Abstract
Almost all the inorganic carbon on Earth is converted into biomass via the Calvin–Benson–Bassham (CBB) cycle. Here, the central carboxylation reaction is catalyzed by ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), which can be found in numerous primary producers including plants, algae, cyanobacteria, and many autotrophic bacteria. Although RubisCO possesses a crucial role in global biomass production, it is not a perfect catalyst. Therefore, research interest persists on accessing the full potential of yet unexplored RubisCOs. We recently developed an activity-based screen suited to seek active recombinant RubisCOs from the environment—independent of the native host’s culturability. Here, we applied this screen to twenty pre-selected genomic fosmid clones from six cultured proteobacteria to demonstrate that a broad range of phylogenetically distinct RubisCOs can be targeted. We then screened 12,500 metagenomic fosmid clones from six distinct hydrothermal vents and identified forty active RubisCOs. Additional sequence-based screening uncovered eight further RubisCOs, which could then also be detected by a modified version of the screen. Seven were active form III RubisCOs from yet uncultured Archaea. This indicates the potential of the activity-based screen to detect RubisCO enzymes even from organisms that would not be expected to be targeted.
Subject terms: Water microbiology, Microbial ecology, Functional genomics
Introduction
Around 200 gigatones of CO2 are annually fixed via the Calvin–Benson–Bassham (CBB) cycle [1]. Its key enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO, EC 4.1.1.39) mediates the actual step of carbon fixation and attaches CO2 to ribulose-1,5-bisphosphate (RuBP) to produce two molecules of 3-D-phosphoglycerate (3-PGA). Despite RubisCO’s major role in Earth’s net primary production, the enzyme is quite an inefficient catalyst (slow turnover and wasteful oxygenase activity) [2]. Catalytic properties of RubisCOs from phylogenetically distinct organisms vary [1] and some in fact may have the potential to transfer superior photosynthetic performance to specific crops. With the world’s population growing rapidly, improvement of agricultural productivity is a major task for meeting humanity’s increasing nutritional requirements. Hence, great efforts have been made for enhancing autotrophic carbon fixation: they range from attempts to modify existing RubisCOs to the development of completely artificial CO2 fixation pathways ([3] and references therein).
Up until now four forms of RubisCO are distinguished. However, only forms I, II, and III have been shown to possess RuBP-dependent CO2 incorporation abilities [2, 4]. It was shown that the properties of different RubisCO forms evolved as a consequence of selective pressure suggesting that each RubisCO form is optimized to its environment (e.g., through its CO2/O2 specificity, its turnover velocity, and its optimum temperature) [5]. All three forms (I, II, and III) have been identified in hydrothermal vent habitats [6–8]. Here hot hydrothermal fluids enriched with reduced inorganic electron donors e.g., hydrogen and sulfide [9, 10] emit from inner earth. As the fluids ascend they become admixed with cold, oxygenated seawater. This chemical disequilibrium is exploited by chemosynthetic microorganisms, which oxidize reduced inorganic compounds and use the gained energy to fix carbon autotrophically.
Nevertheless, our ability to discover RubisCO enzymes from the environment is limited, given that the vast majority of all microorganisms on earth are currently not cultivable. So far, this resource of uncultured microorganisms has been largely untapped, primarily due to difficulties in accessing it. Sequence-based techniques have been used to fish RubisCOs from metagenomes [11] but can only recognize RubisCO genes if a comparable sequence is available in the public databases. In contrast, activity-based tools can expand the spectra of detectable active RubisCO enzymes to those not necessarily originating from known and/or cultured organisms [12]. Even though drawbacks exist when expressing environmental DNA in a surrogate host (troubles are associated with e.g., recognition of intrinsic promoters, codon usage, translation, assembly and folding of enzymes etc.) [13], it is the only way to discover truly novel enzymes [14].
In 2015, we described a new activity-based technique that enables us to recover recombinant active RubisCO enzymes from the environment [12]. In the current study, we first tested the applicability of this screen and investigated the expressability of diverse proteobacterial RubisCOs in our host–vector system. Second, we aimed at seeking novel active RubisCO enzymes from the environment. For this purpose we screened six metagenomic fosmid libraries from hydrothermal vent environments, differing with respect to their (i) geographic origin (north or south Mid Atlantic Ridge) and their (ii) environmental characteristics (basalt vs. ultramafic hosted, temperature, and pH). We also modified the screen to target RubisCOs that are primarily active under anoxic conditions. The identified recombinant RubisCOs were biochemically characterized (temperature and O2 tolerance) and related to local habitat characteristics.
Material and methods
Constructing genomic fosmid libraries
Six genomic fosmid libraries were constructed with DNA isolated from (i) Hydrogenovibrio crunogenus TH-55 (formerly Thiomicrospira crunogena [15]), (ii) Thiobacillus denitrificans AB7, (iii) Rhodovulum sulfidophilum W4, (iv) Thioclava pacifica TL 2, (v) Rhodobacter capsulatus SB 1003, and (vi) Rhodobacter sphaeroides ATH 2.4.1. All strains were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) with the exception of R. capsulatus SB 1003, which was provided by Prof. Gabriele Klug from the University of Giessen. For DSMZ numbers and details on cultivation see Supplementary Material and methods S1. Genomic DNA (gDNA) was isolated as described in Supplementary Material and methods S2. gDNA concentrations and qualities were evaluated as reported in Supplementary Material and methods S3. The genomic libraries were constructed using the CopyControl™ Fosmid Library Production Kit (epicentre®, Madison, WI, USA) according to manufacturer’s protocol. Quality of fosmid inserts were randomly checked using restriction analyses and insert-end sequencing (for further details see Supplementary Material and methods S4).
Sampling of hydrothermal vents
Six hydrothermally influenced environmental samples from along the Mid-Atlantic Ridge were investigated (see Supplementary Fig. S1 for location of sampling sites). These included chimney samples from (i) Sisters Peak at 4°48′S/12°22′W [16], from (ii) Mephisto at 4°47′S/12°22′W [16], and from (iii) Site B at 14°45′N/44°58′W ([9] and references therein) and hydrothermal fluids from (i) the interface zone between hot fluids emanating from Drachenschlund and ambient seawater at 8°18′S/13°30′W [17], from (ii) the mussel bed at Irina II at 14°45′N/44°58′W [10], and from (iii) the Bathymodiolin mussel assemblages at the Lilliput main site at 9°32′S/13°12′W [6]. Physical and chemical parameters specific for the studied hydrothermal vent locations are summarized in Supplementary Table S1. Detailed sampling procedures and further information on the sampling sites are described elsewhere (see Supplementary Materials and methods S5 and [6, 9, 10, 18–21]).
