Abstract
Unlike biologically available nitrogen and phosphorus, which are often at limiting concentrations in surface seawater, sulfur in the form of sulfate is plentiful and not considered to constrain marine microbial activity. Nonetheless, in a model system in which a marine bacterium obtains all of its carbon from co-cultured phytoplankton, bacterial gene expression suggests that at least seven dissolved organic sulfur (DOS) metabolites support bacterial heterotrophy. These labile exometabolites of marine dinoflagellates and diatoms include taurine, N-acetyltaurine, isethionate, choline-O-sulfate, cysteate, 2,3-dihydroxypropane-1-sulfonate (DHPS), and dimethylsulfoniopropionate (DMSP). Leveraging from the compounds identified in this model system, we assessed the role of sulfur metabolites in the ocean carbon cycle by mining the Tara Oceans dataset for diagnostic genes. In the 1.4 million bacterial genome equivalents surveyed, estimates of the frequency of genomes harboring the capability for DOS metabolite utilization ranged broadly, from only 1 out of every 190 genomes (for the C2 sulfonate isethionate) to 1 out of every 5 (for the sulfonium compound DMSP). Bacteria able to participate in DOS transformations are dominated by Alphaproteobacteria in the surface ocean, but by SAR324, Acidimicrobiia, and Gammaproteobacteria at mesopelagic depths, where the capability for utilization occurs in higher frequency than in surface bacteria for more than half the sulfur metabolites. The discovery of an abundant and diverse suite of marine bacteria with the genetic capacity for DOS transformation argues for an important role for sulfur metabolites in the pelagic ocean carbon cycle.
Subject terms: Biogeochemistry, Microbiology
Introduction
The trophic linkage between marine bacteria and phytoplankton in the ocean represents a key step in the global carbon cycle, with phytoplankton lysis products [1] and “dissolved primary production” [the dissolved organic carbon (DOC) released from living phytoplankton [2]] supporting a major fraction of labile carbon flux. Approximately 20 Gt of labile DOC are turned over by heterotrophic marine bacteria each year, an amount more than an order of magnitude higher than the annual turnover from the semi-labile and refractory marine DOC pools combined [3].
Sulfur is a component of amino acids, sulfolipids, and other biomolecules essential to marine bacteria, but the high inorganic sulfur concentration in seawater (~28 mM SO4−2) suggests that its availability is not an important factor governing microbial growth. However, discoveries that members of the ubiquitous SAR11 and SAR86 bacterial clades are incapable of assimilating sulfate and thus dependent on uptake of organic sulfur [4, 5] bring a new perspective to the role of dissolved organic sulfur (DOS) in microbial heterotrophy. Overall, little is currently known about the availability or trophic fate of DOS compounds in the ocean. The one exception is the sulfonium compound dimethylsulfoniopropionate (DMSP), a DOS metabolite synthesized by phytoplankton as an osmoprotectant, possible antioxidant [6], and possible predator deterrent [7] that is readily utilized by heterotrophic marine bacteria [8].
An improved understanding of the participation of labile DOS in the carbon cycle requires better knowledge of the phytoplankton-derived molecules supporting the metabolism of heterotrophic bacteria. These molecules have proven difficult to identify, however, because efficient bacterial scavenging drives their concentrations into the picomolar range [9, 10] and their half-lives in seawater down to minutes [11]. An alternative strategy for discovering the molecular currencies supporting microbial heterotrophy is to extract biological signals from bacterial messenger RNA (mRNA) [12–14]. In this approach, the transcriptional responses of bacteria to phytoplankton-derived metabolites provide indications of substrate uptake and metabolism.
We previously established a model system to investigate how the compounds supporting bacterial heterotrophy varied with changes in the dominant phytoplankton taxon. The transcriptional response of marine bacterium Ruegeria pomeroyi DSS-3 was measured when co-cultured first with the dinoflagellate Alexandrium tamarense and then with the diatom Thalassiosira pseudonana [15]. Bacterial gene expression patterns indicated that both phytoplankton species were releasing labile DOS metabolites into the medium. At a minimum, these included 2,3-dihydroxypropane-1-sulfonate (DHPS), taurine, N-acetyltaurine, and DMSP [15]. In this study, we follow-up on these initial results using data from this co-culture system and metagenomic data from the TARA Oceans expedition [16] to answer two questions: (1) What does bacterial gene expression reveal about the identity of labile phytoplankton-derived DOS metabolites? (2) What is the abundance, taxonomy, and distribution of the ocean bacterioplankton capable of utilizing these metabolites? Our results show that at least seven phytoplankton-derived DOS metabolites can support bacterial heterotrophy, and indicate a more important role for DOS in microbial trophic interactions and carbon flux than previously recognized.
Methods
Experimental setup
Axenic strains of the dinoflagellate A. tamarense CCMP1771 and diatom T. pseudonana CCMP1335 were obtained from the National Center for Marine Algae and maintained with regular checks for bacterial contamination by microscopy and plating [15]. Co-cultures of the phytoplankton with or without the bacterium R. pomeroyi DSS-3 were established in six 20-L LDPE cubitainers (Reliance, Winnipeg, Canada). The cubitainers were first filled with 18 L of autoclaved, 0.2-µm-filtered Guillard’s f/2 medium with vitamin B12 added at f/50 concentration and 2 L of an axenic A. tamarense culture in exponential phase grown in the same medium. The f/2 medium contained 880 μM N as NO3− and 36 μM P as PO4−3 at the time of inoculation. Cubitainers were maintained at 18 °C with a 16–8 h light–dark cycle under ~160 μmol photons m−2 s−1. After 7 days, bacteria grown overnight in ½YTSS medium were harvested in exponential phase, washed five times with sterile phytoplankton medium, and added to three of the cultures at ~105 cells ml−1. Five days later, axenic T. pseudonana in exponential phase grown in the same modified f/2 medium was added to all cubitainers at 200 cells ml−1 along with silicate at 100 µM final concentration. The order in which the phytoplankton were introduced was determined based on T. pseudonana’s ability to outcompete A. tamarense under the culture conditions used. The cultures were maintained for 37 days [15].
Sampling and monitoring
Samples were taken regularly from the cubitainers and streaked onto ½YTSS plates to check for bacterial growth. Plates from bacteria-free phytoplankton controls were checked for growth of any bacteria, while plates from experimental cubitainers were visually inspected for colonies with different morphology from R. pomeroyi. No contamination was detected throughout the 37-day experiment. Bacterial and phytoplankton cell numbers were monitored in the cubitainers by flow cytometry. Triplicate samples were preserved in glutaraldehyde (2% final concentration), stained with SYBR® Green I (final concentration 0.75×; Life Technologies, Waltham, MA, USA), and analyzed on a Cyan instrument (Beckman Coulter, Brea, CA, USA) using the FlowJo software [15]. On days 7, 9, 12, 15, 18, 23, 30, and 37, 1 L samples were collected for chemical analyses and RNA-sequencing (RNA-seq).
Chemical analyses
Duplicate 6 ml samples filtered through 0.45-µm hydrophilic polyethersulfone Acrodisc Supor membranes (Pall Life Sciences, Port Washington, NY, USA) were analyzed for dissolved inorganic nitrogen (nitrate + nitrite) (APHA Standard Method 4500-NO3 F), ammonium (4500-NH3 G), dissolved inorganic phosphorus (4500-P F), and silicate (spectrophotometric silicomolybdate assays). Triplicate samples that were unfiltered or filtered through ashed Whatman GF/F filters were combusted on a Shimadzu TOC-5000A Analyzer to determine total organic carbon (TOC) and dissolved organic carbon (DOC) concentrations, respectively. Analyses were performed at the Center for Applied Isotope Studies (University of Georgia, Athens, GA, USA).
DMSP was the only organic sulfur metabolite measured in the co-culture medium. For total DMSP analysis, 10 ml subsamples were collected in 15 ml polypropylene tubes with plug seal caps (Corning, NY, USA) containing HCl (1.5% final concentration) for preservation. For dissolved DMSP analysis, 10 ml samples were gently poured into a polysulfone filtration tower equipped with a 25 mm GF/F filter, and 4 ml were gravity filtered and preserved in 1.5% HCl. At the time of analysis, DMSP was cleaved into dimethylsulfide (DMS) by the addition of NaOH (1.3 M final concentration). Resulting DMS was quantified using a gas chromatograph (Shimadzu GC-2014 or GC-14A) with a Chromosil 330 column and a flame photometric detector coupled with a purge and trap system [17].
