Abstract
High rates of homologous recombination (HR) in the bacterial plant pathogen Xylella fastidiosa have been previously detected. This study aimed to determine the extent and explore the ecological significance of HR in the genomes of recombinants experimentally generated by natural transformation and wild-type isolates. Both sets of strains displayed widespread HR and similar average size of recombined fragments consisting of random events (2–10 kb) of inter- and intrasubspecific recombination. A significantly higher proportion and greater lengths (>10 kb, maximum 31.5 kb) of recombined fragments were observed in subsp. morus and in strains isolated in Europe from intercepted coffee plants shipped from the Americas. Such highly recombinant strains pose a serious risk of emergence of novel variants, as genetically distinct and formerly geographically isolated genotypes are brought in close proximity by global trade. Recently recombined regions in wild-type strains included genes involved in regulation and signaling, host colonization, nutrient acquisition, and host evasion, all fundamental traits for X. fastidiosa ecology. Identification of four recombinant loci shared between wild-type and experimentally generated recombinants suggests potential hotspots of recombination in this naturally competent pathogen. These findings provide insights into evolutionary forces possibly affecting the adaptive potential to colonize the host environments of X. fastidiosa.
Subject terms: Bacteriology, Phylogenetics, Applied microbiology, Bacterial genomics, Bacterial evolution
Introduction
Horizontal gene transfer, a fundamental mechanism of evolution in bacteria, occurs by conjugation, transduction, and transformation [1]. These processes lead to homologous recombination (HR) that mediates exchange of genetic material among related taxa, including acquisition of novel genetic elements, such as antibiotic resistance and virulence factors, that can result in emergence of new pathotypes [2]. In bacteria, HR is proposed to play a key role in repairing damaged DNA [3] to promote survival under stressful conditions. But HR may also give rise to new variants if the homologous DNA originates from a closely related but genetically distinct donor [4]. New genetic variants can result in profound phenotypic changes, such as evasion of host defenses by creating variation in surface structures [5, 6], facilitate disease emergence [7], or enhance adaptation to new environments [8]. The relative role of HR and point mutations in generating genetic variation differs among bacterial species [9], with HR expected to play a major role in naturally transformable bacteria.
The first observation of HR was achieved by detection of mosaic genes that contained fragments originating from different taxa of the same genus [10]. Use of multilocus sequence typing/analysis (MLST/MLSA), a technique to assess genetic diversity within a species by sequence analysis of 5–11 housekeeping genes scattered across the genome [11], has revealed HR in bacterial genomes. Until recently, MLST was a signature tool for bacterial phylogenetic studies and detection of HR [12–15]. However, examination of only few genes with presumed neutral variation will overlook HR present in other genomic regions, including genes involved in host adaptation and/or virulence [16]. Whole-genome sequence (WGS) comparisons provide a superior approach for detecting HR in bacterial genomes [17–19].
Xylella fastidiosa is a plant pathogenic bacterium that causes massive economic losses and was believed to be limited to the Americas, but recently has emerged as an important pathogen in other regions of the world [20], including Europe [21, 22] and Asia [23]. X. fastidiosa exclusively colonizes two habitats: the plant xylem vessels and foregut of insect vectors (sharpshooter leafhoppers and spittlebugs) [24]. Taxonomically, strains within X. fastidiosa are generally categorized into three subspecies (i.e., fastidiosa, multiplex, and pauca) [25–27], although additional subspecies have been suggested (i.e., morus, sandyi) [28, 29]. While the species has a very broad plant host range [30], this differs among subspecies. However, mechanisms of host specificity are still unknown [31].
Extensive HR has been identified in X. fastidiosa based on genetic diversity and phylogenetic patterns revealed by MLST [29, 31–34]. These studies made notable speculations on the impacts of HR on host range expansion in X. fastidiosa strains, including those strains that infect citrus, mulberry, and blueberry [15, 28, 34]. However, these inferences were made based on the evidence of HR in housekeeping genes, rather than taking advantage of whole genomes.
Experimental studies have demonstrated natural competence and HR in X. fastidiosa both in vitro [35] and in artificial habitats mimicking the natural growth environment of the bacterium [36]. Moreover, intersubspecific HR (IHR) between X. fastidiosa subsp. fastidiosa and multiplex was recently demonstrated experimentally [37]. The objective of this study was to use WGS analysis to identify the extent of intra- and intersubspecific HR in X. fastidiosa. We did this by first focusing on X. fastidiosa recombinant strains experimentally generated in vitro [37], and later by studying multiple strains isolated from field infections around the world. Extensive inter- and intrasubspecific recombination of both ancient and recent origins was detected in the genomes of X. fastidiosa, indicating patterns of gene flow among different subspecies. The increased global movement of strains through trade of plant materials can lead to introduction of novel genotypes into an endemic pathogen population. HR can play a significant role in such cases, where novel genotypes might exchange adaptive alleles with the existing population, resulting in niche-adapted recombinant strains with greater fitness than the existing population.
Materials and methods
Bacterial strains, media, and culture conditions
Strains and mutants used in this study are listed in Table S1. For an explanation on recombinant mutants generated in vitro see Supplementary Methods and Fig. 1. A Temecula1 variant strain, that differed based on whole-genome sequence analysis in this study, was named here as TemeculaL [38]. Temecula1* was also a variant of the Temecula1 strain that showed reduced virulence in our previous study [39]. Strains PD7202 and PD7211 were isolated from coffee plants shipped from Costa Rica and intercepted in the Netherlands [40]. Strains were cultured in PW [41] modified agar without phenol red and with 1.8 g l−1 bovine serum albumin (BSA) (Gibco Life Sciences Technology) for a week at 28 °C from −80 °C stock, re-streaked, and cultured for another week before use. Kanamycin (Km) and chloramphenicol (Cm) were used at 30 and 10 µg ml−1, respectively.
Fig. 1.
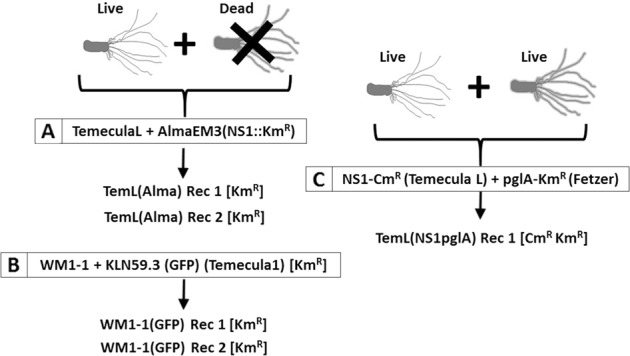
Diagram summarizing the lineage of the recombinant strains used in this study. Parental strains are highlighted in boxes. Recombinant strains were obtained by mixing either one live strain with DNA from another dead strain (a, b), or by co-culturing two live strains (c). Denomination between parenthesis after the strain names corresponds to antibiotic-resistance markers used for selection: Km, kanamycin; Cm, chloramphenicol. Whenever available, selection was also done by GFP production in addition to antibiotic resistance. Parental strains include Xylella fastidiosa subsp. fastidiosa wild-type TemeculaL, WM1-1, and mutants KLN59.3 (GFP) [a KmR and GFP-marked mutant of Temecula1 [75]], NS1-CmR [a CmR cassette inserted in the noncoding region NS1 of TemeculaL [76]], and pglA-KmR [a CmR pglA mutant of strain Fetzer [67]]. X. fastidiosa subsp. multiplex AlmaEm3 (marked with a KmR cassette in the NS1 site) was the only strain classified in a different subspecies. The genomes of all parental and recombinant strains shown here were sequenced for this study. For more details see Supplementary Methods
DNA extraction, library preparation, sequencing, and assembly
In this study, 17 novel X. fastidiosa genome sequences (including nine WT, five in vitro recombinant strains, and three parental strains) were generated (Table S1). Genomic DNA extraction, library preparation and MiSeq/Pacific Biosciences (PacBio) sequencing protocol, quality filtering, trimming, assembly, and annotation details are presented in Supplementary Methods. Briefly, paired-end MiSeq FASTQ files were quality assessed with FastQC (Babraham Bioinformatics) and trimmed with Trimmomatic-0.35 [42]. Trimmed reads (>36 bp) were de novo assembled using SPAdes 3.9.0 [43]. For WM1-1 and AlmaEM3, sequenced by both MiSeq and PacBio, a hybrid assembly of SPAdes 3.9.0 was used. Information for read and assembly statistics, breadth and depth of coverage, and annotation results is summarized in Tables S2 and S3.
