Abstract
Picocyanobacteria make up half of the ocean’s primary production, and they are subjected to frequent viral infection. Viral lysis of picocyanobacteria is a major driving force converting biologically fixed carbon into dissolved organic carbon (DOC). Viral-induced dissolved organic matter (vDOM) released from picocyanobacteria provides complex organic matter to bacterioplankton in the marine ecosystem. In order to understand how picocyanobacterial vDOM are transformed by bacteria and the impact of this process on bacterial community structure, viral lysate of picocyanobacteria was incubated with coastal seawater for 90 days. The transformation of vDOM was analyzed by ultrahigh-resolution mass spectrometry and the shift of bacterial populations analyzed using high-throughput sequencing technology. Addition of picocyanobacterial vDOM introduced abundant nitrogen components into the coastal water, which were largely degraded during the 90 days’ incubation period. However, some DOM signatures were accumulated and the total assigned formulae number increased over time. In contrast to the control (no addition of vDOM), bacterial community enriched with vDOM changed markedly with increased biodiversity indices. The network analysis showed that key bacterial species formed complex relationship with vDOM components, suggesting the potential correspondence between bacterial populations and DOM molecules. We demonstrate that coastal bacterioplankton are able to quickly utilize and transform lysis products of picocyanobacteria, meanwhile, bacterial community varies with changing chemodiverisity of DOM. vDOM released from picocyanobacteria generated a complex labile DOM pool, which was converted to a rather stable DOM pool after microbial processing in the time frame of days to weeks.
Subject terms: Biogeochemistry, Ecology
Introduction
Marine picocyanobacteria, the genera of Synechococcus and Prochlorococcus, are the most abundant and widely distributed photoautotrophs in the World’s Oceans [1, 2]. Picocyanobacteria contribute predominately to global primary production [3]. Picocyanobacteria have been predicted to have large impacts on the future ocean environment, with 10–20% increase of abundance globally by the end of 2100 [4]. Marine viruses are also abundant and play critical roles on microbial mortality in the ocean [5, 6]. About 15% of cyanobacteria are predicted to be infected by viruses in the World’s Oceans [7]. Viruses which infect diverse marine picocyanobacteria (cyanophage) have been isolated from various oceanic environments, ranging from estuarine systems to open ocean environments [8–11]. Metagenomics data revealed that cyanophage-like sequences represented 60–80% of the total identified viral genomes at time from the cellular fractions within the euphotic zone [12], indicating the wide-spread viral infection and lysis of picocyanobacteria. Viral lysis of host cells releases intracellular materials and cell detritus quickly into the environment, thus contributing to the organic matter pool in the ocean [13]. The viral lysis process of marine microbes changes the stoichiometry of organic matter [14] and redistributes the storage and composition of ocean’s carbon [15]. Dissolved organic matter (DOM) released from picocyanobacteria via viral-lysis directly links the carbon fixed by primary producers to the marine DOM pool, hence affecting the biogeochemical cycling of DOM in the ocean [6, 16]. Viral lysis of planktonic microbes releases an estimated 150 Gt DOC to the ocean every year, thereof is a major contributor to the marine DOM pool [5]. However, little is known about the composition and fate of virus-induced DOM (hereinafter referred to as vDOM) from picocyanobacteria.
With recent advances in the last two decades in mass spectrometry (MS), the composition of DOM can now be characterized with ultrahigh resolution and exact molecular formulae can be assigned to the ionizable fraction of desalted or solid-phase extracted DOM. Specifically, ultrahigh-resolution electrospray ionization (ESI) Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) has tremendously advanced the analytical capacity to characterize complex marine DOM molecules [17, 18] and bacterial DOM [19, 20]. FT-ICR MS has revealed abundant nitrogen-containing compounds in Synechococcus DOM [21], which may in part result from degradation of picocyanobacterial phycobilin pigments. Our recent study has shown that vDOM could contribute substantially to fluorescent DOM accumulated in the deep ocean [21].
Allochthonous DOM have been shown to have strong impacts on bacterial communities in the coastal environment [22], and in a culture study [23]. Previous seawater incubation experiments inoculated with simple organic molecules resulted in an increased complexity of DOM composition which appeared to be similar to naturally occurring marine DOM [19, 20]. These earlier studies suggest that natural bacterial communities are able to utilize and transform DOM into complex DOM molecules. Viral lysis of picocyanobacteria occurs frequently in the photic zone, and the question how do bacteria respond to viral lysis products of picocyanobacteria has yet to be addressed.
In this study, we prepared virus-induced DOM, or vDOM by lysing a Synechococcus culture with viruses, and added the vDOM to a natural bacterial community collected from the Atlantic coastal ocean. The goal is to understand: (1) How does the bacterial community in the coastal seawater respond to the enrichment of vDOM? (2) How does the chemical composition of vDOM changes upon bacterial activities? (3) Is there a correspondence between bacterial populations and molecular signatures of DOM?
Methods and materials
vDOM collection
Marine Synechococcus strain CB0101 was grown at 25 °C under constant cool white light (20–30 μE m–2 s–1) in the modified SN15 medium [24], in which no EDTA and vitamin B12 were added to reduce DOC background. The CB0101 culture was challenged by a lytic phage P1 that was isolated in our previous study [25]. The concentration of infectious phage P1 was determined by plaque assay. Viral strain P1 was added into CB0101 culture at the initial MIO = 1. It normally takes about 4–5 days for phage P1 to completely lyse the CB0101 culture. Viral lysate was collected at day 5 in the CB0101 culture challenged with P1 viruses (Supplemental Fig. S1). The lysate was filtered through GF/F (Whatman®) glass fiber filters. The filtrate containing vDOM was stored at 4 °C for 3 days until further use.
Dark incubation experiments
Seawater was collected at the Ocean City Inlet, Maryland, USA (38.324°N, 75.085°W, surface water). Seventy liters of surface seawater was taken with recorded temperature of 7 °C and salinity 32. The water sample was consequently filtered through 0.8-μm PC filters (Millipore®) 2 h after sample collection. A total of six 10 L polycarbonate bottles were used in the incubation experiments (triplicates for the vDOM treatment and triplicates for the control). All the bottles were acid washed and rinsed with MilliQ water prior to the experiment. To set up the incubation, 9 L of filtered seawater was added into each bottle and all six bottles were kept at room temperature in the dark condition for 4 days to lower the presumed background labile DOC level [23] and to stabilize the microbial community. Afterward, 1 L of vDOM filtrate was added into the treatment while the control received only 1 L of sterilized modified SN15 medium (major nutrients NO3−: 75 mg L−1, PO43−:1.586 mg L−1, CO32−:1.04 mg L−1) to balance the nutrients present in the vDOM solution. All bottles were incubated in the dark condition at room temperature (23 ± 2 °C).
Subsampling procedures
Subsamples were collected at day 0, 1, 3, 10, 30, 60, and 90, respectively, for analyses of bacterial abundance, bacterial community structure, nutrients, dissolved organic carbon (DOC), and DOM chemical composition analyses. Additional subsamples were taken at day 5, 7, 16, 22, and 45, respectively, for the analyses of bacterial abundance and DOC. Subsamples were taken from all the triplicates of the vDOM treatment and the control.
