Abstract
The eukaryotic chaperonin TRiC/CCT is a large hetero-oligomeric complex that plays an essential role assisting cellular protein folding and suppressing protein aggregation. It consists of two rings, each composed of eight different subunits; non-native polypeptides bind and fold in an ATP-dependent manner within their central chamber. Here we review recent advances in our understanding of TRiC structure and mechanism enabled by application of hybrid structural methods including the integration of cryo-electron microscopy with distance constraints from crosslinking mass spectrometry. These new insights are revealing how the different TRiC/CCT subunits create asymmetry in its ATP-driven conformational cycle and its interaction with non-native polypeptides, which ultimately underlie its unique ability to fold proteins that cannot be folded by other chaperones.
Keywords: TRiC, CCT, Chaperonin, Archaea, eukaryote, protein folding, chaperone, ATP, allostery, hybrid structural methods, chaperonin, TCP1, actin
Graphical Abstract
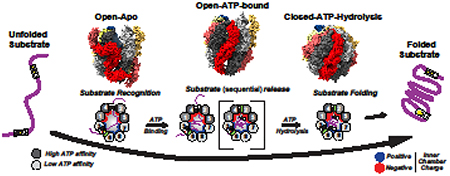
Introduction
All kingdoms of life contain a large double ring chaperonin complex that uses ATP binding and hydrolysis to bind and fold non-native polypeptides within its central chamber. Chaperonins have diverged into two evolutionarily distinct groups, differing substantially in their structure and mechanism. Prokaryotes and organelles of endosymbiotic origin contain Group I chaperonins, including the well characterized E. coli chaperonin GroEL/GroES (reviewed in (Hayer-Hartl et al., 2016)). GroEL consists of two stacked homo-heptameric rings. In response to ATP-binding, the GroEL chamber is capped by the homo-heptameric GroES complex, which acts as a detachable lid required substrate encapsulation and subsequent folding. Eukaryotes and archaea contain Group II chaperonins, which differ significantly from their prokaryotic counterparts in their architecture and mechanisms. First, the two rings of Group II chaperonins are generally eight membered and are stacked in a different inter-ring geometry than bacterial chaperonins. In addition, Group II chaperonins lack a GroES-like cofactor, using instead a flexible protrusion at the top of each subunit to create a built-in-lid that opens and closes in an ATP-dependent manner. Because of their distinct architecture, Group II chaperonins also have diverged in their conformational cycle, allosteric regulation, and in their substrate-recognition and folding mechanisms. In addition, while most prokaryotic chaperonins are homo-oligomeric group II chaperonins have undergone a progressive diversification of subunit composition. In archaea, chaperonins typically consist of 1–3 paralog subunits, while the eukaryotic chaperonin TRiC (TCP1-ring complex, also called CCT for cytosolic chaperonin containing TCP1) has eight different subunits per ring. The underlying causes and consequences of subunit diversification, which have long been enigmatic, are now beginning to emerge, as discussed in this review.
TRiC architecture and overall structure
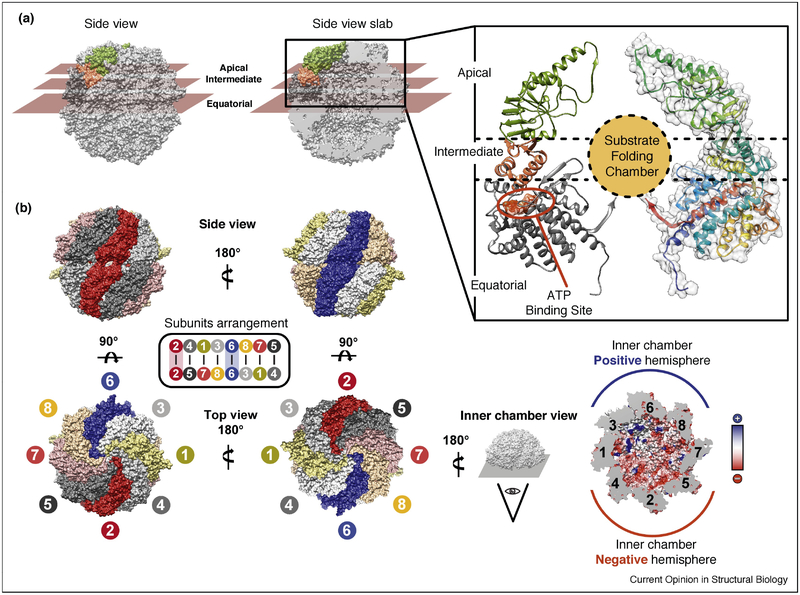
TRiC/CCT is a 1 MDa complex composed of eight paralogous subunits that assemble into a double ring hexadecamer. Each subunit has a characteristic three domains architecture; an apical domain at the top of the ring which contains the substrate recognition site and the lid-forming loop, an equatorial domain that contains the ATP-binding site, and an intermediate domain which contains a conserved Aspartic acid required for ATP hydrolysis and communicates ATP cycling at the equatorial domain to movements of the apical domain. (Fig 1 A)
Figure 1: TRiC Architecture and Subunit Arrangement.