Core-genome prediction
Complete or draft genomes of 55 strains of X. fastidiosa sequenced here or obtained from NCBI (Table S1) were aligned using progressiveMauve [44] using default settings. Core-genome alignment was obtained by removing the accessory regions from the alignment using stripSubsetLCBs script on the aligned locally colinear blocks (LCBs) [44], https://github.com/xavierdidelot/ClonalOrigin/wiki/Usage, after discarding core LCBs shorter than 500 nt. The same steps were followed to obtain the core genome of 57 X. fastidiosa strains when two additional intercepted strains PD7202 and PD7211 [40] were added to our database. Implementing the Mauve approach instead of using alignment of concatenated core genes allowed us to include intergenic regions and contiguous order in recombination analysis. For the model implemented in BratNextGen, contiguously ordered core genome alignment is preferred, which was obtained by concatenation of core LCBs.
Population structure and inference of recombination
Population structure was defined with BAPS 6.0 (hierBAPS module for hierarchical BAPS) using the default settings [45]. The core genome (i.e., LCBs) obtained from Mauve alignment of 55 X. fastidiosa genomes was used as input in BAPS 6.0 to obtain the population structure. A total of 33824 backbone entries were concatenated to obtain core genome alignment of the 55 genomes. In addition, the core genome from Mauve alignment of 13 genomes of in vitro recombinants and their parents was used as input for analyzing population structure for in vitro recombinants. These core genome alignments were used to infer maximum likelihood (ML) phylogeny estimated with RAxML v 8.2.11 under the generalized time-reversible model (GTRGAMMA) with 1000 bootstrap replicates [46]. Mid-point rooted ML trees were visualized and annotated using FigTree (v.1.4.2, http://tree.bio.ed.ac.uk/software/figtree/).
For genealogy reconstruction, recombination was identified from the core genome alignments using BratNextGen algorithm. The cutoff level for obtaining clusters from the proportion of shared ancestry tree (PSA tree) was guided by the number of clusters determined by hierBAPS. Recombination was estimated with 20 iterations of the HMM estimation algorithm and 100 permutations with a 5% significance threshold [18]. To analyze the origin of recombination, we used the fastGEAR algorithm, which employs hierBAPS to identify lineages as clusters of genomes that share a monophyletic signal in at least 50% of the sites. Recombination events between X. fastidiosa lineages that affected a subset of recipient lineages are considered “recent” with the origin of the recombinant sequence assigned to the lineage with the highest probability at that position. Regions affected by recent recombination were removed prior to detection of “ancestral” recombination events that are shared by all strains, which comprise a lineage. The origin of these ancestral recombination events cannot be inferred due to their conserved nature [47]. In identifying both recent and ancestral recombination events, Bayes factor >1 or >10, respectively, was used for testing the statistical significance. We conducted this fastGEAR analysis on the core genome alignments for both sets of 55 and 57 X. fastidiosa strains (including newly intercepted strains PD7202 and PD7211). Next, to obtain recombination-filtered core-genome alignment, positions in the core genome alignment that corresponded to the predicted recent recombination events by fastGEAR were masked using the Perl script, bng-mask_recombination.pl (https://github.com/tseemann/bng-tools/blob/master/bng-mask_recombination.pl). Gaps were then removed from the alignment using trimal (v1.2) with the ‘-nogaps’ option [48]. This recombination-filtered core genome alignment was then used to construct a ML phylogeny as described above. Further analysis of recently recombined regions was carried out to understand ecological significance of intersubspecies recombination. Detailed methodology of extracting coordinates for recently recombined genes as well as annotations, is described in Fig. S1.
Accession numbers
The genomes and the raw reads generated in this study have been deposited in NCBI GenBank under BioProject PRJNA433735 with BioSample numbers SAMN08537137–SAMN08537151, SAMN10499944, and SAMN10499946. GenBank accession numbers are included in Table S1.
Results and Discussion
Recombination by natural competence in vitro leads to multiple events of DNA exchange across the genome
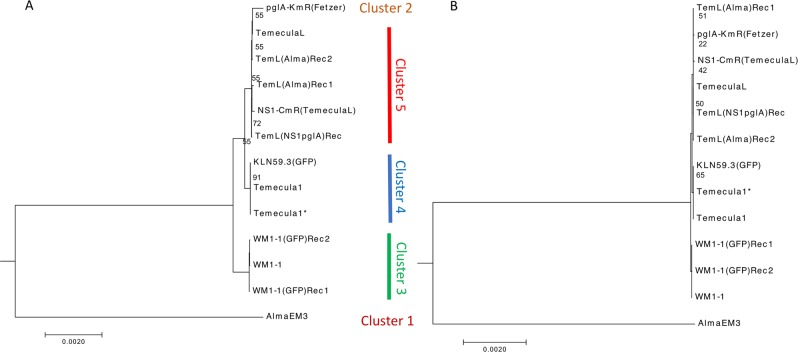
Previously, we performed in vitro natural transformation experiments of X. fastidiosa by co-culturing combinations of either live or heat-killed donor and live recipient strains, generating recombinant strains that were selected based solely on the acquisition of antibiotic-resistance markers [36, 37] (Fig. 1). Complete genomes were analyzed to determine the overall distribution of recombination events that occurred, including the length of DNA flanking the selection marker. Initially, the presence of recombinogenic regions were identified by reference mapping of the reads and alignment of contigs to the parental strain genome (Fig. S2). To further analyze recombinant regions beyond those flanking selection markers, we classified strains used in vitro by hierBAPS that identified five clusters among parental and recombinant strains (Fig. 2a). One interesting observation of this analysis was that strains referred to as “Temecula1” in previous reports are actually different strains that were classified into two different clusters: cluster 5 [TemeculaL, NS1-CmR(TemeculaL)]; and cluster 4 [Temecula1, Temecula1*, KLN59.3(GFP)(Temecula1)]. The NCBI-deposited Temecula1 genome (Table S1) differs from the genome of other strains that have been referred to in the literature by the same name. For instance, clustering showed that NS1-CmR (TemeculaL) has TemeculaL as background and not Temecula1 as previously thought [35]. Recombinant regions shared among Temecula1 and Temecula1* represented different alleles compared with TemeculaL, further highlighting the differences among strains referred to with the same name.
Fig. 2.
Maximum likelihood phylogeny of the core genome alignment of 13 strains of X. fastidiosa in vitro recombinants and their donor and recipient parental strains before (a) and after (b) recombination filtering. Population structure was inferred using nested clustering implemented in hierBAPS. For (b), recombinant regions detected by BratNextGen in the genomes were excluded to build a recombination-free phylogenetic tree. The overall topology remained similar, but the phylogenetic distance decreased among the genomes after recombination filtering. The phylogenetic tree was mid-point rooted, representing an increasing order of nodes. Bootstrap support was >98% for all nodes, except where otherwise indicated
To further investigate variable regions, we used more robust detection tools that detect recombination events at the intrasubspecific level (within subspecies fastidiosa; BratNextGen) and at the intersubspecific level (between subspecies fastidiosa and multiplex; fastGEAR). BratNextGen and fastGEAR identified recombination events in the core genome of X. fastidiosa subsp. fastidiosa WT and in vitro recombinant strains, with prediction of 72 recombination events (Fig. 3). Influence of recombination on phylogenetic relationships among in vitro recombinants and their parents was evident based on recombination-free tree topology that showed increased cohesiveness among clusters within X. fastidiosa subsp. fastidiosa and reduced branch length among subspecies fastidiosa and multiplex strains (Fig. 2b).
Fig. 3.
