Abstract
Symbiotic associations impact and are impacted by their surrounding ecosystem. The association between Burkholderia bacteria and the soil amoeba Dictyostelium discoideum is a tractable model to unravel the biology underlying symbiont-endowed phenotypes and their impacts. Several Burkholderia species stably associate with D. discoideum and typically reduce host fitness in food-rich environments while increasing fitness in food-scarce environments. Burkholderia symbionts are themselves inedible to their hosts but induce co-infections with secondary bacteria that can serve as a food source. Thus, Burkholderia hosts are “farmers” that carry food bacteria to new environments, providing a benefit when food is scarce. We examined the ability of specific Burkholderia genotypes to induce secondary co-infections and assessed host fitness under a range of co-infection conditions and environmental contexts. Although all Burkholderia symbionts intracellularly infected Dictyostelium, we found that co-infections are predominantly extracellular, suggesting that farming benefits are derived from extracellular infection of host structures. Furthermore, levels of secondary infection are linked to conditional host fitness; B. agricolaris infected hosts have the highest level of co-infection and have the highest fitness in food-scarce environments. This study illuminates the phenomenon of co-infection induction across Dictyostelium associated Burkholderia species and exemplifies the contextual complexity of these associations.
Subject terms: Symbiosis, Microbiome
Introduction
Symbiotic interactions can alter the fitness and evolutionary trajectory of both partners [1–4]. Clearly detrimental or mutualistic associations have been investigated for obvious reasons: to eliminate infectious disease, boost health, and restore ecosystems. However, many symbiotic associations evade simple characterization and related mechanisms can underlie opposing outcomes [5, 6]. Invasion and replication strategies employed by mutualists and pathogens often resemble each other, while genotypes and external factors modify subsequent outcomes [7]. Genotype pairing determines the outcome of plant-mycorrhizae interactions [8] and amplification of a genomic region in a normally beneficial Wolbachia symbiont leads to over-replication at the hosts expense [9]. Light mediates pathogenicity of a fungal plant endosymbiont [10], temperature affects reproductive fitness of aphids hosting Buchnera [11], and parasitoid pressure determines whether Hamiltonella defensa is beneficial to host aphids [12]. These examples demonstrate that even canonically beneficial or detrimental associations may produce alternative effects in alternative contexts [4, 13–17].
Eukaryotic microbes, such as amoebae, are attractive models for exploring eukaryote-prokaryote interactions. Amoebae are ubiquitous and efficient phagocytic predators of bacterial prey, making them important shapers of the microbial community [18]. This pressures prey microbes to evolve virulence strategies that enable evasion of phagocytosis or subsequent digestion [19]. Amoebae are thereby potential training grounds and environmental reservoirs for bacterial pathogens. Amoebae phagocytosis also enables bacteria to gain easy access to an attractive intracellular niche, bypassing the requirement for evolving specialized cell-entry mechanisms. After invasion, bacteria can be retained in an environmentally resistant cyst or spore [20]. A number of bacterial pathogens, such as Legionellae pneumophila and others [21, 22], are harbored in different species of amoebae and there is a growing list of recently identified amoebae symbionts [23, 24].
Dictyostelium discoideum has been appreciated as a model host for studying bacterial pathogens for some time [25–27]. Recently, work with wild isolates has emphasized its power for exploring naturally occurring microbial symbioses [28, 29]. As a social amoeba Dictyostelium exhibits a unique life cycle, transitioning between single- and multi-cellular forms. Under favorable conditions, it lives as a unicellular amoeba, consuming bacteria and dividing by binary fission. When bacterial food is depleted, amoebae secrete cAMP, which triggers the transition to multi-cellularity. During this phase, amoebae aggregate to form a multicellular slug that seeks out a location for fruiting body formation (such as the soil surface). Fruiting bodies are comprised of a spherical sorus containing hardy spore cells resting atop a long stalk of dead cells. This positioning of spore cells likely aids in their dispersal [30]. Once dispersed, spores germinate, and the cycle continues.
D. discoideum grown with a variety of bacterial food traditionally form germ-free sori, clearing residual bacteria from the multicellular state during development. Microbial clearance is aided by immune-like sentinel cells, which engulf debris and slough off the migrating slug [31, 32]. However, approximately one third of wild D. discoideum isolates are naturally and stably colonized by Burkholderia bacteria [33, 34]. Burkholderia can be easily eliminated from host populations with antibiotic treatment and new associations can be readily initiated through co-culture. These Burkholderia symbionts establish intracellular infections, which persist through host development, resulting in sori containing both extracellular and intracellular bacteria [34]. Burkholderia symbionts thereby remain associated with host populations during spore dispersal and can be acquired through vertical and horizontal transmission routes. This mixed mode of transmission has interesting implications for the fitness consequences and evolutionary trajectory of the symbiosis.
Burkholderia symbionts of D. discoideum are members of the plant beneficial environmental group within the Burkholderia genus [35]. Symbiont strains are genetically diverse, belonging to three species arising from two independent lineages: B. agricolaris, B. hayleyella, and B. bonniea [36]. Burkholderia differentially impacts host fitness according to host-symbiont genotype combinations and environmental context [33, 34, 37]. Symbionts generally reduce host fitness in food-rich conditions but enhance fitness in food-scarce environments [33, 34, 37]. Fitness benefits are attributed to retention of bacteria within host spores, allowing them to reseed new environments with bacterial food. This trait is called farming and Burkholderia infected hosts are thus referred to as “farmers”. Burkholderia symbionts themselves are poor food sources for their hosts [33, 34]. However, Burkholderia infection appears to increase host susceptibility to secondary bacterial infection, promoting the formation of a mini-microbiome. It is these secondary bacteria that can serve as an amoebae food source and thereby provide the farming benefit.
Given the importance of secondary infection in farming, we sought to explore the underlying dynamics of this interaction. While a commonly used lab food strain, K. pneumoniae, can be identified as an occasional co-infecting partner, co-infection dynamics might vary depending on particular bacterial pairings [34]. Thus, food bacteria identity is an important environmental context that may affect outcomes. Furthermore, the three different Burkholderia symbiont species have divergent evolutionary histories of association with D. discoideum. While they have converged on the farming phenotype, the effects and underlying mechanisms of infection may differ across Burkholderia species [38].
Here, we reveal the density and location of secondary co-infections induced by each Burkholderia species with a collection of secondary bacteria. Next, we clarify the downstream benefits of Burkholderia infection in varied food availability contexts and link these to symbiont genotype and co-infection induction. Specifically, we analyzed co-infection patterns and host outcomes with a variety of secondary bacteria including: laboratory food Klebsiella pneumoniae, Rhizobium, and Serratia isolates that naturally co-occur with D. discoideum, and Agrobacterium tumefaciens and Pseudomonas aeruginosa as pathogens that D. discoideum may encounter in nature. We found that all Burkholderia symbionts induce some degree of secondary infection in host sori but the density and location of secondary infections is dependent on Burkholderia genotype and secondary bacterial identity. Contrary to previous inference, secondary infections are predominantly extracellular with intracellular co-infections only readily visualized in B. agricolaris infected spores. Overall, B. agricolaris induces the highest density of combined co-infection resulting in a higher fitness benefits in food-scarce environments. B. bonniea and B. hayleyella induce lower levels of secondary co-infection but only B. bonniea provides significant host benefits under specific dispersal conditions. This work illuminates the interplay between symbiont genotypes and environmental context in mediating the expression and consequences of novel symbiont-endowed phenotypes.
