Abstract
Currently, liver transplantation is the only available remedy for patients with end-stage liver disease. Conservation of transplanted liver graft is the most important issue as it directly related to patient survival. Carbonyl reductase 1 (CBR1) protects cells against oxidative stress and cell death by inactivating cellular membrane-derived lipid aldehydes. Ischemia-reperfusion (I/R) injury during living-donor liver transplantation is known to form reactive oxygen species. Thus, the objective of this study was to investigate whether CBR1 transcription might be increased during liver I/R injury and whether such increase might protect liver against I/R injury. Our results revealed that transcription factor Nrf2 could induce CBR1 transcription in liver of mice during I/R. Pre-treatment with sulforaphane, an activator of Nrf2, increased CBR1 expression, decreased liver enzymes such as aspartate aminotransferase and alanine transaminase, and reduced I/R-related pathological changes. Using oxygen-glucose deprivation and recovery model of human normal liver cell line, it was found that oxidative stress markers and lipid peroxidation products were significantly lowered in cells overexpressing CBR1. Conversely, CBR1 knockdown cells expressed elevated levels of oxidative stress proteins compared to the parental cell line. We also observed that Nrf2 and CBR1 were overexpressed during liver transplantation in clinical samples. These results suggest that CBR1 expression during liver I/R injury is regulated by transcription factor Nrf2. In addition, CBR1 can reduce free radicals and prevent lipid peroxidation. Taken together, CBR1 induction might be a therapeutic strategy for relieving liver I/R injury during liver transplantation.
Keywords: carbonyl reductase 1, ischemia-reperfusion injury, lipid peroxidation, living-donor liver transplantation, Nrf2, oxidative stress
INTRODUCTION
The blockage and reflow of blood in the liver during transplantation causes ischemia–reperfusion (I/R) injury, which is a challenge for successful outcome. Reoxygenation after oxygen shortage in the liver increases the levels of cellular reactive oxygen species (ROS) and initiates tissue injury (Jaeschke, 2003; Jaeschke and Woolbright, 2012). Carbonyl reductase 1 (CBR1), a member belonging to the family of short-chain dehydrogenase/reductases, catalyzes reactions involving many biologically and pharmacologically active carbonyl substrates (Kassner et al., 2008; Oppermann, 2007). Under oxidative stress during I/R injury, ROS drive lipid peroxidation by producing compounds with carbonyl functional groups including aldehydes and ketones, which are known to be highly reactive and potentially toxic to the cellular environment (Mathews et al., 1994; Oppermann, 2007). Interestingly, several studies report that CBR1 decreases the formation of lipd peroxides, thereby protecting cells from I/R injury (Rashid et al., 2010; Rotondo et al., 2016; Tak et al., 2011).
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a master transcription factor that regulates antioxidant production to maintain cellular redox homeostasis (Klaassen and Reisman, 2010). Under normal circumstances, Nrf2 is located in the cytoplasm and degraded by its repressor Kelch-like erythroid-associated protein 1 (Keap1) via the ubiquitin–proteasome pathway (Kaspar et al., 2009). Under pathological conditions, the interaction between Keap1 and Nrf2 is disrupted, leading to Nrf2 activation. Activated Nrf2 is translocated into the nucleus, where it heterodimerizes with Maf (Itoh et al., 1997). The heterodimer binds to the antioxidant response element (ARE) promoter regions, inducing transcription of many cytoprotective and antioxidative genes, including NAD(P)H:quinone oxidoreductase 1 (NQO1) and heme oxygenase 1 (HO-1) (Amersi et al., 1999; Kensler et al., 2007; Klaassen and Reisman, 2010). While the upregulation of Nrf2 decreases I/R injury, the underlying mechanism is unclear (Kudoh et al., 2014).
Based on our previous finding that Nrf2 directly regulates CBR1 transcription via the ARE region upstream of the CBR1 gene (Miura et al., 2013), we suggest that CBR1 is a potential therapeutic target for attenuation of hepatic I/R injury under the control of Nrf2. The functions of CBR1, in particular its tissue-specific activity and the protective mechanism, remain unknown. Here, we elucidated the function of CBR1 during oxidative stress and I/R using pharmacologic studies in tissue culture, animal experiments, and clinical investigations of living-donor liver biopsies.
MATERIALS AND METHODS
Cell culture
HepaRG (HPRGC10; Life Technologies, USA) cells were cultured in William’s E Medium (12551-032; Life Technologies) containing HepaRG Thaw, Plate, & General Purpose Medium Supplement (HPRG770; Life Technologies) and GlutaMAX Supplement (35050-061; Life Technologies). The cells were maintained in a humidified 37°C incubator (21% O2 and 5% CO2, MCO-15AC; SANYO Electric, Japan).
Oxygen-glucose deprivation and recovery (OGD/R) and hydrogen peroxide treatment
HepaRG cells were subjected to OGD/R using a humidified incubator with oxygen regulation up to 7 h followed by 2 h of reoxygenation. Briefly, the medium of HepaRG cells was replaced with glucose and sodium pyruvate-free DMEM (Thermo Fisher Scientific, USA) prior to inducing low-oxygen culture conditions. The glucose-deprived cells were exposed to hypoxic conditions (1% O2, 5% CO2, and 94% N2) in a multi-gas incubator (MCO-18M; SANYO Electric). The cells were washed with phosphate-buffered saline (PBS), replaced with normal culture medium, and maintained in 21% O2 conditions for 2 h. For hydrogen peroxide (H2O2) treatment, HepaRG cells were treated with hydrogen peroxide (200 μM, H1009; Sigma-Aldrich, USA) up to 7 h.
Immunofluorescence staining
HepaRG cells were seeded onto poly-L-lysine coated glass (GG-18-PLL; Neuvitro, USA) and treated OGD for 7 h followed by 2 h of recovery. After washing with ice-cold PBS 3 times, cells were fixed with 4% formaldehyde. Cells were permeabilized in PBS with 0.2% NP-40, 1% bovine serum albumin (BSA), and 0.1% sodium azide and washed 3 times with ice-cold PBS. The primary antibody, anti-CBR1 (1:100, ab156590; Abcam, UK), was diluted into PBS with 1% BSA and 0.1% NP-40 for 10 min, followed by incubation with the secondary antibody, anti-rabbit IgG-FITC (1:100, sc-2012; Santa Cruz Biotechnology, USA). After washing with PBS 3 times, cells were mounted using DAPI mounting solution (ab104139; Abcam). All images were observed under a fluoroscopic microscope (AxioObserver Z1; Carl Zeiss, Germany).
Luciferase reporter gene assay
HepaRG cells were seeded on a 12-well plate with 1.0 × 106 cells per well and transfected with 1 μg of pRL-SV40 and 3 μg of either a pGL4.20 backbone or similar vectors containing CBR1 promoters. The pGL4.20 Basic, pGL4.20-2062, pGL4.20-412, and pGL4.20-412-mutant vectors were designed as previously described (Miura et al., 2013). Lipofectamine 2000 (11668-019; Thermo Fisher Scientific) was used for transfection. After transfecting luciferase vectors, the cells were exposed to either OGD conditions for 7 h followed by 2 h of recovery or 200 μM of hydrogen peroxide for 7 h. Luciferase assay was conducted using the Promega dual-luciferase reporter assay system (E1910; Promega, USA) following the manufacturer’s instructions. The GloMax 96 microplate luminometer (Promega) was used to detect the activity.
Stable cell line transfection
HepaRG cells were transfected with pcDNA3-CBR1 wild type (CBR1/WT) or pcDNA3-basic using the lipofectamine 2000 system (Thermo Fisher Scientific). Stable cell lines were established as described previously (Tak et al., 2011). Briefly, the transfected HepaRG cells were maintained in a culture medium treated with G418 disulfate salt (600 mg/ml, A1720; Sigma-Aldrich), and G-418 resistant colonies were selected.
RNA interference
Small interfering RNAs (siRNA) specific to either CBR1 (CBR1-siRNA) or a scrambled sequence (scrambled-siRNA) were prepared by Bioneer (Korea). Cells were transfected with siRNA (0.5 μg) using Lipofectamine 2000 (Thermo Fisher Scientific). The siRNA target sequences were as follows: CBR1-siRNA sense, 5′-rCrArCrArGArAUUrArCUrCrCrCUrCUrCUrATT-3′; antisense, 5′-UrA rGrArGrGrArGUrArAUUrCUrGUrGTT-3′; scrambled-siRNA sense, 5′-UCCCAGAUAGAGACUUCAATT-3′; scrambled-siRNA antisense, 5′-UUGAAGUCUCUAUCUGGGATT-3′. The efficiency of siRNA-based interference with CBR1 synthesis was assessed by quantitative real time-polymerase chain reaction (qRT-PCR) and western blot.
