Abstract
The term ‘pathogen emergence’ encompasses everything from novel viruses entering the human population to established pathogens invading new populations, to the evolution of drug resistance. Mathematical models of emergent pathogens allow forecasts of case numbers, investigation of transmission mechanisms, and evaluation of control options. Yet, there are numerous limitations and pitfalls to their use, often driven by data scarcity. Growing availability of data on pathogen genetics and human ecology, coupled with computational and methodological innovations, are amplifying the power of models to inform the public health response to emergence events. Tighter integration of infectious disease models with public health practice, and development of resources at the ready have the potential to increase the timeliness and quality of responses.
Introduction
Public health emergencies driven by emerging infectious diseases are at the forefront of global awareness. From HIV in the 1980s, to Zika virus’ recent invasion of the Americas, models that mathematically capture disease processes have played a role in assessing the risk and framing the response to emerging pathogens. The most prominent, and perhaps most fraught, role of such models is to forecast the course of epidemics (1, 2). Yet, explicit representation of mechanisms of spread and persistence can help us to do far more than forecast incidence. Models can elucidate the properties of emergent pathogens (3, 4), uncover general principles of emergence (5), and compare potential mechanisms of spread and persistence (6).
Models are only as good as the data on which they rely. Data scarcity is the norm when a novel pathogen emerges, amplifying uncertainty and obscuring key drivers of the epidemic. Misrepresentation of core mechanisms can bias inferences and potentially misdirect intervention efforts. The strengths of models must be considered in the context of the limitations and pitfalls of their use.
Here, we focus on emergent viruses, both because of the speed with which they can spread death and disease, and because the dynamics of viral epidemics exemplify key principles in the modeling of infectious diseases. This focus should not, however, detract from the importance of non-viral emergence events, nor from the unique issues involved in modeling non-viral pathogens.
The ‘classical’ dynamic modeling toolkit
The past decade has seen several viral emergence events, including the 2009 pandemic of H1N1 influenza, the emergence of Middle East Respiratory Syndrome associated coronavirus (MERS-CoV) in the Arabian peninsula, the West African Ebola outbreak, and Zika virus’ (ZIKV) invasion of the Americas. These diseases are very different: pandemic H1N1 is spread person-to-person and is closely related to circulating influenza viruses (3); since its emergence MERS-CoV has failed to persist outside of the Middle East and the epidemic appears to be largely driven by zoonotic infections from camels (though a rapidly contained human driven outbreak occurred in South Korea) (6); Ebola is extremely virulent and spread mostly through direct contact with very sick or dead cases (7); and ZIKV is a mosquito transmitted virus, known for decades but recently discovered to be a cause of severe pathogenic disease after emerging in the Americas (8).
Despite these differences, the response to each emerging virus has relied on models grounded in the same dynamic principles and key data that have informed the response to emerging infections since at least the 1980s (Figure 1). Then, Anderson & May used mathematical models to elucidate the key variables required for forecasting the future trajectory and impact of the emerging HIV epidemic (11) (Box 1). Although there have been significant advances in our ability to assess disease threats, ranging from increased statistical rigour enabled by powerful computers, to entirely new methods of inference driven by analyses of pathogen genetics (3, 4), the underlying core principles remain the same.
Figure 1: Classic Zoonotic Emergence to Human-to-Human Transmission.
