Optical coherence tomography-derived measures correlate with clinical outcomes in multiple sclerosis. By conducting genome-wide association studies, Fitzgerald et al. show that variants in early complement pathway genes are associated with changes in structural and functional visual outcomes in multiple sclerosis. Early complement factors may be mediators of neurodegeneration.
Keywords: genome-wide association studies, multiple sclerosis, optical coherence tomography, early complement pathway genes
Abstract
Multiple sclerosis is a heterogeneous disease with an unpredictable course and a wide range of severity; some individuals rapidly progress to a disabled state whereas others experience only mild symptoms. Though genetic studies have identified variants that are associated with an increased risk of developing multiple sclerosis, no variants have been consistently associated with multiple sclerosis severity. In part, the lack of findings is related to inherent limitations of clinical rating scales; these scales are insensitive to early degenerative changes that underlie disease progression. Optical coherence tomography imaging of the retina and low-contrast letter acuity correlate with and predict clinical and imaging-based outcomes in multiple sclerosis. Therefore, they may serve as sensitive phenotypes to discover genetic predictors of disease course. We conducted a set of genome-wide association studies of longitudinal structural and functional visual pathway phenotypes in multiple sclerosis. First, we assessed genetic predictors of ganglion cell/inner plexiform layer atrophy in a discovery cohort of 374 patients with multiple sclerosis using mixed-effects models adjusting for age, sex, disease duration, optic neuritis and genetic ancestry and using a combination of single-variant and network-based analyses. For candidate variants identified in discovery, we conducted a similar set of analyses of ganglion cell/inner plexiform layer thinning in a replication cohort (n = 376). Second, we assessed genetic predictors of sustained loss of 5-letters in low-contrast letter acuity in discovery (n = 582) using multivariable-adjusted Cox proportional hazards models. We then evaluated candidate variants/pathways in a replication cohort. (n = 253). Results of both studies revealed novel subnetworks highly enriched for connected genes in early complement activation linked to measures of disease severity. Within these networks, C3 was the gene most strongly associated with ganglion cell/inner plexiform layer atrophy (P = 0.004) and C1QA and CR1 were top results in analysis of sustained low-contrast letter acuity loss. Namely, variant rs158772, linked to C1QA, and rs61822967, linked to CR1, were associated with 71% and 40% increases in risk of sustained LCLA loss, respectively, in meta-analysis pooling discovery and replication cohorts (rs158772: hazard ratio: 1.71; 95% confidence interval 1.30–2.25; P = 1.3 × 10−4; rs61822967: hazard ratio: 1.40; 95% confidence interval: 1.16–1.68; P = 4.1 × 10−4). In conclusion, early complement pathway gene variants were consistently associated with structural and functional measures of multiple sclerosis severity. These results from unbiased analyses are strongly supported by several prior reports that mechanistically implicated early complement factors in neurodegeneration.
Introduction
Multiple sclerosis, an inflammatory demyelinating and neurodegenerative disorder of the CNS, is the most common cause of progressive disability in young adults. The disease is highly heterogeneous. Certain individuals with multiple sclerosis experience a mild course with little disability progression, while others worsen insidiously and accumulate substantial disability over years. Mechanisms contributing to the observed heterogeneity in disease evolution are poorly understood; discovery of specific pathways associated with risk of disease progression will allow for more accurate individualized prognosis as well as facilitate the discovery of novel therapeutic targets, which could be tested in progressive multiple sclerosis.
Variation in human leukocyte antigen (HLA) genes within the major histocompatibility complex (MHC) is a strong, well-known risk factor for multiple sclerosis, and previous genome-wide association studies (GWAS) have successfully identified over 200 non-MHC variants associated with disease susceptibility (Patsopoulos et al., 2017). However, in comparison, genetic correlates of multiple sclerosis severity are much less well-understood, with no variant(s) being identified consistently with disease course. Major limitations include use of moderate sample sizes without replication and a lack of accurate sensitive measures of disease course. Common clinical outcomes, including the Expanded Disability Status Scale (EDSS) score, are relatively coarse, semi-quantitative measures. They also can be insensitive to change, especially in the early stages of multiple sclerosis when behavioural changes are masked by functional compensation. Other limitations include difficulties in performing longitudinal analyses of advanced/non-conventional MRI outcomes that are acquired from multiple centres.
Advances in quantification of disease progression have allowed for more sensitive and quantitative assessments of the evolution of multiple sclerosis over time. Both optical coherence tomography (OCT) structural imaging of the retina and low-contrast letter acuity (LCLA) are highly reproducible and validated outcome measures, shown to correlate with clinical and imaging-based outcomes in multiple sclerosis patients. Thus, they could serve as sensitive phenotypic measures to discover genetic predictors of multiple sclerosis disease progression (Balcer et al., 2012; Saidha et al., 2015; Martinez-Lapiscina et al., 2016). For example, in patients with multiple sclerosis degeneration of the composite ganglion cell-inner plexiform (GCIP) layer correlates strongly with grey matter atrophy and disability accrual (Saidha et al., 2015), and decline in other OCT-derived measures are associated with progressive visual function loss and risk of disability progression (Talman et al., 2010; Martinez-Lapiscina et al., 2016). Furthermore, increased lesion volume, reductions in brain volume, and increased damage to optic radiations correlate with impaired LCLA in multiple sclerosis (Wu et al., 2007; Maghzi et al., 2014). Hence, the visual pathway may serve as an ideal set of outcome measures to interrogate genetic predictors of multiple sclerosis course.
Herein, we present results of two GWAS (both with independent replication) that capitalize on these recent technological advancements and leverage longitudinal structural and functional changes of visual pathway outcomes in patients with multiple sclerosis followed on average for over 3 years. Using single variant- and pathway-based analyses, we identified subnetworks of potentially associated genes enriched in early complement activation for both visual system outcomes. Our findings are supported by extensive prior work highlighting increased C1q and C3 expression in brain tissues from patients with progressive multiple sclerosis, and recent mechanistic work highlighting the role of early complement pathway factors in mediating neurodegeneration (Stevens et al., 2007; Orsini et al., 2014; Michailidou et al., 2015, 2017; Watkins et al., 2016; Liddelow et al., 2017).
Materials and methods
Genetic predictors of structural retinal thinning using OCT
Discovery: study population and OCT imaging, Johns Hopkins Multiple Sclerosis Center
Patients with a confirmed diagnosis of multiple sclerosis (2010 revised McDonald criteria) were followed beginning in 2010 from the Johns Hopkins (JH) Multiple Sclerosis Center for an ongoing prospective retinal imaging cohort study. Individuals with other known neurological or ophthalmological disorders, diabetes mellitus, uncontrolled hypertension, glaucoma, or refractive errors exceeding ±6 dioptres were excluded from study enrolment. Retinal imaging was performed approximately biannually using spectral domain Cirrus HD-OCT (model 4000, software version 6.0; Carl Zeiss Meditec) as described in detail previously (Syc et al., 2012; Saidha et al., 2015). Briefly, peripapillary and macular retinal layer thicknesses were obtained with the Optic Disc Cube 200×200 protocol and Macular Cube 512×128 protocol, respectively. Scans with signal strength <7/10 or with artefact were excluded in agreement with the OSCAR-IB criteria and were used to compute thicknesses of the composite of the GCIP layers (Tewarie et al., 2012). We selected rates of changes in the composite GCIP layers because in patients with multiple sclerosis the GCIP is less vulnerable to swelling associated with inflammation of the optic nerve (unlike the retinal nerve fibre layer) and GCIP degeneration correlates more strongly with grey matter loss and disability in multiple sclerosis (Saidha et al., 2015). We censored individuals who experienced optic neuritis during follow-up (i.e. stopped updating GCIP). Inflammatory pathology associated with optic neuritis may contribute to accelerated GCIP loss that may be unrelated to the underlying progressive GCIP atrophy of interest in this study (Trip et al., 2005; Saidha et al., 2015).
