Abstract
Anti-oxidative agents hold great potential in osteoarthritis (OA) therapy. However, most radical scavengers have poor biocompatibility and potential cytotoxicity, which limit their applications. Herein we explore dopamine melanin (DM) nanoparticles as a novel scavenger of reactive oxygen species (ROS) and reactive nitrogen species (RNS). DM nanoparticles show low cytotoxicity and a strong ability to sequester a broad range of ROS and RNS, including superoxides, hydroxyl radicals, and peroxynitrite. This translates to excellent anti-inflammatory and chondro-protective effects by inhibiting intracellular ROS and RNS and promoting antioxidant enzyme activities. With an average diameter of 112.5 nm, DM nanoparticles can be intra-articularly (i.a.) injected into an affected joint and retained at the injection site. When tested in vivo in rodent OA models, DM nanoparticles showed diminished inflammatory cytokine release and reduced proteoglycan loss, which in turn slowed down cartilage degradation. Mechanistic studies suggest that DM nanoparticles also enhance autophagy that benefits OA control. In summary, our study suggests DM nanoparticles as a safe and promising therapeutic for OA.
Keywords: osteoarthritis, reactive oxygen species, reactive nitrogen species, dopamine melanin nanoparticles, autophagy
Graphical Abstract
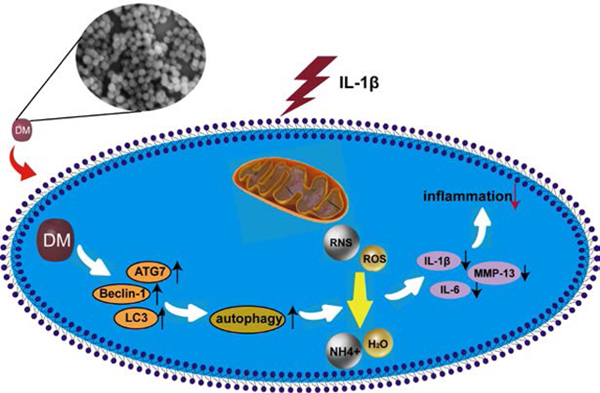
1. Introduction
Osteoarthritis (OA) is a major cause of chronic disability and it affects more than 400 million people worldwide. OA is characterized by the gradual progression of chronic inflammation and cartilage degeneration, leading to stubborn joint pain and joint deformity.1 Current treatments, including analgesics, non-steroidal anti-inflammatory drugs (NSAIDs), and intra-articular injection of hyaluronic acid or corticosteroid,2 aim to ease the symptoms rather than altering the course of disease progression. There is an urgent need for new and safe options for OA.
Recent studies reveal that ROS and RNS play an essential role in OA development and progression.3 ROS and RNS are produced mainly by NADPH oxidase and iNOS, and remain at low levels in normal articular chondrocytes. In OA patients, the antioxidant mechanisms, including superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidise (GPX), are deregulated and insufficient to detoxify ROS and RNS.4–6 This leads to elevated oxidative stress that induces DNA, lipid, and protein damage, as well as cytotoxicity. Reagents that can suppress ROS and RNS in the chondrocytes are therefore promising OA therapeutics. Previously, anti-oxidants like melatonin and N-acetylcysteine (NAC)7, 8 have been tested but these small molecules show short retention in the joint. More recently, nanozymes and metal nanomaterials such as PEGylated bilirubin nanoparticles,9 sialic acid-modified selenium nanoparticles,10 and TEMPO-conjugated gold nanoparticles11 have been explored for OA therapy. But the poor biocompatibility and/or biodegradability have limited their applications.
Herein we investigate melanin nanoparticles as a novel type of radical scavenger for OA therapy. Natural melanin is known to protect the skin from ultraviolet (UV) irradiation. This is in large part attributed to its surface quinone residues that can efficiently scavenge radicals. Artificial melanin nanospheres have been prepared by polymerization of dopamine.12 The Shi group recently investigated the anti-oxidant mechanisms of these melanin nanoparticles, showing that materials can scavenge a broad range of radicals.13 Rageh et al. demonstrated that melanin nanoparticles can protect mice from DNA damage induced by γ-radiation and effectively restore hematopoietic tissues.14 To the best of our knowledge, however, there has been no attempt on exploiting melanin nanoparticles for OA treatment. Unlike small molecule scavengers, DM nanospheres are large in size (~110 nm) and thereby potentially afford much longer retention in the joint, which is beneficial for sustained ROS/RNS suppression and OA management. We hypothesize that i.a. injected DM nanospheres can efficiently suppress cartilage RNS and ROS levels, leading chondrocyte protection and OA suppression. Specifically, we first assessed whether DM nanoparticles can suppress ROS and RNS in IL-1β induced rat chondrocytes, and tested their efficacy in vivo in a rat OA model. The underlying mechanism of action was also investigated.
2. Experimental details
2.1. Synthesis of DM Nanoparticles
2.1.1. Materials
Dopamine hydrochloride (Sigma-Aldrich, H8502), ammonium hydroxide (Sigma-Aldrich, 320145), H2O2 (Fisher Chemical, H325–500), nitrotetrazolium blue chloride (NBT, Sigma-Aldrich, N6876), deoxyribose (Sigma-Aldrich, 121649), ferric chloride (Acros Organics, 423705000), ascorbic acid (Sigma-Aldrich, A5960), riboflavin (Sigma-Aldrich, PHR1054), ethylenediaminetetraacetic acid (EDTA, Sigma-Aldrich, EDS-500G), NaCN (Alfa Aesar, L13278), 2,2 diphenyl-1-picryl hydrazyl (DPPH, Sigma-Aldrich, D9132), sodium nitroprusside (Sigma-Aldrich, PHR1423), HCl (baker analyzed, 0000170466), potassium nitrite (Fisher Scientific, P263–500), diethylenetriaminepentaacetic acid (DTPA, Sigma-Aldrich, D6518), peroxynitrite (Cayman, 81565), Evans Blue (Sigma-Aldrich, E2129), trichloroacetic acid (Fisher Scientific, ICN15259291)
2.1.2. Synthesis of dopamine melanin nanospheres
Two milliliters of ammonium hydroxide (28–30%) was added to a mixed solution containing 40 mL ethanol and 90 mL DI water. The solution was magnetically stirred at 30oC for 30 minutes. 0.5 g of dopamine hydrochloride in 10 mL DI water was dropwise added into the above solution. The color of the solution immediately turned to pale yellow and gradually become dark brown. The solution was magnetically stirred for another 24 hours. The products were collected by centrifugation and purified three times with DI water.
2.2. Characterization of DM Nanoparticles
2.2.1. Physical characterizations
Scanning electron microscopy (SEM) images were acquired on a FEI Teneo field emission SEM. Transmission electron microscopy (TEM) was carried out on a FEI Tecnai 20 transmission electron microscope operating at an accelerating voltage of 200 kV. Particle size distribution was analyzed on a Malvern Zetasizer Nano ZS system. Fourier-transform infrared (FT-IR) spectra were recorded on a Nicolet iS10 FT-IR spectrometer.15 HNMR analysis was performed on a Varian Mercury Plus 400 system. Zeta potential was measured by Zetasizer Nano-ZS.
2.2.2. Radical scavenging assays
DPPH assay:
Free radical-scavenging capacity of DM nanoparticles was also measured by the DPPH scavenging method.16 The test was carried out in a 96 well plate using a total volume of 200 μL methanol containing 0.004 μg DPPH and a series of sample aliquots with the final concentration at 1, 0.5, 0.25, and 0.125 μg/mL. The measurements were done in triplicate. DPPH solutions at the same concentration without the tested samples were used as control. The 520-nm absorbance was read every 5 min for 30 min. Samples were analyzed in triplicate.
