AtNDB2 catalyzes the majority of exogenous NADH oxidation in mitochondria and is required for tolerance of and recovery from environmental stress in Arabidopsis.
Abstract
In addition to the classical electron transport pathway coupled to ATP synthesis, plant mitochondria have an alternative pathway that involves type II NAD(P)H dehydrogenases (NDs) and alternative oxidase (AOX). This alternative pathway participates in thermogenesis in select organs of some species and is thought to help prevent cellular damage during exposure to environmental stress. Here, we investigated the function and role of one alternative path component, AtNDB2, using a transgenic approach in Arabidopsis (Arabidopsis thaliana). Disruption of AtNDB2 expression via T-DNA insertion led to a 90% decrease of external NADH oxidation in isolated mitochondria. Overexpression of AtNDB2 led to increased AtNDB2 protein abundance in mitochondria but did not enhance external NADH oxidation significantly unless AtAOX1A was concomitantly overexpressed and activated, demonstrating a functional link between these enzymes. Plants lacking either AtAOX1A or AtNDB2 were more sensitive to combined drought and elevated light treatments, whereas plants overexpressing these components showed increased tolerance and capacity for poststress recovery. We conclude that AtNDB2 is the predominant external NADH dehydrogenase in mitochondria and together with AtAOX1A forms a complete, functional, nonphosphorylating pathway of electron transport, whose operation enhances tolerance to environmental stress. This study demonstrates that at least one of the alternative NDs, as well as AOX, are important for the stress response.
Mitochondria from higher plants possess a branched electron transport chain (ETC). In addition to the classical ETC, composed of four large protein complexes that oxidize intramitochondrial NADH and succinate, terminating in cytochrome oxidase (COX) and coupled to ATP synthesis, there are a number of type II NAD(P)H dehydrogenases (NDs) located on the inside (NDA and NDC) and outside (NDB) of the inner mitochondrial membrane, as well as an alternative oxidase (AOX). The alternative NDs and AOX constitute what is known as the alternative pathway (AP), which is not coupled to ATP synthesis and therefore is not controlled directly by the cell’s energy status (for review, see Millar et al., 2011).
The AP is present in all higher plants examined to date and is expressed in at least some tissues, but its role in these tissues remains somewhat enigmatic. In thermogenic floral appendages of some plants, the AP, particularly AOX, is expressed in large quantities and contributes to uncontrolled respiration and heat production (Wagner et al., 2008). In many plants, components of the AP are strongly expressed upon exposure to chemical or environmental stresses (Rasmusson et al., 2004; Clifton et al., 2006; Rasmusson and Møller, 2011; Vanlerberghe, 2013). In Arabidopsis (Arabidopsis thaliana), the alternative NDs are encoded in a small gene family of four NDB genes (AtNDB1–AtNDB4), three NDA genes (AtNDA1–AtNDA3), and an NDC gene (AtNDC1), whereas there are five AOX genes (AtAOX1a–AtAOX1d and AtAOX2). Most of these have been heterologously expressed in Escherichia coli and their individual activities explored in some detail, revealing different regulatory features (Djajanegara et al., 1999; Umbach et al., 2002; Selinski et al., 2016, 2017). In the case of the NDs, three genes were functionally expressed and displayed different substrate preferences (Geisler et al., 2007). By contrast, we know much less about the in planta activities and regulation of these enzymes.
In Arabidopsis cell cultures and whole plants, AtNDB2, AtNDA2, and AtAOX1a are concomitantly up-regulated in response to a wide range of treatments, effectively leading to the production of a complete bypass of the classical ETC (Clifton et al., 2005; Ho et al., 2008; Vijayraghavan and Soole, 2010). These and earlier studies (Maxwell et al., 1999; Djajanegara et al., 2002; Gray et al., 2004; Polidoros et al., 2005) have led to the idea that AOX, with or without ND activity, helps to minimize the production of reactive oxygen species (ROS) in mitochondria by keeping the ubiquinone pool in an oxidized state (for review, see Vanlerberghe, 2013). Consistent with such a role, AtAOX1a knockout plants have increased sensitivity to stress (Giraud et al., 2008) and plants with increased AtAOX1a expression produce less ROS (Smith et al., 2009).
