Abstract
To evaluate the prognostic and molecular mechanisms of sex and racial differences in gastric cancer, data from two large centers were used to retrospectively analyze the survival of gastric cancer patients with regard to sex and racial differences. In examining the molecular mechanism of sex in gastric cancer patients of different races, data from The Cancer Genome Atlas database were used to analyze differentially expressed genes (DEGs), and Gene Ontology (GO) enrichment and DNA methylation analyses were performed. Among White gastric cancer patients, it was found that the survival prognosis for females was better than that for males; conversely, among Chinese patients, males had a better prognosis. For African Americans, sex may have an impact on gastric cancer, but this relationship was unclear. The core DEGs between the different sexes included glycogenin 2 pseudogene 1, ribosomal protein S4 Y-linked 1, taxilin-γ and eukaryotic translation initiation factor 1A X-linked among White patients, and GO enrichment analysis revealed that these genes act mainly through RNA binding and transcription pathways. Among Black patients, core DEGs included DnaJ heat shock protein family (Hsp40) member C5, histone deacetylase 10, neogenin 1 and SMG5 nonsense mediated mRNA decay factor, which are mainly related to pathways of cellular structural changes based on GO enrichment analysis. For Asian patients, core DEGs included zinc finger protein Y-linked, thymosin β4 Y-linked, zinc finger protein 787 and ubiquitously transcribed tetratricopeptide repeat containing, Y-linked, participating in cell surface receptor-associated signal transduction and G-protein coupled receptor protein signaling pathways, according to GO. The expression of different core genes and differences in path ways are likely to be the main causes affecting the variation observed among gastric cancer patients of different races and sexes.
Keywords: sex, race, gastric cancer, differentially expressed genes, DNA methylation
Introduction
Gastric cancer is the third leading cause of tumor-related death, killing >730,000 people a year worldwide (1). However, the incidence of gastric cancer ranks only fifth globally, reaching 1,000,000 new cases/year (2). Notably, there are differences in the incidence of gastric cancer in different regions and races. In the United States, the incidence of gastric cancer in White American patients is low, with 25,000 new cases of gastric cancer each year, but it causes 10,700 deaths/year (3). Moreover, 75% of White American patients with gastric cancer have progressive disease, and the 5 year survival rate is only 30.4% (3). For African American patients, the incidence rate for gastric cancer is 11.8 (95% CI, 10.3-13.2) per 100,000 person-years (3). In addition, >70% of new cases of gastric cancer occur in developing countries, with ~50% of cases in eastern Asia, mainly in China, and the occurrence and mortality of gastric cancer in China accounts for 42.6 and 45.0% of the worldwide total, respectively (4). China ranks fifth in gastric cancer incidence and sixth in mortality in the world among 183 countries (4). Overall, there are notable sex differences in the occurrence of gastric cancer, with a previous study indicating that gastric cancer occurs significantly more frequently in male patients than in female patients, with a ratio of 2:1 (5). However, the impact of sex differences on gastric cancer is not clear, nor is it clear whether sex differences occur in gastric cancer patients of different races. Overall, research in this area is still lacking, and further studies are needed to analyze such differences. Clarification of the factors that lead to sex and ethnic differences in gastric cancer may offer new insight into the key pathways involved.
To obtain survival and tumor-related data from gastric cancer patients of different ethnic backgrounds, data from two large centers, the Gastric Cancer Research of Sun Yat-Sen University and the Surveillance Epidemiology and End Results (SEER) databases, were utilized. The SEER database is part of the National Cancer Institute and contains records of the pathogenesis, treatment, pathology and prognosis of millions of patients in the United States from 1973 onwards (6). In addition to analyzing differences in patient survival and prognosis, mRNA and DNA methylation data from gastric cancer tissue samples were obtained from The Cancer Genome Atlas (TCGA) database to identify relevant molecular mechanisms affecting sex differences among different races.
Materials and methods
Study subjects
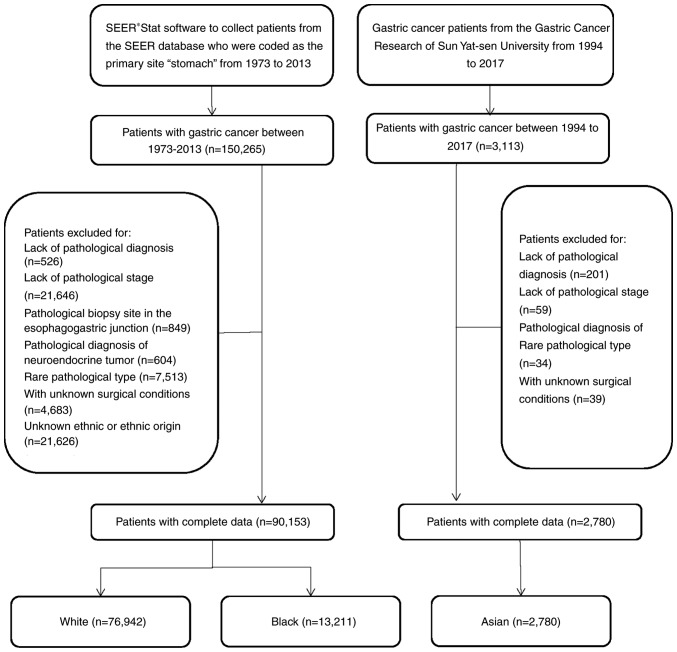
During the period 1973-2013, 150,264 patients with gastric cancer were registered in the SEER database. In the present study, patients with gastric cancer and complete information were included as the study subjects. Patients with no pathological diagnosis (n=526), patients without pathological staging (n=21,646), patients with tumors at the esophagogastric junction (n=849), patients with neuroendocrine tumors (n=604), patients with a rare pathological type (n=7,513), patients with unknown surgical status (n=4,683), patients with unknown radiotherapy (n=2,664) and patients with unknown ethnicity or ethnic origin (n=21,626) were excluded. Ultimately, 90,153 patients with gastric cancer were included. Among them, there were 76,942 White patients and 13,211 Black patients, with 49,614 males and 27,328 females in the former and 7,985 males and 5,226 females in the latter. The data obtained from the SEER database included patient demographics, primary tumor location, tumor morphology, diagnosis phase, first course of treatment and tracked patient follow up (https://seer.cancer.gov/).
The Center for Gastric Cancer Research of Sun Yat-Sen University collected follow up data for patients with gastric cancer between 1994 and 2017, including general demographic data, location of primary tumor, family history, Borrmann classification, surgical status and tumor characteristics. The present study included 3,113 patients with gastric cancer who were enrolled in the Center for Gastric Cancer Research at Sun Yat-Sen University between 1994 and 2017, and had complete follow-up information. Patients with no pathological diagnosis (n=201), patients without pathological staging (n=59), patients with rare pathological types (n=34) and patients with unknown surgical status (n=39) were excluded (2,780 patients; Fig. 1). In the Asian population, there were 1,845 male patients and 935 female patients.
Figure 1.
Flowchart of the patient cohorts.
TCGA contained information for 396 patients with gastric cancer. Patients with unknown ethnicities were excluded (n=55), leading to a total of 341 patients, including 256 White patients, 11 Black patients and 74 Asian patients.
Clinicopathological parameters and ethics statement
Because the American Joint Committee on Cancer (AJCC) staging system was not used before 2004, patients in the SEER data base before 2004 were mainly classified according to SEER staging (7). SEER staging includes localized, regional and distant classifications. For patients after 2004, TNM staging was performed according to the 2004 AJCC standard (7), and this applied to most of the data from Sun Yat-Sen University Center for Gastric Cancer Research (7). Patients from the Sun Yat-Sen University Center for Gastric Cancer Research before 2004 were reclassified according to their data. Age was divided into categories of ≥65 years old and <65 years old. The diagnosis of gastric cancer was based on the standards of the International Classification of Diseases for Oncology (ICD-O). None of the information obtained from the SEER database contains any identifiers, and all information is public. Due to the retrospective nature of the study, informed consent from the patients was not required. This analysis did not involve interaction with human subjects or the use of personally identifiable information. Prior to analysis, the patient records/information was anonymized and deidentified, and these methods were performed in accordance with the study protocol approved by the ethics committee. The Ethics Committee of the First Affiliated Hospital of Sun Yat-Sen University approved the use of the aforementioned databases in the present study.
Screening of differentially expressed genes (DEGs)
The RNA-Seq by Expectation Maximization method was used to normalize the level 3 transcriptome data of the dataset, and all gene expression values were logarithmically trans formed. Approximate data were normally distributed after quantile normalization (8). In the present study, the R program package limma v3.28.14 (https://www.bioconductor.org/packages/devel/bioc/vignettes/limma/inst/doc/users-guide.pdf) was used to analyze gene expression data for male and female patient of different ethnicities, and mRNAs satisfying P<0.01, false discovery rate (FDR) <0.01, and |log2 fold change (FC)|>2 were further investigated, where P<0.05 indicates that the hypothesis test was statistically significant and FDR is a control index for the hypothesis test error rate. As an evaluation index of the selected differential genes, the number of false rejections was proportional to the number of rejected null hypotheses. FC is generally used to describe the degree of change from an initial to a final value. In the present study, the ratio of the tumor tissue gene expression value to the normal tissue gene expression value, also called the difference multiple, was used. Heat maps of the differential genes were constructed in R (https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf) for easy visualcomparison.
Preprocessing of DNA methylation chip data
Whole-genome DNA methylation analysis of the data set was performed using the Illumina Infinium Human Methylation 450 Bead Chip method (Illumina, Inc.), and raw data (primary data in TCGA) were processed using the R package minfi version 1.20.0 (9). Subsequent background subtraction, quantile normalization and quality control were performed. A low-quality probe was removed if it met the following criteria: i) An undetectable rate (P>0.05) in ≥5% of the total sample; ii) a variance coefficient <5%; or iii) single-nucleotide polymorphisms (SNPs) located in the detected CpG dinucleotide (10). β-mixture quantile normalization was used for further Type I and II probe correction (11). The DNA methylation data of the validation set were standardized (12), and the methylation and expression data of the core genes were correlated. VanderWeele's mediation analysis was used to investigate whether the prognostic effects of seven DNA methylation sites were mediated by affecting the corresponding mRNA expression (13).
Functional enrichment and pathway analysis of DEGs
Gene Ontology (GO) analysis was performed on the identified DEGs using the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.8; https://david.ncifcrf.gov/), which provides a comprehensive and systematic annotation tool for elucidating the biological significance of genes. GO analysis includes molecular function, biological process and cellular component aspects (14). DEGs detected using DESeq2 software v3.6 (http://bioconductor.org/packages/stats/bioc/DESeq2/) were used as enrichment prospects, human database reference genes were used as the enrichment analysis background, P<0.05 was used to screen significantly enriched GO terms, and the number of DEGs in each significantly enriched GO term was counted (15). The results were visualized using the R Goplot package v1.0.2 (https://www.rdocumentation.org/packages/GOplot/versions/1.0.2).
Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com/analysis/) is a platform that contains gene expression information for 10 tumors and 1,065 clinical survival data points for patients with gastric cancer. This website was used to obtain core gene expression and patient survival prognosis information in an effort to identify the reasons for differences in the survival of gastric cancer patients of different races and sexes (16). To assess the prognostic value of a particular gene, patient samples were divided into two groups based on the median expression level of the gene (high vs. low expression). Core genes were uploaded to the database to obtain a Kaplan-Meier survival map and this map was used to analyze the overall survival (OS) of patients with gastric cancer. The 95% CI, P-value and hazard ratio (HR) were calculated and are displayed on the graphs.
Statistical analyses
Differences in the clinicopathological parameters between groups were calculated using Pearson's χ2 test. Survival analysis was performed to calculate cancer-specific survival (CSS) and OS. CSS was defined as the time from the initial diagnosis of primary gastric cancer to the death of a patient due to the tumor. OS was defined as the time from the initial diagnosis of primary gastric cancer to death for any reason. The survival rate was statistically examined using the Kaplan-Meier method, and the differences in survival curves were statistically analyzed using the log-rank test. Propensity analysis is a statistical method used to generate matched patients when comparing long term survival in observational nonrandomized controlled trials. A one-to-one match of nonpermutation was performed using the nearest neighbor matching method (17). All factors with an impact on survival prognosis were included in the propensity score matching (Figs. S1-S5). The caliper width was 0.05 times the standard deviation of the logit of the propensity score, and deviations of the co-construction variable of >99% were eliminated (17). Spearman's rank correlation (rs) was used to investigate the relationship between methylation and gene expression. Univariate analysis of continuous and binary categorical dependent variables was performed using simple linear analysis and logistic regression analysis. All statistical analyses were performed using SPSS Statistical Software version 20.0 (IBM Corp.) and R version 3.3.0 (The R Foundation; https://www.rproject.org/foundation/). Two-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological characteristics of the study subjects
The total number of subjects was 93,278, among which 2,857 were Asian, 13,223 were Black and 77,198 were White. In the Asian population, there were 1,893 male patients and 964 female patients, and the male to female ratio was 1.96. In the Black population, there were 7,993 male and 5,241 female patients, and the male to female ratio was 1.53. There were 49,766 male and 27,432 female patients with gastric cancer in the White population, with a male to female ratio of 1.81. Compared with White male patients, White female patients had a lower degree of differentiation (59.43 vs. 63.70%), a higher incidence of signet ring cell carcinoma (9.91 vs. 15.28%), a lower incidence of lymph node metastasis (35.35 vs. 28.22%), T1 stages (23.08 vs. 24.37%) and decreased distant metastasis (34.91 vs. 31.32%) (Table I). Moreover, the primary site of the tumor more frequently occurred in the gastric antrum (13.59 vs. 21.62%), and more female patients underwent radical gastrectomy (46.47 vs. 49.45%) (Tables I and II). Among Black patients, the degree of differentiation, the incidence of signet ring cell carcinoma, lymph node metastasis and T stage were similar to those of White patients. However, the primary site of gastric cancer in Black patients was more likely to be associated with the gastric antrum; additionally, the incidence of cancer in the gastric antrum in male and female patients was similar (27.19 vs. 29.80%), and fewer patients underwent radical gastrectomy (51.68 vs. 49.45%) (Table II). A rare pathological type occurred more frequently in female patients than in male patients (12.40 vs. 14.77%) (Tables I and II). Table III illustrates the clinicopathological features of Asian patients with gastric cancer. The incidence of the primary tumor site in the corpus was higher among females (16.15 vs. 30.70%), as was the incidence of diffuse infiltrative gastric cancer (9.11 vs. 14.97%). The general characteristics of patients with gastric cancer of different ethnic backgrounds showed that the effects of different ethnicities on gastric cancer are varied.
Table I.
Demographics according to gender for White and Black gastric cancer patients (1973-2003).
| Characteristic No. | White gastric cancer patients
|
Black gastric cancer patients
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted
|
Adjusted
|
Unadjusted
|
Adjusted
|
|||||||||||||||
| M | SD/% | F | SD/% | P‑value | M | SD/% | F | SD/% | M | SD/% | F | SD/% | P‑value | M | SD/% | F | SD/% | |
| 28,775 | 63.86 | 16,281 | 36.14 | 9,975 | 50 | 9,975 | 50 | 4,469 | 61.40 | 2,810 | 38.60 | 1,188 | 50 | 1,188 | 50 | |||
| Age, years | ||||||||||||||||||
| ≥65 | 10,184 | 35.39 | 4,290 | 26.35 | <0.01 | 3,028 | 30.36 | 3,028 | 30.36 | 1959 | 43.84 | 953 | 33.91 | <0.01 | 437 | 36.78 | 437 | 36.78 |
| <65 | 18,591 | 64.61 | 11,991 | 73.65 | 6,947 | 69.64 | 6,947 | 69.64 | 2,510 | 56.16 | 1,857 | 66.09 | 751 | 63.22 | 751 | 63.22 | ||
| Tumor grade | ||||||||||||||||||
| 1 | 1,910 | 6.64 | 1,037 | 6.37 | <0.01 | 510 | 5.11 | 510 | 5.11 | 371 | 8.30 | 217 | 7.72 | <0.01 | 49 | 4.12 | 49 | 4.12 |
| 2 | 8,151 | 28.33 | 3,901 | 23.96 | 2461 | 24.67 | 2461 | 24.67 | 1324 | 29.63 | 807 | 28.72 | 334 | 28.11 | 334 | 28.11 | ||
| 3 | 17,101 | 59.43 | 10,371 | 63.70 | 6,597 | 66.14 | 6,597 | 66.14 | 2,575 | 57.62 | 1,664 | 59.22 | 786 | 66.16 | 786 | 66.16 | ||
| 4 | 1,613 | 5.61 | 972 | 5.97 | 407 | 4.08 | 407 | 4.08 | 199 | 4.45 | 122 | 4.34 | 19 | 1.60 | 19 | 1.60 | ||
| Primary site | ||||||||||||||||||
| Cardia | 10,784 | 37.48 | 2,727 | 16.75 | <0.01 | 2,240 | 22.46 | 2,240 | 22.46 | 532 | 11.90 | 227 | 8.08 | <0.01 | 86 | 7.24 | 86 | 7.24 |
| Fundus | 1,257 | 4.37 | 799 | 4.91 | 445 | 4.46 | 445 | 4.46 | 188 | 4.21 | 118 | 4.20 | 31 | 2.61 | 31 | 2.61 | ||
| Body | 1,662 | 5.78 | 1,328 | 8.16 | 719 | 7.21 | 719 | 7.21 | 333 | 7.45 | 238 | 8.47 | 72 | 6.06 | 72 | 6.06 | ||
| Gastric antrum | 4,028 | 14.00 | 3,523 | 21.64 | 2,007 | 20.12 | 2,007 | 20.12 | 1,164 | 26.05 | 829 | 29.50 | 417 | 35.10 | 417 | 35.10 | ||
| Pylorus | 863 | 3.00 | 649 | 3.99 | 321 | 3.22 | 321 | 3.22 | 194 | 4.34 | 158 | 5.62 | 53 | 4.46 | 53 | 4.46 | ||
| Lesser curvature | 2,542 | 8.83 | 1,676 | 10.29 | 971 | 9.73 | 971 | 9.73 | 598 | 13.38 | 343 | 12.21 | 157 | 13.22 | 157 | 13.22 | ||
| Greater curvature | 1,148 | 3.99 | 860 | 5.28 | 442 | 4.43 | 442 | 4.43 | 230 | 5.15 | 125 | 4.45 | 34 | 2.86 | 34 | 2.86 | ||
| Overlapping lesion | 2,422 | 8.42 | 1,737 | 10.67 | 987 | 9.89 | 987 | 9.89 | 494 | 11.05 | 277 | 9.86 | 116 | 9.76 | 116 | 9.76 | ||
| Stomach | 4,069 | 14.14 | 2,982 | 18.32 | 1,843 | 18.48 | 1,843 | 18.48 | 736 | 16.47 | 495 | 17.62 | 222 | 18.69 | 222 | 18.69 | ||
| Histology | ||||||||||||||||||
| Adenocarcinoma | 20,438 | 71.03 | 10,229 | 62.83 | <0.01 | 7,114 | 71.32 | 7,114 | 71.32 | 3,032 | 67.85 | 1,770 | 62.99 | <0.01 | 929 | 78.20 | 929 | 78.20 |
| Signet ring cell carcinoma | 2,852 | 9.91 | 2,487 | 15.28 | 1,403 | 14.07 | 1,403 | 14.07 | 465 | 10.41 | 399 | 14.20 | 147 | 12.37 | 147 | 12.37 | ||
| Mucinous carcinoma | 1,218 | 4.23 | 628 | 3.86 | 214 | 2.15 | 214 | 2.15 | 203 | 4.54 | 109 | 3.88 | 13 | 1.09 | 13 | 1.09 | ||
| Linitis plastica | 523 | 1.82 | 536 | 3.29 | 220 | 2.21 | 220 | 2.21 | ||||||||||
| Adenocarcinoma intestinal | 1,022 | 3.55 | 535 | 3.29 | 217 | 2.18 | 217 | 2.18 | 215 | 4.81 | 117 | 4.16 | 26 | 2.19 | 26 | 2.19 | ||
| Other | 2,722 | 9.46 | 1,866 | 11.46 | 807 | 8.09 | 807 | 8.09 | 554 | 12.40 | 415 | 14.77 | 73 | 6.14 | 73 | 6.14 | ||
| SEER historic stage | ||||||||||||||||||
| Distant | 10,126 | 35.19 | 5,253 | 32.26 | <0.01 | 3,455 | 34.64 | 3,455 | 34.64 | 1,677 | 37.53 | 889 | 31.64 | <0.01 | 422 | 35.52 | 422 | 35.52 |
| Localized | 5,579 | 19.39 | 3,566 | 21.90 | 2,030 | 20.35 | 2,030 | 20.35 | 897 | 20.07 | 650 | 23.13 | 222 | 18.69 | 222 | 18.69 | ||
| Regional | 10,399 | 36.14 | 5,525 | 33.94 | 3,456 | 34.65 | 3,456 | 34.65 | 1,450 | 32.45 | 911 | 32.42 | 404 | 34.01 | 404 | 34.01 | ||
| Unstaged | 2,671 | 9.28 | 1,973 | 12.12 | 1,034 | 10.37 | 1,034 | 10.37 | 445 | 9.96 | 360 | 12.81 | 120 | 10.10 | 120 | 10.10 | ||
| Sequence with surgery | ||||||||||||||||||
| No radiation | 25,482 | 88.56 | 15,077 | 92.60 | <0.01 | 9,273 | 92.96 | 9,273 | 92.96 | 4,088 | 91.47 | 2,577 | 91.71 | 0.94 | 1,121 | 94.36 | 1,121 | 94.36 |
| Radiation after surgery | 690 | 2.40 | 149 | 0.92 | 74 | 0.74 | 74 | 0.74 | 355 | 7.94 | 217 | 7.72 | 66 | 5.56 | 66 | 5.56 | ||
| Radiation prior to surgery | 2,603 | 9.05 | 1,055 | 6.48 | 628 | 6.30 | 628 | 6.30 | 26 | 0.58 | 16 | 0.57 | 1 | 0.08 | 1 | 0.08 | ||
| Gastrectomy | ||||||||||||||||||
| No | 12,089 | 42.01 | 6,781 | 41.65 | 0.46 | 4,192 | 42.03 | 4,192 | 42.03 | 1,915 | 42.85 | 1,179 | 41.96 | 0.45 | 508 | 42.76 | 508 | 42.76 |
| Yes | 16,686 | 57.99 | 9,500 | 58.35 | 5,783 | 57.97 | 5,783 | 57.97 | 2,554 | 57.15 | 1,631 | 58.04 | 680 | 57.24 | 680 | 57.24 | ||
| Radiation | ||||||||||||||||||
| No | 22,022 | 76.53 | 13,783 | 84.66 | <0.01 | 8,532 | 85.53 | 8,532 | 85.53 | 3,701 | 82.81 | 2,381 | 84.73 | 0.03 | 1,074 | 90.40 | 1,074 | 90.40 |
| Yes | 6,753 | 23.47 | 2,498 | 15.34 | 1,443 | 14.47 | 1,443 | 14.47 | 766 | 17.14 | 429 | 15.27 | 114 | 9.60 | 114 | 9.60 | ||
M, male; F, female
Table II.