Ischemia-reperfusion (I/R) animal model
Male C57BL/6 mice (8 weeks of age) were purchased from Orient Bio (Korea). Mice were maintained at 22°C ± 2°C, relative humidity 50% to 60%, under a 12-h/12-h light–dark cycle. To monitor time-dependent changes in I/R injury, mice were divided into 4 groups (sham, 2 h reperfusion, 6 h reperfusion, and 24 h reperfusion; n = 6 per group). All mice were anesthetized via intraperitoneal injection of a mixture of Zoletil 50 (0.15 ml/kg; Virbac, France) and Rompun (0.3 ml/kg; Bayer Korea, Korea) diluted in saline. To induce partial hepatic ischemia, an 11-mm long micro-clamp (00398-02; Fine Science Tools, USA) was used to obstruct the portal vein for 45 min and removed. Mice were sacrificed after reperfusion for 2 h, 6 h, or 24 h.
To observe drug-related phenotypic changes in I/R injury, mice were randomly divided into 8 groups (n = 6–12 per group). Mice were treated with either sulforaphane (SF, Cat#14797; Cayman Chemical Company, USA) or luteolin (LT, Cat#10004161; Cayman Chemical Company). SF was dissolved in saline and administered to mice (50 mg/kg body weight intraperitoneally). As LT did not dissolve in the saline, Polyoxyl 35 castor oil (also known as Kolliphor EL; BASF, Germany) was used as an emulsifier. LT was dissolved in saline containing 8% w/v Kolliphor EL and orally administrated to the mice (100 mg/kg body weight). Sham and I/R groups with SF and LT were treated with corresponding vehicles. Vehicles or SF and LT were administered 48 h and 24 h before starting the hepatic I/R protocol. Mice undergoing hepatic I/R injury were exposed to 45 min of ischemia followed by 6 h of reperfusion using the methods described above. All animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Asan Institute for Life Sciences at the Asan Medical Center (protocol No. 2015-04-172).
Serum chemistry
Whole blood samples were obtained from the inferior vena cava of the mice. All whole blood samples were collected in serum separator tubes (SSTs) (BD, USA) and incubated for 30 min at room temperature (22°C ± 2°C). After coagulation, SSTs were centrifuged at 4,000g for 10 min at 4°C. The clear supernatants were collected and stored as serum samples. Serum aspartate aminotransferase (AST) and alanine transaminase (ALT) were measured using a Hitachi 7180 autoanalyzer (Hitachi, Japan).
Immunohistochemistry
Liver tissues were harvested, fixed in 4% formalin solution, and embeded in paraffin. The paraffin blocks were cut into 2-μm sections and attached to glass slides. After removing the paraffin, the pre-fixed tissues were stained with H&E solution. Damage was scored by three pathologists using the Suzuki method based on the presence and/or severity of sinusoidal congeston, cytoplasmic vacuolization, and necrosis of parnechymal cells (score range, 0–4). Another set of sections (thickness, 4 μm) was procesed using anti-CBR1 antibody (1:500, ab186825; Abcam). Liver tissue was harvested following 6 h of reperfusion and stained as previously described. Sections were stained with H&E. Examination and scoring (Suzuki scoring, 0–4) based on the presence and/or severity of sinusoidal congestion, cytoplasmic vacuolization, and necrosis of parenchymal cells were performed with six representative sections of each liver sample in a blinded fashion.
Patient tissue samples
Liver tissues were obtained from patients undergoing living-donor liver transplantation who visited Asan Medical Center and provided written consent. Liver biopsies (– I/R) were obtained at the end of the cold ischemia time (CIT) during preparation of the donor liver. A second biopsy (+ I/R) was obtained immediately before closure of the abdomen following drain placement. Importantly, the total reperfusion time (RT) was defined as the time from portal vein perfusion to abdominal closure at the conclusion of the procedure. Patient characteristics are described on Table 1. The collection and use of patient samples were approved by the Asan Medical Center Institutional Review Board (AMC IRB) (approval No. 2016-0582).
Table 1.
Patients characteristics
| Patient No. | Donor age (y) | Recipient age (y) | MELD | CIT (min) | WIT (min) | RT (min) |
|---|---|---|---|---|---|---|
| 1 | 26 | 49 | 6 | 96 | 47 | 143 |
| 2 | 26 | 55 | 7 | 75 | 33 | 108 |
| 3 | 27 | 57 | 9 | 92 | 37 | 129 |
| 4 | 28 | 59 | 8 | 99 | 33 | 132 |
| 5 | 29 | 63 | 7 | 105 | 58 | 163 |
| 6 | 33 | 65 | 10 | 63 | 29 | 92 |
| 7 | 32 | 51 | 10 | 64 | 44 | 108 |
| 28.71 ± 1.06 | 58.43 ± 2.03 | 8.14 ± 0.59 | 84.86 ± 6.53 | 40.14 ± 3.83 | 125.00 ± 9.13 |
Values are presented as number only or mean ± SD.
MELD score, model for end-stage liver disease score; CIT, cold ischemia time; WIT, warm ischemia time; RT, reperfusion time.
Western blot analysis
HepaRG cells (2.0 × 106 cells) or 25–30 mg of liver tissues were obtained for further processing. Cytoplasmic and nuclear extracts were collected separately using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) including a protease inhibitor (Roche Diagnostics, Germany) following the manufacturer’s instructions. The protein concentration was measured using the bicinchoninic acid (BCA) method with 2 mg/ml BSA as the standard (Thermo Fisher Scientific). Equal amounts of protein were separated by 10% SDS-PAGE. The separated proteins were transferred onto nitrocellulose membranes. The membranes were blocked with 5% BSA in Tris-buffered saline with 0.1% Tween-20 (TBST, pH 7.6) for 1 h at room temperature (22°C–24°C), followed by incubation with the appropriate primary antibody overnight at 4°C and incubation with a secondary antibody for 1 h at room temperature. All antibodies were diluted in TBST (pH 7.6). Primary antibodies against NRF2 (1:1,000, ab62352; Abcam), TBP (1:1,000, sc-421; Santa Cruz Biotechnology), CBR1 (1:1,000, ab174852; Abcam), HSP70 (1:2,000, 610608; BD Biosciences, USA), and HSP60 (1:2,000, 611562; BD Biosciences) were used. Other compounds such as 4-hydroxynonenal (4HNE, 1:1000; LifeSpan Biosciences, USA), MDA (1:1,000, ALX-210-879; Enzo Life Sciences, USA), and Acrolein (1:1,000, LS-C63521; LifeSpan Biosciences) were also used. The nitrocellulose membranes were washed 3 times with TBST (pH 7.6) for 10 min at room temperature and incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Rabbit-IgG, 1:5,000, sc-2004; Mouse-IgG, 1:5,000, sc-2005; Santa Cruz Biotechnology). The membranes were washed five times with TBST for 10 min at room temperature. Membranes were treated with the chemiluminescence reagent SuperSignal West Femto Substrate (Thermo Fisher Scientific) and detected using the Image Quant LAS-4000 imaging system (GE Healthcare Life Sciences, USA). Western blot image was quantified using Image Studio Lite 5.0 (LI-COR Biosciences, USA). Protein levels were normalized to Actin, except for Nrf2 which was normalized to TBP. The levels of other compounds were normalized to matching control group.
Quantitative real-time PCR
Total RNAs were collected using Qiazol lysis reagent (Qiagen, Germany) and RNeasy mini kit (Qiagen) using a modified protocol. In short, HepaRG cells (2.0 × 106 cells) or 25–30 mg of liver tissues were treated with 0.5 ml of Qiazol lysis buffer and were collected, mixed with 0.1 ml of chloroform and centrifuged in a refrigerated centrifuge at the highest speed for 10 min. The clear supernatants were separately collected and the nucleic acids were precipitated using 70% ethanol. The precipitated total RNAs were bound on the silica column of RNeasy mini kit components, washed twice with RPE buffer (provided in the kit), and eluted with RNase-free distilled water (provided in the kit).
