Abstract
The actinobacterial acI lineage is among the most successful and ubiquitous freshwater bacterioplankton found on all continents, often representing more than half of all microbial cells in the lacustrine environment and constituting multiple ecotypes. However, stably growing pure cultures of the acI lineage have not been established despite various cultivation efforts based on ecological and genomic studies on the lineage, which is in contrast to the ocean from which abundant microorganisms such as Prochlorococcus, Pelagibacter, and Nitrosopumilus have been isolated. Here, we report the first two pure cultures of the acI lineage successfully maintained by supplementing the growth media with catalase. Catalase was critical for stabilizing the growth of acI strains irrespective of the genomic presence of the catalase-peroxidase (katG) gene. The two strains, representing two novel species, displayed differential phenotypes and distinct preferences for reduced sulfurs and carbohydrates, some of which were difficult to predict based on genomic information. Our results suggest that culture of previously uncultured freshwater bacteria can be facilitated by a simple catalase-supplement method and indicate that genome-based metabolic prediction can be complemented by physiological analyses.
Subject terms: Water microbiology, Microbial ecology, Water microbiology, Microbial ecology
Introduction
The acI lineage of the phylum Actinobacteria is the most abundant and cosmopolitan bacterial group in most freshwater environments. Since the acI lineage was first suggested to denote an abundant monophyletic actinobacterial group exclusively found in freshwater environments [1–3], many studies have demonstrated the ubiquity and prevalence of the acI lineage in diverse freshwater ecosystems on all continents [4–10].
The acI lineage of freshwater bacteria has been studied extensively. Studies employing fluorescence in situ hybridization (FISH) and PCR-based 16S rRNA gene sequence profiling showed that the acI lineage and its subgroups exhibit specific distributions depending on the season, depth, and habitat characteristics and that there are >10 monophyletic tribes belonging to three sublineages (acI-A, -B, and -C) [4, 11–13]. Ecophysiological studies using FISH combined with microautoradiography (MAR-FISH) showed that the acI lineage uptakes diverse substrates including leucine, thymidine, glucose, acetate, N-acetylglucosamine, and amino acid mixtures, but the substrate utilization patterns vary depending on the substrates, habitats, and sublineages [14–17]. Shotgun metagenomics of freshwater samples [18, 19] and PCR-based surveys of enrichment cultures and single-amplified genomes (SAGs) of freshwater origin [20, 21] suggested that members of the acI lineage have genomes with low G+C content and carry genes for actinorhodopsin. Subsequent studies of SAGs [22, 23] and metagenome-assembled genomes (MAGs) [24] have suggested that acI genomes are small in size and enriched with genes for acquiring and utilizing carbohydrates and N-rich organic compounds.
As genomic data of the acI lineage has accumulated through culture-independent studies, cultivation of the acI lineage has become increasingly necessary because pure cultures can enable experimental verification of hypotheses deduced from the genomic data and reveal ecophysiological traits relevant to their survival and niche partitioning in natural habitats [25, 26]. The first report on cultivation of the acI lineage was based on a mixed culture obtained from a freshwater lake, but the proportion of acI cells was <6% [27]. Several enrichment cultures containing acI-B strains with higher proportions (up to >50%) were obtained by dilution culturing from freshwater lakes, and MAGs retrieved from these enrichment cultures suggested that the acI lineage depends on co-occurring microorganisms for the supply of various vitamins, amino acids, and reduced sulfur [28–30]. Recently, Kang et al. [31] and Neuenschwander et al. [32] obtained axenic cultures of 20 acI strains in the acI-A, -B, and -C sublineages by dilution-to-extinction culturing. The complete genomes of these acI strains, obtained by whole-genome-amplification of the initial cultures, showed several characteristics of genome streamlining [33] such as small sizes (1.2–1.6 Mb), low G+C content, and high coding density. Metabolic reconstruction based on the genome sequences predicted auxotrophy of all strains for several vitamin B compounds and reduced sulfurs, as well as tribe-specific auxotrophy for some amino acids.
Although rich genomic data have been obtained from mixed or axenic cultures of the acI lineage after various cultivation efforts, all initial cultures failed to become stably-growing pure cultures [31, 32]. This lack of pure cultures prevents further investigations of the physiology and ecology of this lineage, which dominates freshwater bacterioplankton, in contrast to marine environments where stably growing pure cultures of several ubiquitous prokaryotic groups including Prochlorococcus [34], Pelagibacter [35], and Nitrosopumilus [36] are available.
In this study, we report the first establishment of stably growing pure cultures of the acI lineage. Two acI strains belonging to different tribes were successfully cultured and maintained by supplementing the culture medium with catalase. Using these novel pure cultures, we analyzed the phenotypic characteristics of the acI strains and determined metabolic features that are difficult to predict based only on genome information as well as features conforming to previous genome-guided inferences. Several physiological features differed between the two acI strains, providing insight into the niche separation and spatiotemporal dynamics of diverse acI populations.
Results and discussion
Isolation of acI strains and efforts towards culture maintenance
Strain IMCC25003 of the acI-A1 tribe and strain IMCC26103 of the acI-A4 tribe [3, 4] were isolated from Lake Soyang in Korea by dilution-to-extinction culturing [31]. Strains IMCC25003 and IMCC26103 were most closely related to the recently proposed Candidatus species ‘Ca. Planktophila sulfonica’ MMS-IA-56 (100%) and ‘Ca. Planktophila lacus’ MMS-21-148 (99.9%) [32], respectively, based on 16S rRNA gene phylogenetic analyses (Fig. S1). According to the average nucleotide identity [37] with their closest relatives (74–86%) [32], the two strains each represent novel species of the genus ‘Ca. Planktophila’.
Because the two strains are among the few acI isolates and the ability to maintain stably growing acI strains is prerequisite to studying the phenotypic characteristics of this lineage, we first attempted to resuscitate both strains from glycerol stocks using the same media used for isolation. IMCC26103 did not grow for 21 days after revival from the original frozen glycerol stocks but showed an abrupt increase in cell density (Fig. S2a) because of the growth of contaminating strains (Fig. S2b). IMCC25003 began growing without a lag from the original frozen stocks and reached a cell density of 7.6 × 105 cells mL−1 in 5 days (Fig. S2c; upper panel), but showed an atypical growth curve when the revived culture was transferred into the same medium on day 12 (Fig. S2c; middle panel). When the first-transferred culture was inoculated again on day 15, no growth was detected until ~70 days (Fig. S2c; lower panel). Because maintenance of the growing cultures of acI strains was not successful in the media used for their original isolation, further attempts to obtain stable cultures were made by introducing various modifications to the media composition. These attempts were possible only for IMCC25003, as many glycerol stocks were stored using the initially revived cultures of IMCC25003. Modifications to media composition were based on the putative metabolic features and growth requirements of the acI lineage predicted by approaches such as MAR-FISH and single-cell genomics [15, 23]. Culture media contained various supplements of FAMV medium (Table S1 for media composition) such as carbon substrates, proteinogenic amino acids, peptone, and yeast extract and different basal media including artificial freshwater medium [38] and spent medium of Limnohabitans sp. IMCC26003 (Table S2). However, growth of IMCC25003 was not detected under any culture conditions tested. Because cryo-preserved stocks were used as inocula for all the culture trials described above, the growth of acI cells might have been inhibited by the stress of an abrupt change in temperature. However, the acI strains revived from glycerol stocks grew well in the catalase-treated experiments described below, therefore the likelihood of growth inhibition due to temperature stress is very low.
