Abstract
Aggregatibacter and Haemophilus species are relevant human commensals and opportunistic pathogens. Consequently, their bacteriophages may have significant impact on human microbial ecology and pathologies. Our aim was to reveal the prevalence and diversity of bacteriophages infecting Aggregatibacter and Haemophilus species that colonize the human body. Genome mining with comparative genomics, screening of clinical isolates, and profiling of metagenomes allowed characterization of 346 phages grouped in 52 clusters and 18 superclusters. Less than 10% of the identified phage clusters were represented by previously characterized phages. Prophage diversity patterns varied significantly for different phage types, host clades, and environmental niches. A more diverse phage community lysogenizes Haemophilus influenzae and Haemophilus parainfluenzae strains than Aggregatibacter actinomycetemcomitans and “Haemophilus ducreyi”. Co-infections occurred more often in “H. ducreyi”. Phages from Aggregatibacter actinomycetemcomitans preferably lysogenized strains of specific serotype. Prophage patterns shared by subspecies clades of different bacterial species suggest similar ecoevolutionary drivers. Changes in frequencies of DNA uptake signal sequences and guanine–cytosine content reflect phage-host long-term coevolution. Aggregatibacter and Haemophilus phages were prevalent at multiple oral sites. Together, these findings should help exploring the ecoevolutionary forces shaping virus-host interactions in the human microbiome. Putative lytic phages, especially phiKZ-like, may provide new therapeutic options.
Subject terms: Bacteriophages, Bacterial genetics, Phylogenetics, Microbial ecology, Biodiversity
Introduction
Aggregatibacter and Haemophilus species of the family Pasteurellaceae encompass commensals and pathogens of considerable ecological and medical importance [1]. They dominate mucosal surfaces in the oral cavity and pharynx as well as contributing to dental biofilm development [2–5]. Haemophilus parainfluenzae is among the most abundant and transcriptionally active species across all oral and pharyngeal sites [3–8]. Haemophilus influenzae is the most important human opportunistic pathogen among Pasteurellaceae species [1]. Although vaccination against H. influenzae serotype b dramatically reduced the burden of invasive H. influenzae diseases, unvaccinated children and patients with predisposing conditions are still at risk. H. influenzae strains that form no capsule may cause common pediatric infections as well as acute exacerbation of chronic obstructive pulmonary disease in adults. “Haemophilus ducreyi” is the cause of chancroid, a sexually transmitted disease characterized by genital ulcerations, and cutaneous ulcers in children [1]. Aggregatibacter species have been linked to periodontal disease, infective endocarditis, and extra-oral abscesses [1].
Relatively little is known about evolutionary and ecological drivers that control populations of Aggregatibacter and Haemophilus species. Bacteriophages may be candidates for such force [9–12]. These bacterial viruses, in short phages, are either obligatory parasites with strictly lytic lifestyle or they also act as mutualists and exhibit a temperate lifestyle [13]. Phage and bacterium exert reciprocal selective pressures on each other leading to rapid adaptation of both of them [12, 14, 15]. On the one hand, phages may decimate a bacterial population or even bring it to extinction; on the other, they may transfer genetic traits improving ecological fitness [10] or mediate defense against other phages [16]. Therefore, phages infecting Aggregatibacter and Haemophilus species are likely contributors to the control of their host populations and may indirectly influence the human microbiome and potentially human health.
Despite the high medical and ecological importance of Aggregatibacter and Haemophilus species, the population of phages infecting these species has not been widely explored [17, 18]. Although the first Haemophilus phage was reported in early 1960s [19], so far only three types of H. influenzae phages have been reported. These were (i) five P2-like phages: HP1, HP2, S2A, S2B, and S2C, (ii) transposable Mu-like FluMu phage, and (iii) unclassified siphovirus N3 [20, 21]. These P2-like and Mu-like phages represent the family Myoviridae. Exploration of Aggregatibacter actinomycetemcomitans phages started with induction and characterization of lambda-like phiAal7/phiAa phage [22]. Later, dozens of related Aggregatibacter myoviruses were detected and genetics of two, Aaphi23 and S1249, were revealed as reviewed in ref. [18]. Although lysogeny has not been linked to clinical conditions, in vitro studies showed that Aggregatibacter myoviruses can transfer antibiotic resistance cassettes, induce serotype conversion, and potentially increase release of leukotoxin [18]. Aggregatibacter siphoviruses and podoviruses were occasionally observed but have not been characterized. Virulent phages infecting A. actinomycetemcomitans or H. influenzae have not been reported. Phages for the other human-associated Aggregatibacter and Haemophilus species are either not known or are represented by unclassified putative phage sequences. Putative virulent phiKZ-like Haemophilus phages were detected by metagenomics [23] but this type of phages is also known to show pseudolysogeny [24]. Current taxonomy of Aggregatibacter and Haemophilus phages curated by the International Committee on Taxonomy of Viruses is limited to only two species: Haemophilus virus HP1 and HP2 both classified in genus Hp1virus, subfamily Peduovirinae, family Myoviridae and order Caudovirales (see Table S1 in the Supplementary Information for more details). Thus, prevalence, diversity and genetics of Aggregatibacter and Haemophilus phages remained largely unknown, and consequently the impact of these phages on the human microbiome could not be fully explored until now.
In the present study, we explored three main datasets to reveal the prevalence and diversity of Aggregatibacter and Haemophilus phages with focus on temperate, tailed, double-stranded DNA phages. First, we described the prophages found in publicly available genomes of human-associated Pasteurellaceae species [25], and established a classification scheme for these phages. Subsequently, we detected prophages in genomes of clinical isolates of A. actinomycetemcomitans and obtained full-genome sequences for representative prophages. Finally, we detected phages in publicly available human microbiome metagenomes [26] and classified metagenomic phage assemblies from geographically diverse samples [23, 27]. By linking various metadata to classified phage sequences, we could show how phage diversity differs across clades and niches, and how phages co-evolve with their host. Our data laid the foundation for a comprehensive definition of the population structure of phages infecting Aggregatibacter and Haemophilus species that can be further used to explore the ecological role of these phages in the human microbiome and develop new therapeutic approaches.
Results
We screened 276 publicly available genomes of 14 human-associated Aggregatibacter and Haemophilus species for phage-like elements (Fig. S1a). In total, 828 phage-like elements were discovered, 258 of which were predicted to be intact, coding for at least 40 open reading frames (ORFs), and not duplicated within the dataset—hereafter simply referred to as “prophages” (Fig. S1b; Table S2). We chose this ORF number cutoff based on results of alignments and clustering where obviously truncated prophage sequences formed separate groups. Prophages found in 11 of 14 bacterial species, had, on average, the size of 38.8 kb [±1.2, 95% confidence interval (CI)], and coded for 53 (±1, 95% CI) proteins. Evolutionary relationships of prophages were studied at DNA and protein level to cover both close and more distant relations, respectively. We used the presence and position of conserved domains in phage genomes to identify phylotype-specific marker proteins. Multiple complementary analyses gave consistent results and created a framework for phage phylogeny.
Phylogenetic diversity of prophages revealed by comparative genomics
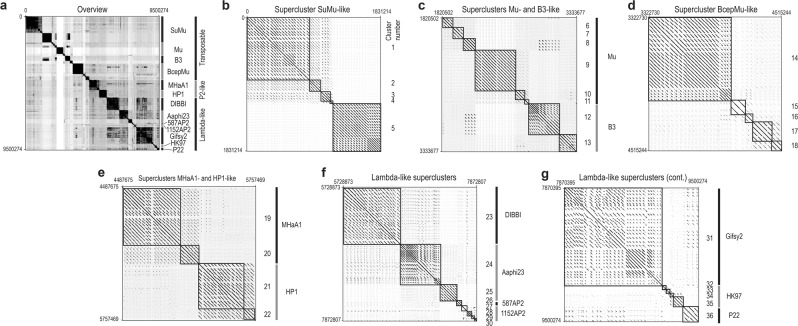
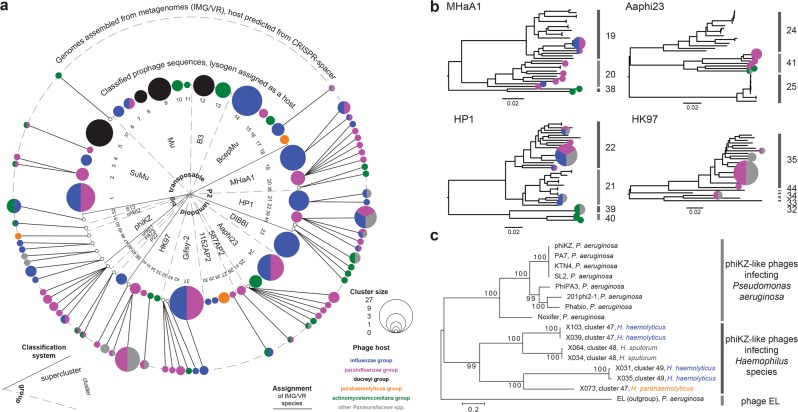
Prophages were first stratified into groups based on two complementary approaches: (i) Genome sequences of prophages were aligned and clustered based on shared DNA content [28]. (ii) Predicted coding sequences from the phage genomes were used as a query against the Prophage/Virus Database (version from Aug 3, 2017) to identify the best matching phages [29]. Both approaches gave consistent groupings. Next, genome sequences of prophages were compared within stratified groups, i.e., individually for each supercluster, using the Genome BLAST Distance Phylogeny method via VICTOR [30], and manually curated. The relationship between genomes was visualized with a dot plot (Fig. 1). In total, 36 clusters of phage genomes were assigned to 13 superclusters and three main groups (Fig. 1a). Transposable phages, that are characterized by transpositional mode of replication [31], were divided into four superclusters (Fig. 1b–d). P2-like phages, recognized by the presence of a Q-P-O-N-M-L gene cluster, in which terminase genes (P and M) are located in reverse order and interspaced with scaffold- and capsid encoded genes [32], were distributed between two superclusters (Fig. 1e). Lambda-like phages, i.e., a diverse group of myo- and siphoviruses that somehow resemble phage lambda and have highly mosaic genomes [33, 34], were grouped into seven superclusters (Fig. 1f, g). Average nucleotide identity of sequences grouped in clusters ranged between 88.5 and 100%, whereas average Mash distances ranged between 0 and 0.082, as assessed by OrthoANI [35] and Mash [36], respectively (Table 1). There was a positive correlation between average Mash distances and (1-ANI), Pearson’s r = 0.83. Cluster sizes differed considerably and exchange of genome fragments between clusters was noticed. As phage mosaicism can pose a problem for whole-genome phylogenies, we looked for marker proteins that can be used to evaluate our classification.
Fig. 1.