Bacterial RNA extraction and RNA-seq
Bacterial cells were collected on 0.2 µm pore-size polycarbonate (PC) membranes after pre-filtration of 1 L samples through 2 µm pore-size membranes to remove eukaryotic cells [15]. Filters were flash frozen in liquid nitrogen and stored at −80 °C. For RNA extraction, filters were incubated at 37 °C for 1 h in TE buffer, sodium dodecyl sulfate (0.6% final concentration), and proteinase K (120 ng µl−1 final concentration, Qiagen, Hilden, Germany). An extraction with acid phenol:chloroform:isoamylalcohol was performed, and RNA was resuspended in RNAse-free water. RNA was precipitated with sodium acetate (0.3 M final concentration) and three volumes of 100% ethanol, and incubated overnight at −20 °C. Pellets were washed twice with 75% ethanol, centrifuged, dried, and resuspended in RNAse-free water [15].
Samples were treated with the Turbo DNA-free kit (Invitrogen, Waltham, MA, USA) to remove DNA, and tests for residual DNA by a 40-cycle PCR targeting the 16S ribosomal RNA (rRNA) gene of R. pomeroyi were negative. rRNA was depleted using custom probes for small and large subunit rRNA genes from all three microbes [18]. Libraries were prepared for two replicate cubitainers at each time point using the KAPA Stranded mRNA-Seq kit (Kapa Biosystems, Wilmington, MA, USA) at the Georgia Genomics and Bioinformatics Core (University of Georgia) and sequenced on a HiSeq Illumina 2500 at the Hudson Alpha Institute for Biotechnology (AL, USA).
RNA-seq analysis
The FASTX toolkit was used for quality control of 249 million 50-bp reads (10 ± 2 million reads per sample; Table S1), imposing a minimum quality score of 20 over 80% of read length. Reads aligning to an in-house rRNA database were removed (blastn, score cutoff ≥50). Bowtie 2 [19] and HTSeq [20] were used to map the remaining reads to the R. pomeroyi genome, conserving strand information and removing reads that mapped to more than one location (Table S1). Counts were converted to transcripts per million [TPM [21]; Table S2] and data deposited in the NCBI BioProject database under accession PRJNA381627. Genes with differential expression between selected time points (representing diatom- and dinoflagellate-dominated phases) were determined with DESeq2 [22]. Although bacterial expression can change more rapidly than the time intervals between samples, genes identified as significantly different were consistent over multiple sample dates within each phase and most likely represented sustained transport/metabolism activities.
Ruegeria pomeroyi expression assays
In a separate experiment, R. pomeroyi was grown in a minimal medium with 12 mM choline-O-sulfate, 20 mM cysteate, or 30 mM acetate as the sole substrate. Overnight cultures were pelleted by centrifugation, resuspended in 1 ml Ambion (Thermo Fisher Scientific) denaturation solution, flash frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. Samples were treated with the TURBO DNA-free kit to remove DNA and Ribo-Zero-Bacteria (Illumina) to remove rRNA. Library preparation, sequencing, and differential expression analysis were as described above.
Mining of the TARA Oceans metagenomes
We analyzed 225 TARA Oceans metagenomes in the bacterial/archaeal size range, either 0.22–1.6 or 0.22–3 µm, from surface (SRF), deep chlorophyll maximum (DCM), and mesopelagic (MES) depths (Table S3). Accounting for replicate samples, 132 unique stations/depths were analyzed. The metagenomes were assembled using SPAdes version 3.9 using k-mers of 21, 33, 55, and 77 after standard adapter trimming and error correction using BFC (https://github.com/lh3/bfc) with the k-mer size set to 19. The reassembly pipeline built contigs within individual samples, allowing coverage calculation by station and depth. Genes were called using Prodigal version 2.6.3 with default parameters. Coverage was obtained using BWA (https://github.com/lh3/bwa) with default parameters to map reads back to contigs and samtools (https://github.com/samtools/samtools) to calculate coverage for each contig.
Hidden Markov Models (HMMs) were used to search the contigs for DOS genes. The HMMs were constructed from reference sequences from bacterial isolates obtained for 11 DOS-related genes that were differentially expressed by R. pomeroyi during the co-culture experiment. Proteins with experimentally verified function were used whenever possible (Table S4). These gene databases were used to build the HMMs using HMMER 3.1b2. From the output from each HMM run against the gene calls in the Tara contigs, only sequences with at least 40% protein identity to a reference gene were kept. A score was then determined where at least 80% of assembled genes were manually annotated as the correct gene based on blastP analysis and tree building (Table S4). Most of the DOS-related genes mediate transformations in catabolic pathways, although transporter substrate binding genes were used in the case of the three C2 sulfonates that have a shared catabolic pathway. We note that our estimates of DOS gene frequencies are conservative, since HMM cutoffs were stringent, and any alternate pathway or transport proteins would not be counted. The Tara dataset was also searched for the single-copy gene recA using the HMM in the Pfam Database (http://pfam.xfam.org/family/PF00154#tabview=tab0). Relative gene abundance information was obtained for each DOS gene and recA based on coverage of the Tara contigs. The proportion of cells harboring a DOS gene at a given location and depth was estimated as: DOS gene counts (normalized to the length of the recA gene)/recA gene counts. A Bray–Curtis dissimilarity analysis based on DOS gene abundances by sample was carried out in R (vegan package, vegdist function) [23].
Taxonomy information was obtained for the gene assemblies that passed the HMM cutoff using Diamond [24] against the May 2018 version of the IMG non-redundant database by keeping the taxonomy of the best hit. In cases of frequent taxonomy assignments to a bacterial group not previously known to possess the genetic capability, manual annotation of gene function and that of its neighboring genes was carried out. Neighboring genes were also classified taxonomically to check for evidence of mis-assembled contigs, but this was not found.
Results and discussion
Co-culture dynamics
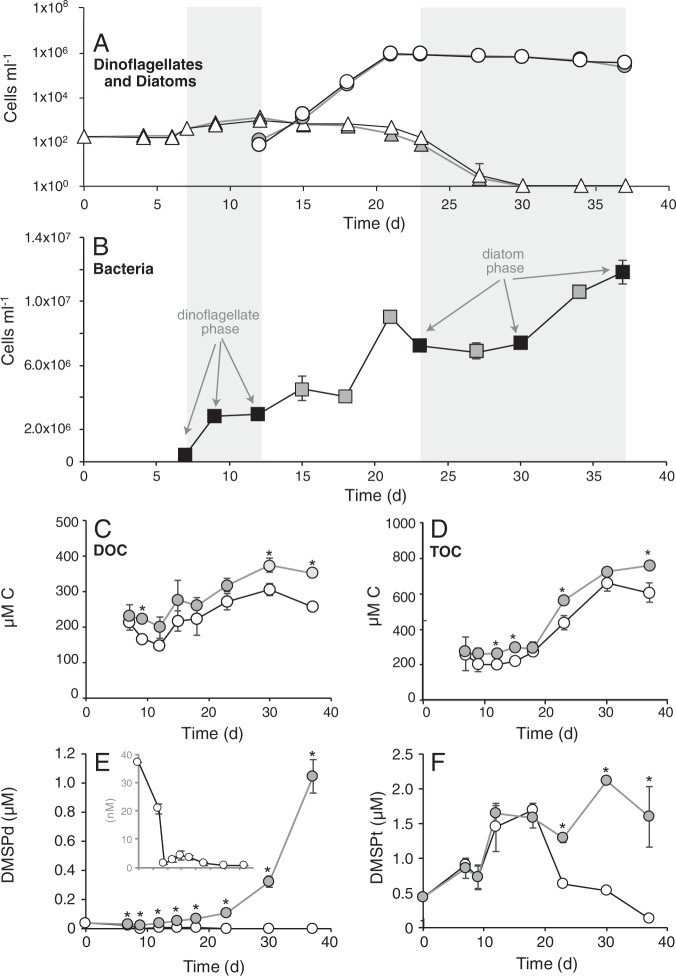
Dinoflagellate A. tamarense was the only phytoplankton species in the co-cultures until day 12, when diatom T. pseudonana was inoculated. Thalassiosira pseudonana became numerically dominant by day 18, and was the only phytoplankton species detected by day 30 (Fig. 1a). As described in Landa et al. [15], the bacterium R. pomeroyi was inoculated into three phytoplankton cultures on day 7 at a concentration of 3.7 × 105 cells ml−1 and increased in abundance by 32-fold by day 37 (Fig. 1b). Bacterial growth was sustained solely by phytoplankton exometabolites since no exogenous carbon or nitrogen sources usable by the bacterium were provided. The remaining three phytoplankton cultures were left bacteria free. DOC and TOC increased over time in both conditions, but were lower in the co-cultures with bacteria (Fig. 1c, d). Inorganic nitrogen and phosphorus were gradually drawn down during the experiment, but remained high enough to ensure that phytoplankton growth was not limited by either nitrogen or phosphate availability (Fig. S1). There was no difference in phytoplankton cell dynamics between treatments with and without bacteria (Fig. 1a).