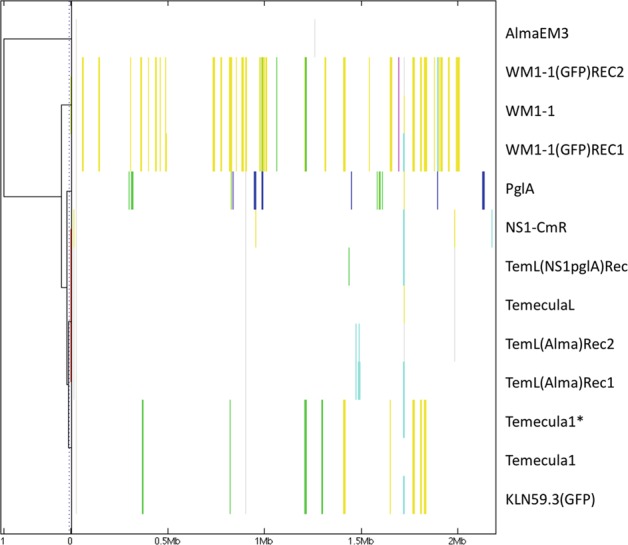
Detection of recombinogenic regions in the core genomes of X. fastidiosa in vitro recombinants and their parental strains using BratNextGen. Core-genome alignment obtained with progressiveMauve was used for genealogy reconstruction, and recombination was identified using BratNextGen algorithm. The estimation of recombination was performed with 20 iterations of the HMM estimation algorithm and 100 permutations with 5% significance threshold. The left panel corresponds to the proportion of shared ancestry (PSA) tree computed by BratNextGen. In PSA tree, strains are clustered according to the proportion of sequence shared, including recombinant regions. In the right panel, recombinant segments identified by BratNextGen for each strain are shown. The colors are arbitrarily assigned. The fragments of the same color present in the same column share the same origin. BratNextGen identified intrasubspecies recombination events (within fastidiosa subspecies), also indicating differences among different “Temecula” strains (see text for more details)
Recombinants representing an intersubspecific strain combination, i.e., heat-killed X. fastidiosa subsp. multiplex strain AlmaEM3 as a donor and X. fastidiosa subsp. fastidiosa strain TemeculaL as a recipient (Fig. 1a), showed recombination only around the Km marker cassette, as identified by both fastGEAR and the stringent method BratNextGen (data not shown). The lengths of recombining regions varied in the case of the two recombinants, with TemL(Alma) Rec1 showing a total of 10-kb region (spanning five recombination events surrounding the Km marker gene) and TemL(Alma) Rec2 showing a 3.5-kb region (spanning one recombination event) flanking the Km marker cassette, as was observed in reference mapping and alignment (Fig. S2). No additional recombinant regions were identified, indicating that recombination in this case was confined to the flanking regions of the Km selection marker insertion site.
On the other hand, in the case of in vitro intrasubspecific recombinants generated by mixing live cells of X. fastidiosa subsp. fastidiosa WM1-1 (hierBAPS cluster 3, Fig. 2a) with heat-killed cells of X. fastidiosa subsp. fastidiosa KLN59.3(GFP) (hierBAPS cluster 4) (Fig. 1b), recombination events were detected away from the GFP marker using BratNextGen and fastGEAR analysis in WM1-1(GFP) Rec1 and WM1-1(GFP) Rec2 (10.5-kb region identified in both recombinants and an additional 6.1 kb in WM1-1(GFP) Rec1). Such random recombination events away from the selection marker were also observed in in vitro recombinants of Helicobacter pylori [49]. We scanned these recombinant regions for polymorphisms in amino acid sequences, and identified polymorphisms in genes encoding lipase/alpha–beta hydrolase and putative Ctpa-like serine protease/peptidase S41 (Table 1). Previously, homologous serine proteases have been linked to virulence of X. fastidiosa [50, 51].
Table 1.
List of recent recombination events in genes/categories identified in WT strains and their possible ecological role in host adaptation
| Recent recombined genesa | Recombined subspecies (number of recombination events) | Recombination origin | Ecological role | References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| morus/ sandyi (4)b | multiplex (12) | fastidiosa (20) | pauca (19) | morus/ sandyi | multiplex | fastidiosa | pauca | Outside | ||||
| 1. Movement | Type IV pili genes | S (31)c | M (52) | F (3) | P (22) | M F P d | S P | S M P | S | P | Xfe: Plant colonization via twitching motility; virulence determinant. | [66] |
| 2. Attachment | Type I pili | - | M (14) | F (3) | P (55) | M F P | P | F | P | Xf: Cell-cell aggregation, attachment to surfaces and biofilm formation; important for colonization of plant and insect hosts; virulence determinant. | [63–65] | |
| Afimbrial adhesin genes | S (14) | M (1) | F (5) | P (18) | F P | S F P | S | M P | ||||
| 3. Exoenzymes | TIISS genes | S (18) | M (29) | F (4) | P (18) | M F P | S P | S M P | S | P | Xf: Hydrolytic enzymes are important for degrading plant structural components and therefore colonization. | [51, 67] |
| Xylan 1,4-xylosidase | S (9) | - | F (7) | - | F | S F | S | S | ||||
| 4. Regulation and signaling | DSF (rpfF/rpfC/rpfG) | S (14) | M (54) | F (2) | P (34) | M F P | S | S M P | P | Xf: Diffusible Signaling Factor (DSF) is a quorum sensing molecule important for plant host specificity and switching between plant and insect environments. | [62,69] | |
| Sensor kinases | S (53) | M (29) | F (6) | P (15) | M F P | S | S M P | P | Part of two-component signal transduction pathways responding to external stimuli for adaptation to different environments. | [77] | ||
| phoP/phoQ | S (14) | M (29) | F (4) | P (5) | M F P | S | S M | S | Xf: Essential for survival in plant host. | [70] | ||
| Vitamin B12 transporter BtuB | S (33) | M (39) | F (30) | P (59) | M F P | S F P | S M P | S | S F P | Regulatory system affecting gene expression, enzyme activity, organism abundance; affects virulence and biofilm formation. | [71–73] | |
| RNA binding Hfq | S (2) | M (6) | - | - | M | S | Small RNAs chaperone, regulates virulence and stress adaptation. | [78] | ||||
| 5. Nutrient acquisition | Tryptophan metabolism | S (10) | M (5) | F (1) | P (9) | M F P | S P | S | Important for interaction host-microflora in humans. | [79] | ||
| Phosphate assimilation | S (39) | M (53) | F (20) | P (22) | M F P | S F | S M | S P | Xf: Indirect evidence suggests phosphorus metabolism important for plant colonization. | [80, 81] | ||
| 6. Adaptation to new conditions | Recombination genes | S (26) | M (11) | F (19) | P (7) | M F P | S F P | S M P | S | P | Xf: Acquisition of new genomic information, evolution. | [35] |
| tRNA biosynthesis / modification genes | S (17) | M (37) | F (8) | P (17) | M F P | S F | S M P | P | Optimization of translation, adaptation to changes. | [74] | ||
| 7. Cell envelope structure | Peptidoglycan biosynthesis genes | S (42) | M (71) | F (12) | P (38) | M F P | S P | S M P | S | S P | Plant host recognition, bacterial growth. | [82, 83] |
| Lipopolysaccharide genes | S (24) | M (24) | F (9) | P (12) | M F P | S F | S M P | S P | Xf: Lipopolysaccharide O-antigen allows to delay initial recognition by plant innate immune system. | [68] | ||
| 3-Oxoacyl [acyl-carrier-proteins] | S (34) | M (48) | F (14) | P (6) | M F | S F | S M P | F | Involved in fatty acid metabolism, influence membrane structure and quorum sensing signaling. | [84–86] | ||
| Recently recombined genes in experimentally-generated recombinants f | ||||||||||||
| WM1-1 (GFP) Rec1f | Lipase/alpha-beta hydrolase (PD1702)g | S (2) | - | - | P (11) | P | P | S P | P | Xf: Homolog (PD1703) identified as pathogenicity effector. | [50] | |
| Putative Ctpa-like serine protease /peptidase S41 (PD0948) g | S (1) | M (15) | F (1) | - | M F | S M | Associated with virulence of other bacterial species. | [52] | ||||
| TemL(NS1-pgla)Rec 1f | Hypothetical proteins (PD1421, PD1422, PD1423)g | - | - | - | - | Unknown. | ||||||
| Extracellular serine protease (PD0950)g | - | M (14) | F (2) | P (1) | M F P | Xf: Serine protease PD0956 identified as pathogenicity effector. | [50] | |||||
| Putative Ctpa-like serine protease/ C-terminal processing protease CtpA/Prc, contains a PDZ domain (PD0949)g | - | M (8) | - | - | M | M | Associated with virulence of other bacterial species. | [52] | ||||
| Translocation and assembly module TamA/ outer membrane protein assembly factor (PD0507)g | - | - | - | - | Secretion of adhesion protein. | [87] | ||||||
a For a complete list of genes considered for each category see Table S6.
b Number in parenthesis indicates the total number of strains considered for each subspecies, see Table S1 for details.
c The number of recent recombination events in parenthesis is categorized by X. fastidiosa subspecies for each gene/category shown in the table. Different letters indicate presence of recombination in each subspecies for each analyzed gene/category. Letters represent each subspecies, i.e., S: subsp. morus/sandyi; M: subsp. multiplex; F: subsp. fastidiosa; P: subsp. pauca.
dPresence of letters in the columns under “Recombination origin” shows which subspecies acted as donor for the recombination event, according to the letter coding of each subspecies used in “Recombined subspecies” column. For instance, if a “M” (subspecies multiplex) is in the “Recombination origin” column of subspecies “fastidiosa”, it means that the latter was the donor for that recombination event(s) in subspecies multiplex.
e Presence of “Xf” means that the ecological role of genes belonging to these categories was functionally studied in X. fastidiosa and references appear in the following column. In the absence of “Xf” the description and corresponding references represent knowledge gained in other bacteria.
f Experimentally-generated recombinants strains where recombined regions were found, and were searched in WT strains.
g Corresponding locus tag from X. fastidiosa Temecula1 for each gene is included in parenthesis.
