Abstract
Liquid biopsy of plasma is a simple and non-invasive technology that holds great promise in biomedical research. It is based on the analysis of nucleic acid-based biomarkers with predictive potential. In the present work, we have combined this concept with the FTA technology for sentinel mussels. We found that hemocytes collected from liquid biopsies can be readily fixed on FTA cards and used for long-term transcriptome analysis. We also showed that liquid biopsy is easily adaptable for metagenomic analysis of bacterial profiles of mussels. We finally provide evidence that liquid biopsies contained circulating cell-free DNA (ccfDNA) which can be used as an easily accessible genomic reservoir. Sampling of FTA-fixed circulating nucleic acids is stable at room temperature and does not necessitate a cold-chain protection. It showed comparable performance to frozen samples and is ideally adapted for sampling in remote areas, most notably in polar regions threatened by anthropogenic activities. From an ethical point of view, this minimally-invasive and non-lethal approach further reduces incidental mortality associated with conventional tissue sampling. This liquid biopsy-based approach should thus facilitate biobanking activities and development of omics-based biomarkers in mussels to assess the quality of aquatic ecosystems.
Introduction
Because of their ability to accumulate xenobiotics in their tissues, their wide distribution, and their ecological and economical importance, blue mussels (Mytilus spp.) have long been recognized as good biological indicators for monitoring the effects of pollution and climate change in marine habitats [1]. Not surprisingly, a relatively large number of biomarkers have been developed, often using a multi-tier approach. They include functional biomarkers based on enzymatic activities or cellular functions, or on measuring concentrations of specific stress indicators [2–4]. In most cases, measure of these biomarkers requires on-site analysis or tissue biopsy performed by trained personnel [5, 6]. It also often requires an uninterrupted cold chain to maintain sample quality. For sampling in remote areas, such as polar regions, this implies complex logistical challenges and considerable risk and cost associated with transportation and storage [7]. The use of formalin-fixed tissue biopsy is an alternative, although formalin is known to affect DNA quality. There are, however, novel methods for collection, storage and shipping of biological samples that offers some solutions to these problems. The Flinders Technology Associates (FTA) cards, for example, can be used to fix and preserve nucleic acids for long-term periods at ambient temperature with high stability [8,9]. This approach has been particularly useful for identification/genotyping and long-term surveillance of infectious pathogens in remote areas at reduced shipping costs [10–11]. The use of FTA cards for monitoring marine habitat with mussels as sentinel species, however, will require concomitant development of nucleic acid-based biomarkers. Fortunately, immense progress in the development has been achieved thanks to new methods in gene sequencing and the need for accurate predictive biomarkers in medicine. Nucleic acid-based biomarkers are particularly well-adapted for liquid biopsies. Liquid biopsy is a concept-based approach that is gaining significant attention in the biomedical area. The concept is that a clinician is able to use a small sample of blood collected from a patient to obtain vital genetic information that can be used: 1) to predict the onset of a disease, 2) to assess the severity of a disease (prognostic), 3) to evaluate and measure with high precision the efficacy of a given treatment, and 4) to determine long-term survival and relapse. It is designed to replace the use of logistically complicated, high cost invasive tissue biopsies to assess ongoing tissue damage. It is largely based on the analysis of circulating cell-free DNA (ccfDNA), which is considered as a genomic reservoir that is easily accessible for studying genetic traits, including genomic alterations [12]. Recent studies in humans have further shown that ccfDNA can be used to identify damages in specific tissues [13]. Liquid biopsies are also compatible for metagenomic data analysis. Microbiome information is now considered an important component of the biomarker schematic [14, 15]. With the help of recent advances in next generation sequencing, microbiome-associated biomarkers, just like ccfDNA, can be used in clinical practice for risk prediction, diagnosis and progression of a disease, or to predict and modulate response to treatment [16]. In marine ecology, microbial biomarkers are also being used for measuring the impact of anthropogenic activities on marine ecosystems [17, 18]. In the present work, we have combined the concept of liquid biopsy and the use of FTA cards to develop a liquid biopsy-based sampling method that involves fixation of nucleic acids on FTA cards. This method is simple, does not require maintaining a cold chain and is compatible with transcriptome analyses. We have also identified, for the first time, circulating cell-free ccfDNA in hemolymph of mussels and performed 16SrDNA gene-based bacterial microbiota profiles of mussels using FTA-fixed bacterial DNA collected from liquid biopsies.