Peroxynitrite assay:
The peroxynitrite scavenging capacity was determined by Evans Blue bleaching assay with slight modifications. The reaction mixture contained 50 mM phosphate buffer (pH 7.4), 0.1 mM DTPA, 90 mM NaCl, 5 mM KCl, 12.5 μM Evans Blue, various concentrations of DM nanoparticles (10–400 μg/mL), and 1 mM peroxynitrite, and the final volume of the solution was 1 mL. The absorbance at 611 nm was measured 30 min later. The percentage of ONOO- that was scavenged was calculated by comparing the results with the blank samples. Gallic acid was used as the standard.
Superoxide radical assay:
The superoxide radical generated from the photo reduction of riboflavin was detected by NBT reduction.17 The reaction mixture in PBS (67 mM, pH 7.8) contained EDTA (0.1 M), 0.0015% NaCN, riboflavin (0.12 mM), NBT (1.5 mM) and various concentrations of DM nanoparticles (10–1000 μg/mL) and the total volume was 3 mL. The tubes were uniformly illuminated by light for 15 min. The optical density at 530 nm was measured before and after the illumination.
Hydroxyl radical assay:
The scavenging capacity against hydroxyl radicals was measured using the deoxyribose test-tube method with minor changes.15 All solutions used as freshly prepared. These include 200 μL of 2.8 mM 2-deoxy-2-ribose, 5 μL of DM nanoparticles (3.125 μg/mL - 400 μg/mL), 400 μL of 200 mM FeCl3, 1.04 mM EDTA, 200 μL H2O2 (1.0 mM), and 200 μL ascorbic acid (1.0 mM). The resulting mixture was incubated for 1 h at 37°C. 1.5 mL aqueous solution containing 2.8% TCA was added, and the solution was first kept at room temperature for 20 min and then at 90°C for 15 min. Afterwards, the solution was cooled down and the absorbance at 532 nm measured and compared.
2.3. Isolation and culture of chondrocytes
Primary chondrocytes were isolated from knee joint cartilage of 3-day-old Sprague-Dawley (SD) rats (Guangxi Medical University, Experimental Animal Center). Articular cartilage tissue was finely minced, rinsed with phosphate-buffered saline (PBS), and digested with 2mg/mL of collagenase II (Solarbio,USA) at 37 °C for 4 h after treatment with 0.25% trypsin/EDTA (Solarbio,USA) for 30 min. And chondrocytes were obtained from articular cartilage tissue and cultured in alpha-modified eagle’s medium (α-MEM, Gibco, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, USA) and 1% (v/v) penicillin/streptomycin (Solarbio, China). Cells were detached from the culture dish using 0.25% trypsin and passaged after reaching confluency. Passage 2 chondrocytes were used for further experiments. To induce OA Chondrocytes, Chondrocytes were cultured with 10 ng/mL of IL-1β for 24h. For therapeutic group, Chondrocytes were cultured with IL-1β and/or different concentration of DM nanoparticles (10 μg/ mL, 30 μg/mL and 60 μg/mL) for indicated time points.
2.4. Cytotoxicity assay
Chondrocytes were cultured in 96-well plates at the concentration of 104 cells/well and added 10 ng/mL of IL-1β (Gibco, USA) and/or 10 different concentrations of DM nanoparticles (ranging from 0 to 180 μg/mL) after 24 hours. MTT reagent (Gibco, USA) was added to culture medium after 24 hours of treatment with IL-1β and DM nanoparticles, cells were then incubated at 37 °C for 4 hours. Absorbance of culture medium was measured using a spectrophotometer (Thermo, USA) at a wavelength of 490 nm. The three best concentrations of DM nanoparticle (10 μg/mL, 30 μg/mL and 60 μg/mL) were selected for further experiments. The test is repeated three times.
2.5. Flow cytometry for detection of apoptosis
1×106 chondrocytes were seeded in a six-well plate and incubated with IL-1β and/or DM nanoparticles for 24 h. After that, cells were digested by trypsin without EDTA and collected by centrifugation. The resuspended chondrocytes were processed by using 10 μL of Annexin V-EGFP (enhanced green fluorescent protein, Key GEN Bio TECH, China) and 5 uL of Propidium Iodide (PI, Key GEN Bio TECH, China) in the dark for 20 minutes. The prepared samples were tested using a flow cytometer.
2.6. Live/dead assay
The chondrocytes incubated with 1 μM calcein-AM and 1 μM PI (Invitrogen, USA) in the dark at 37 °C for 5 minutes after washing three times with PBS. After a quick rinse in PBS, the image of live/death cells were acquired by laser scanning confocal microscopy (Nikon A1, Japan).
2.7. Confocal fluorescence assay
Autophagy and intracellular ROS level of chondrocytes was detected using Cyto-ID Autophagy Detection Kit (green, ENZO Life Science, USA) and MitoSox Red mitochondrial superoxide indicator (red, Eugene, USA), respectively. Cells were incubated with 10 ng/mL of IL-1β and/or 30 μg/mL DM Nanoparticles for indicated times, then the mixture of MitoSox Red, Cyto-ID Green Dye and Hoechst 33342 (green, ENZO Life Science, USA) were used to treat with cells at 37 °C for 15 minutes in the dark after cells were washing by PBS. Images were captured by confocal microscopy (Nikon A1, Japan) immediately after chondrocytes were washed with α-MEM. All the relative fluorescence intensity was measure by Image J (National Institutes of Health, NIH, USA).
2.8. Transmission electron microscopy (TEM)
TEM was used to observe the localization of DM nanoparticles and Autophagosome in chondrocytes. 30 μg/mL of DM-treated cells were collected and centrifuged at 1000 rpm for 3 min. The centrifuged cell agglomerates were fixed by 2.5% (v/v) glutaraldehyde for 24h and 1% (v/v) osmium tetroxide for 2 h to stabilize the organelles and phospholipid membranes of cell. The cells were dehydrated using a concentration gradient of alcohol (30%, 50%, 75%, 90%, 99%,100%) for 15 minutes each and embedded in resin after washing with PBS. Thin sections were sliced from embedded cells and stained with 2% aqueous uranyl acetate for 1h in the dark. Pictures were captured by a TEM (Hitachi H-7650, Japan).
2.9. Quantification of intracellular glycosaminoglycan (GAG)
The content of intracellular DNA was detected using Hoechst 33258 (Sigma, USA) as previous described.18 In briefly, cellular DNA was stained by Hoechst 33258 dye and absorbance was measured at 460 nm by using fluorescence spectrophotometer (Bio-Tek Instruments, USA). Calf thymus DNA was used as the standard. Intracellular GAG secretion was detected using 1,9-dimethylmethylene blue assay (DMMB, Sigma, USA) and the absorption at wavelength of 525 nm was recorded on fluorescence spectrophotometer (Bio-Tek Instruments, USA), and chondroitin sulfate (Sigma, USA) was used as a standard. Finally, intracellular GAG secretion was normalized to the DNA content of the chondrocytes and express as GAG/DNA.
2.10. CAT, GSH-Px and MDA activity assay
After 24h of treatment with three different concentrations of DM nanoparticles (10 μg/mL, 30 μg/mL and 60 μg/mL) and/or 10 ng/mL of IL-1β, the cultured medium of cells were collected and subjected to detection of antioxidant capacity by using Glutathione peroxidase (GSH-Px) and Catalase(CAT) kit (Nanjing Jiancheng Bioengineering Institute, China) and evaluation of oxidative stress level by using Malondialdehyde (MDA) assay kits (Nanjing Jiancheng Bioengineering Institute, China) according to the instructions. Then the absorbance at 450 nm was recorded using a Fluorescence microplate reader (Bio-Tek Instruments, USA). All samples were repeated in triplicate.
2.11. Intracellular ROS measurement
Chondrocytes ROS level were detected by using Reactive Oxygen Species Assay Kit (Beyotime Biotechnology, China), respectively. Cells were washed with serum-free medium and loaded fluorogenic probes 2’,7’-dichlorofluorescein diacetate (DCFH-DA). After incubation with 10 μM/L of DCFH-DA at 37 °C for 30 minutes, the cells were washed three times with serum-free medium. Subsequently, the DCFH- DA-loaded chondrocytes were divided into two aliquots and subjected to determination of relative fluorescent intensity by using a fluorescence spectrophotometer and measurement of ROS levels by using Flow cytometer, respectively. All samples were repeated in triplicate.
