Abstract
Altered genetic features in cancer cells lead to a high rate of aerobic glycolysis and metabolic reprogramming that is essential for increased cancer cell viability and rapid proliferation. Pyruvate kinase muscle (PKM) is a rate-limiting enzyme in the final step of glycolysis. Herein, we report that PKM is a potential therapeutic target in triple-negative breast cancer (TNBC) cells. We found that PKM1 or PKM2 is highly expressed in TNBC tissues or cells. Knockdown of PKM significantly suppressed cell proliferation and migration, and strongly reduced S phase and induced G2 phase cell cycle arrest by reducing phosphorylation of the CDC2 protein in TNBC cells. Additionally, knockdown of PKM significantly suppressed NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) activity by reducing the phosphorylation of p65 at serine 536, and also decreased the expression of NF-κB target genes. Taken together, PKM is a potential target that may have therapeutic implications for TNBC cells.
Keywords: cell cycle, NF-κB, PKM1, PKM2, triple-negative breast cancer cells
INTRODUCTION
Triple-negative breast cancer (TNBC) represents 10% to 15% of all breast cancers and is characterized by the low expression of progesterone receptor, estrogen receptor, and human epidermal growth factor receptor 2 (Lebert et al., 2018). Additionally, it is clinically associated with aggressive behavior, rapid progression of distant metastasis and poor prognosis (Bianchini et al., 2016; Lebert et al., 2018). Despite advanced platinum-based chemotherapy (Tutt et al., 2018), approximately 30% of TNBC patients will die within 5 years (Bonotto et al., 2014). Additionally, patients with metastatic TNBC have a short progression-free survival (Chang et al., 2014). Therefore, challenges remain to identify potential targets in the field of TNBC research for treating this disease.
Cancer cells exhibit metabolic alterations to provide biomaterials and energy that are essential for growth and survival (DeBerardinis et al., 2008; King and Gottlieb, 2009; Vander Heiden et al., 2009). Metabolic changes in cancer cells increase glucose uptake and lactate production (Warburg effect) (Fantin et al., 2006). Pyruvate kinases are crucial rate-limiting glycolytic enzymes that convert phosphoenolpyruvate to pyruvate in the final step of glycolysis (Chaneton and Gottlieb, 2012). Two types of genes encode mammalian pyruvate kinase (pklr and pkm). The PKL and PKR isoforms are encoded by the pklr gene and the pkm gene encodes the PKM1 and PKM2 isoforms (Mazurek, 2007; 2011). PKM1 exists in an active tetrameric form, whereas PKM2 can switch between an active tetrameric form and an inactive dimeric form (Mazurek, 2007; 2011). The PKM2 protein is regulated by several post-translational modifications, including phosphorylation (Gao et al., 2012; Yang et al., 2012b), prolyl hydroxylation (Luo et al., 2011), acetylation (Lv et al., 2011), cysteine oxidation (Anastasiou et al., 2011), and demethylation (Wang et al., 2014). These modifications lead to the suppression of pyruvate kinase activity (Harris et al., 2012) and the resultant dimeric PKM2 is translocated into the nucleus and acts as an active protein kinase to phosphorylate specific nuclear proteins (Gao et al., 2012; Yang et al., 2012a). It also acts as a co-activator of hypoxia-inducible factor (HIF)-1 alpha (Luo et al., 2011) and is heavily involved in tumorigenesis. Additionally, PKM1 promotes tumor growth by activating glucose catabolism and autophagy in pulmonary neuroendocrine tumors (Morita et al., 2018). PKM1 is a therapeutic target in paclitaxel-resistant gastric cancer cells (Okazaki et al., 2018). These findings suggest a requirement for therapeutic drugs that target PKM1 and PKM2 in cancer treatment. Interestingly, PKM2 promotes angiogenesis through the activation of NF-κB/p65 and HIF-1α in hypoxic pancreatic tumors (Azoitei et al., 2016). NF-κB/RelA binds to the PKM promoter and induces the expression of PKM2 in glioblastoma multiforme (Han et al., 2015). Thus, these reports suggest the importance of metabolic cooperation between the NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) pathway and PKM.
The NF-κB family of transcription factors are key regulators of inflammation, immune response, cell differentiation, proliferation, and survival (Hayden and Ghosh, 2008). NF-κB comprises a family of five transcription subunits, p65/RelA, c-Rel, RelB, p50/NF-κB1, and p52/NF-κB2, that form distinct protein complexes, which bind to consensus DNA sequences at promoter regions of responsive genes regulating cellular processes (Nabel and Verma, 1993). Additionally, NF-κB is frequently activated in TNBC and inhibition of NF-κB activity suppresses growth of TNBC cells (Barbie et al., 2014; Yamaguchi et al., 2009). Treatment with STAT3/NF-κB responsive element-driven suicide gene therapy inhibits growth of TNBC cells (Kuo et al., 2017).
The purpose of our study was to identify a promising target that plays crucial roles in TNBC cell growth. Here, we report that knockdown of PKM results in anticancer effects against TNBC cells by reducing NF-κB activation. This might be a potential therapeutic strategy against TNBC cell growth.