(A) Closed TRiC structure with the three domains in a single subunit highlighted: equatorial domain (dark gray) responsible for binding ATP, apical domain (green) responsible for binding substrate and forming the built in lid, and hinge domain (salmon) responsible for relaying ATP changes into movements of the apical domain. Domain distribution indicated by planar sectioning, and also indicated on ribbon diagram of single subunit. (B) Subunit arrangement of TRiC, rings stacked back to back with CCT2 (red) and CCT6 (blue) stacked on top of each other. (B, lower right) Charge asymmetry in the interior of the closed chamber: one hemisphere (blue) is lined with positively charged side chains (+)and the other hemisphere (red) lined with negatively charged side chains (−). Inner chamber electrostatic potential also displayed. (PDB structures used: 4V94)
Structural insight into group II chaperonins was first obtained from crystallographic structures of simpler archaeal chaperonins composed of 1 to 2 subunits (Ditzel et al., 1998; Shomura et al., 2004). For TRiC, crystallography (Dekker et al., 2011; Munoz et al., 2011) and cryo-electron microscopy (cryo-EM) (Cong et al., 2010; Martin-Benito et al., 2007) identified the overall architecture (Fig 1) and the basic features of the ATP-driven conformational cycle (see below, Fig. 2) but the low resolution of the structures and the pseudo-symmetrical appearance of the complex precluded assignment of the subunit arrangement, and limited resolution for structural approaches. TRiC subunits have a very similar three dimensional structure despite having only a 27–39% sequence identity (Archibald et al., 2001). Hybrid structural approaches based on crosslinking/mass spectrometry and combinatorial modelling (Kalisman et al., 2012; Leitner et al., 2012) (Fig 1B), resolved the subunit arrangement ambiguity; and demonstrated that TRiC/CCT has a fixed subunit arrangement, confirmed by in vivo studies in yeast (Leitner et al., 2012). Elucidating the TRiC arrangement enabled application of symmetry-free structural approaches, which together with single subunits eGFP tags (Zang et al., 2016) and antibodies against specific subunits (Balchin et al., 2018), have greatly advanced our understanding of this complex, both in the ATP-induced closed state and in the more flexible open-state.
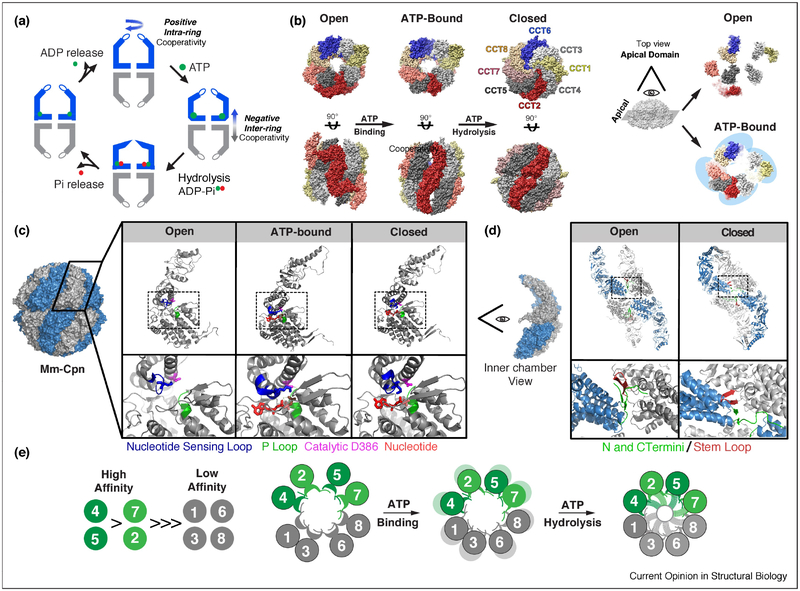
Figure 2: The ATP-driven conformational cycle of TRiC.
(A) Schematic of the ATPase cycle, ATP binding drives a contraction of the complex and ATP hydrolysis leads to further contraction and formation of built-in lid; phosphate release leads to opening of the ring, ADP release resets the cycle. (B) Structures of TRiC in the open, AMPPNP (non-hydrolysable ATP analog) bound, and ATP hydrolysis induced closed state. ATP binding results in dramatic rearrangement of apical domains; their transition into tetramer of dimers is highlighted by top end views. (C) Structural transitions of an individual Mm-Cpn subunit during the ATPase cycle. Inset: changes within the ATP binding pocket. Nucleotide Sensing Loop (blue), P Loop (green), Catalytic residue (magenta), and Nucleotide (red). (D) Chamber view of 4 subunits highlights changes in inter-subunit contacts between the open and closed state. N and C termini (green) interact with the stem loop (red) in the adjacent subunit. (E) Relative ATP affinities of each subunit partition in TRiC into a high affinity lobe (green) and low affinity lobe (grey). PDB structures used for TRiC and Mm-Cpn respectively: Open 5GW4, 3IYF, ATP analog 5GW5, 3RUV, Closed 4V94, 3RUW.