Successful maintenance of stably growing acI cultures by catalase addition
The failure to establish stable acI bacterial cultures by supplementation with defined or undefined nutrients led us to hypothesize that controlling the putative growth inhibitors may be more crucial than supplying growth promoters. Because the growth of Prochlorococcus and ammonia-oxidizing archaea was promoted when H2O2 was removed [39–41], we examined whether catalase, an enzyme that degrades H2O2, could facilitate the cellular growth of IMCC25003, although the IMCC25003 genome contains katG, which codes for a bifunctional catalase-peroxidase. IMCC25003 showed good growth and reached a maximum cell density of 2.7 × 107 cells mL−1 in catalase (10 U mL−1)-amended medium (FAMV+CM+AA, see “Materials and Methods” for media composition), while no growth was detected in the medium devoid of catalase (Fig. S3a). IMCC26103, which does not contain katG, was also successfully revived from the last-remained glycerol stocks in catalase-amended medium, reaching a maximum cell density of 5.2 × 107 cells mL−1 (Fig. S3b). Because both strains exhibited robust and reproducible growth upon repeated transfer into media supplemented with catalase, catalase was considered as a critical supplement in acI strain cultivation.
Before further experiments, dilution-to-extinction culturing of the successfully revived strains was performed twice to ensure the purity of the cultures, resulting in the establishment of stably growing pure cultures. The purity of the established cultures was verified by FISH (Fig. 1a), transmission electron microscopy (Fig. 1b), and whole genome sequencing followed by read mapping. Both strains produced reddish cell pellets because of the presence of carotenoids and actinorhodopsin in 4-L cultures (Fig. S4a), which were subsequently used for genome sequencing. The genome sequences were identical (1 base pair difference) to those obtained by multiple displacement amplification in our previous study [31], with more even sequencing coverages (Fig. S4b). For both strains, more than 99.9% of sequencing reads were mapped to the complete genomes, demonstrating that the cultures were pure.
Fig. 1.
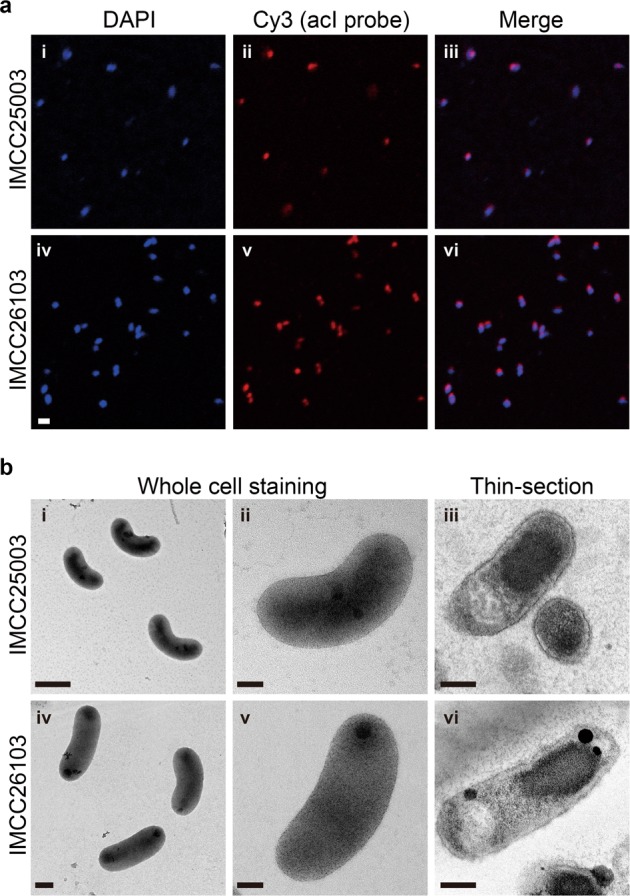
Microscopic examination of the two acI strains for purity evaluation and morphological characterization. a Demonstration of culture purity of strains IMCC25003 (upper; i, ii, and iii) and IMCC26103 (lower; iv, v, and vi) by FISH analysis. The acI cells were dual-stained with DAPI (i and iv) and Cy3-conjugated oligonucleotide probes specific to the acI lineage (AcI-852 and AcI-1214) (ii and v). Merged images are shown in iii and vi. Bar, 1 μm. b Cell morphology of strains IMCC25003 (upper; i, ii, and iii) and IMCC26103 (lower; iv, v, and vi) examined by transmission electron microscopy. The acI cells were stained with uranyl acetate (i, ii, iv, and v) or double stained with uranyl acetate and lead oxide after thin-sectioning (iii and vi). Scale bars; 0.5 μm (i), 0.2 μm (iv), and 0.1 μm (ii, iii, v, and vi)
Characteristics of catalase-dependent growth of acI cultures
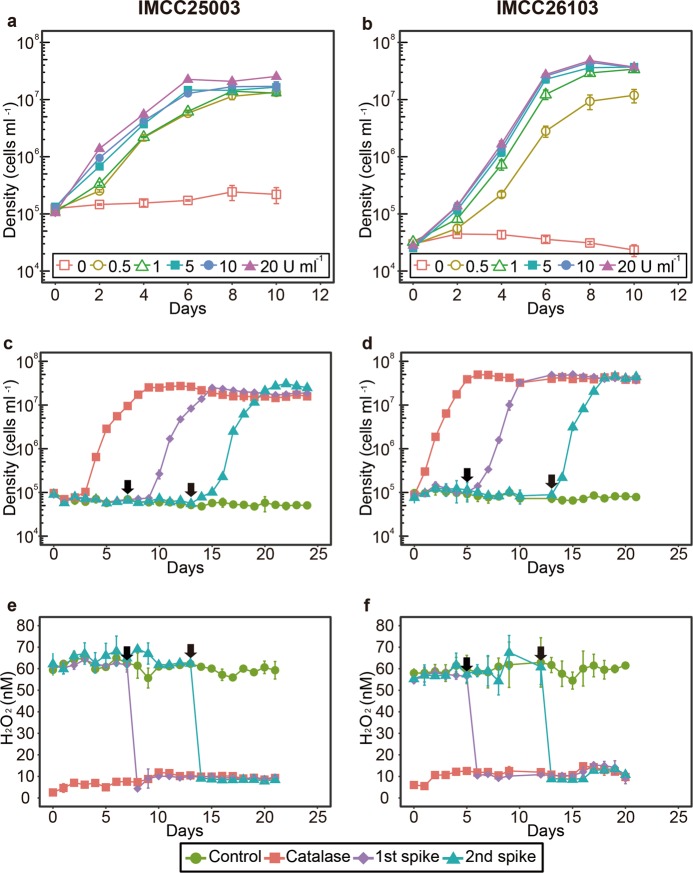
To reconfirm the catalase-dependent growth, the two acI strains were cultivated with various concentrations of catalase (0–20 U mL−1). Growth of the strains was dependent on the presence of catalase, as no growth was detected in the absence of catalase, while 0.5 U mL−1 catalase increased the cell density to ~107 cells mL−1 (Fig. 2a, b). The addition of ferric ion (100 nM; comparable to the concentration obtained by the addition of 10 U mL−1 catalase, a heme-containing enzyme) or autoclaved catalase did not improve the growth of the acI strains, demonstrating that their growth was dependent on the activity of catalase (Fig. S5).