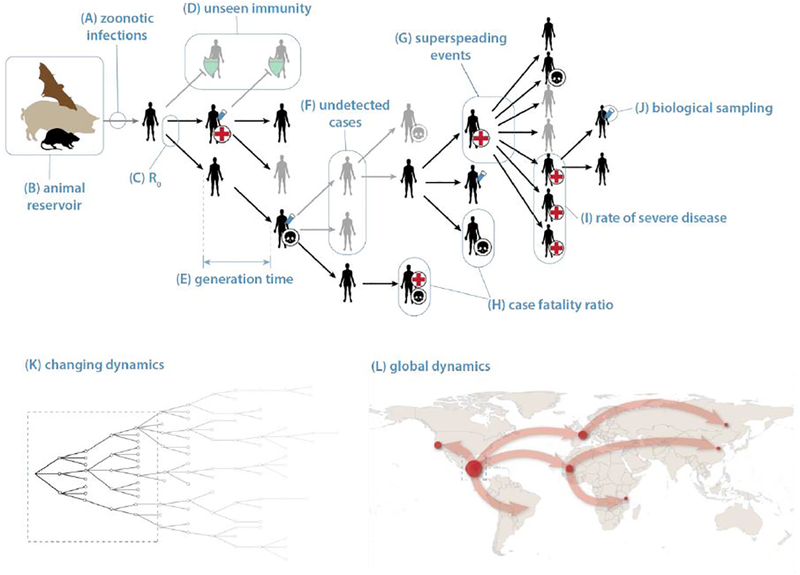
Typically, emergence occurs after a pathogen circulating in an animal reservoir enters the human population through a zoonotic infection event (A-B). The key to understanding whether the pathogen will pose a sustained threat to humans is the average number of cases caused by early infections, i.e., the basic reproductive number R0 (C). Generally, models assume every human is susceptible, but there may be significant unseen immunity (D, green shields) if the pathogen is closely related to a circulating disease (e.g., pandemic H1N1 (3)) or in populations with frequent exposure to the animal reservoir (e.g., antibodies to MERS-CoV in shepherds (9)). The time between subsequent generations of cases, i.e., the generation time (E) combined with R0 determine the speed of epidemic growth. Asymptomatic and undetected cases (F) and superspreading events (G) can have important impacts on disease dynamics and control not obvious from observed aggregate case counts. Superspreading events are often associated with health care facilities (as with MERS-CoV (10)) or other high contact settings (e.g., funerals in Ebola (7)). Reducing or eliminating transmission in these contexts can have a disproportionate impact on reducing R and controlling the epidemic. R0 and the generation time combined with the frequency at which cases die (H, skulls) or have severe outcomes (I, red crosses indicate hospitalization) determine the impact of the disease on the human population. Mechanistic models are both informed and can be used to estimate these values, and the combination of such models with pathogen genetic information obtained from biological sampling of a subset of cases (J, test tube) can allow for inferences when simple observational data does not. Over the medium- to long-term, the reproductive number will change (K) due to depletion of susceptibles, interventions and behaviour changes in response to the emerging threat. The latter two are hard to predict, adding to the difficulties in forecasting over the medium-term. Spread at the global scale (L) may require alternate modeling approaches, with more emphasis on human mobility and environmental suitability than drivers local of pathogen dynamics.
Box 1: Key dynamic quantities estimated early after an emergence event.
There are several key dynamics quantities that determine the course of the epidemic and indicate needs for the structure of the response, that should be identified rapidly after emergence.
Basic Reproductive Number (R0): The number of cases expected to be directly infected by a single index case in an immunologically naive population. Provides an estimate of the transmissibility of an emergent pathogen. If R0<1 the emerging pathogen will die out, while if R0>1 it can spread widely and cause a major epidemic or pandemic. R0 further determines the final size of epidemics in the absence of control measures.
Reproductive number (R): The number of cases expected to be directly infected by a single infected individual case in a population where there is some underlying immunity.
Generation Time: The time between a case becoming infected and that case causing other infections. Combined with R0, determines the speed at which an epidemic spreads through the population.
Incubation period: The time from infection to symptom onset.
Latent Period: The time from infection to becoming infectious.
Infectious period: The length of time that infected individuals can transmit.
Case Fatality Ratio: The proportion of cases that prove fatal.
Hospitalization Rate/Clinical Attack Rate: The proportion of cases where disease is sufficiently severe as to result in hospitalization, potentially affecting detection via passive surveillance.
Asymptomatic Proportion: The percent of infected individuals that do not develop recognizable symptoms.