Fig. 1.
Cell abundance and chemical data from the model system. a Dinoflagellate (triangles) and diatom (circles) cell abundances in co-cultures (white symbols) and axenic cultures (gray symbols). b Bacteria cell numbers in co-cultures, with time points of transcriptional analysis indicated by black symbols. c–f Concentration of dissolved organic carbon (DOC), total organic carbon (TOC), dissolved dimethylsulfoniopropionate (DMSP) (DMSPd), and total DMSP (DMSPt) in co-cultures (white symbols) and axenic cultures (gray symbols). The inset in e shows DMSPd concentrations in the co-culture treatment at a resolved scale. n = 3 for all measurements. Standard deviations falling within the height of the symbol are not shown. Asterisks indicate significant differences (p < 0.05)
Data from this co-culture system served as the basis to investigate the role of phytoplankton-derived DOS metabolites in supporting heterotrophic bacterial growth and activity. An early co-culture phase was defined based on dinoflagellate dominance and encompassed sampling days 7, 9, and 12; and a late phase was defined by diatom dominance and encompassed days 23, 30, and 37 (Fig. 1b). Bacterial genes with experimentally confirmed roles in DOS utilization were tested for differential expression during growth with the two phytoplankton species (Table 1).
Table 1.
Expression of sulfur metabolism genes by Ruegeria pomeroyi in the three-member model system
| Module | Locus tag | Gene name | Function | Reference | Differential expression results | |
|---|---|---|---|---|---|---|
| Fold difference | Adjusted p value | |||||
| DMSP utilization | ||||||
| Regulation | SPO1912 | dmdR | Transcriptional regulator | [52] | 6.2 | < 0.001 |
| Demethylation pathway | SPO1913 | dmdA | Dimethylsulfoniopropionate demethylase | [62] | 7.8 | < 0.001 |
| SPO2045 | dmdB-1 | 3-Methylmercaptopropionyl-CoA ligase | [63] | 0.7 | 0.025 | |
| SPO0677 | dmdB-2 | 3-Methylmercaptopropionyl-CoA ligase | [64] | −1.2 | 0.404 | |
| SPO3804 | dmdC | 3-Methylmercaptopropionyl-CoA dehydrogenase | [63] | −1.2 | 0.747 | |
| SPO3805 | dmdD | Methylthioacryloyl-CoA hydratase | [63] | −1.2 | 0.715 | |
| Methanethiol to methionine | SPOA0318 | megL | Methionine gamma-lyase | [65] | 1.3 | 0.178 |
| Methanethiol oxidation | SPOA0269 | mtoX | Methanethiol oxidase | [45] | 15.7 | < 0.001 |
| Cleavage pathway | SPO0453 | dddW | DMSP lyase | [66] | 25.7 | < 0.001 |
| SPO1703 | dddD | DMSP lyase | [67] | −1.6 | 0.091 | |
| SPO2299 | dddP | DMSP lyase | [68] | 1.7 | 0.312 | |
| SPO1596 | dddQ | DMSP lyase | [69] | 1.1 | 0.724 | |
| SPO2934 | prpE | Acrylate-CoA ligase | [63] | 2.1 | 0.225 | |
| SPO1914 | acuI | Acryloyl-CoA reductase | [47] | 15.0 | < 0.001 | |
| N-acetyltaurine, taurine and isethionate utilization | ||||||
| Regulation | SPO0659 | naaR | Transcriptional regulator | [70] | 3.0 | 0.008 |
| N-acetyltaurine transport | SPO0660 | naaA | N-acetyltaurine ABC transporter, periplasmic | [30] | 1.8 | 0.118 |
| SPO0661 | naaB | N-acetyltaurine ABC transporter, permease | [30] | 1.0 | 0.920 | |
| SPO0662 | naaB' | N-acetyltaurine ABC transporter, permease | [30] | 1.2 | 0.700 | |
| SPO0663 | naaC | N-acetyltaurine ABC transporter, ATP-binding | [30] | 1.5 | 0.371 | |
| SPO0664 | naaC' | N-acetyltaurine ABC transporter, ATP-binding | [30] | 1.9 | 0.108 | |
| N-acetyltaurine metabolism | SPO0657 | naaT | Metallochaperone, putative | [30] | −1.1 | 0.878 |
| SPO0658 | naaS | N-acetyltaurine amidohydrolase | [30] | 1.6 | 0.239 | |
| Taurine transport | SPO0674 | tauA | Taurine ABC transporter, periplasmic | [31] | 1.4 | 0.423 |
| SPO0675 | tauB | Taurine ABC transporter, ATP-binding | [31] | 1.8 | 0.172 | |
| SPO0676 | tauC | Taurine ABC transporter, permease | [31] | 1.3 | 0.554 | |
| Regulation | SPO2355 | iseR | Transcriptional regulator | [32] | 1.8 | 0.056 |
| Isethionate transport | SPO2356 | iseM | Isethionate TRAP transporter, DctM | [32] | 12.5 | < 0.001 |
| SPO2357 | iseL | Isethionate TRAP transporter, small permease | [32] | 6.2 | < 0.001 | |
| SPO2358 | iseK | Isethionate TRAP transporter, periplasmic | [32] | 18.4 | < 0.001 | |
| Isethionate metabolism | SPO2359 | iseJ | Isethionate dehydrogenase | [32] | 20.8 | < 0.001 |
| Regulation | SPO3562 | tauR | Transcriptional regulator | [31] | 5.7 | < 0.001 |
| Taurine/N-acetyltaurine metabolism | SPO0673 | tpa | Taurine-pyruvate aminotransferase | [31] | 4.6 | < 0.001 |
| Taurine/N-acetylaurine/isethionate metabolism | SPO3561 | xsc | Sulfoacetaldehyde acetyltransferase | [31] | 11.7 | < 0.001 |
| SPO3560 | pta | Phosphate acetyltransferase | [31] | 12.0 | < 0.001 | |
| DHPS utilization | ||||||
| Regulation | SPO0590 | hpsR | Transcriptional regulator | [28] | −6.3 | < 0.001 |
| DHPS transport | SPO0591 | hpsK | Dihydroxypropanesulfonate (DHPS) transporter | [28] | −67.7 | < 0.001 |
| SPO0592 | hpsL | Dihydroxypropanesulfonate (DHPS) transporter | [28] | −28.5 | < 0.001 | |
| SPO0593 | hpsM | Dihydroxypropanesulfonate (DHPS) transporter | [28] | −17.5 | < 0.001 | |
| DHPS metabolism | SPO0594 | hpsN | Dihydroxypropanesulfonate-3-dehydrogenase | [28] | −28.7 | < 0.001 |
| SPO0595 | hpsO | R- or S-DHPS-2-dehydrogenase | [28] | −28.9 | < 0.001 | |
| SPO0596 | hpsP | S- or R-DHPS-2-dehydrogenase | [28] | −40.2 | < 0.001 | |
| SPO0598 | slcD | Sulfolactate dehydrogenase | [28] | −21.4 | < 0.001 | |
| Regulation | SPOA0159 | cuyR | Transcriptional regulator | [28] | 1.2 | 0.70 |
| DHPS metabolism | SPOA0158 | cuyA | L-cysteate sulfolyase | [28] | −3.0 | 0.010 |
| Cysteate utilization | ||||||
| Cysteate metabolism | SPO2657 | cuyA-2 | l-Cysteate sulfolyase | This study | −27.8 | < 0.001 |
| Cysteate transport | SPO2658 | ABC transporter, periplasmic | This study | −8.6 | < 0.001 | |
| SPO2659 | ABC transporter, permease | This study | −6.3 | < 0.001 | ||
| SPO2660 | ABC transporter, permease | This study | −3.1 | 0.003 | ||
| SPO2661 | ABC transporter, ATP-binding | This study | −5.0 | < 0.001 | ||
| Choline-O-sulfate utilization | ||||||
| Choline-O-sulfate metabolism | SPO1083 | betC | Choline sulfatase | [35] | 2.6 | 0.015 |
| Choline metabolism | SPO1088 | betA | Choline dehydrogenase | [35] | −2.7 | 0.039 |
| SPO0084 | betB | Betaine-aldehyde dehydrogenase | [35] | −1.9 | 0.065 | |
| SPO3398 | Glycine betaine:homocysteine methyltransferase | [35] | 4.4 | < 0.001 | ||
| SPO3400 | Dimethylglycine dehydrogenase | [35] | 6.0 | < 0.001 | ||
| SPO3396 | Sarcosine dehydrogenase | [35] | 1.7 | 0.008 | ||
Positive fold differences indicate genes with higher relative expression during the dinoflagellate-dominated phase; negative fold differences represent genes with higher relative expression during the diatom-dominated phase. Numbers in bold indicate statistical significance. Experimental verification of gene function in R. pomeroyi is indicated in the Reference column.
