Abstract
Nitrous acid (HONO) is a precursor of the hydroxyl radical (OH), a key oxidant in the degradation of most air pollutants. Field measurements indicate a large unknown source of HONO during the day time. Release of nitrous acid (HONO) from soil has been suggested as a major source of atmospheric HONO. We hypothesize that nitrite produced by biological nitrate reduction in oxygen-limited microzones in wet soils is a source of such HONO. Indeed, we found that various contrasting soil samples emitted HONO at high water-holding capacity (75–140%), demonstrating this to be a widespread phenomenon. Supplemental nitrate stimulated HONO emissions, whereas ethanol (70% v/v) treatment to minimize microbial activities reduced HONO emissions by 80%, suggesting that nitrate-dependent biotic processes are the sources of HONO. High-throughput Illumina sequencing of 16S rRNA as well as functional gene transcripts associated with nitrate and nitrite reduction indicated that HONO emissions from soil samples were associated with nitrate reduction activities of diverse Proteobacteria. Incubation of pure cultures of bacterial nitrate reducers and gene-expression analyses, as well as the analyses of mutant strains deficient in nitrite reductases, showed positive correlations of HONO emissions with the capability of microbes to reduce nitrate to nitrite. Thus, we suggest biological nitrate reduction in oxygen-limited microzones as a hitherto unknown source of atmospheric HONO, affecting biogeochemical nitrogen cycling, atmospheric chemistry, and global modeling.
Subject terms: Biogeochemistry, Biogeochemistry
Introduction
The composition of the Earth’s atmosphere is strongly affected by the biogeochemical cycling of reactive nitrogen species. Nitrous acid (HONO) is a key species due to its effect on hydroxyl free radical (OH) formation and recycling [1, 2]. HONO can also damage the respiratory system of asthmatics. It forms mutagenic and carcinogenic nitrosamines [3, 4], and is thus a health risk and source of indoor air pollution [5].
Atmospheric HONO concentrations range from 5 ppb in cities [6] to about 0.1 ppb in rural areas [7]. The only known significant gas-phase source of HONO is the reaction of NO with OH. The dominant sink during the day time is HONO photolysis [2, 8], which reforms NO and OH. From gas-phase sources and sinks, a photostationary state is expected to be established, but measured HONO concentrations almost always exceed those calculated from known gas-phase chemistry. The heterogeneous disproportionation of NO2 to HONO and HNO3 is a source of HONO that might be able to explain nighttime values, but is too slow to explain day time levels [2, 9]. The reduction of NO2 to HONO is accelerated by organics [10], and chemisorption on mineral particles might take place with regard to HONO [11, 12]. Furthermore, as a day time source was missing, many light or temperature-dependent mechanisms have been postulated, but information on their relevance under ambient conditions is sparse [13]. Results from field experiments showed that the ground surface significantly contributes to HONO concentrations in the lower atmosphere [14, 15].
Soils can emit large amounts of HONO to the atmosphere, potentially explaining the missing HONO source [16–19]. Multiple mechanisms and modeling approaches have been proposed to quantitatively explain the release of HONO from soil. They include chemical equilibrium with soil nitrite [17], surface acidity [19], reactive uptake and displacement [20], and release by ammonia-oxidizing bacteria [18, 21, 22]. HONO emissions from the soil can also involve the heterogeneous hydrolysis of NH2OH [23] when the soils dry out. The maximum HONO flux was reported to occur at 0–40% soil water-holding capacity, WHC [18]. Release of HONO from dried soils has been reported in quantities comparable with NO emissions [17, 18, 24]. At high water-holding capacity, the release of HONO from soils is expected to be low due to low gas diffusion rates and solubility in soil water.
However, denitrification and anaerobic nitrate reduction to ammonia are important biogenic sources of extracellular nitrite [25, 26] under the oxygen-limited or anoxic conditions that frequently occur at high soil water content [26, 27]. Thus, we hypothesize a hitherto undetected source of HONO in soils. To test our hypothesis, we investigated HONO emissions from various soils at high water content (in the following called “wet peak”) using a dynamic chamber system [28]. We measured the wet peak for soils from different ecosystems, and investigated the underlying mechanisms by soil incubations, combined with functional gene expression as well as transcript diversity analyses, pure bacterial culture experiments, and knockout mutant studies.
Materials and methods
Soil samples
Soil samples were taken from the upper layer of the soils. Detailed information about the soils can be found in Supplementary Information. The soil physical and chemical properties were analyzed according to the following standard procedures: pH, DIN ISO 10390 (in water); ammonium/nitrite/nitrate, DIN ISO/TS 14256-1; total carbon and nitrogen, DIN ISO 13878; sand/silt/clay content, DIN ISO 11277. Ammonium, nitrite, and nitrate were measured in extracts with 0.0125 M CaCl2 for most of the soils. Measurements were performed in water extracts for growth media of pure culture studies and mutant experiments, as well as soils S5, S8, and S9. Due to the near-neutral pH and/or high-nitrite concentrations (SI Table S1), a possible underestimation of nitrite, as highlighted in a recent publication by Homyak et al. [29], is unlikely. The physical and chemical properties of the soils, the maximal HONO and NO fluxes and the corresponding WHCs, and ratios of the highest HONO and NO flux of wet peak to dry peak are summarized in SI Table S1.
Flux measurements
Emissions of HONO and NO from the soils were measured with a dynamic chamber system (SI Fig. S1) that has been described in detail in elsewhere [18, 28]. Previous studies showed that our dynamic chamber system can simulate the field fluxes [30–33]. The experiments were conducted in the dark at 25 °C in a temperature-controlled climate chamber. Briefly, 50 g of a homogeneously mixed soil sample was placed in a petri dish (inner diameter = 88 mm) and wetted with ~50 g of ultrapure water. The petri dish was placed into a Teflon chamber (volume 47 L) and flushed with purified dry air (8 L min−1), resulting in low concentrations of HONO inside the chamber and residence times <6 min. The surface of the petri dishes divided by the volume of the Teflon chamber was 0.0016. The petri dishes were filled with the soil material, and the bottom was in direct contact with the chamber. Thus, only a small surface of the petri dishes was exposed to the gas phase of the chamber, providing a very small potentially HONO-reactive surface. These measures minimized the probability of a potential loss of HONO due to heterogeneous reactions in the chamber. Mixing ratios of HONO, NO, NO2, O3, CO2, and H2O in the headspace were continuously monitored as the soil dried out. HONO and NO were detected by a LOPAP (QUMA Elektronik & Analytik GmbH, Germany) and a NOx chemiluminescence analyzer (Model 42i-TL, Thermo Scientific, USA), respectively. The limit of detection was ~5 ppt for HONO and ~80 ppt for NO. Mixing ratios of N2O were determined using the University of Mainz Quantum Cascade Laser (QCL) Absorption spectrometer (UMAQS [34]), which is based on an Aerodyne QCL Mini Monitor [35, 36]. Soil water content (normalized as % WHC), fluxes of HONO, NO, and N2O, and the corresponding errors were calculated based on water loss during the experiment, flow rate and mixing ratios of gases, and Gaussian error propagation, respectively [18, 28]. The WHC was calculated by the mass of water in soil at field capacity and the mass of dry soil [18]. The Gaussian error propagation represents the uncertainty of the fluxes (ΔF), and is calculated as the following [18]:
where F is the flux of HONO, NO, or N2O (ng m−2 s−1, in terms of nitrogen, which is the same as follows), Q is the purging flow rate (m3 s−1), A is the area of soil (m2), and χout and χin are the headspace mixing ratios at the outlet and inlet of the chamber (ppb), respectively.