Constructing metagenomic fosmid libraries
For constructing metagenomic libraries from the six sampled hydrothermal sites, whole metagenomic DNA (mDNA) was isolated as described in Supplementary Material and methods S2. Multiple displacement amplification (MDA) was used to increase mDNA concentrations and improve mDNA purities (see Supplementary Material and Methods S3). Resulting MDA treated mDNA was used for the construction of metagenomic libraries by using the CopyControl™ Fosmid Library Production Kit (epicentre®) according to manufacturer’s protocol. Quality of fosmid inserts was validated performing (i) restriction analysis and (ii) fosmid insert-end sequencing (see Supplementary Material and methods S4, for results see Supplementary Fig. S2). Total clone numbers and average insert sizes of the six metagenomic libraries are summarized in Table 1.
Table 1.
Summary of all tested and identified metagenomic fosmid clones
| sampling sites | Average insert size [kb] | Total number of clones | Number of tested clones | Number of identified clones | |||||
|---|---|---|---|---|---|---|---|---|---|
| Via activity-based screen | Via sequence-based screen | ||||||||
| Form I | Form II | Form III | |||||||
| RubIrF-RubIrR | RubIgF-RubIgR | cbbLF-cbbLR | RuIIF1-RuIIR3 | arubIIIF-arubIIIR | |||||
| Basalt-hosted | |||||||||
| Sisters Peak, 5°S (274 ROV1B, MAR-SUED V) | 39 | 14,000 | 2,496 | 0 | 0 | 0 | 0 | 0 | 7 |
| Mephisto, 5°S (297 ROV 1, MAR-SUED V) | 26 | 20,500 | 2,496 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lilliput, 9°S (324 ROV 6–8, MAR-SUED V) | 23 | 15,000 | 2,496 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ultramafic-hosed | |||||||||
| Site B, 15°N (313 ROV, HYDROMAR VII) | 31 | 17,000 | 2,400 | 0 | 1 | 0 | 1 | 0 | 0 |
| Irina II, 15°N (300 ROV 19–21, HYDROMAR VII) | 31 | 18,000 | 2,688 | 1 | 0 | 0 | 0 | 1 | 0 |
| Drachenschlund, 8°S (314 ROV 7–9, MAR-SUED V) | 37 | 20,200 | 2,496 | 39 | 39 | 0 | 0 | 39 | 0 |
Seeking RubisCOs by activity-based screening
Roughly 12,500 metagenomic fosmid clones from the six constructed metagenomic fosmid libraries (∼2,600 clones each) were screened for clones harboring RubisCO activities by using the recently established high performance liquid chromatography (HPLC) based screening procedure [12, 22] with one modification: crude extracts were stored over night at 4 °C prior to using it as template to perform the RubisCO activity assay (see Supplementary Material and methods S6 for a brief methods description of the used activity-based RubisCO screen).
Measuring RubisCO activities of single clones
Specific RubisCO activities of 20 genomic and 48 metagenomic fosmid clones were measured using HPLC as described for fosmid clone pools (see Supplementary Material and methods S6). Some modifications with respect to culturing and assay performance were applied (see Supplementary Material and methods S7).
Seeking RubisCO structural genes by sequence-based screening
(i) Genomic as well as (ii) metagenomic fosmid libraries were screened on a sequence-based level for clones encoding RubisCO structural genes on their inserts. Primer pairs and corresponding PCR conditions used for the sequence-based screening of (i) genomic fosmid libraries are listed in Supplementary Table S2. The six (ii) metagenomic fosmid libraries were each screened for RubisCO forms I, II, and III structural genes using the following five degenerate primes pairs, respectively: (I) cbbLF and cbbLR [23], (II) RubIgF and RubIgR [24], (III) RubIrF and RubIrR [24], RuIIF1 and RuIIR3 [24] and arubIIIF (5′-GCCGGKAAYGTDTTYGGMATGAAG-3′) and arubIIIR (5′-GYRCTYCCYTCCATCTTGCC-3′). PCR conditions for the latter were: denaturation at 95 °C for 30 s, primer annealing at 62 °C for 30 s, and elongation at 72 °C for 45 s (32 cycles). For all other primer pairs PCR conditions were as described elsewhere [23, 24] with the exception that the annealing temperature of cbbLF and cbbLR was adapted from 48 °C to 46 °C. Amplification was done with the GoTaq® DNA Polymerase (Promega, Mannheim, Germany), according to manufacturer’s instructions with pools of 96 fosmid clones as template. For detailed information on the preparation of PCR suited DNA pools see Supplementary Material and method S8. If a pool of 96 clones was tested positive, it was broken down until the RubisCO harboring clone was identified (see Supplementary Material and method S9). Insert sizes of all identified metagenomic clones were determined using restriction analyses (for results see Supplementary Table S3).
Primer walk, de novo sequencing, and sequence editing
RubisCO-containing DNA inserts of identified (meta)genomic fosmid clones were sequenced starting from both insert ends (Eurofins MWG Operon, Ebersberg, Germany) using T7 promoter primer and pCC1FOS reverse sequencing primer (epicentre®). Sequences were aligned and edited with Lasergene Software SeqMan (DNA Star, Madison, WI, USA). Homologies were evaluated by comparing generated sequences with those deposited in the NCBI database (National Center for Biotechnology Information, Bethesda, MD, USA), using nucleotide BLAST (blastn) search (Basic Local Alignment Search Tool) [25]. For the most active metagenomic clones IrII31F9 and SB29A9 sequencing were continued through primer walking (for details of used primers see Supplementary Tables S4 and S5, respectively). Open reading frames (orfs) were searched using Prodigal with metagenomic settings [26]. Functions of identified orfs were deduced from sequences with highest similarities collected from blastp searches [27].
Sequence access
The nucleotide sequence of partial insert sequences were submitted to the GenBank database of NCBI under following accession numbers: (i) MK204370—Fragment A of IrII31F9, (ii) MK204371—Fragment BC of IrII31F9, (iii) MK204372—Fragment A of SB29A9, (iv) MK204373—Fragment B of SB29A9, (v) MK204374—Fragment C of SB29A9, (vi) MK564616—RubisCO form III of SP52C8, (vii) MK564617—RubisCO form III of SP88G2, (viii) MK564618—RubisCO form III of SP92E10, (ix) MK564619—RubisCO form III of SP96F12, (x) MK564620—RubisCO form III of SP119A5, and (xi) MK564621—RubisCO form III of SP133E7.
Results and discussion
We applied a recently described activity-based screen [12] for recovering recombinant active RubisCO enzymes from metagenomic fosmid libraries. First, we demonstrated the feasibility of the screen to target RubisCOs from phylogenetically distinct Alpha-, Beta-, and Gammaproteobacteria (Fig. 1A) and investigated the role that some flanking genes may play for RubisCO activity. Second, we screened metagenomic libraries constructed with environmental DNA from six hydrothermal habitats to recover RuBP converting enzymes (Fig. 1B). For the latter we additionally conducted the activity-based assay under anoxic conditions and characterized the biochemical properties of identified RubisCO forms to test whether an interconnection between habitat and enzyme specificities exists.
Fig. 1.
