Abstract
CDHR3 (cadherin-related family member 3) is a transmembrane protein that is highly expressed in airway epithelia and the only known receptor for rhinovirus C (RV-C). A CDHR3 SNP (rs6967330) with G to A base change has been linked to severe exacerbations of asthma and increased susceptibility to RV-C infections in young children. The goals of this study were to determine the subcellular localization of CDHR3 and to test the hypothesis that CDHR3 asthma-risk genotype affects epithelial cell function and susceptibility to RV-C infections of the airway epithelia. We used immunofluorescence imaging, Western blot analysis, and transmission electron microscopy to show CDHR3 subcellular localization in apical cells, including expression in the cilia of airway epithelia. Polymorphisms in CDHR3 rs6967330 locus (G→A) that were previously associated with childhood asthma were related to differences in CDHR3 expression and epithelial cell function. The rs6967330 A allele was associated with higher overall protein expression and RV-C binding and replication compared with the rs6967330 G allele. Furthermore, the rs6967330 A allele was associated with earlier ciliogenesis and higher FOXJ1 expression. Finally, CDHR3 genotype had no significant effects on membrane integrity or ciliary beat function. These findings provide information on the subcellular localization and possible functions of CDHR3 in the airways and link CDHR3 asthma-risk genotype to increased RV-C binding and replication.
Keywords: rhinovirus, bronchial epithelium, cadherin-related family member 3, genetics, cilia
There are more than 160 known types of rhinoviruses (RVs), which are classified into A, B, and C species of the Enterovirus genus in the Picornaviridae family. All three RV species frequently cause upper respiratory illnesses, and RV-A and RV-C infections can also cause severe respiratory illnesses such as pneumonia; bronchiolitis; chronic rhinosinusitis; and exacerbations of asthma, COPD, and cystic fibrosis (1–10). The only known cellular receptor for RV-C is CDHR3 (cadherin-related family member 3), which is a transmembrane protein identified by previous studies as a susceptibility factor for early childhood asthma (11, 12). Two CDHR3 alleles, G and A (SNP rs6967330), representing a missense mutation between codons TGT and TAT, result in protein variants Cys529 and Tyr529, respectively. CDHR3-Tyr529 is linked to severe asthma exacerbations (52%) in children (12, 13), and children carrying the A allele of this gene are more susceptible to RV-C infections and illnesses (2, 3).
Human bronchial epithelial cells (BEC) cultured at air–liquid interface (ALI) represent an almost native cell system in which to study RV-C infections of the airways in vitro. Multipotent basal cells extracted from bronchial tissue are cultured in specialized medium in ALI to initiate cell differentiation into mature pseudostratified columnar epithelium consisting of basal cells, ciliated cells, secretory club cells, and mucin-producing goblet cells (14–16). We previously reported that CDHR3 is expressed in FOXJ1-expressing ciliated cells, which are also the targets of RV-C binding (17). In the present study, we conducted experiments to identify the subcellular localization of CDHR3 and to test the hypothesis that CDHR3 genotype impacts epithelial cell function, including the susceptibility of the airway epithelium to RV-C infections in vitro.
Methods
BEC and Nasal Epithelial Cell Culture
Human primary basal BECs were derived from tissue trimmed from healthy donor lungs destined for transplant. BECs extracted from this tissue were stocked in liquid nitrogen for long-term storage or until needed, as previously described (18, 19). Human primary nasal epithelial cells (NECs) were obtained from nasal brushings collected from participants in the Childhood Origins of Asthma (COAST) birth cohort study (20). The families of these children provided informed consent for them to participate in the study, including the collection of nasal brushings. Both BECs and NECs were cultured at ALI in 0.4-μm pore polyester membrane Transwell plates (Corning). The process of cellular differentiation took approximately 28 days to complete.
Immunoblot Analysis
For CDHR3 subcellular compartment analysis, cells from BEC-ALI cultures were lysed and then separated into three cellular protein fractions (Qproteome kit; Qiagen), purified, and quantitated by bicinchoninic acid assay (Pierce Biotechnology). Then, 20 μg of each protein fraction was mixed with 2× SDS gel loading buffer, denatured by boiling, then loaded into Mini-PROTEAN TGX gels (Bio-Rad Laboratories). Proteins were transferred to a polyvinylidene difluoride membrane and blocked with 3% nonfat dry milk in Tris-buffered saline with Tween 20. For whole-cell protein analysis, BEC-ALI cultures were directly lysed with 2× SDS before loading onto gels. After membrane transfer, samples were incubated with monoclonal antibody (mAb)-CDHR3 primary antibody (1:1,000; Abcam) overnight at 4°C before a secondary antimouse IgG-peroxidase (1:5,000; Sigma-Aldrich) was added, followed by incubation for 1 hour at room temperature. The bands were imaged after treatment with chemiluminescent substrate (Pierce Biotechnology).
Rhinovirus Infection
RV-C15 was produced from the pC15-Rz plasmid, and RV-B52 was produced from pB52-Rz plasmid, as previously described (21, 22). BECs with distinct CDHR3 rs6967330 genotypes (A/A, A/G, and G/G) were cultured at ALI in 12-well Transwell inserts (Corning) and infected weekly with RV-C15 (multiplicity of infection, 10). BECs were incubated at 34°C for 1 hour or 24 hours to assess virus binding or yield, respectively. The cells were washed three times with PBS, and the subsequent whole-cell lysates were analyzed for viral RNA levels by real-time PCR.
