Abstract
Microbial communities in arctic–alpine soils show biogeographic patterns related to elevation, but the effect of fine-scale heterogeneity and possibly related temperature and soil moisture regimes remains unclear. We collected soil samples from different micro-topographic positions and elevational levels in two mountain regions of the Scandes, Central Norway. Microbial community composition was characterized by 16S rRNA gene amplicon sequencing and was dependent on micro-topography and elevation. Underlying environmental drivers were identified by integration of microbial community data with a comprehensive set of site-specific long-term recorded temperature and soil moisture data. Partial least square regression analysis allowed the description of ecological response patterns and the identification of the important environmental drivers for each taxonomic group. This demonstrated for the first time that taxa responding to elevation were indeed most strongly defined by temperature, rather than by other environmental factors. Micro-topography affected taxa were primarily controlled by temperature and soil moisture. In general, 5-year datasets had higher explanatory power than 1-year datasets, indicating that the microbial community composition is dependent on long-term developments of near-ground temperature and soil moisture regimes and possesses a certain resilience, which is in agreement with an often observed delayed response in global warming studies in arctic–alpine regions.
Subject terms: Microbial ecology, Biogeography
Introduction
Arctic ecosystems are highly sensitive to elevated temperatures and are therefore often considered as models for global warming response scenarios [1]. The geographic location and elevation of arctic–alpine systems crucially affects environmental variables, such as temperature, soil moisture, solar radiation, and vapor pressure, resulting in spatial alterations concerning the occurrence of plants and animals [2, 3]. An effect of elevation was observed in a couple of microbial studies [4–6], though with different effects. Some studies reported that the community structure, but not alpha diversity, is influenced by elevation [5, 7], while others observed differences also in alpha diversity [8–11]. In these studies, the observed differences were best explained by elevation-related changes in soils, e.g. by altered carbon and nitrogen contents or pH values, but usually not by temperature. Likewise, a global-scale study reported that microbial community composition (MCC) is largely driven by edaphic factors rather than temperature or other variables, which typically predict plant and animal diversity [12]. Only a sediment-mesocosm study along an elevational transect revealed good correlation between MCC and water temperature so far [13]. Thus, the minor relevance of temperature for soil microbial communities in mountain ecosystems remains unexpected.
The apparently minor role of temperature may result from the fact that the seemingly simple task of selecting physiologically and ecologically meaningful measures of temperature has been proven difficult [14]. Mean annual air temperatures derived from synoptic weather stations in combination with grid-based interpolations are often used as explanatory variables, though they are known to be decoupled from the near-surface thermal regime [15, 16]. Moreover, it is especially the multifaceted nature of thermal conditions (including extremes, accumulated heat sums, length of growing season, etc.) that affects, at least for plants, different stages in life cycle and phenology [17]. To approach this multifaceted nature of thermal conditions, the concept of heat units [18] with growing degree days or growing degree hours is often used in plant distribution models [19]. All of these concepts use a certain pre-defined threshold temperature that is quantified by accumulating the respective number of days or hours above this chosen threshold. It can be assumed that different species are determined by different thermal thresholds. Consequently, an inductive approach that screens the entire range of occurring environmental conditions for potential species-specific thresholds might also be useful in microbial ecology to explain variation in MCC and to assess a possible impact of temperature in mountain ecosystems more specifically.
Arctic–alpine ecosystems are further characterized by high micro-topographic heterogeneity, which affects micro-environmental parameters [20], and therefore likely controls microbial activity and MCC. Micro-topographic features provide an explanation for micro-topographic patterns of arctic–alpine plants and arthropods [2, 20, 21]. They influence near-ground air and soil temperatures [15, 16, 22], modulate soil properties such as moisture content, and affect carbon and nitrogen cycling in arctic–alpine soils, as suggested by the observation of a micro-site specific prevalence of the N-fixation gene nifH [23]. Thus, we expect microbial communities to show micro-geographical patterns. Literature about such fine-scaled phenomena in MCC in arctic–alpine soils is scarce and focused mainly on the influence of vegetation patterns [24–27], or on slope exposition [10, 28].
In this study, we evaluated how elevation and micro-topography affect the diversity and biogeography of microbes in a complex terrain and aimed to identify underlying environmental drivers. We used long-term datasets for temporarily highly variable environmental parameters such as temperature and soil moisture, which were recorded at each individual sampling site, instead of using agglomerated variables over whole areas. To relate amplicon datasets with the extensive environmental datasets, we used a statistical approach rather unknown in microbial ecology, partial least square regression (PLSR) analysis. This allowed us to identify the most relevant environmental factors and to describe the ecological response of each microbial taxon.
Material and methods
Sampling sites
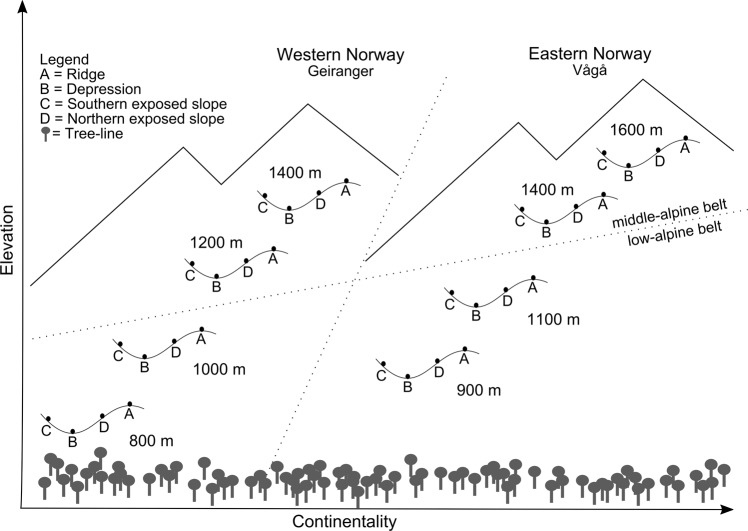
Soil samples were obtained from two arctic–alpine mountain regions in Norway after the first severe frost event in September 2011. The eastern region in Vågå, Oppland (61°53′ N, 9°15′ E) is characterized by a slightly continental climate with low annual precipitation, ranging from 325 to 650 mm a−1. The annual mean temperature is −1.2 °C (min. −29.2 °C, max. 16.7 °C; [20, 29]). The western region near Geiranger, Møre og Romsdal (62°03′ N, 7°15′ E) has an oceanic climate with much higher annual precipitation sums between 1500 and 2000 mm a−1 and a slightly higher mean temperature of 1.9 °C (min. −23.2 °C, max. 17.2 °C; [20]). For this study, we chose four elevations above the treeline, i.e. at 900 m, 1100 m, 1400 m, 1600 m a. s. l. in the Vågå region, which is comparable to 800 m, 1000 m, 1200 m, 1400 m a. s. l. in the Geiranger region (Fig. 1). The two lower elevational levels in each region belong to the low-alpine belt, whereas the two higher levels belong to the middle-alpine belt [30]. At each elevation, samples were taken from four micro-topographic positions, i.e. at ridges, depressions, south-facing, and north-facing slopes. At each site, the upper soil layer directly below the vegetation was sampled in triplicates (for one site a replicate is missing), resulting in 95 samples from 32 sites. Soils were sieved through a 4-mm sieve and stored at −20 °C. On-site near-ground soil and air temperature and soil moisture data were continuously recorded over a period of 5 years (2006–2011) before sampling. Temperatures were measured 15 cm below (soil temperature) and 15 cm above the soil surface (near-ground air temperature). Soil moisture was also measured in 15 cm depth. Soil carbon (C) and nitrogen (N) content, C:N ratio, pH, and soil texture were determined in the soil samples taken in September 2011 (see details in supplementary material Table S1).