Ethanol-treated experiments
Soil S1 was used to check the effects of ethanol treatment on HONO and NO flux. Fifty milliliters of 70% ethanol (Carl Roth GmbH, Germany) were added to a petri dish, which contained 50 g of soil S1, for ~10 h to reduce the numbers of live microbial cells and their activities in the soil. After the soil dried, 50 g of ultrapure water was added to the petri dish. Then, HONO and NO flux were measured by the dynamic chamber system.
Temperature-dependence experiments
Soil S1 was used to check the temperature dependence of the dry and wet peaks. The procedure for the flux measurement was the same as above, except that, the temperature was regulated to 5, 15, 20, 25, 30, 35, 40, and 45 °C in a temperature-controlled climate chamber.
Soil incubation experiments
The soil incubation experiments were conducted under flooded conditions. A sample of 500 g of soil S1 was placed in a glass beaker, and ultrapure water was added to reach ~160% soil water-holding capacity, which is ~100% soil gravimetric water content. Parafilm was used to cover and seal the beaker, which was pierced with seven holes to allow gas exchange between the beaker and the atmosphere. The water loss was negligible during the incubation. The beaker was placed in a dark and constant 25 °C climate chamber (Vötsch Industrietechnik GmbH, Balingen-Frommern, Germany) for incubation. After incubation for 30, 54, and 200 h, 50-g soil subsamples were taken from the beaker, and the fluxes of HONO and NO were measured at 25 °C, as described above. For the experiments with the addition of nitrate, 7.45 mL of potassium nitrate (KNO3) solution with a concentration of 1000 mg L−1, which corresponds to 180 kg N ha−1 fertilizer applied in the field, was added to the soil sample, and then the HONO and NO fluxes were measured.
Strains, culture, and media
All model Proteobacteria were facultative aerobes that were capable of anaerobic nitrate reduction and/or denitrification, hosting a contrasting nitrate reduction and/or denitrification associate gene equipment. The nitrate reducer was Escherichia coli K-12, and the denitrifiers were Pseudomonas G-179, Pseudomonas stutzeri JM-300 (DSM 10701), Bradyrhizobium japonicum (DSM 1755), and Rhodanobacter denitrificans (DSM 23569). These strains were selected to test the effect of nitrate and nitrite on HONO and NO emissions under anoxic conditions.
For anoxic cell incubation, E. coli and B. japonicum were routinely grown in liquid yeast extract (pH 7.0), which contained 10 g L−1 mannitol, 0.5 g L−1 K2HPO4, 0.2 g L−1 MgSO4·7H2O, 0.1 g L−1 NaCl, and 0.4 g L−1 yeast extract. Pseudomonas G-179 and P. stutzeri JM300 were routinely grown in liquid nutrient broth (Sigma-Aldrich Co., USA), which contained 3.0 g L−1 beef extract and 5.0 g L−1 peptone with a pH of 7.0. Rhodanobacter denitrificans was routinely grown in R2A liquid medium (pH 7.2), which contained 0.5 g L−1 casamino acids, 0.5 g L−1 yeast extract, 0.5 g L−1 proteose peptone, 0.5 g L−1 soluble starch, 0.5 g L−1 dextrose, 0.3 g L−1 K2HPO4, 0.05 g L−1 MgSO4·7H2O, and 0.3 g L−1 sodium pyruvate. In addition to the medium used as described above, each medium contained 7.5 mM sodium nitrate and 3 mM glucose during anoxic cell incubation. After inoculation, the cultures were incubated in a glove box (Coy lab products, USA). The box is a vinyl anaerobic airlock chamber, and was filled with 1.5–3.5% of H2 with the balance as N2. All of the bacteria were grown for 12–48 h at 25 °C in the dark. At the stationary phase, 50 mL of the cultures were harvested by centrifugation for 30 min at 3000 rpm and room temperature. The pellet was resuspended in 50 mL of cell-free medium. This procedure was repeated twice to wash out nitrate and nitrite, which might have been present after preculturing. Then 50 mL of the cell culture were transferred into a sterilized petri dish, which contained 50 g of sterilized glass beads (0.25–0.50 mm, Carl Roth GmbH, Germany). Then 1 mL of a 100 mM nitrate or nitrite solution and 0.5 mL of 100 mM glucose solution were added to the petri dish. Thus, the initial concentration was 2 mM for nitrate and nitrite and 1 mM for glucose, respectively. The petri dish containing the cell culture and nutrient solution was placed in the dynamic chamber, and the fluxes of HONO, NO, and N2O were measured by flushing with N2 gas (99.999%) instead of purified air. During the anaerobic measurements, the petri dish was covered with a lid at the beginning. The lid had two holes, one inlet and one outlet, and was flushed with 2 L min−1 N2 gas to exclude potential contamination from oxygen. The total N2 gas flow rate flushed into the chamber was still 8 L min−1. After 30 min of flushing, the lid was removed and the emissions of reactive nitrogen gases were measured.
Mutants
Strains and mutants of E. coli K-12 used in this study have been described in detail elsewhere [37]. Briefly, the parent strain RK4353 is a derivative of MC4100 (∆lacU169 araD139 rpsL gyrA non) [38]. P1 transduction was used to transfer the nirBDC::kan mutation from strain JCB4081a to RK4353 and the nrfAB::cat mutation from strain JCB4053 to RK4353. Strains that were defective in two different proteins were constructed by bacteriophage P1 transduction of a deletion marked with an antibiotic resistance cassette, followed by the pCP20-mediated removal of the kan or cat cassette. Thus, an isogenic RK4353 mutant, JCB5225 (RK4353 ∆nirBDC::kan ∆nrfAB::cat) was available for further experiments. JCB5225 was resistant to chloramphenicol because the cat cassette inserted into nrf has not been “cured”. Bacteria were grown anaerobically in a liquid yeast extract medium as described above, supplemented with 7.5 mM sodium nitrate and 3 mM glucose. After harvesting at the stationary phase, the emissions of HONO, NO, and N2O from different mutants were measured, as described in detail above under anoxic conditions (with N2 gas) in the dynamic chamber.
Extraction of nucleic acids and functional gene and transcript amplification
For nucleic acid analysis, samples were collected at different soil water contents from the dynamic chamber system. Subsamples from the homogenized soil S1 were measured until the desired water content was reached; then the measurements were stopped, and six replicate samples of the soil were immediately stored at −80 °C until use. Total RNA was isolated from three out of the six replicate soil samplings using the RNA PowerSoil Total RNA Isolation Kit (MO BIO Laboratories, Inc., USA). Potential DNA contamination of the RNA was removed by DNA-free DNAse, and polymerase chain reaction (PCR) amplification of 16S rRNA genes from the isolated RNA as a template failed, indicating DNA-free RNA. The DNA-free purified RNA was reversely transcribed with random hexamers and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol [39]. Sequences associated with nitrate reduction (napA and narG) and nitrite reduction (nirK, nirS, and nrfA), and 16S rRNA were amplified from cDNA using the following primer pairs: napA_F1 (CTGGACIATGGGYTTIAACCA)/napA_R1 (CCTTCYTTYTCIACCCACAT), narG1960f (TAYGTSGGSCARGARAA)/narG2650r (TTYTCRTACCABGTBGC), F1aCu (ATCATGGTSCTGCCGCG)/R3Cu (GCCTCGATCAG(A/G)TTGTGGTT), cd3aF (GTSAACGTSAAGGARACSGG)/R3cd (GASTTCGGRTGSGTCTTGA), nrfA_F2aw (CARTGYCAYGTBGARTA)/nrfA_R1 (TWNGGCATRTGRCARTC), and 341F (CCTACGGGAGGCAGCAG)/907RM (CCGTCAATTCMTTTGAGTTT), respectively [40–43]. A total of 100 µL of PCR reactions consisted of 40 µL of 2.5 × 5 Prime Master Mix solution (5 Prime GmbH, Hamburg, Germany), 4 µL of MgCl2 solution (25 mM), 6 µM of each primers (Microsynth AG, Balgach, Switzerland), 7 µl of cDNA template, and 37 µl of deionized water. PCR was performed in a SensoQuest Thermo Cycler (SensoQuest GmbH, Göttingen, Germany) using the following program for the 16S rRNA sequence: initial denaturation at 94 °C for 3 min, then 19 cycles of 94 °C for 30 s, 65 °C for 30 s (touchdown PCR, 0.5 °C per cycle), 72 °C for 90 s, followed by 14 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s. Final extension was at 72 °C for 3 min. PCR conditions for other amplifications were summarized in SI Table S2.