For bacterial cell counting, 2 mL of subsamples was fixed with glutaraldehyde at a final concentration of 1% v/v and kept at −20 °C until flow cytometry analyses. To analyze bacterial community structure, 250 mL of subsample was filtered using 0.22-μm filter (Millipore®) and filters were kept at −20 °C for later DNA extraction. Forty milliliters of subsample was taken and stored at 4 °C for nutrient analysis. To measure DOC, 20 mL subsample was transferred to a pre-cleaned 40-mL glass VOC vial (Thermo ScientificTM). To analyze the chemical composition of DOM, 1 L subsample was filtered through glass fiber (Whatman®) filters and solid-phase extracted using the method described previously [26] (referred to SPE-DOM thereafter). Briefly, filtered samples were acidified to pH 2 using formic acid (Sigma Aldrich, 98%), instead of HCl to avoid chloride adduct formation in the electrospray and then extracted using 1 g Agilent Bond Elut PPL cartridges, the cartridge was then rinsed with acidified pure water, dried, and eluted with 10 mL of methanol (Sigma Aldrich Chromasolv LC-MS®). Triplicate subsamples collected at each time point were mixed as one for further mass spectrometric analysis. Samples were stored at −20 °C prior to MS analyses.
Flow cytometry
Bacterial cells were enumerated using an Epics Altra II flow cytometer (Beckman Coulter, USA) with a 306C-5 argon laser (Coherent, USA). The enumeration was performed following the method of Jiao et al. [27]. Bacterial cells were stained with SYBR Green, sample preparation, and laser settings followed the protocol described by Liang et al. [28].
Chemical analyses
DOC and total dissolved nitrogen (TDN) were analyzed using a Shimadzu TOC-V and aTNM-1 unit. High temperature (680 °C) platinum catalyzed oxidation was used to quantitatively mineralize DOC and TDN in pH 2 acidified (hydrochloric acid, pure, 32%, Sigma Aldrich) samples that were sparged for 2.5 min to remove inorganic carbon. Potassium hydrogen phthalate and potassium nitrate standards were used to quantify DOC and TDN, respectively. All samples were run in triplicates and then averaged. Total dissolved phosphorus (TDP) was measured using the alkaline persulfate digestion of phosphorus to orthophosphate and EPA method 365.1. Dissolved inorganic orthophosphate (PO43−) was measured using EPA method 365.1 and the reaction of phosphate with ammonium molybdate and potassium antimony tartrate to form a blue-colored complex. Inorganic ammonium was quantified using the Bertholet reaction and the formation of a blue-colored compound after its reaction with sodium phenoxide. Nitrite and nitrate were measured using EPA method 353.2 and cadmium reduction of nitrate to nitrite. Nitrite is then quantified colormetrically upon its reaction with sulfanilamide and coupling to N-1-napthylethylenediamine dihydrochloride to form an azo dye. Concentrations of dissolved organic nitrogen (DON) and dissolved organic phosphorus (DOP) were calculated by subtracting inorganic nitrogen (NH4+, NO3−, and NO2−) and inorganic phosphorus from TDN and TDP, respectively.
DNA extraction and high-throughput sequence
Triplicate filters collected at each sampling time point were combined. The DNA was extracted from the combined sample using the PowerSoil Kit (MoBio®) based on the manufacturer’s protocol with some modifications. Briefly, 200 μl of bead solution was replaced with 200 μl of PCI and an extra wash procedure was added when the membrane was stained with a mixture of 100% ethanol and solution C4 (provided in the kit).
The V3–V4 region of bacterial 16S rRNA gene was amplified with PCR using forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. Sequencing was performed using the Illlumina MiSeq platform with MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China). Sequencing data were processed with the Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) pipeline as previously described [29]. Operational taxonomic units (OTUs) were clustered from the assembled high-quality sequences and classified with BLAST in Greengenes Database [30]. OTUs containing <0.001% were discarded. Shannon and Simpson diversity indices were calculated using the OTU table in QIIME.
Sequence data were deposited to the National Center for Biotechnology Information under accession number PRJNA495727 (releasing at May 31st, 2019).
Ultrahigh-resolution MS
A non-targeted ultrahigh-resolution MS approach was used to characterize the SPE–DOM samples from the incubation experiments. All SPE–DOM samples were analyzed at the Helmholtz Center for Environmental Health, Munich, Germany using a 12 Tesla Bruker Solarix Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS) interfaced with negative mode ESI. The soft ESI ionization, in combination with ultrahigh resolution MS, allowed to accurately assign exact molecular formulas to the observed singly charged m/z molecular ions. Mass accuracy exceeded <0.2 ppm at a mass resolution of 400,000 at m/z 500. Samples were directly infused into the ionization source at 2 µL/min and injection tubing was rinsed with ~600 µL of a 50% water/methanol mixture after each sample. All samples were analyzed in triplicates and only m/z ions consistently present in all spectra at comparable relative abundances were considered in this study, but replicate samples showed excellent agreement to each other which is the pre-requisite to assess complex direct infusion ultrahigh-resolution MS data in a semi-quantitative manner. Formula assignments used in this study are based on the following number of atoms of C0–70, O0–25, N0–10, and S0–2. At a mass accuracy of <0.2 ppm, the number of possible atomic combinations is very limited in the mass range of 200–600 Da, but when in doubt and to prevent false assignments, we cross-validated assignments using isotope simulation matching and always applied the seven golden rules [31]. After cross-validation, all formulas presented in this study were unambiguous. Results were plotted using van Krevelen diagrams [32], in which the hydrogen to carbon ratio (H/C) was plotted against the (O/C) ratio of all unambiguously assigned molecular formulas.
Network analyses and statistics
Spearman’s rank correlation coefficient between relative abundance of OTUs and m/z ions in the vDOM treatment were calculated with R (version 3.2.3). Positive strong correlation (r ≥ 0.9, P ≤ 0.05) and negative strong correlation (r ≤ −0.9, P ≤ 0.05) were selected and visualized in network with Cytoscape (version 3.5.1), respectively. OTUs and m/z ions with high correlations were set as nodes in the network, direct edges were made from source (m/z ions) to target (OTUs) based on correlation. Networks were displayed with a Spring Embedded Layout method and analyzed with network analyzer in Cytoscape.
Results
DOM and nutrients released from viral-lysis of Synechococcus
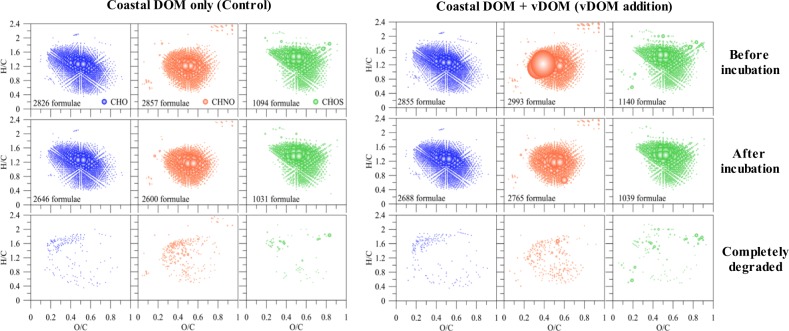
The vDOM from Synechococcus yielded about 2000 more DOM m/z ions compared to the DOM produced from the same Synechococcus culture without being challenged by its phage (Supplemental Fig. S2). This result suggests that vDOM contains more complex chemical composition compared to the DOM exudates from the cyanobacterial cells. Intense and diverse CHON ions were identified in the vDOM produced from Synechococcus [21] and the N-rich components were also visible when vDOM was added into the coastal water at the beginning of the incubation experiment (Fig. 3).
Fig. 3.
