Abstract
Sake-brewing cultivars among varieties of Japanese rice (Oryza sativa L.) have traits adapted to the sake-brewing process, such as a high white-core expression rate (WCE). Our previous study detected putative quantitative trait loci (QTLs) associated with a high WCE derived from Yamadanishiki, a popular brewing rice cultivar. Because the occurrence of white-core grains depends on air temperature and the position of the grain on the panicle, phenotyping of WCE must consider these variable conditions. In this study, qWCE6, a QTL for the WCE on chromosome 6, was validated for the first time, and the phenotyping method examined for its suitability in fine-mapping. A clear tendency towards high WCE was observed in late-heading substituted lines which headed under low daily mean temperature at the experimental location. White-core grains were often expressed by the primary spikelets on the upper panicle, producing a high percentage of superior grains. The segregating population for qWCE6 in late heading revealed a distinct difference in WCE between the Koshihikari and Yamadanishiki homozygous alleles at qWCE6 as determined from that locality. Further, two insertion/deletion markers were developed for the marker-assisted selection of qWCE6. Our results will be useful for informing the breeding of sake-brewing rice cultivars.
Keywords: sake-brewing rice cultivar, QTL, white-core grain, MAS, Yamadanishiki
Introduction
Japanese rice wine, “sake”, has recently become popular internationally, and the export amount has increased by about 2.7 times in the last 10 years (National Tax Agency Japan 2018). To produce sake, grains of rice (Oryza sativa L.) are first polished to a high grade and steamed after water absorption. Next, “koji” is fermented with the steamed rice and Aspergillus oryzae. Finally, sake is brewed from a combination of koji, a large amount of steamed rice, yeast (Saccharomyces cerevisiae), and water. Brewing rice cultivars are used which have been adapted to the brewing process and possess grain traits different from those of cooking rice cultivars. Of these traits, a high white-core expression rate (WCE) is characteristic of brewing rice with respect to grain appearance, and breeding of brewing rice cultivars targets this trait (Ikegami et al. 2003). White-core grain has an opaque structure in the central grain and displays positive effects on water absorption and, in particular, on the fermentation of koji (Horigane et al. 2014, Yanagiuchi et al. 1997). Elucidation of the genetic basis of this trait is therefore of interest for brewing rice cultivar breeding.
The quantitative trait loci (QTLs) associated with WCE in Yamadanishiki, the most popular brewing rice cultivar in Japan, have been previously reported. Yoshida et al. (2002) detected several QTLs on chromosome 12 and at an unknown position, and Okada et al. (2017) detected five QTLs on chromosomes 1 (qWCE1-1 and qWCE1-2), 3 (qWCE3), 6 (qWCE6), and 10 (qWCE10), which increased the WCE of the Yamadanishiki allele. Because the latter study used a greater number of DNA markers and lines than the former, its results should be treated with higher confidence. Of the detected QTLs, qWCE6 was found to have the largest genetic effect, and multiple QTLs for WCE were reported around qWCE6 in non-brewing rice cultivars (Peng et al. 2014, Tan et al. 2000). qWCE6 can thus be regarded as a highly reliable QTL for WCE; however, the genetic effects of all the QTLs associated with WCE have not been validated to date.
Chalky grains, including white-core grains, often result from high temperatures during rice ripening (Morita 2008, Tsukaguchi et al. 2012, Zhao and Fitzgerald 2013), and some heat-induced QTLs and genes associated with chalky grain have been identified (Hakata et al. 2012, Kobayashi et al. 2007, 2013, Miyahara et al. 2017, Nakata et al. 2017, Wada et al. 2015). On the other hand, the WCE of brewing rice cultivars exposed to high temperatures during ripening is lower than under normal conditions (Ishii et al. 2008). This phenomenon is found under both natural and controlled conditions, and was determined to yield larger white-core under high temperatures (Ikegami et al. 2015, Ishii et al. 2008). Therefore, very large size white-core was judged as milky-white grain (Ishii et al. 2008). Increasing the incidence of grain with large white-core also leads to an unwanted increase in the number of cracked grains under high-grade polishing. In addition, Okada et al. (2017) also detected an interaction between QTL and annual environmental conditions for qWCE6, in that the genetic effect of qWCE6 was lower in a year with a higher daily average temperature during the ripening stage. Thus, environmental conditions, in particular temperature, should be considered when evaluating WCE accuracy.
Furthermore, Nagato and Ebata (1958) speculated that there is a relationship between the occurrence of white-core grains and spikelet position on the panicle in the brewing rice cultivars Hattan-Jugo and Hyogo-Omachi. They reported that WCE was higher in the upper versus lower branches, and that the vigorously growing superior grains often expressed a white-core. The present study aimed to verify this phenomenon in Koshihikari, Yamadanishiki, and the chromosomal segment substitution lines selected for investigation.
The purpose of this study is to contribute to the breeding of brewing rice cultivars through validation of the genetic effect of qWCE6 and development of DNA markers for marker-assisted selection (MAS) of qWCE6. To validate qWCE6, we used several backcrossed lines and residual heterozygous lines (RHLs) derived from crossing Koshihikari and Yamadanishiki, as employed by Okada et al. (2017). Koshihikari and Yamadanishiki respectively have early and late heading dates, with a difference of about 20 days. The heading date distinction was largely expressed by qDTH3 (Okada et al. 2017, 2018a), which had very large genetic effects. Two causal genes linked closely have already been established as Hd6 and Hd16 (Hori et al. 2013, Takahashi et al. 2001). We were thus able to control whether plants had an early or late heading date by selecting the genotypes of DNA markers based on the causal variants of Hd6 and Hd16. In the case of rice plants with an early heading date, the daily average temperature during the ripening stage was higher than for those with a late heading date under our cultivation conditions. We therefore used lines with early and late heading date because WCE of Yamadanishiki and the genetic effect of qWCE6 were variable within an environment during the ripening stage (Ishii et al. 2008, Okada et al. 2017).