Gene expression by quantitative PCR
Quantitative kinetic real-time PCRs (qPCRs) were performed in an iQTM5 Real-Time qPCR cycler (Bio-Rad, Munich, Germany) to enumerate the starting quantities of 16S rRNA, napA, narG, nirK, nirS, and nrfA transcripts. All reactions were run in technical triplicates with cDNA as a template utilizing SensiMix (Bioline GmbH, Luckenwalde, Germany) chemistry and external standards [40]. Transcript abundances were normalized to the abundances (g−1 dry soil) of 16S rRNA, which yields expression levels less sensitive to varying RNA extraction efficiencies (i.e., RNA extraction bias) than copy numbers per gram of dry weight soil. nirK expression was below the quantification limit of our qPCR method and thus was not shown. Detailed information can be found elsewhere [39, 40].
Sequencing and transcript diversity analyses
Amplicons (i.e., PCR products) were generated from three replicate soil samples per time point and purified on 1% agarose gels using the MinElute Gel extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. All amplicons per time point and replicates were pooled on a mass basis to yield 18 amplicon pools in total: 16S rRNA : narG : napA : nrfA : nirS : nirK = 46 : 15 : 21 : 10 : 8. Adaptamers including barcodes were ligated to amplicon pools for sequencing library generation according to standard protocols prior to paired-end sequencing on a Illumina MiSeq platform utilizing V2 chemistry (2 × 250 bp). Only sequences that matched primer sequences were further analyzed. Paired-end merging for napA, nrfA, nirS, and nirK, quality filtering (Q > 15), length trimming (Q > 15), dereplication, and clustering was done with the usearch pipeline and Jaguc [44, 45]. For 16S rRNA, narG, napA, nrfA, nirS, and nirK, 43,000 ± 5000 (16S rRNA), 63,000 ± 5100 (narG), 17,500 ± 2300 (napA), 211,000 ± 25,000 (nrfA), 211,000 ± 28,000 (nirS), and 9800 ± 1700 (nirK), respectively, reads were obtained per replicate and transcript (mean ± standard error). OTUs were called at 15% threshold distance for narG and at 3% for all other sequences. Classification of 16S rRNA data was done by RDP [46] and BlastN against the nonredundant nucleotide collection. Classified 16S RNA data at the family level were generated by grouping OTUs called at 3% and presented. Other sequences were classified by BlastX against the nonredundant protein database (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Results and discussion
Large HONO emissions at high-soil moisture
Agricultural soil samples were collected from a wheat field (soil S1, Mainz-Finthen, Germany, Fig. 1a) and from 10 other fields (crops, rice, oasis, and grassland, see SI Fig. S2 and Table S1). Two peaks of HONO and NO emission were found for all of these samples, with a wet peak at 75–140% WHC, as well as the previously reported dry peak at 0–40% of the WHC [18]. The HONO and NO peaks were reproducible in three replicates of the soil S1, with standard errors of the maximum HONO and NO flux within 15% of the average value (SI Fig. S2). The maximum fluxes of HONO emission at 75–140% WHC were in the range of 5–190 ng m−2 s−1 (nitrogen mass-based, see SI Table S1), which is of a similar magnitude to the dry peak emission fluxes reported by Oswald et al. [18]. Sörgel et al. [13] estimated that a surface HONO flux of about 28–70 ng N m−2 s−1 would be required to sustain the measured boundary layer (boundary layer height about 1000 m) values at a rural site. Since most of our observed HONO flux values at high WHC were in the range of 15–85 ng N m−2 s−1, this source could explain the boundary layer values at that rural site. Furthermore, Su et al. [17] calculated HONO fluxes from about 1–3000 ng N m−2 s−1 for different soils and gave a range of ~1–1000 ng N m−2 s−1 for the missing sources calculated for boundary layer heights of 100 and 1000 m. Thus, soil emissions under wet conditions might well explain all or part of the missing HONO source during the day time.
Fig. 1.
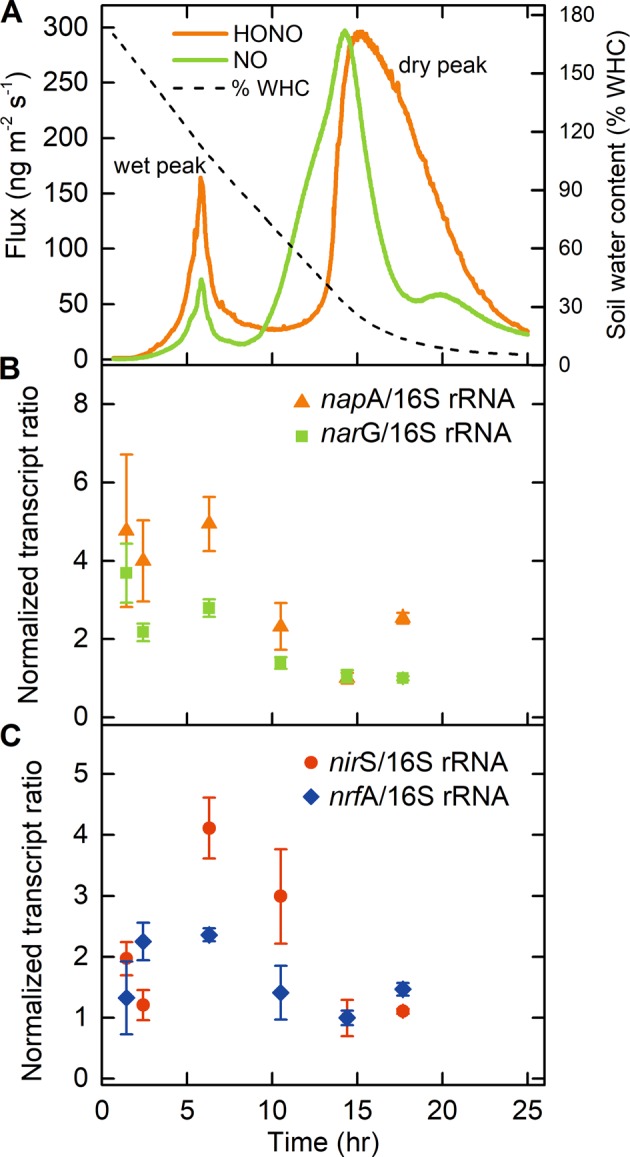
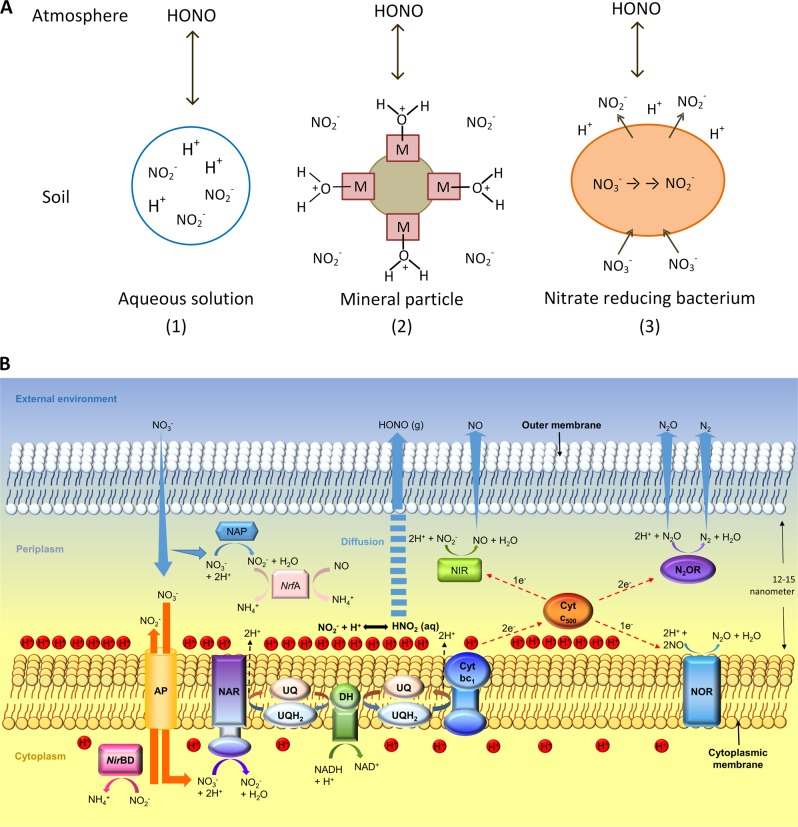
HONO and NO emissions from an agricultural soil sample. a Nitrogen mass-based emission fluxes of HONO (solid orange line) and NO (solid green line) from soil S1 (wheat field, Mainz-Finthen, Germany) plotted against measurement time (hours). Soil water content (percentage of water-holding capacity, % WHC) during the measurements is shown in the right Y axis. b Corresponding expression levels of periplasmic and cytoplasmic nitrate reductase genes (napA and narG, respectively), and c of nitrite reductase genes (nirS and nrfA, respectively). Data points and error bars represent mean values and standard errors of three replicates
For the soil samples investigated in this study, the magnitude of the wet peak maximum flux was 10–90% of the dry peak maximum flux. Interestingly, the HONO wet peak occurred at the highest water content (i.e., the earliest time point) for the moderately acidic soils S3, S5, and S9. The HONO flux peaks under high-moisture content were not as clearly defined and were lower than those in the other soils, suggesting that the low pH constrained the microbial community. However, considering that soils S3, S5, and S9 were regularly flooded, the soil microbial community might be adapted to the changing redox potentials and to nitrate respiration. Thus, the early onset of HONO fluxes might be related to a microbial community prone to react to low-redox potentials/anoxia by nitrate reduction to nitrite.
