Abstract
DNA methylation represents a fundamental epigenetic mark that is associated with transcriptional repression during development, maintenance of homeostasis, and disease. In addition to methylation-sensitive PCR and targeted deep-amplicon bisulfite sequencing to measure DNA methylation at defined genomic loci, numerous unsupervised techniques exist to quantify DNA methylation on a genome-wide scale, including affinity enrichment strategies and methods involving bisulfite conversion. Both affinity-enriched and bisulfite-converted DNA can serve as input material for array hybridization or sequencing using next-generation technologies. In this practical guide to the measurement and analysis of DNA methylation, the goal is to convey basic concepts in DNA methylation biology and explore genome-scale bisulfite sequencing as the current gold standard for assessment of DNA methylation. Bisulfite conversion chemistry and library preparation are discussed in addition to a bioinformatics approach to quality assessment, trimming, alignment, and methylation calling of individual cytosine residues. Bisulfite-converted DNA presents challenges for standard next-generation sequencing library preparation protocols and data-processing pipelines, but these challenges can be met with elegant solutions that leverage the power of high-performance computing systems. Quantification of DNA methylation, data visualization, statistical approaches to compare DNA methylation between sample groups, and examples of integrating DNA methylation data with other –omics data sets are also discussed. The reader is encouraged to use this article as a foundation to pursue advanced topics in DNA methylation measurement and data analysis, particularly the application of bioinformatics and computational biology principles to generate a deeper understanding of mechanisms linking DNA methylation to cellular function.
Keywords: DNA methylation, epigenetics, bisulfite sequencing, next-generation sequencing, bioinformatics
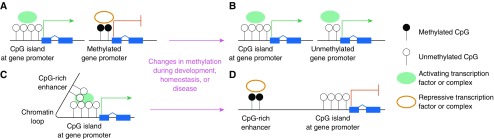
Epigenetic phenomena are heritable changes in cellular phenotype that are not due to mutations in DNA sequence (1, 2). Mechanisms responsible for these phenomena include noncoding RNA species, covalent modifications of histone proteins, and methylation of DNA cytosine residues (3). In animals, DNA methylation predominantly, although not exclusively (4), occurs at cytosine residues that are followed by a guanine residue on their 3′ flank, referred to as cytosine-phospho-guanine (CpG) dinucleotides to distinguish them from CG interstrand base pairing (5). Approximately 4% of cytosines appear in CpG context, and 60–80% of CpG cytosines are methylated depending on the cell type and physiologic or pathologic state. Importantly, CpG residues tend to exhibit a highly nonuniform distribution, clustering together in so-called CpG islands, which are defined as >200-bp regions (typically ∼1 kb) with a GC fraction greater than 0.5 and an observed-to-expected CpG ratio greater than 0.6 (6). These CpG islands localize near gene promoters and other gene-regulatory elements, and tend to be hypomethylated (7) (Figures 1A–1D). In general, CpG methylation is associated with transcriptional repression, and the association is causal in many, but not all, contexts. Moreover, just as the distribution of CpG dinucleotides tends toward nonuniformity, their pattern of methylation is also nonuniform, with methylated residues clustering together in patterns that can vary dramatically between cell types, functional states, and disease conditions.
Figure 1.
The landscape of DNA methylation. (A) Cytosine-phospho-guanine (CpG) islands often occur near gene promoter elements. Hypomethylated CpG islands are associated with active gene transcription, facilitating the binding of activating transcription factors and complexes. In contrast, CpG methylation in gene promoters is associated with transcriptional repression. Methylated CpGs recruit complexes containing methyl-CpG binding domain–containing proteins and other factors that form multiprotein repressive complexes to silence transcription. (B) DNA methylation is a dynamic process, and changes in CpG methylation that occur during development, homeostasis, or disease result in altered gene expression patterns. (C) Many enhancer elements contain CpG residues and islands that facilitate chromatin looping and enhancer–promoter interactions to activate gene expression. (D) Dynamic changes in CpG methylation can alter gene transcription by modifying the three-dimensional chromatin landscape to result in loss of activating enhancer–promoter interactions.
DNA methylation is a chemically stable yet biologically dynamic mark (Figure 2A). A family of DNA methyltransferases (DNMTs) catalyzes the conversion of cytosine to 5-methylcytosine (8). Maintenance of DNA methylation during cell division is a regulated process involving the DNA methyltransferase Dnmt1 and its regulatory adapter protein Uhrf1, among other molecules (9, 10). De novo methylation occurs via Dnmt3a or Dnmt3b activity. DNA demethylation can occur passively during DNA replication or via the catalytic activity of the ten-eleven translocase (TET) family of dioxygenase enzymes (11). Although it was once considered to be a long-lasting epigenetic mark because of its thermodynamic stability, DNA methylation and demethylation are in fact quite dynamic, with rapid kinetics observed in multiple biologic systems (12–14).
Figure 2.
Biochemistry of DNA methylation and methods used to measure DNA methylation in the laboratory (see the data supplement for an expanded figure legend). (A) DNMTs modify the 5-carbon of cytosines in CpG context, a reaction that can be passively reversed during DNA replication or under the activity of a family of TET dioxygenase enzymes. (B) Methyl-DNA immunoprecipitation and MBD methods begin with fragmentation of genomic DNA, followed by enrichment for methylated DNA using anti-5-methylcytosine antibodies or MBD-conjugated beads, respectively. Array hybridization or next-generation sequencing then permits measurement of DNA methylation. (C) The chemical reactions involved in bisulfite treatment convert unmethylated cytosine residues to uracil residues while leaving 5-methylcytosine residues and other residues with 5-carbon modifications unconverted, thus transforming epigenetic information into genetic information. (D) Schematic illustrating how standard PCR chemistry replaces uracils with thymines (now complemented by adenines instead of guanines in the double helix) while cytosines are amplified as cytosines (complemented by guanines in the double helix). (E) Our modified reduced representation bisulfite sequencing method, which is redrawn from Figure 3A in Reference 36. 5 hmC = 5-hydroxymethylcytosine; 5 mC = 5-methylcytosine; α-KG = α-ketoglutarate; BER = base excision repair; CTOB = complement to the original bottom; CTOT = complement to the original top; DNMT = DNA methyltransferase; MBD = methyl-CpG binding domain protein; NaHSO3 = sodium bisulfite; OB = original bottom; OT = original top; SAH = S-adenosylhomocysteine; SAM = S-adenosylmethionine; TDG = thymine DNA glycosylase; TET = ten-eleven translocase.