Discovery: genotyping, quality control, imputation
JH-OCT cohort participants provided blood samples for DNA extraction and were genotyped on the Illumina Multi-Ethnic Global Array (MEGA). We implemented a standard GWAS quality control (QC) pipeline. We excluded individuals with genotype success rates <0.99, related individuals [e.g. those with proportion of identity-by-descent values ≥0.125 (third degree relative)], and those with excess heterozygosity [±4 standard deviation (SD) Fmean]. We tested for potential population outliers using principal components analyses (PCA; –pca option in PLINK); we excluded those who were >2 SD of the mean for PC1 and PC2. Also using PCA, we determined the likely genetic ancestry of our population by mapping the included individuals onto the 1000 Genomes Population (Supplementary Fig. 1). Results suggest our population to be largely of European descent. Using generalized linear regression models, we also confirmed that the identified principle components were not collinear with the included covariates used in subsequent regression models. For example, the identified principle components were not associated with age, sex, history of optic neuritis, or disease duration (all P > 0.05). A total of 474 individuals were genotyped and 458 (97%) remained after sample QC; 374 of 458 individuals also had longitudinal OCT data available and were eligible for the analysis. We also implemented a standard GWAS QC pipeline with respect to exclusion of poor performing variants. We excluded ambiguous variants, variants with minor allele frequencies (MAF) <0.05, those with missing genotype rates >0.05, and those demonstrating deviation from Hardy Weinberg equilibrium (P < 0.001). A total of 1 731 558 variants were genotyped, and 503 678 remained after QC and were included in the analysis. We also imputed classical alleles for HLA class I genes (HLA-A, HLA-B, and HLA-C) and HLA class II genes (HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, and HLA-DRB1), corresponding amino acid sequences and additional MHC variants not genotyped explicitly using SNP2HLA (Jia et al., 2013; Patsopoulos et al., 2017). To do so, we used the MHC-specific reference panel provided by the Type 1 Diabetes Genetics Consortium. Imputed MHC-region and HLA alleles with info scores <0.70 were excluded. For risk scores of interest, our MHC risk score was defined as the linear combination of previously-identified MHC-region multiple sclerosis risk alleles where each risk allele is weighted by its effect on multiple sclerosis susceptibility (Moutsianas et al., 2015). Our non-MHC multiple sclerosis risk score was defined as linear combination of previously-identified non-MHC multiple sclerosis risk alleles where each allele is weighted by its effect on multiple sclerosis susceptibility (Jager et al., 2009). For non-MHC multiple sclerosis susceptibility variants not genotyped explicitly, we imputed allele dosages using the Minimac algorithm as used on the Michigan Imputation Serve with the updated Haplotype Reference Consortium Panel version r1.1 excluding variants with imputation R2 < 0.70 (Howie et al., 2012; Reed et al., 2015; Das et al., 2016; McCarthy et al., 2016). In total, 180 possible non-MHC multiple sclerosis susceptibility variants were genotyped or imputed with high quality and included in our analyses and risk score, and no participants were missing data on non-MHC multiple sclerosis risk alleles.
Discovery: statistical analyses
An overview of our overall analytic approach is provided by Supplementary Fig. 2I. We derived a patient-specific rate of change in GCIP from a multivariable-adjusted mixed effects regression models to use as the dependent variable in genetic analyses. Briefly, we used a linear mixed effects model (also adjusting for age at first OCT, sex, history of optic neuritis and disease duration) incorporating patient-specific intercepts and slopes to model the rate of change in GCIP over time; patient-specific slopes were derived as the sum of the fixed effect and predicted individual random slope. The multivariable-adjusted patient-specific slopes of GCIP decline were used in all subsequent analyses. This approach has been applied previously in GWAS of longitudinal changes in age-related cognitive decline (De Jager et al., 2012). Our analyses did not adjust for multiple sclerosis therapies as (i) multiple sclerosis therapies may act as mediators and potentially attenuate associations between variants and GCIP decline; and (ii) statistical adjustment for mediators in this context may introduce bias in the effect of interest (collider bias) that may stem from unmeasured common factors influencing therapy class decisions and disease severity (e.g. comorbidity or socio-economic factors; Vanderweele et al., 2014). For individual variant- or risk score-based analyses, we regressed multivariable-adjusted patient-specific slopes of GCIP atrophy on allelic dosages of genetic variants (including individual variants, imputed individual MHC susceptibility or non-MHC multiple sclerosis susceptibility alleles, or composite MHC susceptibility/non-MHC multiple sclerosis susceptibility risk scores) in an additive linear regression model also adjusting for genetic ancestry using the first six principal components. Sensitivity analyses additionally adjusted for multiple sclerosis subtype (relapsing-remitting or progressive multiple sclerosis).
Network analysis
Because of the relatively small sample size of our study and previous observations that alterations to biological pathways rather than in individual genes may provide key biological insight to the results of GWAS, we subsequently performed an extensive set of pathway- and network-based analyses using the results of the individual variant analyses, which are described in detail here (Baranzini et al., 2009, 2010; Gourraud et al., 2013; Housley et al., 2015; Matsushita et al., 2015). For these analyses, we calculated a gene-based P-value using the Multi-marker Analysis of GenoMic Annotation (MAGMA) algorithm, which is a fast and flexible method for gene-based analyses of GWAS genotype data (de Leeuw et al., 2015). It applies a multiple regression-based approach that properly incorporates linkage disequilibrium between markers and estimates potential multi-marker effects. We then performed an agnostic pathway analysis using the HotNet2, which is a topology-based algorithm that was originally developed for the analysis of somatic mutations in cancer datasets and has been applied previously to genome-wide association data (Nakka et al., 2016). Briefly, HotNet2 is a method for finding significantly mutated subnetworks within protein-protein interaction networks that applies a directed heat diffusion process through an interaction network. Each gene represents one node in the network and the amount of ‘diffusible heat’ is determined by a heat-score, which we calculated as the negative log-transformed P-value derived from the gene-based tests from the MAGMA analysis. We assessed significance of HotNet2 results for each run by permuting the heat score interaction networks to calculate a P-value for the number of identified subnetworks containing k or more genes, as suggested by HotNet2 developers. We then tested for enrichment of the novel HotNet2-identified subnetworks using pathway information provided by FUMAgwas GENE2FUNC analyses (Kamburov et al., 2013; Watanabe et al., 2017). HotNet2 analyses were performed using the iRefIndex protein interaction database (Razick et al., 2008).
In sensitivity analyses, since HotNet2 may underperform when many heat scores are assigned similar values (as in the case when the majority of P-values are insignificant), we assigned a value of 0 for heat scores, for which we have low confidence in their association with GCIP decline, as in previous analyses (Leiserson et al., 2015; Nakka et al., 2016). We identified low-confidence genes using data-derived threshold based on the local false discovery rate (FDR) of gene-based P-values and used (1 − [local FDR]) as a quantitative metric of ‘confidence’ (interpreting the local FDR as the probability a gene is not associated with GCIP decline given the observed P-value). To identify the gene-score threshold cut-off, we plotted confidence versus gene-based P-values and used the P-value limit corresponding to the first inflection point (e.g. evidence of a sharp drop in confidence), as described previously (Nakka et al., 2016). When the curve did not demonstrate an obvious inflection point, we set gene-score threshold corresponding to a P-value limit of 0.20. We then applied HotNet2 using the confidence-adjusted gene-based heat-scores to derive novel subnetworks and subsequently tested for enrichment, as above.
Further, to ensure the robustness of our network approach in its ability to extract biologically meaningful information from GWAS data, we applied the algorithm to existing summary statistics from Alzheimer’s disease and age-related macular degeneration. In both primary and sensitivity analyses, HotNet2 identified networks of apolipoprotein genes (e.g. APOE) in Alzheimer’s disease and complement pathway genes in age-related macular degeneration (e.g. C3, CFH). Both pathways are highly studied for each respective disease (Supplementary Fig. 3). All statistical tests were two-sided, and analyses were conducted using PLINK, R and Python.
Replication: study population and OCT imaging, University of California at San Francisco Multiple Sclerosis Center
Patients with a confirmed diagnosis of multiple sclerosis (n = 376) were recruited from the University of California at San Francisco (UCSF) Multiple Sclerosis Center for a similar prospective retinal imaging cohort study and similarly met McDonald (2010 criteria) for the diagnosis of multiple sclerosis. They were imaged using Heidelberg Spectralis OCT (Heidelberg Engineering). Similar to the JH-OCT, image sequences included a macular volume scan and have been previously described in detail elsewhere (Balk et al., 2016). Further, similar to the JH-OCT cohort, OSCAR IB criteria were adhered to ensure sufficient data quality standards (Tewarie et al., 2012).
Replication: genotyping
Participants in the UCSF-OCT cohort were genotyped on the Multiple Sclerosis chip, a custom content Illumina Chip containing an exome-chip backbone and all candidate variants derived from a previous meta-analysis of multiple sclerosis-susceptibility variants (International Multiple Sclerosis Genetics Consortium et al., 2011). We used a targeted validation of the selected variants (or viable proxies) we identified from the JH-OCT cohort discovery analyses that were also genotyped in UCSF samples.