The concentration and quality of the extracted RNA were measured using Nanodrop2000 (Thermo Fisher Scientific). Samples with optical density 260/280 value above 1.8 were used for further experiments. The cDNA was generated from the mRNA using the ReverTra Ace qPCR RT Master Mix (Toyobo, Japan). The transcripts were quantified by real-time reverse transcription-polymerase chain reaction (RT-PCR) using the CFX Connect Real-Time PCR Detection System (Bio-Rad, USA) with 5× HOT FIREPol EvaGreen qPCR Supermix (Solis BioDyne, Estonia). The samples were first denatured at 95°C for 15 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 55°C–60°C for 15 s, and elongation at 72°C for 20 s. The data were expressed as the fold change in the treatment groups relative to the control and normalized to GAPDH levels. The primer sequences are shown in Table 2.
Table 2.
Human and mouse primers
| Sense or Antisense | Sequence (5′–3′) | |
|---|---|---|
| Human | ||
| GAPDH | Sense | ACAGTTGCCATGTAGACC |
| Antisense | TTGAGCACAGGGTACTTTA | |
| CBR1 | Sense | ACAAATTTCTTTGGTACCCG |
| Antisense | TAGATACGTTCACCACTCTC | |
| HSPD1 (HSP60) | Sense | CGTCTTGAATAGGCTAAAGG |
| Antisense | TGAACATCTTCAAGATTCAG | |
| HSPA14 (HSP70) | Sense | AGGAACATCCTTATCTCTCAG |
| Antisense | GATCTTTGGAACTCAGAAGC | |
| SOD1 | Sense | GAGCAGAAGGAAAGTAATGG |
| Antisense | GATTAAAGTGAGGACCTGC | |
| SOD2 | Sense | ATCATACCCTAATGATCCCAG |
| Antisense | AGGACCTTATAGGGTTTTCAG | |
| TXN1 | Sense | CTTTGGATCCATTTCCATCG |
| Antisense | GCAACATCATGAAAGAAAGG | |
| TXNRD1 | Sense | AGACAGTTAAGCATGATTGG |
| Antisense | AATTGCCCATAAGCATTCTC | |
| TNFα | Sense | AGGCAGTCAGATCATCTTC |
| Antisense | TTATCTCTCAGCTCCACG | |
| IL-6 | Sequence not provided | Hs_IL6_1_SG QuantiTect Primer Assay (Qiagen; product No. 249900, cat No. QT00083720) |
| IL-1β | Sequence not provided | Hs_IL1B_1_SG QuantiTect Primer Assay (Qiagen; product No. 249900, cat No. QT00021385) |
| Mouse | ||
| Gapdh | Sense | AGGTCGGTGTGAACGGATTTG |
| Antisense | TGTAGACCATGTAGTTGAGGTCA | |
| Cbr1 | Sense | CCTCTAATAAAACCCCAAGG |
| Antisense | CTATTAGGCCAACCTTCTTC | |
| TNFα | Sense | CTATGTCTCAGCCTCTTCTC |
| Antisense | CATTTGGGAACTTCTCATCC | |
| IL-6 | Sequence not provided | Mm_Il6_1_SG QuantiTect Primer Assay (Qiagen; product No. 49900, cat No. QT00098875) |
| IL-1β | Sequence not provided | Mm_Il1b_2_SG QuantiTect Primer Assay (Qiagen; product No. 249900, cat No.QT01048355) |
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software, USA). Liver injury score data were presented as the median and range. All other data were presented as the mean ± SD. Statistical analyses were performed using ANOVA followed by Bonferroni’s multiple comparison test; the exact P value (when P > 0.001 but < 0.05) or the highest P value (P < 0.001) were described.
RESULTS
OGD/R and hydrogen peroxide treatment both induces CBR1 transcription via Nrf2
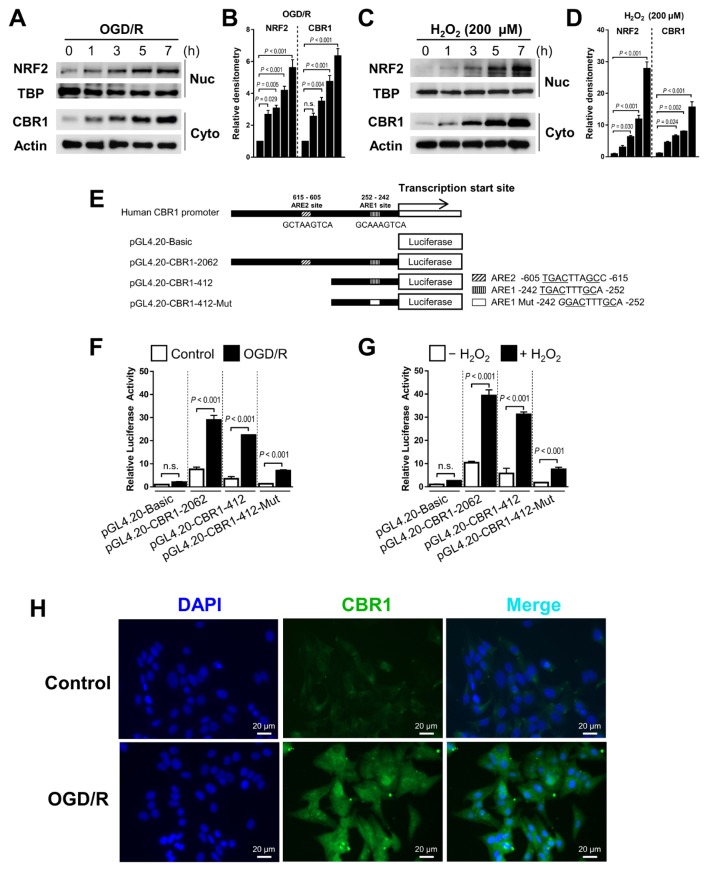
To investigate whether Nrf2 increased the CBR1 transcription, we treated HepaRG, the normal hepatocyte cells OGD/R or hydrogen peroxide for up to 7 h. In response to OGD/R and hydrogen peroxide treatment, the expression of both CBR1 and Nrf2 was increased in quantified western blot analysis (Figs. 1A–1D). To determine whether OGD/R and hydrogen peroxide-induced CBR1 expression requires Nrf2, we investigated the CBR1 promoter region up to −2,000 bp from the transcription start site (Fig. 1E). We found two putative Nrf2-binding regions matching the ARE (TGACXXXGC consensus sequence), located at −615 bp and −252 bp upstream. We found that pGL4.20-CBR1-2062 and pGL4.20-CBR1-412 showed high luciferase activity under OGD/R, while a pGL4.20-CBR1-ARE1 mutant containing 5′-GGACTTTGCA-3′, instead of 5′-TGACTTTGCA-3′ at the ARE exhibited limited luciferase activity (Figs. 1F and 1G). Immunofluorescence staining using a CBR1 monoclonal antibody revealed that CBR1 was strongly induced by OGD/R (Fig. 1H).
Fig. 1. Temporal regulation of CBR1 expression by Nrf2 with OGD/R and hydrogen peroxide.
(A–D) CBR1 is upregulated temporally by Nrf2 during OGD/R and hydrogen peroxide treatment. Western blot images and densitometric quantitation of the blots. (E) Schematic diagram of the promoter regions of human CBR1, including full-length ARE1 and ARE2 and an ARE1 mutant. (F and G) Luciferase reporter assay with OGD/R and hydrogen peroxide treatment. (H) CBR1 immunofluorescent staining. Normal hepatic hepaRG cells were stained with anti-CBR1 monoclonal antibody. Blue, DAPI; Green, human CBR1. Data are representative of five independent experiments. Data are presented as the mean ± SD. Respective comparison was indicated by a line and P value. n.s., not significant.