MATERIALS AND METHODS
Cell culture
All cell lines were purchased from the American Type Culture Collection (ATCC, USA) and were cytogenetically tested and authenticated before the cells were frozen. Each vial of frozen cells was thawed and maintained in culture for a maximum of 8 weeks. MCF10A normal breast cells and 4T1 mouse TNBC cells were cultured in Roswell Park Memorial Institute medium 1640 (RPMI1640) supplemented with 10% fetal bovine serum (FBS; Biological Industries, USA) and 1% penicillin/streptomycin (Biological Industries). HCC1937 TNBC cells were cultured in RPMI1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin (100 μg/ml), non-essential amino acids (NEAA; Thermo Fisher Scientific, China), and sodium pyruvate (Thermo Fisher Scientific). MDA-MB-231 and MDA-MB-436 TNBC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin.
Reagents
The antibodies to detect PKM1 (Cat# 7076S), total PKM2 (Cat# 4053S), -CDC2 (Cat# 28439S), p65 (Cat# 8242), cyclin B1 (Cat# 4135S), phosphorylated PKM2 (Tyr; Cat# 3827), CDC2 (Tyr15; Cat# 4539S), and p65 (Ser536; Cat# 3033) were purchased from Cell Signaling Technology (USA). The antibody to detect β-actin (Cat# KM9001) was from Tianjin Sungene Biotech (China). 2-Deoxy-D-glucose (2-DG; Cat# HY-13966) was purchased from MedChem Express (USA).
Cell proliferation assay
MCF 10A (3 × 103 cells per well) or HCC1937 (3.2 × 103 cells per well) and MDA-MB-231 (2.9 × 103 cells per well) cells were seeded in 96-well plates and incubated for 48 h. Twenty microliters of the MTT solution (Solarbio, China) were added to each well and incubated for 2 h at 37°C in a 5% CO2 incubator. The cell culture medium was removed and then 200 μl of DMSO were added to each well and crystals were dissolved. Absorbance was measured at 570 nm using the Thermo Multiskan plate-reader (Thermo Fisher Scientific).
Anchorage-independent cell growth
Cells (8 × 103 per well) suspended in complete growth medium (Basal Medium Eagle [BME]; Thermo Fisher Scientific) supplemented with 10% FBS, 1% gentamicin and L-glutamine were added to 0.3% agar in a top layer over a base layer of 0.6% agar. The cultures were maintained at 37°C in a 5% CO2 incubator for 3 weeks and then colonies were counted under a microscope using the Image-Pro Plus software (v.6) program (Media Cybernetics, USA).
Migration assay
Transwell chambers (Corning, USA) were coated outside with Matrigel and dried for 30 min at room temperature under sterile conditions. The lower compartment was filled with 600 μl complete medium. Samples containing 5 × 104 cells in 200 μl serum-free medium were added to the upper compartment. The chambers were incubated for 72 h at 37°C in a 5% CO2 atmosphere. Migrated cells were fixed with methanol and stained with hematoxylin and eosin. Photos of the stained cells were captured with the MShot Digital Imaging System (China) under a microscope and analyzed using the Image-Pro Plus software (v.6) program.
Knockdown of PKM by siRNA against PKM1 or PKM2, and overexpression of PKM by cloning into a PKM1 or PKM2 expression vector
A short RNAi sequence against PKM1 (siPKM1-1, 5′-GCGU GGAGGCUUCUUAUAA-3′; siPKM1-2, 5′-CUUGCCUGCUGUGUCGGAG-3) and PKM2 (siPKM2-1, 5′-GUUCGGAGGUUU GAUGAAAUC-3′; siPKM2-2, 5′-CCAUA AUCGUCCUCACCAA-3′) was designed. TNBC cells were transfected with each siRNA and incubated for 48 h. An overexpressing-PKM1 or -PKM2 vector was cloned into the pcDNA3.1-myc/his vector (Kim et al., 2015). MCF10A breast cells were transfected with PKM1, PKM2, or both PKM1 and PKM2 vectors and incubated for 48 h. After selection with G418 (200 μg/ml) for 1 week, cells were used to conduct further studies.
shRNA constructs and lentiviral production
A short hairpin RNA (shRNA) sequence against PKM was designed (5′-CCGGGCCCACCTGAATGTCAATAAACTCGAGTTTATTGACATTCAGGTGGGCTTTTTG-3′) and cloned into the lentiviral vector (pLKO-shPKM). To prepare PKM viral particles, HEK293T cells were transfected with each viral vector and packaging vectors and incubated for 48 h. Viral particles were harvested by filtration using a 0.45 mm sodium acetate syringe filter. After combined with 8 μg/ml of polybrane (Sigma-Aldrich, USA), the viral particles were infected into MCF 10A, HCC1937, or MDA-MB-231 cells. Cell culture medium was replaced at 24 h with fresh complete medium after infection.
Cell cycle analysis
Cells were seeded into 60-mm culture dishes (5 × 104 cells per dish) and incubated for 24 h. Cells were infected with shControl or shPKM virus particles and incubated for 24 h. Cell culture medium was replaced with fresh complete medium and incubated for 48 h. Cells were harvested by trypsinization and washed with cold phosphate buffered saline and then fixed in 1 ml 70% cold ethanol. After rehydration, cells were disrupted in 0.6% Triton X-100 and digested with 100 μg/ml RNase for 1 h. Cells were subsequently stained with 20 μg/ml propidium iodide (Clontech, USA) for 15 min and then cell cycle was analyzed by flow cytometry (Becton, Dickinson and Company, USA).