The TRiC/CCT subunit arrangement is conserved from yeast to mammals (Leitner et al., 2012). Each ring in the hexadecamer has identical subunit order (CCT 2–4–1–3–6–8–7–5) with the two rings related by a two-fold symmetry axis centered on inter-ring CCT2–CCT2’ and CCT6–CCT6’ homotypic dimers (Fig 1B, dark red and blue, respectively). Defining the subunit arrangement illuminated a number of biochemical and biophysical observations, and revealed that this seemingly symmetric complex is deeply asymmetric in many of its functions, including nucleotide binding and hydrolysis as well as substrate binding and folding. Strikingly, upon lid closure, the substrate becomes encapsulated within a folding chamber, which as a result of subunit diversification, is lined with an evolutionarily conserved asymmetric distribution of charges, with CCT 5–2–4– and CCT 3–6–8 subunits creating negative and positive hemispheres, respectively (Balchin et al., 2018; Leitner et al., 2012) (Fig 1B, red and blue, respectively). This unique folding environment likely contributes to TRiC/CCT’s unique ability to fold proteins that cannot be folded by other chaperones.
ATP regulation of the TRiC/CCT conformational cycle
Group II chaperonins use ATP binding and hydrolysis to alternate between an open state, where the lid forming segments are unstructured and the substrate binding sites in each subunit are accessible and a closed state, where the lid segments form a beta-stranded iris (schematic in Fig 2A). The conformational cycle, allosteric regulation and ATP-driven structural changes are largely conserved between TRiC and its simpler archaeal homologues. Many of the early insights into the ATP cycle came from structural and mechanistic studies of the homo-oligomeric archaeal chaperonin from Methanococcus maripaludis, also known as Mm-Cpn; however, these have been confirmed and extended through recent studies of TRiC/CCT (Fig 2B). In the absence of nucleotide, both TRiC and Mm-Cpn reside in an open and highly dynamic state. Cryo-EM revealed that in the homo-oligomeric Mm-Cpn the apical domains alternate between many conformations (Zhang et al., 2011) and for TRiC the apical domains exist in a variety of conformations, the most extreme of which is the apical domain of CCT2 which is flipped out behind CCT5 (Zang et al., 2016). ATP binds to the equatorial domains, which have a universally conserved P-loop motif. ATP binding does not suffice to induce the closed state, but does lead to a ~ 45% rotation of the Mm-Cpn apical domain leading to a more symmetric and compact open state (Zhang et al., 2011). Similarly, ATP-binding to TRiC leads to formation of specific apical domains contacts, generating a more compact open conformation that can be described as a tetramer of dimers (Cong et al., 2012; Zang et al., 2016) (Fig 2B, right). ATP hydrolysis is triggered by a catalytic Aspartic acid in the intermediate domain (D386 in Mm-Cpn). Upon ATP hydrolysis there is a conformational rotation of the apical domains that both closes the lid and releases the bound substrate to the central chamber, which now adopts a highly polar/charged environment (Douglas et al., 2011; Leitner et al., 2012). Mutations in Mm-Cpn showed that both lid closure and release of the substrate to the chamber are required for substrate folding (Douglas et al., 2011). The rate of ATP hydrolysis and the state of the nucleotide are sensed by a loop termed the nucleotide sensing loop (NSL). The NSL contains a lysine (K161 in Mm-Cpn) that interacts with the γ phosphate of ATP in the pre-hydrolysis state and with the α phosphate of the nucleotide following ATP hydrolysis (Fig 2C) (Pereira et al., 2012). ATP hydrolysis causes a shift in the NSL, which is relayed through long alpha helixes at the top of the apical domains to drive the formation of the β stranded lid closing off the chamber (Zhang et al., 2010) (Fig 2A,B). Phosphate and ADP release complete the cycle, triggering reopening of the chamber (Fig 2A) (Bigotti et al., 2006).
TRiC and archaeal chaperonins share their mechanisms of allosteric regulation, even though the rate of ATP hydrolysis is higher for archaea than for TRiC. In vitro, TRiC hydrolyses ~2.4 ATPs per minute*complex (Reissmann et al., 2007), consistent with the reported ~50 s TRiC release halftime for the obligate substrate actin in vivo (Thulasiraman et al., 1999). At low ATP concentrations group II chaperonins exhibit positive cooperativity with a Hill coefficient of 1.3–~2 (Kafri et al., 2001) (Lopez et al., 2017; Reissmann et al., 2007). Positive cooperativity within a ring is regulated through an intersubunit interface formed by four beta-strands, two of which are a beta-stranded hairpin from one subunit and two of which are the N-and C-termini of its neighbor (Fig 2D) (Lopez et al., 2017; Zhang et al., 2011). A network of covarying residues connects the ATP binding pockets of all subunits of a given ring through this interface. Strikingly, chaperonin allostery is tunable through mutations at a key position in this interface (Met47 in Mm-Cpn). This residue regulates the allostery of nucleotide cycling by sensing and communicating nucleotide occupancy within a ring (Lopez et al., 2017). At higher ATP concentrations, negative cooperativity for ATP hydrolysis is observed. By analogy with Group I chaperonins, this negative cooperativity is proposed to reflect the negative inter-ring regulation, leading to an asymmetric ATPase cycle (Kafri et al., 2001) (Reissmann et al., 2007). However, how the two rings of TRiC and archaeal chaperonins communicate is unclear, and other hypotheses have been proposed for the observed negative cooperativity at high ATP concentrations (Jiang et al., 2011).