CBR1 expression increases during murine hepatic I/R in a time-dependent manner
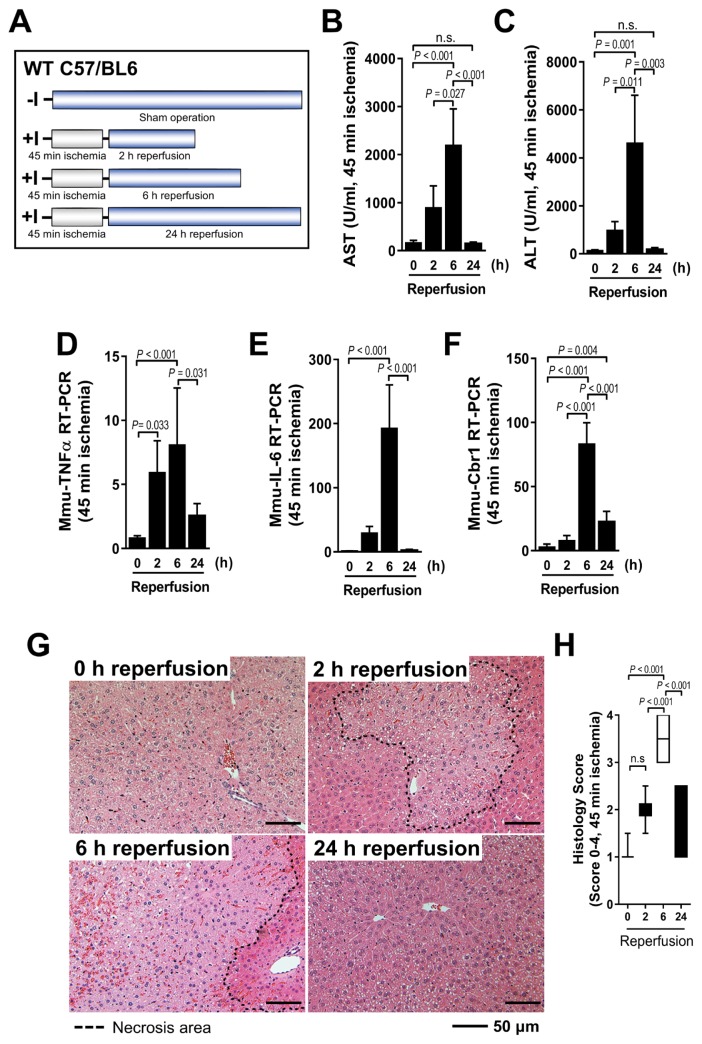
The time course of animal experiment was measured during partial hepatic I/R in WT mice (Fig. 2A). The serum AST and ALT levels peaked after reperfusion for 6 h (Figs. 2B and 2C). Similar patterns of tumor necrosis factor alpha (TNFα) and interleukin 6 (IL-6) transcription were detected in the damaged liver (Figs. 2D and 2E). These results were confirmed by qRT-PCR, with CBR1 expression increasing in a time-dependent manner, peaking after 6 h of reperfusion and decreasing significantly thereafter (Fig. 2F). In addition, histological analysis revealed a significantly greater necrosis (Fig. 2G) and higher Suzuki scores (Fig. 2H) in the sections undergoing 6 h of reperfusion after 45 min of ischemia. Taken together, these findings indicate that CBR1 expression was associated with murine hepatic I/R injury.
Fig. 2. CBR1 expression increased during hepatic I/R in a mouse model.
(A) Schematic outline of mice experiments. (B) AST and (C) ALT after ischemic period for 45 min followed by 2 h, 6 h, and 24 h of reperfusion. Relative expression of (D) murine TNFα, (E) murine IL-6, and (F) murine Cbr1 measured by qRT-PCR. Expression levels are normalized to Gapdh. (G) Liver histology and (H) quantification of histologic tissue injury using the Suzuki Scoring Index (0–4). The necrosis area is outlined by dotted lines. Comparison is indicated by a line and P value. n.s., not significant. n = 6 per group.
Sulforaphane attenuates hepatic I/R injury by increasing CBR1 expression via Nrf2
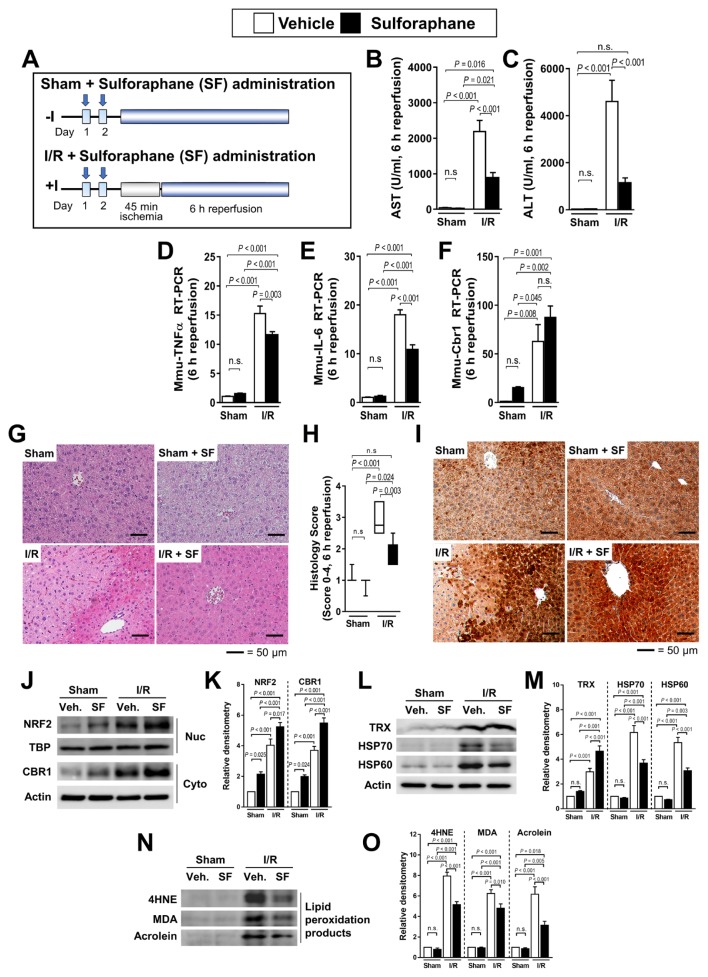
To confirm whether SF, an inducer of Nrf2 target genes, attenuated hepatic I/R injury by increasing CBR1 expression, we administered SF 48 h and 24 h before 45 min of hepatic ischemia and 6 h of reperfusion (Fig. 3A). Following I/R, SF-treated mice showed lower levels of AST and ALT compared with untreated mice (Figs. 3B and 3C). The expression of the inflammatory cytokines TNFα and IL-6 was also lower in treated mice, as revealed by RT-PCR (Figs. 3D and 3E). Liver Cbr1 levels were consistently elevated by SF but not significantly (Fig. 3F). Histological analysis revealed less tissue injury in SF-treated than untreated mice (Figs. 3G and 3H). Immunohistochemical analysis showed that SF administration induces Cbr1 expression (Fig. 3I). Western blot analysis showed that both CBR1 and Nrf2 expression was increased in SF-treated mice (Figs. 3J and 3K). The expression of thioredoxin (TRX) was significantly increased upon exposure to SF or I/R conditions. The expression of TRX peaked upon exposure to I/R and SF together. Expression of the oxidative stress markers HSP70 and HSP60 was increased significantly under I/R conditions and restored when treated with SF (Figs. 3L and 3M). The lipid peroxidation products 4HNE, MDA, and Acrolein increased significantly under I/R conditions, which was attenuated by SF treatment (Figs. 3N and 3O).
Fig. 3. Administration of CBR1 inducer SF attenuates hepatic cell death.
(A) Schematic outline of SF administration of mice. Measurement of (B) ALT and (C) AST. Relative expression of (D) murine TNFα, (E) IL-6, and (F) Cbr1 measured by qRT-PCR. Expression levels are normalized to Gapdh. (G) Liver histology and (H) quantification of liver injury. (I) Immunohistochemistry of murine CBR1. (J and K) Protein expression of Nrf2 and CBR1 and relative density. (L and M) Western blot analysis of oxidative stress markers (TRX, HSP70, and HSP60) and relative density. (N and O) Western blot of lipid peroxidation products (4HNE, MDA, and Acrolein) and relative density. Protein levels were normalized to Actin, except for Nrf2 which was normalized to TBP. The levels of other compounds were normalized to matching control group. Veh., vehicle; White bar, vehicle-treated; Black bar, SF-treated. Comparison is indicated by a line and P value. n.s., not significant. n = 6–12 per group.