Materials and methods
Bacterial strains and culturing
All bacterial strains are described in Supplementary table 1. Briefly, Burkholderia were isolated from D. discoideum stocks. Rhizobium and Serratia were isolated from Dictyostelium grown directly from soil. Pseudomonas aeruginosa PAO1-GFP was provided by R. Fred Inglis [39]. Agrobacterium tumefaciens was provided by Daniel Gage. Klebsiella pneumoniae is the Dictyostelium food strain. We used GFP labeled secondary bacteria for all experiments, with the exception of K. pneumoniae, which is unlabeled when mixed with other bacteria. We grew all bacteria on SM/5 medium (Formedium: 2 g Peptone, 0.2 g yeast extract, 2 g glucose, 1.9 g KH2PO4, 1.3 g K2HPO4.3H20, 0.49 g MgO4.anhydrous, 17 g agar/l) at room temperature. To prepare bacteria for culturing Dictyostelium, we suspended bacterial colonies from SM/5 medium into KK2 (2.2 g KH2PO4 monobasic and 0.7 g K2HPO4 dibasic/l) and set to an OD600nm of 1.5. For K. pneumoniae/secondary bacterial mixtures, we combined bacterial suspensions equally by volume. For Burkholderia infections, we added 5% by volume Burkholderia-RFP to bacterial mixtures.
Construction of fluorescent bacterial strains
We generated RFP labeled Burkholderia by triparental mating with E. coli helper E1354 (pTNS3-asdEc) and donor E2072 (pmini-Tn7-gat-P1-rfp) and confirmed identity of RFP conjugants using a Burkholderia specific PCR as previously described [34, 40, 41]. We GFP labeled Rhizobium, Serratia, A. tumefaciens, and K. pneumoniae through triparental mating with E. coli donor WM3064 (pmini-Tn7-KS-GFP) and helper E1354 (pUXBF13) as previously described [42] and confirmed identity of GFP positive conjugants through 16S rRNA gene sequencing.
Dictyostelium culture conditions
We used D. discoideum clone QS864 (naturally symbiont free) for all experiments. Cultures were initiated by plating spores on SM/5 medium with K. pneumoniae and incubating under lights at room temperature until fruiting bodies developed (4–7 days). For experiments, 105 spores were harvested from developed sori and plated with 200 μL of the appropriate bacterial mixtures. For all experiments (unless otherwise indicated) we analyzed sori 5 days after plating.
For co-infection assays we plated uninfected spores on bacterial mixtures with Burkholderia, uninfected controls were plated without Burkholderia. To compare spore productivity under food variable conditions, we harvested sori from indicated co-infection conditions and plated 105 spores onto SM/5 with K. pneumoniae at an OD600 of 1.5 for food-rich conditions or with heat-killed (30 min at 80 °C) K. pneumoniae at an OD600 of 6 for food-scarce conditions.
Spore production assays
To harvest total spores, we flooded each plate with 5–10 mL KK2 + 0.1% Nonidet P-40 alternative and collected the entire surface contents into 15-mL Falcon tubes. We then diluted samples in KK2 and counted spores on a hemocytometer. At least five replicates were analyzed for each treatment.
Confocal microscopy
We imaged spores by staining with 1% calcofluor in KK2, placing on glass bottom culture dishes (Electron Microscopy Sciences) and overlaying with 2% agarose. We imaged samples on an Olympus Fluoview FV1000 confocal microscope using Plan Apo Oil 1.4NA 60X objective. Z-sections were taken every 0.5 microns at 1024 resolution. Calcofluor was visualized with DAPI, GFP with FITC, and RFP with Cy3 then pseudocolored grey, green, and red, respectively. We imaged at least three individual replicates and counted more than 30 spores for each.
Colony-forming unit quantification
To quantify secondary bacteria, we harvested sori grown from the indicated co-culture conditions from 6- or 14-day incubations. We suspended individual sori in KK2 + 0.05% Nonidet P-40 alternative, counted spores on a hemocytometer, plated serial dilutions on SM/5 medium and incubated plates at room temperature until colony formation (~2 days), and counted GFP colonies using a safe-light imaging system. We performed three or more independent replicates for each treatment.
Streak test
Our streak test assay was initiated from the indicated co-culture conditions by touching individual sori with sterile pipette tips and transferring them to SM/5 plates along a ~1 inch streak. We incubated plates face up under lights at room temperature and examined them 5 days (or 2 weeks) after streaking. We determined the percentage of streaks with bacterial growth, percentage of bacterial positive streaks with fruiting bodies, and number of fruiting bodies in positive streaks. Streaks were photographed on a Cannon Eos7D with a macro-lens. Six sori were streaked for each replicate for at least four individual replicates per condition.
Statistical analysis
We analyzed all data using R (version 3.3.1). For normally distributed data we determined significance using a standard one-way analysis of variance (ANOVA) and a post hoc Tukey HSD test. For non-normally distributed data we performed a Kruskall-wallis test and post hoc analysis with a Dunn test using the dunnTest function in the FSA package [43]. We used Burkholderia status as fixed effects for all conditions.
Results
Burkholderia and secondary bacterial combinations
To investigate the induction of secondary infection by Burkholderia symbionts, we cultured an uninfected natural isolate of D. discoideum with different Burkholderia-RFP and secondary bacteria-GFP combinations. We began with three Burkholderia strains: Ba.70, Bh.11, and Bb.859, each representing one D. discoideum symbiont species B. agricolaris, B. hayleyella, and B. bonniea (Supplementary Fig. 1) [36]. Secondary bacteria consisted of a Klebsiella pneumoniae strain, soil isolated Rhizobium and Serratia, and lab Agrobacterium tumefaciens and Pseudomonas aeruginosa strains. We chose these representatives because: (1) K. pneumoniae is a widely used lab food source for D. discoideum and serves as a starting point for experimental conditions while providing context to other Dictyostelium research. (2) The Rhizobium and Serratia strains where cultured from D. discoideum fruiting bodies that had been plated directly from soil and are thereby ecologically relevant potential co-associates [44]. (3) A. tumefaciens, in addition to its use in plant molecular biology, is an important soil dwelling plant pathogen. As such, amoebae may interact with A. tumefaciens in the environment and this could subsequently impact the surrounding ecosystem. (4) P. aeruginosa is an important opportunistic human pathogen whose association with other bacterial species in biofilms (such as pathogenic Burkholderia cenocepacia) influences infection outcomes [45, 46]. Adding Pseudomonas to the Burkholderia–Dictyostelium system provides a novel approach to explore microbial interactions and virulence.
Host outcomes differ according to Burkholderia and secondary bacteria conditions
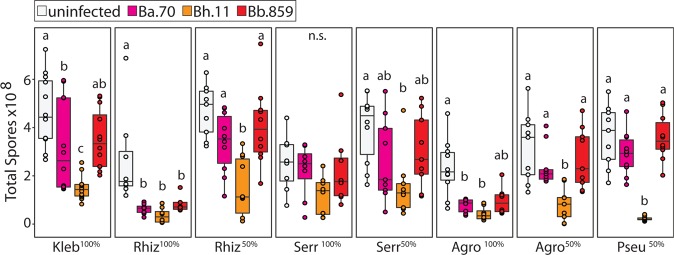
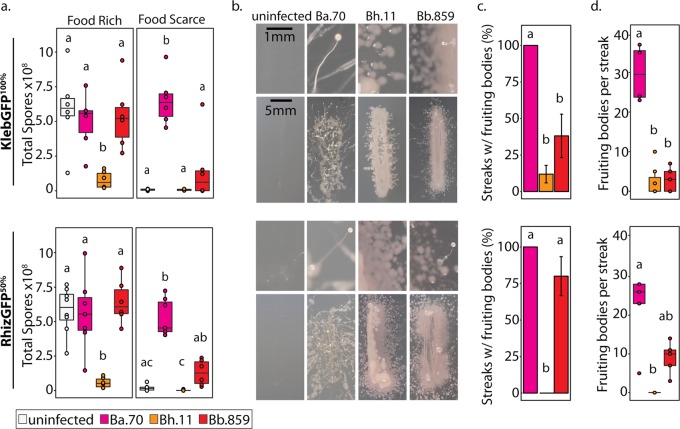
First, we examined host fitness when amoebae were co-cultured with Burkholderia and secondary bacteria. We determined total spore productivity of host amoebae after one social cycle on each labeled secondary bacterium, either alone or in a 50% mixture with K. pneumoniae. Five percent by volume of Burkholderia-RFP was included to establish infections (Fig. 1). D. discoideum was unable to develop on any conditions where P. aeruginosa was the only food source suggesting that this strain was toxic and/or inedible for amoebae. All other conditions supported fruiting body development, but spore productivity varied across conditions (Fig. 1). In line with previous studies, Burkholderia species differentially impact spore productivity on K. pneumoniae [34, 37]. Typically, B. hayleyella was the most detrimental for host fitness with B. agricolaris and B. bonniea being neutral or moderately detrimental. However, these patterns and the degree by which symbiont altered host fitness varied across culture conditions (Fig. 1 and Table 1). These results highlight the variability of fitness outcomes caused by distinct Burkholderia symbionts and suggest that surrounding bacterial communities also impact fitness outcomes.
