Abstract
The plague agent, Yersinia pestis, employs a type III secretion system (T3SS) to selectively destroy human immune cells, thereby enabling its replication in the bloodstream and transmission to new hosts via fleabite. The host factors responsible for the selective destruction of immune cells by plague bacteria were not known. Here we show that LcrV, the needle cap protein of the Y. pestis T3SS, binds N-formylpeptide receptor (FPR1) on human immune cells to promote the translocation of bacterial effectors. Plague infection in mice is characterized by high mortality, however N-formylpeptide receptor deficient animals exhibit increased survival and plague-protective antibody responses. We identified FPR1 p.R190W as a candidate human resistance allele that protects neutrophils from Y. pestis T3SS. These findings reveal the plague receptor on immune cells and show that FPR1 mutations provide for plague survival, which appears to have shaped human immune responses towards other infectious diseases and malignant neoplasms.
Yersinia pestis has caused human disease for more than 5,000 years1. Three pandemics were recorded, including the plague of Justinian (6th-8th), the Black Death (14th-18th) and the Asian Pandemic (19th-20th century)2. The Black Death killed more than half of Europe’s population, suggesting plague must have shaped the human immune system by selecting for mutations that confer resistance3. Carriers of CCR5-Δ32, a gene variant of CC-type chemokine receptor 5 (CCR5), are resistant to infections with human immunodeficiency virus type 14. CCR5 is a member of the 7-transmembrane spanning G protein coupled receptor (GPCR) family that is expressed on immune cells, whereas CCR5-Δ32 is not presented on cell surfaces5. The allele frequency of CCR5-Δ32 is high in Northern Europe and originated 800 years ago, suggesting its selection may be linked to the Black Death6. However, studies in mice did not reveal an impact of CCR5 on plague survival7,8.
Pathogenesis of Y. pestis and of related Yersinia enterocolitica and Yersinia pseudotuberculosis, relies on a conserved type III secretion system (T3SS) that delivers effector proteins into host cells9–11. Y. pestis T3SS targets immune cells for destruction with preferences for neutrophils, macrophages, and dendritic cells12. Immune cell ablation enables bacteria to replicate to high density resulting in high mortality13. Without therapy, approximately half of all bubonic plague victims survive and mount pathogen-specific antibody responses that prevent replication of Y. pestis in blood14. We hypothesized that humans may have acquired mutations in the immune cell receptor for Y. pestis T3SS, thereby diminishing the destruction of immune cells and increasing survival. Here we establish N-formylpeptide receptor (FPR1) as the physiologically relevant plague receptor.
CRISPR-Cas9 screen for T3SS receptor
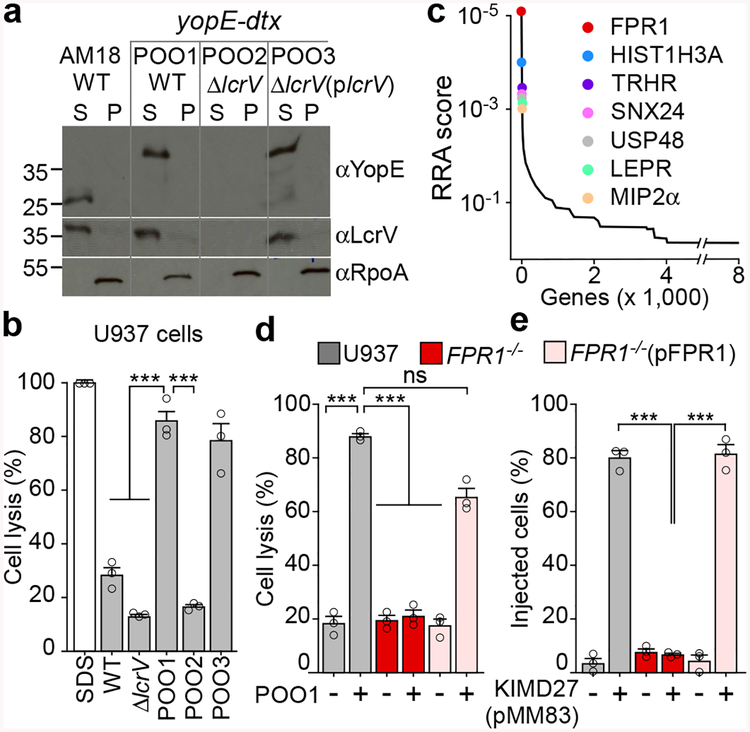
Y. pestis AM18, a ΔyfeAB variant of the vaccine strain EV76, is defective for iron and manganese scavenging15. In broth cultures, Y. pestis AM18 secretes the YopE effector via the lcrV-dependent T3SS pathway (Fig. 1a). Compared with the ΔlcrV variant (AM46), which cannot kill immune cells above control levels, Y. pestis AM18 infection resulted in modest killing of U937 human histiocytic leukemia cells differentiated into macrophages (Fig. 1b). Y. pestis POO1 is a variant of AM18 expressing yopE-dtx, a translational hybrid between the YopE effector and the mature domain of diphtheria toxin16. Dtx is an ADP-ribosyltransferase that modifies elongation factor 2 (EF2) at its diphthamide residue to promote cell death16. Y. pestis POO1 secretes YopE-Dtx and causes death of U937 macrophages in an lcrV-dependent manner (Fig. 1ab). U937 cells were transduced with lentiviral libraries expressing RNA guided endonuclease Cas9 and short guide RNAs (sgRNA – 3 per gene) targeting 19,052 genes. After 12 hours of infection with Y. pestis POO1 or POO2 (yopE-dtx ΔlcrV), resistant cells were cultured with antibiotics to approximately 70% confluence and re-infected. The sgRNA sequences from surviving U937 cells were identified via next-generation sequencing (NGS) and candidate genes ranked based on the number of unique sgRNAs versus NGS reads (Fig. 1c). FPR1 stood out in three independent screens with the most abundant sgRNAs (Supplementary Databases S1–S3). FPR1 is a member of the GPCR family that activates immune cell chemotaxis and cytokine release in response to N-formylpeptides contained in bacteria or mitochondria17. To validate these results, we generated FPR1−/− U937 cells using CRISPR-Cas9 and analyzed mutant cells by sequencing the mutated (frame-shifted) FPR1 alleles (Extended Data Fig. 1a). FPR1−/− cells were transfected with pFPR118. When analyzed for FPR1 expression by immunoblot with αFPR1m, U937 cells, but not their FPR1−/− variants, produced FPR1 (Extended Data Fig. 1b). FPR1 production was restored upon transfection with pFPR1; expression of FPR2 and FPR3, homologs of FPR1 that promote chemotaxis to distinct signals was unaffected (Extended Data Fig. 1b)19. When infected with Y. pestis POO1, FPR1−/− cells were resistant to Y. pestis T3SS-mediated killing (Fig. 1d). This defect was restored in FPR1−/−(pFPR1) cells (Fig. 1d). To validate FPR1-dependent resistance to T3SS, translocation of YopM-Bla into CCF2-AM stained U937 cells was assessed using KIM D27 (pMM83). Increase in blue fluorescence indicates YopM-Bla mediated CCF2-AM cleavage in the cytoplasm of immune cells12. YopM-Bla translocation was abolished in FPR1−/− cells and restored by plasmid-borne expression of FPR1 (Fig. 1e).
Figure 1 |. FPR1 is essential for Y. pestis T3SS into U937 macrophages.
a, Y. pestis AM18 (ΔyfeAB, Δpgm) and its variants POO1 (yopE-dtx), POO2 (ΔlcrV, yopE-dtx) and POO3 (ΔlcrV(plcrV), yopE-dtx) were grown at 37°C with 5 mM EGTA to induce T3SS. Cultures were centrifuged to separate the supernatant (S) from the bacterial pellet (P) and extracts analyzed by immunoblotting with antibodies specific for YopE (αYopE), LcrV (αLcrV) and cytoplasmic RNA polymerase subunit A (αRpoA). b, Y. pestis cells (AM18, POO1, POO2 or POO3) were added at MOI of 10 to U937 for 4 hours at 37°C. Cell lysis was measured as LDH activity in centrifuged supernatants. SDS was used to generate a control sample. c, CRISPR-Cas9 mutagenesis of U937 cells was performed to select for variants resistant to Y. pestis POO1 intoxication as compared to Y. pestis POO2 control. Candidate genes were identified by next generation sequencing and data which are representative of three independent replicates were analyzed using the MaGeCK-based robust rank aggregation (RRA) score analysis. d, Y. pestis POO1 induced cell lysis in U937, FPR1−/− and FPR1−/− (pFPR1) cultures. e, Y. pestis KIM D27 (pMM83) mediated YopM-Bla translocation into U937, FPR1−/− and FPR1−/− (pFPR1) cells. Error bars represent the s.e.m. (n = 3 biological replicates) (b,d,e). One-way ANOVA with Bonferroni Correction was used to identify significant differences: ***, P<0.001; ns= not significant. One of three repeats is shown (a-e).
Diphthamide is a modified histidine residue synthesized by 7 gene products20. As YopE-Dtx is responsible for some, but not all, of the T3SS-mediated killing of U937 cells by strain POO1, one would expect CRISPR-Cas9 mutagenesis to target the diphthamide synthesis pathway. Indeed, sgRNAs targeting DPH1, DPH2, DPH3, DPH5, DPH6 and DPH7 were enriched in U937 macrophages that survived Y. pestis POO1-mediated killing (Supplementary Databases S1–S3). sgRNAs targeting genes that scored even higher than the DPH determinants, were also identified suggesting that these genes may be involved in T3SS-mediated translocation of Yersinia effectors: sorting nexin 24 (SNX24), ubiquitin C-terminal hydrolase (USP48), histone 3.1 (HIST1H3A), thyrotropin-releasing hormone receptor (TRHR), leptin receptor (LEPR) and macrophage inflammatory protein alpha (MIP2α=CXCL2)(Fig. 1c).
Prior work using RNA interference identified CCR5 as contributing to Y. pseudotuberculosis T3SS into 293T cells and into primary murine immune cells21. CCR5 was not identified in our CRISPR-Cas9 screen (Supplementary Databases S1–S3). We used CRISPR-Cas9 and CCR5-specific sgRNAs to generate two independent cell lines with null mutations in CCR5 (Extended Data Fig. 2a). CCR5−/− cells did not exhibit resistance to Y. pestis POO1-mediated killing (Extended Data Fig. 2b). When analyzed for YopM-Bla translocation, Y. pestis infected CCR5−/− cells exhibited a small reduction in T3SS whereas FPR1−/− cells were resistant to effector translocation (Extended Data Fig. 2c). Y. pseudotuberculosis T3S into U937 cells relied in part on CCR5, whereas FPR1 was dispensable for effector translocation (Extended Data Fig. 2d). Thus, Y. pseudotuberculosis and Y. pestis utilize distinct receptors for translocation of effectors into immune cells. Of note, Y. pestis LcrV acquired 10 amino acid substitutions during evolution from its ancestor Y. pseudotuberculosis LcrV, supporting a mechanism for host-cell receptor selectivity.
FPR1 inhibitors block Y. pestis T3SS
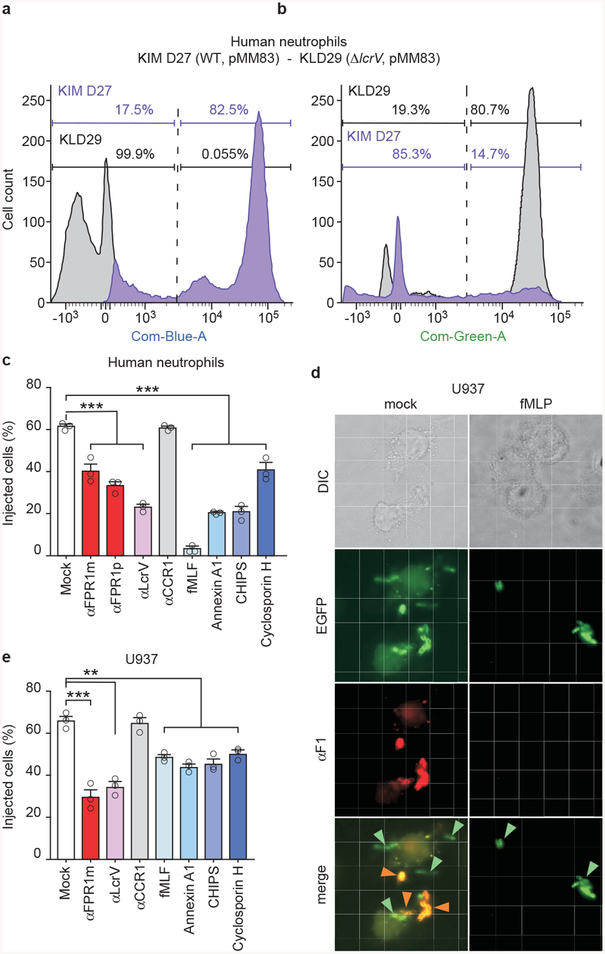
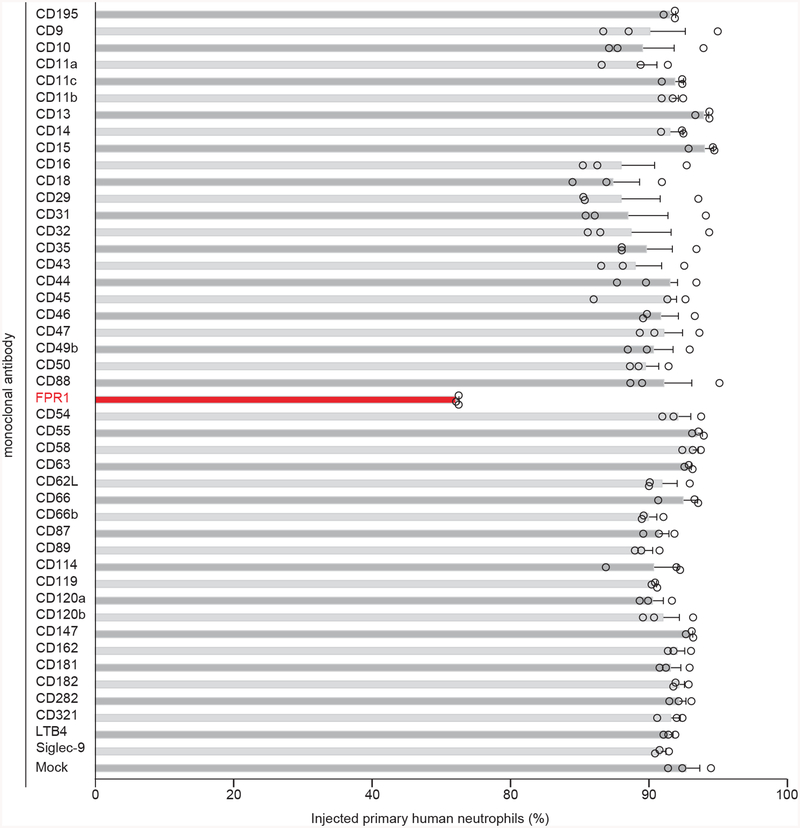
We screened monoclonal antibodies (mAbs) specific for surface proteins of human neutrophils to identify inhibitors of Y. pestis YopM-Bla translocation (Extended Data Fig. 3ab)22. Only the mAb against FPR1 (αFPR1m) inhibited effector translocation (Extended Data Fig. 4). Polyclonal antibodies against FPR1 and LcrV (αLcrV), the needle cap protein of the T3SS23, also inhibited T3SS into neutrophils (Extended Data Fig. 3c). Annexin, a ubiquitous cytosolic protein, is another ligand of FPR124. During cell death, released annexin undergoes Ca2+-dependent rearrangements to expose its A1 peptide for FPR1 recognition25. Addition of N-formylpeptide (fMLF) or annexin A1 to human neutrophils interfered with Y. pestis effector translocation (Extended Data Fig. 3c). FPR1 activation is inhibited by Staphylococcus aureus CHIPS and Tolyplocadium inflatum cyclosporin H26,27; these compounds also inhibited Y. pestis effector translocation (Extended Data Fig. 3c). Fluorescence and differential interference contrast microscopy revealed physical interactions between enhanced green-fluorescent protein (EGFP)-labeled bacteria and U937-derived macrophages. In the absence, but not in the presence of fMLF, Y. pestis (pEGFP) was associated with the cell surface (Extended Data Fig. 3d). Treatment with αFPR1m, αLcrV, fMLF, annexin A1, CHIPS and cyclosporin H also diminished Y. pestis effector translocation into U937 cells (Extended Data Fig. 3e).