The chemical composition of DOM molecules before and after incubation. The van Krevelen diagram showed the assigned formulae in different composition of CHO (blue), CHNO (orange) and CHOS (green) series in the control (left) and the vDOM treatment (right). “Completely degraded” refers to the formulae which were identified at the beginning of incubation but not in the post-incubation samples. Bubble size represents the relative intensity of mass peaks
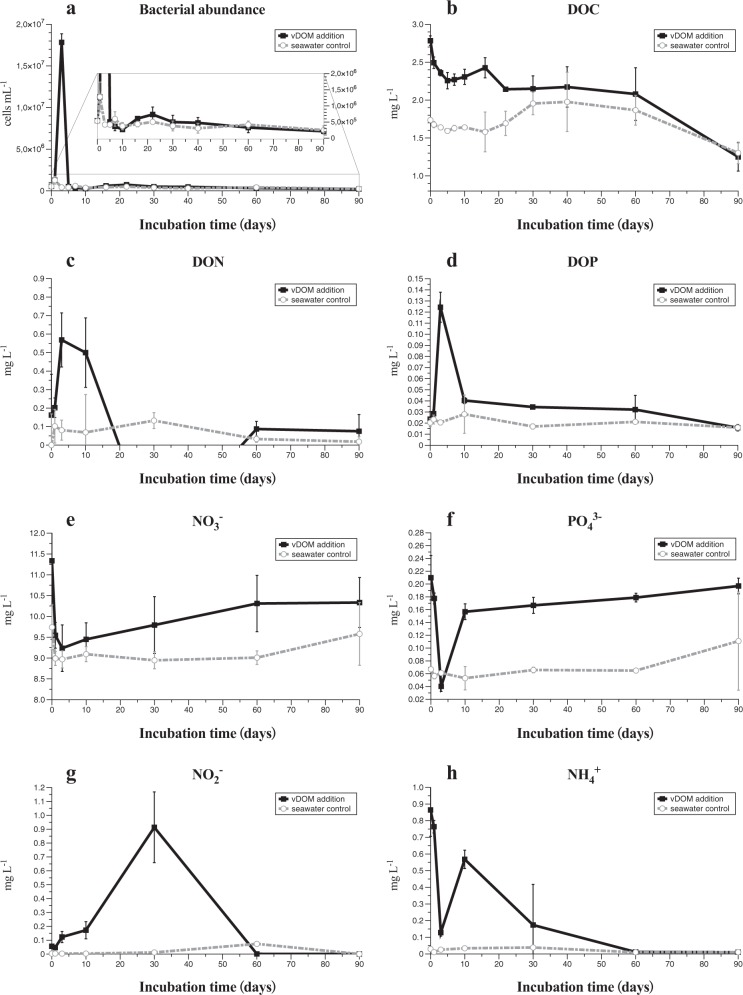
The seawater we used for incubation experiment contained 1.73 mg L−1 DOC. The addition of vDOM to the seawater increased DOC by 1.05 mg L−1, resulting in 2.78 mg L−1 DOC in the final concentration at the beginning of the experiment (Fig. 1b). The DON in the control was below the detection limit at the beginning of experiment, while vDOM treatment contained 0.16 mg L−1 DON (Fig. 1c). This is consistent with the abundant N-containing DOM formulae assigned in the samples from vDOM treatment. The original seawater contained 0.02 mg L−1 DOP, and no extra DOP was brought in with vDOM. The addition of vDOM also increased concentrations of inorganic nitrogen (NO3− and NH4+) and phosphate (PO43−) compared to the control which was compensated for the same volume amount with SN15 medium (Fig. 1e, f, h). vDOM added substantial DOC, DON, inorganic N and P into the incubation system.
Fig. 1.
The dark incubation experiment (total 90 days). The total bacterial counts (a), dissolved organic carbon (DOC) (b), dissolved organic nitrogen (DON) (c) and dissolved organic phosphorus (DOP) (d), phosphate PO43− (e), nitrate NO3− (f), nitrite NO2− (g), and ammonium NH4+ (h) in the vDOM treatment (square in black with solid line) and the control (circle in gray with dashed line)
Bacterial response to the addition of vDOM
The sharp increase of bacterial abundance was observed at day 3 in the community amended with vDOM (Fig. 1a), accompanied by a quick increase of DON and DOP (Fig. 1c, d). However, inorganic nutrients of PO43−, NO3−, and NH4+ (Fig. 1e, f, h) in the vDOM treatment showed an opposite response at day 3. DOC decreased in the vDOM treatment quickly in response to the bacterial dynamic before day 3. Bacterial abundance in the vDOM treatment increased quickly from 5.0 × 105 cells mL−1 to 1.8 × 107 cells mL−1 at day 3, decreased to 4.2 × 105 cells mL−1 at day 5 and maintained ca. 5.0 × 105 cells mL−1 throughout the 90 days’ incubation experiment. The concentration of DOC decreased from 2.78 to 2.30 mg L−1 before day 10, and decreased slightly from day 10 to 60, but decreased sharply from day 60 to 90, finally reaching the same level as the control at day 90 (1.25 mg L−1 in the vDOM treatment and 1.30 mg L−1 in the control). Concentrations of DON and DOP decreased sharply after the sudden increase at day 3, and kept at the low or undetectable level afterward (Fig. 1c, d). This result suggests that microbial community responded to the addition of vDOM by scavenging the available DOC and releasing DON and DOP into the incubating system. It appears that the uptakes of DOC, DON, and DOP were not highly synchronized at the early stage of incubation experiment (within day 10). Our results show that bacteria in the coastal water are able to consume a large portion of DOC in vDOM and increase their abundance by more than 30-fold within the first few days of vDOM addition.
It is intriguing that bacterial cell density decreased quickly to the background level (seawater) at day 5 in the vDOM treatment. The growth of bacteria caused the quick consumption of DOC, resulting in about 50% loss of added DOC and production of DON and DOP within 5 days. vDOM added very little or no DON, DOP, and NO2− to the treatment at the beginning of the incubation experiment. The “bacterial bloom” at day 3 resulted in the production of DON and DOP, which were quickly utilized by bacteria. It is possible that vDOM lost in the first few days was very labile and utilized rapidly by bacteria. vDOM present after day 5 was likely semi-labile and became more recalcitrant in the later stage, as bacterial abundance remained relatively constant and low in the later stages.
The addition of vDOM also brought inorganic N and P (NO3−, PO43−, and NH4+) into the treatment (Fig. 1e, f, h). These three types of inorganic nutrients all decreased drastically in the vDOM treatment from day 0 to 3, suggesting that the microbial community consumed inorganic N and P nutrient introduced with vDOM. It is not clear how bacteria utilized NO3−, PO43−, and NH4+ during the dark incubation period. Concentrations of NO3− and PO43− increased slowly from day 10 to the end of experiment (Fig. 1e, f) indicating little consumption of NO3− and PO43− by the microbial community during this period, likely due to the limitation of other growth factors. The concentration of NH4+ increased from day 3 to 10, and decreased gradually to the undetectable level by day 60 (Fig. 1h). The increase of NH4+ from day 3 to 10 could be associated with the decomposition of largely produced bacterial biomass at day 3. Nitrite increased gradually from day 0 to 30 and decreased to the undetectable level by day 60 (Fig. 1g). We speculate that the relatively fast oxidization of ammonium by ammonium oxidizing bacteria led to the increase of nitrite by day 30 with a slower development of nitrite-oxidizing bacteria and resulting oxidation of nitrite to nitrate. The decrease of nitrite and the slow increase of nitrate at the later stage of incubation are consistent with a slowly developing equilibrium between nitrite production and its oxidation in the later stage of incubation.