Material and methods
Liquid biopsy
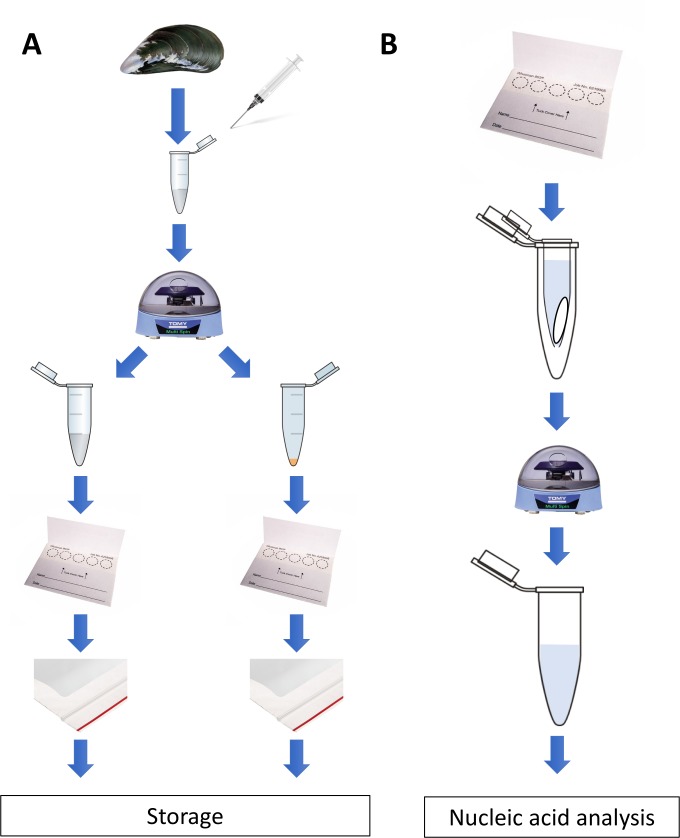
Adult specimens (55–70 mm length) of blue mussels Mytilus spp. were obtained from a commercial supplier (PEI Mussel King Inc., Prince Edward Islands, CANADA) and placed in a temperature-controlled (4°C) aerated aquarium containing 10 L of 32‰ artificial saline water (Reef Crystal artificial marine salt, Instant Ocean, VA, USA). For M. desolationis and Aulacomya ater (A. ater) specimens, they were collected on the intertidal rocky shore of Port-aux-Français (Kerguelen Islands, France) as previously described [19]. In all cases, hemolymph was collected from single individuals, without pooling and centrifugation. For routine preparation of liquid biopsies on FTA cards, the intervalvar liquid was removed with the tip of a knife and hemolymph was withdrawn from the adductor muscle using a syringe fitted with a 25 gauge needle. This method has been shown to be minimally invasive and non-lethal for freshwater bivalvles [20]. Samples (<1ml of hemolymph) were immediately transferred into sterile 1.5 ml Eppendorf tube and centrifuged for 3 minutes at maximum speed (approx. 3000 x g) at room temperature using a battery-powered mini-centrifuge (TOMY, Japan) (Fig 1). After centrifugation, aliquots (50 μl) of supernatants were spotted on individual discs (10 mm diameter) on Whatman 903™ FTA cards (Whatman, Z694827, Sigma-Aldrich, Oakville, ON, Canada). The remaining supernatant was discarded and hemocyte pellets gently resuspended and spotted on FTA cards. In some experiments, Whatman FTA Gene Cards (GE Healthcare, Marlborough, MA) were used. The cards were dried for 15 min at room temperature and stored individually in small (3” x 4”) clear 2 mil plastic ziplock baggies containing one silica gel desiccant (1gr/bag) moisture absorber. When indicated, hemolymph were pooled, transferred into a sterile 50 mL and centrifuged for 15 min at 3000 x g on a table low speed centrifuge (Beckman Coulter Allegra 6R). Unless otherwise indicated, all FTA cards were kept at room temperature in the dark.
Fig 1. Diagrammatic summary of the sampling protocol.
Left, for routine preparation of liquid biopsies on FTA cards, hemolymph was withdrawn from the adductor muscle. Samples (approx. 1ml of hemolymph) were immediately transferred into sterile 1.5 ml and centrifuged at room temperature using a battery-powered mini-centrifuge (TOMY, Japan). After centrifugation, aliquots (50 μl) of supernatants were spotted on individual discs (10 mm diameter) on FTA® cards. The remaining supernatant was discarded and hemocyte pellets gently resuspended and spotted on FTA cards. The cards were dried for 15 min at room temperature and stored individually in small plastic Ziploc baggies containing a silica gel desiccant moisture absorber. Right, to recover nucleic acids, discs were carefully placed at the bottom of a sterile microcentrifuge tube with a hole punctured in its bottom. The tube was then transferred into a larger sterile microcentrifuge tube before isolation of nucleic acids as described in details in the material and methods section.
Isolation of ccfDNA from hemolymph
ccfDNA from hemolymph (5 ml) was carried out using the NucleoSpin Plasma XS kit (Macherey Nagel) with optimized manufacturer’s protocols. Purified DNA was quantified by the PicoGreen assay according to the manufacturer’s recommendations [21].
Isolation of nucleic acid
For RNA isolation and purification, individual 12 mm2 disks from FTA cards were carefully placed at the bottom of a sterile 0.6 ml microcentrifuge tube with a hole punctured in its bottom by a 18G needle. The tube was then transferred into a sterile 1.5 ml microcentrifuge tube before addition of TRIzol (800 μl) to the upper tube. The cards were soaked with TRIzol for 10 min at room temperature and centrifuged for 3 min at high speed (8000 rpm) in a table centrifuge (BioFuge Fresco, Heraeus Instrument, Grimsby, ON, Canada). The TRIzol solution was transferred and topped with 0.2 ml of chloroform. Tubes were incubated 3 min at room temperature and centrifuged at 12000 x g for 15 min at 4°C. The aqueous phase was used to perform RNA extraction following a standard protocol. The RNA was dissolved in RNase/DNase free water and quantified using the Thermo Scientific™ NanoDrop™ spectrophotometer. DNA was isolated using an adapted the prepGEM™ Universal kit for Blood storage cards (Zygem, Charlottesville, VA). Briefly, 3 mm disc were placed on an eppendorf tube in washed with 100 μl of DNA-free water and DNA recovered with the prepGEM solution following 5 min incubations at 75°C and 95°C. Nucleic acid samples were stored at -20°C until further analysis. Purified DNA was quantified by the PicoGreen assay (Lumiprobe, Hunt Valley, MD) according to the manufacturer’s recommendations [21].
Thermal stress
Following acclimatization, groups of ten mussels of similar length were transferred into tank containing oxygenated seawater at 30°C or at 4°C (control). After 30 min, liquid biopsies were collected. Hemocytes pellets and hemolymph were either fixed on FTA cards or kept frozen at -20°C.
RT-PCR analysis
Total cellular RNA was isolated from cell pellets fixed on FTA cards using the TRIzol reagent (Life Technologies) as described above. First-strand cDNA was prepared from 2 μg of cellular RNA in a total reaction volume of 20 μl using the High Capacity cDNA Reverse Transcription Kit from Thermo-Fisher. qPCR specificity was carried out with a Rotor-Gene® 3000 (Corbett Research) with primers listed in Table 1. All qPCR reactions were carried out in 20 μl with the QuantiFast SYBR Green PCR Kit (Qiagen). qPCRs were amplified using the following conditions: 95°C for 5 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s with a single acquisition. A final melting curve analysis was performed using at 95°C for 5 s, 65°C for 60 s and 97°C with a continuous acquisition. To ensure primer specificity, melting temperature curves were determined for each amplicon. PCR products were run on a 1.5% agarose gel to visualize the bands and ensure a unique band is amplified. The expression level (log2 of the fold change) was expressed 1) calculating the delta CT (ΔCT) by subtracting the averaged threshold cycle (CT) value of the three housekeeping genes (EF1γ, EF1α and SNX14) from the CT value of the target gene and 2) comparing the fold change of non-exposed mussels versus thermal-exposed mussels [22].