Fig. 2.
Catalase-dependent growth of the two acI strains. a and b The effect of various concentrations (0–20 U mL−1) of catalase on the growth of strains IMCC25003 (a) and IMCC26103 (b). c-f The effect of catalase addition on cell densities (c and d) and H2O2 concentrations (e and f) during cultivation of strains IMCC25003 (c and e) and IMCC26103 (d and f). Catalase (10 U mL−1) was added to the culture media at day 0 (‘Catalase’) or days indicated by black arrows (‘1st spike’ and ‘2nd spike’). No catalase was added to the cultures designated as ‘Control’. All experiments were performed in triplicate using the culture medium FAMV+CM+AA (Table S1). Error bars indicate standard deviation. Note that error bars shorter than the size of the symbols are hidden
Cell densities and H2O2 concentrations were simultaneously monitored in a catalase-spiked growth experiment to determine whether the pivotal role of catalase in promoting growth was mediated by degradation of H2O2. Without catalase, no cellular growth was detected (Fig. 2c, d) and the H2O2 concentration was maintained at the initial level of ~60 nM (Fig. 2e, f). In contrast, when 10 U mL−1 catalase was added at the time of inoculation, the H2O2 concentration rapidly decreased to less than 10 nM and the acI strains began growing, reaching maximum cell densities in 6–9 days. When catalase was spiked at 7 or 13 days for IMCC25003 and 5 or 12 days for IMCC26103, the H2O2 concentration rapidly decreased from ~60 to ~10 nM (Fig. 2e, f). The acI strains, which showed no growth before catalase addition, began growing following the decrease in H2O2 concentration immediately after catalase addition. This suggests that H2O2 removal by catalase may be important for maintaining the growth of the acI strains.
To ascertain whether H2O2 removal was important for the growth of acI strains, they were grown in media containing various concentrations (0–5 mM) of pyruvate, a chemical H2O2 scavenger [42, 43]. Increasing pyruvate concentration led to a decrease in H2O2 concentration, and resulted in an H2O2 concentration of <40 nM at a pyruvate concentration of ≥0.5 mM (Fig. S6e, f). The growth of acI strains was enhanced slightly in the media containing high concentrations (≥0.5 mM) of pyruvate, increasing the maximum cell density of IMCC25003 and IMCC26103 by up to ~2-fold and ~5-fold, respectively (Fig. S6a, b). However, pyruvate addition caused a lower level of growth enhancement than catalase supplementation. Assuming that high pyruvate concentrations inhibit cell growth regardless of H2O2 removal, we tested the growth of the acI strains in media containing catalase and high concentrations of pyruvate. When catalase (10 U mL−1) was added to media containing a high concentration of pyruvate, the growth of acI strains was increased and the strains reached cell densities of ~107 cells mL−1 (Fig. S7), indicating that there was negligible growth inhibition by pyruvate. These results suggest that catalase is essential for the growth of acI strains via mechanisms involving, but not restricted to, H2O2 decomposition. Currently, it is unclear why the acI strains did not grow efficiently under low H2O2 conditions created by pyruvate, yet grew to a density of ~107 cells mL−1 in catalase-supplemented culture media. These observations may be due to differences in the H2O2 degradation rates of catalase- and pyruvate-based reactions or the non-canonical activities of catalase, including its peroxidase activity for the decomposition of small compounds such as methanol and formate [44], which should be further studied.
The euphotic zones of freshwater lakes are known to contain levels of H2O2 that vary spatially and temporally between nanomolar and micromolar concentrations [45, 46]. The H2O2 concentrations at the surface (depth ~1 m) of Lake Soyang where the two acI strains were isolated were approximately 60 nM throughout the year when measured after long-term storage at −20 °C (Fig. S8a). During the preparation of the culture medium, the H2O2 concentration varied from ~120 nM in the freshly collected water sample, to ~600 nM in the autoclaved medium, and ~50 nM in the culture medium containing 50 µM pyruvate (Fig. S8b). We showed that the acI strains could not replicate in the basal culture media without added catalase; however, diverse acI populations are known to prevail in most freshwater lakes at H2O2 concentrations higher than that of the basal culture media used in this study (>50 nM) [47]. The reasons for the discrepancy in the growth of acI bacteria in their natural habitats versus laboratory conditions are unclear, although microenvironments within the water or help from co-occurring organisms [40] might affect the prevalence of the acI lineage in freshwater environments.
Characterization of KatG of the acI lineage
It was questioned whether katG was functional in IMCC25003 during the experiments. Although katG is only found in IMCC25003, the two acI strains showed similar growth responses to the reduction in H2O2 concentration caused by catalase or pyruvate addition, suggesting that katG or its gene product, bifunctional catalase-peroxidase (KatG), did not decompose H2O2 properly in IMCC25003. Therefore, we first examined the expression of katG in IMCC25003 by qPCR. The qPCR results showed that katG was expressed and the relative expression level increased slightly with increasing H2O2 concentrations (Fig. S9). Next, katG of IMCC25003 was cloned and expressed in E. coli and the gene product was purified by affinity chromatography (Fig. S10). The native molecular weight of IMCC25003 KatG was ~165 kDa (Fig. S10d) and molecular weight of the subunit analyzed by SDS-PAGE was ~83 kDa (Fig. S10a), indicating that IMCC25003 KatG formed a homodimer composed of two subunits. In an in-gel enzyme activity assay, IMCC25003 KatG showed both catalase and peroxidase activities but its catalase activity was much lower than that of bovine catalase (KatE), which belongs to the monofunctional catalase group and was supplemented into the culture media in this study (Fig. S11). To more accurately assay the catalase activity of IMCC25003 KatG, decomposition of H2O2 was plotted using varying amounts of enzymes (Fig. S12). All enzymatic parameters including specific activity (U mg−1) and catalytic efficiency (kcat/Km) showed that IMCC25003 KatG displayed lower catalase activities than those of other microbial KatGs reported previously, as well as KatE (Table S3). The low catalase activity of IMCC25003 KatG raises an interesting point. The level of cellular resources required to synthesize low-activity proteins would not differ greatly from that required to synthesize normal-activity proteins. Therefore, it is difficult to propose a mechanism(s) how the possession of a low-activity catalase, not the loss of the catalase gene studied with respect to the Black Queen Hypothesis [48], could benefit the cells, especially for the oligotrophic acI lineage with streamlined genomes.