Nitrate-dependent biotic HONO emissions
Soil S1 was treated with 70% ethanol to reduce the numbers of live microbial cells and their activities. HONO and NO fluxes at high- and low-soil WHC (wet and dry peak, respectively) were significantly decreased (~40 and 10-fold, respectively) relative to non-ethanol-treated soil (SI Fig. S3a), as had been already shown with a different inhibition method for the dry peak [18], indicating that biotic processes dominated HONO emissions at high soil moisture.
Biotic HONO and NO emissions are known to depend on soil temperature [18, 47]. We found that HONO and NO emissions increased with temperature increasing from 5 to 45 °C (SI Fig. S3b and c), which is in agreement with a biotic process [48].
After incubation for 54 h at ~160% WHC (flooded conditions), HONO and NO fluxes from soil S1 diminished, but recovered after the addition of nitrate (SI Fig. S4). These data indicated (1) nitrate-dependent HONO and NO emissions and suggested that (2) nitrate reduction might significantly contribute to HONO and NO emissions at high soil moisture.
Microbial nitrate reduction is associated with HONO emissions from soil
We measured concentrations of nitrate and nitrite, levels of mRNA for genes involved in nitrate reduction and denitrification, and transcript diversity in the soil sample S1. The nitrate concentration decreased concomitantly with an increase in nitrite concentration at the HONO wet peak, while changes in soil pH were negligible (SI Fig. S5a). Thus, HONO emissions were linked to increased nitrate-derived nitrite concentrations at ~110% WHC. The HONO wet peak also correlated with the highest levels of the expression of napA and narG, both encoding nitrate reductases, and nitrite reductase genes nirS and nrfA, encoding cytochrome cd1-dependent denitrifier and DNRA-related nitrite reductases, respectively (Fig. 1b, c). Expression of the nitrate reduction associated gene was higher than that of the nitrite reductases prior to the wet peak. Expression of nirK (encoding a Cu-dependent denitrifier nitrite reductase) was below the detection limit. These data suggest the ongoing microbial anaerobic nitrate reduction to ammonia and/or denitrification as a source of HONO emissions due to a temporal nitrite accumulation.
Proteobacteria as key players for HONO emission
By producing high local nitrite concentrations right at the outer side of the cytoplasmic membrane, which is a low-pH environment, HONO formation is suggested to be due to pH-dependent nitrite loss. Active microbial key players of the nitrate reducers were identified by 16S rRNA and functional gene transcript sequencing. The dominant active microorganisms in soil S1 during the HONO measurements (Fig. 1a) were related to Telluria mixta (OTU 14), Stenotrophomonas maltophilia (OTU 999), Yersinia kristensenii (OTU 15), Ochrobactrum anthropi (OTU 39), Rhodanobacter D206a (OTU 314), and Y. kristensenii (OTU 2220), as indicated by 16S rRNA, narG, napA, nirK, nirS, and nrfA transcript diversity analyses, respectively (SI Fig. S6 and Table S3). Interestingly, only one 16S rRNA-based OTU with a mean relative abundance of 0.39% (range: 0.0–0.8%) was affiliated with a potential nitrifier (i.e., Nitrosospira sp.), suggesting a minor activity/importance of nitrifiers and potentially nitrifier denitrification during our experiments. The relative abundance of Serratia sp. (OTU 470) and Y. kristensenii (OTU 15) related nitrate reductase transcripts (narG and napA) was increased by ~6 and 2%, respectively at the wet peak. The relative abundances of Achromobacter sp. (OTU 32), Gammaproteobacteria bacterium SG8_30 (OTU 306), and Shigella sonnei (OTU 2226) related nitrite reductase transcripts (nirK, nirS, and nrfA, respectively) were increased by ~19, 14, and 12%, respectively, at the wet peak. Thus, diverse anaerobic nitrate reducing Proteobacteria were active during HONO emissions.
Model anaerobic nitrate reducing Proteobacteria emits HONO
The gammaproteobacterial model nitrate reducer Escherichia coli reduces nitrate to nitrite catalyzed by the nitrate reductase NarG and NapA. E. coli produced HONO and NO under anoxic conditions in the presence of initial nitrate or nitrite (Fig. 2, Table 1). HONO and NO formation was marginal in the absence of E. coli. HONO and NO fluxes of E. coli cultures were similar for nitrate and nitrite-supplemented media, suggesting that nitrate was completely reduced to nitrite in the nitrate-supplemented cultures.
Fig. 2.
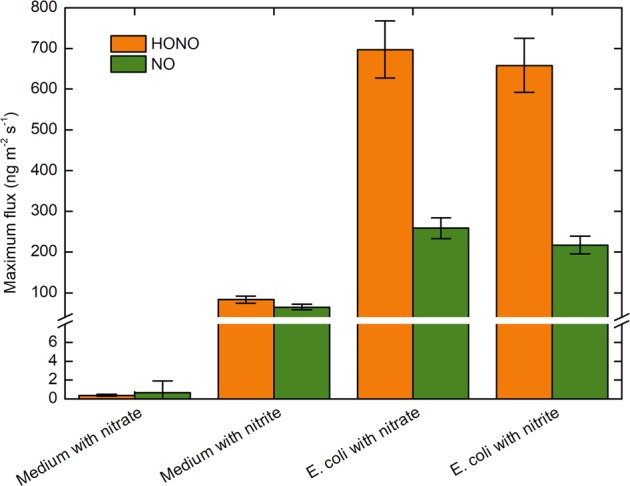
Maximum anaerobic emissions of HONO and NO from Escherichia coli K-12. Nitrogen mass-based maximum fluxes of HONO and NO from the nitrate reducer, E. coli K-12, incubated with nitrate or nitrite and measured under anoxic conditions. Data bars represent the maximum values, and error bars represent relative errors that were calculated based on Gaussian error propagation
Table 1.