Differentiation Markers
BECs with distinct CDHR3 rs6967330 genotypes (A/A, A/G, and G/G) were cultured at ALI and collected weekly in RLT buffer (Qiagen). Their mRNA for CDHR3 and differentiation gene markers was quantified by real-time PCR. Gene markers detected for distinct cell subpopulations were 1) C-MYB for intermediate cilia-fated cells, 2) FOXJ1 for ciliated cells, 3) MUC5AC for goblet cells, and 4) SCGB1A1 for club cells.
Immunofluorescence and Confocal Imaging
Human bronchial tissue or BEC-ALI cultures were fixed in 4% paraformaldehyde and embedded in paraffin blocks. The samples were sectioned and mounted on charged glass slides, then deparaffinized and treated with 10 mM sodium citrate for antigen retrieval. After blocking with 5% BSA (1 h, room temperature), the slides were incubated overnight at 4°C with primary antibody (mAb CDHR3, Abcam; mAb acetylated α-tubulin, Cell Signaling Technology; mAb E-cadherin, Abcam; mAb β-actin, Sigma-Aldrich). The following day, the slides were washed with PBS with Tween 20 and then incubated (1 h, room temperature) with fluorophore-conjugated secondary antibody (Alexa Fluor fluorophores; Pierce Biotechnology) before counterstaining with DAPI and mounting (EMS mounting medium; Electron Microscopy Sciences). Fluorescence images were captured with a Nikon Ti Eclipse microscope. For Z-stack imaging, BEC-ALI cultures were fixed, then permeabilized with 0.2% Triton X-100 before serum block and antibody incubation. Cell specimens were counterstained with DAPI and then placed on glass-bottomed plates in mounting medium.
Transmission Electron Microscopy
We followed modified AURION’s protocol for immunogold labeling of preembedded samples to prepare cells for transmission electron microscopy (www.aurion.nl). Differentiated primary human BECs were fixed in 4% paraformaldehyde with 0.5% glutaraldehyde overnight at 4°C and then permeabilized with 0.2% Triton X-100 for 1 hour at room temperature. Specimens were blocked and then incubated overnight with primary antibody (1–5 μg/ml) at 4°C. Specimens were then immunolabeled with F(ab′)2-conjugated ultrasmall gold particles, silver enhanced (AURION R-Gent SE-EM), postfixed with 1% osmium tetroxide, and embedded in Poly/Bed 812 resin (Polysciences Inc.). These were sectioned into 10-μm slices before 4% uranyl acetate staining and examination on a Philips CM120 transmission electron microscope.
Fluorescent Bead Tracking
Cells were washed with warm PBS and then treated apically with 1 × 106 of 1-μm fluorescent polystyrene beads (Thermo Fisher Scientific). Time-lapse images were collected over 5 minutes, and fluorescent bead movement over the apical surface of BEC-ALI culture was recorded using a fluorescence microscope (Nikon Ti Eclipse). The relative velocity was measured by quantifying distance traveled by individual beads in TrackMate (Fiji, ImageJ) software.
Colocalization Quantification
Using NIS-Elements AR software (Nikon Instruments), multiple regions in the cilia were selected, and overlap of red (acetylated α-tubulin) and green (CDHR3) signals was analyzed using Pearson’s colocalization coefficient and Mander’s overlap coefficient measurements.
Statistical Analysis
The comparative RV binding to cells and virus replication yields in infected cells were evaluated using Wilcoxon’s rank-sum test, as implemented in R (R Foundation for Statistical Computing).
Cytokine Quantification
RV-C–induced CXCL10 and IFN-λ (1/3) were measured in NEC culture media 24 hours postinfection by ELISA according to the manufacturer’s protocol (R&D Systems).
Results
CDHR3 Localizes in the Apical Cells of Differentiated Bronchial Epithelia
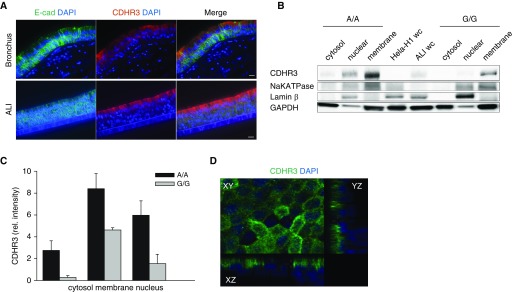
To identify the cellular distribution of CDHR3 expression within the epithelium, we differentiated primary human BECs at ALI for 28 days and imaged sections under a fluorescence microscope. For comparison, we imaged human bronchial tissue sections from normal lung donors. Immunofluorescent staining of BEC-ALI culture and human bronchial tissue sections indicated that CDHR3 is expressed predominantly in the differentiated apical cells of the bronchial epithelia. Expression of CDHR3 in cultured BEC-ALI was similar to that in bronchial tissue (Figure 1A).
Figure 1.
CDHR3 localizes in the apical cells of human airway epithelial cells. (A) Immunofluorescent staining of E-cadherin (E-cad; green), CDHR3 (red), and nucleus (DAPI; blue) in bronchial tissue (top) and human bronchial epithelial cell air–liquid interface (BEC-ALI) culture (bottom). (B) Immunoblot analysis of three subcellular compartments extracted from BEC-ALI cultures (n = 3) with A/A and G/G genotype: cytosol, membrane, and nuclear, stained for CDHR3 (∼100 kD). (C) Quantification of CDHR3 protein band signal intensity of subcellular compartments (n = 3). (D) Z-stack slice view of BEC-ALI culture stained for CDHR3 (green) and nucleus (DAPI). Scale bars: 10 μm. CDHR3 = cadherin related family member 3; rel. = relative; wc = whole-cell lysate.