Demographics according to gender for White and Black GC patients (2004-2013).
| Characteristic No. | White GC patients
|
Black GC patients
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted
|
Adjusted
|
Unadjusted
|
Adjusted
|
|||||||||||||||
| M | SD/% | F | SD/% | P‑value | M | SD/% | F | SD/% | M | SD/% | F | SD/% | P‑value | M | SD/% | F | SD/% | |
| 20,839 | 65.24 | 11,047 | 34.76 | 2,704 | 50 | 2,704 | 50 | 3,516 | 59.24 | 2,416 | 40.76 | 183 | 50 | 183 | 50 | |||
| Age, years | ||||||||||||||||||
| ≥65 | 8,022 | 38.50 | 3,567 | 32.29 | <0.01 | 812 | 30.03 | 812 | 30.03 | 2,522 | 71.73 | 1,739 | 71.98 | 0.89 | 155 | 84.70 | 155 | 84.70 |
| <65 | 12,871 | 61.76 | 7,480 | 67.71 | 1,892 | 69.97 | 1,892 | 69.97 | 994 | 28.27 | 680 | 28.15 | 28 | 15.30 | 28 | 15.30 | ||
| Tumor grade | ||||||||||||||||||
| 1 | 1,017 | 4.88 | 600 | 5.43 | <0.01 | 105 | 3.88 | 105 | 3.88 | 175 | 4.98 | 129 | 5.34 | <0.01 | 4 | 2.19 | 4 | 2.19 |
| 2 | 6,025 | 28.91 | 2,533 | 22.93 | 732 | 27.07 | 732 | 27.07 | 1,079 | 30.69 | 653 | 27.03 | 58 | 31.69 | 58 | 31.69 | ||
| 3 | 13,327 | 63.95 | 7,633 | 69.10 | 1,859 | 68.75 | 1,859 | 68.75 | 2,189 | 62.26 | 1,576 | 65.23 | 121 | 66.12 | 121 | 66.12 | ||
| 4 | 524 | 2.51 | 281 | 2.54 | 8 | 0.30 | 8 | 0.30 | 73 | 2.08 | 61 | 2.52 | 0 | 0.00 | 0 | 0.00 | ||
| Primary site | ||||||||||||||||||
| Cardia | 9,778 | 46.92 | 2,620 | 23.72 | <0.01 | 1,087 | 40.20 | 1,087 | 40.20 | 579 | 16.47 | 252 | 10.43 | <0.01 | 15 | 8.20 | 15 | 8.20 |
| Fundus | 847 | 4.06 | 507 | 4.59 | 67 | 2.48 | 67 | 2.48 | 139 | 3.95 | 94 | 3.89 | 2 | 1.09 | 2 | 1.09 | ||
| Body | 1,515 | 7.27 | 1,246 | 11.28 | 221 | 8.17 | 221 | 8.17 | 365 | 10.38 | 289 | 11.96 | 25 | 13.66 | 25 | 13.66 | ||
| Gastric antrum | 2,833 | 13.59 | 2,388 | 21.62 | 598 | 22.12 | 598 | 22.12 | 956 | 27.19 | 720 | 29.80 | 71 | 38.80 | 71 | 38.80 | ||
| Pylorus | 448 | 2.15 | 352 | 3.19 | 32 | 1.18 | 32 | 1.18 | 145 | 4.12 | 125 | 5.17 | 1 | 0.55 | 1 | 0.55 | ||
| Lesser curvature | 1,244 | 5.97 | 821 | 7.43 | 131 | 4.84 | 131 | 4.84 | 376 | 10.69 | 241 | 9.98 | 15 | 8.20 | 15 | 8.20 | ||
| Greater curvature | 618 | 2.97 | 499 | 4.52 | 53 | 1.96 | 53 | 1.96 | 141 | 4.01 | 100 | 4.14 | 2 | 1.09 | 2 | 1.09 | ||
| Overlapping lesion | 1,368 | 6.56 | 965 | 8.74 | 134 | 4.96 | 134 | 4.96 | 309 | 8.79 | 218 | 9.02 | 10 | 5.46 | 10 | 5.46 | ||
| Stomach | 2,242 | 10.76 | 1,849 | 16.74 | 381 | 14.09 | 381 | 14.09 | 506 | 14.39 | 380 | 15.73 | 42 | 22.95 | 42 | 22.95 | ||
| Histology | ||||||||||||||||||
| Adenocarcinoma | 12,774 | 61.30 | 5,445 | 49.29 | <0.01 | 1,842 | 68.12 | 1,842 | 68.12 | 2,006 | 57.05 | 1,211 | 50.12 | <0.01 | 127 | 69.40 | 127 | 69.40 |
| Signet ring cell carcinoma | 3,262 | 15.65 | 2,672 | 24.19 | 534 | 19.75 | 534 | 19.75 | 502 | 14.28 | 502 | 20.78 | 20 | 10.93 | 20 | 10.93 | ||
| Mucinous carcinoma | 1,338 | 6.42 | 834 | 7.55 | 54 | 2.00 | 54 | 2.00 | 119 | 3.38 | 74 | 3.06 | 1 | 0.55 | 1 | 0.55 | ||
| Adenocarcinoma, intestinal type | 1,782 | 8.55 | 975 | 8.83 | 158 | 5.84 | 158 | 5.84 | 438 | 12.46 | 283 | 11.71 | 8 | 4.37 | 8 | 4.37 | ||
| Other | 1,737 | 8.34 | 1,121 | 10.15 | 116 | 4.29 | 116 | 4.29 | 451 | 12.83 | 349 | 14.45 | 27 | 14.75 | 27 | 14.75 | ||
| T stage | ||||||||||||||||||
| T1 | 4,810 | 23.08 | 2692 | 24.37 | <0.01 | 787 | 29.11 | 787 | 29.11 | 857 | 24.37 | 649 | 26.86 | <0.01 | 68 | 37.16 | 68 | 37.16 |
| T2a | 1,585 | 7.61 | 868 | 7.86 | 177 | 6.55 | 177 | 6.55 | 265 | 7.54 | 181 | 7.49 | 9 | 4.92 | 9 | 4.92 | ||
| T2b | 4,669 | 22.41 | 2,174 | 19.68 | 527 | 19.49 | 527 | 19.49 | 789 | 22.44 | 486 | 20.12 | 28 | 15.30 | 28 | 15.30 | ||
| T3 | 2,803 | 13.45 | 1,566 | 14.18 | 270 | 9.99 | 270 | 9.99 | 410 | 11.66 | 294 | 12.17 | 10 | 5.46 | 10 | 5.46 | ||
| T4 | 2,400 | 11.52 | 1,342 | 12.15 | 199 | 7.36 | 199 | 7.36 | 499 | 14.19 | 324 | 13.41 | 10 | 5.46 | 10 | 5.46 | ||
| TX | 4,626 | 22.20 | 2,405 | 21.77 | 744 | 27.51 | 744 | 27.51 | 696 | 19.80 | 485 | 20.07 | 58 | 31.69 | 58 | 31.69 | ||
| N stage | ||||||||||||||||||
| N0 | 8,187 | 39.29 | 4,827 | 43.70 | <0.01 | 1,325 | 49.00 | 1,325 | 49.00 | 1,464 | 41.64 | 4,827 | 43.70 | <0.01 | 106 | 57.92 | 106 | 57.92 |
| N1 | 7,366 | 35.35 | 3,118 | 28.22 | 754 | 27.88 | 754 | 27.88 | 1,142 | 32.48 | 3,118 | 28.22 | 35 | 19.13 | 35 | 19.13 | ||
| N2 | 1,660 | 7.97 | 898 | 8.13 | 120 | 4.44 | 120 | 4.44 | 317 | 9.02 | 898 | 8.13 | 6 | 3.28 | 6 | 3.28 | ||
| N3 | 649 | 3.11 | 386 | 3.49 | 31 | 1.15 | 31 | 1.15 | 121 | 3.44 | 386 | 3.49 | 0 | 0.00 | 0 | 0.00 | ||
| NX | 3,031 | 14.54 | 1,818 | 16.46 | 474 | 17.53 | 474 | 17.53 | 472 | 13.42 | 1,818 | 16.46 | 36 | 19.67 | 36 | 19.67 | ||
| M stage | ||||||||||||||||||
| M0 | 12,327 | 59.15 | 6,727 | 60.89 | <0.01 | 1,698 | 62.80 | 1,698 | 62.80 | 2,091 | 59.47 | 6,727 | 60.89 | <0.01 | 106 | 57.92 | 106 | 57.92 |
| M1 | 7,274 | 34.91 | 3,460 | 31.32 | 805 | 29.77 | 805 | 29.77 | 1,218 | 34.64 | 3,460 | 31.32 | 62 | 33.88 | 62 | 33.88 | ||
| MX | 1,292 | 6.20 | 860 | 7.78 | 201 | 7.43 | 201 | 7.43 | 207 | 5.89 | 860 | 7.78 | 15 | 8.20 | 15 | 8.20 | ||
| Tumor size | ||||||||||||||||||
| ≤1 cm | 11,071 | 53.13 | 5,608 | 50.76 | <0.01 | 1,415 | 52.33 | 1,415 | 52.33 | 1,870 | 53.19 | 5,608 | 50.76 | <0.01 | 88 | 48.09 | 88 | 48.09 |
| >1 cm | 9,822 | 47.13 | 5,439 | 49.24 | 1,289 | 47.67 | 1,289 | 47.67 | 1,646 | 46.81 | 5,439 | 49.24 | 95 | 51.91 | 95 | 51.91 | ||
| Sequence with surgery | ||||||||||||||||||
| No radiation | 17,489 | 83.92 | 9,622 | 87.10 | <0.01 | 2,406 | 88.98 | 2,406 | 88.98 | 2,997 | 85.24 | 9,622 | 87.10 | <0.01 | 173 | 94.54 | 173 | 94.54 |
| Radiation after surgery | 2,117 | 10.16 | 1,203 | 10.89 | 202 | 7.47 | 202 | 7.47 | 465 | 13.23 | 1,203 | 10.89 | 9 | 4.92 | 9 | 4.92 | ||
| Radiation prior to surgery | 1,287 | 6.18 | 222 | 2.01 | 96 | 3.55 | 96 | 3.55 | 54 | 1.54 | 222 | 2.01 | 1 | 0.55 | 1 | 0.55 | ||
| Gastrectomy | ||||||||||||||||||
| No | 11,210 | 53.79 | 5,584 | 50.55 | <0.01 | 1,414 | 52.29 | 1,414 | 52.29 | 1,699 | 48.32 | 5,584 | 50.55 | <0.01 | 83 | 45.36 | 83 | 45.36 |
| Yes | 9,683 | 46.47 | 5,463 | 49.45 | 1,290 | 47.71 | 1,290 | 47.71 | 1,817 | 51.68 | 5,463 | 49.45 | 100 | 54.64 | 100 | 54.64 | ||
| Radiation | ||||||||||||||||||
| No | 14,062 | 67.48 | 8,518 | 77.11 | <0.01 | 2,173 | 80.36 | 2,173 | 80.36 | 2,616 | 74.40 | 8,518 | 77.11 | <0.01 | 171 | 93.44 | 171 | 93.44 |
| Yes | 6,831 | 32.78 | 2,529 | 22.89 | 531 | 19.64 | 531 | 19.64 | 900 | 25.60 | 2,529 | 22.89 | 12 | 6.56 | 12 | 6.56 | ||
M, male; F, female.