Net anaerobic HONO and NO emissions from different Proteobacteria in the presence of initial nitrite or nitrate
| Proteobacteria | Strains | Genes encoding nitrogen oxide reductases converting | Maximum flux (ng m−2 s−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NO3− to NO2− | NO2− to NO | NO2− to NH4+ | NO to N2O | N2O to N2 | Nitrite | Nitrate | ||||||
| narG | napA | nirK | nirS | nirBDC/nrfAB | norBC | nosZ | HONO | NO | HONO | NO | ||
| Nitrate reducer | Escherichia coli K-12 | + | + | − | − | +a | − | − | 575 ± 115 | 165 ± 33 | 697 ± 139 | 258 ± 52 |
| Denitrifiers | Pseudomonas G-179 | − | + | + | − | − | + | − | −12 ± 2 | 96 ± 19 | 11 ± 2 | 39 ± 8 |
| Bradyrhizobium japonicum | − | + | + | − | − | + | + | −31 ± 6 | −13 ± 3 | 14 ± 3 | 16 ± 3 | |
| Rhodanobacter denitrificans | + | − | + | − | − | + | + | 4 ± 1 | 5 ± 1 | 0.1 ± 0 | 0 ± 0.1 | |
| Pseudomonas stutzeri JM-300 | + | + | − | + | − | + | + | −37 ± 7 | −31 ± 6 | 0.7 ± 0.1 | 0.6 ± 0.1 | |
+ and − indicate that the bacteria have or do not have the specific denitrification-associated gene, respectively. Nitrogen mass-based maximum fluxes of HONO and NO were calculated by the differences between the maximum flux from pure cell culture and sterilized background medium, and can be attributed to the direct effect of the bacteria. Values are the maximum fluxes ± relative errors. Relative errors were calculated based on 20% of the flux value, which were derived from four replications of sterilized background measurement.
aAlternative nitrite reductases, see Fig. 3
HONO and NO fluxes from denitrifying Proteobacteria differing in their set of denitrification-associated genes were minimal relative to the nitrate reducer E. coli hosting a nrfA-encoded nitrite reductase (Table 1). This suggests that the NirK and NirS nitrite reductases of denitrifying bacteria efficiently prevent the accumulation of nitrite because nitrite reduction rates were similar to nitrate reduction rates. Indeed, N2O peaks with a maximum flux of ~100 and ~40,000 ng m−2 s−1 were detected for Pseudomonas G-179 and P. stutzeri, respectively, in the presence of nitrite (SI Fig. S7). High rates of N2O emission by P. stutzeri were associated with the depletion of NO2−–N from 24.4 mg kg−1 determined for the sterilized control to 0.3 mg kg−1 at the end of the measurement, suggesting complete denitrification and loss of nitrogen as N2, N2O, and NO due to high rates of denitrification by P. stutzeri compared with other denitrifying bacteria [49]. Thus, the data suggest that (i) nitrate reducers are more prone to accumulate nitrite and emit HONO than actively denitrifying cultures, and (ii) active nitrite reduction mitigates HONO formation.
Absence of nitrite reductase genes increases HONO emissions from E. coli K-12
A double mutant, strain JCB5225, defective in the nirBDC and nrfAB nitrite reductase genes is unable to reduce nitrite to ammonia. This strain reduces nitrate quantitatively to nitrite, which accumulates in the growth medium. Compared with the parent strain RK4353, HONO and NO emissions increased significantly when the strain JCB5225 was grown in the presence of supplemental nitrite or nitrate (Fig. 3). As expected, nitrite concentrations for the strain without nirBDC and nrfAB genes were much higher than for the wild-type strain (SI Table S4), suggesting that nirBDC and nrfAB-encoded nitrite reductases impacted nitrite concentrations [50] and thus HONO and NO emissions.
Fig. 3.
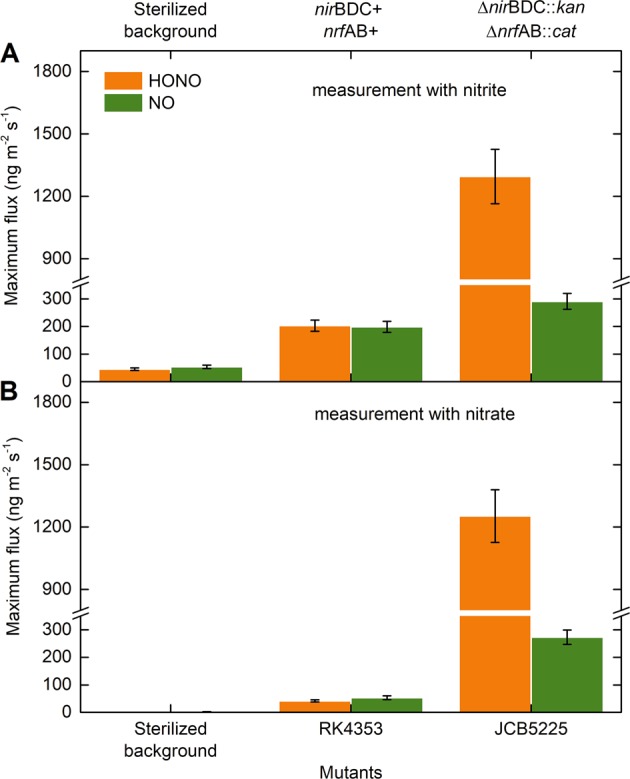
Maximum anaerobic emissions of HONO and NO from mutant strains of Escherichia coli K-12. Nitrogen mass-based maximum fluxes of HONO and NO incubated with a nitrite or b nitrate at an initial concentration of 2 mM. Investigated strains include RK4353 (parent strain, with both functional nirBDC and nrfAB genes) and JCB5225 (RK4353 ∆nirBDC::kan ∆nrfAB::cat, without both functional nirBDC and nrfAB genes). Data bars represent the maximum values, and error bars represent relative errors that were calculated based on Gaussian error propagation
Nitrate reducer-driven HONO emissions
We calculated HONO fluxes based on the acid–base equilibrium in solution and the volatilization of HONO according to Henry’s law [17] (Fig. 4a, pathway 1). The results were compared with the measured fluxes from the E. coli wild-type and mutant experiments (SI Table S4). The model was able to account for about 30–45% of the HONO flux from the heat-sterilized background (autoclaved medium with filter-sterilized nitrate or nitrite added after autoclaving), but less than 5% of the measured HONO flux from experiments with pure cultures. Except for very dry conditions (<1% WHC), the calculated HONO fluxes for soil S1 were also much lower than the measured fluxes (SI Fig. S5b). There are several reasons why application of Henry’s law might lead to an underestimation of HONO fluxes. A recent study showed that surface acidity in soil particles rather than bulk pH controls HONO uptake and release from soil [19] (Fig. 4a, pathway 2). Nitrite and HONO might be highly concentrated in water and biofilms rather than evenly distributed. Indeed, nitrate reducers release nitrite generated in their cytoplasm into their surroundings, supporting the view that nitrate reducers represent such local “hot spots” of high-nitrite concentrations. Second, a proton motive force is generated during nitrate reduction, resulting in an accumulation of positive charges at the outside of the bacterial cytoplasmic membrane relative to the cytoplasm or the external environment [51, 52]. Acidification at the outside of the bacterial cytoplasmic membrane might accelerate HONO emissions as well (Fig. 4a, pathway 3, and Fig. 4b). Based upon acid–base equilibria alone, a pH of 1.0–2.5 units lower than that of the bulk fluid would be required to explain the observed rates of the HONO efflux. This is within the range that typically occurs across cytoplasmic membranes [51–54]. HONO might leave the soil immediately once formed on the surface. Soil is very heterogeneous and even at high soil moisture, a certain fraction of the soil surface (including internal surfaces connected by pores) is exposed to the atmosphere. Indeed, soil pore networks are complex, and control gas transport in soil [55].