Finally, we assessed the extent of recombination in another intrasubspecific in vitro recombinant. TemL(NS1pglA) Rec1 was obtained by co-culturing a live donor and recipient parents NS1-CmR (TemeculaL) and pglA-KmR, belonging to clusters 5 and 2, respectively (Figs. 1c, 2a). Upon analyzing the flanking region of the Km marker cassette in the recombinant genome, a 6-kb region upstream of the cassette was identical to the region from pglA-KmR, indicating recombination at this region. A 5-kb region downstream of the Km cassette was identical in both pglA-KmR as well as NS1-CmR. In addition to recombination near the Km cassette, we identified three recombination events (320 bp, 1.5, and 3 kb) away from the Km cassette, similar to what was described above. Scanning these regions for changes in amino acid polymorphisms in protein-coding genes indicated recombinant genes encoding extracellular serine protease, Ctpa-like serine protease, hypothetical proteins and translocation, and assembly module TamA/surface antigen/autotransporter domain (Table 1). Except for the last two, all the other proteins were identified also as recombinants in WT strains (Table 1). Interestingly, a recombinant gene encoding Ctpa-like serine protease, identified in this in vitro recombinant, is present upstream of the recombinant gene (also encoding Ctpa-like serine protease) [52] in WM1-1(GFP) Rec1 (see above). The presence of recombination in similar regions of the genome in two independent recombinants arising from different parents is surprising. Ctpa-like serine protease was also identified to be a recombinant locus in morus/sandyi, multiplex, and fastidiosa subspecies in WT strains (Table 1, see below), suggesting this region as a possible recombination hotspot. Potential role of these genes in the interaction of X. fastidiosa with its host plants is described in Table 1.
Our results indicate that in vitro recombination by natural competence is not limited to a small fraction of the genome, and it is variable, even when considering the same pair of strains under controlled conditions in the same experiment [37], as seen in the case of two TemL(Alma) recombinants that vary in the extent of integration of transformed DNA. While this study analyzed recombinants obtained after a single cycle of transformation, frequency of recombination upon natural competence is expected to increase dramatically after several cycles [49]. Thus, co-infection of plants [53] or insects with different strains of a single or multiple subspecies is expected to result in unpredictable widespread exchanges of genetic material. Interestingly, we found that in the case of intersubspecific recombination (Fig. 1a), only the region of the selection marker was exchanged, while in intrasubspecific recombination (Fig. 1b, c), additional regions away from the selection marker were also exchanged. This may indicate that in nature, intrasubspecific HR may be more widespread in the genome than intersubspecific recombination, where it could be inhibited by sequence divergence [54], or possibly by different restriction-modification systems [49]. Testing this hypothesis would require extensive sampling and genome analysis of strains belonging to each subspecies from wild populations followed by genome sequencing.
Ancient and recent recombinogenic regions are common among X. fastidiosa strains
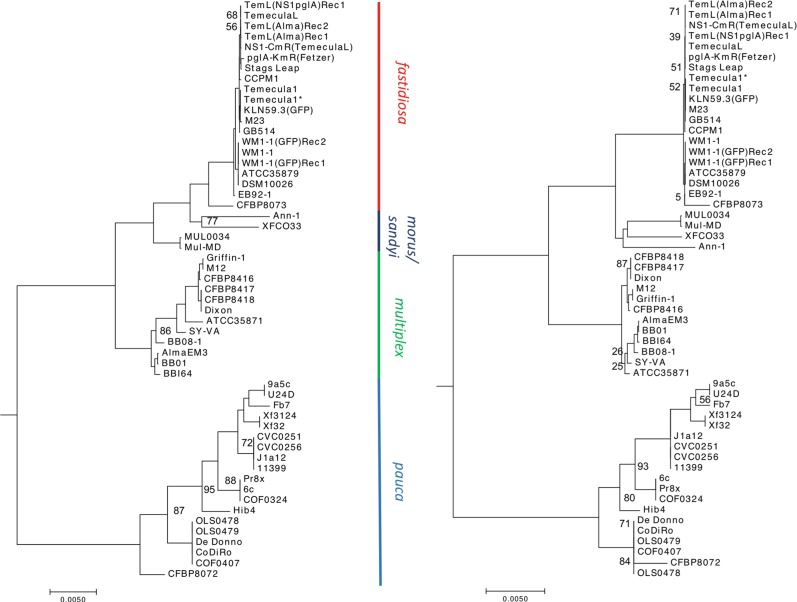
Analysis of in vitro transformation detected multiple recombination events within and between subspecies multiplex and fastidiosa. To estimate the influence of recombination on the evolution of X. fastidiosa at the intersubspecific level, we analyzed 55 genomes belonging to all X. fastidiosa subspecies described/available to date [i.e., subsp. fastidiosa, multiplex, pauca, morus, and sandyi (15 sequenced in this study and 40 obtained from NCBI GenBank)] (Table S1). Population structure of all the genomes inferred using nested clustering implemented in hierBAPS identified five clusters: one cluster each representing subsp. fastidiosa and multiplex, two clusters within subsp. pauca, and a single cluster representing subsp. morus and sandyi (Fig. 4a). The secondary level clustering further divided the cluster containing subspecies morus and sandyi into individual subspecies clusters, and a cluster containing multiplex into two clusters (as seen by two subclades within subspecies multiplex). The core genome alignment of 1.79 MB was used for the construction of a phylogenetic tree (Fig. 4a). Subspecies morus and sandyi were positioned in a “hybrid” subclade between fastidiosa and multiplex; this is in agreement with the phylogeny based on MLST [20].
Fig. 4.
Population structure of X. fastidiosa. Maximum likelihood phylogeny of core genome alignment of 1.79 Mb of 55 X. fastidiosa strains analyzed in this study, before (a) and after (b) filtering for the recombinant regions. The phylogenetic tree was mid-point rooted, representing an increasing order of nodes. For (a), a total of 33,824 backbone entries were concatenated to obtain core genome alignments of 1.79 MB. Population structure was inferred using nested clustering implemented in hierBAPS. For (b), recombinant regions detected by fastGEAR in all subspecies of X. fastidiosa were excluded to build a recombination-free phylogenetic tree. Bootstrap support was >98% for all nodes, except where otherwise indicated
While BratNextGen allowed us to detect recombination within a single lineage (subsp. fastidiosa), it did not provide enough resolution when diverse lineages belonging to a subspecies were included, with no recombination identified in subsp. multiplex or pauca (Fig. S3). To address this limitation, we used fastGEAR software that inferred recombination between lineages as well as from external origins [47]. As evident from Fig. 5, subsp. morus and sandyi showed mosaic/chimeric genomes due to a relatively high degree of recent (Table S4) as well as ancient (Table S5) recombination events, thus, confirming previous observations [28].
Fig. 5.