T3SS triggers immune cell chemotaxis
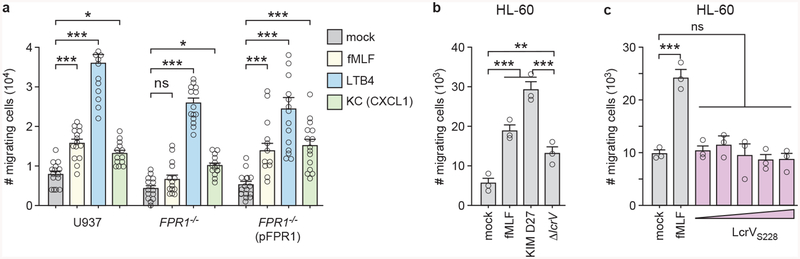
Using a transwell migration assay, we observed chemotaxis of U937 cells towards fMLF, Y. pestis KIM D27, Leukotriene B4 (LTB4) and Keratinocyte Chemoattractant (KC) (Fig. 2a; Extended Data Fig. 5a). Chemotaxis toward fMLF and Y. pestis, but not toward LTB4 and KC, was abolished in FPR1−/− monocytes, and restored following transfection with pFPR1 (Fig. 2a; Extended Data Fig. 5a). U937 monocyte chemotaxis was also diminished when Y. pestis KLD29 (ΔlcrV) was analyzed as a chemoattractant, suggesting that the N-formylpeptide signal is amplified by secretion via the bacterial T3SS (Fig. 2a). To test whether other immune cells migrate towards Y. pestis in a T3SS-dependent manner, we analyzed primary human neutrophils and differentiated HL-60 cells, neutrophils derived from acute promyelocytic leukemia cells. Compared to wild-type Y. pestis, migration of primary human neutrophils and HL-60 cells was diminished towards the ΔlcrV mutant (Fig. 2b; Extended Data Fig. 5b). Granulocytes from wild-type mice, but not N-formylpeptide receptor deficient mice, mFpr1−/−, migrate towards fMLF peptide and Y. pestis wild type for lcrV (Fig. 2c). Chemotaxis towards recombinant LcrVS228, a strep-tagged variant that functions identically to wild-type LcrV28 was not observed suggesting that LcrV is not a chemoattractant signal (Extended Data Fig. 5c). Addition of LcrVS228 to differentiatd HL-60 cells blocked chemotaxis toward fMLF (data not shown), an activity previously reported for the V antigen of plague (LcrV)29.
Figure 2 |. Immune cell chemotaxis towards Y. pestis is mediated by the T3SS and FPR1.
Numbers of migrating immune cells were quantified in a transwell assay primed with mock, Y. pestis KIM D27 (WT) or KLD29 (ΔlcrV) (107 CFU/ml). Chemotaxis toward fMLF is shown as a control: U937, FPR1−/− and FPR1−/− (pFPR1) cells (a); human neutrophils (b); granulocytes from wild-type (C57BL/6) and mFpr1−/− mice (c). One of three repeats is shown and error bars represent the s.e.m. (n = 3 biological replicates). One-way ANOVA with Bonferroni Correction (a, b) and un-paired Student’s t-test (c) were used to identify significant differences: ***, P<0.001; **, P <0.01; *, P <0.05; ns, not significant.
The T3SS cap protein LcrV binds FPR1
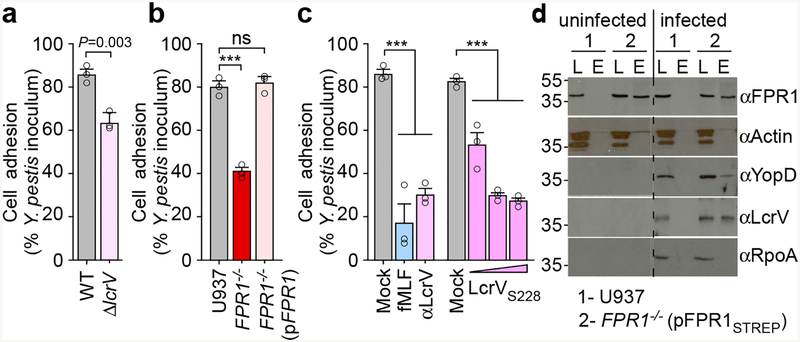
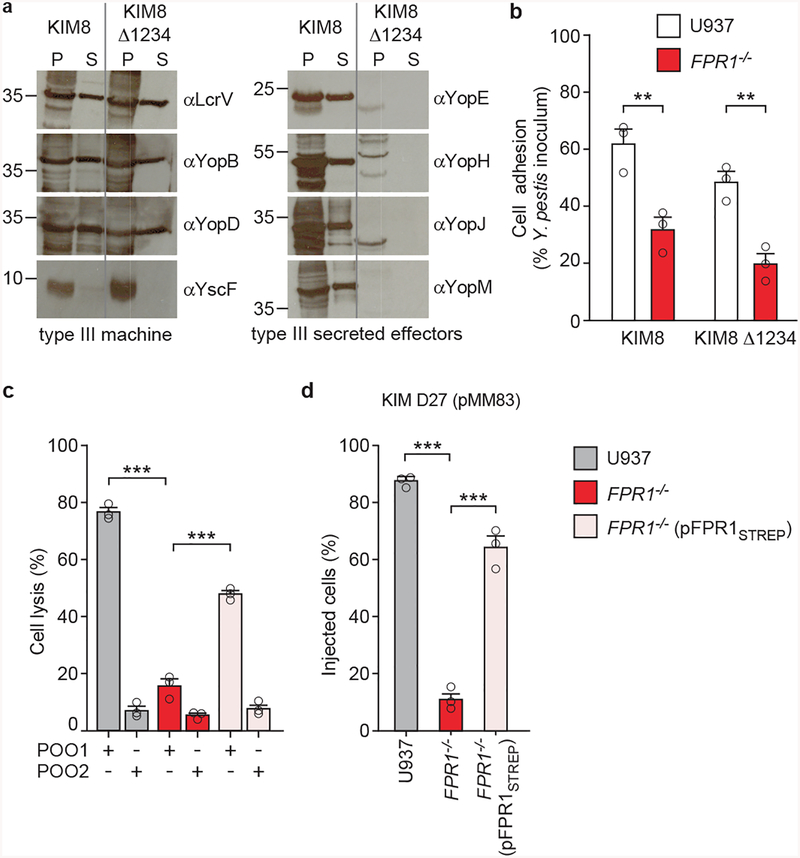
Y. pestis KIM D27 adhesion to U937-derived macrophages was diminished for the ΔlcrV variant (Fig. 3a). FPR1 is required for Y. pestis adherence and plasmid-borne expression of FPR1 in FPR1−/− (pFPR1) macrophages restored the adhesion of plague bacteria (Fig. 3b). Addition of fMLF, αLcrV or purified LcrVS228 each caused a reduction in bacteria adhesion to macrophages (Fig. 3c). To exclude the possibility that secreted bacterial effectors may impact adhesion, U937 macrophages were inoculated with Y. pestis KIM8, a plasminogen activator protease mutant, or with KIM8 Δ1234, an isogenic variant lacking segments of the pCD1 virulence plasmid encoding YopE, YopH, YopJ, YopK, YopM, YopT, and YpkA30 (Extended Data Fig. 6a). Similar levels of adhesion were observed albeit that as expected KIM8 Δ1234 was unable to intoxicate immune cells (Extended Data Fig. 6b). When transfected into FPR1−/− cells, pFPR1STREP encoding FPR1 with C-terminal Strep-tag II restored Y. pestis POO1 killing of, and YopM-Bla cleavage of CCF2-AM (Extended Data Fig. 6cd). When subjected to pull-down experiments, FPR1STREP from detergent lysate of FPR1−/− (pFPR1STREP) cells, but not FPR1 from U937 macrophages, was retained on StrepTactin-sepharose (Fig. 3d). Y. pestis T3SS needle complex components, LcrV and YopD28, co-purified with FPR1STREP (Fig. 3d). As control, actin from U937 macrophages and RNA polymerase A (RpoA) from bacterial cells did not associate with FPR1STREP (Fig. 3d).
Figure 3 |. Adhesion of Y. pestis to human macrophages involves LcrV binding to N-formylpeptide receptor.
a, Wild-type Y. pestis KIM D27 or its ΔlcrV variant (KLD29) were added to U937 cells (MOI of 10) and adherence quantified as percent inoculum. b, Adherence of Y. pestis KIM D27 to U937, FPR1−/− and FPR1−/− (pFPR1) cells. c, Treatment of U937 cells with 10 μM fMLF or αLcrV or LcrVS228 (10–103 ng/ml) reduces Y. pestis KIM D27 adherence. One of three repeats is shown and error bars represent the s.e.m. (n = 3 biological replicates) (a-c). Significant differences were measured with the two tailed t-test (a), and one-way ANOVA with Bonferroni post-hoc analysis (b, c): ***, P<0.001; ns= not significant. d, Cleared detergent lysates of U937 or FPR1−/− (pFPR1STREP) cells were incubated without (left panel) or with 10 MOI Y. pestis KIM D27 (right panel) and subjected to affinity chromatography on StrepTactin-sepharose. Load (L) and eluate (E) samples were analyzed by immunoblotting with IgG specific for FPR1, actin, YopD, LcrV, and RpoA. One of three repeats is shown.
mFpr1−/− mice exhibit plague resistance
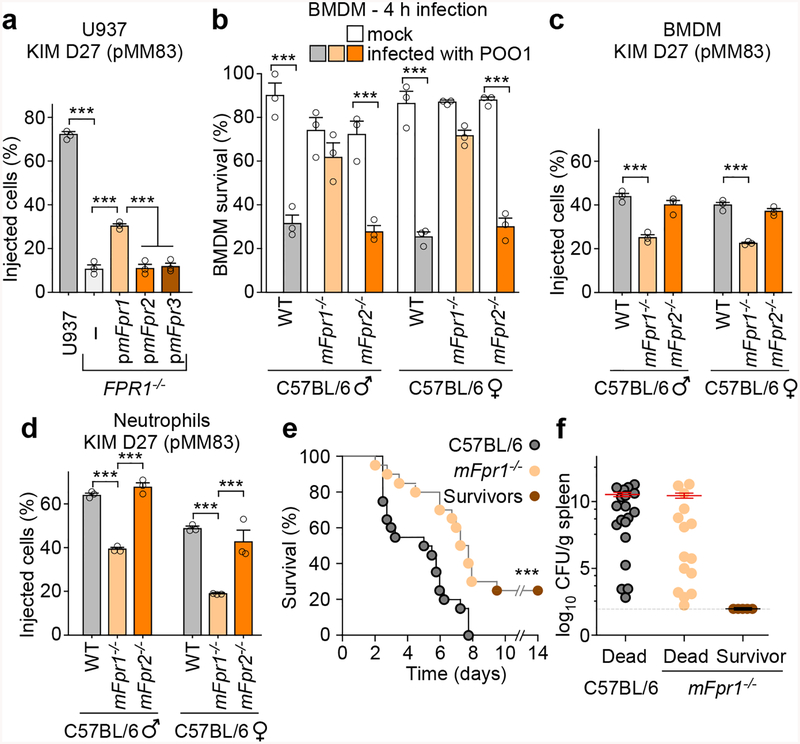
Rodents play important roles in the epidemiology of plague as highly-susceptible epizootic reservoirs for pathogen amplification and flea-borne disease transmission31. The mouse chemotaxis locus harbors three genes (mFpr1, mFpr2, and mFpr3) whose products are known to function as GPCR, promoting immune cell chemotaxis towards N-formylpeptides (mFpr1 and to a lesser degree mFpr2) or other bacterial products (mFpr3)32,33. mFpr1, mFpr2, and mFpr3 were cloned into pBGSA and transfected into FPR1−/− macrophages for infection with Y. pestis KIM D27 (pMM83). Expression of mFpr1, but not of mFpr2 and mFpr3, restored in part Y. pestis T3SS effector translocation into FPR1−/− macrophages (Fig. 4a). C57BL/6 mice lacking mFpr1 that are reared under pathogen-free conditions develop normally32. However, when infected with bacteria and compared to wild-type animals, mFpr1−/− mice exhibit reduced survival (accelerated time-to-death), increased pathogen burden in the spleen and liver and their neutrophils display diminished chemotaxis towards fMLF32,34. When infected with Y. pestis POO1, bone marrow derived macrophages (BMDM) of wild-type animals (male and female) were rapidly killed, whereas mFpr1−/− BMDM exhibited partial resistance to Y. pestis T3SS-induced killing (Fig. 4b). mFpr2−/− BMDM did not exhibit resistance to Y. pestis POO1 induced killing (Fig. 4b). Y. pestis KIMD27 (pMM83) infection of macrophages demonstrated reduced translocation of YopM-Bla into mFpr1−/− BMDM, when compared to wild-type and mFpr2−/− BMDMs (Fig. 4c). Peritoneal granulocytes from mFpr1−/− mice also displayed a reduction in Y. pestis effector translocation, whereas neutrophils from mFpr2−/− mice did not (Fig. 4d).