Variations of the bacterial community
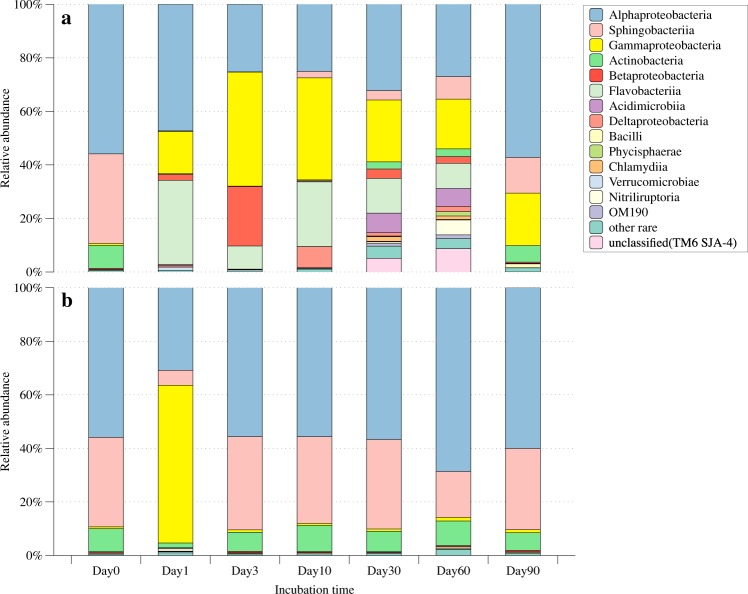
The bacterial community responded differently to the addition and no addition of vDOM (Fig. 2a, b). The original bacterial community (day 0) in seawater was dominated with Alphaproteobacteria, Sphingobacteria, and Actinobacteria, which kept steady to the temperature shift to laboratory and filtration process (Supplemental Fig. S3). The bacterial community enriched with vDOM responded quickly and drastically when compared to the controls (Fig. 2a). At day 1 and 3, Gammaproteobacteria, Betaproteobacteria, and Flavobacteria became more abundant compared to day 0. Such a community shift corresponded to the rapid increase of bacterial abundance (ca. 30-folds) during this period. At day 10, Alphaproteobacteria, Gammaproteobacteria, and Flavobacteria were the dominant bacteria in the vDOM sample. The complexity of the bacterial communities in the vDOM samples increased in day 30 and 60, reflected by the increased diversity indices (Table 1). Many different groups of bacteria (i.e., Acidimicrobiia, Chlamydiia, and Nitriliruptoria) appeared between day 30 and 60. Despite these changes, Gammaproteobacteria, Betaproteobacteria, and Flavobacteria were the three abundant groups, making up more than half of bacterial community throughout the experiment period. At day 90, these three groups of bacteria made up 90% of bacterial community. Gammaproteobacteria were present in the vDOM sample throughout the 90 days’ incubation, and they were an important group of bacteria responding to the addition of vDOM. In contrast, the bacterial community remained relatively stable in the control and was similar to the original seawater community which mainly consisted of Alphaproteobacteria, Sphingobacteria, and Actinobacteria (Fig. 2b).
Fig. 2.
The relative abundance of major bacterial taxa at different sampling time points during the incubation, showing the dynamics of bacterial community structure in the vDOM treatment (a) and control (b). Other rare: sum of taxa relative abundance below 1%. Unclassified: unclassified in the GreenGene database including unclassified groups of TM6 and SJA4. No triplicates were sequenced separately; samples at each sampling time point were combined together before DNA isolation
Table 1.
Simpson and Shannon biodiversity indices for the vDOM treatment and the control at different sampling times
| Time | Day 0 | Day 1 | Day 3 | Day 10 | Day 30 | Day 60 | Day 90 |
|---|---|---|---|---|---|---|---|
| vDOM | |||||||
| Simpsona | 0.65 | 0.92 | 0.89 | 0.92 | 0.98 | 0.98 | 0.70 |
| Shannona | 2.45 | 5.11 | 4.24 | 5.29 | 6.83 | 6.75 | 3.11 |
| Control | |||||||
| Simpson | 0.58 | 0.89 | 0.65 | 0.66 | 0.66 | 0.61 | 0.63 |
| Shannon | 2.19 | 4.65 | 2.50 | 2.54 | 2.58 | 2.85 | 2.48 |
aSignificant different from control, double tailed t-test, P < 0.05
Chemical composition of vDOM
The chemical composition of DOM analyzed by FT-ICR MS was displayed in van Krevelen space. The addition of vDOM increased the number of molecular formulae for all the CHO, CHNO, and CHOS formulae classes (Fig. 3). The numbers of different CHO, CHNO, and CHOS formulae increased from 2826, 2857, and 1094 to 2855, 2993, and 1140, respectively, at the time when vDOM was added into the coastal water, suggesting that the addition of vDOM increased the chemical complexity of DOM, even though it is expected that an unknown portion of this vDOM is not visible in negative mode ESI-FT-ICR MS due to weak ionization and possible strong ion suppression (e.g., alcohols and saccharides). In particular, abundant N-containing formulae were identified in the samples with vDOM addition and at the beginning of the experiment (Fig. 3b). After 90 days’ incubation time, the relative abundance of these vDOM formulae was decreased but not completely degraded, which might be due to a presumed large number of isomers of which some are readily available whereas others might be resistant to fast microbial degradation. Regardless, a clear decrease in intensity of m/z ions assigned to nitrogen-containing molecular formulae were observed at the end of 90-day incubation, implying that the N-rich DOM produced from the lysis of picocyanobacteria were effectively utilized by bacteria. The overall molecular formula patterns in the vDOM treatment and control became similar in both groups after 90 days, but the numbers of identified CHON and CHOS formulae in the vDOM addition treatments were still higher compared to the control. Hence, the addition of vDOM increased the diversity of DOM chemical composition of coastal seawater not only at the beginning of the experiment but also after 90 days of dark incubation.
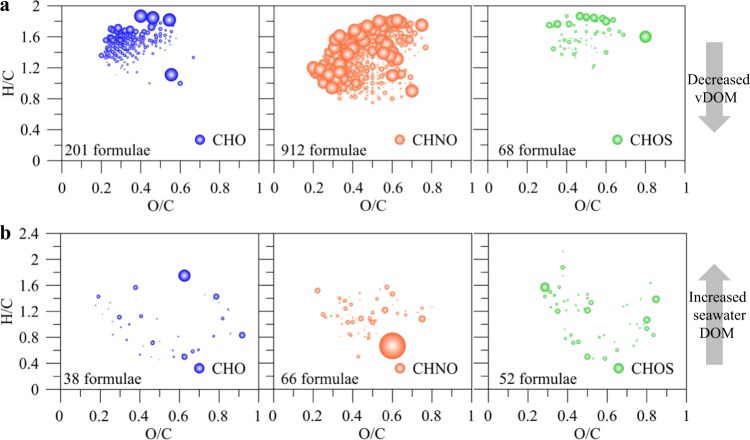
To further assess the different components of vDOM before and after the 90 days’ incubation, we separated the formulae in samples from the vDOM treatment into two parts: First, isolated vDOM m/z ions which are common with the initial vDOM composition before addition to seawater (Fig. 4a) and second: coastal DOM m/z ions present in the controls. We compared the relative changes in abundance of m/z ions before and after the incubation experiments. The majority of these isolated vDOM m/z ions decreased more than 10%, but a few also increased. There were also some coastal DOM m/z ions which increased >10%. Relative changes in abundance are typically associated with high variability, because slight changes in the sample matrix and composition may result in differences in abundance of individual m/z ions, meaning that relative changes should be taken as indicators but not used for quantifying distinct changes.
Fig. 4.