Diversity of prophages assessed by DNA similarity matrix analysis. a Overview dot plot of 243 prophage sequences arranged in groups, superclusters and clusters. Dot plot compared sorted and merged sequences of prophages (combined length of 9.5 megabase) on the x-axis, and the same collection of sequences on the y-axis of a plot. When the DNA residues of both sequences match at the same location on the plot, a dot is drawn at the corresponding position. Once the dots have been plotted, they will combine to form lines, and dense groups of lines will form black squares that correspond to clusters of similar genomes. The bigger the cluster, the higher the prevalence of prophages belonging to that cluster. The main diagonal represents the sequence’s alignment with itself; lines off the main diagonal but around it represent similar patterns within the closely related phage genomes. Signals found more far from the diagonal represents similar patterns within more distantly related phage genomes. Both halves of the graph created by the diagonal provide the identical information but we decided to keep them both for plot clarity. Enlarged sections for phages assigned to supercluster SuMu-like (b), Mu- and B3-like (c), BcepMu-like (d), MHaA1- and HP1-like (e), and lambda-like supercluster (f, g) are shown. Clusters (numbers 1–36) were highlighted with black frames. Superclusters and main groups are labeled and indicated by black or gray stripes. DNA coordinates for merged sequence are given in plots corners
Table 1.
Characteristics of phage clusters
| Supercluster | Cluster number | Prophages | Assemblies | OrthoANI (mean ± s.d.) | Mash distance (mean ± s.d.) | Predicted tail morphology | Taxonomy of lysogens | Most related phages | Reported phages | Comments | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SuMu-like | 1 | 20 | 0 | 93.9 ± 3.3 | 0.048 ± 0.02 | Contractile | H. aegyptius; H. haemolyticus; H. influenzae; H. parainfluenzae | Glaesserella phage SuMu; Mannheimia phage 3927AP2 | |||
| 2 | 4 | 2 | 92.4 ± 3.7 | 0.05 ± 0.025 | Contractile | H. parainfluanzae | Glaesserella phage SuMu; Mannheimia phage 3927AP2 | ||||
| 3 | 3 | 0 | 100 ± 0.0 | 0.002 ± 0.002 | Contractile | H. influenzae | Glaesserella phage SuMu; Mannheimia phage 3927AP2 | ||||
| 4 | 1 | 1 | – | – | Contractile | H. parainfluenzae | Glaesserella phage SuMu; Mannheimia phage 3927AP2 | ||||
| 5 | 18 | 0 | 90.9 ± 0.1 | 0.01 ± 0.008 | Flexible | H. ducreyi | Glaesserella phage SuMu; Mannheimia phage 3927AP2 | DNA homology with phages from clusters 8 and 12 | Gangaiah et al., [70] | ||
| 37 | 0 | 1 | – | – | Flexible | – | Glaesserella phage SuMu; Mannheimia phage 3927AP2 | Paez-Espino et al., [27] | |||
| Mu-like | 6 | 3 | 0 | 95.1 ± 4.3 | 0.038 ± 0.033 | Contractile | H. haemolyticus; H. influenzae | Enterobacteria phages Mu and SfMu; Escherichia phage D108 | |||
| 7 | 3 | 0 | 93.9 ± 3.3 | 0.05 ± 0.041 | Contractile | H. influenzae; H. parainfluenzae | Enterobacteria phages Mu and SfMu; Escherichia phage D108 | FluMu | Morgan et al., [21] | ||
| 8 | 4 | 0 | 100 ± 0.0 | 0.001 ± 0.001 | Flexible | H. ducreyi | Enterobacteria phages Mu and SfMu; Escherichia phage D108 | DNA homology with phages from clusters 5 and 12 | Gangaiah et al., [70] | ||
| 9 | 13 | 0 | 99.9 ± 0.1 | 0.001 ± 0.001 | Flexible | H. ducreyi | Enterobacteria phages Mu and SfMu; Escherichia phage D108 | Gangaiah et al., [70] | |||
| 10 | 3 | 0 | 91.3 ± 6.7 | 0.067 ± 0.052 | Flexible | A. actinomycetemcomitans | Enterobacteria phages Mu and SfMu; Escherichia phage D108 | ||||
| 11 | 1 | 0 | – | – | Flexible | A. aphrophilus | Enterobacteria phages Mu and SfMu; Escherichia phage D108 | ||||
| B3-like | 12 | 11 | 0 | 93.7 ± 6.1 | 0.058 ± 0.05 | Flexible | H. ducreyi | Pseudomonas phages B3 and PM105 | DNA homology with phages from clusters 5 and 8 | Gangaiah et al., [70] | |
| 13 | 6 | 0 | 90.0 ± 6.2 | 0.082 ± 0.051 | Flexible | A. actinomycetemcomitans; A. sp. HMT-458 | Pseudomonas phages B3 and PM105 | ||||
| BcepMu-like | 14 | 22 | 0 | 98.7 ± 1.7 | 0.021 ± 0.021 | Contractile | H. influenzae | Burkholderia phages BcepMu and φE255 | |||
| 15 | 2 | 0 | 100 | – | Contractile | H. parainfluenzae | Burkholderia phages BcepMu and φE255 | ||||
| 16 | 2 | 0 | 88.5 | – | Contractile | A. aphrophilus/segnis | Burkholderia phages BcepMu and φE255 | ||||
| 17 | 4 | 2 | 99.2 ± 0.6 | 0.008 ± 0.005 | Contractile | H. influenzae | Burkholderia phages BcepMu and φE255 | ||||
| 18 | 2 | 0 | 100 | – | Contractile | H. paraphrohaemolyticus | Burkholderia phages BcepMu and φE255 | ||||
| MHaA1-like | 19 | 16 | 5 | 93.7 ± 2.4 | 0.054 ± 0.02 | Contractile | H. aegyptius; H. haemolyticus; H. influenzae | Mannheimia phages 587AP1 and φMHaA1 | |||
| 20 | 5 | 7 | 92.3 ± 2.8 | 0.054 ± 0.018 | Contractile | H. parainfluenzae | Mannheimia phages 587AP1 and φMHaA1 | ||||
| 38 | 0 | 2 | – | – | Contractile | – | Mannheimia phages 587AP1 and φMHaA1 | Paez-Espino et al., [27] | |||
| HP1-like | 21 | 10 | 3 | 95.5 ± 2.5 | 0.043 ± 0.018 | Contractile | H. influenzae | Haemophilus phages HP1 and HP2 | HP1, HP2 | Harm and Rupert, [19]; Williams et al., [20] | |
| 22 | 3 | 16 | 90.9 ± 1.2 | 0.066 ± 0.019 | Contractile | H. parainfluenzae | Haemophilus phages HP1 and HP2 | One of phages has head morphogenesis protein resembling one from Pasteurella phage F108 | |||
| 39 | 0 | 3 | – | – | Contractile | – | Haemophilus phages HP1 and HP2 | Paez-Espino et al., [27] | |||
| 40 | 0 | 3 | – | – | Contractile | – | Haemophilus phages HP1 and HP2 | Paez-Espino et al., [27] | |||
| DIBBI-like | 23 | 17 | 0 | 97.3 ± 1.3$ | 0.043 ± 0.017 | – | H. aegyptius; H. influenzae | Aggregatibacter phages Aaphi23 and S1249 | OrthoANI indicated three outliers, DNA homology with some phages from cluster 31 | ||
| Aaphi23-like | 24 | 17 | 0 | 94.2 ± 3.5 | 0.07 ± 0.038 | – | H. aegyptius; H. haemolyticus; H. influenzae; H. parainfluenzae | Aggregatibacter phages Aaphi23 and S1249 | |||
| 25 | 4 | 0 | 97.1 ± 1.6 | 0.029 ± 0.015 | Contractile | A. actinomycetemcomitans | Aggregatibacter phages Aaphi23 and S1249 | Aaphi23, S1249 | Resch et al., [34]; Chen et al., [62] | ||
| 41 | 0 | 8 | – | – | – | Aggregatibacter phages Aaphi23 and S1249 | Two phages have similar head morphogenesis protein like phages from cluster 24 | Paez-Espino et al., [27] | |||
| 587AP2-like | 26 | 2 | 1 | 99.9 | 0.000 | – | H. parainfluenzae | Mannheimia phage 587AP2 | |||
| 1152AP2-like | 27 | 1 | 2 | – | – | – | H. parainfluenzae | Mannheimia phage 1152AP2 | |||
| 28 | 3 | 0 | 95.8 ± 3.8 | 0.06 ± 0.051 | Flexible | H. paraphrohaemolyticus; H. parahaemolyticus | Mannheimia phage 1152AP2 | ||||
| 29 | 1 | 0 | – | – | – | H. haemolyticus | Mannheimia phage 1152AP2 | ||||
| 30 | 1 | 0 | – | – | – | H. haemolyticus | Mannheimia phage 1152AP2 | ||||
| Gifsy2-like | 31 | 28 | 2 | 93.6 ± 4.7 | 0.057 ± 0.036 | Flexible | H. haemolyticus; H. influenzae; H. paraphrohaemolyticus; H. parainfluenzae | Multiple phages | Some show DNA homology with phages from cluster 23 | ||
| 42 | 0 | 1 | – | – | Flexible | – | Mannheimia phages 587AP2 and 535AP2 | Paez-Espino et al., [27] | |||
| 43 | 0 | 5 | – | – | Flexible | – | Mannheimia phages 587AP2 and 1152AP2 | Paez-Espino et al., [27] | |||
| HK97-like | 32 | 1 | 0 | – | – | Flexible | A. aphrophilus | Mannheimia phage 1152AP2 | |||
| 33 | 1 | 1 | – | – | Flexible? | A. segnis | Mannheimia phage 535AP2 | ||||
| 34 | 1 | 0 | – | – | Flexible | H. parainfluenzae | Mannheimia phage 1152AP2 | ||||
| 35 | 3 | 18 | 93.8 ± 4.7 | 0.046 ± 0.003 | Flexible | H. parainfluenzae | Mannheimia phage 587AP2 | ||||
| 44 | 0 | 1 | – | – | Flexible | – | Pseudomonas phages PA73 and SCH_Ab26 | Paez-Espino et al., [27] | |||
| P22-like | 36 | 3 | 0 | – | 0.024 ± 0.019 | – | H. influanzae | Mannheimia phage 587AP2 | |||
| PA73-like | 45 | 0 | 2 | – | – | Flexible | – | Mannheimia phage 587AP2 | Potentially defective | Paez-Espino et al., [27] | |
| unknown 1 | 46 | 0 | 6 | – | – | – | – | Mannheimia phage 587AP2 | Potentially defective | Paez-Espino et al., [27] | |
| phiKZ-like | 47 | 0 | 4 | – | – | – | – | Pseudomonas phage phiKZ | Genome size of 279 kbp, likely lytic, pseudolysogeny possible | Paez-Espino et al., [27] | |
| 48 | 0 | 2 | – | – | – | – | Pseudomonas phage phiKZ | Genome size of 241–254 kbp, likely lytic, pseudolysogeny possible | Paez-Espino et al., [27] | ||
| 49 | 0 | 2 | – | – | – | – | Pseudomonas phage phiKZ | Genome size of 220 kbp, likely lytic, pseudolysogeny possible | Paez-Espino et al., [27] | ||
| 50 | 0 | 1 | – | – | – | – | Pseudomonas phage phiKZ | Genome size of 211 kbp, likely lytic, pseudolysogeny possible | Paez-Espino et al., [27] | ||
| unknown 2 | 51 | 0 | 1 | – | – | – | – | Serratia phage phiMAM13 | Genome size of 190 kbp | Paez-Espino et al., [27] | |
| S13-like | 52 | 0 | 5 | – | – | – | – | Cronobacter phage S13 | Genome size of 151–152 kbp, likely lytic | Paez-Espino et al., [27] |
Phylogenetic diversity of prophages assessed using marker proteins
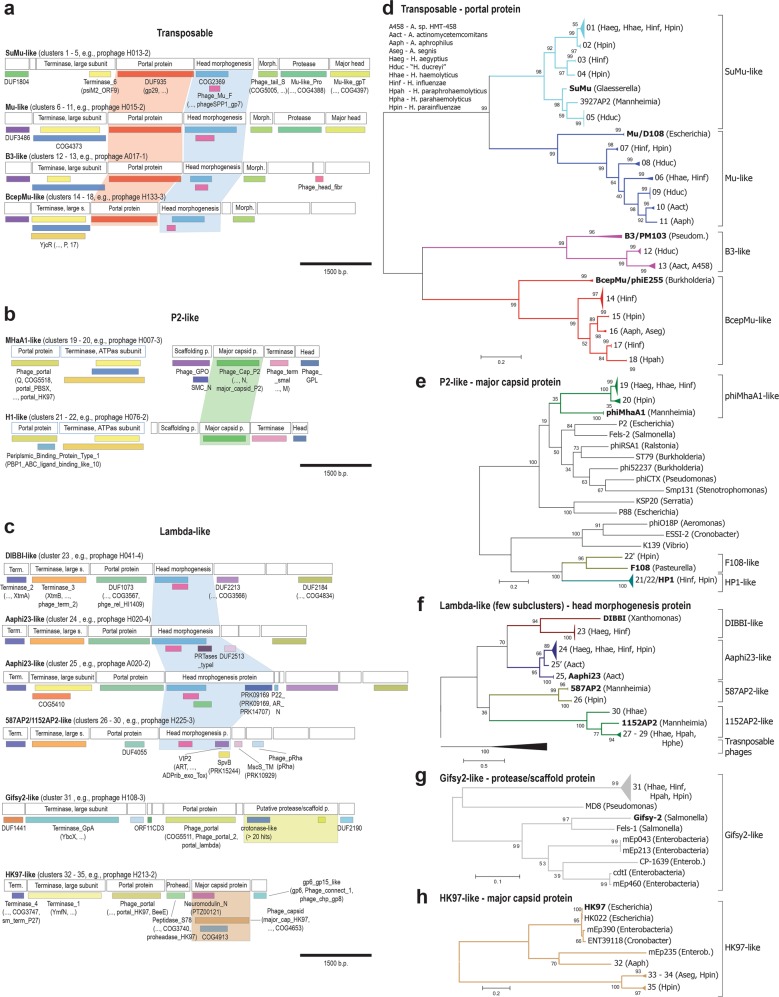