Sulfonate metabolism
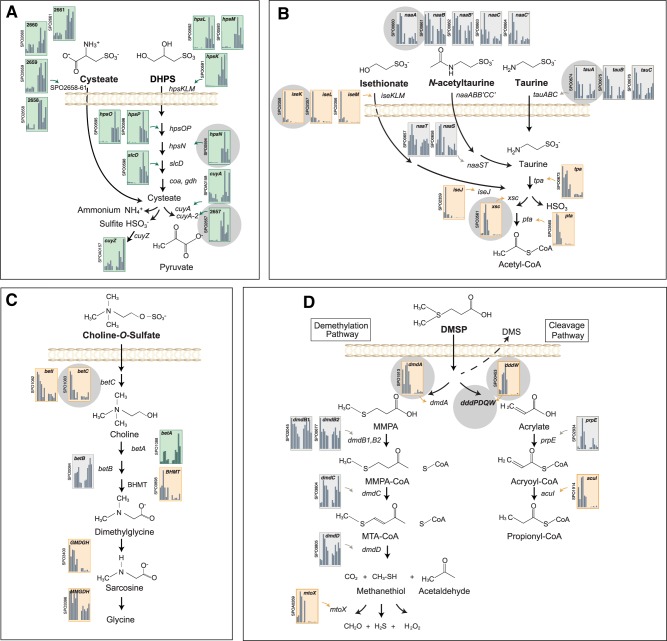
Expression patterns indicated that five sulfonates, a class of organic sulfur compounds characterized by a R-SO3− functional group, were likely serving as substrates for R. pomeroyi growth in the co-cultures. Transcripts for the transport and catabolism of the C3 sulfonate DHPS were enriched during the diatom-dominated phase of the co-culture, including those encoding uptake (hpsKLM) and catabolism to cysteate (hpsOPN, slcD) and pyruvate plus sulfite (cuyA) (Fig. 2a; Table 1). DHPS was previously identified as an abundant metabolite in T. pseudonana [25–27] and was also found in the dissolved organic matter pool associated with a marine diatom bloom [27].
Fig. 2.
Expression of organic sulfur pathways by R. pomeroyi. Histograms indicate relative expression (TPM) for genes encoding transformation of a C3 sulfonates, b C2 sulfonates, c a sulfate ester, and d a sulfonium compound. The first six bars in each represent expression during the dinoflagellate phase (sampling days 7, 9, and 12) and the last six bars represent expression during the diatom phase (sampling days 23, 30, and 37), with replicates (n = 2) shown separately. Orange backgrounds indicate genes with significant enrichment in the dinoflagellate phase and green indicate significant enrichment in the diatom phase (DESeq2, p < 0.05). Genes used in the Tara Oceans bioinformatic analysis are marked with gray circles
We noticed enrichment of transcripts from an operon containing an unannotated ABC transporter (SPO2658-2661) and an adjacent gene (SPO2657) having high identity to cuyA, the gene that encodes l-cysteate sulfolyase in the DHPS catabolism pathway [28] (Table 1). Based on the function of l-cysteate sulfolyase, we hypothesized that the C3 sulfonate cysteate was also available as a bacterial substrate during the diatom-dominated co-culture, and transported into the cell by the upregulated unannotated transporter system (Fig. 2a). Transcriptional analysis of R. pomeroyi grown on cysteate compared to acetate showed strong upregulation of the ABC transporter components and adjacent cuyA-like gene (cuyA-2; Fig. S2), supporting a role in cysteate utilization. As is the case for cuyA in the DHPS pathway, cuyA-2 is predicted to oxidize cysteate to pyruvate and sulfite, although cysteate catabolism provides a net source of ammonium to the bacterium, whereas DHPS does not (Fig. 2a). While sulfonate concentrations were not measured in this study, previous studies showed cysteate to be a component of diatom metabolomes [25, 29].
Co-culture transcription patterns indicated that R. pomeroyi growth was likely supported by three other sulfonates, in this case C2 sulfonates released during the dinoflagellate-dominated phase (Fig. 2b). Seventeen genes in the R. pomeroyi genome have been experimentally confirmed to transport and catabolize the structurally related C2 sulfonates taurine, N-acetyltaurine, and isethionate [30–32] (Fig. 2b). These are catabolized through a shared lower pathway leading from sulfoacetaldehyde to acetyl-CoA (via genes xsc and pta), and this pathway was enriched in the bacterial transcriptome. The isethionate transporter components (iseKLM) and upper pathway genes for isethionate, taurine, and N-acetyltaurine (iseJ and tpa; Fig. 2b) were also significantly enriched during the dinoflagellate phase (Table 1). These bacterial expression results are supported by previous findings of both taurine [33] and isethionate [34] in metabolomes of marine diatoms. Here, the higher expression of R. pomeroyi C2 sulfonate genes during the dinoflagellate phase suggests greater release by the dinoflagellate than by the diatom. N-acetyltaurine has not yet been identified in phytoplankton metabolomes.
Sulfate ester metabolism
We noted enrichment of transcripts from betC (choline sulfatase) and betI (choline catabolism transcriptional regulator) during growth of R. pomeroyi on dinoflagellate-derived metabolites. betC was previously experimentally verified as an essential gene in the catabolism of the sulfate ester choline-O-sulfate by R. pomeroyi [35]. Transcription analysis of the bacterium growing on choline-O-sulfate compared to acetate identified betC and betI among the most highly enriched, along with transcripts from several downstream genes that process choline to glycine betaine, sarcosine, and glycine (Fig. 2c and Fig. S2). This DOS metabolite was previously identified in the metabolome of dinoflagellate Amphidinium carteri [36, 37].
Sulfonium metabolism
Ruegeria pomeroyi harbors two degradation pathways for the sulfonium compound DMSP, each with different physiological and ecological fates. DMSP-sulfur processed through the demethylation pathway is incorporated into cellular material or oxidized for energy, while DMSP-sulfur processed through the cleavage pathway is largely lost from the cells in the form of DMS [38] (Fig. 2d). In the axenic controls, dissolved DMSP accumulated to 1050 ± 117 nM (Fig. 1e), while in the R. pomeroyi co-cultures accumulation was only 1.4 ± 0.4 nM, indicative of highly efficient bacterial uptake. Transcription of the first genes in each of the pathways (dmdA for demethylation, dddW for cleavage) was significantly higher during the dinoflagellate phase (Table 1). This is consistent with previous data showing that although T. pseudonana also synthesizes DMSP, A. tamarense is the higher producer of the two [39]. We checked whether this was the case under the specific growth conditions used here and found 772 ± 3 nM DMSP in the A. tamarense exometabolome vs. 101 ± 18 nM in the T. pseudonana exometabolome after 1 week of axenic growth. Although R. pomeroyi has other genes that can also mediate the first step in the DMSP cleavage pathway [dddD, dddP, dddQ [40]], only dddW was expressed to any appreciable levels.
The timing of peak expression of the gatekeeper genes for demethylation (dmdA) vs. cleavage (dddW) was offset in the co-culture by 3–5 days (Fig. S3). Variations in the ratio of DMSP routed through the two pathways have been detected in natural marine bacterial communities as well [8, 41], although the controls over this differential routing are still not understood [42]. Because DMSP demethylation produces hydrogen peroxide [43–45] and cleavage produces acrylate/acryloyl-CoA [46, 47], one hypothesis views differential regulation as a mechanism for bacteria to balance the different physiological stresses each pathway imposes [48]. In the co-culture experiment, expression of the R. pomeroyi catalase gene (katG) was significantly correlated with expression of demethylation pathway genes (dmdA and mtoX; r = 0.73 and 0.89, p ≤ 0.01), but not with the cleavage pathway gene dddW (r = 0.27; Fig. S3), consistent with hydrogen peroxide stress linked to demethylation. Acryloyl-CoA reductase (acuI) from the cleavage pathway is co-located and co-regulated with dmdA from the demethylation pathway (Fig. S3), an unusual arrangement that, upon upregulation of the operon, would protect the bacterium both by metabolizing acryloyl-CoA generated in the cleavage pathway and routing DMSP into the demethylation pathway. We checked whether acryloyl-CoA accumulation switches gene expression to the demethylation pathway by adding acrylate, the precursor of acryloyl-CoA, to R. pomeroyi cultures growing on DMSP. As hypothesized from the genome organization, the relative DMSP flux through demethylation pathway increased significantly upon acrylate addition (Fig. S4).
