Abstract
Bladder cancer (BC) is a potentially life-threatening malignancy. Due to a high recurrence rate, frequent surveillance strategies and intravesical drug therapies, BC is considered one of the most expensive tumors to treat. As a fundamental evolutionary catabolic process, autophagy plays an important role in the maintenance of cellular environmental homeostasis by degrading and recycling damaged cytoplasmic components, including macromolecules and organelles. Scientific studies in the last two decades have shown that autophagy acts as a double-edged sword with regard to the treatment of cancer. On one hand, autophagy inhibition is able to increase the sensitivity of cancer cells to treatment, a process known as protective autophagy. On the other hand, autophagy overactivation may lead to cell death, referred to as autophagic cell death, similar to apoptosis. Therefore, it is essential to identify the role of autophagy in cancer cells in order to develop novel therapeutic agents. In addition, autophagy may potentially become a novel therapeutic target in human diseases. In this review, the current knowledge on autophagy modulation in BC development and treatment is summarized.
Keywords: autophagy, bladder cancer, autophagosome, lysosome, autophagic cell death
1. Introduction
Bladder cancer (BC) is a potentially life-threatening malignancy that is considered one of the most expensive tumors in terms of treatment and medical care (1–3). After prostate cancer, it is the second most common type of urological cancer and ranks 10th among the most common types of cancer around the globe (4). It has been estimated that there were 549,393 new cases of BC and 199,922 deaths resulting from this disease worldwide in 2018, according to a report from the International Agency for Research on Cancer (4). The primary histological subtype of human BC is transitional cell carcinoma, which occurs at a high rate of >90% (5). Moreover, 70–80% of new cases are diagnosed as non-muscle invasive BC (NMIBC) (6–8). Despite undergoing transurethral resection of bladder tumor, up to 50% of patients with NMIBC can experience relapse, and 20% continue to progress within 5 years (9). The risk factors for NMIBC progression include tumor stage, grade, size, number and recurrence rate (10). A multidisciplinary approach for the reduction of the tumor recurrence rate, involving surgical intervention combined with radiotherapy, chemotherapy or immunotherapy, is a therapeutic option in patients with BC (10,11). Nevertheless, such a treatment tactic still has unsatisfactory clinical effects. Thus, the development of novel therapies and enhancement of responses to current therapies is urgently required, in order to improve clinical outcomes.
Autophagy, a fundamental evolutionary catabolic process, plays an important role in the maintenance of cellular environmental homeostasis by degrading and recycling damaged cytoplasmic components, including macromolecules and organelles (12). This process can be activated by various cellular stress conditions, including nutrient deprivation (13,14), organelle damage (15), radiotherapy or chemotherapy (15,16), to satisfy cellular needs and promote cell survival. The role of autophagy in cancer development and therapy appears to be paradoxical depending on the context. During the initial stage of cancer development, autophagy serves a major role in tumor suppression by maintaining genomic integrity and preventing proliferation and inflammation (17). However, after the establishment of cancer, cancer cells may utilize autophagy to survive cellular stresses in the adverse microenvironment (18). In addition, autophagy is considered a double-edged sword with regard to the treatment of cancer. Cellular protective autophagy induced by anticancer therapy plays an important role in therapy resistance among cancer cells. Thus, autophagy inhibition can increase the sensitivity of cancer cells to anticancer therapy. On the contrary, autophagy promotion may induce type II programmed cell death, which is referred to as autophagic cell death, similar to apoptosis (19). In this review, the current knowledge on autophagy modulation in BC development and treatment is summarized, with the aim of exploring novel and potential therapeutic targets.
2. Autophagy overview
Autophagy is a lysosomal degradation process in which damaged, long-lived cytoplasmic proteins and organelles are swallowed by double-membrane autophagic vesicles termed autophagosomes (20). The fusion of an autophagosome with a lysosome results in the formation of an autolysosome, which provides an acidic environment for hydrolytic enzymes to degrade the internalized cellular components (21). Autophagy is a biological phenomenon widely occurring in eukaryotic cells. In addition, it is a fascinating process regulated by multiple autophagy-related proteins.
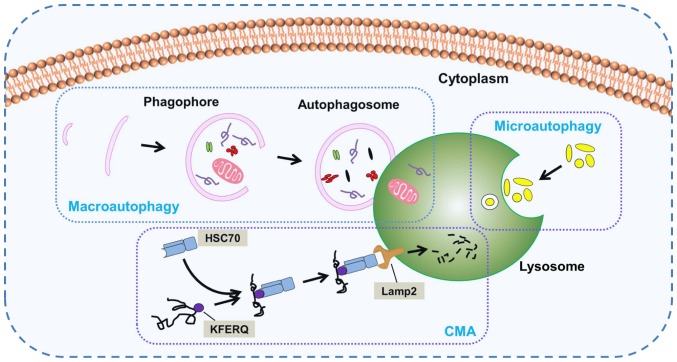
According to the mode of transport for intracellular components to the lysosome, the following three important subtypes of autophagy in mammals have been identified: Macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) (Fig. 1). Macroautophagy, which is usually referred to as autophagy, is the main autophagy pathway. During this pathway, the cell forms a double-membrane structure termed a phagophore, which develops into an autophagosome. Subsequently, cytoplasmic components are engulfed into autophagosomes and delivered to lysosomes for fusion and degradation. Nevertheless, there is a fundamental difference between micro- and macroautophagy. In microautophagy, the lysosome membrane autonomously changes shape via invagination or bulging to engulf cytoplasmic components directly. CMA is a selective and unique autophagic process that selectively discerns and degrades substrate proteins containing the specific KFERQ pentapeptide sequence. Heat shock cognate protein of 70 kDa, a cytosolic chaperone protein, is a key component in CMA able to discern the KFERQ motif and deliver it to lysosome associated membrane protein type 2 for degradation (22,23). Owing to its easily recognizable double-membrane vesicles, macroautophagy is the best-characterized variant of autophagy. The unique morphological features of macroautophagy have been the focus of numerous studies since 1950 (24). Moreover, macroautophagy is the central hub of autophagy-lysosomal pathways. In consideration of the large volume of studies existing on macroautophagy in mammals, the rest of this review will focus on macroautophagy, hereafter referred to as autophagy.
Figure 1.