2.12. Intracellular RNS measurement
Chondrocytes RNS level were detected by using Reactive Nitrogen Species Assay Kit (BestBio, China). Cells were washed and suspended with serum-free medium and loaded fluorogenic probes BBoxiProbeTM R21F. After incubation with 10 μM/L of BBoxiProbeTM R21F at 37 °C for 30 minutes, the cells were washed three times with PBS. RNS levels were quantified by using a fluorescence microplate reader (Bio-Tek Instruments, USA) with excitation wavelength of 488 nm and emission wavelength of 516 nm. All samples were repeated in triplicate.
2.13. Quantitative real-time PCR
Total RNA was extracted from the chondrocytes using RNA extraction kit (Tiangen Biotech Co., Ltd., China) and digested with DNase to remove any contaminating genomic DNA in accordance with the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized using reverse transcription kit (Fermentas Company, USA). The qRT-PCR was performed using FastStart Universal SYBR Green Master (Roche) by quantitative PCR detection system (Realplex 4, Eppendorf Corporation). The primer sequences were listed in Table 1 (Supporting Information). Relative gene expression levels were calculated using the 2-ΔΔCT method and normalized to GAPDH. All samples were repeated in triplicate.
2.14. Western blotting
Protein extracts were obtained from chondrocytes by RIPA lysis buffer (BOSTER, China) with 1 mM phenylmethanesulfonyl fluoride (PMSF, BOSTER, China). 50 μg of protein extracts were subjected to electrophoresis on 10% SDS-PAGE gel, and transferred to a polyvinylidene fluoride (PVDF, Millipore, US) membrane after concentration of protein was detected by BCA protein assay kit (Beyotime, China). The membranes were incubated with specific antibodies, including anti-IL-6 (BOSTER, China, 1:200), anti-IL-1β (BOSTER, China, 1:200), anti-TNF-α(BOSTER, China, 1:200), anti-MMP-13 (BOSTER, China, 1:200), anti-COX-2 (BOSTER, China, 1:200), anti-iNOS (BOSTER, China, 1:200) anti-LC3 (BOSTER, China, 1:200), anti-ATG7 (BOSTER, China, 1:200), anti-Beclin-1 (BOSTER, China, 1:200) and anti-GAPDH ( BOSTER, China, 1:200) overnight at 4 °C. Subsequently, membranes were incubated with horseradish peroxidase-labelled secondary antibody (Invitrogen, USA) for 2 hours after washing. The signals were visualized using Odyssey Infrared Imaging System (LI-COR Biotechnology, USA).
2.15. OA model establishment and treatment in vivo
Thirty 7-week-old female SD rats were provided by the Experimental Animal Center of Guangxi Medical University. The use of animals was approved by Animal Ethics Committee from the Experimental Animal Center of Guangxi Medical University and all animal manipulations were carried out strictly according to the National Institutions of Health Guide for the Care and Use of Laboratory Animals. To obtained rat model of OA, we used surgically induced method. After anesthesia with 10% chloral hydrate, anterior cruciate ligament transection (ACLT) was performed on the right knee of rat, meanwhile, sham surgery was performed with the same skin incision on the left knee of rat. Four weeks after the ACLT surgery, the OA rats were ready for test. The rats were randomly divided into five groups as following: normal group (receiving no treatment); OA group (i.a. injected with 0.2 mL of saline three times a week for 4 weeks); DM nanoparticles therapeutic group (i.a. injected with 0.2 mL of DM nanoparticles three times a week for 4 weeks). The therapeutic group was divided into 3 subgroups base on different concentration of DM nanoparticles (10 μg/mL, 30 μg/mL and 60 μg/mL), which were represented by DM+10, DM+30 and DM+60, respectively.
After 4 weeks, the rats were sacrificed by overdose of chloral hydrate, and the cartilages of knee joint were harvested for further investigation. The severity of OA lesions was evaluated based on integer score for cartilage structural changes (score range 0–6, 0= normal cartilage structure and 5=erosion of the cartilage down to the subchondral bone), cellular changes (score range 0–3), H&E and Safranin-O fast green staining (score range 0–4) and tidemark integrity (scored rang 0–1).
2.16. ROS detection of articular cartilage
50 mg of fresh cartilage tissue from knee joint was made into homogenate with 1 mL of buffer, and homogenate was using and centrifuged at 4 °C for 10 minutes. 190 μL of supernatant was collected and incubated with 10 μL of BBcellProbeTM O11 ROS probe (BestBio, China) in a 96-well plate at 37 °C in the dark for 30 minutes. ROS levels were quantified by fluorescence microplate reader (Bio-Tek Instruments, USA) with excitation wavelength of 488 nm and emission wavelength of 530 nm. All samples were repeated in triplicate.
2.17. RNS detection of articular cartilage
Fifty mg of fresh cartilage tissue from knee joint was made into homogenate with 1 mL of buffer, and homogenate was using and centrifuged at 4 °C for 10 minutes. 190 μL of supernatant was collected and incubated with 10 μL of BBcellProbeTM O52F RNS probe (BestBio, China) in a 96-well plate at 37 °C in the dark for 30 minutes. RNS levels were quantified by fluorescence microplate reader (Bio-Tek Instruments, USA) with excitation wavelength of 488 nm and emission wavelength of 516 nm. All samples were repeated in triplicate.
2.18. Histological staining
The knee tissues from rat were fixed in 4% paraformaldehyde for 48hours. After decalcification in 10% (w/v) Tris-EDTA for 4 weeks on a shaker, the fixed knee tissues were embedded in paraffin. The embedded tissues were then cut into 5μm frontal sagittal sections. Slides of femur and tibia were stained with Hematoxylin and Eosin (H&E) (Solarbio, China) and Safranin-O fast green (Solarbio, China). The morphological manifestations of cartilage and subchondral bone were observed in a double-blind manner using a microscope (Olympus, Japan), and cartilage degeneration of knee joint was evaluated using OA Research Society International scoring system (OARSI) as described previously.19
2.19. Immunohistochemical analysis
Immunohistochemical staining was performed to detect protein expression of MMP-13(BOSTER, China) in articular cartilage. Briefly, the slices were incubated with primary antibody for MMP-13 (1:100) (BOSTER, China, 1:200) at 4°C overnight after deparaffinage of paraffin-embedded sections. After extensive washes with PBS, the sections were incubated successively with the second antibody (ZSGb Bio, China) for 15 minutes and biotin-labeled horse radish peroxidase (ZSGb Bio, China) for 15 minutes. Subsequently, the sections were counter-stained with hematoxylin (Solarbio, China) after processing with a 3, −3’-diaminobenzidine tetrahydrochloride (DAB) kit (BOSTER, China) according to the protocol. Neutral resin sealed-slices were prepared for observation and pictures were captured by an Inverted microscope (Olympus, Japan).
2.20. Statistical Analysis
All tests are performed at least triplicate. Statistical analysis of all data (mean ± S.D, n = biological replicates) was analyzed by SPSS 64.0 (SPSS Inc., Chicago, Illinois, USA). Comparison between OA and treatment groups was examined by one-way analysis of variance (ANOVA). P<0.05 was considered statistically significant. Bars with different letters are significantly different from each other at P<0.05, and those with the same letter exhibit no significant difference.
3. Results
3.1. DM nanoparticles synthesis and characterizations
DM nanoparticles were synthesized following a published protocol with minor modifications.12 Briefly, we dissolved dopamine hydrochloride in an ethanol/water mixed solution and added into the solution ammonium hydroxide. After 24 h reaction in the open air, the nanoparticle products were collected by centrifugation and purified with water. Scanning emission microscopy (SEM) and transmission emission microscopy (TEM) showed that the products were uniform nanospheres, with an average diameter of ~112.5 nm (Figure 1a, b). Dynamic light scattering (DLS) found that the hydrodynamic size of the nanoparticles was ~230 nm (Figure 1c). Zeta potential analysis showed that the surface of the nanoparticles was negatively charged (−13.5 mV, Figure 1d). This is attributed to the multiple catechol groups on the nanoparticle surface, which was confirmed by FR-IR (Figure S1a). No benzene ring 1H atom was detected in DM nanoparticles by 1H NMR (Figure S1b), suggesting successful polymerization.