Table III.
Demographics according to gender for Asian GC patient (1994-2017).
| Characteristic | Unadjusted
|
Adjusted
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M | SD/% | F | SD/% | P-value | M | SD/% | F | SD/% | P-value | |
| No. | 1,845 | 66.37 | 935 | 33.63 | 293 | 50 | 293 | 50 | ||
| Age, years | ||||||||||
| ≥65 | 1,213 | 65.75 | 711 | 76.04 | <0.001 | 233 | 79.52 | 233 | 79.52 | 1.000 |
| <65 | 632 | 34.25 | 224 | 23.96 | 60 | 20.48 | 60 | 20.48 | ||
| Family history | ||||||||||
| No | 1,670 | 90.51 | 852 | 91.12 | 0.834 | 283 | 96.59 | 283 | 96.59 | |
| Yes | 168 | 9.11 | 83 | 8.88 | 10 | 3.41 | 10 | 3.41 | ||
| Tumor grade | ||||||||||
| 1 | 93 | 5.04 | 103 | 11.02 | <0.001 | 3 | 1.02 | 3 | 1.02 | 1.000 |
| 2 | 527 | 28.56 | 137 | 14.65 | 51 | 17.41 | 51 | 17.41 | ||
| 3 | 1,149 | 62.28 | 667 | 71.34 | 223 | 76.11 | 223 | 76.11 | ||
| 4 | 76 | 4.12 | 28 | 2.99 | 16 | 5.46 | 16 | 5.46 | ||
| Ascites | ||||||||||
| No | 1,630 | 88.35 | 815 | 87.17 | 0.329 | 289 | 98.63 | 289 | 98.63 | 1.000 |
| Serous fluid | 186 | 10.08 | 109 | 11.66 | 4 | 1.37 | 4 | 1.37 | ||
| Hemorrhagic | 29 | 1.57 | 11 | 1.18 | 0 | 0.00 | 0 | 0.00 | ||
| Primary site | ||||||||||
| Cardia | 630 | 34.15 | 227 | 24.28 | <0.001 | 68 | 23.21 | 68 | 23.21 | 1.000 |
| Corpus | 298 | 16.15 | 287 | 30.70 | 75 | 25.60 | 75 | 25.60 | ||
| Pylorus | 759 | 41.14 | 402 | 42.99 | 150 | 51.19 | 150 | 51.19 | ||
| Stomach | 58 | 3.14 | 19 | 2.03 | 0 | 0.00 | 0 | 0.00 | ||
| Histology | ||||||||||
| Adenocarcinoma | 1,413 | 76.59 | 688 | 73.58 | <0.001 | 261 | 89.08 | 261 | 89.08 | 1.000 |
| Signet ring cell carcinoma | 179 | 9.70 | 127 | 13.58 | 29 | 9.90 | 29 | 9.90 | ||
| Mucinous carcinoma | 72 | 3.90 | 32 | 3.42 | 1 | 0.34 | 1 | 0.34 | ||
| Other | 181 | 9.81 | 88 | 9.41 | 2 | 0.68 | 2 | 0.68 | ||
| T stage (AJCC, 2004) | ||||||||||
| T1 | 204 | 11.06 | 132 | 14.12 | 0.137 | 40 | 13.65 | 40 | 13.65 | |
| T2 | 237 | 12.85 | 119 | 12.73 | 25 | 8.53 | 25 | 8.53 | ||
| T3 | 848 | 45.96 | 414 | 44.28 | 170 | 58.02 | 170 | 58.02 | ||
| T4 | 556 | 30.14 | 270 | 28.88 | 58 | 19.80 | 58 | 19.80 | ||
| N stage (AJCC, 2004) | ||||||||||
| N0 | 414 | 22.44 | 212 | 22.67 | 0.532 | 68 | 23.21 | 68 | 23.21 | 1.000 |
| N1 | 770 | 41.73 | 403 | 43.10 | 157 | 53.58 | 157 | 53.58 | ||
| N2 | 645 | 34.96 | 316 | 33.80 | 68 | 23.21 | 68 | 23.21 | ||
| N3 | 16 | 0.87 | 4 | 0.43 | 0 | 0.00 | 0 | 0.00 | ||
| M stage (AJCC, 2004) | ||||||||||
| M0 | 1,389 | 75.28 | 713 | 76.26 | 0.492 | 271 | 92.49 | 271 | 92.49 | 1.000 |
| M1 | 419 | 22.71 | 209 | 22.35 | 22 | 7.51 | 22 | 7.51 | ||
| MX | 37 | 2.01 | 13 | 1.39 | 0 | 0.00 | 0 | 0.00 | ||
| Borrmann | ||||||||||
| Massive type | 102 | 5.53 | 32 | 3.42 | <0.001 | 5 | 1.71 | 5 | 1.71 | 1.000 |
| Localized type | 436 | 23.63 | 211 | 22.57 | 69 | 23.55 | 69 | 23.55 | ||
| Infiltrating ulcerative type | 1,023 | 55.45 | 484 | 51.76 | 199 | 67.92 | 199 | 67.92 | ||
| Diffuse infiltrative type | 168 | 9.11 | 140 | 14.97 | 14 | 4.78 | 14 | 4.78 | 1.000 | |
| Mixed type | 116 | 6.29 | 68 | 7.27 | 6 | 2.05 | 6 | 2.05 | ||
| Gastrectomy | ||||||||||
| No | 207 | 11.22 | 120 | 12.83 | 0.213 | 1 | 0.34 | 1 | 0.34 | 1.000 |
| Yes | 1,638 | 88.78 | 815 | 87.17 | 292 | 99.66 | 292 | 99.66 | ||
M, male; F, female.
Survival analysis of subgroups according to race and sex
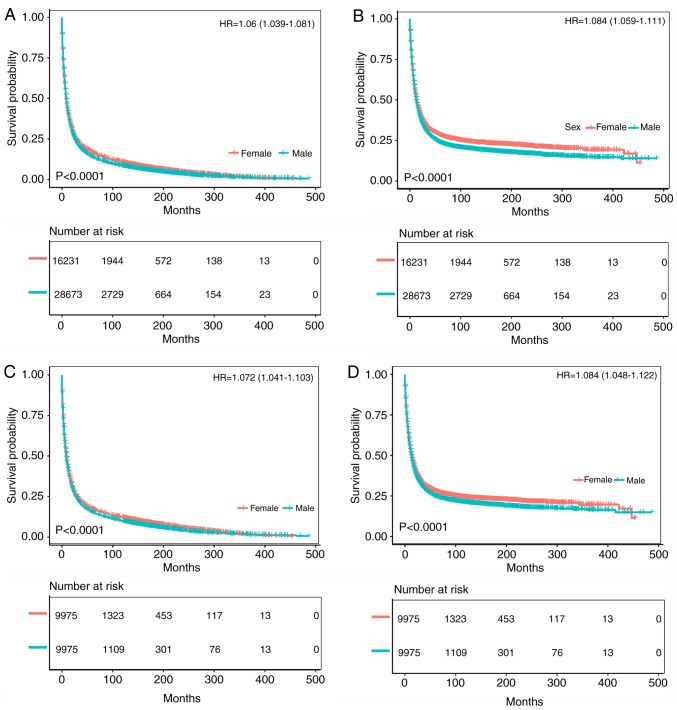
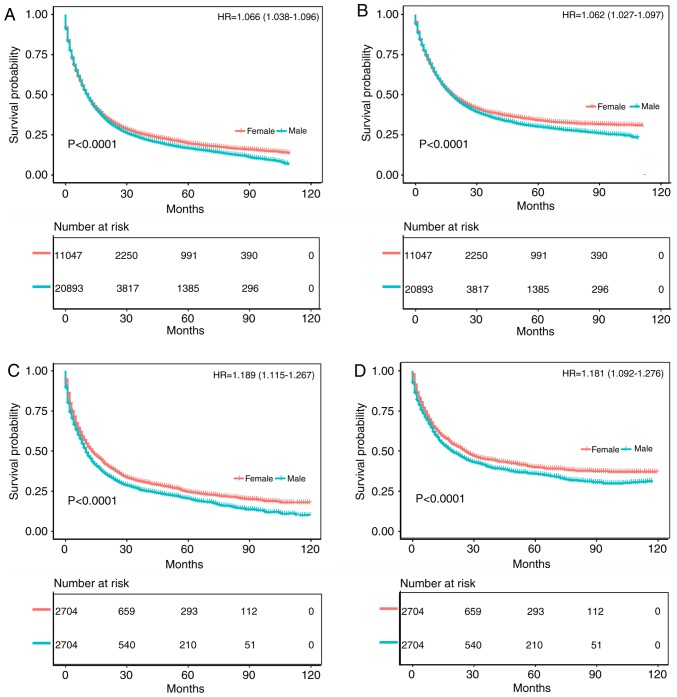
As shown in Figs. 2 and 3, the survival prognosis of female gastric cancer patients was better than that of male patients among Whites. Fig. 2A and B compare the OS and CSS of patients with different gastric cancer types between 1973 and 2003. The survival time of male patients was significantly lower than that of female patients (OS: HR of death=1.06, 95% CI=1.039-1.081, P<0.001; CSS: HR of death=1.084, 95% CI=1.059-1.111, P<0.001), which remained after matching (OS: HR of death=1.072, 95% CI=1.041-1.103, P<0.001; CSS: HR of death=1.084, 95% CI=1.048-1.122, P<0.001) (Fig. 2C and D). Fig. 3A and B compare the OS and CSS of patients with different gastric cancer types between 2004 and 2013, revealing that the survival time of male patients was significantly lower than that of female patients (OS: HR of death=1.066, 95% CI=1.038-1.096, P<0.001; CSS: HR of death=1.062, 95% CI=1.027-1.097, P<0.001), which again remained after matching (OS: HR of death=1.189, 95% CI=1.115-1.267, P<0.001; CSS: HR of death=1.181, 95% CI=1.092-1.276, P<0.001) (Fig. 3C and D). Fig. S1A and B displays the trend in HRs for OS and CSS among male and female patients (P<0.0001), with significantly better results for the latter.