When responding to an emerging virus, perhaps the first priority is measuring the distributions of R0 and generation time (Box 1). R0 is of particular interest, as it determines if the disease will die out after introduction. For example, early estimates of R0 for MERS-CoV were well below one (4, 12), while estimates for pandemic H1N1 were in the neighborhood of 1.5 (3, 13). The former remains confined to the Arabian peninsula, and apparently requires continuous re-seeding into the human population from camels to persist, while the latter has established itself globally. Knowing the generation time allows us to estimate R0 from the growth in case numbers during the early (exponential growth) phase of an epidemic. Likewise, using these two values, relatively accurate short-term forecasts can be made early on with simple models (Figure 2).
Figure 2: Phases of the emergence process.
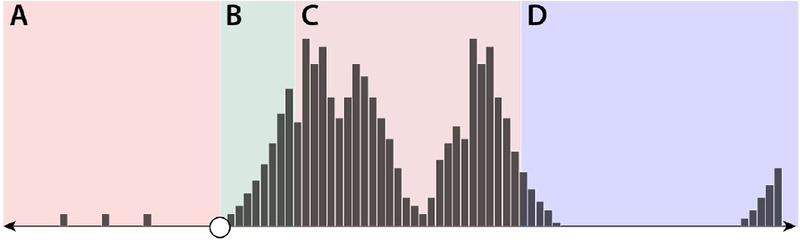
(A) Pre-emergence period: can last years to decades, and can feature occasional zoonotic transmission events. (B) Short-term post-emergence: the first several generations (lasting months to years depending on the pathogen’s generation time), characterized by exponential growth. (C) Medium-term post-emergence: patterns driven by hard to predict aspects of pathogen ecology and human behavior. (D) Long-term post-emergence period: general trends dictated by pathogen properties and basic epidemic theory.
Moving from forecasting cases to forecasting disease burden requires estimates of the risk of severe illness and mortality following infection. Dynamic aspects of both the disease and reporting processes, and potentially large numbers of unobserved infections (Figure 1), mean that models of both are often necessary to estimate these quantities (14). Biases can go both ways; models of the time-course of individual infections have been used to correct for under estimates of the case fatality rate for both SARS-CoV and Ebola early in each epidemic (14, 15); while statistical models that account for the structure of reporting have been used to adjust for upward biases due to underreporting of less severe cases of MERS-CoV (16).
Subsequent to characterizing the growth and potential health impact of an emerging epidemic, modelling efforts shift to evaluating control measures. The design and effectiveness of interventions, such as isolation and quarantine depend critically on the distribution of the disease’s incubation period and latent period, as well as the frequency of asymptomatic infection (5, 17). More detailed models require more data or assumptions, but can be used for strategic evaluation of particular interventions. In response to the Ebola epidemic, models were used to compare the impact of case isolation, contact-tracing with quarantine, and sanitary funeral practices (18); and to evaluate the impact of travel bans, and exit and entry screening at airports (19). Such models may not accurately forecast the exact number of cases prevented by each intervention: their value is in providing an assessment of relative impact. Importantly, such models rely not only on accurate characterization of the epidemic process, but modeling of logistics, health systems and human behaviors.
Information on R0, human demographics, and the distribution and duration of immunity also allow us to make general predictions about an emergent pathogen’s long-term impact (Figure 2). For instance, basic models of the post invasion dynamics of an immunizing infection predict a lull in ZIKV transmission in the Americas starting a few years after its introduction, potentially lasting decades (2). The presence of pre-existing immunity can also profoundly affect both short- and long-term epidemic patterns, and is a critical, usually missing, component when forecasting epidemics.
Limitations of the classical toolkit and pitfalls in model based approaches
Basic infectious disease models can forecast short term incidence, and broadly characterize long-term trends (Figure 2); and can do so with increasing accuracy as new data becomes available. However, medium-term forecasts may require unattainably detailed information about biological, ecological and social systems. Variations in patterns of infectious contact, arising from local contact structure to regional variation in mobility are driven by changing human behavior. For example, even in the absence of formal directives, people naturally change their behavior in the presence of Ebola cases. This phenomenon helped control previous outbreaks in the Democratic Republic of Congo and elsewhere, and may have contributed to mismatches between pessimistic forecasts of the 2014 West African Ebola outbreak and its observed trajectory (20). Control efforts may also change the trajectory of an epidemic, invalidating forecasts that leave them out. Variation in environmental suitability for transmission at small and large spatial scales may also shape spread in unanticipated ways. This is particularly true for vector borne diseases, even when nuances of biological mechanism (e.g., the effect of temperature on mosquito survival) are well known (21, 22).
