Abstract
The DNA sequencing technologies in use today produce either highly accurate short reads or less-accurate long reads. We report the optimization of circular consensus sequencing (CCS) to improve the accuracy of single-molecule real-time (SMRT) sequencing (PacBio) and generate highly accurate (99.8%) long high-fidelity (HiFi) reads with an average length of 13.5 kilobases (kb). We applied our approach to sequence the well-characterized human HG002/NA24385 genome and obtained precision and recall rates of at least 99.91% for single-nucleotide variants (SNVs), 95.98% for insertions and deletions <50 bp (indels) and 95.99% for structural variants. Our CCS method matches or exceeds the ability of short-read sequencing to detect small variants and structural variants. We estimate that 2,434 discordances are correctable mistakes in the ‘genome in a bottle’ (GIAB) benchmark set. Nearly all (99.64%) variants can be phased into haplotypes, further improving variant detection. De novo genome assembly using CCS reads alone produced a contiguous and accurate genome with a contig N50 of >15 megabases (Mb) and concordance of 99.997%, substantially outperforming assembly with less-accurate long reads.
Introduction
DNA sequencing technologies have improved at rates eclipsing Moore’s law1 revolutionizing biological sciences. Beginning in the 1970s, Sanger sequencing2, and subsequent automation3 facilitated large scale DNA sequencing projects and paved the way for modern genomic research4–7. The first reference genomes were followed by the advent of several high-throughput sequencing technologies (next-generation sequencing or NGS) including 454™, Solexa/Illumina®, ABI® Solid™, Complete Genomics™, and Ion Torrent™. These technologies employed a range of chemistries and detection strategies8–13. All produce relatively accurate reads but are limited in read length, typically to less than 300 basepairs (bp). These accurate short reads are well-suited for calling single-nucleotide variants (SNVs) and small insertions and deletions (indels), but are less useful for de novo assembly, haplotype phasing and structural variant detection, all of which require information across longer sequence spans.
In order to detect structural variants, phase haplotypes and assemble genomes, superior results14–18 could be obtained with technologies such as PacBio® SMRT Sequencing19 and Oxford Nanopore sequencing20 both of which produce long reads (>10 kb). These technologies rely on single-molecule detection and are characterized by reduced read accuracy (75–90%)19,20. High consensus accuracy can be achieved by read-to-read error correction, but it is computationally intensive, and errors remain from mis-mapping reads or mixing haplotypes during correction15,21. As a result of the error rate, long-read technologies are rarely used to detect SNVs or indels. Although human genomes can be sequenced at population scales, it remains necessary to combine sequencing technologies to detect all the different types of genetic variation. This increases cost and adds complexity to projects. A sequencing technology with long read length and high accuracy could enable comprehensive variant discovery in a single experiment.
Circular consensus sequencing (CCS) derives a consensus sequence from multiple passes of a single template molecule, producing accurate reads from noisy individual subreads22,23. The length and accuracy of CCS (also known as “HiFi”) reads is limited by the number of passes required to achieve the desired accuracy and the overall (“polymerase”) read length of the sequencing platform. To date, CCS has primarily been applied to DNA inserts shorter than 2 kb24, and has not been used to produce a deep-coverage dataset for an entire human genome.
We devised an approach to produce long CCS reads and applied our method to sequence the well-characterized human male HG002/NA2438525,26. HG002/NA24385 is one of the benchmark samples from the Genome in a Bottle (GIAB) Consortium. GIAB provides physical reference materials along with detailed characterization of the sample genome, defining “benchmark regions” at which the sequence of the sample is known and “benchmark variants” within those regions at which the sample differs from the human reference genome. We chose this sample as an exemplar for studying sequencing accuracy and variant detection. Using CCS reads, we identify small and large sequence variation and phase haplotypes in HG002/NA24385 more accurately than existing technologies. We also use CCS reads to assemble the genome with similar contiguity and 5.9× the accuracy of the most recently reported PacBio human genome assembly (GCA_001542345.1).
Results
CCS Library Preparation and Sequencing
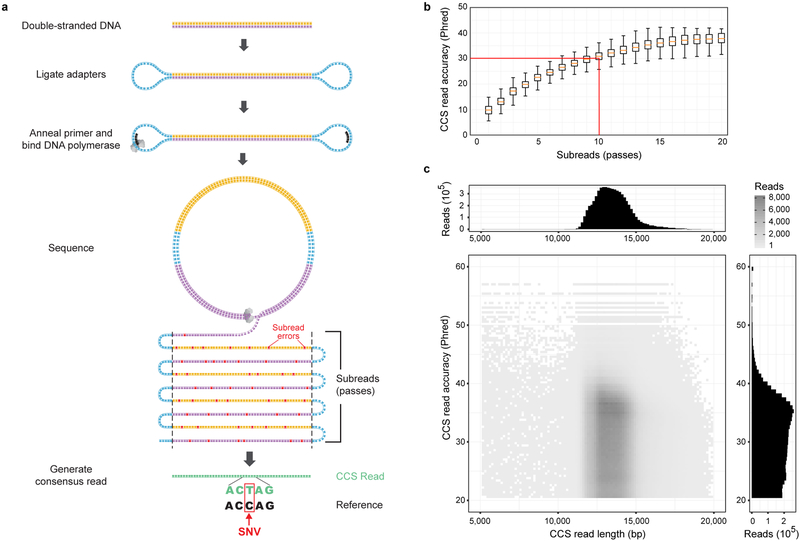
An opportunity to produce long CCS reads was suggested by a 16-fold increase in the fraction of polymerase reads longer than 100 kb for a control E. coli 10 kb amplicon library compared to a long-insert (>30 kb) library of sheared E. coli genomic DNA sequenced under identical conditions with 10 hour collections (Supplementary Figure 1a). This suggested that polymerases on newly-synthesized, shorter, discrete-sized inserts have better survival, which we hypothesized was due to DNA damage on long inserts that terminates the polymerase reaction. To evaluate this hypothesis, a library was prepared from BsaAI-digested lambda DNA to examine the effect of pre-extension, in which the polymerase extends (without laser illumination) prior to sequence data collection to effectively eliminate damaged templates (which terminate during the pre-extension period) and select for surviving polymerases. The DNA loading concentration was increased with pre-extension to compensate for the polymerases lost due to damaged templates. With pre-extension and with 4 hour collections to detect early polymerase survival, the fraction of reads of an 8 kb fragment from the digested lambda DNA that survive to 40 kb polymerase read length increased by 4.5-fold, confirming that pre-extension improves read length for discrete-sized inserts (Supplementary Figure 1b). Insert size was then evaluated, with adjustments in collection time, to maximize the yield of high-accuracy reads (Supplementary Table 1).
Ultimately, a SMRTbell library tightly distributed at 15 kb was chosen for high-coverage circular consensus sequencing (Figure 1a, Supplementary Figure 1c–f) based on estimates of 150 kb polymerase read length and a requirement of 10 passes to achieve Q30 (Phred quality score 30) read accuracy (Figure 1b). CCS reads with a predicted accuracy of at least Q20 (99%) were retained (Supplementary Figure 2a). The total CCS read yield was 89 Gb (mean 2.3 ± standard deviation of 0.4 Gb over 39 SMRT Cells 1M), with read length of 13.5±1.2 kb (Figure 1c). The predicted accuracy of the CCS reads has a median of Q30 (99.9%) and a mean of Q27 (99.8%) (Figure 1c). Predicted accuracy matches well with concordance to the GIAB HG002 benchmark (average [Qpredicted – Qconcordance] = −1.2), which indicates that the predicted accuracy is well calibrated (Supplementary Figure 2b–c). Average mapped coverage of the genome is 28-fold, with minimal difference across [GC] content (Supplementary Figure 2d–e).
Figure 1. Sequencing HG002 with highly-accurate, long reads.