Fig. 1.
Location of sampling sites and experimental design. Samples were collected from two regions (Geiranger and Vågå), four micro-topographic positions and four elevational levels representing two alpine belts. Both regions are located above the treeline. The vegetation is characterized by dwarf shrubs, graminoids, lichens, and mosses growing on siliceous bedrock and differs strongly due to micro-topographic features (e.g. Dahl [71])
DNA extraction, PCR amplification, and Illumina sequencing
DNA was isolated from 0.25 g of soil using the PowerSoil® DNA Isolation Kit (MoBio Laboratories, Inc., Carlsbad, CA). Cell lysis was performed with a tissue lyzer (10 min, 30 Hz). DNA was eluted in 100 µL PCR-water. 16S rRNA genes were amplified in triplicate reactions per sample using the fivefold diluted DNA extracts and non-barcoded primers [31]. One microliter of the amplification product was used in a second PCR with barcoded forward primers (Table S2). Each PCR assay contained 1× Herculase II Fusion buffer, 0.25 µM of each primer, 0.25 mM of each dNTP, 0.8 mg mL−1 BSA, 1 mM MgCl2, 1 U Herculase II Fusion Polymerase, and 1 µL template. The PCR program consisted of 26 cycles in the first PCR and 5 cycles in the second PCR (initial denaturation: 2 min at 95 °C; cycles: 20 s at 95 °C, 20 s at 52 °C, 20 s at 72 °C; final elongation: 3 min at 72 °C). The triplicate PCR products per sample were pooled and purified with CleanNA beads (GC-Biotech, Alphen aan den Rijn, Netherlands). The purified amplicons were quantified using the QuantiFluor® dsDNA system (Promega, Mannheim, Germany) and combined at equimolar ratios into two pools. Library preparation and sequencing on an Illumina HiSeq system was performed by the Max Planck-Genome-centre Cologne and resulted in 5.4 million 250 bp paired-end raw reads. After assembly and quality control, 2.8 million reads were used in QIIME [32] to build a 97% consensus OTU table for diversity and MCC analyses (for further details about sequence processing see supplementary material). Raw sequence reads have been deposited in the European Nucleotide Archive (PRJEB25202).
Analysis of MCC and diversity
Alpha and beta diversity were assessed based on a rarefied subset of the 97% OTU table (9132 reads per sample). Principal coordinates analysis (PCoA) plots and analysis of similarity (ANOSIM) were calculated based on a weighted UniFrac distance matrix using tools of QIIME [33, 34]. ANOSIM was performed to test for significant differences between samples with regard to mountain region, elevation, and microtopography. Because of the nested experimental design and the significant effect of elevation (but not mountain region), we separated the dataset according to elevation before evaluating differences due to micro-topography by ANOSIM.
Observed OTUs, Chao1, and Pilou’s Evenness were calculated in RStudio with the package vegan [35, 36]. Non-parametric Kruskal–Wallis tests were used to identify significant differences between samples in dependence on elevation and micro-topography. Mann–Whitney U tests with Bonferroni–Holm correction were used to further assess differences between the four micro-topographic expositions.
STAMP [37] was used to identify taxonomic groups (TG) responding to elevation or micro-topography. Therefore, the 97% OTU table was used to set up a TG table using the summarize_taxa.py command, which summarizes TGs at the last known phylogenetic level, followed by data rarefaction. The same dataset was used for PLSR, which allowed to integrate results from both analyses. Significant differences in relative abundances of the TGs were proven by Kruskal–Wallis tests and Scheffé post-hoc tests corrected with a False Discovery Rate calculation after Benjamini–Hochberg.
Analysis of microbial community data in relation to environmental parameters by PLSR
In addition to data on soil carbon (C) and nitrogen (N) content, C:N ratio, pH, and texture as potential explanatory variables for MCC, we used long-term on-site temperature and soil moisture measurements to calculate different temperature and soil moisture estimates (Table S1). This includes mean values, minimum and maximum values, the amplitude, and the sum of hours for which a specific temperature and moisture threshold were exceeded. For temperature, thresholds were set to 0.1 °C intervals from −25 to +25 °C, and for soil moisture, intervals of 0.01 m³ H2O m−³ soil were used within the range of 0–0.5 m³ H2O m−³ soil. The sum of hours above these thresholds was calculated based on 1-year, 2-year and 5-year datasets. This enabled us to test the relative effect of long-term versus short-term environmental conditions on MCC.
Overall, this approach resulted in 3000 environmental variables to be considered (Table S1), leading to the challenging statistical problem that there are many more variables (p) than observations (n). Moreover, the explanatory variables are not independent of each other but correlated, introducing complex interactions, and redundancies. The n < p problem makes common multiple regression approaches inapplicable because the degrees of freedom are equal to zero. An accepted shortcut to overcome this problem is to reduce the multidimensionality in the explanatory variables by multivariate reduction methods, such as principal component analysis (PCA). However, the PCA-obtained components maximize the covariation among the predictor variables regardless of the response variable. Thus, this approach might yield components lacking any ecological justification and consequently hinders the interpretation of the results [38]. To overcome these limitations, we used PLSR, a statistical method that is widely and successfully used in other scientific disciplines dealing with high-dimensional data (e.g., bioinformatics, genomics, spectrometrics). An introduction into PLSR is given in several references [39–42], and additional background information about the usefulness of PLSR for the purpose of this study is given in the supplementary material. To identify the most important variables out of the entire set of environmental variables, we applied different algorithms that are available for PLSR, as reviewed in Mehmood et al. [43]. These include Variable Importance in Projection (VIP) [44], which is likely to be the most popular, as well as the Selectivity Ratio (SR) [45, 46] and the significance Multivariate Correlation (sMC) approach [47], which were both introduced more recently.
To relate our set of environmental variables to the relative abundances of each TG using PLSR, mean relative abundance values were calculated for each TG from the three replicate samples per site using the same rarefied TG table as for STAMP. Variables (both dependent and independent) exhibiting near-zero variance were removed prior to further analyses. To assess the robustness of the PLSR results, a centered log-ratio transformed (clr) dataset, generated from the summarized TG table, was used instead of the rarefied TG table, thus taking into account the compositional nature of the data by a dedicated normalization procedure [48]. The PLSR was set up as single response (PLSR1) model (i.e., one model per taxonomic group) using the SIMPLS algorithm [49], implemented in the package mdatools [50] for R 3.3.1 [35]. The optimal number of components in the PLSR model was found using Wold’s R criterion [51]. We applied SR, VIP, and sMC (for the latter the function being adapted from the package plsVarSel [43]) to assess variable importance. For the SR approach, we defined explanatory variables with a SR of ≥ 2 as trustworthy, meaning that the explained variance by the variable was twice as high as the residual variance in the data.
