Abstract
The genus of Parasutterella has been defined as a core component of the human and mouse gut microbiota, and has been correlated with various health outcomes. However, like most core microbes in the gastrointestinal tract (GIT), very little is known about the biology of Parasutterella and its role in intestinal ecology. In this study, Parasutterella was isolated from the mouse GIT and characterized in vitro and in vivo. Mouse, rat, and human Parasutterella isolates were all asaccharolytic and producers of succinate. The murine isolate stably colonized the mouse GIT without shifting bacterial composition. Notable changes in microbial-derived metabolites were aromatic amino acid, bilirubin, purine, and bile acid derivatives. The impacted bile acid profile was consistent with altered expression of ileal bile acid transporter genes and hepatic bile acid synthesis genes, supporting the potential role of Parasutterella in bile acid maintenance and cholesterol metabolism. The successful colonization of Parasutterella with a single environmental exposure to conventional adult mice demonstrates that it fills the ecological niche in the GIT and contributes to metabolic functionalities. This experiment provides the first indication of the role of Parasutterella in the GIT, beyond correlation, and provides insight into how it may contribute to host health.
Subject terms: Symbiosis, Bacterial host response
Introduction
Parasutterella, a genus of Betaproteobacteria, has been defined as a member of the healthy fecal core microbiome in the human gastrointestinal tract (GIT) [1]. Members of the genus Parasutterella have also been found in a variety of host species, including mice, rats, dogs, pigs, chickens, turkeys, and calves according to reported sequences available in the Ribosomal Database Project (RDP). The genus of Parasutterella contains two type strains, Parasutterella excrementihominis YIT11859 and Parasutterella secunda YIT12071, which were first isolated from human feces [2, 3]. 16S ribosomal RNA (rRNA) gene sequence similarities indicate that the Parasutterella sequences from mice are most closely related to P. excrementihominis. In humans, the genus of Parasutterella has a unique phylogenetic classification [4] as it stands out as one of the most frequently reported taxa within the class Betaproteobacteria in the gut, and is largely represented by a single species, P. excrementihominis. The relative abundance of this species has been associated with different host health outcomes such as inflammatory bowel disease, obesity, diabetes, and fatty liver disease [4–6].
A significant reduction of Parasutterella in response to high-fat diet (HFD) treatment has been observed in multiple animal models and human studies, indicating a negative correlation between Parasutterella abundance and HFD-induced metabolic phenotypes including hypothalamic inflammation [7–10]. Parasutterella has also been identified to respond to antibiotic administration and other dietary interventions such as prebiotic and resistance starch supplementation in human studies and animal trials [11–13]. In addition, patients with Clostridium difficile infection (CDI) exhibited a dramatic increase in the abundance of Proteobacteria in the gut, however, within the phylum of Proteobacteria, Parasutterella was significantly lower in CDI patients and asymptomatic carriers than in healthy controls [14]. An in vitro fermentation study, which investigated how microbial composition of fecal donors impacted the fermentation properties of dietary fiber, showed that the abundance of Parasutterella decreased during the fermentation and positively correlated with ammonia production [15]. These results provide evidence that Parasutterella has a role in impacting microbial activities and host responses, however, beyond correlative work, our knowledge of Parasutterella’s physiological characteristics is extremely limited. With a growing number of studies correlating Parasutterella with diverse outcomes, including beneficial and detrimental, it becomes essential that we begin to understand the basic role of this microbe and its uncharacterized metabolites in the microbial ecosystem and host physiology.
Alteration of microbially derived metabolites is an important mechanism through which changes in gut microbial activity generate functional consequences for host health outcomes [16, 17]. Several microbially generated metabolites derived from substrates including carbohydrates, aromatic amino acids, bile acids, and choline have been identified as regulatory molecules. For instance, short-chain fatty acids (SCFAs) play a crucial role in linking diet, gut microbiota, and host immune response [18], and aromatic amino-acid metabolites such as indole derivatives enhance epithelial barrier integrity [19]. Additionally, the modulatory effects of microbial-derived bile acid metabolites on the farnesoid X receptor (FXR) signaling pathway are recognized to influence bile acid profile and host lipid metabolism [20]. The identification of these molecules provides new insights into the microbe–microbe and microbe–host interactions, though there are still a considerable number of yet-to-be described microbial-derived metabolites. Specifically, when pursuing the functionalities of a gut commensal bacterium that has not been well characterized, the assessment of their contribution to gut metabolite profiling becomes a sufficient and valid approach to deepen our understanding of lifestyle and physiological characteristics.
Different experimental models and detection methodologies have been established to unravel the metabolic functionalities of the gut microbiota including in vitro fermentation models, human clinical trials, humanized germ-free mouse models, and monocolonization of germ-free mice in combination with targeted or untargeted metabolomic approaches [21, 22]. In vitro characterization of metabolic capabilities of gut commensal bacteria is a simpler approach, however, it is limited by culturability and required growth conditions. For example, many gut bacteria will only grow to pinpoint colonies and have limited ability to grow in broth culture [23]. The animal model integrates the interactions between the microbe and host to improve the understanding of the holobiont. In the current study, the murine Parasutterella strain was successfully isolated and physiological characteristics were identified based on the draft genome and in vitro culture results. The isolate was subsequently introduced to Parasutterella-free mice harboring a complex microbiota, to investigate the capability of Parasutterella to colonize the mouse GIT and its effects on microbial community, intestinal metabolite profiling, and host physiology. The results provide the first indication of the role of Parasutterella in the GIT and improve our understanding of the mechanism through which it may influence host health outcomes.
Materials and methods
Bacterial strains
The mouse Parasutterella strain, Parasutterella mc1, was isolated from feces of healthy phosphatidylethanolamine N-methyltransferase knockout mice using two different types of selective media including the Gifu anaerobic medium (GAM) agar at pH value of 6.0 and GAM supplemented with 4 μg/ml oxacillin, as modified from previously published methods [2, 3]. Feces were collected immediately after defecation into 1× Phosphate-buffered saline (PBS) (pH = 7.4) supplemented with 0.05% (w/v) l-cysteine HCl, transferred into an anaerobic chamber (Sheldon, Cornelius, Oregon) containing the anaerobic gas mixture of 85% N2, 10% CO2, and 5% H2, homogenized, diluted, spread on selective agar, and incubated at 37 °C for 3 days.
The methodology of whole-genome sequencing, Parasutterella-targeted primer (Paraf/Parar) design and real-time PCR assay, as well as quantification of metabolites in bacterial culture are described in the supplementary methods.
Mice
Six- to 8-week-old female C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were housed in the animal facilities at the University of Alberta. Mice were kept under specific pathogen-free (SPF) conditions with sterilized filter-topped cages and standard chow diet (LabDiet®, 5053). All the mice were screened with designed primers (Paraf/Parar, Table S1) and 16 mice that did not harbor Parasutterella were selected. Mice were randomly grouped into four cages with four mice per cage by a blinded lab animal technician and balanced for average body weight. The cages were allocated into two treatments: control (CON) and Parasutterella colonization (PARA). Preliminary results showed that the exposure to Parasutterella by oral gavage or administration to the bedding resulted in a successful colonization of the mouse intestine. Therefore, in the current study, the PARA group was exposed to Parasutterella mc1 by administering 1 ml of the bacterial suspension with the concentration of 107 colony forming unit (CFU)/ml to the bedding while the CON group received PBS (Figure S1). Body weight was recorded weekly and mice were euthanized by CO2 asphyxiation 6 weeks after Parasutterella/PBS treatment. Contents from ileum, cecum, and colon were collected and liver, intestinal tissue, and blood were harvested. The protocols employed were approved by the University of Alberta’s Animal Care Committee and in accordance with the guideline of the Canadian Council on the Use of Laboratory Animals. The colonization experiment was repeated three times with the total sample size of 24 for each treatment and the untargeted metabolomic analysis was repeated twice with the total sample size of 16 for each treatment.