Replication: statistical analysis
An overview of our overall analytic approach is provided by Supplementary Fig. 2(II). Similar to the discovery JH-OCT, we created a patient-specific rate of change in OCT outcomes that were estimated from linear mixed-effects model to use as a continuous phenotype in subsequent analyses. Estimated outcomes included inner plexiform layer, ganglion cell layer and the composite ganglion cell complex. Also similar to JH-OCT analyses, we regressed multivariable-adjusted patient-specific slopes of the measures of retinal layer thinning atrophy on these specified genetic variants. Slopes were adjusted for age at first OCT, sex, disease duration and a history of optic neuritis.
Genetic predictors of changes in visual function
Discovery: study population and visual function, CombiRx trial
The CombiRx Trial including 1008 patients with relapsing-remitting multiple sclerosis, was a 3-arm, phase III, randomized, double-blinded, and placebo-controlled trial of combination therapy employing a partial 2 × 2 factorial design and a 2:1:1 randomization allocation to combination interferon β-1a and glatiramer acetate or each single therapy (and matching placebo) on annualized relapse rate. Full details of the trial and results have been described in detail elsewhere (Lindsey et al., 2012; Lublin et al., 2013). Briefly, eligible participants had relapsing-remitting multiple sclerosis, were treatment naïve, and had to have at least two clinical relapses in the prior 3 years or at least one clinical relapse with subsequent MRI activity in that time frame. Patients were followed for a minimum of 3 years (or up to 7 years if he or she entered the extension study). At baseline and annually thereafter, binocular visual acuity was assessed using Sloan letter charts with 2.5% and 1.25% contrast in a room illuminated to 80–100 cd/m2 at 2-m distance. The number of letters correctly identified (out of 60) was recorded, and patients were instructed to wear usual distance glasses or contacts. In patients with multiple sclerosis, changes in LCLA at 1.25% and at 2.5% contrast are each associated with disability progression. As CombiRx trial participants were relatively early in their disease course and had experienced little disability progression (Table 1), we a priori defined a sustained change in LCLA as a loss of five letters (confirmed at a subsequent visit 1-year later) at 1.25% contrast for our primary analysis. We selected a sustained loss of 5-letters as the primary outcome as (i) such criteria have been applied in ophthalmology clinical trials and in multiple sclerosis; and (ii) assessments of LCLA were relatively infrequent in CombiRx; some previous studies of LCLA changes included testing every 12 weeks (Beck et al., 2007; Balcer et al., 2012, 2017).
Table 1.
Characteristics of OCT cohorts
| Cohort | ||
|---|---|---|
| JH-OCT (n = 374; 50%) | UCSF-OCT (n = 376; 50%) | |
| Mean follow-up time, years (SD) | 3.6 (2.1) | 3.9 (2.0) |
| Number of OCT scans (SD) | 4.6 (3.5) | 3.1 (1.2) |
| Mean age at baseline, years (SD) | 42.7 (11.0) | 47.7 (10.6) |
| Female sex, n (%) | 292 (78) | 263 (68) |
| Relapsing-remitting multiple sclerosis (%) | 319 (85.3) | 309 (80) |
| History of optic neuritis, n (%) | 148 (39.5) | 179 (46) |
| Mean disease duration (SD) | 9.0 (8.2) | 14.0 (9.1) |
Discovery: genotyping, quality control and HLA imputation
At baseline, a subset of CombiRx participants provided blood samples (n = 608; 60%) for DNA extraction and were genotyped on the genome-wide Human Illumina Bead Chip. We again implemented a standard GWAS QC pipeline. We excluded individuals with genotype success rates <0.99, related individuals (e.g. those with proportion of identity-by-descent values ≥0.125) and those with excess heterozygosity (±4 SD Fmean). In total, 582 participants were eligible for the analysis. As in the JH-based analyses, we tested for potential population outliers PCA (–pca option in PLINK). We removed individuals who were >2 SD of the mean for PC1 and PC2. Similar to JH-based analyses, we also determined the likely genetic ancestry of CombiRx trial participants by mapping included individuals onto the 1000 Genomes Population (Supplementary Fig. 4). As in JH-based analyses, results suggest our population to be largely of European descent. The identified principle components were not collinear with covariates used in subsequent regression models (e.g. principle components were not associated with age, sex, history of optic neuritis, or disease duration). We also excluded ambiguous variants, those with missing genotype rates >0.05, and those with deviations from Hardy Weinberg equilibrium (P < 0.001), as in the JH-OCT cohort.
To facilitate replication and meta-analyses between results from discovery and replication studies of visual function outcomes (both CombiRx and JH cohorts had available genome wide genotyping data), we imputed CombiRx genotypes using the Minimac algorithm as used on the Michigan Imputation Server with the updated Haplotype Reference Consortium version r1.1 reference panel (Howie et al., 2012; Reed et al., 2015; Das et al., 2016; McCarthy et al., 2016). Following imputation, we restricted all analyses to variants with MAF > 0.05, those with imputation R2 values > 0.70, as outlined by Verma et al. (2014) and performed subsequent analyses using the estimated allele dosages. We also imputed classical alleles for HLA class I and II genes using SNP2HLA and similarly created MHC and non-MHC multiple sclerosis susceptibility scores, as described in the JH-OCT cohort.
Discovery: statistical analysis
We assessed the association between additive allele dosages (imputed and genotyped) of 5 258 576 variants and time to a sustained 5-letter loss in LCLA using a Cox proportional hazards model adjusted for age, sex, disease duration, history of optic neuritis and genetic ancestry (using first six principle components). We subsequently conducted a similar set of HotNet2 pathway- and network-based analyses (including sensitivity analyses using the results of the individual variant analyses in CombiRx).
Replication and meta-analysis: study population and visual function Johns Hopkins Multiple Sclerosis Center (replication)
A subset of participants (n = 253; 66%) from the JH-OCT cohort also underwent visual function testing (denoted JH-LCLA). Standardized visual function testing was performed with retro-illuminated eye charts in a darkened room prior to OCT examination. To assess visual acuity 2.5% and 1.25% contrast we used LCLA Sloan letter charts (at 2 m). Similar to CombiRx, the number of letters identified correctly were recorded. Approximate biannual testing was performed binocularly, and participants also used their habitual glasses or contact lenses. Relative to CombiRx trial participants, JH cohort members were older and had greater disease duration (Table 1). In the JH-LCVA cohort we similarly defined a priori a sustained change in LCLA as a loss of five letters (confirmed at a subsequent visit) at 2.5% contrast for our primary analysis to avoid any floor-type effects potentially relevant for changes in 1.25% in this population with more advanced disease (changes in LCLA at 2.5% may be more sensitive among patients with greater disease duration).
Replication and meta-analysis: genotyping, quality control and HLA imputation
JH-LCLA participants were genotyped as part of the JH-OCT Cohort (e.g. on the Illumina MEGA). We performed an identical genotype imputation procedure as in the CombiRx discovery cohort using the Minimac algorithm as used on the Michigan Imputation Server and the updated Haplotype Reference Consortium version r1.1 reference panel (McCarthy et al., 2016).
Replication and meta-analysis: statistical analysis
Initial analyses evaluated the effect of genetic variants identified from the CombiRx discovery cohort on time to a sustained change in LCLA in the JH-LCLA replication cohort using a Cox proportional hazards model adjusting for age, sex, disease duration, history of optic neuritis and genetic ancestry (using first six principle components). We then pooled results from analyses of high-quality variants measured in both studies (variants with MAF > 0.05 and imputation R2 > 0.7) using fixed effects meta-analysis (Willer et al., 2010). We performed a fixed-effects meta-analysis as we assume an underlying common study population and underlying common genetic effect, and our study prespecified discovery and replication cohorts. For variants demonstrating potential heterogeneity, we also conducted random-effects meta-analysis as an additional sensitivity analysis. We assessed potential inflation of test statistics (estimated by comparing the median test statistic to that expected by chance) and corrected subsequent results using a genomic control correction. We then performed network and pathway-based analyses (and corresponding sensitivity analyses), as the JH-OCT study, using the P-values from the meta-analysis. We also excluded variants demonstrating potential statistical heterogeneity between the two cohorts (P < 0.10) from subsequent network and pathway analyses, as these variants are likely variants which did not replicate between the two studies.
Data availability
Data collected for this study are part of a long-term research project; access to de-identified subject level data may be granted to those who meet criteria for confidential access and have the necessary data-sharing agreements in place.