Changes of specific DOM molecular formulae in the vDOM treatment after 90 days’ incubation. The isolated vDOM formulae decreased (a), and the common formulae of the seawater background increased (b). Bubble size was set proportional with the changed percentage of particular mass peaks, cutoff = 10%
Associations between bacterial species and vDOM formulae
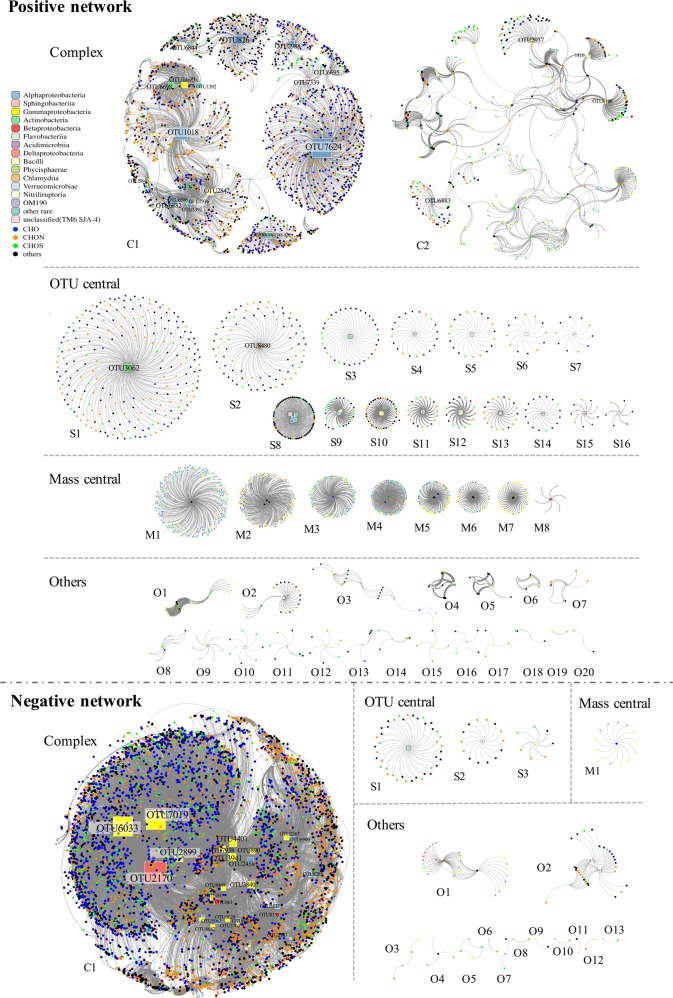
In order to explore the coherence between bacterial species and DOM molecular signature, the network analysis was used to visualize the complex interactions between bacteria and DOM at species to molecule level. Sub-datasets were selected when the spearman’s ranking correlation coefficient is strong (r ≥ 0.9 or r ≤ −0.9) between each m/z ion and OTU identified in the vDOM treatment. Direct connections were made from selected circle nodes (m/z ions, representing the DOM molecular signatures) to selected square nodes (OTUs, representing the bacterial species) as edges in the networks. The strong positive correlations (r ≥ 0.9) and strong negative correlations (r ≤ −0.9) were defined as the positive and negative networks, respectively (Fig. 5).
Fig. 5.
Network layout showing the association between bacterial species and DOM formulae in the vDOM treatment. Bacterial species are shown in square nodes, and different bacterial classes are labeled with different colors (same color bar, in Fig. 2). Circle nodes represent assigned molecular formulae of CHO (blue), CHON (orange), CHOS (green) and others (dark gray, including CHONS formulae and not assigned ions). Direct connections were made between mass formulae and OTUs when the Spearmen’s rank correlation coefficient is strong r ≥ 0.9 (Positive network, upper) and r ≤ −0.9 (Negative network, lower), P ≤ 0.05. Size of square nodes are proportional to the in-connected degree. The OTU name tags were shown when square nodes with connections >100 in the positive network and >200 in the negative network (the others were listed in supplemental Tables 1 and 2). Assigned molecular formulae in each subnetwork were shown in supplemental Tables 3 and 4
In the vDOM treatment, a total of 1829 OTUs and 11,210 assigned m/z ions were selected. Among them, 1430 OTUs and 3491 m/z ions were selected based on their high positive (r ≥ 0.9) spearman’s correlation coefficients, forming the positive networks. They consisted of complex networks with 20,455 edges which were divided into 47 individual subnetworks. Among these 47 independent subnetworks, there were two large complex subnetworks with more than 500 nodes and 2000 edges (Fig. 5 positive network C1 and C2, Complex). The positive subnetwork C1 is the largest one among all the subnetworks. Positive subnetwork C1 contains 52 OTU nodes, 2298 m/z nodes and 3578 edges in total forming several OTU centers. It is noteworthy that the OTU label size is proportional to the rank of OTU nodes (number of connections), some of them may not be readable if they do not occur frequently. Positive subnetwork C2 contained 254 OTU nodes, 302 m/z nodes and 2023 edges forming an evenly connected complex network without large connection center. Positive subnetworks S1–7 contained a single OTU node (Fig. 5, positive network S1–7, OTU central), while S8–16 contained multi-OTU nodes centrally connected (Fig. 5, positive network S8–16, OTU central). There were eight mass central subnetworks with more OTU nods connected with several m/z ions nodes as central (Fig. 5, positive network M1–16, mass central). The small subnetworks which were hard to define as OTU central or mass central and which contained less than ten total nodes were shown in the “Other” category (Fig. 5, positive network O1–20, others).
In the negative network, a total of 232 OTU nodes and 5291 m/z ions nodes were selected based on their high negative (r ≤ −0.9) spearman’s correlation coefficient. These selected nodes consisted of complex networks with 22,022 edges which divided into 18 individual subnetworks. The negative network contained 1 large complex subnetwork (Fig. 5, negative network C1, Complex), 3 OTU central subnetworks (Fig. 5, negative network S1–3, OTU central), 1 mass central subnetwork (Fig. 5, negative network M1, mass central), and 12 others (Fig. 5, negative network O1–12, others). Except for negative subnetwork C1, none of the subnetworks contained more than 50 total nodes. Negative subnetwork C1 contains 161 OTU nodes, 4948 m/z nodes and 21,755 edges. In general, the positive network contained less nodes and edges, but was divided into more numbers of subnetworks than the negative network.
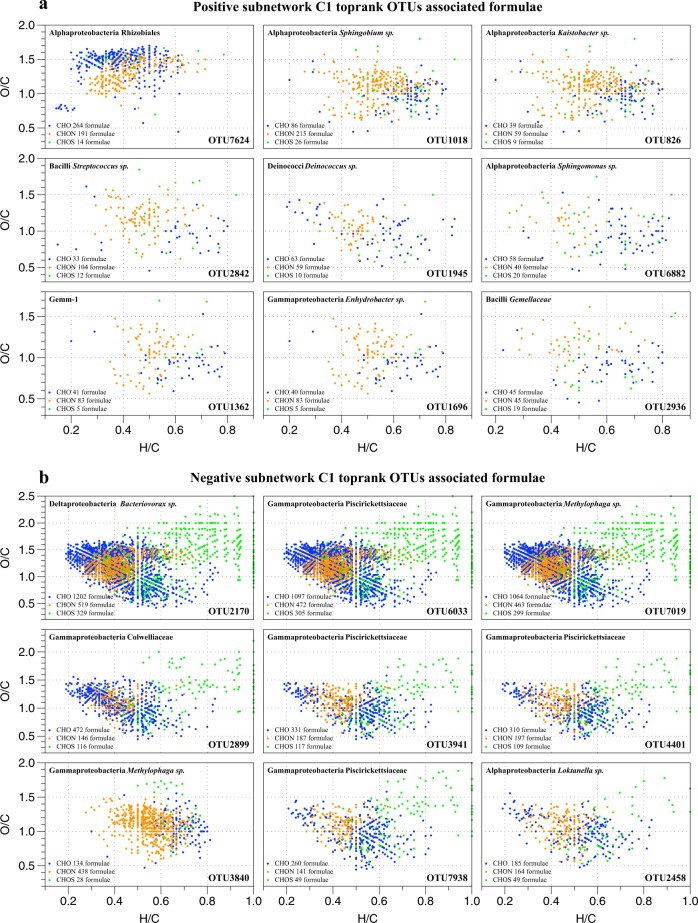
The m/z ions associated with top-rank OTUs in positive subnetwork C1 and negative subnetwork C1 were assigned to chemical formulae in groups of CHO, CHON, and CHOS (Fig. 6). The formulae patterns were consistent in the same subnetwork but distinct between the positive and the negative networks. The formulae associated with the top one ranking OTU7624 had relatively higher O/C ratio but lower H/C ratio than the other top-rank OTUs in the positive subnetwork C1 (Fig. 6a). All the top-rank OTUs in the negative subnetwork C1 except for OTU3840 were associated high O/C and H/C ratio CHOS formulae (Fig. 6b). The top-rank OTU nodes with high connections in the positive subnetwork C1 were OTUs from Alphaproteobacteria (Rhizobiales, Sphingobium sp., Kaistobacter sp., Sphingomonas sp., Hyphomicrobium sp., and Ochrobactrum sp.) Bacilli (Streptococcus sp. and Gemellaceae), Actinobacteria (Kocuria sp. and Mycobacterium sp.), Gammaproteobacteria (Enhydrobacter sp. and Stenotrophomonas sp.), Betaproteobacteria (Achromobacter sp. and Delftia sp.), and others (Deinococci, Clostridia, and Gemm-1). The top-rank OTU nodes with high connections in the negative subnetwork C1 were OTUs from Deltaproteobacteria (Bacteriovorax sp.), Gammaproteobacteria (Piscirickettsiaceae, Methylophaga sp., and Amphritea sp.), Alphaproteobacteria (Loktanella sp., Rhodobacteraceae and Tropicibacter sp.), and Betaproteobacteria (Methylotenera sp.). Only the top nine OTUs in positive and negative subnetwork C1 were shown in Fig. 6, whereas the others can be found in Supplemental Tables 1–4.