Numerous developmental, physiologic, and pathologic processes exhibit specific DNA methylation patterns (15). These processes include the development of myriad cell types and tissues, the plasticity of immune cell identity and function, and malignancy. Because of the power inherent in epigenetic control mechanisms, researchers have developed sophisticated tools to investigate DNA methylation in both animal models and human subjects. My goal here is to provide a focused overview of technologies and computational strategies to measure and analyze DNA methylation, highlighting bisulfite sequencing-based methods and pipelines, and using some of my group’s techniques and informatics procedures to illustrate key concepts. This review is not intended to be comprehensive, but rather to serve as a practical guide to explore DNA methylation measurement and data analysis. Excellent reviews and guidelines on next-generation sequencing have been published recently (16–18). The reader is encouraged to review these references for a thorough background on next-generation sequencing, control of batch effects, and other issues relevant to the design and analysis of sequencing-based studies.
Methods of Measuring DNA Methylation
Numerous technologies permit measurement of DNA methylation. Each has its own advantages and disadvantages, and these are reviewed in depth in Reference 19 and summarized in Table 1. Most common methods involve a treatment that distinguishes unmethylated from methylated cytosines, followed by a step that leverages this identification strategy to generate a DNA methylation data set. Although most of this review will focus on methods that use a chemical strategy to distinguish unmethylated from methylated cytosines followed by next-generation sequencing (bisulfite sequencing), it is important to discuss other common techniques, such as affinity enrichment methods. Strategies that exploit methylation-sensitive restriction endonucleases coupled with array hybridization (e.g., comprehensive high-throughput arrays for relative methylation [CHARM] [20]) and next-generation sequencing [Methyl-seq] [21]), as well as single-cell approaches (22) and single-molecule nanopore sequencing (23), may be of interest but are not reviewed further here.
Table 1.
Common Techniques Used to Measure DNA Methylation
| Technique | Advantages | Disadvantages | Notes |
|---|---|---|---|
| Affinity enrichment based (e.g., MeDIP and MBD-based methods) | Low cost relative to bisulfite sequencing. | Low resolution relative to bisulfite sequencing. Bias due to copy number variation, GC content, and CpG density. Higher input requirements than bisulfite conversion–based methods. | Straightforward for laboratories already facile with chromatin immunoprecipitation, sequencing chemistry, and bioinformatics. |
| Bisulfite conversion based (e.g., WGBS, RRBS, and Infinium) | Low input requirements (pg–ng scale). Single-nucleotide resolution. Can provide non-CpG information. | Labor and computation intensive compared with affinity enrichment techniques. Susceptible to bias from incomplete bisulfite conversion and bisulfite PCR artifacts. | Current gold standard. Requires specialized chemistry and computational platforms. Oxidative bisulfite sequencing permits identification of 5-hydroxymethylcytosine (see text for alternatives). |
Definition of abbreviations: CpG = cytosine-phospho-guanine; GC = guanine-cytosine; MBD = methyl-CpG binding domain protein; MeDIP = methyl-DNA immunoprecipitation; RRBS = reduced representation bisulfite sequencing; WGBS = whole-genome bisulfite sequencing.
Affinity Enrichment Strategies
Common affinity enrichment strategies include methyl-DNA immunoprecipitation (MeDIP (24–26)) and methyl-CpG binding domain protein (MBD [27, 28]) methods. Figure 2B illustrates the basic steps involved in these techniques. Both MeDIP- and MBD-based methods have the advantage of being low-cost and straightforward for laboratories that are already skilled in chromatin immunoprecipitation sequencing (ChIP-seq) or related techniques. The disadvantages include relatively low resolution, susceptibility to copy number variation bias, and a bias toward observations of methylated DNA compared with bisulfite-based protocols (19).
Bisulfite-based Methods
Although bisulfite-based methods are more labor and computation intensive than other approaches, many consider them to be the gold standard for measuring DNA methylation because of their single-nucleotide resolution, flexibility across organisms and model systems, and very low input requirements (we have successfully performed bisulfite sequencing on 10–100 pg of genomic DNA). As detailed in Figure 2C, treatment of genomic DNA with sodium bisulfite transforms epigenetic information into genetic information that can then be assessed with the use of strategies detailed below. The fundamental result of the bisulfite conversion reaction is rapid transformation of unmethylated cytosine residues to uracil residues—a reaction from which 5-methylcytosine residues are thermodynamically protected (29, 30). It is critical to achieve very high cytosine-to-uracil conversion rates to satisfy the assumptions of bisulfite-based analysis discussed below; our conversion rates are routinely greater than 99%, as measured by the observed frequency of unmethylated CpGs in an unmethylated λ-bacteriophage genome spiked into every sample. Bisulfite-converted DNA is fragmented because of the harsh chemical treatment. It is also single stranded because the supermajority of non-CpG cytosines are unmethylated and thus are converted to uracils, resulting in loss of CG interstrand base pairing. Importantly, when these bisulfite-converted fragments are subjected to standard PCR amplification, thymines replace uracils (Figure 2D), creating a DNA sequence that can be compared with a reference unconverted sequence to determine whether individual cytosines were methylated or not in the original sample. The PCR amplification can be locus specific, in which case the amplified fragments are cloned and sequenced by standard Sanger-based methods or pyrosequencing, or subjected to targeted deep-amplicon bisulfite sequencing. Methylation-specific PCR assays can also be designed to amplify only converted (i.e., thymine-ated) sequences, thus distinguishing methylated from unmethylated genomic regions of interest. Along with mass spectrometry–based methods such as the EpiTYPER platform (31), these locus-specific techniques can also be used to validate findings obtained from genome-wide methods.