Analysis of purified subcellular compartments (cytosolic, plasma membrane, and nuclear fractions) from fully differentiated BECs that were homozygous for either the CDHR3 asthma-risk allele (rs6967330 A/A) or the low-risk allele (G/G) revealed that CDHR3 is present in all three subcellular compartments (Figure 1B). BECs with the A/A genotype had greater overall protein expression, whereas the proportion of CDHR3 distribution between subcellular compartments was similar (Figure 1C). The highest proportion of CDHR3 was detected in the plasma membrane fraction, followed by the nuclear fraction, and the least proportion was in the cytosolic fraction (Figures 1B–1D). Confocal image analysis confirmed that CDHR3 localization is highest in the plasma membrane (Figure 1D).
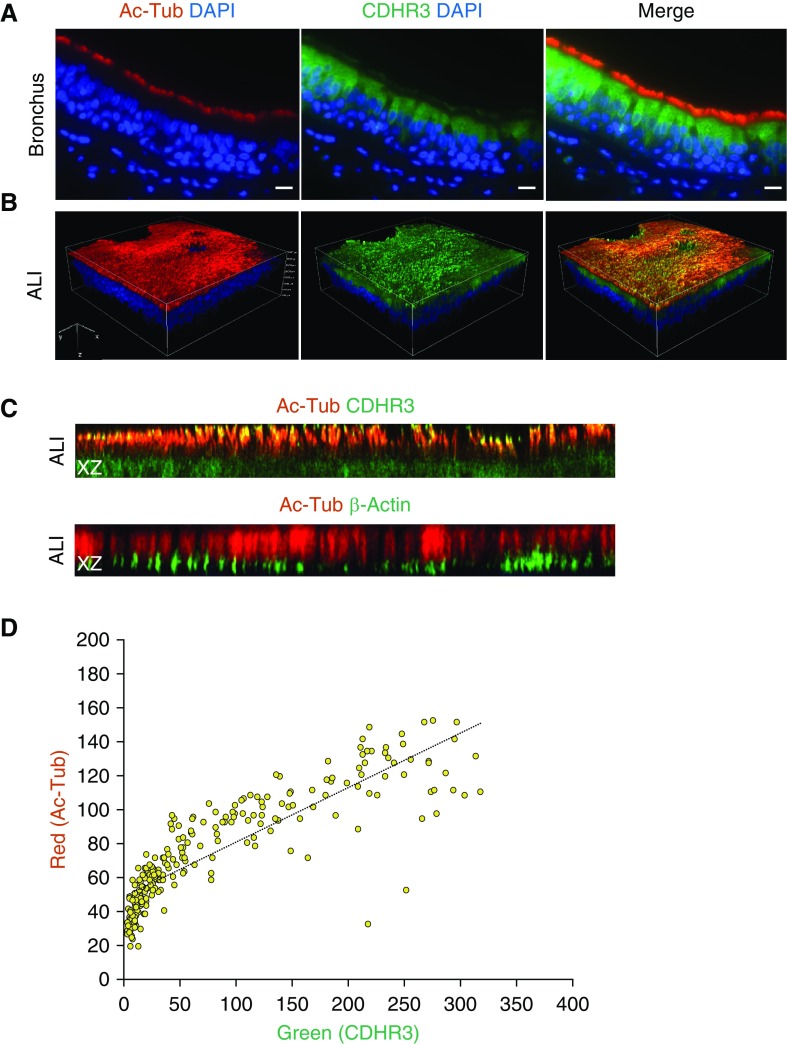
CDHR3 Is Expressed in Cilia
Using flow cytometry, we previously demonstrated that ciliated cells of human epithelia expressed CDHR3 (17). Given the apical expression pattern of CDHR3, we next tested whether this protein was expressed in conjunction with cilia by fluorescence imaging. We found that apical cells which stained positive for cilia also expressed CDHR3, whereas apical cells that were negative for cilia were also negative for CDHR3 (Figure 2A). Furthermore, most cells that were negative for CDHR3 were positive for MUC5AC, a marker for mucin secreted by goblet cells (Figure E1 in the data supplement).
Figure 2.
CDHR3 expression in cilia of bronchial epithelial cells. (A) Immunofluorescent staining profiles of acetylated α-tubulin (Ac-Tub; red), CDHR3 (green), and nucleus (DAPI) in the epithelium of human bronchial tissue. (B) Three-dimensional volume view of Z-stack confocal images of immunofluorescent Ac-Tub (red), CDHR3 (green), nucleus (DAPI), and merged signals. Regions of red and green signal overlap are shown in yellow. (C) Z-stack confocal XZ slice view comparison of two separate cultures stained for (top) Ac-Tub (red) with CDHR3 (green) and (bottom) Ac-Tub (red) with β-actin (green). Regions of signal overlap are shown in yellow. (D) Scatterplot quantification of colocalization correlation (Pearson’s correlation = 0.72; Mander’s overlap = 0.88) of Ac-Tub and CDHR3 signals in the cilia region of XZ slice image in C. Scale bars: 10 μm.