Phage genome sequences were screened for open reading frames. Genetics of these prophages will be described in detail elsewhere; here we screened the genomes for gene arrangements that were conserved for supercluster members and for marker proteins that could be used for phylogenetic profiling. We found the genome segment coding for proteins involved in head morphogenesis and DNA packaging (e.g., portal protein) to be the most suitable (Fig. 2a–c) because it is easy to identify and essential for the functionality of virions (i.e., entire phage particles). Our selection is in good agreement with previous comprehensive phylogenetic studies [31–33, 37]. Multiple markers were used because there is no single phage protein that has homologs in all phage genomes [38]. Next, we used the marker proteins to study the evolutionary relationship between newly reported and reference phages (Fig. 2d–h). Well-supported clusters and superclusters were observed. The findings were generally consistent with the results of the DNA-based full-genome analysis. One of the very rare exceptions was, for example, a recombination event where a HP1-like capsid gene (cluster 22) was replaced by one of F108-type (Fig. 2e). Such recombination events disrupt correlation between biomarker and cluster relationship (Table 1). To control for gene shuffling, we performed comparative analyses applying multiple biomarkers independently (data not shown). Occasionally, closely related clusters were grouped together (Fig. 2e, f, h), indicating that evolutionary relationship at fine phylogenetic level is better captured by analysis at whole-genome DNA level. We then compared our phylogenetic dataset with publicly available sequences of phages infecting Aggregatibacter and Haemophilus species [27, 39, 40] and discovered that only 10% of our phage clusters contain phage species that have been reported before (Table 1, for more details see Table S1).
Fig. 2.
Diversity of prophages assessed by marker protein-based phylogeny. Genome segments coding for head morphogenesis and DNA packaging are shown for transposable (a), P2-like (b), and lambda-like (c) phages. Conserved domains are indicated and conserved marker proteins are highlighted. Blue frames in b indicate transcription in opposite direction (from right to left). The phylogenetic trees for transposable (d), P2-like (e), and lambda-like (f, g, h) phages were constructed. Reference sequences were included [32, 33, 99]. The evolutionary history was inferred by using the Maximum Likelihood method. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Trees collapsed at cluster level. Number and length of studied sequences were as follows: 127 sequences with 324 amino acid positions (d), 55 with 300 aa (e), 63 with 131 aa (f), 37 with 554 aa (g), and 11 with 354 aa (h). Branches were colored to highlight superclusters. Abbreviated host taxonomy is given in brackets for each cluster. Full species names are listed in d
Diversity patterns of phages across species and subspecies clades
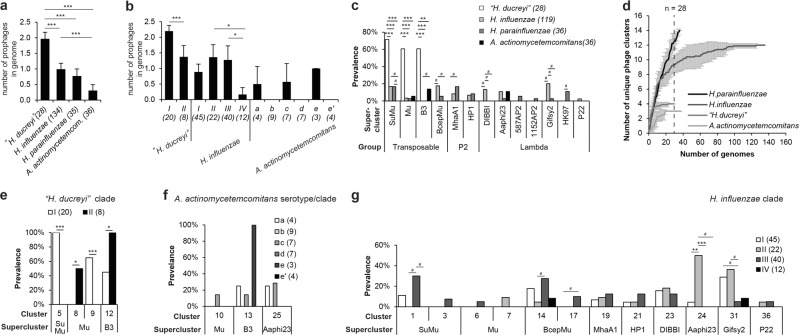
We uncovered the quantitative distribution of prophages across different host phylotypes. Because of the unequal distribution of available genomes, we focused on four well-represented species, i.e., A. actinomycetemcomitans, “H. ducreyi”, H. influenzae, and H. parainfluenzae as well as the subspecies clades of the first three species. Clades, i.e., discrete population structure at subspecies resolution (Table S2), were previously inferred from whole-genome sequence data [41–43]. Genomes of “H. ducreyi” harbor, on average, 2–6 times more prophages than other species (Fig. 3a). H. influenzae and H. parainfluenzae showed intermediate values. Differences in prophage prevalence were observed at subspecies resolution as well (Fig. 3b).
Fig. 3.
Prevalence and diversity patterns of phages across species and subspecies clades. a Number of prophages per genome plotted for four species that were best represented in genome database. b Number of prophages per genome plotted for subspecies clades of “H. ducreyi”, H. influenzae, and A. actinomycetemcomitans. Discrete population structure at subspecies resolution was inferred from whole-genome sequences [41–43]. c Prevalences of phage superclusters were plotted across four species. d Rarefaction curves were constructed to assess phage cluster richness from the results of sampling the genomes. Mean values are plotted and errors bars represent 95% CIs. The smallest sample size is indicated by a vertical dashed line. Prevalence of phage clusters across subspecies clades was plotted for “H ducreyi” (e), A. actinomycetemcomitans (f), and H. influenzae (g). Bar graph and error bars show mean value and 95 % CI, respectively. Number of observations per group is given in brackets. In a and b, significant differences between group means was detected by one-way ANOVA with post hoc Tukey test. In c and e–g, Fisher’s exact test (two-sided) was used to analyze the significance of the association between the phage presence of phage (grouped in superclusters or clusters) and host clades. Bonferroni correction was applied to compensate for multiple comparisons. The number of observations is given in brackets following the clade name. Symbols ***, **, *, and # indicated p < 0.001, p < 0.01, p < 0.05, and significant p value prior to Bonferroni correction, respectively
We then explored the qualitative distribution of phage groups, superclusters, and clusters across bacterial species and subspecies (Fig. 3c–g). Phages from all three main groups (i.e., transposable, P2-like, and lambda-like) lysogenized H. influenzae and H. parainfluenzae (Fig. 3c). A. actinomycetemcomitans was not lysogenized by P2-like phages, while “H. ducreyi” was exclusively lysogenized by transposable prophages. We observed interesting patterns in prophage distribution at supercluster level across bacterial species (Fig. 3c). The most striking observation was a very high prevalence of prophages from three transposable superclusters in genomes of “H. ducreyi”. The profiles for H. influenzae and H. parainfluenzae were generally similar, except for the distribution of DIBBI- and Gifsy2-like prophages, which were less prevalent in H. parainfluenzae. The Aaphi23-like supercluster was the only lambda-like supercluster represented in A. actinomycetemcomitans strains in contrast to H. influenzae and H. parainfluenzae strains that hosted prophages from multiple superclusters.
Subsequently, we looked at patterns of phage clusters and bacterial species at finer phylogenetic resolution. We first constructed rarefaction curves to assess phage richness from the results of sampling the genomes of selected species (Fig. 3d). This curve is a plot of the number of phage clusters as a function of the number of bacterial genomes studied. We decided to use genus-like clusters (as opposed to species-like viral operational taxonomic units, in short vOTUs) to effectively remove redundancy, although at a cost of diversity underestimation. Rarefaction curves generally grew rapidly at first, as the most common phage clusters were found, but plateaued (for all species but H. parainfluenzae) as only the rarest phage clusters remained to be sampled. Phage cluster richness (which we take as an approximate equivalent for phage genus richness) was high for H. influenzae and H. parainfluenzae and low for “H. ducreyi” and A. actinomycetemcomitans. H. parainfluenzae was lysogenized by the most taxonomically diverse group of phages, which was far from being fully explored in this dataset because of the limited number of available genome sequences.
Subspecies bacterial clades showing divergent traits are a sign of ongoing speciation. We wondered if, at such a fine phylogenetic resolution, we would be able to see additional patterns of phage diversity, which could shed light on a potential role of prophages in speciation processes. Indeed, we observed differences in the prevalences of phage clusters across such bacterial subspecies-like clades. Two clades of “H. ducreyi” were colonized by different transposable phage clusters (Fig. 3e). The effect was even more pronounced if truncated prophages were taken into account (Fig. S2). Clade-specific association was supported by balanced distribution of phage clusters across diseases and geographical origin (Fig. S2). Only half of A. actinomycetemcomitans serotypes/clades were lysogenized (Fig. 3f) but a high prevalence of cluster 13 prophages in serotype/clade e strains was noticed. H. influenzae clade II was associated with cluster 24-lysogeny (Fig. 3g). Strains from clade III were more often hosting transposable prophages from clusters 1, 14, and 17.
In summary, clear quantitative and qualitative differences in prophage profiles obtained for different clades suggest that either (i) phages piggybacked on hosts that outcompeted other variants in the process of natural selection, or (ii) phages were enriched due to either biological characteristics of their host clade (e.g., presence of specific receptor) and/or environmental factors shaping the occupied niche (e.g., favored specific phage variant that is able to co-exist with its host in a specific niche), or (iii) phages conferred a trait that is advantageous for their host, consequently contributing to their host’s success and, potentially, divergence, or (iv) a combination of factors favors the observed phage-host associations. Prophage patterns observed for A. actinomycetemcomitans strains could not be fully evaluated using the main dataset because of the small sample size. To address this problem, we included a second dataset built on a wide collection of clinical isolates.