Figure 1.
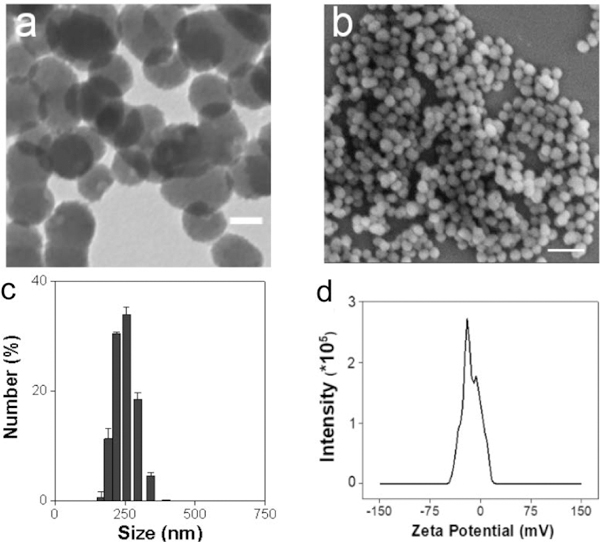
Physical characterizations of DM nanoparticles. (a) TEM of DM nanoparticles. (b) SEM of DM nanoparticles. (c) DLS of DM nanoparticles. (d) Zeta potential of DM nanoparticles.
We then analyzed the capacity of DM nanoparticles for scavenging radicals (Figure 2). We first assessed free radical scavenging by DPPH assay. Our studies showed effective and concentration dependent DPPH suppression (Figure 2a). Compared with PBS control, DM nanoparticles (80 μg/mL) inhibited the DPPH fluorescence by 64%. Similarly, Evans Blue bleaching, NBT reduction and deoxyribose test-tube method assays showed significant sequestration of peroxynitrite, superoxide, and hydroxyl radicals (Figure 2b, c, d). These results confirm the capacity of melanin nanoparticles to efficiently scavenge a broad range of ROS and RNS.
Figure 2.
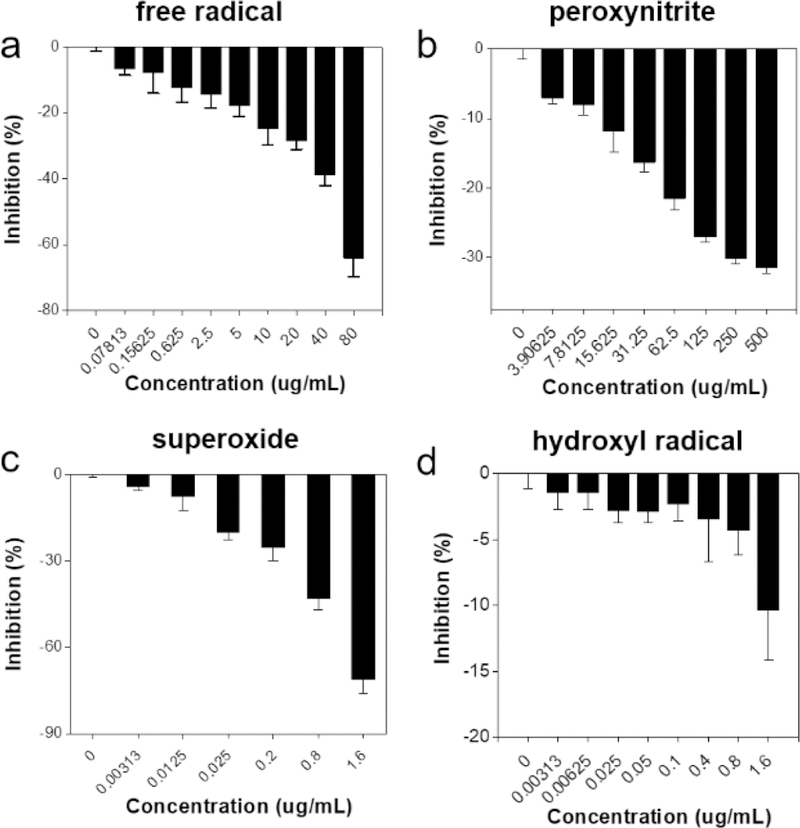
DM nanoparticles as a radical scavenger. (a) Free radical inhibition by DM nanoparticles. (b) The capacity of DM nanoparticles to scavenge peroxynitrite radicals. (c) Superoxides inhibition by DM nanoparticles. (d) The capacity of DM nanoparticles to scavenge hydroxyl radicals.
3.2. Anti-inflammatory and chondro-protective effects of DM nanoparticles on IL-1β-induced chondrocytes
We then studied the chondro-protective effects of DM nanoparticles. This was investigated with rat chondrocytes with IL-1β. As shown in Figure 3a, DM nanoparticles showed no cytotoxicity below 120 μg/mL but rather increased the chondrocytes viability. In particular, DM nanoparticles of 10, 30 and 60 μg/mL significantly promoted cell growth compared with the IL-1β control (P<0.001). Thus, DM nanoparticles of 10, 30 and 60 μg/mL were chosen for further investigation.
Figure 3.
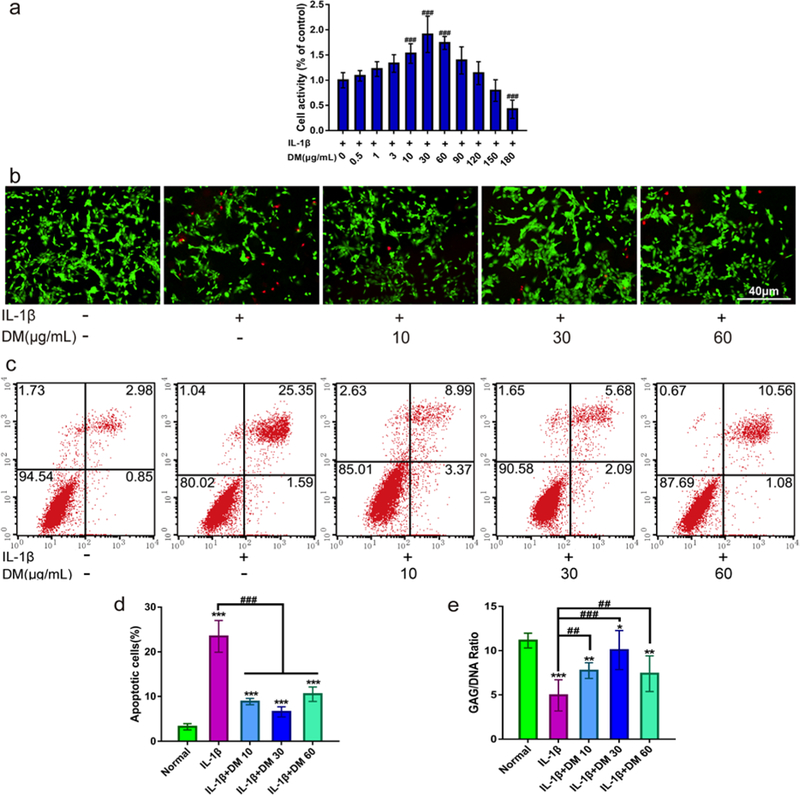
Chondro-protective effects of DM nanoparticles on IL-1β-induced chondrocytes. MTT assay was to detect the cytotoxicity of DM nanoparticles. (Control: only with 10ng/mL IL-1β). (b-e) Chondrocytes were treated with IL-1β (10 ng/mL) and/or various concentrations of DM nanoparticles (10, 30, 60 μg/mL) for 24 hours. (b) FDA//PI stained for cell viability. (c) Flow cytometry for cell apoptosis. (d) Quantitative flow cytometry for apoptosis. (e) Quantification of matrix production of GAG (n=6) for cell proliferation. Normal (without IL-1β); IL-1β (with 10ng/mL IL-1β); IL-1β +DM 10 (with 10ng/mL IL-1β and 10 μg/ml DM nanoparticles); IL-1β+DM 30 (with 10ng/mL IL-1β and 30 μg/ml DM nanoparticles); IL-1β+DM 60 (with 10 ng/mL IL-1β and 60 μg/ml DM nanoparticles). Values are presented as means ± SD, n=6. *, P <0.05; **, P <0.01; ***, P <0.001, relative to the normal group; #, P <0.05; ##, P <0.01; ###, P <0.001, relative to the IL-1β group. Scale bar, 40 μm.