Overview of the experimental setup. The overview summarizes the experimental setup. A) An existing activity-based RubisCO screen was evaluated using six phylogenetically distinct bacteria, namely Alpha-, Beta-, and Gammaproteobacteria. B) The validated activity-based screen was applied to six distinct metagenomic deep-sea vent fosmid libraries. Color-schemes used in A), B) re-occur in Figs. 3 and 4, respectively. The photo of the hydrothermal deep-sea vent chimney is protected by copyright of the ROV Kiel 6000, GEOMAR
Genomic fosmid clones of cultured representatives
Feasibility of the activity-based screen for seeking RubisCOs from distinct Alpha-, Beta-, and Gammaproteobacteria
RubisCO genes were heterologously expressed from six microorganisms in our host–vector system (Fig. 1A). These were from (i) the Gammaproteobacterium H. crunogenus (forms I and II), (ii) the Betaproteobacterium T. denitrificans (forms I and II), and the four Alphaproteobacteria (iii) R. sulfidophilum (form I), (iv) R. capsulatus (form II), (v) T. pacifica (form I), and (vi) R. sphaeroides (form I and form II) (for RubisCO classification see Fig. 2, for gene arrangements see Supplementary Fig. S3). Structural RubisCO genes alone are often not enough to express a fully functional RubisCO and lacking gene products of adjacent genes may cause a dramatic loss of RubisCO activity [12, 22]. Hence, we worked with genomic fosmid clones carrying structural RubisCO genes and adjacent flanking gene regions. For each isolate recombinant RubisCO activities of at least three genomic fosmid clones were measured. Clones comprising most contrasting flanking regions were selected. Overall, 20 genomic fosmid clones were tested of which 15 exhibited heterologous RubisCO activity in our host–vector system. Gene arrangements of selected genomic fosmid clones and respective RubisCO activities are summarized in Fig. 3. Fosmid clones with gDNA inserts from T. pacifica (Tpac14A2) and H. crunogenus (Hcrun6F3, Hcrun9C4, and Hcrun9E6) had activities between 166 and 455 nmol 3-PGA mg−1 min−1 and would have been found in the pools of 24 clones (pool size commonly used for screening). Activities of fosmid clones with gDNA fragments from R. sphaeroides (Rsph1C7), R. capsulatus (Rcap18B6), and T. pacifica (Tpac5B3) were with 37 ± 8, 35 ± 7 and 33 ± 9 nmol 3-PGA mg−1 min−1 below the detection limit of our used HPLC based RubisCO screen (≥50 nmol 3-PGA mg−1 min−1) and would only have been detected if the pool size of clones was reduced to 8 instead of 24 clones. The fosmid clones exhibiting RubisCO activities below 20 nmol 3-PGA mg−1 min−1—including that of T. denitrificans (Tden7G11 and Tden10F2) or R. sulfidophilum (Rsul7B4, Rsul10E8, and Rsul9H7)—would have only been identified if clones had been screened individually.
Fig. 2.
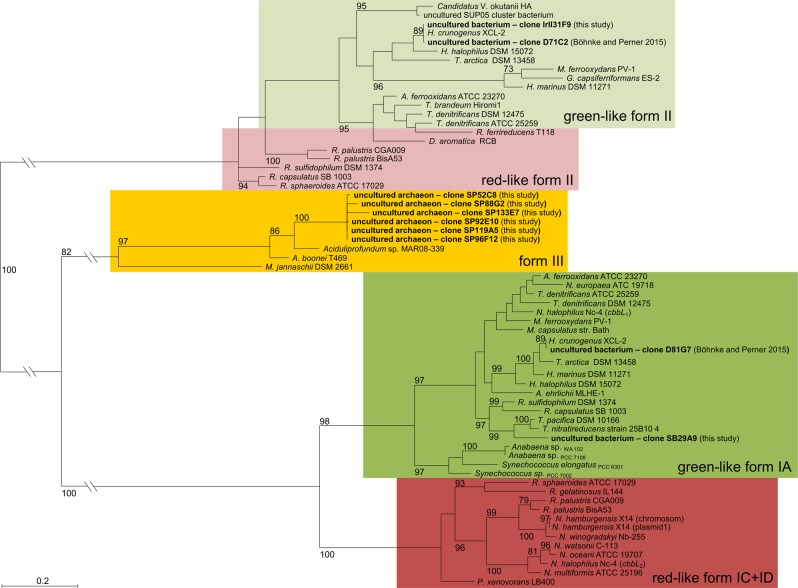
Classification of red-type and green-type forms I and II RubisCO genes. The classification of identified metagenome derived RubisCO genes are shown together with red-type and green-type form I (cbbL) and form II (cbbM) RubisCO genes of representative microorganisms. The phylogenetic tree was calculated based on the amino-acid sequences using Maximum-Likelihood analyses (accession numbers of included genes are listed in Supplementary Table S8). Bootstrap values, calculated for 100 replicates, are presented as percentages at the node and are indicated only when above 70%. The scale bar represents the expected number of changes per amino acid position
Fig. 3.
Evaluating the spectra of the applied activity-based RubisCO screen. Gene arrangements and recombinant RubisCO activities of different fosmid clones isolated from genomic libraries of: A Hydrogenovibrio crunogenus XCL-2 (NCBI accession: NC_007520.2), B Thiobacillus denitrificans AB 7 (NCBI accession: NZ_AQWL01000006.1), C Rhodovulum sulfidophilum W4 (NCBI accession: NZ_DF260912), D Thioclava pacifica TL 2 (NCBI accession: NZ_AUND01000034.1), EI, II Rhodobacter capsulatus SB 1003 (NCBI accession: NC_014034.1), and FI, II Rhodobacter sphaeroides ATH 2.4.1 (NCBI accession: Chr1 - NC_007493.2 and Chr2 - NC_007494.2). The order of genes and corresponding functions are deduced from blastn annotations. Arrows indicate orfs in the direction of transcription. Used gene abbreviations are as follows: cbbQ-m/cbbQ-1—AAA + protein, encoding prokaryotic RubisCO activase of green-type RubisCO form IA; cbbO-m/cbbO-1—von Willebrand factor type A operating as adaptor between RubisCO and CbbQ; cbbM—ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, form II; cbbR1/cbbRm—LysR type transcriptional RubisCO regulators; cbbL—ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, form I; cbbS—ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit, cbbA—fructose-1,6-bisphosphate aldolase; cbbT—transketolase; cbbF—fructose-1,6-phosphosphatase; cbbZ—phospoglycolate phosphatase; cbbG—glyceraldehyd-3-phosphate dehydrogenase; cbbP—phosphoribulokinase; cbbE—ribulose-phosphate-3-epimerase; cbbY—CbbY family protein with predicted function in carbohydrate transport and metabolism; moxR—MoxR-like ATPase; vWA domain—Von Willebrand domain containing protein; luxR—LuxR family transcriptional regulator
Impact of flanking regions on RubisCO activity from tested proteobacteria
Depending on which of the flanking genes were present or absent, the RubisCO activities of the three fosmid clones from the same isolate could differ significantly (p-value < 0.05). Affected were the genomic fragments from H. crunogenus, T. pacifica, R. capsulatus, and R. sphaeroides (Fig. 3A, D, EII, and FII, respectively). Indeed, we have shown before that the degree of RubisCO activity appears to relate to the presence of distinct adjacent genes [12].