Three main subtypes of autophagy. Long-lived and damaged cytoplasmic components are degraded via different autophagic processes. The diagram presents the three main subtypes: Macroautophagy, microautophagy and CMA. In macroautophagy, a double-membrane vesicle (phagophore) surrounds degradation components to form an autophagosome, which fuses with a lysosome for subsequent hydrolysis. In microautophagy, the lysosomal membrane alters its shape via invagination or bulging to engulf cytoplasmic components for degradation. In CMA, the adapter molecule HSC70 discerns and binds to the specific KFERQ motif of substrate proteins for translocation to the lysosome and ensures binding to Lamp2 for degradation. CMA, chaperone-mediated autophagy; HSC70, heat shock cognate protein of 70 kDa; Lamp2, lysosome associated membrane protein type 2.
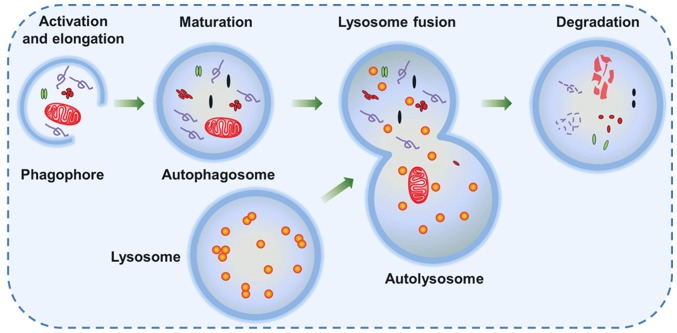
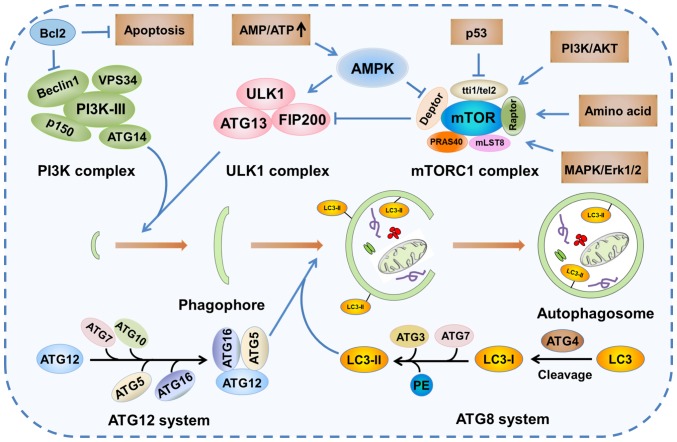
Autophagy is a dynamic process, and is referred to as autophagic flux. The complete process of autophagic flux can be divided into the following four steps: i) Activation and elongation, ii) maturation, iii) lysosome fusion and iv) degradation (Fig. 2) (25). The initial stage of phagophore formation is the most complex step in the process of autophagic flux, in which various functional units are involved, including the mammalian uncoordinated-51-like protein kinase (ULK1/ATG1) complex, the PI3K complex and two ubiquitin-like conjugation systems [autophagy-related protein 12 homolog (ATG12) and ATG8] (Fig. 3) (26,27). Presumably, ATG8-phosphatidylethanolamine (PE), a useful marker of autophagic membranes, is involved in almost all steps of the autophagic flux, particularly the later steps, including phagophore expansion and lysosome fusion (26).
Figure 2.
Steps of the autophagic flux. Autophagy is activated in response to various cellular stress conditions. A double-membrane vesicle (phagophore) begins to form and elongate into an autophagosome in order to engulf intracellular degradation components, including mitochondria, damaged organelles and lipid droplets. The mature autophagosome with intracellular degradation components then fuses with the lysosome and forms an autolysosome, which provides an acidic environment for hydrolytic enzymes to hydrolyze the engulfed components.
Figure 3.
Signaling pathways of autophagy. mTOR kinase is a pivotal molecule in the mTORC1 complex that plays an important role in the regulation of autophagy. Autophagy activation is triggered by decreased activity of the mTORC1 complex due to the activation of AMPK or p53 signaling. The decreased activity of mTORC1, an inhibitor of the mammalian ULK1 complex, leads to the increase the activity of the ULK1 complex, which subsequently initiates the formation of phagophore in conjunction with the PI3K complex. The elongation and maturation of the phagophore is dependent on two ubiquitin-like conjugation systems (ATG12 and ATG8), which involve multiple autophagy proteins, including ATG5, ATG16 and LC3. ATG, autophagy-related protein homolog; mTORC1, mTOR complex 1; AMPK, AMP-activated protein kinase; ULK1, uncoordinated-51-like protein kinase; LC3, microtubule-associated protein light chain 3; PE, phosphatidylethanolamine; TTI1, Tel2-interacting protein 1; TEL2, telomere length regulation protein TEL2; DEPTOR, DEP domain-containing mTOR-interacting protein; RAPTOR, regulatory-associated protein of mTOR; PRAS40, proline-rich Akt substrate of 40 kDa; MLST8, mTOR-associated protein LST8 homolog; MAPK, mitogen-activated protein kinase; FIP200, fusion-inhibiting peptide 200.
In addition, more than 30 autophagy-related genes and homologous proteins have been identified to be essential for autophagy (28). Therefore, the activation of autophagy in response to various stresses leads to alterations in the expression levels of autophagy-related proteins, including microtubule-associated protein light chain 3 (LC3), p62 and Beclin1. As a member of the ATG8 family in mammals, LC3 is a specific protein marker of autophagosome formation. In the ATG8 ubiquitin-like conjugation system, LC3 is first cleaved by ATG4, a specific cysteine protease, to expose the C-terminal cysteine. Subsequently, LC3 is conjugated to PE by ATG7 and ATG3, facilitating the conversion of cytosolic LC3-I to membrane-bound LC3-II (Fig. 3) (29). p62, also referred to as sequestosome 1, is a multi-functional signaling protein implicated in cell proliferation, survival and death. Recent studies have revealed that p62 plays a critical role in the autophagic proteolytic cascade by binding to and delivering ubiquitinated contents to autophagosomes. p62 interacts with membrane protein LC3 via its LC3-interacting region motif (30–32). Therefore, p62 degradation occurs in autolysosomes. On the contrary, autophagy inhibition is often accompanied by upregulation of p62 expression. Beclin1, a multi-domain protein, regulates the cross talk between autophagy and apoptosis. Additionally, the class III PI3K complex has been identified as an important signaling hub that regulates the autophagy-flux and balances the processes of autophagy and apoptosis. Therefore, as a PI3K-III regulatory subunit, Beclin1 plays an important role in the regulation of autophagy through phosphorylation and ubiquitination (33).