Luciferase reporter assay
Transient transfection was conducted using Lipofectamine 2000 (Invitrogen, USA) and assays to determine firefly luciferase and Renilla activities were performed according to the manufacturer’s manual (Promega, USA). Cells (1 × 104 cells per well) were seeded in 12-well culture plates and incubated for 24 h. Cells were co-transfected with an NF-κB or AP1 reporter plasmid (250 ng) and an internal control vector (pCMV-Renilla, 50 ng) and incubated for 24 h. Cells were infected with shControl or shPKM virus particles and incubated for 24 h. Cell culture medium was replaced with fresh complete medium and incubated for 48 h. Luciferase activities were measured using the substrates provided in the reporter assay system (Promega) using a Luminoskan Ascent plate reader (Thermo Fisher Scientific). The luciferase activity was normalized to Renilla luciferase activity.
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
RNA extraction from cells was performed according to the manufacturer’s manual (AXYGEN, China). One microgram of each RNA sample was reverse transcribed using reverse transcriptase (TaKaRa Bio, China) according to the manufacturer’s guidelines. The primers used in this study are as follows: PKM1-F, 5′-CAGCCAAAG GGGACTATCCT-3′, PKM1-R 5′-GAGGCTCGCACAAGTTCTTC-3′; PKM2-F 5′-C TATCCTCTGGAGGCTGTGC-3′, PKM2-R 5′-GTGGGGTCGCTGGTAATG-3′; IL6-F 5′-TCCACAAGCGCCTTCGGTC-3′, IL6-R 5′-CAGGCTGGCATTTGTGGTTG 3′; CXCL1-F 5′-CCCAAACCGAAGTCATAGCCA-3′, CXCL1-R 5′-TGTTGCAGG C TCCTCAGAAAT-3′; tumor necrosis factor alpha (TNFα)-F 5′-CAAGGACAGCAGAGGACCAG -3′, TNFα-R 5′-GGGTTTGCTACAACATGGGC-3′ and β-actin-F 5′-CCTCGCCTTTGCCGATC C-3′, β-actin-R 5′-TGAAGGTCTCAAACATGATCTGG-3′.
Statistical analysis
All quantitative results are expressed as mean ± SD. Statistically significant differences were obtained using the Student’s t-test or by one-way ANOVA (IBM SPSS Statistics ver. 24.0; IBM, USA). A value of P < 0.05 was considered to be statistically significant.
RESULTS
PKM1 and PKM2 are highly expressed in TNBC tissues and cells
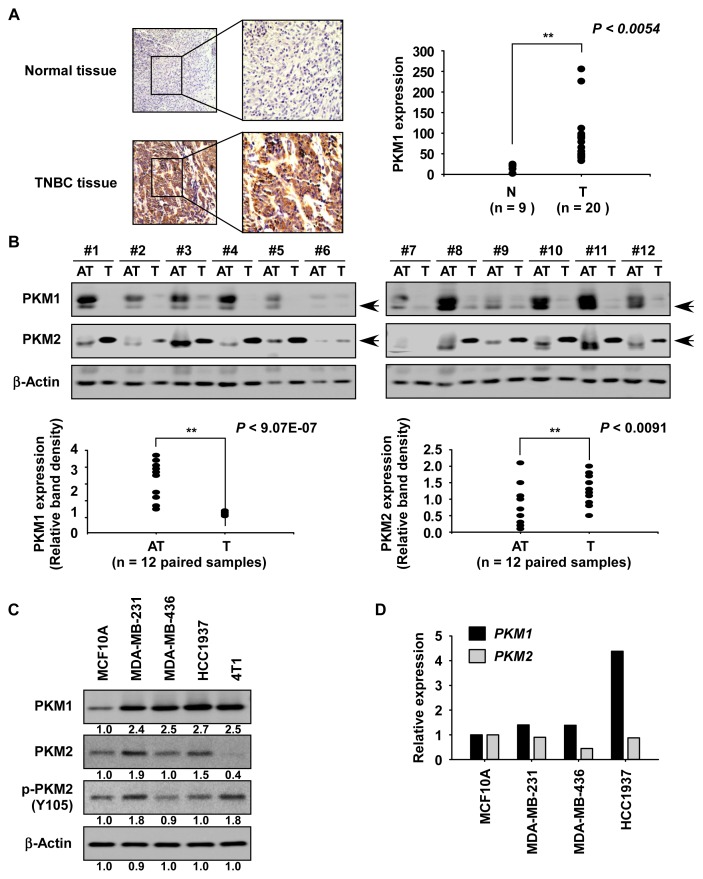
To determine whether the PKM1 protein is differentially expressed, normal or TNBC tissues were analyzed by immunohistochemistry. Results showed that the PKM1 protein was highly expressed in TNBC tissues compared to normal tissues (Fig. 1A). We also examined the expression of PKM1 and PKM2 by Western blotting in 12 paired TNBC tisseus. Results indicated that the PKM1 protein was highly expressed in adjacent tissues, whereas the PKM2 protein was strongly expressed in tumor tissues (Fig. 1B). Additionally, we examined the expression of PKM1 or PKM2 by Western blotting or reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in MCF10A normal breast and human TNBC (MDA-MB-231, MDA-MB-436, and HCC1937) and mouse TNBC (4T1) cells. Results showed that the protein and mRNA levels of PKM1 are highly expressed in TNBC cells compared to MCF10A normal breast cells (Figs. 1C and 1D). The PKM2 protein is overexpressed in MDA-MB-231 and HCC1937 TNBC cells, and the phosphorylated PKM2 protein is highly expressed in MDA-MB-231 and 4T1 TNBC cells (Fig. 1C).