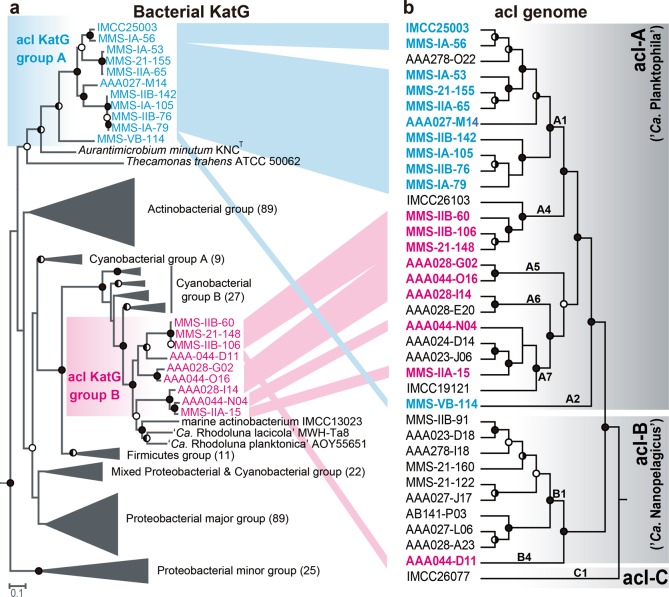
We next investigated the distribution of katG among available acI genomes, as only one (IMCC25003) of four acI strains isolated from Lake Soyang contains katG [31]. katG was detected in 14 of 20 complete acI genomes (Table S4). When SAGs were included, 20 of 35 genomes carried katG. The distribution of katG in the acI lineage was biased toward acI-A compared to acI-B. Of the 25 acI-A genomes, 19 carried katGs, while only one of 10 acI-B genomes contained katG (Table S4); thus, we analyzed the phylogenetic features of acI KatG proteins. Unexpectedly, phylogenetic analyses including diverse bacterial KatG proteins [49] indicated that KatGs of the acI lineage belonged to two distinct groups that are separated widely in the entire KatG tree (Fig. 3a). Further, this KatG phylogeny of the acI lineage was not consistent with a phylogenomic tree of the lineage based on conserved proteins (Fig. 3b). KatGs of the acI-A sublineage were divided into two different KatG groups depending on the tribes. acI-A1 and A2 KatGs formed group A, while acI-A4, A5, A6, and A7 KatGs formed group B together with KatG of acI-B4 (Fig. 3). These results indicate that complicated evolutionary processes occurred for KatG in the acI lineage. Since the KatG of IMCC25003 belonging to group A showed low catalase activity, it would be interesting to determine whether the KatG proteins of group B also exhibit low catalase activity.
Fig. 3.
Comparison between a phylogenetic tree of bacterial KatG proteins and phylogenomic tree of members of the acI lineage. a A maximum-likelihood tree of bacterial KatG protein sequences including those predicted in the acI genomes. The acI KatG proteins are indicated in color (blue and red for acI KatG group A and B, respectively), while other bacterial KatG proteins were grouped following the scheme proposed by Zamocky et al. [49]. The number of proteins included in each KatG group are indicated within parentheses at the end of group names. The KatG sequences of Rhodopirellula baltica and Pirellula staleyi (both from the phylum Planctomycetes) were used as the outgroup. b A phylogenomic tree of members of the acI lineage constructed using concatenated alignment of conserved marker proteins. The acI strains containing katG are indicated in color according to the grouping in the left KatG tree. The SAG AAA027-D23 (belonging to the acSTL-A1 tribe) [30] was set as an outgroup. Only the tree topologies are shown, and branch lengths do not represent phylogenetic distances. For both trees, bootstrap values (from 100 and 250 replicates for the KatG tree and phylogenomic tree, respectively) are shown at the nodes as filled circles (≥90%), half-filled circles (≥70%), and empty circles (≥50%)
Phenotypic characterization of the acI strains
Establishment of stable pure cultures by catalase addition enabled the first phenotypic characterization of the acI lineage, including detailed morphology (Fig. 1b), temperature preference (Fig. S13), fatty acid profile (Table S5), growth requirement, and substrate utilization (Fig. 4), leading to the proposal of two novel species named as ‘Ca. Planktophila rubra’ (ru’bra. L. fem. adj. rubra reddish, pertaining to the reddish color of cells) for IMCC25003 and ‘Ca. Planktophila aquatilis’ (a.qua.ti′lis. L. fem. adj. aquatilis living, growing, or found, in or near water, aquatic) for IMCC26103 (see Supplementary Information Text for detailed description). Actively growing cells of the two strains were curved rods of very small sizes and biovolumes of 0.041 (for IMCC25003) and 0.061 µm3 (for IMCC26103), representing some of the smallest ultramicrobial (<0.1 µm3) cells among known cultivated freshwater bacteria, but 1.6–2.4-fold larger cell volumes than those of other acI bacteria estimated by epifluorescence microscopy [32].
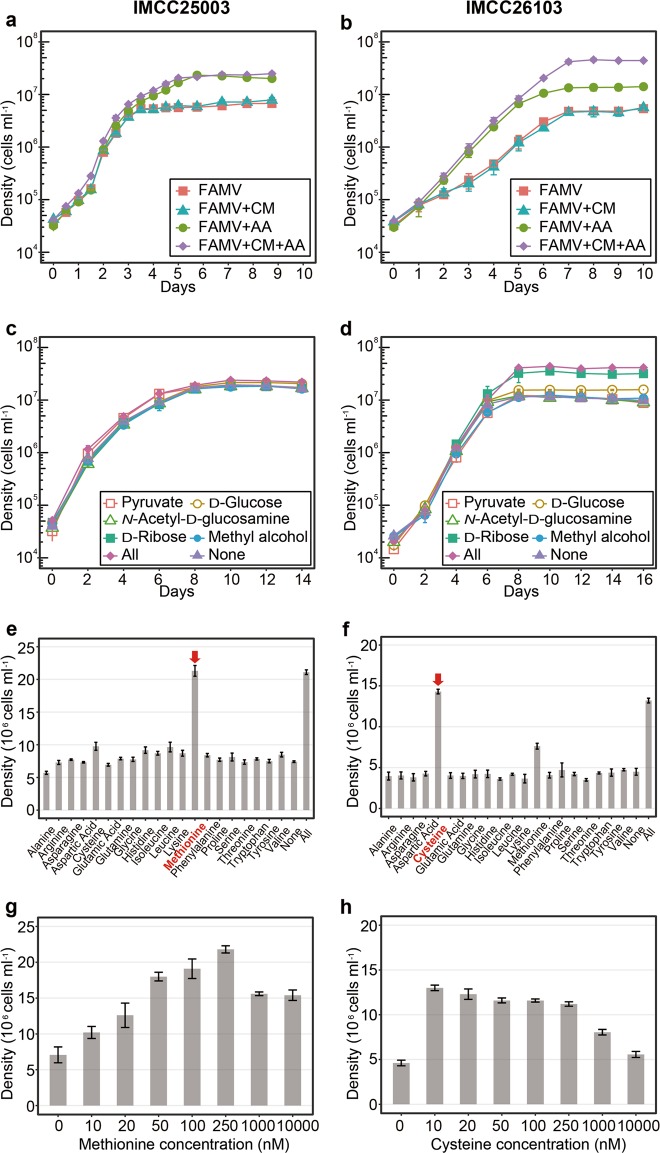
Fig. 4.