3. Autophagy signaling pathways
Autophagy is induced under conditions of cellular stress, including hypoxia, oxidative stress, nutrient deprivation, organelle damage and radiotherapy or chemotherapy in order to meet cellular needs and promote cell survival. Moreover, autophagy levels are regulated by these cellular stresses through different signaling pathways. Subsequently, once these stresses are eliminated by autophagy upregulation, levels return to normal.
mTOR signaling pathway
mTOR is a 300-kDa serine/threonine protein kinase present in mTOR complex 1 (mTORC1) (34). It plays a crucial role in regulating cellular growth, proliferation and protein synthesis. mTORC1 lies upstream of the ULK1 complex and negatively regulates this complex, resulting in autophagy suppression. Specifically, ULK1 is phosphorylated at Ser757 by mTORC1 and then inactivated under nutrient-enriched conditions. On the contrary, nutrient deficiency or mTORC1 inhibitors can lead to the activation of ULK1. Activated ULK1 phosphorylates ATG14 at Ser29 and Beclin1 at Ser14 to stimulate the kinase activity of the class III PI3K, resulting in phagophore and autophagosome formation (35). In addition, numerous upstream signaling pathways that regulate mTORC1 activity may affect the intracellular autophagy levels (Fig. 3).
Mitogen stimulation signaling pathway
The mitogen stimulation signal pathway is characterized by its dependence on serine/threonine kinase Akt. Class I PI3K is stimulated by multiple mitogen signals, including activated receptor tyrosine kinases, activated oncogene Ras and G protein-coupled receptors (36). Activated class I PI3Ks phosphorylate phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 recruits Akt and phosphoinositide-dependent kinases-1 (PDK1), which bind to the cell membrane. Akt serine/threonine kinases are phosphorylated by PDK1 and mTORC2 at Thr308 and Ser473, respectively, resulting in their activation. The downstream effector of the PI3K/Akt pathway is mTOR, which forms two macromolecular complexes, mTORC1 and mTORC2. Akt activates mTORC1 via the inhibition of tuberous sclerosis complex proteins 1/2 (TSC1/2), thereby promoting Rheb (37). Subsequently, active mTORC1 inhibits autophagy by hindering ULK1.
Energy signaling pathway
AMP-activated protein kinase (AMPK), a heterotrimer comprising of an α-catalytic subunit and regulatory β- and γ-subunits, is an evolutionarily conserved regulator of cellular energy homeostasis (38). When the cellular AMP/ATP ratio increases under conditions of cellular stress, such as hypoxia and nutrient deprivation, AMP binds directly to the γ-subunit, thereby activating AMPK. TSC2 is phosphorylated by activated AMPK at Ser1345 and Ser1227, subsequently leading to the inhibition of mTORC1 activity (39).
Amino acid signaling pathway
Amino acids are not only substrates of various metabolic pathways, but also signal molecules that regulate signal transduction pathways. The regulation of mTOR by amino acids is associated with the PI3K/Akt independent pathway. Mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3), a Ste20-related kinase, has been identified as an upstream regulator of mTORC1 in response to amino acids (40). Amino acids can upregulate the activity of MAP4K3, which is essential for the activation of S6 kinase and induce the phosphorylation of eukaryotic initiation factor 4E-binding protein, an mTOR-regulated inhibitor (40). Amino acids are capable of regulating autophagy via several signaling pathways, including the Ras/Raf-1/ERK1/2 pathway (41), the integrin/p38 mitogen-activated protein kinase pathway (42), and the mTOR signaling pathway, which is dominant compared with the other pathways.
p53 signaling pathway
Numerous studies have shown that p53, an important tumor suppressor, plays an important role in regulating glucose homeostasis (43,44). In the case of glucose deficiency, p53 is able to induce autophagy through the mTOR and damage regulated autophagy modulator (DRAM) pathways (45). Further studies have indicated that p53 activation is able to suppress mTOR activity and that this regulation involves p53-dependent AMPK activation with the subsequent activation of the TSC1/TSC2 complex (46–48). Moreover, DRAM is a p53 target gene and encodes a lysosomal protein that induces autophagy. Crighton et al (49) showed that p53 was able to induce autophagy in a DRAM-dependent manner.
Beclin 1-related regulatory pathways
At present, mTOR and Beclin1 are considered significant signaling hubs in the context of autophagy. Beclin1, which is described as the mammalian homolog of yeast ATG6, plays an important role in the process of autophagosome nucleation. It recruits class III PIK3/vacuolar sorting protein-34 to form a regulated complex that generates phosphatidylinositol 3-phosphate [PI(3)P]. Subsequently, certain proteins, including ATG8 and ATG12 complex, bind with PI(3)P-binding domains to modulate autophagosome formation (50). Anti-apoptotic protein Bcl-2 is able to bind to the N-terminal Bcl-2 homology 3 domain of Beclin1, thus inhibiting autophagy (51).
4. Autophagy regulation in BC
During tumor formation, autophagy plays a major role in suppressing tumor initiation and development by maintaining genomic integrity and preventing proliferation and inflammation (17). In the present review, we hypothesize that this is also the case for BC. To the best of our knowledge, no studies that prove this hypothesis have been published to date. Once a tumor has become established, tumor cells can utilize autophagy to survive cellular stresses in the adverse microenvironment. Zhu et al (52) demonstrated that ATG7 was notably overexpressed in invasive BC and knockdown of this protein was able to markedly inhibit BC invasion, suggesting that ATG7 was involved in the regulation of BC development.
The role of autophagy appears to be paradoxical in cancer therapy depending on the context. On one hand, inhibition of autophagy may be employed to increase the cytotoxic effect of treatments, including chemotherapy and radiotherapy (53–55). On the other hand, excessive activation of autophagy may lead to autophagic cell death, also known as programmed cell death type II, which is similar to apoptosis and is defined as cell death in the presence of lysosomes (56–60). Therefore, it is essential to identify the role of autophagy in cancer cells in order to develop new therapeutic agents.