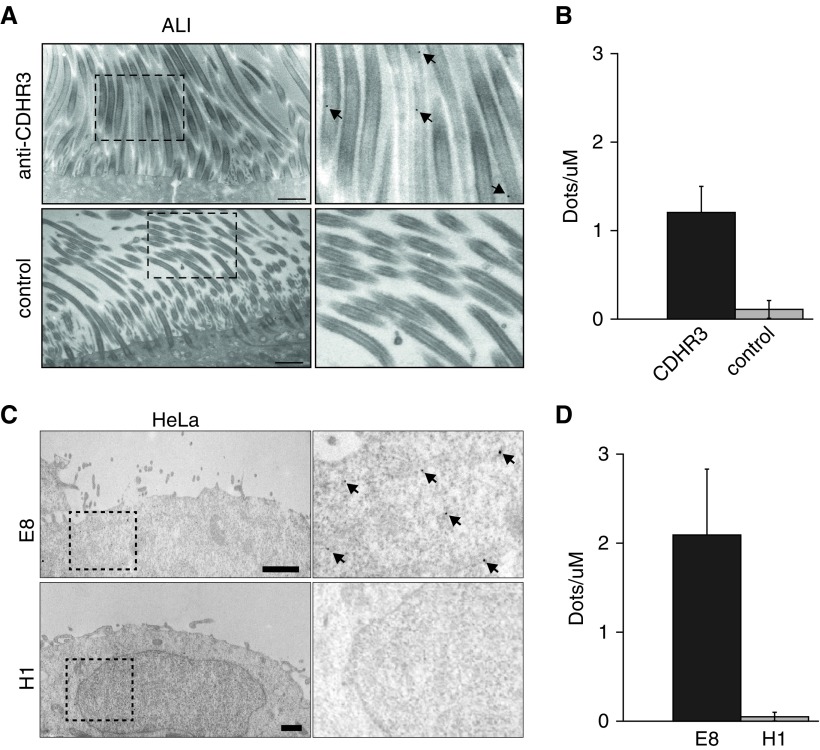
We noted positive fluorescent staining for CDHR3 within the cilia by confocal Z-stack imaging (Figures 2B and E2). The fluorescent staining for acetylated α-tubulin expression overlapped with CDHR3 but not with β-actin (Figure 2C). Furthermore, there was a close correlation between CDHR3 and acetylated α-tubulin expression by image analysis (Figure 2D). Transmission electron microscopy confirmed CDHR3 expression in cilia of airway epithelial cells and also in HeLa-E8 cells transduced with CDHR3 (Figure 3).
Figure 3.
Transmission electron microscopy micrographs of immunolabeled CDHR3 in human BEC-ALI and HeLa cells. (A) BEC-ALI cultures were incubated with immunogold-labeled anti-CDHR3 monoclonal antibody (black dots, top row) or a control antibody (bottom row). Right column shows labeled CDHR3 (arrows) at a higher magnification within inset boxes. (B) Quantification of immunogold labeling in BEC-ALI cultures incubated with anti-CDHR3 or control antibody. (C) HeLa-E8 (transduced with CDHR3) and native HeLa-H1 cells were incubated with immunogold-labeled anti-CDHR3 monoclonal antibody (black dots). Right column shows labeled CDHR3 (arrows) at a higher magnification within inset boxes. (D) Quantification of immunolabeling in HeLa-H8 and HeLa-H1. Scale bars: 1 μm.
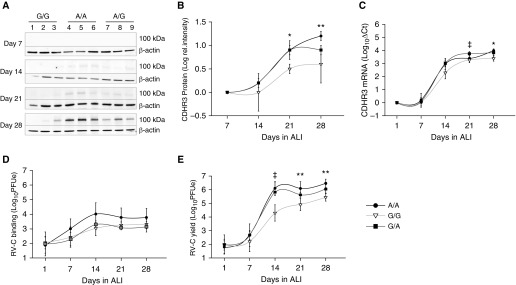
CDHR3 rs6967330 Genotype Affects Protein Expression and RV-C Infection
We next analyzed effects of CDHR3 rs6967330 genotype (A/A, G/G, and G/A) on CDHR3 expression during cellular differentiation at ALI. CDHR3 protein and mRNA were first detected between 7 and 14 days of cellular differentiation (Figures 4A–4C). From Day 14 to Day 28, the G/A and A/A genotypes were associated with higher overall CDHR3 protein expression than the G/G genotype (Figures 4A and 4B). This difference was most significant during cellular differentiation at or before Day 21. There were similar trends, although less robust, for CDHR3 mRNA (Figure 4C). BECs carrying just one CDHR3 A allele displayed similar protein and mRNA expression phenotypes as cells having homozygous A/A alleles (Figures 4A–4C).
Figure 4.
CDHR3 rs6967330 genotype impacts protein expression and rhinovirus C (RV-C) infection in human BEC-ALI cultures. (A) CDHR3 (∼100 kD) protein expression in A/A, A/G, and G/G genotypes of differentiating BEC-ALI cultures on Days 7, 14, 21, and 28. (B) Quantification of CDHR3 protein expression signal intensity from Western blots shown in A. (C) CDHR3 mRNA expression in BEC-ALI cultures with A/A, A/G, and G/G genotypes over a 28-day differentiation time period (n = 3). (D) RV-C15 binding (1 h postinfection) and (E) RV-C15 RNA progeny yield (24 h postinfection) in A/A, A/G, and G/G genotypes of ALI cultures over 28-day differentiation period (n = 3). ‡P < 0.05 between donors that have A/A and G/G genotypes only; *P < 0.05 and **P < 0.01.