Fig. 1. Expression of PKM1 and PKM2 in TNBC tissues and cells.
(A) The expression of the PKM1 protein in normal and TNBC tissues was analyzed by immunohistochemistry. The expression of PKM1 was determined using a microscope and the Image-Pro Plus software (v.6) program. N, normal tissue; T, TNBC tissue. (B) The expressioin of PKM1 and PKM2 proteins in 12 paried TNBC tissues was analyzed by Western blotting. AT, adjacent tissue; T, cancer tissue. β-Actin was used to verify equal protein loading. Band density was measured using the ImageJ software program (National Institutes of Health, USA). Arrows indicate the PKM1 or PKM2 protein. The results of the expression of PKM1 (left panel) and PKM2 (right panel) proteins are shown as a graph. **P < 0.01. (C and D) The expression of PKM1 or PKM2 in normal breast cells and TNBC cells. The expression of PKM1, PKM2, or phosphorylated PKM2 was analyzed by Western blotting (C) or RT-qPCR (D). Similar results were obtained from 3 independent experiments and representative blots are shown.
Knockdown of PKM suppresses growth and migration of TNBC cells
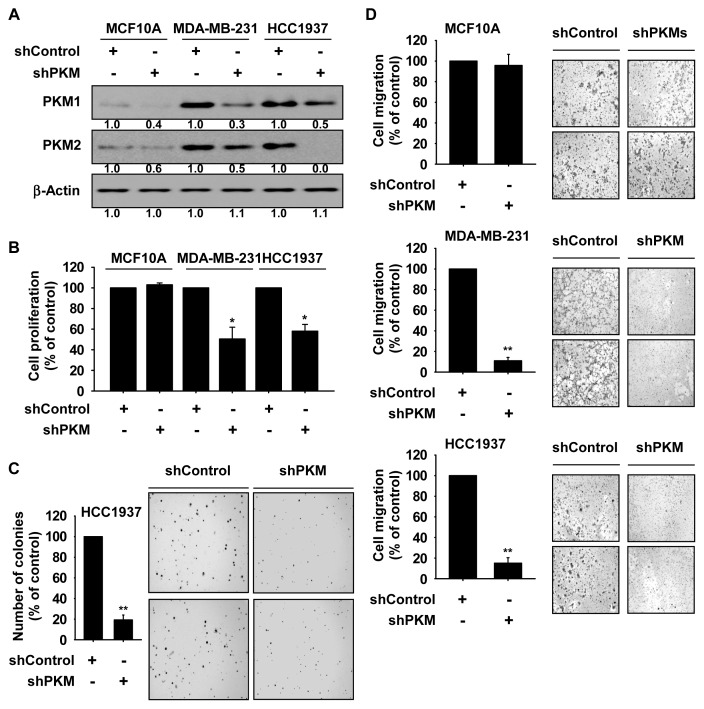
To study the influence of PKM1 or PKM2 expression on cell growth, specific siRNAs against PKM1 or PKM2 were used (Supplementary Figs. 1A and 1B). Cell growth was analyzed by MTT or soft agar assay and results indicated that knockdown of PKM1 or PKM2 significantly suppressed growth of TNBC cells (Supplementary Figs. 1C and 1D). Additionally, the phosphorylated p65 protein was strongly inhibited by knockdown of PKM1 or PKM2 (Supplementary Fig. 1E). Therefore, we established stable knockdown of PKM in MCF10A, MDA-MB-231, or HCC1937 cells (Fig. 2A and Suppplementary Fig. 2), and these cells were used to conduct further studies. Cell growth was analyzed by MTT or soft agar assay. Results showed that growth of TNBC cells was significantly inhibited by knockdown of PKM, whereas growth was not affected in MCF10A normal breast cells (Figs. 2B and 2C). We next examined whether knockdown of PKM could affect cell migration. Results indicated that migration of TNBC cells was significantly inhibited by knockdown of PKM but not changed in MCF10A normal breast cells (Fig. 2D).
Fig. 2. Knockdown of PKM1/2 suppresses growth and migration of TNBC cells.