Characterization of gut microbial composition
DNA was extracted from ileal, cecal, and colonic contents after Parasutterella/PBS treatment, as well as from feces before the treatment. The DNA extraction, amplicon library construction, paired-end sequencing, and data analysis were performed using the protocols and pipelines published previously [24].
Ultrahigh performance liquid chromatography-electrospray ionization/Fourier transform mass spectrometry (UPLC-ESI/FTMS)
Frozen cecal contents were lyophilized and homogenized in 80% aqueous methanol (25 μl/mg) and supernatants were subsequently collected after centrifugation at 15,000 rpm for 20 min. The supernatants were profiled by UPLC-ESI/FTMS in positive and negative ionization mode with a pooled quality control sample injected for every eight samples. Samples were analyzed in a random way using a computerized list of random numbers. The UPLC-FTMS systems consisted of an acquity UPLC system (Walters, MA, USA) coupled with an LTQ-Orbitrap Fusion mass spectrometer (Thermo Scientific, Nepean, ON), which was operated in a survey scan and Fourier transform MS detection mode.
To maximize the chromatographic separations, the analytes were separated on a C4 column, a C18 column, and a HILIC column, respectively. The chromatographic conditions, sample preparation techniques, and data process are described in the supplementary methods.
Data analysis and visualization
For microbial composition analysis, the comparison of individual taxa/operational taxonomic units (OTUs) between groups were performed using the Mann–Whitney U-test. Nonparametric multivariate analysis of variance was used to identify the difference between groups using the adonis function in the vegan package (R v3.4.4). The principal coordinate analysis (PCoA) based on the Bray–Curtis dissimilarity metric was plotted using the phyloseq package (R v3.4.4) [25]. For metabolomic data analysis, the Metaboanalyst [26] was used to conduct the data normalization, univariate, and multivariate analysis. The quality of UPLC-FTMS data was controlled by filtering the missing values and interquartile range (IQR). The quality-controlled data were normalized by log transformation and proceeded to statistical analyses. The fold change values between the two groups were calculated using the datasets before normalization. The corrections of P-values generated from the Student’s t-test or Mann–Whitney U-test were conducted for multiple testing by the Benjamini–Hochberg procedure (false discovery rate, q-value). Features with a q-value < 0.05 were considered as statistically significant indicated as follows: ##q-value < 0.01; #q-value < 0.05. For determination of body weight, gene expression, 16S rRNA gene copy numbers (log transformed) and cytokine levels, the Shapiro–Wilk test was performed to check the normality of data distribution and the Student’s t-test was used to compare the difference between two treatments. P-values indicate statistical significance as follows: **P < 0.01; *P < 0.05. R (v3.4.4) and GraphPad Prism were used for visualizing the results.
Results
Parasutterella can be successfully isolated from mouse intestine using the selective medium
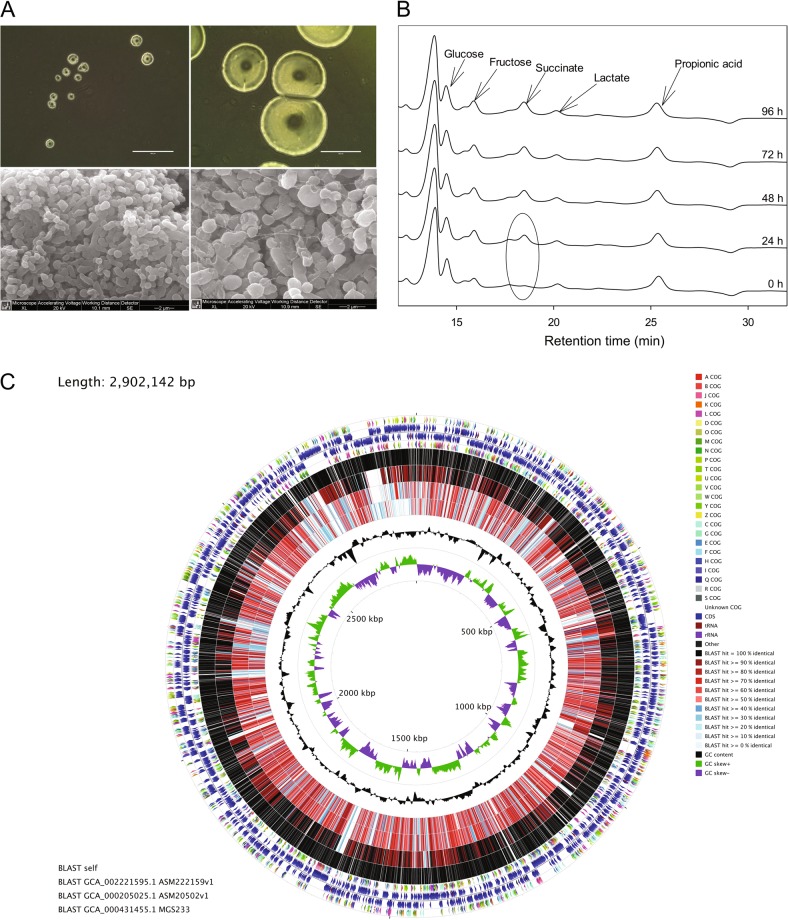
Parasutterella mc1 was isolated from the selective medium including GAM (pH = 6.0) and GAM supplemented with 4 μg/ml oxacillin inoculated with a 10−6 serially diluted fecal sample. Colonies of Parasutterella appeared entire, circular, convex, and translucent with a diameter of 0.5–0.7 mm after 72 h of incubation on GAM agar at 37 °C (Fig. 1a). Cells of Parasutterella were Gram-negative, obligately anaerobic cocci, or coccobacilli (0.5–0.8 × 1.0–1.5 μm in size) (Fig. 1a). When grown in GAM broth for upto 120 h at 37 °C anaerobically, Parasutterella produced no visible turbidity and no changes in glucose, fructose, or lactate were detected in the broth culture. However, the Parasutterella strain produced approximately 2.5 mM succinate after 72 h of growth, indicating that Parasutterella is a succinate-producing bacterium (Fig. 1b).
Fig. 1.
