdenakker G. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992;176:59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara H, Hamaoka T. Coordination of chemokine and adhesion systems in intratumoral T cell migration responsible for the induction of tumor regression. Int Immunopharmacol. 2001;1:613–623. doi: 10.1016/S1567-5769(00)00049-7. [DOI] [PubMed] [Google Scholar]
- 23.Lacalle RA, Blanco R, Carmona-Rodriguez L, Martin-Leal A, Mira E, Manes S. Chemokine receptor signaling and the hallmarks of cancer. Int Rev Cell Mol Biol. 2017;331:181–244. doi: 10.1016/bs.ircmb.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Dite P, Hermanova M, Trna J, Novotny I, Ruzicka M, Liberda M, Bartkova A. The role of chronic inflammation: Chronic pancreatitis as a risk factor of pancreatic cancer. Dig Dis. 2012;30:277–283. doi: 10.1159/000336991. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahimi S, Mukherjee S, Alhyari L, Rubin E, Aljumaily R. Spontaneous regression of metastatic pancreatic cancer: A role for recurrent inflammation. Pancreas. 2019;48:e4–e6. doi: 10.1097/MPA.0000000000001193. [DOI] [PubMed] [Google Scholar]
- 26.Incio J, Liu H, Suboj P, Chin SM, Chen IX, Pinter M, Ng MR, Nia HT, Grahovac J, Kao S, et al. Obesity-Induced inflammation and desmoplasia promote pancreatic cancer progression and resistance to chemotherapy. Cancer Discov. 2016;6:852–869. doi: 10.1158/2159-8290.CD-15-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes JB, Sircy LM, Heusinkveld LE, Ding W, Leander RN, McClelland EE, Nelson DE. Modulation of macrophage inflammatory nuclear Factor κB (NF-κB) signaling by intracellular cryptococcus neoformans. J Biol Chem. 2016;291:15614–15627. doi: 10.1074/jbc.M116.738187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, Paschall AV, Shi H, Savage N, Waller JL, Sabbatini ME, Oberlies NH, Pearce C, Liu K. The MLL1-H3K4me3 Axis-Mediated PD-L1 expression and pancreatic cancer immune evasion. J Natl Cancer Inst. 2017:109. doi: 10.1093/jnci/djw283. doi: 10.1093/jnci/djw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui K, Zou H, Shi M, Ou Y, Han L, Zhang B, Hu D, Li S. Gene expression profiles in chemokine (C-C Motif) Ligand 21-Overexpressing pancreatic cancer cells. Pathol Oncol Res. 2018 Apr 23; doi: 10.1007/s12253-018-0390-z. doi: 10.1007/s12253-018-0390-z (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson L. Pancreatic cancer: Disrupting the chemokine axis in PDAC. Nat Rev Clin Oncol. 2016;13:330. doi: 10.1038/nrclinonc.2016.62. [DOI] [PubMed] [Google Scholar]
- 31.Marchesi F, Grizzi F, Laghi L, Mantovani A, Allavena P. Molecular mechanisms of pancreatic cancer dissemination: The role of the chemokine system. Curr Pharm Des. 2012;18:2432–2438. doi: 10.2174/13816128112092432. [DOI] [PubMed] [Google Scholar]
- 32.Nakata B, Fukunaga S, Noda E, Amano R, Yamada N, Hirakawa K. Chemokine receptor CCR7 expression correlates with lymph node metastasis in pancreatic cancer. Oncology. 2008;74:69–75. doi: 10.1159/000139126. [DOI] [PubMed] [Google Scholar]
- 33.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gebauer F, Tachezy M, Effenberger K, von Loga K, Zander H, Marx A, Kaifi JT, Sauter G, Izbicki JR, Bockhorn M. Prognostic impact of CXCR4 and CXCR7 expression in pancreatic adenocarcinoma. J Surg Oncol. 2011;104:140–145. doi: 10.1002/jso.21957. [DOI] [PubMed] [Google Scholar]
- 35.Lee YH, Song GG. Association between chemokine receptor 5 delta32 polymorphism and susceptibility to cancer: A meta-analysis. J Recept Signal Transduct Res. 2015;35:509–515. doi: 10.3109/10799893.2014.960934. [DOI] [PubMed] [Google Scholar]