Results
Genetic predictors of structural retinal thinning using OCT
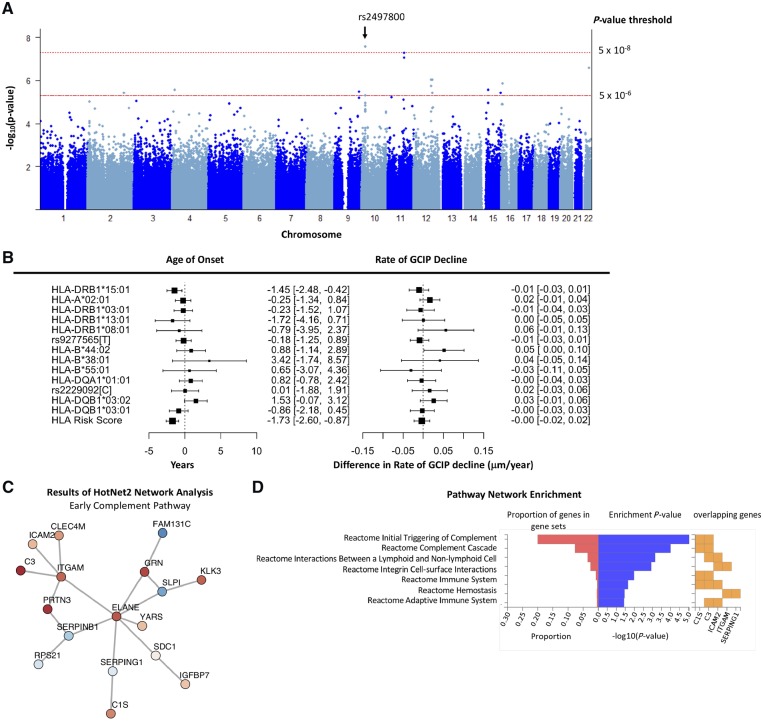
A total of 374 participants (78% female; mean age 42.7 years) were included in OCT discovery analysis. Mean follow-up time for JH-OCT was 3.6 years (SD: 2.1 years) and the average rate of decline in GCIP at the population level was −0.32 μm/year [95% confidence interval (CI): −0.39 to −0.24 μm/year; P < 0.0001]. Using multivariable-adjusted patient-specific slopes of GCIP atrophy as a continuous phenotype, we found one intergenic variant near SLC39A12 (rs2497800; hg19: chr10:18351061) that achieved genome-wide significance with faster rates of GCIP atrophy (additive difference in rate of GCIP = −0.08 μm/year increase in effect allele; 95% CI: −0.11 to −0.06; P = 2.65 × 10−8). We also detected suggestive associations (P < 5 × 10−6) between six other variants and GCIP thinning (Fig. 1A and Supplementary Table 1). However, these associations remain to be confirmed, as replication cohorts to further interrogate them were not available.
Figure 1.
Summary of results for GWAS of GCIP decline in multiple sclerosis. (A) Manhattan plot of P-values from analyses assessing the association between genetic variants and rate of GCIP decline in the discovery JHU OCT cohort. One variant (rs2497800) achieved genome-wide significance. (B) Results for known multiple sclerosis HLA susceptibility variants. The left panel demonstrates the association between a given HLA allele and age of onset of multiple sclerosis. Values are displayed in years. The right panel demonstrates the association between a given HLA allele and difference in rate of change in GCIP. Values are displayed as difference in rate of change in µm/year. The HLA risk score is calculated as the linear combination of previously identified HLA risk alleles where each allele is weighted by its effect on multiple sclerosis susceptibility. (C) Composition of identified networks from HotNet2 analyses that were identified in both primary and secondary analyses. (D) Enrichment results of the HotNet2 network analyses of networks identified in both primary and secondary analyses. The plotted network reflects the identified network from the sensitivity analyses (where low-confidence genes are assigned a heat score of 0). The identified network was highly enriched in early complement pathway genes (P = 1.38 × 10−8). The left (red) portion of the plot denotes the proportion of a given pathway that is composed of the genes in the network. The middle (blue) portion of the plot denotes the −log(P-value) of the tests for enrichment in the pathway using the identified network of genes. The right (orange) portion of the plot denotes the genes in the network which are included in the given pathway.
Consistent with previous GWAS in multiple sclerosis, burden of MHC risk alleles was strongly associated with earlier age of multiple sclerosis onset (per 1-unit increase in genetic risk score: −1.73 years earlier; 95% CI: −2.60 to −0.87 years; P < 0.001; Fig. 1C). However, individual MHC alleles and MHC risk burden were not associated with differential rates of GCIP atrophy (per 1-unit increase in MHC risk score: −0.003 μm/year; 95% CI: −0.02 to 0.02 μm/year; P = 0.73; Fig. 1B). Several non-MHC multiple sclerosis susceptibility variants also displayed nominal evidence of a potential association with GCIP thinning. Our weighted non-MHC multiple sclerosis risk score (e.g. an individual’s burden of multiple sclerosis susceptibility alleles) was not associated with GCIP decline (per 1-unit: −0.002 μm/year; 95% CI: −0.02 to 0.01 μm/year; P = 0.81; Supplementary Table 2).
Table 2.
Characteristics of discovery (CombiRx) and replication (JHU LCVA) cohorts
| Cohort (total n = 835) | ||
|---|---|---|
| CombiRx (n = 582; 69%) | JHU (n = 253; 31%) | |
| Sustained 5-letter change in visual acuity at 1.25%, n (%) | 180 (31) | 58 (23) |
| Sustained 5-letter change in visual acuity at 2.5%, n (%) | 136 (23) | 88 (35) |
| Annualized rate of change in LCLA at 1.25% | −0.35 (−0.49, −0.21) | −0.07 (−0.19, 0.05) |
| Annualized rate of change in LCLA at 2.5% | −0.16 (−0.28, −0.05) | −0.30 (−0.42, −0.18) |
| Mean follow-up time (SD) | 3.1 (1.7) | 4.22 (3.22) |
| Female sex, n (%) | 416 (72) | 200 (79) |
| Mean age at baseline (SD) | 38.1 (9.4) | 41.9 (10.9) |
| Median EDSS (IQR) at baseline | 2.0 (1.0–2.5) | - |
| Relapsing-remitting multiple sclerosis | 582 (100) | 216 (85) |
| Disease duration | ||
| <1 year, n (%) | 372 (64) | 41 (16) |
| 1–2 years, n (%) | 153 (26) | 23 (9) |
| ≥2–10 years, n (%) | 54 (9) | 111 (44) |
| ≥10 years, n (%) | 0 (0) | 78 (31) |
| History of optic neuritis, n (%) | 122 (21) | 116 (46) |
EDSS = Expanded Disability Status Scale.
In analyses to derive novel clusters of connected genes associated with GCIP thinning, we identified one network of genes in primary and in sensitivity analyses (Fig. 1C and D). We observed potentially highly biologically-relevant enrichment in early complement system genes (Penrichment = 1.38 × 10−8). Though the network size-based P-value derived from the distribution of gene-based heat scores was not significant (possibly related to the relatively small sample size and resultant proportion of non-contributory gene-based P-values; Pnetwork = 0.19), the complement pathway has substantial biological relevance to neurodegeneration that has been extensively described in prior work. For example, evidence from mechanistic studies demonstrate a role of early complement regulation of synaptic pruning (notably first described in a retinal model) (Stevens et al., 2007). Further, more recent evidence suggesting that variation in C3 is associated with lower brain volumes and high C3 expression in neurotoxic astrocytes (A1) (Liddelow et al., 2017; Roostaei et al., 2019). As a result, we focused subsequent analyses on validating the complement pathway and C3 related findings. To perform the validation of this network, we attempted to identify whether variants identified in C3 were associated with GCIP atrophy in an independent cohort, since this gene has (i) strong biological plausibility; and (ii) demonstrated the strongest association with GCIP decline in preliminary analyses. Twenty-three variants were mapped to C3 and were included in this analysis; rs11569523 was the leading variant at this locus (Supplementary Table 3; chr19:6689042; difference in rate of change per increase in 1 risk allele: −0.08; 95% CI: −0.12 to −0.04; FDR-adjusted P = 2.0 × 10−3).