Fig. 1.
Culture Conditions Modify the Impact of Burkholderia Infections on Host Fitness. Total spore productivity for D. discoideum after one round of development on the indicated GFP labeled bacterial species (Kleb = K. pneumoniae, Rhiz = Rhizobium, Serr = Serratia, Agro = A. tumefaciens, and Pseu = P. aeruginosa. Fifty percentage cultures are mixed with 50% unlabeled K. pneumoniae by volume). D. discoideum failed to develop on 100% Pseudomonas conditions and is thereby not plotted. Fuschia, orange, and red boxes indicate cultures wherein Burkholderia infections are initiated by inclusion of 5% B. agricolaris.70-RFP, B. hayleyella.11-RFP, and B. bonniea.859-RFP, respectively. Points within boxes indicate individual replicates. Letters indicate post hoc significance within panels
Table 1.
Burkholderia infections significantly alter spore productivity in most (but not all) bacterial culture conditions. Statistical analysis of spore fitness from Fig. 1
| Condition | χ 2 | p Value |
|---|---|---|
| K. pneumoniae-GFP100% | 23.537 | >0.001 |
| Rhizobium-GFP100% | 20.81 | >0.001 |
| Rhizobium-GFP50% | 18.759 | >0.001 |
| Serratia-GFP100% | 6.411 | 0.09 |
| Serratia-GFP50% | 11.292 | 0.01 |
| Agrobacterium-GFP100% | 18.76 | >0.001 |
| Agrobacterium-GFP50% | 16.267 | >0.001 |
| Pseudomonas-GFP50% | 23.36 | >0.001 |
All df values = 3
Intracellular co-infection is rare and depends on Burkholderia and secondary bacterial combinations
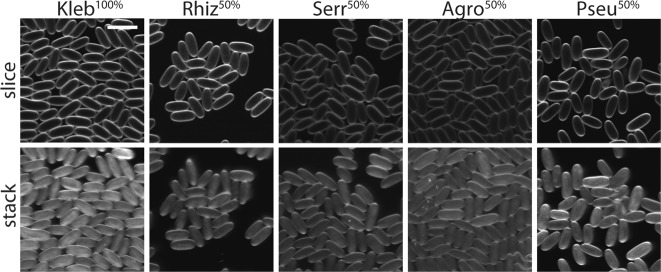
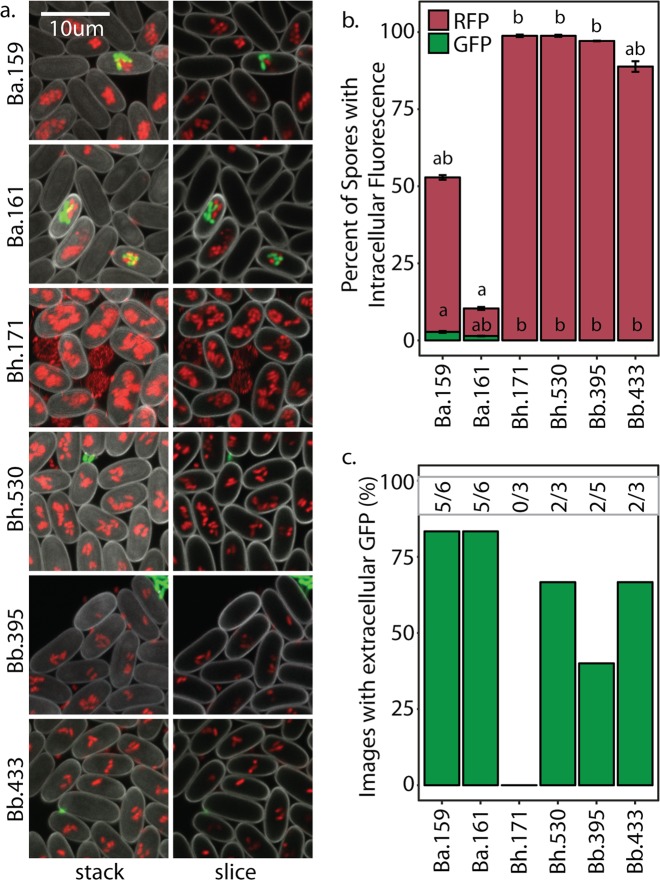
To investigate induction of secondary infection we imaged D. discoideum sori after development on Burkholderia and secondary bacteria. We used 50/50 K. pneumoniae/secondary bacteria-GFP conditions as they resulted in better amoebae development than secondary bacteria-only conditions. We also imaged sori grown from K. pneumoniae-GFP. Importantly, we do not detect any secondary bacteria in sori in the absence of Burkholderia (Fig. 2). Thus, these bacteria are not capable of infecting D. discoideum on their own. In contrast, we can detect secondary-GFP cells in sori from amoebae co-exposed to Burkholderia (Fig. 3). To determine their prevalence in host spore populations, we quantified the percent of spores intracellularly infected with Burkholderia-RFP and with secondary bacteria-GFP. First, the percent of spores infected with each Burkholderia species significantly differs (χ2 = 44.02, df = 2, p < 0.001). In aggregate, B. hayleyella infects the most (89.2%), B. bonniea infects an intermediate (68.6%), and B. agricolaris infects the fewest (33%) percent of spores. However, we only readily observe intracellular secondary co-infections in B. agricolaris host spores (average of 5.5% across conditions). We very rarely observe intracellular secondary bacteria-GFP in B. hayleyella and B. bonniea hosts (0.01 and 0.05%, respectively) (Fig. 3). We did not observe intracellular secondary bacteria-GFP in the absence of intracellular Burkholderia-RFP, suggesting that secondary bacteria are only retained in Burkholderia co-infected spores.
Fig. 2.
Secondary bacteria do not infect symbiont free spores. Representative confocal micrographs of sori contents after growth on the indicated GFP labeled bacterial cultures. Scale bar = 10 μm
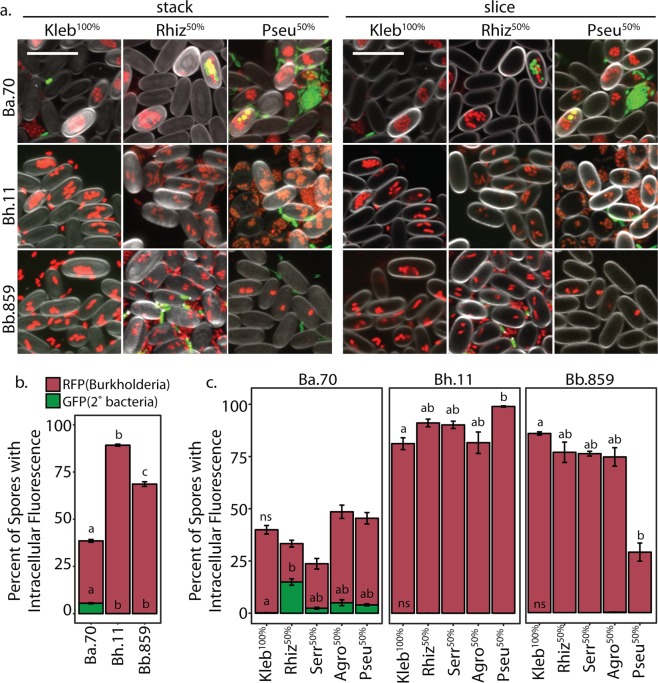
Fig. 3.