Genome-scale interrogation of methylation status at single-nucleotide resolution can be performed via array hybridization of bisulfite-converted DNA using site-specific, bead-ligated probes that distinguish methylated and unmethylated loci based on their differential sequence after bisulfite treatment. The most recent iteration of the commonly used Illumina Infinium methylation assay uses this approach to measure methylation at up to 850,000 sites (32) and is popular for large-scale human studies. Comprehensive methylation profiling can be performed with whole-genome bisulfite sequencing (WGBS), which represents the current gold standard for DNA methylation assessment (33). In WGBS, strategies such as random PCR priming are used to amplify DNA without respect to any specific loci. Adapter ligation and indexing (barcoding) can occur before or after bisulfite conversion, and these adapter-ligated fragments are then sequenced using next-generation technologies. After the data-processing steps outlined below, a computer algorithm assigns an unmethylated value to sequenced thymines occurring at positions for which the reference genome contains a cytosine. Conversely, a methylated value is assigned to sequenced cytosines occurring at positions for which the reference genome contains a cytosine. For some so-called nondirectional protocols, including ours, all four strands that result from bisulfite treatment—the original top, original bottom, complement to the original top, and complement to the original bottom—are sequenced and provide methylation information (see Figure 2D). Thus, in addition to detecting cytosine-to-thymine conversions, sophisticated algorithms can detect unmethylated residues by recording guanine-to-adenine conversions (i.e., the result of a cytosine-to-thymine conversion on the complementary strand), as illustrated in Figure 2D and discussed below.
WGBS provides the most comprehensive assessment of cytosine methylation, although knowing the methylation status of almost every genomic cytosine in any context (not just CpG) is unnecessary for most studies. Moreover, as cytosines tend to display locally conserved methylation status, it is also not typically necessary to measure the methylation status of every CpG because the methylation status of nearby cytosines can be inferred. Accordingly, our group and many others perform reduced representation bisulfite sequencing (RRBS), which implements an initial unsupervised enrichment step for CpG-rich regions of the genome (34–38). Our modified RRBS (mRRBS) protocol is illustrated in Figure 2E. Although the technical details vary, most RRBS procedures measure 10–20% of all genomic CpGs (upwards of 2–4 million CpGs in mice or humans) while sequencing only 1–2% of the total genome because of the critical digestion and enrichment steps. This approach produces cost savings in terms of sequencing expenses and enables multiplexing of multiple indexed (barcoded) samples into a sequencing run to limit batch effects. For comparison, the NIH Roadmap Epigenomics Project’s guidelines for WGBS (http://www.roadmapepigenomics.org/protocol) suggest a 30× depth at the whole-genome scale and a minimum of 100-bp reads (>800–1,000 million aligned reads in total), whereas we target ∼50 million aligned reads per mRRBS sample. Accordingly, we multiplex four to six samples per run using single-end 75-bp reads on an Illumina NextSeq 500 instrument with a V2 High-Output Reagent Kit (∼400 million reads/sequencing run). This flexibility allows the incorporation of additional biological replicates, which increases the statistical power of bisulfite sequencing studies. Additional replicates and increased sequencing depth improve the detection rate of differentially methylated loci for a given difference in methylation, and implementation guidelines can be found in Reference 39. It is important to note that standard bisulfite-based techniques cannot distinguish 5-methylcytosine from other 5-carbon cytosine modifications, including 5-hydroxymethylation. Techniques such as oxidative bisulfite sequencing (40), Tet-assisted bisulfite sequencing (41), hydroxy-MeDIP (which is similar to MeDIP but uses anti-5-hydroxymethylcytosine antibodies) (42), and selective 5-hydroxymethylcytosine labeling techniques, such as a combined glycosylation restriction analysis (43, 44), are available to measure 5-hydroxymethylation but are beyond the scope of this review.
Building a Bisulfite Sequencing Data-Processing Pipeline
The goal of bioinformatics pipelines is to provide reproducible processing of sequencing data, generating the same output for a given raw data set, pipeline components, and input variables. Many pipelines and pipeline components for processing and analyzing DNA methylation data have been published (45–49). In this section, my objective is to illustrate the general contours of a bisulfite-based processing pipeline by reviewing the steps we use to process our WGBS and mRRBS data. Our pipeline, written for command line, contains modules for demultiplexing, quality assessment, trimming, alignment to reference genomes, and finally methylation extraction (also known as methylation calling) (Figure 3A). Pipelines for processing bisulfite sequencing data are computation, memory, and storage-space intensive; use of a high-performance computing cluster or cloud-based computing system is recommended. Example commands from our pipeline are included in the data supplement.
Figure 3.
Outline and principles of bisulfite sequencing data-processing pipelines (see the data supplement for an expanded figure legend and the text for details and abbreviations). (A) Our pipeline begins with raw base call (*.bcl) files from Illumina sequencers. It then generates multiple intermediate files and reports, and ends with output files for use in visualization and analysis procedures. (B) The Bismark alignment procedure performs a series of in silico residue conversions to align bisulfite-converted sample fragments to a reference genome. (C) The Bismark methylation extraction (calling) procedure. CHH = cytosine followed by any two noncytosine bases; DSS = Dispersion Shrinkage for Sequencing.
Demultiplexing
The standard output of Illumina sequencers consists of base call (*.bcl) files. Particularly when multiple uniquely indexed samples are sequenced together, it is necessary to create quality-annotated sequence files (*.fastq files) for each sample. Unlike the other steps of our pipeline, demultiplexing bisulfite sequencing data requires no special modifications to standard packages such as Illumina’s BCL2FASTQ software (https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html). After running BCL2FASTQ, it is useful to review the demultiplexing quality statistics, including the relative proportion of each indexed library to ensure even representation of each library in the pool.