Sulfur fate in Ruegeria pomeroyi
The S moiety in sulfonates is more oxidized than in DMSP and therefore less valuable as a reduced sulfur source for bacterial biosynthesis. For the inorganic sulfur derived from the three dinoflagellate-derived sulfonates (taurine, N-acetyltaurine, and isethionate), sulfur has been shown to be exported from R. pomeroyi via the soe system in the form of sulfate [49, 50]. Consistent with this, there was higher average expression of soeABC during the dinoflagellate phase, and expression of soe genes had significant positive correlations with tauR, xsc, and pta (Fig. S5). Inorganic sulfur from the two C3 diatom-derived sulfonates (DHPS and cysteate) is exported by R. pomeroyi instead in the form of sulfite via the cuyZ gene [28, 51]. cuyZ had peak expression in the diatom phase (Fig. S5). Finally, inorganic sulfur from DMSP metabolism is exported by R. pomeroyi in the form of sulfate using a third system, soxABCDXYZ [52]. Expression of sox genes peaked during the dinoflagellate phase and was significantly correlated with expression of DMSP demethylation gene dmdA (Fig. S5).
DOS transformation by ocean bacteria
We used knowledge of the sulfur metabolites important in this model system to characterize the capacity for DOS utilization by ocean bacteria. Thirteen bacterial genes indicative of catabolism of phytoplankton-derived organic sulfur molecules were analyzed in the Tara Oceans metagenomic database [16]. DNA sequences representing the bacterial/archaeal size class of marine plankton (0.22–1.6 or 0.22–3 µm filter pore-size ranges; Table S3) from surface (100 samples from 5 m depth), DCM (72 samples from 17 to 188 m depths), and mesopelagic (53 samples from 250–1000 m depths) collections (Fig. S6) were assembled into contigs by individual sample (see Methods), which allowed us to link reads back to the station and depth where they were collected. HMM searches were conducted for genes characteristic of the model system sulfur metabolites, using diagnostic catabolic genes or, in the case of the C2 sulfonates that share catabolic genes, using transporter binding proteins (Fig. 2). The percent of genomes harboring the genes was estimated based on length-normalized ratios to recA, a housekeeping gene present in single copy in all bacterial and archaeal genomes. The sum of recA sequences across all samples (Table S3) indicated that 1.4 million bacterial/archaeal genome equivalents were surveyed in these Tara Oceans collections, of which ~650,000 were from the surface ocean, ~500,000 from the DCM, and ~230,000 from the mesopelagic.
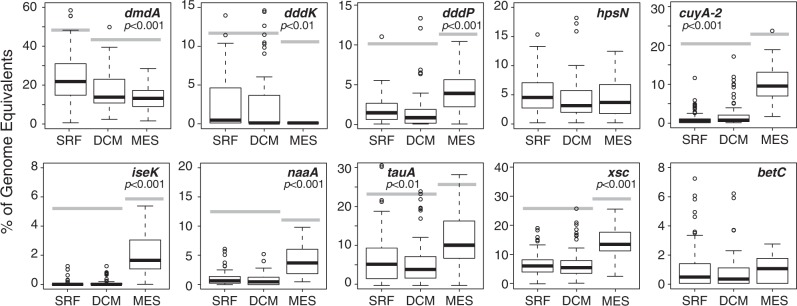
The frequency of genes for DOS utilization averaged across all locations and depths is highest for DMSP, and this is the only DOS metabolism capability identified in all 225 Tara samples. Twenty percent of bacterial genomes harbored dmdA, the diagnostic gene for DMSP demethylation, while 3.8%, 2.5%, and 2.4% of genomes had dddK, dddP, and dddD, the most abundant DMSP cleavage genes (Table S3). Although dddW was the most highly transcribed of the R. pomeroyi DMSP cleavage genes in the co-culture, it is extremely rare in the Tara dataset—present in only 92 of the 1.4 million genome equivalents we surveyed (0.01% of genomes; Table S3). Cleavage genes dddL and dddQ were found in 0.06% and 2.3% of genomes; whether DMSP cleavage is the native function of dddQ has been questioned [53]. A recent analysis of DMSP cleavage gene expression in the Tara Oceans metatranscriptome data shows they are actively expressed, with dddP and dddK having the highest transcription levels [54].
Genes for utilization of sulfoacetaldehyde (xsc), taurine (tauA), and DHPS (hpsN) are the next most frequent in Tara bacterioplankton (estimated to be in 8.4%, 8.2%, 4.8% of genomes). Genes for transforming choline-O-sulfate (betC) and isethionate (iseK) are not frequent (0.9% and 0.5% of genomes) (Table S3). Notably, bacterial DOS genes are not distributed evenly by depth. DMSP demethylation gene dmdA and cleavage gene dddK had distributions significantly biased toward surface ocean bacterial genomes. The DMSP cleavage gene dddP and genes for taurine (tauA), N-acetyltaurine (naaA), cysteate (cuyA-2), isethionate (iseK), and sulfoacetaldehyde (xsc) utilization had distributions significantly biased toward mesopelagic bacterial genomes (Fig. 3).
Fig. 3.
Dissolved organic sulfur (DOS) metabolite genes in the ocean. Relative abundance of 10 DOS metabolite genes in marine bacterial/archaeal genomes was estimated by normalizing to recA counts for 100 surface, 72 deep chlorophyll maximum (DCM), and 58 mesopelagic samples from the Tara Oceans database. For genes with significant differences in relative abundance by depth zone (analysis of variance (ANOVA)), gray bars connect zones that are not statistically different (Tukey’s honestly significant difference (HSD) multiple comparison)
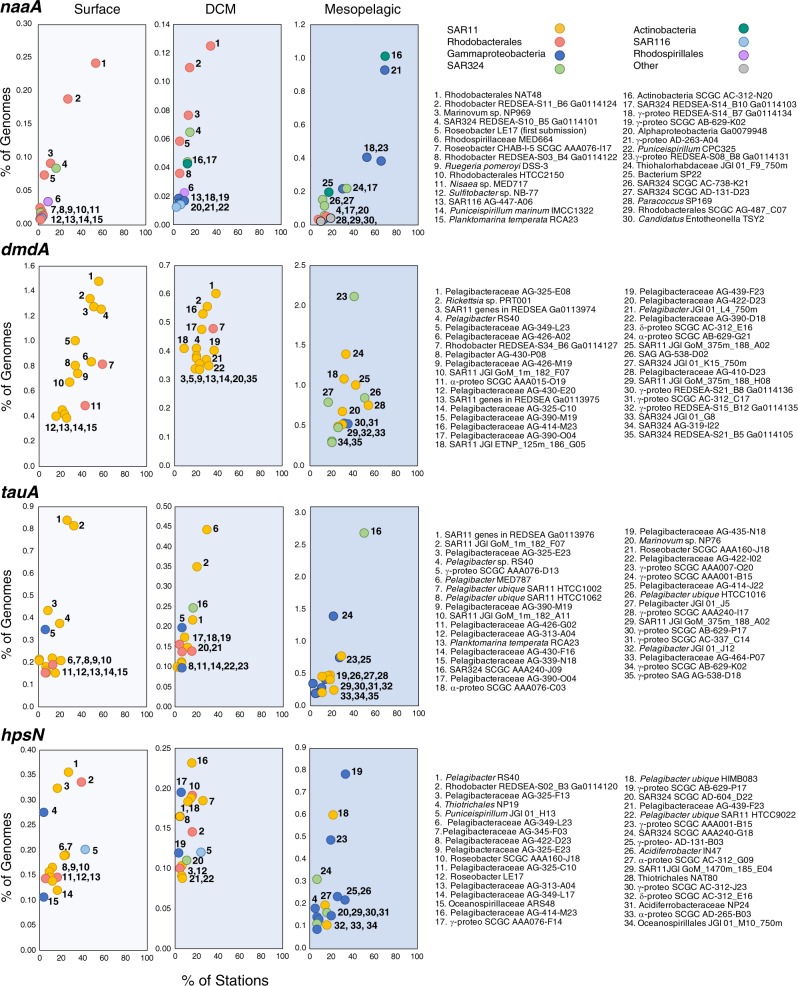
To learn which bacterial taxa harbor DOS genes, we assigned taxonomy to the Tara genes based on blast analysis against the Integrated Microbial Genomes non-redundant database. For all DOS genes except dddK, taxonomic assignments include bacteria belonging to the Rhodobacterales, an expected consequence of Rhodobacterales member R. pomeroyi being the sensor bacterium that generated the DOS metabolite list. Because of the numerical importance of SAR11 cells in the ocean, they dominate the taxonomic assignments for any DOS genes found in the group. Thus, SAR11 genomes account for >80% of bacterial genes for DMSP utilization (dmdA and dddK), >55% of genes for DHPS utilization (hpsN), and >60% of genes for taurine utilization (tauA). Other marine Alphaproteobacteria from the SAR116, Rhodospirillales, and Rhizobacterales clades also harbor DOS genes, but to a lesser extent than Rhodobacterales and SAR11 (Fig. 4).