Comparing the ATP regulation of archaeal chaperonins with TRiC revealed a surprising consequence of subunit diversification in eukaryotes, namely a highly asymmetric ATPase cycle. While all subunits in the eukaryotic chaperonin maintain the P-loop and the catalytic Asp residue in their ATP binding site, many of the element of ATPase regulation are asymmetrically distributed in the eukaryotic chaperonin. This includes the residues in the NSL that control the ATPase rate (Pereira et al., 2012), the identity of residue at position 47 controlling positive cooperativity (Lopez et al., 2017) and most dramatically, the equatorial nucleoside contacts that control affinity for ATP (Reissmann et al., 2012). Both biochemical, genetic and single molecule experiments have demonstrated that of the eight subunits in TRiC, only four bind ATP with any appreciable affinity (Jiang et al., 2011; Reissmann et al., 2012). Single molecule studies showed TRiC binds ~7–8 nucleotides at nearly all ATP concentrations, which could be the results of 8 nucleotides bound to a single ring, or 4 nucleotides per ring (Jiang et al., 2011). Crosslinking studies then indicated that four paralog subunits of TRiC bind ATP with significant affinity (CCT5,4,1 and 2), while the others bound ATP very poorly. Careful titration also indicated a hierarchy of ATP affinity with CCT4/5>CCT1/2>>>CCT7,8,6,3 (Reissmann et al., 2012). The measured affinities are consistent with the degree of conservation of nucleoside contacts in the high affinity subunits, suggesting an evolutionarily conserved hierarchy of ATP binding and hydrolysis. Recently, it was proposed that the “low” affinity subunits have a very slow ADP off-rate (Zang et al., 2016). Importantly, mutations in the low affinity subunits (CCT7,8,6,3) that impair ATP binding, i.e. in the P-loop motif, or ATP hydrolysis, i.e. the catalytic Asp acid, have no phenotypic consequences in yeast, indicating their interaction with nucleotide is dispensable for function. In contrast, these same mutations in the high affinity subunits (CCT5,4,1,2) exhibit in vivo phenotypes with a severity that mirrors their affinity for ATP, with CCT4 mutations in either ATP binding or hydrolysis being lethal (Reissmann et al., 2012) When mapped onto the TRiC arrangement, these findings suggest the ATPase cycle is driven by a high-affinity hemisphere in the ring consisting of CCT5,4,1 and 2 (Fig 2E).
The asymmetric ATP utilization by TRiC has led to a model of sequential closure initiating on the high affinity hemisphere and progressing through the other subunits. Early structural work found different levels of closure of the ring based on ATP concentrations (Rivenzon-Segal et al., 2005). Structural work using H/D exchange MS and cryo-EM support the notion that the ATP-dependent conformational change initiates in the “high” ATP affinity lobe of TRiC (CCT5,2,4,1) (Balchin et al., 2018; Zang et al., 2016).
The cellular functions and substrates of TRiC/CCT
TRiC is essential for viability as many essential proteins, such as actin, tubulin and cell cycle regulators, exhibit an obligate TRiC requirement to achieve the native state. TRiC binds co- and post-translationally to ~10% of the proteome (Hein et al., 2015; Yam et al., 2008). Why TRiC selects these particular set of proteins as substrates and how it facilitates their folding are key unanswered questions for future research. TRiC substrates tend to be topologically complex with aggregation prone β-sheet folds, both properties are predicted to cause slow folding kinetics and increase aggregation propensity (Yam et al., 2008). These include proteins with a WD40 Beta propeller domain, such as Gβ(Wells et al., 2006), CSA (Pines et al., 2018), and WDR68 (Miyata et al., 2014) as well as oncoproteins acting at the transcriptional level, such as p53 (Trinidad et al., 2013), AML1-ETO (Roh et al., 2016) and STAT3 (Kasembeli et al., 2014). TRiC has also been shown to suppress aggregation of amyloid proteins linked to age related diseases, including aggregation of the polyQ expanded HTT protein responsible for Huntingtin’s disease (Shen and Frydman, 2012; Sontag et al., 2013); as well as synphilin and alpha-synuclein, linked to aging-related Alzheimers’ and Parkinson’s disease, respectively (Chen et al., 2018; Sot et al., 2017).