Morphology, metabolite production, and whole-genome visualization of Parasutterella mc1. a Top: colonies of Parasutterella mc1 formed on fastidious anaerobic agar (FAA, left) and Gifu anaerobic medium (GAM) agar (right), respectively. Bright field, magnification, ×100; Scale bars, 400 μm. Bottom: scanning electron microscopy (SEM) micrographs of Parasutterella mc1 grown on GAM agar, showing ×15,000 (left) and ×20,000 (right) magnification, respectively. b HPLC chromatogram showing metabolite profile in GAM broth with Parasutterella growth collected at 0 h, 24 h, 48 h, 72 h, and 96 h. c Circular genome map of Parasutterella mc1 generated with CGView comparison tools. From the outer to inner circles: the first (outermost) and fourth rings depicting COG categories of protein coding genes on the forward and reverse strands, respectively. The second and third rings show the locations of protein coding, tRNA, and rRNA genes on the forward and reverse strands, respectively. The fifth, sixth, seventh, and eighth rings depict BLAST comparisons (expected threshold = 0.1) between Parasutterella mc1 CDS translations and the translations from Parasutterella mc1, Turicimonas muris YL45 (ASM222159v1), P. excrementihominis YIT11859 (ASM20502v1), and P. excrementihominis CAG:233 (MGS233), respectively. The ninth ring (black plot) depicts GC content and the innermost plot represents GC skew. Both base composition plots were generated using a sliding window of 10,000 nt
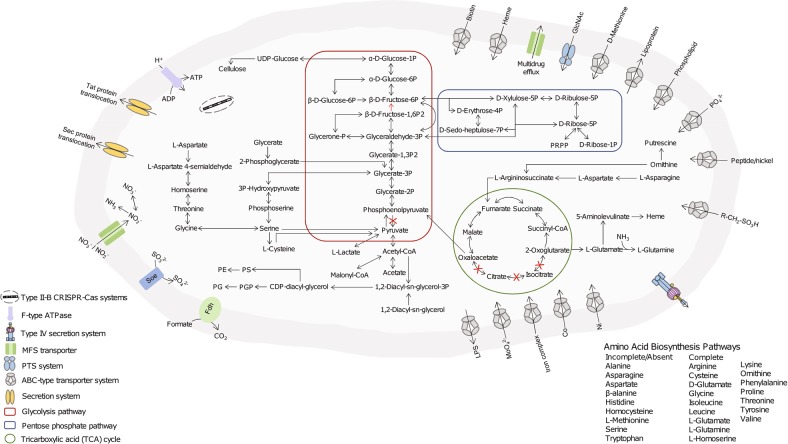
Parasutterella mc1 shared 93% 16S rRNA sequence identity with the type strain of P. excrementihominis YIT11859, which was isolated from human feces (Figure S1). The complete genome size of Parasutterella is 2.8 Mb with a G + C content of 44.1 mol% (Fig. 1c). Detailed information including the numbers of total raw reads, contigs, N50, and the accession number of deposited sequence data is provided in Table S2. There are 2648 unique genes predicted and the absence of the genes for transporting and metabolizing exogenous sugars was consistent with the asaccharolytic characteristic of Parasutterella shown in broth culture [2, 3]. In accordance with the presence of genes encoding l-asparaginase, aspartate ammonia-lyase, and putative aspartate dehydrogenase, asparagine was the most rapid and preferred amino acids metabolized by Parasutterella in GAM broth (Fig. 2; Figure S2). Supplementing l-asparagine and l-aspartate at 2 mM in the GAM broth culture of Parasutterella at 48 h of growth increased bacterial counts at 72 h compared with that in the GAM broth control (P < 0.05) (Figure S3). In addition, there were no identified hits of toxin virulence factor (VF)-related genes in the genome of Parasutterella mc1, which suggests that Parasutterella is either a commensal or symbiotic member of the gut microbiota.
Fig. 2.
Selected genomic features of Parasutterella mc1. Predicted metabolic pathways and physiological capabilities are shown based on the annotations of draft genome. Periplasma (gray) and cytoplasma (light gray) are shown bounded by outer and inner membranes, respectively. PRPP phosphoribosyl pyrophosphate, PGP phosphatidylglycerophosphate, PS phosphatidylserine, PE phosphatidylethanolamine, PG phosphatidylglycerol, Fdh formate dehydrogenase, Soe sulfite dehydrogenase (quinone)
Parasutterella isolate readily colonized the mouse GIT without shifting the microbial structure
SPF C57BL/6J mice were screened using a set of designed PCR primers targeting the 16S rRNA gene of the Parasutterella isolate to select 16 Parasutterella-free individuals. We were able to confidently detect the presence of Parasutterella down to the 106 CFU/g of feces using the real-time PCR system. With a single environmental exposure, Parasutterella rapidly and stably colonized the mouse GIT through the duration of the study, representing an average of 9.31 ± 0.16 log10 16S rRNA gene copies per gram of feces (Figure S3). Parasutterella were successfully isolated using selective medium from the mouse feces 6 weeks after environmental inoculation, verifying the stable colonization of Parasutterella. The colonization of the Parasutterella isolate in the mouse GIT did not significantly affect body weight (Figure S1). The success of Parasutterella to colonize the mature mouse gut after a single exposure demonstrated that the microbe fills the ecological niche in the GIT.
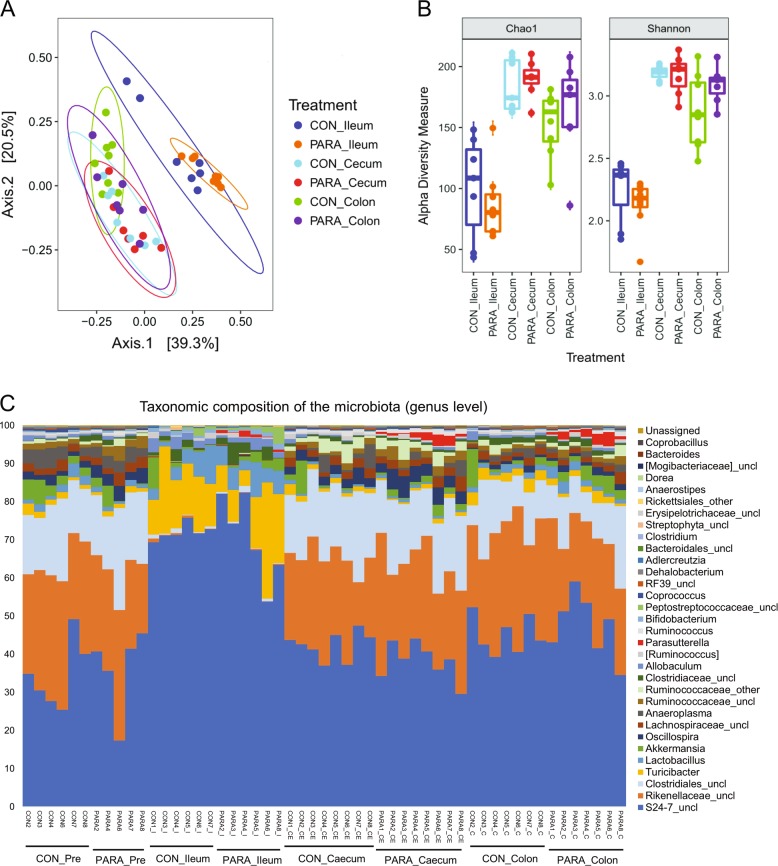
The gut microbiota of the CON and PARA groups was characterized by sequencing 16S rRNA gene amplicons from ileal, cecal, and colonic contents. The sequencing obtained 20,086 ± 7581 (mean ± standard deviation) quality-controlled and chimera-checked reads per sample. The successful colonization of Parasutterella did not cause a significant shift in gut microbiota structure as revealed by the permutational multivariate analysis of variance (Fig. 3a). Specifically, there was no significant difference in the structural pattern between the CON and PARA group at all three intestinal sites, which was evaluated using Bray–Curtis and unweighted UniFrac distance matrices with 999 random permutations (adonis, P > 0.05). Instead, the microbial community was segregated by intestinal sites, which explained 39.3% variation in bacterial composition (adonis, P < 0.001, 999 permutations). The richness and evenness of the microbial community, indicated by alpha-diversity index (Chao1 and Shannon index), did not vary between the CON and PARA groups at each intestinal site (Fig. 3b).
Fig. 3.
Microbial structural analysis of contents collected from different intestinal segments. a Principal coordinates analysis (PCoA) plots of the bacterial communities based on the Bray–Curtis dissimilarity metric. Colors represent intestinal sites and within the same intestinal site, each point represents an individual mouse. For all treatment groups, n = 8. b Alpha-diversity analysis of bacterial communities in ileal, cecal, and colonic contents of mice. All the contents were harvested 6 weeks after Parasutterella/PBS administration. Data are shown as mean ± s.e.m. c Bar chart indicating microbial community profiles between groups, summarized down to the genus level. Microbial composition of the two groups before experimental treatment are labeled as CON_PRE and PARA_PRE (color figure online)
To assess the abundance of different gut microbes in the CON and PARA group, all sequences were assigned to taxonomy using RDP classifier. In accordance with the separation of the overall microbial structure by intestinal sites, the regional variation played a key role in shaping the microbial community. However, at each intestinal site, there were no notable changes in microbial abundance profile between the two treatments. Parasutterella was the only significantly changed genus between the CON and PARA group, contributing the average of 0.64, 1.88, and 1.71% of the 16S rRNA gene reads in the ileum, cecum, and colon, respectively (Fig. 3c).