- 36.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yip SH, Wang P, Kocher JA, Sham PC, Wang J. Linnorm: Improved statistical analysis for single cell RNA-seq expression data. Nucleic Acids Res. 2017;45:e179. doi: 10.1093/nar/gkx828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43 (Database Issue) 2015:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao X, Huang K, Huang R, Liu X, Han C, Yu L, Yu T, Yang C, Wang X, Peng T. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther. 2017;10:4493–4506. doi: 10.2147/OTT.S142557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao X, Zhu G, Huang R, Yang C, Wang X, Huang K, Yu T, Han C, Su H, Peng T. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803. doi: 10.2147/CMAR.S161334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francois O, Martins H, Caye K, Schoville SD. Controlling false discoveries in genome scans for selection. Mol Ecol. 2016;25:454–469. doi: 10.1111/mec.13513. [DOI] [PubMed] [Google Scholar]
- 47.Glickman ME, Rao SR, Schultz MR. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol. 2014;67:850–857. doi: 10.1016/j.jclinepi.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 48.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 49.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget. 2013;4:2171–2185. doi: 10.18632/oncotarget.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao G, Wang X, Yu Y. CXCR4/Let-7a Axis regulates metastasis and chemoresistance of pancreatic cancer cells through targeting HMGA2. Cell Physiol Biochem. 2017;43:840–851. doi: 10.1159/000481610. [DOI] [PubMed] [Google Scholar]
- 51.Gao Z, Wang X, Wu K, Zhao Y, Hu G. Pancreatic stellate cells increase the invasion of human pancreatic cancer cells through the stromal cell-derived factor-1/CXCR4 axis. Pancreatology. 2010;10:186–193. doi: 10.1159/000236012. [DOI] [PubMed] [Google Scholar]
- 52.Shakir M, Tang D, Zeh HJ, Tang SW, Anderson CJ, Bahary N, Lotze MT. The chemokine receptors CXCR4/CXCR7 and their primary heterodimeric ligands CXCL12 and CXCL12/high mobility group box 1 in pancreatic cancer growth and development: Finding flow. Pancreas. 2015;44:528–534. doi: 10.1097/MPA.0000000000000298. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Liu C, Mo X, Shi H, Li S. Mechanisms by which CXCR4/CXCL12 cause metastatic behavior in pancreatic cancer. Oncol Lett. 2018;15:1771–1776. doi: 10.3892/ol.2017.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Wang H, Cai J, Du S, Xin B, Wei W, Zhang T, Shen X. Artemin regulates CXCR4 expression to induce migration and invasion in pancreatic cancer cells through activation of NF-κB signaling. Exp Cell Res. 2018;365:12–23. doi: 10.1016/j.yexcr.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 55.Sleightholm RL, Neilsen BK, Li J, Steele MM, Singh RK, Hollingsworth MA, Oupicky D. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol Ther. 2017;179:158–170. doi: 10.1016/j.pharmthera.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 56.Little EC, Kubic JD, Salgia R, Grippo PJ, Lang D. Canonical and alternative transcript expression of PAX6 and CXCR4 in pancreatic cancer. Oncol Lett. 2017;13:4027–4034. doi: 10.3892/ol.2017.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aravindan S, Ramraj S, Kandasamy K, Thirugnanasambandan SS, Somasundaram DB, Herman TS, Aravindan N. Hormophysa triquerta polyphenol, an elixir that deters CXCR4- and COX2-dependent dissemination destiny of treatment-resistant pancreatic cancer cells. Oncotarget. 