Luteolin increases hepatic ischemia–reperfusion injury via the Nrf2–CBR1 signaling pathway
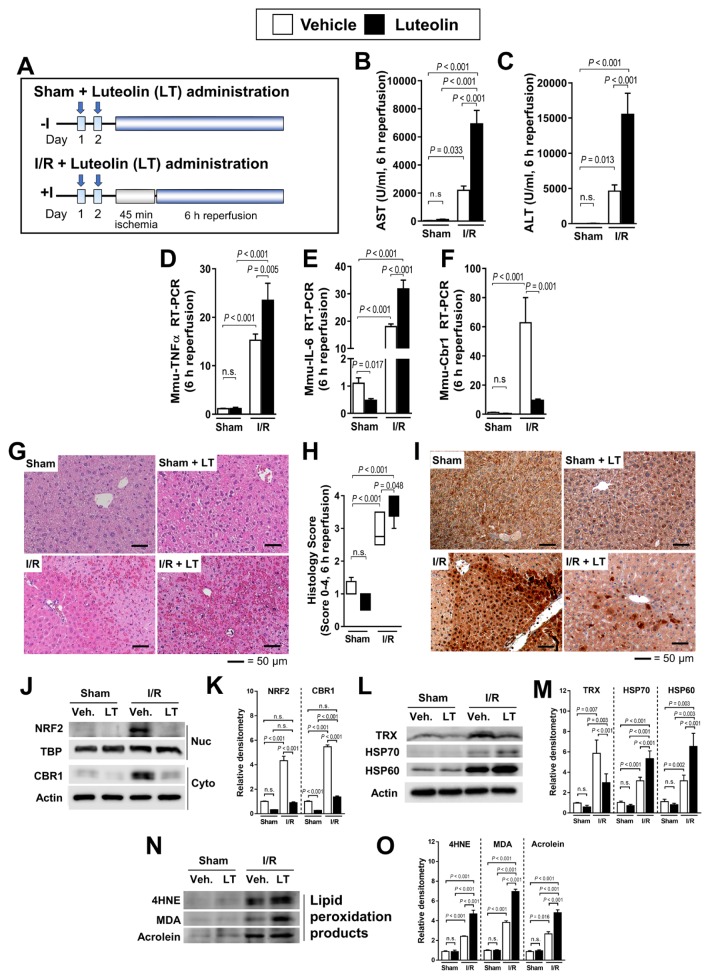
The administration of LT, an inhibitor of CBR1 expression, 48 h or 24 h before 45 min of hepatic ischemia and 6 h of reperfusion injury (Fig. 4A) resulted in elevated AST and ALT levels in treated than in untreated mice (Figs. 4B and 4C). RT-PCR revealed that TNFα and IL-6 expression also was higher in LT-treated mice (Figs. 4D and 4E). The CBR1 expression was dramatically lower in LT-treated mice (Fig. 4F). Additionally, histological analysis revealed a more severe damage in LT-treated than untreated mice (Figs. 4G and 4H). Immunohistochemical analysis (Fig. 4I) and western blot results showed that LT administration before I/R decreased CBR1 expression (Figs. 4J and 4K). The levels of HSP70 and HSP60, which are the oxidative stress markers, increased under I/R exposure in individuals treated with LT, while the expression of TRX was decreased by LT (Figs. 4L and 4M). Lipid peroxidation products (4HNE, MDA, and Acrolein) were elevated in all mice after I/R, with greater elevation in LT-treated mice (Figs. 4N and 4O).
Fig. 4. Administration of the CBR1 reducer LT increases hepatic ischemia–reperfusion injury and hepatic cell death.
(A) Schematic outline of LT administration of mice. Serum measurement of (B) AST and (C) ALT. Relative expression of (D) murine TNFα, (E) IL-6, and (F) Cbr1 measured by qRT-PCR. Expression levels are normalized to Gapdh. (G) H&E staining of liver injury and (H) histology score of LT-treated mice. (I) Immunohistochemistry of CBR1. (J and K) Protein expression of Nrf2 and CBR1 and relative density. (L and M) Western blot analysis of oxidative stress markers (TRX, HSP70, and HSP60) and relative density. (N and O) Western blot of lipid peroxidation products (4HNE, MDA, and Acrolein) and relative density. Protein levels were normalized to Actin, except for Nrf2 which was normalized to TBP. The levels of other compounds were normalized to matching control group. Veh., vehicle; White bar, vehicle-treated; Black bar, LT-treated. Comparison is indicated by a line and P value. n.s., not significant. n = 6–12 per group.
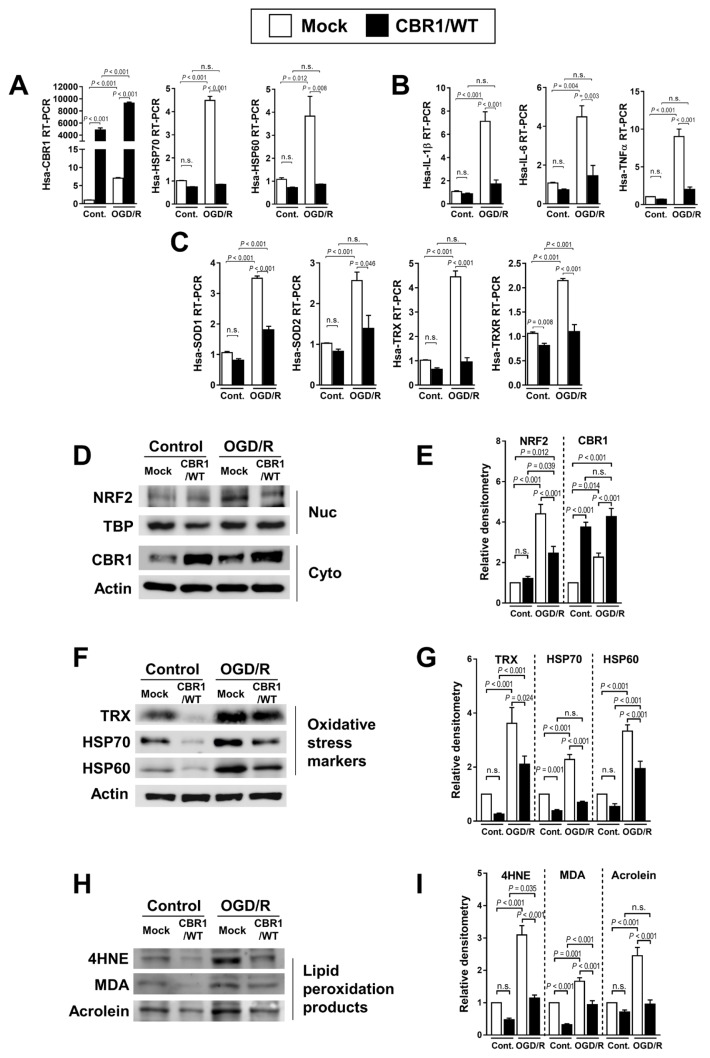
Overexpressed or knockdown CBR1 is related to lipid peroxidation downstream products
To further investigate the role of CBR1 expression in attenuating I/R injury, we used hepaRG cells to create a cell line overexpressing CBR1, which exhibited stable expression of CBR1. CBR1-overexpressing cells (CBR1/WT) showed a significantly high expression of CBR1. The cells with control vectors (Mock) and CBR1-overexpressing cell lines both expressed higher transcriptional levels of CBR1 under OGD/R conditions (Fig. 5A). RT-PCR analysis of this cell line after 7 h exposure to OGD/R revealed that the levels of the stress markers HSP70 and HSP60 were lower than in the parent cell line (Fig. 5A). The expression of interleukin 1 beta (IL-1β), IL-6, and TNFα also was lower in CBR1-overexpressing cells (Fig. 5B), similar to the expression of SOD1, SOD2, TRX, and TRX reductase (TRXR) (Fig. 5C). The protein levels of Nrf2 were significantly increased under OGD/R while the expression of CBR1 was diminished (Figs. 5D and 5E). We also observed a lower expression of a variety of stress-response markers (TRX, HSP70, and HSP60) in CBR1-overexpressing cells (Figs. 5F and 5G). Similar trends were also observed for the lipid peroxidation products, 4HNE, MDA, and Acrolein (Figs. 5H and 5I).
Fig. 5. Gain-of-function study of CBR1 in normal hepatocyte cells under hydrogen peroxide treatment.