Aggregatibacter phages lysogenize strains of specific serotype/clade and show different host range
A. actinomycetemcomitans is an opportunistic pathogen that can devote most of its transcriptional effort to produce virulence factors, including adhesins and toxins specific for human neutrophils [44]. We explored prophage diversity patterns using strain collections [45–48] that encompass 157 isolates representing different serotypes, countries of origin, and clinical conditions (Table S3 and Fig. S3a). We developed PCR tests for detection of prophages in A. actinomycetemcomitans genomes (Fig. S3b, c). In A. actinomycetemcomitans strains, we detected 45 prophages from cluster 25, eight from cluster 13, and 11 from cluster 10 (Fig. S3d). No prophages were detected in genomes of related Pasteurellaceae species: A. aphrophilus, A. segnis, H. influenzae, H. parahaemolyticus, H. parainfluenzae, and H. pittmaniae.
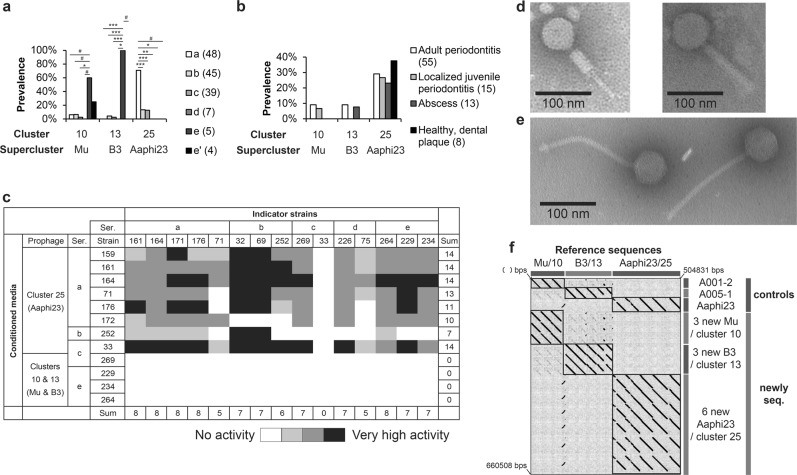
We then looked at phage patterns across Aggregatibacter serotypes/clades (Fig. 4a). Cluster 25 prophages were highly prevalent in serotype/clade a strains. All five serotype/clade e strains were lysogenized with cluster 13 phages, and three of them were co-infected by cluster 10 phages (Fig. 4a). No lysogens could be detected among seven serotype/clade d strains. Next, we searched for phage patterns across clinical conditions. Cluster 25 prophages were highly prevalent in strains isolated from various conditions as well as from dental biofilms of healthy individuals (Fig. 4b). In contrast, transposable phages (i.e. from clusters 10 and 13) lysogenized only strains isolated from diseases such as periodontitis.
Fig. 4.
Characterization of Aggregatibacter phages. Prevalence of phages representing three clusters in different A. actinomycetemcomitans serotypes/clades (a) and clinical groups (b). Fisher’s exact test (two-sided) was used to analyze the significance of the association between the presence of phages and either host lineages or diseases. Bonferroni correction was applied to compensate for multiple comparisons. The number of observations is given in brackets following the clade name. Symbols ***, **, *, and # indicated p < 0.001, p < 0.01, p < 0.05, and significant p value prior to Bonferroni correction, respectively. c Antimicrobial activity of conditioned media from A. actinomycetemcomitans cultures treated with Mitomycin C. Results of drop spot assay are presented. Sources of conditioned media are listed in rows while the indicator strains are given in columns. Strains were grouped by prophage type and serotype/clade. Ser. is an abbreviation for serotype/clade. Electron micrographs of Aaphi23-like phage with a contractile tail and transposable phage with flexible tail induced from A. actinomycetemcomitans are shown in d and e, respectively. f Overview dot plot of newly sequenced genomes arranged in clusters. Dot plot compared sorted and merged sequences of reference prophages on the x-axis, and the control and new sequences on the y-axis of the plot. More details about the dot plot method can be found in legend of Fig. 1. Control and new sequences are indicated by black and gray stripes, respectively. DNA coordinates for merged sequence are given in the corners
Subsequently, we explored the host range of Aggregatibacter phages. Prophages were induced from lysogens using Mitomycin C, and filtered conditioned media were tested for antimicrobial activity on A. actinomycetemcomitans indicator strains representing five serotypes (Fig. 4c). Conditioned media originating from strains harboring cluster 25 prophages in their genome showed broad antimicrobial activity. In contrast, samples from strains harboring transposable prophages from clusters 10 and 13 showed no activity. Super-sensitive indicator strains (SPS 32 and SPS 69) and super-resistant strain SPS 33 of A. actinomycetemcomitans were observed. Intriguingly, superinfection exclusion was observed in strains SPS 33 and SPS 71, but not fully functional in strains SPS 161 and SPS 164 and even absent in strain SPS 176. This may be a sign of genetic variation. Alternatively, lysis was caused indirectly, e.g., via bacteriocins or toxin induction. To address this problem, reference strains lysogenized with different phages have to be studied [49]. Virions characterized by contractile and flexible tails were detected in conditioned media from induced lysogens harboring cluster 25 and cluster 10/13 prophages, respectively (Fig. 4d, e). Full-genome sequencing performed for selected lysogens confirmed the presence of prophages (Fig. 4f; prophage sequences are provided in File S1) and identified mismatches in some of the PCR primers (Fig. S3e, f).
In summary, Aggregatibacter phages preferably lysogenize specific phylogenetic lineages [41, 46] and are not strongly correlated with specific clinical conditions. These phages show either an extremely narrow or a broad host range.
Phage patterns revealed across phylogenetic lineages made us wonder about their biogeographical distribution, i.e., niche tropism. We speculated that phages should undergo environmental selection and could, potentially, drive niche-association of their hosts. Therefore, we investigated the prevalence of Aggregatibacter and Haemophilus phages in human-associated microbiomes.
Aggregatibacter and Haemophilus phages are prevalent in oral metagenomes and show niche specificity
The National Institutes of Health Human Microbiome Project (HMP) has provided one of the broadest characterizations of human-associated microbiomes to date [8, 26]. We used the HMP metagenome dataset to study the prevalence of Aggregatibacter and Haemophilus species and their temperate phages in different sites of the human body. The human oral cavity was a major reservoir of Pasteurellaceae species. H. parainfluenzae was highly abundant and prevalent (Fig. S4a, b). Few other species were prevalent, but usually at low abundance.
By using marker protein sequences, the prevalence of phages (grouped to superclusters and clusters) was profiled at different sites of the human body. Aggregatibacter and Haemophilus phages were found exclusively in the oral cavity and pharynx. Data for the three best represented oral sites, i.e., buccal mucosa, gingiva, and tongue, is shown as, for other sites, only few samples were available (Fig. 5). Representatives of transposable superclusters SuMu and BcepMu were highly prevalent at all three sites (Fig. 5a), while Mu-like phages were rarely encountered. Among P2-like phages, both MhaA-like and HP1-like phages were quite prevalent. HK97-like phages were the most prevalent phages among lambda-like superclusters.
Fig. 5.
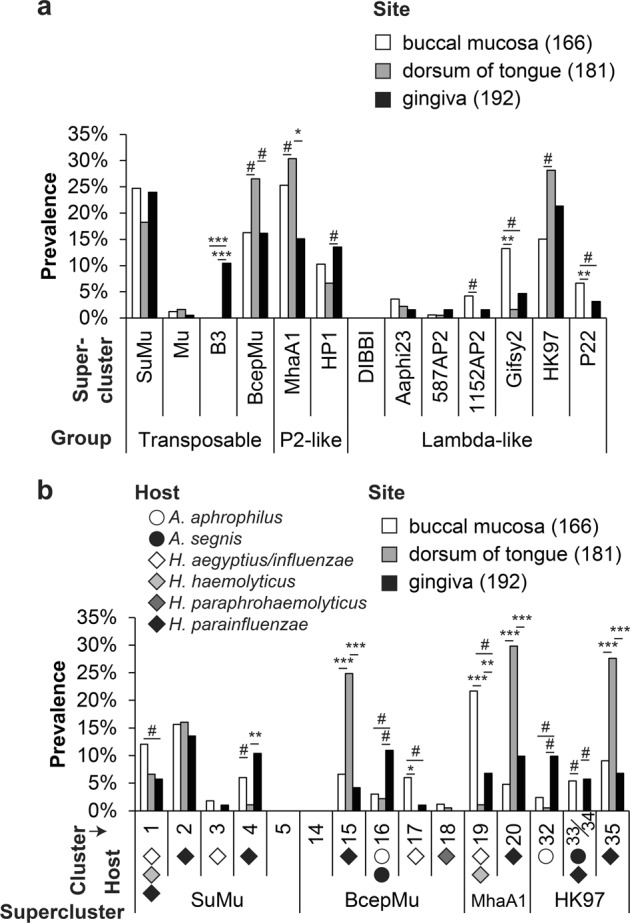
Prevalence of phage clusters among human microbiomes. Prevalence of phages grouped to superclusters (a) and clusters (b) at three oral sites, as assessed by metagenome analysis. Dataset from [26] was used. Predicted host of the phage is indicated in b. Fisher’s exact test (two-sided) was used to analyze the significance of the association between the phages and sites. Bonferroni correction was applied to compensate for multiple comparisons. The number of observations is given in brackets following the site name. Symbols ***, **, *, and # indicated p < 0.001, p < 0.01, p < 0.05, and significant p value prior to Bonferroni correction, respectively
Subsequently, we studied the phages of the four most prevalent superclusters, i.e., SuMu, BcepMu, MHaA1, and HK97, at higher phylogenetic resolution (Fig. 5b). SuMu-like cluster 2 phages were uniformly prevalent at all sites. In contrast, the representatives of BcepMu-like clusters 15 showed strong preference for the microbiota of the tongue. Among MhaA1-like clusters (19 and 20) and HK97-like clusters [35], a clear preference for either buccal mucosa or tongue was observed. As expected, most of the detected phages belong to clusters whose members infect H. parainfluenzae, H. haemolyticus, commensal Aggregatibacter species or have broader host range (Fig. 5b).
In summary, we measured the high prevalence of SuMu-, BcepMu-, MhaA1-, and HK97-like phages in human oral metagenomes. Niche tropism was commonly observed for phages at higher phylogenetic resolution. A diverse group of phages infecting H. parainfluenzae was observed.
Phage genomes assembled from metagenomes expanded the classification
The number and diversity of phage sequences extracted from metagenomes far exceeds that of phage isolates and could complement the prophage sequences retrieved from genomic databases [50]. High prevalence of phages observed in different oral niches prompted us to search the metagenomes for new sequences from Aggregatibacter and Haemophilus phages. IMG/VR is a database of cultured and uncultured DNA viruses and retroviruses that integrates the sequences with associated metadata, e.g., predicted host [27]. It contains over 760,000 viral genomes or genome fragments from: (i) publicly available isolate genomes, (ii) curated prophages, and, most important for us, (iii) ~8000 assembled metagenomes. We retrieved the metagenomic genome assemblies (version 2.0 from June 2018, accessed in February 2019) for all phages predicted to infect Aggregatibacter and/or Haemophilus species and classified them in the same way as the prophage sequences above.