(a) Circular consensus sequencing (CCS) derives a consensus or CCS read from multiple passes of a single template molecule, producing accurate reads from noisy individual subreads (passes). (b) Accuracy – predicted by CCS software – of reads with different numbers of passes, for sequencing of the human male HG002. At 10 passes, the median read achieves Q30 predicted accuracy. Orange lines are medians; boxes extend from lower to upper quartiles; whiskers extend 1.5 interquartile distances; n=1,000 CCS reads for each number of passes. (c) Length and predicted accuracy of CCS reads.
Quality Evaluation of CCS Reads
To characterize the few residual errors in CCS reads, discordances between the reads and the GIAB HG002 benchmark were tallied. The average read concordance is 99.8%, comparable to the concordance of short reads from the Illumina NovaSeq (99.9% for 2×151 bp reads) and HiSeq 2500 (99.5% for 2×250 bp reads) (Supplementary Table 2). The large majority of CCS read discordances are indels in homopolymer contexts: 3.4% are mismatches, 4.6% are indels in non-homopolymer contexts, and 92.0% are indels in homopolymers. This equates to a mismatch every 13,048 bp in CCS reads, a non-homopolymer indel every 9,669 bp, and a homopolymer indel every 477 bp (Supplementary Table 2). The mismatch rate is 17× lower than reads from the Illumina NovaSeq, while the indel rate is 181× higher (Supplementary Table 2).
To confirm independently the high quality of CCS reads, error rates were measured through read-to-read alignments27. Consistent with the reference-based methods, the average read accuracy is estimated at 99.8%. A putative large artifact is detected in 0.6% of reads: 0.5% are molecular chimeras, likely due to ligation of DNA fragments during library construction, 0.1% contain a “low quality” run of bases, anecdotally in microsatellites, and 0.03% have a missing SMRTbell adapter on one end. Overall, the read-to-read comparison supports the predicted quality of the CCS reads.
Increased Mappability of CCS Reads
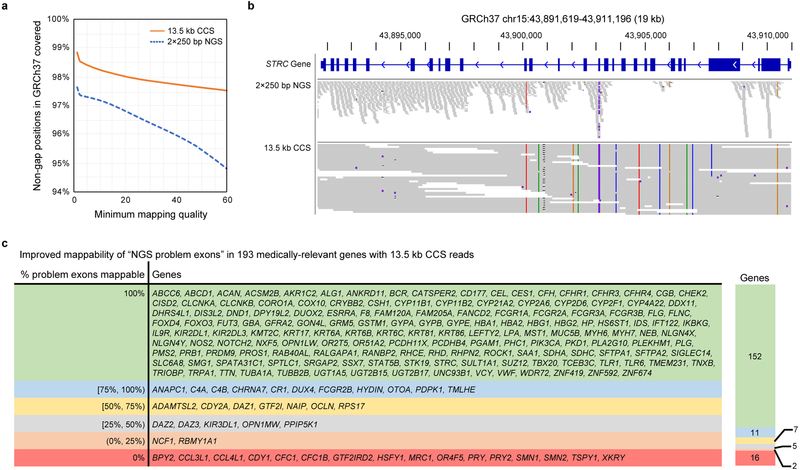
To evaluate increases in mappability with long reads, the 13.5 kb CCS reads and a coverage-matched number of 2×250 bp NGS short reads were mapped to GRCh37. A genomic position was considered to be mappable if it is covered by at least ten reads. The CCS reads cover more of the genome at all mapping quality values. At the highest reported value (60), 97.5% of the non-gap GRCh37 is mappable with 13.5 kb CCS reads, while 94.8% is mappable with NGS short reads (Figure 2a).
Figure 2. Mappability of the human genome with CCS reads.
(a) Percentage of the non-gap GRCh37 human genome covered by at least 10 reads from 28-fold coverage NGS (2×250 bp, HiSeq 2500) and CCS (13.5 kb) datasets at different mapping quality thresholds. (b) Coverage of the congenital deafness gene STRC in HG002 with 2×250 bp NGS reads and 13.5 kb CCS reads at a mapping quality threshold of 10. (c) Improvement in mappability with 13.5 kb CCS reads for 193 human genes previously reported as medically-relevant and problematic to map with NGS reads28.
The additional regions that are now accessible with longer CCS reads include numerous medically-relevant genes which have been previously reported as recalcitrant to NGS sequencing28. Of the 193 reported medically-relevant genes with at least one NGS problem exon, 152 are fully mappable with the CCS reads, including CYP2D6, GBA, PMS2, and STRC (Figure 2b–c).
The 13.5 kb CCS reads also resolve complex regions, like the HLA class 1 and 2 genes, which are fully phased and typed to four-field resolution29 (Supplementary Figure 3).
Small Variant Detection with CCS Reads
GATK30 was used to call SNVs and small indels (<50 bp) with CCS reads. Evaluated against the GIAB benchmark26, genome-wide precision for SNVs is 99.468% and recall is 99.559%. For indels, precision is 78.977% and recall is 81.248%. While GATK performance with CCS reads is comparable to NGS for SNVs, it is lower for indels (Table 1). Unlike NGS read errors, which are mostly mismatches that are modeled well by base quality scores from the read, CCS read errors are mostly indels for which GATK uses fixed models designed for NGS reads (Supplementary Table 2), likely contributing to the low indel precision and recall of GATK for CCS reads.
Table 1. Performance of small variant calling with CCS reads.
Precision, recall, and F1 of small variant calling measured against the Genome in a Bottle v3.3.2 benchmark using hap.py. Bold indicates the highest value in each column. Underline indicates a value higher than the GATK HaplotypeCaller run on 30-fold Illumina NovaSeq reads. Coverage is 28-fold for PacBio CCS and 30-fold for Illumina NovaSeq. Rows are sorted (“^”) based on F1 for SNVs.
| Variant caller (training model) | SNVs | Indels | |||||
|---|---|---|---|---|---|---|---|
| Platform | Precision | Recall | F1 ^ | Precision | Recall | F1 | |
| Illumina (NovaSeq) | DeepVariant (Illumina model) | 99.960% | 99.940% | 99.950% | 99.633% | 99.413% | 99.523% |
| PacBio (CCS) | DeepVariant (CCS model) | 99.914% | 99.959% | 99.936% | 96.901% | 95.980% | 96.438% |
| PacBio (CCS) | DeepVariant (haplotype-sorted CCS model) | 99.904% | 99.963% | 99.934% | 97.835% | 97.141% | 97.486% |
| Illumina (NovaSeq) | GATK HaplotypeCaller (no filter) | 99.852% | 99.910% | 99.881% | 99.371% | 99.156% | 99.264% |
| PacBio (CCS) | GATK HaplotypeCaller (hard filter) | 99.468% | 99.559% | 99.513% | 78.977% | 81.248% | 80.097% |
Variant callers based on deep learning have an inherent ability to adapt to the error profiles of new data types31,32. To evaluate variant calling with a deep learning framework, Google DeepVariant32 was used to call SNVs and indels from CCS reads, with chromosome 20 held out during model training and selection. Using a model trained on Illumina reads, precision on chromosome 20 is 99.533% and recall is 99.793% for SNVs, and precision is 23.991% and recall is 81.692% for indels (Supplementary Table 3). Training a model on CCS reads provides a large boost in precision and recall for both SNVs and indels. For SNVs, DeepVariant achieves genome-wide precision of 99.914% and recall of 99.959%. For indels, DeepVariant achieves 96.901% precision and 95.980% recall (Figure 3a, Table 1). Performance is similar on the held-out chromosome 20 (Supplementary Table 3). Most discordant indels (90.33%) occur in homopolymer runs, matching the most common discordance in CCS reads (Supplementary Table 2). The callset includes 1,969 SNVs and 62 indels in exons of 193 medically-relevant genes previously reported as recalcitrant to NGS sequencing.
