Abstract
Lactation persistency (LP), defined as the ability of a cow to maintain milk production at a high level after milk peak, is an important phenotype for the dairy industry. In this study, we used a targeted genotyping approach to scan for potentially functional single nucleotide polymorphisms (SNPs) within 57 potential candidate genes derived from our previous genome wide association study on LP and from the literature. A total of 175,490 SNPs were annotated within 10-kb flanking regions of the selected candidate genes. After applying several filtering steps, a total of 105 SNPs were retained for genotyping using target genotyping arrays. SNP association analyses were performed in 1,231 Holstein cows with 69 polymorphic SNPs using the univariate liner mixed model with polygenic effects using DMU package. Six SNPs including rs43770847, rs208794152, and rs208332214 in ADRM1; rs209443540 in C5orf34; rs378943586 in DDX11; and rs385640152 in GHR were suggestively significantly associated with LP based on additive effects and associations with 4 of them (rs43770847, rs208794152, rs208332214, and rs209443540) were based on dominance effects at P < 0.05. However, none of the associations remained significant at false discovery rate adjusted P (FDR) < 0.05. The additive variances explained by each suggestively significantly associated SNP ranged from 0.15% (rs43770847 in ADRM1) to 5.69% (rs209443540 in C5orf34), suggesting that these SNPs might be used in genetic selection for enhanced LP. The percentage of phenotypic variance explained by dominance effect ranged from 0.24% to 1.35% which suggests that genetic selection for enhanced LP might be more efficient by inclusion of dominance effects. Overall, this study identified several potentially functional variants that might be useful for selection programs for higher LP. Finally, a combination of identification of potentially functional variants followed by targeted genotyping and association analysis is a cost-effective approach for increasing the power of genetic association studies.
Keywords: Candidate genes, Canadian Holstein cows, lactation persistency, SNPs
Introduction
Lactation persistency (LP) refers to the ability of cows to maintain milk production at a high level after milk peak (Swalve, 1995; Dekkers et al., 1998). The lactation curve has a direct impact on the total milk yield of a cow and LP is thus an important economic trait for the dairy industry. Lactation persistency is influenced by many factors including management practice, disease, lactation number, physiological status, and genetics (Dekkers et al., 1998; Stefanon et al., 2002; Muir et al., 2004). Genetic factors contribute substantially to the variation in LP and reported heritability estimates for LP range from 0.10 to 0.34 (Swalve, 1995; Jakobsen et al., 2002; Muir et al., 2004; Cole and VanRaden, 2006). Although a number of studies have detected quantitative trait loci (QTL) and candidate genes related to LP (Harder et al., 2006; Sharma et al., 2006; Kolbehdari et al., 2009; Verbyla and Verbyla, 2009; Pryce et al., 2010; Do et al., 2017; Nayeri et al., 2017; Bissonnette, 2018), the biology of LP is still poorly understood. Using a candidate gene approach, single nucleotide polymorphisms (SNPs) in the TLR4, IL10, and SPP1 genes were found to be associated with LP in Canadian Holstein cattle (Sharma et al., 2006; Verschoor et al., 2011; Bissonnette, 2018). Kolbehdari et al. (2009) performed a genome-wide scan on a small set of samples (462 bulls and 1,536 SNP markers) and identified only one genome-wide significant SNP (rs41634436) on Bovine chromosome (BTA) 15 for LP and suggested CD44 (harbors the associated SNP) as a positional candidate gene for LP. Using a larger population and high density SNP chip (3,729 cows and 602,095 SNPs), Nayeri et al. (2017) identified several genomic regions with highly significant SNPs for LP on BTA6, 13, 20, and 27 and suggested MYT1, SLC2A4RG, and SLC17A9 (genes on BTA13) and THRB (gene on BTA27) as potential novel candidate genes for LP. Recently, we performed a genome-wide association study (GWAS) on 3,796 cows using BovineSNP50 Genotyping BeadChip and identified 2 important regions containing multiple significantly associated SNPs for LP on BTA5 (106 to 108 Mb) and BTA20 (29.3 to 31.3 Mb) (Do et al., 2017).
Several studies have identified the targeted genotyping approach as a powerful and cost-effective approach to enhance the identification of variants for complex traits (Li et al., 2016b; Cirera et al., 2018). For the present study, we adapted a targeted array genotyping approach to identify new functional SNPs for LP in potential candidate genes identified in our GWAS (Do et al., 2017) and from the literature (Sharma et al., 2006; Kolbehdari et al., 2009; Verschoor et al., 2011; Nayeri et al., 2017).
Materials and Methods
Animal Resource, DNA Sampling, and Isolation
A total of 1,500 Holstein cows including 340 cows from our previous study (Do et al., 2017) were used. Cows were from 27 herds in Quebec. Official estimated breeding values (EBVs) for LP were provided by Canadian Dairy Network (Guelph, ON, Canada, www.cdn.ca). Deregressed EBVs (deEBVs) of LP for cows were calculated according to the deregression procedure of Garrick et al. (2009). In brief, deEBV adjusts for ancestral information; therefore, only own and descendant’s information was retained for each animal. Moreover, the deregression procedure also removes shrinkage associated with EBV, thereby avoiding the double counting problem encountered when EBV is used as a response variable in association studies (Garrick et al., 2009). The weight factors for dEBV of the ith animal were estimated as
where c is the part of the genetic variance that is assumed not to be explained by markers (c = 0.1), h2 is the heritability of the trait, and ri2 is the reliability of the deEBV of the ith animal.
The population mean of deEBVs of LP was 99.10 and standard deviation was 9.92. Moreover, the pedigree of cows was traced back to 5 generations which included 5,421 animals.
Milk sampling (50 mL from each cow) was coordinated by Valacta laboratories (Sainte-Anne-de-Bellevue, QC, Canada, www.valacta.com). Samples were centrifuged at 4,500 × g for 20 min at 4 °C. After centrifugation, the topmost layer (fat) and whey (middle layer) were discarded and the cells at the bottom were resuspended in 1 × PBS (phosphate buffered saline) and homogenized by vortexing for about 30 s followed by centrifugation at 4,500 × g for 20 min at 4 °C. The supernatant was discarded and milk cells transferred into 2 mL Eppendorf tube and stored at −20 °C. The DNA was isolated from milk cells using Nucleospin Blood kit (Macherey-Nagel GmbH & Co., KG Düren, Germany) adapted for milk cells (Ibeagha-Awemu et al., 2014). For each sample, 30 µL of DNA (5 ng/µL) was used for SNP genotyping.