Effects of carbon sources and amino acids on the growth of the acI strains IMCC25003 (figures on the left: a, c, e, and g) and IMCC26103 (figures on the right: b, d, f, and h). a and b Effects of amino acid mixture (AA) and/or carbon source mixture (CM). AA and CM were added to the FAMV medium separately or together. Detailed medium composition is presented in Table S1. c and d Effects of individual carbon compound. Each of 5 carbon compounds [5 μM, except for pyruvate (50 μM)] was added to the FAMV+AA medium. All, 5 carbon substrates were added together; None, no carbon source was added. e and f Effects of individual amino acids. Each of 20 proteinogenic amino acids was added at a concentration of 100 nM to the basal medium FAMV. All, 20 amino acids were added together; None, no amino acid was added. g and h Optimal concentrations of l-methionine (for IMCC25003) and l-cysteine (for IMCC26103). Various concentrations (0–10,000 nM) of l-methionine or l-cysteine were added to the basal medium FAMV. All experiments were performed in triplicate. Error bars indicate standard deviation. Note that error bars shorter than the size of the symbols are hidden
The cell densities of IMCC25003 were increased by amino acids but not by carbon mixtures (Fig. 4a). Growth of IMCC26103 was enhanced by both amino acids and carbon mixtures, but the enhancement by carbon mixtures was observed only in the presence of amino acids (Fig. 4b). These results indicate that the growth of both strains was mainly limited by at least one of 20 amino acids and that IMCC26103 (but not IMCC25003) utilized at least one of the carbon sources added. In each carbon compound-amended experiment, no single carbon compound affected the growth of IMCC25003 (Fig. 4c), while ribose and glucose enhanced the growth of IMCC26103 (Fig. 4d). These results were supported by genomic information showing that the IMCC26103 genome contained genes for d-ribose pyranase, ribokinase, and glucose ABC transporter, while these genes were not found in the IMCC25003 genome. In each amino acid-amended test, methionine and cysteine increased the growth of both strains (Fig. 4e, f), supporting the genome-based metabolic prediction that acI strains lack an assimilatory sulfate reduction pathway and thus require reduced sulfur compounds for growth [31]. Interestingly, the two strains showed different preferences for reduced sulfur compounds. IMCC25003 preferred methionine (optimum at 250 nM) over cysteine (Fig. 4e, g), while IMCC26103 preferred cysteine (optimum at 10 nM) over methionine (Fig. 4f, h), which was difficult to predict from genome information. Although a limited number of substrates was tested, these differential preferences of different acI tribes on growth substrates may lead to niche differentiation, underlying the tribe-specific spatiotemporal dynamics of the acI lineage found in several cultivation-independent studies [6, 30, 47, 50].
Conclusion
We successfully maintained actively-growing pure cultures of the acI lineage, the most abundant freshwater bacterioplankton group, by supplementing catalase into the culture media to lower H2O2 concentration, enabling further analysis of the physiological properties that could not be inferred from genome sequences alone. This simple catalase-supplement method may accelerate the cultivation of bacterioplankton with streamlined genomes and thus contribute to studies of the ecological roles of ubiquitous and abundant freshwater oligotrophic bacteria.
Materials and methods
Please see Supplementary Information for experimental details on “Measurement of bacterial cell densities”, “Measurement of katG expression by qPCR”, “Expression, purification, and characterization of KatG from IMCC25003”, and “Phylogenetic analyses based on 16 S rRNA gene, whole genome, and KatG protein”.
Initial isolation of the acI stains, medium preparation, and revival experiments
Freshwater collection, initial isolation, and identification of strains IMCC25003 and IMCC26103 were described in our previous study [31]. Briefly, both strains were initially isolated as liquid cultures by high-throughput culturing method based on dilution-to-extinction in 0.2 μm-filtered and autoclaved freshwater media supplemented with very low levels of carbon compounds, amino acids mixture, and vitamin mixture. Two cryovials each containing 200 μL of initial high-throughput cultures suspended in 10% (v/v) sterile glycerol were stored at −80 °C and used for further revival experiments. As shown in Fig. S2, the axenic culture of IMCC25003 was obtained from the revival experiments, which was further used in the following experiments to establish stably maintained pure cultures of IMCC25003.
Culture media for reviving IMCC25003 were prepared based on natural freshwater (see Table S1 for culture media ingredients). Natural freshwater collected at a depth of 1 m from the Dam station (37°56ʹ50.6″ N, 127°49ʹ7.9″ E) of Lake Soyang in February 2016 was filtered through a 0.2-μm pore-size membrane filter (Supor, Pall Corporation), autoclaved for 1.5 h, and aerated for 3 h. The filtered-autoclaved-aerated medium was supplemented with 10 μM NH4Cl, 10 μM KH2PO4, and trace metal mixture [51], which was designated as FAM (filtered and autoclaved freshwater medium) (Table S1). The FAM medium supplemented with the vitamin mixture [52], designated as FAMV (Table S1), was used as a basal medium that did not contain extra-amended organic compounds. FAMV supplemented with a carbon mixture (CM; 50 μM pyruvate, 5 μM d-glucose, 5 μM N-acetyl-d-glucosamine, 5 μM d-ribose, and 5 μM methyl alcohol) and amino acid mixture (AA; 20 proteinogenic amino acids, 100 nM each) were prepared and used as basal heterotrophic growth media.
As the first attempt to obtain a stable culture of IMCC25003 (Table S2), a 100-μL glycerol stock of IMCC25003 was inoculated into 20 mL of FAMV, FAMV amended with CM (FAMV+CM), FAMV amended with AA (FAMV+AA), and FAMV amended with CM and AA (FAMV+CM+AA). Different concentrations (0.5×, 1×, 5×, and 10×) of CM were added to FAMV+AA and the growth of IMCC25003 was tested. A 0.1-μm-filtered but non-autoclaved freshwater medium (FM) and artificial freshwater medium [38] each supplemented with vitamin mixture, CM, and AA were also tested. In the second attempt, the following organic nutrients were individually supplemented into FAMV+CM+AA: 20 μM acetate, 20 μM oxaloacetate, 20 μM putrescine, 20 μM glycerol, 20 μM xylose, 1 mg L−1 proteose peptone, and 1 mg L−1 yeast extract. In the third attempt, the spent medium of the genus Limnohabitans which is often considered as a co-occurring bacterium with acI bacteria, was added to the growth test medium. Limnohabitans sp. IMCC26003 grew to 3.1 × 106 cells mL−1 in 100 mL of FAMV+CM+AA at 25 °C for 2 weeks. After the Limnohabitans culture was filtered through a 0.2-μm pore-size membrane followed by a 0.1-μm pore-size membrane, 1 mL of the filtrate of the spent medium and 100 μL of glycerol stock of IMCC25003 were inoculated into 20 mL of FAMV+CM+AA. In the fourth attempt, catalase (from bovine liver, C9322, Sigma-Aldrich) stock solution (105 U mL−1 in 10 mM PBS, pH 7.4) was added to FAMV+CM+AA at a final concentration of 10 U mL−1 and 100 μL glycerol stock of IMCC25003 was inoculated. All reviving tests were performed at 18 °C for 5 weeks.