In total, there were 115 IMG/VR singletons and 140 IMG/VR clusters encompassing 1090 viral contigs that were predicted to originate from phages infecting Aggregatibacter and Haemophilus species. Seven IMG/VR singletons and 53 IMG/VR clusters have at least one representative with a high quality draft genome (in total there were 108 such viral contigs). We assigned these high quality draft genomes to 14 superclusters and 28 clusters (Table 1, Table S1). Next, we compared the prophage dataset with the metagenome dataset (Fig. 6). The two approaches complemented each other: from a total of 52 clusters only 12 were shared, while 24 and 16 were unique for the prophage and metagenome datasets, respectively. Phages infecting important pathogens like A. actinomycetemcomitans, H. influenzae, and “H. ducreyi” were almost exclusively found in the prophage dataset (Fig. 6a, b), likely, because metagenomic studies poorly covered corresponding infections. Transposable phages were overrepresented in the prophage dataset (Fig. 6a), possibly, because metagenomic assemblies of transposable phages were less often categorized as high quality draft. On the other hand, the metagenome dataset was superior in providing sequences of phages infecting oral commensals. Aggregatibacter sp. HMT-458, H. haemolyticus, and H. parainfluenzae are poorly represented in the genome database but well covered by the HMP metagenomic study (Table S3). Finally, we found putative lytic phages characterized by big genome size (ranging from 151 to 279 kbp) exclusively in the metagenome dataset. Phages resembling Pseudomonas phage phiKZ (Fig. 6c) drew our special attention due to their potential therapeutic use [51].
Fig. 6.
Classification of viral assemblies from metagenomes. a Comparison between prophage dataset and metagenome dataset. The following information is provided (starting from inner ring): phage group, phage supercluster, phage cluster, size of clusters and corresponding host species in prophage dataset, size of clusters and corresponding host species in metagenome dataset [27]. Assignment of IMG/VR assemblies to clusters is indicated by solid line linkers. b Dendrograms based on full-genome comparison for selected clusters constructed with VICTOR [30]. New sequences introduced by metagenomes are indicated by circle graphs [same as in a]. c The phylogenetic tree for phiKZ-like phages was constructed using sequences of major capsid protein. The evolutionary history was inferred by using the Maximum Likelihood method. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site
In summary, metagenomic data complemented prophage screening. Addition of metagenomic assemblies to our classification system increased the number of phage genomes by 45%, created new clusters, and allowed cataloguing of putative lytic phages with therapeutic potential. Moreover, comparative analyses identified the bacterial species that are underrepresented in genome database and infectious diseases that have not been widely studied with metagenomics.
Phage-host coevolution revealed by frequencies of DNA uptake signal sequences (USSs) and guanine–cytosine (GC) content
Uncovering the vast diversity of phages made us wonder if we could track phage-host coevolution. In this context, we came across a unique property of Pasteurellaceae species and phages, namely the presence of DNA uptake signal sequences (USSs) in their genomes.
Pastuerallaceae species exhibit strong preferences for genomic DNA from very close or somehow close relatives, a self-specificity likely arising from the combined effects of biases in the DNA uptake machinery and genomic overrepresentation of the USSs that this machinery prefers [52–54]. USSs have multiple “dialects” with two main clades within the Pasteurellaceae family preferring two distinct sequence variants (i.e., Hin-USS and Apl-USS, standing for H. influenza-like and Actinobacillus pleuropneumoniae-like, respectively) and having corresponding variants enriched in their genomes. Notably, in genome sequences of H. influenzae isolates, the uptake sequences have a higher density in core genes than in accessory genes [55]. This suggests that uptake sequences accumulate slowly, since recently acquired accessory genes would not yet have accumulated them [52]. Consequently, the frequency of USSs in a prophage may be a good proxy for phage-host coevolution time. This hypothesis is further supported by the existence of both USS-rich and USS-poor prophages/phage-like elements [56, 57].
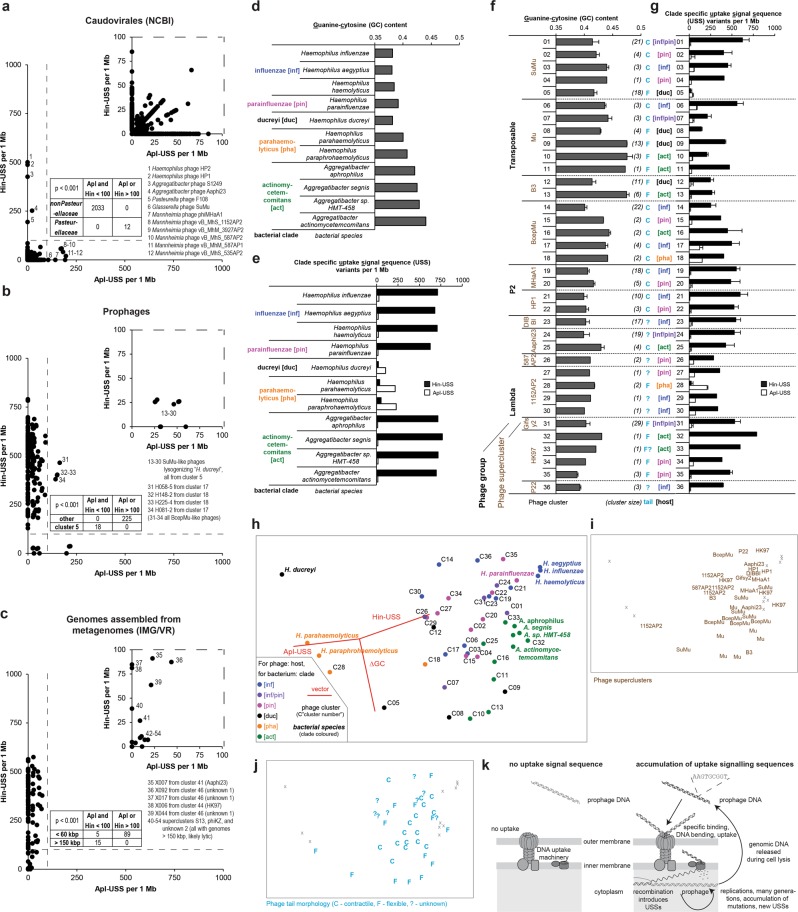
If the hypothesis holds true, phages known to lysogenize Pasteurellaceae species should have accumulated USSs in their genomes, all other phages not. Indeed, this is the case, as shown in Fig. 7a: USS-profiling of 2045 Caudovirales phage genomes [[39], accessed in February 2019)] correctly identified all the 12 phages lysogenizing Pasteurellaceae species (with a cutoff of either Hin- or Apl-USS frequency of 100, calculated per 1 megabase). Next, we profiled prophages identified in this study. As expected, almost all of them accumulated USSs (Fig. 7b). The only exception was the group of very closely related phages from cluster 5, suggesting that these phages were acquired by Pasteurellaceae more recently. Generally, Hin-dialect predominated, probably because the species using the Apl-dialect, e.g., H. parahaemolyticus (see below), were underrepresented in our study. Interestingly, four BcepMu-like phages (representing solely closely related clusters, either 17 or 18) seem to be “bilingual”, i.e., to accumulate both Hin- and Apl-USSs, which may suggest phage interaction with diverse hosts. Subsequently, we profiled high quality draft genomes [27] of Aggregatibacter and Haemophilus phages assembled from metagenomes (Fig. 7c). Again, almost all phages accumulated USSs and the Hin-dialect predominated. Among USS-poor phages we observed two distinct groups: (i) five phages located at boarder line mostly from supercluster Unknown 1, and (ii) all the phages with big (>150 kbp) genomes, all likely lytic.
Fig. 7.
Frequencies of DNA uptake signal sequences (USSs) and guanine–cytosine content (GC%) in genome sequences of Pasteurellaceae species and their phages. Frequencies of clade-specific uptake signal sequence (USS) variants in both orientations and guanine–cytosine content (GC%) were measured for genomes from diverse phages and human-associated Pasteurellaceae species. Frequencies are given per 1 Mb (i.e., 106 nt) of genome sequence. a Scatter plot showing frequencies of H. influenzae USS (Hin-USS) and Actinobacillus pleuropneumoniae USS (Apl-USS) in genomes of phages from order Caudovirales [25] on y- and x-axis, respectively. Enlarged fragment of plot covering low values is shown in top right corner. Contingency table summarizing studied groups is shown in the middle. Fisher’s exact test (two-sided) was used to analyze the significance of the association between the phage groups and presence of USSs at given cutoff. Phages characterized by noteworthy values are numbered and labeled. b like a but prophages classified in this study are presented. c like a but IMG/VR assemblies [27] are presented. d GC content was depicted for selected species grouped in clades based on genetic relationship inferred from concatenated nucleotide sequences (~2650 nt) of 16 S rRNA and three housekeeping (infB, pgi, recA) genes [1]. The subfamily clade labels were colored and abbreviated in square brackets throughout the figure. Genome sequences from either the representative National Center for Biotechnology Information (NCBI) strains or the type strains were retrieved from NCBI genome database. e Frequencies of Hin- and Apl-USS in the same genomes as in d. f GC content (mean ± s.d.) was depicted for phage clusters gathered in groups and superclusters (latter in brown throughout the figure). g Frequencies (mean ± s.d., per 1 Mb) of Hin-USS and Apl-USS in prophage sequences grouped like in f. For clusters depicted in f and g additional information is provided: the cluster size, which is the number of studied phages and written in parentheses, the predicted or confirmed morphology of phage tail written in light blue throughout the figure (C—contractile, F—flexible,?—unknown), and the abbreviated clade name for the phage host. h Ordination of bacterial species and phage clusters was constructed based on GC% and USS frequencies from d to g. The Bray-Curtis coefficient was calculated between every pair of samples using three variables: ΔGC (i.e., GC content reduced by the minimal GC in studied dataset), frequencies of Hin-USS, frequencies of Apl-USSs, each standardized by maximum (i.e., values were scaled so that their maxima across these three variables were always 100). Non-metric multidimensional scaling (nMDS) was used to represent the samples in two-dimensional space. Points were colored based on bacterial clades and phage host (labels starting with “C”). Superimposed is a vector plot for three variables (in red), with the vector direction for each variable reflecting the Pearson correlations of their values with the ordination axes, and length giving the multiple correlation coefficient from this linear regression on the ordination points. 2D-stress of 0.12 was observed. Same ordination was used in i and j. i Supercluster assignment was plotted for all phage clusters. Location of bacterial species is indicated by gray “x”. j Phage tail morphology for all phage clusters is given. k Mechanism of USS accumulation in prophages. Picture adapted from [52]
To get better resolution of phage-host coevolution and to track host change, we coupled USS-profiling with guanine–cytosine content measure (GC%). Bacterial hosts (grouped to subfamily clades for the sake of simplicity) showed a broad range of GC% (Fig. 7d) and used either Hin- or Apl-dialects (Fig. 7e). It is unclear what the source of considerable variation of GC% among members of the same family, in the majority sharing the same habitat, is. Likewise, for phages grouped in clusters, we observed a variation in both GC% (Fig. 7f) and USS-profiles (Fig. 7g, for USS-profiles with single nucleotide mismatch see Fig. S5a). Generally, Hin-dialect predominated except for clusters 5, 17, and 18, as well as 28, where prophages lack any USS sequences, used both dialects, or favored the Apl-dialect, respectively (Fig. 7g). Frequencies of Hin-USS reached saturation at ~750, while maximal Apl-USS values were much smaller (Fig. 7e, g). To juxtapose phages with hosts, we created a 2D-ordination based on both USS-profiles and GC% (Fig. 7h, for 3D plot go to Fig. S5b). For phages, we observed a tendency to group with their corresponding hosts suggesting long-term coevolution. “H. ducreyi” phages were the noteworthy exception, and they likely originated either from Aggregatibacter-like hosts (cluster 9, high GC%, Hin-dialect) or potentially from species outside the Pasteurellaceae family (cluster 5, USS-poor). Traces of phage-host coevolution were also visible at phage supercluster level (Fig. 7d–g, i). Although phage clusters from some superclusters showed similar profiles, for most phage clusters (e.g., those grouped in Mu, BcepMu, HP1, and MHaA1 superclusters) clear shifts towards hosts were usually observed. For example, Aggregatibacter phages showed elevated GC%, or phages infecting the parahaemolyticus clade accumulated Apl-USS. In some cases, the cause of the shift remained unclear: e.g., why cluster 14 is very different from all the other BcepMu-like clusters, including cluster 17 that encompasses phages infecting the same host species, i.e., H. influenzae. Finally, we used tail morphologies as ordination labels (Fig. 7j), and observed that clusters containing phages with flexible tail showed more extreme profiles, when compared to clusters with myoviruses (characterized by contractile tail). This might suggest that certain types of phages are more prone to the observed shifts.