In addition to facilitating folding, TRiC may assist complex assembly, as many of its substrates are part of larger complexes (Kaisari et al., 2017). For example, α and β tubulin, form a dimer downstream of TRiC (Lewis et al., 1997) and tumor suppressor VHL assembles with binding partners Elongin B/C (Spiess et al., 2006). TRiC also plays a role in viral protein folding, including roles in capsid folding/assembly(Hong et al., 2001; Knowlton et al., 2018) and replication(Zhou et al., 2008), making it a possible target for antiviral therapies.
TRiC substrate interactions: how subunit diversity impacts polypeptide binding and folding
Substrate folding is coordinated with the ATPase cycle. In the open state TRiC binds unfolded polypeptides through the apical domains; ATP-induced lid formation releases the substrate to the closed chamber where folding is thought to occur. While a detailed understanding of how TRiC assists substrate folding has remained elusive, recent studies revealed a key role of subunit diversity in both substrate binding and folding.
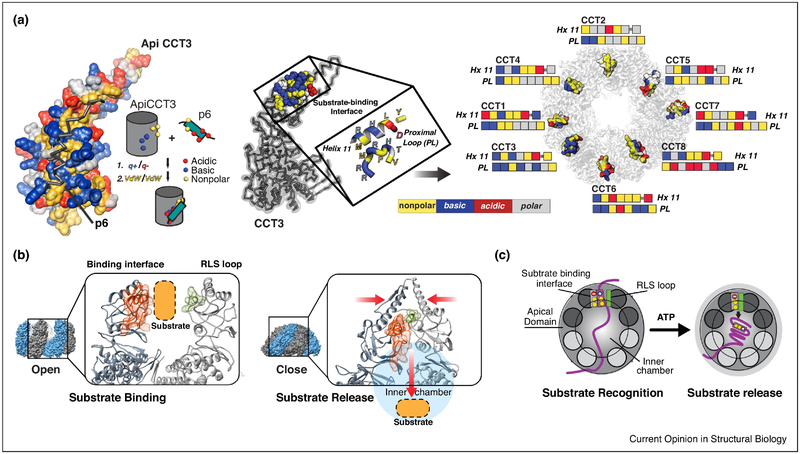
The important question of substrate recognition, and how TRiC can recognize so many different substrates was recently addressed using a hybrid structural approach that combined NMR, biophysical assays, modeling and crosslinking-mass spectrometry (XL-MS) approaches to determine the substrate-binding interface of the apical domains in TRiC subunits and obtain a structural model of the apical domain-substrate interaction. These experiments revealed that specific motifs within the substrate polypeptide bind to an apical domain interface created by Helix 11 and a flexible proximal loop (H11-PL) (Fig. 3A, left) (Joachimiak et al., 2014). Strikingly, and unlike the hydrophobic binding interfaces of other chaperones, the H11-PL interface of each apical domain has a unique combination of polar, charged and hydrophobic residues, which should result in a diversification of the type of substrate motifs recognized in its substrates. As a result of the plasticity of the TRiC apical domain-substrate motif interaction, different substrates can bind in different configurations to the same apical domain, while the unique substrate-binding motifs enable different substrates that share no sequence similarly to bind, allowing for recognition of a broader set of substrate sequences (Fig.3A, right). Substrates recruited to the apical domain surfaces through the interaction of specific sequence or structural elements with a specific combination of polar and nonpolar contacts across the eight subunits will also orient the substrate polypeptide within the chamber and promote specific topologies that may be productive for folding. The partitioning of the substrate binding surface by charge and nonpolar contacts may also play a role in substrate recruitment through modulation of association and dissociation rates. The eight subunits thus are thought to coordinate multivalent interaction with a substrate which prevent aggregation and prime the substrate conformation for subsequent folding.
Figure 3: Role of TRiC subunit diversity in substrate recognition and binding.
(A) Structural model of apical domain of TRiC subunit CCT3 bound to substrate HIV protein p6. Helix 11 (and Proximal Loop make up binding domain, amino acid characteristics indicated by color: Nonpolar (yellow), Basic (blue), Acidic (red) and Polar (grey). (B) Top view of TRiC open conformation, which exposes the substrate binding regions of each apical domain. The Hx11/PL determinant is shown in spacefill following coloring scheme in (A), their differing characteristics allow polyvalent binding and diversity of substrate recognition. (C) The substrate binding region of apical domain (red) clashes with the RLS domain of its neighboring subunit (green) in the closed conformations, facilitates substrate release into the chamber.