Figure 4 |. Contribution of mouse N-formylpeptide receptors to plague disease.
a, Transfection with pmFpr1, but not pmFpr2 or pmFpr3, restores Y. pestis KIM D27 (pMM83) translocation of YopM-Bla into U937 FPR1−/− cells. One of three repeats is shown and error bars represent the s.e.m. (n = 3 biological replicates). b, Compared to C57BL/6 mice, BMDM from mFpr1−/−, but not mFpr2−/− mice, exhibit increased survival when infected with Y. pestis POO1 (yopE-dtx); one of three repeats is shown and error bars represent the s.e.m (n = 3 biological replicates). Y. pestis KIM D27 (pMM83) translocation of YopM-Bla in mouse BMDM (c) and neutrophils (d) requires mFpr1; one of three repeats is shown and error bars represent the s.e.m. (n = 3 biological replicates). e, Survival of wild-type C57BL/6 and mFpr1−/− mice (n=10 males and 10 females) following subcutaneous inoculation with (600 CFU) Y. pestis CO92. f, Bacterial loads (CFU) in spleen tissues of dead and surviving animals. Horizontal bars denote the mean and error bars represent the s.e.m. One of two repeats is shown (e, f). Significant differences were determined using one-way ANOVA and Bonferroni’s post-hoc analysis (a-d) and the log-rank (Mantel-Cox) test (e): ***, P<0.001.
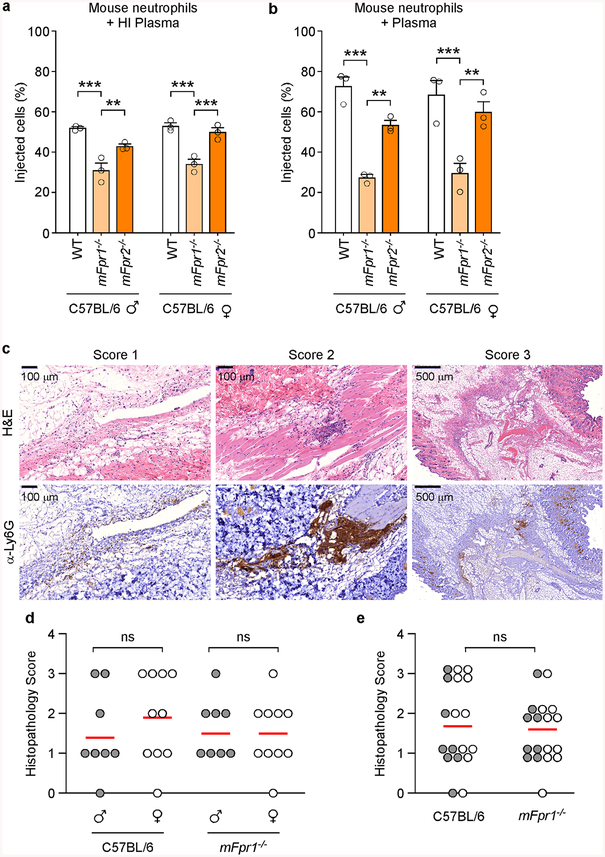
To assess whether mFpr1 functions as the plague receptor in mice, wild-type C57BL/6 and mFpr1−/− animals were infected by subcutaneous inoculation with 600 CFU Y. pestis CO92. Wild-type mice succumbed 3–8 days post-inoculation and mFpr1−/− animals exhibited delayed time-to-death (2.25 days) and a small, but significant increase in survival, suggesting that another GPCR may function as a plague receptor (Fig. 4e). Unlike with human macrophages and FPR1 (Fig. 1e), mFpr1 is not absolutely essential for effector translocation into mouse macrophages and neutrophils (Fig. 4cd). To rule out the possibility that complement coating of Y. pestis during infection may have bypassed the requirement for mFpr1 binding, bacteria were preincubated with mouse plasma and effector injection measured using peritoneal granulocytes from C57BL/6, mFpr1−/− and mFpr2−/− mice (Extended Data Fig. 7ab). Although plasma enhanced T3SS-mediated translocation into wild-type and mFpr2−/− granulocytes relative to heat-inactivated plasma, preincubation of bacteria with plasma did not restore the translocation of YopM-Bla into mFpr1−/− granulocytes.
In mouse models for pneumonic and bubonic plague, Y. pestis triggers neutrophil influx to the site of infection35,36. Neutrophil-mediated tissue damage is thought to prime the devastating pathology associated with plague35. To determine whether knockout mutation in mFpr1 affects predominantly chemotaxis of immune cells towards Y. pestis rather than blocking T3SS-mediated effector injection, we asked whether neutrophil influx into plague infected tissues is reduced in mFpr1−/− mice as compared to wild-type animals. Similar numbers of anti-Ly6G+ immune cells were detected in inguinal tissues of mFpr1−/− and wild-type animals four hours following intradermal inoculation with Y. pestis (Extended Data Fig. 7c–e). Thus, during bubonic plague infection, the mFpr1−/− mutation predominantly blocks the injection of bacterial effectors without abolishing the neutrophil influx into infected tissues.
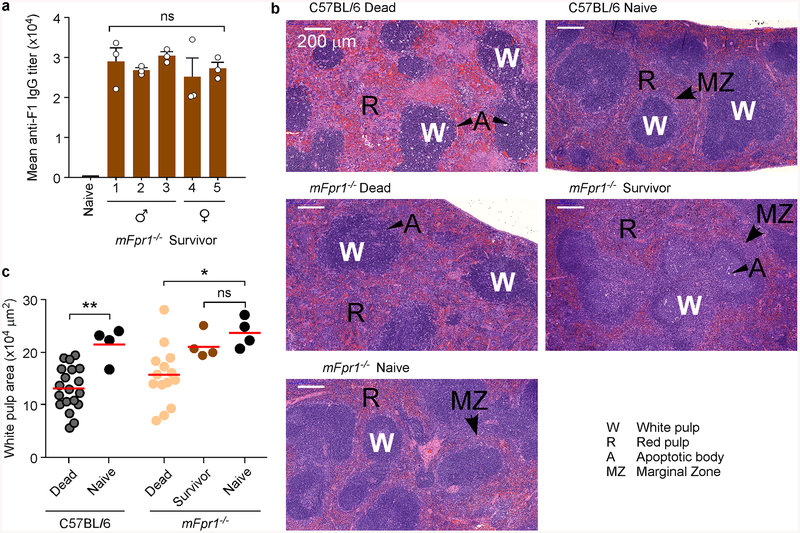
All animals that died of plague displayed high bacterial loads in spleens, but Y. pestis bacteria were not recovered from tissues of surviving mFpr1−/− animals (Fig. 4f). mFpr1−/− survivors developed high-titer pathogen-specific F1-antibodies (Extended Data Fig. 8a), which provide protective immunity against plague37. Plague infection results in the massive depletion of immune cells in the spleen i.e. the destruction of white pulp and its replacement by fibrinoid necrosis lesions (Extended Data Fig. 8b; label A). Loss of white pulp can be visualized by H&E staining of thin-sectioned spleen samples and can be quantified by measuring areas of white pulp (Extended Data Fig. 8c). Unlike wild-type and mFpr1−/− animals that succumbed to plague, spleens of mFpr1−/− survivors displayed histology similar to that of naïve animals with larger white pulp area (Extended Data Fig. 8bc).
FPR1 R190W is a plague resistance allele
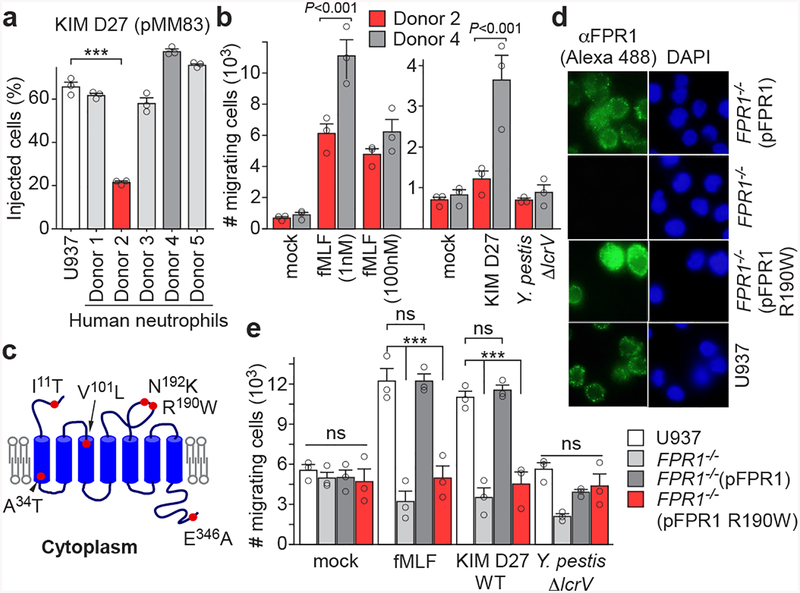
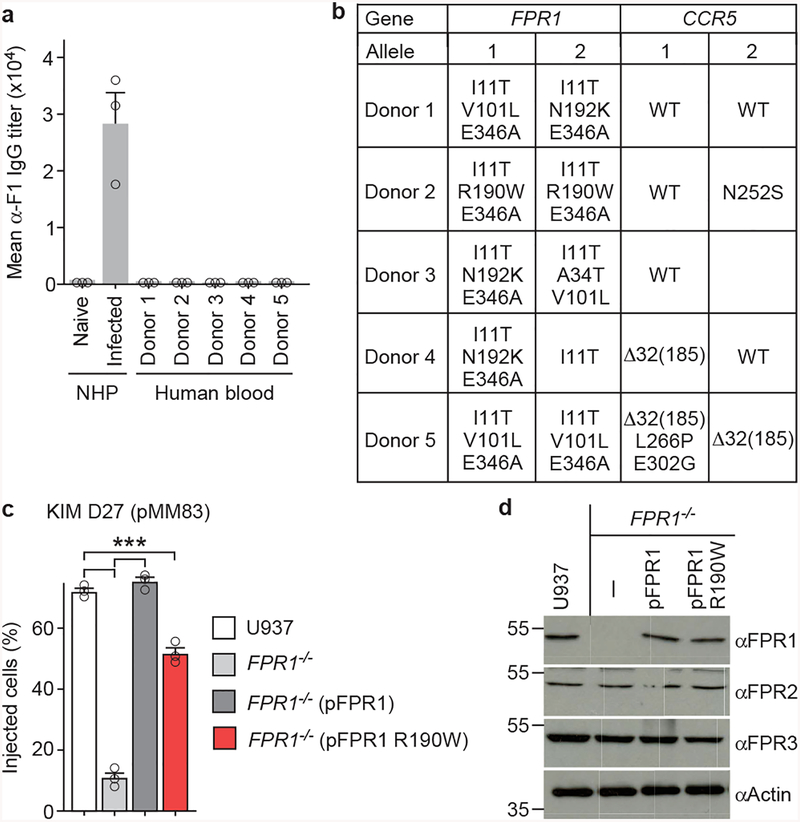
Neutrophils from human volunteers were infected with Y. pestis KIM D27 (pMM83) to measure YopM-Bla effector translocation. Neutrophils of donor 2 exhibited decreased T3SS translocation and reduced chemotaxis towards fMLF and Y. pestis (Fig. 5ab). ELISA measurement of specific F1-antibodies ruled out the possibility of plague immunity in volunteers (Extended Data Fig. 9a). FPR1 and CCR5 genes were PCR amplified from leukocyte DNA and sequenced to identify single nucleotide polymorphisms (SNPs) in each proband (Supplementary Table 1 & Extended Data Fig. 9b). FPR1 genes from each donor carried between 3–4 SNPs, however only donor 2 carried rs5030880 (A>T, p.R190W), with a single amino acid substitution, Arg190Trp, located in extracellular loop 2 between transmembrane domains 4 and 5 (Fig. 5c). rs5030880 occurs in 11–13% of Europeans and Americans with European descent, 6–9% of Africans and Americans with African descent, and 20% of Asians. Two donors were carrier or homozygote for CCR5-Δ32, whereas donor 2 did not harbor the CCR5-Δ32 deletion (Supplementary Table 1). To test whether rs5030880 is sufficient to confer diminished chemotaxis towards Y. pestis and reduced T3SS, we generated FPR1−/− cells expressing pFPR1 R190W. Compared with U937 and FPR1−/− (pFPR1) cells, FPR1−/− (pFPR1 R190W) cells exhibited reduced effector translocation during Y. pestis infection (Extended Data Fig. 9c). Immunoblotting and fluorescence microscopy experiments documented that FPR1−/− (pFPR1 R190W) macrophages produce and deposit N-formylpeptide receptor on the cell surface similar to U937 or FPR1−/− (pFPR1) macrophages (Fig. 5d; Extended Data Fig. 9d). Chemotaxis experiments revealed reduced migration of FPR1−/− (pFPR1 R190W) monocytes towards fMLF and Y. pestis as compared to U937 or FPR1−/− (pFPR1) monocytes (Fig. 5e).