H. crunogenus—forms I and II RubisCO
RubisCO activities from the different genomic H. crunogenus fosmid clones Hcrun9C4 and Hcrun9E6 were 32% and 63% lower than activities measured for Hcrun6F3 (Fig. 3A). The three genes TCR_RS02240, TCR_RS02245, and TCR_RS02265 are missing on the genomic fragment of clone Hcrun9C4 but are present on the Hcrun6F3 fosmid insert (Fig. 3A). Gene products of these three genes have been previously demonstrated to affect RubisCO activity in another metagenome derived H. crunogenus related DNA fragment [12]. Thus, at least one of these three genes seems to be responsible for the different RubisCO activities. Although Hcrun9E6 possesses all orfs that are posited to influence RubisCO activity, RubisCO activity of Hcrun9E6 was reduced threefold relative to Hcrun6F3 (Fig. 3A). However, on the Hcrun9E6’s genomic fragment an additional gene, namely TCR_RS02100 encoding an ATP-dependent protease subunit HsIV, is present. This gene product appears to directly or indirectly affect RubisCO in its functioning.
T. pacifica—form I RubisCO
The clone Tpac14A2 with genomic DNA form T. pacifica had eight- and five-fold higher RubisCO activities than the two genomic fosmid clones Tpac4C12 and Tpac5B3, respectively (Fig. 3D). Unlike the other two genomic fragments, Tpac14A2 lacks the genes TP2_RS12655, an elongation factor, and TP2_RS12660, a LuxR family transcriptional regulator. LuxR regulators are known to work both as activators and repressors [28]. As the RubisCO activity increased when the LuxR gene is absent (Fig. 3D), it here may directly or indirectly function as a repressor for RubisCO.
R. capsulatus—form I and form II RubisCO
Genomic R. capsulatus fosmid clones Rcap12D9 and Rcap12G12 had comparable form I RubisCO activities although their DNA inserts are very contrasting (additional 16.7 kb DNA upstream in Rcap12G12 and instead 20.6 kb downstream on Rcap12D9) (Fig. 3EI). The RubisCO form II activity of the R. capsulatus genomic fosmid clone Rcap18B6 was, on the contrary, significantly higher (<0.001 p-value) than that of Rcap20G4 (Fig. 3EII). Relative to Rcap18B6, the fosmid clone Rcap20G4 has additional 23 orfs (26.1 kb) upstream of the form II RubisCO gene cluster and misses 11 orfs (15.1 kb) downstream. Specifying “the one” orf responsible for the varying RubisCO activity is hardly possible. However, one promising gene missing on the genomic fragment of Rcap18B6 encodes a LuxR gene. Its gene product may function here as a repressor.
R. sphaeroides—form I and form II RubisCO
No difference in form I RubisCO activities from R. sphaeroides genomic fosmid clones Rsph3A12 and Rsph9D6 was observed, although flanking gene regions are very diverging: i.e., Rsph3A12 had 25 additional orfs upstream of the form I RubisCO gene cluster (22.4 kb) but lacks 14 orfs downstream (17.4 kb), relative to Rsph9D6 (Fig. 3FI). In case of R. sphaeroides fosmid clones, Rsph1C7’s RubisCO form II activity is significantly higher (p-value < 0.001) than that of Rsph9E12 (Fig. 3FII). Relative to the fosmid clone Rsph9E12, Rsph1C7 lacks genes at the 5′ end which encode transporters. It includes additional genes at the 3′ end of the fragment encoding a (i) MoxR-like AAA + ATPase (RSP_3275), (ii) some hypothetical proteins as well as a (iii) von Willebrand factor A (VWA) domain containing protein (RSP_3278) (Fig. 3FII). The MoxR AAA + family is a large group of ATPases and are often associated with a protein containing a VWA domain [29]. In RubisCO gene clusters the genes cbbQ, belonging to the MoxR group of AAA + ATPases, and the cbbO, containing a VWA domain, have been shown to interact with RubisCO enzymes and enhance their activity [22, 30]. The presence of these genes in Rsph1C7 as well as their product’s putative function may explain why the RubisCO activity of Rsph1C7 is higher than in Rsph9E12.
In summary, although some of the recombinant RubisCO activities are very low and these fosmid clones would only have been found if the pool size of the screened fosmid clones were reduced, our work demonstrates that the activity-based screen can detect different types of RubisCO forms I and II from the tested Alpha-, Beta-, and Gammaproteobacteria. Thus, the screen is useful for targeting proteobacterial RubisCOs from the environment and provides an opportunity to learn of the role some orfs play for RubisCO functioning.
Metagenomic fosmid clones of yet uncultured microorganisms
Seeking recombinant RubisCOs from six deep-sea hydrothermal vent habitats
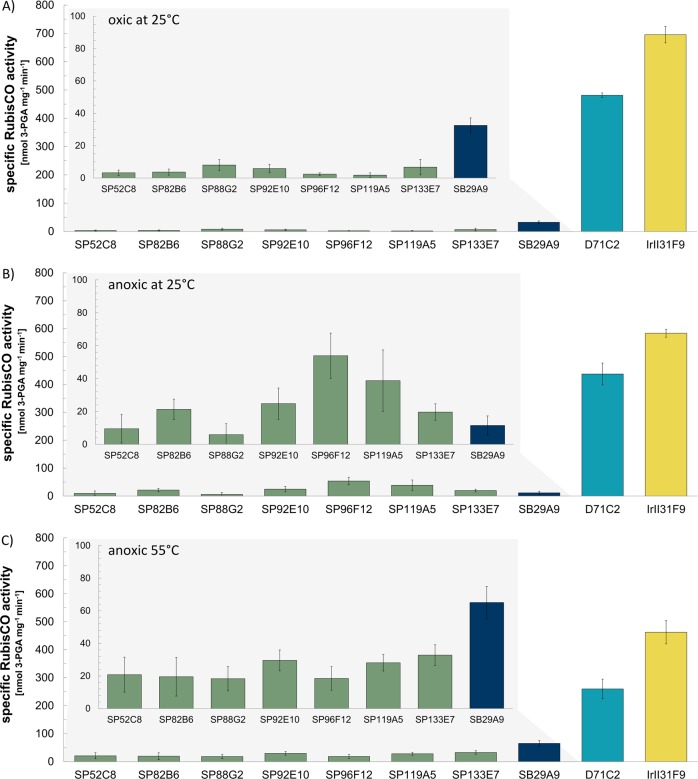
For each of the six metagenomic fosmid libraries we screened between 2,400 and 2,688 fosmid clones (Table 1). We identified forty fosmid clones with RubisCO activity using the activity-based approach (Table 1, Fig. 4). Via the sequence-based screening, all these 40 as well as 8 additional fosmid clones encoding RubisCO genes were identified (Table 1, Supplementary Table S6). The eight fosmid clones detected only via sequence-based screening included (i) one from the Site B library (SB29A9) encoding a Thioclava atlantica relative RubisCO form I (90% aa similarity, Fig. 2 and Supplementary Table S7) and (ii) seven fosmid clones from the Sisters Peak library (SP52C8, SP96F12, SP119A5, SP88G2, SP133E7, SP92E10, and SP82B6) comprising RubisCO form III encoding genes related to that of Aciduliprofundum sp. MAR06-339 (85% to 89% aa similarities, Fig. 2). All of these clones exhibited RubisCO activities on the single clone level (Fig. 4A). Clone SB29A9 had 32.5 ± 4.6 nmol 3-PGA mg−1 min−1 and the RubisCO form III encoding fosmid clones had 2.3 ± 0.7 to 8.0 ± 3.4 nmol 3-PGA mg−1 min−1 RubisCO activities. They would have been detected in the pool via activity-based searches if pools were smaller, i.e., eight clones or less.