Burkholderia Differentially Induce Intracellular Co-Infections. a Representative confocal micrographs of Burkholderia-RFP infected sori contents after development on the indicated GFP labeled secondary bacteria. Scale bar = 10 μm. b Percent of spores infected with Burkholderia-RFP (brick) and co-infected with secondary bacteria-GFP (green) averaged across all replicates of all secondary bacterial conditions. c Average percent of spores infected (as in b) for each secondary culture condition. Error bars ± SE
The identity of the secondary bacterium also plays a role in the prevalence of intracellular infections for both Burkholderia and secondary bacteria. For instance, slightly fewer spores are infected with B. bonniea when cultured with P. aeruginosa-GFP (29%) than with all other bacteria (85.9–75.6%) (χ2 = 10.843, df = 4, p = 0.028). In contrast, significantly more spores are infected with B. hayleyella when cultured with P. aeruginosa-GFP (98.9%) than with all other bacteria (81.2–91%) (χ2 = 10.217, df = 4, p = 0.036). For B. agricolaris hosts, the degree of secondary co-infections significantly varied by secondary bacteria (χ2 = 15.019, df = 4, p = 0.004). K. pneumoniae-GFP is localized in only 0.2% of total spores while Rhizobium-GFP was localized in 14.9%. We observe similar co-infection patterns for each Burkholderia species when Rhizobium is used as the sole food source (Supplementary Fig. 2). When secondary bacterial infections are considered as a percentage of spores co-infected with B. agricolaris, Rhizobium-GFP is co-localized in almost half of total infected spores. This suggests that should B. agricolaris infection levels increase in conditions that promote higher infection titers, secondary infections may correspondingly increase.
Burkholderia symbionts induce extracellular secondary infections
Although we found minimal intracellular co-infections in most conditions, the farming phenotype may instead be explained by extracellular secondary infections. To get an initial indication of extracellular co-infections, we determined the percent of confocal images in which any extracellular GFP could be visualized (Fig. 4a). We found that all Burkholderia symbionts induced at least some level of extracellular co-infections, as we could visualize external GFP in each condition. Similar to our observations for intracellular co-infections, extracellular secondary bacteria appeared most frequently in B. agricolaris host sori (Fig. 4a).
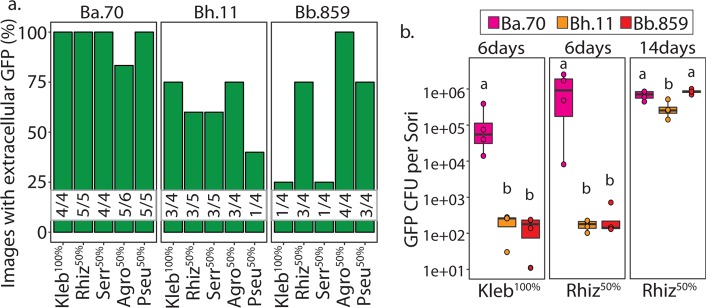
Fig. 4.
Burkholderia Induce Extracellular Co-infections. a Percent of confocal micrographs wherein secondary bacterial-GFP cells were visualized extracellularly within sori contents. Numbers nested within bars indicate the number of images with visible GFP over the number of total images collected per condition. b Number of GFP colony-forming units (log 10) from Burkholderia infected sori contents when grown on K. pneumoniae-GFP and Rhizobium-GFP. Individual sori contents were harvested either 6 or 14 days after plating on the Burkholderia/Secondary bacterial culture conditions. These numbers may represent both extracellular and intracellularly derived secondary bacteria. Points represent the GFP-CFU count for individual replicates
To quantify overall secondary co-infection, we counted GFP colony-forming units per sori for K. pneumoniae-GFP100% and Rhizobium-GFP50% conditions six days after co-culturing (Fig. 4b). No bacterial colonies were recovered from sori grown without Burkholderia, again indicating that these bacteria do not by themselves infect D. discoideum. We found that B. agricolaris induces the highest level of secondary infection, with B. agricolaris host sori colonized by an average of 1.29 × 105 K. pneumoniae-GFP cfu’s and 1.17 × 106 Rhizobium-GFP cfu’s (Fig. 4b). We recover notable, albeit far fewer, secondary bacterial colonies from B. hayleyella and B. bonniea hosts. B. hayleyella sori host an average of 206 and 169, and B. bonniea an average of 153 and 276, K. pneumoniae-GFP and Rhizobium-GFP cfu’s, respectively.
To explore whether secondary bacteria could further amplify within fruiting bodies over time, we also quantified Rhizobium-GFP colony-forming units 14 days after plating. We found that cfu’s did not increase for B. agricolaris hosts, but dramatically increased for B. hayleyella and B. bonniea hosts, which produced 2.87 × 105 and 8.37 × 105 GFP cfu’s, respectively. This brought the number of cfu’s in all Burkholderia infected sori up to fairly similar levels, perhaps representing a peak carrying capacity. However, we noticed that the number of spores per sori for B. hayleyella and B. bonniea infected hosts appeared to decrease over time (not shown). Replication of secondary bacteria within these sori could be damaging to spores, counter-acting potential benefits of hosting more food bacteria.
Burkholderia symbionts benefit hosts in food-scarce conditions in relation to co-infection induction
Farmers have been shown to have an advantage when dispersed to food-scarce environments [33, 34]. This is attributed to the induction of secondary bacterial food carriage, enabling host spores to reseed new environments with edible bacteria. However, this benefit has previously only been measured as an average fitness outcome across hosts infected with genotypically diverse Burkholderia symbionts [34]. Whether or how specific Burkholderia genotypes correspond with this beneficial outcome remained unknown. Given our results demonstrating that B. agricolaris induces the highest co-infection levels, we speculated that B. agricolaris hosts would have the highest reproductive fitness after dispersal to food-scarce environments.
To compare host fitness under different food availability conditions, we first plated uninfected spores with Burkholderia and secondary bacteria (K. pneumoniae-GFP100% and Rhizobium-GFP50%) under the same conditions employed previously. After 5 days of incubation we harvested developed sori and transferred 105 spores with rich (live K. pneumoniae-GFP) or scarce (heat-killed K. pneumoniae) food on nutrient medium. After 5 days of incubation in these conditions, we measured total spore productivity (Fig. 5a). For food-rich conditions, we again found significant differences according to infection status (F = 7.41, df = 3, p = 0.0015, and χ2 = 13.95, df = 3, p = 0.0029 for K. pneumoniae-GFP100% and Rhizobium-GFP50% conditions respectively). However, in this experiment only B. hayleyella hosts produced significantly fewer spores than the uninfected control (Fig. 5a). For food-scarce conditions, spore productivity was also significantly different according to Burkholderia infection status (χ2 = 11.87 and 20.616, df = 3, p = 0.0078 and <0.001 for K. pneumoniae-GFP100% and Rhizobium-GFP50% conditions, respectively). Here, B. agricolaris hosts had the highest spore productivity for both secondary conditions (Fig. 5a). B. bonniea also resulted in slightly higher, but not significantly different, spore productivity compared to the uninfected control (Fig. 5a). Thus, B. agricolaris infections endow a benefit for their amoeba host when dispersed to food-scarce environments. B. agricolaris-RFP and Rhizobium-GFP could again be found co-infecting spores after development on food-scarce conditions (Supplementary Fig. 3).
Fig. 5.