Table 1. List of primers used in this study.
| Gene | Sense primers | Antisense primers | Product size |
|---|---|---|---|
| EF1γ | 5’-AAA GCT CAA ATT GCA GCG GA-3’ | 5’-ATG cAA TCT cCT CCA GCT GT-3’ | 158 bp |
| SXN14 | 5’-AGA GGC GAG GAT CAG ATG TG-3’ | 5’-ATC TTC ACC ACC ACC AGA GG-3’ | 158 bp |
| HSP70 | 5’-ACA GAG AGA CTT GTT GGC GA-3’ | 5’-TTC CGA TCA GTC TCT TGG CA-3’ | 88 bp |
| SOD | 5’-ACC ATG TGC GTC TAC AGG AT-3’ | 5’-TTG ATC AGG TTT CCG ATG GC-3’ | 95 bp |
Taxonomic analysis of hemolymph microbiota
Following fixation on FTA cards, DNA was extracted from hemocyte pellets, as described above. The whole 16S bacterial DNA V2-V3 region was targeted by the primers 28F‐519R primers and pyrosequenced by the Illumina Mi-Seq Next Generation Sequencer at Research & Testing Laboratory (http://www.researchandtesting.com/, Texas, USA). A detailed description of bioinformatic filters that were used for analysis can be found at http://www.rtlgenomics.com/docs/Data_Analysis_Methodology.pdf). Briefly, post-sequencing processing of the raw data for both types of samples (frozen and FTA-fixed cell pellets) was carried out by standard methods to eliminate low quality readings, contaminants and chimeras. The remaining filtered sequences were then grouped into Operational Taxonomy Units (OTUs) at a similarity level ≥ 95%. For each sample, we have obtained approximately 10 000–20 000 OTUs. For taxonomic classification, we used the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/). The metagenomic data from this publication have been deposited to the NCBI Sequence Read Archives with the assigned the identifier PRJNA564645. To take into account the problem of contamination, we used, as controls, template-free FTA cards that were processed with the same DNA extraction and PCR amplification kits as the real samples, as previously recommended [23].
Statistics
All datasets were validated for normality (with Shapiro-Wilk test) and homoscedasticity (with Bartlett and F of Fisher test). Student's t-test was used as parametric test to compare data set with two groups and a two-way analysis of variance (ANOVA) was used to compare datasets with more than two groups. For non-parametric data, Kruskall Wallis test was used to compare datasets. All data are expressed as the mean ± S.D. Results were considered statistically significant at P≤0.05.
Results
Isolation and analysis of RNA from hemocyte pellets fixed on FTA cards
Because hemocytes are implicated in several physiological functions, such as phagocytosis of pathogens, homeostasis, and reproduction, there is increasing interest in studying how their transcriptome is modulated by environmental stress factors [24–28]. We thus developed a general framewok procedure for collecting hemolymph samples using FTA cards without the need of a cold chain is shown and detailed in Fig 1A. Other than a battery-operated centrifuge, it does not require specialized equipment and can be easily incorporated into a small, light-weight portable unit (S1 Fig).
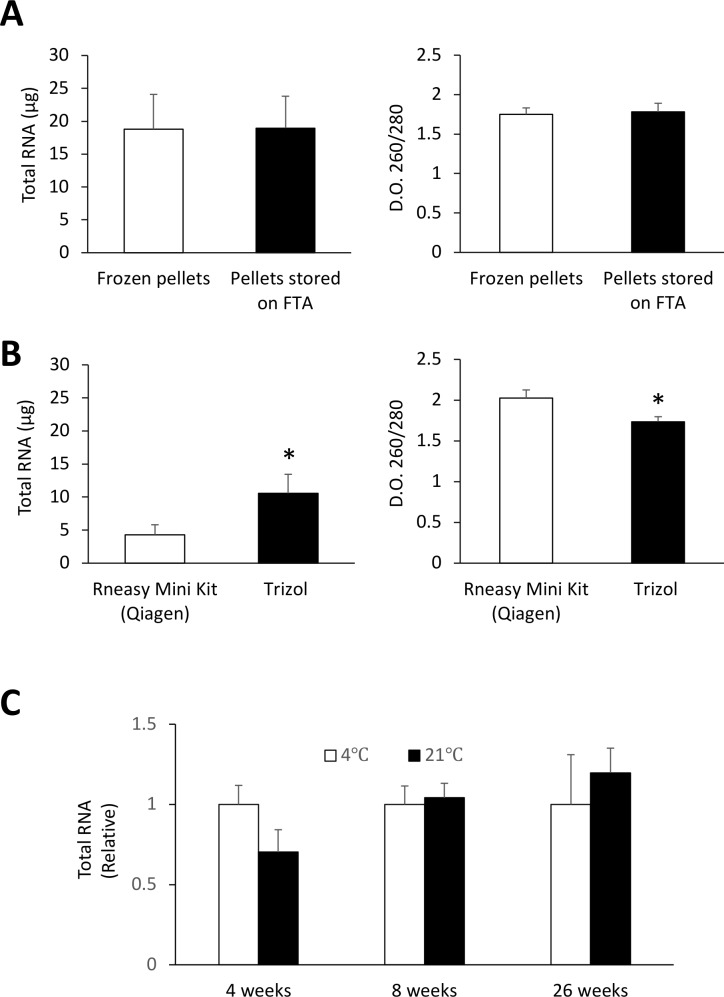
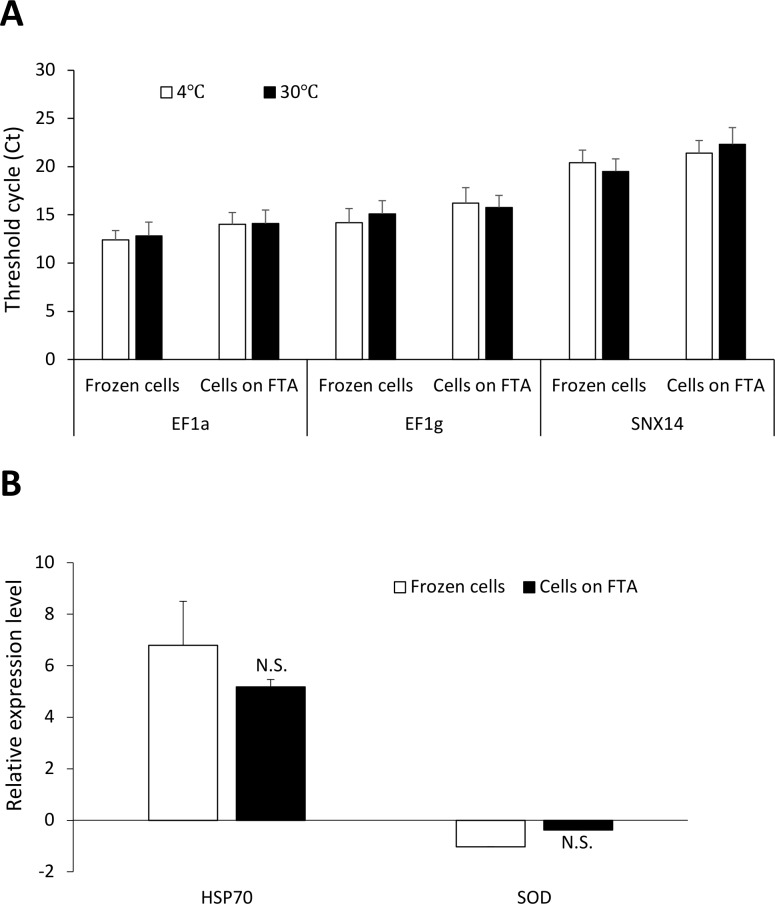