The lack of data that makes mechanistic models essential in the response to an emergent pathogen, also handicaps these models. Numerous modeling exercises, and hard experience show that appropriate control measures must be implemented early in an emergent epidemic to be successful (23, 24). This sets up a Catch-22 scenario because the time when good models are most needed is when they are the hardest to make. The potential to publish high profile results and a desire to have a meaningful impact on the public health response can lead to an explosion of modeling studies (e.g., a pubmed search for ‘mathematical model Ebola’ since 2014 yields over 300 results). These may be based on data and methods of varying quality, and are rarely followed by any attempt to synthesize results and reach consensus.
Efforts to “get ahead of the game” by collecting data and creating models prior to disease emergence may be doomed, as emergent pathogens rarely conform to our expectations. For example, despite extensive work preparing for a flu pandemic (e.g., (23, 25, 26)), pandemic H1N1 violated model assumptions on nearly every point: it arose in a different part of the world, it was the same major subtype as an already circulating strain, and it failed to displace all circulating influenza A subtypes. Likewise, prior to the start of the 2014 outbreak, Ebola was considered a well-characterized threat, of significant concern in limited parts of East and Central Africa, and causing only small epidemics. Though models suggest the size of the West African outbreak was not inconsistent with previous knowledge (27), its scale and location were completely unanticipated.
One solution to the lack of data is increased reliance on our understanding of biological mechanism. However, models, particularly complex models, heavily based on prior knowledge or mechanistic assumptions have the potential to be (sometimes spectacularly) wrong. Forecasts of the range of ZIKV provide a prime example. Several models driven by vector ecology, and in some cases human sexual behavior, suggested a risk of significant ZIKV outbreaks in the US outside of the southern-most counties (28–30); a prediction inconsistent both with our experience with other arboviruses, and observations from the ZIKV epidemic thus far. Models based on assumed mechanism may be useful, revealing potential deviations from past experience and galvanizing public health action, but should be paired with an examination of the data and explorations of why predictions deviate from experience.
Appropriately capturing and communicating uncertainty is a constant challenge when modeling emergent pathogens. Classical approaches that rely heavily on systems of differential equations are powerful, but can easily neglect to account for statistical uncertainties arising from limited data, and uncertainties rising from the role of chance events in the epidemic process (i.e., process uncertainty). The best recent work takes advantage of methods built on growing computational resources to capture both, and also attempts to account for uncertainties driven by knowledge gaps. For instance, early work on pandemic H1N1 and MERS-CoV estimated R0 using multiple approaches and assumptions, calculating confidence intervals for each, and reporting the full spectrum of results (3, 4). Epidemiologists must carefully evaluate which types of uncertainty are critical to capture given the goals of their analysis, e.g., process uncertainty may be critical when forecasting incidence, but less so when comparing hypothetical interventions.
Communication of results from modeling exercises can be difficult. The media and other laypersons tend to focus on a model’s most dire predictions. In the fall of 2014, researchers at the CDC projected that, in the absence of control measures, over a half-million Ebola cases would be reported in Liberia and Sierra Leone by early 2015 (1). The actual numbers were an order of magnitude lower (7). While these projections may have indeed been unduly dire, the fact is that control measures were implemented in the region, and the CDC forecasts accounting for the potential impact of interventions (e.g., increasing use of specialized Ebola treatment units) were far closer to what actually occurred. However, the media almost exclusively focused on the extreme values, contributing to an impression that the forecast was a failure (31). A focus on prediction exacerbates the problem; models may be successful in their aims of identifying planning scenarios or evaluating hypothetical interventions, but still produce unrealistic or unrealized epidemic forecasts. Media headlines tend to miss the nuanced difference between forecasts and planning scenarios, as occurred with early pandemic H1N1 planning exercises, where attention focussed almost exclusively on the high end of the worst case scenarios (32).