Quality Assessment
We perform a multidimensional quality assessment of *.fastq files both before and after the trimming procedure outlined below. Our pipeline uses FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to measure multiple aspects of *.fastq file quality. It is valuable to review the total number of reads obtained per sample; again, we aim for a minimum of 50 million aligned reads per sample for mRRBS. The per-base sequence quality graph is also useful to ensure good quality (average quality score >28–30 across read positions). The per-base sequence content metric, which reports the relative frequency of each DNA base across read positions, will invariably fail because of the bisulfite treatment, which disproportionately increases thymines (and adenines in nondirectional libraries) in comparison with other bases. Although it is reported as a failure, the observation of a disproportionate frequency of bases across read positions is a coarse indicator of the success of bisulfite conversion. GC ratios and content measurements are likewise confounded by bisulfite treatment, and sequence duplication levels are often higher than expected in RRBS due to the enrichment for CpG-rich portions of the genome. Post-trimming FastQC reports are useful to determine the number of reads that remain after trimming, ensure removal of adapters, and verify that any variability in per-base sequence content imparted by random priming has been removed (see below). Example pre- and post-trimming FastQC reports are available in the data supplement.
Trimming
Our pipeline uses Trim Galore! (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), a wrapper around Cutadapt (https://github.com/marcelm/cutadapt/) and FastQC, which has useful features for trimming *.fastq files generated from bisulfite sequencing experiments. For example, in contrast to many other trimming packages, Trim Galore! allows us to specify that our mRRBS libraries are generated from MspI-digested fragments. After adapter trimming, this option instructs the software to remove another 2 bp from the 3′ end to avoid an artifact introduced during preparation of MspI-digested libraries. We also specify the nondirectional nature of our libraries (i.e., they must include all four possible strands resulting from bisulfite treatment and amplification, not just the original top and original bottom). In addition to standard adapter trimming and trimming of low-quality bases, we also trim bases from the 5′ end of each read to remove the artifacts added by random priming (note the difference in the per-base sequence content before and after trimming in the example FastQC reports found in the data supplement).
Alignment
The challenge with aligning bisulfite sequencing reads comes from the fact that every sequenced thymine could represent either a genuine genomic thymine or a bisulfite-converted cytosine. Likewise, on the complementary strand, every adenine could represent either a genuine genomic adenine or the complement to a thymine that resulted from bisulfite conversion of an unmethylated cytosine. Therefore, alignment and methylation extraction of bisulfite sequencing data require not only a standard reference genome but also an in silico bisulfite-converted genome, including both cytosine-to-thymine and guanine-to-adenine versions to account for the original and complementary strands, respectively (Figure 3B). We use the popular Bismark package (50) for multiple steps of our pipeline, including genome conversion, which must be completed once before alignment. We selected Bismark, which uses the Bowtie 2 alignment algorithm (51), as our standard aligner because of its integrated features and relative resistance to error across ranges of methylation levels compared with other packages (52).
Our pipeline executes two alignment scripts for each sample, creating aligned, sorted, and indexed *.bam files: one for alignment to the genome corresponding to the experiment (usually mouse or human) and one to the ∼48-kb λ-bacteriophage genome added to every sample before bisulfite conversion. The result is a Bismark alignment report, which summarizes numerous important parameters, including the mapping rate, which is typically lower in bisulfite sequencing than other sequencing technologies due to the complexities of alignment as discussed above, and an estimate of the methylation frequency in each possible cytosine context (CpG, CHG, and CHH, where H is any noncytosine base). The frequency of CpG methylation in the λ-bacteriophage genome gives an estimate of the bisulfite conversion efficiency, as the λ-bacteriophage genome is grown in methylase-negative Escherichia coli, which cannot methylate cytosines in CpG context. Thus, for example, if the observed CpG methylation frequency in the λ-bacteriophage is 0.2%, the cytosine-to-thymine (bisulfite) conversion in the sample is estimated to be 99.8% efficient.
Methylation Extraction
The final step in our processing pipeline also uses Bismark to perform methylation extraction. The principle is straightforward: assign a methylated call when a cytosine is observed at a position showing a cytosine in the reference genome, and assign an unmethylated call when a thymine is observed at a position showing a cytosine in the reference genome (Figure 3C). This process is iterated across the genome, generating a number of outputs, including raw methylation call files for each cytosine context and strand (CpG, CHG, and CHH for the two original and two complementary strands), *.bedgraph tracks for visualization of methylation at each position in standard genome browsers, a methylation call report with associated M-bias plots, and a methylation coverage file. M-bias plots are useful to determine whether any substantial bias exists in methylation calls across reads (see example in the data supplement). The methylation coverage (*.cov) file is the most useful format for analysis, as it lists the methylation percentage in addition to the total number of methylated and unmethylated calls for each CpG positon.