Inhibiting protective autophagy in BC
BC is a malignant tumor associated with high morbidity and mortality, and a significant economic burden associated with it (4). A comprehensive treatment approach involving surgery combined with chemoradiotherapy or immunotherapy is a therapeutic option to reduce the tumor recurrence rate in patients with BC (10,11). Yet, in spite of effective therapy, the majority of patients still experience disease relapse and ultimately die of tumor metastasis (61). Poor prognosis is often attributed to resistance to various therapeutic interventions, which is a distinguishing feature of cancer. Numerous studies suggest that cancer cells may achieve resistance through a wide variety of mechanisms, including cell intrinsic and extrinsic factors, such as genetic heterogeneity (62), autophagy (19,53), tumor microenvironment (63) and cancer stem cells factors (64). Furthermore, autophagy may affect the tumor microenvironment by supplying cellular energy demands and preventing cytotoxicity under stressful conditions such as hypoxia, oxidative stress, inflammation and cytokine release. In addition, autophagy may have an impact on the regulation of cancer stem cell homeostasis by contributing to the maintenance of stemness (65). Given the importance of these mechanisms, increasing interest has arisen in the development of efficient therapeutic approaches based on the autophagy regulation.
Autophagy may be inhibited at any stage of the autophagic flux. In the last decade, many studies involving autophagy mechanisms have been performed to identify chemical inhibitors of autophagy, including chloroquine (CQ) and 3-methyladenine (3-MA). Numerous studies have revealed that inhibition of protective autophagy via various approaches, including pharmaceutical inhibitors (53,55,66,67), RNA-interference agents (66,68) and natural bioactive compounds (69,70) (Table I), is able to increase the sensitivity of BC to therapeutic interventions.
Table I.
Autophagy inhibitors in bladder cancer.
| Inhibitor | Mechanism of action | Treatments combined with inhibitor | Bladder cell line | (Refs.) |
|---|---|---|---|---|
| Chloroquine | Lysosomal lumen alkalizer | Cisplatin, radiotherapy, lapatinib or gefitinib | EJ, T24, RT-112, 5637, J82 | (53,55,66,68) |
| 3-Methyladenine | PI3K inhibitor | Cisplatin, Fangchinoline, lapatinib or gefitinib | RT-112, T24, J82 | (55,67,68) |
| Icaritin | Protein synthesis inhibitor | Epirubicin | 5637, T24 | (69) |
| Frondoside A | Protein synthesis inhibitor | Cisplatin and gemcitabine | RT112 | (70) |
| shRNA | Knockdown of Beclin1 and ATG7/ATG12 | Cisplatin | 5637, T24 | (66) |
| siRNA | Suppression of ATG12 | Lapatinib or gefitinib | T24, J82 | (68) |
shRNA, short hairpin RNA; siRNA, small interfering RNA; ATG, autophagy-related protein.
CQ, an anti-malarial drug, is the most frequently used and proficient agent for the inhibition of autophagy. Currently, CQ and its derivative hydroxychloroquine are the only clinically available autophagy inhibitors approved by the US Food and Drug Administration (71). These drugs are weak lipophilic bases that have the ability to accumulate in lysosomes, increasing lysosomal pH. Subsequently, the alkalization of lysosomes prevents their fusion with autophagosomes and inactivates lysosomal acidic proteases, thereby preventing cargo degradation in autolysosomes. Studies have indicated that CQ-mediated lysosomal dysfunction is able to sensitize cancer cells to chemotherapy and radiotherapy, thus enhancing the anticancer effect of this therapeutic method. A recent study in which BC cells were treated with CQ in combination with cisplatin, demonstrated that inhibition of autophagy enhanced the cytotoxicity of cisplatin (55). Another study revealed the same result that inhibition of cisplatin-induced autophagy using CQ significantly increased BC cell sensitivity to cisplatin, hence enhancing its cytotoxicity (66). These findings suggest that autophagy is induced by cisplatin as a protective mechanism in BC cells and that inhibition of autophagy is able to significantly enhance chemosensitivity in cisplatin-resistant cells. Moreover, it has also been proved that autophagy inhibition by CQ is able to enhance the radiosensitization efficiency in BC cells. Wang et al (53) reported that radiotherapy activated autophagy in BC cells and that subsequent protective autophagy was strongly associated with radioresistance. In addition, the combination of radiation and CQ induced synergistic anticancer effects, as confirmed by evidence of enhanced apoptosis rate, indicating that inhibition of autophagy contributes to the enhancement of radiosensitization (53).
The second most widely used autophagy inhibitor is 3-MA, a class III PI3K inhibitor. PI3Ks are a diverse family of lipid kinases that play important roles in cellular processes, including cell proliferation, metabolism and autophagy regulation (72,73). Class III PI3K, which represents one of the three classes of PI3Ks in mammalian cells, is an activator of autophagy that plays an important role in the early stages of autophagosome formation (25). Fan et al (67) performed a study that aimed to assess the antitumor effects of fangchinoline (Fcn), which is a natural product found in Stephania tetrandra, on BC. They reported that Fcn was able to induce autophagy and apoptosis in BC cells and that inhibition of autophagy by 3-MA resulted in the enhancement of Fcn-induced apoptosis, evident by the increased cleavage of caspase-3 (67). These results suggest that inhibition of protective autophagy may enhance the apoptotic efficiency of anticancer drugs.
In addition to the pharmaceutical inhibitors listed in Table I, natural bioactive compounds have also been proved to possess synergistic anticancer effects with other agents by inhibiting autophagy. Icaritin, a flavonol glycoside isolated from the genus Epimedium, is a hydrolysate of the traditional Chinese herb icariin (69). Previous studies have demonstrated that icaritin has the potential to be an effective anticancer agent by promoting apoptosis, inhibiting cell proliferation and inducing the cell cycle. Pan et al (69) treated human BC cells with icaritin and/or epirubicin (EPI) to investigate how icaritin plays a synergistic role in suppressing BC development. The results indicated that the half maximal inhibitory concentration values with regard to the inhibition of both BT5637 and T24 cell proliferation were significantly higher with icaritin or EPI alone than in combination. Western blot analysis revealed that icaritin not only downregulated the expression levels of major autophagy proteins (ATG3, ATG5, ATG7 and ATG12), but also induced a significant decrease in the LC3-II/LC3-I ratio, suggesting that icaritin was able to enhance BC cell sensitivity to EPI through the inhibition of EPI-induced protective autophagy (69). In addition, another bioactive compound, marine triterpene glycoside frondoside A, extracted from the sea cucumber Cucumaria frondosa, has been demonstrated to exert an anticancer effect. Dyshlovoy et al (70) used frondoside A in combination with cisplatin and gemcitabine and demonstrated that frondoside A was able to enhance the anticancer capabilities of the two standard chemotherapeutic agents in BC RT112 cells by inhibiting protective autophagy.