Fig. 4.
Potential pathways of HONO emission from soil (a), and illustration of HONO production during denitrification and anaerobic nitrate reduction (b). a Pathway 1: acid–base equilibrium in soil aqueous solution and the volatilization of nitrous acid according to Henry’s law [17]; Pathway 2: nitrous acid formation and release controlled by soil mineral particle surface acidity [19]; Pathway 3: nitrite accumulation at the outside of the cytoplasmic membrane of nitrate-reducing microbes, resulting in locally increased nitrite concentrations and nitrous acid formation enhanced by proton motive force-dependent acidification. b Nitrite is produced by anaerobic nitrate reductases close to the cytoplasmic membrane, either at the outer or inner side of the membrane. Nitrite produced at the inner side of the membrane is subsequently transported to the outer side via nitrate–nitrite antiporters (AP). The outer side of the membrane is positively charged due to accumulation of protons (proton motive force). Nitrous acid is formed due to the high-proton concentrations from nitrite (acid–base equilibrium) and diffused out of the outer membrane through porin channels to the external environment. HONO (g) and HNO2 (aq) represent molecular nitrous acid in the gas and aqueous phase, respectively
In conclusion, soil HONO emissions can account for the missing HONO source during the day time. Temperatures are higher during the day than at nighttime, and all dominating processes with respect to HONO formation and transport correlated with temperature, e.g., HONO solubility decreases, fluxes from soil (SI Fig. S3) increase, and turbulent transport processes are more efficient. In accordance with Su et al. [17], such processes associated with HONO fluxes from soil can indeed account for the missing source. HONO emissions under conditions of high soil water content contribute to soil HONO fluxes and can be explained by nitrite accumulation that is driven by nitrate reducers. This process is an additional significant source of atmospheric HONO that has previously not been taken into account.
Emissions under “dry conditions” likewise contribute and could be attributed to the activity of ammonia oxidizers [18, 21] and ammonia-oxidizing Archaea [21, 23]. At least some of the HONO emissions at the dry peak originate from the heterogeneous hydrolysis of NH2OH [23], which is an intermediate in ammonium oxidation. In contrast, we now showed that the wet peak arises from nitrite accumulation during nitrate reduction. Our data suggest that HONO formation during the wet peak is due to the microbial formation of nitrite by nitrate reducers under anaerobic conditions in anoxic or oxygen-reduced microsites in soil.
Agricultural soils receive large amounts of nitrogen fertilizer, adding nitrate to the soils. It is well known that soils host anoxic microsites within aggregates [56], and that N2O emissions are strongly stimulated by fertilization events [57]. This stimulation is considered to be due to denitrification releasing nitrite as an intermediate [58]. Rain events, irrigation, or fertilization with manure will increase soil moisture and thus the extent of anoxic microsites in soil. Thus, our results and conclusions apply to many diverse soils, including agricultural upland soils (SI Table S1). If we assume 14.2 × 1012 m2 of arable land (data from FAO, global arable land area in 2016), 5 irrigation events per year, and 20 precipitation events higher than 2.0 mm h−1 per year with substantial regional variations [59], then ~112 and 94 Gg y−1 of HONO–N and NO–N might be released at high soil water content from global arable land soils [22]. If, however, only 3.3 × 1012 km2 of arable land equipped for irrigation (data from FAO, global arable land area in 2016) is taken into account, with 5 irrigation events per year and 10 precipitation events higher than 2.0 mm h−1 per year, then only ~16 and 13 Gg y−1 of HONO–N and NO–N might be released at high soil water content from global arable land soils. According to these calculations, the emissions of reactive nitrogen (HONO–N and NO–N) from global arable land soils at high soil water content may range between 29 and 210 Gg y−1, corresponding to around 0.8–5.6% of NOx emissions from agricultural soil (3.7 Tg y−1) according to the IPCC report [60]. The impact of HONO emissions from these soils on the chemistry within the atmospheric boundary layer, nitrogen use efficiency, and climate change should be further quantified and included in regional and global models of atmospheric chemistry and air quality.
Supplementary information
Acknowledgements
We thank J. Cui and M. Badawy for helping to collect the soil samples. We also thank G. Braker and L.R. Bakken for providing pure bacteria cultures. We are grateful to J. Kesselmeier, F.X. Meixner, and H.L. Drake for allowing us to complete experiments in their laboratories. We are also grateful to M. Welling, D. Plake, U. Parchatka, A. Moravek, A. Dallinger, S. Hetz, M. Morawe, N. Roßbach, R. Mertel, and A. Wieczorek for supporting techniques in our experiments. This work was supported by the Max Planck Society and the Chinese Academy of Sciences. D. Wu was sponsored by the National Natural Science Foundation of China (41807449), Shanghai Pujiang Program (18PJ1403500), and “the Fundamental Research Funds for the Central Universities”. M. Liu was supported by the National Natural Science Foundation of China (41761144062). B. Weber would like to thank Paul Crutzen for awarding her a Nobel Laureate Fellowship (2013–2015).
Author contributions
This study was co-initiated by D.W., I.T., T.B., Y.C., and H.S. The development of the experimental design and investigations were guided by M.A.H., M.S., B.W., and U.P. The measurements were performed by D.W., M.A.H., S.M., and J.L. The mutants were constructed by J.A.C. The soil samples and the corresponding nutrient data were supported by T.B., B.X., and X.J. Equilibrium model calculations were accomplished by D.W., M.S., U.P., Y.C., H.S., and G.L. The paper was written by D.W. and M.A.H. with the major input from M.S., J.A.C., U.P., and B.W., and further input from all the other authors.
Data availability
The sequence data reported in this paper have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics, Chinese Academy of Sciences (http://bigd.big.ac.cn/gsub/) with accession number CRA000459. All data needed to evaluate the conclusions are present in the paper and/or the Supplementary Information. Additional data related to this paper may be requested from the authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dianming Wu, Marcus A. Horn
Contributor Information
Dianming Wu, Phone: +86 (0)21-54341171, Email: dmwu@geo.ecnu.edu.cn.
Marcus A. Horn, Phone: +49 (0)511-762-17980, Email: horn@ifmb.uni-hannover.de
Bettina Weber, Phone: +49(0)6131-305-7501, Email: b.weber@mpic.de.
Matthias Sörgel, Phone: +49(0)6131-305-4432, Email: m.soergel@mpic.de.
Supplementary information
The online version of this article (10.1038/s41396-019-0379-y) contains supplementary material, which is available to authorized users.