Fig. 4.
Specific RubisCO activities of metagenomic fosmid clones. RubisCO activities of 15 identified fosmid clones measured at A 25 °C under oxic conditions, B 25 °C under anoxic conditions, and C 55 °C under anoxic conditions. The bars of clones sought from one vent site are indicated in the same color: (I) ‘Sisters Peak’ clones are olive green-colored, (II) the ‘Site B’ clone is dark blue-colored, (III) the ‘Drachenschlund’ clone is petrol blue-colored, and (IV) the ‘Irina II’ clone is yellow-colored
The 40 fosmid clones identified via activity-based screening included (i) one clone detected from the Irina II metagenomic library (IrII31F9) comprising one form I and one form II RubisCO on its insert (Supplementary Table S7 and Supplementary Fig. S3) and (ii) 39 clones isolated from the Drachenschlund library possessing both RubisCO forms I and II encoding genes on their inserts (Supplementary Fig. S3). The metagenomic fosmid inserts of the 39 Drachenschlund clones share a 100% similarity, likely due to a methodological bias caused prior to or during library construction. RubisCO forms I and II encoding genes found in the Irina II and in the Drachenschlund fosmid libraries show highest similarities with Hydrogenovibrio crunogenus XCL-2 (Fig. 2). However, the sequences of the Irina II clone IrII31F9 share only around 92% similarities with sequences of the Drachenschlund clones. Although H. crunogenus is known to encode three different RubisCO forms on its genome (one cytoplasmic form I, one carboxysomal form I, and one form II) [31], carboxysomal RubisCOs could not be detected via sequence-based or via activity-based screening approaches. This likely indicates that carboxysomal RubisCO gene clusters may not have been covered when constructing the libraries due to technical troubles as has been noted before for other gene regions [32].
Activity-based screen under anoxic conditions—effects of O2
All fosmid clones were measured under oxic conditions at 25 °C (Fig. 4A) and additionally under anoxic conditions at 25 and at 55 °C (Fig. 4B, C). Significant differences (p-value < 0.05) in the RubisCO activities, were measured for clones SP82B6, SP92E10, SP96F12, SP119A5, SP133E7, SB29A9, and IrII31F9 under oxic versus anoxic conditions.
In case of the five Sisters Peak clones (SP82B6, SP92E10, SP96F12, SP119A5, SP133E7) resembling Aciduliprofundum sp. MAR08-339 (Fig. 2, Supplementary Tables S6) RubisCO activities increased 3- to 23-fold at 25 °C under anoxic conditions. Archaeal form III RubisCOs are highly sensitive against molecular O2 often resulting in reversible or even in an irreversible inhibition of RubisCO activity [33, 34]. This might explain why measured RubisCO activities of identified metagenomic RubisCO form III encoding fosmid clones were hardly detectable when measuring RubisCO activity via the unmodified HPLC based assay, i.e., under atmospheric O2 conditions.
The activity of the Thioclava relative form I RubisCO (clone SB29A9) measured at 25 °C under anoxic conditions decreased 3-fold (Fig. 4A, B). Form I RubisCO has been shown to possess lowest sensitivities against O2 [4] and O2 concentrations were shown to influence the diversity of form I RubisCO encoding cbbL genes [6, 35]. Nevertheless, up until now no mechanism has ever been described which allows O2 to enhance RubisCO form I activity. It thus remains enigmatic why RubisCO activities measured with the anoxic version of our assay decrease relative to those measured under atmospheric O2 levels. Particularly, as one would rather expect that due to the discontinued wasteful oxygenase reaction form I activities measured under anoxic conditions would still be higher than those measured under oxic conditions [36]. Fosmid clones for the oxic and anoxic assay were both grown under fully oxic conditions and only the enzyme activity measurement itself was performed under oxic or anoxic settings. Consequently, the observed activity loss under anoxic assay conditions could not be occurring on the transcriptional level, but instead is likely associated with posttranslational processes. CbbO and CbbQ are such proteins that have been shown to be involved in posttranslational activation of RubisCO [22, 30] and homologous genes are encoded on the SB29A9’s metagenomic insert. Yet, to our knowledge, the influence of O2 depletion on posttranslational RubisCO activation has not been investigated and experiments are needed to look into this phenomenon.
Total RubisCO activity of the Hydrogenovibrio relative Irina II clone (IrII31F9) decreased if the assay was performed under O2 exclusion (83% residual activity, Fig. 4B). On the IrII31F9’s metagenomic insert both form I and form II RubisCOs are encoded. Generally, it has been assumed that form I RubisCO is adapted to functioning in oxic environments, while form II RubisCO is better adapted to low O2 levels [4]. Form II RubisCO is also catalytically much faster than form I RubisCO [37]. Assuming that both RubisCO forms are present in Ir31F9’s crude extract in equal or almost equal amounts, CO2 fixation rates should be higher when prevailing conditions support RubisCO form II functioning under anoxic conditions and forms I and II RubisCO activities add up. As already mentioned, fosmid clones were autoinduced under atmospheric conditions and only the activity measurement was performed anoxically. As form II RubisCO is not that good in discriminating between CO2 and O2, one might assume that it would be wasteful to express form II RubisCO under atmospheric O2 levels when a more efficient enzyme could be expressed instead. Indeed, previous investigations of another metagenomic fragment also possessing both RubisCO forms (I and II), showed that under oxic conditions the RubisCO form I transcript levels are 29-fold higher than those of form II RubisCO [22]. Another study investigating the expression of distinct RubisCO forms under different redox conditions in polluted groundwater even showed that form II RubisCO was not expressed at all in groundwater supplied with O2 [35]. Even if we would assume that form II RubisCO is not expressed when fosmid clone IrII31F9 is grown under atmospheric O2 levels, the reasons why RubisCO form I activity decreases if measured under anoxic conditions remains unknown.