Culture maintenance and confirmation of culture purity
Bacterial cultures of strains IMCC25003 and IMCC26103 revived first from glycerol stocks by catalase amendment were purified twice by dilution-to-extinction culturing using FAMV+CM+AA containing 10 U mL−1 of catalase. Cells were diluted to 5, 1, or 0.1 cells mL−1 and dispensed into 48-well plates (1 mL per each well). After incubation, growth-positive wells (>105 cells mL−1) from the most diluted inoculum (0.1 cells mL11) were used for next round of dilution-to-extinction, resulting in the establishment of stably-growing pure cultures.
To confirm the purity of the established pure cultures by FISH, 5 mL of exponentially grown cells were fixed with 2% paraformaldehyde (in PBS, pH 7.4) and filtered through 0.2-μm polycarbonate filters (Isopore, Millipore). Hybridization was performed at 35 °C for 6 h in hybridization buffer [900 mM NaCl, 20 mM Tris (pH 7.4), 0.01% SDS, 15% formamide] with Cy3-labeled oligonucleotide probes (AcI-852 and AcI-1214; 2 ng μL−1 each) and helper probes (AcI-852-H1 and AcI-852-H2; 2 ng μL−1 each) targeting the acI lineage [53]. After washing the membranes twice with washing buffer [150 mM NaCl, 20 mM Tris (pH 7.4), 6 mM EDTA, and 0.01% SDS] at 55 °C for 10 min followed by DAPI (4,6-diamidino-2-phenylindole) staining, Cy3-positive and DAPI-positive cells were visualized with a confocal laser scanning microscope (LSM 510 META, Carl Zeiss).
For purity confirmation by genome sequencing, the two strains were cultivated in 4 L of FAMV+CM+AA supplemented with 10 U mL−1 of catalase. Cells were harvested by centrifugation at 20,000 × g for 120 min and the cell pellets (Fig. S4a) were used for genomic DNA extraction. Sequencing libraries were constructed using the Nextera library preparation kit (Illumina). Genome sequencing was performed on an Illumina MiSeq platform (2 × 300 bp) by ChunLab, Inc (Seoul, Republic of Korea). Raw sequencing data were assembled using SPAdes 3.9.0 [54], resulting in a circularly closed complete genome for both strains. Comparison of the genome sequences obtained in this study to those obtained by multiple displacement amplification [31] was performed using BLASTn. Calculation of sequencing coverages and estimation of culture purity based on reads mapping were performed as described by Kang et al. [31], using the ‘depth’ and ‘flagstat’ options in samtools.
After purification, both strains were stored as 10% (v/v) glycerol suspensions at −80 °C and working cultures were maintained in FAMV+CM+AA supplemented with 1 U mL−1 of catalase at 25 °C. Throughout this study, the peak shape of the cultures in the flow cytometry, morphology of SYBR Green I-stained cells in epifluorescence microscopy, FISH images obtained using acI-specific probes, and electropherograms from 16S rRNA gene sequencing were routinely examined to evaluate culture purity.
Characterization of catalase-dependent growth properties
The effects of the catalase concentration on the growth of strains IMCC25003 and IMCC26103 were tested in triplicate in FAMV+CM+AA supplemented with 0.5, 1, 5, 10, and 20 U mL−1 of catalase at 25 °C. Bacterial cell densities were measured by flow cytometry every 2 days. The effects of ferric ion (100 nM; supplied by FeCl3) and catalase autoclaved for 2 h were tested in the same media, and cell density was measured daily.
To confirm that catalase enhanced cellular growth and decreased H2O2 concentration, 10 U mL−1 of catalase was added to IMCC25003-inoculated culture medium (FAMV+CM+AA) that had been maintained for 0, 7, and 13 days without catalase treatment. Similarly, for strain IMCC26103, 10 U mL−1 of catalase was spiked at 0, 5, and 12 days. Cultures maintained without catalase were used as controls. The effect of pyruvate concentration was tested using a medium with the same composition as FAMV+CM+AA, without pyruvate. Various final concentrations (0, 50, 200, 500, 1000, and 5000 μM) of pyruvate were then added to this medium, with or without catalase (10 U mL−1), before inoculation. For all cultures, cell densities were monitored every day by flow cytometry. At the same time, the concentration of H2O2 in the culture medium was measured using the modified acridinium ester chemiluminescence method [55] described below. Inocula for these experiments were grown in FAMV+CM+AA supplemented with 1 U mL−1 of catalase, and were transferred to the test media during stationary phase at a dilution ratio of 1:1000.
Determination of hydrogen peroxide concentration
A modified acridinium ester chemiluminescence method [55] was used to determine H2O2 concentrations in the culture medium. To measure H2O2 concentrations at the nanomolar level, acridinium NHS ester (200200; Cayman Chemical) was used as an indicator and the resulting chemiluminescence was estimated using a SpectraMax L microplate Reader (Molecular Devices). A standard curve for H2O2 was prepared in the range of 0.02–2.0 μM by diluting 30% (v/v) of H2O2 solution (H1009; Sigma-Aldrich) with ddH2O. The acridinium ester-H2O2 response signal was integrated for 13 s after sequential injections of 20 μL 2 M Na2CO3 (pH 11.3) followed by 80 μL of 2.2 mg L−1 acridinium ester (in 1 mM phosphate buffer, pH 3) at 300 μL s−1 injection flow speed and 0.1-s interval. All measurements were conducted in triplicate at 20 °C.
For the H2O2 concentration measurements, water samples from Lake Soyang were collected monthly from 2016–2017 at a depth of 1 m and stored at −20 °C in the dark. On the day of measurement, the water samples were thawed at room temperature and used immediately. The water sample used to measure the change in H2O2 concentration during media preparation was collected from the same depth in February 2019. The water sample was processed on the day of collection following the procedure for the preparation of FAMV+CM+AA, with a few modifications; the sample was not aerated and pyruvate was added separately as a final component. Aliquots were collected at each step of media preparation and were frozen at −80 °C until the H2O2 concentration was assayed.
Morphological and physiological characterization of acI strains
Cell morphology was examined by transmission electron microscopy (CM200, Philips) using whole cell staining and thin-sectioning. For whole cell staining, 20-mL cultures were gently filtered using a 0.2-μm pore size polycarbonate membrane on which formvar/carbon-coated copper grids were placed, followed by staining of the grids with 0.5% uranyl acetate. To prepare thin-sectioned samples, cell pellets were harvested by centrifugation of 2-L cultures at 20,000 × g for 120 min. Cell pellets that were primary fixed with 2.0% Karnovsky’s fixative and secondary fixed with 1.0% osmium tetroxide were dehydrated using ethanol series from 35% to 100% and Epon 812 resin was infiltrated. Ultrathin sections (70-nm) were prepared with an ultramicrotome using a diamond knife, placed on formvar/carbon-coated copper grids, and double-stained with 2% uranyl acetate and 0.5% lead oxide.