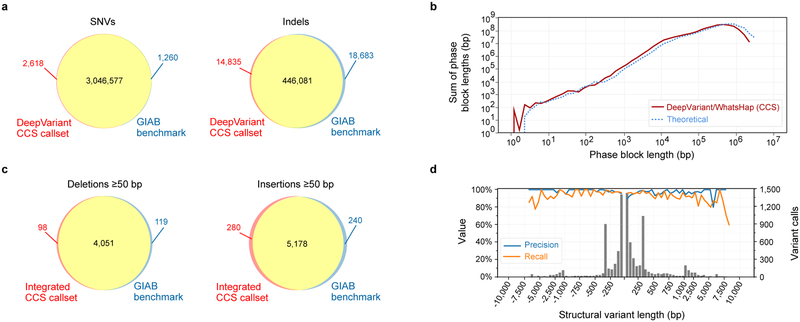
Figure 3. Variant calling and phasing with CCS reads.
(a) Agreement of DeepVariant SNV and indel calls with Genome in a Bottle v3.3.2 benchmark measured with hap.py. (b) Phasing of heterozygous DeepVariant variant calls with WhatsHap, compared to theoretical phasing of HG002 with 13.5 kb reads. (c) Agreement of integrated CCS structural variant calls with the Genome in a Bottle v0.6 structural variant benchmark measured with Truvari, (d) by variant length. Negative length indicates a deletion; positive length indicates an insertion. The histogram bin size is 50 bp for variants shorter than 1 kb, and 500 bp for variants >1 kb. All comparisons to GIAB are for the benchmark subset of the genome.
Phasing Small Variants with CCS Reads
To determine whether CCS reads could provide both highly-accurate variant calls and long-range information needed to generate haplotypes, we used WhatsHap33 to phase the DeepVariant variant calls. Nearly all (99.64%) autosomal heterozygous variants were phased into 19,215 blocks with an N50 of 206 kb (Supplementary Table 4). The phase block length distribution closely matches the theoretical limit estimated by creating breaks between variants that are separated by more than the average CCS read length of 13.5 kb. This suggests that the phase block length is limited by read length and the amount of variation in HG002, not by coverage or the quality of the variant calls (Figure 3b, Supplementary Figure 4). Evaluated against the GIAB benchmark phase set, the switch error rate is 0.37% and the Hamming error rate is 1.91% (Supplementary Table 4).
Improving Small Variant Detection with Haplotype Phasing
GATK and DeepVariant do not directly incorporate long-range haplotype phase information when calling variants. To evaluate whether phase information from reliable SNV calls improves results, particularly for less-reliable indel calls, CCS reads were haplotype-tagged based on trio-phased variants from GIAB (which tags 84.55% of reads) and a DeepVariant model was then trained on reads passed in haplotype-sorted order. The haplotype-sorted model performs similarly to the original DeepVariant CCS model for SNVs, but it reduces indel false negatives and false positives by around 30%, achieving precision of 97.835% and recall of 97.141% (Table 1).
Structural Variant Detection with CCS Reads
Insertion and deletion structural variants ≥50 bp were called using two read mapping-based tools, pbsv (https://github.com/PacificBiosciences/pbsv) and Sniffles34. The callsets show similar precision (>94%) and recall (>91%) against the GIAB benchmark (Supplementary Table 5). Precision is consistent across variant length, but recall is lower for variants ≥3kb (Supplementary Figure 5a–b). To increase recall for larger variants, haplotype-resolved de novo assemblies were analyzed with paftools35 (see “De Novo Assembly of CCS Reads”), with precision >93% and recall >89% (Supplementary Figure 5c–d, Supplementary Table 5).
An integrated callset includes 12,091 insertions and 8,432 deletions. Precision is 96.13% and recall is 95.99% (Figure 3c, Supplementary Table 5), with similar performance for insertions as deletions and for variants less than 1kb as for greater than or equal to 1kb (Figure 3d), indicating the complementarity of mapping- and assembly-based structural variant calling. The callset has 143 insertions and 163 deletions that intersect exons.
For comparison, structural variants were called in PacBio continuous (“noisy”) long reads (CLR) (with pbsv and Sniffles), Illumina 2×250 bp short reads25 (with Manta36 and Delly37), and 10X Genomics linked reads25 (with LongRanger38). The CCS callset exceeds all others in both precision and recall. The closest in performance is the pbsv CLR callset, which has precision of 94.64% and recall of 94.48%. The Manta callset has precision of 85.34% and recall of 55.88%, with much worse recall for insertions (39.65%) than deletions (76.90%). The LongRanger callset has precision of 83.79% and recall of 39.83%, again with worse recall for insertions (16.41%) than deletions (70.18%). A callset from paftools run on a linked-read SuperNova assembly has precision of 64.52% and recall of 52.74%. All considered short- and linked-read callsets have worse performance than all CCS and CLR callsets in both precision and recall (Supplementary Table 5, Supplementary Figure 5).
De Novo Assembly of CCS Reads
Three different algorithms – FALCON39, Canu40, and wtdbg241 – were used to assemble the full CCS read set, which is a mix of paternal and maternal reads. By skipping the initial read-to-read error correction step, the algorithms completed 10–100× faster than is typical for long-read assemblies21 (Supplementary Table 6). All assemblies have high contiguity with a contig N50 from 15.43 to 28.95 Mb. The total assembly size is near the expected human genome size for FALCON and wtdbg2. The Canu assembly has a total genome size of 3.42 Gb, larger than the expected haploid human genome, because it resolves some heterozygous alleles into separate contigs (Table 2, Supplementary Figure 6).
Table 2. Statistics for de novo assembly of CCS reads.
The “mixed” haplotype assemblies use all reads. The “maternal” and “paternal” assemblies use parent-specific reads from trio binning plus unassigned reads. HG002 concordance is measured against the Genome in a Bottle benchmark. BUSCO gene completeness uses the Mammalia ODB9 gene set. Ensembl genes is the percentage of genes from Ensembl R94 that are full-length, single-copy in the assembly relative to the full-length, single-copy count for GRCh38. Contigs shorter than 13 kb were excluded from genome size and contiguity measurements; contigs shorter than 100 kb were excluded from the concordance measurement. “*” indicates polishing with Arrow.
| Canu | 3.42 | 18,006 | 22.78 | 25.02 | 108.46 | 30.16 | 31.1 | 92.3% | 93.2% |
| FALCON | 2.91 | 2,541 | 28.95 | 24.51 | 110.21 | 38.04 | 25.8 | 87.6% | 97.6% |
| wtdbg2 | 2.79 | 1,554 | 15.43 | 12.62 | 84.67 | 22.61 | 44.6 | 94.2% | 96.1% |
| Canu* | 3.04 | 5,854 | 18.02 | 17.04 | 48.81 | 19.78 | 47.2 | 94.1% | 98.1% |
| FALCON* | 2.80 | 924 | 19.99 | 15.54 | 74.33 | 24.07 | 43.5 | 95.1% | 97.8% |
| wtdbg2 | 2.75 | 2,637 | 12.10 | 9.29 | 66.34 | 16.55 | 43.5 | 93.8% | 95.6% |
| Canu* | 2.96 | 6,868 | 16.14 | 14.90 | 64.83 | 20.19 | 47.7 | 93.4% | 98.2% |
| FALCON* | 2.70 | 1,489 | 16.40 | 14.06 | 95.34 | 25.61 | 43.5 | 93.6% | 97.7% |
Short reads from the parents of HG002 were used to identify k-mers unique to one parent and then partition (“trio bin”) the CCS reads by haplotype42. Three different k-mer sizes were evaluated: 21 bp (previously reported for trio binning) and longer k-mers of 51 bp and 91 bp enabled by the accuracy of CCS reads. The 21-mer binning assigns 35.3% of reads to the mother and 33.6% to the father (68.9% binned). The 51-mer binning is more complete at 78.5% binned; using longer 91-mers provides only a small additional gain to 79.2% binned. The 51-mer binning was selected for assembly (Supplementary Table 7).