In silico SNP Selection for Genotyping
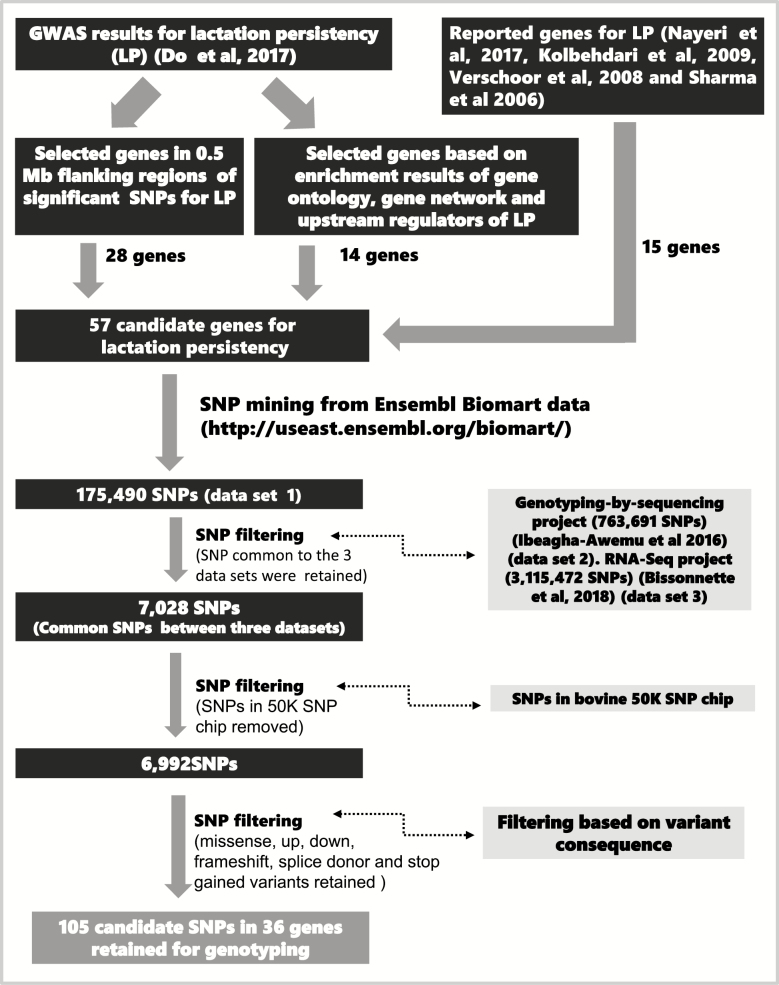
To select potential candidate SNPs for genotyping, we integrated SNPs for LP in potential candidate genes identified in our GWAS (Do et al., 2017) and published data on candidate genes for LP (Figure 1). Based on the GWAS and due to linkage disequilibrium of SNPs in the population, 28 positional candidate genes within 0.5 Mb flanking regions of SNPs significantly associated with LP, 4 genes most enriched (P < 0.05) in biological processes gene ontology terms for LP, 5 hub genes in molecular networks enriched for LP, and 5 most enriched relevant upstream regulators of LP were selected (Supplementary Table S1a). Moreover, 15 genes (ADRM1, AGPAT6, CD44, DGAT1, IL10, IRF2, ITGB5, LAMA5, MYT1, OSBPL2, PAPPA, SLC17A9, SLC2A4RG, THRB, and TLR4) were identified from the literature for their potential roles in LP (Supplementary Table S1a). In total, 57 genes were selected for querying SNPs from Ensembl Biomart database (Ensembl 83, Bos taurus UMD3.1, http://useast.ensembl.org/Bos_taurus/Info/Index, accessed in September, 2017). A total of 175,490 SNPs were identified within selected genes and their 10-Kb flanking upstream regions (Supplementary Table S1b). The upstream 10-Kb flanking region was selected to capture SNPs in the promoter region of genes. Predicted consequence of selected SNPs were obtained via Variant Effect Predictor (http://useast.ensembl.org/info/docs/tools/vep/index.html). Moreover, the potential effects of SNPs on protein function were determined with SIFT (Sorting Intolerant From Tolerant) score (Ng and Henikoff, 2003) via manual query of Ensembl database using the Cow Short Variants (ARS-UCD1.2) option (http://useast.ensembl.org/biomart/martview/ accessed in September, 2017). SIFT-predicted substitution scores less than 0.05 are considered deleterious and scores from 0.05 to 1 are considered tolerated (Ng and Henikoff, 2003).
Figure 1.
Schematic illustration of the procedure for SNP selection.
To ensure that selected SNPs are present in Canadian Holstein populations, the 175,490 SNPs (data set 1) were queried against SNPs obtained from Canadian Holstein cows by the method of genotyping-by-sequencing (GBS) (763,691 SNPs) (data set 2) (Ibeagha-Awemu et al., 2016) and by RNA-sequencing (3,115,472 SNPs) (data set 3) (Bissonnette et al., 2018) (Figure 1). The GBS data were generated from 1,246 Canadian Holstein cows and detailed information on data processing is reported in Ibeagha-Awemu et al. (2016). A total of 7,028 SNPs common to the three data sets were retained for further analysis (Figure 1). To avoid genotyping a SNP twice, we removed SNPs that are present in the BovineSNP50 v3 BeadChip used in our previous GWAS (Do et al. 2017). A total of 6,992 SNPs were retained and further filtered based on their potential functional activity or variant consequence (Supplementary Table S1c). Finally, a total of 105 SNPs that are missense variants or located in regions with potential effects on post-transcriptional regulation (up- and down-stream untranslated region variants) as well as frameshift, splice donor, and stop gained mutations were prioritized for genotyping (Table 1, Figure 1). In silico prediction indicated that among the genotyped SNPs, 82 are missense variants including 15 SNPs with potential deleterious effects on protein variants (SFIT score < 0.05) while 67 might cause tolerated changes in protein variants (SFIT score ≥ 0.05) (Table 1).
Table 1.