Materials and Methods
Plant materials
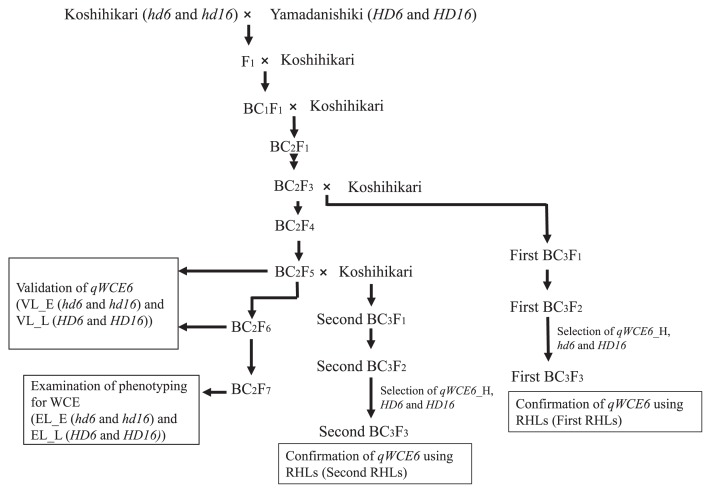
Fig. 1 shows the development process of materials used in this study. We used lines that featured a substituted chromosomal segment of Yamadanishiki on a Koshihikari genetic background. In total, two pairs of backcrossed lines and two RHLs were employed.
Fig. 1.
Development process of the materials used in this study. In the first validation of qWCE6, we used two lines from the BC2F5 and BC2F6 generations (validation lines with early and late heading dates; VL_E and VL_L). In the next experiment, we used the first residual heterozygous line (First RHLs). Two lines of the BC2F7 generation were used to verify the relationship between WCE and the spikelet position on the panicle (examination lines with early and late heading dates; EL_E and EL_L). Finally, the second BC3F3 population (Second RHLs) was used in the experiment segregating qWCE6. We selected for genotypes of qWCE6, Hd6 and Hd16 in the first and second RHLs using DNA markers. qWCE6_H indicates heterozygosity at qWCE6. The genotypes of the heading date genes are represented as hd6 and hd16 (Koshihikari alleles) or HD6 and HD16 (Yamadanishiki alleles).
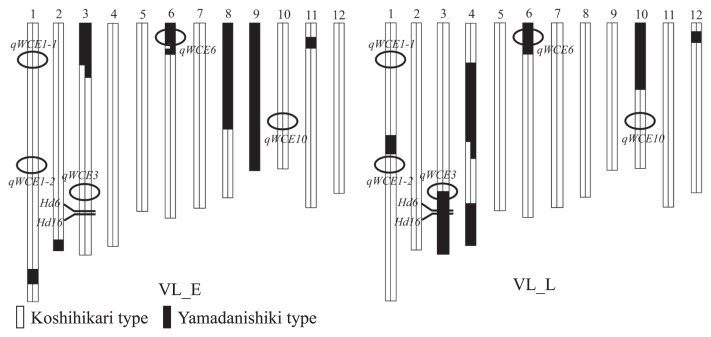
A total of the four backcrossed lines was derived from population increasing fixation by selfing BC2F1, generated by successively backcrossing Koshihikari twice followed by a F1 crossing between Koshihikari and Yamadanishiki. These lines were genotyped with 106 DNA markers in the BC2F4 generation using DNA extracted from the leaves following the approach of Dellaporta et al. (1983) with minor modifications (Supplemental Table 1). Two of the lines were used to verify the genetic effects of qWCE6 on the BC2F5 and BC2F6 generation and treated as validation lines with early or late heading date (VL_E and VL_L; Fig. 1). The genetic structure of the other two lines, also selected under early or late heading date conditions, had only qWCE6 at the Yamadanishiki allele of QTLs, as determined by our previous study, although a genotype of qWCE3 in VL_L remained unclear (Fig. 2).
Fig. 2.
Graphical genotypes of VL_E and VL_L in the BC2F4 generation. The white and black bars indicate the Koshihikari and Yamadanishiki chromosomal segments, respectively. The five circles represent the QTLs for WCE, and Hd6 and Hd16 are the genes associated with the heading date.
The other backcrossed lines were utilized to examine the WCE evaluation method focusing on the position of spikelets on a panicle in the BC2F7 generation, and to confirm whether a backcrossed line on Koshihikari genetic background also tended to express white-core on superior grains (Fig. 1; Nagato and Ebata 1958). These two backcrossed lines (examination line with early or late heading date; EL_E and EL_L) were homozygous at the qWCE6 Yamadanishiki allele and had similar WCEs and heading date to VL_E and VL_L, respectively.
Two RHLs populations were used to verify the genetic effect of qWCE6 under segregation of qWCE6 and the heading date control, because VL_E and VL_L had residual chromosomal segments of Yamadanishiki on regions except for qWCE6. These populations were selected by MAS with RM1369 and RM2615 as the markers enclosing qWCE6, a cleaved amplified polymorphic sequence (CAPS) marker targeted at a functional nucleotide polymorphism (FNP) of Hd6, and a PCR-confronting two-pair primer (PCR-CTPP) marker targeted at an FNP of Hd16 (Supplemental Table 1). One RHLs population (First RHLs) was derived from backcrossing a BC2F3 generation plant with Koshihikari (Fig. 1). From the First BC3F2 generation, we selected a rice plant for which qWCE6 was heterozygous and Hd16 was homozygous at the Yamadanishiki allele. Hd6 was already fixed as homozygous at the Koshihikari allele. The other RHLs population (Second RHLs) was developed by backcrossing a BC2F5 generation plant with Koshihikari and self-pollinating (Fig. 1). In this population, we selected a plant that was qWCE6 heterozygous and Hd6 and Hd16 homozygous at the Yamadanishiki allele from the Second BC3F2 generation.
Validation of qWCE6
VL_E, VL_L, and the parents were cultivated in an experimental field at Kobe University, Food Resources Education and Research Center (Kasai City, Hyogo Prefecture Japan; 34.88°N, 134.86°E) in 2013 and 2014. After harvesting, the grains were air-dried for three days and the WCE was evaluated. After dehulling, 100 grains were selected, omitting immature and cracked grains. The grains were visually inspected with a light box, white-core grains were counted, the WCE was calculated, and the occurrence rates of other chalky-grain types were also measured. In VL_E, seven (2013) and four (2014) plants were evaluated, and in VL_L, six (2013) and four (2014) plants were evaluated. Parents were measured using six plants in each year.