Mechanistic models play an essential role in the response to emergence events. However, given the unknowns inherent in the situation, accurately characterizing an emerging pathogen is hard and always will be. Even when researchers are sensitive to limitations of their models, these limitations may be very hard to communicate. A focus on forecasting, intended or not, can obscure the broader value of mechanistic approaches: their ability to synthesize multiple complex data streams in an informative way, leveraging our understanding of biologic and epidemic process to improve situational awareness and reveal properties of pathogen transmission.
Improving modeling of emergent pathogens: trends and opportunities
A clear trend over the past decade is the increasing availability of data on pathogen genetics. Whole-genome sequencing is ever more affordable, and novel analytic techniques are developing a-pace. Phylodynamic methods can characterize the timing of pathogen introduction, for example indicating earlier ZIKV introduction into the Americas than originally thought (33), and suggesting both the single spillover origin, and subsequent determinants of Ebola spread (34). Phylodynamic methods can also be paired with disease models to estimate R0 (3, 4), providing an alternative to analysis of case data for estimating this important quantity. However, the benefits of obtaining an estimate that is not subject to the same biases as case data should not obscure the fact that phylogenetic approaches have their own biases and limitations, and are not a panacea.
Modeling of emergent pathogens has also benefitted from the “big data” revolution driven by the ever expanding pool of large datasets created through automated data collection. High resolution (sub-kilometer) satellite based measures of environmental variables (e.g., land surface temperature) is one novel big data stream, that can be combined with other data sources via machine learning algorithms to determine, for example, the likely range and local transmission intensity of vector borne infections like ZIKV (21, 22). Similar approaches have been used to disaggregate census data yielding high-resolution maps of population density and demographics (35), thus tackling the perennial problem in epidemiology of determining the population at risk. Another trend is increasing availability of data-streams characterizing mobility, such as air-travel flows (36). Novel data-streams on mobility are also becoming increasingly important: mobile phone-call records yield unprecedented temporal and spatial resolution on human mobility and aggregation (37). As ever, potential biases and limitations must be considered carefully, e.g., spatial locations can only be mapped when a call is made, and mobile phone ownership is not necessarily representative of populations of interest.
Both phylodynamic and ‘big-data’ techniques have been enabled by increasing availability of affordable, high-performance computing resources. These resources also allow for implementation of statistical and modeling techniques that were once prohibitively computationally expensive, and have improved the rigor of models of emergent pathogens, particularly the quantification of uncertainty. Computationally intensive techniques that integrate across multiple predictive models (e.g., (38)) are leading to clear improvements in forecasting of established pathogens, and may provide similar benefits for emergent pathogens. Likewise, with new techniques and enough computational power (though sometimes more than is currently available), essentially any probabilistic model construction can be fit to data. This allows researchers to combine often complex representations of the transmission process with techniques of statistical inference to estimate critical transmission properties while taking into account the large scale uncertainty in the underlying transmission tree (39).
As models become more flexible and easier to fit, there is the promise of updating results in ‘real time’ as the response to an emerging pathogen develops. Realizing this potential requires improvements in the way data flows through health systems, as well as how it is combined and processed by models. Data must be updated in a timely manner, and forecasts and inferences must be sensibly adjusted as new data arrives and old data is modified. Rapid modeling exercises can be critical in making timely decisions and guiding interventions and field studies in a rapidly changing environment. For instance, models played a critical role in design of vaccine trials during the Ebola outbreak (40). However, such real-time efforts remain sporadic and ad-hoc.