An increasing number of studies focusing on RNA-interference have been performed to further explore the effects of protective autophagy inhibition on anticancer therapy. Kang et al (68) demonstrated that genetic inhibition of autophagy by disabling ATG12, which is one of the key transcription genes for autophagosome formation and completion, was able to potentiate the anticancer effects of epidermal growth factor receptor (EGFR) inhibitors on BC cells. Small interfering RNA (siRNA) was transfected into BC cells to suppress autophagy through blockage of ATG12. Subsequently, the transfected cells were treated with the EGFR inhibitors lapatinib or gefitinib. The results indicated that inhibition of autophagy by ATG12-siRNA synergistically increased apoptotic cell death when combined with EGFR inhibitors, as confirmed by flow cytometry analysis, suggesting that autophagy acted as a protective mechanism in BC cells (68). In addition, Lin et al (66) combined the autophagy inhibitor short hairpin RNA (shRNA)-based lentivirus with cisplatin and demonstrated that inhibition of autophagy through knockdown of ATG7/ATG12 or Beclin1 using shRNA was able to synergistically enhance the anticancer capabilities of cisplatin in BC 5637 and T24 cells. The results showed that inhibition of autophagy by shRNA increased cisplatin-induced apoptosis, which was evident by the enhanced cleavage of caspase-3, indicating that the combination of cisplatin with autophagy inhibitors was able to increase the sensitivity of BC cells to cisplatin, thus enhancing the cytotoxicity of this chemotherapeutic drug (66).
In summary, autophagy has been identified as a critical mechanism contributing to cancer therapy resistance. Moreover, autophagy may be regarded as a potential target for therapies involving autophagy inhibitors in combination with conventional therapeutics. The numerous studies mentioned above indicate that inhibition of protective autophagy may be able to increase the sensitivity of BC to chemotherapy or radiotherapy, providing important information on the effective regulation of autophagy during cancer treatment.
Activation of autophagic cell death in BC
Autophagy appears to play a contradictory role in cancer therapy depending on the context. Apart from cytoprotective autophagy, the other primary and opposing form of autophagy, which may facilitate cell death either alone or in association with apoptosis, is cytotoxic autophagy. Functionally, cytotoxic autophagy is capable of decreasing the number of viable cells and/or reducing clonogenic survival upon treatment (74). Gewirtz (74) hypothesized that the contradictory functions of autophagy may be associated with specific signaling pathways and/or substrates for the autophagic machinery. Moreover, it has been widely acknowledged that high autophagy levels may induce type II programmed cell death, which is also known as autophagic cell death (56). On the basis of this concept, a number of studies have been performed to investigate the therapeutic potential of autophagic cell death activation in different fields of cancer treatment. In BC, autophagic cell death has been noted in chemotherapy with pazopanib (57), biological therapy with Cheliensisine A-fluoride (ChlA-F) (58) and with alternative therapies (59,60,75) (Table II).
Table II.
Activators of autophagic cell death in bladder cancer.
| Activator | Mechanism of action | Signaling pathways involved | Bladder cell line | (Refs.) |
|---|---|---|---|---|
| Pazopanib | Increasing cathepsin B activity | ERK1/2 | 5637, J82 | (57) |
| ChlA-F | Upregulating Sestrin-2 expression | Sestrin-2 | RT4, T24T, UMUC3 | (58) |
| Ubenimex | Akt agonist | Akt | 5637, RT112 | (75) |
| Salidroside | Suppressing PI3K and p-Akt | Autophagy/PI3K/Akt | T24 | (59) |
| Tetrandrine | Upregulating p-AMPK and downregulating p-mTOR | AMPK/mTOR | T24, 5637 | (60) |
ChlA-F, Cheliensisine A-fluoride; AMPK, AMP-activated protein kinase; p-, phosphorylated.
A better understanding of the potential role of vascular endothelial growth factor (VEGF) in tumor formation and progression has led to the addition of more effective agents to the therapeutic field of multiple tumor types. Among these anti-angiogenic agents, pazopanib, an oral tyrosine kinase inhibitor that targets the VEGF receptor, has been approved for metastatic renal cell carcinoma and soft tissue sarcoma (57). However, to the best of our knowledge, the development of pazopanib for BC is still in the initial stages of clinical research. A previous study, in which BC cells were treated with pazopanib, indicated that this anti-angiogenic agent was able to induce autophagy by increasing ERK1/2 phosphorylation, as evaluated by LC3-II/LC3-I ratio increase, acidic vesicular organelle formation, p62 protein degradation and autophagic flux (57). In addition, pazopanib has been proven to induce autophagic cell death, which is markedly reverted by 3-MA. The aforementioned study confirmed that autophagic cell death induced by pazopanib was associated with increased cathepsin B activity. Finally, comparative gene expression analysis in BC cells at the molecular level indicated that pazopanib induced cytotoxic autophagy by affecting the expression of autophagic genes, such as the upregulation of ATG9B and downregulation of the tumor protein p73 gene (57).