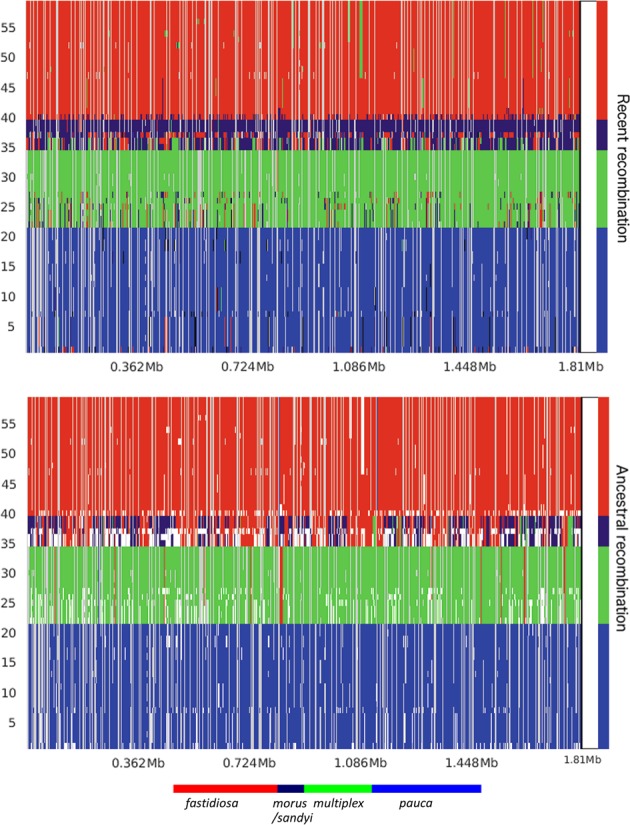
Interlineage recombination. Distribution, origin, and proportion of recent (a) and ancestral (b) recombination events across the X. fastidiosa core genome as predicted by fastGEAR. The lineage predictions are based on hierBAPS analysis and are color-coded, and clade designations are labeled accordingly. The presence of differently colored markings within a given lineage indicates a recombination event relative to the genomic position shown in the X axis. Predicted recent recombination events were removed prior to the analysis of ancestral recombination, which are represented by white gaps in (b). Subspecies morus and sandyi showed a high degree of both ancestral and recent recombination, indicating mosaicism in these two subspecies
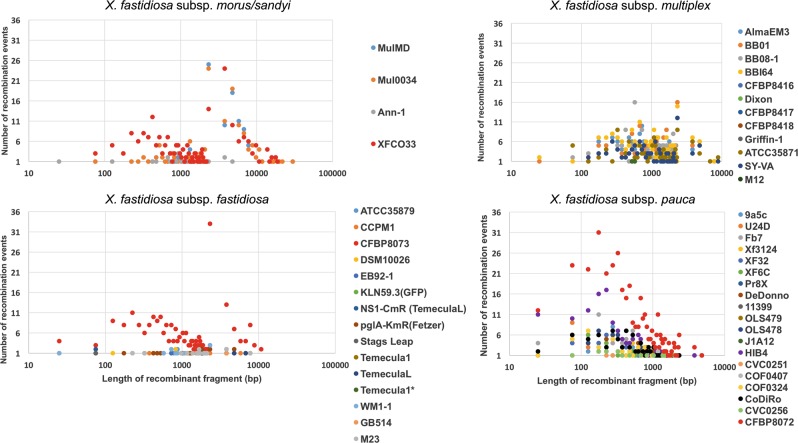
Next, we assessed the influence of recent recombination events on different subspecies. Subspecies pauca has experienced recent recombination events originating from all three lineages, morus/sandyi (27% of the events), multiplex (16% events), and fastidiosa (16% of the events) subspecies, as well as from possibly other bacteria (40% of the total recombination events) (Table S4). There are multiple reports of coexistence in the same geographic areas of subsp. pauca and other subspecies, that can explain the exchange of DNA leading to recombination [55, 56]. The total size of recombination events varied from 530 bp to ~250 kb, representing ~0.02–10% of the genome (Table S4). We identified CFBP8072 as highly recombinant among pauca strains, showing a total of 376 recombination events, with origins in all other subspecies of X. fastidiosa and possible outside sources. This strain, isolated in France from intercepted coffee plants imported from Ecuador, was previously identified as a recombinant strain based on MLSA [57]. Analysis of length distribution of recombination events across genomes of subsp. pauca indicated an average size of 1 kb, ranging from 25 to 3791 bp, with the exception of 4423 bp for CFBP8072 (Fig. 6). A plot of the frequency of specific lengths of recombinant segments showed a normal distribution, and a small peak at 25 bp followed by a large peak at 300 bp (without the outlier CFBP8072), indicating two frequently recombined lengths across pauca subspecies. This bimodal distribution model is similar to what has been observed in other bacteria [49, 58], but the mechanism(s) behind it is still unclear.
Fig. 6.
Distribution of the length of recombinant fragments and the number of recombination events for each subspecies of X. fastidiosa. The number of recent recombination events identified by fastGEAR were plotted on the Y axis, with the lengths of recombination events plotted on the X axis using a logarithmic scale. The average length of a recombined fragment was around 1000 bp for all subspecies, with 10,000 bp being a higher limit for the recombined fragment length (subsp. morus as an exception). Higher incidence of recombination as well as the greater lengths of recombinant fragments were observed in subsp. morus (31,554 bp as the longest recombinant fragment) as well as in strains isolated from intercepted coffee plants (XFCO33, CFBP8072, and CFBP8073, marked in red, 19,280 bp as the longest recombinant fragment)
Subspecies fastidiosa has experienced relatively low levels of recombination events compared with other subspecies, except for an outlier strain, CFBP8073, which is highly recombinant (Fig. 6). The overall number of recombination events experienced by subsp. fastidiosa strains, except CFBP8073, were 235 in total, of which 87% of the events originated in subsp. multiplex, 11% originated in subsp. morus/sandyi, and 2% originated from outside species, with none from subsp. pauca. A highly recombinant strain CFBP8073 contains recombinant regions with 83% of the events originating in morus/sandyi subspecies. This is in contrast with other subsp. fastidiosa strains that contained recombination origins in subsp. multiplex. Interestingly, strain CFBP8073 was also intercepted in Europe in coffee plants shipped from Mexico [57]. Despite a comparatively low rate of recombination in subsp. fastidiosa, two frequently recombined segments included 75 and 2325 bp, indicating a similar pattern as observed above, including a normal distribution. The maximum length of a recombinant segment was 10,909 bp (Fig. 6). These lengths are comparable with the recombinant segment lengths observed in experimentally generated recombinants.
A population structure using hierBAPS indicated that subspecies multiplex is diverse (Fig. 4a), with at least five subclades. A sublineage within multiplex consisting of Dixon and Griffin-1 strains was minimally affected by recombination (Fig. 5, Table S4). However, sublineages containing susbp. multiplex ATCC35871, SY-VA, and BB08-1-containing subclades were significantly affected by recombination, with donor lineages being morus/sandyi (51% of recombination events) and fastidiosa (48% of recombination events) (Fig. 5, Table S4). The length distribution of recombinant events also followed a normal curve for subsp. multiplex, with the two most frequently recombined fragments of 649 and 2318 bp.
In addition to ancient recombination, subsp. morus and sandyi have also undergone recent recombination. It is interesting to note that these two subspecies have different origins of recombination, with morus showing a higher proportion of recombination originating in multiplex (56% in multiplex compared with 41% recombination originating in fastidiosa), and with sandyi showing a higher proportion of recombination originating in fastidiosa (64% of the events originating in fastidiosa and 20% originating in multiplex). Strain XFC033 belonging to subspecies sandyi can be referred to as a hybrid strain, with origins in pauca, fastidiosa, and multiplex. Interestingly, this strain was identified from symptomatic coffee leaves originating in Costa Rica and intercepted in northern Italy [59]. Although similar to other subspecies, fragments of ~1 kb were frequently recombined in morus/sandyi strains; subsp. morus contains the largest recombined fragment of 31.5 kb. The two subsp. also contained at least nine fragment sizes >10 kb, reiterating the highly recombinant nature of these subspecies. The widespread recombination found here is in concordance with X. fastidiosa being a γ proteobacteria, since this group is considered to have the “most chamaleonic-like evolutionary history” [60].
Phylogenetic relationships are influenced by recombination events between X. fastidiosa subspecies
To understand the influence of recombination on the overall topology of the X. fastidiosa phylogeny, we filtered core genome alignments to exclude the regions identified as recent recombinant events predicted by fastGEAR. The remaining nonrecombinant regions were then used to build a recombination-free phylogenetic tree. As shown in Fig. 4b, the overall topology of the tree appeared similar to that of the unfiltered tree, showing similar major clades and subclades. Recombinant filtering resulted in a strongly increased cohesiveness within subsp. fastidiosa, multiplex, and pauca. The most notable observation after recombinant filtering was the shift of subclades morus and sandyi with respect to their relatedness to fastidiosa and multiplex (Fig. 4b). When we compared branch-length distances from both trees, the average distance between morus and sandyi decreased (from 0.015 to 0.010), whereas that between morus and multiplex significantly increased (from 0.014 to 0.019) upon recombinant filtering. This led to a shift in the position of subsp. morus closer to sandyi in the recombinant-filtered phylogeny. These observations indicate that recombination plays an important role in X. fastidiosa evolution.