To test whether such sampling framework is compatible for multi-omics analysis of hemolymph samples, we first compared RNA yields obtained from frozen hemocytes and hemocytes that were fixed on FTA cards. Isolation of RNA from FTA discs was carried out using a simple TRIzol-based protocol which consisted of soaking FTA discs in TRIzol for 10 min at room temperature (Fig 1B). Nucleic acids were then eluted from the discs by brief centrifugation and RNA purified using standard phase separation with chloroform and subsequent precipitation with isopropyl alcohol [29]. Our results showed that similar amount of RNA was recovered from pellets fixed on FTA discs as compared to pellets frozen at -20°C (Fig 2A). Both methods yielded similar ratio of the absorbance at 260 and 280 nm. RNA recovery from FTA discs could also be achieved using a commonly-used silica-based RNA extraction kit, which resulted in lower yield compared to the TRIzol-based method but provided purer RNA (Fig 2B). We also found that the RNA yield from hemocyte pellets fixed on FTA was not significantly affected by long term storage (up to 6 months) at 4°C or ambient temperatures (Fig 2C). We next compared the performance RT-qPCR carried out using frozen RNA and RNA fixed on FTA cards and found that both conservation methods performed equally when measuring expression of three commonly used housekeeping genes (EF1α, EF1γ, and SNX14) (Fig 3A). Both methods were also equally efficient measuring the de novo expression of stress-related genes (Hsp70 and SOD) in hemocytes collected from mussels subjected to an acute thermal stress (Fig 3B).
Fig 2. RNA extraction from FTA cards.
(A) Recovery of total RNA from hemocytes stored frozen at -20°C or on FTA cards using a TRIzol-based protocol. (B) Comparative RNA recovery from FTA cards using TRIzol or the RNeasy kit. (C) Stability of RNA fixed on FTA cards and kept at different temperatures. * p < 0.05. Values are expressed as the total amount of RNA recovered from a 10 mm diameter sample disc FTA cards.
Fig 3. RT-qPCR analysis using frozen and FTA-fixed RNA.
(A) Cycle tresholds (Ct) values obtained for expression analysis of three housekeeping genes in pooled hemocytes pellets that were either kept frozen or fixed on FTA cards. (B) Relative gene expression of Hsp70 and superoxide dismutase (SOD) in hemocytes following an acute thermal stress at 30°C for 30 min. Expression levels are relative to controls (4°C). No significant (N.S.) differences were found using both methods.
Bacterial microbiome analysis
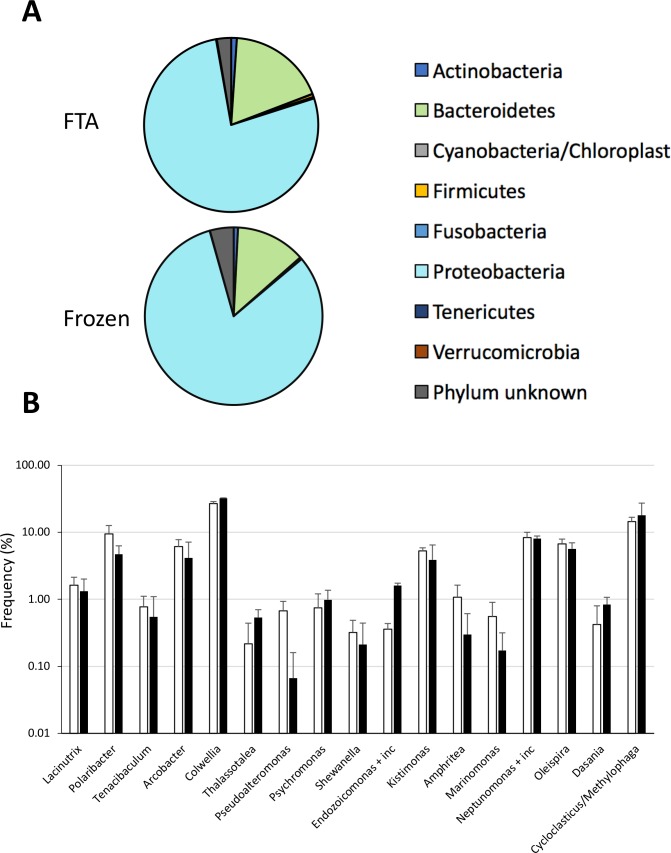
Bacteria microbiome-derived profiles are emerging as potent biomarkers not only in the biomedical field, but also in monitoring environmental stress in a vast number of species, including marine organisms [30, 31]. For these experiments, we compared the 16SrDNA gene-based bacterial profiles from frozen and FTA-conserved DNA from cellular pellets. Our results showed that both methods generated similar bacterial profiles with regards to phylum (Fig 4A). We found no significant differences in genus-level bacterial groups that were most frequently found in hemolymph (Fig 4B). The most common bacteria found in the hemolymph was from the genus Colwellia. This genius is commonly found in marine habitats, including the hemolymph of blue mussels [18]. Taken together, these findings show that FTA-fixed DNA is perfectly compatible with metagenomic analysis of circulating microbiota.