2017;8:5717–5734. doi: 10.18632/oncotarget.13900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakraborty K, Bose A, Chakraborty T, Sarkar K, Goswami S, Pal S, Baral R. Restoration of dysregulated CC chemokine signaling for monocyte/macrophage chemotaxis in head and neck squamous cell carcinoma patients by neem leaf glycoprotein maximizes tumor cell cytotoxicity. Cell Mol Immunol. 2010;7:396–408. doi: 10.1038/cmi.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jo H, Zhang R, Zhang H, McKinsey TA, Shao J, Beauchamp RD, Ballard DW, Liang P. NF-kappa B is required for H-ras oncogene induced abnormal cell proliferation and tumorigenesis. Oncogene. 2000;19:841–849. doi: 10.1038/sj.onc.1203392. [DOI] [PubMed] [Google Scholar]
- 60.Wong M, Fish EN. RANTES and MIP-1alpha activate stats in T cells. J Biol Chem. 1998;273:309–314. doi: 10.1074/jbc.273.1.309. [DOI] [PubMed] [Google Scholar]
- 61.Manes S, Mira E, Colomer R, Montero S, Real LM, Gomez-Mouton C, Jimenez-Baranda S, Garzon A, Lacalle RA, Harshman K, et al. CCR5 expression influences the progression of human breast cancer in a p53-dependent manner. J Exp Med. 2003;198:1381–1389. doi: 10.1084/jem.20030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datar I, Qiu X, Ma HZ, Yeung M, Aras S, de la Serna I, Al-Mulla F, Tan TZ, Thiery JP, Trumbly R, et al. Correction: RKIP regulates CCL5 expression to inhibit breast cancer invasion and metastasis by controlling macrophage infiltration. Oncotarget. 2016;7:26925. doi: 10.18632/oncotarget.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Velasco-Velazquez M, Pestell RG. The CCL5/CCR5 axis promotes metastasis in basal breast cancer. Oncoimmunology. 2013;2:e23660. doi: 10.4161/onci.23660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava A, Pandey SN, Choudhuri G, Mittal B. CCR5 Delta32 polymorphism: Associated with gallbladder cancer susceptibility. Scand J Immunol. 2008;67:516–522. doi: 10.1111/j.1365-3083.2008.02097.x. [DOI] [PubMed] [Google Scholar]
- 65.Li K, Xu B, Xu G, Liu R. CCR7 regulates Twist to induce the epithelial-mesenchymal transition in pancreatic ductal adenocarcinoma. Tumour Biol. 2016;37:419–424. doi: 10.1007/s13277-015-3819-y. [DOI] [PubMed] [Google Scholar]
- 66.Feng R, Morine Y, Ikemoto T, Imura S, Iwahashi S, Saito Y, Shimada M. Nab-paclitaxel interrupts cancer-stromal interaction through C-X-C motif chemokine 10-mediated interleukin-6 downregulation in vitro. Cancer Sci. 2018;109:2509–2519. doi: 10.1111/cas.13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lanca T, Costa MF, Goncalves-Sousa N, Rei M, Grosso AR, Penido C, Silva-Santos B. Protective role of the inflammatory CCR2/CCL2 chemokine pathway through recruitment of type 1 cytotoxic γδ T lymphocytes to tumor beds. J Immunol. 2013;190:6673–6680. doi: 10.4049/jimmunol.1300434. [DOI] [PubMed] [Google Scholar]
- 68.Gonzalez-Arriagada WA, Lozano-Burgos C, Zuniga-Moreta R, Gonzalez-Diaz P, Coletta RD. Clinicopathological significance of chemokine receptor (CCR1, CCR3, CCR4, CCR5, CCR7 and CXCR4) expression in head and neck squamous cell carcinomas. J Oral Pathol Med. 2018;47:755–763. doi: 10.1111/jop.12736. [DOI] [PubMed] [Google Scholar]
- 69.Farrow B, Evers BM. Inflammation and the development of pancreatic cancer. Surg Oncol. 2002;10:153–169. doi: 10.1016/S0960-7404(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 70.Greer JB, Whitcomb DC. Inflammation and pancreatic cancer: An evidence-based review. Curr Opin Pharmacol. 2009;9:411–418. doi: 10.1016/j.coph.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 71.Hausmann S, Kong B, Michalski C, Erkan M, Friess H. The role of inflammation in pancreatic cancer. Adv Exp Med Biol. 2014;816:129–151. doi: 10.1007/978-3-0348-0837-8_6. [DOI] [PubMed] [Google Scholar]