Majority of strains from intercepted plants are highly recombinant
The findings that (i) different degrees of ancestral and recent recombination have shaped the population structure of X. fastidiosa, and (ii) each lineage contains different degrees of recombination with origins in other subspecies, show that a shared ecological niche [55, 56] of these diverse lineages could have contributed to the emergence of the recombinant strains. Interestingly, the highly recombinant strains CFBP8072, CFBP8073, and XFC033, were strains isolated from intercepted infected coffee plants, with their origin in American countries. We further included two additional strains belonging to subspecies pauca, PD7202 (CFBP8495, belonging to Sequence Type 53, ST53) and PD7211 (CFBP8498, belonging to ST73) (Table S1), that were isolated from asymptomatic coffee plants intercepted in the Netherlands with their origin in Central America. Based on our findings, we hypothesized that these two strains are highly recombinant. Upon analyzing recombination using fastGEAR, we found PD7211 to contain in total 99 recent recombination events, with recombination regions shared with CFBP8072. On the contrary, strain PD7202 contained 49 recent recombination events, similar to other pauca strains, indicating that not all intercepted strains were highly recombinant (Fig. S4). Maximum likelihood phylogeny of the core genome indicated that PD7202 belongs to the same clade as subsp. pauca strains, containing CoDiRo and DeDonno, while PD7211 with a higher degree of recombination formed a distinct clade (Fig. S5). The presence of all subspecies has been identified in countries of the American continents in different hosts [55, 56]. It is likely that recombination events are rampant in the case of plants co-infected by multiple strains [53]. The presence of multiple subspecies in certain hosts or vectored by insects can foster recombination among these subspecies, resulting in emergence of recombinant strains. The importance of these recombinant regions in the epistatic contribution of each strain background [61] in terms of host range expansion, or aggressiveness of strains, remains to be investigated. Movement of these highly recombinant strains, that have acquired genomic regions from diverse subspecies, across the globe via shipment of asymptomatic plant material, is a major concern. Such introduction of novel variants to a new geographic area could further foster generation of new recombinants. Acquisition of novel traits by endemic pathogenic species could lead to increasing losses due to changes in host adaptation.
Recombinant regions shared among subspecies inform eco-evolutionary dynamics of X. fastidiosa–host interactions
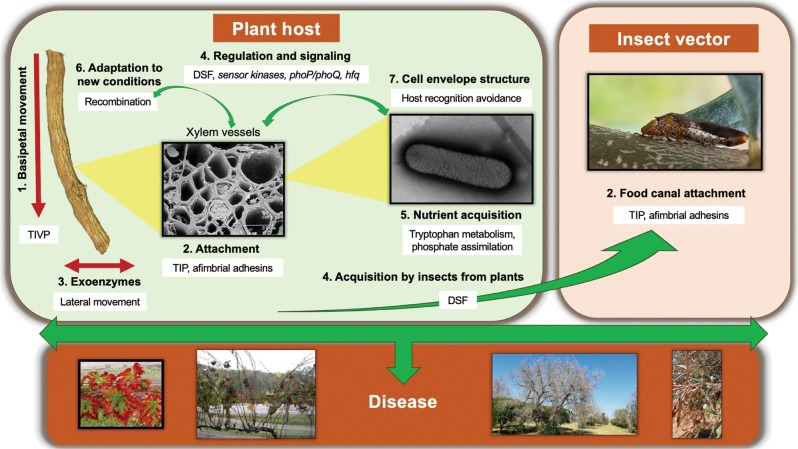
Functional significance of 1026 genes (~40% of the X. fastidiosa genome), that were identified in recombinant regions, was analyzed. We focused on recently recombined annotated loci that were included among the top 10% in terms of the frequency of recombination events (105 genes with 19 recombination events or higher, see Table S7 genes highlighted in yellow). The predicted functions of genes encoded in these loci can be considered to play roles in adjustment to and/or transition between the two ecological niches experienced by the pathogen during its life cycle, i.e., plant xylem and insect foregut [62] (Table 1, S6, S7, and Fig. 7). Thus, they have a major impact on the ecology of X. fastidiosa. Cell attachment [63] is a fundamental feature needed to live in physically restricted environments with strong liquid flow, including xylem and insect foregut [64, 65], while movement against flow (twitching motility) [66] and secretion of degrading enzymes [67] are crucial for colonization of xylem vessels. Although it is not completely understood how the plant host recognizes X. fastidiosa, lipopolysaccharide (LPS) [68] has been suggested as a key target, and other cell envelope structures could be involved in evading recognition by the host innate immune system [68]. Recombinant alleles for genes encoding utilization of specific nutrients, such as mineral elements [39] or amino acids, indicate adaptation of the pathogen to the nutritionally constrained environment. A great number of genes involved in regulatory and signaling cascades were detected as recombinants. Interestingly, genes involved in production (rpfF) and sensing (rpfC and rpfG) of the quorum-sensing molecule diffusible signal factor (DSF), which is critical for acquisition of X. fastidiosa by insects from plants [62], and regulates plant host specificity [69], were detected in high frequencies. Other regulatory genes detected here, have been shown to be important for virulence of X. fastidiosa; interestingly, phoP/phoQ [70] were shown to be needed for survival in plants.
Fig. 7.
Diagram illustrating the ecological role of genes found to be under recent recombination in Xylella fastidiosa. X. fastidiosa is only found in two very specific ecological niches: xylem vessels of host plants and food canal of xylem-feeding insects. Genes that were identified through our analysis as having the highest rate of recent intersubspecific recombination are listed in Table 1, S6, and S7. Based on the functional classification of these genes (Table 1), we placed these functions in the context of the ecology of X. fastidiosa during its life cycle. Briefly, in plant hosts (left panel), X. fastidiosa needs twitching motility [1] and exoenzymes [3] to colonize the xylem system inside plants, where nutrients are very limited [5]. While colonizing xylem vessels, they need attachment [2] to withstand constant liquid flow in the vascular system and to form biofilms. Proteins in the cell envelope are fundamental for recognition by the plant [7]. When colonizing a new host, X. fastidiosa most likely needs to acquire new genetic information for adaptation to new conditions [6]. Several regulatory cascades detected to be under recombination, have been implicated in regulating bacterial colonization and virulence [4]. DSF quorum-sensing molecule has a role in causing an increase in X. fastidiosa attachment and biofilm formation that is important to increase cell stickiness and therefore insect acquisition [4]. In the food canal of the xylem-feeding insect (right panel), attachment by TIP and afimbrial adhesins [2] is important to sustain population under high liquid flow. TIVP: type IV pili; TIP: type I pili; T2SS: type two secretion system; DSF: diffusible signal factor. Numbers of the highlighted X. fastidiosa traits from 1 to 7 correlate to the categories described in Table 1. For more information refer to the text and Table 1. Photo credits: plant host 1, 2: Harvey Hoch (Cornell University); insect vector 2: Phil Brannen (University of Georgia). All other pictures are credited to the authors
Notably, the vitamin B12 transporter BtuB was the single gene with the highest recombination events (165) across subspecies, and was among the most frequently recombined genes for each subspecies (Table S7). Interestingly, vitamin B12 regulates gene expression, enzyme activity, and abundance of microorganisms [71], and has been related to virulence [72] and biofilm formation [73] in bacterial human pathogens, but has not been studied in X. fastidiosa. The presence of a greater number of recombination events (collectively 2631 events) in genes encoding “hypothetical proteins”, brings attention to functionally uncharacterized proteins. Four genes encoding lipase, Ctpa-like serine proteases, and extracellular serine protease, were common recombinant loci in intrasubspecific recombination (in vitro recombinants), as well as in intersubspecific recombination in WT strains (Table 1). Genes identified to be highly recombinant across different subspecies and encoding functions not previously shown to be important in X. fastidiosa, are of great interest to further understand the ecological adaptation of X. fastidiosa to specific environmental niches. Recombination in tRNA modification genes and recombination machinery indicates adaptability to receive and successfully integrate foreign DNA (from different subspecies) in order to obtain adaptive alleles that provide overall fitness to this naturally competent pathogen [35, 74].
Conclusions
In this study, we assessed the effect of HR on the diversity of the bacterial plant pathogen X. fastidiosa. Through our comparison of both experimentally generated recombinants and WT strains, we conclude that recombination occurs randomly and with varying lengths of DNA recombination across the genome. While the extent of recombination in different subspecies varies among WT strains, all subspecies were affected by recent intersubspecific recombination at common loci, encoding important functions, such as host colonization, nutrient acquisition, and gene regulation/signaling. Four of such recombinant loci were also identified under intrasubspecific recombination in experimentally generated recombinants, further supporting the importance of in vitro studies to understand recombination potential of these naturally competent strains. These findings underline the prominent ability of this pathogen to evolve and adapt to new conditions by acquiring new DNA, which is facilitated by the lifestyle of this bacterium that is restricted to xylem vessels of plants or mouthparts of insect vectors [62], where it can easily encounter other bacteria in very confined spaces.
Supplementary information
Acknowledgements
This project was funded by Agriculture and Food Research Initiative competitive grant no. 2015-67014-23085 from the USDA National Institute of Food and Agriculture, and HATCH AAES (Alabama Agricultural Experiment Station) program provided to N.P and L.D, and partly supported by research grant OS 2015330 of the Ministry of Economic Affairs in the Netherlands. We thank the Alabama Supercomputing Authority for granting access to their high-performance computing platform.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Neha Potnis, Prem Kandel, Marcus Merfa
Supplementary information
The online version of this article (10.1038/s41396-019-0423-y) contains supplementary material, which is available to authorized users.