(A) Transcription levels of CBR1 and oxidative stress markers (HSP70 and HSP60) in mock transfected cells and a CBR1-overexpressing stable cell line under OGD/R. (B) Fold change in inflammatory cytokines (IL-1β, IL-6, and TNFα) in mock control vs CBR1-overexpressing cells. (C) Expression of antioxidant enzymes (SOD1, SOD2, TRX, and TRXR) in CBR1-overexpressing cells under OGD/R. (D and E) Western blot analysis of Nrf2 and CBR1 with relative density. (F and G) Oxidative stress markers (TRX, HSP70, and HSP60) with relative density. (H and I) Lipid peroxidation products (4HNE, MDA, and Acrolein) with relative density. Protein levels were normalized to Actin, except for Nrf2 which was normalized to TBP. The levels of other compounds were normalized to matching control group. Cont., control; White bar, Mock-treated; Black bar, CBR1/WT. Comparison is indicated by a line and P value. n.s., not significant. n = 10 per group.
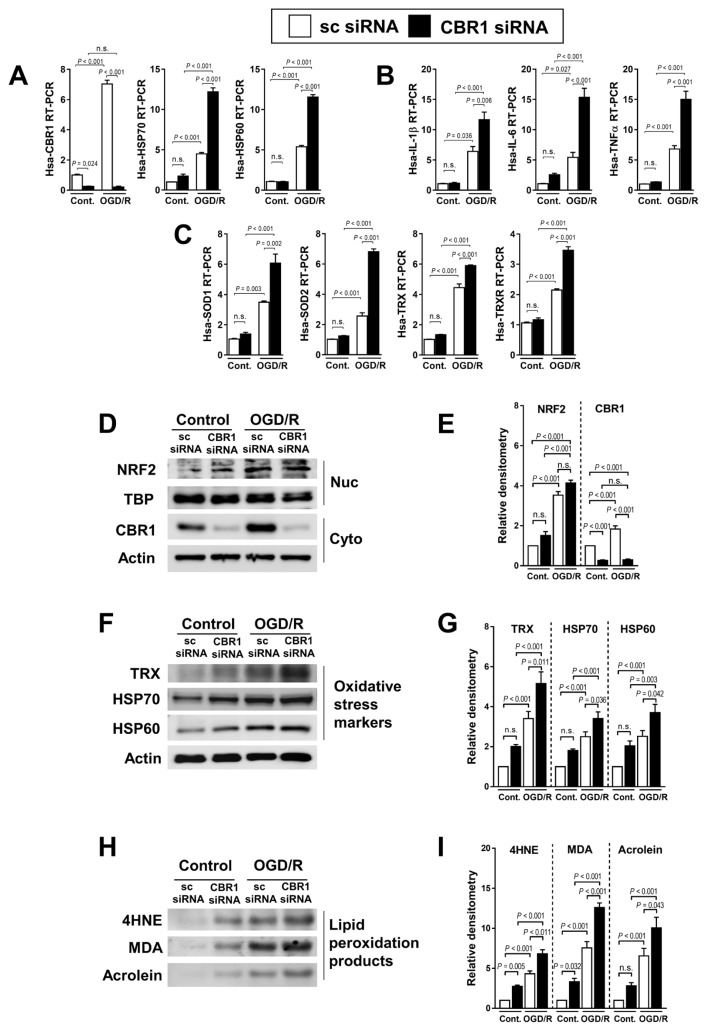
Conversely, we constructed a CBR1-knock down cell line with CBR1 siRNA, resulting in a profoundly decreased CBR1. The RT-PCR analysis revealed that cells with a lower CBR1 expression showed higher levels of damage markers (HSP70 and HSP60; Fig. 6A), pro-inflammatory cytokines (IL-1β, IL-6, and TNFα; Fig. 6B), and oxidative stress markers (SOD1, SOD2, TRX, and TRXR; Fig. 6C). The nuclear levels of Nrf2 were not changed by CBR1 siRNA; however, they were increased by OGD/R (Figs. 6D and 6E). The expression of oxidative stress markers (TRX, HSP70, and HSP60; Figs. 6F and 6G) and lipid peroxidation products (4HNE, MDA, and Acrolein) was elevated in CBR knockdown cells (Figs. 6H and 6I).
Fig. 6. Loss-of-function study of CBR1 in normal hepatocyte cells under hydrogen peroxide treatment.
(A) Transcription levels of CBR1 and oxidative stress markers with control siRNA and CBR1 siRNA under OGD/R. (B) Fold change in inflammatory cytokine expression in control and CBR1 siRNA cells. (C) Expression of antioxidant enzymes in CBR1 siRNA cells under OGD/R. (D and E) Western blot analysis of Nrf2 and CBR1 with relative density. (F and G) Oxidative stress markers (TRX, HSP70, and HSP60) with relative density. (H and I) Lipid peroxidation products (4HNE, MDA, and Acrolein) with relative density. Protein levels were normalized to Actin, except for Nrf2 which was normalized to TBP. The levels of other compounds were normalized to matching control group. Cont., control; White bar, scrambled siRNA-treated; Black bar, CBR1 siRNA-treated. Comparison is indicated by a line and P value. n.s., not significant. n = 10 per group.
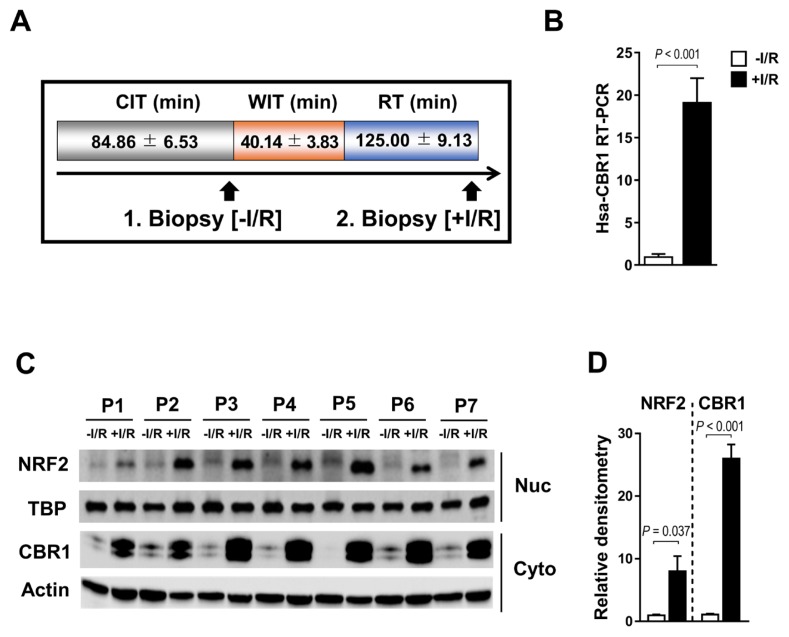
Human CBR1 expression is upregulated by Nrf2 during orthotopic living-donor liver transplantation
To investigate the role of CBR1 in I/R injury in humans, we harvested two liver biopsies from each of the seven patients; one immediately after incision for use as a control and the other after reperfusion (Fig. 7A). Consistent with our previous results in mice, human CBR1 was upregulated after reperfusion compared with the control (Fig. 7B). Western blot analysis of the nuclear and cytosolic fractions of cell lysates revealed elevated expression of Nrf2 and CBR1 during living-donor liver transplantation (Fig. 7C). Quantitative western blot analysis of Nrf2 and CBR1 expression showed consistent results (Fig. 7D).
Fig. 7. CBR1 expression in biopsies of transplanted human liver.
(A) Liver specimens were harvested at the conclusion of CIT during the preparation of living-donor liver allograft and during reperfusion after warm ischemia and reperfusion, immediately before suturing of the abdomen following drain placement. WIT, warm ischemia time; RT, reperfusion time. (B) CBR1 mRNA level by RT-PCR. (C) Nrf2 and CBR1 proteins in 7 patients (P1–P7; one representative blot of 3 is shown) as assessed by western blot analysis and (D) quantification of CBR1 expression by density. Protein levels were normalized to Actin, ecxept for Nrf2 which was normalized to TBP. White bar, before I/R; Black bar, after I/R. Comparison is indicated by a line and P value. n.s., not significant.
DISCUSSION
Hepatic I/R injury leads to severe liver damage and is a significant cause of liver transplantation failure (Serracino-Inglott et al., 2001). Prevention and amelioration of I/R injury is an unmet clinical need. The current paradigm of hepatic I/R injury is based on two apparently separate events involving cell damage in the ischemic phase and inflammation during the reperfusion phase (Kensler et al., 2013). While ischemic insult is readily tolerated by the liver, the reperfusion-induced inflammatory response is the main culprit in liver destruction (Kalogeris et al., 2011; 2012). Thus, recent efforts to develop therapeutic agents have focused on the direct inhibition of inflammation and cell death during the reperfusion stage. However, the limited therapeutic effects observed in rodent models cannot be translated to humans, and the clinical benefits of this approach have yet to be established unequivocally.