FALCON, Canu, and wtdbg2 were run separately on the paternal and maternal reads, with the unassigned reads included in both sets. All algorithms produce highly contiguous and nearly complete assemblies for the parental genomes, with N50 from 12.10 to 19.99 Mb and genome size from 2.67 to 3.04 Gb (Table 2). From 95.3% to 98.2% of human genes are identified as single-copy in each parental assembly (Table 2). Assembly-based structural variant calls have high precision and recall, suggesting few large-scale mis-assemblies (Supplementary Table 5). Furthermore, analysis of the phase-consistency43 of maternal and paternal haplotigs shows the assemblies are phased properly (Supplementary Figure 7).
All mixed and parental assemblies are high quality with concordance to the HG002 benchmark ranging from Q44-Q48 for polished44 and Q26-Q45 for unpolished assemblies (Table 2, Supplementary Table 8). This greatly exceeds that of previously published and accessioned assemblies at Q40 (6× worse) for PacBio noisy long reads and Q29 (77× worse) for Oxford Nanopore reads with Illumina polishing (Figure 4a, Supplementary Table 8).
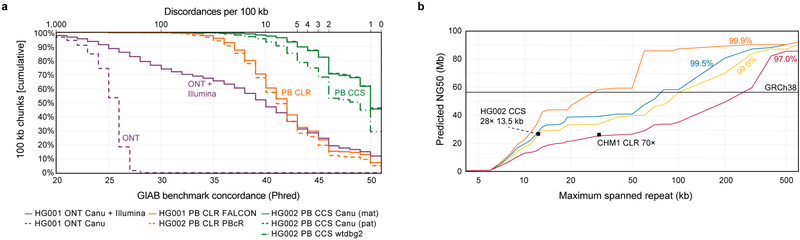
Figure 4. Impact of read accuracy on de novo assembly.
(a) The concordance of seven assemblies to the Genome in a Bottle (GIAB) v3.3.2 benchmark (Supplementary Table 8). Contigs longer than 100 kb were segmented into 100 kb chunks and aligned to GRCh37. Concordance was measured per chunk, and chunks with no discordances were assigned concordance of Q51. PB=PacBio, ONT=Oxford Nanopore, CLR=continuous (“noisy”) long reads. (b) Predicted contiguity of a human assembly based on ability to resolve repeats of different lengths (x-axis) and percent identities (colored lines)21. The solid line indicates the contiguity of GRCh38. The 97.0% identity line is representative of CLR assemblies using standard read-to-read error correction. The points show example CCS and CLR46 assemblies using Canu. Repeat identity and length are proxies for read accuracy and length.
Large segmental duplications often result in contig breaks in de novo assemblies, and assemblies of noisy long reads typically span less than 50 of 175 Mb of segmental duplications in the human genome15,18,45. The most contiguous assemblies of CCS reads span over 60 Mb of segmental duplications, a 20% improvement (Supplementary Table 9). A model of assembly contiguity based on large repeat resolution suggests that the current assemblies of CCS reads resolve 15 kb repeats of 99 to 99.5% identity (Figure 4b).
Coverage Requirements for Variant Calling and De Novo Assembly
To evaluate the depth of CCS read coverage required for variant calling and assembly, we randomly subsampled from the full dataset. For SNVs, precision and recall with DeepVariant remain above 99.5% for coverage down to 15-fold; performance decays steeply below 10-fold (Supplementary Figure 8a). For indels, DeepVariant remains comparable to typical NGS performance (>90%) down to 17-fold coverage (Supplementary Figure 8b). For structural variants, precision with pbsv is above 95% for all evaluated coverage levels. Recall is above 90% down to 15-fold coverage, and decays steeply below 10-fold (Supplementary Figure 8c). For phasing with WhatsHap, the phase block N50 remains above 150 kb down to 10-fold coverage (Supplementary Figure 8d). Mixed-haplotype wtdbg2 assemblies have consistent size above 2.7 Gb, contig N50 around 15 Mb, and concordance above Q42 until coverage falls below 15-fold (Supplementary Figure 8e–g).
Revising and Expanding Genome in a Bottle Benchmarks
High-quality callsets from CCS reads provide an opportunity to identify mistakes in the GIAB benchmarks, particularly for structural variants where the benchmark is still in draft form. Sixty small variant and 40 structural variant discrepancies between the GIAB benchmark (small variant v3.3.2, structural variant v0.6) and the CCS callsets (DeepVariant haplotype-sorted, structural variant integrated) were selected for manual curation. Selected variants were spread across variant types, discrepancy types, and both inside and outside homopolymers and tandem repeats.
For small variants, 29 of 31 discrepancies in homopolymers were classified as correct in the benchmark. Outside of homopolymers, 19 of 29 were classified as errors in the benchmark. Most of these benchmark errors (13 of 19) are true variants in L1 elements called homozygous reference in GIAB (Supplementary Figure 9a–b, Supplementary Table 10). The identified benchmark errors overlap with putative errors in a DeepVariant Illumina whole genome case study (https://github.com/google/deepvariant). Of 745 putative false positive (i.e. in callset but not benchmark) SNVs in the case study, 344 agree with the CCS callset, with 282 (82.0%) falling within large interspersed repeats. Fewer of the false negative (i.e. in benchmark but not callset) SNVs (8%), false negative indels (25%), and false positive indels (19%) from the case study agree with the CCS callset. Extrapolating from manual curation, we estimate that 2,434 (1,313–2,611; 95% confidence interval) errors in the current GIAB benchmark could be corrected using the CCS reads.
For structural variants, curator classification was unclear for 11 of 40 discrepancies, typically because of tandem repeat structure that permits multiple representations of a variant. For the remainder, 15 of 16 false negative discrepancies were classified as correct in the benchmark. However, for false positive discrepancies, 11 of 13 were classified as errors in the benchmark (Supplementary Figure 9c–d, Supplementary Table 11). This suggests that the GIAB structural variant benchmark set is precise but incomplete.
The high-quality CCS callsets also provide an opportunity to expand the benchmarks into repetitive and highly polymorphic regions that have been difficult to characterize with confidence using short reads. Adding the CCS DeepVariant callset to the existing GIAB small variant integration pipeline would expand the benchmark regions by up to 1.3% and 418,875 variants (210,184 SNVs and 208,691 indels). For structural variants, only 9,232 of 18,832 autosomal variant calls overlap benchmark regions, which means that the number of variants in the benchmark would more than double if all CCS variants calls were incorporated.
Discussion
We present a protocol for producing highly-accurate long reads using circular consensus sequencing (CCS) on the PacBio Sequel System. We apply the protocol to sequence the human HG002 to 28-fold coverage with average read length of 13.5 kb and an average read accuracy of 99.8%. We analyze the CCS reads to call SNVs, indels, and structural variants; to phase variants into haplotype blocks; and to de novo assemble the HG002 genome. We demonstrate that CCS reads from a single library approach the accuracy of short reads for small variant detection, while accessing more of the genome including in medically-relevant genes. CCS reads also enable structural variant detection and de novo assembly at similar contiguity and markedly higher concordance than noisy long reads.
The CCS performance for SNV and indel calling rivals that of the commonly-used pairing of BWA and GATK on 30-fold short-read coverage. Interestingly, though the overall accuracy of CCS reads is similar to short reads, direct application of the GATK pipeline to CCS reads produces inferior results, especially for indels. The major residual error in CCS reads – indels in homopolymers – is not as frequent in short reads. We suspect that the current GATK, which was designed for short reads, does not properly model the CCS error profile, and thus performance lags for indels. This is supported by results with DeepVariant. When a DeepVariant model trained on Illumina reads is run on CCS reads, the performance is poor for indels. When DeepVariant is trained on CCS reads, performance improves dramatically. As more CCS datasets are made available, both model-based callers like GATK and learning-based callers like DeepVariant will have the opportunity to improve on the performance reported here, including by incorporating haplotype phase information and evaluating and training against updated GIAB benchmarks that correct errors using CCS reads. Further, advances in sequencing chemistry or consensus base calling (such as the application of deep learning) that reduce the residual indel errors in CCS reads also could improve variant calling performance.