Figure 2.
Kaplan-Meier analysis after gastric cancer diagnosis among White patients. The mean survival time of female patients was significantly prolonged compared with that of male patients (P<0.001). (A) Kaplan-Meier curve for OS in female patients and male patients from 1973-2003. (B) Kaplan-Meier curve for CSS in female patients and male patients from 1973-2003. (C) Kaplan-Meier curve for OS in female patients and male patients from 1973-2003, adjusted via PSM. (D) Kaplan-Meier curve for CSS in female patients and male patients from 1973-2003, adjusted via PSM. HR, hazard ratio; PSM, propensity score matching; OS, overall survival; CSS, cancer-specific survival.
Figure 3.
Kaplan-Meier analysis after gastric cancer diagnosis among White patients. The mean survival time of female patients was significantly prolonged compared with that of male patients (P<0.001). (A) Kaplan-Meier curve for OS in female patients and male patients from 2004-2013. (B) Kaplan-Meier curve for CSS in female patients and male patients from 2004-2013. (C) Kaplan-Meier curve for OS in female patients and male patients from 2004- 2013, adjusted via PSM. (D) Kaplan-Meier curve for CSS in female patients and male patients from 2004-2013, adjusted via PSM. HR, hazard ratio; PSM, propensity score matching; OS, overall survival; CSS, cancer-specific survival.
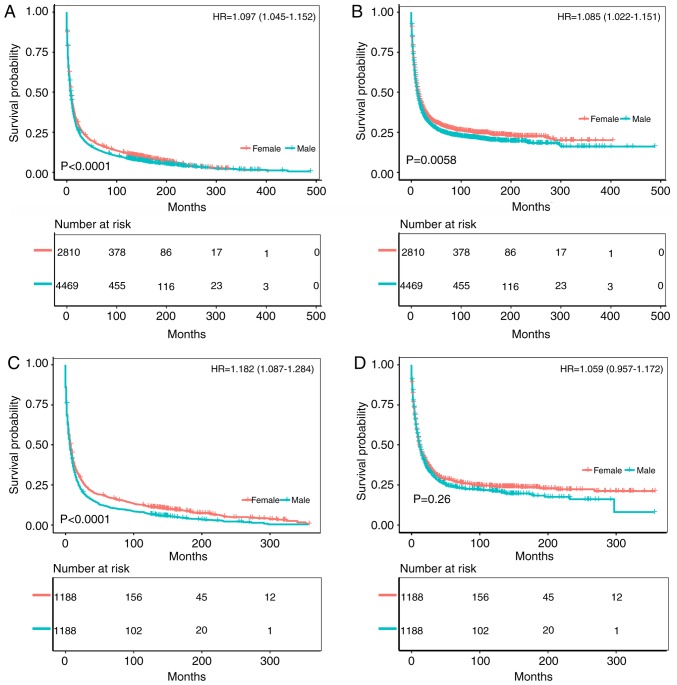
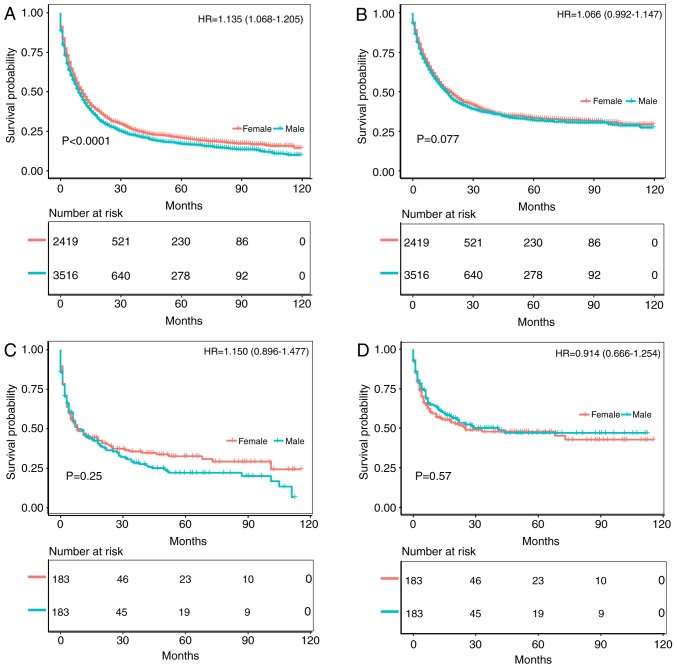
The survival prognosis for female gastric cancer patients was not greatly different from that of male patients among Black patients (Figs. 4 and 5). Fig. 4A and B compare the OS and CSS of patients with different gastric cancer types between 1973 and 2003, wherein the survival time of male patients was significantly lower than that of female patients (OS: HR of death=1.097, 95% CI=1.045-1.152, P<0.001; CSS: HR of death=1.085, 95% CI=1.022-1.151, P=0.0058). After matching, the OS of male patients was still significantly lower than that of female patients (HR of death=1.182, 95% CI=1.087-1.284, P<0.001), but there was no significant difference in CSS (HR of death=1.059, 95% CI=0.957-1.172, P=0.260) (Fig. 4C and D). Fig. 5A and B compare the OS and CSS of patients with different gastric cancer types between 2004 and 2013. Although the OS of male patients was significantly lower than that of female patients (HR of death=1.135, 95% CI=1.068-1.205, P<0.001), there was no significant difference in CSS (HR of death=1.066, 95% CI=0.992-1.147, P=0.077). Furthermore, there was no significant difference in survival between the two groups after matching (OS: HR of death=1.15, 95% CI=0.896-1.477, P=0.25; CSS: HR of death=0.914, 95% CI=0.666-1.254, P=0.570) (Fig. 5C and D). Fig. S1C and D illustrates the trend in HRs for OS and CSS among male and female patients, with no significant differences between the groups.
Figure 4.
Kaplan-Meier analysis after gastric cancer diagnosis among Black patients. The mean survival time of female patients was significantly prolonged compared with that of male patients (P<0.001). (A) Kaplan-Meier curve for OS in female patients and male patients from 1973-2003. (B) Kaplan-Meier curve for CSS in female patients and male patients from 1973-2003. (C) Kaplan-Meier curve for OS in female patients and male patients from 1973-2003, adjusted via PSM. (D) Kaplan-Meier curve for CSS in female patients and male patients from 1973-2003, adjusted via PSM. HR, hazard ratio; PSM, propensity score matching; OS, overall survival; CSS, cancer-specific survival.
Figure 5.
Kaplan-Meier analysis after gastric cancer diagnosis among Black patients. The mean survival time of female patients was significantly prolonged compared with that of male patients (P<0.001). (A) Kaplan-Meier curve for OS in female patients and male patients from 2004-2013. (B) Kaplan-Meier curve for CSS in female patients and male patients from 2004-2013. (C) Kaplan-Meier curve for OS in female patients and male patients from 2004-2013, adjusted via PSM. (D) Kaplan-Meier curve for CSS in female patients and male patients from 2004-2013, adjusted via PSM. HR, hazard ratio; PSM, propensity score matching; OS, overall survival; CSS, cancer-specific survival.
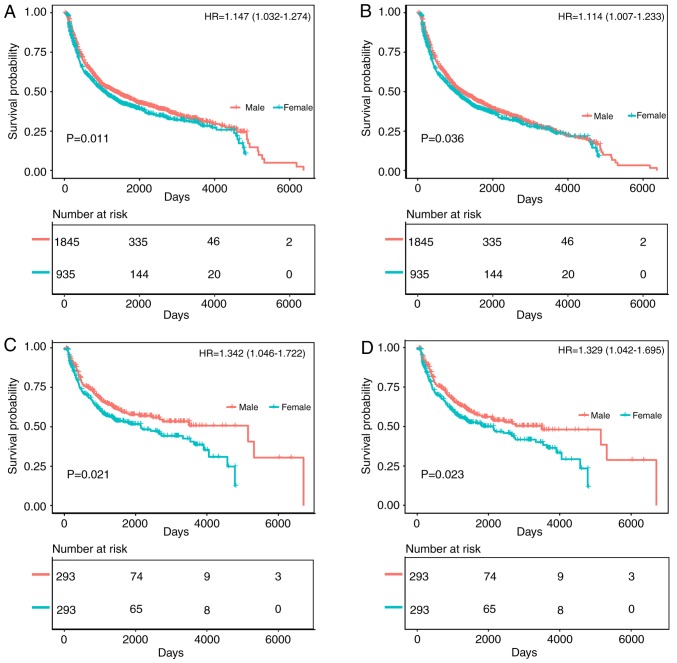
Among Asian patients, the survival prognosis of female gastric cancer patients was worse than that of male patients (Fig. 6). The comparison of the OS and CSS of patients with different gastric cancer types between 1994 and 2017 is shown in Fig. 6A and B. The survival time of female patients was significantly lower than that of male patients (OS: HR of death=1.147, 95% CI=1.032-1.274, P=0.011; CSS: HR of death=1.114, 95% CI=1.007-1.233, P=0.036), and the survival time of female patients was significantly lower than that of male patients after matching (OS: HR of death=1.342, 95% CI=1.046-1.722, P=0.021; CSS: HR of death=1.329, 95% CI=1.042-1.695, P=0.023) (Fig. 6C and D). The trend in HRs for OS and CSS among male and female patients is illustrated in Fig. S1E and F (P<0.0001), with significantly better results for men than women.
Figure 6.
Kaplan-Meier analysis after gastric cancer diagnosis among Asian patients. The mean survival time of male patients was significantly prolonged compared with that of female patients (P<0.05). (A) Kaplan-Meier curve for OS in female patients and male patients from 1994-2017. (B) Kaplan-Meier curve for CSS in female patients and male patients from 1994-2017. (C) Kaplan-Meier curve for OS in female patients and male patients from 1994-2017, adjusted via PSM. (D) Kaplan-Meier curve for CSS in female patients and male patients from 1994-2017, adjusted via PSM. HR, hazard ratio; PSM, propensity score matching; OS, overall survival; CSS, cancer-specific survival.