Results
Biogeographic patterns in microbial communities
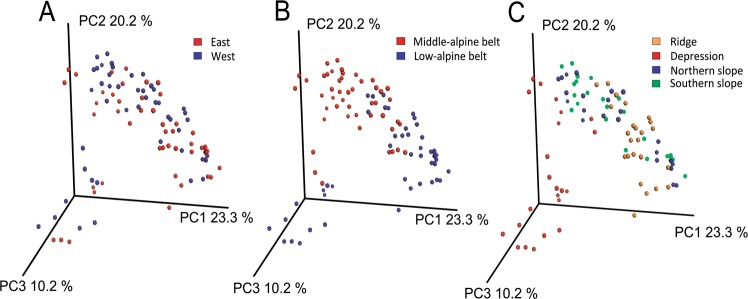
PCoA and ANOSIM revealed the variation in MCC to be unaffected by the mountain region (R = 0.08, P ≤ 0.001, Fig. 2a). In contrast, we observed a significant effect of elevation (R = 0.31, P ≤ 0.001, Fig. 2b). Likewise, micro-topography affected the MCC in both elevational belts significantly (low-alpine belt: R = 0.44, P ≤ 0.001, middle-alpine belt: R = 0.34, P ≤ 0.001).
Fig. 2.
Principal coordinate analysis (PCoA) of the microbial community compositional data based on a weighted UniFrac distance matrix. Samples were colored according to a mountain region, b alpine belt, and c micro-topography
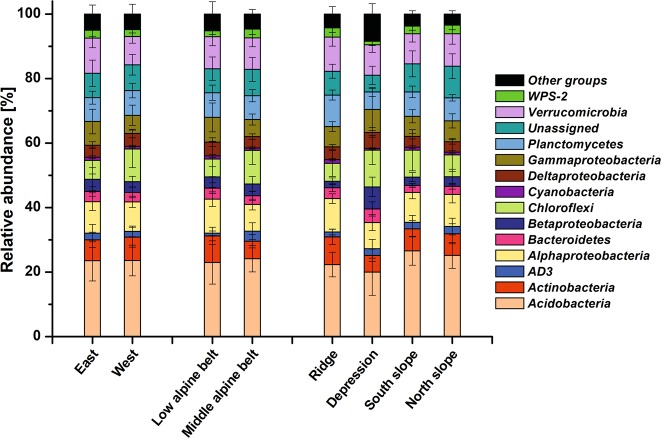
The most abundant phyla in the analyzed soils were Acidobacteria (23.5% ± 5.5) and Proteobacteria (23.3% ± 5.9), followed by Verrucomicrobia (9.8% ± 2.8), Chloroflexi (7.9% ± 5.5), Planctomycetes (7.5% ± 2.1), and Actinobacteria (6.8% ± 3.6; Fig. 3). Lower abundant phyla were Bacteroidetes, WPS-2 (“Candidatus Eremiobacteraeota”), and AD3 (“Candidatus Dormibacteraeota”). Within the Proteobacteria, Alpha- and Gammaproteobacteria were the dominant groups (9.4% ± 2.3 and 6.5% ± 3.6, respectively), while Beta- and Deltaproteobacteria showed abundances between 3 and 4%. In the low-alpine belt, families belonging to the phyla Actinobacteria, Alphaproteobacteria, Bacteroidetes, as well as Deltaproteobacteria, and Gammaproteobacteria showed significantly higher relative abundances, while members of the phyla Chloroflexi, Gemmatimonadetes, “Candidatus Dormibacteraeota”, and “Candidatus Eremiobacteraeota” were more prevalent in the middle-alpine belt (Fig. 3, Table S3). Microtopography also influenced MCC significantly, especially in depressions, where members of Bacteroidetes, Betaproteobacteria, Chlorobi, and Chloroflexi showed higher relative abundances, whereby an enrichment of freshwater-associated and (facultative) anaerobic TGs was found, e.g. Holophagaceae, Spirochaetaceae, Geobacteraceae, Myxococcales, Methylococcales, Comamonadaceae, Rhodocyclaceae, and Syntrophobacterales (Table S4). In contrast, soils from ridges harbored more Actinobacteria, Armantimonadetes, and Planctomycetes. A comparison of the alpha-diversity between samples revealed that elevation did not affect richness or evenness of the communities, while microtopography had a strong impact on all diversity indices (P < 0.001, Table 1), with slopes supporting a lower microbial richness and evenness than depressions and ridges. The mountain region influenced only Chao1 richness, showing higher values in the western region.
Fig. 3.
Stacked bar charts presenting the relative abundance of the predominant phyla and proteobacterial classes in samples grouped by eastern and western region, elevation according to alpine belt or micro-topography. Illustrated are differences in mean relative abundance and the corresponding standard deviation
Table 1.
Differences in diversity indices in dependence on mountain region, elevational belt, and micro-topography calculated by using Kruskal–Wallis tests
| Test parameters | Sample grouping | Observed | Chao | Pilou’s evenness |
|---|---|---|---|---|
| Mountain region | East | 1686 ± 237 | 3221 ± 476*** | 0.823 ± 0.029 |
| West | 1768 ± 229 | 3543 ± 536 | 0.829 ± 0.026 | |
| Elevational belt | Low-alpine | 1708 ± 270 | 3367 ± 553 | 0.822 ± 0.031 |
| Middle-alpine | 1746 ± 195 | 3395 ± 511 | 0.829 ± 0.024 | |
| Micro-topography*** | Ridge | 1764 ± 118 | 3370 ± 252 | 0.845 ± 0.014 |
| Depression | 1906 ± 275 | 3695 ± 629 | 0.829 ± 0.036 | |
| South slope | 1675 ± 217 | 3372 ± 562 | 0.817 ± 0.024 | |
| North slope | 1571 ± 181 | 3099 ± 456 | 0.813 ± 0.023 | |
| Mann–Whitney U test results for micro-topography | ||||
| Observed | Chao | Pilou’s evenness | ||
| Ridge | Depression | n.s. | n.s. | n.s. |
| Ridge | South slope | n.s. | n.s. | *** |
| Ridge | North slope | *** | * | *** |
| Depression | South slope | ** | n.s. | n.s. |
| Depression | North slope | *** | ** | n.s. |
| South slope | North slope | n.s. | n.s. | n.s. |
Significant differences are indicated as *P < 0.05, **P < 0.01, **P < 0.001, n.s. non-significant. Significant differences between the four individual micro-topographic positions were evaluated using Mann–Whitney U tests.
Relevance of different temperature and moisture variables for MCC
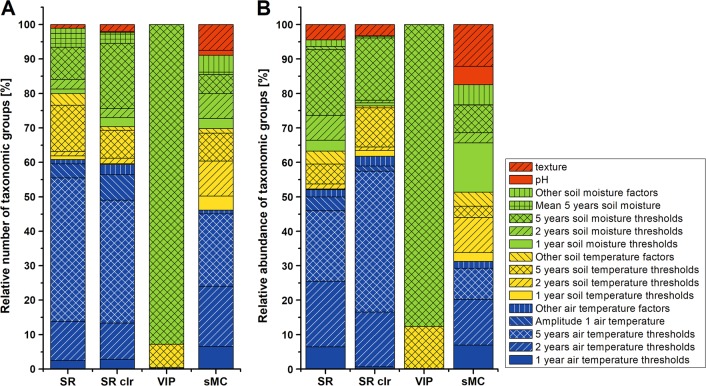
To identify environmental variables that can explain the variation in MCC we performed PLSR in combination with the SR, VIP, and sMC algorithm. Since PLSR analysis does not perform well for very low abundant taxa (read abundance ≤ 0.001%), presented results are derived from 470 TGs, which were obtained from the rarefied TG dataset upon zero-variance filtering, which precedes the PLSR procedure. Focusing on SR data, we observed that most TGs showed the highest SR values for near-ground air temperature (61%; Fig. 4a), soil temperature (19%), or soil moisture variables (17.5%), but rarely for other soil characteristics (2.5%). When taking into account the mean relative abundance of each taxon across the study sites, soil moisture was found to be the more important factor (32%; Fig. 4b), while the relevance of air temperature (52%) and soil temperature (11%) decreased slightly, i.e. several of the dominant taxa at the sites were air temperature or soil moisture driven. A clr-transformed TG table instead of a rarefied dataset as input in combination with the SR approach gave similar results (Fig. 4). The use of sMC as an alternative approach to identify the most important variables resulted in similar findings, i.e. soil temperature, soil texture, and pH became slightly more relevant based on this algorithm, but air temperature and soil moisture remained the predominant factors (Fig. 4). In contrast, VIP identified soil moisture as the major driver for 93% of the TGs. Interestingly, soil characteristics such as pH, carbon and nitrogen content, or soil texture were seldom found to be the most important factor (2% for the SR algorithm, 10% for sMC; Fig. 4a). Upon focusing on the type of measure, most microorganisms showed a better relation to specific temperature or moisture thresholds than to mean, maximum or minimum values, or the amplitude, regardless of the algorithm used for factor identification (SR or sMC; Fig. 4). Furthermore, the 2-year and 5-year air temperature datasets were better related to variations in microbial relative abundance across sites than the 1-year data.