To determine the effect of CDHR3 asthma-risk genotype on RV-C infection, we cultured primary BECs selected by their CDHR3 genotype and infected them weekly for 28 days with RV-C15 while the cells were differentiating. Undifferentiated cells at Day 1 had minimal RV-C binding and replication. From Day 14 onward, the A/A genotype was associated with a trend toward higher virus binding than the G/G genotype (Figure 4D). The viral yield 24 hours later was significantly higher (up to 2 logs; P < 0.01) in A/A BECs than in G/G BECs from Day 14 to Day 28 (Figure 4E). BECs carrying just one CDHR3 A allele displayed an RV-C replication phenotype similar to that of cells with the homozygous A/A genotype (Figure 4E).
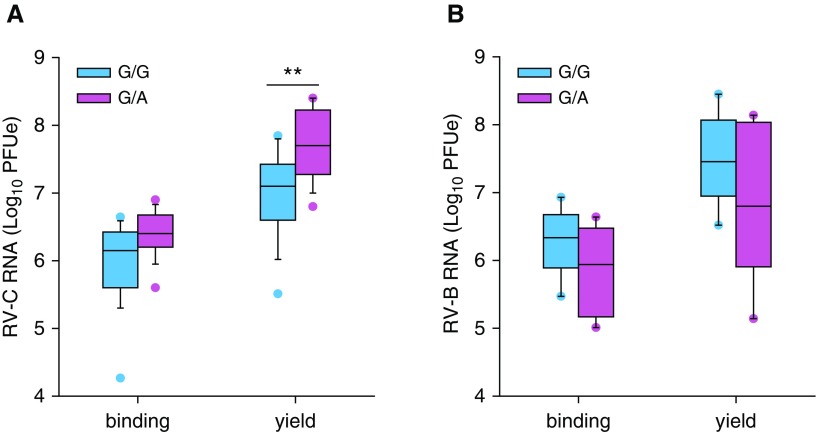
We next analyzed effects of CDHR3 rs6967330 genotype on RV-C infection of NEC cultures obtained from 15- to 17-year-old children of known genotype (16 G/A, 30 G/G). Similarly to BECs, we observed a trend for G/A heterozygote NECs to bind slightly greater amounts of RV-C (Figure 5A). However, G/A genotype NECs gave significantly greater RV-C15 progeny yields than the G/G genotype NECs (median, 7.8 vs. 7.2 log plaque-forming unit equivalents (PFUe); P < 0.01) (Figure 5A). As a control, we infected NEC cultures of known genotype (6 G/A, 6 G/G) with RV-B52, a rhinovirus that uses ICAM-1 (intercellular adhesion molecule 1) as its cell receptor (23). There was no effect of CDHR3 genotype on RV-B52 binding and replication (Figure 5B).
Figure 5.
Rhinovirus (RV) infection of nasal epithelial cells from donors with different CDHR3 rs6967330 genotypes. Nasal epithelial cells from volunteers were differentiated at the air–liquid interface and infected with (A) RV-C15 or (B) RV-B52. Viral RNA at 4 hours postinfection and 24 hours postinfection was used to quantify virus binding and virus RNA progeny yield, respectively. Vertical box plots show mean RV binding and progeny yield in differentiated cells with CDHR3 homozygous G/G genotype (cyan; n = 30 [A], n = 6 [B]) and heterozygous G/A genotype (magenta; n = 16 [A], and n = 6 [B]). **P < 0.01. PFUe = plaque-forming unit equivalents.
To examine the airway epithelium cytokine response to RV-C infection relative to CDHR3 rs6967330 genotype, we measured virus-induced secretions of CXCL10 and IFN-λ (1/3) in RV-C15–infected NEC cultures at 24 hours postinfection. There were trends toward higher CXCL10 and IFN-λ protein secretions in the asthma-risk genotype (G/A) NECs (Figure E3).
CDHR3 Genotype Effects on Epithelial Differentiation and Barrier Function
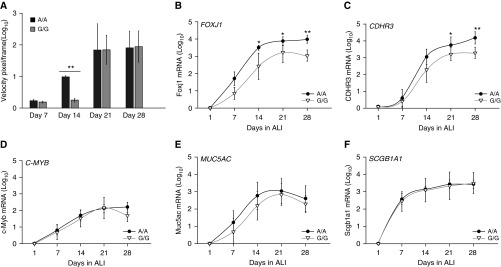
During cellular differentiation, CDHR3 rs6967330 homozygote BECs (A/A) develop cilia more quickly than low-risk homozygotes (G/G) by visual inspection (data not shown). This was confirmed by tracking the motion of fluorescent beads as an indicator of ciliary function (Figure 6A). There was little or no bead motion until Day 14, when the A/A genotype was associated with significantly greater bead velocity (fivefold; P < 0.001) than the G/G genotype. Genotype did not affect the relative bead velocity at later time points (Days 21–28).
Figure 6.
CDHR3 rs6967330 genotype affects ciliogenesis during differentiation of human BEC-ALI cultures. (A) Bar graph represents fluorescent bead tracking velocity (pixels/frame) as a measurement of cilia beat function in differentiating BECs with A/A or G/G genotype at Days 14, 21, and 24 of ALI culture. (B–F) Curves illustrate mRNA expression of FOXJ1 (B), CDHR3 (C), C-MYB (myeloblastosis family transcription factor) (D), MUC5AC (mucin 5AC, oligomeric mucus/gel-forming) (E), and SCGB1A1 (secretoglobin family 1A member 1) (F) in A/A and G/G genotype–differentiating BEC-ALI cultures over 28 days. *P < 0.05 and **P < 0.001.