Farming Benefits Vary by Symbiont and Dispersal Strategy. For all figures, the top panel represents data from sori pre-grown on K. pneumoniae-GFP100% and the bottom panel represents sori pre-grown on Rhizobium-GFP50%, both with (fuschia, orange, and red) and without (white) supplementation of 5% Burkholderia. a Quantification of total spores harvested from food-rich and food-scarce plating conditions after transfer from K. pneumoniae-GFP100% (top) or Rhizobium-GFP50% (bottom) plating conditions. Points represent data from each individual replicate. b Representative images of individual sori streaks from fruiting bodies developed on K. pneumoniae-GFP100% (top) or Rhizobium-GFP50% (bottom) plates. Top panels are magnified sections of bottom panels. c Percentage of bacterial positive sori streaks with observable fruiting bodies. Error bars ± SE. d Number of fruiting bodies per fruiting body positive streaks for each individual replicate. All letters indicated post hoc significance within panels. For streak tests, all sori where streaked five days after plating on K. pneumoniae-GFP100% (top panels) or Rhizobium-GFP50% (bottom panels) and all data was collected five days after streaking
For the above assay, we evenly distributed spores on plates. If secondary bacteria are less numerous in host sori (as for B. hayleyella and B. bonniea), they might be spread too far from germinating spores to access and thus benefit from. Further, this assay might not best simulate spore dispersal in nature, where spores might be deposited in smaller denser patches by passing soil inhabitants. Therefore, we examined host fitness in food-scarce conditions using a “streak” dispersal strategy. Here, we deposited individual sori from fruiting bodes grown on K. pneumoniae-GFP100% or Rhizobium-GFP50% in small patches (~1 inch streaks) on nutrient medium. After a week of incubation, we measured the percent of fruiting body positive streaks and the number of fruiting bodies per each streak (Fig. 5b–d).
Streaks from uninfected sori did not produce bacterial colonies nor fruiting bodies. Over 95% of Burkholderia infected sori produced streaks with bacteria, however, the percent of streaks with fruiting bodies growing from these bacterial colonies significantly varies across Burkholderia species (χ2 = 13.728 and 12.127, df = 2, p = 0.001 and 0.0023 for K. pneumoniae-GFP100% and Rhizobium-GFP50% conditions, respectively). Burkholderia species also significantly influences the number of fruiting bodies per streak (χ2 = 14 and 11.24, df = 2, p < 0.001 and 0.0036 for K. pneumoniae-GFP100% and Rhizobium-GFP50% conditions, respectively). B. agricolaris infected sori generated significantly more fruiting bodies than B. hayleyella hosts from both conditions and B. bonniea hosts from K. pneumoniae-GFP100% conditions. The number of fruiting bodies also increased the longer streak plates were left to incubate (Fig. 6). Since fruiting bodies often developed from B. bonniea host sori, this suggests they gain better access to food under this dispersal strategy.
Fig. 6.
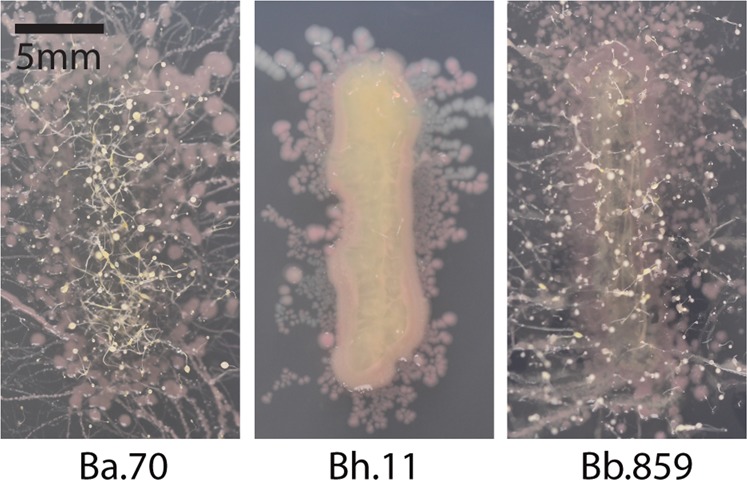
Fruiting Bodies Amplify in Burkholderia Infected Sori Streaks. Images of sori streaks two weeks after streak-testing. Sori were harvested from individual fruiting bodies that developed 5 days after plating spores on Rhizobium-GFP50% culture conditions with 5% of the indicated Burkholderia-RFP strain
We rarely witnessed fruiting bodies from B. hayleyella host sori streaks, providing an interesting counterpart to B. bonniea. Both symbionts induce similar densities of co-infection (Fig. 4) yet differ in downstream benefits (Fig. 5). This may be explained by the relative detriment each species exerts on its host. B. hayleyella reduces host fitness compared to B. bonniea, whereas B. bonniea hosts are often indistinguishable from uninfected counterparts (Figs. 1 and 5). Thus, B. hayleyella toxicity may inhibit host development despite food availability.
Co-infections and conditional benefits are consistent across Burkholderia species members
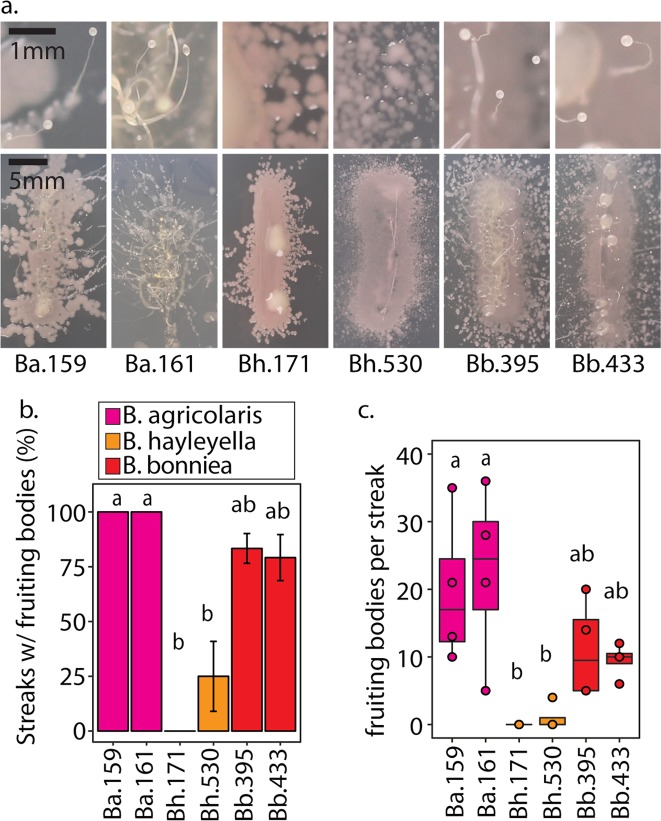
Our representative Burkholderia symbiont species significantly differed in their induction of co-infection and host impacts. We next asked whether these phenotypes were similar across strains of the same Burkholderia species. We imaged (Fig. 7) and streak tested (Fig. 8) host sori for additional Burkholderia-RFP strains after growth on Rhizobium-GFP50%. We again found noticeable, but low, levels of intracellular Rhizobium in B. agricolaris infected spores, with co-infection rare or absent in B. hayleyella and B. bonniea infected spores (Fig. 5). The percent of spores infected by Burkholderia was significantly different depending on genotype (χ2 = 22.65, df = 5, and P < 0.001), with B. agricolaris strains infecting fewer spores than B. hayleyella and B. bonniea (Fig. 5b). Despite low levels of intracellular Rhizobium co-infection, we again frequently observed extracellular GFP in sori (Fig. 7c).
Fig. 7.
Co-infection Patterns Correspond to Burkholderia Species. a Representative confocal micrographs of sori after five days post plating with 5% of the indicated Burkholderia-RFP strain and Rhizobium-GFP50%. b Average percent of spores visualized with intracellular Burkholderia-RFP (brick) and intracellular Rhizobium-GFP (green) for each of the indicated Burkholderia species. Letters indicate post hoc significance. c Percent of images in which external GFP was visualized out of the total number of independent images acquired. Numbers above bars represent the raw number of external GFP positive images over the number of images acquired for each Burkholderia infection condition
Fig. 8.