Figure 5 |. Single-nucleotide polymorphism in human FPR1 associated with neutrophil resistance to Y. pestis T3SS.
a, Quantification of Y. pestis KIM D27 (pMM83) translocation of YopM-Bla into neutrophils from five different donors (1–5) as compared to U937 cells. b, Quantification of migrating neutrophils from donors 2 and 4 following addition of fMLF (1–100 nM) or priming with mock, Y. pestis KIM D27 (WT) or KLD29 (ΔlcrV) (2×107 CFU/ml). c, Model illustrating the position of amino acid substitutions in human FPR1. d, Surface display of FPR1 revealed by immunofluorescence microscopy using Alexa 488 labeled anti-FPR1 antibodies (αFPR1) (left). DAPI staining showing nuclei of U937 cells and variants (right). One of three repeats is shown. e, Quantification of migrating U937, FPR1−/−, FPR1−/− (pFPR1) and FPR1−/− (pFPR1 R190W) cells in a transwell assay primed with mock, fMLF, Y. pestis KIM D27 (WT) or KLD29 (ΔlcrV). One of three repeats is shown (a, b, d, e). Error bars represent the s.e.m. (n = 3 biological replicates) (a, b, e). One-way ANOVA and Bonferroni’s post-hoc analyses (a, e) and two-tailed t-test (b) were used to identify significant differences: ***, P<0.001; ns, not significant.
Discussion
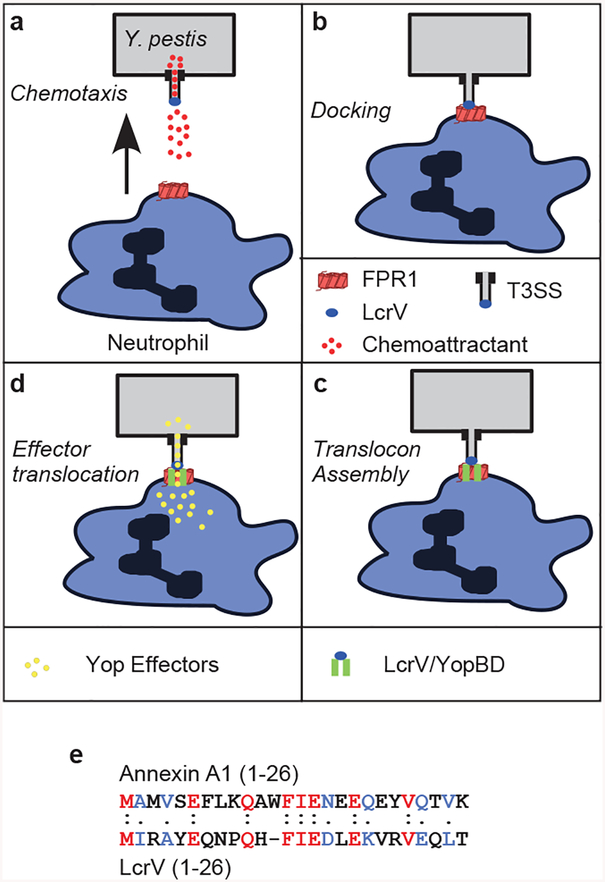
Our findings suggest that chemotaxis of immune cells towards Y. pestis is enhanced by the pathogen’s T3SS, releasing N-formylpeptides to activate FPR1 signaling (Extended Data Fig. 10a). Following chemotaxis, the T3SS cap protein, LcrV, binds to FPR1 (Extended Data Fig. 10b). The N-terminal domain of LcrV with homology to annexin A1 (Extended Data Fig. 10e) but without N-formyl modification15,24 may contribute to this docking event. Docking enables the assembly of the T3SS translocon, composed of LcrV, YopB, and YopD (Extended Data Fig. 10c)38. LcrV and YopD co-purified with FPR1STREP, demonstrating interaction between FPR1 and the T3SS needle. Translocon assembly promotes the formation of a conduit between the Y. pestis T3SS and the cytoplasm of immune cells, a low-calcium environment activating Yop effector translocation (Extended Data Fig. 10d)39. Yop effectors then inhibit host cell signaling, block actin-polymerization and activate immune cell apoptosis40. The concurrent release of formylated peptides from mitochondria (DAMPs, damage-associated molecular patterns) may further exacerbate infection through FPR1-mediated recruitment of neutrophils, and subsequent acute inflammation41,42.
Phylogenetic analyses revealed that early duplication of FPR1 generated FPR2 in the chemotaxis locus of mammals43. This was followed by a late duplication of FPR2 to yield FPR3 near the evolutionary origin of primates43. FPR1 is the high affinity receptor for N-formylpeptides, whereas FPR2 functions as a low affinity receptor for these compounds yet also recognizes other types of chemoattractants44. Rodents generally harbor FPR1 and an expanded family of FPR2-derived receptors that are expressed in vomeronasal neurons and respond to olfactory stimuli. In contrast, canidae (wolves, dogs, ferrets, coyotes, foxes) have lost FPR1 and the ability to respond to N-formylpeptides, harboring duplicated FPR2 in their chemotaxis locus45. Y. pestis causes plague in a wide range of different rodent species (rats, mice, prairie dogs, rabbits, guinea pigs and gerbils), however canidae are resistant to plague disease46. Although coyotes are frequently infected with Y. pestis through the consumption of plague-infected prey, these animals generate plague-protective F1 antibodies without developing disease symptoms47. Thus, plague resistance in canidae may be related to the loss of FPR1 in these species. More than 200 SNPs, many of which cause amino acid substitutions, have been described for FPR1. FPR1 SNPs have been associated with macular degeneration48, gastric cancer49 and the reduced survival of cancer patients receiving chemotherapy24. Of note, the ability of dendritic cells to physically associate with dying cancer cells and promote neoplasm-specific T cell immune responses requires expression of functional FPR1 on dendritic cells and annexin A1 on tumor cells24. Thus, while myeloid cell chemotaxis towards dying cancer cells is an important element of anti-tumor immune responses, annexin A1 binding to FPR1 enhances the presentation of cancer neoantigens by antigen-presenting cells and the stimulation of T lymphocytic responses that promote the killing of tumor cells24. We propose that the most abundant FPR1 SNPs may have been selected to enhance human survival from infectious diseases, including plague. The consequences of such selection may be the association of FPR1 SNPs with human diseases, most importantly malignant neoplasms. Of note, the onset of cancer occurs predominantly late in life, and may therefore not exert positive selection on FPR1.
METHODS
Ethics statement.
The University of Chicago’s Institutional Review Board (IRB) reviewed, approved, and supervised the protocols used for all experiments utilizing blood from human volunteers and informed consent forms were obtained from all participants. Animal research was performed in accordance with institutional guidelines following experimental protocol review, approval, and supervision by the Institutional Animal Care and Use Committee at The University of Chicago. Experiments with Y. pestis were performed in biosafety level 3 (BSL3)/animal BSL3 (ABSL3) containment at the Howard Taylor Ricketts Regional Biocontainment Laboratory. The University of Chicago Select Agent Program is approved and routinely inspected by both Institutional Biosafety Committee and Centers for Disease Control and Prevention (CDC) officials. Based on information provided to the CDC Select Agent Program and the APHIS Agriculture Select Agent Program, the University of Chicago is authorized to possess, use and transfer select agents and toxins under the conditions specified in the University of Chicago registration application, in accordance with 42 CFR part 73, 9 CFR part 121 and 7 CFR part 331.
Biosafety and Dual Use Research of Concern.
This manuscript describes experiments with Y. pestis AM18 (Δpgm, ΔyfeAB), a non-virulent variant of the plague vaccine strain Y. pestis EV76 (Δpgm), which was genetically modified to express the yopE-dtx hybrid (a fusion between full length yopE and the coding sequence for the mature domain of diphtheria toxin), generating Y. pestis POO1 (Δpgm, ΔyfeAB, yopE-dtx). To construct Y. pestis POO1, the investigators (O.S. and D.M.) submitted a proposal that was reviewed by The University of Chicago’s Institutional Biosafety Committee DURC Task Force (DTF) for the possibility of generating a strain with enhanced virulence. As mandated by DTF, the investigators analyzed the virulence attributes of strains Y. pestis EV76, Y. pestis AM18 and Y. pestis POO1 in a mouse model of bubonic plague infection. The data demonstrated that Y. pestis POO1 is avirulent in the mouse model of plague infection, whereas Y. pestis EV76 exhibits residual virulence, which is known to be required for eliciting a protective immune response against plague50,51. The analysis was reviewed by The University of Chicago’s DTF. DTF agreed with the investigators’ assessment that the findings presented in this proposal do not present DURC potential in that the yopE-dtx allele attenuates Y. pestis virulence. The DURC analysis allowed the investigators to use Y. pestis POO1 for CRISPR-Cas9 screening to isolate mutations in cell lines that confer resistance to Y. pestis T3SS effector translocation.
Bacterial strains and media.
Y. pestis KIM D27, an attenuated variant of the Y. pestis Mediaevalis strain KIM lacking the 102-kb pgm locus52, AM18, a ΔyfeAB variant of Y. pestis EV7615, the corresponding ΔlcrV mutant strains KLD2953 and AM46, (constructed in KIM D27 and AM18 backgrounds, respectively)15, and strains KIM8 and KIM8 Δ123430 were propagated on Heart Infusion Agar (HIA) plates at 26°C for two days. Overnight cultures were grown in Heart Infusion Broth (HIB) or thoroughly modified Higuchi’s (TMH) medium at 26°C15,54. Antibiotics were added as appropriate to a final concentration of 20 μg/ml chloramphenicol, 50 μg/ml kanamycin or 50 μg/ml ampicillin. Y. pestis CO92, a fully virulent isolate of the biovar Orientalis, was propagated on HIA plates supplemented with 0.2% galactose and 0.01% Congo Red at 26°C for two days55. Overnight cultures were grown in HIB at 26°C. Escherichia coli DH5α was propagated on Luria-Bertani (LB) agar plates at 37°C for 18–24 hours56. Overnight cultures were grown in Luria-Bertani (LB) broth at 37°C. Antibiotics were added as appropriate to a final concentration of 20 μg/ml chloramphenicol or 100 μg/ml ampicillin. Y. pseudotuberculosis YPIII (PB1+) was propagated on LB or Tryptic Soy agar (TSA) plates at 26°C for 2 days, overnight cultures were grown in LB or Tryptic Soy broth (TSB) at 26°C57. Chloramphenicol was added as required to a final concentration of 20 μg/ml.
Yersinia pestis strains and plasmid construction.
Construction of Y. pestis POO1 and POO2 was performed as follows. Upstream and downstream 1 kb flanking fragments of yopE were PCR amplified (5’- AAATCTAGAGGTAAACATTAATATTTGCCCGACAGGATGCTCTG-3’ in combination with 5’-AAAGTCGACCATCAATGACAGTAATTTCTGCATCTGTTGCGCC-3’ and 5’-AAAGTCGACTATGGATAAAAACAAGGGGATAGTGTTTCCCCC-3’ in combination with 5’-AAAGAGCTCCTCTGTTGAGCATTACACACTCCACAGTTGGG-3’) using the pCD1 plasmid from Y. pestis AM18 as template. The fragments were digested with XbaI/SalI and SalI/SacI and cloned into XbaI/SacI digested pUC19 in a three-way ligation yielding pUC19::yopE (all restriction enzymes were from New England Biolabs, Ipswich, Massachusetts, USA). A DNA fragment containing the codons 33–225 of diphtheria toxin fragment A (dtxA) was amplified by PCR (5’-AAAGTCGACTCTAAATCTTTTGTGATGGAAAAC-3’ and 5’-AAAGTCGACTGACACTGAGCTACCTACTGATCGCCTG-3’) using chromosomal DNA from Corynebacterium diphtheriae NCTC1312958. The PCR products encoding 579 base pairs of dtxA was digested with SalI and cloned into the corresponding site of pUC19::yopE yielding pUC19::yopE-dtxA. pUC19::yopE-dtxA was digested with Sacl/Xbal and the resulting products sub-cloned into Sacl/Xbal digested pCVD442 for allelic replacement. The resulting plasmid pCVD442::yopE-dtxA was electroporated into Y. pestis AM18 (to yield POO1) or AM46 (to yield POO2) and single-crossover events were selected by plating on HIA supplemented with 50 μg ml−1 ampicillin. Resolution of replication-defective plasmid co-integrates of pCVD442 were achieved by plating on HIA supplemented with 5% sucrose as counter-selection for sacB58. Ampicillin-sensitive and sucrose-resistant colonies were examined by PCR and DNA sequencing with 5’-AAATCTAGAGGTAAACATTAATATTTGCCCGACAGGATGCTCTG-3’ and 5’-AAAGAGCTCCTCTGTTGAGCATTACACACTCCACAGTTGGG-3’to verify mutants. Plasmid transformation of Y. pestis KIM D27 and its variants with pEGFP (Clontech, Palo Alto, California, USA), in addition to the plasmids described above, was performed by electroporation at 25 μF, 100 Ω, 1.8 kV cm−1. Y. pseudotuberculosis was transformed with pMM83 and pMM91 by electroporation at 25 μF, 200 Ω, 2.5 kV cm−1. Plasmid encoding FPR1 (FPR1 sgRNA resistant allele), which expresses human FPR1 from the SRα promoter18 was obtained from Addgene (plasmid #62600). The FPR1 insert is encoded in the pBGSA vector backbone18. pFPR1STREP, which carries a Strep-tag II (residues WSHPQFEK) appended to the C-terminal end of FPR1, and pFPR1 R190W were constructed by inverse PCR, as described in QuikChange® II Site-Directed Mutagenesis Kit. Briefly, PCR was performed using pFPR1 as template and two oligonucleotide primers (5’-TGGAGTTACAGGCAAAGAGGAGCCACCCGCAGTTCGAGAAATAATAGAATTC-3’ and 5’-GGGCTGCAGGAATTCTATTTTTCTCGAACTGCGGGTGGCT CCTCTTTGCCTG-3’ for pFPR1STREP and 5’-GGACCAACGACCCTAAAGAGTGGATAAATGTGGCCG-3’ and 5’-CATGGCAACGGCCACATTTATCCACTCTTTAGGGTCG-3’ for pFPR1 R190W) using PfuUltra HF DNA polymerase (Agilent, Santa Clara, California, USA). The PCR products were treated with DpnI endonuclease for 1 hour to digest the parental DNA template and subsequently transformed into E. coli DH5α super-competent cells. Clones were screened using the primer pair 5’-CAGCTGGTGAACAGTCCAGGAGCAGACAAG ATGGAG-3’ and 5’-CCCCCGGGCTGCAGGAATTCTATTACTTTGCCTGTA-3’. The plasmids were transfected into U937 FPR1−/− cells as described below. Human-codon-optimized sequences encoding mFPR1, mFPR2 and mFPR3 were synthesized by Integrated DNA Technologies (Skokie, Illinois) and amplified in E. coli DH5α. The genes were excised with EcoRI (New England Biolabs, Ipswich, Massachusetts, USA) and ligated into the parent vector pBGSA. Clones were screened for directionality using the following primer pairs: 5’-CAGGACGCAGATACAGCGGTC-3’ in combination with 5’-CAGCTGGTGAACAGTCCAGGAGCAGACAAGATGGAG-3’ (mFPR1, 2 and 3), or 5’-CCCCCGGGCTGCAGGAATTCTATTACTTTGCCTGTA-3’ combination with 5’-GAAGGTGGCCGTTACCATGTTGAC-3’, 5’-GAAGCTGAATACCGCCATCACCTTCG-3’, or 5’-CTGAATACCGCCATCACCTTCGTG-3’ (mFPR1, 2 and 3, respectively). The plasmids were transfected into U937 FPR1−/− cells as described below.