Fig. 6.
The assigned chemical formulae associated with top-rank OTUs in the positive subnetwork C1 (a) and negative subnetwork C1 (b). The van Krevelen diagram showed the CHO (blue), CHON (orange), and CHOS (green) formulae associated with each OTU and the taxonomic information of these top rank OTUs
Discussion
Viral lysis of Synechococcus releases intracellular materials to produce not only large amount of diverse DOM but also substantial quantities of inorganic nutrients. vDOM of picocyanobacteria brought in N-rich DOM (Fig. 3). Our result also shows that the abundant N-containing components introduced by vDOM were largely degraded during the 90 days’ incubation experiment, suggesting the important contribution of nitrogen sources from vDOM to natural bacterial community and their fast utilization. Viral lysis from Synechococcus were proved to be novel and distinct from intracellular materials [33]. Viral lysate contained unique components compared to the exudate from Synechococcus cells, since vDOM contain products released from the lysis of whole cells and viral particles. The viral particles themselves contributed insignificantly to the amount of total DOC or DON during the viral lysis process of Synechococcus or during the vDOM incubation (Supplemental Fig. S4). The most possible source of these abundant N-containing components is the unique phycobilisome complexity in Synechococcus cells. The degradation products of phycobilisome is consistent with the labile peptides derived from phycoerythrin protein [33] and relatively stable pyrrolic materials degraded from phycobilin pigments [21].
The DOM chemical signatures of the vDOM treatment became similar to that of seawater at the end of incubation (90 days). It seems that bacteria are capable of using and transforming the vDOM produced from viral lysis of Synechococcus into DOM products similar to those in the natural seawater. Long-term bioassays with simple substrate additions such as glucose [19], glutamic acid or a mixture of oligosaccharides and oligopeptides [20] showed that DOM composition after incubation experiments shared similar signature with seawater DOM. A recent study based on FT-ICR MS showed that molecular composition was similar throughout the water column [34]. Does this imply that the majority of DOM in natural aquatic systems are relatively stable and contain similar DOM species which cannot be easily utilized by bacteria? Although FT-ICR MS differentiate the chemical compositional diversity of DOM, it cannot predict the chemical structure of each formula. The limitations of the current analysis of DOM include extraction biases [26] and machine reproducibility [18].
In our study, the coastal bacterial community responded quickly to the addition of vDOM, both in terms of bacterial biomass and population structure. The re-assembling of original bacterial community and DOM signature at the end of 90 days’ incubation suggests that bacterial community is able to transform vDOM into less labile or more recalcitrant DOM in 3 months. The turnover of labile DOM to refractory DOM by natural bacterial community may occur in a relatively fast time frame. Although more studies are needed to confirm this hypothesis, our study provides preliminary evidence supporting the microbial carbon pump concept [35, 36], which emphasizes the importance of microbial activities in transformation of labile DOM into refractory DOM.
The addition of vDOM changed bacterial community during the incubation period, increased not only the chemodiversity of DOM but also the bacterial diversity. However, the bacterial community seemed to resume the original assemblage at the end of incubation. A marked increase in bacterial biomass was also observed after the addition of vDOM. The increased bacterial diversity and abundance within the first few days of vDOM addition is likely due to the presence and consumption of labile vDOM. High bacterial diversity was observed in the vDOM sample at day 30 and 60, suggesting that more bacterial species were involved in degrading diverse DOM molecules during this period. The initial bacterial community was re-established after the exogenous perturbation was over (90 days in this study). The recovery of bacterial community echoes the turnover of vDOM to seawater DOM at the end of 90 days’ incubation. We assume that there was no further bacterial transformation of the additional DOM in this incubation system after 3 months when the environmental condition tended to be stable. Under this circumstance, microbial transformation of vDOM resulted in the environmental context-dependent recalcitrant DOC (referred as RDOCt [36]). RDOCt can be considered as a type of RDOC which is present in nature due to the lack of microbial metabolic process or enzymes. The formation of RDOCt is the consequence of microbial carbon sequestration [36].
Gammaprotebacteria, Betaproteobacteria, and Flavobacter thrived in response to the enrichment of vDOM. Occurrence of these bacterial groups reduced the relative abundance of Alphaproteobacteria. It is generally believed that bacterial community in the natural environment is limited by the availability of DOM [37]. Alphaproteobacteria are known to be active in utilizing low-molecular weight DOM, such as glucose and amino acids, while Bacteroidetes (including Flavobacter) can actively take up high-molecular weight DOM such as chitin, N-acetyl glucosamine, and protein [38, 39]. Gammaproteobacteria and Flavobacter became more abundant in the vDOM treatment, particularly in the first 10 days. Their relative abundance decreased gradually after day 30. Alphaproteobacteria regained their niche at day 90 (similar to the pre-incubation period). Prevalence of Gammaproteobacteria and Flavobacter in the vDOM treatment could be triggered by the enrichment of complex DOM substrates. Viral lysis of picocyanobacteria is a frequent ecological event in the ocean. Unlike extracellular excretion, burst of cyanobacterial cells as result of viral infection releases diverse DOM components including cell wall glycans, phycobilisomes, lipid, carbohydrate, protein, etc. Gammaproteobacteria and Flavobacter could break down large DOM molecules which are less preferred by Alphaproteobacteria. Bacteriotes (Flavobacter) also dominate the human gut microbiome [40]. Bacteriotes are active members in human gut and they are able to catabolize diverse dietary plan glycans substrates including cellulose and hemicellulose [41]. Bacteriotes from different environments employ a similar strategy for glycan metabolism [41]. Cyanobacterial cell walls contain peptidoglycans which are thicker and more cross-linked compared to bacterial peptidoglycans [42, 43]. Occurrence of Flavobacter in the vDOM treatment could be related to the degradation of cyanobacterial cell wall components. Betaproteobacteria were only abundant at day 3, and remained relatively insignificant during the incubation. Gammaprotebacteria and Betaproteobacteria can be found in the coastal water [44], but Betaproteobacteria tend to prevail in the freshwater environment [45]. While Gammaproteobacteria are able to use amino acids, which are labile DOM to many bacteria, they also utilize extracellular polysacharides, proteins, and DOM released from phytoplankton [39, 46]. It is noteworthy that even within Gammaproteobacteria, the uptake of DOM can be different [47]. Some Gammaproteobacteria rely more on exogenous amino acids because they have fewer metabolic capability for synthesizing common amino acids [48].