Statistical Hypothesis Testing for Differential DNA Methylation
Quantification of CpG Methylation
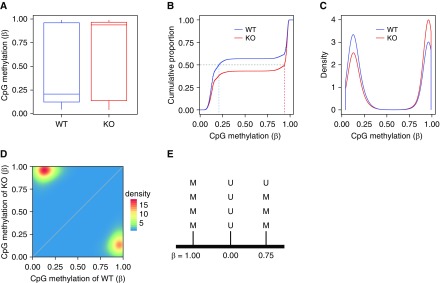
A useful parameter known as β represents the average methylation at unique cytosines measured in the population of cells that make up a sample (Figures 4A–4D). If a cytosine residue is completely unmethylated in the population, then β = 0 (or 0%); if it is completely methylated, then β = 1 (or 100%). Fundamentally, in a single cell on one allele, an individual cytosine is either unmethylated or methylated, prompting the question of how β can range continuously from 0 to 1. There are at least three explanations. First, β is calculated by summing the methylated calls from the methylation extraction procedure divided by the total number of reads at that position. For example, if three methylated calls and one unmethylated call are observed at a position covered by four reads, then β = 0.75 (Figure 4E). Second, incomplete bisulfite conversion will result in intermediate β scores as an artifact of uneven bisulfite conversion. Third, there may be heterogeneity in methylation due to mixtures of cell types or cell states within the population used as a sample. If a sample contains 50% cells that are methylated at a certain cytosine position and 50% cells that are unmethylated at that position, then β will be 0.5 if all other variables are equal. Flow-cytometric enrichment for cell types of interest can reduce this heterogeneity, although fixation protocols can degrade DNA and increase the heterogeneity of DNA methylation (53). It is important to note that although it is the most useful parameter to describe cytosine methylation, β can demonstrate substantial heteroscedasticity (i.e., unequal variability across its range) and does not account for read depth (i.e., observability) at a position (54). A few approaches can mitigate the observability issue, including filtering for positions that have at least a minimal depth (e.g., 3×, 5×, etc.) and using algorithms that account for read depth when comparing cytosines between samples. Heteroscedasticity is a more complicated issue that can be addressed by modeling procedures to apply mathematical transforms to β. We use the bisulphite feature methylation pipeline within the SeqMonk package (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) to perform raw quantification of β scores from *.cov files generated in the final step of our processing pipeline. SeqMonk also serves as a genome browser and contains helpful visualization, data-handling, and statistical procedures for bisulfite sequencing data. Our statistical hypothesis-testing procedure using the Dispersion Shrinkage for Sequencing (DSS) R/Bioconductor package discussed below also generates methylation values based on a modeling approach (55–57). Although it is computationally intensive, we selected the DSS procedure because of its ability to account for both technical (coverage) and biological (methylation level) variability in an algorithmic, unsupervised manner that does not require the setting of an arbitrary read depth cutoff (36, 58). DSS also uses mathematical transforms to limit the heteroscedasticity inherent in raw β scores.
Figure 4.
Quantification of DNA methylation (see the data supplement for an expanded figure legend). Different approaches for displaying data reveal multiple aspects of DNA methylation data. Each graph shows the same data, comparing the CpG methylation profile of regulatory T cells from either chimeric wild-type (WT) or chimeric mitochondrial complex III knockout (KO) mice, as originally reported in Figure 3 from Reference 38; raw data are available in the Gene Expression Omnibus database under accession number GSE120452. The figure shows 17,588 differentially methylated CpGs. (A) A standard Tukey box-and-whisker plot. (B) The empirical cumulative distribution function. The median β score for each group is shown, corresponding to the median displayed in A. (C) Density histogram. (D) Scatter/density plot. (E) Schematic representation of β score calculation, showing how β can be an intermediate value between 0 and 1 based on the average of methylated (M) and unmethylated (U) calls. For the three cytosines shown in the figure, .
Comparing Methylation between Samples
Once the raw or transformed β scores are calculated, statistical hypothesis testing can be performed at single-CpG resolution to identify CpGs that are differentially methylated between groups of samples—so-called differentially methylated cytosines (DMCs). The null hypothesis for these tests is that there is no difference in β between groups at a given position. Many methods are available for statistical hypothesis testing, and each has its own strengths and weaknesses (58). The most straightforward approach involves Fisher’s exact test or the chi-square test; however, these tests do not take into account biological variability or variability due to sequencing depth, and demonstrate skewing of P values toward lower-than-expected values when tested against the null condition. A different approach that accounts for read depth and biological dispersion is based on the commonly used edgeR method for RNA sequencing (RNA-seq) and demonstrates reasonable performance in test settings (59).
Because methylation data are inherently bimodal (i.e., most β scores are near 0 or 1, as explored in Figures 4A–4D), methods that use the binomial or β-binomial distribution tend to exhibit better performance for methylation data than statistical tests that use other distributions. We use the DSS package to generate P values and then a standard Benjamini-Hochberg correction for multiple comparisons to generate false discovery rate (FDR) q-values at well-observed CpG positions as defined by the DSS modeling procedure. A DMC can then be defined as a CpG with an FDR q value less than a desired threshold, typically 0.05 or 0.01. A Δ value can be assigned to obtain a list of DMCs that are different by a defined magnitude in β score—for example, a β difference of 0.10 or 0.25 (i.e., 10% or 25%). The DSS procedure for pairwise comparisons involves a Bayesian hierarchical model that approximates and shrinks CpG site-specific dispersions, which accounts for both biological variability and sequencing depth (55–57). DSS then solves the β-binomial model and tests the null hypothesis of equal mean methylation between two groups using Wald tests. DSS can also perform hypothesis testing using F tests in a general experimental design, which allows comparison of multiple groups, factors, or other variables using a β-binomial regression model. Because of the manner in which regression coefficients are calculated in DSS, the general experimental design procedure does not quantitate β scores, but it does generate a list of well-observed positions. Accordingly, we often use raw β scores at these positions when comparing multiple groups, again avoiding the need to use an arbitrary read depth cutoff that results in loss of information.
A list of DMCs then permits the generation of a set of differentially methylated regions (DMRs). The definition of a DMR is not standardized, and there are no well-validated procedures for generating an unsupervised set of DMRs. For example, the default DSS approach defines a DMR as a region with a minimum length of 50 bp that contains at least three CpGs, 50% of which meet an arbitrary P value threshold. These regions are merged when they occur within 50 bp of one another, creating larger DMRs without an upper bound. Consistent with the arbitrary definition of a DMR, the DSS package documentation states, “It is very difficult to select a natural and rigorous threshold for defining DMRs. We recommend users try different thresholds to obtain satisfactory results.” Our general approach is to define regions of interest based on prior annotations of promoters, enhancers, and other functional genomic elements, and then interrogate these areas for DMCs. In a converse approach, we also test the likelihood of DMCs appearing in a selected set of regions (e.g., promoters, enhancers, etc.) above that expected by chance using hypergeometric testing (35). Machine learning approaches to DMR identification hold promise for the future of unsupervised DNA methylation analysis (60).