Final list of 105 SNPs selected for genotyping from a total of 175,490 SNPs
| Genes | SNP | Chromosome | SNP position | Variant Consequence1 | SIFT score2 | SIFT prediction | Protein Allele (one letter code) | Protein allele (three letter code) |
|---|---|---|---|---|---|---|---|---|
| ADRM1 | rs208332214 | 13 | 55418416 | missense | 0 | deleterious | T/N | Thr/Asn |
| rs208566185 | 13 | 55427615 | missense | 0.84 | tolerated | Q/K | Gln / Lys | |
| rs208794152 | 13 | 55431510 | missense | 0.07 | tolerated | R/W | Arg /Trp | |
| rs209179785 | 13 | 55410932 | missense | 0.02 | deleterious | E/Q | Glu/ Gln | |
| rs211087002 | 13 | 55431496 | missense | 0.91 | tolerated | R/K | Arg/ Lys | |
| rs378318832 | 13 | 55407771 | missense | 1 | tolerated | S/P | Ser/Pro | |
| rs383024237 | 13 | 55407768 | missense | 1 | tolerated | C/R | Cys /Arg | |
| rs384785350 | 13 | 55407786 | missense | 0.82 | tolerated | S/A | Ser/Ala | |
| rs41696753 | 13 | 55424454 | missense | 0.04 | deleterious | R/Q | Arg/ Gln | |
| rs43770847 | 13 | 55415352 | missense | 0.29 | tolerated | S/G | Ser/ Gly | |
| rs446082190 | 13 | 55418674 | missense | 0.08 | tolerated | I/M | Ile/Met | |
| rs475020220 | 13 | 55421182 | missense | 0.01 | deleterious | K/T | Lys/Thr | |
| AGPAT6 | rs110454169 | 27 | 36212557 | 5_prime_UTR | ||||
| rs208675276 | 27 | 36212352 | 5_prime_UTR | |||||
| rs379137591 | 27 | 36212515 | 5_prime_UTR | |||||
| rs477793555 | 27 | 36212766 | missense | 0.06 | tolerated | K/N | Lys/Asn | |
| APOA1 | rs134430767 | 15 | 27932525 | missense | 0.05 | tolerated | V/A | Val/Ala |
| rs472309323 | 15 | 27932366 | missense | 0.48 | tolerated | V/A | Val/Ala | |
| APOA4 | rs210304158 | 15 | 27907074 | missense | 1 | tolerated | H/R | His /Arg |
| rs210921720 | 15 | 27907105 | missense | 0.08 | tolerated | Q/E | Gln / Glu | |
| APOA5 | rs208297208 | 15 | 27866581 | missense | 0.55 | tolerated | E/D | Glu/Asp |
| rs463483753 | 15 | 27868126 | splice_donor | |||||
| BCL2 | rs462904013 | 24 | 62105166 | missense | 0.64 | tolerated | A/S | Ala/Ser |
| C5orf34 | rs208163974 | 20 | 31327150 | missense | 0.34 | tolerated | H/R | His /Arg |
| rs209443540 | 20 | 31326810 | missense | 0.85 | tolerated | G/S | Gly /Ser | |
| CD44 | rs210543167 | 15 | 66525817 | missense | 0.37 | tolerated | H/R | His /Arg |
| rs42309927 | 15 | 66540981 | missense | 1 | tolerated | Q/P | Gln /Pro | |
| DDX11 | rs109173661 | 5 | 107486607 | missense | 0.49 | tolerated | V/A | Val/Ala |
| rs378943586 | 5 | 107487273 | missense | 0.29 | tolerated | M/I | Met/ Ile | |
| rs379141637 | 5 | 107497280 | downstream_gene | |||||
| rs482813209 | 5 | 107491924 | missense | 0.54 | tolerated | R/Q | Arg/ Gln | |
| DGAT1 | rs109286048 | 14 | 1787761 | missense | 0.44 | tolerated | D/N | Asp /Asn |
| rs109326954 | 14 | 1802266 | missense | 0.95 | tolerated | A/E | Ala/ Glu | |
| rs523413537 | 14 | 1802265 | frameshift | -/X | (-/X | |||
| EPOR | rs207986369 | 7 | 17003689 | missense | 1 | tolerated | A/P | Ala /Pro |
| rs209618019 | 7 | 16984076 | 5_prime_UTR | |||||
| rs466628285 | 7 | 16984149 | 5_prime_UTR | |||||
| ESRRA | rs477471785 | 29 | 43226583 | downstream_gene | ||||
| FKBP4 | rs109367585 | 5 | 107451448 | missense | 0.78 | tolerated | M/T | Met/Thr |
| rs110664228 | 5 | 107451447 | missense | 0.18 | tolerated | M/I | Met/ Ile | |
| rs134534391 | 5 | 107430855 | missense | 0.68 | tolerated | L/S | Leu/Ser | |
| rs449158440 | 5 | 107432580 | missense | 0.56 | tolerated | L/H | Leu/His | |
| rs476205399 | 5 | 107430347 | missense | 0.49 | tolerated | P/L | Pro/ Leu | |
| FOXM1 | rs134093880 | 5 | 107375702 | upstream_gene | ||||
| rs135216344 | 5 | 107377538 | upstream_gene | |||||
| GHR | rs109212162 | 20 | 31891025 | missense | 1 | tolerated | T/I | Thr/ Ile |
| rs109300983 | 20 | 31891050 | missense | 0.09 | tolerated | S/G | Ser/ Gly | |
| rs110265189 | 20 | 31891130 | missense | 0.02 | deleterious | N/T | Asn /Thr | |
| rs385640152 | 20 | 31909478 | missense | 0.02 | deleterious | F/Y | Phe /Tyr | |
| HMGCS1 | rs433464233 | 20 | 31472618 | missense | 0 | deleterious | Y/N | Tyr/Asn |
| IL10 | rs464052754 | 16 | 4406316 | missense | 0.62 | tolerated | T/S | Thr/Ser |
| IL1B | rs109004886 | 11 | 46417272 | missense | 0.07 | tolerated | G/D | Gly /Asp |
| rs449928032 | 11 | 46411289 | missense | 0.62 | tolerated | I/V | Ile/Val | |
| rs477020822 | 11 | 46411299 | stop_gained | Y/* | Tyr/* | |||
| IQSEC3 | rs385440401 | 5 | 107639460 | missense | 0.39 | tolerated | P/L | Pro/ Leu |
| rs432450632 | 5 | 107622167 | missense | 0 | deleterious | I/F | Ile/Prohe | |
| ITGB5 | rs135061845 | 1 | 69889500 | missense | 0.23 | tolerated | K/R | Lys/Arg |
| rs208109365 | 1 | 69899627 | missense | 1 | tolerated | A/E | Al Ala / Glu | |
| rs458450978 | 1 | 69832625 | missense | 0 | deleterious | D/G | Asp /Gly | |
| ITGB5 | rs474996003 | 1 | 69852928 | missense | 0 | deleterious | H/P | His /Pro |
| LDLRAP1 | rs211106371 | 2 | 128082792 | missense | 0.44 | tolerated | P/L | Pro/ Leu |
| MAN1C1 | rs448142609 | 2 | 128007912 | missense | 0.47 | tolerated | A/G | Ala / Gly |
| rs463343368 | 2 | 127909836 | missense | 0.02 | deleterious | S/R | Ser/Arg | |
| rs469242409 | 2 | 128007906 | missense | 0.57 | tolerated | V/G | Val/ Gly | |
| rs477509557 | 2 | 127915414 | missense | 0.42 | tolerated | V/M | Val/Met | |
| rs479080803 | 2 | 128007651 | missense | 0.35 | tolerated | H/P | His /Pro | |
| rs482632351 | 2 | 128007909 | missense | 0.3 | tolerated | Q/P | Gln /Pro | |
| rs482762432 | 2 | 127888951 | missense | 0.02 | deleterious | L/P | Leu/Pro | |
| MAP3K5 | rs109478031 | 9 | 75560882 | missense | 0.45 | tolerated | D/N | Asp /Asn |
| MRPS30 | rs135415772 | 20 | 30081437 | missense | 1 | tolerated | T/A | Thr/Ala |
| NNT | rs449852282 | 20 | 31204527 | missense | 0.08 | tolerated | A/P | Ala /Pro |
| rs478597655 | 20 | 31257968 | 5_prime_UTR | |||||
| OSBPL2 | rs383712095 | 13 | 55450613 | missense | 0.23 | tolerated | N/K | Asn / Lys |
| rs41696759 | 13 | 55450615 | missense | 1 | tolerated | N/D | Asn /Asp | |