ChlA-F, a novel derivative of Cheliensisin A isolated from Goniothalamus cheliensis Hu, has been identified as a potential anticancer drug due to its enhanced water solubility and chemical stability. Hua et al (58) treated human BC cells and the normal urothelial cell line UROtsa with ChlA-F to investigate the anticancer activity and molecular mechanisms of its biological effects. The results revealed that ChlA-F significantly inhibited BC cell growth and led to a significant increase in the LC3-II/LC3-I ratio, indicating that ChlA-F was able to activate autophagy in human BC cells and may possess antitumor properties. Interestingly, the inhibition of human BC cell growth by ChlA-F was significantly reversed by combining treatment with bafilomycin A1, a fusion inhibitor of autophagosomes and lysosomes (58). Moreover, Hua et al (58) evaluated the effects of ChlA-F on autophagy-related protein expression to illustrate the molecular mechanisms involved in ChlA-F-induced autophagy. They demonstrated that ChlA-F treatment notably increased Sestrin-2 (SESN2) protein expression; however, no prominent effects on the expression levels of other autophagy-related proteins were reported. Further studies confirmed that ChlA-F treatment specifically induced SESN2 expression by increasing its transcription and mRNA stability (58). These findings suggest that the activation of cytotoxic autophagy via specific signaling pathways contributes to the anticancer effects of ChlA-F, therefore providing new information for therapeutic alternatives against human BC.
With the exception of pazopanib and ChlA-F, a number of alternative BC therapies have also been proven to provide anticancer effects through autophagy induction. One example is ubenimex, a broad-spectrum antitumor agent that has been used in adjuvant therapy. Aminopeptidase N (APN), known as the cell surface molecule CD13, is involved in several cell life activities, including cell survival, blood pressure regulation, angiogenesis, invasion and metastasis of tumor cells (76). Therefore, as an APN inhibitor, ubenimex is a promising agent for cancer treatment. Ubenimex has been shown to inhibit the proliferation, migration and invasion of BC cells by downregulating APN expression levels and inducing autophagy through inhibition of the Akt signaling pathway (75). On the contrary, autophagy inhibition with 3-MA reversed the antiproliferative properties of ubenimex, indicating that ubenimex was capable of inducing autophagic cell death in BC cells (75).
Salidroside, a bioactive tyrosine-derived phenolic compound isolated from Rhodiola rosea, possesses properties against fatigue, anoxia, cardiovascular disease and cancer (77). Furthermore, salidroside has been proven to induce autophagic cell death along with apoptosis in BC cells. It has been demonstrated that salidroside causes apoptosis through the autophagy/PI3K/Akt and matrix metalloproteinase-9 signaling pathways (59). Kou et al (60) indicated that autophagy induced by tetrandrine may synergistically enhance apoptosis in human BC cells by regulating the AMPK/mTOR signaling pathway.
5. Conclusions
Although a large number of studies have been performed to confirm novel treatment methods for BC over the past few decades, the management and long-term survival rate of patients with BC have remained relatively stagnant without any significant improvement in clinical outcomes. Autophagy has been shown to be a complex cellular process with contrasting effects in the treatment of BC. Undoubtedly, the aforementioned findings prove that the application of autophagy activators and inhibitors provides further insight for the development of novel therapeutic options against human BC. To the best of our knowledge, since the multidisciplinary approach of surgery combined with radiotherapy, or chemotherapy is usually considered in patients with BC, autophagy inhibitors, including CQ and 3-MA, may be more beneficial in BC treatment due to their ability to increase cancer cell sensitivity to chemotherapy or radiotherapy. Therefore, a better understanding of the role of autophagy in BC treatment is crucial for the selection of effective drugs to target the autophagic pathway.
Acknowledgments
Not applicable.
Funding
This work was supported by Graduate Innovation Fund of Jilin University (grant no. 101832018C068).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
FL and HZ conceived and designed the study and prepared the manuscript. HG, MF and XR were responsible for the literature search, data visualization and analysis. YY and BL retrieved the relevant literature and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Perlis N, Krahn MD, Boehme KE, Alibhai SMH, Jamal M, Finelli A, Sridhar SS, Chung P, Gandhi R, Jones J, et al. The bladder utility symptom scale: A novel patient reported outcome instrument for bladder cancer. J Urol. 2018;200:283–291. doi: 10.1016/j.juro.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Steurer S, Singer JM, Rink M, Chun F, Dahlem R, Simon R, Burandt E, Stahl P, Terracciano L, Schlomm T, et al. MALDI imaging-based identification of prognostically relevant signals in bladder cancer using large-scale tissue microarrays. Urol Oncol. 2014;32:1225–1233. doi: 10.1016/j.urolonc.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DC, Greene PS, Nielsen ME. Surgical advances in bladder cancer: At what cost? Urol Clin North Am. 2015;42:235–252. doi: 10.1016/j.ucl.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Hu L, Chen Y, Hou J. The prognostic value of histological subtype in patients with metastatic bladder cancer. Oncotarget. 2017;8:28408–28417. doi: 10.18632/oncotarget.16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrasekar T, Evans CP. Autophagy and urothelial carcinoma of the bladder: A review. Investig Clin Urol. 2016;57(Suppl 1):S89–S97. doi: 10.4111/icu.2016.57.S1.S89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly JD, Tan WS, Porta N, Mostafid H, Huddart R, Protheroe A, Bogle R, Blazeby J, Palmer A, Cresswell J, et al. BOXIT-A randomised phase III placebo-controlled trial evaluating the addition of celecoxib to standard treatment of transitional cell carcinoma of the bladder (CRUK/07/004) Eur Urol. 2019;75:593–601. doi: 10.1016/j.eururo.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Tan WS, Tan WP, Tan MY, Khetrapal P, Dong L, deWinter P, Feber A, Kelly JD. Novel urinary biomarkers for the detection of bladder cancer: A systematic review. Cancer Treat Rev. 2018;69:39–52. doi: 10.1016/j.ctrv.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Cambier S, Sylvester RJ, Collette L, Gontero P, Brausi MA, van Andel G, Kirkels WJ, Silva FC, Oosterlinck W, Prescott S, et al. EORTC nomograms and risk groups for predicting recurrence, progression, and disease-specific and overall survival in non-muscle-invasive stage Ta-T1 urothelial bladder cancer patients treated with 1–3 years of maintenance bacillus calmette-guérin. Eur Urol. 2016;69:60–69. doi: 10.1016/j.eururo.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 10.Anderson B. Bladder cancer: Overview and management. Part 2: Muscle-invasive and metastatic bladder cancer. Br J Nurs. 2018;27:S8–S20. doi: 10.12968/bjon.2018.27.18.S8. [DOI] [PubMed] [Google Scholar]