The greater part of the difference in heading date between Koshihikari (Aug. 3 in both years) and Yamadanishiki (Aug. 20 in 2013 and Aug. 17 in 2014) was driven by the tight linkage between Hd6 and Hd16 alleles (Fig. 2; Okada et al. 2017, 2018a).
Confirmation of qWCE6 genetic effects using RHLs
The First RHLs were cultivated in 72 plants. We sampled leaves from all plants and extracted DNA. To derive the qWCE6 genotype with the two markers (RM1369 and RM2615), we measured the WCE of nine (Koshihikari homozygous), 21 (heterozygous) and six (Yamadanishiki homozygous) plants, respectively. The WCE of the First RHLs was calculated by measuring the number of white-core grains in 100 grains using the same method as described above.
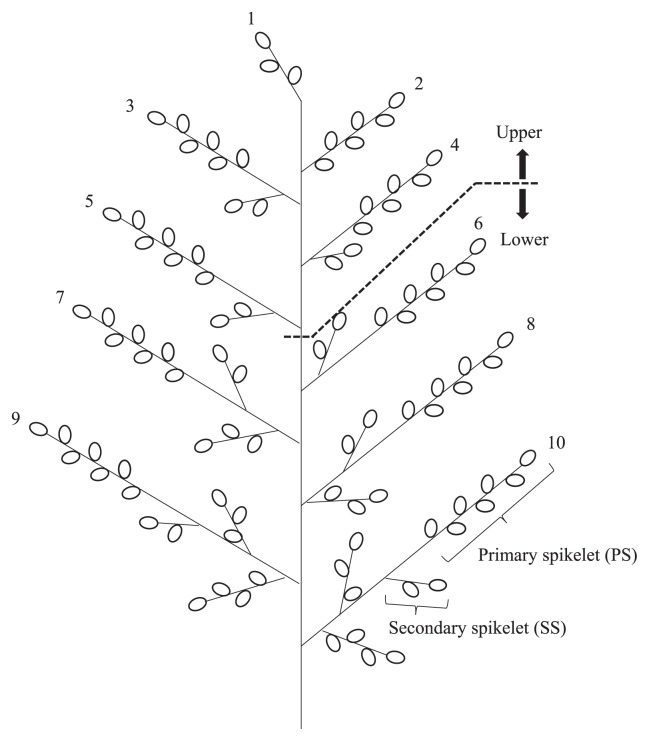
The Second RHLs were cultivated in 96 plants, and the qWCE6 genotype was identified with the same markers as above. We selected 19 (Koshihikari homozygous), 23 (heterozygous) and 15 (Yamadanishiki homozygous) plants, respectively. The WCE evaluation method was determined by the results of the experiment described below (see next section). We thus measured each WCE using 20 grains sampled from the primary spikelets (PS) on the upper part of one panicle per plant (Fig. 3). During sampling, superior grains were selected preferentially. Furthermore, dominance degree value (DDV) of qWCE6 was calculated according to Ukai (2000), namely the absolute value of dominance effect divided by additive effect of the QTL. In addition, in five samples selected randomly from each genotype population of qWCE6 we measured grain length and width using SmartGrain 1.0 (Tanabata et al. 2012).
Fig. 3.
Sampling panicle position. We classified positions on the panicle into “upper” and “lower” as follows: half the number of total primary branches counted from the top of the panicle were designated as “upper”, and the other half was designated as “lower”.
Examination of the WCE evaluation method focusing on the position of spikelets on a panicle
Heading dates of EL_E and EL_L were also similar to VL_E and VL_L, respectively (EL_E: Aug. 3, 2015 and EL_L: Aug. 26, 2015). For five panicles of each line and parent, the position of the spikelets was registered, and we checked whether each spikelet had a white-core. Next, the WCE was calculated for each part of the panicle, including PS, secondary spikelet (SS), upper-panicle PS and lower-panicle PS (Fig. 3).
Statistical analysis
We used R for the statistical analyses (ver. 3.4.1, R Core Team 2017). A Dunnett’s multi-comparison test was used for first validation. A Steel-Dwass test was conducted for experiments except for the first validation, using the NSM3 package (Schneider et al. 2018). Differences in grain length and width of Second RHLs were assessed using a Tukey-Kramer test.
Development of insertion/deletion (INDEL) markers
First, we searched INDEL sites using rice TASUKE (Kumagai et al. 2013, https://ricegenomes.dna.affrc.go.jp/), restricting the search to sequence depth values >5 between 1,534,301 bp and 3,662,323 bp on chromosome 6 on Koshihikari (ID: SAMD00045947) and Yamadanishiki (ID: SAMD00010633), using IRGSP-1.0 (Kawahara et al. 2013). The candidate region of qWCE6 was selected based on a previous QTL analysis of WCE (Okada et al. 2017, 2018b). We developed two INDEL markers, verified under the following PCR conditions: 1 cycle at 95°C for 2 min, 35 cycle at 95°C for 30 s, at 57°C for 30 s and 68°C for 1 min, post-extension at 72°C for 5 min, and stop at 4°C; using a reaction medium of 2 μl of crude extracted DNA, 3 μl of 2 μM mixed primer and 5 μl of Quick TaqTM HS DyeMix (Toyobo Life Science). Amplified products were separated by electrophoresis on a 3 % agarose gel and stained with ethidium bromide.
Results
Validation of the genetic effect of qWCE6
The graphical genotypes of VL_E and VL_L were shown in Fig. 2. VL_E had Koshihikari homozygous on both Hd6 and Hd16 and headed on Jul. 28, 2013 and Jul. 26, 2014, while VL_L had Yamadanishiki homozygous at both Hd6 and Hd16 of and headed on Aug. 21, 2013 and Aug. 20, 2014 (Table 1, Fig. 2).The WCE of VL_E did not significantly differ from that of Koshihikari, while the WCE of VL_L was significantly higher in 2013 (Table 1). In contrast, in 2014 the WCEs of both lines were significantly higher than that of Koshihikari (Table 1). This confirmed the genetic effect of qWCE6 but left a need for additional experiments, as both lines still contained large chromosomal segments of Yamadanishiki. In addition, the WCE of VL_E was variable in both years, while the WCE of VL_L was comparatively stable (Table 1).