| PAPPA | rs43580134 | 8 | 107186321 | missense | 1 | tolerated | M/T | Met/Thr |
| rs43580135 | 8 | 107186299 | missense | 0.52 | tolerated | T/A | Thr/Ala | |
| rs43580136 | 8 | 107186285 | missense | 0.15 | tolerated | A/V | Ala /Val | |
| PARP11 | rs476004287 | 5 | 106624500 | missense | 0.05 | deleterious | P/R | Pro/Arg |
| PPARG | rs109613657 | 22 | 57367375 | missense | 1 | tolerated | Q/H | Gln /His |
| rs132979274 | 22 | 57402368 | stop_gained | Q/* | Gln /* | |||
| rs42016945 | 22 | 57468357 | 5_prime_UTR | |||||
| RHNO1 | rs210028143 | 5 | 107353582 | missense | 0.73 | tolerated | A/T | Ala /Thr |
| SLC17A9 | rs440569152 | 13 | 54923249 | missense | 0.05 | tolerated | Y/S | Tyr/Ser |
| rs472517979 | 13 | 54917820 | missense | 0.02 | deleterious | S/R | Ser/Arg | |
| TEAD4 | rs133238643 | 5 | 107296229 | 5_prime_UTR | ||||
| rs384581336 | 5 | 107325259 | 5_prime_UTR | |||||
| TEX14 | rs109393617 | 19 | 9775707 | missense | 0.22 | tolerated | T/A | Thr/Ala |
| rs110989707 | 19 | 9775272 | missense | 0.23 | tolerated | T/A | Thr/Ala | |
| rs134356175 | 19 | 9742447 | missense | 0.64 | tolerated | P/L | Pro/ Leu | |
| rs134816249 | 19 | 9777840 | missense | 0.21 | tolerated | N/S | Asn /Ser | |
| rs208759383 | 19 | 9788622 | missense | 1 | tolerated | M/L | Met/ Leu | |
| rs378060964 | 19 | 9767815 | missense | 0.13 | tolerated | G/R | Gly /Arg | |
| rs380796779 | 19 | 9789924 | missense | 0 | deleterious | G/R | Gly /Arg | |
| rs383432764 | 19 | 9792619 | missense | 0.19 | tolerated | R/W | Gly /Trp | |
| rs460269897 | 19 | 9775646 | missense | 0.2 | tolerated | H/P | His /Pro | |
| TLR4 | rs29017188 | 8 | 108829143 | 5_prime_UTR | ||||
| rs516362864 | 8 | 108829405 | frameshift | -/X | -/X | |||
| rs8193069 | 8 | 108838685 | missense | 0.28 | tolerated | T/I | Thr/ Ile | |
| TRPS1 | rs381800291 | 14 | 50862411 | missense | 0.09 | tolerated | D/N | Asp /Asn |
| rs468711225 | 14 | 50863245 | splice_donor | |||||
| TSPAN11 | rs110381847 | 5 | 106978486 | 5_prime_UTR | ||||
| rs110799905 | 5 | 106978562 | 5_prime_UTR | |||||
| rs134575400 | 5 | 106976010 | 5_prime_UTR | |||||
| rs135745883 | 5 | 107005431 | missense | 1 | tolerated | I/V | Ile/Val | |
| rs210073944 | 5 | 107030616 | missense | 0.12 | tolerated | D/N | Asp /Asn |
1SNP consequence obtained using Ensembl Biomart database (Ensembl 83, Bos taurus UMD3.1, http://useast.ensembl.org/Bos_taurus/Info/Index).
2SIFT score: Sorts intolerant from tolerant algorithm score and the SIFT score ranges from 0.0 (deleterious) to 1.0 (tolerated).
SNP Genotyping and Filtering
Array genotyping was performed using PlexSeq Genotyping method (Agriplex Genomics, Cleveland, OH). Primers were designed to amplify regions surrounding each SNP using a multiplexed approach. Each primer design included an additional sequence at the 5′ end that was then used to anneal universal barcoded Illumina primers in a secondary amplification reaction. All samples (including negative and positive controls) were uniquely barcoded and sequenced simultaneously using Illumina NextSeq system (Illumina, San Diego, CA). Fastq files were analyzed using PlexCall software (Agriplex Genomics) which provides genotype calls for all SNPs in each sample. Arrays for 96 of 105 SNPs were successfully designed and genotyped. Moreover, 27 of 96 SNPs were removed before association test due to their lower marker call rates (<90%) or monomorphism. A total of 1,231 cows having both genotypic (SNPs) and phenotypic (dEBVs for LP) records were used in the association analysis.
Statistical Analyses
Principal component analysis (PCA) was performed with prcomp() function in R to examine potential population structure in the data (Supplementary Figure S1). The association analyses, using additive and dominance effects model, were performed using a univariate linear mixed model in which each SNP was analyzed individually as follows:
where μ is the overall mean, y is the vector of deregressed EBVs (deEBVs) for LP, Z is an incidence matrix relating phenotypes to the corresponding random polygenic effect, a is a vector of the random polygenic effect ~ (where A is the additive relationship matrix and is the polygenic variance, m is a vector with coded genotypes (2, 1, or 0 for genotypes AA, AB, and BB) for each animal, β 1 is the additive effect of the SNP, g is a vector with coded genotype (2, 1, or 0 for genotypes AA, AB, and BB) for each animal, β 2 is the dominance effect of the SNP, and e is a vector of random environmental deviates ~ N(0,W-1σe2) (where is the general error variance and W is the diagonal matrix containing weights of the deEBVs). All association analyses were fitted by restricted maximum likelihood (REML) using the DMU software (Madsen and Jensen, 2013; http://dmu.agrsci.dk). A Wald test was used to test the null hypothesis H0: β 1 = 0 or β 2 = 0 that was used to determine the significantly associated SNPs for additive and dominance effects, respectively. Significance was established at false discovery rate adjusted P (FDR) < 0.05 and the suggestive threshold was considered at uncorrected P < 0.05. The proportion of total phenotypic variance explained by the additive genetic variance of a SNP was calculated as 2p(1−p)α 2/σ 2p, where p is the allele frequency of one of the alleles of the SNP, α is the estimated allele substitution effect as α = β 1 + β 2[(1 − p) − p], and σ 2p is the phenotypic variance. The proportion of total phenotypic variance of the trait explained by a dominance genetic variance of the SNP was calculated as (2p(1 − p)β 2)2/σ 2p, where β 2 is the estimated dominance effect, and σ 2p is the total phenotypic variance. Finally, linkage disequilibrum (LD) block analysis was performed for the chromosomal region(s) containing SNPs significantly or suggestively associated with LP. The LD block was defined according to Gabriel et al. (2002) as a region with over 95% of informative SNP pairs showing strong LD (strong LD is when the one-sided upper 95% confidence bound on D′ is >0.98 and the lower bound is >0.7) and was detected and visualized with Haploview software (Barrett et al., 2004).