- 72.McKay CJ, Glen P, McMillan DC. Chronic inflammation and pancreatic cancer. Best Pract Res Clin Gastroenterol. 2008;22:65–73. doi: 10.1016/j.bpg.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 73.Momi N, Kaur S, Krishn SR, Batra SK. Discovering the route from inflammation to pancreatic cancer. Minerva Gastroenterol Dietol. 2012;58:283–297. [PMC free article] [PubMed] [Google Scholar]
- 74.Padoan A, Plebani M, Basso D. Inflammation and pancreatic cancer: Focus on metabolism, cytokines, and immunity. Int J Mol Sci. 2019;20(pii):E676. doi: 10.3390/ijms20030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shadhu K, Xi C. Inflammation and pancreatic cancer: An updated review. Saudi J Gastroenterol. 2019;25:3–13. doi: 10.4103/sjg.SJG_390_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duell EJ, Casella DP, Burk RD, Kelsey KT, Holly EA. Inflammation, genetic polymorphisms in proinflammatory genes TNF-A, RANTES, and CCR5, and risk of pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:726–731. doi: 10.1158/1055-9965.EPI-05-0797. [DOI] [PubMed] [Google Scholar]
- 77.Dorgham K, Abadie V, Iga M, Hartley O, Gorochov G, Combadiere B. Engineered CCR5 superagonist chemokine as adjuvant in anti-tumor DNA vaccination. Vaccine. 2008;26:3252–3260. doi: 10.1016/j.vaccine.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 78.Song Y, Gan Y, Wang Q, Meng Z, Li G, Shen Y, Wu Y, Li P, Yao M, Gu J, Tu H. Enriching the housing environment for mice enhances their Nk cell antitumor immunity via sympathetic Nerve-Dependent regulation of NKG2D and CCR5. Cancer Res. 2017;77:1611–1622. doi: 10.1158/0008-5472.CAN-16-2143. [DOI] [PubMed] [Google Scholar]
- 79.Taub DD, Turcovski-Corrales SM, Key ML, Longo DL, Murphy WJ. Chemokines and T lymphocyte activation: I. Beta chemokines costimulate human T lymphocyte activation in vitro. J Immunol. 1996;156:2095–2103. [PubMed] [Google Scholar]
- 80.Tan MC, Goedegebuure PS, Belt BA, Flaherty B, Sankpal N, Gillanders WE, Eberlein TJ, Hsieh CS, Linehan DC. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J Immunol. 2009;182:1746–1755. doi: 10.4049/jimmunol.182.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubie C, Frick VO, Ghadjar P, Wagner M, Grimm H, Vicinus B, Justinger C, Graeber S, Schilling MK. CCL20/CCR6 expression profile in pancreatic cancer. J Transl Med. 2010;8:45. doi: 10.1186/1479-5876-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Masai K, Iwashita Y, Tominaga M, Hirano S, Shibata K, Matsumoto T, Sasaki A, Ohta M, Kitano S. mRNA expression of chemokine receptors in hepatic and pancreatic tumor cell lines. Gan To Kagaku Ryoho. 2004;31:1261–1263. (In Japanese) [PubMed] [Google Scholar]
- 83.Mehta SA, Christopherson KW, Bhat-Nakshatri P, Goulet RJ, Jr, Broxmeyer HE, Kopelovich L, Nakshatri H. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: Implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene. 2007;26:3329–3337. doi: 10.1038/sj.onc.1210120. [DOI] [PubMed] [Google Scholar]
- 84.Shiraishi K, Fukuda S, Mori T, Matsuda K, Yamaguchi T, Tanikawa C, Ogawa M, Nakamura Y, Arakawa H. Identification of fractalkine, a CX3C-type chemokine, as a direct target of p53. Cancer Res. 2000;60:3722–3726. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.