Fig. 4.
Most important environmental drivers as obtained by partial least-square regression (PLSR) for each individual taxonomic group (TG). Relative numbers in the respective categories were obtained by counting the number of TGs falling into each category (a) or considering the mean relative abundance of each TG across all samples (b). Different algorithms were used to identify the most important environmental driver for each TG: selectivity ratio (SR), variable importance in projection (VIP), and Significance Multivariate Correlation (sMC). In addition, PLSR was conducted based on a centered logratio (clr) transformed OTU dataset in combination with SR for variable selection (SR clr). Specific temperature and moisture threshold values were calculated as sum of hours for which a specific temperature and moisture threshold was exceeded over 1, 2, or 5 years
Responses of individual microbial taxa to environmental variables
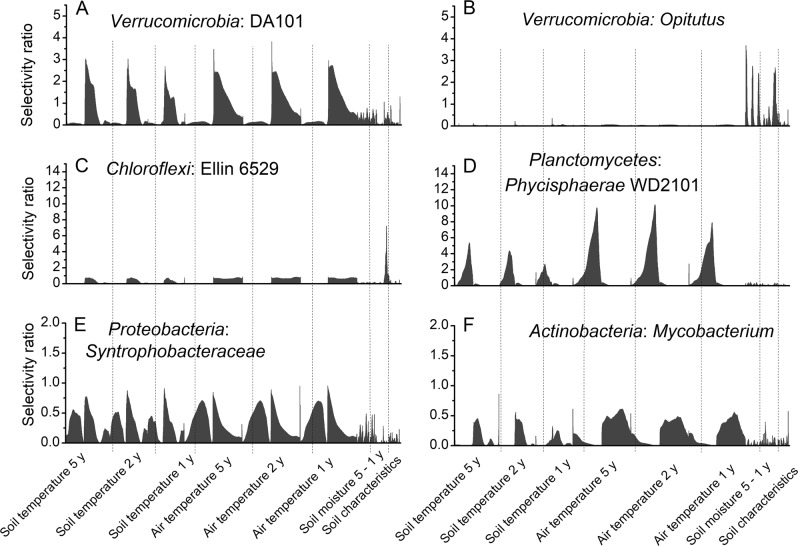
The obtained SR data were used to set up ecological response graphs, which display the importance of specific temperature or moisture regimes for a TG and describe the range in which it has its ecological response (Fig. 5). Remarkably, more than 84% of all analyzed TGs had at least one SR ≥ 2, indicating that relevant environmental variables were identified in most cases. SR profiles showed for some taxa a broad and for others a narrow ecological response pattern, suggesting a broad or narrow ecological niche. Interestingly, soil and air temperature profiles were often quite similar within a TG, although soil temperature ranges appeared sharper and less broad than air temperature thresholds (e.g. Fig. 5a, d and f). TGs with a narrow profile showed often very high SRs, while broader response patterns resulted in lower SRs (e.g. Fig. 5c versus e). In comparison to the SR approach, sMC led to very narrow response profiles, while a clr-transformation of the data resulted in SR profiles that were largely comparable to those of the rarefied SR dataset, though the specific shape and the identified most important factor differed for several TGs (Fig. S1; Table S4).
Fig. 5.
Examples of ecological response graphs showing the ecological response patterns of different taxonomic groups: a genus DA101, b genus Opitutus, c class Ellin 6529, d order Phycisphaerae WD2101, e family Synthrophobacteraceae, and f genus Mycobacterium. Plots display the selectivity ratios (SRs) of all environmental variables (Table S1). The first three sections, separated by dotted lines, represent soil temperature measures over 1, 2, and 5 years, respectively. Within each temperature section, stepwise increasing threshold values ranging from −16.9 °C to +22.4 °C are shown, followed by single values for amplitude, maximum, minimum, and mean. The same pattern is applied to present air temperature in a range from −25 °C to +25 °C and soil moisture ranging from 0 to 0.5 m3 H2O m−3 soil. The last section named soil characteristics includes soil texture, C, N, C:N ratio, and pH values
TGs dependent on temperature with SR values ≥ 2 were identified within the Acidobacteria, Actinobacteria, Alphaproteobacteria, Bacteriodetes, Firmicutes, Verrucomicrobia, and Planctomycetes (Table S4). The taxonomic rank at which TGs responded to temperature differed within the phyla. For example, responses were observed at order or class level within the Acidobacteria for Holophagae, Solibacteriales and the class iii1-8, or within the Actinobacteria for diverse though not all Actinomycetales (Table S4). The exact threshold air or soil temperature was specific for each genus. TGs that were predominantly influenced by soil moisture were also identified in different phyla, but also not exclusively in specific lineages, but rather scattered within these phyla. Soil moisture responsive TGs were found within Beta-, Delta- and Gammaproteobacteria, Chlorobi, Planctomycetes, and Spirochaeta, often with the strongest response to 5-year data (Table S4). These groups included members well-known to live under anoxic conditions, e.g. Desulfobacterales, Synthrophobacterales, Rhodocyclales, Anaerolineae.
Combination of biogeographic community patterns and ecological response patterns
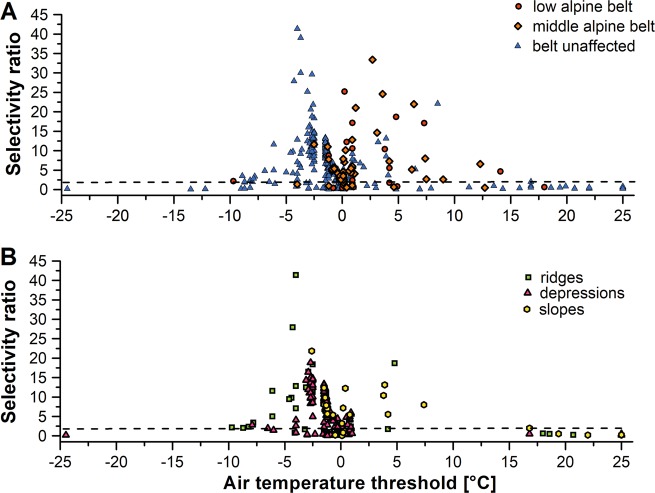
Based on STAMP, approximately half of the 470 TGs, which were analyzed by PLSR, were significantly influenced by micro-topography (50%), elevation (9%), or both factors (7%, Table S4). This knowledge about significantly affected taxa was combined with the information about the ecological response of each taxon obtained by PLSR. Analysis of the taxa being responsive to elevation and their respective maximum SRs for temperatures revealed no specific temperature threshold. Instead, data scattered in a range of −10 to +10 °C air temperature (or −4 to +8 °C soil temperature) with an accumulation between 2.5 and 0 °C air temperature (Fig. 6a and S3). Applying the SR cut-off of ≥ 2, the number of TGs responding to temperature was very high in the middle-alpine belt with 95% of the significantly enriched TGs being soil or air temperature affected. In comparison, 77% of the low alpine belt-affected and 78% of the belt-unaffected TGs had SRs ≥ 2 for soil or air temperature. This suggests that the occurrence of microbes being controlled by temperature increases with increasing elevation. Further, we evaluated the influence of temperature on TGs that were responsive to micro-topography rather than to elevation. A major part of the responsive TGs had air temperature thresholds between −2 and +2 °C (or 0 °C soil temperature; Fig. 6b and S3) and most of these were enriched in depressions. Actually, this accounted for 82% of all enriched taxa in depressions, indicating that temperature is also an important factor for microorganisms in water-filled depressions, with the freezing point of water being determinative. In contrast, temperature-responsive microorganisms at ridges (88%) and slopes showed broader air and soil temperature ranges (Fig. S2).