The anoxic version of our activity-based screen allows detection of RubisCOs that exhibit higher activities under anoxic conditions and elevated temperatures. Some modifications of our procedure could improve activities from such enzymes: (i) Working cultures could be grown under anaerobic conditions. However, autoinduction could become challenging if O2 were excluded because O2 limitation triggers the expression of more than 200 genes to adjust E. coli’s (the host’s) metabolism [38–40]. This results in non-optimal growth paralleled with low or even no protein production. (ii) Crude extracts could be prepared under anoxic conditions. (iii) One further purification step might significantly increase RubisCO form III activity, as has been shown before for crude extracts of cultured representatives [33]. (iv) As active form III RubisCO enzymes of selected cultured representatives are most active between 83 and 93 °C [33, 41], the assay temperatures could be further increased from 55 to 85 °C.
Linking properties from recombinant enzymes with environmental parameters from the hydrothermal vents
We screened six deep-sea hydrothermal vent metagenomes for active RubisCO enzymes using sequence-based and activity-based approaches (Table 1). Specific RubisCO activities of all identified RubisCOs were measured at 25 °C under oxic and at 25 and 55 °C under anoxic conditions. It is of interest to link enzyme biochemical properties to habitat characteristics. The environmental factors of the habitats from which the hydrothermal vent samples, i.e., Sisters Peak, Mephisto, Lilliput, Site B, Irina II, and Drachenschlund, were collected are summarized in Supplementary Table S1. Although the number of identified active RubisCO enzymes is not high enough for statistically relevant conclusions, they do depict trends between the taxonomy and expected physiology of organisms encoding the identified RubisCO and the environments they were recovered from.
Sisters Peak and Aciduliprofundum RubisCO form III
The Sisters Peak chimney hosts some of the currently hottest measured venting temperatures and highly reduced endmember conditions [21]. A large portion of genes including RubisCO form III genes resembling those of the thermophilic Aciduliprofundum (23%) were recognized, indicative of its good adaptation to local chimney conditions [21]. Of the three known bona fide RubisCO forms only archaeal form III has been shown to operate at (hyper)thermophilic temperatures [33, 41]. It exhibits unusually high sensitivities to molecular O2, which results in low capabilities to discriminate between CO2 and O2 [5]. The RubisCO form III detected in the chimney matches the local habitat properties, namely hot and reduced conditions.
Site B and Thioclava RubisCO form I
The Site B vent at the Logatchev hydrothermal field is characterized by high sulfide contents (1,241 µM, Supplementary Table S1), providing a habitat for chemolithoautotrophic sulfur-oxidizers to thrive. In fact, the one active RubisCO that we detected from this environment (clone SB29A9) resembled that of Thioclava atlantica (90% aa similarity, Fig. 2). T. atlantica is known to yield energy by oxidizing reduced sulfur compounds [42]. Most members of the Thioclava genus have been described as facultatively autotrophic bacteria that assimilate inorganic carbon via the CBB cycle ([43], for exceptions see [44]). We showed that the metagenome derived Thioclava relative RubisCO is functional (32.5 ± 4.6 nmol 3-PGA mg−1 min−1, Fig. 4A) likely indicating that the original host organism is also able to fix carbon using the CBB cycle. The activity of this Thioclava relative form I RubisCO can be doubled by raising assay temperatures to 55 °C (Fig. 4C). Only focused hydrothermal emissions have been observed at Site B and maximum measured fluid temperatures were 350 °C (Supplementary Table S1). Hence, enzyme characteristics are in line with the hotter nature of this hydrothermal site. The identified RubisCO appears to be better adapted to moderate thermophilic rather than to mesophilic conditions. Similar temperature adaptations of form I RubisCO to hotter environments have been shown before for members of the cyanobacterial Synechococcus clade [45] and might apply for this Thioclava relative form I RubisCO, too. Although cultured representatives of the genus Thioclava have, for now, only been isolated from psychrophilic and mesophilic, but never from (hyper)thermophilic environments [46–48]. The higher RubisCO activity measured at elevated assay temperatures suggests that the original host of the identified Thioclava relative form I RubisCO is likely a moderately thermophilic organism.
Drachenschlund and Irina II and Hydrogenovibrio RubisCO forms I and II
The hydrothermal fluids emitting from the crater Drachenschlund are quickly mixed with ambient seawater. Low O2 levels and high fluid temperatures stem from measurements taken directly where the fluids emit from within the crater (Supplementary Table S1). In contrast, the fluid sample used for library construction was collected from the interface zone between hot fluids emanating from Drachenschlund and ambient seawater. The sampled Drachenschlund mixed fluids are thus expected to be oxic and mesophilic rather than anoxic and thermophilic. Indeed, total RubisCO activity of the Drachenschlund clone D71C2 decreased by 17 and 40% when O2 is absent (Fig. 4A, B) and the assay temperature is raised from 25 to 55 °C, respectively (Fig. 4B, C). The original host of the investigated RubisCO forms I and II encoding metagenomic fragment thus seems to be better adapted to O2 presence and moderate temperatures [49, 50]. This is in line with the intense mixing processes occurring at Drachenschlund.
Total RubisCO activity of the H. crunogenus relative Irina II clone (IrII31F9) is also highest at 25 °C and oxygenated conditions (83% and 64% residual activity, if measured under anoxic conditions at 25 and 55 °C, respectively). Hence, the organism reflected by this metagenomic RubisCO forms I and II comprising fragment is likely better adapted to mesophilic and oxic conditions than to thermophilic and anoxic conditions. This reflects the parameters specific for the Irina II site (relatively low temperatures: 6.4 to 11 °C and high O2 contents 208–235 µM, compared to other sites; see Supplementary Table S1).
All of the identified H. crunogenus relative fosmid clones have two different RubisCO forms encoded on their metagenomic insert (forms I and II, Supplementary Fig. S3). Form I RubisCO is highly specific for CO2 albeit being quite a slow catalyst [5, 51]. Form II RubisCO is catalytically much faster but its ability to discriminate between O2 and CO2 is comparably low [5, 51]. Organisms possessing only one RubisCO form are thus limited by the imperfection of their RubisCO enzyme. The combination of both RubisCO forms in one organism enables a rapid response to changing CO2 and O2 levels. At hydrothermal deep-sea systems, where CO2 and O2 concentrations can dynamically change over time ([19] and references therein), this flexibility likely entails a great advantage for local microorganisms and might explain why Hydrogenovibrio species are so successful in colonizing chemically changing hydrothermal vent environments [20, 52–54].