Subsequently, we assessed the association between a priori-selected variants annotated to C3 (a total of six variants either overlapped or were proxies) in an independent replication study conducted in 386 multiple sclerosis patients followed on average for 3.9 years (SD: 2.0 years) at the UCSF Multiple Sclerosis Center. Similar to the JH-OCT cohort, patient-specific rates of change in OCT outcomes adjusted for age, sex, disease duration, and history of optic neuritis were used as continuous phenotypes in the analysis. UCSF cohort participants were relatively comparable to JH-OCT cohort participants; they were 32% male, on average aged 47.6 years (SD: 10.6 years) and had an average disease duration of 14.1 years (SD: 9.1 years). Of the six a priori-selected candidate C3 variants tested, rs11569523 was also associated with the inner plexiform layer thinning, which is a component of the composite GCIP layers (difference in rate of change: −0.70; 95% CI: −1.30 to −0.09; Supplementary Table 4). Several of the other a priori-selected candidate variants mapping to the C3 gene were also associated with the ganglion cell layer thinning (another component of the composite GCIP) including rs17030 (chr19:6677989; difference in rate of change: −0.08; 95% CI: −0.15 to −0.001) and rs1047286 (chr19:6713262; difference in rate of change: −0.18; 95% CI: −0.34 to −0.02); rs1047286 was also marginally associated with loss of the composite ganglion cell complex. Other candidate C3 variants: rs428543 (chr19:6702157), rs2230205 (chr19:6709704), rs2230201 (chr19:6713291) were not associated with OCT phenotypes.
Our top individual variant (rs2497800; chr10:18351061) was not available in the UCSF replication genotyping cohort and did not have suitable proxy variants, so we could not evaluate its potential association with loss of ganglion cell layer components in an independent cohort.
Genetic predictors of changes in visual function
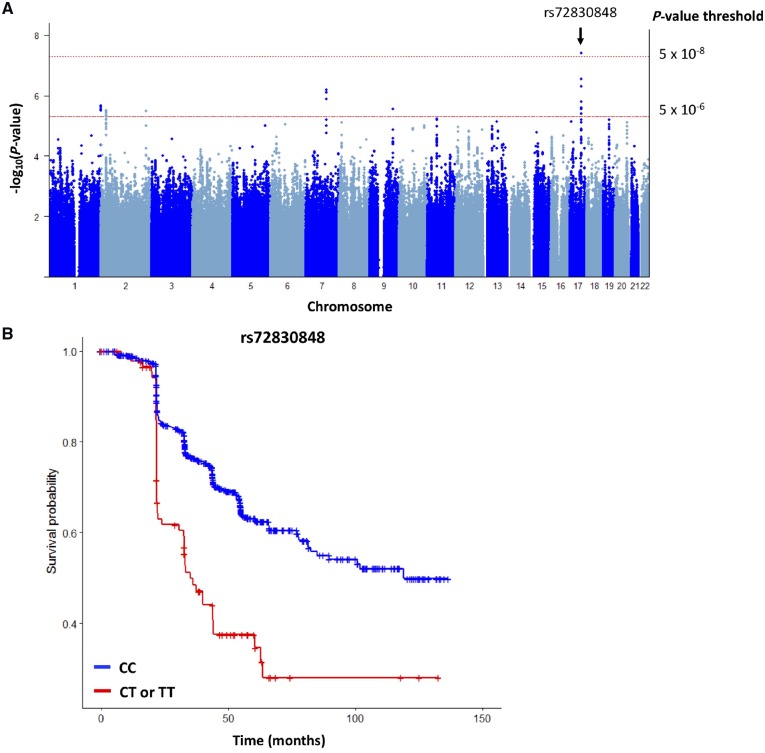
We detected potential associations between six variants and a sustained LCLA loss in CombiRx discovery cohort (P < 5 × 10−6; Supplementary Table 5). Of these, the MSI2 variant (rs72830848; chr17:55398579) replicated in JH-LCLA participants (P = 0.0028) and together yield a joint statistic of P = 3.98 × 10−8. The pooled hazard ratio (HR) for sustained LCLA loss associated with a 1-allele increase was 2.25 (95% CI: 1.78 to 2.85).
In a secondary analysis, six other variants demonstrated suggestive evidence (P < 5 × 10−6) of association with risk of sustained LCLA loss in a meta-analysis pooling data from the CombiRx and JH-LCLA (ntotal = 835; Table 3 and Fig. 2). Similar to analyses of GCIP thinning, we did not detect an association between individual MHC susceptibility or non-MHC multiple sclerosis susceptibility variants or the composite susceptibility scores and sustained LCLA loss (Supplementary Tables 6–8).
Table 3.
Variants demonstrating potential association with risk of LCLA changes in CombiRx (discovery) and in JHU (replication) samples with pooled P-value ≤ 5 × 10−6
| SNP | Chr | Position | Effect allele | Mapped genes | CombiRx (n = 582) | JHU (n = 253) | Meta-analysis (n = 835) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | Imputation R2b | HR (95% CI) | P-value | Freq | Imputation R2 | HR (95% CI) | P-value | HR(95% CI)c | Pooled P-value | I2 | P for heterogeneity | |||||
| rs72830848 | 17 | 55398579 | T | MSI2 | 0.06 | 0.94 | 2.24 (1.61, 3.11) | 1.76 × 10−6 | 0.05 | 0.94 | 2.27 (1.33, 3.89) | 2.78 × 10−3 | 2.25 (1.78, 2.85) | 3.98 × 10−8 | 4.2 | 0.84 |
| rs13241771 | 7 | 98432607 | A | TMEM130 | 0.16 | 0.98 | 1.76 (1.36, 2.27) | 1.65 × 10−5 | 0.16 | 0.98 | 1.77 (1.21, 2.6) | 3.45 × 10−3 | 1.77 (1.47, 2.12) | 6.59 × 10−7 | 1.5 | 0.90 |
| rs10157709 | 1 | 249154567 | C | ZNF672, ZNF692, PGBD2 | 0.05 | 1.00 | 1.99 (1.4, 2.85) | 1.38 × 10−4 | 0.06 | 1.00 | 2.57 (1.36, 4.87) | 3.82 × 10−3 | 2.26 (1.76, 2.91) | 2.12 × 10−6 | 7.2 | 0.79 |
| rs2888449 | 2 | 217861522 | C | 0.29 | 0.99 | 0.67 (0.54, 0.83) | 2.39 × 10−4 | 0.29 | 0.98 | 0.61 (0.44, 0.83) | 2.09 × 10−3 | 0.64 (0.55, 0.74) | 3.25 × 10−6 | 31.4 | 0.58 | |
| rs10980055 | 9 | 112570896 | G | PALM2-AKAP, AKAP, PALM2 | 0.15 | 1.00 | 1.59 (1.24, 2.04) | 2.39 × 10−4 | 0.15 | 0.99 | 1.95 (1.26, 3.03) | 2.85 × 10−3 | 1.76 (1.48, 2.10) | 2.87 × 10−6 | 23.8 | 0.63 |
| rs12713638 | 2 | 25915978 | G | DTNB, ASXL2 | 0.37 | 1.00 | 0.67 (0.54, 0.83) | 2.91 × 10−4 | 0.36 | 0.98 | 0.59 (0.42, 0.82) | 1.48 × 10−3 | 0.63 (0.54, 0.73) | 3.06 × 10−6 | 44.9 | 0.50 |
Bolded variant was significant in discovery and replication analyses.
bImputation R2 after application of the Minimac imputation algorithm (Howie et al., 2012).
cPooled HRs are adjusted for age, disease duration, history of optic neuritis and genetic ancestry (first six principal components). Results are only pooled after testing for heterogeneity.
Figure 2.
Summary of results for GWAS of loss of visual function in multiple sclerosis. (A) Manhattan plot depicting P-values from analyses assessing the pooled association between genetic variants and risk of sustained LCLA from the discovery (CombiRx) and replication (JHU-LCLA). One variant (rs72830848) was potentially associated with sustained LCLA at the 5 × 10−8 significance level in the pooled analysis. (B) Kaplan Meier plot comparing rate of sustained LCLA for individuals with the CC genotype relative to individuals with TC or TT genotype. Individuals with the CT or TT genotype were at an over 2-fold increase risk of sustained LCLA change relative to individuals with the CC genotype (HR: 2.25; 95% CI: 1.70 to 2.98; P-value = 3.98 × 10−8).