Bacterial population and DOM composition in the ocean are both highly diverse. Although it is challenging to bridge complex microbial and chemical species, we intended to use the network analysis to explore the potential relationship between major bacterial and DOM species. In the network analysis, strong positive and negative correlations indicate the increased and decreased monotonic trends, respectively. In the positive networks, top-rank OTU nodes were mostly the connected with CHON formulae, which were accumulated after 90 days of incubation. The top-rank OTUs in the positive network were affiliated with bacterial taxa which were dominant at the beginning and end of incubation experiment (Alphaproteobacteria), contributing to the accumulation of stable DOM pool. In the negative networks, more OTU nodes were connected with the CHO formulae which were degraded during the incubation time and the CHOS formulae with high H/C and O/C ratios. The top-tank OTUs in the negative network were affiliated with the bacterial taxa which were dominant in the middle of incubation experiment (mainly Gammaproteobacteria and Betaproteobacteria) (Figs. 2 and 6), correlated to the degradation and transformation of relatively active DOM compounds. Both methods (network analysis and distribution frequency) concur that different bacterial taxa were responsible for the accumulation and degradation of DOM during the incubation period. The key species or taxa could serve as targets for further functional study of bacteria involved in transformation of vDOM. The network patterns also implicated the possible interactions between bacteria and DOM. The subnetworks were clustered into three major groups of the complex (containing the largest number of nodes), the mass central (single mass node connected with multiple OTU nodes), and the OTU central (single OTU node connected with multiple mass nodes). We proposed that the OTUs in the OTU central networks are supposed to be “generalist” which can utilize multiple DOM substrates and the OTUs in the mass central networks are “specialist” related to limited DOM species. Meanwhile, the chemicals in the OTU central networks would be more complex in structure than which in the mass central networks, since they were found by limited bacterial spices other than the ones connected with multiple bacterial spices. However, this preliminary hypothesis needs to be verified with more evidence from bacterial activities and functions, as well as the structure analysis of the DOM spices.
The consumption and accumulation of DOM are a result of interactions between the diverse bacterial community and DOM molecules. One of interesting questions in marine microbial ecology is whether there is a specific link between bacterial populations and chemical compounds. Network analyses visualized the complex correlations between thousands of bacterial species and DOM molecules, and help us to find the key species or the pattern of interactions. In this study, by focusing on the substantial changes of bacterial species and DOM molecules during the incubation experiment, we were able to identify strong correlations between bacterial OTUs and m/z ions of DOM. To date, only a few studies attempted to unveil the link between microbial biodiversity and DOM chemical diversity. Network analyses were used to describe the coherence between bacterial species and DOM molecular signatures in the former field study [36]. Using the combination of high throughput sequencing and ultrahigh-resolution MS, the study analyzed samples across a latitudinal transect across the North Sea showed that bacterial communities were mainly influenced by local events and correlated with specific labile DOM components. Strong correlations were identified between certain and limited OTUs and DOM components in a short-term in situ observation (20 days) [36], indicating the gap of community functional analyses and bacterial taxa and their connections to specific DOM molecules. Here in this study, the combination of high-throughput sequencing and ultra-high resolution mass spectra provided the unprecedented diversity and complexity of both microbial community and DOM. The discussion of interaction between them has been deepened to the level of “species” to “molecules”. The correlation and interaction were amplified and focused in the close incubation system, thus we identified a large number of OTUs and m/z ions with strong correlation in the vDOM treatment and revealed the complex interaction between the two complementary complexities.
Bacteria play key roles on transforming organic matter, and thousands of bacterial and DOM species are present in the ocean. The ability to link bacterial diversity with chemical diversity of DOM will deepen our understanding of DOM transformation by specific bacterial groups or populations. The network analysis we applied here is just a first step toward this goal. A major challenge here is the complexity and the semi-quantitative data of both microbial and DOM species. Deep sequencing of bacterial community is now possible and enables us to explore the rare species in the community. However, the universal primers based on the 16S rRNA gene regions (V3–V4) tend to overestimate some specific taxa such as Gammaproteobacteria [49]. Ultrahigh MS provides the highly diverse data on DOM chemical composition but is not able to identify single molecules without the information of chemical structures and is also relying on a specific analytical window, which may not be representative for the entire DOM pool. The application of advanced analytical approaches coupling with combination analyses of data sets will further explore the detailed interactions between bacteria and DOM in the ocean to better understand the processes and mechanisms that drive biogeochemical cycles. We used coastal water for this experiment because the Synechococcus and phage were isolated from the coastal estuary. The water was collected during the winter time, mainly to obtain a water sample with low DOM. This will allow the final concentration of vDOM to be higher than the background DOM. This may not be an issue if vDOM is added to oceanic water where the background DOM is much lower compared to the coastal water. The incubation at room temperature could potentially change the composition of bacterial community collected from the cold season. Ideally, the incubation temperature should be similar to in situ temperature for the incubation experiment. We monitored the bacterial community when temperature increased from in situ to room temperature, and found little change on the bacterial community (Supplemental Fig. S3). However, bacterial community responded quickly and strongly to the addition of vDOM. In our study, the added vDOM was mainly the viral lysis products but contained Synechococcus exudates as well. For the future study, additional control such as cyanobacterial exudates should be considered, as Synechococcus cells can release more than 20% of organic carbon via excretion [50].
Supplementary information
Acknowledgements
We would like to acknowledge the major supports from the Ministry of Science and Technology(2018YFA0605800, support Z.Z.) and the Seed Grant from University of Maryland Center for Environmental Science, Institute of Marine and Environmental Technology (awarded to F.C. and M.G. in 2015). This project was partially supported by the Maryland Sea Grant REU program in summer 2015 and National Science Foundation of the United States of America (Award #1829888). This is contribution 5640 of the University of Maryland Center for Environmental Science. This publication is partially supported by grants to N.J. from the Ministry of Science and Technology (2013CB955700), the Natural Science Foundation of China (91428308) and State Ocean Administration (GASI-03-01-02-05), to Z.Z. from the National Postdoctoral Program for Innovative Talents (BX20180388), to L.X. from the Ministry of Science and Technology (2016YFA0601103), and to Q.Z. from the Research Grants Council of the Hong Kong Special Administrative Region, China (16103414) and the Natural Science Foundation of China (41476147).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhao Zhao, Michael Gonsior
Contributor Information
Nianzhi Jiao, Email: jiao@xum.edu.cn.
Feng Chen, Email: chenf@umces.edu.
Supplementary information
The online version of this article (10.1038/s41396-019-0449-1) contains supplementary material, which is available to authorized users.