All 12 clinicopathological factors, including T stage, N stage, M stage, tumor grade, marital status, primary tumor location, pathological type, ethnicity, age group, SEER historic stage, ascites status and Borrmann classification, were significant in the univariate analysis (Figs. S2 S6). Figs. S2 and S3 indicate that for White patients, female sex was an independent protective factor for survival. However, as shown in Figs. S4 and S5, the prognosis of women between 1973 and 2003 was better than that of men among Black patients, whereas sex-related survival was not significant between 2004 and 2013. In Asians, male sex was an independent protective factor for survival in gastric cancer (Fig. S6).
Functional enrichment and pathway analysis of DEGs
Regarding White patients, the mRNA expression data of 161 male and 95 female gastric cancer tissue samples were summarized after pretreatment and the R package limma was used for differential expression analysis according to the top 50 log2 FC values. Genes were mapped using heat maps, in which 50 mRNAs were upregulated and 50 mRNAs downregulated. The specific genes are shown in Fig. S7. Among them, gray indicates male, and yellow indicates female. To further investigate the role of differential genes in White patients with gastric cancer, DAVID was used for GO enrichment analysis. According to the results, in addition to enrichment of the sex differentiation pathway, the DEGs were mainly enriched in ‘RNA binding', ‘transcription', ‘oxidoreductase activity incorporation of two atoms of oxygen', and ‘rRNA binding' (Fig. S8). The mRNA expression data for Black patients were also summarized, comprising 8 male and 3 female gastric cancer tissue samples. The specific genes are shown in Fig. S9. As above, gray indicates male, and yellow indicates female. Based on the GO enrichment analysis, DEGs were mainly enriched in ‘cornified envelope', ‘endopeptidase inhibitor activity', ‘structural molecule activity', ‘extracellular region' and ‘extracellular space', in addition to enrichment of the sex differentiation pathway (Fig. S10). Similarly, the mRNA expression data for Asian patients involved 48 male and 26 female gastric cancer tissue samples. The specific genes are shown in Fig. S11, where gray indicates male and yellow female. In addition to enrichment of the sex differentiation pathway, DEGs were mainly enriched in ‘RNA binding', ‘extracellular region', ‘acute phase response', ‘cell surface receptor linked signal transduction' and ‘G protein coupled receptor protein signaling pathway', according to the GO enrichment analysis (Fig. S12).
Relationship between CpG methylation, gene expression and prognosis
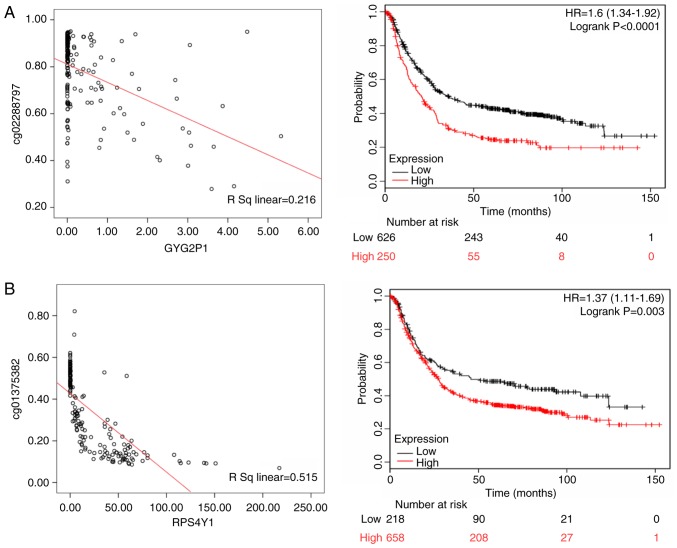
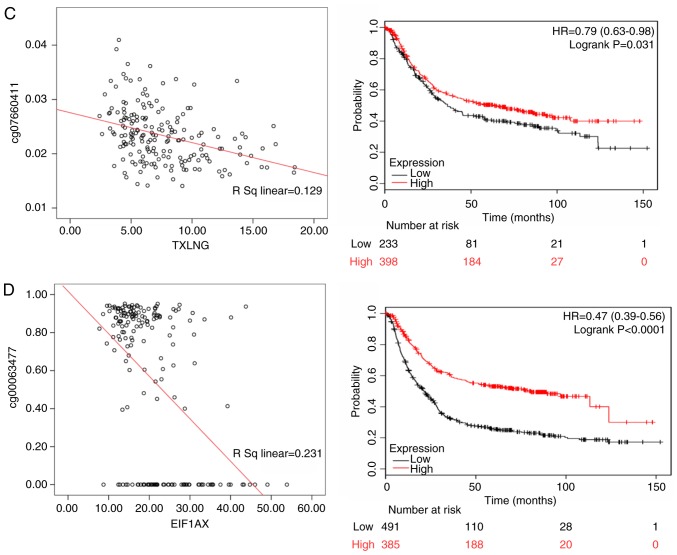
Methylation of seven CpG loci and expression of quantitative trait loci were assessed in the gene set. Because promoter hypermethylation plays an important role in the inactivation of cancer associated genes, there was particular interest in genes with low methylation and high expression levels. In subsequent analyses, these genes may be used to screen candidate diagnostic biomarkers (18). Among White patients, the methylation level of the CpG locus in the promoter and first exon regions was inversely correlated with the expression of corresponding genes, such as glycogenin 2 pseudogene 1 (GYG2P1; rs=0.216, P<0.05), ribosomal protein S4 Y-linked 1 (RPS4Y1; rs= 0.515, P<0.05), taxilin-γ (TXLNG; rs=0.129, P<0.05) and eukaryotic translation initiation factor 1A X-linked (EIF1AX; rs=0.231, P<0.05). In the KMPlot database, OS data were available for 876 patients with gastric cancer, and GYG2P1 (HR of death=1.600, 95% CI=1.340-1.920, P<0.001), RPS4Y1 (HR of death=1.370, 95% CI=1.110-1.690, P=0.003), TXLNG (HR of death=0.790, 95% CI=0.630-0.980, P=0.031) and EIF1AX (HR of death=0.470, 95% CI=0.390-0.560, P<0.001) were differentially expressed. GYG2P1, RPS4Y1, TXLNG and EIF1AX were indicated to be upregulated by DNA hypomethylation. GYG2P1 and RPS4Y1 were highly expressed in male patients and were significantly associated with a poor prognosis. By contrast, high expression of TXLNG and EIF1AX in female patients was associated with a favor able prognosis (Fig. 7).
Figure 7.
The left panels shows the correlation between methylated sites and gene expression in White gastric cancer patients. The right panels represent the Kaplan-Meier OS of gene expression in White gastric cancer patients. (A) GYG2P1; (B) RPS4Y1; (C) TXLNG; and (D) EIF1AX. GYG2P1, glycogenin 2 pseudogene 1; RPS4Y1, ribosomal protein S4 Y-linked 1; TXLNG, taxilin-γ; EIF1AX, eukaryotic translation initiation factor 1A X linked.
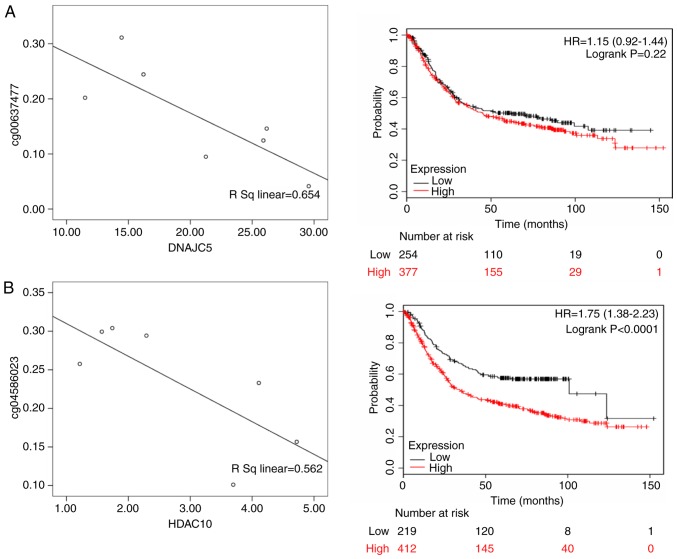
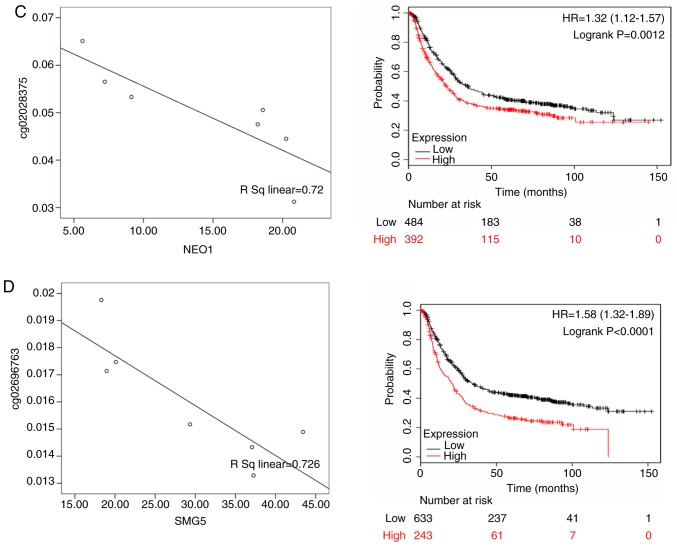
For Black patients, DnaJ heat shock protein family (Hsp40) member C5 (DNAJC5; rs=0.654, P<0.05), histone deacetylase 10 (HDAC10; rs=0.562, P<0.05), neogenin 1 (NEO1; rs= 0.720, P<0.05) and SMG5 nonsense mediated mRNA decay factor (SMG5; rs=0.726; P<0.05) were differentially expressed. According to the KMPlot database, 876 patients with gastric cancer had OS data, including those with differential expression of DNAJC5 (HR of death=1.15, 95% CI= 0.920-1.440, P= 0.220), HDAC10 (HR of death=1.750, 95% CI=1.380-2.230, P<0.001), NEO1 (HR of death=1.320, 95% CI=1.120-1.570, P=0.001) and SMG5 (HR of death=1.580, 95% CI=1.320-1.890, P<0.001). Among these genes, DNAJC5, HDAC10, NEO1 and SMG5 were highly expressed in male patients and were significantly associated with a poor prognosis (Fig. 8).
Figure 8.
The left panels shows the correlation between methylated sites and gene expression in Black gastric cancer patients. The right panels represent the Kaplan-Meier OS of gene expression in Black gastric cancer patients. (A) DNAJC5; (B) HDAC10; (C) NEO1; and (D) SMG5. DNAJC5, DnaJ heat shock protein family (Hsp40) member C5; HDAC10, histone deacetylase 10; NEO1, neogenin 1; SMG5, SMG5 nonsense mediated mRNA decay factor.