We next tested whether CDHR3 genotype affected differentiation rates of BECs, as indicated by cellular expression of specific cell-type markers: C-MYB (intermediate progenitor cell), FOXJ1 (ciliated cells), MUC5AC (goblet cells), and SCGB1A1 (club cells). Compared with the rs6967330 G/G genotype, the A/A genotype was associated with 10-fold higher FOXJ1 mRNA expression starting at Day 14 (P < 0.05) (Figure 6B). This suggests a possible regulatory role of CDHR3 upstream of FOXJ1. These experiments also confirmed the findings shown in Figure 4C in that A/A versus G/G BECs expressed 10-fold higher CDHR3 mRNA (P < 0.05) (Figure 6C). Furthermore, FOXJ1 mRNA expression was closely correlated with CDHR3 mRNA expression (R = 0.85; P < 0.0001). In contrast, the CDHR3 genotype did not affect the relative phenotypes of C-MYB, MUC5AC, and SCGB1A1 mRNA expression (Figures 6D–6F). CDHR3 genotype also did not influence the degree of epithelial cell tight-junction formation, as measured by transepithelial resistance (Figure E4).
Discussion
Understanding of what causes severe RV-C infections is partly limited by incomplete information on the native biology of its only known receptor, CDHR3. CDHR3 asthma-risk genotype (rs6967330-A) has been associated with early childhood asthma exacerbations (12), bronchiolitis (24), chronic rhinosinusitis (25), and early-onset adult asthma (26) triggered by RV infections. The goals of this study were to determine CDHR3 localization within primary airway epithelial cells and to test for effects of rs6967330-A on epithelial cell function and susceptibility to RV-C infections. We show that CDHR3 is highly expressed in the apical ciliated airway epithelial cells and associates with the cilia of these cells. The rs6967330-A genotype had several effects. First, epithelial cells carrying at least one A allele had greater overall CDHR3 protein expression and correspondingly greater RV-C binding and replication. In addition, genotype effects on CDHR3 expression were most pronounced during cellular differentiation. We further found that rs6967330-A was associated with faster differentiation, as indicated by the expression of ciliogenesis transcription factor FOXJ1 and more rapid development of functional cilia.
Our findings are consistent with previous reports that CDHR3 is highly expressed in differentiated ciliated cells and not in basal cells (11, 12, 17). The present findings demonstrate that CDHR3 localization is not restricted to the plasma membrane but is also found at lower concentrations in the cytosolic and nuclear subcellular compartments. Subcellular localization in multiple compartments suggests that CDHR3 may have multiple diverse cellular functions. In contrast with E-cadherin, our findings suggest that CDHR3 does not contribute to epithelial structural integrity. Rather, like some protocadherins, CDHR3 may be working to link extracellular contacts to intracellular signaling (27–31). CDHR3 expression in conjunction with cilia in apical cells suggests that RV-C binding occurs in this location. The length of a cilium from the basal body to the tip ranges between 1 and 10 μm in length, depending on the region of the airways. With up to seven of eight healthy airway epithelial cells displaying cilia at approximately 200 cilia per cell, this organization pattern must clearly provide an accessible receptor-docking platform for RV-C (32, 33). In fact, many other respiratory viruses, such as influenza A and B, likewise bind to receptors localized on the cilia of airway epithelia (34, 35). We speculate that RV-C may preferentially use CDHR3 as a cell receptor because of its apical abundance and easy accessibility on the cilia.
Notably, our findings demonstrate that rs6967330-A is associated with higher protein expression levels and correspondingly higher RV-C replication in cultured primary epithelial cells of both the upper and lower respiratory airways. This finding supports previous clinical studies in two separate birth cohorts showing association of rs6967330-A with increased risk of RV-C infections and illnesses in young children (36), as well as with severe bouts of wheezing illnesses in children with asthma (12, 37). Furthermore, rs6967330-A had greater effects on replication than binding, which indicates that the CDHR3-Y529 protein variant may promote both RV-C binding and entry.
Our study demonstrates that rs6967330-A is associated with increased CDHR3 protein expression and to a lesser extent with CDHR3 mRNA. Previous studies have demonstrated that the cleaved cytoplasmic domains of cadherins self-activate expression of their own genes in the nucleus, which decreases as the cells mature and protein–protein interactions increase (27, 38–46). Perhaps the predicted structural stability and cell surface presentation of the CDHR3-Y529 protein variant (47) confers more efficient self-gene activation via the cytoplasmic domain than the CDHR3-C529 variant. Similarly, the stronger genotype-related difference in protein versus mRNA expression could be due to differential mRNA translation or protein stability, although we have not yet studied these aspects of the phenotypes (47).
Our findings show that CDHR3 rs6967330 genotype affects the rate of ciliated airway epithelial cell differentiation. This is in agreement with a previous report of linkage between CDHR3 and FOXJ1 expression in a gene network analysis conducted in sputum cells (48). The faster differentiation of ciliated airway cells associated with the A allele suggests that epithelial recovery after injury may be more rapid in hosts with this genotype. Notably, rs6967330-A represents the ancestral form of the CDHR3 gene. The rs6967330-G allele is not present in the genomes of ancient hominids or in any other mammals, including nonhuman primates (47). Only modern humans encode the virus-resistant genotype, possibly suggesting that balancing selective pressures may have contributed to the current prevalence of the G allele (49).