Table 1.
Validation of the genetic effects of qWCE6
| WCE (%)a | Days to headingb | |||
|---|---|---|---|---|
|
|
|
|||
| 2013 | 2014 | 2013 | 2014 | |
| VL_E | 9.4 ± 9.1 | 29.0 ± 6.8** | 94 ± 1 | 95 ± 1 |
| VL_L | 18.7 ± 5.3** | 34.3 ± 1.9** | 118 ± 0 | 120 ± 1 |
| Koshihikari | 5.0 ± 2.8 | 9.7 ± 2.7 | 100 ± 1 | 102 ± 1 |
| Yamadanishikic | 70.5 ± 4.2 | 70.3 ± 4.1 | 117 ± 0 | 116 ± 1 |
Phenotypic data indicate “mean ± standard deviation (SD)”, and the astarisks represented significant difference between Koshihikari and each line (P < 0.01).
Significant difference test was not conducted for the data.
Yamadanishiki’s data are supplemental values.
First segregation test of qWCE6
The heading date of the First RHLs was from Aug. 14–24, and the population included late or slightly late heading lines. The heading date data for each genotype population of qWCE6 were Aug. 19–22 (Koshihikari homozygous), Aug. 15–23 (heterozygous) and Aug. 14–16 (Yamadanishiki homozygous). This distribution of heading dates resulted from segregating Hd17 in the candidate region of qWCE6 (Matsubara et al. 2012, Okada et al. 2017, 2018a). WCE of First RHLs was evaluated using the conventional method, but no statistically significant difference was detected among qWCE6 genotypes (Fig. 4). The lines that were homozygous at the qWCE6 Yamadanishiki allele appeared to have a higher WCE, but this tendency was not found to be statistically significant (Fig. 4). It was therefore necessary to consider improving the method used to evaluate WCE for qWCE6 validation and fine-mapping. As white-core grains generally express at superior grains on a panicle (Nagato and Ebata 1958), this was done by focusing on the spikelet position on a panicle, as described in the next section.
Fig. 4.
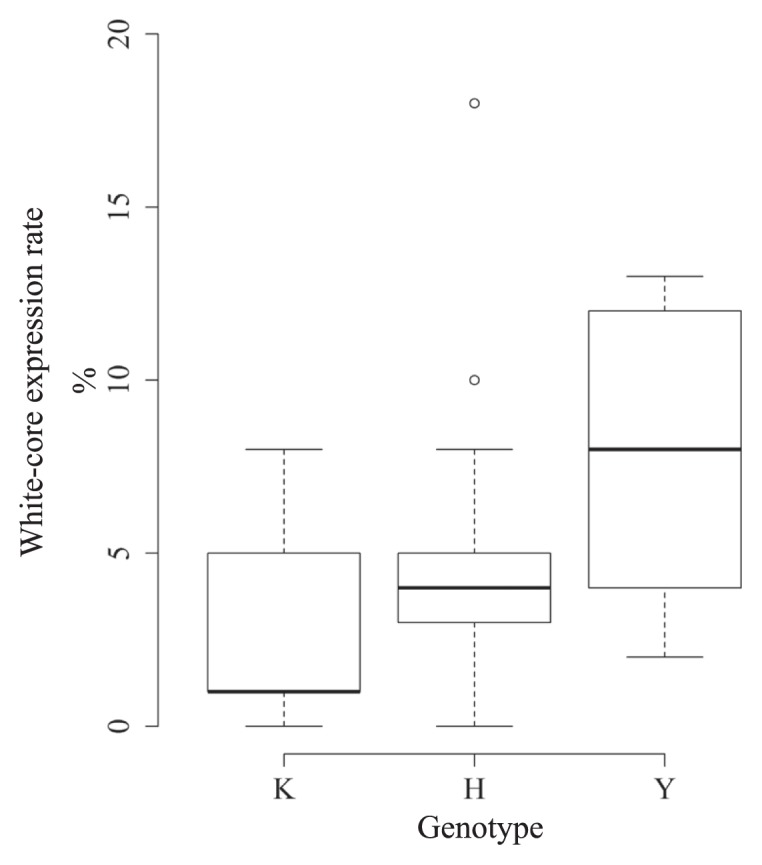
WCE classified the First RHLs into qWCE6 genotypes. On the horizontal axis, K, H, and Y represent the qWCE6 genotype for Koshihikari homozygous, Koshihikari heterozygous, and Yamadanishiki homozygous, respectively.
Relationship between WCE and spikelet position on a panicle
We first inspected the WCE of PS and SS (Table 2). PS WCE was higher than that of SS for all materials, but the WCEs of spikelets in both EL_E and EL_L were not higher than in Koshihikari (Table 2). We next focused on the PS, which had a high WCE, and divided the measure into PS on the upper panicle and lower panicle, respectively. EL_L was found to have a significantly increased PS WCE on the upper panicle (Table 2). This finding suggests that it is important to evaluate PS WCE on the upper panicle in late-heading material, sampling as many superior grains as possible.
Table 2.
Relationship between WCE and spikelet position on the panicle
| WCE of partial panicle (%)† | ||||
|---|---|---|---|---|
|
|
||||
| PS‡ | SS‡ | Upper PS | Lower PS | |
| EL_E | 30.2 ± 13.2a | 7.7 ± 3.4a | 33.5 ± 12.3ab | 26.2 ± 19.1a |
| EL_L | 28.7 ± 5.4a | 8.1 ± 5.3a | 45.0 ± 9b | 10.4 ± 10.1a |
| Koshihikari | 19.9 ± 7.6a | 5.6 ± 2.7a | 20.4 ± 9.3a | 19.5 ± 9.7a |
| Yamadanishiki | 85.2 ± 5b | 65.7 ± 7.2b | 85.4 ± 7.3c | 85.1 ± 8b |
WCE data show mean ± SD; the defferent letters in supertext represent statistical difference between groups (Steel-Dwass method, P < 0.05).