Antibodies and reagents.
The following monoclonal antibodies, sourced from R&D Systems (Minneapolis, Minnesota), were used to investigate inhibition of Y. pestis type III injection of human neutrophils, as described below: Siglec-9, LTB4, CD321, CD282, CD182, CD181, CD162, CD147, CD120b, CD120a, CDw119, CD114, CD89, CD88, CD87, CD66b, CD66, CD62L, CD63, CD58, CD55, CD54, CD50, CD49b, CD47, CD46, CD45, CD44, CD43, CD35, CD32, CD31, CD29, CD18, CD16, CD15, CD14, CD13, CD11b, CD11c, CD11a, CD10, CD9 and CD195. The rabbit polyclonal antibodies targeting FPR1 and FPR2 were purchased from Thermo Fisher Scientific, while the rabbit polyclonal antibody to FPR3 was purchased from Abcam (Cambridge, Massachusetts, USA). Mouse monoclonal antibodies against FPR1 and CCR1 were sourced from R&D Systems. The rabbit polyclonal to β-actin was purchased from Abcam. The mouse monoclonal anti-Y. pestis F1 antigen antibody was purchased from Abcam. Polyclonal anti-sera from rabbits were previously raised against LcrV and YopE59. Goat anti-rabbit IgG HRP-conjugated secondary antibodies were purchased from Cell Signaling Technology (Danvers, Massachusetts) or Abcam. Horse anti-mouse IgG HRP-conjugated secondary antibody was purchased from Cell Signaling Technology. Goat anti-mouse Alexa Fluor488-conjugated secondary antibody and goat anti-Rabbit Alexa Fluor594-conjugated secondary antibody were purchased from Thermo Fisher Scientific. The peptide N-formyl-methionine-leucine-phenylalanine (fMLF), mouse KC (CXCL1) and Tolyplocadium inflatum cyclosporin H were purchased from Sigma Aldrich (St. Louis, Missouri). The annexin A1 peptide (residues 1–25) was sourced from VWR Scientific (Radnor, Pennsylvania) and Leukotriene B4 from Cayman Chemical Company (Ann Arbor, Michigan).
Cell culture.
Human U937 histiocytic leukemia cells were propagated in Roswell Park Memorial Institute medium (RPMI) 1640 (Gibco) supplemented with 10% fetal calf serum (FCS, Sigma Aldrich or Gemini Bio-Products, CA)60. Human HL-60 acute promyelocytic leukemia cells were propagated in Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco) supplemented with 25 mM HEPES, l-Glutamine and 20% FCS61. The human HEK293T embryonic kidney cells were propagated in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% FCS. For transiently transfected cell lines, plasmids were maintained by the addition of 200 μg/ml G-418 (Invitrogen) to the media. U937 cells were differentiated into macrophages with 10 nM phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich) in RPMI 1640 containing 10% FCS for 48 hours and into monocytes with 1 mM N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate (Sigma Aldrich) in RPMI 1640 containing 10% FCS for 48 hours, replacing the medium every 24 hours. HL-60 cells were differentiated into neutrophils in IMDM containing 20% FCS, supplemented with 0.8% DMSO for 5–7 days.
Lentiviral production and isolation.
Lentivirus for transduction of U937 cells were produced by transfection of HEK293T cells. One hour prior to transfection, 80–90% confluent HEK293T cell cultures were washed with PBS (Corning Cellgro) and placed under 13 ml of reduced serum Opti-MEM media (Life Technologies) per T225 flask used. For each flask, 200 μl of Lipofectamine 2000+ reagent (Life Technologies) was diluted in 7.5 ml Opti-MEM and incubated for 5 minutes at room temperature. Separately, 10 μg of each lentiviral CRISPR vector (LentiCRISPRv2) and 50 μg of ViraPower lentiviral packaging mix (Invitrogen) was suspended in 7.5 ml of Opti-MEM media per flask. The following LentiCRISPRv2 plasmids were sourced from Genscript; for inactivation of FPR1 pLentiCRISPRv2::FPR1guideRNA#1 and pLentiCRISPRv2::FPR1guideRNA#2 were used, while pLentiCRISPRv2::CCR5guideRNA#1 and pLentiCRISPRv2::CCR5guideRNA#2 were used for inactivation of CCR5. Alternatively, 20 μg of the pooled human GeCKO v2.0 library, obtained from Addgene (Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA)62 was used in place of the individual LentiCRISPRv2 plasmids. The solutions were then mixed and incubated at room temperature for 20 minutes. The lipofectamine-DNA complex solution was added to the HEK293T cells, which were incubated overnight at 37°C, 5% CO2. The media was then replaced with RPMI 1640 containing 10% FCS, without antibiotics. Lentivirus containing supernatants were removed 72 hours post-transfection and debris removed by centrifugation at 2000 ×g, 15 minutes, 4°C. Viruses were concentrated by ultra-centrifugation at 45,700 ×g, 2 hours, 4°C, filtered through a 0.45 μm PVDF filter (Millex) and stored at −80°C. Lentivirus concentration was determined by titration against U937 cells prior to use, infecting 1×106 cells with 0 to 100 μl of lentiviral stocks, as described below.
Cell transduction using the GeCKO library.
The human CRISPR-Cas9 v2 library was a gift from Feng Zhang and obtained via Addgene (Cambridge, Massachusetts, USA; Catalogue Number 1000000048)62. U937 cells were transduced with the GeCKO library via spinfection. To find optimal virus volumes for achieving a multiplicity of infection (MOI) of 0.3–0.5, each cell type and new virus batch was tested by spinfecting 3×106 cells with a dilution series of the lentivirus. Briefly, 3×106 cells per well were plated into a 12-well plate in RPMI 1640 containing 10% FCS, supplemented with 8 μg/ml polybrene (Sigma Aldrich). Each well received a different titrated virus amount, between 0 to 50 μl. The 12-well plate was centrifuged at 1,500 ×g for 2 hours at 37°C. Cell pellets were suspended in fresh RPMI 1640 containing 10% FCS incubated for 48 hours at 37°C under 5% CO2. Cells were centrifuged, counted, and samples split in two wells with one well containing 2.5 μg/ml puromycin (Gibco). After 3 days, cells were counted, and the transduction efficiency was calculated as cell count from wells containing puromycin divided by cell count from wells without puromycin, and multiplied by 100.
Genomic DNA sequencing.
U937 cells were transduced at MOI of 0.3–0.5 with the GeCKO lentiviral library. Transduced U937 cells were infected with Y. pestis POO1 or POO2 for 12 hours at MOI of 100. Following infection, resistant cells were cultured in the presence of 2.5 μg/ml puromycin until reaching approximately 70% confluence and then reseeded and re-infected. Following incubation for 12 hours, the resistant cells were cultured in the presence of 2.5 μg/ml puromycin. Genomic DNA was extracted with a Blood & Cell Culture Midi kit (Qiagen) and used to prepare a sgRNA library with a two-step PCR protocol62. The first PCR was used to amplify the sgRNA containing cassette. Amplified products were subjected to a second PCR with a primer pair encoding a unique 8-bp barcode required for multiplexing along with a stagger sequence to increase library complexity62. The resulting amplicons from the second PCR were gel extracted, quantified, mixed and sequenced using a HiSeq 2500 (Illumina). The raw sequencing data were processed and analyzed using customized CRISPR-Cas9 library screen pipelines. Briefly, sequencing reads were first de-multiplexed by using the barcode in the reverse primer, and processed by Cutadapt to remove sequences from beginning to sgRNA priming site primers. Trimmed reads were used to map sgRNA sequences to pooled GeCKO v2 libraries A. Read counts of sgRNA for each sample were quantified by MAGeCK v5.6.0. Count data were filtered and normalized, and essential sgRNA and genes were ranked by MAGeCK. The data have been deposited in NCBI’s Gene Expression Omnibus63 and are accessible though GEO Series accession number GSE133826 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133826).
Targeted CRISPR-Cas9 mutagenesis.
U937 cells were suspended in RPMI 1640 containing 10% FCS, supplemented with 8 μg/ml polybrene (Sigma Aldrich), to a density of 1×107 cells/ml. Cells were transduced with CRISPR Lentivirus, produced as described above, at MOI of 10 and the cell-virus mixture centrifuged at 680 ×g for 2 hours at room temperature. The supernatant containing virus was removed and the cells re-suspended in RPMI 1640 containing 10% FCS to a density of 1×107 cells/ml, seeded in 12 well tissue culture plates (Corning) and incubated at 37°C, 5% CO2 for 3 days. The cells were sedimented by centrifugation (200 ×g, 5 minutes, room temperature), the media replaced with RPMI 1640 containing 10% FCS supplemented with 2.5 μg/ml puromycin and the cells incubated at 37°C, 5% CO2 for another 3 days. The cells were pooled, sedimented by centrifugation (200 ×g) for 5 minutes at room temperature, re-suspended in RPMI 1640 containing 10% FCS supplemented with 2.5 μg/ml puromycin and seeded into T225 flasks for expansion. Following expansion, the transduced cells were enumerated and diluted to approximately 5 cell/ml and 96 well plates seeded with 100 μl per well. Wells containing a single cell were identified by microscopy. DNA lesions in the sgRNA target region were identified by PCR as described below. Cell lines harboring deletions in the target gene were expanded in RPMI 1640 containing 10% FCS, without antibiotic supplementation, prior to use.
Isolation of murine bone marrow derived macrophages (BMDMs).
C57BL/6J, mFpr1−/− and mFpr-rs2−/− mice were euthanized by compressed CO2 inhalation. The femur and tibia were removed and stored at 4°C in RPMI 1640 supplemented with penicillin/streptomycin (Gibco) for transportation, as needed. Femur and tibia were surface sterilized with 70% ethanol, washed with phosphate buffered saline (PBS, Sigma Aldrich), separated and ends removed to flush out the bone marrow with 2×10 ml of RPMI 1640 containing penicillin/streptomycin. The suspension was passed through a 40 μm nylon tissue culture strainer (BD) and the cells were sedimented by centrifugation (10 minutes, 200 ×g, room temperature). The supernatant was aspirated and the pellet washed with 10 ml of RPMI 1640 containing penicillin/streptomycin (10 minutes, 200 ×g, room temperature) and re-suspended in 10 ml RPMI 1640 without penicillin/streptomycin. The cells were enumerated using a hemocytometer and diluted with BMDM media (RPMI 1640 (Gibco), supplemented with 20% FCS, 10% filter sterilized supernatant from colony-stimulating-factor-transfected cells, 2 mM glutamine, 1 mM pyruvate and 0.55 mM β-mercaptoethanol) to a density of 3×105 cells/ml. The cells were seeded in 150 mm bacteriological dishes and fed with an additional 30 ml of BMDM medium 3 days post-extraction. The growth medium was replaced 6 days post-extraction. Any non-adherent cells were sedimented by centrifugation (10 minutes, 200 ×g, room temperature), re-suspended in BMDM medium and seeded in new 150 mm bacteriological dishes. Bone marrow derived macrophages were used at day 7 to 9 post extraction.
Isolation of murine peritoneal granulocytes.
C57BL/6J, mFpr1−/− and mFpr-rs2−/− mice were hand restrained and 1 ml of Brewer thioglycollate medium (Sigma Aldrich) injected into the peritoneal cavity. The inflammatory response was allowed to develop for 3 hours before animals were euthanized by compressed CO2 inhalation. The abdomen of each mouse was surface sterilized with 70% ethanol and the peritoneal wall exposed. 5 ml of sterile PBS (Corning Cellgro) containing 0.02% sodium ethylenediaminetetraacetic acid (Na2 EDTA) was injected into the peritoneal cavity and withdrawn. This step was repeated and the peritoneal fluids combined for each mouse. The pooled peritoneal fluids were centrifuged (10 minutes at 200 ×g, room temperature), and the peritoneal exudate washed three times with 10 ml PBS (10 minutes, 200 ×g, room temperature). The washed pellet was re-suspended in 1 ml PBS, enumerated and diluted to 3–5×107 peritoneal exudate cells/ml. The cell suspension was mixed 1:9 (vol/vol) with Percoll gradient solution [1 volume 10 × PBS, pH 7.2 to 9 volumes sterile Percoll (Sigma Aldrich)] and separated by ultracentrifugation (20 minutes, 60,650 ×g, 4°C). The granulocyte containing layer was collected and washed with 10 ml PBS (5 minutes, 200 ×g, room temperature). The granulocytes were re-suspended in the appropriate medium and enumerated.
Isolation of human neutrophils.