(A) Expression of PKM1 and PKM2 proteins in knockdown PKM cells (shPKM) or control cells (shControl) was analyzed by Western blotting. Band density was measured using the Image J (NIH) software program. (B) Effect of PKM knock-down on cell growth. Cells were seeded and then incubated for 72 h. Cell growth were measured at an absorbance of 570 nm. (C) Effect of PKM knockdown on anchorage-independent cell growth. Cells were seeded and incubated for 3 weeks. Colonies were counted using a microscope and the Image-Pro Plus software (v.6) program. (D) Effect of PKM knockdown on cell migration. Cells were seeded in transwells and incubated for 48 h. Migrated cells were counted. (B–D) Data are shown as mean ± SD of triplicate values from 3 independent experiments and the asterisks indicate a significant difference between shPKM cells and shControl cells (*P < 0.05, **P < 0.01).
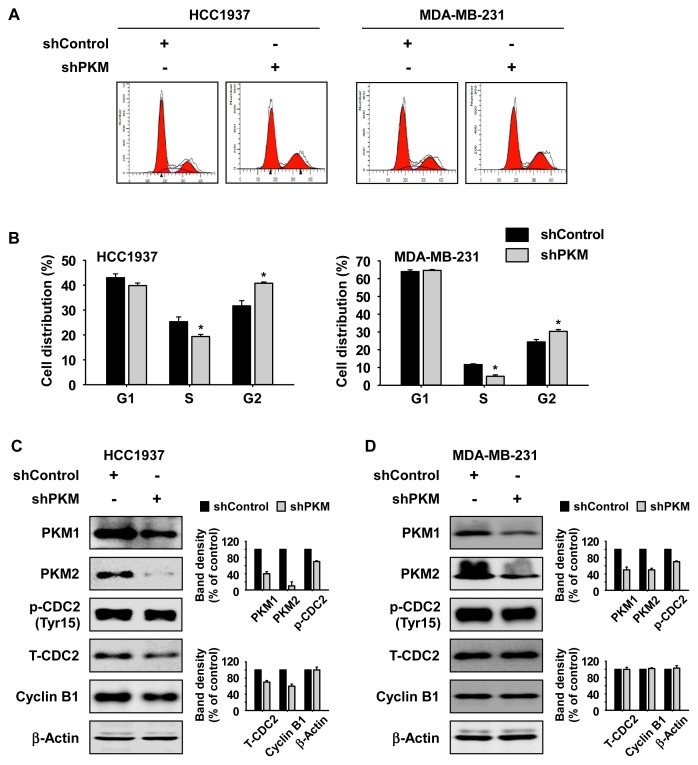
Knockdown of PKM induces G2/M phase cell cycle arrest
To determine the effect of PKM knockdown on cell cycle in TNBC cells, we performed flow cytometry analysis. Cells were infected with shPKM virus particles and then incubated for 72 h. The results indicated that knockdown of PKM significantly reduced S phase and induced G2/M phase cell cycle arrest (Figs. 3A and 3B). Additionally, we investigated whether knockdown of PKM could affect the expression of G2 phase marker proteins. Cells were infected with shPKM virus particles for 72 h and cell cycle marker proteins were analyzed by Western blotting. Results indicated that knockdown of PKM consistently attenuated the expression of the phosphorylated CDC2 protein (Fig. 3C and 3D). Additionally, the expression of total CDC2 and cyclin B1 was decreased in HCC1937 TNBC cells (Fig. 3C and 3D).
Fig. 3. Knockdown of PKM reduces S phase and induces G2 phase cell cycle arrest in TNBC cells.
(A and B) Effect of PKM knockdown on cell cycle. Cells were seeded and incubated for 48 h in 10% serum-supplemented medium and then stained with propidium iodide. Cell cycle was analyzed using Fluorescence Activated Cell Sorting (FACS). Data are shown as mean ± SD of triplicate values from 3 independent experiments and the asterisk indicates a significant difference between shPKM cells and shControl cells (*P < 0.05). (C and D) Effect of PKM knockdown on cell cycle marker proteins. Cells were synchronized by serum starvation for 24 h and incubated for 48 h in 10% serum-supplemented medium and analyzed by Western blotting. Band density was measured using the ImageJ software program and all results of Western blotting are shown as mean ± SD for 3 independent experiments and as a bar graph.