To sum up, USS frequencies in combination with GC% can be used to study phage-host coevolution if bacterial taxa (or their ancestors) exhibit specificity in DNA uptake. The longer a temperate phage co-evolves with its host, the likelier accumulation of USSs (Fig. 7k) and a GC% similar to the host’s is. An important unanswered question here is what exactly drives evolution of GC content in these phages.
Discussion
Prophage diversity has been resolved in microbial genomes and metagenomes at coarse taxonomic resolutions to improve understanding of phage genetics, ecology, and evolution [11, 58–60]. Detailed analyses at finer resolution levels, i.e., for host genera or species, not only extended the understanding of phage biology but also provided a valuable reference for further explorations [16, 32, 33, 61]. Yet, comprehensive updating of the sequence resources for phages, from very biased and incomplete resources to one that is representative, ordered, characterized, and easily accessible has not been performed for most bacterial taxa. Advances in the understanding of prophage diversity are hampered by many factors: lack or unequal distribution of genomes/metagenomes for host clades/microbiomes, laborious processes of data acquisition and curation, rapidly changing taxonomy, lack of representative strain collections, absence of strain metadata, and simply the vast amount of understudied species to choose from. Here, we overcome these hurdles to study phages infecting members of the genera Aggregatibacter and Haemophilus, relevant human commensals and opportunistic pathogens. As phages infecting these species are largely unknown, we first aimed at revealing their taxonomic diversity. After establishing a phage classification scheme, we described the patterns of phage diversity across clades and niches and traced phage-host coevolution. Our findings advocate for more detailed studies exploring multiple ecological roles of these phages in the human microbiome.
Our study creates the first comprehensive classification scheme for Aggregatibacter and Haemophilus phages based on shared similarities in genome and marker-protein sequences (for comparison with existing classifications see Table S1). One of the most significant findings to emerge from prophage screening is that only three of our 33 phage clusters contain phage species previously reported in public databases, although many of these phages are highly prevalent in human oral microbiomes. In raw numbers, we analyzed 258 sequences of complete or almost complete prophages, five of which (Aaphi23, FluMu, HP1, HP2, and S1249) were described previously [19–21, 34, 62]. Classification of metagenomic assemblies yielded 107 additional sequences that form 16 new clusters, six of which likely contain lytic phages with therapeutic potential. Therefore, former microbiome studies focusing on oral phages likely suffered from incomplete reference datasets and propagation of erroneous annotations [63]. We hope that our phage reference catalogue can help others to overcome such and similar problems. We provide the genome and marker-protein sequences (Files S1 and S2) in a phylogenetic context (Table S2) that can be used to study Aggregatibacter and Haemophilus prophages at broad resolution as exemplified by analysis of the HMP dataset.
Another important issue for future research is to establish publicly available databases that provide easy access to well characterized and constantly updated sequences for oral phages. For example, data from recent work on the genomics of Aggregatibacter and Haemophilus species will broaden our catalogue [42, 43, 64]. Aggregatibacter and Haemophilus phages can also be found in metagenomics studies focused either on whole microbiomes or on viruses only (i.e., virome) [23, 65–67]. The latter are attractive since extracellular particles of phages are enriched and therefore low abundant phages can be detected and characterized [50]. On the other hand, analyzing datasets without enrichment has several advantages: it can detect lytic, temperate, and persistent infections, as well as overcome the biases arising from the enrichment procedure [50]. In all metagenomic studies the prediction of phage host(s) poses a challenge, but in silico estimations are often reliable and important guides for subsequent analyses, e.g., in vivo studies [50].
We refrained from assigning the full taxonomic affiliation to phages since the taxonomy of tailed phages is currently undergoing a major reorganization [68]. We are in a process of evaluating superclusters one by one, including sequencing additional phage genomes and screening for new members that infect hosts outside the genera Aggregatibacter and Haemophilus. We plan to gradually integrate our findings within current taxonomy.
Our results show that the quantitative and qualitative distribution of prophages across Aggregatibacter and Haemophilus species is not random. These patterns are signatures of co-evolutionary processes that maintain phage-host interaction. Very high prevalence (common presence) but low phylogenetic diversity (limited number of phage clusters) characterized prophages of the strict human pathogen “H. ducreyi”. These findings suggest either piggybacking where the host strain carrying these phages developed into a successful clone, or a mutualistic relationship, where transposable prophages confer a strong fitness benefit to “H. ducreyi” lysogens. Independent emergence of two similar prophage-rich lineages (clades I and II, see Fig. 3 and Fig. S2) put commensalism forward for consideration. There are several potential mechanisms that could drive such interactions between a pathogen and its prophages. It may be speculated that a prophage allows production of a factor that enhances adhesion, growth, or resistance of the host. Alternatively, specific disruption of a protein-coding gene by prophage integration may be advantageous during host’s adaptation [69]. Conserved locations of “H. ducreyi” prophages may suggest the latter scenario, as transposable prophage formation is believed to occur by integration into nearly random chromosomal locations [42, 70]. Another possible explanation is that these temperate bacteriophages act on the one hand as allelopathic agents, facilitating competition with other populations, and on the other hand as efficient host protectors against such an assault [16, 71]. In this scenario, phages that leak from a lysogenic population kill susceptible cells from a competitor population at a cost of small fraction of the lysogenic population but also protect its host from a secondary infection through superinfection exclusion. Positive frequency-dependent selection occurs because the more phage-releasing lysogens exist, the more beneficial it is to become a phage-resistant lysogen. Finally, the spontaneous or triggered phage-induced lysis of “H. ducreyi” strains may elicit a severe immune response due to release of endotoxins. Extracellular DNA produced in this process can be beneficial for the surviving subpopulation as an access to the key nutrients, source of DNA for genome repair or extension, or by conferring protection as a structural part of a biofilm matrix [72].
Mutualistic relationship implies that there are also benefits for the temperate phage to undergo the lysogenic cycle. One of the current paradigms for establishing lysogeny is low host abundance under stress or nutrient-limited conditions such that the phage is able to wait for more favorable host growth conditions (or host adaptation) in order to switch to lytic propagation. The alternative Piggyback-the-Winner model explains prevalence of lysogeny within nutrient rich environments (presumably such as mucous layers) by reduced phage predation control on bacterial abundance and superinfection exclusion, i.e., preventing closely related phages from infecting the same bacterial cell [73]. Additionally, if extracellular conditions may be physically detrimental to the phage virion (e.g., extreme pH), lysogeny would confer protection. Moreover, additional/alternative modes (i.e., as prophage) of dissemination, replication, and evolution may favor phage survival. Finally, lysogeny may enable broader host range by decreasing host-use trade-offs, e.g., by increasing total burst size calculated per single absorbed virion [74].
In contrast to “H. ducreyi” lysogens, H. influenzae and H. parainfluenzae strains are lysogenized by highly diverse populations of phages. A probable explanation is that these phages infect bacterial populations that are prevalent, abundant, active, biogeographically widespread, dynamic and diverse across multiple sites of the human mouth and pharynx [3–8, 75]. Diverse phage populations may favor host divergence, creating a positive feedback. Such coevolution can then maintain phenotypic and genetic diversity of both phages and their hosts [12]. Importantly, this process is driven by abiotic and other biotic components of the environment. The human mouth is a dynamic environment colonized by a complex and diverse microbiota, and the oral cavity is unique in providing distinct niches with physicochemical gradients, exposure to outer environment, and constant mixing by saliva flow [76, 77]. Consequently, both in situ coevolution of relevant adaptations, and colonization by phages that come in from sites in which these adaptations have already evolved, can take place. Combined, this provides an immense interaction space for diverse phages and host strains. In contrast to Haemophilus species, members of only three phage clusters infect strains of other oral species such as A. actinomycetemcomitans. However, this bacterial specialist is far less prevalent, low abundant (Fig. S4) and preferably inhabits the buccal epithelium and gingival sulcus [8, 78, 79].
Analysis of patterns at finer phylogenetic resolution of the bacterial hosts (i.e., at subspecies-like clades) gave further insights into the parasite-host ecology. Prophages varied remarkably between lineages, indicating either preserving forces or barriers in phage dissemination. Previously, a similar phenomenon was reported for siphoviruses (phages with flexible tail) that preferentially lysogenize Staphylococcus aureus strains from specific clonal complexes [80]. The authors suggested that spread of phages must be restricted potentially by restriction-modification systems and gave a number of examples how lysogeny can be preserved. Strikingly, in our study, similar patterns where shared by clades of different species, suggesting common ecoevolutionary driving forces.
As a first example, we noticed an extremely high prevalence of simultaneous infections with multiple transposable prophages in “H. ducreyi” strains from clades I and II but also in serotype/clade e strains of A. actinomycetemcomitans. These were co-infections of SuMu- Mu- and B3-like phages. Interestingly, in two clades of “H. ducreyi”, either Mu-like cluster 8 or Mu-like cluster 9 reached 100% prevalence when truncated prophages were included. On the other hand, B3-like phages where universally present in both clade II “H. ducreyi” strains and in serotype e strains of A. actinomycetemcomitans. All these bacterial strains originated from highly inflamed tissue—either genital or cutaneous ulcers or chronic gum disease. This would suggest a potential role of transposable phages in pathogenesis or in evasion of the immune response. The potential mechanisms were discussed in the previous section. Alternatively, we observed piggybacking, i.e., host strain carrying these phages developed into a highly infective clone by acquiring non-phage element(s) carrying advantageous gene(s). Conserved polylysogeny, the carriage of multiple prophages, additionally implies that interactions between prophages may be beneficial for a host, e.g., by reducing the rate of spontaneous lysis or regulation of gene expression.