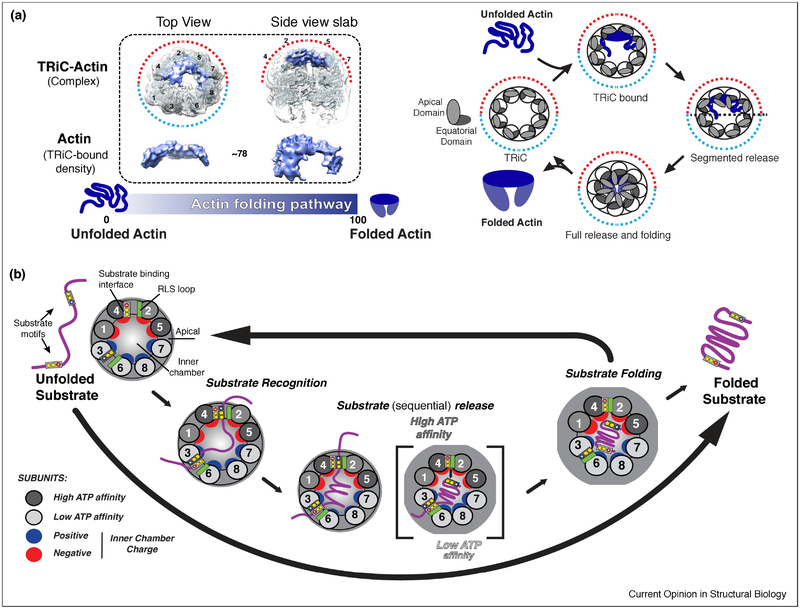
The next step in the folding cycle is ATP-dependent substrate release into the chamber. The mechanism for substrate release was revealed by studies of the archaeal Mm-Cpn, which showed that the conformational change induced by ATP hydrolysis brings the H11-PL into close proximity with a loop in the adjacent subunit, which packs against the binding interface during ring closure. This loop, termed RLS for release loop for substrate, causes a steric clash that pushes the substrate into the chamber (Fig. 3B,C) (Douglas et al., 2011). Importantly, mutations in the RLS abrogate substrate folding, even if the chamber is closed and the substrate encapsulated. Recent work combining H/D-exchange MS and cryo-EM provided new evidence supporting the importance of substrate binding configuration and substrate release to a productive folding cycle. This study found that when the denatured canonical TRiC substrate, Actin binds TRiC in the open state, it has already adopted a significant degree of secondary and tertiary structure. TRiC, however, keeps the two lobes of Actin in an extended conformation and prevents formation of a misfolded state. Binding of ATP and subsequent hydrolysis drive a conformational change that starts in the high ATP–affinity hemisphere, allowing the segmental release of actin, which enhances productive formation of the long range contact bringing the two lobes together (Balchin et al., 2018) (Fig 4A). This data provided strong evidence for ATP binding and hydrolysis initiating a conformational cascade that promotes release of the substrate in a sequential manner dictated by the hierarchical ATP usage of each subunit to facilitate folding. Since many TRiC substrates have complex topologies (Yam et al., 2008), it is likely that one key aspect of TRiC assisted folding is to promote the formation of high contact order interactions which are difficult to form with simpler chaperones, or with chaperonins that expose an array of identical hydrophobic binding sites.
Figure 4: Role of TRiC subunit diversity in substrate folding and release.
(A) TRiC-Actin folding cycle adapted from (Balchin et al., 2018). Denatured Actin binds to apical domains on the high ATP affinity side of TRiC in a largely structured conformation. ATP binding and hydrolysis drives the sequential release of substrate into chamber allowing folding to occur; opening of the lid allows release of Actin into solution. (B) Overall model of TRiC mediated folding. Unique H11/PL binding interfaces allow TRiC to bind substrate (Purple) in a subunit specific conformation during initial recognition. The asymmetry of the ATPase cycle (high to low affinity; dark to light grey), initiates a sequential closure of the TRiC complex. Proximal interactions of the RLS loop (green) and the H11/PL binding interface facilitates sequential substrate release into the folding chamber. In the chamber, substrate is subjected to an asymmetric distribution of charges with the surface of one hemisphere positively (blue), and the other negatively (red) charged.
We propose that the emergence of the unique mechanism of TRiC, with its subunit specific substrate interactions and the segmental release into the chamber, arose as a solution to the problem of folding of topologically complex domains and proteins. This raises the interesting question of how simpler archaeal counterparts evolved into such a complex hetero-oligomeric folding machine in eukaryotes. Strikingly, comparing all organisms containing TRiC-like chaperonins found a positive correlation between subunit diversity and the size of its proteome (Joachimiak et al., 2014), suggesting the subunit diversification of TRiC is linked to the expansion of the proteome in eukaryotes. This raises the possibility that the folding machinery contributes to dictate proteome size.
Once substrate is released into the chamber, a variety of mechanisms have been proposed that could assist folding. The asymmetric charge distribution within the chamber (Leitner et al., 2012) likely assists folding, for instance, by separating charged regions of a peptide that are normally surface exposed, while allowing the formation of a collapsed hydrophobic core. Confinement within the GroEL-ES chamber has been shown to enhance folding perhaps by providing geometric constraints on possible folding pathways (Tang et al., 2006). Theoretical work also suggests that confined water within a polarized chamber will cause an increase in the hydrophobic effect to drive productive folding (England and Pande, 2008). As the above mechanisms mostly arise from studies of the bacterial chaperonin, the specific mechanisms driving folding within the TRiC chamber remain to be determined.