Fig. 6.
Plots displaying the maximum selectivity ratio (SR) in dependence on the 5-year air temperature threshold values of each taxonomic group (TG). Different colors indicate TGs that were significantly affected or unaffected by elevation (a) or affected by different micro-topographic expositions (b) according to STAMP. The dotted line indicates the SR cut-off at the value of 2, below which variables were considered to have no explanatory effect
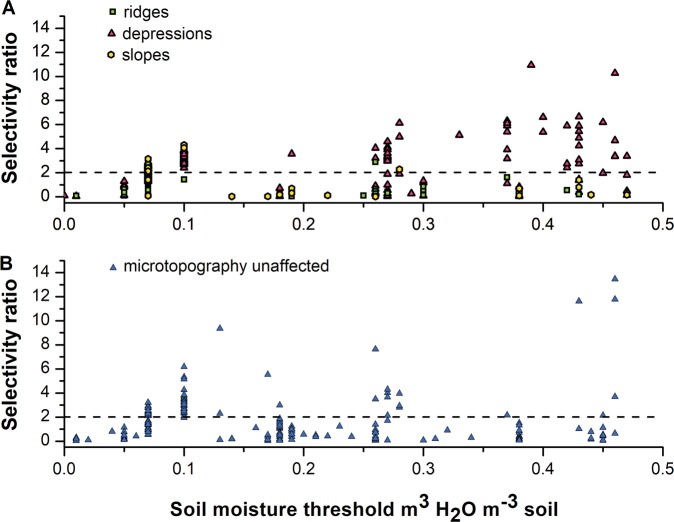
The threshold values for the 5-year moisture data were widely scattered (Fig. 7), but especially the TGs specifically enriched in depressions showed SR values ≥ 2 in 69% of all cases. In contrast, only 23% of TGs in other micro-topographic sites and 37% of all micro-topography-unaffected TGs responded to soil moisture thresholds with SR values ≥ 2 (Table S4). Further, an accumulation of depression enriched taxa was found > 0.25 m³ H2O m³ (Figs. 7b and S4). In fact, 22% of the microbial taxa enriched in depressions showed maximum SR values when moisture increased above 0.25 m³ H2O m−³ soil, while the percentage values were much lower for the other micro-topographic sites, as well as for the TGs being unaffected by micro-topography (3% and 8%, respectively).
Fig. 7.
Plots displaying the maximum selectivity ratio (SR) in dependence on the 5-year soil moisture threshold values of each taxonomic group (TG). Plot A shows TGs that were significantly enriched in depressions, at ridges or at slopes according to STAMP results, while plot B includes all TGs that were unaffected by micro-topography. The color code indicates taxa responding to different microtopographic expositions. The dotted line indicates the SR cut-off at the value of 2, below which variables were considered to have no explanatory effect
Discussion
MCC along gradients of elevation and micro-topography
Multivariate analysis of amplicon data revealed strong effects of elevation and micro-topography on the MCC in arctic–alpine soils, whereby ~50% of all TGs showed a significant shift in relative abundance due to one of these factors (Table S4). However, only a few TGs were affected by both parameters, suggesting that different microbial taxa may be primarily controlled by either elevation or micro-topography. Among the TGs that increased in relative abundance with elevation were “Candidatus Dormibacteraeota”, “Candidatus Eremiobacteraeota”, and Chloroflexi, taxa that have been detected in other arctic and high alpine studies [10, 28, 52, 53] and may thus represent typical colonizers of cold ecosystems. Similar to our findings, it was observed that the micro-topographic position in the landscape influences MCC besides elevation [10]. In our study, the permanently wet depressions differed clearly in soil moisture levels and provide oxic and anoxic microniches, which probably supported the development of distinct microbial communities, with members being characteristic for anoxic aquatic habitats or flooded soils, e.g. Geobacteraceae, Comamonadaceae, or Myxococcales (Table S4; [54–56]).
Alpha diversity was only affected by micro-topography, but not by elevation, which is in agreement with some earlier studies [7, 11, 57]. Micro-topography leads to a lower alpha diversity in soil microbial communities from slopes compared to those from ridges and depressions, implying a probable impoverishment at slopes.
Application of PLSR to combine amplicon data with comprehensive long-term environmental data records
In this study, spatially and temporarily resolved long-term temperature and soil moisture data were used to explain biogeographic patterns of the bacterial community. PLSR analysis allowed us to combine the extensive environmental datasets with amplicon data, which would not be possible with commonly used methods such as linear regression due to the higher number of explanatory variables than response variables [38]. We used different algorithms to determine the importance of environmental variables (SR, VIP, and sMC). Our results demonstrate that SR and sMC are well suited for the identification of the most relevant drivers, but not VIP, which gives a different result for the compiled data of all TGs (Fig. 4). VIP is known to generate false-positive results more likely and is therefore considered as least reliable here [46, 47]. The overall good agreement of sMC and SR data was partially supported when comparing results for individual taxa (Table S4 and Fig. S1), i.e. a clear response to temperature or soil moisture was often consistent, but the most important driver was not necessarily identical for the individual taxa (Table S4). This is explained by the fact that SR-values and sMC values are often quite similar for differently calculated parameters within the same category, e.g. for 2-year versus 5-year temperature data, so that slight variations in the data due to the applied algorithm will lead to differences concerning the most important factor (Fig. 5 and S1). However, the robustness of our findings in sum across all analyzed TGs allows general conclusions about major responses of the MCC to the identified environmental factors.
The ecological response patterns for individual taxa derived based on sMC versus SR were in general similar for many taxa (Fig. S1). SR is known to detect the most influential variables, though it tends to underestimate the total number of important variables, where sMC was described to perform better [58]. This is reflected in the finding that a broader range of different environmental factors were identified as most relevant based on sMC (Fig. 4). Considering that it was a major aim to find the most influential factors that affect MCC and that SR has been described as the most conservative approach, providing the least false positive rates, we focused on SR for the further analyses [46, 47].