PS and SS represent primary and secondary spikelet, respectively.
Second segregation test of qWCE6
The heading dates of the Second RHLs were from Aug. 23 to Sep. 1, and late-heading was a characteristic of the heading date for this population. The heading date data for each genotype population of qWCE6 was from Aug. 25 to Sep. 1 (Koshihikari homozygous), Aug. 23–27 (heterozygous) and Aug. 23–27 (Yamadanishiki homozygous). The segregation of Second RHLs also was due to Hd17 (Matsubara et al. 2012, Okada et al. 2017, 2018a). We evaluated WCE using 20 grains from the PS on the upper panicle (superior grains as priority) based on the results of the above experiment. We found that the WCE of the population that was homozygous at the Yamadanishiki allele of qWCE6 was significantly higher than that of the Koshihikari-homozygous population (Fig. 5). This indicates a better performance of this evaluating method that of the method used for the First RHLs, although rice plants with a low WCE also existed in the Yamadanishiki-homozygous population. At the same time, the DDV of qWCE6 was 0.22 (Fig. 5). Grain length and width did not change with qWCE6 genotype (Supplemental Fig. 1).
Fig. 5.
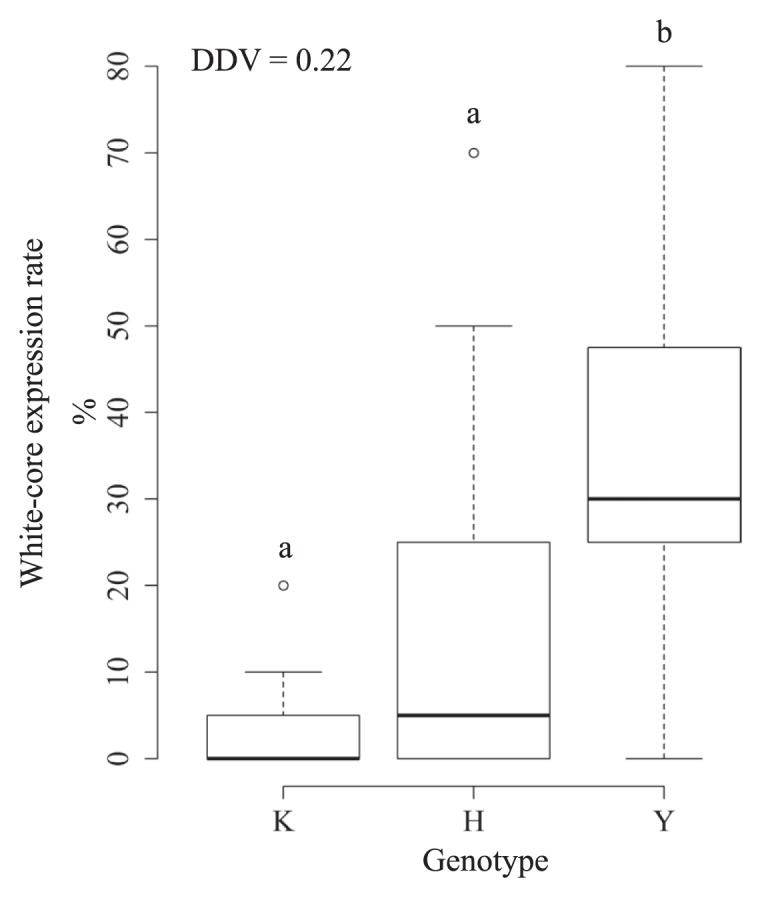
WCE classified the Second RHLs into qWCE6 genotypes. On the horizontal axis, K, H, and Y represent the qWCE6 genotype for Koshihikari homozygous, Koshihikari heterozygous, and Yamadanishiki homozygous, respectively. The different letters above each boxplot indicate the statistical differences between groups (Steel-Dwass test, P < 0.05). DDV represents dominance degree value calculated as an absolute value of dominance effect divided by additive effect of a QTL. When the value is DDV = 0, 0 < DDV < 1, DDV = 1 or 1 < DDV, inheritance manner of a QTL is defined as no dominance, incomplete dominance, complete dominance or overdominace, respectively (Ukai 2000).
Development of INDEL markers
A total of 14 INDEL sites was searched, to which one site at 3,652,857 bp was added due to its close proximity to one upstream INDEL site (Supplemental Table 2). Among these 15 sites, we targeted INDELs of 2,376,346 bp and both 3,652,851 bp and 3,652,857 bp because of relatively large INDELs. Two INDEL markers (INDEL_KY06_1 and INDEL_KY06_2) were established (Table 3, Fig. 6).
Table 3.
Primers for insertion/deletion markers for marker-assisted selection
| Marker name | Target position on chr.6 | Forward | Reverse | Amplicon size (bp)a | AT (°C)b | |
|---|---|---|---|---|---|---|
|
| ||||||
| Koshihikari | Yamadanishiki | |||||
| INDEL_KY06_1 | 2,376,346 | AAGCCAGCAGTTTGTTACACATC | AAACATTAGCACACGACGAGTC | 430 | 502 | 57 |
| INDEL_KY06_2 | 3,652,851 and 3,652,857 | ATCCCGAGTTCTCTTCCGAAAC | CGCCCAGAAAGAGAGAAGAGAAG | 321 | 382 | 57 |
Each PCR product size expected from rice TASUKE (Kumagai et al. 2013, https://ricegenomes.dna.affrc.go.jp/).
Annealing temperature.