References
- 1.Alicke B, Geyer A, Hofzumahaus A, Holland F, Konrad S, Pätz HW, et al. OH formation by HONO photolysis during the BERLIOZ experiment. J Geophys Res. 2003;108:8247. doi: 10.1029/2001JD000579. [DOI] [Google Scholar]
- 2.Kleffmann J, Gavriloaiei T, Hofzumahaus A, Holland F, Koppmann R, Rupp L, et al. Daytime formation of nitrous acid: a major source of OH radicals in a forest. Geophys Res Lett. 2005;32:L05818. doi: 10.1029/2005GL022524. [DOI] [Google Scholar]
- 3.Sleiman M, Gundel LA, Pankow JF, Jacob P, Singer BC, Destaillats H. Formation of carcinogens indoors by surface-mediated reactions of nicotine with nitrous acid, leading to potential thirdhand smoke hazards. Proc Natl Acad Sci. 2010;107:6576–81. doi: 10.1073/pnas.0912820107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen TR, Brauer M, Kjaergaard S. Effects of nitrous acid exposure on human mucous membranes. Am J Respir Crit Care Med. 1995;151:1504–11. doi: 10.1164/ajrccm.151.5.7735607. [DOI] [PubMed] [Google Scholar]
- 5.Gligorovski S, Abbatt JPD. An indoor chemical cocktail. Science. 2018;359:632–33. doi: 10.1126/science.aar6837. [DOI] [PubMed] [Google Scholar]
- 6.Alicke B, Platt U, Stutz J. Impact of nitrous acid photolysis on the total hydroxyl radical budget during the limitation of oxidant production/pianura padana produzione di ozono study in Milan. J Geophys Res. 2002;107:8196. doi: 10.1029/2000JD000075. [DOI] [Google Scholar]
- 7.Acker K, Moller D, Wieprecht W, Meixner FX, Bohn B, Gilge S, et al. Strong daytime production of OH from HNO2 at a rural mountain site. Geophys Res Lett. 2006;33:L02809. doi: 10.1029/2005GL024643. [DOI] [Google Scholar]
- 8.Sörgel M, Regelin E, Bozem H, Diesch JM, Drewnick F, Fischer H, et al. Quantification of the unknown HONO daytime source and its relation to NO2. Atmos Chem Phys. 2011;11:10433–47. doi: 10.5194/acp-11-10433-2011. [DOI] [Google Scholar]
- 9.Kleffmann J. Daytime sources of nitrous acid (HONO) in the atmospheric boundary layer. ChemPhysChem. 2007;8:1137–44. doi: 10.1002/cphc.200700016. [DOI] [PubMed] [Google Scholar]
- 10.Gutzwiller L, Arens F, Baltensperger U, Gäggeler HW, Ammann M. Significance of semivolatile diesel exhaust organics for secondary HONO formation. Environ Sci Technol. 2002;36:677–82. doi: 10.1021/es015673b. [DOI] [PubMed] [Google Scholar]
- 11.Gustafsson RJ, Orlov A, Griffiths PT, Cox RA, Lambert RM. Reduction of NO2 to nitrous acid on illuminated titanium dioxide aerosol surfaces: implications for photocatalysis and atmospheric chemistry. Chem Commun. 2006;0:3936–38. [DOI] [PubMed]
- 12.Finlayson-Pitts BJ. Reactions at surfaces in the atmosphere: integration of experiments and theory as necessary (but not necessarily sufficient) for predicting the physical chemistry of aerosols. Phys Chem Chem Phys. 2009;11:7760–79. doi: 10.1039/b906540g. [DOI] [PubMed] [Google Scholar]
- 13.Sörgel M, Trebs I, Wu D, Held A. A comparison of measured HONO uptake and release with calculated source strengths in a heterogeneous forest environment. Atmos Chem Phys. 2015;15:9237–51. doi: 10.5194/acp-15-9237-2015. [DOI] [Google Scholar]
- 14.Michoud V, Colomb A, Borbon A, Miet K, Beekmann M, Camredon M, et al. Study of the unknown HONO daytime source at a European suburban site during the MEGAPOLI summer and winter field campaigns. Atmos Chem Phys. 2014;14:2805–22. doi: 10.5194/acp-14-2805-2014. [DOI] [Google Scholar]
- 15.Kleffmann J, Kurtenbach R, Lörzer J, Wiesen P, Kalthoff N, Vogel B, et al. Measured and simulated vertical profiles of nitrous acid—Part I: field measurements. Atmos Environ. 2003;37:2949–55. doi: 10.1016/S1352-2310(03)00242-5. [DOI] [Google Scholar]
- 16.Kubota M, Asami T. Source of nitrous acid volatilized from upland soils. Soil Sci Plant Nutr. 1985;31:35–42. doi: 10.1080/17470765.1985.10555215. [DOI] [Google Scholar]
- 17.Su H, Cheng Y, Oswald R, Behrendt T, Trebs I, Meixner FX, et al. Soil nitrite as a source of atmospheric HONO and OH radicals. Science. 2011;333:1616–18. doi: 10.1126/science.1207687. [DOI] [PubMed] [Google Scholar]
- 18.Oswald R, Behrendt T, Ermel M, Wu D, Su H, Cheng Y, et al. HONO emissions from soil bacteria as a major source of atmospheric reactive nitrogen. Science. 2013;341:1233–35. doi: 10.1126/science.1242266. [DOI] [PubMed] [Google Scholar]
- 19.Donaldson MA, Bish DL, Raff JD. Soil surface acidity plays a determining role in the atmospheric-terrestrial exchange of nitrous acid. Proc Natl Acad Sci. 2014;111:18472–77. doi: 10.1073/pnas.1418545112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VandenBoer TC, Young CJ, Talukdar RK, Markovic MZ, Brown SS, Roberts JM, et al. Nocturnal loss and daytime source of nitrous acid through reactive uptake and displacement. Nat Geosci. 2015;8:55–60. doi: 10.1038/ngeo2298. [DOI] [Google Scholar]
- 21.Scharko NK, Schütte UME, Berke AE, Banina L, Peel HR, Donaldson MA, et al. Combined flux chamber and genomics approach links nitrous acid emissions to ammonia oxidizing bacteria and archaea in urban and agricultural soil. Environ Sci Technol. 2015;49:13825–34. doi: 10.1021/acs.est.5b00838. [DOI] [PubMed] [Google Scholar]
- 22.Weber B, Wu D, Tamm A, Ruckteschler N, Rodríguez-Caballero E, Steinkamp J, et al. Biological soil crusts accelerate the nitrogen cycle through large NO and HONO emissions in drylands. Proc Natl Acad Sci. 2015;112:15384–89. doi: 10.1073/pnas.1515818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ermel M, Behrendt T, Oswald R, Derstroff B, Wu D, Hohlmann S, et al. Hydroxylamine released by nitrifying microorganisms is a precursor for HONO emission from drying soils. Sci Rep. 2018;8:1877. doi: 10.1038/s41598-018-20170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maljanen M, Yli-Pirilä P, Hytönen J, Joutsensaari J, Martikainen PJ. Acidic northern soils as sources of atmospheric nitrous acid (HONO) Soil Biol Biochem. 2013;67:94–97. doi: 10.1016/j.soilbio.2013.08.013. [DOI] [Google Scholar]
- 25.Knowles R. Denitrification. Microbiol Mol Biol Rev. 1982;46:43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim NY, Frostegård Aring, Bakken LR. Nitrite kinetics during anoxia: the role of abiotic reactions versus microbial reduction. Soil Biol Biochem. 2018;119:203–09. doi: 10.1016/j.soilbio.2018.01.006. [DOI] [Google Scholar]
- 27.Betlach MR, Tiedje JM. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl Environ Microbiol. 1981;42:1074–84. doi: 10.1128/aem.42.6.1074-1084.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu D, Kampf CJ, Pöschl U, Oswald R, Cui J, Ermel M, et al. Novel tracer method to measure isotopic labeled gas-phase nitrous acid (HO15NO) in biogeochemical studies. Environ Sci Technol. 2014;48:8021–27. doi: 10.1021/es501353x. [DOI] [PubMed] [Google Scholar]
- 29.Homyak PM, Vasquez KT, Sickman JO, Parker DR, Schimel JP. Improving nitrite analysis in soils: drawbacks of the conventional 2 M KCl extraction. Soil Sci Soc Am J. 2015;79:1237–42. doi: 10.2136/sssaj2015.02.0061n. [DOI] [Google Scholar]
- 30.van Dijk SM. Biogenic NO emissions from forest and pasture soils: Relating laboratory studies to field measurements. J Geophys Res. 2002;107: LBA 25-1-LBA 25–11.