Fig 4. Bacterial profiles of hemolymph.
Upper histogram, pie chart representation of the relative abundance (in percentage) of 16SrRNA of the most common phyla found in liquid biopsies of M. edulis. Lower histogram, bacterial species differentially abundant in frozen and FTA-fixed liquid biopsies.
Circulating cell-free DNA
Analysis of ccfDNA from liquid biopsies is another promising avenue for the development of predictive biomarkers. In humans, it serves as a genomic reservoir that is easily accessible with a minimally-invasive procedure. To our knowledge, however, the existence of ccfDNA in hemolymph of invertebrates, and in mussels in particular, has not been reported. Using the same method that is commonly used for isolating ccfDNA (see material and methods section for details) from human plasma, we have thus investigated the presence of ccfDNA in mussels. Our results revealed that we indeed found ccfDNA in hemolymph of Mytilus edulis (Table 2) at in a range of concentrations (nanograms/ml) that were similar to that found in humans [32]. Similar results were obtained for M. desolationis and Aulacomya ater (A. ater). To validate the integrity of the ccfDNA fragments in liquid biopsies (cell-free hemolymph) fixed on FTA cards, we carried out real-time qPCR analysis using primers that generated small amplicons and that were specific for EF1γ, HSP70 and SNX14 genes [33] (Table 3). Our results showed that we could indeed find evidence of ccfDNA fragments encoding SNX14 and HSP70 genes but not EF1γ, suggesting that ccfDNA fragments are generated in a gene-specific manner, as observed in humans (Table 3). Indeed, ccfDNA fragments have been shown to derive from cell-specific nucleosome footprinting from different cell types [13]. These results reveal for the first time the presence of ccfDNA in mussels and open the door for its use as a biomarker in liquid biopsies collected from mussels.
Table 2. Concentrations of ccfDNA.
| Samples | ccfDNA (ng/mL) |
|---|---|
| Human plasma | 8,72 ± 1,43 |
| M. edulis | 4,23 ± 0,18 |
Table 3. qPCR analysis of ccfDNA fixed on FTA cards.
| Gene | Template | CT | MT | % of detection |
|---|---|---|---|---|
| EF1γ | ccfDNA | 25,31 ± 0,85 | 78,50 ± 1,71 | 0% |
| NTC | 25,79 ± 0,67 | 79,65 ± 0,21 | ||
| SNX14 | ccfDNA | 24,64 ± 2,44* | 83,84 ± 0,47 | 56% |
| NTC | 31,79 ± 0.01 | N.D. | ||
| HSP70 | ccfDNA | 21,25 ± 1,55 * | 81,51 ± 0,14 * | 89% |
| NTC | 32,16 ± 0 | 80 ± 0 |
NTC, No template control. CT, threshold cycle, MT, melting temperature, and % of amplicon detection obtained for EF1γ, SNX14 and HSP70 (n = 9 mussels, with 3 replicates for each sample. ND = no detected.
* p < 0.05).
Discussion
Given the growing concerns over the anthropogenic impact on marine ecosystems, it is increasingly critical to develop biomarkers that can be used as diagnostic and predictive tools, as well as for monitoring the success of the remediation efforts. The use of multi-omics biomarkers is a step in that direction. It obliges us, however, to go back to the drawing board to re-examine our sampling strategy not only for logistical reasons, but for economical reasons as well. This is particularly true for polar regions where the logistic complexity of tissue sampling is a major obstacle. In the present work, we have combined the use of FTA cards and the concept of liquid biopsies for the development of novel omics-based biomarkers in mussels, a widely used sentinel species. More specifically, we have shown that: 1) hemocytes pellets can be readily fixed on FTA cards and used for gene expression analysis; 2) FTA-fixed cell pellets are stable for long term periods when stored at room temperature, facilitating the biobanking activities and shipment to third parties at lower cost when compared to frozen samples; 3) our sampling strategy is easily adaptable for microbiome analysis of bacterial profiles, and 4) ccfDNA is present in mussel hemolymph and can be used as a genomic reservoir for the development of novel biomarkers. Taken together, this study lays the foundation for a logistically-friendly, omics-based multi-biomarker approach using mussels to assess the quality of marine ecosystems.
Until now, the concept of liquid biopsies has been almost exclusively used for guiding clinical practice, especially in oncology, prenatal screening, transplantation and presence of non-human ccfDNA (virus-derived sequences, for example) [12, 34, 35]. One of the main benefits of non-invasive liquid biopsies is that it overcomes many of the drawbacks associated with tissue biopsies. It also provides an accurate snapshot of the genomic landscape, bypassing many problems associated with tissue heterogeneity. In fact, to our knowledge, this is the first report of the presence of ccfDNA in invertebrates. In humans, the normal plasma concentration of ccfDNA is generally below 10–20 ng/ml, which is relatively similar to what we found in hemolymph of mussels. while it can increase by 5–10 times in patients with malignant disease or stress conditions [36]. Future studies will be needed to determine whether measuring ccfDNA levels in mussels can be used as a rapid stress biomarker as it does in humans [32, 36, 37]. Analysis of ccfDNA is also perfectly adaptable for serial samplings of DNA abnormalities, gene expression analysis, point mutations, chromosome translocations, miRegulome, epigenetic changes, etc. For example, analysis of ccfDNA can theoretically be used for measuring mutations in genes associated with hematopoietic neoplasia (such as p53), for rapid identification of mussel species, or for any genomic traits. It could also be used to detect the presence of parasitic and viral DNA to reduce the risk for public human health. From an ethical point of view, it reduces incidental mortality associated with conventional tissue biopsies. To develop the full potential of this approach, we are currently using next-generation sequencing to perform a genomic analysis of the entire ccfDNA found in different mussel populations. The results of these analyses will be reported in a separate manuscript.