Host Benefits Correspond to Burkholderia Species and their Induction of Co-infections. a Images of representative sori streaks from fruiting bodies grown on Rhizobium-GFP50% with 5% of the indicated Burkholderia strain. Top panels are magnified sections of bottom panels. b Percentage of bacterial positive sori streaks with observable fruiting bodies. Error bars ± SE. c Number of fruiting bodies per fruiting body positive streaks for each individual. All letters indicated post hoc significance within panels. All sori where streaked five days after plating on Rhizibium-GFP50% and all data was collected five days after streaking
Next, we investigated the benefits of infection by these strains in food-scarce environments (Fig. 8). We found that Burkholderia genotype significantly influences both the percentage of sori that generate fruiting bodies and the number of fruiting bodies in positive streaks (χ2 = 12.127, df = 2, p = 0.002 and χ2 = 11.24, df = 2, p = 0.003, respectively). In accordance with our previous pattern B. agricolaris infections result in high, B. bonniea intermediate, and B. hayleyella low, levels of fruiting body production (Fig. 8). These results demonstrate that species members similarly induce co-infections and result in similar subsequent host benefits.
Discussion
Since elucidating the link between farming and Burkholderia, it has been assumed that Burkholderia symbionts allow for secondary co-infections via intracellular co-infection events. Intracellular co-infections of B. agricolaris with K. pneumoniae had previously been identified [34]. Co-phagocytosis with Burkholderia and subsequent inhibition of phagocytic digestion could be a parsimonious mechanistic explanation for this phenomenon. Burkholderia can be visualized within intracellular vacuoles that appear similar to phagosomes [37] but how Burkholderia invades and survives within amoebae is not resolved. Nonetheless, in this scenario secondary bacteria must first be liberated from infected amoebae or spores (via regurgitation or host cell lysis) so that surrounding amoebae may reap the benefits of farming. However, here we found that co-intracellular infection is quite rare, occurring with some frequency only for B. agricolaris, and occurring differentially across secondary bacteria. In contrast, all Burkholderia symbiont species generate extracellular secondary infections that can be visualized for all secondary species. The induction of secondary infection and corresponding downstream benefits is significantly different across symbionts. B. agricolaris induces the highest levels of secondary infection (both intracellularly and overall) with B. hayleyella and B. bonniea generating almost exclusively extracellular infections at low levels. This suggests that Burkholderia symbionts could induce secondary infections by multiple mechanisms that qualitatively and quantitatively differ between genotypes.
Overall, these results indicate that the predominant route by which different Burkholderia symbionts induce farming leans more towards extracellular bacterial carriage than intracellular co-infections. Susceptibility to secondary infections could be due to Burkholderia symbionts compromising the primitive immune system of their multicellular hosts. Sentinel cells serve as immune-like cells in multicellular slugs by trapping unwanted cargo through phagocytosis and/or neutralization by DNA nets [31, 32]. When sentinel cells have accumulated cargo, they drop out the slug, thereby cleansing it of potential toxic entities [31]. A gene deletion that reduces sentinel cells leads to retention of secondary bacteria through the slug stage and into the sorus [31]. Burkholderia host slugs have fewer sentinel cells than uninfected counterparts and this defect goes away when hosts are cured of their Burkholderia symbiont via antibiotic treatment [47]. Thus, the induction of secondary infections could be an indirect consequence of Burkholderia symbiosis resulting in sentinel cell reduction. This scenario may be comparable to the phenomenon of secondary infections in mammalian systems whereby primary infectious agents compromise the immune system of their hosts. Despite this, we cannot rule out the possibility that extracellular secondary infection originates from intracellular co-infection. It is possible that co-infected cells are more susceptible to lysis, rupturing and spewing their secondary bacterial passengers into the extracellular matrix. In either situation, secondary bacteria might then amplify within sori.
Intracellular co-infections are most frequent with B. agricolaris and the soil Rhizobium strain. This might reflect an ecologically relevant association between these species in nature. Burkholderia and Rhizobium are both ubiquitous in soil and contain several important symbiont species which have been found co-colonizing the same hosts [48–51]. Predation by amoebae in soil and aquatic systems shapes microbial community assembly and overall food webs [52]. Given the likely co-occurrence of soil amoebae with Burkholderia and other soil microbes it’s tempting to speculate on how these multipartite interactions influence overall microbial communities and higher trophic levels. Here, we show that amoebae co-disperse Burkholderia symbionts and secondary bacterial hitchhikers to new environments. Thus, the impact of amoebae on their surrounding microbial network can go well beyond predator-prey dynamics. Finally, our observation of Burkholderia and P. aeruginosa co-infection amplifies the concern that soil amoebae can serve as reservoirs for bacterial pathogens. These results suggest that Burkholderia symbionts can increase the suite of potential pathogenic partners hosted by amoebae.
Burkholderia–fungal associations have been well recognized for their importance in the soil ecosystem and for their bio-restoration potential [53–55]. There are compelling parallels between Burkholderia–Dictyostelium and Burkholderia–fungal associations. Some Burkholderia (notably B. terrae) are capable of adhering to and migrating with growing fungal hyphae through soil [56]. Similar to our system, some of these fungal associates assist in the co-migration of other (non-migrating) bacteria [57]. Several mechanisms have been proposed to underlie these interactions, such as direct receptor binding and indirect biofilm co-aggregation [58]. B. terrae extracellularly colonizes fungal hyphae but many other Burkholderia symbionts of diverse hosts persist intracellularly [13, 59]. An interesting example is the Rhizopus microsporus endosymbiont B. rhizoxinica fungi, which produces the rice seedling blight toxin [60]. Recently, secretion systems have been shown to be important for the active invasion of B. rhizoxinica across the fungal cell wall and into the host cytoplasm [59]. Secretion systems have also been implicated in B. psuedomallei infections [61, 62]. However many plant mutualistic Burkholderia species, which are closer relatives to Burkholderia symbionts of Dictyostelium, appear to lack some of these systems [61–63] The hypothesized portal of entry into Dictyostelium is via phagocytosis, which could circumvent the need for invasion specific mechanisms. Overall, Burkholderia symbionts of other hosts can help inform our understanding of the Burkholderia–amoebae symbiosis and vice versa.
Biofilm formation is intriguing to consider as a mechanistic explanation of secondary infection. Burkholderia adherence to secondary bacteria would increase the likelihood of co-phagocytosis or extracellular co-colonization. Different adhesive capacities of Burkholderia and secondary species could explain differences in the extent of secondary infections across bacterial combinations. Interestingly, recent work implicates Dictyostelium lectins in the farming phenomenon, higher lectin expression was detected in farmer D. discoideum clones and addition of endogenous lectins induced bacterial carriage [64]. Although this work did not consider the presence or impact of Burkholderia, we think Burkholderia symbionts play a key role. Burkholderia could induce farming via induction of lectin expression in amoebae or more simplistically, Burkholderia lectins may mediate co-adherence of secondary bacteria. Indeed, lectin expression by B. cenocepacia is an important component of biofilm formation and lectin aids in adherence of B. cepacia to host tissues [45, 65]. Future exploration into lectin expression and adhesion mechanisms will be helpful for clarifying these themes.
In addition to elucidating the phenomenon of secondary infections our results exemplify the context dependency of symbiotic outcomes in this system. We found that the costs and benefits of this symbiosis can be modified by different bacterial conditions and spore dispersal processes. The nature and extent of farming induction by Burkholderia symbionts differs across symbiont species and so do their corresponding contextual fitness outcomes. Previously, all Burkholderia symbionts were thought to benefit their hosts under food-scarce conditions. Here, B. hayleyella strains, though similar in secondary infection patterns to B. bonniea, are more detrimental in food-rich, and not beneficial in food-scarce, conditions. Perhaps B. hayleyella strains encode virulence genes that B. bonniea strains lack. We also find that Burkholderia symbionts display different population wide infectivity patterns that appear roughly similar across strains of the same species. How symbiont density is regulated and how it influences the parasitism to mutualism continuum are compelling questions in infectious biology [66, 67]. The variation of infection metrics and outcomes in this system, and the ease in which they can be explored, makes it well poised for investigating these questions. Ultimately, further research into the mechanisms, consequences, and ecological framework of the Burkholderia–Dictyostelium symbiosis will help illuminate microbial interaction dynamics relevant to infection biology and microbial ecology.