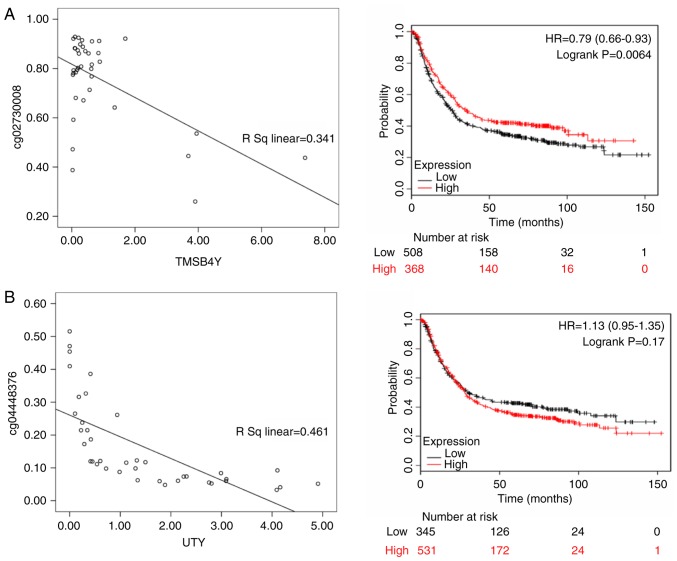
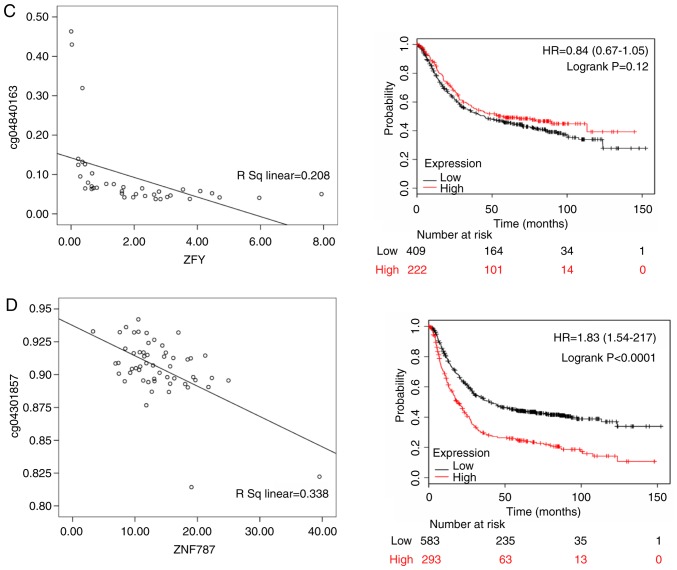
Thymosin β4 Y-linked (TMSB4Y; rs=0.341, P<0.05), ubiquitously transcribed tetratricopeptide repeat containing, Y-linked (UTY; rs= 0.461, P<0.05), zinc finger protein Y-linked (ZFY; rs=0.208, P<0.05) and zinc finger protein 787 (ZNF787; rs=0.338, P<0.05) were differentially expressed in Asian patients, and there were 876 gastric cancer patients with OS data from KMPlot database. ZFY (HR of death=0.840, 95% CI=0.670-1.050, P=0.12), TMSB4Y (HR of death=0.790, 95% CI=0.660-0.930, P=0.006), UTY (HR of death=1.130, 95% CI= 0.950-1.350, P= 0.170) and ZNF787 (HR of death=1.830, 95% CI=1.540-2.170, P<0.001) were differentially expressed. ZFY, TMSB4Y and UTY were highly expressed in male patients, and TMSB4Y was significantly associated with a better prognosis; ZNF787 was highly expressed in female patients and associated with a poor prognosis (Fig. 9).
Figure 9.
The left panels shows the correlation between methylated sites and gene expression in Asian gastric cancer patients. The right panels represent the Kaplan-Meier OS of gene expression in Asian gastric cancer patients. (A) TMSB4Y; (B) UTY; (C) ZFY; and (D) ZNF787. TMSB4Y, thymosin β4 Y-linked; UTY, ubiquitously transcribed tetratricopeptide repeat containing, Y-linked; ZFY, zinc finger protein Y-linked; ZNF787, zinc finger protein 787.
Discussion
The present study analyzed 93,278 gastric cancer patients in the SEER and Sun Yat-Sen University Center for Gastric Cancer Research databases, with 2,857 Asian, 13,223 Black and 77,198 White patients. In the Asian population, the male to female ratio was 1.96; that of the Black population was 1.53, and that of the White population was 1.81. This ratio is similar to that in a previous study, indicating that the incidence of gastric cancer in men is almost twice that in women (3). Based on the general tumor characteristics of gastric cancer patients of the different ethnic groups, it was found that for White patients, the tumor grade of male patients was worse than that of female patients; this difference was not significant for Black patients. Conversely, the tumor grade of Asian female patients was worse than that of Asian male patients. This may be an important factor leading to the effect of sex on the prognosis of gastric cancer. Despite the few studies on the tumor character istics of gastric cancer in different sexes, Aron et al (19) studied patients with renal cell carcinoma in the SEER database, and found that tumor size and tumor stage were lower among females compared to males, which is similar to the results for gastric cancer in the present study. Furthermore, there are few studies on the prognosis of gastric cancer in the different sexes. However, Kim et al (20) analyzed 4,722 patients undergoing radical gastrectomy, including 3,136 males and 1,586 females, and found that younger female patients had a higher incidence of signet ring cell carcinoma than did men, and that the prognosis of gastric cancer among women was worse than that among men. Li et al (21) also investigated 96,501 gastric cancer patients, including 61,639 males and 34,862 females, reporting that the former presented with larger tumors, higher stages and higher grade gastric cancer; in addition, OS and CSS were better in women, with a significant difference. These studies suggested that sex has a significant effect on gastric cancer.
To date, the impact of ethnicity on gastric cancer has been assessed in a considerable number of reports. Dong et al (3) conducted a retrospective cohort study of the comprehensive healthcare system in southern California between 2008 and 2014 by assessing the incidence of gastric cancer in different races, and found that race is indeed a risk factor. Non-Hispanic Black patients had an odds ratio (OR) of 1.500, White Hispanic patients had an OR of 1.400 and Asian patients had an OR of 1.500 (3). The above data showed that the incidence of gastric cancer in Asian populations is high. In fact, 75% of gastric cancer patients in the world are of Asian descent (5). In addition to these racial differences, the impact of sex on gastric cancer is also worthy of further study. Globally, the incidence of gastric cancer in women is almost half that in men. However, a study from South Korea found that although the incidence in women is lower than that in men, the prognosis of women is generally poor and the average age of onset is low (55.0 years for females and 57.9 years for males) (20). A study in the United States between 2010 and 2014 reported that the incidence of gastric cancer in male Asian patients was 14/100,000, and that in female patients was 8/100,000, with mortality rates of 7.1/100,000 and 4.3/100,000 for males and females, respectively. The incidence of gastric cancer in males of the African American population was 13.6/100,000 and that in females was 7.8/100,000, with mortality rates of 8.6/100,000 and 4.1/100,000 for males and females, respectively. Lastly, the incidence of gastric cancer in males of the White American population was 8.2/100,000 and that in females was 3.7/100,000; the mortality rate was 3.4/100,000 for males and 1.7/100,000 for females (22). In fact, the effects of sex on gastric cancer in patients of different ethnic backgrounds vary considerably. The present study showed that the prognosis of White female patients with gastric cancer in the United States was better than that of White male patients. By contrast, the difference between men and women may not be significant among Black gastric cancer patients, but for Chinese patients, the prognosis of females was worse than that of males. These conclusions are consistent with the conclusions of the previous studies mentioned above.
Different populations carry different gene mutations, and gene expression also varies, which may explain why different populations have differences in cancer risk. For example, Loh et al (23) studied polymorphisms in gastric cancer genes in different ethnic groups by evidence-based medicine. A total of 203 related studies were included, and 225 polymorphisms in 95 genes were evaluated; the results showed that race can have a marked impact on the risk of gastric cancer. The present study also evaluated differential gene expression between ethnicities and found significant differences among White, Black and Asian patients. Among them, the DEGs between White male and female patients were mainly enriched in RNA binding and transcription pathways. A total of four core genes were identified, GYG2P1, RPS4Y1, TXLNG and EIF1AX, according to the relationship between gene expression and DNA methylation (Table IV). GYG2P1 (24), RPS4Y1 (25), TXLNG (26) and EIF1AX (26) are mainly involved in tumor RNA binding and transcriptional path ways, as well as the survival prognosis of patients with different sexes of gastric cancer. In general, the impact of these core genes on tumors may be an important factor leading to poor prognosis in males. The DEGs between Black male and female patients were mainly enriched in cell structural changes, and DNAJC5, HDAC10, NEO1 and SMG5 were identified by the same means described above (Table IV). However, there is little research on the relationship between DNAJC5 and SMG5 in tumors, and tumor-related effects are not yet clear (27). Although studies have shown that high expression of HDAC10 can improve prognosis, high expression of this gene was reported to be associated with a poor prognosis (28). Increased expression of NEO1 increases the sensitivity of gastric cancer cells to carboplatin (29). Overall, these genes may play a role by altering the structural function of cells, though the influence of these genes on the survival prognosis of gastric cancer in different sexes remains unclear. For Asian patients, the DEGs between the sexes were mainly enriched in cell surface receptor-associated signal transduction and the G protein coupled receptor protein signaling, and four core genes were screened, TMSB4Y, UTY, ZFY and ZNF787, based on the relationship between gene expression and DNA methylation (Table IV). TMSB4Y is significantly related to the inhibition of tumor migration, suggesting that it may be a tumor suppressor gene (30). In this study, it was also found that elevated expression of TMSB4Y can improve the prognosis of patients. UTY and ZFY are mainly involved in apoptosis in stem cells in males (31), yet their impact on cancer is still unclear. ZNF can promote the migration and invasion of gastric cancer (32). The present study found that elevated expression of ZNF787 affects the prognosis of patients. The impact of these core genes on tumors may be an important factor leading to poor prognosis in females.
Table IV.