Another benefit of combining both FTA-based technology and liquid biopsy is its compatibility with microbiome analysis. In humans, although blood is normally considered a sterile environment, there is increasing studies showing the circulating cells harbor a rich bacterial microbiome that can be used as disease biomarkers [38, 39]. This is also true for bivalves. The microbiome of hemolymph samples collected from the adductor muscle from Pacific oysters, for example, has been shown to undergo significant changes following abiotic and biotic stress factors [40]. A recent study showing that bacterial diversity and richness in mussels were higher in the hemolymph compared to other tissues support the idea that liquid biopsies is a valid approach for microbiome analysis in mussels [18]. Indeed, Vezzulli and colleagues have shown that the alpha diversity of the micriobiome of blue mussels was higher in the hemolymph compared to the digestive gland, in both [18]. Our results further showed that the bacterial profiles obtained from DNA of cells fixed on FTA was not significantly different from that obtained from frozen cells (S2 Fig). We found no significant differences in those bacteria that are commonly found in marine ecosystems, such as Alteromonadales or Oceanospirillales [41, 42]. In fact, our results showed that DNA from cells fixed on FTA cards was more prone to detect those rare bacterial DNA, consistent with the ability of the chemical matrix of FTA cards to stabilize and protect nucleic acids from environmental degradation. Not surprisingly, one of the most abundant bacterial DNA that we found in both FTA cards and frozen samples derived from the Polaribacter genus, which species are almost always found in polar or marine environments [43]. The other most frequently found genus were Colwellia, Oleispira, Cycloclastricus and Neptunomonas. These bacteria are well known to degrade polycyclic aromatic hydrocarbons (PAH) or short-chain alkans. Cycloclasticus bacterial DNA has been recently found in Bathymodiolus heckerae mussels where they have established a symbiosis at a 3000m depth in oil-rich habitats near asphalt volcanoes in the Gulf of Mexico [44]. They have also been shown to bloom in marine habitats following the Deep Horizon oil spill [44]. Interestingly, the Oleispira genus, another bacterium that has bloomed following the Deep Horizon oil spill, was also among the most frequently found bacterium in M. edulis. The third most frequent bacteria found was from the genus Neptunomonas, another bacterium implicated in degradation of PAHs and normally found in contaminated sediments [45]. Whether the presence of this microbiota reflects a symbiotic relationship with M. edulis is unclear at the present time. Overall, our findings indicate that microbiome analysis in liquid biopsies collected from cellular pellets of mussels is an interesting avenue to study dysbiosis in mussels following exposure to environmental stress.
Our study showed that combining liquid biopsies and the FTA technology could be useful for long-term biobanking and retesting of omics biomarkers collected from sentinel mussels, particularly in remote regions. This approach is also economically viable, especially when using the Whatman 903™ FTA cards. Although we have used for this study 10 mm diameter circular FTA discs, we obtained similar RNA yield using smaller discs (S3 Fig). If necessary, one could use the the Whatman FTA Gene Cards, which have been adapted for cature on nucleic acids. Although more expensive than regular Whatman 903™ cards, we found that they are more performant for DNA recovery (S4 Fig). It is important to note, however, that there are multiple options for isolating nucleic acid from FTA cards. In our hands, we have found that TRIzol was an adequate, low cost, and versatile method for routine RT-qPCR analysis. It can also be combined with commercial kits to obtain purer RNA preparations (S1 Table). TRIzol is also suitable for isolation of small RNAs (<200 nucleotides) as compared to the silica-based methods which do not retain small RNAs [46]. This has to be taken into consideration for future studies aimed at testing the possibility of using non-coding RNAs as putative biomarkers in hemolymph of mussels. In humans, non-coding RNAs have shown great promise as potential cancer biomarkers for liquid biopsies.
Another clear benefit of combining liquid biopsy and the FTA technology is from a logistically-point of view. We can now collect samples at remote site with a minimal amount of equipment (S1 Fig). Moreover, this approach addresses increasing concerns in field work ethics [47]. Liquid biopsies require sampling of a very small volume of hemolymph and does not lead to animal death. We believe that such approach could also be extended to other marine and freshwater organisms and offers many advantages for the development of long-term ecological observatories in polar regions threatened by anthropogenic activities.
Supporting information
The case contains a battery powered minicentrifuge and a portable solar panel battery charger for long term expeditions. The case also contains all necessary reagents and equipment to collect biopsies on site.
(TIFF)
Data are shown as mean ± SD from duplicates.
(TIFF)
(A) Recovery of total RNA from a 10 mm disc (100%) and a quarter of a disc (25%). (B) RNA recovery following soaking for 10 min versus overnight at room temperature before centrifugation. Results are representative of two independent experiments. RNA recovery was carried out using the TRIzol-based method.
(TIFF)
DNA recovery from hemocyte pellets that were fixed on the Whatman 903™ cards or the Whatman Gene FTA cards. Values are expressed as the total amount of DNA recovered from a 3 mm diameter sample disc using the PropGEM-based protocol. Results are representative of two independent experiments (n = 12/fgroup). * p < 0.05.
(TIFF)
(DOCX)
Acknowledgments
The authors would like to thank Ms. Marlène Fortier for her technical help, all the personnel from the French Polar Institute Paul Emile Victor (IPEV) and the Terres Australes et Antarctiques Françaises (TAAF) for their help and hospitality during our stay in the Kerguelen archipelago.
Abbreviations
- FTA
Flinders Technology Associates
- ccfDNA
circulating cell-free DNA
- EF1α
Elongation factor 1-alpha
- EF1γ
Elongation factor 1-gamma
- SNX14
Sorting Nexin 14
- Hsp70
heat shock protein 70
- SOD
superoxide dismutase
Data Availability
The metagenomic data from this publication have been deposited to the NCBI Sequence Read Archive with the assigned the identifier PRJNA564645.