This chondro-protective effect was confirmed by FDA/PI live/dead assay. As shown in Figure 3b, a significantly reduced number of live cells (green) and an increased number of dead cells (red) were found in chondrocytes treated with IL-1β. This toxicity was largely inhibited when DM nanoparticles were co-incubated, with 30 μg/ml DM showing the most prominent viability improvement. The cyto-protective effects of DM nanoparticles were further analyzed by flow cytometry. As shown in Figure 3c, d, IL-1β induced remarkable chondrocyte death with the apoptosis level increased by 7.51 fold relative to the PBS control. On the contrary, co-incubation with DM nanoparticles greatly attenuated IL-1β induced cell apoptosis, decreasing the population by 64.50%, 77.59% and 58.34%, respectively, when the DM concentration was 10, 30, 60 μg/mL.
During cartilage development, GAGs play an important role in the integrity of the cartilage matrix. Loss of GAG is a hallmark for early-stage OA.20 To evaluate the impact of DM nanoparticles on GAG expression, we used DMMB assay. As shown in Figure 3e, a significant loss of GAGs in chondrocytes was induced by IL-1β (up to 58.39%). However, DM nanoparticles rescued IL-1β induced GAG loss, restoring its contents by 50.98 %, 56.86% and 47.06% for 10, 30 and 60 μg/mL DM nanoparticles respectively. 30 μg/ml DM in particular promoted the GAG production to nearly a normal level.
3.3. Protective effects of DM nanoparticles on IL-1β-induced inflammation
In order to further explore the effects of DM nanoparticles on mRNA levels of inflammatory factors such as IL-6, IL-1β, TNF-α, MMP-13, COX-2 and iNOS, chondrocytes pretreated with 10 ng/ml IL-1β were cultured with or without DM nanoparticles of different concentrations for 24 hours. As shown in Figure 4a, the expression of inflammatory markers was remarkably elevated after IL-1β treatment. As a comparison, DM nanoparticles led to significant decreased regulation of all tested mRNA. Particularly, 30 μg/ml DM nanoparticles induced the most prominent decline, with all inflammatory factors approximating normal chondrocytes. This was further confirmed by Western blot analysis (Figure 4b), which showed that DM nanoparticles at 30 μg/mL greatly inhibited IL-1β induced upregulation of the inflammatory factors at the molecular levels.
Figure 4.
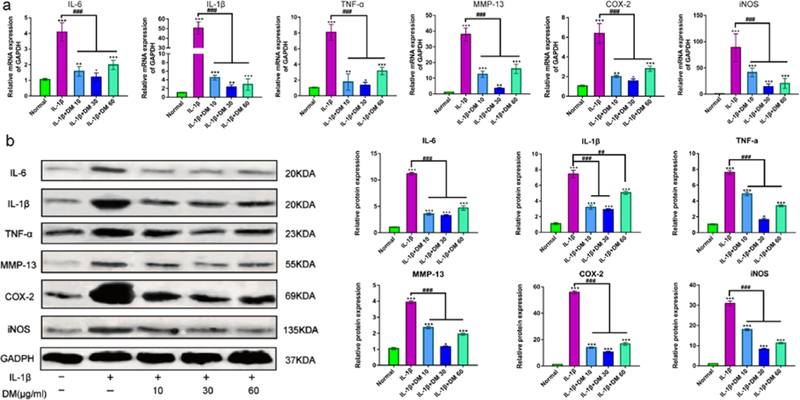
Effect of DM nanoparticles on the treatment of OA. (a) QRT-PCR was used to analyze the gene expression levels of IL-1β, TNF-α, IL-6, MMP-13, COX-2 and iNOS in vitro. (b) Western Blot was used to analyze the protein expression of IL-1β, TNF-α, IL-6, MMP-13, COX-2 and iNOS. Values are presented as means ± SD, n=6. *, P <0.05; **, P <0.01; ***, P <0.001, relative to the normal group; #, P <0.05; ##, P <0.01; ###, P <0.001, relative to the IL-1β group.
3.4. DM nanoparticles suppressed IL-1β-induced free radicals
Intracellular ROS generation induced by IL-1β was investigated by flow cytometry. As shown in Figure 5a, b, increased ROS was produced in IL-1β-mediated chondrocytes over time. Nevertheless, administration of DM nanoparticles remarkably reduced the ROS production. Especially at the time point of 24 h, ROS was reduced greatly, close to the level of normal cells. Of note, there was a slight increase of ROS production ranged from 3 h to 6 h, followed by a drop afterwards. These results indicated DM nanoparticles have powerful ability to scavenge intracellular ROS.
Figure 5.
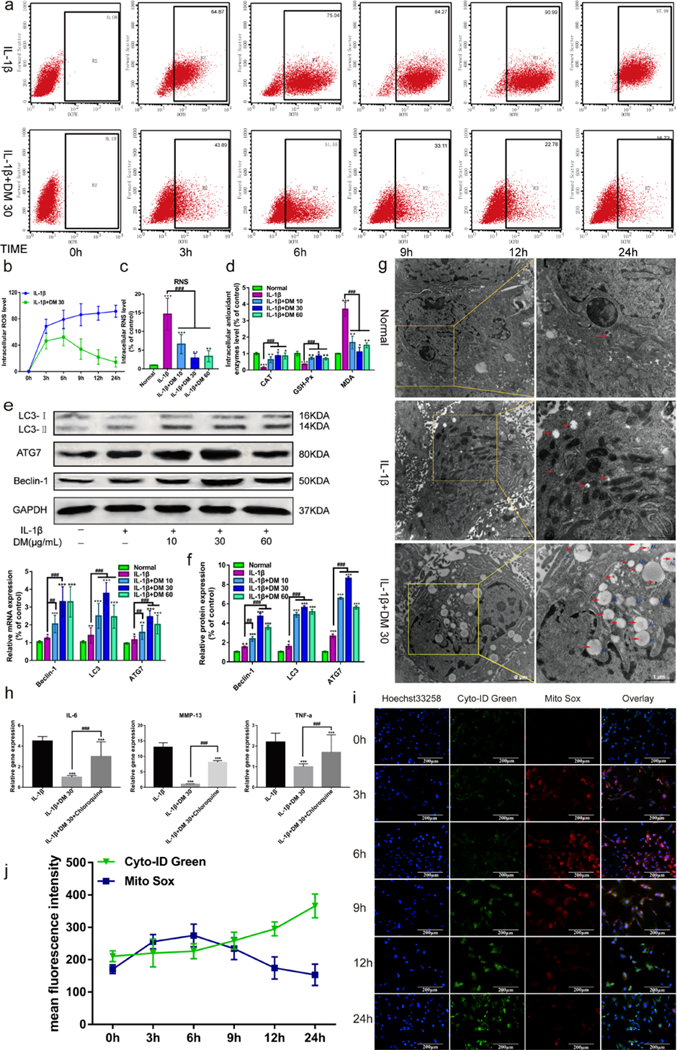
Effect of DM nanoparticles on the oxygen free radicals and autophagy. The chondrocytes stained with DCFH 30 minutes, followed by analyzed using flow cytometry. Upper-right corner region represents Chondrocyte ROS levels. (b) Quantifying the production of ROS in chondrocytes from flow cytometry. (c) Quantifying the production of RNS in chondrocytes. (d) Production of CAT, GSH-Px, MDA was assayed by a microplate fluorescence reader. (e) Western Blot was used to analyze the protein expression of LC3, ATG7 and Beclin-1. (f) QRT-PCR was used to analyze the gene expression of LC3, ATG7 and Beclin-1 in vitro. (g) TEM was used to analyze the uptake of DM nanoparticles and intracellular autophagy. (h) QRT-PCR was used to analyze the gene expression of MMP-13, IL-6 and TNF-α after Chondrocytes Treated with Autophagy Inhibitor Chloroquine. IL-1β (with 10 ng/mL IL-1β); IL-1β +DM (with 10 ng/mL IL-1β and 30 μg/ml DM nanoparticles); IL-1β+DM+Chloroquine (with 10 ng/mL IL-1β, 30 μg/ml DM nanoparticles and 1 μmol/L Chloroquine ). (i) After treated with DM nanoparticles (30 μg/mL) and IL-1β (10 ng/mL) for 0h, 3h, 6h, 9h, 12h and 24h, cell samples concurrently stained with Cyto-ID Green dye and MitoSox Red dye were analyzed by confocal microscopy. (j) The fluorescence intensity of chondrocytes was analyzed by Image J. Values are presented as means ± SD, n=6. *, P <0.05; **, P <0.01; ***, P <0.001, relative to the normal group; #, P <0.05; ##, P <0.01; ###, P <0.001, relative to the IL-1β group. Scale bars, 200 μM.