Fig. 4.
Taxonomic assignments of the 15 most abundant bacterial taxa at each depth zone harboring DOS metabolite genes naaA, dmdA, tauA, and hpsN. Taxonomy was obtained for Tara Oceans genes based on the best hit against the May 2018 version of the Integrated Microbial Genomes non-redundant database
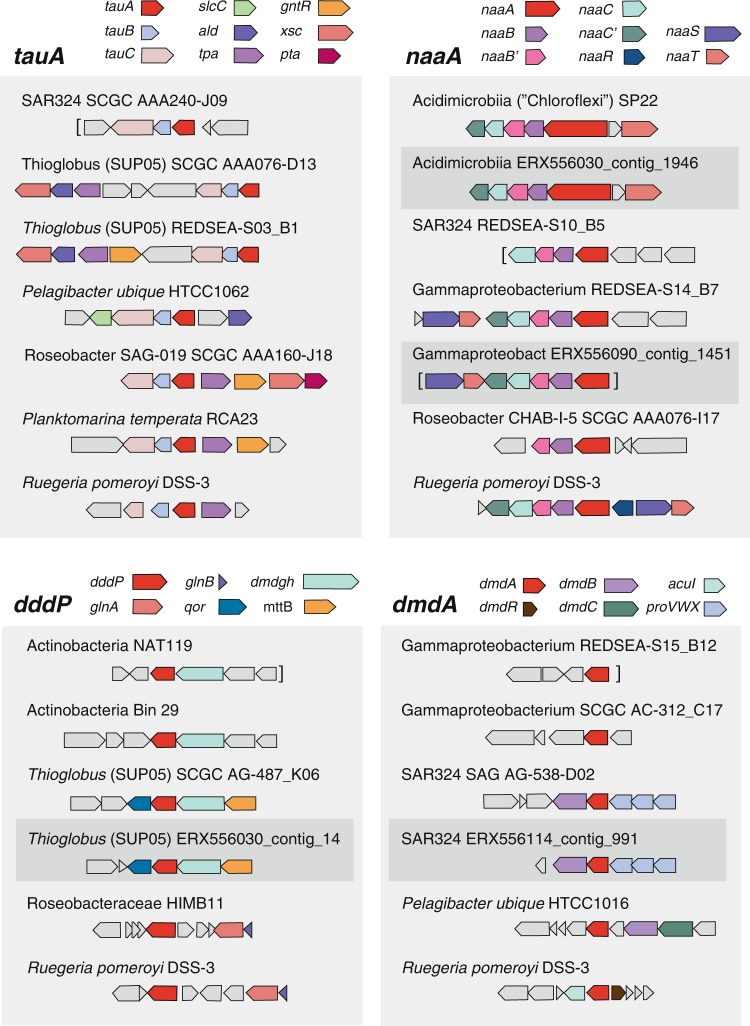
In the mesopelagic, novel groups not previously recognized for organic sulfur metabolism were prominent (Fig. 4). One such group is SAR324, whose genomes contain orthologs for utilization of DMSP (dmdA and dddP), DHPS (hpsN), taurine (tauA), N-acetyltaurine (naaA), isethionate (iseK), and choline-O-sulfate (betC). For example, SAR324 genomes account for 41% of dmdA, 92% of iseK, and 29% of tauA in bacteria sampled from mesopelagic depths. A second novel taxon potentially participating in DOS metabolite catabolism at depth is the Acidimicrobiia group of the Actinobacteria, whose genomes contain orthologs for utilization of N-acetyltaurine (30% of mesopelagic naaA), taurine (1% of mesopelagic tauA), and DMSP cleavage (46% of mesopelagic dddP). The Acidimicrobiia tauA and naaA genes in the Tara data are uncharacteristically long (~680 amino acids compared to ~300 in other bacteria), which was due to two substrate binding domains encoded in each. Two substrate binding domains were also found in tauA and naaA in some marine Acidimicrobiia reference genomes (Fig. 5), an arrangement that could improve scavenging for scarce substrates. A third novel group at DCM and mesopelagic depths is Thioglobus/SUP05, whose genomes contain orthologs for DMSP cleavage (21% of DCM dddP, 7% of mesopelagic dddP) and taurine utilization (11% of DCM tauA, 33% of mesopelagic tauA). Lastly, DOS genes that mapped to a novel Gammaproteobacteria group with 16S rRNA sequences most similar to Thiohalorhabdaceae bacterium JGI 01_F9_750m (95% identity) included those for DMSP demethylation (13% of mesopelagic dmdA) and N-acetyltaurine utilization (51% of mesopelagic naaA) (Fig. 4).
Fig. 5.
Gene neighborhoods of dissolved organic sulfur (DOS) metabolite genes in Tara Oceans contigs (dark gray shading, ERX556xxx labels), the closest reference sequences to these genes, and model bacterium R. pomeroyi. tauA, taurine transport; naaA, N-acetyltaurine transport; dddP, DMSP cleavage; dmdA, DMSP demethylation. Genes colored gray are unrelated to organic sulfur metabolism. Brackets indicate contig ends
Two catabolism genes used in the Tara HMM searches, cuyA-2 and xsc, are not specific for a single DOS metabolite. cuyA-2 encodes a second copy of l-cysteate sulfolyase for cysteate metabolism in R. pomeroyi (Fig. 2a), but might not be distinguishable from the canonical cuyA from the DHPS pathway in homology-based searches. xsc encodes a sulfoacetaldehyde acetyltransferase that in R. pomeroyi mediates a step in the shared lower pathway through which C2 sulfonates taurine, N-acetyltaurine, and isethionate are degraded (Fig. 2b), but this gene is also used in an alternate pathway for DHPS catabolism by some marine bacteria [28]. Taxonomic assignments of cuyA-2 and xsc are dominated by SAR11, Rhodobacter, SAR324, and Gammaproteobacteria, and their frequency and distribution are consistent with other sulfonate utilization genes (Table S3).
In cases where HMMs identified orthologs in taxa not previously recognized to transform DOS, we carried out synteny-based checks on the annotations by examining gene neighborhoods in the Tara assemblies and in their closest reference genomes. The co-location of genes with annotations consistent with DOS metabolism corroborated the HMM annotations (Fig. 5). The HMMs did not recover any orthologs originating from archaeal genomes, although we note that there were no characterized marine archaeal DOS genes to include in the HMM reference sequences.
Insights into DOS processing and fate
Bacteria capable of DMSP utilization are the most frequent DOS degraders in the Tara Oceans database, an expected result given both that ~10% of marine net primary production is directed to DMSP synthesis [8, 55] and phytoplankton intracellular concentrations can reach 400 mM [39, 56]. Marine bacteria serving as model organisms for DMSP gene discovery, including R. pomeroyi, can harbor up to four different DMSP cleavage genes in addition to dmdA [40, 42]. Yet, based on the frequency of cleavage genes in the Tara dataset compared to demethylation gene dmdA, it appears that most oceanic DMSP-degrading bacteria are capable only of demethylation. Among DMSP-degrading SAR11 cells, for example, frequencies of dmdA and dddK indicate that no more than 25% can harbor both pathways. Although community gene frequency is not a measure of activity level, the lower frequency of cleavage genes accords with estimates that <2% of the DMSP synthesized by marine phytoplankton is ultimately released from the ocean surface as DMS [42]. Nonetheless, this small DMS flux relative to DMSP production accounts for over 40% of Earth’s atmospheric sulfur burden [41], and even minor changes in bacterial routing between the two pathways could impact atmospheric sulfur concentrations.