Structural variant calling and de novo genome assembly with CCS reads match or exceed that reported for noisy long reads. The CCS reads have an advantage of high accuracy, which eliminates the need for read correction, allows more stringent criteria to be used in variant calling or read overlapping, and ultimately produces more accurate assemblies and variant calls. Noisy long reads have an advantage of longer maximum read length, but increased accuracy of CCS reads compensates for the length required for highly contiguous assembly. Modeling (Figure 4b) suggests modest advances in accuracy (to 99.9%) at 15 kb read length would double the current contiguity, which already matches the best published de novo assemblies46.
Evaluation of variant calls and assemblies relies on high-quality benchmarks and supporting tools47. For small variants, CCS reads provide an opportunity to improve the GIAB benchmark by expanding into difficult genomic regions and correcting errors, primarily in large interspersed repeats. For structural variants, both the GIAB set and benchmarking tools need to be further developed. It remains a challenge to reconcile when two structural variant calls describe the same allele, particularly in tandem repeats. Moreover, the GIAB structural variant set is not as complete or accurate as for small variants. Including structural variant calls from CCS reads offers to improve the GIAB set.
The CCS read approach alleviates some challenges of long-read sequencing. First, aiming for fragments in the 10–20 kb size range relaxes the need to isolate ultra-long genomic DNA. Second, increased accuracy allows for more stringent alignment and overlap comparisons, greatly reducing the compute time and cost while improving assembly results by recognizing fine-grained repeat and haplotype phase information. Third, familiar tools like GATK that were developed for accurate short reads are readily applied to CCS reads. Fourth, manual interpretation in genome browsers is easier with accurate reads. Finally, increased accuracy could enable calling of low-frequency somatic variants.
Variant calling and assembly with CCS reads perform well down to 15-fold coverage, which offsets the current reduced throughput per run compared to noisy long reads. The newer Sequel II System provides more CCS reads per run (8× compared to this study), simplifying the workflow, and reducing data collection to 2–3 SMRT Cells for 15-fold coverage of a human genome (Supplementary Table 12). Improved CCS consensus calling algorithms and increased polymerase read length will enable longer inserts and further increase throughput and quality. Together, this will facilitate rapid, population-scale analysis of full genomes with CCS reads to improve human health.
Online Methods
Library Preparation
All sequencing libraries were prepared using SMRTbell Template Prep Kit 1.0 (Pacific Biosciences Ref. No. 100-259-100). To minimize ligation chimeras, hairpin adapters were ligated overnight at 500× molar excess to the genomic fragment molecules. Sequencing primers were conditioned by heating to 80°C for 2 minutes and rapidly cooled to 4°C . The sequencing primer was annealed to the template at a molar ratio of 20:1 (primer:template) for 30 minutes at 20°C. After primer annealing, polymerase was bound to the primed template at a molar ratio of 10:1 (polymerase:template) for 4 hours at 30°C. Polymerase-bound samples were then kept at 4°C prior to use. Prior to sequencing, excess unbound polymerase was removed by incubating the complexes for 5 minutes with 0.6× (vol:vol) AMPure PB beads (Pacific Biosciences Ref. No. 100-265-900) at room temperature. Beads were not washed with 80% ethanol. After removing the free polymerase in the supernatant, the polymerase-bound complexes were eluted with MagBead Binding Buffer v2 (Pacific Biosciences Ref. No. 101-046-400). Modifications to this protocol are listed below for the amplicon, E. coli large insert, lambda digest, and human sequencing libraries.
10 kb E. coli PCR Amplicon Library Preparation
Library input DNA was generated by amplification of a 10 kb region of E. coli strain K12 (CP032667.1:871,407–881,407) using genomic DNA as the template. Products were purified using Sage BluePippin with a 9 kb high-pass cutoff on a 0.75% agarose cassette (Sage Science Product No. PAC30KB) and the library was constructed as described above.
30 kb E. coli Large-Insert Library Preparation
A SMRTbell library was prepared using unsheared, high molecular weight (> 50 kb) genomic DNA from E. coli strain K12. Prior to annealing primers, the library was size-selected on the Sage BluePippin using a 0.75% agarose cassette (Sage Science Product No. PAC30KB) and a 30 kb high-pass cutoff.
Lambda-Digest Library Preparation
Lambda DNA (New England BioLabs Product No. N3011L) was digested with BsaAI (New England BioLabs Product No. R0531L) and the fragments purified with AMPure and constructed into SMRTbells as described above.
Human CCS Library Preparation
Library preparation was performed on the human reference genome sample HG002 obtained from NIST. Genomic DNA was sheared using the Megaruptor from Diagenode with a long hydropore cartridge and a 20 kb shearing protocol. Prior to library preparation, the size distribution of the sheared DNA was characterized on the Agilent 2100 BioAnalyzer System using the DNA 12000 kit. In order to tighten the size distribution of the SMRTbell library, the sample was separated into 3 kb fractions on the SageELF System from Sage Science using the SageELF Native Agarose for DNA 0.75% 1–18kb cassette (Sage Science Product No. ELD7510). Fractions having the desired size distributions were identified on the Agilent 2100 BioAnalyzer using the DNA 12000 kit (Supplementary Figure 1c–f). Fractions centered at 10 kb, 15 kb, and 18 kb were used for sequencing with the 15 kb fraction selected for high-coverage sequencing.
Sequencing
All sequencing reactions were performed on the PacBio Sequel System with the Sequel Sequencing Kit 3.0 chemistry (Pacific Biosciences Ref. No. 101-500-400 and 101-427-800). The 10 kb E. coli PCR amplicon and 30 kb E. coli large-insert libraries were sequenced with no pre-extension and 10-hour collection time. The lambda-digest library was sequenced with 0 and 5-hour pre-extension and 4-hour collection time. The HG002 human libraries were sequenced with 4- or 12-hour pre-extension and 20-, 24-, or 30-hour collection depending on insert length.
Consensus Read Generation
Consensus reads (“CCS reads”) were generated using the ccs software version 3.0.0 (https://github.com/pacificbiosciences/unanimity/) with --minPasses 3 --minPredictedAccuracy 0.99 --maxLength 21000. For the 13.5 kb human library, average run time is 3,035 CPU core hours per SMRT Cell (118,365 total). Total CCS read yield was 89 Gb (2.3±0.4 Gb per SMRT Cell), with read length of 13.5±1.2 kb.
Read Mapping
Reads were mapped to the GRCh37 human reference genome, specifically the hs37d5 build from the 1000 Genome Project48. CCS reads were mapped using pbmm2 version 0.10.0 (https://github.com/PacificBiosciences/pbmm2) with --preset CCS. CLR reads were mapped using pbmm2 with --preset SUBREAD. NGS reads were mapped with minimap235 version 2.14-r883 with -x sr.
Measuring HG002 Concordance
To measure concordance to HG002, alignments to GRCh37 were evaluated at positions within GIAB v3.3.2 benchmark regions that have no variant call26. Concordance = M/(M+X+D+I) where M is the number of matches, X is the number of mismatches, D is the number of deletion basepairs, and I is the number of insertion basepairs. Phred = min[−10×log10(1-Concordance), −10×log10(1/(1+ReadLength))]
A deleted basepair is considered a homopolymer deletion when it matches the preceding or following basepair in the reference genome. An insertion is considered a homopolymer insertion when the basepairs of the insertion are identical and match either the preceding or following basepair in the reference genome.