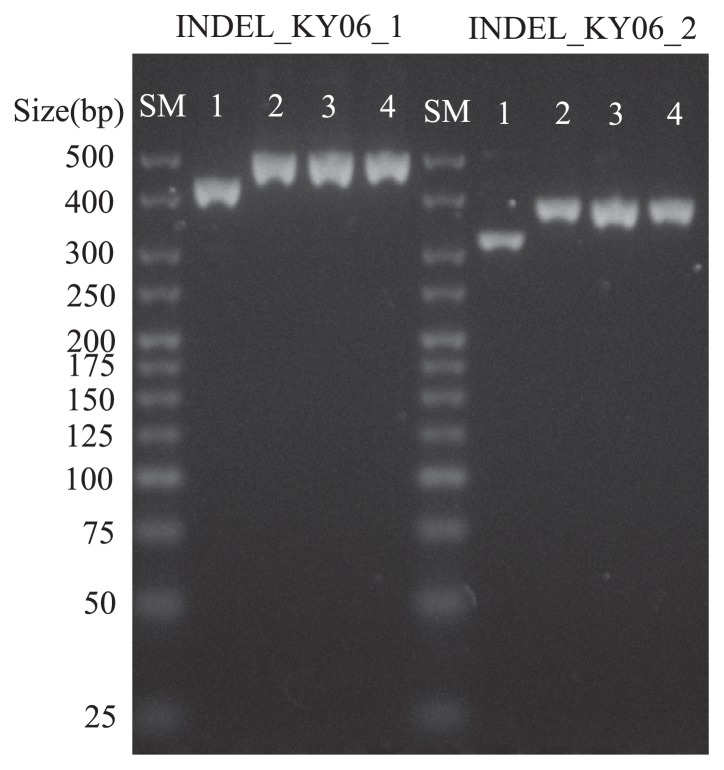
Fig. 6.
Electrophoregram of insertion/deletion (INDEL) markers developed in this study. The lanes of SM, 1, 2, 3 and 4 represented size marker, Koshihikari, Yamadanishiki, VL_E and VL_L.
Discussion
In this study, a line with the QTL for WCE showed WCE variation across two years; the early heading line (VL_E) exhibited variation in both years (2013 and 2014; Table 1), similar to the trend observed by Okada et al. (2017). In 2013 and 2014, the daily average temperature in the middle of August 2013 was higher than that in 2014 (Supplemental Fig. 2, cited from Okada et al. 2017). On the other hand, the average daily temperature from the end of August tended to show lower and lower transition in both years. This suggests that WCE variation in VL_E resulted from differences in average daily temperatures during the middle of August, which corresponds to the ripening stage of the VL_E. On the other hand, it is likely that the WCE of VL_L was stable because the ripening stages of VL_L in 2013 and 2014 were at lower temperature and under better conditions for the expression of white-core.
High temperatures during ripening have been shown to cause larger white-core size in a brewing rice cultivar (Ikegami et al. 2015, Ishii et al. 2008). Under the same conditions, the incidence of milky-white and white-back grains also increased in cooking rice cultivars (Ikegami et al. 2015). In the present study, VL_E in 2013 showed an increased occurrence rate over VL_L of other types of chalky grain through heat-stress, because its ripening period was at high daily mean air temperature (Supplemental Table 3, Supplemental Figs. 2, 3). We also suggest that distinguishing between white-core and chalky grains induced by heat-stress is difficult because part of the white-core fuses with other grain components in chalky grains.
We investigated the relationship between spikelet position on the panicle and WCE. As we found that there generally were many superior grains in the PS of the upper panicle, our results are consistent with those of Nagato and Ebata (1958), and we confirmed the same phenomenon for backcrossed lines on a Koshihikari genetic background. However, white-core grains found in Koshihikari and EL_E may result from high temperatures during the ripening stage due to the difficulties in distinguishing them from other chalky grain types, as noted above. Moreover, Yamadanishiki also had a higher WCE than the other lines in the SS (Table 2). It may be considered that high WCE in the SS resulted from having plural factors for occurring white-core because reported QTLs for WCE had low phenotypic variance explained and frequency of a line having WCE as high as Yamadanishiki was very low (Okada et al. 2017). Therefore, it is possible for a line with a high WCE in the SS to be developed, although the correlation between WCE in the SS and WCE evaluated with the conventional method needs to be confirmed using a segregating population.
The WCE of the First RHLs displayed no significant differences among the qWCE6 genotypes, while the WCE of the Second RHLs was clearly distinguished between populations that were homozygous at the Koshihikari and Yamadanishiki alleles by qWCE6 (Figs. 4, 5). In the experiment using the First RHLs, we measured WCE in 100 normal grains (randomly selected ignoring panicle position), and the genetic effect of qWCE6 was unclear. In addition, the WCE of the population that was qWCE6 homozygous at the Yamadanishiki allele was lower than that of VL_E and VL_L (Table 1, Fig. 4). To account for this phenomenon, we propose that the rate of Yamadanishiki chromosomal segments in the First RHLs was lower than in VL_E and VL_L due to triple backcrossing. Heterozygous regions in the population would thus still exist, as the genes increasing WCE are considered to be of slightly recessive inheritance (Kamijima et al. 1981). Okada et al. (2017) reported four QTLs that exclude qWCE6; however the genetic variation of the WCE population used in that study was not fully expressed by these QTLs, suggesting that other QTLs exist and have very small genetic effects.
The genetic effect of qWCE6 was thus clearly validated under the improving sampling method employed with the Second RHLs, and white-core grain associated with qWCE6 was found to be independent of grain size (Fig. 5, Supplemental Fig. 1). From the DDV of qWCE6, the mode of inheritance appears to be incomplete dominance (Fig. 5). However, some plants were found to have a low WCE despite being homozygous at the qWCE6 Yamadanishiki allele, probably because we calculated the WCE using only 20 grains in the Second RHLs. WCE should therefore be evaluated using a greater number of grains to attain a more accurate measurement.
The developed INDEL markers were able to distinguish between Koshihikari and Yamadanishiki (Fig. 6). These INDELs at 2,376,346 bp and 3,652,851 bp on chromosome 6 showed the different alleles from Yamadanishiki in more than 80% of 215 accessions of all temperate japonica in the rice TASUKE. This indicates that these markers will be of ready use for MAS of qWCE6.