- 11.Anderson B. Bladder cancer: Overview and disease management. Part 1: Non-muscle-invasive bladder cancer. Br J Nurs. 2018;27:S27–S37. doi: 10.12968/bjon.2018.27.18.S8. [DOI] [PubMed] [Google Scholar]
- 12.Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177:1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower CM, Wills CA, Frisch SM, Wang HG. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy. 2018;14:1110–1128. doi: 10.1080/15548627.2018.1450020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long M, McWilliams TG. Monitoring autophagy in cancer: From bench to bedside. Semin Cancer Biol. 2019 Jul 15; doi: 10.1016/j.semcancer.2019.05.016. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 15.Zachari M, Gudmundsson SR, Li Z, Manifava M, Shah R, Smith M, Stronge J, Karanasios E, Piunti C, Kishi-Itakura C, et al. Selective autophagy of mitochondria on a ubiquitin-endoplasmic-reticulum platform. Dev Cell. 2019 doi: 10.1016/j.devcel.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Ren X, Hait WN, Yang JM. Therapeutic targeting of autophagy in disease: Biology and pharmacology. Pharmacol Rev. 2013;65:1162–1197. doi: 10.1124/pr.112.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop E, Bradshaw TD. Autophagy modulation: A prudent approach in cancer treatment? Cancer Chemother Pharmacol. 2018;82:913–922. doi: 10.1007/s00280-018-3669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizumura K, Cloonan S, Choi ME, Hashimoto S, Nakahira K, Ryter SW, Choi AM. Autophagy: Friend or foe in lung disease? Ann Am Thorac Soc. 2016;13(Suppl 1):S40–S47. doi: 10.1513/AnnalsATS.201507-450MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang M, Park R, Kim H, Namkoong S, Jo D, Huh YH, Jang IS, Lee JI, Park J. AMPK contributes to autophagosome maturation and lysosomal fusion. Sci Rep. 2018;8:12637. doi: 10.1038/s41598-018-30977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee A, Patel B, Koga H, Cuervo AM, Jenny A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016;12:1984–1999. doi: 10.1080/15548627.2016.1208887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinković M, Šprung M, Buljubašić M, Novak I. Autophagy modulation in cancer: Current knowledge on action and therapy. Oxid Med Cell Longev. 2018;2018:8023821. doi: 10.1155/2018/8023821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor MA, Das BC, Ray SK. Targeting autophagy for combating chemoresistance and radioresistance in glioblastoma. Apoptosis. 2018;23:563–575. doi: 10.1007/s10495-018-1480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang S, Shin KD, Kim JH, Chung T. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018;37:653–664. doi: 10.1007/s00299-018-2258-9. [DOI] [PubMed] [Google Scholar]
- 27.Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 28.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Döring T, Zeyen L, Bartusch C, Prange R. Hepatitis B virus subverts the autophagy elongation complex Atg5-12/16L1 and does not require Atg8/LC3 lipidation for viral maturation. J Virol. 2018;92 doi: 10.1128/JVI.01513-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, Ebner M, Peterbauer T, Sztacho M, Trapannone R, Tarafder AK, et al. Phasing out the bad-How SQSTM1/p62 sequesters ubiquitinated proteins for degradation by autophagy. Autophagy. 2018;14:1280–1282. doi: 10.1080/15548627.2018.1462079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cha-Molstad H, Yu JE, Feng Z, Lee SH, Kim JG, Yang P, Han B, Sung KW, Yoo YD, Hwang J, et al. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat Commun. 2017;8:102. doi: 10.1038/s41467-017-00085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon DH, Park OH, Kim L, Jung YO, Park Y, Jeong H, Hyun J, Kim YK, Song HK. Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat Commun. 2018;9:3291. doi: 10.1038/s41467-018-05825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menon MB, Dhamija S. Beclin 1 phosphorylation-at the center of autophagy regulation. Front Cell Dev Biol. 2018;6:137. doi: 10.3389/fcell.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grunwald DS, Otto NM, Park JM, Song D, Kim DH. GABARAPs and LC3s have opposite roles in regulating ULK1 for autophagy induction. Autophagy. 2019:1–15. doi: 10.1080/15548627.2019.1632620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol Biosyst. 2015;11:1946–1954. doi: 10.1039/C5MB00101C. [DOI] [PubMed] [Google Scholar]
- 37.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dite TA, Ling NXY, Scott JW, Hoque A, Galic S, Parker BL, Ngoei KRW, Langendorf CG, O'Brien MT, Kundu M, et al. The autophagy initiator ULK1 sensitizes AMPK to allosteric drugs. Nat Commun. 2017;8:571. doi: 10.1038/s41467-017-00628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W, Pan Y, Wang S, Liu Y, Chen G, Zhou L, Ni W, Wang A, Lu Y. Cryptotanshinone activates AMPK-TSC2 axis leading to inhibition of mTORC1 signaling in cancer cells. BMC Cancer. 2017;17:34. doi: 10.1186/s12885-016-3038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 42.Prick T, Thumm M, Köhrer K, Häussinger D, Vom Dahl S. In yeast, loss of Hog1 leads to osmosensitivity of autophagy. Biochem J. 2006;394:153–161. doi: 10.1042/BJ20051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 44.Goldstein I, Yizhak K, Madar S, Goldfinger N, Ruppin E, Rotter V. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer Metab. 2013;1:9. doi: 10.1186/2049-3002-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itahana Y, Itahana K. Emerging roles of p53 family members in glucose metabolism. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19030776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dando I, Cordani M, Donadelli M. Mutant p53 and mTOR/PKM2 regulation in cancer cells. IUBMB Life. 2016;68:722–726. doi: 10.1002/iub.1534. [DOI] [PubMed] [Google Scholar]