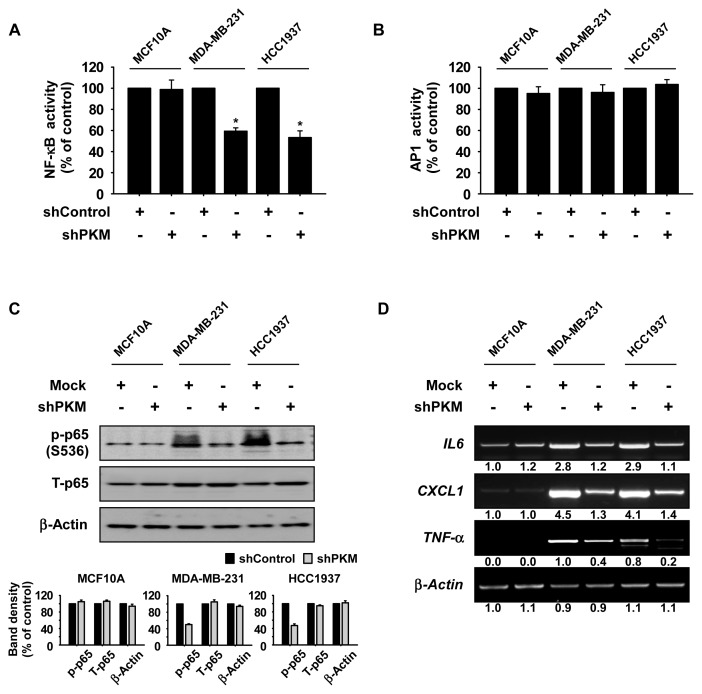
Knockdown of PKM inhibits NF-κB activity in TNBC cells
PKM2 was shown to induce NF-κB activation in fibroblasts (Chiavarina et al., 2011) and AP1 activation is essential for TNFα-mediated TNBC progression (Qiao et al., 2016). Therefore, we investigated the effect of PKM knockdown on NF-κB or AP1 transcritional activation in MCF10A or TNBC cells. Cells were transfected with an NF-κB or AP1-luciferase reporter vector and then infected with shPKM virus particles for 72 h, and the luciferase activity was measured from the cell lysates. Results showed that NF-κB reporter activity was significantly decreased in PKM knockdown TNBC cells compared to control TNBC cells, whereas NF-κB reporter activity was not changed in MCF10A normal breast cells (Fig. 4A). Additionally, AP1 reporter activity was not affected by knockdown of PKM (Fig. 4B). The decreased NF-κB reporter activity was verified by assessing the expression of phosphorylated p65 and total p65 proteins as well as target gene expression of NF-κB signaling. Results showed that the expression of the phosphorylated p65 protein and the transcriptional expression of IL6, CXCL1, and TNF-α were strongly inhibited by knockdown of PKM in TNBC cells, but not changed in MCF10A normal breast cells (Figs. 4C and 4D).
Fig. 4. Knockdown of PKM inhibits NF-κB activity in TNBC cells.
The effect of PKM knockdown on NF-κB (A) and AP-1 (B) reporter activity. Cells were transfected with an NF-κB or AP-1 reporter plasmid and CMV-renilla and then incubated for 48 h. Reporter activity was measured using substrates included in the reporter assay system. Data are shown as mean ± SD of triplicate values from 3 independent experiments and the asterisks indicate a significant difference between shPKM cells and shControl cells (*P < 0.05). (C) Effect of PKM knockdown on the phosphorylation of the p65 protein. Cells were analyzed by Western blotting. All results of Western blotting are shown as mean ± SD for 3 independent experiments and as a bar graph. (D) Effect of PKM knockdown on the expression of NF-κB target genes. Gene expression was analyzed by RT-PCR. (C and D) β-Actin was used to verify equal protein loading and band density was measured using the ImageJ software program. Similar results were obtained from 3 independent experiments and representative blots are shown.
DISCUSSION
The metabolic function of pyruvate kinase and the mechanism of its regulation of cancer growth have been reported for varous cancer types (Christofk et al., 2008; Luo and Semenza, 2011). Therefore, it could be useful for the targeting of cellular metabolism for responsiveness to cancer therapy (Dang, 2012; Kroemer and Pouyssegur, 2008; Tennant et al., 2010). Here, we examined PKM as a metabolic target contributing to the growth of TNBC cells. We found that growth of TNBC cells was sensitive to the inhibitory effect of PKM knockdown compared to growth of MCF10A normal breast cells (Figs. 2B and 2C). Additionally, our results consistently indicated that high doses of 2-DG as a glucose analogue weakly suppressed growth of MCF10A cells (Supplementary Fig. 3A), whereas 2-DG dose-dependently inhibited growth of HCC1937 cells (Supplementary Figs. 3B and 3C). This result suggested that cancer cells exhibit alternative metabolic requirements, including glycolysis, compared to normal cells (DeBerardinis et al., 2008), which are essential for supporting tumor cell growth and survival (Tennant et al., 2009). Therefore, PKM is a potential metabolic target of the glycolytic signaling pathway in TNBC cells. Additionally, the expression of PKM1 and PKM2 was differentially regulated in TNBC tissues and cells (Fig. 1). Therefore, we also suggest that the individual function of PKM1 and PKM2 may be involved in tumorigenesis or progression of TNBC, respectively. Elucidating the relationship between the expression of the PKMs and metabolic functions during TNBC tumorigenesis is essential. Therefore, we next investigated whether overexpression of PKM could affect tumorigenesis of normal breast cells. MCF10A cells were transfected with PKM1, PKM2, or both PKM1 and PKM2. Expression of PKM1 and PKM2 was analyzed by Western blotting (Supplementary Fig. 4A). Cell growth was analyzed by MTT, soft agar, or foci formation assay. Results showed that growth of cells stably overexpressing PKM1, PKM2, or both PKM1 and PKM2 was little changed compared with control cells (Supplementary Figs. 4B–4D). Additionally, we examined the effect of PKM overexpression on NF-κB activity. Results indicated that overexpression of PKM could not affect the activity of NF-κB and the phosphorylation of p65 (Supplementary Figs. 4E and 4F). Theses results indicated that stably overexpressing PKM1, PKM2, or both PKM1 and PKM2 cannot induce tumorigenesis of normal breast cells and activity of NF-κB. Oncogenic tyrosine kinases are reportedly activated in TNBC cells, but not in normal cells (Wu et al., 2015). Among them, 9 different tyrosine kinases induce the phosphorylation of PKM2 at tyrosine 105 (Zhou et al., 2018). Furthermore, phosphomimetic PKM2 (Y105D), as a dimeric form, induces colony formation in MCF10A normal breast cells, but not wildtype PKM2 expressing cells (Zhou et al., 2018). Therefore, we suggest that overexpression of PKM2 combined with the activation of tyrosine kinase signaling leads to the dimeric form of PKM2 and can promote tumorigenesis of normal breast cells. Additionally, overexpression of PKM1 might provide biomaterials through metabolic alterations during tumorigenesis.