UPLC-FTMS revealed the effects of Parasutterella colonization on the cecal metabolome
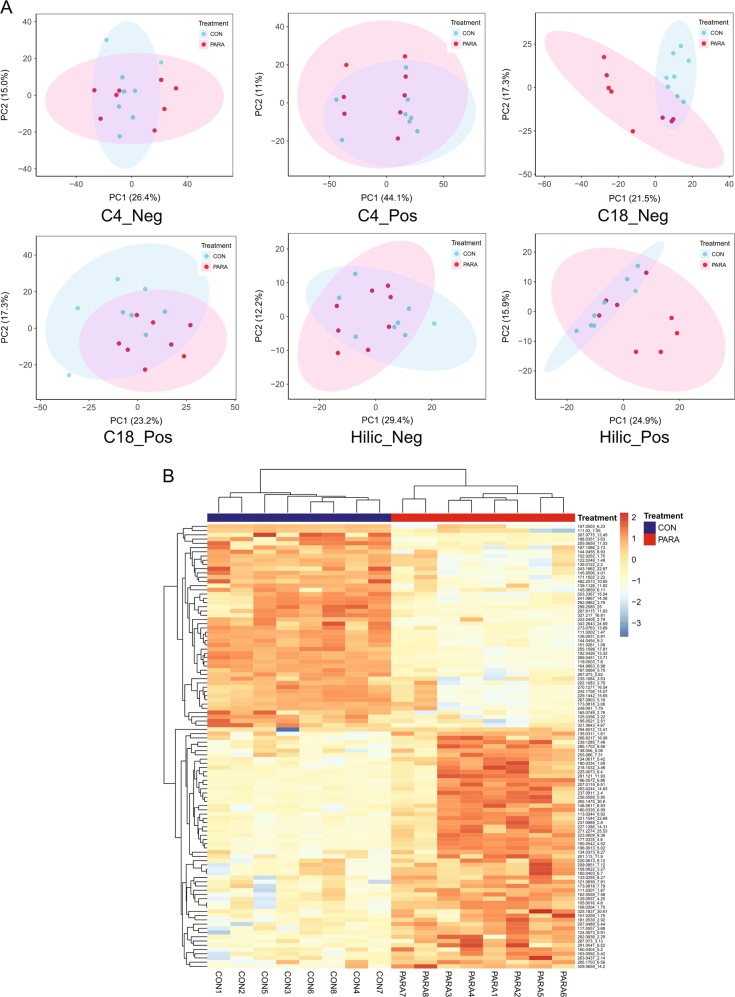
The UPLC-FTMS methodology was optimized to determine the metabolomic profile of cecal contents from the CON and PARA group collected 6 weeks after Parasutterella exposure. To detect as many metabolites as possible, the UPLC-FTMS-based untargeted approach was used, which contained three types of chromatographic conditions (C4, C18, and HILIC column) with two ionization modes (ESI in positive and negative mode, + /–) to generate six possible combinations. The reverse-phase UPLC-FTMS on C18 column with negative ionization mode generated the largest number of features (2165 features) and characterized the most significantly different features (105 features) between the CON and PARA group (Table S3). Principle component analysis (PCA) of all detected features revealed a distinct clustering between the CON and PARA group based on the C18-ESI– dataset (Fig. 4a). The separation was also displayed using the features that were differently presented between treatment groups analyzed by Student’s t-test (n = 8 per treatment, P < 0.05) (Figure S4). In total, 132 of 8990 detected features differentiated the CON and PARA group and the colonization of Parasutterella significantly impacted the features detected by C18-ESI– approach to the greatest extent, indicating the changed profile of non-phospholipid metabolites in cecal contents (Fig. 4b; Table S4, S5). The results showed that the presence of Parasutterella changed the cecal metabolomic profile even though the colonization did not result in a significant shift in microbial community structure.
Fig. 4.
Parasutterella colonization altered the cecal metabolite profile. a PCA plot of features that were detected from the CON and PARA group. Cecal metabolite features were generated from six chromatographic conditions (n = 8 per treatment). Each dot represents an individual mouse. b Heatmap of changed features detected from cecal samples based on the C18-ESI– dataset. Features significantly increased or decreased in the PARA group compared with the CON group are shown, q-value < 0.05. Each column represents an individual mouse (n = 8 per treatment)
The colonization of Parasutterella altered levels of identifiable metabolites
The untargeted metabolomic analysis approach allowed us to detect metabolite features broadly and extensively. However, the identification of metabolites based on detected mass spectra remains challenging due to limited standard reference materials and difficulties in distinguishing the biological source for commonly shared metabolites across the host and gut microbiota [22, 27, 28]. In the current study, the univariate and multivariate approaches were performed on the features detected from the CON and PARA group, and features with a q-value < 0.05 were selected for metabolite identification. The pure standards including certain bile acids were obtained and analyzed under the identical chromatographic condition and ionization mode to validate the spectra from the samples.
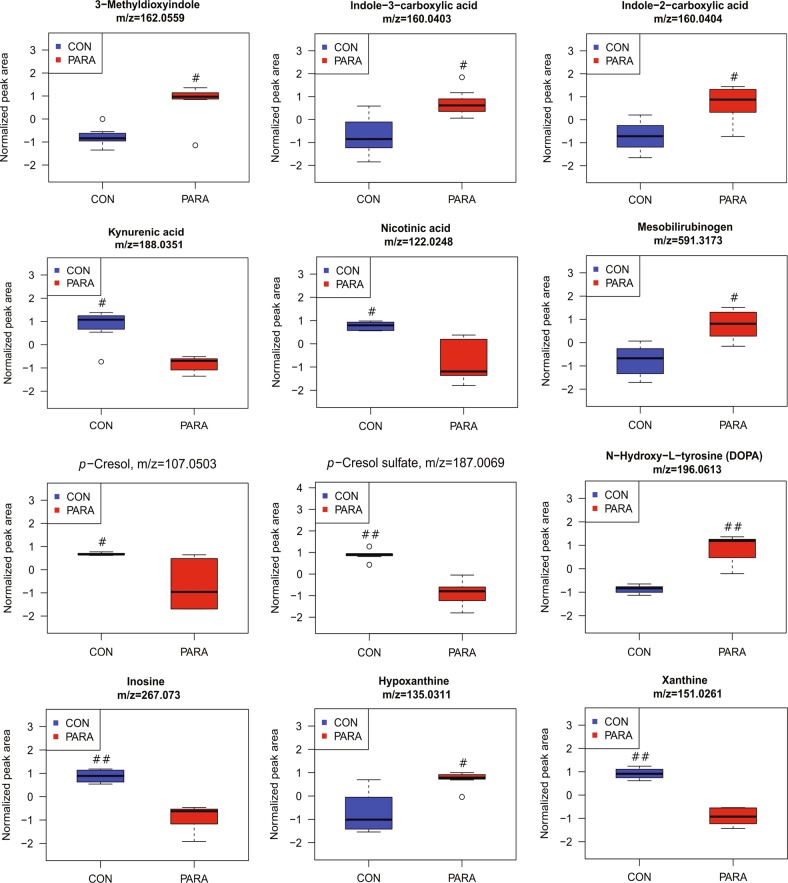
Approximately 40% of the features within the 132 significantly differentiated features remained unidentified, which revealed a substantial proportion of yet-to-be described microbial-associated metabolites. Though the identification of significant metabolites was incomplete, certain metabolites and pathways consistently associated with gut microbial activity were observed. With the colonization of Parasutterella, the level of tryptophan metabolites, 3-Methyldioxyindole (m/z 162.0559, ESI–), indole-2-carboxylic acid (m/z 160.0404, ESI–), and indole-3-carboxylic acid (m/z 160.0403, ESI–) were increased, whereas the abundance of kynurenic acid (m/z 188.0351, ESI–) and nicotinic acid (m/z 122.0248, ESI–) were decreased (q-value < 0.05, Fig. 5). 3-Methyldioxyindole exhibited an 8.3-fold increase and kynurenic acid showed an 11.2-fold decrease in the PARA group compared with the CON group, which were the most notable changes in tryptophan metabolism. The metabolism of another aromatic amino acid, tyrosine, was also altered by Parasutterella colonization, as indicated by reduced levels of p-Cresol (m/z 107.0503, ESI–) and p-Cresol sulfate (m/z 187.0069, ESI–), but elevated ethylphenol (m/z 121.0659, ESI–) and a tyrosine derivative predicted as N-hydroxy-l-tyrosine or DOPA (m/z 196.0613, ESI-) (q-value < 0.05, Fig. 5). p-Cresol and p-Cresol sulfate were decreased 5.6-fold and 43.4-fold in PARA group compared with that in the CON group, whereas the ethylphenol and the tyrosine derivative in PARA group were increased 2.1-fold and 84.3-fold than that in the CON group, respectively. Overall, the results indicate that the presence of Parasutterella significantly changed aromatic amino-acid metabolism in the gut including tryptophan and tyrosine catabolism, and the changes in microbial-associated metabolites indicated an altered functionality of the gut microbial community to utilize these aromatic amino acids.