Supplementary information
Acknowledgements
We thank Kyle Skottke for fruitful discussions and manuscript review, Joan Strassmann and David Queller for initial guidance in the system, and all members of the DiSalvo lab at SIUE, particularly Jacob W. Miller for general laboratory support.
Funding
This study was supported by SIUE start-up funds from the DiSalvo lab.
Author contributions
NK and SD designed the study. NK and SD performed experiments with assistance from MNE, RAN and TSH. SD and TSH wrote the manuscript.
Data availability
All raw data supporting the conclusions of this manuscript is available at 10.6084/m9.figshare.7547834.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0419-7) contains supplementary material, which is available to authorized users.
References
- 1.Kiers ET, West SA. Evolving new organisms via symbiosis. Science. 2015;348:392–4. doi: 10.1126/science.aaa9605. [DOI] [PubMed] [Google Scholar]
- 2.Margulis L, Fester R. Bellagio conference and book. Symbiosis as source of evolutionary innovation: speciation and morphogenesis. Conference—June 25–30, 1989, Bellagio Conference Center, Italy. Symbiosis. 1991;11:93–101. [PubMed] [Google Scholar]
- 3.Douglas AE. Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6:a016113. doi: 10.1101/cshperspect.a016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas AE. Symbiotic interactions. Oxford: Oxford University Press, Oxford science publications Press; 2002. p. 148.
- 5.Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–9. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 6.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–65. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Yang S, Tang F, Zhu H. Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbio. 2012;14:334–42. doi: 10.1111/j.1462-5822.2011.01736.x. [DOI] [PubMed] [Google Scholar]
- 8.Klironomos JN. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology. 2003;84:2292–301. doi: 10.1890/02-0413. [DOI] [Google Scholar]
- 9.Chrostek Ewa, Teixeira Luis. Mutualism Breakdown by Amplification of Wolbachia Genes. PLOS Biology. 2015;13(2):e1002065. doi: 10.1371/journal.pbio.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Álvarez-Loayza P, Jr, Torres JFW, Balslev MS, Kristiansen H, Svenning J-C T, et al. Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, iriartea deltoidea. PLOS ONE. 2011;6:e16386. doi: 10.1371/journal.pone.0016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar HE, Wilson ACC, Ferguson NR, Moran NA. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLOS Biol. 2007;5:e96. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver KM, Campos J, Moran NA, Hunter MS. Population dynamics of defensive symbionts in aphids. Proc R Soc B Biol Sci. 2008;275:293–9. doi: 10.1098/rspb.2007.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonfante P, Anca IA. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol. 2009;63:363–83. doi: 10.1146/annurev.micro.091208.073504. [DOI] [PubMed] [Google Scholar]
- 14.Bronstein JL. Conditional outcomes in mutualistic interactions. Trends Ecol Evol. 1994;9:214–7. doi: 10.1016/0169-5347(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 15.Hussa EA, Goodrich-Blair H. It takes a village: ecological and fitness impacts of multipartite mutualism. Annu Rev Microbiol. 2013;67:161–78. doi: 10.1146/annurev-micro-092412-155723. [DOI] [PubMed] [Google Scholar]
- 16.Newton AC, Fitt BDL, Atkins SD, Walters DR, Daniell TJPathogenesis. parasitism and mutualism in the trophic space of microbe-plant interactions. Trends Microbiol. 2010;18:365–73. doi: 10.1016/j.tim.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Brocal Vicente, Latorre Amparo, Moya Andrés. Between Pathogenicity and Commensalism. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. Symbionts and Pathogens: What is the Difference? pp. 215–243. [Google Scholar]
- 18.Rønn R, McCaig AE, Griffiths BS, Prosser JI. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl Environ Microbiol. 2002;68:6094. doi: 10.1128/AEM.68.12.6094-6105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paquet Valérie E., Charette Steve J. Amoeba-resisting bacteria found in multilamellar bodies secreted byDictyostelium discoideum:social amoebae can also package bacteria. FEMS Microbiology Ecology. 2016;92(3):fiw025. doi: 10.1093/femsec/fiw025. [DOI] [PubMed] [Google Scholar]
- 20.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–8. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor-Mulneix Dawn L., Bendor Liron, Linz Bodo, Rivera Israel, Ryman Valerie E., Dewan Kalyan K., Wagner Shannon M., Wilson Emily F., Hilburger Lindsay J., Cuff Laura E., West Christopher M., Harvill Eric T. Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLOS Biology. 2017;15(4):e2000420. doi: 10.1371/journal.pbio.2000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horn M, Wagner M. Bacterial endosymbionts of free-living amoebae. J Eukaryot Microbiol. 2004;51:509–14. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 24.Rubeniņa I, Kirjušina M, Bērziņš A, Valciņa O, Jahundoviča I. Relationships between free-living amoeba and their intracellular bacteria. Proc Latv Acad Sci Sect B Nat Exact Appl Sci. 2017;71:259–65. [Google Scholar]
- 25.Bozzaro S, Eichinger L. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets. 2011;12:942–54. doi: 10.2174/138945011795677782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bozzaro S, Bucci C, Steinert M. Phagocytosis and host-pathogen interactions in Dictyostelium with a look at macrophages. Int Rev Cell Mol Biol. 2008;271:253–300. doi: 10.1016/S1937-6448(08)01206-9. [DOI] [PubMed] [Google Scholar]
- 27.Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–6. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Nasser W, Santhanam B, Miranda ER, Parikh A, Juneja K, Rot G, et al. Bacterial discrimination by dictyostelid amoebae reveals the complexity of ancient interspecies interactions. Curr Biol. 2013;23:862–72. doi: 10.1016/j.cub.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skriwan C, Fajardo M, Hägele S, Horn M, Wagner M, Michel R, et al. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol. 2002;291:615–24. doi: 10.1078/1438-4221-00177. [DOI] [PubMed] [Google Scholar]
- 30.smith jeff, Queller DC, Strassmann JE. Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC Evol Biol. 2014;14:105. doi: 10.1186/1471-2148-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–81. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Zhuchenko O, Kuspa A, Soldati T. Social amoebae trap and kill bacteria by casting DNA nets. Nat Commun. 2016;7:10938. doi: 10.1038/ncomms10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brock DA, Douglas TE, Queller DC, Strassmann JE. Primitive agriculture in a social amoeba. Nature. 2011;469:393–6. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 34.DiSalvo S, Haselkorn TS, Bashir U, Jimenez D, Brock DA, Queller DC, et al. Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. Proc Natl Acad Sci U S A. 2015;112:E5029–37. doi: 10.1073/pnas.1511878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol. 2012;63:249–66. doi: 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- 36.Brock DA, Hubert AM, Noh S, DiSalvo S, Geist KS, Haselkorn TS, et al. Endosymbiotic adaptations in three new bacterial species associated with Dictyosteliumdiscoideum: Burkholderiaagricolaris sp. nov., Burkholderiahayleyella sp. nov., and Burkholderiabonniea sp. nov. 2018. http://biorxiv.org/lookup/doi/10.1101/304352. [DOI] [PMC free article] [PubMed]
- 37.Shu L, Brock DA, Geist KS, Miller JW, Queller DC, Strassmann JE, et al. Symbiont location, host fitness, and possible coadaptation in a symbiosis between social amoebae and bacteria. eLife. 2018. https://elifesciences.org/articles/42660. [DOI] [PMC free article] [PubMed]
- 38.Haselkorn Tamara S., DiSalvo Susanne, Miller Jacob W., Bashir Usman, Brock Debra A., Queller David C., Strassmann Joan E. The specificity of Burkholderia symbionts in the social amoeba farming symbiosis: Prevalence, species, genetic and phenotypic diversity. Molecular Ecology. 2019;28(4):847–862. doi: 10.1111/mec.14982. [DOI] [PubMed] [Google Scholar]
- 39.Inglis RF, Biernaskie JM, Gardner A, Kümmerli R. Presence of a loner strain maintains cooperation and diversity in well-mixed bacterial communities. Proc R Soc B Biol Sci. 2016;283:20152682. doi: 10.1098/rspb.2015.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norris MH, Kang Y, Lu D, Wilcox BA, Hoang TT. Glyphosate resistance as a novel select-agent-compliant, non-antibiotic-selectable marker in chromosomal mutagenesis of the essential genes asd and dapb of Burkholderia pseudomallei. Appl Environ Microbiol. 2009;75:6062–75. doi: 10.1128/AEM.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su S, Bangar H, Saldanha R, Pemberton A, Aronow B, Dean GE, et al. Construction and characterization of stable, constitutively expressed, chromosomal green and red fluorescent transcriptional fusions in the select agents, Bacillus anthracis, Yersinia pestis, Burkholderia mallei, and Burkholderia pseudomallei. MicrobiologyOpen. 2014;3:610–29. doi: 10.1002/mbo3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kikuchi Y, Fukatsu T. Live imaging of symbiosis: spatiotemporal infection dynamics of a GFP-labelled Burkholderia symbiont in the bean bug Riptortus pedestris. Mol Ecol. 2014;23:1445–56. doi: 10.1111/mec.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogle DH, Wheeler P, Dinno A. Fsa: fisheries stock analysis [Internet]. 2018. https://github.com/droglenc/FSA.