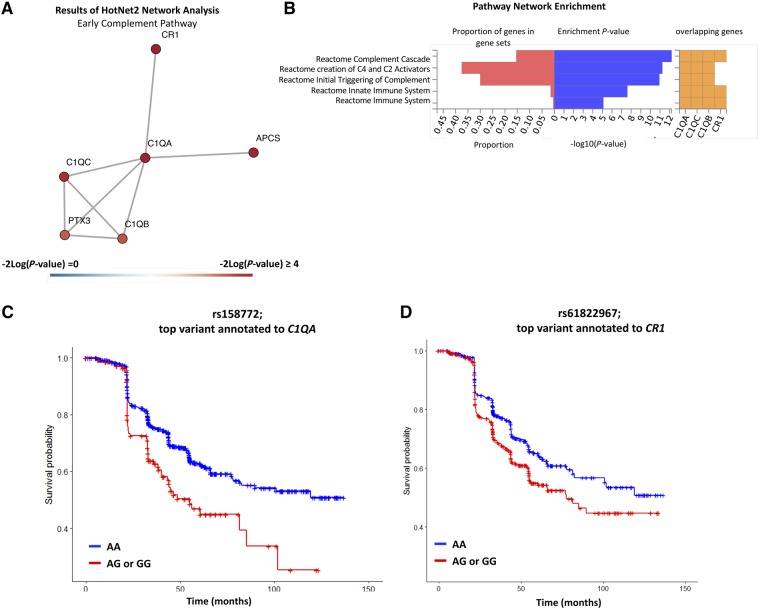
In network analyses of gene-based P-values (derived from the P-values for individual variant meta-analyses) associated with LCLA loss, we identified a significant subnetwork of interest (Pnetwork = 0.03), which remained significant after controlling for false discovery (Fig. 3A). Consistent with the structural retinal thinning results, we observed enrichment in early complement genes (Penrichment = 9.15 × 10−6). We noted variants at the C1QA locus and at the CR1 locus as associated with sustained LCLA loss (Fig. 3). Specifically, rs158772 (chr1:22945619) linked to C1QA and rs61822967 (chr1:207694165) linked to CR1 and were associated with a 71% and 40% increase in risk of sustained LCLA loss, respectively (rs158772: HR: 1.71; 95% CI: 1.30 to 2.25; P = 1.31 × 10−4, rs61822967: HR: 1.40; 95% CI: 1.16 to 1.68; P = 3.83 × 10−4).
Figure 3.
Summary of results of the network analyses related to loss of visual function in multiple sclerosis. (A) Composition of identified networks from HotNet2 analyses that were identified in both primary and secondary analyses. (B) Enrichment results of the HotNet2 network analyses of networks identified in both primary and secondary analyses. The identified network was highly enriched in early complement pathway genes (P = 9.15 × 10−16). The plotted network reflects the identified network from the sensitivity analyses (where low-confidence genes are assigned a heat score of 0). (C) Kaplan Meier plot comparing rate of sustained LCLA for individuals with the CC genotype relative to individuals with TC or TT genotype for the variant rs158772, which is the top variant annotated to C1QA. Individuals with the CT or TT genotype were at a 71% increased risk of sustained LCLA change relative to individuals with the CC genotype (HR: 1.71; 95% CI: 1.30 to 2.25; P-value = 10−4). For CombiRx, the imputation R2 for rs158772 is 0.98; for JHU, the imputation R2 for rs158772 is 0.99. (D) Kaplan Meier plot comparing rate of sustained LCLA for individuals with the AA genotype relative to individuals with AG or GG genotype for the variant rs61822967, which is the top variant annotated to CR1. Individuals with the AG or GG genotype were at a 40% increased risk of sustained LCLA change relative to individuals with the AA genotype (HR: 1.40; 95% CI: 1.16 to 1.68; P-value = 4 × 10−4). For CombiRx, the imputation R2 for rs61822967is 0.96; for JHU, the imputation R2 for rs61822967 is 0.99.
Discussion
New technologies that allow a more intricate, quantitative and objective study of large populations of patients with multiple sclerosis may help to provide insight into disease. Using relatively recent imaging technology that allows a quantitative and real time surrogate measurement of disease progression (OCT), exhaustive genome wide screens, and functional outcomes, we identified one of the strongest associations with disease progression reported to date. The results of this study provide guidance to novel studies elucidating mechanisms of disease course in multiple sclerosis. Network analyses used herein offer a structured approach to identify coordinated signals in a functionally related set of genes rather than depending on a single variant. These findings are consistent with the notion that consideration of variant sets (or genes), often with modest associations, rather than individual genes, may aid in identifying pivotal signalling cascades relevant to disease biology, a key component to interpreting GWAS results in a biological context (Baranzini et al., 2009; Housley et al., 2015).
Our analyses suggest that there are genetic variants, particularly in early complement pathway genes that predispose some patients with multiple sclerosis to develop more rapid retinal neurodegeneration (measured by OCT) or increased susceptibility to visual function loss, which are validated outcomes previously linked with EDSS and MRI progression. While many complement system genes are critical for immune function, non-immune functions of complement have been more recently discovered in the CNS (Scolding et al., 1998; Wing et al., 1999; Stevens et al., 2007; Liddelow et al., 2017; Morgan et al., 2017). The discovery of expression of early classical complement pathway molecules (C1q and C3) on neuronal synapses or in neurotoxic glia has led to new hypotheses that aberrant complement expression may play a role in mediating neurodegeneration in several neurodegenerative diseases (Hong et al., 2016; Liddelow et al., 2017). In a process first described in a murine retinal-development model, astrocyte-induced production of neuronal C1q and C3 was found to tag extraneous synapses for removal by C3r expressing macrophages and microglia. In the mature brain, synapses remain stable (in the absence of complement), while in neurodegenerative disease, this mechanism is aberrantly reactivated (Stevens et al., 2007; Hong et al., 2016). Liddelow et al. (2017) recently demonstrated that C1q is also secreted by activated microglia and can induce a subtype of reactive astrocytes (A1 astrocytes, demarcated by C3), which become unable to promote neuronal survival or synaptogenesis as they do in a healthy environment, but rather mediate neurotoxicity. Variants within CR1 (in moderate linkage disequilibrium with variants identified here) are consistently identified in Alzheimer’s disease susceptibility studies (Lambert et al., 2013). Finally, recent studies have also demonstrated that variants in complement component 4 (C4) may be risk factors for neuromyelitis optica (Estrada et al., 2018).
In the experimental autoimmune encephalitis (EAE) model of multiple sclerosis, astrocyte-specific gene expression analyses have indicated complement pathway enrichment with particularly high C3 expression (Itoh et al., 2018; Tassoni et al., 2018). Similarly, post-mortem tissues from multiple sclerosis brains demonstrate enrichment of C3-expressing A1 astrocytes in white matter and in acute demyelinating lesions lending to speculation that within demyelinating lesions A1 astrocytes may contribute to remyelination failure (Liddelow et al., 2017). Roostaei et al. (2019) also found that in a smaller sample of multiple sclerosis patients, a common coding variant in C3 (rs2230199) is associated with cognitive impairment and lower grey matter volume in cross-sectional analyses. Our study adds considerably to these findings as ours is longitudinal, larger, and included replication. While specific variants identified differ, several identified variants are in moderate linkage disequilibrium with rs2230199 and provide corroborative support for a role of early complement genes in multiple sclerosis course. Other pathological studies showed increased expression of C1q and C3 in brains of multiple sclerosis patients with progressive multiple sclerosis as well (Michailidou et al., 2017).
Our exploratory single variant analyses of genetic predictors of retinal atrophy identified several variants of interest. Namely, we identified a variant near SLC39A12 potentially associated with accelerated GCIP atrophy. SLC39A12 is highly expressed in human brains and enhanced cortical expression has been observed in pathology studies of schizophrenia patients (Scarr et al., 2016). However, while intriguing, independent validation of these variants in other cohorts is necessary. In analyses of LCLA, we also identified a variant near MSI2 that is associated with sustained LCLA loss. The MSI2 protein may regulate alternative splicing in mouse retinal photoreceptors and neural stem cells; however, exact mechanisms are not well-described (Katz et al., 2014; Murphy et al., 2016). Notably, whether variants linked SLC39A1 or MSI2 have functional consequences remains unknown, as neither appear to be cis expression quantitative trait loci (eQTLs). Another important observation is that we did not find that known multiple sclerosis risk alleles or the global burden of susceptibility variants are predictors of severity as measured by visual pathway outcomes in either OCT or LCLA analyses. These findings are consistent with previous studies demonstrating MHC and non-MHC susceptibility variants influence the age of onset of multiple sclerosis but have had no definitive effect on multiple sclerosis course or severity to date (Isobe et al., 2016).
Primary strengths of our study include its longitudinal design; for both structural and functional visual phenotypes, individuals were followed for on average of over 3 years, with follow-up extending for some to over 7 years. Other innovative features include use of novel phenotypic outcome measures to evaluate the effects of genetic variants on multiple sclerosis disease course over time. Both GCIP thickness measured using OCT and visual function testing using LCLA have excellent reproducibility and are each sensitive, inexpensive measures that correlate with more-conventional imaging and clinical measures of disability in patients with multiple sclerosis (Saidha et al., 2015; Balcer et al., 2017). Measures of ganglion cell layer thicknesses using OCT, in particular, may serve as an ideal phenotypic outcome in larger, more comprehensive multi-site follow-up studies, as this technique is highly sensitive and reproducible, while structural MRIs are often difficult to compare across sites and traditional clinical outcomes (e.g. EDSS) may lack sensitivity in determining small changes in disability. Another strength is that both analyses of structural retinal thinning as well as visual dysfunction included replication cohorts.