CBR1 is known to increase cell survival under oxidative stress by preventing the formation of lipid peroxidation products (Ellis, 2007; Rashid et al., 2010). In this study, our results indicate that Nrf2 induced CBR1 transcription during both OGD/R and hydrogen peroxide treatment, and hepatic I/R injuries in mice and humans. Previous studies showed the chemically induced Nrf2 promoted CBR1 expressions, but not in physiological conditions, especially under liver I/R injury. We showed that pre-treatment with SF, an inducer of CBR1 expression (Thimmulappa et al., 2002) decreased the expression of the liver function enzymes (AST and ALT) and significantly decreased I/R injury-related pathological changes in liver tissues. On the other hand, LT, a known inhibitor of Nrf2 decreased the CBR1 expression in liver tissues exposed to more severe I/R injury. As TRX is a known target for Nrf2, the protein levels of TRX were increased when mice were treated with SF. Conversely, LT exposure decreased the levels TRX suggesting that SF and LT worked as Nrf2 regulators in the mouse model of liver I/R injury.
Western blot analysis showed that the expression of oxidative stress markers (TRX, HSP70, and HSP60) and lipid peroxidation products (4HNE, MDA, and Acrolein) was significantly lower in cells overexpressing CBR1. CBR1 knockdown elevated the levels of oxidative stress proteins compared with the parental cell line. We also observed that human CBR1 was overexpressed during orthotropic living-donor liver transplantation via Nrf2.
Studies showed that overexpressed CBR1 attenuated hepatic cell death against oxidative stress by reducing ROS within cells and such mechanisms were partially regulated by Nrf2 system (Higdon et al., 2012; Marinho et al., 2014; Ray et al., 2012). In addition, we confirmed that CBR1 transcription was increased during hepatic I/R injury via Nrf2 following the binding of Nrf2 to the ARE sequence 5′-TGACXXXGC-3′ within the CBR1 promoter region. The human ARE sequence containing the Nrf2 binding site is 5′-TGACTTTGCA-3′, which is located −252 bp upstream of the CBR1 start codon (Miura et al., 2013). Nrf2 stabilization and activation of the transcription factors were observed under OGD/R conditions as well as hydrogen peroxide-induced stress. The results indicated that physiological conditions of oxidative stress during I/R injuries also stabilized Nrf2 and increased the cellular levels of CBR1.
LT is known as an antioxidant, but it is also known as an inhibitor of Nrf2. In other study, LT inhibited Nrf2/ARE pathways resulting in a negative regulation of cancer cells (Tang et al., 2011). In addition, several cancer studies suggested LT acted as a chemotherapeutic agent by suppressing Nrf2 pathways in cancer and facilitated the therapeutic suppression of tumor growth (Chian et al., 2014). Apparently, LT reduces oxidative stress not by directly increasing the antioxidant proteins but via regulation of inflammation (Funakoshi-Tago et al., 2011; Ziyan et al., 2007). Indeed, our data showed that treating LT under basal conditions significantly decreased the level of inflammatory cytokine IL-6. Therefore, LT can be partially beneficial in restoring liver injuries by suppressing the pro-inflammatory signals even though it suppresses Nrf2-regulated antioxidant genes discussed in our study.
We confirmed that CBR1 expression is regulated by Nrf2 in human liver. Numerous studies have provided substantial evidence supporting the critical role of ROS in hepatic I/R injury (Clavien et al., 2001; Fondevila et al., 2003; Jaeschke, 2003; Serracino-Inglott et al., 2001). Therefore, strategies to inhibit ROS production and enhance ROS scavenging have been suggested as therapeutic approaches. Nrf2, a transcription factor mediating the expression of many endogenous antioxidants, is known to play an essential role in protecting cells against oxidative stress (Jaiswal, 2004; Kaspar et al., 2009; Kobayashi and Yamamoto, 2005; Leonard et al., 2006; Motohashi and Yamamoto, 2004; Nguyen et al., 2009). Recent studies have shown that Nrf2 also exerts anti-inflammatory response through regulation of NADPH oxidase (Kovac et al., 2015) and pro-inflammatory signaling (Kong et al., 2010). Several reports have revealed that Nrf2-null mice are highly susceptible to hepatic injuries induced by chemicals (Liu et al., 2013), alcohols (Lamlé et al., 2008), high-fat diet (Meakin et al., 2014), methionine/choline-deficient diet (Chowdhry et al., 2010), and cytokines (Kong et al., 2010), indicating that Nrf2 protects the liver via multiple cytoprotective pathways.
Our study demonstrated that Nrf2 increases CBR1 expression and attenuates hepatic cell death under I/R conditions. The attenuated cell death is attributed to the ability of CBR1 to reduce oxidative stress, including lipid peroxidation (Ellis, 2007). Therefore, we propose that CBR1 is one of the anti-oxidative and anti-apoptotic molecules regulated by Nrf2.
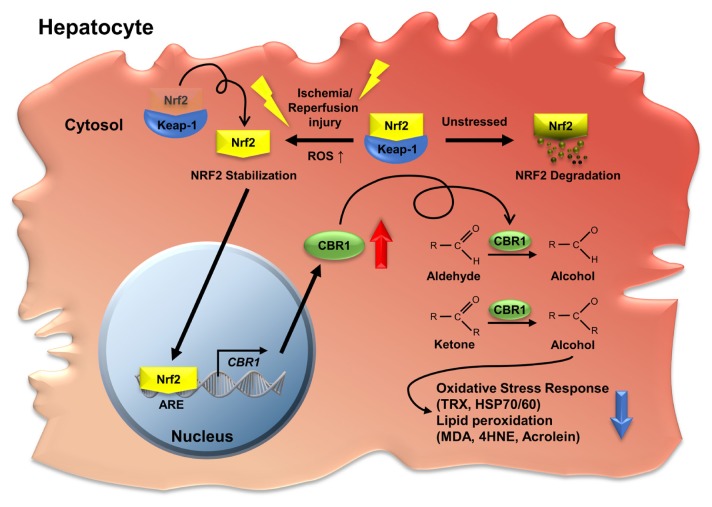
Oxidative-stress–induced hepatic senescence is one of the primary causes of severe liver I/R injury (Teoh and Farrell, 2003). CBR1 deficiency aggravates oxidative stress, inflammatory response, and liver damage. We observed that overexpression of CBR1 before ischemia significantly suppressed the expression of oxidative stress enzymes and the production of lipid peroxidation products. The development of a specific and practical CBR1 activator represents a possible strategy for protection against hepatic I/R injury (Fig. 8).
Fig. 8. Graphic summary of the CBR1 function under hepatic ischemia reperfusion injury.
CBR1 is transcriptionally induced by Nrf2 during liver I/R injury. Higher expression of CBR1 relieves oxidative stress and lipid peroxidation in the liver during I/R injury while suppressed CBR1 increases the damage. As human CBR1 is upregulated by Nrf2 during liver transplantation, CBR1 induction can be a therapeutic strategy for relieving liver I/R injury during liver transplantation.
In this study, we found that Nrf2-CBR1 regulation was robustly activated under oxidative stress conditions in normal hepatocyte cell lines, hepatic I/R animal model, and clinical samples. We found that the up-regulation of Nrf2-CBR1 system attenuated I/R injury in the liver while the down-regulation of Nrf2-CBR1 system severely damaged the liver. These findings suggest that Nrf2-CBR1 plays a protective role in living-donor liver transplantation.