- 44.Brock DA, Haselkorn TS, Garcia JR, Bashir U, Douglas TE, Galloway J, et al. Diversity of free-living environmental bacteria and their interactions with a bactivorous amoeba. Front Cell Infect Microbiol. 2018;8:411–411. doi: 10.3389/fcimb.2018.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. Regulation of biofilm formation in Pseudomonas and Burkholderia species: regulation of biofilm formation. Environ Microbiol. 2014;16:1961–81. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 46.Tomlin KL, Coll OP, Ceri H. Interspecies biofilms of Pseudomonas aeruginosa and Burkholderia cepacia. Can J Microbiol. 2001;47:949–54. doi: 10.1139/w01-095. [DOI] [PubMed] [Google Scholar]
- 47.Brock Debra A., Callison W. Éamon, Strassmann Joan E., Queller David C. Sentinel cells, symbiotic bacteria and toxin resistance in the social amoebaDictyostelium discoideum. Proceedings of the Royal Society B: Biological Sciences. 2016;283(1829):20152727. doi: 10.1098/rspb.2015.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Compant S, Nowak J, Coenye T, Clément C, Ait Barka E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev. 2008;32:607–26. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 49.Estrada-De Los Santos P, Bustillos-Cristales R, Caballero-Mellado J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl Environ Microbiol. 2001;67:2790–8. doi: 10.1128/AEM.67.6.2790-2798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen NH, Bruns TD. The microbiome of Pinus muricata ectomycorrhizae: community assemblages, fungal species effects, and Burkholderia as important bacteria in multipartnered symbioses. Microb Ecol. 2015;69:914–21. doi: 10.1007/s00248-015-0574-y. [DOI] [PubMed] [Google Scholar]
- 52.Edgcomb V. Marine protist associations and environmental impacts across trophic levels in the twilight zone and below. Curr Opin Microbiol. 2016;31:169–75. doi: 10.1016/j.mib.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Banitz T, Wick LY, Fetzer I, Frank K, Harms H, Johst K. Dispersal networks for enhancing bacterial degradation in heterogeneous environments. Environ Pollut. 2011;159:2781–8. doi: 10.1016/j.envpol.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Ellegaard-Jensen L, Knudsen BE, Johansen A, Albers CN, Aamand J, Rosendahl S. Fungal–bacterial consortia increase diuron degradation in water-unsaturated systems. Sci Total Environ. 2014;466–467:699–705. doi: 10.1016/j.scitotenv.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 55.Rashid MI, Mujawar LH, Shahzad T, Almeelbi T, Ismail IMI, Oves M. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol Res. 2016;183:26–41. doi: 10.1016/j.micres.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Nazir R, Zhang M, de Boer W, van Elsas JD. The capacity to comigrate with Lyophyllum sp. strain Karsten through different soils is spread among several phylogenetic groups within the genus Burkholderia. Soil Biol Biochem. 2012;50:221–33. doi: 10.1016/j.soilbio.2012.03.015. [DOI] [Google Scholar]
- 57.Warmink JA, Nazir R, Corten B, van Elsas JD. Hitchhikers on the fungal highway: The helper effect for bacterial migration via fungal hyphae. Soil Biol Biochem. 2011;43:760–5. doi: 10.1016/j.soilbio.2010.12.009. [DOI] [Google Scholar]
- 58.Vila T, Nazir R, Rozental S, dos Santos GMP, Calixto ROR, Barreto-Bergter E, et al. The role of hydrophobicity and surface receptors at hyphae of Lyophyllum sp. strain karsten in the interaction with Burkholderiaterrae BS001—implications for interactions in soil. Front Microbiol. 2016. 10.3389/fmicb.2016.01689/full. [DOI] [PMC free article] [PubMed]
- 59.Moebius N, Üzüm Z, Dijksterhuis J, Lackner G, Hertweck C. Active invasion of bacteria into living fungal cells. eLife. 2014. https://elifesciences.org/articles/03007. [DOI] [PMC free article] [PubMed]
- 60.Partida-Martinez LP, Monajembashi S, Greulich KO, Hertweck C. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr Biol. 2007;17:773–7. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 61.Gong Lan, Lai Shu-Chin, Treerat Puthayalai, Prescott Mark, Adler Ben, Boyce John D., Devenish Rodney J. Burkholderia pseudomallei Type III Secretion System Cluster 3 ATPase BsaS, a Chemotherapeutic Target for Small-Molecule ATPase Inhibitors. Infection and Immunity. 2015;83(4):1276–1285. doi: 10.1128/IAI.03070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stone JK, DeShazer D, Brett PJ, Burtnick MN. Melioidosis: molecular aspects of pathogenesis. Expert Rev Anti Infect Ther. 2014;12:1487–99. doi: 10.1586/14787210.2014.970634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Angus Annette A., Agapakis Christina M., Fong Stephanie, Yerrapragada Shailaja, Estrada-de los Santos Paulina, Yang Paul, Song Nannie, Kano Stephanie, Caballero-Mellado Jésus, de Faria Sergio M., Dakora Felix D., Weinstock George, Hirsch Ann M. Plant-Associated Symbiotic Burkholderia Species Lack Hallmark Strategies Required in Mammalian Pathogenesis. PLoS ONE. 2014;9(1):e83779. doi: 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dinh C, Farinholt T, Hirose S, Zhuchenko O, Kuspa A. Lectins modulate the microbiota of social amoebae. Science. 2018;361:402–6. doi: 10.1126/science.aat2058. [DOI] [PubMed] [Google Scholar]
- 65.Šulák Ondřej, Cioci Gianluca, Lameignère Emilie, Balloy Viviane, Round Adam, Gutsche Irina, Malinovská Lenka, Chignard Michel, Kosma Paul, Aubert Daniel F., Marolda Cristina L., Valvano Miguel A., Wimmerová Michaela, Imberty Anne. Burkholderia cenocepacia BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity. PLoS Pathogens. 2011;7(9):e1002238. doi: 10.1371/journal.ppat.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–95. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cunnington Aubrey J. The Importance of Pathogen Load. PLoS Pathogens. 2015;11(1):e1004563. doi: 10.1371/journal.ppat.1004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data supporting the conclusions of this manuscript is available at 10.6084/m9.figshare.7547834.