The temperature range for growth of the two acI strains was monitored at 10–35 °C in FAMV+CM+AA supplemented with 10 U mL−1 of catalase. After the optimum temperature had been determined, all growth experiments were performed in triplicate at 25 °C in different growth media supplemented with 10 U mL−1 of catalase. The growth curves in FAMV, FAMV+CM, FAMV+AA, and FAMV+CM+AA were determined to evaluate the effects of carbon compound mixtures and amino acids mixtures on cellular growth. To identify which amino acids are required for growth, 20 individual amino acids were supplemented in the FAMV medium at 100 nM and cellular growth was monitored. Similarly, five individual carbon sources (50 μM pyruvate, 5 μM d-glucose, 5 μM N-acetyl-d-glucosamine, 5 μM d-ribose, and 5 μM methyl alcohol) were supplemented into FAMV+AA and the growth curves were determined. To determine the optimal concentration of reduced sulfur compounds, growth of the acI strains was measured at 0–10 μM of methionine (for IMCC25003) and cysteine (for IMCC26103).
Cellular fatty acid profiles were determined using cells harvested from 2-L cultures grown in FAMV+CM+AA supplemented with 10 U mL−1 of catalase. Fatty acid methyl esters were extracted from cell pellets using the standard protocol for the Sherlock Microbial Identification System (MIDI, Inc.) and analyzed by gas chromatography (Agilent 7890 GC) based on MIDI version 6.1 with the TSBA6 database [56].
Supplementary information
Acknowledgements
The authors are grateful to Dr. Sung-Keun Rhee and Dr. Jong-Geol Kim for their help in measuring H2O2 concentration. This study was supported by the Mid-Career Research Program (to J-CC, No. NRF-2016R1A2B2015142) and Science Research Center grant (to J-CC, No. NRF-2018R1A5A1025077) through the National Research Foundation (NRF) funded by the Ministry of Sciences and ICT, and by the Basic Science Research Program funded by the Ministry of Education, Republic of Korea (to IK, No. NRF-2016R1A6A3A11934789; to J-HS, No. NRF-2016R1A6A3A11935361).
Data availability
The complete genome sequences of strains IMCC25003 and IMCC26103 obtained from cell pellets using the Illumina MiSeq platform have been deposited in GenBank with the accession numbers CP029557 for IMCC25003 and CP029558 for IMCC26103.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Suhyun Kim, Ilnam Kang
Supplementary information
The online version of this article (10.1038/s41396-019-0432-x) contains supplementary material, which is available to authorized users.
References
- 1.Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, et al. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol. 2000;66:5053–65. doi: 10.1128/AEM.66.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwart G, Crump BC, Agterveld MPKv, Hagen F, Han SK. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–55. doi: 10.3354/ame028141. [DOI] [Google Scholar]
- 3.Warnecke F, Amann R, Pernthaler J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol. 2004;6:242–53. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- 4.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parfenova VV, Gladkikh AS, Belykh OI. Comparative analysis of biodiversity in the planktonic and biofilm bacterial communities in Lake Baikal. Microbiology. 2013;82:91–101. doi: 10.1134/S0026261713010128. [DOI] [Google Scholar]
- 6.Newton RJ, McLellan SL. A unique assemblage of cosmopolitan freshwater bacteria and higher community diversity differentiate an urbanized estuary from oligotrophic Lake Michigan. Front Microbiol. 2015;6:1028. [DOI] [PMC free article] [PubMed]
- 7.Li J, Zhang J, Liu L, Fan Y, Li L, Yang Y, et al. Annual periodicity in planktonic bacterial and archaeal community composition of eutrophic Lake Taihu. Sci Rep. 2015;5:15488. doi: 10.1038/srep15488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henson Michael W., Hanssen Jordan, Spooner Greg, Fleming Patrick, Pukonen Markus, Stahr Frederick, Thrash J. Cameron. Nutrient dynamics and stream order influence microbial community patterns along a 2914 kilometer transect of the Mississippi River. Limnology and Oceanography. 2018;63(5):1837–1855. doi: 10.1002/lno.10811. [DOI] [Google Scholar]
- 9.Satinsky B, Fortunato C, Doherty M, Smith C, Sharma S, Ward N, et al. Metagenomic and metatranscriptomic inventories of the lower Amazon River, May 2011. Microbiome. 2015;3:39. doi: 10.1186/s40168-015-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savio D, Sinclair L, Ijaz UZ, Parajka J, Reischer GH, Stadler P, et al. Bacterial diversity along a 2600 km river continuum. Environ Microbiol. 2015;17:4994–5007. doi: 10.1111/1462-2920.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allgaier M, Grossart HP. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol. 2006;72:3489–97. doi: 10.1128/AEM.72.5.3489-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allgaier M, Brückner S, Jaspers E, Grossart HP. Intra- and inter-lake variability of free-living and particle-associated Actinobacteria communities. Environ Microbiol. 2007;9:2728–41. doi: 10.1111/j.1462-2920.2007.01385.x. [DOI] [PubMed] [Google Scholar]
- 13.Newton RJ, Jones SE, Helmus MR, McMahon KD. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol. 2007;73:7169–76. doi: 10.1128/AEM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert EM, Salcher MM, Posch T, Eugster B, Pernthaler J. Rapid successions affect microbial n-acetyl-glucosamine uptake patterns during a lacustrine spring phytoplankton bloom. Environ Microbiol. 2012;14:794–806. doi: 10.1111/j.1462-2920.2011.02639.x. [DOI] [PubMed] [Google Scholar]
- 15.Salcher MM, Posch T, Pernthaler J. In situ substrate preferences of abundant bacterioplankton populations in a prealpine freshwater lake. ISME J. 2013;7:896–907. doi: 10.1038/ismej.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez MT, Hörtnagl P, Sommaruga R. Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ Microbiol. 2010;12:74–82. doi: 10.1111/j.1462-2920.2009.02043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck U, Grossart HP, Amann R, Pernthaler J. Substrate incorporation patterns of bacterioplankton populations in stratified and mixed waters of a humic lake. Environ Microbiol. 2009;11:1854–65. doi: 10.1111/j.1462-2920.2009.01910.x. [DOI] [PubMed] [Google Scholar]
- 18.Ghai R, Rodriguez-Valera F, McMahon KD, Toyama D, Rinke R, Cristina Souza de Oliveira T, et al. Metagenomics of the water column in the rristine upper course of the Amazon River. PLoS ONE. 2011;6:e23785. doi: 10.1371/journal.pone.0023785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghai R, McMahon KD, Rodriguez-Valera F. Breaking a paradigm: cosmopolitan and abundant freshwater actinobacteria are low GC. Environ Microbiol Rep. 2012;4:29–35. doi: 10.1111/j.1758-2229.2011.00274.x. [DOI] [PubMed] [Google Scholar]