Highlights:
The eukaryotic chaperonin TRiC/CCT is a large hetero-oligomeric ring-shaped complex
TRiC uses ATP to fold many essential cellular proteins within its central chamber
TRiC undergoes a complex conformational cycle driven by ATP binding and hydrolysis
Unfolded substrates bind TRiC through polyvalent subunit-specific contacts
TRiC subunit diversity creates an asymmetric cycle essential for folding activity
Perspectives.
The wealth of new structural and mechanistic insight emerging in recent years has completely changed our view of TRiC/CCT. Initially thought to be a variation on the theme of GroEL-ES, this chaperonin has emerged as a highly asymmetrical and complex machine, where subunit diversification has created unique substrate binding and folding capabilities and a unique ATP-driven cycle unlike that of any other chaperone. This must clearly be linked to TRiC’s unique ability to fold eukaryotic proteins. Clearly, many exciting new questions are now open. These include fundamental questions such as how folding occurs within the chamber, how the two rings communicate and, importantly, what are the folding intermediates that TRiC recognized in vivo, during translation. Another important and very poorly understood aspect of TRiC biology relates to its function in the cell, its interaction with cofactors and other chaperones and its regulation and assembly. The next years will likely bring light into this very important and until now enigmatic folding machine.
Acknowledgements
Work in the Frydman lab is supported by NIH (R01GM074074 to JF; F32GM103124 to DG). We thank members of the Frydman lab for discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
5 References
**Outstanding interest/*Special interest
- Archibald JM, Blouin C, and Doolittle WF (2001). Gene duplication and the evolution of group II chaperonins: implications for structure and function. J Struct Biol 135, 157–169. [DOI] [PubMed] [Google Scholar]
- Balchin D, Milicic G, Strauss M, Hayer-Hartl M, and Hartl FU (2018). Pathway of Actin Folding Directed by the Eukaryotic Chaperonin TRiC. Cell 174, 1507–1521 e1516. [DOI] [PubMed] [Google Scholar]; ** This paper examines the folding pathway of Actin on TRiC. Using H/D exchange MS and cryo-EM, the authors provide evidence for binding to specific apical domains, and segmented release of Actin that allows a stepwise folding of a long range contact that simulations predict would be an otherwise rare event.
- Bigotti MG, Bellamy SR, and Clarke AR (2006). The asymmetric ATPase cycle of the thermosome: elucidation of the binding, hydrolysis and product-release steps. J Mol Biol 362, 835–843. [DOI] [PubMed] [Google Scholar]
- Chen XQ, Fang F, Florio JB, Rockenstein E, Masliah E, Mobley WC, Rissman RA, and Wu C (2018). T-complex protein 1-ring complex enhances retrograde axonal transport by modulating tau phosphorylation. Traffic 19, 840–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. (2010). 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A 107, 4967–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Schroder GF, Meyer AS, Jakana J, Ma B, Dougherty MT, Schmid MF, Reissmann S, Levitt M, Ludtke SL, et al. (2012). Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. EMBO J 31, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, and Willison KR (2011). The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J 30, 3078–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, and Steinbacher S (1998). Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93, 125–138. [DOI] [PubMed] [Google Scholar]
- Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, and Frydman J (2011). Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell 144, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Identifies mechanism for substrate release into TRiC cavity during lid closure
- England JL, and Pande VS (2008). Potential for modulation of the hydrophobic effect inside chaperonins. Biophys J 95, 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer-Hartl M, Bracher A, and Hartl FU (2016). The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem Sci 41, 62–76. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723. [DOI] [PubMed] [Google Scholar]
- Hong S, Choi G, Park S, Chung AS, Hunter E, and Rhee SS (2001). Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J Virol 75, 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Douglas NR, Conley NR, Miller EJ, Frydman J, and Moerner WE (2011). Sensing cooperativity in ATP hydrolysis for single multisubunit enzymes in solution. Proc Natl Acad Sci U S A 108, 16962–16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimiak LA, Walzthoeni T, Liu CW, Aebersold R, and Frydman J (2014). The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper uses a variety of techniques to identify where substrates initially bind TRiC apical domains. They uncover a small region in apical domains, that is conserved across homologs and diverse among paralogs that allows TRiC to form specific polyvalent interactions with substrate.
- Kafri G, Willison KR, and Horovitz A (2001). Nested allosteric interactions in the cytoplasmic chaperonin containing TCP-1. Protein Sci 10, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisari S, Sitry-Shevah D, Miniowitz-Shemtov S, Teichner A, and Hershko A (2017). Role of CCT chaperonin in the disassembly of mitotic checkpoint complexes. Proc Natl Acad Sci U S A 114, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisman N, Adams CM, and Levitt M (2012). Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc Natl Acad Sci U S A 109, 2884–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasembeli M, Lau WC, Roh SH, Eckols TK, Frydman J, Chiu W, and Tweardy DJ (2014). Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol 12, e1001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton JJ, Fernandez de Castro I, Ashbrook AW, Gestaut DR, Zamora PF, Bauer JA, Forrest JC, Frydman J, Risco C, and Dermody TS (2018). The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat Microbiol 3, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, et al. (2012). The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using a crosslinking/mass spectrometry approach, this paper finally identifies the conserved arrangement of TRiC subunits. This arrangement reveals that the subunits are segregated asymmetrically into a (+) and (−) charged hemisphere and also a high ATP affinity and low ATP affinity hemisphere.