Results and Discussion
The PCA results showed no genetic cluster or population structure in the current data (Supplementary Figure S1). The maximum proportion of variance explained by a principal component was 6.1% (Supplementary Figure S1). The association analyses of 69 polymorphic SNPs indicated that SNPs rs43770847, rs208794152, and rs208332214 in ADRM1; rs209443540 in C5orf34; rs378943586 in DDX11; and rs385640152 in GHR were suggestively associated (P < 0.05) with LP (Table 2). The most significant association was reported for SNP rs43770847 in ADRM1 (P = 0.008, FDR = 0.25). Among the significantly associated SNPs, the highest value in additive effects was reported for rs209443540 in C5orf34 (11.75 kg). The additive variance of significantly associated SNPs explained from 0.15% (rs43770847 in ADRM1) to 5.69% (rs209443540 in C5orf34) of total phenotypic variance of LP (Table 2). The LD analysis for SNPs on chromosome 20 indicated that SNP rs385640152 in GHR is in a LD block with SNP rs110265189 in GHR (Supplementary Figure S2). Four SNPs (rs43770847, rs208794152, and rs208332214 in ADRM1, and rs209443540 in C5orf34) also had suggestive dominance effects on LP (P < 0.05) and dominance variance of the SNPs accounted for 0.24% to 1.35% of total phenotypic variance of LP (Table 3). Two SNPs in ADRM1 (rs209179785) and DDX11 (rs378943586) tended to have suggestive dominance effects on LP (P < 0.1) (Table 3) and these dominance effects, however, explained a very small proportion of phenotypic variance of LP (Table 3).
Table 2.
Significantly associated SNPs with lactation persistency based on additive effects
| Genes | SNP | Genotype | Genotype count | Minor allele frequency | P value | FDR1 | Additive effect (kg) | Phenotypic variance explained (%) by additive variance |
|---|---|---|---|---|---|---|---|---|
| ADRM1 | rs43770847 | G/G | 146 | 0.40 | 0.008 | 0.26 | –1.07 | 0.15% |
| A/G | 671 | |||||||
| A/A | 399 | |||||||
| C5orf34 | rs209443540 | A/A | 5 | 0.08 | 0.008 | 0.26 | 11.75 | 5.69% |
| G/G | 1031 | |||||||
| G/A | 194 | |||||||
| DDX11 | rs378943586 | G/G | 926 | 0.13 | 0.016 | 0.41 | 6.10 | 2.23% |
| A/A | 12 | |||||||
| G/A | 285 | |||||||
| GHR | rs385640152 | A/T | 328 | 0.16 | 0.029 | 0.49 | 2.68 | 0.52% |
| T/T | 30 | |||||||
| A/A | 862 | |||||||
| ADRM1 | rs208332214 | C/A | 195 | 0.08 | 0.039 | 0.49 | –10.15 | 4.27% |
| C/C | 1030 | |||||||
| A/A | 5 | |||||||
| ADRM1 | rs208794152 | T/T | 5 | 0.08 | 0.039 | 0.49 | –10.15 | 4.27% |
| C/T | 196 | |||||||
| C/C | 1011 | |||||||
| DDX11 | rs109173661 | C/T | 536 | 0.25 | 0.099 | 0.76 | −2.12 | 0.46% |
| T/T | 45 | |||||||
| C/C | 650 |
1False discovery rate adjusted P-values.
Table 3.
Significantly associated SNPs with lactation persistency based on dominance effect
| Genes | SNP | Genotype | Genotype count | Minor allele frequency | P value | FDR1 | Dominance Effect, kg | Phenotypic variance explained (%) by dominance variance |
|---|---|---|---|---|---|---|---|---|
| C5orf34 | rs209443540 | A/A | 5 | 0.08 | 0.007 | 0.26 | 5.01 | 1.04% |
| G/G | 1031 | |||||||
| G/A | 194 | |||||||
| ADRM1 | rs43770847 | G/G | 146 | 0.40 | 0.008 | 0.26 | 1.48 | 0.28% |
| A/G | 671 | |||||||
| A/A | 399 | |||||||
| ADRM1 | rs208794152 | T/T | 5 | 0.08 | 0.033 | 0.49 | −5.65 | 1.35% |
| C/T | 196 | |||||||
| C/C | 1011 | |||||||
| ADRM1 | rs208332214 | C/A | 195 | 0.08 | 0.034 | 0.49 | −5.58 | 1.29% |
| C/C | 1030 | |||||||
| A/A | 5 | |||||||
| ADRM1 | rs209179785 | G/C | 624 | 0.33 | 0.071 | 0.76 | 1.42 | 0.24% |
| C/C | 92 | |||||||
| G/G | 514 | |||||||
| DDX11 | rs378943586 | G/G | 926 | 0.13 | 0.073 | 0.76 | 3.03 | 0.55% |
| A/A | 12 | |||||||
| G/A | 285 |
1False discovery rate adjusted P-values.
Targeted SNP genotyping provides a cost-effective approach for post-GWAS validation or for genotyping limited genomic regions of interest. Targeted genotyping could be performed by either using custom SNPs array or by targeted resequencing depending on the purpose of the study. The custom SNP array genotyping allows a focus on biologically meaningful variants via the genotyping of specific sets of targeted SNPs (Sõber et al., 2009; Cirera et al., 2018), while targeted sequencing supports SNP discovery, validation, and screening of genetic variants in genome or gene regions of interest (De Donato et al., 2013; Jiang et al., 2014; Gorjanc et al., 2015; Ibeagha-Awemu et al., 2016; Li et al., 2016a; Brouard et al., 2017). The approach of targeted arrays used in this study was to get deeper information about potential functional SNPs of LP which could influence the expression of proteins or downstream transcriptional regulation. This approach is potentially very powerful to obtain deeper information on informative SNPs within candidate genes (LaFramboise, 2009; Sõber et al., 2009; Jiang et al., 2014; Cirera et al., 2018).
Although many recent studies have shown that noncoding variants (synonymous variants and variants located in intergenic and intronic regions of genes) can significantly contribute to variance of complex human (Yang et al., 2011) and livestock traits (Morota et al., 2014; Do et al., 2015), it is still important to study missense variants since they directly change the amino acids in proteins with potential effects on phenotypic variance (Hindorff et al., 2009; Kindt et al., 2013). Therefore, about 80% of SNPs selected for genotyping in this study were missense variants (Table 1).
In fact, none of the studied SNPs was significantly associated with LP after multiple testing correction was applied and might be due to our limited sample size. Moreover, multiple testing procedure is based on the hypothesis that all test are independent which may not be true for association test with SNPs because of probable existence of linkage disequilibrium among markers. Notably, 3 SNPs in ADRM1 gene suggestively associated with LP by both additive and dominance effects (Tables 2 and 3). Previously, ADRM1 was identified as a potential candidate gene for LP through a GWAS on bulls (Nayeri et al., 2017). Since ADRM1 was not identified as a candidate gene in our previous study (Do et al., 2017), these 3 SNPs in ADRM1 are considered novel markers associated with LP. The protein coded by ADRM1 is a putative cell adhesion regulating protein that plays important roles in the maintenance of protein homeostasis (Jørgensen et al., 2006). The SNP rs43770847 in ADRM1 was the most associated SNP (P = 0.008) based on additive effects but it explained only a small proportion of LP variance (0.15%) (Table 2). SNP rs43770847 allele substitution influenced the protein sequence by changing serine to glycine (Ser1322Gly). Substantial dominance variance has been reported for different production traits in dairy cows which might be useful in genomic selection (Ertl et al., 2014; Aliloo et al., 2016; Jiang et al., 2017; Varona et al., 2018). For example, total phenotypic variance of milk, fat, and protein yield traits explained by dominance effects ranged from 5% to 7% (Sun et al., 2014). The dominance effect of ADRM1 rs43770847 was suggestively significant (P < 0.05) and explained a small proportion of the phenotypic variance of the trait (0.028%). To the best of our knowledge, this is the first study to estimate dominance effects and phenotypic variance explained by SNPs for LP.