The 2×250 bp Illumina HiSeq 2500 reads were obtained from GIAB25 (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/NIST_Illumina_2×250bps/ or https://bit.ly/2WjH7tF) and mapped to GRCh37 with minimap235 version 2.14-r883 with -x sr. The 2×151 bp Illumina NovaSeq reads were obtained from SRX5648942.
Coverage by [GC] Content
To measure coverage by local [GC] content, bedtools49 version 2.27.1 was used to divide the GRCh37 reference genome into 500 bp windows (bedtools makewindows -w 500) and then to calculate the [GC] content (bedtools nuc) and average coverage (bedtools coverage -mean) of each window.
Reference-independent Quality Evaluation
The Dazzler suite (https://dazzlerblog.wordpress.com/) was used to evaluate the accuracy of the CCS reads without relying on a reference genome. Briefly, daligner27 commit 381fa920 was used to align pairs of CCS reads and produce all local alignments longer than 1 kb with less than 5% difference in sequence. Each CCS subject read was partitioned into 100 bp panels, within which its coverage by and concordance to other reads aligning to it was calculated. Panels with a concordance in the worst 0.1% of all panels were considered low quality. Abrupt ends in the alignment of 5 or more reads to a given panel along the CCS subject read were used to estimate library artifacts like chimeric molecules and missing adapters.
Mappability of CCS and NGS Reads
To compare with the mappability of 13.5 kb CCS reads, a coverage-matched (89 Gb) set of 2×250bp Illumina HiSeq 2500 reads for HG002 was obtained from GIAB25 (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/NIST_Illumina_2×250bps/ or https://bit.ly/2WjH7tF) and mapped to GRCh37 with minimap235 version 2.14-r883 with -x sr.
A genome position is considered mappable if it is covered by alignments for at least ten reads at a specified mapping quality or higher, which was evaluated using bedtools bamtobed and bedtools genomecov -bga. Gaps (“N” basepairs in the reference) were excluded.
Previously-reported NGS problem exons28 were considered mappable if every basepair in the exon is covered by a read at mapping quality of 60.
HLA Typing
The HLA-A and HLA-DPA1 genes were typed by comparing the sequence of CCS reads that span the genes to entries in the IMGT database50 version 3.19.0.
Small Variant Detection and Benchmarking
To develop a workflow for calling variants in CCS reads with GATK30 HaplotypeCaller v4.0.6.0, different values of the HaplotypeCaller parameter --pcr-indel-model and VariantFiltration parameter --filter-expression were considered to maximize SNV and indel F1 without excessive complication, starting from the GATK best practices for hard filtering. In the end, HaplotypeCaller was run on reads with a minimum mapping quality of 60 using allele-specific annotations (--annotation-group AS_StandardAnnotation) and --pcr_indel_model AGGRESSIVE. Autosomes and the pseudo-autosomal regions (PARs) on chromosome X were called with --ploidy 2; chromosome Y and the non-PAR regions of chromosome X were called with --ploidy 1. Multi-allelic variant sites were split into separate entries for filtration with a custom script. SNVs were filtered using GATK VariantFiltration with --filter_expression of AS_QD < 2.0 for SNVs and indels longer than 1bp, and AS_QD < 5.0 for 1 bp indels. A similar pipeline was used to call variants in coverage-matched 2×151 bp Illumina NovaSeq reads with a few differences: a minimum mapping quality of 20, --pcr-indel-model NONE, --standard_min_confidence_threshold_for_calling 2.0, and no variant filtration.
A Google DeepVariant model for CCS reads was generated as previously reported32 using DeepVariant version 0.7.1. Briefly, models were trained using CCS reads for chromosomes 1–19 and the HG002 GIAB v3.3.2 benchmark. A single model was selected based on performance in chromosomes 21 and 22 to avoid overfitting. Neither training nor model selection considers chromosome 20, which is available for accuracy evaluations. To support long reads, local reassembly is disabled for DeepVariant with CCS reads. The wgs_standard model version 0.7.1 was used to call variants in NovaSeq reads and to apply a model trained on Illumina reads to CCS reads.
To incorporate long-range haplotype information, DeepVariant was modified to produce pileups with reads sorted by the BAM haplotype (“HP”) tag. Haplotype information was added to the pbmm2 CCS alignments using WhatsHap v0.17 (whatshap haplotag) with the trio-phased variant calls from GIAB (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/NIST_MPI_whatshap_08232018/RTG.hg19.10x.trio-whatshap.vcf.gz or https://bit.ly/2R73grR). A new DeepVariant model then was trained as described above.
Small variant callsets were benchmarked against the GIAB v3.3.2 HG002 set26 by vcfeval51 (https://github.com/RealTimeGenomics/rtg-tools) with no partial credit run through hap.py version 0.3.10 (https://github.com/Illumina/hap.py). Only PASS calls were considered.
Phasing Small Variants
Small variant calls were phased using WhatsHap v0.17 (whatshap phase). The number of switch and Hamming errors was computed against trio-phased variant calls from GIAB using whatshap compare.
To model the phase blocks achievable with a given read length, cuts were introduced between heterozygous variants in the GIAB trio-phased variant callset that are separated by more than the read length, which effectively assumes that adjacent heterozygous variants separated by less than the read length can be phased.
Structural Variant Detection
pbsv version 2.1.0 (https://github.com/PacificBiosciences/pbsv) was run on pbmm2 CCS read alignments. The pbsv discover stage was run separately per chromosome with tandem repeat annotations (https://github.com/PacificBiosciences/pbsv/tree/master/annotations) passed with --tandem-repeats. The pbsv call stage was run on the full genome.
Sniffles version 1.0.10 was run on pbmm2 CCS reads alignments with -s 3 --skip_parameter_estimation and with the variant sequence obtained from reads.
Structural variants in the maternal and paternal Canu and FALCON assemblies from CCS reads (see “De novo Assembly”) were called using a previously described workflow52. Briefly, contigs were mapped to GRCh37 using minimap2 --paf-no-hit -cx asm5 --cs -r 2k; variants were called with paftools.js call35; maternal and paternal variants were concatenated; and indel calls at least 30 bp were retained.
An integrated callset was produced from the pbsv, Sniffles, and paftools/Canu callsets using SURVIVOR53 and custom scripts. Two calls were considered supporting if the calls had the same structural variation type, a start position within 1 kb, and a difference in length less than 5%. One call from each matching set was retained with precedence given to pbsv, then Sniffles, and then paftools. Because pbsv and Sniffles have poor sensitivity for calls larger than 1 kb, all non-matched calls from paftools that are larger than 1 kb were retained. The callset integration is robust to more stringent definitions of supporting calls (99.1% identical when requiring start position within 100 bp instead of 1 kb).
Structural variants were called in pbmm2 alignments of PacBio CLR reads (SRX5590586) with pbsv version 2.1.0 exactly as for the CCS reads, and with Sniffles version 1.0.10 with default parameters.
NovoAlign (http://www.novocraft.com) alignments to GRCh37 of 300-fold coverage of HG002 with 2×250bp Illumina HiSeq 2500 reads were obtained from GIAB. Structural variants were called with Manta36 version 1.4.0 with all coverage, and Delly37 version 0.7.6 with coverage subsampled to 30-fold using samtools view -b -s 0.1.
Structural variant callsets on 10X Genomics reads from LongRanger version 2.2 were obtained from GIAB (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/10XGenomics_ChromiumGenome_LongRanger2.2_Supernova2.0.1_04122018/ or https://bit.ly/2Mtj084). Insertion and deletion variants at least 30 bp were combined from the sequence-resolved indels and large deletion calls (NA24385_LongRanger_snpindel.vcf.gz, NA24385_LongRanger_sv_deletions.vcf.gz). Another callset was produced using paftools on the diploid Supernova 2.0.1 assembly as described above.