Fig. 5.
Parasutterella colonization induced changes in cecal metabolites. Changed metabolites associated with tryptophan metabolism (3-Methyldioxyindole, indole-3-carboxylic acid, indole-2-carboxylic acid, kynurenic acid, and nicotinic acid), tyrosine metabolism (p-Cresol, p-Cresol sulfate, and N-hydroxy-l-tyrosine or DOPA), and purine metabolism (inosine, hypoxanthine, and xanthine). Data are presented as box plots, where the boxes represent the 25th to 75th percentiles and the lines within the boxes represent the median. Circles represent values beyond 1.5 times the interquartile range. Significance was indicated as follows, ##q-value < 0.01;#q-value < 0.05. Eight biological replicates from the CON and PARA mice were plotted
Microbial-associated metabolites, mesobilirubinogen (m/z 591.3173, ESI–), and purine metabolites including inosine (m/z 267.073, ESI–), hypoxanthine (m/z 135.0311, ESI–), and xanthine (m/z 151.0261, ESI–) were altered in Parasutterella colonized mice. The bilirubin-derived compound, mesobilirubinogen, was significantly increased (q-value < 0.05, 2.4-fold change) in the PARA group, indicating enhanced metabolism of conjugated bilirubin by gut microbiota (Fig. 5). Altered purine metabolites were observed in PARA group as indicated by remarkably decreased inosine (74.4-fold difference) and xanthine (9.3-fold difference), and increased hypoxanthine (3.4-fold change) (q-value < 0.05, Fig. 5). The alterations in these microbial-derived metabolites revealed a distinct metabolomic signature between the CON and PARA group.
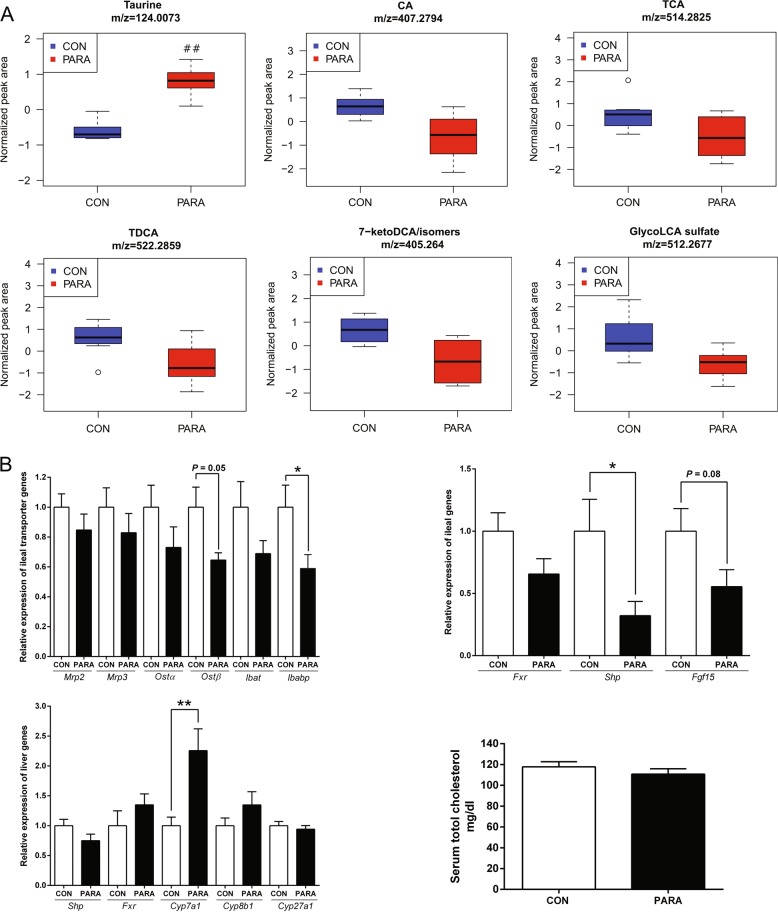
Noteworthy changes in bile acids were consistent between repeated experiments and were identified as significant before fasle discovery rate correction, indicating that Parasutterella may actively participate in bile acid metabolism and homeostasis. A reduction in the level of cholic acid (CA, m/z 407.2794, ESI–, 2.9-fold change), taurocholic acid (TCA, m/z 514.2825, ESI–, 2.2-fold change), taurodeoxycholic acid (TDCA, m/z 522.2859, ESI+, 2.0-fold change), 7-ketodeoxycholic acid or its isomers (7-keto DCA, m/z 405.2640, ESI–, 2.8-fold change), and glycolithocholic acid sulfate (glycol-LCA sulfate, m/z 512.2677, ESI–, 2.3-fold change) was observed in the PARA group, indicating alterations in bile acid metabolism (Fig. 6a). Though the changes in bile acids between the CON and PARA group were not significant based on the q-value, the consistent decreases in these bile acid metabolites clearly suggested that the presence of Parasutterella influenced bile acid profiling in cecal contents. The concomitant increase in taurine (m/z 124.0073, ESI–, 2.0-fold change) concentration further indicated changes in bile acid metabolism, including the process of deconjugating primary bile acids (Fig. 6a). Overall, the bile acid components detected by the current chromatographic conditions were consistently impacted by Parasutterella colonization.
Fig. 6.