Next, we detected intracellular levels of RNS using BBoxiProbeTM R21 fluorescent probe. As shown in Figure 5c, IL-1β mediated the increase of RNS production, which was 12.43 times higher than that of the normal group. The treatment of DM greatly reduced the production of RNS in a dose-dependent manner. Administration of 30 μg/mL DM led to a decrease of 75.86 % in RNS levels compared with IL-1β group.
Further, we detected intracellular antioxidant enzymes CAT and GSH-Px, which are key indicators of oxidative stress. As shown in Figure 5d, the levels of CAT and GSH-Px in chondrocytes were notably down-regulated by IL-1β, while the downregulation was reversed by DM nanoparticles treatment. 30 μg/mL of DM nanoparticles, in particularly, upregulated the levels of CAT and GSH-Px the most in all the groups. MDA, which is one of compounds produced in the process of lipid peroxidation was also investigated. As shown in Figure 5d, MDA activity was dramatically activated in the IL-1β group compare with normal cells, but suppressed after DM treatment. Especially in the IL-1β+30 group, MDA level comes up to that in normal cells. These results revealed that DM nanoparticles act as potent ROS scavengers protect chondrocytes from damage by reducing excessive intracellular oxidative stress.
3.5. DM nanoparticles stimulate autophagy for chondrocyte protection
Autophagy as a protective process in the pathogenesis of OA has been demonstrated.21 It was also shown that enhanced autophagy is linked to consumption of excessive ROS and thus protection of chondrocytes.22 Quantitative RT-PCR analysis showed that IL-1β led to a marginal increase of specific autophagy markers, including LC3-Ⅱ, ATG7 and Beclin-1, compared with normal chondrocytes. By contrast, DM nanoparticles drastically elevated the expression of these autophagy markers in IL-1β-pretreated chondrocytes (Figure 5f). In particular, 30 μg/ml DM nanoparticles showed the most prominent induction. Western blot analysis also confirmed this, finding that LC3, ATG7 and Beclin-1 protein levels were significantly increased upon DM nanoparticles treatment (Figure 5e). Conversion of LC3-Ⅰ to LC3-Ⅱ is an initiating step in autophagy. It is noteworthy that the production of LC3-Ⅱ in OA chondrocytes was facilitated by DM nanoparticles, indicating autophagy activation. Further, a large number of autophagosomes were observed in chondrocytes treated with 30 μg/ml DM nanoparticles using TEM (Figure 5g).
For further validation, we co-incubated chondrocytes with chloroquine, an autophagy inhibitor. As shown in Figure 5h, quantitative RT-PCR results showed increased levels of inflammatory factors (MMP-13, IL-6, and TNF-α) when IL-1β-induced chondrocytes were incubated with DM nanoparticles in the presence of chloroquine. This indicated that autophagy inhibition attenuates the anti-inflammatory and chondro-protective effects of DM nanoparticle. Taken together, these findings demonstrate that DM nanoparticles may suppress IL-1β-induced oxidative stress by activating autophagy.
3.6. DM nanoparticles mediated ROS scavenging followed by autophagy activation
In order to verify the sequential events of DM nanoparticle-mediated autophagy and ROS scavenging in IL-1β-induced OA chondrocytes, we used immunofluorescence assay with cyto-ID Green probe for autophagy and MitoSox Red dye for ROS at different time point. At the first 6h, oxidative fluorescence (red) intensified gradually, while the intensity of autophagic fluorescence (green) remained weak (Figure 5i, j). Interestingly, the autophagic fluorescence was enhanced dramatically in a time-dependent manner between 6 to 24 h. In contrast, the intensity of oxidative fluorescence decreased sharply. These indicated that ROS was cleared while autophagy was activated.
3.7. Therapy studies in ACLT-induced rat OA models
To assess the effect of DM nanoparticles on pathological manifestations of OA, we established an OA model of rats by conducting an anterior cruciate ligament transaction (ACLT) surgery at the lower end of the femur and the upper end of the tibia in the knee-joint. And i.a. injection of DM nanoparticles with three different doses (10, 30, and 60 μg/mL, 0.1mL) was performed. Degeneration of cartilage was evaluated by macroscopic observation. As shown in Figure 6a, characteristics of OA, such as erosion and osteophyte formation was observed in the femoral condyles and tibial plateau in the ACLT surgery group (OA), while the normal cartilage was presented with a glistening, smooth surface without any defects and osteophyte formation (sham-operation group). However, 4-week injection of DM nanoparticles led to markedly reduced cartilage damage with the glistening cartilage surfaces resembling normal cartilage. Among all the treatment groups, 30 μg/mL DM nanoparticles showed the best performance. Consistent with the macroscopic observation, the macroscopic scores were ranked in the order of OA, DM 10, DM 60 and DM 30 (Figure 6b).
Figure 6.
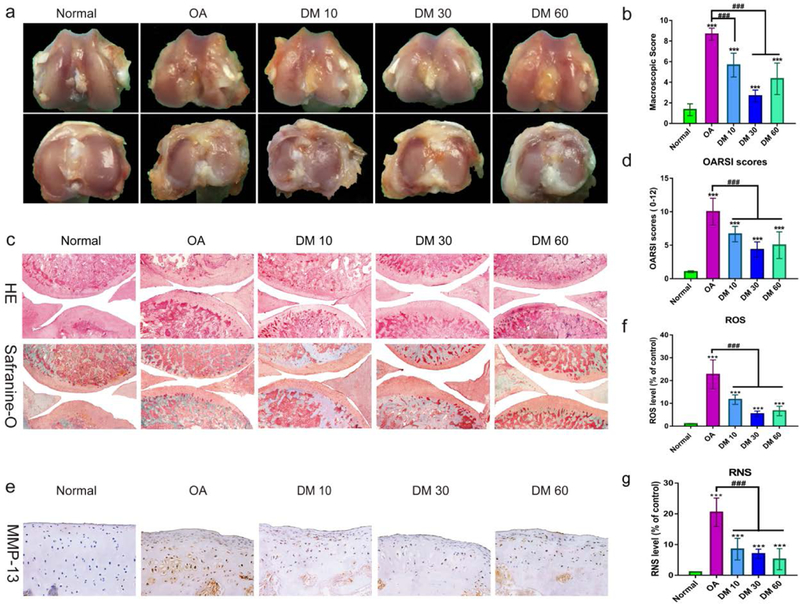
Effect of DM nanoparticles on ACLT-induced OA. SD rats were randomly divided into five groups: Normal group, OA group, DM 10 group, DM 30 group, DM 60 group. Normal group underwent sham operation, DM 10 group, DM 30 group and DM 60 group respectively received different concentrations of DM nanoparticles(10, 30, and 60 μg/mL), while OA group received with nothing in OA rats. All the samples were harvested after 8 weeks. (a) Macroscopic appearance. (b) Macroscopic scores of femoral condyles from normal and OA rats. (c) Histological analysis of OA was evaluated by Hematoxylin-Eosin staining and Safranin O staining. (d) OARSI score of articular cartilage was determined. (e, f) Quantification of the ROS production in articular cartilage. (g) Quantifying the production of RNS in articular cartilage. Values are presented as means ± SD. *, indicates P <0.05; **, P <0.01; ***, P <0.001, relative to the normal group. #, P <0.05; ##, P <0.01; ###, P <0.001, relative to the OA group.