While these strengths add a high level of rigour, JH and UCSF-OCT cohorts were relatively inhomogeneous clinical populations of patients with respect to stage of their disease, whereas CombiRx was a trial of patients relatively early in disease course. However, as we had access to primary data for each cohort, we were able to standardize analytic methodology across cohorts and minimize heterogeneity in analyses, allowing us to optimally combine information from cohorts. For example, for analyses of LCLA, we selected phenotypic changes a priori (1.25% in CombiRx versus 2.5% in JH-LCLA), maximizing sensitivity to detect progression due to the differences in disease stage between the two cohorts (1.25% charts are more sensitive in early patients while 2.5% charts are more sensitive to changes later in disease). However, despite our relatively modest sample size (and heterogeneous OCT cohort populations), remarkably convergent findings demonstrating complement pathway enrichment from each cohort support our results. A key limitation of our study is the lack of genome-wide genotyping in the UCSF-OCT cohort. As participants were genotyped on a more targeted genotyping array, we were unable to validate findings for some candidate variants beyond validation of candidate genes from our network analyses. A critical next step is to independently validate the variants of high interest identified in this study. Furthermore, our gene-based P-values are derived from a statistical algorithm and are not cell-type or expression specific, so functional effects of the observed genetic variation may be heterogeneous within different cells and upon immune activation. However, previous studies have generated highly biologically relevant observations and new hypotheses from the implementation of such statistical methodology (Baranzini et al., 2009; Nakka et al., 2016). It is also possible that our network analyses were underpowered; results are based from single-variant analyses that are derived from our modest sample size. Still, for both visual system outcomes, we performed extensive sensitivity analyses where we tried to identify and exclude low confidence genes as an attempt to mitigate this concern. Variants in a number of complement pathway genes are associated with age-related macular degeneration and partial overlapping pathophysiology may be relevant to visual system changes observed. To counteract this possibility, we excluded patients from our cohort with other ophthalmological or neurological conditions that may influence visual pathway-based outcomes.
Conclusion
Results of our longitudinal GWAS analysis suggest variations in early complement pathway genes are associated with changes to previously validated structural and functional visual outcomes in patients with multiple sclerosis. Early complement proteins C1q and C3 are highly expressed in progressive multiple sclerosis brain tissues and may be mediators of neurodegeneration. This novel genetic discovery has implications both for the potential of developing methods to predict progression of disease as well as for providing new insight into the mechanisms of pathology in multiple sclerosis.
Supplementary material
Supplementary material is available at Brain online.
Funding
This study was supported by National Institutes of Health (NIH) grants R01NS082347 to P.A.C. and R01NS088155 to S.E.B. and by support from the Race to Erase. K.C.F. is supported by a postdoctoral fellowship from the National Multiple Sclerosis Society. Parts of this study were enabled by the collaboration with CombiRx Trial Investigators. The CombiRx study was funded by the NIH National Institute of Neurological Disorders and Stroke (phase III study grant UO1NS045719, planning grant R21NS41986) and is listed on www.clinicaltrials.gov as NCT00211887.
Competing interests
K.C.F., K.K., M.D.S., S.A.A., N.F., A.M.R., D.K., P.B., K.A.W. report no competing interests. S.K. has served on consulting or advisory boards for Acorda, Bayer, Biogen, Celgene, EMD Serono, Genentech, Genzyme, Mallinckrodt, Novartis, and Teva. S.S.C. has received personal compensation for activities with Orthotech Biotech, the American Academy for Orthopedic Surgery, Oxford University Press, Department of Defense, and Medimmune. S.S. has received consulting fees from Medical Logix for the development of CME programs in neurology and has served on scientific advisory boards for Biogen-Idec, Genzyme, Genentech Corporation, EMD Serono and Novartis. He is the PI of investigator-initiated studies funded by Genentech Corporation and Biogen Idec, and received support from the Race to Erase multiple sclerosis foundation. He has received equity compensation for consulting from JuneBrain LLC, a retinal imaging device developer. He is also the site investigator of a trial sponsored by MedDay Pharmaceuticals. A.J.G. reports receiving research grants from the National Multiple Sclerosis Society, NIH, Novartis and Inception 5 Sciences. He has served on an end point adjudication committee for MedImmune and a steering committee for OCTiMS and has provided expert witness for Mylan and Anneal. He has also served on then Scientific Advisory Board of Bionure and Inception Sciences. E.M.M. reports receiving free glatiramer acetate for the investigator-initiated vitamin D trial, of which she is the PI from Teva Neuroscience provides. She is also the PI of investigator-initiated studies funded by Biogen, Sanofi-Genzyme. She is also a site investigator of trials sponsored by Sun Pharma, Biogen and royalties from Up-to-date. G.R.C. serves on Data and Safety Monitoring Boards: AMO Pharmaceuticals, Biolinerx, Horizon Pharmaceuticals, Merck, Merck/Pfizer, Opko Biologics, Neurim, Ophazyme, Sanofi-Aventis, Reata Pharmaceuticals, Receptos/Celgene, Teva pharmaceuticals, NHLBI (Protocol Review Committee), NICHD (OPRU oversight committee). He also serves on Consulting or Advisory Boards forAtara Biotherapeutics, Axon, Biogen, Biotherapeutics, Argenix, Brainstorm Cell Therapeutics, Charleston Labs Inc, Click Therapeutics, Genzyme, Genentech, GW Pharma, Klein-Buendel Incorporated, Medimmune, Medday, Novartis, Roche, Scifluor, Somahlution, Teva pharmaceuticals, TG Therapeutics, UT Houston. G.R.C. is employed by the University of Alabama at Birmingham and President of Pythagoras, Inc. a private consulting company located in Birmingham AL. F.D.L. has received funding for research grants Novartis Pharmaceuticals Corp, Teva Neuroscience, Actelion, Transparency Life Sciences, and the National multiple sclerosis Society. He has received consulting fees, served on advisory boards, or data safety and monitoring boards for Bayer HealthCare Biogen, EMD Serono, Novartis, Teva, Actelion, Sanofi/Genzyme, Acorda, Roche/Genentech, MedImmune, Receptos/Celgene, Forward Pharma, TG Therapeutics, Abbvie, Regeneron, Medday, Atara Biotherapeutics, Polpharma, Mapi Pharma, Innate Immunotherapeutic, Apitope. S.E.B. has received consulting fees or participated in Scientific Advisory Boards from Novartis Pharma, Sanofi-Aventis, Teva, and Biogen-Idec. P.L.D. has received honoraria from Genzyme and Celgene. He is the principal investigator on grants from Biogen and Roche Pharmaceuticals. P.A.C. has received personal honorariums for consulting from Biogen and Disarm Therapeutics. He is PI on research grants to Johns Hopkins from MedImmune, Annexon, Biogen, and Genzyme.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
Abbreviations
- GCIP
ganglion cell-inner plexiform
- GWAS
genome-wide association studies
- HLA
human leukocyte antigen
- LCLA
low-contrast letter acuity
- MHC
major histocompatibility complex
- OCT
optical coherence tomography
References
- Balcer LJ, Galetta SL, Polman CH, Eggenberger E, Calabresi PA, Zhang A et al. Low-contrast acuity measures visual improvement in phase 3 trial of natalizumab in relapsing multiple sclerosis. J Neurol Sci 2012; 318: 119–24. [DOI] [PubMed] [Google Scholar]