Percentage of phenotypic variance explained by dominance effect of rs43770847 (ADRM1) was approximately 5 times lower than by additive effect which follows the same trend reported in previous studies on milk traits in dairy cows (Ertl et al., 2014; Aliloo et al., 2016; Jiang et al., 2017; Varona et al., 2018). Further notable significantly associated SNPs in ADRM1 (rs208332214 and rs208794152) explained a considerable amount of the phenotypic variance of LP (4.27%); therefore, these SNPs might be important for genetic selection for enhanced LP.
The second most associated SNP with LP was rs209443540 in C5orf34. This mutation is characterized by a substitution of G by A causing a change from glycine to serine in the protein sequence (Tables 1 and 2). This SNP is located at position 31,326,810 bp in chromosome 20 and 66,383 bp away from a significant SNP (rs109823394, position: 31,393,193 bp) reported in our previous study (Do et al., 2017). Moreover, this SNP was not present in a LD block in the current data set; therefore, it might be considered a novel marker for LP. Interestingly, both additive and dominance variances of this SNP explained a considerable amount of the variation in LP (5.69% and 1.04%, respectively). Recently, C5orf34 was suggested to have roles in lung cancer development and progression by regulating MAPK signaling pathway (He et al., 2019). Its function in bovine is unknown.
SNP rs385640152 is a missense variant within exon 8 (ENSBTAT00000001758.2:c.836T>A) of GHR gene predicted to cause a potentially deleterious amino acid substitution from phenylalanine to tyrosine (Phe279Tyr) in the GHR protein. GHR is considered a potential candidate gene for LP in our previous study (Do et al., 2017) since it was located close to 3 significantly associated SNPs with LP on BTA 20. Interestingly, Nayeri et al. (2017) also associated several SNPs (rs41639260, rs110482506, and rs41639261) located in intronic regions of the GHR gene with LP in bulls. Growth hormone is well known for its galactopoietic action in bovine (Burton et al., 1994). Some studies have associated polymorphisms in the GHR gene with milk yield and lactation (Moisio et al., 1998; Rahmatalla et al., 2011). In particularly, a T to A nucleotide substitution (SNP rs385640152) in exon 8 of the GHR gene resulting in a phenylalanine (GHR279Phe) to tyrosine (GHR279Tyr) change in the transmembrane domain of the GHR protein was associated with a major effect on milk yield (Blott et al., 2003). Furthermore, cows of the GHR279Phe protein variant produced about 200 kg more milk annually than cows carrying the GHR279Tyr variant (Blott et al., 2003). In the present study, the substitution of allele A by T of this mutation (rs385640152) only accounted for 0.52% of the phenotypic variation in LP (Table 2). However, Fontanesi et al. (2007) indicated that the use of the GHR Phe279Tyr allele in marker-assisted selection may not have a significant impact on selection for milk yield and milk components due to the high frequency of the putative positive allele for milk protein percentage.
SNP rs378943586 in DDX11 was also suggestively associated with LP based on additive effects (Table 2). DDX11 is important in breast cancer regulation and mammary cell development (Callari et al., 2011) but its functionality in cows is unknown. The additive and dominance variances of this SNP explained 2.23% and 0.55% of the phenotypic variance in LP, respectively; therefore, this SNP might also be considered in selection for LP with the inclusion of both additive and dominance effects.
Since more than 100 QTLs for milk yield and milk component traits have been reported around the centromeric region of BTA14 where the DGAT1 gene is located (Hu et al., 2015), we also genotyped three functionally informative SNPs in the DGAT1 gene (Table 1). However, none of the SNPs in the DGAT1 gene was significantly associated with LP in this study, suggesting that the DGAT1 gene might not be important for LP. Other GWAS studies did not also report associated SNPs for LP within the DGAT1 gene region (Kolbehdari et al., 2009; Pryce et al., 2010; Nayeri et al., 2017). Finally, it is important to note that none of the SNPs in candidate genes derived by gene ontology, network, or pathways analyses (Do et al., 2017) were significantly associated with LP in this study, suggesting that the relationship of these genes with LP may be through indirect relationships with other genes.
Conclusions
We analyzed associations between 69 potential functional variants located in 30 candidate genes and dEBVs of LP in Canadian Holstein cows. SNPs rs43770847, rs208794152, and rs208332214 in ADRM1, rs209443540 in C5orf34, rs378943586 in DDX11, and rs385640152 in GHR might be important for understanding the biology of LP as well as for selection of cows with higher LP. The amount of phenotypic variance explained by dominance effects for each associated SNPs was substantial; therefore, it might be important to include dominance effects in genetic selection for enhanced LP. Functional validation of the suggestively associated SNPs and SNPs on further genes like C5orf34, ADRM1, DDX11, and GHR might further understanding of additive and nonadditive effects on LP and for inclusion in a genomic selection for LP.
Supplementary Material
Acknowledgments
We thank participating farmers for animal management and Valacta (Valacta Laboratories, Ste-Anne-de-Bellevue, QC, Canada, www.valacta.com) for assistance in collecting milk samples. Funding for this research was provided by the Dairy Farmers of Canada, Agriculture and Agri-Food Canada, the Canadian Dairy Network and the Canadian Dairy Commission.
Conflict of interest statement
None declared.