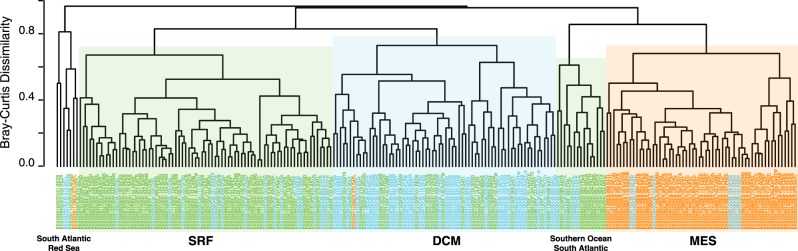
The fact that DMSP genes dmdA and dddK have frequencies biased toward surface ocean genomes suggests a close coupling between phytoplankton synthesis and bacterial transformation for this compound. Yet, in the case of sulfonate genes, with frequencies mostly biased toward mesopelagic genomes, the proximate source of labile DOS is less clear. Sulfonate-sulfur is more oxidized than DMSP-sulfur and may be less preferred as a substrate in surface waters. Sinking phytoplankton biomass could also provide a source of sulfonates at depth, as supported by evidence of DHPS production during bacterial degradation of sulfolipids from photosynthetic membranes [57]. DOS metabolites could also be available from viral lysates and zooplankton detritus. For example, taurolipids have been identified in protist membranes [58], and taurine release by crustacean zooplankton has been observed in surface and upper mesopelagic waters [59]. When the Tara Ocean samples are clustered based on gene patterns, collection depth is the main correlate (Fig. 6), with depth-related variation in both sources of DOS and the bacterial taxa that process DOS likely to be driving this pattern. Only minor groupings were observed based on oceanic region, although a separate cluster of 15 surface and DCM samples with high DOS gene abundance emerged, driven primarily by high frequencies of genomes with dddK, hpsN, and tauA (Fig. 6).
Fig. 6.
Dendrogram of a Bray–Curtis dissimilarity matrix constructed from the relative abundance (as % of genome equivalents) of 10 dissolved organic sulfur (DOS) metabolite genes in 225 Tara Oceans bacterial/archaeal size fraction datasets (Table S3). Sample names are colored based on collection depth (green = surface; blue = DCM; orange = mesopelagic) and indicate sample number (“ERX55xxxx”), followed by depth zone (XXX), station number (123), and the Tara Oceans biome code (XXXX). For two smaller clusters containing samples from mixed depths, the main ocean regions of origin are indicated
Our important findings are, first, that a diverse suite of DOS metabolites have the potential to support bacterial heterotrophy in the ocean. Membership in this suite may well expand further, particularly when viewed through a more comprehensive lens than just a single model bacterium’s gene expression. The recent finding that the structurally unusual DOS compound dimethylsulfoxonium propionate is an exometabolite of marine phytoplankton and readily taken up by marine bacteria, including R. pomeroyi [60], supports this prediction. The biosynthetic apparatus and physiological roles of these DOS metabolites in marine phytoplankton remain largely unknown. Second, in addition to the bacterial taxa already recognized to play roles in organic sulfur transformation (Rhodobacter, SAR11, and SAR116), we discovered that other taxonomic groups can process DOS metabolites and have gene frequencies biased toward the mesopelagic. These include SAR324, Acidimicrobiia, Thioglobus/SUP05, and Thiohalorhabdaceae-like cells with varying capabilities for utilization of DMSP, taurine, N-acetyltaurine, isethionate, and choline-O-sulfate. Non-labile marine DOS (defined as the semi-polar molecules captured on solid-phase extraction resin) in the ocean’s dissolved organic matter reservoir has been found to degrade at a higher long-term rate than bulk DOC, indicating selective removal of organic sulfur over time and depth [61]. Here, an analysis window targeting the labile components of marine DOS similarly implicates organic sulfur turnover as a key process in the global carbon cycle.
Supplementary information
Acknowledgements
We appreciate assistance from C Smith and S Roth, and thank N Ivanova for help with IMG database access. Computing resources and technical expertise were provided by the Georgia Genomics and Bioinformatics Core. This work was funded by NSF grants OCE-1342694, OCE-1342699, IOS-1656311 and The Gordon and Betty Moore Foundation grant #5503. This research used resources of the Joint Genome Institute (JGI) and the National Energy Research Scientific Computing Center (NERSC), US Department of Energy Office of Science User Facilities operated under Contract No. DE-AC02-05CH11231.
Author contributions
ML and MAM conceived of the study and wrote the paper with input from all authors. ML, ASB, BPD, KE, RPK, BN, and AV performed experiments or conducted analyses; TN and SS assembled and analyzed sequence data; ML, RPK, and MAM analyzed data and interpreted findings.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0455-3) contains supplementary material, which is available to authorized users.
References
- 1.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 2.Morán XAG, Ducklow HW, Erickson M. Carbon fluxes through estuarine bacteria reflect coupling with phytoplankton. Mar Ecol Prog Ser. 2013;489:75–85. doi: 10.3354/meps10428. [DOI] [Google Scholar]
- 3.Hansell DA. Recalcitrant dissolved organic carbon fractions. Ann Rev Marine Sci. 2013;5:421–45. doi: 10.1146/annurev-marine-120710-100757. [DOI] [PubMed] [Google Scholar]
- 4.Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–99. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JW, Wilhelm LJ, Giovannoni SJ. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–4. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 6.Sunda W, Kieber D, Kiene R, Huntsman S. An antioxidant function for DMSP and DMS in marine algae. Nature. 2002;418:317–20. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- 7.Strom S, Wolfe G, Slajer A, Lambert S, Clough J. Chemical defense in the microplankton II: inhibition of protist feeding by beta-dimethylsulfoniopropionate (DMSP) Limnol Oceanogr. 2003;48:230–7. doi: 10.4319/lo.2003.48.1.0230. [DOI] [Google Scholar]
- 8.Kiene RP, Linn LJ, Bruton JA. New and important roles for DMSP in marine microbial communities. J Sea Res. 2000;43:209–24. doi: 10.1016/S1385-1101(00)00023-X. [DOI] [Google Scholar]
- 9.Kaiser K, Benner R. Organic matter transformations in the upper mesopelagic zone of the North Pacific: chemical composition and linkages to microbial community structure. J Geophys Res Oceans. 2012;C10123:117.
- 10.Mopper K, Schultz CA, Chevolot L, Germain C, Revuelta R, Dawson R. Determination of sugars in unconcentrated seawater and other natural waters by liquid chromatography and pulsed amperometric detection. Environ Sci Technol. 1992;26:133–8. doi: 10.1021/es00025a014. [DOI] [Google Scholar]
- 11.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 12.McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poretsky RS, Sun S, Mou X, Moran MA. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12:616–27. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satinsky BM, Crump BC, Smith CB, Sharma S, Zielinski BL, Doherty M, et al. Microspatial gene expression patterns in the Amazon River Plume. Proc Natl Acad Sci USA. 2014;111:11085–90. doi: 10.1073/pnas.1402782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landa M, Burns AS, Roth SJ, Moran MA. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 2017;11:2677–90. doi: 10.1038/ismej.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 17.Kiene RP, Service SK. Decomposition of dissolved DMSP and DMS in estuarine waters: dependence on temperature and substrate concentration. Mar Ecol Prog Ser. 1991;76:1–11. doi: 10.3354/meps076001. [DOI] [Google Scholar]
- 18.Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 22.Love M, Anders S, Huber W. Differential analysis of count data—the DESeq2 package. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. vegan: Community Ecology Package. 2017. https://CRAN.R-project.org/package=vegan.
- 24.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using Diamond. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 25.Busby WF. Sulfopropanedial and cysteinolic acid in the diatom. Biochim Biophys Acta. 1966;121:160–1. doi: 10.1016/0304-4165(66)90360-6. [DOI] [PubMed] [Google Scholar]
- 26.Busby WF, Benson AA. Sulfonic acid metabolism in the diatom Navicula pelliculosa. Plant Cell Physiol. 1973;14:1123–32. [Google Scholar]
- 27.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer J, Huhn T, Habeck M, Denger K, Hollemeyer K, Cook AM. 2, 3-Dihydroxypropane-1-sulfonate degraded by Cupriavidus pinatubonensis JMP134: purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology. 2010;156:1556–64. doi: 10.1099/mic.0.037580-0. [DOI] [PubMed] [Google Scholar]
- 29.Shibuya I, Yagi T, Benson AA. In Japanese Society of Plant Physiologists, editor. Studies on microalgae and photosynthetic bacteria. Tokyo: University of Tokyo Press; 1963. p. 627–36.