Bile acid metabolites and host gene expression. a Changes in bile acid derivatives in cecal contents of the CON and PARA group. Normalized values are presented as box plots, where the boxes represent the 25th to 75th percentiles and the lines within the boxes represent the median. Circles represent values beyond 1.5 times the interquartile range. Eight biological replicates from the CON and PARA mice were plotted for taurine, CA, TCA, TDCA, 7-keto DCA, or its isomers, and Glyco-LCA sulfate. ##q-value < 0.01. b Quantitative RT-PCR results of gene expression. Top left: ileal bile acid transporters including Mrp2, Mrp3, Ostα, Ostβ, Ibat, and Ibabp. Top right: ileal FXR signaling pathway including Fxr, Shp, and Fgf15. Bottom left: liver FXR and bile acid synthesis genes including Shp, Fxr, Cyp7a1, Cyp8b1, and Cyp27a1. Bottom right: serum cholesterol concentration of the CON and PARA mice. For all treatment group, n = 8. Data are shown as mean ± s.e.m. **P < 0.01; *P < 0.05
The alterations in bile acid profile were consistent with hepatic and ileal gene expression
Previous studies have indicated correlations between the abundance of Parasutterella and alterations of bile acid metabolites in the gut [9, 29–31]. To follow-up on differences in bile acid profiles observed, we evaluated host gene expression of bile acid transporters in the ileum and the FXR signaling pathway. Gene expression of ileal bile acid-binding protein (Ibabp) in the PARA group was significantly lower than that in the CON group (P < 0.05), and the organic solute transporter β (Ostβ) tended to be reduced in the PARA group (P = 0.05), which was consistent with the reductions in bile acid metabolites. The mRNA levels of ileal genes in the FXR signaling pathway, including the small heterodimer partner (Shp, P < 0.05) and the fibroblast growth factor 15 (Fgf15, P = 0.08) were decreased in the PARA group. The regulatory effects of gut microbiota on bile acid synthesis through modulating the FXR signaling pathway has been demonstrated [20]. Therefore, we further measured the associated liver gene encoding the rate-limiting enzyme in bile acid synthesis, cholesterol 7α-hydroxylase (Cyp7a1). The expression of Cyp7a1 was significantly higher in Parasutterella colonized mice than that in control mice, which indicated an enhanced bile acid synthesis in the PARA group. The other liver genes related to bile acid synthesis including Cyp8b1 and Cyp27a1 were not affected by Parasutterella colonization, supporting the role of FXR signaling in the Cyp7a1 regulation. The Cyp7a1 expression has been correlated with decreased serum cholesterol in human and mouse studies [32, 33], accordingly, serum cholesterol was measured. Despite a numerical decrease consistent with the hypothesis, no significant difference in serum cholesterol concentration was observed (Fig. 6b).
The presence of Parasutterella did not impact the colonic cytokine expression
To investigate if the colonization of Parasutterella affects the immune response in the gut, a panel of colonic cytokines were measured by enzyme-linked immunosorbent assay. Parasutterella colonization did not alter the level of detected colonic cytokines except for a tendency to reduce the IL-1β expression (P = 0.092, Figure S5), which indicates that the colonization of Parasutterella did not induce the host innate immune responses and further supports the role of Parasutterella as commensal or symbiotic gut microbe.
Discussion
The Parasutterella genus has been recognized for approximately 10 years and increasing studies have shown its relationship with different health outcomes. However, beyond the correlations between the abundance change and host phenotype, our understanding of the metabolic features and functionalities of Parasutterella remains limited. In the current study, the murine Parasutterella was successfully isolated and characterized for its genomic features and metabolic characteristics. A tractable model was subsequently used by adding a single bacterium, Parasutterella mc1, to a complex microbial community to investigate the role Parasutterella plays in affecting the microbial ecosystem and host physiology. Parasutterella successfully thrived in the mouse GIT without shifting microbial structure, but significantly altered the cecal metabolome including multiple biological processes and pathways.
It has been described that the genus of Parasutterella was largely unreactive in biochemical characterization and only limited end products of metabolism have been identified [4]. The type strains of genus Parasutterella, P. excrementihominis YIT11859 and P. secunda YIT12071, were asaccharolytic in the analytical profile index test systems and exhibited weak growth in peptone–yeast extract medium, which could not be enhanced by adding glucose, succinate, or lactate to the broth [2, 3]. In the current study, the mouse isolate showed no turbidity when grown in GAM broth for upto 120 h, which was consistent with previously reported biochemical characteristics. The bacterial density reached 107 CFU/ml in the initial 48 h of growth, however, failed to significantly increase in the following 72 h of culture. The rapid growth of bacterial cells in the first 48 h was accompanied by the depletion of asparagine and a slight decrease of aspartate in the medium, as validated by nuclear magnetic resonance spectroscopy (Figure S2). The supplementation of asparagine and aspartate in the bacterial culture stimulated the growth in the following 24 h (Figure S3). Therefore, in agreement with the presence of genes encoding amino-acid-utilizing enzymes in the genome, the asaccharolytic genus, Parasutterella, is likely to rely on amino acids such as asparagine, aspartate, and serine to support its metabolic activities and physiological functions. In addition, the preferential catabolism of the non-essential amino acids (NEAAs) for the host may indicate the adaptation of Parasutterella to the gut environment, which contains readily available NEAAs.
Importantly, it is the first study showing Parasutterella produces succinate as a fermentative end-product. Succinate production in feces has been positively correlated with the relative abundance of genera including Bacteroides, Lactobacillus, and Parasutterella in a dextran sulfate sodium (DSS)-induced colitis mouse model [34]. The current study provided direct evidence for succinate accumulation in the broth culture of Parasutterella. Additionally, despite the fact that succinate is an intermediate compound in intestinal metabolism, we observed elevated levels of succinate in Parasutterella colonized mice when measured on a C8 column (ESI–, m/z 117.01934, Figure S3). When grown in GAM broth for 72 h, Parasutterella produced about 2.5 mM succinate, whereas Bacteroides fragilis, one of the well-identified succinate producers, was able to produce 100 mM succinate under the same conditions. However, the amount of succinate generated by Parasutterella was substantial when the significant differences in growth rate and bacterial density between Parasutterella and Bacteroides were taken into consideration. Succinate, as one of the key intermediate metabolites produced by gut microbiota, plays an important role in cross-feeding metabolic pathways [35]. The capacity of Parasutterella to produce succinate indicates one potential way Parasutterella supports interspecies metabolic interactions within the gut ecosystem.
Surprisingly, with a single environmental exposure, the Parasutterella isolate successfully colonized the mature mouse gut without causing significant shifts in bacterial composition and host innate immune response. Although Parasutterella has been described as an obligate anaerobe, it was able to persist sufficiently long to colonize the mouse intestine. It has been reported that some strict anaerobic bacteria such as members of Bacteroides can survive and even grow in the presence of low oxygen concentration [36]. We hypothesize that the defined obligate anaerobe, Parasutterella, could protect itself from certain level of oxygen through yet-to-be characterized mechanisms. It is possible that the colonization of Parasutterella led to strain-level differences in gut microbiota that was not detected by 16S rRNA sequencing analysis, however, a lack of changes in the overall structure demonstrated that Parasutterella fills the ecological niche and persists in the gut microbial community. The transmission of Parasutterella has been observed between mother and vaginally born infants, with relative abundance gradually increasing to 12 months of age, indicating that Parasutterella is one of the early colonizers in the newborn gut and increases in response to dietary change and host development [37]. A study using a germ-free mouse model reconstituted with neonatal microbiota indicated that bacterially derived succinate promoted the colonization of strict anaerobic bacteria, Clostridiales, to protect mice from infection [38]. It was assumed that succinate enhanced the growth of Clostridiales through an indirect mechanism, which boosted the consumption of oxygen by aerobic or facultative anaerobic bacteria [38]. Being one of the early colonizers, as well as succinate-producing commensal bacteria, Parasutterella may play a role in microbial interactions and infection resistance especially early in life.