Among the novel aspects of our study is the use of fresh donor tissue as the source of airway epithelial cultures, providing abundant never-passaged cells from donors with defined CDHR3 genotypes. These were used to show that protein expression and virus susceptibility from bronchial and nasal cells were similar. This at least was true for the virus used in this study, RV-C15, a recombinant virus cloned and sequenced from a clinical isolate. However, a technical limitation of our approach was the limited availability of rs6967330-A homozygote BECs, which are present in only approximately 3% of the population. Another limitation was that the anti-CDHR3 antibodies we had available target a cytoplasmic domain. Anti-CDHR3 antibodies capable of detecting extracellular domains or recombinant CDHR3 are not currently available but would be valuable in subsequent direct tests for cell surface expression of CDHR3 protein variants.
In conclusion, this study presents new information on the biology of CDHR3, including localization and effects of genotype on protein expression, RV-C infections, and epithelial cell function. A better understanding of the native function of CDHR3 and the role of genotype on RV-C infection severity could help to inform new approaches to RV-C prevention and treatment in young children at increased risk for more severe RV-C illnesses.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the University of Wisconsin Optical Imaging Core and the University of Wisconsin Medical School Electron Microscope Facility for technical assistance and Michael D. Evans for assistance in statistical analysis. The authors also thank the children and families of the COAST cohorts for their support and commitment to the study, as well as the COAST research teams for their efforts.
Footnotes
Supported by National Institutes of Health (NIH) grants U19AI104317 and P01 HL070831 (J.E.G.) and by the Clinical and Translational Science Awards (CTSA) program through NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR000427.
Author Contributions: Conception and design: S.B. and J.E.G.; data acquisition: S.B., Y.A.B., R.A.B.-S., I.K., and C.O.; analysis and interpretation: S.B., Y.A.B., S.W.A., D.J.J., R.F.L., C.O., A.C.P., and J.E.G.; drafting of the manuscript: S.B. and J.E.G.; and revision and editing of the manuscript: S.B., Y.A.B., R.A.B.-S., I.K., S.W.A., D.J.J., R.F.L., A.C.P., and J.E.G.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0220OC on March 27, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Heymann PW, Carper HT, Murphy DD, Platts-Mills TA, Patrie J, McLaughlin AP, et al. Viral infections in relation to age, atopy, and season of admission among children hospitalized for wheezing. J Allergy Clin Immunol. 2004;114:239–247. doi: 10.1016/j.jaci.2004.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwane MK, Prill MM, Lu X, Miller EK, Edwards KM, Hall CB, et al. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis. 2011;204:1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 3.Annamalay AA, Jroundi I, Bizzintino J, Khoo SK, Zhang G, Lehmann D, et al. Rhinovirus C is associated with wheezing and rhinovirus A is associated with pneumonia in hospitalized children in Morocco. J Med Virol. 2017;89:582–588. doi: 10.1002/jmv.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soto-Quiros M, Avila L, Platts-Mills TA, Hunt JF, Erdman DD, Carper H, et al. High titers of IgE antibody to dust mite allergen and risk for wheezing among asthmatic children infected with rhinovirus. J Allergy Clin Immunol. 2012;129:1499–1505, e5. doi: 10.1016/j.jaci.2012.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain S, Self WH, Wunderink RG CDC EPIC Study Team. Community-acquired pneumonia requiring hospitalization. N Engl J Med. 2015;373:2382. doi: 10.1056/NEJMc1511751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieninger E, Singer F, Tapparel C, Alves MP, Latzin P, Tan HL, et al. High rhinovirus burden in lower airways of children with cystic fibrosis. Chest. 2013;143:782–790. doi: 10.1378/chest.12-0954. [DOI] [PubMed] [Google Scholar]
- 7.McManus TE, Marley AM, Baxter N, Christie SN, O’Neill HJ, Elborn JS, et al. Respiratory viral infection in exacerbations of COPD. Respir Med. 2008;102:1575–1580. doi: 10.1016/j.rmed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox DW, Bizzintino J, Ferrari G, Khoo SK, Zhang G, Whelan S, et al. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Respir Crit Care Med. 2013;188:1358–1364. doi: 10.1164/rccm.201303-0498OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller EK, Khuri-Bulos N, Williams JV, Shehabi AA, Faouri S, Al Jundi I, et al. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol. 2009;46:85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Møller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 13.Loisel DA, Du G, Ahluwalia TS, Tisler CJ, Evans MD, Myers RA, et al. Genetic associations with viral respiratory illnesses and asthma control in children. Clin Exp Allergy. 2016;46:112–124. doi: 10.1111/cea.12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randell SH. Airway epithelial stem cells and the pathophysiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:718–725. doi: 10.1513/pats.200605-117SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawlins EL, Hogan BL. Epithelial stem cells of the lung: privileged few or opportunities for many? Development. 2006;133:2455–2465. doi: 10.1242/dev.02407. [DOI] [PubMed] [Google Scholar]