Primary neutrophils from healthy human volunteers were isolated from Na2 EDTA-anticoagulated venous blood samples. The blood was layered 1:1 (vol/vol) onto PolymorphPrep and separated by centrifugation (35 minutes, 500 ×g, 20°C). The neutrophil containing layer was removed, mixed 1:1 (vol/vol) with 50% HBSS/water, and centrifuged (10 min, 350 ×g, room temperature). Sedimented neutrophils were washed with 10 ml Hank’s Balanced Salt Solution (HBSS, Corning Cellgro) (10 min, 350 ×g, room temperature) and re-suspended in 5 ml of red blood cell lysis buffer (Roche). Neutrophils were sedimented by centrifugation (5 min, 250 ×g, room temperature) and washed with 10 ml HBSS (10 min, 350 ×g, room temperature). The neutrophils were suspended in 2 ml RPMI 1640 supplemented with 2% bovine serum albumin (BSA, Fisher Scientific). Neutrophil viability and purity were determined by Trypan blue (MP Biomedicals, LLC) exclusion and Wright/Giemsa staining, respectively.
Isolation of DNA for sequencing of FPR1 and CCR5.
Genomic DNA was isolated from clonal transductant cells (following CRISPR-Cas9 mutagenesis) or from Na2 EDTA-anticoagulated venous blood obtained from healthy human volunteers using the Wizard Genomic DNA Purification kit (Promega), according to the manufacturer’s instructions. For CRISPR-Cas9 derived cells, primers 5’-CCTGGTAAAACGGGGACAGTAGC-3’ and 5’-CTGTAACTCCACCTCTGCAGAAGG-3’ were used to amplify the sgRNA target region of FPR1 and primers 5’-CTGCTTGTCATGGTCATCTGCTAC-3’ and 5’-CTTAGACCCTCTATAACAGTAACTTCC-3’ were used for CCR5. The entire coding region of both FPR1 and CCR5 was then amplified from candidate mutant cell lines, or genomic DNA donor using primer pairs 5’-GATCAAGCTTGAGCAGACAAGATGGAGACAAATTC-3’ and 5’-GATCGAATTCCTCAAGGTGAGACGAAGCTGG-3’ and 5’-GATCAAGCTTGCACAGGGTGGAACAAGATGG-3’ and 5’-GATCGAGCTCGCACAACTCTGACTGGGTCAC-3’, respectively. PCR products digested with HindIII/EcoRI (FPR1) or HindIII/SacI (CCR5) were cloned into pUC19. Plasmids were transformed into E. coli DH5α and clonal isolates of pUC19::FPR1 and pUC19::CCR5 Sanger sequenced by the University of Chicago Cancer Sequencing Center, using the primers described above as well as the plasmid targeting primers 5’-GTAAAACGACGGCCAGT-3’ and 5’-CACACAGGAAACAGCTATGACCAT-3’. A minimum of four reads per sequence were used for alignment and SNP analysis, performed using Geneious 6.0.6 (Biomatters Ltd).
Type III injection assay.
Overnight Y. pestis cultures were diluted 1:20 into fresh HIB supplemented with 20 μg/ml chloramphenicol, incubated for 1.5 hours at 26°C and then incubated for 1.5 hours at 37°C. Target cells were suspended at a density of 1×106 cell/ml in 500 μl of RPMI 1640 containing 2% BSA and 50% mouse plasma in RPMI when indicated. Cells (U937 cells, mouse BMDM and neutrophils, and human neutrophils) were pre-incubated with fMLF, FPR1 inhibitors, antibodies or LcrVS228 for 30 minutes at 37°C, 5% CO2 prior to infection, if required. The target cells were infected at MOI of 10 in a final volume of 600 μl, at 37°C, 5% CO2. Infection with Y. pestis KIMD27 YopM-Bla (pMM83) and, as a negative control, Y. pestis KLD29 (ΔlcrV) YopM-Bla (pMM83) was allowed to proceed for 3 hours. Alternatively, infection with Y. pseudotuberculosis YPIII (PB1+) YopM-Bla (pMM83), with Y. pseudotuberculosis YPIII (PB1+) GST-Bla (pMM91) serving as the negative control, was allowed to proceed for 1 hour. Cells were sedimented by centrifugation (3 minutes, 1,500 ×g, room temperature) and re-suspended in 100 μl of RPMI 1640 containing 2% BSA, with 50 μg/ml kanamycin to abrogate Yersinia growth and type III injection. The cells were stained with 1 × CCF2-AM (Invitrogen) for 1 hour in the dark at room temperature. Cells were collected by centrifugation (3 minutes, 1,500 ×g, room temperature), washed with 500 μL of HBSS (Corning Cellgro) (3 minutes, 1,500 ×g, room temperature), and re-suspended in 500 μl HBSS Flow (HBSS supplemented with 0.5 mM EDTA, 25 mM HEPES and 2% bovine serum albumin, pH 7.4). To discriminate between live and dead cells, propidium iodide was added to each sample at a final concentration of 0.5 μg/ml immediately prior to flow cytometry analysis. A BD FACSCanto flow cytometer was used to analyze at least 10,000 cells per sample, and data were analyzed using FlowJo v10.0.7 (FlowJo, LLC, USA) and FACSDiva (BD Biosciences, San Jose, California, USA). Samples were first gated with the forward and side scatter for the population of singlet cells and subsequently gated for live cells by negative PI selection. PI-negative cells were analyzed for blue fluorescence indicative of type III injection of YopM-Bla.
Trans-well migration assay.
HL-60 cells were seeded at a density of 2×105 cell/ml, 6 days prior to the assay and differentiated as described above. U937 cells were seeded at a density of 3×105 cell/ml, 2 days prior to the assay and differentiated into monocytes as described above. Human neutrophils and murine granuloctes were prepared as described above and used directly for the trans-well migration (chemotaxis) assay. The cells were sedimented by centrifugation (5 minutes, 200 ×g, room temperature). For chemotaxis towards fMLF, the cells were re-suspended in 1 ml HBSS with calcium, magnesium and glucose (Gibco) and 1% BSA; HBSS without calcium and magnesium (Corning Cellgro) was used for U937 derived monocytes. For chemotaxis towards Y. pestis, the cells were re-suspended in the same medium as present in the bottom chamber, typically thoroughly modified Higuchi’s (TMH) medium. The cells were counted using a hemocytometer and adjusted to 2×106 cell/ml; where required the cells were pre-incubated with LcrVS228 for 30 minutes at 37°C, 5% CO2 prior to setting up the trans-well plates. Trans-well plates with polycarbonate inserts of a 5 μm pore size (Corning Costar) were used. Appropriate medium (600 μl) was placed in the lower wells, containing the chemoattractant fMLF (Sigma Aldrich) at a concentration of 1 nM for HL-60 cells and primary neutrophils and 10 nM for U937 monocytes. The chemoattractants LTB4 and KC (CXCL1) were used at 10 nM and 100 ng ml−1, respectively. To measure chemotaxis towards Y. pestis, 2×107 CFUs of KIMD27 or its derivatives were re-suspended in 600 μl HBSS containing 1% BSA and placed in the bottom chamber. 100 μl of the cell suspension was added to the top chamber and the trans-well plates incubated for 2 hours at 37°C, 5% CO2. Following migration, the media and un-translocated cells were aspirated from the top well and discarded. The trans-well inserts were treated with trypsin at 37°C, 5% CO2 for 5 minutes and then discarded. The medium from the lower chamber was combined with the trypsin and the translocated cells recovered by centrifugation (5 minutes, 200 ×g, room temperature). The cells were suspended in HBSS containing 0.04% Trypan blue (MP Biomedicals, LLC) and enumerated using a hemocytometer.
Cell adhesion assay.
For adhesion assays, 2×105 U937 cells were seeded into 24-well plates and differentiated into macrophages for 48 hours with 100 ng/ml of PMA in RPMI containing 10% FCS. Fresh medium was added without PMA and the cells allowed to rest for 24 hours. U937 cells were infected for 2 hours at a MOI of 10 with Y. pestis. Un-inoculated medium was used for mock infection. Supernatants were collected without disturbing the cells, followed by serial dilution and plating of bacteria on HIA. The adherent cells were washed with PBS and lysed with 0.1% Triton X-100 in PBS for 30 min. The cells were serially diluted and plated on HIA and incubated at 26°C for 48 hours. Bacteria attached to the cells were counted and calculated as a percentage of inoculum.
Immunofluorescence microscopy.
U937 cells were seeded in 12-well plates on 18-mm glass coverslips. Cells were fixed with 10% formalin for 20 minutes at room temperature, washed with 1 × PBS three times and permeabilized with 0.1% Triton X-100 in PBS for 30 minutes. For human peripheral neutrophils, 18-mm glass coverslips were coated with 50 μl fresh human serum for 1 hour at 37°C. One ml of neutrophils (1×106 cells) was placed on coated coverslips and incubated for 1 hour at 37°C. The neutrophils were stimulated with 200 nM PMA for 30 minutes, washed with PBS, fixed with 10% formalin for 30 minutes and permeabilized with 0.1% Triton X-100 in PBS for 30 minutes. Cells were blocked in 5% goat serum in PBS (blocking buffer) for 1 hour, followed by overnight incubation with primary antibodies (as described above) at 4°C. Next, the cells were washed three times with PBS followed by incubation with fluorescently labeled secondary antibody for 1 hour at room temperature. Cells were counterstained with Alexa 488 and DAPI (4′,6-diamidino-2-phenylindole). For extracellular binding of Alexa 488-tagged FPR1, cells were not permeabilized; for intracellular binding, cells were permeabilized and stained as described above.
FPR1STREP affinity purification.
U937 and U937 FPR1−/−(pFPR1STREP) cells (10×106) grown in 100 mL RPMI 1640 containing 10% FCS were centrifuged, washed in PBS and suspended in 100 mL serum free RPMI. Each sample was split into two flasks (5×106 cells) and one aliquot infected with 5×107 Y. pestis KIM D27 for 2 hours at 37°C (MOI 10), while the other aliquot was left uninfected. Y. pestis KIM D27 was pre-grown in HIB for 1.5 hours at 37°C. Both samples were sedimented by centrifugation at 300 ×g for 5 minutes at 25°C. Permeabilization buffer with Halt Protease Inhibitor Cocktail was added to the cell pellets, vortexed briefly and incubated 10 minutes at 25°C. A 50% slurry StrepTactin sepharose (100 μl) was added to fresh tubes, centrifuged at 3,000 ×g for 1 minute and washed with permeabilization buffer. Permeabilized cells were centrifuged for 15 minutes at 16,000 ×g and 0.7 ml supernatant containing proteins was transferred to a new tube. Protein samples from U937 and U937 FPR1−/−(pFPR1STREP) were added to the washed Strep-Tactin sepharose and rotated for 30 minutes at 25°C. Samples were centrifuged for 1 minute at 3,000 ×g. The supernatant was transferred to a fresh tube and sample loading buffer added to the beads. Samples were vortexed and heated to 90°C and analyzed by 15% SDS-PAGE and immunoblotting for FPR1, Actin, LcrV, YopD and RpoA.
LDH release assay/cell killing assay.
LDH release was measured using a Pierce LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Briefly, U937 cells were infected with Y. pestis POO1 or POO2 at MOI of 10 for 4 hours in a 96-well plate. At the end of incubation, the plate was centrifuged at 250 ×g for 3 minutes and 50 μl of each supernatant was transferred to a 96-well flat bottom plate in triplicate wells. Reaction Mixture (50 μl) was added to each sample well and mixed by gentle tapping. The plate was incubated at room temperature for 30 minutes protected from light. Stop Solution (50 μl) was added to each sample well and mixed by gentle tapping and the absorbance measured at 490 nm and 680 nm. The 680 nm absorbance values were subtracted from the 490 nm absorbance value to determine LDH activity.
Trypan blue exclusion assay.
The Trypan blue exclusion assay was used to measure killing of U937 cells and murine BMDMs. Briefly, cells were suspended at a density of 5×105 cell/ml in 1 ml of RPMI 1640 supplemented with 2% BSA. The target cells were infected at MOI of 10 by the addition of Y. pestis KIMD27, KLD29 (ΔlcrV) or POO1 (yopE-DTA) in 10 μl PBS (Corning Cellgro) and incubated at 37°C, 5% CO2 for 4 or 18 hours. At the conclusion of the infection, Trypan blue solution was added to a final concentration of 0.04% and the number of live and dead cells counted using a hemocytometer. The number of live cells was calculated as a percentage of the total number of live (Trypan blue negative) and dead (Trypan blue positive) cells per grid square. Three biological repeats were performed per assay, with each replicate being counted in technical triplicate.
U937 cell lysis.
FPR1 containing extracts were prepared from U937 cells and their derivatives using the M-PER plus membrane protein extraction reagent (Thermo Fisher Scientific), according to manufacturer’s instructions. Briefly, 1×108 cells were washed with PBS by centrifugation (5 minutes, 300 ×g, room temperature). The cell pellet was lysed using 2 ml of permeabilization buffer, supplemented with 1×HALT protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific), for 10 minutes at 4°C. Insoluble material was removed by centrifugation (15 minutes, 16,000 ×g, 4°C). The supernatant was removed, mixed 1:1 (vol/vol) with 2×loading buffer and boiled at 90°C prior to separation by SDS-PAGE and visualization of proteins by immuno-blotting.
Animal breeding.
C57BL/6J, mFpr1−/− and mFpr2−/− mice were sourced from the National Institutes of Health (NIH) (28) and bred in a barrier facility at the University of Chicago. All mFpr1−/− and mFpr2−/− mice were genotyped by PCR prior to use. DNA was extracted from tissue samples taken by ear punch, using the Hot-SHOT DNA extraction method. Briefly, tissue samples were placed under 100 μl of alkaline lysis reagent (25 mM sodium hydroxide, 0.2 mM sodium EDTA) and boiled at 90°C for 30 minutes. The samples were neutralized with 100 μl of 40 mM Tris-HCl, the tissue sedimented by centrifugation (3 minutes, 1,500 ×g, room temperature) and the DNA containing supernatant removed. One μl of the DNA extract was used as a template for PCR, using KOD polymerase (Novagen) and primers 5’-CTGATGATTCCTCTGACAGTGAGC-3’ and 5’-GCAACGGGCTCGTGATCTGG-3’ to amplify mFpr1 and 5’-CGGGGTGTGTAGTGTGACTTTTC-3’ and 5’-CCTGACAAATGCTATGATTATAGGCATG-3’ to amplify mFpr2. PCR products were analyzed by agarose gel electrophoresis, in comparison to products generated from wild-type C57BL/6J mouse tissue.