Due to the limited ability to characterize Parasutterella using a culture-based approach, we propose that colonization of Parasutterella in a controlled animal model provides greater insight into its functionality. Despite limited effects on microbial composition, notable changes in cecal metabolic profile including tryptophan, tyrosine, bilirubin, purine, and bile acid metabolism were observed. The presence of Parasutterella influenced the microbial metabolism of aromatic amino acids, as indicated by changes in the abundance of metabolites generated from tryptophan and tyrosine. The decreased level of p-Cresol and p-Cresol sulfate and increased level of ethylphenol, as well as a tyrosine derivative demonstrated an altered tyrosine metabolism. Various species of gut microbiota have been characterized to be capable to convert tyrosine to p-Cresol and ethylphenol including Clostridium, Bifidobacterium, Bacteroides fragilis, and Escherichia coli [39]. The impact on tryptophan metabolism by Parasutterella colonization was indicated by the altered abundance of tryptophan metabolites including kynurenic acid, nicotinic acid, 3-Methyldioxyindole, indole-2-carboxylic acid, and indole-3-carboxylic acid in cecal contents. Tryptophan, as one of the essential amino acids in mammals, can be co-metabolized by the host and gut microbiota through multiple pathways in the GIT. The representative microbial metabolites produced from tryptophan in the intestine include tryptamine, indole, indole acetic acid, indole-propionic acid, and 3-methylindole (skatole) [40, 41]. 3-Methyldioxyindole has been considered as a potential in vivo oxidation product of skatole [42, 43], which perform as a marker of altered tryptophan metabolism [44, 45]. Indole-3-carboxylic acid is a side-chain shortened product of indole-3-acetic acid that has been reported to be produced in the culture medium by certain gut bacterial species [41]. The increased abundance of 3-Methyldioxyindole and indole-3-carboxylic acid in the cecal content suggests that the colonization of Parasutterella enhanced the deamination and chain shortening procedure of tryptophan metabolism in the gut. On the other hand, the concentration of kynurenic acid and nicotinic acid generated from the kynurenine pathway were reduced by the presence of Parasutterella, indicating the impact of Parasutterella colonization on pathway interactions in tryptophan metabolism (Figure S3).
In addition, purine derivatives were identified to be differentiated between the CON and PARA group, including inosine, hypoxanthine, and xanthine. Purine-derived metabolites have been reported to modulate immune responses of immune cells and gut mucosal barrier [46, 47]. A study using mouse DSS-induced colitis model identified that hypoxanthine potentially modulates the energy balance of intestinal epithelium and performs a critical role in maintaining intestinal barrier function [47]. Therefore, in the current study, the elevated level of hypoxanthine resulting from Parasutterella colonization may exert potential beneficial effects on the intestinal mucosal homeostasis. Inosine has been reported to inhibit the Treg cell deficiency-induced autoimmunity in mice and the administration of Lactobacillus reuteri restored the reduction of plasma inosine [48]. The restored plasma inosine was associated with decreased levels in feces, implying that L. reuteri may promote the absorption of inosine in the intestine [48]. In the current study, the cecal abundance of inosine was significantly reduced (74.4-fold change) by the presence of Parasutterella, which may indicate a potential alteration in plasma inosine level and immunomodulatory effects. Future work integrating blood metabolomics approach will be helpful to underpin the effect of Parasutterella colonization on systematic metabolism and function.
Bile acids, in addition to facilitate the absorption of lipid-soluble nutrients in the intestine, have been recognized as important endocrine signaling molecules that impact host physiology, including metabolic processes [49, 50]. In the current study, bile acid metabolism was changed in Parasutterella colonized mice as indicated by the reduced abundance of detected primary bile acids (CA and TCA) and certain secondary bile acids (TDCA, 7-keto DCA, and glycol-LCA) in cecal contents. The genus Parasutterella has been inversely correlated with fecal deoxycholic acid (DCA) and lithocholic acid (LCA) levels in a non-alcoholic steatohepatitis-hepatocellular carcinoma mouse model [9]. Another study using a humanized microbiota mouse model showed a positive association between Parasutterella and beta-muricholic acid as well as 7-keto DCA in feces [29]. Patients with recurrent CDI who were cured by autologous fecal microbiota transplantation had increased relative abundance of Clostridium XIVa members, Holdemania, and Parasutterella. The authors hypothesized that these taxa may play an active role in bile acid metabolism, especially the secondary bile acid biosynthesis [30]. In addition, a remarkable reduction of P. excrementihominis in alcoholic hepatitis patients suggested that Parasutterella species may exert protective effects on liver health [31]. Interestingly, the secondary bile acid ursodeoxycholic acid was identified as a hepatoprotective molecule in the healthy control group [31], which may indicate a correlative relationship between the presence of Parasutterella and increased secondary bile acid in healthy people. In agreement with these previous reports, our results showed that Parasutterella participated in bile acid metabolism, however, due to the limitation of chromatographic conditions in the current study we were unable to demonstrate changes in all bile acid species. In addition, bile acid metabolism is a complex biochemical process including multiple reactions performed by diverse gut microbes. Even though our study used a well-controlled system by adding one single bacterium to a mature microbial community, we could not eliminate the microbial interactions in bile acid transformation. The changes in detected bile acid metabolites may be attributed to the colonization of Parasutterella itself or indirect consequences such as altering the activities of other bacteria by Parasutterella. Due to a paucity of information regarding the capability of Parasutterella to utilize bile acids, future work will focus on the mechanism driving altered bile acid metabolism by Parasutterella colonization.
Consistent with bile acid modifications, gene expression of ileal bile acid transporters, including Ibabp and Ostβ, was decreased. The transporter encoded by Ibabp actively interacts with FXR signaling pathway to modulate FXR expression in the small intestine [51] and the Ost-α/β complex can be induced by FXR, as well as bile acids [52]. Moreover, FXR has been reported to be antagonized by certain bile acids such as the tauro-conjugated muricholic acid in mice, and consequently modulate the expression of Cyp7a1 in the liver [20, 50]. In the current study, the reductions in gene expression of bile acid transporters, as well as the inhibition of FXR signaling pathway matched the elevated expression of liver Cyp7a1 gene, which revealed that the colonization of Parasutterella modified the bile acid metabolites and thereby impacted bile acid transport and synthesis. These effects on bile acid transporters and FXR signaling pathway may be exerted directly by Parasutterella or indirectly via microbial interactions. Additionally, there was no significant difference in serum cholesterol between the CON and PARA group, which may be explained by the normal physiological state of the mice. Future studies incorporating a high-fat dietary treatment will be helpful to investigate the effects of Parasutterella-induced Cyp7a1 expression on host physiology.
In summary, the results show the potential metabolic functionality of Parasutterella as a member of gut microbiota. It is the first study to reveal the chemical “fingerprint” of Parasutterella and highlight the potential to impact the host physiology via modulating the abundance or the function of commensal bacteria in the gut. Additionally, the controlled model system of adding a single commensal microorganism to a complex community can be applied to investigate other commensal species of interest.
Supplementary information
Acknowledgements
We thank the Metabolomics Innovation Centre (TMIC) and UVic Genome BC Proteomic Centre for the support of untargeted metabolomics analysis. We are also grateful to Dr. Michael Gänzle (University of Alberta, Edmonton, Canada) and his assistance for the HPLC system. TJ was supported by a Graduate Student Scholarship from the Alberta Innovates-Technology Futures, Alberta, Canada. BPW is supported by the Canada Research Chair Program.
Funding
This research was supported by the Natural Sciences and Engineering Research Council Discovery Grant # 436154 held by BPW.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary material
The online version of this article (10.1038/s41396-019-0364-5) contains supplementary material, which is available to authorized users.