References
- 1.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–21. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 2.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2(1):376–81. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michod RE, Bernstein H, Nedelcu AM. Adaptive value of sex in microbial pathogens. Infect, Genet Evol: J Mol Epidemiol Evolut Genet Infect Dis. 2008;8:267–85. doi: 10.1016/j.meegid.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Guttman DS. Recombination and clonality in natural populations of Escherichia coli. Trends Ecol Evol. 1997;12:16–22. doi: 10.1016/s0169-5347(96)10057-4. [DOI] [PubMed] [Google Scholar]
- 5.Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–90. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- 6.Nelson K, Selander RK. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci USA. 1994;91:10227–31. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan S, Liu H, Mohr TJ, Jenrette J, Chiodini R, Zaccardelli M, et al. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a ver, atypical tomato strain. Appl Environ Microbiol. 2008;74:3171–81. doi: 10.1128/AEM.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baltrus DA, Guillemin K, Phillips PC, Travisano M. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution. 2008;62:39–49. doi: 10.1111/j.1558-5646.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- 9.Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98:182–7. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spratt BG, Bowler LD, Zhang QY, Zhou J, Smith JM. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol. 1992;34:115–25. doi: 10.1007/BF00182388. [DOI] [PubMed] [Google Scholar]
- 11.Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 12.Freel KC, Millán-Aguiñaga N, Jensen PR. Multilocus sequence typing reveals evidence of homologous recombination linked to antibiotic resistance in the genus Salinispora. Appl Environ Microbiol. 2013;79:5997–6005. doi: 10.1128/AEM.00880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong Y, Ma JH, Warren K, Tsang RS, Low DE, Jamieson FB, et al. Homologous recombination drives both sequence diversity and gene content variation in Neisseria meningitidis. Genome Biol Evol. 2013;5:1611–27. doi: 10.1093/gbe/evt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timilsina S, Jibrin MO, Potnis N, Minsavage GV, Kebede M, Schwartz A, et al. Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl Environ Microbiol. 2015;81:1520–9. doi: 10.1128/AEM.03000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nunney L, Hopkins DL, Morano LD, Russell SE, Stouthamer R. Intersubspecific recombination in Xylella fastidiosa strains native to the United States: infection of novel hosts associated with an unsuccessful invasion. Appl Environ Microbiol. 2014;80:1159–69. doi: 10.1128/AEM.02920-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker JK, Havird JC, De La, Fuente L. Differentiation of Xylella fastidiosa strains via multilocus sequence analysis of environmentally mediated genes (MLSA-E) Appl Environ Microbiol. 2012;78:1385–96. doi: 10.1128/AEM.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D. Efficient inference of recombination hot regions in bacterial genomes. Mo Biol Evol. 2014;31:1593–605. doi: 10.1093/molbev/msu082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida RP, Nunney L. How do plant diseases caused by Xylella fastidiosa emerge? Plant Dis. 2015;99:1457–67. doi: 10.1094/PDIS-02-15-0159-FE. [DOI] [PubMed] [Google Scholar]
- 21.Saponari M, Boscia D, Nigro F, Martelli GP. Identification of DNA sequences related to Xylella fastidiosa in Oleander, Almond and Olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy) J Plant Pathol. 2013;95:668. [Google Scholar]
- 22.Strona G, Carstens CJ, Beck PS. Network analysis reveals why Xylella fastidiosa will persist in Europe. Sci Rep. 2017;7:71. doi: 10.1038/s41598-017-00077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su CC, Deng WL, Jan FJ, Chang CJ, Huang H, Shih HT, et al. Xylella taiwanensis sp. nov., causing pear leaf scorch disease. Int J Syst Evolut Microbiol. 2016;66:4766–71. doi: 10.1099/ijsem.0.001426. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins DL, Purcell AH. Xylella fastidiosa: cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 2002;86:1056–66. doi: 10.1094/PDIS.2002.86.10.1056. [DOI] [PubMed] [Google Scholar]
- 25.Marcelletti S, Scortichini M. Genome-wide comparison and taxonomic relatedness of multiple Xylella fastidiosa strains reveal the occurrence of three subspecies and a new Xylella species. Arch Microbiol. 2016;198:803–12. doi: 10.1007/s00203-016-1245-1. [DOI] [PubMed] [Google Scholar]
- 26.Schaad NW, Postnikova E, Lacy G, Fatmi M, Chang CJ. Xylella fastidiosa subspecies: X. fastidiosa subsp piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst Appl Microbiol. 2004;27:763. doi: 10.1078/0723-2020-00263. [DOI] [PubMed] [Google Scholar]
- 27.Schaad NW, Postnikova E, Lacy G, Fatmi MB, Chang CJ. Xylella fastidiosa subspecies: X-fastidiosa subsp piercei, subsp nov., X-fastidiosa subsp multiplex subsp. nov., and X-fastidiosa subsp pauca subsp nov. Syst Appl Microbiol. 2004;27:290–300. doi: 10.1078/0723-2020-00263. [DOI] [PubMed] [Google Scholar]
- 28.Nunney L, Schuenzel EL, Scally M, Bromley RE, Stouthamer R. Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl Environ Microbiol. 2014;80:3025–33. doi: 10.1128/AEM.04112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scally M, Schuenzel EL, Stouthamer R, Nunney L. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl Environ Microbiol. 2005;71:8491–9. doi: 10.1128/AEM.71.12.8491-8499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.EFSA. Update of the Xylella spp. host plant database. EFSA Journal. 2018;16:5408. [DOI] [PMC free article] [PubMed]
- 31.Nunney L, Yuan X, Bromley RE, Stouthamer R. Detecting genetic introgression: high levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Appl Environ Microbiol. 2012;78:4702–14. doi: 10.1128/AEM.01126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida RP, Nascimento FE, Chau J, Prado SS, Tsai CW, Lopes SA, et al. Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Appl Environ Microbiol. 2008;74:3690–701. doi: 10.1128/AEM.02388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coletta-Filho H, Francisco C, Lopes J, Muller C, Almeida R. Homologous recombination and Xylella fastidiosa host-pathogen associations in South America. Phytopathology. 2017;107:305–312. doi: 10.1094/PHYTO-09-16-0321-R. [DOI] [PubMed] [Google Scholar]
- 34.Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astua M, Moreira L, et al. Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce’s disease of grapevine in the U.S. PLoS One. 2010;5:e15488. doi: 10.1371/journal.pone.0015488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kung SH, Almeida RP. Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl Environ Microbiol. 2011;77:5278–84. doi: 10.1128/AEM.00730-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandel PP, Lopez SM, Almeida RP, De La Fuente L. Natural competence of Xylella fastidiosa occurs at a high frequency inside microfluidic chambers mimicking the bacterium’s natural habitats. Appl Environ Microbiol. 2016;82:5269–77. doi: 10.1128/AEM.01412-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kandel PP, Almeida RPP, Cobine PA, De La, Fuente L. Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex. Mol Plant-Microbe Interact. 2017;30:589–600. doi: 10.1094/MPMI-02-17-0053-R. [DOI] [PubMed] [Google Scholar]
- 38.Kandel PP, Chen HY, De La Fuente L. A short protocol for gene knockout and complementation in Xylella fastidiosa shows that one of the type IV pilin paralogs (PD1926) is needed for twitching while another (PD1924) affects pilus number and location. Appl Environ Microbiol. 2018;84:e01167–18. [DOI] [PMC free article] [PubMed]
- 39.Oliver JE, Sefick SA, Parker JK, Arnold T, Cobine PA, De La Fuente L. Ionome changes in Xylella fastidiosa-infected Nicotiana tabacum correlate with virulence and discriminate between subspecies of bacterial isolates. Mol Plant-Microbe Interact. 2014;27:1048–58. doi: 10.1094/MPMI-05-14-0151-R. [DOI] [PubMed] [Google Scholar]
- 40.Bergsma-Vlami M, van de Bilt JLJ, Tjou-Tam-Sin NNA, Helderman CM, Gorkink-Smits P, Landman NM, et al. Assessment of the genetic diversity of Xylella fastidiosa in imported ornamental Coffea arabica plants. Plant Pathol. 2017;66:1065–74. [Google Scholar]