Known function/phenotype of Protein-protein interaction analysis of differential genes between of gender in different races of patients with gastric cancer.
| Author, year | Protein | Protein function | Expression | (Refs.) |
|---|---|---|---|---|
| Loh et al, 2009 | GYG2P1 | Glycogenin 2 pseudogene 1. The expression of pseudogenes is significantly related to the development and prognosis of tumors. | White male | (23) |
| Lou et al, 2019 | RPS4Y1 | RPS4Y1 is a full name of ribosomal protein S4 Y-linked 1, which is a member. of the ribosomal protein S4E family. The role of RPS4Y1 is similar to that of RPS4X; its high level of expression is associated with poor prognosis of intrahepatic cholangiocarcinoma and non mucinous carcinoma | White male | (24) |
| Jung et al, 2011 | TXLNG | Located in Xp22.2, the full name of taxilin-γ, which encodes a member of the paclitaxel family. The encoded protein binds to the c terminal coil region of the diamine family members 1a, 3a and 4a and may play a role in intracellular vesicle transport, which is mediated by lipopolysaccharide and may be involved in Cell cycle regulation. | White female | (25) |
| Yu et al, 2005 | EIF1AX | EIF1AX is located at Xp22.12 and is called the eukaryotic translation initiation. factor 1A X-linked. This gene encodes an important eukaryotic translation initiation factor. The expression of EIF1AX is an important factor in translation which affect tumor progression and prognosis | White female | (26) |
| Benitez et al, 2017 | DNAJC5 | Located at 20q13.33, the full name of DnaJ heat shock protein family (Hsp40) member C5, which is a member of the J protein family. The relationship between this gene and tumors, and the tumor-related effects are not yet clear. | Black male | (27) |
| Tao et al, 2017 | HDAC10 | Located at 22q13.33, the full name of histone deacetylase 10, which encodes a protein belonging to the family of histone deacetylases, members of which deacetylate lysine residues at the n terminal portion of the core histone. | Black male | (28) |
| Qu et al, 2018 | NEO1 | Located at 15q24.1, the full name of neogenin 1, which encodes a cell surface protein that is a member of the immunoglobulin superfamily. Increased expression of NEO1 increases the sensitivity of gastric cancer cells to carboplatin. | Black male | (29) |
| Benitez et al, 2017 | SMG5 | Located in 1q22, the full name is nonsense mediated mRNA decay factor, and SMG5 is involved in non-sensory-mediated mRNA decay. | Black male | (27) |
| Wong et al, 2015 | TMSB4Y | Located in Yq11.221, the full name of thymosin beta 4 Y-linked, which is located in a specific region of the Y chromosome and encodes an actin-separating protein. The expression of TMSB4Y is significantly related to the inhibition of tumor migration. | Asian male | (30) |
| Tricoli et al, 1993 | UTY | Located in Yq11.221, the full name of ubiquitously transcribed tetratricopeptide repeat containing, Y-linked, which encodes a protein containing a repeat of tetra quartin, thought to be involved in protein protein-interactions. | Asian male | (31) |
| Tricoli et al, 1993 | ZFY | Located in Yp11.2, the full name zinc finger protein Y-linked, which encodes a zinc-containing finger protein that acts as a transcription factor and is mainly involved in apoptosis of male stem cells. | Asian male | (31) |
| Li et al, 2014 | ZNF787 | Located at 19q13.43, the full name of zinc finger protein 787. Some abnormalities in ZNF protein can increase the expression of MMP 2, MMP-9 and ICAM-1, reduce the expression of timp-1, and promote the migration and invasion of gastric cancer. | Asian female | (32) |
GYG2P1, glycogenin 2 pseudogene 1; RPS4Y1, ribosomal protein S4 Y-linked 1; TXLNG, taxilin-γ; EIF1AX, eukaryotic translation initiation factor 1A X-linked; DNAJC5, DnaJ heat shock protein family (Hsp40) member C5; HDAC10, histone deacetylase 10; NEO1, neogenin 1; SMG5, SMG5 nonsense mediated mRNA decay factor; TMSB4Y, thymosin β4 Y-linked; UTY, ubiquitously transcribed tetratricopeptide repeat containing, Y-linked; ZFY, zinc finger protein Y-linked; ZNF787, zinc finger protein 787; MMP, matrix metal loproteinase; ICAM-1, intercellular adhesion molecule 1; timp-1, TIMP metallopeptidase inhibitor 1.
Although the present study provided a comprehensive analysis of the prognosis of patients with different ethnic back grounds based on the world's largest database and data from China's largest center for gastric cancer research, there are several limitations. First, because the staging standards were different at different periods, SEER data were divided into 1973-2003 and 2004-2013 periods to include more patients and bolster the results. Due to the limitations of follow up in the databases, some data were not available, such as the situation of patients with respect to chemotherapy, which may have an impact on the conclusion. Second, as the classification of patients in the SEER database does not distinguish between Asians and Indians and South Americans, analysis of data for the Asian population from the SEER database is not possible. Third, this study was a retrospective study, and thus there may be some bias in the statistical results. Fourth, a period of 40 years was examined because of differences in treatment and diet, and potentially variable environmental factors. These factors could affect the accuracy of the results. Given that the present study was retrospective, the data collection was not strictly randomized; therefore, the presence of confounding factors is unavoidable. Furthermore, whether sex differences are the cause of differences in the prognosis of patients with gastric cancer cannot be directly determined.
In the present study, it was found that the effects of sex on the prognosis of gastric cancer patients of different ethnicities differed. It was concluded that differences in molecular mechanisms between the two sexes among gastric cancer patients of different ethnic groups are the main reasons for the observed variability.
Supplementary Data
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CZ and HL conceived and designed the study. HL, CW, ZW, ZG, WC and YH performed the data analysis. HL, CW and ZW wrote the paper. All authors read and approved the manuscript.
Ethics approval and consent to participate
The information of for White and Black patients with gastric cancer obtained from the SEER database does not contain any identifiers and is public. Due to the retrospective nature of the study, informed consent was not required. All SEER database information was deidentified by the United States National Cancer Institute review board approval. This analysis did not involve interaction with human subjects or the use of person ally identifiable information. The research on the Asian cohort was approved by the ethics committee of the First Affiliated Hospital of Sun Yat-Sen University, and written informed consent was obtained from all of these patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD -3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Dong E, Duan L, Wu BU. Racial and Ethnic minorities at increased risk for gastric cancer in a regional US population study. Clin Gastroenterol Hepatol. 2017;15:511–517. doi: 10.1016/j.cgh.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang KT, Kim J, Jung J, Chang JH, Chai YJ, Oh SW, Oh S, Kim YA, Park SB, Hwang KR. Impact of breast cancer subtypes on prognosis of women with operable invasive breast cancer: A population-based study using SEER database. Clin Cancer Res. 2019;25:1970–1979. doi: 10.1158/1078-0432.CCR-18-2782. [DOI] [PubMed] [Google Scholar]
- 7.Greene FL, Compton CC, Fritz AG, Shah JP, Winchester DP. AJCC Cancer Staging Atlas. Springer; New York, NY: 2006. [DOI] [Google Scholar]
- 8.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: A flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona FJ, Sayols S, Moran S, Heyn H, Vizoso M, Gomez A, et al. A prog nostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013;31:4140–4147. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 11.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langevin SM, Butler RA, Eliot M, Pawlita M, Maccani JZ, McClean MD, Kelsey KT. Novel DNA methylation targets in oral rinse samples predict survival of patients with oral squamous cell carcinoma. Oral Oncol. 2014;50:1072–1080. doi: 10.1016/j.oraloncology.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böhnke JR. Explanation in causal inference: Methods for mediation and interaction. Q J Exp Psychol (Hove) 2016;69:1243–1244. doi: 10.1080/17470218.2015.1115884. [DOI] [PubMed] [Google Scholar]
- 14.Gene Ontology Consortium The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 17.Austin PC. Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies. Pharm Stat. 2011;10:150–161. doi: 10.1002/pst.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 19.Aron M, Nguyen MM, Stein RJ, Gill IS. Impact of gender in renal cell carcinoma: An analysis of the SEER database. Eur Urol. 2008;54:133–140. doi: 10.1016/j.eururo.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Kim HW, Kim JH, Lim BJ, Kim H, Kim H, Park JJ, Youn YH, Park H, Noh SH, Kim JW, Choi SH. Sex disparity in gastric cancer: Female sex is a poor prognostic factor for advanced gastric cancer. Ann Surg Oncol. 2016;23:4344–4351. doi: 10.1245/s10434-016-5448-0. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Wei Z, Wang C, Chen W, He Y, Zhang C. Gender differences in gastric cancer survival: 99,922 cases based on the SEER database. J Gastrointest Surg. 2019 Jul 25; doi: 10.1007/s11605-019-04304-y. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 22.Ashktorab H, Kupfer SS, Brim H, Carethers JM. Racial disparity in gastrointestinal cancer risk. Gastroenterology. 2017;153:910–923. doi: 10.1053/j.gastro.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh M, Koh KX, Yeo BH, Song CM, Chia KS, Zhu F, Yeoh KG, Hill J, Iacopetta B, Soong R. Meta-analysis of genetic polymorphisms and gastric cancer risk: Variability in associations according to race. Eur J Cancer. 2009;45:2562–2568. doi: 10.1016/j.ejca.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Lou W, Ding B, Fan W. High expression of pseudogene PTTG3P indicates a poor prognosis in human breast cancer. Mol Ther Oncolytics. 2019;14:15–26. doi: 10.1016/j.omto.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, Seo J, Kim B, Jung Y, Kim WK, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 26.Yu VW, Ambartsoumian G, Verlinden L, Moir JM, Prud'homme J, Gauthier C, Roughley PJ, St-Arnaud R. FIAT represses ATF4-mediated transcription to regulate bone mass in transgenic mice. J Cell Biol. 2005;169:591–601. doi: 10.1083/jcb.200412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benitez BA, Sands MS. Primary fibroblasts from CSPα mutation carriers recapitulate hallmarks of the adult onset neuronal ceroid lipofuscinosis. Sci Rep. 2017;7:6332. doi: 10.1038/s41598-017-06710-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao X, Yan Y, Lu L, Chen B. HDAC10 expression is associated with DNA mismatch repair gene and is a predictor of good prognosis in colon carcinoma. Oncol Lett. 2017;14:4923–4929. doi: 10.3892/ol.2017.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu H, Sun H, Wang X. Neogenin-1 promotes cell proliferation, motility, and adhesion by up-regulation of zinc finger E-box binding homeobox 1 via activating the Rac1/PI3K/AKT pathway in gastric cancer cells. Cell Physiol Biochem. 2018;48:1457–1467. doi: 10.1159/000492255. [DOI] [PubMed] [Google Scholar]
- 30.Wong HY, Wang GM, Croessmann S, Zabransky DJ, Chu D, Garay JP, Cidado J, Cochran RL, Beaver JA, Aggarwal A, et al. TMSB4Y is a candidate tumor suppressor on the Y chromosome and is deleted in male breast cancer. Oncotarget. 2015;6:44927–44940. doi: 10.18632/oncotarget.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tricoli JV, Bracken RB. ZFY gene expression and retention in human prostate adenocarcinoma. Genes Chromosomes Cancer. 1993;6:65–72. doi: 10.1002/gcc.2870060202. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Tan BB, Zhao Q, Fan LQ, Wang D, Liu Y. ZNF139 promotes tumor metastasis by increasing migration and invasion in human gastric cancer cells. Neoplasma. 2014;61:291–298. doi: 10.4149/neo_2014_037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.