Funding Statement
Supported in part by the Fondation Armand-Frappier (YSP), the French Polar Institute Paul Émile Victor (IPEV, Project N°409 IMMUNOTOXKER) (SB), and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-06607). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Beyer J, Green NW, Brooks S, Allan IJ, Ruus A, Gomes T, et al. Blue mussels (Mytilus edulis spp.) as sentinel organisms in coastal pollution monitoring: a review. Marine environmental research. 2017. September 1;130:338–65. 10.1016/j.marenvres.2017.07.024 [DOI] [PubMed] [Google Scholar]
- 2.Bodin N, Burgeot T, Stanisiere JY, Bocquené G, Menard D, Minier C, et al. Seasonal variations of a battery of biomarkers and physiological indices for the mussel Mytilus galloprovincialis transplanted into the northwest Mediterranean Sea. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology. 2004. August 1;138(4):411–27. [DOI] [PubMed] [Google Scholar]
- 3.Dondero F, Dagnino A, Jonsson H, Caprì F, Gastaldi L, Viarengo A. Assessing the occurrence of a stress syndrome in mussels (Mytilus edulis) using a combined biomarker/gene expression approach. Aquatic Toxicology. 2006. June 1;78:S13–24. 10.1016/j.aquatox.2006.02.025 [DOI] [PubMed] [Google Scholar]
- 4.Farcy E, Burgeot T, Haberkorn H, Auffret M, Lagadic L, Allenou JP, et al. An integrated environmental approach to investigate biomarker fluctuations in the blue mussel Mytilus edulis L. in the Vilaine estuary, France. Environmental Science and Pollution Research. 2013. February 1;20(2):630–50. 10.1007/s11356-012-1316-z [DOI] [PubMed] [Google Scholar]
- 5.Berg DJ, Haag WR, Guttman SI, Sickel JB. Mantle biopsy: a technique for nondestructive tissue-sampling of freshwater mussels. Journal of the North American Benthological Society. 1995. December 1;14(4):577–81. [Google Scholar]
- 6.Mioduchowska M, Kaczmarczyk A, Zając K, Zając T, Sell J. Gender-associated mitochondrial DNA heteroplasmy in somatic tissues of the endangered freshwater mussel Unio crassus (Bivalvia: Unionidae): implications for sex identification and phylogeographical studies. Journal of Experimental Zoology Part A: Ecological Genetics and Physiology. 2016. November;325(9):610–25. [DOI] [PubMed] [Google Scholar]
- 7.Lacoursière‐Roussel A, Howland K, Normandeau E, Grey EK, Archambault P, Deiner K, et al. eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecology and evolution. 2018. August;8(16):7763–77. 10.1002/ece3.4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Cunha Santos G. FTA Cards for Preservation of Nucleic Acids for Molecular Assays: A Review on the Use of Cytologic/Tissue Samples. Archives of pathology & laboratory medicine. 2018. March;142(3):308–12. [DOI] [PubMed] [Google Scholar]
- 9.Muthukrishnan M, Singanallur NB, Ralla K, Villuppanoor SA. Evaluation of FTA® cards as a laboratory and field sampling device for the detection of foot-and-mouth disease virus and serotyping by RT-PCR and real-time RT-PCR. Journal of virological methods. 2008. August 1;151(2):311–6. 10.1016/j.jviromet.2008.05.020 [DOI] [PubMed] [Google Scholar]
- 10.Liang X, Chigerwe M, Hietala SK, Crossley BM. Evaluation of Fast Technology Analysis (FTA) Cards as an improved method for specimen collection and shipment targeting viruses associated with Bovine Respiratory Disease Complex. Journal of virological methods. 2014. June 15;202:69–72. 10.1016/j.jviromet.2014.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam KI, Esona MD, Williams A, Ndze VN, Boula A, Bowen MD. Evaluation of BBL™ Sensi-Discs™ and FTA® cards as sampling devices for detection of rotavirus in stool samples. Journal of virological methods. 2015. September 15;222:41–6. 10.1016/j.jviromet.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences. 2008. October 21;105(42):16266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016. January 14;164(1–2):57–68. 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016. July;535(7610):94 10.1038/nature18850 [DOI] [PubMed] [Google Scholar]
- 15.Guglielmi G. How gut microbes are joining the fight against cancer. Nature. 2018. May;557(7706):482 10.1038/d41586-018-05208-8 [DOI] [PubMed] [Google Scholar]
- 16.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018. January 5;359(6371):97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apprill A. Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Frontiers in Marine Science. 2017. July 18;4:222. [Google Scholar]
- 18.Vezzulli L, Stagnaro L, Grande C, Tassistro G, Canesi L, Pruzzo C. Comparative 16SrDNA gene-based microbiota profiles of the Pacific oyster (Crassostrea gigas) and the Mediterranean mussel (Mytilus galloprovincialis) from a shellfish farm (Ligurian Sea, Italy). Microbial ecology. 2018. February 1;75(2):495–504. 10.1007/s00248-017-1051-6 [DOI] [PubMed] [Google Scholar]
- 19.Caza F, Betoulle S, Auffret M, Brousseau P, Fournier M, St-Pierre Y. Comparative analysis of hemocyte properties from Mytilus edulis desolationis and Aulacomya ater in the Kerguelen Islands. Marine environmental research. 2015. September 1;110:174–82. 10.1016/j.marenvres.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 20.Gustafson LL, Stoskopf MK, Bogan AE, Showers W, Kwak TJ, Hanlon S, et al. Evaluation of a nonlethal technique for hemolymph collection in Elliptio complanata, a freshwater bivalve (Mollusca: Unionidae). Diseases of aquatic organisms. 2005. June 30;65(2):159–65. 10.3354/dao065159 [DOI] [PubMed] [Google Scholar]
- 21.Chiminqgi M, Moutereau S, Pernet P, Conti M, Barbu V, Lemant J, et al. Specific real-time PCR vs. fluorescent dyes for serum free DNA quantification. Clinical Chemical Laboratory Medicine. 2007. August 1;45(8):993–5. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001. December 1;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 23.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC biology. 2014. December;12(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanguy M, McKenna P, Gauthier-Clerc S, Pellerin J, Danger JM, Siah A. Sequence analysis of a normalized cDNA library of Mytilus edulis hemocytes exposed to Vibrio splendidus LGP32 strain. Results in immunology. 2013. January 1;3:40–50. 10.1016/j.rinim.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreira R, Pereiro P, Canchaya C, Posada D, Figueras A, Novoa B. RNA-Seq in Mytilus galloprovincialis: comparative transcriptomics and expression profiles among different tissues. BMC genomics. 2015. December;16(1):728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Detree C, Núñez-Acuña G, Roberts S, Gallardo-Escárate C. Uncovering the complex transcriptome response of Mytilus chilensis against saxitoxin: implications of harmful algal blooms on mussel populations. PloS one. 2016. October 20;11(10):e0165231 10.1371/journal.pone.0165231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong W, Chen Y, Lu W, Wu B, Qi P. Transcriptome analysis of Mytilus coruscus hemocytes in response to Vibrio alginnolyficus infection. Fish & shellfish immunology. 2017. November 1;70:560–7. [DOI] [PubMed] [Google Scholar]