- 20.Sharma AK, Sommerfeld K, Bullerjahn GS, Matteson AR, Wilhelm SW, Jezbera J, et al. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J. 2009;3:726–37. doi: 10.1038/ismej.2009.13. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Garcia Manuel, Swan Brandon K, Poulton Nicole J, Gomez Monica Lluesma, Masland Dashiell, Sieracki Michael E, Stepanauskas Ramunas. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. The ISME Journal. 2011;6(1):113–123. doi: 10.1038/ismej.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, Stepanauskas R, et al. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J. 2013;7:137–47. doi: 10.1038/ismej.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghylin TW, Garcia SL, Moya F, Oyserman BO, Schwientek P, Forest KT, et al. Comparative single-cell genomics reveals potential ecological niches for the freshwater acl Actinobacteria lineage. ISME J. 2014;8:2503–16. doi: 10.1038/ismej.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol Ecol. 2014;23:6073–90. doi: 10.1111/mec.12985. [DOI] [PubMed] [Google Scholar]
- 25.Salcher MM, Šimek K. Isolation and cultivation of planktonic freshwater microbes is essential for a comprehensive understanding of their ecology. Aquat Microb Ecol. 2016;77:183–96. doi: 10.3354/ame01796. [DOI] [Google Scholar]
- 26.Rappé MS. Stabilizing the foundation of the house that ‘omics builds: the evolving value of cultured isolates to marine microbiology. Curr Opin Microbiol. 2013;16:618–24. doi: 10.1016/j.mib.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Jezbera J, Sharma AK, Brandt U, Doolittle WF, Hahn MW. ‘Candidatus Planktophila limnetica’, an actinobacterium representing one of the most numerically important taxa in freshwater bacterioplankton. Int J Syst Evol Microbiol. 2009;59:2864–9. doi: 10.1099/ijs.0.010199-0. [DOI] [PubMed] [Google Scholar]
- 28.Garcia SL, McMahon KD, Grossart HP, Warnecke F. Successful enrichment of the ubiquitous freshwater acI Actinobacteria. Environ Microbiol Rep. 2014;6:21–7. doi: 10.1111/1758-2229.12104. [DOI] [PubMed] [Google Scholar]
- 29.Garcia SL, Buck M, McMahon KD, Grossart HP, Eiler A, Warnecke F. Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol Ecol. 2015;24:4449–59. doi: 10.1111/mec.13319. [DOI] [PubMed] [Google Scholar]
- 30.Garcia SL, Stevens SLR, Crary B, Martinez-Garcia M, Stepanauskas R, Woyke T, et al. Contrasting patterns of genome-level diversity across distinct co-occurring bacterial populations. ISME J. 2018;12:742–55. doi: 10.1038/s41396-017-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang I, Kim S, Islam MR, Cho JC. The first complete genome sequences of the acI lineage, the most abundant freshwater Actinobacteria, obtained by whole-genome-amplification of dilution-to-extinction cultures. Sci Rep. 2017;7:42252. doi: 10.1038/srep42252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2018;12:185–98. doi: 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–65. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–7. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 35.Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–3. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 36.Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 37.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–72. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Kang I, Cho JC. Genomic analysis of a freshwater actinobacterium, “Candidatus Limnosphaera aquatica” strain IMCC26207, isolated from Lake Soyang. J Microbiol Biotechnol. 2017;27:825–33. doi: 10.4014/jmb.1701.01047. [DOI] [PubMed] [Google Scholar]
- 39.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “Helper” heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530–4. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One. 2011;6:e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JG, Park SJ, Damste JSS, Schouten S, Rijpstra WIC, Jung MY, et al. Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc Natl Acad Sci USA. 2016;113:7888–93. doi: 10.1073/pnas.1605501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desagher S, Glowinski J, Premont J. Pyruvate protects neurons against hydrogen peroxide-induced toxicity. J Neurosci. 1997;17:9060–7. doi: 10.1523/JNEUROSCI.17-23-09060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varma SD, Hegde KR. Lens thiol depletion by peroxynitrite. Protective effect of pyruvate. Mol Cell Biochem. 2007;298:199–204. doi: 10.1007/s11010-006-9352-y. [DOI] [PubMed] [Google Scholar]
- 44.Glorieux C, Calderon PB. Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol Chem. 2017;398:1095–108. doi: 10.1515/hsz-2017-0131. [DOI] [PubMed] [Google Scholar]
- 45.Cory RM, Davis TW, Dick GJ, Johengen T, Denef VJ, Berry MA, et al. Seasonal Dynamics in Dissolved Organic Matter, Hydrogen Peroxide, and Cyanobacterial Blooms in Lake Erie. Front Mar Sci. 2016;3:54. doi: 10.3389/fmars.2016.00054. [DOI] [Google Scholar]
- 46.Rusak SA, Richard LE, Peake BM, Cooper WJ, Bodeker GE. The influence of solar radiation on hydrogen peroxide concentrations in freshwater. Mar Freshw Res. 2010;61:1147–53. doi: 10.1071/MF10001. [DOI] [Google Scholar]
- 47.Berry MA, Davis TW, Cory RM, Duhaime MB, Johengen TH, Kling GW, et al. Cyanobacterial harmful algal blooms are a biological disturbance to Western Lake Erie bacterial communities. Environ Microbiol. 2017;19:1149–62. doi: 10.1111/1462-2920.13640. [DOI] [PubMed] [Google Scholar]
- 48.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio. 2012;3:e00036–12. [DOI] [PMC free article] [PubMed]
- 49.Zamocky M, Gasselhuber B, Furtmuller PG, Obinger C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch Biochem Biophys. 2012;525:131–44. doi: 10.1016/j.abb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okazaki Y, Nakano SI. Vertical partitioning of freshwater bacterioplankton community in a deep mesotrophic lake with a fully oxygenated hypolimnion (Lake Biwa, Japan) Environ Microbiol Rep. 2016;8:789–788. doi: 10.1111/1758-2229.12439. [DOI] [PubMed] [Google Scholar]
- 51.Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 2013;7:592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis HC, Guillard RRL. Relative value of ten genera of micro-organisms as food for oyster and clam larvae. Fish Bull. 1958;58:293–304. [Google Scholar]
- 53.Warnecke F, Sommaruga R, Sekar R, Hofer JS, Pernthaler J. Abundances, identity, and growth state of actinobacteria in mountain lakes of different UV transparency. Appl Environ Microbiol. 2005;71:5551–9. doi: 10.1128/AEM.71.9.5551-5559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King DW, Cooper WJ, Rusak SA, Peake BM, Kiddle JJ, O’Sullivan DW, et al. Flow injection analysis of H2O2 in natural waters using acridinium ester chemiluminescence: Method development and optimization using a kinetic model. Anal Chem. 2007;79:4169–76. doi: 10.1021/ac062228w. [DOI] [PubMed] [Google Scholar]
- 56.Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Newark, DE: MIDI Inc 1990.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequences of strains IMCC25003 and IMCC26103 obtained from cell pellets using the Illumina MiSeq platform have been deposited in GenBank with the accession numbers CP029557 for IMCC25003 and CP029558 for IMCC26103.