The immunological landscape on which a pathogen emerges can have profound effects on its spread, with the immunologic imprint of related viruses potentially providing protection (41) or increasing disease severity in affected subpopulations (42). Immunological signatures also provide a marker of previous exposure, and can reveal whether a pathogen is truly novel, or has circulated undetected in human populations before. However, this immunological landscape has historically been part of the “dark matter” of epidemiologic information; serologic laboratory and analytic techniques have lagged behind developments in the molecular analysis of genomic data, and there are few sources of data on the pre-emergence immune status of populations. Establishment of a global serum bank, combined with improved methods for efficiently testing for a broad range of immunological markers could provide an invaluable resource for responding to emergence events (43). However, commensurate improvements to how such information is incorporated into disease models is also needed.
The ultimate achievement in modeling emergent pathogens would be to develop models of sufficient biological and ecological sophistication to identify emergent disease threats before they entered the human population. Identifying and sequencing novel viruses is a necessary first step, but “virus hunting” activities are of limited utility without some way to assess which viruses pose a threat. We know generalities, for example that viruses are more likely to jump between closely related species (44), or that RNA viruses’ rapid rate of mutation may make them more prone to emergence events than DNA viruses (45); and we can theoretically assess how the relationship between introduction frequency, R0, and the number of mutations needed to efficiently transmit among humans impacts emergence rates (45). Yet we lack the depth of understanding of the relation between genotype and phenotype to assess which viruses will spread and cause disease in the human population, and which will not (46). However, our ability to observe “viral chatter” between human and animal populations is ever increasing, and may soon lead the breakthroughs needed to identify likely emerging threats.
Future emergence events
Global biosecurity depends on our ability to effectively confront emerging infectious disease threats. Mechanistic models, which capture our scientific understanding of disease processes, will continue to play an important role in assessing and responding to pathogen emergence. While these methods have numerous limitations and pitfalls, and it may sometimes be difficult to tell good work from bad, they provide vital information to the global health response that is unavailable through other means. Continued methodological improvements that take advantage of new sources of data will increase the range and accuracy of inferences that can be made from leveraging infectious disease models. Tighter integration with public health practice and development of resources at the ready may increase the timeliness and quality of analyses to inform the public health response.
REFERENCES
- 1.Meltzer MI et al. , Estimating the future number of cases in the Ebola epidemic—Liberia and Sierra Leone, 2014−−2015. MMWR Surveill. Summ 63, 1–14 (2014). [PubMed] [Google Scholar]
- 2.Ferguson NM et al. , EPIDEMIOLOGY. Countering the Zika epidemic in Latin America. Science. 353, 353–354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser C et al. , Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 324, 1557–1561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cauchemez S et al. , Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect. Dis 14, 50–56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser C, Riley S, Anderson RM, Ferguson NM, Factors that make an infectious disease outbreak controllable. Proc. Natl. Acad. Sci. U. S. A 101, 6146–6151 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cauchemez S et al. , Unraveling the drivers of MERS-CoV transmission. Proc. Natl. Acad. Sci. U. S. A 113, 9081–9086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Ebola Response Team et al. , Exposure Patterns Driving Ebola Transmission in West Africa: A Retrospective Observational Study. PLoS Med 13, e1002170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessler J et al. , Assessing the global threat from Zika virus. Science, aaf8160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller MA et al. , Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect. Dis 15, 559–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assiri A et al. , Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med 369, 407–416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May R M And Anderson, R M, COMMENTARY~ Transmission dynamics of HIV infection. Nature. 326, 137 (1987). [DOI] [PubMed] [Google Scholar]