- 31.Remde A, Ludwig J, Meixner FX, Conrad R. A study to explain the emission of nitric oxide from a marsh soil. J Atmos Chem. 1993;17:249–75. doi: 10.1007/BF00694400. [DOI] [Google Scholar]
- 32.Rummel U, Ammann C, Gut A, Meixner FX, Andreae MO. Eddy covariance measurements of nitric oxide flux within an Amazonian rain forest. J Geophys Res. 2002;107:LBA 17-1–9. doi: 10.1029/2001JD000520. [DOI] [Google Scholar]
- 33.Plake D, Stella P, Moravek A, Mayer JC, Ammann C, Held A, et al. Comparison of ozone deposition measured with the dynamic chamber and the eddy covariance method. Agric Meteorol. 2015;206:97–112. doi: 10.1016/j.agrformet.2015.02.014. [DOI] [Google Scholar]
- 34.Müller S, Hoor P, Berkes F, Bozem H, Klingebiel M, Reutter P, et al. In situ detection of stratosphere–troposphere exchange of cirrus particles in the midlatitudes. Geophys Res Lett. 2015;42:949–55. doi: 10.1002/2014GL062556. [DOI] [Google Scholar]
- 35.McManus JB, Zahniser MS, Nelson JDD, Shorter JH, Herndon S, Wood E, et al. Application of quantum cascade lasers to high-precision atmospheric trace gas measurements. Opt Eng. 2010;49:111124. doi: 10.1117/1.3498782. [DOI] [Google Scholar]
- 36.McManus JB, Zahniser MS, Nelson DD. Dual quantum cascade laser trace gas instrument with astigmatic Herriott cell at high pass number. Appl Opt. 2011;50:A74–A85. doi: 10.1364/AO.50.000A74. [DOI] [PubMed] [Google Scholar]
- 37.Vine CE. Escherichia coli response to nitrosative stress. PhD thesis, University of Birmingham, 2012.
- 38.Stewart V, MacGregor CH. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982;151:788–99. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu YJ, Zaprasis A, Liu SJ, Drake HL, Horn MA. The earthworm Aporrectodea caliginosa stimulates abundance and activity of phenoxyalkanoic acid herbicide degraders. ISME J. 2011;5:473–85. doi: 10.1038/ismej.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer K, Biasi C, Horn MA. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 2012;6:1058–77. doi: 10.1038/ismej.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dallinger A, Horn MA. Agricultural soil and drilosphere as reservoirs of new and unusual assimilators of 2,4-dichlorophenol carbon. Environ Microbiol. 2014;16:84–100. doi: 10.1111/1462-2920.12209. [DOI] [PubMed] [Google Scholar]
- 42.Alcántara-Hernández RJ, Valenzuela-Encinas C, Marsch R, Dendooven L. Respiratory and dissimilatory nitrate-reducing communities from an extreme saline alkaline soil of the former lake Texcoco (Mexico) Extremophiles. 2009;13:169–78. doi: 10.1007/s00792-008-0207-1. [DOI] [PubMed] [Google Scholar]
- 43.Welsh A, Chee-Sanford J, Connor L, Löffler F, Sanford R. Refined NrfA phylogeny improves PCR-based nrfA gene detection. Appl Environ Microbiol. 2014;80:2110–19. doi: 10.1128/AEM.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nebel ME, Wild S, Holzhauser M, Hüttenberger L, Reitzig R, Sperber M, et al. JAguc—a software package for environmental diversity analyses. J Bioinform Comput Biol. 2011;09:749–73. doi: 10.1142/S0219720011005781. [DOI] [PubMed] [Google Scholar]
- 45.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–98. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–67. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gödde M, Conrad R. Immediate and adaptational temperature effects on nitric oxide production and nitrous oxide release from nitrification and denitrification in two soils. Biol Fertil Soils. 1999;30:33–40. doi: 10.1007/s003740050584. [DOI] [Google Scholar]
- 48.Keeney DR, Fillery IR, Marx GP. Effect of temperature on the gaseous nitrogen products of denitrification in a silt loam soil. Soil Sci Soc Am J. 1979;43:1124–28. doi: 10.2136/sssaj1979.03615995004300060012x. [DOI] [Google Scholar]
- 49.Carlson CA, Ingraham JL. Comparison of denitrification by Pseudomonas stutzeri, Pseudomonas aeruginosa, and Paracoccus denitrificans. Appl Environ Microbiol. 1983;45:1247–53. doi: 10.1128/aem.45.4.1247-1253.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Gunsalus RP. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J Bacteriol. 2000;182:5813–22. doi: 10.1128/JB.182.20.5813-5822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calamita HG, Ehringer WD, Koch AL, Doyle RJ. Evidence that the cell wall of Bacillus subtilis is protonated during respiration. Proc Natl Acad Sci USA. 2001;98:15260–63. doi: 10.1073/pnas.261483798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konhauser KO (ed). Introduction to Geomicrobiology. (John Wiley & Sons, Malden, USA, 2009).
- 53.Padan E, Zilberstein D, Rottenberg H. The proton electrochemical gradient in Escherichia coli Cells. Eur J Biochem. 1976;63:533–41. doi: 10.1111/j.1432-1033.1976.tb10257.x. [DOI] [PubMed] [Google Scholar]
- 54.Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol. 2007;189:5601–07. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katuwal S, Arthur E, Tuller M, Moldrup P, de Jonge LW. Quantification of soil pore network complexity with X-ray computed tomography and gas transport measurements. Soil Sci Soc Am J. 2015;79:1577–89. doi: 10.2136/sssaj2015.06.0227. [DOI] [Google Scholar]
- 56.Keiluweit M, Gee K, Denney A, Fendorf S. Anoxic microsites in upland soils dominantly controlled by clay content. Soil Biol Biochem. 2018;118:42–50. doi: 10.1016/j.soilbio.2017.12.002. [DOI] [Google Scholar]
- 57.Liu C, Wang K, Meng S, Zheng X, Zhou Z, Han S, et al. Effects of irrigation, fertilization and crop straw management on nitrous oxide and nitric oxide emissions from a wheat–maize rotation field in northern China. Agric Ecosyst Environ. 2011;140:226–33. doi: 10.1016/j.agee.2010.12.009. [DOI] [Google Scholar]
- 58.Ruser R, Flessa H, Russow R, Schmidt G, Buegger F, Munch JC. Emission of N2O, N2 and CO2 from soil fertilized with nitrate: effect of compaction, soil moisture and rewetting. Soil Biol Biochem. 2006;38:263–74. doi: 10.1016/j.soilbio.2005.05.005. [DOI] [Google Scholar]
- 59.Trenberth KE, Zhang Y. How often does it really rain? Bull Am Meteorol Soc. 2018;99:289–98. doi: 10.1175/BAMS-D-17-0107.1. [DOI] [Google Scholar]
- 60.Ciais P, Sabine C, Bala G, Bopp L, Brovkin V, Canadell J, et al. Carbon and other biogeochemical cycles. In: Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, et al., editors. Climate change 2013: the physical science basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, UK: Cambridge University Press; 2014. pp. 465–570.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data reported in this paper have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics, Chinese Academy of Sciences (http://bigd.big.ac.cn/gsub/) with accession number CRA000459. All data needed to evaluate the conclusions are present in the paper and/or the Supplementary Information. Additional data related to this paper may be requested from the authors.