The second pattern was a very high prevalence of Aaphi23-like prophages in serotype/clade a strains of A. actinomycetemcomitans and strains of H. influenzae grouped in clade II. This is in agreement with our previous findings for A. actinomycetemcomitans obtained by Southern blot hybridization [45]. In contrast to serotype/clade b and c strains, serotype/clade a strains of A. actinomycetemcomitans are usually genetically competent [81]. High prevalence of Aaphi23-like prophages in naturally competent linages suggests that DNA uptake may be a preserving force [52]. As high amounts of extracellular DNA can usually be found in oral biofilms, these prophages may protect competent lysogenic strains from harmful excessive uptake of foreign DNA by abortive killing induced via SOS response [82]. Phage mediated allopathy is another potential mechanism as we observed that Aaphi23-like phages showed broad activity spectrum and tended to have higher activity against serotype/clade b than a strains. A potential explanation is that the loss of competence in serotype/clade b and c strains was shown to be followed by the loss of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs), bacterial adaptive immune systems that protect against parasitic DNA [83].
The third pattern was absence or very low incidence of prophages in serotype d strains of A. actinomycetemcomitans and clade IV strains of H. influenzae. This may be caused by a lack of efficient cell receptors or integration sites. Multiple phage resistance mechanisms may also form a barrier to lysogenization [14, 84]. We previously observed a high resistance of serotype d strains of A. actinomycetemcomitans to Aaphi23-like phages supporting this notion [45]. Alternatively, these strains thrive in particular microenvironments that might hinder phage dispersal or favor inactivation of the virions. Further experimental evaluation is required to identify the relevant explanation.
The spatial ecology of the microbial communities that inhabit the human body, in particular those of the mouth, deserves greater attention [76]. Important questions include which factors influence the size, boundary and spatial structure of microbial populations in a given niche. The influence of populations from different sites on each other is also not known. Here we profiled the prevalence of Aggregatibacter and Haemophilus phages in oral microbiomes and observed that members of related phage clusters have preferences for different oral niches. Putative clades of H. parainfluenzae showed niche adaptation as well [8]. Different niches have unique physical, chemical, and biological (i.e., microbiome) profiles that likely promote speciation in both phages and their bacterial hosts. Mucin, a heavily glycosylated protein, may play a key role in this, since Pasteurellaceae species preferentially colonize mucosal surfaces. It was reported that bacteria (including Pasterurellaceae species) and phages express receptors for specific mucins (e.g., outer membrane proteins or proteins containing Ig-like domains) [85–87]. We rarely observed Ig-fold in proteins from Pasteurellaceae phages (data not shown), so likely other protein domains contribute to mucin binding. On the one hand, immobilization and/or subdiffusive motion of phages and bacteria in mucosal layers increase the frequency of their encounters [88]. On the other hand, dispersion between niches, enabled by salivary and air flow, drags invaders. Phages may provide protection from such invading species [89]. Recently, spatially structured lytic to lysogenic switches were hypothesized to reconcile Piggyback-the-Winner and ‘Bacteriophage adherence to mucus’ models [90].
Discovery of high phage diversity prompted us to study the dynamics of phage-host coevolution. By measuring the frequencies of clade-specific DNA uptake signaling sequences (USSs) and guanine–cytosine content (GC%) we could trace phage-host coevolution and host shift events. We hope that future studies on the evolutionary ecology of prokaryotic immune mechanisms [91] and viral counter strategies [14, 15] reveal mechanisms behind observed dynamics and assign a proper time scale.
Pasteurellaceae species and phages provide a unique subject to study ecology and evolution. These bacteria and viruses are niche-specific and strongly host-associated, therefore barrier formation, dissemination, rate of speciation, and adaptive radiation, can be studied across different scales. They usually inhabit mucous layers that are easily accessible; for this reason, parameters like population size or host range can be obtained easier than for other habitats. Finally, Pasteurellacae species are usually genetically competent but some clades lost this property, consequently, the relationship between phage infection and DNA uptake can be dissected. Here we are providing a robust classification framework for phages that should be a good basis for such explorations.
Conclusion
We established a classification scheme for Aggregatibacter and Haemophilus prophages and described the patterns of their diversity across clades and niches. Niche-specific coevolution may favor development of mutualistic relationships between members of specific bacterial clades and phage phylotypes. Further work is needed on the potential mechanistic underpinnings of prophage patterns and therapeutic use of lytic phages. In summary, Aggregatibacter and Haemophilus phages are prevalent, diverse, likely play a significant ecoevolutionary role in the human microbiome, and are therefore interesting candidates for further research.
Experimental procedures
Information about used bacterial strains, culture conditions, species taxonomy, clades phylogeny, DNA isolation, Mitomycin C induction, drop spot assay, electron microscopy, genome sequencing, classification of metagenomic assemblies, USS-profiling, and statistical analyses can be found in Supplementary Information.
Identification and characterization of prophages infecting Aggregatibacter and Haemophilus species
The PHASTER (PHAge Search Tool—Enhanced Release) web server [29] was used to identify and annotate prophage sequences within publicly available genomes of human-associated Aggregatibacter and Haemophilus spp. [1] retrieved from the NCBI genome database (March 2017). Defective and incomplete prophages are commonly encountered in bacterial genomes; therefore, we focused on non-duplicated, putatively intact prophages (PHASTER score >90) with genomes encoding a minimum of 40 ORFs (if not otherwise stated). We identified duplications by manual screening. Highly similar sequences were identified, metadata was retrieved when possible, and potential pseudoreplication (e.g., re-sequencing under different name, strains isolated from same clinical site or patient, colony variants of same isolate) was judged. Phage genome alignments were built using progressive Mauve [28]. Genome-based taxonomic classification was performed with VICTOR’s formula d4 [30]. Similarity matrix analysis was performed with Gepard using word length 10 [92]. Average nucleotide identity was calculated with OrthoANI [35]. Mash was used (k-mer size: 21; sketch-size: 1000) to estimate genome distance using MinHash [36]. Phage genome sequences were screened for open reading frames using Glimmer [93]. Translated sequences of proteins were annotated using three variants of BLAST against the following databases: (i) Prophage/Virus Database [29], (ii) Subsystem Technology [94], and (iii) Conserved Domain Database [95]. Multiple alignments of amino acid sequences were performed with MUSCLE [96]. Phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis v. 6 [97]. Evolutionary histories were inferred using the Maximum Likelihood method based on the Jones Taylor Thorton (JTT) matrix-based model [98]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. The trees were drawn to scale, with branch lengths measuring the number of substitutions per site. Phage richness from the results of genome sampling was assessed by rarefaction (custom spreadsheet, 10 replicates).
Identification of phages and their hosts in human-associated biofilm metagenomes
Metagenomic taxon abundances (MetaPhlAn2) were plotted for Aggregatibacter and Haemophilus species [8]. A species was regarded as present if its abundance was higher than 0.1%. To identify phages, metagenomic whole-genome assembly fasta files originating from the Human Microbiome Project WGS production phase I (HMP, study WGS-PP1) were downloaded via the HMP portal (https://portal.hmpdacc.org/) [26]. Files marked as “scaffolds” were removed from the present analysis, resulting in a database of 1021 contig files. Using a custom perl script, all contigs were combined into one file in which the fasta headers contained the unambiguous name of the original file in addition to the original contig identifier. All contigs were combined into one file, processed into a BLAST database, and the list of maker protein sequences (File S2) was searched against this database using tblastn with NCBI stand-alone BLAST (v. 2.5.0+). The BLAST E-value cutoff for this search was 1E-80. Based on the BLAST results table, an identity score was calculated as the percentage of query sequence covered by the BLAST hit, multiplied by the identity value of the BLAST hit. Only hits with an identity score of at least 95% were further analyzed. Prevalences of phage clusters and superclusters were plotted for different body sites.
Detection of prophages by PCR
Used strains and primers are listed in Table S4 and Table S5, respectively. Detailed information can be found in Supplementary Information.
Supplementary information
Acknowledgements
Work of SPS has been supported by HiLF (Hochschulinterne Leistungsförderung), an intramural program of Hannover Medical School. MS would like to acknowledge the Ministry of Lower Saxony and VolkswagenStiftung (both BIOFABRICATION FOR NIFE: VWZN2860) and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2155 “RESIST”, Project ID 39087428) for funding. We thank S. N. Stumpp for lab management, M. Dalton, and R. Schreeb for technical assistance, D. P. Melidis for running programs on Linux, A. Hoffmann for providing access to a centrifuge, R. Mutters for bacterial strains used for preliminary work, E. Kulik for providing access to old reprints, E. Rubalskii for discussion, and two anonymous reviewers for comments and suggestions. We are grateful to the research community for publicly available genome sequences of phages and Pasteurellaceae species. We would like to thank the HMP Consortium for publicly available human-associated biofilm metagenomes and colleagues from Joint Genome Institute for viral assemblies publically available in IMG/VR database.
Author contributions
SPS, MK, and MS designed the study; SPS and JH performed the experiments; SPS, IY, and GBdW analyzed the data. SPS wrote the manuscript with input from MK, IY, GBdW, AW, JH, and MS. All the authors approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0450-8) contains supplementary material, which is available to authorized users.