Histopathological examination by H&E and Safranin-O fast green staining showed severe pathologic changes in the OA group, as evidenced by fissures and fibrillation, a loss of cartilage superficial zone and osteophyte proliferation (Figure 6c). On the contrary, DM nanoparticles significantly slowed down cartilage degeneration, manifested in a remarkable reduction in the severity of histologic lesions and an increased GAG content. The OARSI Score was used to assess cartilage degeneration and damage (Figure. 6d). Relative to the OA control, 10, 30 and 60 μg/mL DM nanoparticles reduced theOARSI score by 27.28%, 52.52% and 45.46 % respectively, which is consistent with the macroscopic findings.
As a marker of OA, MMP-13 is commonly used in the evaluation of OA. As shown in Figure 6e and Figure S2, intense positive staining of MMP-13 was observed in the cartilage layer of the OA group. As a comparison, much less positive staining was shown in DM nanoparticles groups, particularly the 30μg/mL group. In addition, we examined ROS production in the articular cartilage using BBcellProbeTM probe. As shown in Figure 6f, g, ROS and RNS levels were increased by 24.35 and 28.87 times, respectively, in OA group. With DM nanoparticles therapy, on the other hand, the ROS and RNS levels in cartilage were dramatically reduced. In summary, these results suggest that DM nanoparticles could effectively scavenge free radicals in the articular cartilage and as a result alleviate pathological progression of OA.
4. Discussion
Under physiological conditions, minute concentrations of ROS and RNS have important roles in various aspects of intracellular signaling and metabolic regulation.23 Excessive ROS and RNS produced by injured chondrocytes accelerate cell death and cartilage degradation in OA progression.24–28 ROS scavengers have been demonstrated as effective OA therapeutics, but many are associated with poor biocompatibility, low degradation, or short drug retention. In this study, we are the first to explore DM nanoparticles as a safe therapeutic for OA treatment. We found that DM nanoparticles prevented elevated oxidative stress and autophagy, both contributing to inhibited cartilage remodeling in inflammatory diseases.
It has been reported that high density of extracellular matrix (ECM) of avascular cartilage hinders drug penetration and that nanoparticles with a size between 100 to 300 nm are favorable with regard to entering the cartilage ECM while staying long in the joint.29 DM nanoparticles have an average diameter of 112.5 nm (Figure 1a, b) and a hydrodynamic size of 230 nm (Figure 1c), and are well within this optimal nanoparticle size range. The nanoparticles’ colloidal stability comes from their negatively charged surface (−13.5 mV, Figure 1d).30
In this study, we found that DM nanoparticles exhibited the capacity of scavenging multiple types of ROS and RNS (Figure 2), which is beneficial for protecting chondrocytes from oxidative stress, inflammatory reactions, and cartilage degeneration (Figure 3, 4, 6). RNS and ROS play an essential role in OA development and progression,3 and are potential therapy targets. NO inhibition may maintain the articular phenotype by maintaining Dickkopf-1 (DKK1) and Frizzled related protein (FRZB)31 and suppressing the release of NO, ROS, PGE2 and MMP-13, thus attenuating chondrocyte death in OA.32 Salerno et al. reported that iNOS knockout mice have lower rates of OA compared with the wild type.33 DM nanoparticles serve as a trap for potentially harmful radicals because they contains quinone residues34 that can react with ROS.35 DM nanoparticles as a dopamine derivative can also react with RNS and thus inhibiting their nitrating effects.36 This includes reduced NO production and protection against LPS-induced cell damage.37 Previous studies showed that increased levels of ROS and RNS, can modify the Cys residues at the DNA-binding sites and influence the binding between DNA and transcription factors,38 leading to proteoglycan loss in chondrocytes39–41 and thus cartilage degradation.42
Interestingly, we found that DM nanoparticles exert anti-inflammatory and chondro-protective effects by autophagy activation, which was demonstrated by the increased production of LC3-Ⅱ/LC3-Ⅰ, ATG7 and Beclin-1 and induced autophagic vesicles formation in chondrocytes (Figure 5e, f, g). Treatment of an autophagy inhibitor, chloroquine, reversed the anti-inflammatory effect of DM nanoparticles, evidenced by increased production of MMP-13, IL-1β and TNF-α (Figure 5h). These results suggest that ROS and RNS scavenging effects of DM nanoparticles might be related to its autophagy activation (Figure S3). Autophagy is a vital cellular homeostatic mechanism for eliminating damaged cellular organelles and macromolecules.43 It was found that autophagic activity decreases with aging, meaning better cyto-protective effects in young cartilage44 to prevent damage caused by ROS and inflammatory cytokines.45, 46 Pharmacological activation of autophagy is wildly used in OA therapy.21, 47, 48 Over-expression of iNOS inhibits autophagosome formation49 and NO inhibits autophagy by downregulating JNK1 by S-nitrosylation and activating the mTOR complex1.50 As a ligand of DRD3 (dopamine receptor D3), dopamine could induce the DRD3 internalization and LC3B upregulation, activating autophagy51 through the PI3K/Akt and mTOR pathways.52
5. Conclusion
In conclusion, as a novel ROS and RNS scavengers, DM nanoparticles alleviated the pathological process of OA and cartilage degeneration. DM nanoparticles exert excellent anti-inflammatory effects by inhibiting intracellular ROS and RNS generation and activating antioxidant enzymes by autophagy, thereby inhibiting cartilage degradation and OA progression. This study provides a novel insight for therapy of OA by nanoparticulated ROS and RNS scavengers.
Supplementary Material
Acknowledgements
This work was supported by the National key research and development program of China (2018YFC1105900). We thank the funding support by the National Science Foundation (CAREER grant no. NSF1552617 to J.X.), the National Institute of Biomedical Imaging and Bioengineering (grant no. R01EB022596 to J.X.), the National Heart, Lung, and Blood Institute (grant no. R01NS093314). We also thank the Distinguished Young Scholars Program of Guangxi Medical University. Gang Zhong, Xueyuan Yang, and Xianfang Jiang contributed equally to this work.