References
- 1.Scanlan DJ, West NJ. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol. 2002;40:1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 2.Agawin NSR, Duarte CM, Agustí S, McManus L. Abundance, biomass and growth rates of Synechococcus sp. in a tropical coastal ecosystem (Philippines, South China Sea) Estuar Coast Shelf Sci. 2003;56:493–502. doi: 10.1016/S0272-7714(02)00200-7. [DOI] [Google Scholar]
- 3.Li WKW. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnol Oceanogr. 2003;39:169–75. doi: 10.4319/lo.1994.39.1.0169. [DOI] [Google Scholar]
- 4.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao NZ, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110:9824–9. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 6.Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 7.Suttle CA. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol. 1994;28:237–43. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- 8.Suttle CA, Chan AM. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol. 1994;60:3167–74. doi: 10.1128/aem.60.9.3167-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marston MF, Sallee JL. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island’s coastal waters. Appl Environ Microbiol. 2003;69:4639–47. doi: 10.1128/AEM.69.8.4639-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millard AD, Mann NH. A temporal and spatial investigation of cyanophage abundance in the Gulf of Aqaba, Red Sea. J Mar Biolog Assoc UK. 2006;86:507. doi: 10.1017/S0025315406013415. [DOI] [Google Scholar]
- 11.Wang K, Wommack KE, Chen F. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl Environ Microbiol. 2011;77:7459–68. doi: 10.1128/AEM.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong EF. Community genomics among stratified microbial assemblages in the Ocean’s Interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 13.Proctor LM, Fuhrman JA. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–62. doi: 10.1038/343060a0. [DOI] [Google Scholar]
- 14.Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Microbiol. 2014;12:519–28. doi: 10.1038/nrmicro3289. [DOI] [PubMed] [Google Scholar]
- 15.Weinbauer MG, Chen F. Virus-mediated redistribution and partitioning of carbon in the global oceans. In: Jiao N, et al., editors. Microbial carbon pump in the ocean. Washington, DC: SCIENCE/AAAS; 2011. p. 54–6.
- 16.Zhang R, Wei W, Cai LL. The fate and biogeochemical cycling of viral elements. Nat Rev Microbiol. 2014;12:850–1. doi: 10.1038/nrmicro3384. [DOI] [PubMed] [Google Scholar]
- 17.Gonsior M, et al. Characterization of dissolved organic matter across the subtropical convergence off the South Island, New Zealand. Mar Chem. 2011;123:99–110. doi: 10.1016/j.marchem.2010.10.004. [DOI] [Google Scholar]
- 18.Hertkorn N, Harir M, Koch BP, Michalke B, Schmitt-Kopplin P. High-field NMR spectroscopy and FTICR mass spectrometry: powerful discovery tools for the molecular level characterization of marine dissolved organic matter. Biogeosciences. 2013;10:1583–624. doi: 10.5194/bg-10-1583-2013. [DOI] [Google Scholar]
- 19.Koch BP, Kattner G, Witt M, Passow U. Molecular insights into the microbial formation of marine dissolved organic matter: recalcitrant or labile? Biogeosciences. 2014;11:4173–90. doi: 10.5194/bg-11-4173-2014. [DOI] [Google Scholar]
- 20.Lechtenfeld OJ, Hertkorn N, Shen Y, Witt M, Benner R. Marine sequestration of carbon in bacterial metabolites. Nat Commun. 2015;6:6711.. doi: 10.1038/ncomms7711. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Gonsior M, Luek J, Timko S, Ianiri H, Hertkorn N, et al. Picocyanobacteria and deep-ocean fluorescent dissolved organic matter share similar optical properties. Nat Commun. 2017;8:1–10. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traving SJ, Rowe O, Jakobsen NM, Sørensen H, Dinasquet J, Stedmon CA, et al. The effect of increased loads of dissolved organic matter on estuarine microbial community composition and function. Front Microbiol. 2017;8:1272–15. doi: 10.3389/fmicb.2017.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landa M, Cottrell MT, Kirchman DL, Kaiser K, Medeiros PM, Tremblay L, et al. Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ Microbiol. 2014;16:1668–81. doi: 10.1111/1462-2920.12242. [DOI] [PubMed] [Google Scholar]
- 24.Waterbury JB, Watson SW, Valois FW, Franks DG. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can Bull Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 25.Wang K, Chen F. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ Microbiol. 2008;10:300–12. doi: 10.1111/j.1462-2920.2007.01452.x. [DOI] [PubMed] [Google Scholar]
- 26.Dittmar T, Koch B, Hertkorn N. A simple and efficient method for the solid‐phase extraction of dissolved organic matter (SPE‐DOM) from seawater. Limnol Oceanogr Methods. 2008;6:230–6. doi: 10.4319/lom.2008.6.230. [DOI] [Google Scholar]
- 27.Jiao NZ, Yang YH, Koshikawa H, Watanabe M. Influence of hydrographic conditions on picoplankton distribution in the East China Sea. Aquat Microbial Ecol. 2002;30:37–48. doi: 10.3354/ame030037. [DOI] [Google Scholar]
- 28.Liang Y, Li L, Luo TW, Zhang Y, Zhang R, Jiao NZ. Horizontal and vertical distribution of marine virioplankton: a basin scale investigation based on a global cruise. PLoS ONE. 2014;9:e111634–12. doi: 10.1371/journal.pone.0111634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kind T, Fiehn O. Seven golden rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Krevelen DW. Graphical-statistical method for the study of structure and reaction processes of coal. Fuel. 1950;29:269–84. [Google Scholar]
- 33.Ma X, Coleman ML, Waldbauer JR. Distinct molecular signatures in dissolved organic matter produced by viral lysis of marine cyanobacteria. Environ Microbiol. 2018;20:3001–11. doi: 10.1111/1462-2920.14338. [DOI] [PubMed] [Google Scholar]
- 34.Hansman RL, Dittmar T, Herndl GJ. Conservation of dissolved organic matter molecular composition during mixing of the deep water masses of the northeast Atlantic Ocean. Mar Chem. 2015;177:288–97. doi: 10.1016/j.marchem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao N, Azam F. Microbial carbon pump and its significance for carbon sequestration in the ocean. In: Jiao N, et al., editors. Microbial carbon pump in the ocean. Washington, DC: Science/AAAS; 2011.
- 36.Lucas J, Koester I, Wichels A, Niggemann J, Dittmar T, Callies U, et al. Short-term dynamics of North Sea bacterioplankton-dissolved organic matter coherence on molecular level. Front Microbiol. 2016;7:321. doi: 10.3389/fmicb.2016.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson CA, Ducklow HW. Growth of bacterioplankton and consumption of dissolved organic carbon in the Sargasso Sea. Aquat Microbial Ecol. 1996;10:69–85. doi: 10.3354/ame010069. [DOI] [Google Scholar]
- 38.Cottrell MT, Kirchman DL. Natural assemblages of marine Proteobacteria and members of the cytophaga-flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–7. doi: 10.1128/AEM.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elifantz H, Malmstrom RR, Cottrell MT, Kirchman DL. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware estuary. Appl Environ Microbiol. 2005;71:7799–805. doi: 10.1128/AEM.71.12.7799-7805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79:2012–20. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the bacteroidetes sus-like paradigm. J Biol Chem. 2009;284:24673–7. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glauner B, Höltje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–95. [PubMed] [Google Scholar]
- 43.Hoiczyk E, Hansel A. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriolog. 2000;182:1191–9. doi: 10.1128/JB.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappé MS, Kemp PF, Giovannoni SJ. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–26. doi: 10.4319/lo.1997.42.5.0811. [DOI] [Google Scholar]
- 45.Glöckner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–6. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarmento H, Gasol JM. Use of phytoplankton‐derived dissolved organic carbon by different types of bacterioplankton. Environ Microbiol. 2012;14:2348–60. doi: 10.1111/j.1462-2920.2012.02787.x. [DOI] [PubMed] [Google Scholar]
- 47.Nikrad MP, Cottrell MT, Kirchman DL. Uptake of dissolved organic carbon by gammaproteobacterial subgroups in coastal waters of the West Antarctic Peninsula. Appl Environ Microbiol. 2014;80:3362–8. doi: 10.1128/AEM.00121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grzymski JJ, Carter BJ, DeLong EF, Feldman RA, Ghadiri A, Murray AE. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl Environ Microbiol. 2006;72:1532–41. doi: 10.1128/AEM.72.2.1532-1541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 50.Becker JW, Berube PM, Follett CL, Waterbury JB, Chisholm SW, DeLong EF, et al. Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol. 2014;5:111. doi: 10.3389/fmicb.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.