Structural variant callsets were benchmarked against the GIAB v0.6 HG002 structural variant set (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/NIST_SVs_Integration_v0.6 or https://bit.ly/2T7iLBX) using Truvari (https://github.com/spiralgenetics/truvari) commit 600b4ed7 modified to allow a single variant in the benchmark set to support multiple variants in the callset. Truvari was run with -r 1000 -p 0.01 --multimatch --includebed HG002_SVs_Tier1_v0.6.bed -c HG002_SVs_Tier1_v0.6.vcf.gz. The -p 0 option was used to disable sequence checks for callsets that report symbolic alleles instead of sequence-resolved calls (LongRanger, Delly).
De novo Assembly
Mixed haplotype assemblies were produced using all CCS reads. Canu40 version 1.7.1 was run with -p asm genomeSize=3.1g correctedErrorRate=0.015 ovlMerThreshold=75 batOptions=“-eg 0.01 -eM 0.01 -dg 6 -db 6 -dr 1 -ca 50 -cp 5” -pacbio-corrected. FALCON39 kit version 1.2.0 was run with ovlp_HPCdaligner_option = -v -B128 -M24 -k24 -h1024 -e.97 -l2500 -s100, ovlp_DBsplit_option = -s400, and overlap_filtering_setting = --max-diff 90 --max-cov 120 --min-cov 2. Wtdbg2 (https://github.com/ruanjue/wtdbg2) version 2.2 was run with -k 0 -p 21 -AS 4 -s 0.5 -e 2 -K 0.05 and followed by wtdbg2-cns.
CCS reads from HG002 were “trio binned” as maternal, paternal, or unassigned as previously described42. Briefly, 2×250 bp Illumina HiSeq 2500 reads for the father (HG003/NA24149) and mother (HG004/NA24143) of HG002 were obtained from GIAB (HG003: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG003_NA24149_father/NIST_Illumina_2×250bps/ or https://bit.ly/2TADePc; HG004: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG004_NA24143_mother/NIST_Illumina_2×250bps/ or https://bit.ly/2uw7joJ). Sequence k-mers unique to the mother or father were identified and used to categorize CCS reads (https://github.com/skoren/triobinningScripts), using k-mer size of 21, 51, and 91 and excluding k-mers that occur 25 times or fewer. The maternal and unassigned reads were used for the “maternal” assemblies; paternal and unassigned reads were used for the “paternal” assemblies.
The maternal and paternal assemblies were generated with Canu and wtdbg2 using the same software version and same options as for the mixed haplotype assembly. For the maternal and paternal assemblies, FALCON version 0.7 was run with length_cutoff_pr = 2000, ovlp_HPCdaligner_option = -k24 -e.95 -s100 -l1000 -h600 -mdust -mrep8 -mtan -M21, ovlp_DBsplit_option = -x2000 -s400, falcon_sense_option = --min_idt 0.70 --min_cov 4 --max_n_read 200, and overlap_filtering_setting = --max_diff 40 --max_cov 80 --min_cov 2 --min_len 500.
The maternal and paternal Canu assemblies were polished with Arrow version 2.2.2 run through ArrowGrid (https://github.com/skoren/ArrowGrid) using subreads that correspond to the CCS reads used for each assembly. The maternal and paternal FALCON assemblies were polished with Arrow version 2.2.2 using all subreads.
Assembly Evaluation
For each assembly, contigs were broken into 100 kb chunks with remainders shorter than 100 kb ignored. The chunks were aligned to GRCh37 using minimap2 --eqx -x asm5, and primary alignments that span at least 50 kb in the reference at higher than 50% identity were retained. The concordance of each chunk was evaluated just as for CCS reads (see “Measuring HG002 Concordance”). The overall assembly concordance was calculated as the average concordance of the 100 kb chunks.
Gene completeness was measured using BUSCO54 version 3.0.2 using the Mammalia ODB9 gene set. The single plus duplicated gene count in the BUSCO summary is reported. For a human-specific measure of completeness, we calculated the fraction of single-copy human genes that remain single-copy in each assembly. The human transcript sequences from Ensembl55 build r94 were mapped to each assembly with minimap2 -cx splice -B 4 -O 4,34 -C9 -uf --cs and evaluated with paftools.js asmgene -i 0.98, which retains the longest of overlapping transcripts and counts a transcript hit if 99% of the transcript sequence maps at 98% identity or higher. A single-copy transcript has exactly one hit. Counts are normalized to the number of transcripts that are considered single-copy by these criteria in GRCh38 (GCA_000001405.15).
To measure the number of segmental duplications spanned by each assembly, the assemblies were processed with segDupPlots (https://github.com/mvollger/segDupPlots), which maps contigs to GRCh38 and considers a segmental duplication to be spanned by the assembly if a contig alignment extends fully through the segmental duplication and into at least 50 kb of unique sequence on each flank45.
Model of Assembly Contiguity
To predict assembly contiguity at different read lengths and read accuracies, a previously described model21 was updated with improvements for high-accuracy reads. Briefly, all repeat annotations for GRCh38 were downloaded from the UCSC Genome Browser. Repeat identity was defined as by each track except for: the nested repeat track where identity was 50+50×(score/1000), RepeatMasker where identity was 1-((mismatches + deleted + inserted)/1000), and microsat and windowmasker/sdust which does not define identity and thus was treated as 100%. Gaps were included as 100% identity repeats. Additional repeats were added from self-matches using MashMap56 (https://github.com/marbl/MashMap).
The assembly contiguity was predicted based on the ability to resolve repeats. At a given percent identity, repeats below that identity were excluded and remaining repeats separated by 15 bp or fewer were merged. Then, cuts were introduced at each repeat of a given length, and assembly NG50 was calculated assuming that contigs end at each cut.
Coverage Titration
To evaluate the performance of variant calling and assembly at different coverage levels, CCS reads were downsampled from the 28-fold dataset and processed. For small variant calling, alignments were subsampled in DeepVariant version 0.7.1 from 4% to 100% in steps of 3%. Variants were called on each subsample using the DeepVariant CCS model. Precision and recall for SNVs and indels were evaluated with hap.py as described above (see “Small Variant Detection and Benchmarking“). For phasing, alignments were subsampled (samtools view -s) at rates from 10% to 100% in steps of 10%. The DeepVariant callset from the full 28-fold coverage data was phased using WhatsHap v0.17 (whatshap phase) with the subsampled alignments. For structural variants, alignments were subsampled (samtools view -s) at rates from 10% to 100% in steps of 10%. Variants were called on the subsampled alignments with pbsv version 2.1.0 and benchmarked with Truvari as described above (see “Structural Variant Detection“). For assembly, reads were subsampled at rates from 10% to 100% in steps of 10%. Sampling was performed based on read name (10% sample is reads that end in 0, 20% is reads that end in 0–1, and so on). Assembly of subsampled reads was performed with wtdbg2 version 2.2 and benchmarked as described above (see “De novo Assembly” and “Assembly Evaluation”).
Revising and Expanding Genome in a Bottle Benchmarks
Discrepancies between the GIAB v3.3.2 small-variant benchmark and the DeepVariant callset from haplotype-sorted CCS reads were identified with vcfeval from RTG Tools 3.8.2 and hap.py 0.3.10. Discrepancies between the GIAB v0.6 structural variant benchmark and the integrated structural variant callset from CCS reads were identified with Truvari. A sample of 60 small variant and 40 structural variant discrepancies were selected for manual curation by random sampling across discrepancy types (false positive, false negative, genotype difference), variant types (SNV, indel, insertion structural variant, and deletion structural variant), both inside and outside homopolymers and tandem repeats. Variants were curated as previously described26. Briefly, curators evaluated variants in IGV along with tracks showing segmental duplications, interspersed and simple repeats, and alignments of CCS reads, 10X Genomics reads (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/10XGenomics_ChromiumGenome_LongRanger2.2_Supernova2.0.1_04122018/ or https://bit.ly/2Mtj084), Illumina short reads (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/NIST_Illumina_2×250bps/ or https://bit.ly/2WjH7tF) , and Illumina reads from a 6 kb mate pair library (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/NIST_Stanford_Illumina_6kb_matepair/ or https://bit.ly/2I2Zdw1), all obtained from GIAB. At each variant site, the reliability of alignments for each read set was evaluated by examining depth of coverage compared to genome-wide average, evenness of coverage around the variant, mapping quality of alignments, density of variants in the region, and clipping of aligned reads while considering genomic repeat context. The correct call was determined as the one supported by the reliable technologies with normal coverage, consistent haplotype structure, and consistency of forward and reverse reads. The benchmark error rate was estimated by variant type and discrepancy type and used to extrapolate from the sample to the number of errors in the full GIAB benchmark. Confidence intervals were calculated assuming a binomial distribution.