- Balcer LJ, Raynowska J, Nolan R, Galetta SL, Kapoor R, Benedict R et al. Validity of low-contrast letter acuity as a visual performance outcome measure for multiple sclerosis. Mult Scler 2017; 23: 734–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk LJ, Cruz-Herranz A, Albrecht P, Arnow S, Gelfand JM, Tewarie P et al. Timing of retinal neuronal and axonal loss in multiple sclerosis: a longitudinal OCT study. J Neurol 2016; 263: 1323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet 2009; 18: 2078–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini SE, Srinivasan R, Khankhanian P, Okuda DT, Nelson SJ, Matthews PM et al. Genetic variation influences glutamate concentrations in brains of patients with multiple sclerosis. Brain 2010; 133: 2603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck RW, Maguire MG, Bressler NM, Glassman AR, Lindblad AS, Ferris FL. Visual acuity as an outcome measure in clinical trials of retinal diseases. Ophthalmology 2007; 114: 1804–9. [DOI] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A et al. Next-generation genotype imputation service and methods. Nat Genet 2016; 48: 1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging 2012; 33: 1017.e1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Whelan CW, Zhao F, Bronson P, Handsaker RE, Sun C et al. A whole-genome sequence study identifies genetic risk factors for neuromyelitis optica. Nat Commun 2018; 9: 1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourraud P-A, Sdika M, Khankhanian P, Henry RG, Beheshtian A, Matthews PM et al. A genome-wide association study of brain lesion distribution in multiple sclerosis. Brain 2013; 136: 1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 2016; 352: 712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housley WJ, Fernandez SD, Vera K, Murikinati SR, Grutzendler J, Cuerdon N et al. Genetic variants associated with autoimmunity drive NFκB signaling and responses to inflammatory stimuli. Sci Transl Med 2015; 7: 291ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Sawcer S, Hellenthal G, Pirinen M, Spencer CCA et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011; 476: 214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe N, Keshavan A, Gourraud P-A, Zhu AH, Datta E, Schlaeger R et al. Association of HLA genetic risk burden with disease phenotypes in multiple sclerosis. JAMA Neurol 2016; 73: 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Itoh Y, Tassoni A, Ren E, Kaito M, Ohno A et al. Cell-specific and region-specific transcriptomics in the multiple sclerosis model: focus on astrocytes. PNAS 2018; 115: E302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager PLD, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol 2009; 8: 1111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Han B, Onengut-Gumuscu S, Chen W-M, Concannon PJ, Rich SS et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013; 8: e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 2013; 41: D793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Li F, Lambert NJ, Sokol ES, Tam W-L, Cheng AW et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. Elife 2014; 3: e03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MDM, Vandin F, Wu H-T, Dobson JR, Eldridge JV, Thomas JL et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015; 47: 106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017; 541: 481–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey J, Scott T, Lynch S, Cofield S, Nelson F, Conwit R et al. The CombiRx trial of combined therapy with interferon and glatiramer cetate in relapsing remitting multiple sclerosis: design and baseline characteristics. Mult Scler Relat Disord 2012; 1: 81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin FD, Cofield SS, Cutter GR, Conwit R, Narayana PA, Nelson F et al. Randomized study combining interferon & glatiramer acetate in multiple sclerosis. Ann Neurol 2013; 73: 327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghzi A-H, Revirajan N, Julian LJ, Spain R, Mowry EM, Liu S et al. Magnetic resonance imaging correlates of clinical outcomes in early multiple sclerosis. Mult Scler Relat Disord 2014; 3: 720–7. [DOI] [PubMed] [Google Scholar]
- Martinez-Lapiscina EH, Arnow S, Wilson JA, Saidha S, Preiningerova JL, Oberwahrenbrock T et al. Retinal thickness measured with optical coherence tomography and risk of disability worsening in multiple sclerosis: a cohort study. Lancet Neurol 2016; 15: 574–84. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Madireddy L, Sprenger T, Khankhanian P, Magon S, Naegelin Y et al. Genetic associations with brain cortical thickness in multiple sclerosis. Genes Brain Behav 2015; 14: 217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature Genetics 2016; 48: 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou I, Naessens DMP, Hametner S, Guldenaar W, Kooi E, Geurts JJG et al. Complement C3 on microglial clusters in multiple sclerosis occur in chronic but not acute disease: implication for disease pathogenesis. Glia 2017; 65: 264–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou I, Willems JGP, Kooi E-J, van Eden C, Gold SM, Geurts JJG et al. Complement C1q-C3-associated synaptic changes in multiple sclerosis hippocampus. Ann Neurol 2015; 77: 1007–26. [DOI] [PubMed] [Google Scholar]
- Morgan BP, Boyd C, Bubeck D. Molecular cell biology of complement membrane attack. Semin Cell Dev Biol 2017; 72: 124–32. [DOI] [PubMed] [Google Scholar]
- Moutsianas L, Jostins L, Beecham AH, Dilthey AT, Xifara DK et al. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nature Genetics 2015; 47: 1107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D, Cieply B, Carstens R, Ramamurthy V, Stoilov P. The Musashi 1 controls the splicing of photoreceptor-specific exons in the vertebrate retina. PLoS Genet 2016; 12: e1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakka P, Raphael BJ, Ramachandran S. Gene and network analysis of common variants reveals novel associations in multiple complex diseases. Genetics 2016; 204: 783–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F, De Blasio D, Zangari R, Zanier ER, De Simoni M-G. Versatility of the complement system in neuroinflammation, neurodegeneration and brain homeostasis. Front Cell Neurosci 2014; 8: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsopoulos N, Baranzini SE, Santaniello A, Shoostari P, Cotsapas C, Wong G et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv 2017; 143933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed E, Nunez S, Kulp D, Qian J, Reilly MP, Foulkes AS. A guide to genome-wide association analysis and post-analytic interrogation. Stat Med 2015; 34: 3769–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. iRefIndex: A consolidated protein interaction database with provenance. BMC Bioinform 2008; 9: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostaei T, Sadaghiani S, Mashhadi R, Falahatian M, Mohamadi E, Javadian N et al. Convergent effects of a functional C3 variant on brain atrophy, demyelination, and cognitive impairment in multiple sclerosis. Mult Scler 2019; 25: 532–40. [DOI] [PubMed] [Google Scholar]
- Saidha S, Al-Louzi O, Ratchford JN, Bhargava P, Oh J, Newsome SD et al. Optical coherence tomography reflects brain atrophy in multiple sclerosis: a four-year study. Ann Neurol 2015; 78: 801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr E, Udawela M, Greenough MA, Neo J, Suk Seo M, Money TT et al. Increased cortical expression of the zinc transporter SLC39A12 suggests a breakdown in zinc cellular homeostasis as part of the pathophysiology of schizophrenia. npj Schizophrenia 2016; 2: 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolding NJ, Morgan BP, Compston DA. The expression of complement regulatory proteins by adult human oligodendrocytes. J Neuroimmunol 1998; 84: 69–75. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N et al. The classical complement cascade mediates CNS synapse elimination. Cell 2007; 131: 1164–78. [DOI] [PubMed] [Google Scholar]
- Syc SB, Saidha S, Newsome SD, Ratchford JN, Levy M, Ford E et al. Optical coherence tomography segmentation reveals ganglion cell layer pathology after optic neuritis. Brain 2012; 135: 521–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talman LS, Bisker ER, Sackel DJ, Long DA, Galetta KM, Ratchford JN et al. Longitudinal study of vision and retinal nerve fiber layer thickness in multiple sclerosis. Ann Neurol 2010; 67: 749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassoni A, Itoh Y, Itoh N, Farkhondeh V, Sofroniew M, Voskuhl R. Astrocyte specific transcriptomics reveal an A1 profile and complement activation in optic nerve during EAE. 2018. https://actrims.confex.com/actrims/2018/meetingapp.cgi/Paper/3096 (9 March 2018, date last accessed) [Google Scholar]
- Tewarie P, Balk L, Costello F, Green A, Martin R, Schippling S et al. The OSCAR-IB consensus criteria for retinal OCT quality assessment. PLoS One 2012; 7: e34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trip SA, Schlottmann PG, Jones SJ, Altmann DR, Garway-Heath DF, Thompson AJ et al. Retinal nerve fiber layer axonal loss and visual dysfunction in optic neuritis. Ann Neurol 2005; 58: 383–91. [DOI] [PubMed] [Google Scholar]
- Vanderweele TJ, Vansteelandt S, Robins JM. Effect decomposition in the presence of an exposure-induced mediator-outcome confounder. Epidemiology 2014; 25: 300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SS, de Andrade M, Tromp G, Kuivaniemi H, Pugh E, Namjou-Khales B et al. Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet 2014; 5: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8: 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LM, Neal JW, Loveless S, Michailidou I, Ramaglia V, Rees MI et al. Complement is activated in progressive multiple sclerosis cortical grey matter lesions. J Neuroinflamm 2016; 13: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing MG, Seilly DJ, Nicholas RS, Rahman S, Zajicek J, Lachmann PJ et al. Comparison of C1q-receptors on rat microglia and peritoneal macrophages. J Neuroimmunol 1999; 94: 74–81. [DOI] [PubMed] [Google Scholar]
- Wu GF, Schwartz ED, Lei T, Souza A, Mishra S, Jacobs DA et al. Relation of vision to global and regional brain MRI in multiple sclerosis. Neurology 2007; 69: 2128–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data collected for this study are part of a long-term research project; access to de-identified subject level data may be granted to those who meet criteria for confidential access and have the necessary data-sharing agreements in place.