Due to the fact that community compositional data are meanwhile more frequently transformed [48], we also evaluated potential effects of transformed input data by using exemplarily a clr-transformed dataset instead of the rarefied TG table. The overall ecological response patterns were very similar (Fig. 4), indicating that data preprocessing does not affect PLSR considerably in our case. We attribute this to the presence of clear differences between our samples and the fact that soil harbors highly complex microbial communities, where the limitations of compositional data analyses are discussed to be less relevant [59]. This will probably be different for less complex communities, especially when harboring few highly responsive taxa. Again, differences between rarefied and clr-transformed data were seen in individual response patterns (Fig. S1, Table S4), but the overall results leading to the identification of the most important drivers for the arctic alpine soil microbial communities remained consistent (Fig. 4).
Taken together, the PLSR approach couples state-of-the-art MCC data with comprehensive microenvironmental observations, as it has been done to investigate temperature effects on plants, arthropods, phytomass, or primary productivity [21, 58]. This is to our knowledge a new approach in microbial ecology, providing new opportunities to identify important environmental drivers for microbial communities. Combining the results for all TGs, we obtained robust results, although we observed some variation in the ecological response graphs for individual taxa, depending on the applied algorithm. Further in-depth studies will help to identify those methodological approaches that provide the most robust results also for individual taxa.
Near-ground air temperature and soil moisture as most important factors describing the ecological response of microbes
The PLSR analysis in combination with the calculation of SR values revealed that long-term temperature and soil moisture data were best related to the MCC. We found slightly higher SR values between MCC and near-ground air temperatures than with soil temperatures, indicating that short-term shifts, e.g. frost events, which are more pronounced 15 cm above ground than 15 cm below ground, are of superior importance for microorganisms thriving close below the soil surface. Our study also demonstrated that strongly agglomerated temperature or moisture values (mean, minimum, maximum, and amplitude) were not very well suited to explain MCC, while summing up hours of specific temperature or soil moisture thresholds resulted in better statistical relations. This may explain why studies using mean annual temperature values did not identify temperature as a major explanatory factor [5, 7, 10, 12]. In contrast to these earlier studies, different soil characteristics (pH, total C, total N, C:N ratio, and soil texture) showed weaker relations with the MCC. However, long-term records of dynamic soil parameters or additional variables may be further important explanatory variables for the biogeographic patterns of some TGs, especially for those that showed SRs < 2 for all measured parameters. In summary, the high SR values for temperature and soil moisture indicate that these factors provide the main environmental framework for the MCC in our study and should thus be considered as relevant factors for microbial communities in arctic–alpine ecosystems.
Variation in MCC is better explained by long-term than short-term temperature and moisture regimes
The 5-year temperature and soil moisture data often generated the highest SR values for most TGs, while the responses were weaker for 1-year data (Fig. 4). This suggests that the spatial patterns we observed in MCC are the result of long-term rather than short-term site-specific temperature or soil moisture regimes. Conversely, the better relation to long-term conditions indicates a certain resilience of microbial communities to year-to-year variations. This suggests that short-term dynamics in the MCC due to e.g. seasonality or snow melt [53, 60, 61] lead to a certain amplitude of microbial reactions without changing the whole system [62]. Our observation of a well-adapted MCC to long-term temperature and soil moisture conditions provides an explanation why global warming studies investigating the MCC in arctic–alpine regions found only little effects or only after long-term treatments over several years [63–67].
Identification of main drivers for elevational and micro-topographic differences in MCC
Microorganisms responding to elevation were often dependent on temperature variables, i.e., 83% of the enriched TGs in the middle-alpine belt and 87% in the low-alpine belt showed SRs ≥ 2 (Fig. 7). In comparison, 73% of the elevation-unaffected TGs had SRs ≥ 2. This temperature effect is in agreement with studies reporting elevation-dependent changes in animal and plant communities [2, 3, 7], in which temperature is the most evident underlying environmental factor. However, it contradicts the results of several previous microbial studies, which reported only a minor influence of temperature on elevation-dependent MCC shifts [5, 12]. Most studies identified soil properties such as pH, continentality, soil moisture, or carbon or nitrogen contents as underlying environmental drivers for elevation-dependent MCC shifts [7, 8, 11, 57]. The use of long-term near-ground temperature data with high spatial resolution instead of mean annual temperature values might be the key to resolve this contradiction. This would be in line with vegetation studies, which demonstrate that relations to mean atmospheric air temperatures should be used with caution due to their decoupling from the complex micro-environmental conditions [15, 16, 22]. A distinct threshold temperature for elevation-dependent taxa was not evident, even though the responsiveness to temperature increased with increasing elevation. It appears that each TG is affected by a slightly different threshold temperature ranging between −10 °C and +10 °C, indicating taxon-specific microbial responses to temperature. Responsive genera or species were in some cases bot not necessarily members of the same family, class or order. This is in agreement with the observation of Martiny et al. [68], who reported that temperature adaptation is not a conserved trait at higher taxonomic ranks.
MCC in depressions was strongly affected by soil and air temperature (0 °C and −2 to +1.5 °C, respectively). A temperature threshold around 0 °C, the freezing point of water, appears reasonable for organisms living in environments with high soil moisture levels. In agreement, a temperature of 0 °C has been reported to induce low soil respiration rates in permafrost soils, which is explained by the combination of cold, wet conditions, and low oxygen availability [69]. Thus, temperature seems to play a major role for microbial activity and community composition in water-logged environments, likewise as observed by Wang et al. in their mesocosm study [13]. We identified a specific soil moisture threshold > 0.25 m³ H2O m−³ of depression-enriched TGs (Fig. 7). Along with the detection of (facultative) anaerobic bacteria with SR ≥ 2, we conclude that this volumetric threshold value influences microbial life in the studied soils by providing anoxic microniches, which are known to affect microbial life substantially [70].
Conclusion
This study shows that not only elevation but also micro-topography is an important parameter, determining the biogeography of microbes in arctic–alpine ecosystems. To identify the most important underlying environmental factors contributing to the observed biogeographic pattern, a statistical approach new to the field of microbial ecology was used. PLSR allowed us to combine comprehensive long-term temperature and soil moisture datasets with 16S rRNA amplicon data and provided information about the ecological response for individual taxa. We identified for the first time temperature as a relevant factor for elevation-dependent soil microbial patterns in mountain ecosystems and soil moisture as factor being closely linked to micro-topographic differences. A specific soil moisture threshold was identified for microorganisms being affected by micro-topography, and linked to it a shift to anaerobic microorganisms in depressions. Overall, the microbial communities showed a stronger response to 5-year temperature or soil moisture conditions than to 1-year data, indicating a certain resilience of microbial communities to temperature variation in arctic–alpine ecosystems. This is in line with several global warming studies in arctic–alpine ecosystems, which report MCC responses only after long-term warming treatments. Thus, our study revealed that temporarily highly dynamic environmental factors such as temperature or soil moisture contribute more importantly to MCC variation in arctic–alpine ecosystems than previously thought. This becomes evident when proxies for these parameters are derived from long-term datasets with high temporal and spatial resolution to account for microscale heterogeneity.
Supplementary information
Acknowledgments
We like to thank Merle Noschinski-Reetz for excellent technical assistance.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0409-9) contains supplementary material, which is available to authorized users.
References
- 1.Arctic Monitoring and Assessment Programme. AMAP 2009 update on selected climate issues of concern (observations, short-lived climate forcers, arctic carbon cycle, predictive capability). Oslo: AMAP; 2009.