Mephisto and Lilliput and no detected RubisCOs
RubisCO activities were not detected from metagenomic fragments when screening 2,496 fosmid clones from the Mephisto and the Lilliput metagenomic fosmid libraries each. This is surprising as members of the genus Hydrogenovibrio are considerable parts of the microbial community in the Lilliput fluids [20]. Possible reasons for not recognizing active RubisCOs could be related to (i) difficulties in expressing their metagenomic DNA in the surrogate host E. coli ([13] and references therein), (ii) not covering the whole metagenome when constructing the libraries as previously described [32] (iii) methodological shortcomings associated with the utilized RubisCO screen, e.g., overnight incubations of crude extracts at 4 °C may lead to the exclusion of particularly unstable RubisCOs, or (iv) the fact that not enough clones were screened.
Conclusion
The activity-based screen utilized here for seeking RubisCOs from the environment is currently the only opportunity to identify and study active RubisCOs from the majority of yet uncultured microorganisms. RubisCO enzymes from a broad phylogenetic spectrum can be detected with this screen including proteobacterial forms I and II RubisCOs as well as some archaeal form III RubisCOs. This activity-based RubisCO screen, thus, grants access to phylogenetically distinct RubisCOs from the environment, circumventing the bottleneck of cultivation. Here, we identified new RubisCOs of yet uncultured strains. We characterized some biochemical properties (temperature and O2 tolerance) of these recombinant enzymes and showed that they reflect the predominant environmental conditions of investigated hydrothermal vent sites. Hence, the activity-based RubisCO screen allows us not only to seek active RubisCOs form the environment but also enables studying the biochemistry of active RubisCO enzymes from uncultured organisms.
Supplementary information
Acknowledgements
We thank the captain and crews of the RV Meteor, the Maria S. Merian as well as the ROV Kiel6000 (GEOMAR, Kiel) for helping us to obtain deep-sea vent samples. We thank Wenke Bahnsen, Dagmar Svensson, Anna Ulatowski, and Nicole Adam for excellent technical assistance in the laboratory. The work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (PE1549/5-1) and the DFG priority program 1144 “From Mantle to Ocean: Energy-, Material- and Life-cycles at Spreading Axes”.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0439-3) contains supplementary material, which is available to authorized users.
References
- 1.Yokota A. Revisiting RuBisCO. Biosci Biotechnol Biochem. 2017;81:2039–49. doi: 10.1080/09168451.2017.1379350. [DOI] [PubMed] [Google Scholar]
- 2.Erb TJ, Zarzycki J. A short history of RubisCO: the rise and fall (?) of nature’s predominant CO2 fixing enzyme. Curr Opin Biotechnol. 2018;49:100–7. doi: 10.1016/j.copbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwander T, Schada von Borzyskowski L, Burgener S, Cortina NS, Erb TJ. A synthetic pathway for the fixation of carbon dioxide in vitro. Science. 2016;354:900–4. doi: 10.1126/science.aah5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badger MR, Bek EJ. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot. 2008;59:1525–41. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 5.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perner M, Seifert R, Weber S, Koschinsky A, Schmidt K, Strauss H, et al. Microbial CO2 fixation and sulfur cycling associated with low-temperature emissions at the Lilliput hydrothermal field, southern Mid-Atlantic Ridge (9°S) Environ Microbiol. 2007;9:1186–201. doi: 10.1111/j.1462-2920.2007.01241.x. [DOI] [PubMed] [Google Scholar]
- 7.Hügler M, Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci. 2011;3:261–89. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol. 2008;65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- 9.Perner M, Kuever J, Seifert R, Pape T, Koschinsky A, Schmidt K, et al. The influence of ultramafic rocks on microbial communities at the Logatchev hydrothermal field, located 15 degrees N on the Mid-Atlantic Ridge. FEMS Microbiol Ecol. 2007;61:97–109. doi: 10.1111/j.1574-6941.2007.00325.x. [DOI] [PubMed] [Google Scholar]
- 10.Perner M, Hansen M, Seifert R, Strauss H, Koschinsky A, Petersen S. Linking geology, fluid chemistry, and microbial activity of basalt- and ultramafic-hosted deep-sea hydrothermal vent environments. Geobiology. 2013;11:340–55. doi: 10.1111/gbi.12039. [DOI] [PubMed] [Google Scholar]
- 11.Witte B, John D, Wawrik B, Paul JH, Dayan D, Tabita FR. Functional prokaryotic RubisCO from an oceanic metagenomic library. Appl Environ Microbiol. 2010;76:2997–3003. doi: 10.1128/AEM.02661-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Böhnke S, Perner M. A function-based screen for seeking RubisCO active clones from metagenomes: novel enzymes influencing RubisCO activity. ISME J. 2014;9:735–45. doi: 10.1038/ismej.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perner M, Ilmberger N, Köhler HU, Chow J, Streit WR. Emerging fields in functional metagenomics and its industrial relevance: Overcoming limitations and redirecting the search for novel biocatalysts. In: de Bruijn FJ, editor. Handbook of molecular microbial ecology II. New Jersey: Wiley-Blackwell; 2011. pp. 484–5. [Google Scholar]
- 14.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–85. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boden R, Scott KM, Williams J, Russel S, Antonen K, Rae AW, et al. An evaluation of Thiomicrospira, Hydrogenovibrio and Thioalkalimicrobium: reclassification of four species of Thiomicrospira to each Thiomicrorhabdus gen. nov and Hydrogenovibrio, and reclassification of all four species of Thioalkalimicrobium to Thiomicrospira. Int J Syst Evol Microbiol. 2017;67:1140–51. doi: 10.1099/ijsem.0.001855. [DOI] [PubMed] [Google Scholar]
- 16.Haase KM, Petersen S, Koschinsky A, Seifert R, Devey CW, Keir R, et al. Young volcanism and related hydrothermal activity at 5°S on the slow-spreading southern Mid-Atlantic Ridge. Geochem Geophys Geosyst. 2007;8:1–17. doi: 10.1029/2006GC001509. [DOI] [Google Scholar]
- 17.Melchert B, Devey CW, German CR, Lackschewitz KS, Seifert R, Walter M, et al. First evidence for high-temperature off-axis venting of deep crustal/mantle heat: The Nibelungen hydrothermal field, southern Mid-Atlantic Ridge. Earth Planet Sci Lett. 2008;275:61–9. doi: 10.1016/j.epsl.2008.08.010. [DOI] [Google Scholar]
- 18.Perner M, Bach W, Hentscher M, Koschinsky A, Garbe-Schönberg D, Streit WR, et al. Short-term microbial and physico-chemical variability in low-temperature hydrothermal fluids near 5°S on the Mid-Atlantic Ridge. Environ Microbiol. 2009;11:2526–41. doi: 10.1111/j.1462-2920.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 19.Perner M, Gonnella G, Hourdez S, Böhnke S, Kurtz S, Girguis P. In situ chemistry and microbial community compositions in five deep-sea hydrothermal fluid samples from Irina II in the Logatchev field. Environ Microbiol. 2013;15:1551–60. doi: 10.1111/1462-2920.12038. [DOI] [PubMed] [Google Scholar]