Functional Enrichment Analysis and Integration with Other –Omics Data Sets
Functional Enrichment Analysis
Before integration with other –omics data sets, such as those generated by ChIP-seq or RNA-seq, we perform a functional enrichment analysis on a list of DMCs or DMRs using tools such as the Genomic Regions Enrichment of Annotations Tool (61). Originally designed for ChIP-seq data, this tool uses stringent control for false positives to associate cis-regulatory regions with input genomic coordinates, drawing from an extensive set of annotated ontologies. As with any functional enrichment tool, we are cautious about interpreting its output because of the inherently biased nature of functional enrichment due to the human-annotated databases from which these tools draw their biological associations.
Integration with ChIP-Seq Data
DNA methylation does not exist in a vacuum, and the power of DNA methylation sequencing lies in integration with data sets generated by other –omics technologies. Integration with ChIP-seq can be performed by examining DNA methylation at well-observed CpGs across putative enhancers identified by occupancy of histone 3 lysine 4 monomethylation (H3K4 me1) and other DNA-bound proteins. For example, we recently conducted a study in which we deleted TET2 in a breast cancer cell line and queried the effect on histone modifications, DNA methylation, and transcription factor binding (37). Our approach began with clustering of estrogen receptor-α ChIP-seq binding peaks to identify regions with differential binding upon loss of TET2. After confirming TET2 occupancy at these sites in wild-type cells, we used mRRBS and the DSS procedure to determine that loss of TET2 resulted in DNA hypermethylation at these estrogen receptor-α/TET2–regulated regions. This integration analysis allowed us to establish a model in which TET2 coactivates gene expression through DNA demethylation of critical enhancer elements.
Integration with RNA-Seq Data
Perhaps the most common integration occurs between DNA methylation and RNA-seq (transcriptional profiling) data, as transcription represents the proximate readout of epigenetic control mechanisms, including DNA methylation. One straightforward approach is to examine the DNA methylation status of the promoters of differentially expressed genes. We took this approach in a recent study examining the effect of loss of mitochondrial complex III on regulatory T cell–suppressive function (38). Using a Metagene analysis and display, we determined that loss of complex III resulted in hypermethylation of the promoters of downregulated genes, particularly those with an associated CpG island. These data supported our model in which loss of complex III results in an increase in the metabolite L-(S)-2-hydroxyglutarate, which inhibits demethylase enzymes, including the TET family of DNA demethylases (62).
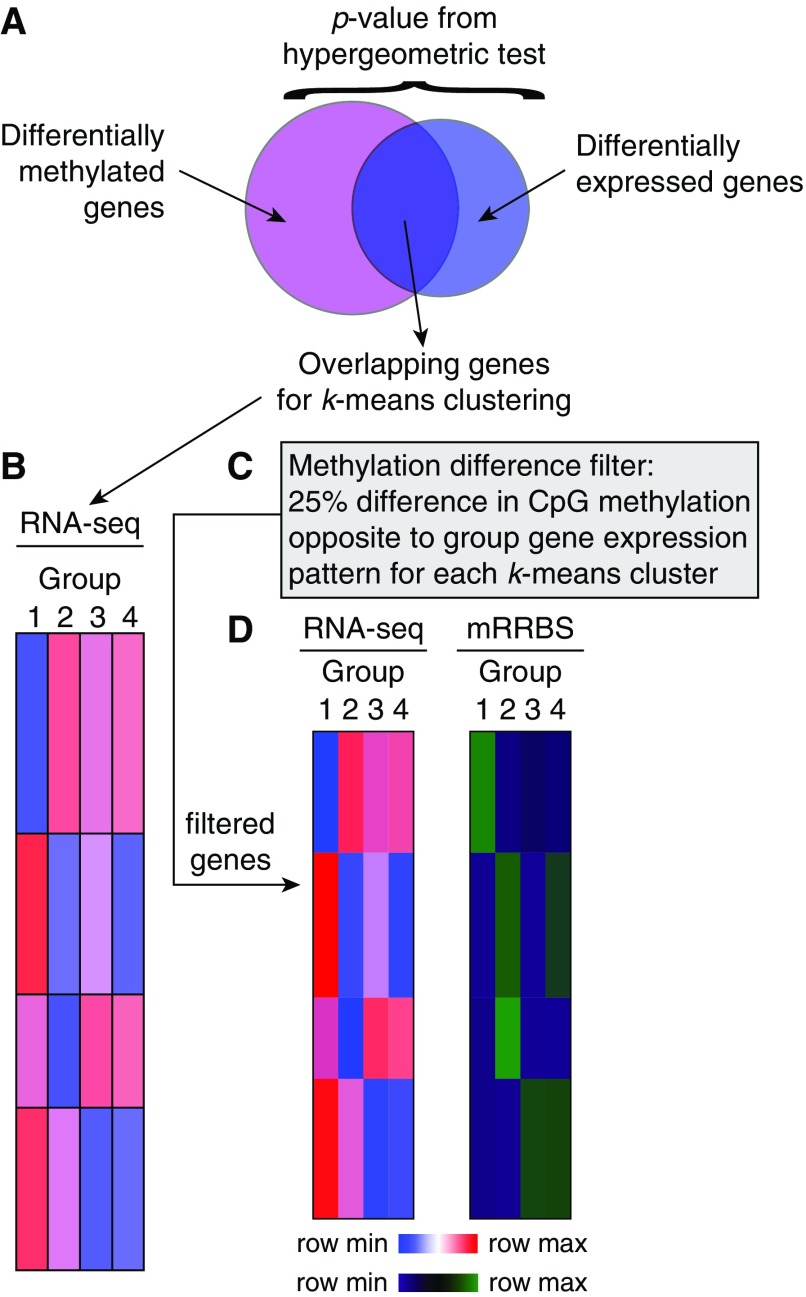
The above approach works well with pairwise comparisons, in which relative hyper- or hypomethylation can be easily defined between two groups. A challenge arises when multiple groups are examined, as was the case in our study of differential DNA methylation and transcription within sorted lung CD4+ T cells during neonatal pneumonia in mice (35). In that study, we examined four groups in a crossed experimental design: neonatal and juvenile mice exposed to either PBS (control) or E. coli bacteria (pneumonia). For the analysis we created a semisupervised DNA methylation difference-filtering algorithm, which is explored in Figures 5A–5D. Conceptually, the algorithm begins by determining the genes that are 1) differentially expressed in the RNA-seq data set (by an ANOVA-like test in edgeR [63]) and 2) differentially methylated in the mRRBS data set (liberally defined as genes with at least one DSS general experimental design-defined DMC within 2 kb of their gene bodies, inclusive). The overlap between these two gene lists is evaluated using a hypergeometric test and visualized using a simple Venn diagram. If the overlap is greater than that expected by chance, the list of overlapping genes is then subjected to k-means clustering using standard procedures (16). Based on the assumption that DNA methylation in promoters is a repressive mark, the algorithm then selects (filters for) CpGs within gene promoters that are hypermethylated within the lower-expressed groups (and therefore hypomethylated in the higher-expressed groups) by an arbitrary difference in the β score, usually 0.1 or 0.25 (i.e., 10% or 25%). This step is repeated for each k-means cluster based on the observed pattern of expression particular to that cluster. The result is a subset of genes passing the methylation filter whose promoters display a methylation pattern that is anticorrelated with gene expression, conforming to the biologic assumption of methylation as a repressive mark. This final list of candidate genes has a high statistical probability of being regulated by DNA methylation. In the neonatal pneumonia study, we used this procedure to determine a list of genes that form a core regulatory signature within lung CD4+ T cells along both developmental (time) and response-to-inflammation (pneumonia) axes. In summary, although only a few standardized, validated approaches are available to perform unsupervised integration of DNA methylation and other –omics data sets (64), creative strategies can be used to combine these data sets in biologically informative ways.