- 48.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 50.Qian X, Li X, Cai Q, Zhang C, Yu Q, Jiang Y, Lee JH, Hawke D, Wang Y, Xia Y, et al. Phosphoglycerate kinase 1 phosphorylates Beclin1 to induce autophagy. Mol Cell. 2017;65:917.e6–931.e6. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Zhu J, Tian Z, Li Y, Hua X, Zhang D, Li J, Jin H, Xu J, Chen W, Niu B, et al. ATG7 promotes bladder cancer invasion via autophagy-mediated increased ARHGDIB mRNA stability. Adv Sci (Weinh) 2019;6:1801927. doi: 10.1002/advs.201801927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang F, Tang J, Li P, Si S, Yu H, Yang X, Tao J, Lv Q, Gu M, Yang H, Wang Z. Chloroquine enhances the radiosensitivity of bladder cancer cells by inhibiting autophagy and activating apoptosis. Cell Physiol Biochem. 2018;45:54–66. doi: 10.1159/000486222. [DOI] [PubMed] [Google Scholar]
- 54.Piya S, Andreeff M, Borthakur G. Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy. 2017;13:214–215. doi: 10.1080/15548627.2016.1245263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlütermann D, Skowron MA, Berleth N, Böhler P, Deitersen J, Stuhldreier F, Wallot-Hieke N, Wu W, Peter C, Hoffmann MJ, et al. Targeting urothelial carcinoma cells by combining cisplatin with a specific inhibitor of the autophagy-inducing class III PtdIns3K complex. Urol Oncol. 2018;36:160.e1–160.e13. doi: 10.1016/j.urolonc.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 56.Chiao MT, Cheng WY, Yang YC, Shen CC, Ko JL. Suberoylanilide hydroxamic acid (SAHA) causes tumor growth slowdown and triggers autophagy in glioblastoma stem cells. Autophagy. 2013;9:1509–1526. doi: 10.4161/auto.25664. [DOI] [PubMed] [Google Scholar]
- 57.Santoni M, Amantini C, Morelli MB, Liberati S, Farfariello V, Nabissi M, Bonfili L, Eleuteri AM, Mozzicafreddo M, Burattini L, et al. Pazopanib and sunitinib trigger autophagic and non-autophagic death of bladder tumour cells. Br J Cancer. 2013;109:1040–1050. doi: 10.1038/bjc.2013.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hua X, Xu J, Deng X, Xu J, Li J, Zhu DQ, Zhu J, Jin H, Tian Z, Huang H, et al. New compound ChlA-F induces autophagy-dependent anti-cancer effect via upregulating Sestrin-2 in human bladder cancer. Cancer Lett. 2018;436:38–51. doi: 10.1016/j.canlet.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li T, Xu K, Liu Y. Anticancer effect of salidroside reduces viability through autophagy/PI3K/Akt and MMP-9 signaling pathways in human bladder cancer cells. Oncol Lett. 2018;16:3162–3168. doi: 10.3892/ol.2018.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kou B, Liu W, Xu X, Yang Y, Yi Q, Guo F, Li J, Zhou J, Kou Q. Autophagy induction enhances tetrandrine-induced apoptosis via the AMPK/mTOR pathway in human bladder cancer cells. Oncol Rep. 2017;38:3137–3143. doi: 10.3892/or.2017.5988. [DOI] [PubMed] [Google Scholar]
- 61.Alfred Witjes J, Lebret T, Compérat EM, Cowan NC, De Santis M, Bruins HM, Hernández V, Espinós EL, Dunn J, Rouanne M, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71:462–475. doi: 10.1016/j.eururo.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 62.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21:1258–1266. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun Y. Tumor microenvironment and cancer therapy resistance. Cancer Lett. 2016;380:205–215. doi: 10.1016/j.canlet.2015.07.044. [DOI] [PubMed] [Google Scholar]
- 64.Carnero A, Garcia-Mayea Y, Mir C, Lorente J, Rubio IT, LLeonart ME. The cancer stem-cell signaling network and resistance to therapy. Cancer Treat Rev. 2016;49:25–36. doi: 10.1016/j.ctrv.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Sharif T, Martell E, Dai C, Kennedy BE, Murphy P, Clements DR, Kim Y, Lee PW, Gujar SA. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy. 2017;13:264–284. doi: 10.1080/15548627.2016.1260808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin JF, Lin YC, Tsai TF, Chen HE, Chou KY, Hwang TI. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des Devel Ther. 2017;11:1517–1533. doi: 10.2147/DDDT.S126464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan B, Zhang X, Ma Y, Zhang A. Fangchinoline induces apoptosis, autophagy and energetic impairment in bladder cancer. Cell Physiol Biochem. 2017;43:1003–1011. doi: 10.1159/000481698. [DOI] [PubMed] [Google Scholar]
- 68.Kang M, Lee KH, Lee HS, Jeong CW, Kwak C, Kim HH, Ku JH. Concurrent autophagy inhibition overcomes the resistance of epidermal growth factor receptor tyrosine kinase inhibitors in human bladder cancer cells. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan XW, Li L, Huang Y, Huang H, Xu DF, Gao Y, Chen L, Ren JZ, Cao JW, Hong Y, Cui XG. Icaritin acts synergistically with epirubicin to suppress bladder cancer growth through inhibition of autophagy. Oncol Rep. 2016;35:334–342. doi: 10.3892/or.2015.4335. [DOI] [PubMed] [Google Scholar]
- 70.Dyshlovoy SA, Madanchi R, Hauschild J, Otte K, Alsdorf WH, Schumacher U, Kalinin VI, Silchenko AS, Avilov SA, Honecker F, et al. The marine triterpene glycoside frondoside A induces p53-independent apoptosis and inhibits autophagy in urothelial carcinoma cells. BMC Cancer. 2017;17:93. doi: 10.1186/s12885-017-3085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu X, Long YC, Shen HM. Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy. Autophagy. 2015;11:1711–1728. doi: 10.1080/15548627.2015.1043076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gewirtz DA. The four faces of autophagy: Implications for cancer therapy. Cancer Res. 2014;74:647–651. doi: 10.1158/0008-5472.CAN-13-2966. [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Liu Y, Liu W, Zhang Y, Guo F, Zhang L, Cui M, Liu S, Wu R. Ubenimex, an APN inhibitor, could serve as an anti-tumor drug in RT112 and 5637 cells by operating in an Akt-associated manner. Mol Med Rep. 2018;17:4531–4539. doi: 10.3892/mmr.2018.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amin SA, Adhikari N, Jha T. Design of aminopeptidase N inhibitors as anti-cancer agents. J Med Chem. 2018;61:6468–6490. doi: 10.1021/acs.jmedchem.7b00782. [DOI] [PubMed] [Google Scholar]
- 77.Torrens-Spence MP, Pluskal T, Li FS, Carballo V, Weng JK. Complete pathway elucidation and heterologous reconstitution of rhodiola salidroside biosynthesis. Mol Plant. 2018;11:205–217. doi: 10.1016/j.molp.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.