NF-κB activation can promote tumor growth, proliferation, and invasion of TNBC (House et al., 2018; Ito-Kureha et al., 2015). Additionally, positive crosstalk between NF-κB and PKM has been reporoted (Azoitei et al., 2016; Han et al., 2015). Therefore, we investigated the effect of PKM knockdown on NF-κB activity and results indicated that knockdown of PKM significantly inhibited NF-κB activity by reducing phosphorylation of p65 in TNBC cells (Figs. 4A and 4C). However, how PKM can regulate NF-κB activity through the phosphorylation of p65 is not clear. IKKβ plays an essential role in the phosphorylation of RelA/p65 on serine 536 and regulates NF-κB activation (Israel, 2010). Dimeric PKM2 acts an active protein kinase that phosphorylates specific nuclear proteins (Gao et al., 2012; Yang et al., 2012a). Therefore, in future studies we will investigate whether the expression of the PKM2 protein is differentially localized in the cytoplasm or nucleus, and whether dimeric PKM2 can phosphorylate the p65 protein at serine 536 or affect IKKβ signaling molecules.?
In conclusion, PKM is a novel metabolic target of TNBC cells that inhibits growth and migration by reducing NF-κB activation. Future studies will focus on providing additional mechanisms and metabolic changes of PKM in TNBC cells. These studies may assist in the successful development of therapeutics against TNBC.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Henan Joint Fund, National Natural Science Foundation China (NSFC) (grant Nos. U1804196 and 81572812) and Key program of Henan Province, China (grant No. 161100510300).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Anastasiou D., Poulogiannis G., Asara J.M., Boxer M.B., Jiang J.K., Shen M., Bellinger G., Sasaki A.T., Locasale J.W., Auld D.S., et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoitei N., Becher A., Steinestel K., Rouhi A., Diepold K., Genze F., Simmet T., Seufferlein T. PKM2 promotes tumor angiogenesis by regulating HIF-1alpha through NF-kappaB activation. Mol Cancer. 2016;15:3. doi: 10.1186/s12943-015-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie T.U., Alexe G., Aref A.R., Li S., Zhu Z., Zhang X., Imamura Y., Thai T.C., Huang Y., Bowden M., et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J Clin Invest. 2014;124:5411–5423. doi: 10.1172/JCI75661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonotto M., Gerratana L., Poletto E., Driol P., Giangreco M., Russo S., Minisini A.M., Andreetta C., Mansutti M., Pisa F.E., et al. Measures of outcome in metastatic breast cancer: insights from a real-world scenario. Oncologist. 2014;19:608–615. doi: 10.1634/theoncologist.2014-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B., Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37:309–316. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Chang B., Sokhn J., James E., Abu-Khalaf M. Prolonged progression-free survival in a patient with triple-negative breast cancer metastatic to the liver after chemotherapy and local radiation therapy. Clin Breast Cancer. 2014;14:e61–e64. doi: 10.1016/j.clbc.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Chiavarina B., Whitaker-Menezes D., Martinez-Outschoorn U.E., Witkiewicz A.K., Birbe R., Howell A., Pestell R.G., Smith J., Daniel R., Sotgia F., et al. Pyruvate kinase expression (PKM1 and PKM2) in cancer-associated fibroblasts drives stromal nutrient production and tumor growth. Cancer Biol Ther. 2011;12:1101–1113. doi: 10.4161/cbt.12.12.18703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., Cantley L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Dang C.V. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Gao X., Wang H., Yang J.J., Liu X., Liu Z.R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Wei W., Chen X., Zhang Y., Wang Y., Zhang J., Wang X., Yu T., Hu Q., Liu N., et al. NF-kappaB/RelA-PKM2 mediates inhibition of glycolysis by fenofibrate in glioblastoma cells. Oncotarget. 2015;6:26119–26128. doi: 10.18632/oncotarget.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris I., McCracken S., Mak T.W. PKM2: a gatekeeper between growth and survival. Cell Res. 2012;22:447–449. doi: 10.1038/cr.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- House C.D., Grajales V., Ozaki M., Jordan E., Wubneh H., Kimble D.C., James J.M., Kim M.K., Annunziata C.M. IKKε cooperates with either MEK or non-canonical NF-kB driving growth of triple-negative breast cancer cells in different contexts. BMC Cancer. 2018;18:595. doi: 10.1186/s12885-018-4507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb Perspect Biol. 2010;2:a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Kureha T., Koshikawa N., Yamamoto M., Semba K., Yamaguchi N., Yamamoto T., Seiki M., Inoue J. Tropomodulin 1 expression driven by NF-kappaB enhances breast cancer growth. Cancer Res. 2015;75:62–72. doi: 10.1158/0008-5472.CAN-13-3455. [DOI] [PubMed] [Google Scholar]