Figure 5.
A DNA methylation difference-filtering algorithm (see the data supplement for an expanded figure legend). (A) The algorithm begins by examining the intersection of two candidate gene lists: one list of genes containing a differentially methylated cytosine within 2 kb of their gene body (inclusive), and one list of differentially expressed genes. A hypergeometric test evaluates the statistical significance of the overlap. (B) Genes that demonstrate both differential methylation and expression are then subjected to k-means clustering based on their gene expression level. (C) The methylation difference filter is then applied to each k-means cluster in turn based on the assumption that DNA methylation and gene transcription are anticorrelated. Genes with no CpGs that meet the filter criteria do not pass the filter; remaining genes pass the filter. (D) Gene expression by RNA sequencing and (unfiltered) promoter methylation by mRRBS are then evaluated for the genes that pass the filter. The diagrams in this figure are schematized versions of Figures 5G and 6 from Reference 35; raw data are available in the Gene Expression Omnibus database under accession number GSE106807. mRRBS = modified reduced representation bisulfite sequencing.
Conclusions and General Recommendations
DNA methylation is a fundamental, dynamic epigenetic mark that is involved in myriad developmental, homeostatic, and pathologic processes. A detailed mechanistic understanding of the biology of DNA methylation as a biomarker or causal substrate requires methods to measure and analyze DNA methylation using low-bias and high-resolution techniques. Although many approaches can be used to accomplish these goals, in this review I have highlighted bisulfite sequencing as the current gold standard, and outlined a biochemical and analytical strategy to measure and analyze DNA methylation in a comprehensive, single-nucleotide-resolution, unsupervised manner. These techniques, particularly the computational methods, may seem daunting for junior and senior investigators alike. Nevertheless, I would encourage interested readers to attempt these techniques in their own laboratory, beginning at a small scale. Numerous online tutorials and resources can be accessed to start exploring DNA methylation data. The SeqMonk platform discussed above is a user-friendly graphical user interface accompanied by a helpful tutorial that can provide an easy entrance into the world of DNA methylation data analysis. Going forward as technologies and computational resources evolve, adherence to sound scientific, mathematical, and computational principles will be important to facilitate discoveries involving DNA methylation in the context of other epigenetic phenomena and the numerous regulatory levels linking genome, environment, and cellular function.
Supplementary Material
Acknowledgments
Acknowledgment
The author thanks Shang-Yang (Sam) Chen and Kishore Anekalla for their work in developing the initial code sets for the DNA methylation processing and analysis pipelines, as well as the members of his laboratory and research group for their helpful comments on the manuscript. This work was supported in part by the computational resources and staff contributions provided by the Genomics Compute Cluster, which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University’s high-performance computing facility, with the purpose to advance research in genomics.
Footnotes
Supported by National Institutes of Health grants K08HL128867 and U19AI135964, and a Parker B. Francis Research Opportunity Award.
B.D.S. has a pending patent application—U.S. Patent App. 15/542,380, “Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation.”
