The plant cell wall is a complex structure composed of cellulose embedded in a matrix of polysaccharides (hemicellulose and pectin), phenolic compounds, and proteins. The primary cell wall is a thin layer formed in growing cells and is present ubiquitously, whereas the secondary cell wall is a rigid and thick structure formed after cell growth in specialized cell types, such as xylem vessels, fiber cells, and anther endothecium (Zhong and Ye, 2015). Secondary cell walls originated when vascular plants appeared on land around 430 million years ago (Zhong et al., 2010). They provide rigidity to the upright plant body, enable water and nutrient transport through vascular tissues, and protect cells from microbial infection. The main components of the secondary cell wall are cellulose, hemicellulose (xylan and glucomannan), and lignin (Zhong and Ye, 2015). Secondary wall biosynthesis is regulated by an orchestra of transcription factors (TFs), including NAC (NAM, NO APICAL MERISTEM; ATAF, ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR; CUC, CUP-SHAPED COTYLEDON), MYB, KNOX (KNOTTED1-like homeobox), and BEL1-LIKE TFs (Rao and Dixon, 2018). In this issue of Plant Physiology, Wang et al. (2019) characterized the rice (Oryza sativa) class II KNOX TF KNOX ARABIDOPSIS THALIANA7 (KNAT7) that coordinates secondary wall thickening and cell expansion in stem and rice grains.
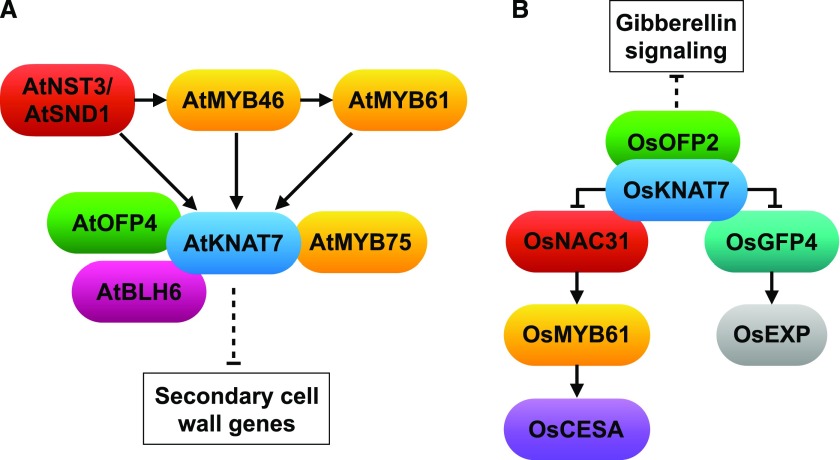
KNAT7 was first designated as IRREGULAR XYLEM11 based on its role in xylem development and secondary wall regulation in Arabidopsis (Arabidopsis thaliana; Brown et al., 2005). The knat7 knockout mutant has thicker secondary cell walls and increased lignin content in fiber cells (Li et al., 2012). KNAT7 is a direct target of the secondary cell wall master regulator NAC SECONDARY WALL THICKENING PROMOTING3 (NST3)/SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1), MYB46, and MYB61 (Rao and Dixon, 2018). KNAT7 physically interacts with a number of transcription factors including OVATE FAMILY PROTEIN4 (OFP4), BELL1-LIKE HOMEODOMAIN6 (BLH6), and MYB75, and the protein complexes suppress the expression of secondary wall biosynthetic genes (Li et al., 2012; Nakano et al., 2015; Fig. 1A).
Figure 1.
A model comparing the gene network of KNAT7 in Arabidopsis and rice. A, Regulatory network of Arabidopsis KNAT7. NAC SECONDARY WALL THICKENING PROMOTING3 (AtNST3)/SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (AtSND1), AtMYB46, and AtMYB61 directly bind to the promoter of AtKNAT7. AtKNAT7 physically interacts with OVATE FAMILY PROTEIN4 (AtOFP4), BELL1-LIKE HOMEODOMAIN6 (AtBLH6), and AtMYB75. AtKNAT7 and its interacting partners act as transcriptional suppressors of downstream secondary cell wall gene expression. B, Regulatory network of rice KNAT7. OsKNAT7 physically interacts with OsNAC31, GROWTH-REGULATING FACTOR4 (OsGRF4), and OsOFP2. OsKNAT7 suppresses the function of OsNAC31 in activating OsMYB61, which further activates the expression of cellulose synthase CESA. Similarly, OsKNAT7 suppresses the function of OsGFP4 in activating EXPANSIN (EXP) expression. OsOFP2 is a negative regulator of GA signaling. → denotes promoter binding and activation. –| denotes physical interaction and suppression. ---| denotes indirect suppression.