References
- 1.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zeng Z, et al. A pyrosequencing study in twins shows that GI microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 2.Nagai F, Morotomi M, Sakon H, Tanaka R. Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces. Int J Syst Evol Microbiol. 2009;59:1793–7. doi: 10.1099/ijs.0.002519-0. [DOI] [PubMed] [Google Scholar]
- 3.Morotomi M, Nagai F, Watanabe Y. Parasutterella secunda sp. nov., isolated from human faeces and proposal of Sutterellaceae fam. nov. in the order Burkholderiales. Int J Syst Evol Microbiol. 2011;61:637–43. doi: 10.1099/ijs.0.023556-0. [DOI] [PubMed] [Google Scholar]
- 4.Morotomi M. The family Sutterellaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin, Heidelberg: Springer; 2014. p. 1005-12.
- 5.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Blasco-Baque V, Coupé B, Fabre A, Handgraaf S, Gourdy P, Arnal JF, et al. Associations between hepatic miRNA expression, liver triacylglycerols and gut microbiota during metabolic adaptation to high-fat diet in mice. Diabetologia. 2017;60:690–700. doi: 10.1007/s00125-017-4209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreutzer C, Peters S, Schulte DM, Fangmann D, Türk K, Wolff S, et al. Hypothalamic inflammation in human obesity is mediated by environmental and genetic factors. Diabetes. 2017;66:2407–15. doi: 10.2337/db17-0067. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012;6:1848–57. doi: 10.1038/ismej.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie G, Wang X, Huang F, Zhao A, Chen W, Yan J, et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int J Cancer. 2016;139:1764–75. doi: 10.1002/ijc.30219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danneskiold-Samsøe Niels Banhos, Andersen Daniel, Radulescu Ilinca Daria, Normann-Hansen Ann, Brejnrod Asker, Kragh Marie, Madsen Tobias, Nielsen Christian, Josefsen Knud, Fretté Xavier, Fjaere Even, Madsen Lise, Hellgren Lars I., Brix Susanne, Kristiansen Karsten. A safflower oil based high-fat/high-sucrose diet modulates the gut microbiota and liver phospholipid profiles associated with early glucose intolerance in the absence of tissue inflammation. Molecular Nutrition & Food Research. 2017;61(5):1600528. doi: 10.1002/mnfr.201600528. [DOI] [PubMed] [Google Scholar]
- 11.Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GM, Neyrinck AM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–86. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzler-Zebeli BU, Schmitz-Esser S, Mann E, Grüll D, Molnar T, Zebeli Q. Adaptation of the cecal bacterial microbiome of growing pigs in response to resistant starch type 4. Appl Environ Microbiol. 2015;81:8489–99. doi: 10.1128/AEM.02756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staley C, Kaiser T, Beura LK, Hamilton MJ, Weingarden AR, Bobr A, et al. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome. 2017;5:87. doi: 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Dong D, Jiang C, Li Z, Wang X, Peng Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe. 2015;34:1–7. doi: 10.1016/j.anaerobe.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Brahma S, Martínez I, Walter J, Clarke J, Gonzalez T, Menon R, et al. Impact of dietary pattern of the fecal donor on in vitro fermentation properties of whole grains and brans. J Funct Foods. 2017;29:281–9. doi: 10.1016/j.jff.2016.12.042. [DOI] [Google Scholar]
- 16.Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26:110–30. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan S, Alden N, Lee K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr Opin Biotechnol. 2015;36:137–45. doi: 10.1016/j.copbio.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Di Yu, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA. 2010;107:228–33. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–35. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013;27:701–18. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–43. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–60. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ju T, Shoblak Y, Gao Y, Yang K, Fouhse J, Finlay BB, et al. Initial gut microbial composition as a key factor driving host response to antibiotic treatment, as exemplified by the presence or absence of commensal Escherichia coli. Appl Environ Microbiol. 2017;83:e01107–17. doi: 10.1128/AEM.01107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted metabolomics strategies-challenges and emerging directions. J Am Soc Mass Spectrom. 2016;27:1897–905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alonso A, Marsal S, Julia A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3:23. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey N, Wagner VE, Blanton LV, Cheng J, Fontana L, Haque R, et al. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell. 2015;163:95–107. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staley C, Kelly CR, Brandt LJ, Khoruts A, Sadowsky MJ. Complete microbiota engraftment is not essential for recovery from recurrent Clostridium difficile infection following fecal microbiota transplantation. mBio. 2016;7:e01965–16. doi: 10.1128/mBio.01965-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830–9. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 32.Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, et al. Human cholesterol 7α-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110:109–17. doi: 10.1172/JCI0215387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin JZ, Martagón AJ, Hsueh WA, Baxter JD, Gustafsson JAring, Webb P, et al. Thyroid hormone receptor agonists reduce serum cholesterol independent of the LDL receptor. Endocrinology. 2012;153:6136–44. doi: 10.1210/en.2011-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osaka T, Moriyama E, Arai S, Date Y, Yagi J, Kikuchi J, et al. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients. 2017;9:E1329. doi: 10.3390/nu9121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–47. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–4. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 37.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG, Pudlo NA, et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science. 2017;356:315–9. doi: 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 40.Hubbard TD, Murray IA, Perdew GH. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab Dispos. 2015;43:1522–35. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523–35. doi: 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- 42.Albrecht CF, Chorn DJ, Wessels PL. Detection of 3-hydroxy-3-methyloxindole in human urine. Life Sci. 1989;45:1119–26. doi: 10.1016/0024-3205(89)90169-0. [DOI] [PubMed] [Google Scholar]
- 43.Skiles GL, Adams JD, Jr, Yost GS. Isolation and identification of 3-hydroxy-3-methyloxindole, the major murine metabolite of 3-methylindole. Chem Res Toxicol. 1989;2:254–9. doi: 10.1021/tx00010a007. [DOI] [PubMed] [Google Scholar]
- 44.Zheng X, Xie G, Zhao A, Zhao L, Yao C, Chiu NH, et al. The footprints of gut microbial-mammalian co-metabolism. J Proteome Res. 2011;10:5512–22. doi: 10.1021/pr2007945. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y, Men L, Pi Z, Wei M, Song F, Zhao C, et al. Fecal metabolomics of type 2 diabetic rats and treatment with Gardenia jasminoides Ellis based on mass spectrometry technique. J Agric Food Chem. 2018;66:1591–9. doi: 10.1021/acs.jafc.7b06082. [DOI] [PubMed] [Google Scholar]
- 46.Haskó G, Sitkovsky MV, Szabó C. Immunomodulatory and neuroprotective effects of inosine. Trends Pharmacol Sci. 2004;25:152–7. doi: 10.1016/j.tips.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Lee JS, Wang RX, Alexeev EE, Lanis JM, Battista KD, Glover LE, et al. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J Biol Chem. 2018;293:6039–51. doi: 10.1074/jbc.RA117.000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He B, Hoang TK, Wang T, Ferris M, Taylor CM, Tian X, et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med. 2017;214:107–23. doi: 10.1084/jem.20160961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. 2011;108:4523–30. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Nakahara M, Furuya N, Takagaki K, Sugaya T, Hirota K, Fukamizu A, et al. Ileal bile acid-binding protein, functionally associated with the farnesoid X receptor or the ileal bile acid transporter, regulates bile acid activity in the small intestine. J Biol Chem. 2005;280:42283–9. doi: 10.1074/jbc.M507454200. [DOI] [PubMed] [Google Scholar]
- 52.Landrier JF, Eloranta JJ, Vavricka SR, Kullak-Ublick GA. The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-α and -β genes. Am J Physiol Gastrointest Liver Physiol. 2006;290:G476–85. doi: 10.1152/ajpgi.00430.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.