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0150TR on July 2, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Waddington CH. The epigenotype: 1942. Int J Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 6.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 7.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 9.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 10.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 12.Pacis A, Tailleux L, Morin AM, Lambourne J, MacIsaac JL, Yotova V, et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015;25:1801–1811. doi: 10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallner S, Schröder C, Leitão E, Berulava T, Haak C, Beißer D, et al. Epigenetic dynamics of monocyte-to-macrophage differentiation. Epigenetics Chromatin. 2016;9:33. doi: 10.1186/s13072-016-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer BD, Mock JR, Aggarwal NR, Garibaldi BT, Sidhaye VK, Florez MA, et al. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am J Respir Cell Mol Biol. 2015;52:641–652. doi: 10.1165/rcmb.2014-0327OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales-Nebreda L, McLafferty FS, Singer BD. DNA methylation as a transcriptional regulator of the immune system. Transl Res. 2019;204:1–18. doi: 10.1016/j.trsl.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch CM, Chiu SF, Akbarpour M, Bharat A, Ridge KM, Bartom ET, et al. A beginner’s guide to analysis of RNA sequencing data. Am J Respir Cell Mol Biol. 2018;59:145–157. doi: 10.1165/rcmb.2017-0430TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter DR. Thinking BIG rheumatology: how to make functional genomics data work for you. Arthritis Res Ther. 2018;20:29. doi: 10.1186/s13075-017-1504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hersh CP, Adcock IM, Celedón JC, Cho MH, Christiani DC, Himes BE, et al. High-throughput sequencing in respiratory, critical care, and sleep medicine research: an official American Thoracic Society workshop report. Ann Am Thorac Soc. 2019;16:1–16. doi: 10.1513/AnnalsATS.201810-716WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelsey G, Stegle O, Reik W. Single-cell epigenomics: recording the past and predicting the future. Science. 2017;358:69–75. doi: 10.1126/science.aan6826. [DOI] [PubMed] [Google Scholar]
- 23.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 24.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 25.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 26.Hsu HK, Weng YI, Hsu PY, Huang TH, Huang YW. Detection of DNA methylation by MeDIP and MBDCap assays: an overview of techniques. Methods Mol Biol. 2014;1105:61–70. doi: 10.1007/978-1-62703-739-6_5. [DOI] [PubMed] [Google Scholar]
- 27.Jørgensen HF, Adie K, Chaubert P, Bird AP. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aberg KA, Chan RF, Xie L, Shabalin AA, van den Oord EJCG. Methyl-CpG-binding domain sequencing: MBD-seq. Methods Mol Biol. 2018;1708:171–189. doi: 10.1007/978-1-4939-7481-8_10. [DOI] [PubMed] [Google Scholar]
- 29.Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis—a personal account. Proc Jpn Acad, Ser B, Phys Biol Sci. 2008;84:321–330. doi: 10.2183/pjab/84.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang RY, Gehrke CW, Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980;8:4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrath-Morrow SA, Ndeh R, Helmin KA, Chen SY, Anekalla KR, Abdala-Valencia H, et al. DNA methylation regulates the neonatal CD4+ T-cell response to pneumonia in mice. J Biol Chem. 2018;293:11772–11783. doi: 10.1074/jbc.RA118.003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter JM, Helmin KA, Abdala-Valencia H, Wunderink RG, Singer BD. Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight. 2018;3:e123287. doi: 10.1172/jci.insight.123287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Ozark PA, Smith ER, Zhao Z, Marshall SA, Rendleman EJ, et al. TET2 coactivates gene expression through demethylation of enhancers. Sci Adv. 2018;4:eaau6986. doi: 10.1126/sciadv.aau6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martínez-Reyes I, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziller MJ, Hansen KD, Meissner A, Aryee MJ. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods. 2015;12:230–232, 1 p following 232. doi: 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 41.Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nestor CE, Meehan RR. Hydroxymethylated DNA immunoprecipitation (hmeDIP) Methods Mol Biol. 2014;1094:259–267. doi: 10.1007/978-1-62703-706-8_20. [DOI] [PubMed] [Google Scholar]
- 43.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song CX, Yu M, Dai Q, He C. Detection of 5-hydroxymethylcytosine in a combined glycosylation restriction analysis (CGRA) using restriction enzyme Taq(α)I. Bioorg Med Chem Lett. 2011;21:5075–5077. doi: 10.1016/j.bmcl.2011.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adusumalli S, Mohd Omar MF, Soong R, Benoukraf T. Methodological aspects of whole-genome bisulfite sequencing analysis. Brief Bioinform. 2015;16:369–379. doi: 10.1093/bib/bbu016. [DOI] [PubMed] [Google Scholar]
- 46.Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 47.Kangeyan D, Dunford A, Iyer S, Stewart C, Hanna M, Getz G, et al. A (fire)cloud-based DNA methylation data preprocessing and quality control platform. BMC Bioinformatics. 2019;20:160. doi: 10.1186/s12859-019-2750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang KYY, Huang YJ, Chen PY. BS-Seeker3: ultrafast pipeline for bisulfite sequencing. BMC Bioinformatics. 2018;19:111. doi: 10.1186/s12859-018-2120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graña O, López-Fernández H, Fdez-Riverola F, González Pisano D, Glez-Peña D. Bicycle: a bioinformatics pipeline to analyze bisulfite sequencing data. Bioinformatics. 2018;34:1414–1415. doi: 10.1093/bioinformatics/btx778. [DOI] [PubMed] [Google Scholar]
- 50.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JH, Park SJ, Kenta N. An integrative approach for efficient analysis of whole genome bisulfite sequencing data. BMC Genomics. 2015;16(Suppl 12):S14. doi: 10.1186/1471-2164-16-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singer BD, Mock JR, D’Alessio FR, Aggarwal NR, Mandke P, Johnston L, et al. Flow-cytometric method for simultaneous analysis of mouse lung epithelial, endothelial, and hematopoietic lineage cells. Am J Physiol Lung Cell Mol Physiol. 2016;310:L796–L801. doi: 10.1152/ajplung.00334.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng H, Conneely KN, Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42:e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, Xu T, Feng H, Chen L, Li B, Yao B, et al. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 2015;43:e141. doi: 10.1093/nar/gkv715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park Y, Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 2016;32:1446–1453. doi: 10.1093/bioinformatics/btw026. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Baheti S, Sun Z. Statistical method evaluation for differentially methylated CpGs in base resolution next-generation DNA sequencing data. Brief Bioinform. 2018;19:374–386. doi: 10.1093/bib/bbw133. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Pal B, Visvader JE, Smyth GK. Differential methylation analysis of reduced representation bisulfite sequencing experiments using edgeR. F1000 Res. 2017;6:2055. doi: 10.12688/f1000research.13196.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srivastava A, Karpievitch YV, Eichten SR, Borevitz JO, Lister R. HOME: a histogram based machine learning approach for effective identification of differentially methylated regions. BMC Bioinformatics. 2019;20:253. doi: 10.1186/s12859-019-2845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singer BD, Chandel NS. Immunometabolism of pro-repair cells. J Clin Invest. 2019;130:124613. doi: 10.1172/JCI124613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bersanelli M, Mosca E, Remondini D, Giampieri E, Sala C, Castellani G, et al. Methods for the integration of multi-omics data: mathematical aspects. BMC Bioinformatics. 2016;17(Suppl 2):15. doi: 10.1186/s12859-015-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.