- Lewis SA, Tian G, and Cowan NJ (1997). The alpha- and beta-tubulin folding pathways. Trends Cell Biol 7, 479–484. [DOI] [PubMed] [Google Scholar]
- Lopez T, Dalton K, Tomlinson A, Pande V, and Frydman J (2017). An information theoretic framework reveals a tunable allosteric network in group II chaperonins. Nat Struct Mol Biol 24, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Benito J, Grantham J, Boskovic J, Brackley KI, Carrascosa JL, Willison KR, and Valpuesta JM (2007). The inter-ring arrangement of the cytosolic chaperonin CCT. EMBO Rep 8, 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Shibata T, Aoshima M, Tsubata T, and Nishida E (2014). The molecular chaperone TRiC/CCT binds to the Trp-Asp 40 (WD40) repeat protein WDR68 and promotes its folding, protein kinase DYRK1A binding, and nuclear accumulation. J Biol Chem 289, 33320–33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IG, Yebenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Carcer G, Malumbres M, et al. (2011). Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol 18, 14–19. [DOI] [PubMed] [Google Scholar]
- Pereira JH, Ralston CY, Douglas NR, Kumar R, Lopez T, McAndrew RP, Knee KM, King JA, Frydman J, and Adams PD (2012). Mechanism of nucleotide sensing in group II chaperonins. EMBO J 31, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines A, Dijk M, Makowski M, Meulenbroek EM, Vrouwe MG, van der Weegen Y, Baltissen M, French PJ, van Royen ME, Luijsterburg MS, et al. (2018). TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A. Nat Commun 9, 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S, Joachimiak LA, Chen B, Meyer AS, Nguyen A, and Frydman J (2012). A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Rep 2, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Identifies relative ATP affinities for each TRiC subunit, and correlate ATP affinity with in vivo phenotypes
- Reissmann S, Parnot C, Booth CR, Chiu W, and Frydman J (2007). Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Mol Biol 14, 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, and Horovitz A (2005). Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol 12, 233–237. [DOI] [PubMed] [Google Scholar]
- Roh SH, Kasembeli M, Galaz-Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, and Tweardy DJ (2016). Chaperonin TRiC/CCT Modulates the Folding and Activity of Leukemogenic Fusion Oncoprotein AML1-ETO. J Biol Chem 291, 4732–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, and Frydman J (2012). The Interplay Between the Chaperonin TRiC and N-terminal Region of Huntingtin Mediates Huntington’s Disease Aggregation and Pathogenesis In Protein Quality Control in Neurodegenerative Disease, Morimoto RI, and Christen Y, eds. (Springer Verlag; ). [Google Scholar]
- Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, and Miki K (2004). Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J Mol Biol 335, 1265–1278. [DOI] [PubMed] [Google Scholar]
- Sontag EM, Joachimiak LA, Tan Z, Tomlinson A, Housman DE, Glabe CG, Potkin SG, Frydman J, and Thompson LM (2013). Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc Natl Acad Sci U S A 110, 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sot B, Rubio-Munoz A, Leal-Quintero A, Martinez-Sabando J, Marcilla M, Roodveldt C, and Valpuesta JM (2017). The chaperonin CCT inhibits assembly of alpha-synuclein amyloid fibrils by a specific, conformation-dependent interaction. Sci Rep 7, 40859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Miller EJ, McClellan AJ, and Frydman J (2006). Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell 24, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, and Hayer-Hartl M (2006). Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914. [DOI] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, and Frydman J (1999). In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J 18, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, and Vousden KH (2013). Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell 50, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CA, Dingus J, and Hildebrandt JD (2006). Role of the chaperonin CCT/TRiC complex in G protein betagamma-dimer assembly. J Biol Chem 281, 20221–20232. [DOI] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, and Frydman J (2008). Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol 15, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Jin M, Wang H, Cui Z, Kong L, Liu C, and Cong Y (2016). Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM. Nat Struct Mol Biol 23, 1083–1091. [DOI] [PubMed] [Google Scholar]; ** This paper has the first high resolution cryo-EM structures of the eukaryotic TRiC complex form Saccharomyces cerevisiae. Using GFP inserts, the authors confirm the arrangement of TRiC subunits. By solving structures for multiple nucleotide states, the authors identify massive transitions of the TRiC apical domains, and also find that one hemisphere has a very slow off-rate for ADP.
- Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, et al. (2010). Mechanism of folding chamber closure in a group II chaperonin. Nature 463, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma B, DiMaio F, Douglas NR, Joachimiak LA, Baker D, Frydman J, Levitt M, and Chiu W (2011). Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure. Structure 19, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. (2008). Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4, 495–504. [DOI] [PubMed] [Google Scholar]