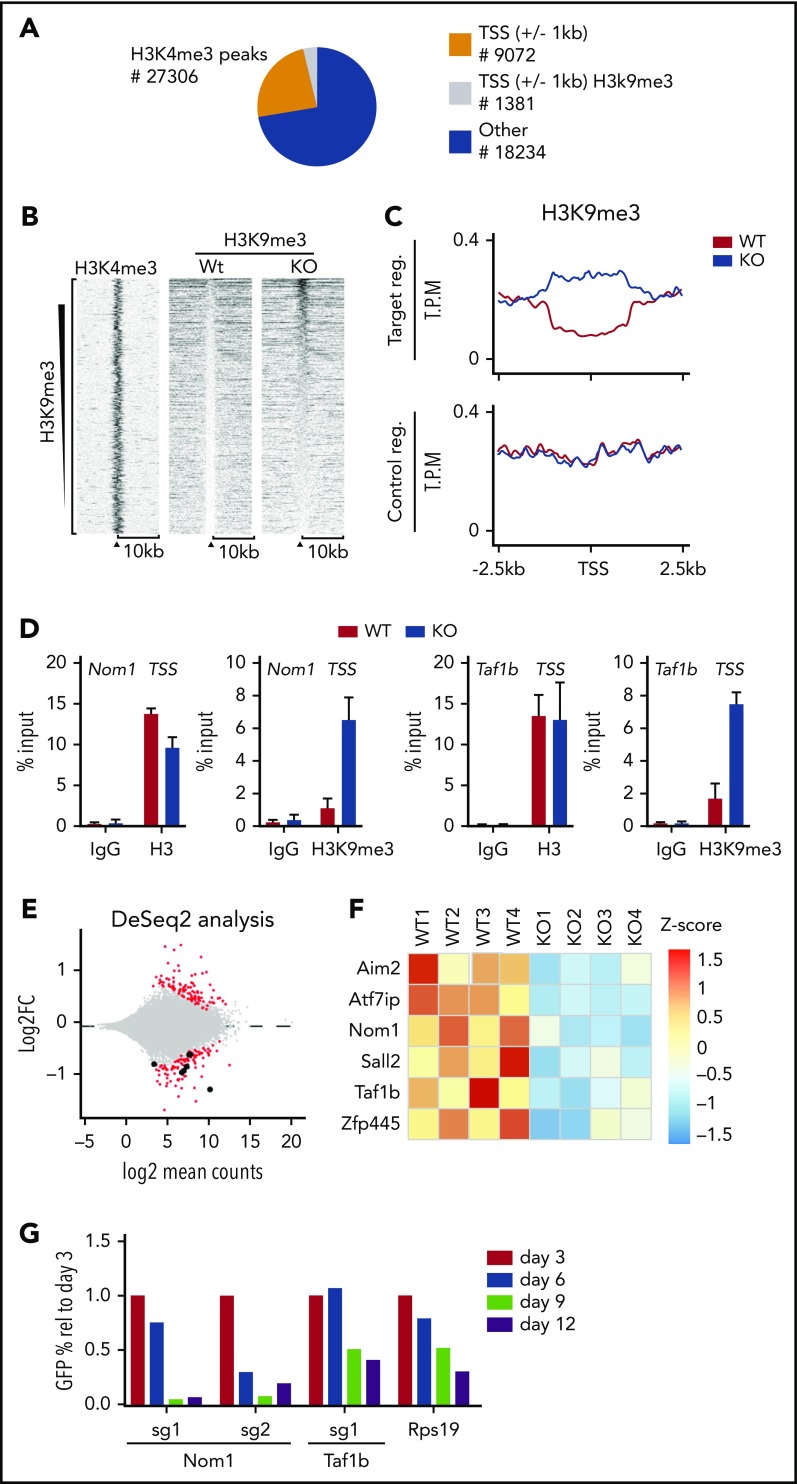
Figure 2.
Loss of KDM4A-C leads to accumulation of H3K9me3 at TSS on a subset of genes in LSK cells. (A) Pie chart indicating the position of H3K4me3 peaks in LSK (Lin−Sca−c-Kit+) cells from C57BL6 mice.12 Orange and gray represent peaks localized ±1 kb of TSS, peaks falling outside of these regions are represented with blue. H3K4me3+ regions ±1 kb of TSS that experience a significant change in H3K9me3 levels are indicated with gray. (B) Heat map of H3K9me3 ChIP-seq read counts ±10 kb of TSS. Data have been filtered to only include regions in which a significant change in H3K9me3 occurs. (C) Summary of the data in panel A zoomed in to ±2.5 kb of TSS. (D) Direct ChIP-qPCR validation on selected target genes using cells sorted in an independent experiment. (E) RNA-seq analysis of HSCs sorted from CreER:Kdm4abcfl/fl or CreER mice 2 weeks after tamoxifen injection (n = 4 in each group). (F) Heat map showing the distribution of normalized counts for the listed genes in the RNA-seq data set. (G) LSK cells were sorted from C57BL6 mice and transduced with lentiviruses expressing GFP, Cas9, and single guide RNA (sgRNA) against Taf1b and Nom1 as well as positive (Rps19) and negative controls (nontargeting sgRNA). The percentage of GFP+ cells was followed over time by FACS. The percentages have been normalized to the negative control and plotted relative to the value at day 3. FC, fold change; IgG, immunoglobulin G; reg., region; rel, relative; sg, sgRNA; T.P.M., tags per million; Wt/WT, wild type.