References
- 1.Norskov-Lauritsen N. Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans. Clin Microbiol Rev. 2014;27:214–40. doi: 10.1128/CMR.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilian M, Prachyabrued W, Theilade E. Haemophili in developing dental plaque. Scand J Dent Res. 1976;84:16–9. doi: 10.1111/j.1600-0722.1976.tb00456.x. [DOI] [PubMed] [Google Scholar]
- 4.Kilian M, Schiott CR. Haemophili and related bacteria in the human oral cavity. Arch oral biol. 1975;20:791–6. doi: 10.1016/0003-9969(75)90055-2. [DOI] [PubMed] [Google Scholar]
- 5.Kuklinska D, Kilian M. Relative proportions of Haemophilus species in the throat of healthy children and adults. Euro J Clin Microbiol. 1984;3:249–52. doi: 10.1007/BF02014895. [DOI] [PubMed] [Google Scholar]
- 6.Szafranski SP, Deng ZL, Tomasch J, Jarek M, Bhuju S, Meisinger C, et al. Functional biomarkers for chronic periodontitis and insights into the roles of Prevotella nigrescens and Fusobacterium nucleatum; a metatranscriptome analysis. NPJ Biofilms Microbiom. 2015;1:15017. doi: 10.1038/npjbiofilms.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szafranski SP, Wos-Oxley ML, Vilchez-Vargas R, Jauregui R, Plumeier I, Klawonn F, et al. High-resolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Appl Environ Microbiol. 2015;81:1047–58. doi: 10.1128/AEM.03534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–6. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roux S, Hallam SJ, Woyke T, Sullivan MB. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. eLife. 2015;4:1–20. Epub 2015/07/23 [DOI] [PMC free article] [PubMed]
- 12.Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38:916–31. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs Z, Abedon ST. Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’. FEMS Microbiol Lett. 2016;363:fnw047. doi: 10.1093/femsle/fnw047. [DOI] [PubMed] [Google Scholar]
- 14.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 15.Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11:675–87. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 16.Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol. 2017;2:16251. doi: 10.1038/nmicrobiol.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol BiolRev. 2003;67:238–76. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szafranski SP, Winkel A, Stiesch M. The use of bacteriophages to biocontrol oral biofilms. J Biotechnol. 2017;250:29–44. doi: 10.1016/j.jbiotec.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Harm W, Rupert CS. Infection of Transformable Cells of Haemophilus Influenzae by Bacteriophage and Bacteriophage DNA. Zeitschrift fur Vererbungslehre. 1963;94:336–48. doi: 10.1007/BF00897593. [DOI] [PubMed] [Google Scholar]
- 20.Williams BJ, Golomb M, Phillips T, Brownlee J, Olson MV, Smith AL. Bacteriophage HP2 of Haemophilus influenzae. J Bacteriol. 2002;184:6893–905. doi: 10.1128/JB.184.24.6893-6905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan GJ, Hatfull GF, Casjens S, Hendrix RW. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J Mol Biol. 2002;317:337–59. doi: 10.1006/jmbi.2002.5437. [DOI] [PubMed] [Google Scholar]
- 22.Stevens RH, Hammond BF, Lai CH. Characterization of an inducible bacteriophage from a leukotoxic strain of Actinobacillus actinomycetemcomitans. Infect Immun. 1982;35:343–9. doi: 10.1128/iai.35.1.343-349.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature. 2016;536:425–30. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 24.Pletnev EA, Krylov SV, Shaburova OV, Burkal’tseva MV, Miroshnikov KA, Krylov VN. [Pseudolysogeny of Pseudomonas aeruginosa bacteria infected with phiKZ-like bacteriophages] Genetika. 2010;46:26–32. [PubMed] [Google Scholar]
- 25.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paez-Espino D, Roux S, Chen IA, Palaniappan K, Ratner A, Chu K, et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2019;47(D1):D678–D86. doi: 10.1093/nar/gky1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meier-Kolthoff JP, Goker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toussaint A, Rice PA. Transposable phages, DNA reorganization and transfer. Curr Opin Microbiol. 2017;38:88–94. doi: 10.1016/j.mib.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Casjens SR, Grose JH. Contributions of P2- and P22-like prophages to understanding the enormous diversity and abundance of tailed bacteriophages. Virology. 2016;496:255–76. doi: 10.1016/j.virol.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grose JH, Casjens SR. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468-470:421–43. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resch G, Kulik EM, Dietrich FS, Meyer J. Complete genomic nucleotide sequence of the temperate bacteriophage Aa Phi 23 of Actinobacillus actinomycetemcomitans. J Bacteriol. 2004;186:5523–8. doi: 10.1128/JB.186.16.5523-5528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbio. 2016;66:1100–3. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 36.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 38.Rohwer F, Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–35. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43(Database issue):D571–7. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46(D1):D708–D17. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittichotirat W, Bumgarner RE, Chen C. Evolutionary Divergence of Aggregatibacter actinomycetemcomitans. J Dent Res. 2016;95:94–101. doi: 10.1177/0022034515608163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks M, Fookes M, Wagner J, Ghinai R, Sokana O, Sarkodie YA, et al. Direct whole-genome sequencing of cutaneous strains of Haemophilus ducreyi. Emerg Infect Dis. 2018;24:786–9. doi: 10.3201/eid2404.171726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettigrew MM, Ahearn CP, Gent JF, Kong Y, Gallo MC, Munro JB, et al. Haemophilus influenzae genome evolution during persistence in the human airways in chronic obstructive pulmonary disease. Proc Natl Acad Sci USA. 2018;115:E3256–E65. doi: 10.1073/pnas.1719654115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szafranski SP, Deng ZL, Tomasch J, Jarek M, Bhuju S, Rohde M, et al. Quorum sensing of Streptococcus mutans is activated by Aggregatibacter actinomycetemcomitans and by the periodontal microbiome. BMC Genom. 2017;18:238. doi: 10.1186/s12864-017-3618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haubek D, Willi K, Poulsen K, Meyer J, Kilian M. Presence of bacteriophage Aa phi 23 correlates with the population genetic structure of Actinobacillus actinomycetemcomitans. Euro J Oral Sci. 1997;105:2–8. doi: 10.1111/j.1600-0722.1997.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 46.Poulsen K, Theilade E, Lally ET, Demuth DR, Kilian M. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology. 1994;140(Pt 8):2049–60. doi: 10.1099/13500872-140-8-2049. [DOI] [PubMed] [Google Scholar]
- 47.Haubek D, Poulsen K, Asikainen S, Kilian M. Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1995;33:395–401. doi: 10.1128/jcm.33.2.395-401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Steenbergen TJ, Bosch-Tijhof CJ, van Winkelhoff AJ, Gmur R, de Graaff J. Comparison of six typing methods for Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1994;32:2769–74. doi: 10.1128/jcm.32.11.2769-2774.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willi K, Sandmeier H, Meyer J. Temperate bacteriophages of Actinobacillus actinomycetemcomitans associated with periodontal disease are genetically related. Med Microbiol Lett. 1993;2:419–26. [Google Scholar]
- 50.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat Biotechnol. 2019;37:29–37. Epub 2018/12/18 [DOI] [PMC free article] [PubMed]
- 51.Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, et al. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J Mol Biol. 2002;317:1–19. doi: 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- 52.Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity. J Bacteriol. 2014;196:1471–83. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakkali M, Chen TY, Lee HC, Redfield RJ. Evolutionary stability of DNA uptake signal sequences in the Pasteurellaceae. Proc Nal Acad Sci USA. 2004;101:4513–8. doi: 10.1073/pnas.0306366101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol Biol. 2006;6:82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogg JS, Hu FZ, Janto B, Boissy R, Hayes J, Keefe R, et al. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 2007;8:R103. doi: 10.1186/gb-2007-8-6-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danner DB, Deich RA, Sisco KL, Smith HO. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene. 1980;11:311–8. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- 57.Smith HO, Tomb JF, Dougherty BA, Fleischmann RD, Venter JC. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–40. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 58.Mavrich TN, Hatfull GF. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol. 2017;2:17112. doi: 10.1038/nmicrobiol.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–93. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 60.Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISMEJ. 2016;10:2744–54. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife. 2015;4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, Kittichotirat W, Si Y, Bumgarner R. Genome sequence of Aggregatibacter actinomycetemcomitans serotype c strain D11S-1. J Bacteriol. 2009;191:7378–9. doi: 10.1128/JB.01203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Gao Y, Zhao F. Phage-bacteria interaction network in human oral microbiome. Environ Microbio. 2016;18:2143–58. doi: 10.1111/1462-2920.12923. [DOI] [PubMed] [Google Scholar]
- 64.Murra M, Lutzen L, Barut A, Zbinden R, Lund M, Villesen P, et al. Whole-Genome Sequencing of Aggregatibacter Species Isolated from Human Clinical Specimens and Description of Aggregatibacter kilianii sp. nov. Journal of Clin Microbiol. 2018;56:e00053–18. doi: 10.1128/JCM.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paez-Espino D, Chen IA, Palaniappan K, Ratner A, Chu K, Szeto E, et al. IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 2017;45(D1):D457–D65. doi: 10.1093/nar/gkw1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, et al. Human oral viruses are personal, persistent and gender-consistent. ISMEJ. 2014;8:1753–67. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA, 3rd, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISMEJ. 2012;6:915–26. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barylski J, Enault F, Dutilh BE, Schuller MB, Edwards RA, Gillis A, et al. Analysis of Spounaviruses as a Case Study for the Overdue Reclassification of Tailed Bacteriophages. Sys Biol. 2019. [DOI] [PMC free article] [PubMed]
- 69.Davies EV, James CE, Williams D, O’Brien S, Fothergill JL, Haldenby S, et al. Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proc Natl Acad Sci USA. 2016;113:8266–71. doi: 10.1073/pnas.1520056113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gangaiah D, Webb KM, Humphreys TL, Fortney KR, Toh E, Tai A, et al. Haemophilus ducreyi cutaneous ulcer strains are nearly identical to class I genital ulcer strains. PLoS Negl Trop Dis. 2015;9:e0003918. doi: 10.1371/journal.pntd.0003918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISMEJ. 2016;10:2854–66. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen M, Yang Y, Shen W, Cen L, McLean JS, Shi W, et al. A linear plasmid-like prophage of Actinomyces odontolyticus Promotes Biofilm Assembly. Appl Environ Microbiol. 2018;84:e01263–18. doi: 10.1128/AEM.01263-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobian-Guemes AG, et al. Lytic to temperate switching of viral communities. Nature. 2016;531:466–70. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 74.Sieber M, Gudelj I. Do-or-die life cycles and diverse post-infection resistance mechanisms limit the evolution of parasite host ranges. Ecol Lett. 2014;17:491–8. doi: 10.1111/ele.12249. [DOI] [PubMed] [Google Scholar]
- 75.Mukundan D, Ecevit Z, Patel M, Marrs CF, Gilsdorf JR. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J ClinMicrobiol. 2007;45:3207–17. doi: 10.1128/JCM.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Proctor DM, Relman DA. The landscape ecology and microbiota of the human nose, mouth, and throat. Cell host Microbe. 2017;21:421–32. doi: 10.1016/j.chom.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edlund A, Santiago-Rodriguez TM, Boehm TK, Pride DT. Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol. 2015;7:27423. doi: 10.3402/jom.v7.27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. 2013;51:2850–61. doi: 10.1128/JCM.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fine DH, Markowitz K, Furgang D, Velliyagounder K. Aggregatibacter actinomycetemcomitans as an early colonizer of oral tissues: epithelium as a reservoir? J Clin Microbiol. 2010;48:4464–73. doi: 10.1128/JCM.00964-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, et al. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009;191:3462–8. doi: 10.1128/JB.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujise O, Lakio L, Wang Y, Asikainen S, Chen C. Clonal distribution of natural competence in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2004;19:340–2. doi: 10.1111/j.1399-302x.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 82.Setlow JK, Boling ME, Allison DP, Beattie KL. Relationship between prophage induction and transformation in Haemophilus influenzae. J Bacteriol. 1973;115:153–61. doi: 10.1128/jb.115.1.153-161.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jorth P, Whiteley M. An evolutionary link between natural transformation and CRISPR adaptive immunity. mBio. 2012;3:e00309–12. doi: 10.1128/mBio.00309-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359:eaar4120. doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 87.Reddy MS, Bernstein JM, Murphy TF, Faden HS. Binding between outer membrane proteins of nontypeable Haemophilus influenzae and human nasopharyngeal mucin. Infect Immun. 1996;64:1477–9. doi: 10.1128/iai.64.4.1477-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barr JJ, Auro R, Sam-Soon N, Kassegne S, Peters G, Bonilla N, et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc Natl Acad Sci SA. 2015;112:13675–80. doi: 10.1073/pnas.1508355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quistad SD, Grasis JA, Barr JJ, Rohwer FL. Viruses and the origin of microbiome selection and immunity. ISMEJ. 2017;11:835–40. doi: 10.1038/ismej.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Silveira CB, Rohwer FL. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiom. 2016;2:16010. doi: 10.1038/npjbiofilms.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Houte S, Buckling A, Westra ER. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol Mol Biol Rev. 2016;80:745–63. doi: 10.1128/MMBR.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–8. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 93.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D3. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences. CABIOS. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 99.Toussaint Ariane, Van Gijsegem Frédérique. Extension of the transposable bacterial virus family: two genomic organisations among phages and prophages with a Tn552-related transposase. Research in Microbiology. 2018;169(9):495–499. doi: 10.1016/j.resmic.2017.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.