- 28.de Boissel PG, Fournier M, Rodriguez-Lecompte JC, McKenna P, Kibenge F, Siah A. Functional and molecular responses of the blue mussel Mytilus edulis' hemocytes exposed to cadmium-An in vitro model and transcriptomic approach. Fish & shellfish immunology. 2017. August 1;67:575–85. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nature protocols. 2006. August;1(2):581 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- 30.Bourlat SJ, Borja A, Gilbert J, Taylor MI, Davies N, Weisberg SB, et al. Genomics in marine monitoring: new opportunities for assessing marine health status. Marine pollution bulletin. 2013. September 15;74(1):19–31. 10.1016/j.marpolbul.2013.05.042 [DOI] [PubMed] [Google Scholar]
- 31.Carugati L, Corinaldesi C, Dell'Anno A, Danovaro R. Metagenetic tools for the census of marine meiofaunal biodiversity: an overview. Marine Genomics. 2015. December 1;24:11–20. 10.1016/j.margen.2015.04.010 [DOI] [PubMed] [Google Scholar]
- 32.Dawson SJ. Characterizing the Cancer Genome in Blood. Cold Spring Harbor perspectives in medicine. 2018. May 29:a026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cubero-Leon E, Ciocan CM, Minier C, Rotchell JM. Reference gene selection for qPCR in mussel, Mytilus edulis, during gametogenesis and exogenous estrogen exposure. Environmental Science and Pollution Research. 2012. August 1;19(7):2728–33. [DOI] [PubMed] [Google Scholar]
- 34.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. Journal of clinical oncology. 2014. February 20;32(6):579 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proceedings of the National Academy of Sciences. 2015. October 27;112(43):13336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huttunen R, Kuparinen T, Jylhävä J, Aittoniemi J, Vuento R, Huhtala H, et al. Fatal outcome in bacteremia is characterized by high plasma cell free DNA concentration and apoptotic DNA fragmentation: a prospective cohort study. PloS one. 2011. July 1;6(7):e21700 10.1371/journal.pone.0021700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forsblom E, Aittoniemi J, Ruotsalainen E, Helmijoki V, Huttunen R, Jylhävä J, et al. High cell-free DNA predicts fatal outcome among Staphylococcus aureus bacteraemia patients with intensive care unit treatment. PloS one. 2014. February 10;9(2):e87741 10.1371/journal.pone.0087741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS microbiology reviews. 2015. May 4;39(4):567–91. 10.1093/femsre/fuv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian Y, Yang X, Xu S, Wu C, Qin N, Chen SD, et al. Detection of Microbial 16 S rRNA Gene in the Blood of Patients With Parkinson’s Disease. Frontiers in aging neuroscience. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lokmer A, Wegner KM. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. The ISME journal. 2015. March;9(3):670 10.1038/ismej.2014.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans FF, Egan S, Kjelleberg S. Ecology of type II secretion in marine gammaproteobacteria. Environmental microbiology. 2008. May;10(5):1101–7. 10.1111/j.1462-2920.2007.01545.x [DOI] [PubMed] [Google Scholar]
- 42.Jensen S, Duperron S, Birkeland NK, Hovland M. Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS microbiology ecology. 2010. December 1;74(3):523–33. 10.1111/j.1574-6941.2010.00981.x [DOI] [PubMed] [Google Scholar]
- 43.Gosink JJ, Woese CR, Staley JT. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. International Journal of Systematic and Evolutionary Microbiology. 1998. January 1;48(1):223–35. [DOI] [PubMed] [Google Scholar]
- 44.Rubin-Blum M, Antony CP, Borowski C, Sayavedra L, Pape T, Sahling H, et al. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nature microbiology. 2017. August;2(8):17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hedlund BP, Geiselbrecht AD, Bair TJ, Staley JT. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 1999. January 1;65(1):251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sultan M, Amstislavskiy V, Risch T, Schuette M, Dökel S, Ralser M, et al. Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC genomics. 2014. December;15(1):675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costello M, Beard KH, Corlett RT, Cumming GS, Devictor V, Loyola R, et al. 2016. Field work ethics in biological research. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The case contains a battery powered minicentrifuge and a portable solar panel battery charger for long term expeditions. The case also contains all necessary reagents and equipment to collect biopsies on site.
(TIFF)
Data are shown as mean ± SD from duplicates.
(TIFF)
(A) Recovery of total RNA from a 10 mm disc (100%) and a quarter of a disc (25%). (B) RNA recovery following soaking for 10 min versus overnight at room temperature before centrifugation. Results are representative of two independent experiments. RNA recovery was carried out using the TRIzol-based method.
(TIFF)
DNA recovery from hemocyte pellets that were fixed on the Whatman 903™ cards or the Whatman Gene FTA cards. Values are expressed as the total amount of DNA recovered from a 3 mm diameter sample disc using the PropGEM-based protocol. Results are representative of two independent experiments (n = 12/fgroup). * p < 0.05.
(TIFF)
(DOCX)
Data Availability Statement
The metagenomic data from this publication have been deposited to the NCBI Sequence Read Archive with the assigned the identifier PRJNA564645.