- 2.Hein N, Pape R, Finch OD, Löffler J. Alpine activity patterns of Mitopus morio (Fabricius, 1779) are induced by variations in temperature and humidity at different scales in central Norway. J Mt Sci. 2014;11:644–55. doi: 10.1007/s11629-013-2913-0. [DOI] [Google Scholar]
- 3.Körner C. Alpine plant life: functional plant ecology of high mountain ecosystems. 2nd ed. Berlin, Heidelberg: Springer; 2003. [Google Scholar]
- 4.Löffler UCM, Cypionka H, Löffler J. Soil microbial activity along an arctic-alpine altitudinal gradient from a seasonal perspective. Eur J Soil Sci. 2008;59:842–54. doi: 10.1111/j.1365-2389.2008.01054.x. [DOI] [Google Scholar]
- 5.Lanzén A, Epelde L, Blanco F, Martín I, Artetxe U, Garbisu C. Multi-targeted metagenetic analysis of the influence of climate and environmental parameters on soil microbial communities along an elevational gradient. Sci Rep. 2016;6:28257. doi: 10.1038/srep28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang LM, Wang M, Prosser JI, Zheng YM, He JZ. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol Ecol. 2009;70:208–17. doi: 10.1111/j.1574-6941.2009.00775.x. [DOI] [PubMed] [Google Scholar]
- 7.Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, et al. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology. 2011;92:797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- 8.Peay KG, von Sperber C, Cardarelli E, Toju H, Francis CA, Chadwick OA, et al. Convergence and contrast in the community structure of Bacteria, Fungi and Archaea along a tropical elevation– climate gradient. FEMS Microbiol Ecol. 2017;93:fix045. doi: 10.1093/femsec/fix045. [DOI] [PubMed] [Google Scholar]
- 9.Shen C, Ni Y, Liang W, Wang J, Chu H. Distinct soil bacterial communities along a small-scale elevational gradient in alpine tundra. Front Microbiol. 2015;6:582. doi: 10.3389/fmicb.2015.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Anderson BJ, Buckley HL, Lewis G, Lear G. Aspect has a greater impact on alpine soil bacterial community structure than elevation. FEMS Microbiol Ecol. 2017;93:1–11. doi: 10.1093/femsec/fiw253. [DOI] [PubMed] [Google Scholar]
- 11.Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, et al. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem. 2013;57:204–11. doi: 10.1016/j.soilbio.2012.07.013. [DOI] [Google Scholar]
- 12.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Pan F, Soininen J, Heino J, Shen J. Nutrient enrichment modifies temperature-biodiversity relationships in large-scale field experiments. Nat Commun. 2016;7:13960. doi: 10.1038/ncomms13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Körner C, Hiltbrunner E. The 90 ways to describe plant temperature. Perspect Plant Ecol Evol Syst. 2018;30:16–21. doi: 10.1016/j.ppees.2017.04.004. [DOI] [Google Scholar]
- 15.Pape R, Wundram D, Löffler J. Modelling near-surface temperature conditions in high mountain environments: an appraisal. Clim Res. 2009;39:99–109. doi: 10.3354/cr00795. [DOI] [Google Scholar]
- 16.Scherrer D, Körner C. Topographically controlled thermal-habitat differentiation buffers alpine plant diversity against climate warming. J Biogeogr. 2011;38:406–16. doi: 10.1111/j.1365-2699.2010.02407.x. [DOI] [Google Scholar]
- 17.Graae BJ, Vandvik V, Armbruster WS, Eiserhardt WL, Svenning JC, Hylander K, et al. Stay or go – how topographic complexity influences alpine plant population and community responses to climate change. Perspect Plant Ecol Evol Syst. 2018;30:41–50. doi: 10.1016/j.ppees.2017.09.008. [DOI] [Google Scholar]
- 18.Wang JY. A critique of the heat unit approach to plant response studies. Ecology. 1960;41:785–90. doi: 10.2307/1931815. [DOI] [Google Scholar]
- 19.Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC. Climate change threats to plant diversity in Europe. Proc Natl Acad Sci. 2005;102:8245–50. doi: 10.1073/pnas.0409902102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löffler J. Micro-climatic determination of vegetation patterns along topographical, altitudinal, and oceanic-continental gradients in the high mountains of Norway. Erdkunde. 2003;57:232–49. doi: 10.3112/erdkunde.2003.03.05. [DOI] [Google Scholar]
- 21.Beckers N, Hein N, Vanselow KA, Löffler J. Effects of microclimatic thresholds on the activityabundance and distribution patterns of alpine Carabidae species. Ann Zool Fenn. 2018;55:25–44. doi: 10.5735/086.055.0104. [DOI] [Google Scholar]
- 22.Löffler J, Pape R, Wundram D. The climatologic significance of topography, altitude and region in high mountains—a survey of oceanic-continental differentiations of the scandes. Erdkunde. 2006;60:15–24. doi: 10.3112/erdkunde.2006.01.02. [DOI] [Google Scholar]
- 23.Stewart KJ, Coxson D, Siciliano SD. Small-scale spatial patterns in N2-fixation and nutrient availability in an arctic hummock-hollow ecosystem. Soil Biol Biochem. 2011;43:133–40. doi: 10.1016/j.soilbio.2010.09.023. [DOI] [Google Scholar]
- 24.Shi Y, Grogan P, Sun H, Xiong J, Yang Y, Zhou J, et al. Multi-scale variability analysis reveals the importance of spatial distance in shaping Arctic soil microbial functional communities. Soil Biol Biochem. 2015;86:126–34. doi: 10.1016/j.soilbio.2015.03.028. [DOI] [Google Scholar]
- 25.Shi Y, Xiang X, Shen C, Chu H, Neufeld JD, Walker VK, et al. Vegetation-associated impacts on arctic tundra bacterial and microeukaryotic communities. Appl Environ Microbiol. 2015;81:492–501. doi: 10.1128/AEM.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker JKM, Egger KN, Henry GHR. Long-term experimental warming alters nitrogen-cycling communities but site factors remain the primary drivers of community structure in high arctic tundra soils. ISME J. 2008;2:982–95. doi: 10.1038/ismej.2008.52. [DOI] [PubMed] [Google Scholar]
- 27.Chu H, Neufeld JD, Walker VK, Grogan P. The influence of vegetation type on the dominant soil bacteria, archaea, and fungi in a low Arctic tundra landscape. Soil Sci Soc Am J. 2011;75:1756–65. doi: 10.2136/sssaj2011.0057. [DOI] [Google Scholar]
- 28.Frey B, Rime T, Phillips M, Stierli B, Hajdas I, Widmer F, et al. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol Ecol. 2016;92:fiw018. doi: 10.1093/femsec/fiw018. [DOI] [PubMed] [Google Scholar]
- 29.Löffler J. Snow cover dynamics, soil moisture variability and vegetation ecology in high mountain catchments of central Norway. Hydrol Process. 2005;19:2385–405. doi: 10.1002/hyp.5891. [DOI] [Google Scholar]
- 30.Dahl E. Zonation in arctic and alpine tundra and fellfield ecobiomes. Ecosystem theory application. London, Chichester: Wiley; 1986.
- 31.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team. R: a language and environment for statistical computing [Computer software]. Vienna: R Foundation for Statistical Computing. 2016
- 36.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. Vegan: Community Ecology Package. R package version 2.0-10. 2013.