- 17.Griggs TF, Bochkov YA, Basnet S, Pasic TR, Brockman-Schneider RA, Palmenberg AC, et al. Rhinovirus C targets ciliated airway epithelial cells. Respir Res. 2017;18:84. doi: 10.1186/s12931-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashraf S, Brockman-Schneider R, Gern JE. Propagation of rhinovirus-C strains in human airway epithelial cells differentiated at air-liquid interface. Methods Mol Biol. 2015;1221:63–70. doi: 10.1007/978-1-4939-1571-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakiela B, Brockman-Schneider R, Amineva S, Lee WM, Gern JE. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am J Respir Cell Mol Biol. 2008;38:517–523. doi: 10.1165/rcmb.2007-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemanske RF., Jr The Childhood Origins of Asthma (COAST) study. Pediatr Allergy Immunol. 2002;13:38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 21.Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, et al. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagome K, Bochkov YA, Ashraf S, Brockman-Schneider RA, Evans MD, Pasic TR, et al. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014;134:332–341. doi: 10.1016/j.jaci.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basnet S, Palmenberg AC, Gern JE.Rhinoviruses and their receptors Chest[online ahead of print] 17 Jan 2019DOI: 10.1016/j.chest.2018.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husby A, Pasanen A, Waage J, Sevelsted A, Hodemaekers H, Janssen R, et al. CDHR3 gene variation and childhood bronchiolitis. J Allergy Clin Immunol. 2017;140:1469–1471, e7. doi: 10.1016/j.jaci.2017.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang EH, Willis AL, McCrary HC, Noutsios GT, Le CH, Chiu AG, et al. Association between the CDHR3 rs6967330 risk allele and chronic rhinosinusitis. J Allergy Clin Immunol. 2017;139:1990–1992, e2. doi: 10.1016/j.jaci.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanazawa J, Masuko H, Yatagai Y, Sakamoto T, Yamada H, Kaneko Y, et al. Genetic association of the functional CDHR3 genotype with early-onset adult asthma in Japanese populations. Allergol Int. 2017;66:563–567. doi: 10.1016/j.alit.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Haas IG, Frank M, Véron N, Kemler R. Presenilin-dependent processing and nuclear function of γ-protocadherins. J Biol Chem. 2005;280:9313–9319. doi: 10.1074/jbc.M412909200. [DOI] [PubMed] [Google Scholar]
- 28.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 29.Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, et al. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci USA. 2016;113:7160–7165. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redies C, Vanhalst K, van Roy F.δ-Protocadherins: unique structures and functions Cell Mol Life Sci 2005622840–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maître JL, Heisenberg CP. Three functions of cadherins in cell adhesion. Curr Biol. 2013;23:R626–R633. doi: 10.1016/j.cub.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomashefski JF, Jr, Farver CF.Anatomy and histology of the lung Tomashefski JF.JrCagle PT, Farver CF, Fraire AE.Dail and Hammar’s pulmonary pathology New York: Springer; 200820–48. [Google Scholar]
- 33.Randell SH, Burns K, Boucher RC. Epithelial cells. In: Barnes PJ, Drazen J, Rennard SI, Thomson NC, editors. Asthma and COPD: basic mechanisms and clinical management. 2nd ed. San Diego, CA: Academic Press; 2009. pp. 201–210. [Google Scholar]
- 34.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carson JL, Collier AM, Hu SS. Acquired ciliary defects in nasal epithelium of children with acute viral upper respiratory infections. N Engl J Med. 1985;312:463–468. doi: 10.1056/NEJM198502213120802. [DOI] [PubMed] [Google Scholar]
- 36.Bønnelykke K, Coleman AT, Evans MD, Thorsen J, Waage J, Vissing NH, et al. Cadherin-related family member 3 genetics and rhinovirus C respiratory illnesses. Am J Respir Crit Care Med. 2018;197:589–594. doi: 10.1164/rccm.201705-1021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stenberg Hammar K, Niespodziana K, van Hage M, Kere J, Valenta R, Hedlin G, et al. Reduced CDHR3 expression in children wheezing with rhinovirus. Pediatr Allergy Immunol. 2018;29:200–206. doi: 10.1111/pai.12858. [DOI] [PubMed] [Google Scholar]
- 38.Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, et al. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. J Biol Chem. 2008;283:12691–12700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hambsch B, Grinevich V, Seeburg PH, Schwarz MK.γ-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression J Biol Chem 200528015888–15897. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Pimplikar SW. The γ-secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc Natl Acad Sci USA. 2001;98:14979–14984. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, et al. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto I, Kawano Y, Murakami D, Sasayama T, Araki N, Miki T, et al. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J Cell Biol. 2001;155:755–762. doi: 10.1083/jcb.200108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tissir F, Qu Y, Montcouquiol M, Zhou L, Komatsu K, Shi D, et al. Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat Neurosci. 2010;13:700–707. doi: 10.1038/nn.2555. [DOI] [PubMed] [Google Scholar]
- 44.Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- 45.Ni CY, Murphy MP, Golde TE, Carpenter G.γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase Science 20012942179–2181. [DOI] [PubMed] [Google Scholar]
- 46.Buchanan SM, Schalm SS, Maniatis T. Proteolytic processing of protocadherin proteins requires endocytosis. Proc Natl Acad Sci USA. 2010;107:17774–17779. doi: 10.1073/pnas.1013105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmenberg AC. Rhinovirus C, asthma, and cell surface expression of virus receptor CDHR3. J Virol. 2017;91:e00072-17. doi: 10.1128/JVI.00072-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones AC, Troy NM, White E, Hollams EM, Gout AM, Ling KM, et al. Persistent activation of interlinked type 2 airway epithelial gene networks in sputum-derived cells from aeroallergen-sensitized symptomatic asthmatics. Sci Rep. 2018;8:1511. doi: 10.1038/s41598-018-19837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Neill MB, Laval G, Teixeira J, Palmenberg A, Pepperell C.Evolutionary genetics of a disease susceptibility locus in CDHR3 [preprint] bioRxiv 2017[accessed 2019 Apr 1]. Available from: https://www.biorxiv.org/content/10.1101/186031v1
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.