Bubonic plague mouse model.
For bubonic plague challenge, mFpr1−/− mice (n = 20, 6- to 8-week old, 10 males and 10 females) were compared to age and sex matched C57BL/6J mice. All mice were infected by subcutaneous injection in the left inguinal fold with Y. pestis strain CO92 in 100 μl of PBS, at a dose of 600 CFU. Animals were monitored every 12 hours, increasing to every 6 hours following the onset of severe disease; deaths were recorded. When animals were deemed moribund (laterally recumbent and lethargic with rapid respiration) they were euthanized by compressed CO2 inhalation. After 14 days post-infection, mFpr1−/− mice that survived the initial challenge were euthanized. Cardiac puncture was performed to obtain sera for analysis of IgG antibody titers. For all animals the draining lymph nodes, liver and spleen were removed during necropsy, a section was taken for histopathological analysis and the remainder analyzed for bacterial load by CFU enumeration. Antibody titers were determined by ELISA as previously described64. All experiments were performed in BSL3 and ABSL3 laboratories at the Howard Taylor Ricketts Laboratory. Samples for histopathology were fixed in Formalin (Fisher Scientific) and hematoxylin and eosin stained slides were prepared by the Human Tissue Resource Center, University of Chicago.
Neutrophil infiltration.
To measure neutrophil infiltration, 20 mFpr1−/− mice (4 groups, n = 5 per group, 6- to 8-week old, 5 males and 5 females, 2 experimental replicates) were compared to age and sex matched C57BL/6J mice. All mice were anaesthetized by intra-peritoneal injection with ketamine/xylazine (100 mg kg−1 and 5 mg kg−1, respectively), shaved and infected by injection into the dermis of the left inguinal region with Y. pestis strain CO92 in 50 μl of PBS, at a dose of 1000 CFU. Four hours post-challenge, all animals were euthanized by compressed CO2 inhalation. The dermis surrounding the injection site was removed during necropsy and fixed in Formalin (Fisher Scientific) for histopathological analysis. Consecutive thin section slides were stained with hematoxylin and eosin, or stained by immunohistochemistry using the neutrophil specific marker anti-Ly6G (1A8). Slides were prepared by the Human Tissue Resource Center, University of Chicago and analyzed by a blinded investigator who assigned each thin-sectioned tissue sample one of four pathology scores: 0 = no neutrophil influx, 1 = local infiltration, 2 = moderate local infiltration and 3 = widespread infiltration (Extended Data Figure 8). Datasets were analyzed for statistical significant differences in neutrophil influx between plague infected wild-type and mFpr1−/− mice.
Murine plasma preparation.
Whole blood was collected by cardiac puncture of CD-1 mice (Charles River Laboratories) and immediately anti-coagulated with 10 μg ml−1 desirudin (Marathon Pharmaceuticals). Plasma was generated by centrifugation of desirudin-treated blood at 1,000 ×g for 5 minutes at 4°C for removal of blood cells, followed by a second centrifugation at 10,000 ×g for 3 minutes at 4°C. Heat inactivation was achieved by incubating plasma at 56°C for 30 minutes.
LcrVS228 purification.
Recombinant LcrVS228 was purified from a 2-liter culture of E. coli DH5α carrying plasmid pKG4828. The culture was grown in LB broth to mid-exponential phase (OD600 ~ 0.6) prior to addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG, Fisher Scientific). The culture was incubated at 37°C for another 4 hours and the bacteria sedimented by centrifugation (10 minutes, 5,000 ×g, 4°C). The cell pellet was washed with PBS (10 minutes, 5,000 ×g, 4°C) and re-suspended in 20 ml of Buffer A (0.1 M Tris (pH 8.0), 0.15 M NaCl, 1 mM EDTA). The cells were lysed in a French pressure cell at 14,000 psi and insoluble material removed by ultra-centrifugation (2 hours, 40,000 ×g, 4°C). The supernatant was subjected to chromatography over 1 ml of Strep-Tactin Sepharose resin (IBA BioTAGnology). The protein-bound resin was washed with 20 ml of strep-purification buffer and the bound LcrVS228 eluted with 10×1 ml of strep-purification buffer, supplemented with 5 mM desthiobiotin (IBA BioTAGnology). The elution fractions were analyzed by 15% SDS-PAGE and those containing LcrVS228 pooled. The protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce, Thermo Fisher Scientific).
Statistical analyses.
All statistical analyses were performed using GraphPad Prism, version 5.0 (GraphPad Software, Inc., La Jolla USA). Datasets featuring one group were analyzed by an un-paired Student’s t-test or Mann Whitney test while a one-way ANOVA with Bonferroni’s post-test or Kruskal-Wallis test was used for datasets containing more than one group. For Trypan blue exclusion, LDH release, cell adhesion, type III injection and trans-well migration assays, a minimum of 3 repeats were performed. In all cases, p is defined as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Data availability statement.
All data generated as part of this study are included in this article or its Extended Data and Supplementary Materials. Data for the CRISPR-Cas9 screen have been deposited in NCBI’s Gene Expression Omnibus and are accessible though GEO Series accession number GSE133826 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133826).
Extended Data
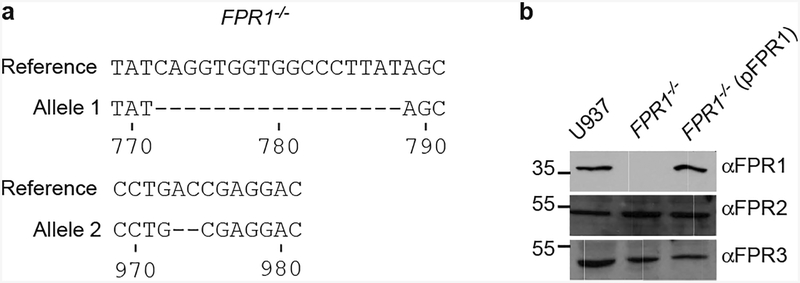
Extended Data Figure 1 |. Generation of FPR1−/− U937 cells using CRISPR-Cas9 and genetic complementation.
a, Sequencing results for alleles cloned from CRISPR-Cas9 derived FPR1−/− U937 macrophages. b, Immunoblot analysis for the production of FPR1, FPR2 and FPR3 in U937 macrophages, CRISPR-Cas9 derived FPR1−/− cells and FPR1−/− cells transfected with pFPR1. Numbers to the left of blots indicate migration of molecular weight markers. One of three repeats is shown (a, b).
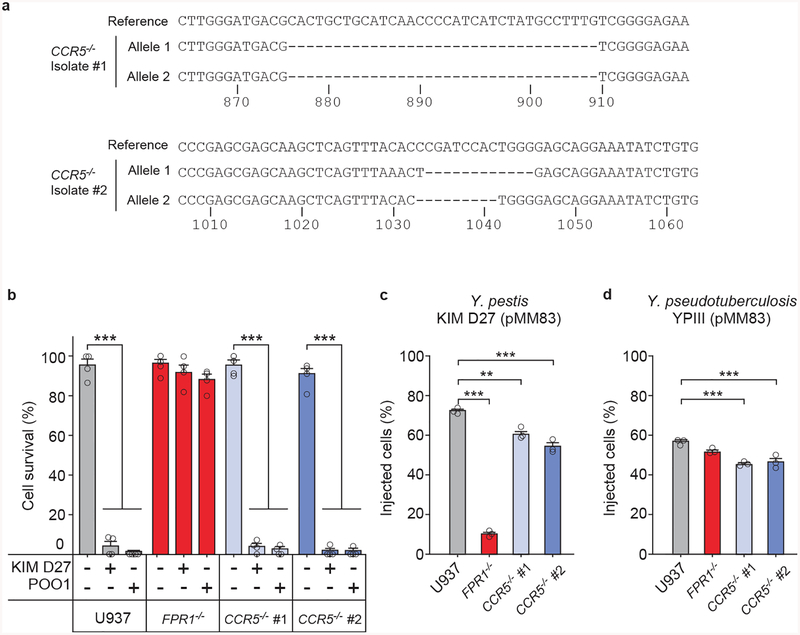
Extended Data Figure 2 |. Contribution of CCR5 to Yersinia spp. intoxication and injection by the T3SS.
a, Sequencing results for the alleles of two CCR5−/− U937 isolates obtained using CRISPR-Cas9 mutagenesis. b, Cell survival following incubation with strains Y. pestis KIM D27 and POO1 was measured using the Trypan blue exclusion assay; error bars represent the s.e.m. (n = 4 biological replicates). T3SS injection into U937, FPR1−/−, and CCR5−/− by Y. pestis KIM D27 (c) and Y. pseudotuberculosis YPIII carrying pMM83 (YopM-Bla) (d); error bars represent the s.e.m. (n = 3 biological replicates). One of three repeats is shown (b-d). Statistical analysis was performed using one-way ANOVA with Bonferroni Correction: ***, P <0.001; **, P <0.01.
Extended Data Figure 3 |. Antibodies and ligands of N-formylpeptide receptor (FPR1) inhibit Y. pestis type III secretion into human neutrophils.
Human neutrophils were stained with the β-lactamase substrate CCF2-AM, infected with wild-type Y. pestis KIM D27 (WT, pMM83) or KLD29 (ΔlcrV, pMM83) variant defective for the type III secretion system (T3SS), and analyzed for (a) blue fluorescence (YopM-Bla translocation into neutrophils and CCF2-AM cleavage) or (b) green fluorescence to derive the percent of stained cells injected with T3SS effector. c, Inhibition of Y. pestis T3SS into human neutrophils by monoclonal (αFPR1m) and polyclonal antibodies against FPR1 (αFPR1p), bacterial LcrV, N-formylpeptide (fMLF), annexin A1 peptide, staphylococcal CHIPS and cyclosporin H. d, Differential interference contrast (DIC) and fluorescence microscopy of mock or fMLF treated differentiated U937 cells infected with green-fluorescent Y. pestis KIM D27 (pEGFP) and stained with F1-specific antibody (red) to reveal extracellular bacteria in merged images of mock, but not in merged images of fMLF-treated cells. Orange and green arrows point to extra- and intracellular bacteria, respectively. e, Antibodies and ligands of FPR1 inhibit Y. pestis T3SS of YopM-Bla into U937 macrophages. One of three repeats is shown (a-e). Error bars represent the s.e.m. (n = 3 biological replicates) (c, e). One-way ANOVA with Bonferroni Correction was used to identify significant differences (c, e): ***, P <0.001; **, P <0.01.
Extended Data Figure 4 |. Screening 45 monoclonal antibodies for inhibition of Y. pestis T3SS into human neutrophils.
Human neutrophils were stained with the β-lactamase substrate CCF2-AM (green fluorescence), and infected with wild-type Y. pestis KIM D27 (pMM83). Translocation of YopM-Bla into neutrophils results in CCF2-AM cleavage (blue fluorescence) allowing for quantification of percent of stained cells injected with T3SS effector in the absence (Mock) or presence of specific monoclonal antibody. Error bars represent the s.e.m. (n = 3 biological replicates). A representative of three independent experiments is shown.
Extended Data Figure 5 |. FPR1−/− cells migrate towards chemoattractants other than formylated peptides and differentiated HL-60 cells migrate towards Y. pestis.
a, Numbers of migrating immune cells were quantified in a transwell assay primed with mock, 10 nM fMLF, 10 nM LTB4 or 100 ng ml−1 KC (CXCL1) for U937, FPR1−/− and FPR1−/− (pFPR1) cells. b, Numbers of migrating HL-60 cells were quantified in a transwell assay primed with mock, Y. pestis KIM D27 (WT) or KLD29 (ΔlcrV) (107 CFU/ml). Chemotaxis toward fMLF is shown as a control. c, Increasing concentrations of LcrVS228 (10−1–103 ng/ml) were added to the transwell assay and number of migrating HL-60 cells recorded. Error bars represent the s.e.m. (n = 3 biological replicates); one-way ANOVA with Bonferroni Correction was used to identify significant differences: ***, P<0.001; **, P <0.01; *, P<0.05; ns, not significant. A representative of three independent experiments is shown.
Extended Data Figure 6 |. Adhesion of Y. pestis to human macrophages does not require effector Yops and loss of FPR1 in human macrophages can be complemented with pFPR1STREP.
a, Y. pestis KIM 8 and its variant KIM 8 Δ1234 (Yop-less) were grown at 37°C in TMH to induce T3SS. Cultures were centrifuged to separate the supernatant (S) from the bacterial pellet (P) and extracts were analyzed by immunoblotting with antibodies specific for YopB (αYopB), YopD (αYopD), YopE (αYopE), LcrV (αLcrV), YscF (αYscF), YopH (αYopH), YopM (αYopM) and YopJ (αYopJ). One of three repeats is shown. b, Wild-type Y. pestis KIM 8 or its Yop-less variant, KIM8 Δ1234, were added to U937 and FPR1−/− cells (MOI of 10) and adherence quantified as percent inoculum. Error bars represent the s.e.m. (n = 3 biological replicates). One-way ANOVA with Bonferroni Correction was used to identify significant differences: **, P<0.01. A representative of three independent experiments is shown. c-d, Genetic complementation in FPR1−/− cells using pFPR1STREP. c, Y. pestis POO1 (yopE-dtx) but not POO2 (ΔlcrV, yopE-dtx) induced cell lysis is restored in FPR1−/− cells transfected with plasmid expressing C-terminal Strep-II tag FPR1 (pFPR1STREP). d, Y. pestis KIM D27 (pMM83) mediated YopM-Bla translocation into U937, FPR1−/− and FPR1−/− (pFPR1STREP) cells. One of three repeats is shown and error bars represent the s.e.m. (n = 3 biological replicates) (c, d). Significant differences were measured with one-way ANOVA with Bonferroni post-hoc analysis: ***, P<0.001.