- 12.Breban R, Riou J, Fontanet A, Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 382, 694–699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y et al. , The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 326, 729–733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghani AC et al. , Methods for estimating the case fatality ratio for a novel, emerging infectious disease. Am. J. Epidemiol 162, 479–486 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucharski AJ, Edmunds WJ, Case fatality rate for Ebola virus disease in west Africa. Lancet. 384, 1260 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Lessler J et al. , Estimating the Severity and Subclinical Burden of Middle East Respiratory Syndrome Coronavirus Infection in the Kingdom of Saudi Arabia. Am. J. Epidemiol 183, 657–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peak CM, Childs LM, Grad YH, Buckee CO, Comparing nonpharmaceutical interventions for containing emerging epidemics. Proc. Natl. Acad. Sci. U. S. A 114, 4023–4028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey A et al. , Strategies for containing Ebola in West Africa. Science. 346, 991–995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Read JM, Diggle PJ, Chirombo J, Solomon T, Baylis M, Effectiveness of screening for Ebola at airports. Lancet. 385, 23–24 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Funk S, Knight GM, Jansen VAA, Ebola: the power of behaviour change. Nature. 515, 492 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Alex Perkins T, Siraj AS, Ruktanonchai CW, Kraemer MUG, Tatem AJ, Model-based projections of Zika virus infections in childbearing women in the Americas. Nature Microbiology. 1, 16126 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Messina JP et al. , Mapping global environmental suitability for Zika virus. Elife. 5 (2016), doi: 10.7554/eLife.15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson NM et al. , Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 437, 209–214 (2005). [DOI] [PubMed] [Google Scholar]
- 24.WHO Ebola Response Team et al. , Ebola virus disease in West Africa--the first 9 months of the epidemic and forward projections. N. Engl. J. Med 371, 1481–1495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson NM et al. , Strategies for mitigating an influenza pandemic. Nature. 442, 448–452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longini IM Jr et al. , Containing pandemic influenza at the source. Science. 309, 1083–1087 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Camacho A et al. , Potential for large outbreaks of Ebola virus disease. Epidemics. 9, 70–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shacham E, Nelson EJ, Hoft DF, Schootman M, Garza A, Potential High-Risk Areas for Zika Virus Transmission in the Contiguous United States. Am. J. Public Health 107, 724–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogoch II et al. , Anticipating the international spread of Zika virus from Brazil. Lancet. 387, 335–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manore CA, Ostfeld RS, Agusto FB, Gaff H, LaDeau SL, Defining the Risk of Zika and Chikungunya Virus Transmission in Human Population Centers of the Eastern United States. PLoS Negl. Trop. Dis 11, e0005255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stobbe M, CDC’s overblown estimate of Ebola outbreak draws criticism. The Seattle Times (2015), (available at http://www.seattletimes.com/nation-world/cdcs-overblown-estimate-of-ebola-outbreak-draws-criticism/).
- 32.Swine Flu Could Infect Half of U.S., Panel Estimates (2009), (available at http://www.washingtonpost.com/wp-dyn/content/article/2009/08/24/AR2009082401733.html).
- 33.Faria NR et al. , Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature (2017), doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudas G et al. , Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature (2017), doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatem AJ, WorldPop, open data for spatial demography. Sci Data. 4, 170004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao L, Wu X, Huang Z, Tatem AJ, Modeling monthly flows of global air travel passengers: An open-access data resource. J. Transp. Geogr 48, 52–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wesolowski A et al. , Quantifying travel behavior for infectious disease research: a comparison of data from surveys and mobile phones. Sci. Rep 4, 5678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Karspeck A, Shaman J, Comparison of filtering methods for the modeling and retrospective forecasting of influenza epidemics. PLoS Comput. Biol 10, e1003583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cauchemez S et al. , Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc. Natl. Acad. Sci. U. S. A 108, 2825–2830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho A et al. , Real-time dynamic modelling for the design of a cluster-randomized phase 3 Ebola vaccine trial in Sierra Leone. Vaccine. 35, 544–551 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO, Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 354, 722–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dejnirattisai W et al. , Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol 17, 1102–1108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metcalf CJE et al. , Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet. 388, 728–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM, The evolution and genetics of virus host shifts. PLoS Pathog 10, e1004395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antia R, Regoes RR, Koella JC, Bergstrom CT, The role of evolution in the emergence of infectious diseases. Nature. 426, 658–661 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pepin KM, Lass S, Pulliam JRC, Read AF, Lloyd-Smith JO, Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat. Rev. Microbiol 8, 802–813 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]