Many QTLs associated with chalky rice grain have been reported around qWCE6. These were detected under either ambient or high temperature conditions. The QTLs for white-back and basal-white grains induced by heat-stress were identified by Kobayashi et al. (2007) and Kobayashi et al. (2013). In contrast, Tan et al. (2000) and Peng et al. (2014) reported QTLs for white-core and milky-white grain around qWCE6 under ambient temperature conditions. This indicates that qWCE6 may correspond to the latter QTLs due to the similar temperature environment. Zhao et al. (2016), Gong et al. (2017), and Qiu et al. (2017) detected the QTLs for the chalky grain rate around qWCE6 and suggested that qWCE6 is also related to these QTLs, although they did not classify chalky grain types. It has been shown that the mutation of genes associated with starch synthase induces chalky grains (Fujita et al. 2007, Nishi et al. 2001). For example, a mutation of Isoamylase 1 on chromosome 8 was shown to increase the chalky grain rate (Sun et al. 2015). Some genes associated with starch synthase are present in the endosperm around qWCE6, such as Waxy and starch synthase I (SSI; Yamakawa et al. 2008). Rice TASUKE demonstrated that two variants of Waxy, one each for Koshihikari and Yamadanishiki, existed in promotor region and 3′ UTR as one SNP and one INDEL, respectively. On the other hand, SSI is invariant between Koshihikari and Yamadanishiki. The Waxy variants are unlikely to represent causal variants of qWCE6 because Nipponbare (a cooking rice used as the reference sequence) and Yamadanishiki had the same allele. Further studies employing methodologies such as fine-mapping are therefore needed.
In conclusion, the present study for the first time validated the genetic effects of a white-core grain QTL, qWCE6, which constitutes a very important factor for Yamadanishiki. The INDEL markers developed in this study allow for MAS of qWCE6. We also improved WCE evaluation based on temperature and spikelet position on the panicle for use in future fine-mapping. Our findings suggest that late-heading lines should be used for evaluation at our study location, and that WCE should be measured using on the order of 100 superior grains. However, late-heading lines may only be suitable for locations comparable to that used in this study. Daily mean air temperature during the ripening stage in our location was normally below 26°C (Supplemental Fig. 2), which likely prevents white-cores from attaining great size and makes the expression of other types of chalky grains difficult (Ikegami et al. 2015, Ishii et al. 2008). In other regions, adjustments should be made to the heading date based on the environment (such as temperature) and the grain quality of the brewing rice cultivar in question. The heading date decision is thus also a key factor for breeding brewing rice cultivars. In future studies, an element of the genetic basis of white-core development will be elucidated by map-based cloning of qWCE6. Moreover, QTLs associated with white-core grain were also detected using mapping populations crossing between cooking rice cultivars around qWCE6 (Peng et al. 2014, Tan et al. 2000). Therefore, an allele decreasing WCE of qWCE6 may be available at breeding of a cooking rice cultivar having high-quality grain.
Supplementary Information
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 17J01082 and the Japan Science and Technology Agency (JST) CREST Grant Number JPMJCR17O3. We would like to thank Miki Suehiro and Wakana Yokoyama for their help with sampling.
Literature Cited
- Dellaporta, S.L., Wood, J. and Hicks, J.B. (1983) A plant DNA minipreparation version II. Plant Mol. Biol. Rep. 1: 19–21. [Google Scholar]
- Fujita, N., Yoshida, M., Kondo, T., Saito, K., Utsumi, Y., Tokunaga, T., Nishi, A., Satoh, H., Park, J.H., Jane, J.L.et al. (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144: 2009–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J., Miao, J., Zhao, Y., Zhao, Q., Feng, Q., Zhan, Q., Cheng, B., Xia, J., Huang, X., Yang, S.et al. (2017) Dissecting the genetic basis of grain shape and chalkiness traits in hybrid rice using multiple collaborative populations. Mol. Plant 10: 1353–1356. [DOI] [PubMed] [Google Scholar]
- Hakata, M., Kuroda, M., Miyashita, T., Yamaguchi, T., Kojima, M., Sakakibara, H., Mitsui, T. and Yamakawa, H. (2012) Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 10: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Hori, K., Ogiso-Tanaka, E., Matsubara, K., Yamanouchi, U., Ebana, K. and Yano, M. (2013) Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigane, A.K., Suzuki, K. and Yoshida, M. (2014) Moisture distribution in rice grains used for sake brewing analyzed by magnetic resonance imaging. J. Cereal Sci. 60: 193–201. [Google Scholar]
- Ikegami, M., Yoshida, S., Nakamura, C. and Kamijima, O. (2003) Heritability estimates of white-core expression in a sake-brewing rice (Oryza sativa L.) cultivar Yamadanishiki based on F2 variance and selection response in the F2 generation. Breed. Res. 5: 9–15. [Google Scholar]
- Ikegami, M., Fujimoto, H., Ogawa, T., Miyoshi, A., Yano, Y., Tsuchida, T. and Hirakawa, K. (2015) The relationships between temperature conditions and brown rice quality of a brewer’s rice cultivar “Yamadanishiki” in Hyogo prefecture. Jpn. J. Crop Sci. 84: 295–302. [Google Scholar]
- Ishii, K., Oba, K., Maruyama, A. and Katano, M. (2008) Effect of high temperature at grain filling period in TGC on grain texture of brewers’ rice “Yamada-nishiki”. Rep. Kyushu Br. Crop Sci. Soc. Japan 74: 24–26. [Google Scholar]
- Kamijima, O., Yamamoto, J. and Nakanishi, K. (1981) Studies on breeding rice for sake brewery II. Segregations of occurrence frequency of white-core kernels, kernel weight and culm length in F2 populations and interrelationships of these characters. Sci. Rept. Fac. Agr. Kobe Univ. 14: 265–272. [Google Scholar]
- Kawahara, Y., de la Bastide, M., Hamilton, J.P., Kanamori, H., McCombie, W.R., Ouyang, S., Schwartz, D.C., Tanaka, T., Wu, J., Zhou, S.et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (NY) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, A., Genliang, B., Shenghai, Y. and Tomita, K. (2007) Detection of quantitative trait loci for white-back and basal-white kernels under high temperature stress in japonica rice varieties. Beed. Sci. 57: 107–116. [Google Scholar]