Extended Data Figure 7 |. T3SS injection of mFpr1−/− neutrophils in vitro and in vivo.
a-b, Murine plasma does not affect mFpr1−/− neutrophil resistance to T3SS injection. Y. pestis KIM D27 (pMM83) translocation of YopM-Bla into mouse neutrophils, incubated in murine plasma with (a) or without (b) prior heat inactivation (HI). One of three repeats is shown. Error bars represent the s.e.m. (n = 3 biological replicates); one-way ANOVA with Bonferroni Correction was used to identify significant differences: ***, P<0.001; **, P<0.01. c-e, The mFpr1−/− mutation does not abolish the influx of neutrophils into Y. pestis infected tissues. Age and sex matched C57BL/6J and mFpr1−/− mice (4 groups, n = 5 per group, 6- to 8-week old, 5 males and 5 females, 2 experimental replicates) were anesthetized and infected by injection of 1,000 CFU Y. pestis CO92 into the inguinal region. Four hours post-challenge, euthanized animals were necropsied, the dermis surrounding the injection site was removed and fixed in formalin for histopathological analysis. Consecutive thin sectioned slides were stained with hematoxylin and eosin (H&E), or stained by immunohistochemistry with the neutrophil marker anti-Ly6G (α-Ly6G). (c) Slides were analyzed by a blinded investigator and assigned one of four pathology scores: 0 = no neutrophil influx, 1 = local infiltration, 2 = moderate local infiltration and 3 = widespread infiltration. Analysis of neutrophil influx was performed in male (n=8) and in female (n= 10) mice (d) or in all mice (male and female, n=18; 2 animals were excluded from the analysis owing to unclear histology)(e). One-way ANOVA with Bonferroni Correction was used to analyze differences: ns, not significant (b-c).
Extended Data Figure 8 |. Contribution of mouse N-formylpeptide receptor 1 to plague disease.
a, Serum derived from naïve or Y. pestis infected mFpr1−/− mice (experiment shown in Fig. 4e) was analyzed for IgG specific for capsular fraction 1 antigen (αF1) via ELISA. One of three repeats is shown and error bars represent the s.e.m. (n = 3 biological replicates). b, Representative spleen sections stained with H&E from naïve or Y. pestis infected animals shown in Fig. 4e. c, Measurements of white pulp areas in spleens. Each dot represents the average white pulp surface area in each animal (n=19–101 per animal, data quantified using ImageJ). Horizontal bars denote the average. Significant differences were determined using one-way ANOVA and Bonferroni’s post-hoc analysis (a) and the Mann-Whitney and Kruskal-Wallis tests (c): **, P <0.01; *, P<0.05; ns, not significant.
Extended Data Figure 9 |. Genetic and functional analyses of FPR1 and CCR5 genes from five human donors.
a, Serum derived from the blood of naïve or Y. pestis infected non-human primates (NHP, Cynomolgous macaque) or human blood donors described in Fig. 5a (donors 1–5) was analyzed for IgG specific for F1 antigen (αF1) via ELISA. b, List of amino acids changes deduced following cloning and sequencing of FPR1 and CCR5 alleles from human blood neutrophils (donors 1–5). c, Quantification of Y. pestis KIM D27 (pMM83) translocation of YopM-Bla into U937 or FPR1−/− macrophages transfected with plasmids pFPR1 and pFPR1 R190W. d, Immunoblot analysis for the production of FPR1, FPR2, FPR3 and actin in U937 macrophages, and derived FPR1−/− cells un-transfected or transfected with pFPR1 and pFPR1 R190W, respectively. One of three repeats is shown (a, c). Error bars represent the s.e.m. (n = 3 biological replicates) (a, c). One-way ANOVA and Bonferroni’s post-hoc analyses (c) was used to identify significant differences: ***, P<0.001.
Extended Data Figure 10 |. Model summarizing Y. pestis interactions with the plague receptor on human immune cells.
a, Y. pestis releases N-formylpeptides via its type III secretion system (T3SS) in order to attract human neutrophils by activating N-formylpeptide receptor (FPR1) signaling and chemotaxis. b, The Y. pestis T3SS docks on the plague receptor (FPR1) via the LcrV needle cap protein. c, Docking promotes assembly of the membrane translocon (including LcrV, YopD and YopB), which provides a conduit for low-calcium signaling to the bacterial T3SS. d, Low-calcium signaling activates T3SS transport of Yop effectors into the cytoplasm, thereby killing host immune cells. e, LcrV shares homology with the annexin A1 peptide. Alignment performed using Clustal Omega.
Supplementary Material
Acknowledgements
We thank Jos A. van Strijp for providing CHIPS, Gregory V. Plano and Deborah Anderson for sharing and providing strains Y. pestis KIM8 and KIM8 Δ1234, Christina Tam and Derek Elli for experimental assistance, Jorge Andrade and Yan Li for assistance with bioinformatics analysis of sequence data, Carole Ober, Tatyana Golovkina, and members of our laboratory for discussion. O.S. dedicated this paper to the memory of Peter Model. This project has been supported by funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under awards AI107792, AI057153, and AI042797.
Footnotes
Online Content. Methods, along with any additional Extended Data display items and Supplementary Materials source data are available in the online version of the paper.
The authors declare no competing financial interests.
References:
- 1.Andrades Valtueña A et al. The stone age plague and its persistence in Eurasia. Curr Biol. 27, 3683–3691 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Stenseth NC et al. Plague: past, present, and future. PLoS Med. 5, e3(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedictow OJ The Black Death 1346–1353: The Complete History. (Boydell Press, 2004). [Google Scholar]
- 4.Samson M et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Liu R et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86, 367–377 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Stephens JC et al. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am. J. Hum. Genet 62, 1507–1515 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mecsas J et al. Evolutionary genetics: CCR5 mutation and plague protection. Nature 427, 606(2004). [DOI] [PubMed] [Google Scholar]
- 8.Elvin SJ et al. Evolutionary genetics: ambiguous role of CCR5 in Y. pestis infection. Nature 430, 417(2004). [DOI] [PubMed] [Google Scholar]
- 9.Galan JE, Lara-Tejero M, Marlovits TC & Wagner S Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol 68, 415–438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Achtman M et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96, 14043–14048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis GR The Yersinia deadly kiss. J. Bacteriol 180, 5495–5504 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marketon MM, DePaolo RW, DeBord KL, Jabri B & Schneewind O Plague bacteria target immune cells during infection. Science 309, 1739–1741 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inglesby TV et al. Plague as a biological weapon: medical and public health management. JAMA 283, 2281–2290 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Quenee LE & Schneewind O Plague vaccines and the molecular basis of immunity against Yersinia pestis. Hum. Vaccin 5, 817–823 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Mitchell A et al. Glutathionylation of Yersinia pestis LcrV and Its efects on plague pathogenesis. MBio 8, e00646–00617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier RJ Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39, 1793–1803 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Boulay F, Tardif M, Brouchon L & Vignais P The human N-formylpeptide receptor. Characterization of two cDNA isolates and evidence for a new subfamily of G-protein-coupled receptors. Biochemistry 29, 11123–11133 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Miettinen HM et al. The ligand binding site of the formyl peptide receptor maps in the transmembrane region. J. Immunol 159, 4045–4054 (1997). [PubMed] [Google Scholar]
- 19.Le Y, Murphy PM & Wang JM Formyl-peptide receptors revisited. Trends Immunol. 23, 541–548 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Schaffrath RSM Decoding the biosynthesis and function of diphthamide, an enigmatic modification of translation elongation factor 2 (EF2). Microb. Cell 1, 203–205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheahan KL & Isberg RR Identification of mammalian proteins that collaborate with type III secretion system function: involvement of a chemokine receptor in supporting translocon activity. MBio 6, e02023–02014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardoel BW et al. Identification of an immunomodulating metalloprotease of Pseudomonas aeruginosa (IMPa). Cell. Microbiol 14, 902–913 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Mueller CA et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310, 674–676 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Vacchelli E et al. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science 350, 972–978 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Perretti M & D’Acquisto F Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat. Rev. Immunol 9, 62–70 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Postma B et al. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol 172, 6994–7001 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Wenzel-Seifert K, Grünbaum L & Seifert R Differential inhibition of human neutrophil activation by cyclosporins A, D, and H. Cyclosporin H is a potent and effective inhibitor of formyl peptide-induced superoxide formation. J. Immunol 147, 1940–1946 (1991). [PubMed] [Google Scholar]
- 28.Ligtenberg KG, Miller NC, Mitchell A, Plano GV & Schneewind O LcrV mutants that abolish Yersinia type III injectisome function. J. Bacteriol 195, 777–787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welkos S, Friedlander A, McDowell D, Weeks J & Tobery S V antigen of Yersinia pestis inhibits neutrophil chemotaxis. Microb Pathog 24, 185–196, doi: 10.1006/mpat.1997.0188 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Bartra SS, Jackson MW, Ross JA & Plano GV Calcium-regulated type III secretion of Yop proteins by an Escherichia coli hha mutant carrying a Yersinia pestis pCD1 virulence plasmid. Infect Immun 74, 1381–1386, doi: 10.1128/IAI.74.2.1381-1386.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gage KL & Kosoy MY Natural history of plague: perspectives from more than a century of research. Annu. Rev. Entomol 50, 505–528 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Hartt JK, Barish G, Murphy PM & Gao JL N-formylpeptides induce two distinct concentration optima for mouse neutrophil chemotaxis by differential interaction with two N-formylpeptide receptor (FPR) subtypes. Molecular characterization of FPR2, a second mouse neutrophil FPR. J. Exp. Med 190, 741–747 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stempel H et al. Strain-specific loss of formyl peptide receptor 3 in the murine vomeronasal and immune systems. J. Biol. Chem 291, 9762–9275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M et al. Formylpeptide receptors are critical for rapid neutrophil mobilization in host defense against Listeria monocytogenes. Sci. Rep 2, 786(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pechous RD, Sivaraman V, Price PA, Stasulli NM & Goldman WE Early host cell targets of Yersinia pestis during primary pneumonic plague. PLoS Pathog 9, e1003679, doi: 10.1371/journal.ppat.1003679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon JG et al. Yersinia pestis subverts the dermal neutrophil response in a mouse model of bubonic plague. MBio 4, e00170–00113, doi: 10.1128/mBio.00170-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quenee L, Cornelius CA, Ciletti NA, Elli D & Schneewind O Yersinia pestis caf1 (F1) variants and the limits of plague vaccine protection. Infect. Immun 76, 2025–2036 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakansson S, Bergman T, Vanooteghem J-C, Cornelis G & Wolf-Watz H YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect. Immun 61, 71–80 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torruellas J, Jackson MW, Pennock JW & Plano GV The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol. Microbiol 57, 1719–1733 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Cornelis GR Yersinia type III secretion: send in the effectors. J. Cell Biol 158, 401–408 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464, 104–107, doi: 10.1038/nature08780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorward DA et al. The role of formylated peptides and formyl peptide receptor 1 in governing neutrophil function during acute inflammation. Am J Pathol 185, 1172–1184, doi: 10.1016/j.ajpath.2015.01.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muto Y, Guindon S, Umemura T, Kőhidai L & Ueda H Adaptive evolution of formyl peptide receptors in mammals. J. Mol. Evol 80, 130–141 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Ye RD et al. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev 61, 119–161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quignon P, Rimbault M, Robin S & Galibert F Genetics of canine olfaction and receptor diversity. Mamm. Genome 23, 132–143 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Perry RD & Fetherston JD Yersinia pestis - etiologic agent of plague. Clin. Microbiol. Rev 10, 35–66 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baeten LA et al. Immunological and clinical response of coyotes (Canis latrans) to experimental inoculation with Yersinia pestis. J. Wildl. Dis 49, 932–939 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Liang XY et al. FPR1 interacts with CFH, HTRA1 and smoking in exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Eye 28, 1502–1510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otani T et al. Polymorphisms of the formylpeptide receptor gene (FPR1) and susceptibility to stomach cancer in 1531 consecutive autopsy cases. Biochem. Biophys. Res. Commun 405, 356–361 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Girard G Plague. Annu. Rev. Microbiol 9, 253–277 (1955). [DOI] [PubMed] [Google Scholar]
- 51.Meyer KF, Smith G, Foster L, Brookman M & Sung MH Live, attenuated Yersinia pestis vaccine: virulent in non-human primates, harmless to guinea pigs. J. Infect. Dis 129, S85–S112 (1974). [DOI] [PubMed] [Google Scholar]
- 52.Brubaker RR Mutation rate to non-pigmentation in Pasteurella pestis. J. Bacteriol 98, 1404–1406 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeBord KL et al. Immunogenicity and protective immunity against bubonic and pneumonic plague by immunization of mice with the recombinant V10 antigen, a variant of LcrV. Infect. Immun 74, 4910–4914 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higuchi K An improved chemically defined culture medium for strain L mouse cells based on growth responses to graded levels of nutrients including iron and zinc ions. J Cell Physiol 75, 65–72 (1970). [DOI] [PubMed] [Google Scholar]
- 55.Doll JM et al. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg 51, 109–114 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Hanahan D Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol 166, 557–572 (1983). [DOI] [PubMed] [Google Scholar]
- 57.Forsberg A, Bolin I, Norlander L & Wolf-Watz H Molecular cloning and expression of calcium-regulated, plasmid-coded proteins of Y. pseudotuberculosis. Microb. Pathogen 2, 123–137 (1987). [DOI] [PubMed] [Google Scholar]
- 58.Ton-That H & Schneewind O Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol 50, 1429–1438 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Anderson DM & Schneewind O A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278, 1140–1143 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Sundström C & Nilsson K Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17, 565–577 (1976). [DOI] [PubMed] [Google Scholar]
- 61.Gallagher R et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood 54, 713–733 (1979). [PubMed] [Google Scholar]
- 62.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210, doi: 10.1093/nar/30.1.207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quenee LE, Ciletti NA, Elli D, Hermanas T & Schneewind O Prevention of pneumonic plague in mice, rats, guinea pigs and non-human primates with clinical grade rV10, rV10–2 or F1-V vaccines. Vaccine 29, 6572–6583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated as part of this study are included in this article or its Extended Data and Supplementary Materials. Data for the CRISPR-Cas9 screen have been deposited in NCBI’s Gene Expression Omnibus and are accessible though GEO Series accession number GSE133826 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133826).