- 20.Perner M, Hentscher M, Rychlik N, Seifert R, Strauss H, Bach W. Driving forces behind the biotope structures in two low-temperature hydrothermal venting sites on the southern Mid-Atlantic Ridge. Environ Microbiol Rep. 2011;3:727–37. doi: 10.1111/j.1758-2229.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 21.Perner M, Gonnella G, Kurtz S, LaRoche J. Handling temperature bursts reaching 464 °C: different microbial strategies in the Sisters Peak hydrothermal chimney. Appl Environ Microbiol. 2014;80:4585–98. doi: 10.1128/AEM.01460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böhnke S, Perner M. Unraveling RubisCO form I and form II regulation in an uncultured organism from a deep-sea hydrothermal vent via metagenomic and mutagenesis studies. Front Microbiol. 2017;8:1303. doi: 10.3389/fmicb.2017.01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell BJ, Cary SC. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl Environ Microbiol. 2004;70:6282–9. doi: 10.1128/AEM.70.10.6282-6289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiridonova EM, Berg IA, Kolganova TV, Ivanovsky RN, Kuznetsov BB, Tourova TP. An oligonucleotide primer system for amplification of the ribulose-1,5-bisphosphate carboxylase/oxygenase genes of bacteria of various taxonomic groups. Microbiology. 2004;73:316–25. doi: 10.1023/B:MICI.0000032243.93917.30. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. MBio. 2013;4:e00378–13. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snider J, Houry WA. MoxR AAA+ATPases: a novel family of molecular chaperones? J Struct Biol. 2006;156:200–9. doi: 10.1016/j.jsb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Tsai YC, Lapina MC, Bhushan S, Mueller-Cajar O. Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat Commun. 2015;6:8883. doi: 10.1038/ncomms9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA, et al. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLOS Biol. 2006;4:2196–212. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam KN, Cheng J, Engel K, Neufeld JD, Charles TC. Current and future resources for functional metagenomics. Front Microbiol. 2015;6:1196. doi: 10.3389/fmicb.2015.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn MW, Tabita FR. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J Bacteriol. 2003;185:3049–59. doi: 10.1128/JB.185.10.3049-3059.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreel NE, Tabita FR. Substitutions at methionine 295 of Archaeoglobus fulgidus ribulose-1,5-bisphosphate carboxylase/oxygenase affect oxygen binding and CO2/O2 specificity. J Biol Chem. 2007;282:1341–51. doi: 10.1074/jbc.M609399200. [DOI] [PubMed] [Google Scholar]
- 35.Alfreider A, Schirmer M, Vogt C. Diversity and expression of different forms of RubisCO genes in polluted groundwater under different redox conditions. FEMS Microbiol Ecol. 2012;79:649–60. doi: 10.1111/j.1574-6941.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- 36.Trudeau DL, Edlich-Muth C, Zarzycki J, Scheffen M, Goldsmith M, Khersonsky O, et al. Design and in vitro realization of carbon-conserving photorespiration. Proc Natl Acad Sci USA. 2018;115(49):E11455–E64. doi: 10.1073/pnas.1812605115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, Ramya RCS, Mueller-Cajar O. Surveying the expanding prokaryotic Rubisco multiverse. FEMS Microbiol Lett. 2017;fnx156:364. 10.1093/femsle/fnx156. [DOI] [PubMed]
- 38.epicentre. Instruction Manual: CopyControl™ Fosmid Library Production Kit with pCC1FOS™ Vector. 2012;EPILIT171 Rev. A:1–27.
- 39.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009;18:936–48. doi: 10.1002/pro.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreel NE, Tabita FR. Serine 363 of a hydrophobic region of archaeal ribulose 1,5-bisphosphate carboxylase/oxygenase from Archaeoglobus fulgidus and Thermococcus kodakaraensis affects CO2/O2 substrate specificity and oxygen sensitivity. PLoS ONE. 2015;10:e0138351. doi: 10.1371/journal.pone.0138351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai Q, Li S, Xu H, Jiang L, Zhang R, Shao Z. Thioclava atlantica sp. nov., isolated from deep sea sediment of the Atlantic Ocean. Antonie Van Leeuwenhoek. 2014;106:919–25. doi: 10.1007/s10482-014-0261-x. [DOI] [PubMed] [Google Scholar]
- 43.Sorokin DY, Tourova TP, Spiridonova EM, Rainey FA, Mulyzer G. Thioclava pacifica gen. nov., sp nov., a novel facultatively autotrophic, marine, sulfur-oxidizing bacterium from a near-shore sulfidic hydrothermal area. Int J Syst Evol Microbiol. 2005;55:1069–75. doi: 10.1099/ijs.0.63415-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhang R, Lai Q, Wang W, Li S, Shao Z. Thioclava dalianensis sp. nov., isolated from surface seawater. Int J Syst Evol Microbiol. 2013;63:2981–5. doi: 10.1099/ijs.0.046094-0. [DOI] [PubMed] [Google Scholar]
- 45.Miller SR, McGuirl MA, Carvey D. The evolution of RuBisCO stability at the thermal limit of photoautotrophy. Mol Biol Evol. 2013;30:752–60. doi: 10.1093/molbev/mss327. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Lai Q, Shao Z. A multilocus sequence analysis scheme for phylogeny of Thioclava bacteria and proposal of two novel species. Front Microbiol. 2017;8:1321. doi: 10.3389/fmicb.2017.01321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thongphrom C, Kim JH, Bora N, Kim W. Thioclava arenosa sp. nov., isolated from sea sand. Int J Syst Evolut Microbiol. 2017;67:1735–9. doi: 10.1099/ijsem.0.001853. [DOI] [PubMed] [Google Scholar]
- 48.Chang R, Bird L, Barr C, Osburn M, Wilbanks E, Nealson K, et al. Thioclava electrotropha sp. nov., a versatile electrode and sulfur-oxidizing bacterium from marine sediments. Int J Syst Evol Microbiol. 2018;68:1652–8. doi: 10.1099/ijsem.0.002723. [DOI] [PubMed] [Google Scholar]
- 49.Feller G, Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol. 2003;1:200–8. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 50.Unsworth LD, van der Oost J, Koutsopoulos S. Hyperthermophilic enzymes – stability, activity and implementation strategies for high temperature applications. FEBS J. 2007;274:4044–56. doi: 10.1111/j.1742-4658.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 51.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59:1515–24. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 52.Gonnella G, Böhnke S, Indenbirken D, Garbe-Schönberg D, Seifert R, Mertens C, et al. Endemic hydrothermal vent species identified in the open ocean seed bank. Nat Microbiol. 2016;1:16086. doi: 10.1038/nmicrobiol.2016.86. [DOI] [PubMed] [Google Scholar]
- 53.Hansen M, Perner M. A novel hydrogen oxidizer amidst the sulfur-oxidizing Thiomicrospira lineage. ISME J. 2015;9:696–707. doi: 10.1038/ismej.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brazelton WJ, Baross JA. Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS ONE. 2010;5:e13530. doi: 10.1371/journal.pone.0013530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.