- 37.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:btu494-. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrascal LM, Galván I, Gordo O. Partial least squares regression as an alternative to current regression methods used in ecology. Oikos. 2009;118:681–90. doi: 10.1111/j.1600-0706.2008.16881.x. [DOI] [Google Scholar]
- 39.Geladi P, Kowalski BR. Partial least-squares regression: a tutorial. Anal Chim Acta. 1986;185:1–17. doi: 10.1016/0003-2670(86)80028-9. [DOI] [Google Scholar]
- 40.LlE Frank, Friedman JH. A statistical view of some chemometrics regression tools. Technometrics. 1993;35:109–35. doi: 10.1080/00401706.1993.10485033. [DOI] [Google Scholar]
- 41.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58:109–30. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- 42.Abdi H. Partial least square regression (PLS regression). In: Salkind NJ, editor. Encyclopedia of measurement and statistics. Thousand Oaks, CA: Sage; 2007.
- 43.Mehmood T, Liland KH, Snipen L, Sæbø S. A review of variable selection methods in partial least squares regression. Chemom Intell Lab Syst. 2012;118:62–69. doi: 10.1016/j.chemolab.2012.07.010. [DOI] [Google Scholar]
- 44.Wold S, Johansson E, Cocchi M. PLS—partial least squares projections to latent structures. In: Kubinyi H, editor. 3D QSAR in Drug Design. KLUWER ESCOM Science Publisher; Leiden 1993.
- 45.Rajalahti T, Arneberg R, Kroksveen AC, Berle M, Myhr KM, Kvalheim OM. Discriminating variable test and selectivity ratio plot: quantitative tools for interpretation and variable (biomarker) selection in complex spectral or chromatographic profiles. Anal Chem. 2009;81:2581–90. doi: 10.1021/ac802514y. [DOI] [PubMed] [Google Scholar]
- 46.Farrés M, Platikanov S, Tsakovski S, Tauler R. Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J Chemom. 2015;29:528–36. doi: 10.1002/cem.2736. [DOI] [Google Scholar]
- 47.Tran TN, Afanador NL, Buydens LMC, Blanchet L. Interpretation of variable importance in partial least squares with significance multivariate correlation (sMC) Chemom Intell Lab Syst. 2014;138:153–60. doi: 10.1016/j.chemolab.2014.08.005. [DOI] [Google Scholar]
- 48.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:2224. [DOI] [PMC free article] [PubMed]
- 49.De Jong S. SIMPLS: an alternative approach to partial least squares regression. Chemom Intell Lab Syst. 1993;18:251–63. doi: 10.1016/0169-7439(93)85002-X. [DOI] [Google Scholar]
- 50.Kucheryavskiy S. Mdatools: multivariate data analysis for chemometrics. R package version 0.7.0. 2015.
- 51.Wold S. Cross-validatory estimation of the number of components in factor and principal components models. Technometrics. 1978;20:397–405. doi: 10.1080/00401706.1978.10489693. [DOI] [Google Scholar]
- 52.Ferrari BC, Bissett A, Snape I, van Dorst J, Palmer AS, Ji M, et al. Geological connectivity drives microbial community structure and connectivity in polar, terrestrial ecosystems. Environ Microbiol. 2016;18:1834–49. doi: 10.1111/1462-2920.13034. [DOI] [PubMed] [Google Scholar]
- 53.Lazzaro A, Hilfiker D, Zeyer J. Structures of microbial communities in alpine soils: seasonal and elevational effects. Front Microbiol. 2015;6:1–13. doi: 10.3389/fmicb.2015.01330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber KA, Urrutia MM, Churchill PF, Kukkadapu RK, Roden EE. Anaerobic redox cycling of iron by freshwater sediment microorganisms. Environ Microbiol. 2006;8:100–13. doi: 10.1111/j.1462-2920.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- 55.Frindte K, Allgaier M, Grossart HP, Eckert W. Redox stability regulates community structure of active microbes at the sediment-water interface. Environ Microbiol Rep. 2016;8:798–804. doi: 10.1111/1758-2229.12441. [DOI] [PubMed] [Google Scholar]
- 56.Randle-Boggis RJ, Ashton PD, Helgason T. Increasing flooding frequency alters soil microbial communities and functions under laboratory conditions. Microbiologyopen. 2017;7:e00548. doi: 10.1002/mbo3.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–20. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pape R, Löffler J. Determinants of arctic-alpine pasture resources: the need for a spatially and functionally fine-scaled perspective. Geogr Ann Ser A, Phys Geogr. 2017;99:353–70. doi: 10.1080/04353676.2017.1368833. [DOI] [Google Scholar]
- 59.Tsilimigras MCB, Fodor AA. Compositional data analysis of the microbiome: fundamentals, tools, and challenges. Ann Epidemiol. 2016;26:330–5. doi: 10.1016/j.annepidem.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 60.McMahon SK, Wallenstein MD, Schimel JP. A cross-seasonal comparison of active and total bacterial community composition in Arctic tundra soil using bromodeoxyuridine labeling. Soil Biol Biochem. 2011;43:287–95. doi: 10.1016/j.soilbio.2010.10.013. [DOI] [Google Scholar]
- 61.Buckeridge KM, Banerjee S, Siciliano SD, Grogan P. The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol Biochem. 2013;65:338–47. doi: 10.1016/j.soilbio.2013.06.012. [DOI] [Google Scholar]
- 62.Allison S. D., Martiny J. B. H. Resistance, resilience, and redundancy in microbial communities. Proceedings of the National Academy of Sciences. 2008;105(Supplement 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamb EG, Han S, Lanoil BD, Henry GHR, Brummell ME, Banerjee S, et al. A High Arctic soil ecosystem resists long-term environmental manipulations. Glob Chang Biol. 2011;17:3187–94. doi: 10.1111/j.1365-2486.2011.02431.x. [DOI] [Google Scholar]
- 64.Frey SD, Drijber R, Smith H, Melillo J. Microbial biomass, functional capacity, and community structure after 12 years of soil warming. Soil Biol Biochem. 2008;40:2904–7. doi: 10.1016/j.soilbio.2008.07.020. [DOI] [Google Scholar]
- 65.Kuffner M, Hai B, Rattei T, Melodelima C, Schloter M, Zechmeister-Boltenstern S, et al. Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing. FEMS Microbiol Ecol. 2012;82:551–62. doi: 10.1111/j.1574-6941.2012.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deslippe JR, Hartmann M, Simard SW, Mohn WW. Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol Ecol. 2012;82:303–15. doi: 10.1111/j.1574-6941.2012.01350.x. [DOI] [PubMed] [Google Scholar]
- 67.Sistla SA, Moore JC, Simpson RT, Gough L, Shaver GR, Schimel JP. Long-term warming restructures Arctic tundra without changing net soil carbon storage. Nature. 2013;497:615–8. doi: 10.1038/nature12129. [DOI] [PubMed] [Google Scholar]
- 68.Martiny J. B. H., Jones S. E., Lennon J. T., Martiny A. C. Microbiomes in light of traits: A phylogenetic perspective. Science. 2015;350(6261):aac9323–aac9323. doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 69.Mu C, Zhang T, Zhang X, Cao B, Peng X. Sensitivity of soil organic matter decomposition to temperature at different depths in permafrost regions on the northern Qinghai-Tibet Plateau. Eur J Soil Sci. 2016;67:773–81. doi: 10.1111/ejss.12386. [DOI] [Google Scholar]
- 70.Brune A, Frenzel P, Cypionka H. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev. 2000;24:691–710. doi: 10.1111/j.1574-6976.2000.tb00567.x. [DOI] [PubMed] [Google Scholar]
- 71.Dahl E. Rondane mountain vegetation in south Norway and its relation to the environment. Det Nor Vid Akad Skr Mat Naturv Klasse. 3 (Skrifter utgitt av Det Norske Videnskaps-Akademi i OsloI, Aschehoug, Oslo) 1956.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.