- 30.Denger K, Lehmann S, Cook AM. Molecular genetics and biochemistry of N-acetyltaurine degradation by Cupriavidus necator H16. Microbiology. 2011;157:2983–91. doi: 10.1099/mic.0.048462-0. [DOI] [PubMed] [Google Scholar]
- 31.Gorzynska AK, Denger K, Cook AM, Smits TH. Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3T. Arch Microbiol. 2006;185:402. doi: 10.1007/s00203-006-0106-8. [DOI] [PubMed] [Google Scholar]
- 32.Weinitschke S, Sharma PI, Stingl U, Cook AM, Smits TH. Gene clusters involved in isethionate degradation by terrestrial and marine bacteria. Appl Environ Microbiol. 2010;76:618–21. doi: 10.1128/AEM.01818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson AE, Ayer SW, Laycock MV. The effect of salinity on growth and amino acid composition in the marine diatom Nitzschia pungens. Can J Bot. 1992;70:2198–201. doi: 10.1139/b92-272. [DOI] [Google Scholar]
- 34.Boroujerdi AF, Lee PA, DiTullio GR, Janech MG, Vied SB, Bearden DW. Identification of isethionic acid and other small molecule metabolites of Fragilariopsis cylindrus with nuclear magnetic resonance. Anal Bioanal Chem. 2012;404:777–84. doi: 10.1007/s00216-012-6169-2. [DOI] [PubMed] [Google Scholar]
- 35.Lidbury I, Kimberley G, Scanlan DJ, Murrell JC, Chen Y. Comparative genomics and mutagenesis analyses of choline metabolism in the marine Roseobacter clade. Environ Microbiol. 2015;17:5048–62. doi: 10.1111/1462-2920.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikawa M, Taylor RF. In Martin D, Padilla G, editors. Marine pharmacognosy. Choline and related substances in algae. New York: Academic Press; 1973. p. 203–40.
- 37.Taylor RF, Ikawa M, Sasner JJ, Jr, Thurberg FP, Andersen KK. Occurrence of choline esters in the marine dionflagellate Amphidinium carteri. J Phycol. 1974;10:279–83. [Google Scholar]
- 38.Kiene RP, Linn LJ, González J, Moran MA, Bruton JA. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl Environ Microbiol. 1999;65:4549–58. doi: 10.1128/aem.65.10.4549-4558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller MD, Bellows WK, Guillard RR. In Saltzman ES, Cooper WJ, editors. Biogenic sulfur in the environment. Dimethyl sulfide production in marine phytoplankton. Washington D.C.: ACS Publications; 1989. p. 167–82.
- 40.Curson AR, Todd JD, Sullivan MJ, Johnston AW. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol. 2011;9:849. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- 41.Simó R. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol Evol. 2001;16:287–94. doi: 10.1016/S0169-5347(01)02152-8. [DOI] [PubMed] [Google Scholar]
- 42.Moran MA, Reisch CR, Kiene RP, Whitman WB. Genomic insights into bacterial DMSP transformations. Annu Rev Mar Sci. 2012;4:523–42. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- 43.Eyice Ö, Schäfer H. Culture-dependent and culture-independent methods reveal diverse methylotrophic communities in terrestrial environments. Arch Microbiol. 2016;198:17–26. doi: 10.1007/s00203-015-1160-x. [DOI] [PubMed] [Google Scholar]
- 44.Suylen G, Large P, Van Dijken J, Kuenen J. Methyl mercaptan oxidase, a key enzyme in the metabolism of methylated sulphur compounds by Hyphomicrobium EG. Microbiology. 1987;133:2989–97. doi: 10.1099/00221287-133-11-2989. [DOI] [Google Scholar]
- 45.Eyice Ö, Myronova N, Pol A, Carrión O, Todd JD, Smith TJ, et al. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 2017;12:145. doi: 10.1038/ismej.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González JM, Kiene RP, Moran MA. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class proteobacteria. Appl Environ Microbiol. 1999;65:3810–9. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todd JD, Curson AR, Sullivan MJ, Kirkwood M, Johnston AW. The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of E. coli and other bacteria in conferring resistance to acrylate. PLoS ONE. 2012;7:e35947. doi: 10.1371/journal.pone.0035947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varaljay VA, Robidart J, Preston CM, Gifford SM, Durham BP, Burns AS, et al. Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J. 2015;9:1677–86. doi: 10.1038/ismej.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann S. Sulfite dehydrogenases in organotrophic bacteria: enzymes, genes and regulation. Konstanz, Germany: University of Konstanz; 2013.
- 50.Lenk S, Moraru C, Hahnke S, Arnds J, Richter M, Kube M, et al. Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J. 2012;6:2178–87. doi: 10.1038/ismej.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denger K, Smits TH, Cook AM. l-Cysteate sulpho-lyase, a widespread pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3T. Biochem J. 2006;394:657–64. doi: 10.1042/BJ20051311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reisch CR, Crabb WM, Gifford SM, Teng Q, Stoudemayer MJ, Moran MA, et al. Metabolism of dimethylsulphoniopropionate by Ruegeria pomeroyi DSS‐3. Mol Microbiol. 2013;89:774–91. doi: 10.1111/mmi.12314. [DOI] [PubMed] [Google Scholar]
- 53.Lei L, Cherukuri KP, Alcolombri U, Meltzer D, Tawfik DS. The dimethylsulfoniopropionate (DMSP) lyase and lyase-like cupin family consists of bona fide DMSP lyases as well as other enzymes with unknown function. Biochemistry. 2018;57:3364–77. doi: 10.1021/acs.biochem.8b00097. [DOI] [PubMed] [Google Scholar]
- 54.Curson AR, Williams BT, Pinchbeck BJ, Sims LP, Martínez AB, Rivera PPL, et al. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat Microbiol. 2018;3:430. doi: 10.1038/s41564-018-0119-5. [DOI] [PubMed] [Google Scholar]
- 55.Galí M, Devred E, Levasseur M, Royer S-J, Babin M. A remote sensing algorithm for planktonic dimethylsulfoniopropionate (DMSP) and an analysis of global patterns. Remote Sens Environ. 2015;171:171–84. doi: 10.1016/j.rse.2015.10.012. [DOI] [Google Scholar]
- 56.Stefels J. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J Sea Res. 2000;43:183–97. doi: 10.1016/S1385-1101(00)00030-7. [DOI] [Google Scholar]
- 57.Denger K, Weiss M, Felux A-K, Schneider A, Mayer C, Spiteller D, et al. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature. 2014;507:114–7. doi: 10.1038/nature12947. [DOI] [PubMed] [Google Scholar]
- 58.Saidha T, Stern AI, Schiff JA. Taurine conjugates in the lipid fraction of Euglena cells and their mitochondria. Microbiology. 1993;139:251–7. [Google Scholar]
- 59.Clifford EL, Hansell DA, Varela MM, Nieto‐Cid M, Herndl GJ, Sintes E. Crustacean zooplankton release copious amounts of dissolved organic matter as taurine in the ocean. Limnol Oceanogr. 2017;62:2745–58. doi: 10.1002/lno.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thume Kathleen, Gebser Björn, Chen Liang, Meyer Nils, Kieber David J., Pohnert Georg. The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature. 2018;563(7731):412–415. doi: 10.1038/s41586-018-0675-0. [DOI] [PubMed] [Google Scholar]
- 61.Ksionzek KB, Lechtenfeld OJ, McCallister SL, Schmitt-Kopplin P, Geuer JK, Geibert W, et al. Dissolved organic sulfur in the ocean: Biogeochemistry of a petagram inventory. Science. 2016;354:456–9. doi: 10.1126/science.aaf7796. [DOI] [PubMed] [Google Scholar]
- 62.Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–52. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 63.Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature. 2011;473:208–11. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- 64.Bullock HA, Reisch CR, Burns AS, Moran MA, Whitman WB. Regulatory and functional diversity of methylmercaptopropionate coenzyme A ligases from the dimethylsulfoniopropionate demethylation pathway in Ruegeria pomeroyi DSS-3 and other proteobacteria. J Bacteriol. 2014;196:1275–85. doi: 10.1128/JB.00026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wirth JS. Phylogenomics and the metabolism of sulfur compounds in the Roseobacter group. Athens, GA, USA: University of Georgia; 2019.
- 66.Todd JD, Kirkwood M, Newton-Payne S, Johnston AW. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 2012;6:223–6. doi: 10.1038/ismej.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science. 2007;315:666–9. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- 68.Todd J, Curson A, Dupont C, Nicholson P, Johnston A. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ Microbiol. 2009;11:1376–85. doi: 10.1111/j.1462-2920.2009.01864.x. [DOI] [PubMed] [Google Scholar]
- 69.Todd JD, Curson AR, Kirkwood M, Sullivan MJ, Green RT, Johnston AW. DddQ, a novel, cupin‐containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ Microbiol. 2011;13:427–38. doi: 10.1111/j.1462-2920.2010.02348.x. [DOI] [PubMed] [Google Scholar]
- 70.Brüggemann C, Denger K, Cook AM, Ruff J. Enzymes and genes of taurine and isethionate dissimilation in Paracoccus denitrificans. Microbiology. 2004;150:805–16. doi: 10.1099/mic.0.26795-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.