ACKNOWLEDGMENTS
The present study was partially supported by the Asan Institute for Life Sciences (Tak E, 15-662; Tak E, 17-662; and Yoon YI, 18-IT0622), the National Research Foundation of Korea (NRF-2015K1A4A3046807); the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B04032429); and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HI15C0972). We thank the core facilities of Confocal Microscopy Core and Comparative Pathology Core at the ConveRgence mEDIcine research center (CREDIT), Asan Medical Center for the use of their shared equipment, services, and expertise.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Amersi F., Buelow R., Kato H., Ke B.B., Coito A.J., Shen X.D., Zhao D.L., Zaky J., Melinek J., Lassman C.R., et al. Upregulation of heme oxygenase-1 protects genetically fat Zucker rat livers from ischemia/reperfusion injury. J Clin Invest. 1999;104:1631–1639. doi: 10.1172/JCI7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chian S., Thapa R., Chi Z., Wang X.J., Tang X. Luteolin inhibits the Nrf2 signaling pathway and tumor growth in vivo. Biochem Biophys Res Commun. 2014;447:602–608. doi: 10.1016/j.bbrc.2014.04.039. [DOI] [PubMed] [Google Scholar]
- Chowdhry S., Nazmy M.H., Meakin P.J., Dinkova-Kostova A.T., Walsh S.V., Tsujita T., Dillon J.F., Ashford M.L., Hayes J.D. Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic Biol Med. 2010;48:357–371. doi: 10.1016/j.freeradbiomed.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Clavien P.A., Rüdiger H.A., Selzner M. Mechanism of hepatocyte death after ischemia: apoptosis versus necrosis. Hepatology. 2001;33:1555–1556. doi: 10.1053/jhep.2001.0103306le02. [DOI] [PubMed] [Google Scholar]
- Ellis E.M. Reactive carbonyls and oxidative stress: potential for therapeutic intervention. Pharmacol Ther. 2007;115:13–24. doi: 10.1016/j.pharmthera.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Fondevila C., Busuttil R.W., Kupiec-Weglinski J.W. Hepatic ischemia/reperfusion injury—a fresh look. Exp Mol Pathol. 2003;74:86–93. doi: 10.1016/S0014-4800(03)00008-X. [DOI] [PubMed] [Google Scholar]
- Funakoshi-Tago M., Nakamura K., Tago K., Mashino T., Kasahara T. Anti-inflammatory activity of structurally related flavonoids, Apigenin, Luteolin and Fisetin. Int Immunopharmacol. 2011;11:1150–1159. doi: 10.1016/j.intimp.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Higdon A., Diers A.R., Oh J.Y., Landar A., Darley-Usmar V.M. Cell signalling by reactive lipid species: new concepts and molecular mechanisms. Biochem J. 2012;442:453–464. doi: 10.1042/BJ20111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2 small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Jaeschke H. Molecular mechanisms of hepatic ischemia-reperfusion injury and preconditioning. Am J Physiol Gastroint Liver Physiol. 2003;284:G15–G26. doi: 10.1152/ajpgi.00342.2002. [DOI] [PubMed] [Google Scholar]
- Jaeschke H., Woolbright B.L. Current strategies to minimize hepatic ischemia-reperfusion injury by targeting reactive oxygen species. Transplant Rev. 2012;26:103–114. doi: 10.1016/j.trre.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- Kalogeris T., Baines C.P., Krenz M., Korthuis R.J. Ischemia/reperfusion. Compr Physiol. 2011;7:113–170. doi: 10.1002/cphy.c160006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeris T., Baines C.P., Krenz M., Korthuis R.J. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317. doi: 10.1016/B978-0-12-394309-5.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassner N., Huse K., Martin H.J., Godtel-Armbrust U., Metzger A., Meineke I., Brockmoller J., Klein K., Zanger U.M., Maser E., et al. Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab Dispos. 2008;36:2113–2120. doi: 10.1124/dmd.108.022251. [DOI] [PubMed] [Google Scholar]
- Kensler T.W., Egner P.A., Agyeman A.S., Visvanathan K., Groopman J.D., Chen J.G., Chen T.Y., Fahey J.W., Talalay P. Keap1–nrf2 signaling: a target for cancer prevention by sulforaphane. Top Curr Chem. 2013;329:163–177. doi: 10.1007/128_2012_339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler T.W., Wakabayash N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Klaassen C.D., Reisman S.A. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol. 2010;244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- Kong X., Thimmulappa R., Kombairaju P., Biswal S. NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J Immunol. 2010;185:569–577. doi: 10.4049/jimmunol.0902315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac S., Angelova P.R., Holmström K.M., Zhang Y., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim Biophys Acta. 2015;1850:794–801. doi: 10.1016/j.bbagen.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh K., Uchinami H., Yoshioka M., Seki E., Yamamoto Y. Nrf2 activation protects the liver from ischemia/reperfusion injury in mice. Ann Surg. 2014;260:118–127. doi: 10.1097/SLA.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamlé J., Marhenke S., Borlak J., Von Wasielewski R., Eriksson C.P., Geffers R., Manns M.P., Yamamoto M., Vogel A. Nuclear factor-eythroid 2–related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159–1168.e2. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Leonard M.O., Kieran N.E., Howell K., Burne M.J., Varadarajan R., Dhakshinamoorthy S., Porter A.G., O’Farrelly C., Rabb H., Taylor C.T. Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J. 2006;20:2624–2626. doi: 10.1096/fj.06-5097fje. [DOI] [PubMed] [Google Scholar]
- Liu J., Wu K.C., Lu Y.F., Ekuase E., Klaassen C.D. Nrf2 protection against liver injury produced by various hepatotoxicants. Oxid Med Cell Longev. 2013;2013 doi: 10.1155/2013/305861. 305861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews W.R., Guido D.M., Fisher M.A., Jaeschke H. Lipid peroxidation as molecular mechanism of liver cell injury during reperfusion after ischemia. Free Radic Biol Med. 1994;16:763–770. doi: 10.1016/0891-5849(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Meakin P.J., Chowdhry S., Sharma R.S., Ashford F.B., Walsh S.V., McCrimmon R.J., Dinkova-Kostova A.T., Dillon J.F., Hayes J.D., Ashford M.L. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol Cell Biol. 2014;34:3305–3320. doi: 10.1128/MCB.00677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Taketomi A., Nishinaka T., Terada T. Regulation of human carbonyl reductase 1 (CBR1, SDR21C1) gene by transcription factor Nrf2. Chem Biol Interact. 2013;202:126–135. doi: 10.1016/j.cbi.2012.11.023. [DOI] [PubMed] [Google Scholar]
- Motohashi H., Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Nioi P., Pickett C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann U. Carbonyl reductases: The complex relationships of mammalian carbonyland quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- Rashid M.A., Lee S., Tak E., Lee J., Choi T.G., Lee J.W., Kim J.B., Youn J.H., Kang I., Ha J., et al. Carbonyl reductase 1 protects pancreatic beta-cells against oxidative stress-induced apoptosis in glucotoxicity and glucolipotoxicity. Free Radic Biol Med. 2010;49:1522–1533. doi: 10.1016/j.freeradbiomed.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Ray P.D., Huang B.W., Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotondo R., Moschini R., Renzone G., Tuccinardi T., Balestri F., Cappiello M., Scaloni A., Mura U., Del-Corso A. Human carbonyl reductase 1 as efficient catalyst for the reduction of glutathionylated aldehydes derived from lipid peroxidation. Free Radic Biol Med. 2016;99:323–332. doi: 10.1016/j.freeradbiomed.2016.08.015. [DOI] [PubMed] [Google Scholar]
- Serracino-Inglott F., Habib N.A., Mathie R.T. Hepatic ischemia-reperfusion injury. Am J Surg. 2001;181:160–166. doi: 10.1016/S0002-9610(00)00573-0. [DOI] [PubMed] [Google Scholar]
- Tak E., Lee S., Lee J., Rashid M.A., Kim Y.W., Park J.H., Park W.S., Shokat K.M., Ha J., Kim S.S. Human carbonyl reductase 1 upregulated by hypoxia renders resistance to apoptosis in hepatocellular carcinoma cells. J Hepatol. 2011;54:328–339. doi: 10.1016/j.jhep.2010.06.045. [DOI] [PubMed] [Google Scholar]
- Tang X., Wang H., Fan L., Wu X., Xin A., Ren H., Wang X.J. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic Biol Med. 2011;50:1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Teoh N.C., Farrell G.C. Hepatic ischemia reperfusion injury: pathogenic mechanisms and basis for hepatoprotection. J Gastroenterol Hepatol. 2003;18:891–902. doi: 10.1046/j.1440-1746.2003.03056.x. [DOI] [PubMed] [Google Scholar]
- Thimmulappa R.K., Mai K.H., Srisuma S., Kensler T.W., Yamamato M., Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Ziyan L., Yongmei Z., Nan Z., Ning T., Baolin L. Evaluation of the anti-inflammatory activity of luteolin in experimental animal models. Planta Med. 2007;73:221–226. doi: 10.1055/s-2007-967122. [DOI] [PubMed] [Google Scholar]