Life Sciences Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability Statement
Data is available in NCBI BioProject PRJNA529679. CCS reads are available on NCBI SRA with accession SRX5327410. Small variant calls are available on NCBI dbSNP with accessions ss3783301452-ss3798736595. Structural variant calls are available on NCBI dbVar with accession nstd167. The trio binned Canu assemblies are available on NCBI Assembly with accessions GCA_004796485.1 (maternal) and GCA_004796285.1 (paternal). Alignments to GRCh37 are available at ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/PacBio_CCS_15kb/ or https://bit.ly/2RW1b3I. Additional data, including all assemblies and a track hub for the UCSC Genome Browser, is available at https://downloads.pacbcloud.com/public/publications/2019-HG002-CCS.
Code Availability Statement
Custom scripts are available at https://github.com/PacificBiosciences/hg002-ccs/. Google DeepVariant, a model trained on PacBio CCS reads, and instructions for use are available at https://github.com/google/deepvariant.
Supplementary Material
Acknowledgements
We would like to thank J. Harting for assistance with HLA typing, K. Robertshaw for figure generation, J. Wilson and J. Ziegle for providing PacBio CLR datasets, and J. Puglisi for critical reading of the manuscript.
S.K. and A.M.P. were supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
This work was supported by NIH grant 1R01HG010040 to H.L. and NSFC grants 31571353 and 31822029 to J.R. M.C.S. is funded by the National Science Foundation (DBI-1350041) and National Institutes of Health (R01-HG006677). F.J.S. and M.M. are funded by NIH grant UM1 HG008898. T.M. acknowledges funding from the German Research Foundation (DFG) (391137747 and 395192176).
N.D.O. and J.M.Z. were supported by intramural funding from the National Institute of Standards and Technology and an interagency agreement with the U.S. Food and Drug Administration.
This work utilized computational resources of DNAnexus and Google to apply DeepVariant to CCS reads.
Certain commercial equipment, instruments, or materials are identified to specify adequate experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose.
Footnotes
Competing Financial Interests Statement
A.M.W., A.T., D.R.R., G.T.C., M.W.H., P.P., R.J.H., W.J.R., and Y.Q. are employees and shareholders of Pacific Biosciences. A.C., A.K., M.A.D., and P.C. are employees and shareholders of Google. A.F. and C-S.C. are employees and shareholders of DNAnexus. A.C. is a shareholder and was an employee of DNAnexus for a portion of this work.
References
- 1.DNA Sequencing Costs: Data. National Human Genome Research Institute (NHGRI) Available at: https://www.genome.gov/27541954/dna-sequencing-costs-data/. (Accessed: 7th December 2018) [Google Scholar]
- 2.Sanger F, Nicklen S & Coulson AR DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A 74, 5463–5467 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith LM et al. Fluorescence detection in automated DNA sequence analysis. Nature 321, 674–679 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Venter JC et al. The sequence of the human genome. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Ronaghi M, Karamohamed S, Pettersson B, Uhlén M & Nyrén P Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem 242, 84–89 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Bentley DR et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shendure J et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005). [DOI] [PubMed] [Google Scholar]
- 11.McKernan KJ et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 19, 1527–1541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drmanac R et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Rothberg JM et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Sedlazeck FJ, Lee H, Darby CA & Schatz MC Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet 19, 329–346 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Chaisson MJP et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo J-S et al. De novo assembly and phasing of a Korean human genome. Nature 538, 243–247 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Cretu Stancu M et al. Mpping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun 8, 1326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eid J et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Mikheyev AS & Tin MMY A first look at the Oxford Nanopore MinION sequencer. Mol. Ecol. Resour 14, 1097–1102 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Jain M et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Travers KJ, Chin C-S, Rank DR, Eid JS & Turner SW A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 38, e159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loomis EW et al. Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 23, 121–128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hebert PDN et al. A Sequel to Sanger: amplicon sequencing that scales. BMC Genomics 19, 219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zook JM et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data 3, 160025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zook JM et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol 37, 561–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers G Efficient Local Alignment Discovery amongst Noisy Long Reads in Algorithms in Bioinformatics (eds. Brown D & Morgenstern B) 52–67 (Springer Berlin Heidelberg, 2014). [Google Scholar]
- 28.Mandelker D et al. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet. Med 18, 1282–1289 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Ambardar S & Gowda M High-Resolution Full-Length HLA Typing Method Using Third Generation (Pac-Bio SMRT) Sequencing Technology. Methods Mol. Biol 1802, 135–153 (2018). [DOI] [PubMed] [Google Scholar]
- 30.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo R, Sedlazeck FJ, Lam T-W & Schatz MC A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun 10, 998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poplin R et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol 36, 983–987 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Patterson M et al. WhatsHap: Weighted Haplotype Assembly for Future-Generation Sequencing Reads. J. Comput. Biol 22, 498–509 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Sedlazeck FJ et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Rausch T et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia S et al. Linked-Read sequencing resolves complex structural variants. bioRxiv 231662 (2017). doi: 10.1101/231662 [DOI] [Google Scholar]
- 39.Chin C-S et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koren S et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan J & Li H Fast and accurate long-read assembly with wtdbg2. bioRxiv 530972 (2019). doi: 10.1101/530972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koren S et al. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol 36, 1174–1182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fungtammasan A & Hannigan B How well can we create phased, diploid, human genomes?: An assessment of FALCON-Unzip phasing using a human trio. bioRxiv 262196 (2018). doi: 10.1101/262196 [DOI] [Google Scholar]
- 44.Chin C-S et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Vollger MR et al. Long-read sequence and assembly of segmental duplications. Nat. Methods 16, 88–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider VA et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27, 849–864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krusche P et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol 37, 555–560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinforma 47, 11.12.1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson J, Soormally AR, Hayhurst JD & Marsh SGE The IPD-IMGT/HLA Database - New developments in reporting HLA variation. Hum. Immunol 77, 233–237 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Cleary JG et al. Comparing Variant Call Files for Performance Benchmarking of Next-Generation Sequencing Variant Calling Pipelines. bioRxiv 023754 (2015). doi: 10.1101/023754 [DOI] [Google Scholar]
- 52.Li H et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat. Methods 15, 595–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeffares DC et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun 8, 14061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV & Zdobnov EM BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Zerbino DR et al. Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain C, Koren S, Dilthey A, Phillippy AM & Aluru S A fast adaptive algorithm for computing whole-genome homology maps. Bioinformatics. 34, i748–i756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available in NCBI BioProject PRJNA529679. CCS reads are available on NCBI SRA with accession SRX5327410. Small variant calls are available on NCBI dbSNP with accessions ss3783301452-ss3798736595. Structural variant calls are available on NCBI dbVar with accession nstd167. The trio binned Canu assemblies are available on NCBI Assembly with accessions GCA_004796485.1 (maternal) and GCA_004796285.1 (paternal). Alignments to GRCh37 are available at ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/PacBio_CCS_15kb/ or https://bit.ly/2RW1b3I. Additional data, including all assemblies and a track hub for the UCSC Genome Browser, is available at https://downloads.pacbcloud.com/public/publications/2019-HG002-CCS.