Literature Cited
- Aliloo H., Pryce J. E., González-Recio O., Cocks B. G., and Hayes B. J.. . 2016. Accounting for dominance to improve genomic evaluations of dairy cows for fertility and milk production traits. Genet. Sel. Evol. 48:8. doi: 10.1186/s12711-016-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. D. M. J., and Daly M. J.. . Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, no. 2 (2004): 263–265. doi:10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bissonnette N. 2018. Short communication: Genetic association of variations in the osteopontin gene (SPP1) with lactation persistency in dairy cattle. J. Dairy Sci. 101:456–461. doi: 10.3168/jds.2017-13129. [DOI] [PubMed] [Google Scholar]
- Bissonnette N., Brouard J.-S., Olivier Ariel N. G., Ibeagha-Awemu E., and Miglior F.. Discovery of expression quantitative trait locus associated with Johne’s disease using both RNA-seq and DNA variants World Congress on Genetics Applied to Livestock Production – NewZeeland Feb 2018; 2018. [Google Scholar]
- Blott S., Kim J.-J., Moisio S., Schmidt-Küntzel A., Cornet A., Berzi P., Cambisano N., Ford C., Grisart B., and Johnson D.. . 2003. Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics 163(1):253–266. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462408/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard J. S., Boyle B., Ibeagha-Awemu E. M., and Bissonnette N.. . 2017. Low-depth genotyping-by-sequencing (GBS) in a bovine population: strategies to maximize the selection of high quality genotypes and the accuracy of imputation. BMC Genet. 18(1):32. doi: 10.1186/s12863-017-0501-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J. L., McBride B. W., Block E., Glimm D. R., and Kennelly J. J.. . 1994. A review of bovine growth hormone. Can. J. Anim. Sci. 74(2):167–201. doi:10.4141/cjas94-027 [Google Scholar]
- Callari M., Cappelletti V., De Cecco L., Musella V., Miodini P., Veneroni S., Gariboldi M., Pierotti M. A., and Daidone M. G.. . 2011. Gene expression analysis reveals a different transcriptomic landscape in female and male breast cancer. Breast Cancer Res. Treat. 127:601–610. doi: 10.1007/s10549-010-1015-8. [DOI] [PubMed] [Google Scholar]
- Campbell S., Swann H. R., Seif M. W., Kimber S. J., and Aplin J. D.. . 1995. Cell adhesion molecules on the oocyte and preimplantation human embryo. Hum. Reprod. 10:1571–1578. doi: 10.1093/humrep/10.6.1571. [DOI] [PubMed] [Google Scholar]
- Cirera S., Clop A., Jacobsen M. J., Guerin M., Lesnik P., Jorgensen C. B., Fredholm M., and Karlskov-Mortensen P.. . 2018. A targeted genotyping approach enhances identification of variants in taste receptor and appetite/reward genes of potential functional importance for obesity-related porcine traits. Anim. Genet. doi: 10.1111/age.12641 [DOI] [PubMed] [Google Scholar]
- Cole J. B., and VanRaden P. M.. . 2006. Genetic evaluation and best prediction of lactation persistency. J. Dairy Sci. 89:2722–2728. doi: 10.3168/jds.S0022-0302(06)72348-7. [DOI] [PubMed] [Google Scholar]
- De Donato M., Peters S. O., Mitchell S. E., Hussain T., and Imumorin I. G.. . 2013. Genotyping-by-sequencing (GBS): a novel, efficient and cost-effective genotyping method for cattle using next-generation sequencing. PLoS One 8:e62137. doi: 10.1371/journal.pone.0062137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J. C. M., Ten Hag J. H., and Weersink A.. . 1998. Economic aspects of persistency of lactation in dairy cattle. Livest. Prod. Sci. 53(3):237–252. doi: 10.1016/s0301-6226(97)00124-3 [DOI] [Google Scholar]
- Do D. N., Bissonnette N., Lacasse P., Miglior F., Sargolzaei M., Zhao X., and Ibeagha-Awemu E. M.. . 2017. Genome-wide association analysis and pathways enrichment for lactation persistency in Canadian Holstein cattle. J. Dairy Sci. 100:1955–1970. doi: 10.3168/jds.2016-11910. [DOI] [PubMed] [Google Scholar]
- Do D. N., Janss L. L., Jensen J., and Kadarmideen H. N.. . 2015. SNP annotation-based whole genomic prediction and selection: an application to feed efficiency and its component traits in pigs. J. Anim. Sci. 93:2056–2063. doi: 10.2527/jas.2014-8640. [DOI] [PubMed] [Google Scholar]
- Ertl J., Legarra A., Vitezica Z. G., Varona L., Edel C., Emmerling R., and Götz K.-U.. . 2014. Genomic analysis of dominance effects on milk production and conformation traits in Fleckvieh cattle. Genet. Sel. Evol. 46(1):40–40. doi: 10.1186/1297-9686-46-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feugang J. M., Kaya A., Page G. P., Chen L., Mehta T., Hirani K., Nazareth L., Topper E., Gibbs R., and Memili E.. . 2009. Two-stage genome-wide association study identifies integrin beta 5 as having potential role in bull fertility. BMC Genomics 10:176. doi: 10.1186/1471-2164-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L., Scotti E., Tazzoli M., Beretti F., Dall’Olio S., Davoli R., and Russo V.. . 2007. Investigation of allele frequencies of the growth hormone receptor (GHR) F279Y mutation in dairy and dual purpose cattle breeds. Ital. J. Anim. Sci. 6(4):415–420. doi:10.4081/ijas.2007.415 [Google Scholar]
- Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., . et al. 2002. The structure of haplotype blocks in the human genome. Science 296, 5576(2002):2225–2229. doi:10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Garrick D. J., Taylor J. F., and Fernando R. L.. . 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41(1):1. doi:10.1186/1297-9686-41-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanc G., Cleveland M. A., Houston R. D., and Hickey J. M.. . 2015. Potential of genotyping-by-sequencing for genomic selection in livestock populations. Genet. Sel. Evol. 47(1):12. doi:10.1186/s12711-015-0102-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M. P., Yan W., Wu M.-H., Lin Y.-N., Agno J. E., Sharma M., Braun R. E., Rajkovic A., and Matzuk M. M.. . 2006. TEX14 is essential for intercellular bridges and fertility in male mice. Proc. Natl. Acad. Sci. U. S. A. 103(13):4982–4987. doi:10.1073/pnas.0505123103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder B., Bennewitz J., Reinsch N., Thaller G., Thomsen H., Kühn C., Schwerin M., Erhardt G., Förster M., Reinhardt F., . et al. 2006. Mapping of quantitative trait loci for lactation persistency traits in German Holstein dairy cattle. J. Anim. Breed. Genet. 123:89–96. doi: 10.1111/j.1439-0388.2006.00577.x. [DOI] [PubMed] [Google Scholar]
- He D., Wu Z., He J., Wang Y., Li Z., and Gao S.. . 2019. Up-regulation of C5orf34 promotes lung adenocarcinoma migration and is correlated with worse prognosis. Gene 696:47–53. doi: 10.1016/j.gene.2019.02.019. [DOI] [PubMed] [Google Scholar]
- Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., Collins F. S., and Manolio T. A.. . 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. of Sci. 106(23):9362–9367. doi:10.1073/pnas.0903103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. L., Park C. A., and Reecy J. M.. . 2016. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 44(D1):D827–D833. doi: 10.1093/nar/gkv1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeagha-Awemu E. M., Akwanji K. A., Beaudoin F., and Zhao X.. . 2014. Associations between variants of FADS genes and omega-3 and omega-6 milk fatty acids of Canadian Holstein cows. BMC Genet. 15:25. doi: 10.1186/1471-2156-15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeagha-Awemu E. M., Peters S. O., Akwanji K. A., Imumorin I. G., and Zhao X.. . 2016. High density genome wide genotyping-by-sequencing and association identifies common and low frequency SNPs, and novel candidate genes influencing cow milk traits. Sci. Rep. 6:31109. doi: 10.1038/srep31109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen J. H., Madsen P., Jensen J., Pedersen J., Christensen L. G., and Sorensen D. A.. . 2002. Genetic parameters for milk production and persistency for Danish Holsteins estimated in random regression models using REML. J. Dairy Sci. 85:1607–1616. doi: 10.3168/jds.S0022-0302(02)74231-8. [DOI] [PubMed] [Google Scholar]
- Jiang J., Shen B., O’Connell J. R., VanRaden P. M., Cole J. B., and Ma L.. . 2017. Dissection of additive, dominance, and imprinting effects for production and reproduction traits in Holstein cattle. BMC Genomics 18(1):425. doi:10.1186/s12864-017-3821-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Liu X., Yang J., Wang H., Jiang J., Liu L., He S., Ding X., Liu J., and Zhang Q.. . 2014. Targeted resequencing of GWAS loci reveals novel genetic variants for milk production traits. BMC Genomics 15:1105. doi: 10.1186/1471-2164-15-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen J. P., Lauridsen A.-M., Kristensen P., Dissing K., Johnsen A. H., Hendil K. B., and Hartmann-Petersen R.. . 2006. Adrm1, a Putative Cell Adhesion Regulating Protein, is a Novel Proteasome-associated Factor. J. Mol. Biol. 360(5):1043–1052. doi: 10.1016/j.jmb.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Kindt A. S., Navarro P., Semple C. A., and Haley C. S.. . 2013. The genomic signature of trait-associated variants. BMC Genomics 14(1):108. doi:10.1186/1471-2164-14-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbehdari D., Wang Z., Grant J. R., Murdoch B., Prasad A., Xiu Z., Marques E., Stothard P., and Moore S. S.. . 2009. A whole genome scan to map QTL for milk production traits and somatic cell score in Canadian Holstein bulls. J. Anim. Breed. Genet. 126:216–227. doi: 10.1111/j.1439-0388.2008.00793.x. [DOI] [PubMed] [Google Scholar]
- LaFramboise T. 2009. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res. 37:4181–4193. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Sun D., Zhang S., Liu L., Alim M. A., and Zhang Q.. . 2016a. A post-GWAS confirming the SCD gene associated with milk medium- and long-chain unsaturated fatty acids in Chinese Holstein population. Anim. Genet. 47:483–490. doi: 10.1111/age.12432. [DOI] [PubMed] [Google Scholar]
- Li C., Sun D., Zhang S., Yang S., Alim M. A., Zhang Q., Li Y., and Liu L.. . 2016b. Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese Holstein cattle population based on a post genome-wide association study. BMC Genet. 17:110. doi: 10.1186/s12863-016-0418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisio S., Elo K., Kantanen J., and Vilkki J.. . 1998. Polymorphism within the 3’ flanking region of the bovine growth hormone receptor gene. Anim. Genet. 29:55–57. doi:10.1046/j.1365-2052.1998.00254.x [DOI] [PubMed] [Google Scholar]
- Morota G., Abdollahi-Arpanahi R., Kranis A., and Gianola D.. . 2014. Genome-enabled prediction of quantitative traits in chickens using genomic annotation. BMC Genomics 15:109. doi: 10.1186/1471-2164-15-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir B. L., Fatehi J., and Schaeffer L. R.. . 2004. Genetic relationships between persistency and reproductive performance in first-lactation Canadian holsteins. J. Dairy Sci. 87:3029–3037. doi: 10.3168/jds.S0022-0302(04)73435-9. [DOI] [PubMed] [Google Scholar]
- Nayeri S., Sargolzaei M., Abo-Ismail M. K., Miller S., Schenkel F., Moore S. S., and Stothard P.. . 2017. Genome-wide association study for lactation persistency, female fertility, longevity, and lifetime profit index traits in Holstein dairy cattle. J. Dairy Sci. 100:1246–1258. doi: 10.3168/jds.2016-11770. [DOI] [PubMed] [Google Scholar]
- Pryce J. E., Haile-Mariam M., Verbyla K., Bowman P. J., Goddard M. E., and Hayes B. J.. . 2010. Genetic markers for lactation persistency in primiparous Australian dairy cows. J. Dairy Sci. 93:2202–2214. doi: 10.3168/jds.2009-2666. [DOI] [PubMed] [Google Scholar]
- Rahmatalla S. A., Müller U., Strucken E. M., Reissmann M., and Brockmann G. A.. . 2011. The F279Y polymorphism of the GHR gene and its relation to milk production and somatic cell score in German Holstein dairy cattle. J. Appl. Genet. 52:459–465. doi: 10.1007/s13353-011-0051-3. [DOI] [PubMed] [Google Scholar]
- Romanska H. M., and Berditchevski F.. . 2011. Tetraspanins in human epithelial malignancies. J. Pathol. 223(1):4–14. doi: 10.1002/path.2779 [DOI] [PubMed] [Google Scholar]
- Sharma B. S., Leyva I., Schenkel F., and Karrow N. A.. . 2006. Association of toll-like receptor 4 polymorphisms with somatic cell score and lactation persistency in Holstein bulls. J. Dairy Sci. 89:3626–3635. doi: 10.3168/jds.S0022-0302(06)72402-X. [DOI] [PubMed] [Google Scholar]
- Sõber S., Org E., Kepp K., Juhanson P., Eyheramendy S., Gieger C., Lichtner P., Klopp N., Veldre G., and Viigimaa M.. . 2009. Targeting 160 candidate genes for blood pressure regulation with a genome-wide genotyping array. PloS One 4(6):e6034. doi:10.1371/journal.pone.0006034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanon B., Colitti M., Gabai G., Knight C. H., and Wilde C. J.. . 2002. Mammary apoptosis and lactation persistency in dairy animals. J. Dairy Res. 69(01):37–52. doi: 10.1017/S0022029901005246 [DOI] [PubMed] [Google Scholar]
- Sun C., VanRaden P. M., Cole J. B., and O’Connell J. R.. . 2014. Improvement of prediction ability for genomic selection of dairy cattle by including dominance effects. PLoS One 9:e103934. doi: 10.1371/journal.pone.0103934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalve H. 1995. Genetic relationship between dairy lactation persistency and yield. J. Anim. Breed. Genet. 112(1‐6):303–311. [Google Scholar]
- Varona L., Legarra A., Toro M. A., and Vitezica Z. G.. . 2018. Non-additive Effects in Genomic Selection. Front. Genet. 9:78-78. doi: 10.3389/fgene.2018.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla K. L., and Verbyla A. P.. . 2009. Estimated breeding values and association mapping for persistency and total milk yield using natural cubic smoothing splines. Genet. Sel. Evol. 41:48. doi:10.1186/1297-9686-41-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschoor C. P., Pant S. D., Biggar G. A., Schenkel F. S., Sharma B. S., and Karrow N. A.. . 2011. Identification of SNPs in interferon gamma, interleukin-22, and their receptors and associations with health and production-related traits in Canadian Holstein bulls. Anim. Biotechnol. 22:7–15. doi: 10.1080/10495398.2011.536078. [DOI] [PubMed] [Google Scholar]
- Yang J., Manolio T. A., Pasquale L. R., Boerwinkle E., Caporaso N., Cunningham J. M., de Andrade M., Feenstra B., Feingold E., Hayes M. G., . et al. 2011. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43:519. doi: 10.1038/ng.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.