- Kobayashi, A., Sonoda, J., Sugimoto, K., Kondo, M., Iwasawa, N., Hayashi, T., Tomita, K., Yano, M. and Shimizu, T. (2013) Detection and verification of QTLs associated with heat-induced quality decline of rice (Oryza sativa L.) using recombinant inbred lines and near-isogenic lines. Breed. Sci. 63: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, M., Kim, J., Itoh, R. and Itoh, T. (2013) TASUKE: a web-based visualization program for large-scale resequencing data. Bioinformatics 29: 1806–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K., Ogiso-Tanaka, E., Hori, K., Ebana, K., Ando, T. and Yano, M. (2012) Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53: 709–716. [DOI] [PubMed] [Google Scholar]
- Miyahara, K., Wada, T., Sonoda, J., Tsukaguchi, T., Miyazaki, M., Tsubone, M., Yamaguchi, O., Ishibashi, M., Iwasawa, N., Umemoto, T.et al. (2017) Detection and validation of QTLs for milky-white grains caused by high temperature during the ripening period in Japonica rice. Breed. Sci. 67: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, S. (2008) Prospect for developing measures to prevent high-temperature damage to rice grain ripening. Jpn. J. Crop Sci. 77: 1–12. [Google Scholar]
- Nagato, K. and Ebata, M. (1958) Studies on white-core rice kernel I. On the occurrence of white core. Jpn. J. Crop Sci. 27: 49–51. [Google Scholar]
- Nakata, M., Fukamatsu, Y., Miyashita, T., Hakata, M., Kimura, R., Nakata, Y., Kuroda, M., Yamaguchi, T. and Yamakawa, H. (2017) High temperature-induced expression of rice α-amylases in developing endosperm produces chalky grains. Front. Plant Sci. 8: 2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National tax agency JAPAN (2018) Sake report. https://www.nta.go.jp/taxes/sake/shiori-gaikyo/shiori/2018/pdf/000.pdf
- Nishi, A., Nakamura, Y., Tanaka, N. and Satoh, H. (2001) Biochemical and genetic analysis of the effects of Amylose-extender mutation in rice endosperm. Plant Physiol. 127: 459–472. [PMC free article] [PubMed] [Google Scholar]
- Okada, S., Suehiro, M., Ebana, K., Hori, K., Onogi, A., Iwata, H. and Yamasaki, M. (2017) Genetic dissection of grain traits in Yamadanishiki, an excellent sake-brewing rice cultivar. Theor. Appl. Genet. 130: 2567–2585. [DOI] [PubMed] [Google Scholar]
- Okada, S., Onogi, A., Iijima, K., Hori, K., Iwata, H., Yokoyama, W., Suehiro, M. and Yamasaki, M. (2018a) Identification of QTLs for rice grain size using a novel set of chromosomal segment substitution lines derived from Yamadanishiki in the genetic background of Koshihikari. Breed. Sci. 68: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, S., Sasaki, M. and Yamasaki, M. (2018b) A novel rice QTL qOPW11 associated with panicle weight affects panicle and plant architecture. Rice (NY) 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, B., Wang, L., Fan, C., Jiang, G., Luo, L., Li, Y. and He, Y. (2014) Comparative mapping of chalkiness components in rice using five populations across two environments. BMC Genet. 15: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Chen, K., Lv, W., Ou, X., Zhu, Y., Xing, D., Yang, L., Fan, F., Yang, J., Xu, J.et al. (2017) Examining two sets of introgression lines reveals background-independent and stably expressed QTL that improve grain appearance quality in rice (Oryza sativa L.). Theor. Appl. Genet. 130: 951–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/ [Google Scholar]
- Schneider, G., Chicken, E. and Becvarik, R. (2018) NSM3: Functions and Datasets to Accompany Hollander, Wolfe, and Chicken—Nonparametric Statistical Methods, Third Edition. https://CRAN.R-project.org/package=NSM3
- Sun, W., Zhou, Q., Yao, Y., Qiu, X., Xie, K. and Yu, S. (2015) Identification of genomic regions and the isoamylase gene for reduced grain chalkiness in rice. PLoS ONE 10: e0122013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Shomura, A., Sasaki, T. and Yano, M. (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 98: 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y.F., Xing, Y.Z., Li, J.X., Yu, S.B., Xu, C.G. and Zhang, Q. (2000) Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 101: 823–829. [DOI] [PubMed] [Google Scholar]
- Tanabata, T., Shibaya, T., Hori, K., Ebana, K. and Yano, M. (2012) SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaguchi, T., Yamamura, T., Inoue, H., Nakagawa, H., Murakami, K. and Kita, E. (2012) The response of the occurrence of milky white kernels with different cross-sectional patterns of chalkiness in the endosperm to grain-filling temperature and to assimilate supply in Koshihikari. Jpn. J. Crop Sci. 81: 267–274. [Google Scholar]
- Ukai, Y. (2000) Genetic analysis at the genomic level: Map and QTL. University of Tokyo Press, p. 241. [Google Scholar]
- Wada, T., Miyahara, K., Sonoda, J., Tsukaguchi, T., Miyazaki, M., Tsubone, M., Ando, T., Ebana, K., Yamamoto, T., Iwasawa, N.et al. (2015) Detection of QTLs for white-back and basal-white grains caused by high temperature during ripening period in japonica rice. Breed. Sci. 65: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, H., Ebitani, T. and Terao, T. (2008) Comparison between locations of QTLs for grain chalkiness and genes responsive to high temperature during grain filling on the rice chromosome map. Breed. Sci. 58: 337–343. [Google Scholar]
- Yanagiuchi, T., Yamamoto, H., Miyazaki, N., Nagano, T., Mizuma, T. and Wakai, Y. (1997) Influence of grain type on suitability of rice for sake brewing. Seibutsu-kogaku 75: 169–176. [Google Scholar]
- Yoshida, S., Ikegami, M., Kuze, J., Sawada, K., Hashimoto, Z., Ishii, T., Nakamura, C. and Kamijima, O. (2002) QTL analysis for plant and grain characters of Sake-brewing rice using a doubled haploid population. Breed. Sci. 52: 309–317. [Google Scholar]
- Zhao, X. and Fitzgerald, M. (2013) Climate change: implications for the yield of edible rice. PLoS ONE 8: e66218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X.Q., Daygon, V.D., McNally, K.L., Hamilton, R.S., Xie, F.M., Reinke, R.F. and Fitzgerald, M.A. (2016) Identification of stable QTLs causing chalk in rice grains in nine environments. Theor. Appl. Genet. 129: 141–153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.