References
- 1.Varela-Eirin M, Loureiro J, Fonseca E, Corrochano S, Caeiro JR, Collado M and Mayan MD, Ageing Res. Rev, 2018, 42, 56–71. [DOI] [PubMed] [Google Scholar]
- 2.Roman-Blas JA, Bizzi E, Largo R, Migliore A and Herrero-Beaumont G, Expert Opin. Pharmaco, 2016, 17, 1745–1756. [DOI] [PubMed] [Google Scholar]
- 3.Mazzetti I, Grigolo B, Pulsatelli L, Dolzani P, Silvestri T, Roseti L, Meliconi R and Facchini A, Clin. Sci. (Lond), 2001, 101, 593–599. [PubMed] [Google Scholar]
- 4.Lepetsos P and Papavassiliou AG, BBA-Mol. Basis Dis, 2016, 1862, 576–591. [DOI] [PubMed] [Google Scholar]
- 5.Erturk C, Altay MA, Selek S and Kocyigit A, Scand J. Clin. Lab Invest, 2012, 72, 433–439. [DOI] [PubMed] [Google Scholar]
- 6.Altindag O, Erel O, Aksoy N, Selek S, Celik H and Karaoglanoglu M, Rheumatol Int, 2007, 27, 339–344. [DOI] [PubMed] [Google Scholar]
- 7.Lim HD, Kim YS, Ko SH, Yoon IJ, Cho SG, Chun YH, Choi BJ and Kim EC, J Pineal Res, 2012, 53, 225–237. [DOI] [PubMed] [Google Scholar]
- 8.Schreck R, Rieber P and Baeuerle PA, EMBO J, 1991, 10, 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim MJ, Lee Y, Jon S and Lee DY, Biomaterials, 2017, 133, 242–252. [DOI] [PubMed] [Google Scholar]
- 10.Yin T, Yang L, Liu Y, Zhou X, Sun J and Liu J, Acta Biomater, 2015, 25, 172–183. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Zhang J, Chen Y, Kawazoe N and Chen G, ACS Appl. Mater. Inter, 2017, 9, 35683–35692. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Ai K, Liu J, Deng M, He Y and Lu L, Adv. Mater, 2013, 25, 1353–1359. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Ai K, Ji X, Askhatova D, Du R, Lu L and Shi J, Journal of the American Chemical Society, 2017, 139, 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rageh MM, El-Gebaly RH, Abou-Shady H and Amin DG, Mol Cell Biochem, 2015, 399, 59–69. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell B, Gutteridge JM and Aruoma OI, Anal Biochem, 1987, 165, 215–219. [DOI] [PubMed] [Google Scholar]
- 16.Brand-Williams W, Cuvelier M and Berset C, Lwt Food Sci Technol, 1995, 28, 25–30. [Google Scholar]
- 17.McCord JM and Fridovich I, J Biol Chem, 1969, 244, 6049–6055. [PubMed] [Google Scholar]
- 18.Meng F, He A, Zhang Z, Zhang Z, Lin Z, Yang Z, Long Y, Wu G, Kang Y and Liao W, J Biomed Mater Res A, 2014, 102, 2725–2735. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Chen LK, Zhu DC, Zhang GR, Guo C, Qi YY and Ouyang HW, Tissue Eng Part A, 2010, 16, 1621–1632. [DOI] [PubMed] [Google Scholar]
- 20.Adams ME, Billingham ME and Muir H, Arthritis Rheum, 1983, 26, 69–76. [DOI] [PubMed] [Google Scholar]
- 21.Mi B, Wang J, Liu Y, Liu J, Hu L, Panayi AC, Liu G and Zhou W, Front Pharmacol, 2018, 9, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goutas A, Syrrou C, Papathanasiou I, Tsezou A and Trachana V, Free Radic Biol Med, 2018, 126, 122–132. [DOI] [PubMed] [Google Scholar]
- 23.Koskenkorva-Frank TS, Weiss G, Koppenol WH and Burckhardt S, Free Radic Biol Med, 2013, 65, 1174–1194. [DOI] [PubMed] [Google Scholar]
- 24.Henrotin YE, Bruckner P and Pujol JP, Osteoarthritis Cartilage, 2003, 11, 747–755. [DOI] [PubMed] [Google Scholar]
- 25.Nemirovskiy OV, Radabaugh MR, Aggarwal P, Funckes-Shippy CL, Mnich SJ, Meyer DM, Sunyer T, Rodney Mathews W and Misko TP, Nitric Oxide, 2009, 20, 150–156. [DOI] [PubMed] [Google Scholar]
- 26.Ersoy Y, Ozerol E, Baysal O, Temel I, MacWalter RS, Meral U and Altay ZE, Ann Rheum Dis, 2002, 61, 76–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakurai H, Kohsaka H, Liu MF, Higashiyama H, Hirata Y, Kanno K, Saito I and Miyasaka N, J Clin Invest, 1995, 96, 2357–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karan A, Karan MA, Vural P, Erten N, Tascioglu C, Aksoy C, Canbaz M and Oncel A, Clin Rheumatol, 2003, 22, 397–399. [DOI] [PubMed] [Google Scholar]
- 29.Jiang T, Kan HM, Rajpura K, Carbone EJ, Li Y and Lo KW, J Nanosci Nanotechnol, 2018, 18, 2310–2317. [DOI] [PubMed] [Google Scholar]
- 30.Jain A, Mishra SK, Vuddanda PR, Singh SK, Singh R and Singh S, Nanomedicine, 2014, 10, 1031–1040. [DOI] [PubMed] [Google Scholar]
- 31.Zhong L, Schivo S, Huang X, Leijten J, Karperien M and Post JN, Int J Mol Sci, 2017, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bentz M, Zaouter C, Shi Q, Fahmi H, Moldovan F, Fernandes JC and Benderdour M, J Cell Biochem, 2012, 113, 2256–2267. [DOI] [PubMed] [Google Scholar]
- 33.Salerno L, Sorrenti V, Di Giacomo C, Romeo G and Siracusa MA, Curr Pharm Des, 2002, 8, 177–200. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson ES, Clin Microbiol Rev, 2000, 13, 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Ai K, Ji X, Askhatova D, Du R, Lu L and Shi J, J Am Chem Soc, 2017, 139, 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S, Geddes TJ, Javitch JA and Kuhn DM, J Biol Chem, 2003, 278, 28736–28742. [DOI] [PubMed] [Google Scholar]
- 37.Yoshioka Y, Sugino Y, Tozawa A, Yamamuro A, Kasai A, Ishimaru Y and Maeda S, J Pharmacol Sci, 2016, 130, 51–59. [DOI] [PubMed] [Google Scholar]
- 38.O’Brian CA and Chu F, Free Radic Res, 2005, 39, 471–480. [DOI] [PubMed] [Google Scholar]
- 39.Starkman BG, Cravero JD, Delcarlo M and Loeser RF, Biochem J, 2005, 389, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin W, Park JI and Loeser RF, J Biol Chem, 2009, 284, 31972–31981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole LB, Karplus PA and Claiborne A, Annu Rev Pharmacol Toxicol, 2004, 44, 325–347. [DOI] [PubMed] [Google Scholar]
- 42.Hosseinzadeh A, Kamrava SK, Joghataei MT, Darabi R, Shakeri-Zadeh A, Shahriari M, Reiter RJ, Ghaznavi H and Mehrzadi S, J Pineal Res, 2016, 61, 411–425. [DOI] [PubMed] [Google Scholar]
- 43.Frudd K, Burgoyne T and Burgoyne JR, Nat Commun, 2018, 9, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang J, Wang W, Zhang H, Hu Y, Wang M and Yin Z, Int J Mol Med, 2013, 32, 1311–1318. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki H, Takayama K, Matsushita T, Ishida K, Kubo S, Matsumoto T, Fujita N, Oka S, Kurosaka M and Kuroda R, Arthritis Rheum, 2012, 64, 1920–1928. [DOI] [PubMed] [Google Scholar]
- 46.Shen C, Yan J, Erkocak OF, Zheng XF and Chen XD, Rheumatology (Oxford), 2014, 53, 1022–1033. [DOI] [PubMed] [Google Scholar]
- 47.Shen C, Cai GQ, Peng JP and Chen XD, Osteoarthritis Cartilage, 2015, 23, 2279–2287. [DOI] [PubMed] [Google Scholar]
- 48.Sun MM, Beier F and Pest MA, Curr Opin Rheumatol, 2017, 29, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales CR, Pedrozo Z, Lavandero S and Hill JA, Antioxid Redox Signal, 2014, 20, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, Garcia-Arencibia M, Rose C, Luo S, Underwood BR, Kroemer G, O’Kane CJ and Rubinsztein DC, Mol Cell, 2011, 43, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, Ji X, Liu J, Li Z and Zhang X, Int J Mol Sci, 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gimenez-Xavier P, Francisco R, Santidrian AF, Gil J and Ambrosio S, Neurotoxicology, 2009, 30, 658–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