- Kim D.J., Park Y.S., Kang M.G., You Y.M., Jung Y., Koo H., Kim J.A., Kim M.J., Hong S.M., Lee K.B., et al. Pyruvate kinase isoenzyme M2 is a therapeutic target of gemcitabine-resistant pancreatic cancer cells. Exp Cell Res. 2015;336:119–129. doi: 10.1016/j.yexcr.2015.05.017. [DOI] [PubMed] [Google Scholar]
- King A., Gottlieb E. Glucose metabolism and programmed cell death: an evolutionary and mechanistic perspective. Curr Opin Cell Biol. 2009;21:885–893. doi: 10.1016/j.ceb.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Kuo W.Y., Hwu L., Wu C.Y., Lee J.S., Chang C.W., Liu R.S. STAT3/NF-kappaB-regulated lentiviral TK/GCV suicide gene therapy for Cisplatin-resistant triple-negative breast cancer. Theranostics. 2017;7:647–663. doi: 10.7150/thno.16827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebert J.M., Lester R., Powell E., Seal M., McCarthy J. Advances in the systemic treatment of triple-negative breast cancer. Curr Oncol. 2018;25(Suppl 1):S142–S150. doi: 10.3747/co.25.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Hu H., Chang R., Zhong J., Knabel M., O’Meally R., Cole R.N., Pandey A., Semenza G.L. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Semenza G.L. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2:551–556. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., Zha Z., Liu Y., Li Z., Xu Y., et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S. Pyruvate kinase type M2: a key regulator within the tumour metabolome and a tool for metabolic profiling of tumours. Ernst Schering Found Symp Proc. 2007;(4):99–124. doi: 10.1007/2789_2008_091. [DOI] [PubMed] [Google Scholar]
- Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Morita M., Sato T., Nomura M., Sakamoto Y., Inoue Y., Tanaka R., Ito S., Kurosawa K., Yamaguchi K., Sugiura Y., et al. PKM1 confers metabolic advantages and promotes cell-autonomous tumor cell growth. Cancer Cell. 2018;33:355–367.e7. doi: 10.1016/j.ccell.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Nabel G.J., Verma I.M. Proposed NF-kappa B/I kappa B family nomenclature. Genes Dev. 1993;7:2063. doi: 10.1101/gad.7.11.2063. [DOI] [PubMed] [Google Scholar]
- Okazaki M., Fushida S., Tsukada T., Kinoshita J., Oyama K., Miyashita T., Ninomiya I., Harada S., Ohta T. The effect of HIF-1alpha and PKM1 expression on acquisition of chemoresistance. Cancer Manag Res. 2018;10:1865–1874. doi: 10.2147/CMAR.S166136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., He H., Jonsson P., Sinha I., Zhao C., Dahlman-Wright K. AP-1 is a key regulator of Proinflammatory cytokine TNFalpha-mediated triple-negative breast cancer progression. J Biol Chem. 2016;291:5068–5079. doi: 10.1074/jbc.M115.702571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant D.A., Duran R.V., Boulahbel H., Gottlieb E. Metabolic transformation in cancer. Carcinogenesis. 2009;30:1269–1280. doi: 10.1093/carcin/bgp070. [DOI] [PubMed] [Google Scholar]
- Tennant D.A., Duran R.V., Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Tutt A., Tovey H., Cheang M.C.U., Kernaghan S., Kilburn L., Gazinska P., Owen J., Abraham J., Barrett S., Barrett-Lee P., et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.J., Hsieh Y.J., Cheng W.C., Lin C.P., Lin Y.S., Yang S.F., Chen C.C., Izumiya Y., Yu J.S., Kung H.J., et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1alpha-mediated glucose metabolism. Proc Natl Acad Sci U S A. 2014;111:279–284. doi: 10.1073/pnas.1311249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zahari M.S., Ma B., Liu R., Renuse S., Sahasrabuddhe N.A., Chen L., Chaerkady R., Kim M.S., Zhong J., et al. Global phosphotyrosine survey in triple-negative breast cancer reveals activation of multiple tyrosine kinase signaling pathways. Oncotarget. 2015;6:29143–29160. doi: 10.18632/oncotarget.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Ito T., Azuma S., Ito E., Honma R., Yanagisawa Y., Nishikawa A., Kawamura M., Imai J., Watanabe S., et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer Sci. 2009;100:1668–1674. doi: 10.1111/j.1349-7006.2009.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W.K., Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012a;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., Lyssiotis C.A., Aldape K., Cantley L.C., Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012b;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li M., Zhang L., Zhao H., Sahin O., Chen J., Zhao J.J., Songyang Z., Yu D. Oncogenic kinase-induced PKM2 tyrosine 105 phosphorylation converts nononcogenic PKM2 to a tumor promoter and induces cancer stem-like cells. Cancer Res. 2018;78:2248–2261. doi: 10.1158/0008-5472.CAN-17-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.