In this issue, Wang et al. (2019) showed that the KNAT7 ortholog in rice has a conserved function in regulating secondary cell wall development but may accomplish this through different or additional interacting partners and downstream factors. The Osknat7 mutant has a thicker wall in fiber cells than the wild type, similar to the Arabidopsis knat7 mutant. However, the increased secondary wall is due to a higher content of cellulose and xylan, rather than lignin as in Arabidopsis (Li et al., 2012). The Osknat7 mutant also exhibits a larger grain size and larger cells in spikelet bracts (Wang et al., 2019), which has not been reported in Arabidopsis. The phenotype in both cell wall thickness and cell size suggests that OsKNAT7 may be a coordinator of secondary cell wall biosynthesis and cell expansion. Indeed, Wang et al. (2019) showed that OsKNAT7 physically interacts with both the secondary wall regulator NAC31 and the cell expansion regulator GROWTH-REGULATING FACTOR4 (GRF4). Similar to the transcriptional repressor activity of AtKNAT7, OsKNAT7 suppresses the function of NAC31 and GRF4, which otherwise activate MYB61 and expansin gene expression, respectively (Wang et al., 2019; Fig. 1B). In addition, OsKNAT7 interacts with OsOFP2 in a yeast two-hybrid assay; the latter regulates both grain shape and vascular bundle development via gibberellin signaling (Schmitz et al., 2015). The OsKNAT7-OsOFP2 complex could be another mechanism coordinating secondary wall thickening and cell growth (Fig. 1B).
In general, the transcriptional regulation of secondary cell wall biosynthesis is conserved in evolution. Numerous studies have shown that the ectopic expression of NAC or MYB master regulators from poplar (Populus trichocarpa), grasses, or gymnosperms is able to rescue the phenotype in mutants of Arabidopsis orthologs (Zhong et al., 2010; Rao and Dixon, 2018). However, the history of duplication in these TF families enables subfunctionalization and neofunctionalization, which may result in diverse regulatory mechanisms and cell wall compositions in different plant species and families. Indeed, the cell wall of the grass species is different from that of many dicots, in that the grass cell wall has an increased amount of xylan, and that lignin and other cell wall components are often cross linked with ferulic acid and p-coumaric acid (Hatfield et al., 2017). The finding in OsKNAT7 provides evidence that the gene network of KNAT7 may differ between Arabidopsis and rice. For instance, in Arabidopsis, MYB61 directly binds to the promoter of KNAT7 and thus acts upstream of KNAT7 (Fig. 1A), while in rice, KNAT7 suppresses NAC31 activation of MYB61 expression (Fig. 1B), suggesting that the order of signal transduction has changed during evolution. Alternatively, there could be a feedback loop between KNAT7 and MYB61 in both species. Further mechanistic studies will uncover the evolution of the KNAT7 network in balancing secondary cell wall formation with cell division and expansion.
References
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield RD, Rancour DM, Marita JM (2017) Grass cell walls: A story of cross-linking. Front Plant Sci 7: 2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ (2012) The class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 194: 102–115 [DOI] [PubMed] [Google Scholar]
- Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Dixon RA (2018) Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front Plant Sci 9: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz AJ, Begcy K, Sarath G, Walia H (2015) Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci 241: 177–188 [DOI] [PubMed] [Google Scholar]
- Wang S, Yang H, Mei J, Liu X, Wen Z, Zhang L, Xu Z, Zhang B, Zhou Y (2019) A Rice homeobox protein KNAT7 integrates the pathways regulating cell expansion and wall stiffness. Plant Phys 181: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H (2010) Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 15: 625–632 [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H (2015) Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56: 195–214 [DOI] [PubMed] [Google Scholar]