- 41.Davis MJ, French WJ, Schaad NW. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr Microbiol. 1981;6:309–14. [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darling Aaron E., Mau Bob, Perna Nicole T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 2013;30:1224–8. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol. 2017;34:1167–82. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bubendorfer S, Krebes J, Yang I, Hage E, Schulz TF, Bahlawane C, et al. Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat Commun. 2016;7:11995-. doi: 10.1038/ncomms11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Chakrabarty PK, Fleites LA, Rayside PA, Hopkins DL, Gabriel DW. Three new Pierce’s disease pathogenicity effectors identified using Xylella fastidiosa biocontrol strain EB92-1. PLoS One. 2015;10:e0133796. doi: 10.1371/journal.pone.0133796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouran H, Gillespie H, Nascimento R, Chakraborty S, Zaini PA, Jacobson A, et al. The secreted protease PrtA controls cell. Growth, Biofilm Form Pathog Xylella Fasti Sci Rep. 2016;6:31098. doi: 10.1038/srep31098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carroll RK, Rivera FE, Cavaco CK, Johnson GM, Martin D, Shaw LN. The lone S41 family C-terminal processing protease in Staphylococcus aureus is localized to the cell wall and contributes to virulence. Microbiol-Sgm. 2014;160:1737–48. doi: 10.1099/mic.0.079798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denance N, Legendre B, Briand M, Olivier V, de Boisseson C, Poliakoff F, et al. Several subspecies and sequence types are associated with the emergence of Xylella fastidiosa in natural settings in France. Plant Pathol. 2017;66:1054–64. [Google Scholar]
- 54.Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–80. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coletta HD, Francisco CS, Lopes JRS, Muller C, Almeida RPP. Homologous recombination and Xylella fastidiosa host-pathogen associations in South America. Phytopathology. 2017;107:305–12. doi: 10.1094/PHYTO-09-16-0321-R. [DOI] [PubMed] [Google Scholar]
- 56.Nunney Leonard, Ortiz Beatriz, Russell Stephanie A., Ruiz Sánchez Rebeca, Stouthamer Richard. The Complex Biogeography of the Plant Pathogen Xylella fastidiosa: Genetic Evidence of Introductions and Subspecific Introgression in Central America. PLoS ONE. 2014;9(11):e112463. doi: 10.1371/journal.pone.0112463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacques MA, Denance N, Legendre B, Morel E, Briand M, Mississipi S, et al. New coffee plant-infecting Xylella fastidiosa variants derived via homologous recombination. Appl Environ Microbiol. 2016;82:1556–68. doi: 10.1128/AEM.03299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mostowy Rafal, Croucher Nicholas J., Hanage William P., Harris Simon R., Bentley Stephen, Fraser Christophe. Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution. PLoS Genetics. 2014;10(5):e1004300. doi: 10.1371/journal.pgen.1004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giampetruzzi A, Loconsole G, Boscia D, Calzolari A, Chiumenti M, Martelli GP, et al. Draft genome sequence of CO33, a coffee-infecting isolate of Xylella fastidiosa. Genome Announcements. 2015;3:e01472-15. [DOI] [PMC free article] [PubMed]
- 60.Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–74. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnold BJ, Gutmann MU, Grad YH, Sheppard SK, Corander J, Lipsitch M, et al. Weak epistasis may drive adaptation in recombining bacteria. Genetics. 2018;208:1247–60. doi: 10.1534/genetics.117.300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatterjee S, Almeida RPP, Lindow S. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol. 2008;46:243–71. doi: 10.1146/annurev.phyto.45.062806.094342. [DOI] [PubMed] [Google Scholar]
- 63.De La Fuente L, Montanes E, Meng YZ, Li YX, Burr TJ, Hoch HC, et al. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl Environ Microbiol. 2007;73:2690–6. doi: 10.1128/AEM.02649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feil H, Feil WS, Lindow SE. Contribution of fimbrial and afimbrial adhesins of Xylella fastidiosa to attachment to surfaces and virulence to grape. Phytopathology. 2007;97:318–24. doi: 10.1094/PHYTO-97-3-0318. [DOI] [PubMed] [Google Scholar]
- 65.Killiny N, Almeida RPP. Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Appl Environ Microbiol. 2014;80:420–6. doi: 10.1128/AEM.03156-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng YZ, Li YX, Galvani CD, Hao GX, Turner JN, Burr TJ, et al. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187:5560–7. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roper MC, Greve LC, Warren JG, Labavitch JM, Kirkpatrick BC. Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol Plant-Microbe Interact. 2007;20:411–9. doi: 10.1094/MPMI-20-4-0411. [DOI] [PubMed] [Google Scholar]
- 68.Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P, et al. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat Commun. 2018;9:390. doi: 10.1038/s41467-018-02861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Killiny N, Almeida RPP. Gene regulation mediates host specificity of a bacterial pathogen. Environ Microbiol Rep. 2011;3:791–7. doi: 10.1111/j.1758-2229.2011.00288.x. [DOI] [PubMed] [Google Scholar]
- 70.Pierce BK, Kirkpatrick BC. The PhoP/Q two-component regulatory system is essential for Xylella fastidiosa survival in Vitis vinifera grapevines. Physiol Mol Plant Pathol. 2015;89:55–61. [Google Scholar]
- 71.Rosnow JJ, Hwang S, Killinger BJ, Kim YM, Moore RJ, Lindemann SR, et al. A cobalamin activity-based probe enables microbial cell growth and finds new cobalamin-protein interactions across domains. Appl Environ Microbiol. 2018;84:e00955–18. [DOI] [PMC free article] [PubMed]
- 72.Cordonnier C, Le Bihan G, Emond-Rheault JG, Garrivier A, Harel J, Jubelin G. Vitamin B12 uptake by the gut commensal bacteria Bacteroides thetaiotaomicron limits the production of shiga toxin by enterohemorrhagic Escherichia coli. Toxins. 2016;8:14. doi: 10.3390/toxins8010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee KM, Go J, Yoon MY, Park Y, Kim SC, Yong DE, et al. Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect Immun. 2012;80:1639–49. doi: 10.1128/IAI.06161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bjork GR, Ericson JU, Gustafsson CED, Hagervall TG, Jonsson YH, Wikstrom PM. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–87. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- 75.Newman KL, Almeida RP, Purcell AH, Lindow SE. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl Environ Microbiol. 2003;69:7319–27. doi: 10.1128/AEM.69.12.7319-7327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matsumoto A, Young GM, Igo MM. Chromosome-based genetic complementation system for Xylella fastidiosa. Appl Environ Microbiol. 2009;75:1679–87. doi: 10.1128/AEM.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Capra Emily J., Laub Michael T. Evolution of Two-Component Signal Transduction Systems. Annual Review of Microbiology. 2012;66(1):325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kavita K, de Mets F, Gottesman S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol. 2018;42:53–61. doi: 10.1016/j.mib.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 80.De La Fuente L, Parker JK, van Santen E, Oliver JE, Brannen P, Granger S, et al. The bacterial pathogen Xylella fastidiosa affects the leaf ionome of plant hosts during infection. PLoS One. 2013;8:e62945. doi: 10.1371/journal.pone.0062945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rock CD. Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development? Trends Plant Sci. 2013;18:601–10. doi: 10.1016/j.tplants.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Antolín-Llovera Meritxell, Ried Martina K., Binder Andreas, Parniske Martin. Receptor Kinase Signaling Pathways in Plant-Microbe Interactions. Annual Review of Phytopathology. 2012;50(1):451–473. doi: 10.1146/annurev-phyto-081211-173002. [DOI] [PubMed] [Google Scholar]
- 83.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–36. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu Z, Dong HJ, Ma JC, Yu YH, Li KH, Guo QQ, et al. Novel Xanthomonas campestris long-chain-specific 3-oxoacyl-acyl carrier protein reductase involved in diffusible signal factor synthesis. Mbio. 2018;9:e00596–18. [DOI] [PMC free article] [PubMed]
- 85.Mao YH, Li F, Ma JC, Hu Z, Wang HH. Sinorhizobium meliloti functionally replaces 3-oxoacyl-acyl carrier protein reductase (FabG) by overexpressing NodG during fatty acid synthesis. Mol Plant-Microbe Interact. 2016;29:458–67. doi: 10.1094/MPMI-07-15-0148-R. [DOI] [PubMed] [Google Scholar]
- 86.Filip’echeva YA, Shelud’ko AV, Prilipov AG, Burygin GL, Telesheva EM, Yevstigneyeva SS, et al. Plasmid AZOBR_p1-borne fabG gene for putative 3-oxoacyl-acyl-carrier protein reductase is essential for proper assembly and work of the dual flagellar system in the alphaproteobacterium Azospirillum brasilense Sp245. Can J Microbiol. 2018;64:107–18. doi: 10.1139/cjm-2017-0561. [DOI] [PubMed] [Google Scholar]
- 87.Selkrig J, Belousoff MJ, Headey SJ, Heinz E, Shiota T, Shen HH, et al. Conserved features in TamA enable interaction with TamB to drive the activity of the translocation and assembly module. Sci Rep. 2015;5:12905. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.