Whereas there have been extensive studies on the role of AOX using a reverse genetics approach, there are limited studies where ND expression has been manipulated. This is further complicated by the dual targeting of some of the NDs (NDAs, NDC, and NDB1) to other organelles in the cell as well as the mitochondrion (Carrie et al., 2008). Recent studies have assessed the effect of reducing the expression of AtNDB1 (Wallström et al., 2014a), AtNDA1 and AtNDA2 (Wallström et al., 2014b), and AtNDC1 (Fatihi et al., 2015) in Arabidopsis. Low-level expression of NDAs resulted in delayed growth and a shift to fermentation but apparently had no effect on photosynthesis, whereas lower levels of AtNDB1 resulted in slower growth and altered NADPH/NADP ratios not linked to photosynthetic function. A knockout of AtNDC1 linked the chloroplast-targeted version of the protein to a role in vitamin K1 biosynthesis, and these plants were very sensitive to high light (Fatihi et al., 2015). Knockdown of AtNDB4 by RNA interference resulted in better or unaltered growth under standard and stress growth conditions, but these lines consistently had higher AtNDB2 and AtAOX1a protein levels (Smith et al., 2011). To our knowledge, there have been no studies published assessing the impact of altered levels of the mitochondria-specific AtNDB2.
In this study, we used transgenic Arabidopsis to further explore the role of AtNDB2. A key role for AtNDB2 in external NADH oxidation was identified through analysis of a T-DNA insertion line that showed increased sensitivity to drought and high-light stress. When AtNDB2 was overexpressed, mitochondria showed enhanced levels of AtNDB2 protein, but only small increases in external NADH oxidation were seen. It was only when AtNDB2 was overexpressed together with AtAOX1a that external NADH oxidation rates increased significantly, a change that was linked mainly to AOX activity. These dual overexpression plants had substantially enhanced tolerance to drought and high-light stress.
RESULTS
Generation and Characterization of Transgenic Plants
To explore the effects of altered expression of AtNDB2 on plant growth, we used a T-DNA insertion line and transgenic lines overexpressing AtNDB2 and AtAOX1a.
Atndb2 T-DNA Insertion Lines
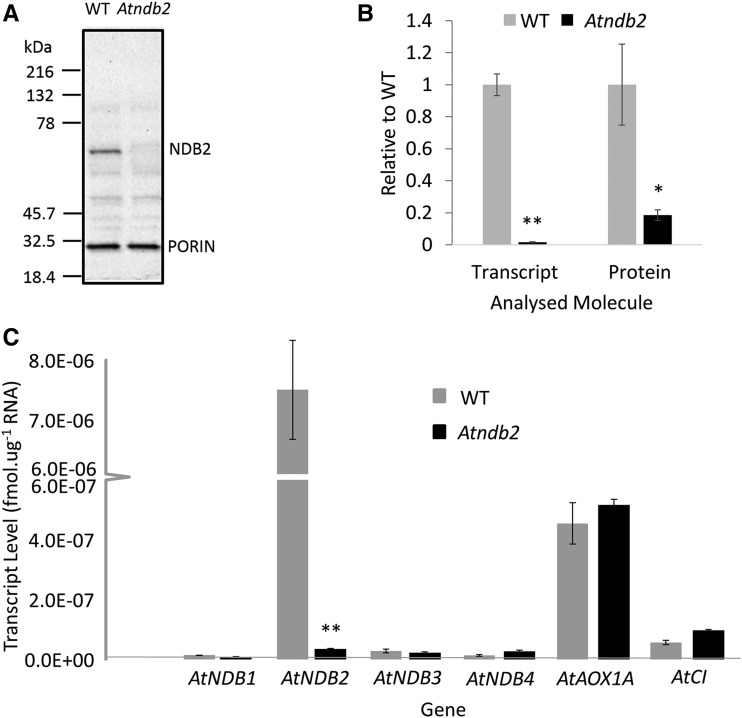
The AtNDB2 (At4g05020) gene contains 10 exons and nine introns (Supplemental Fig. S1). To investigate the effect of a reduction in AtNDB2 expression, a T-DNA insertion line (SALK_036330) for AtNDB2 was obtained with an insertion in intron 7 of the gene. Sequence analysis of the Atndb2 line showed that the insertion had caused a 10-bp deletion around the insertion site, 220 bp upstream of the stop codon of the AtNDB2 gene (Supplemental Fig. S1). Virtually no AtNDB2 transcript could be detected in plants homozygous for the insertion (Fig. 1B), and AtNDB2 protein was below detection limits in isolated mitochondria from the plants (Fig. 1, A and B). Transcript levels of other NDB genes were unaffected in plants grown under normal conditions (Fig. 1C).
Figure 1.
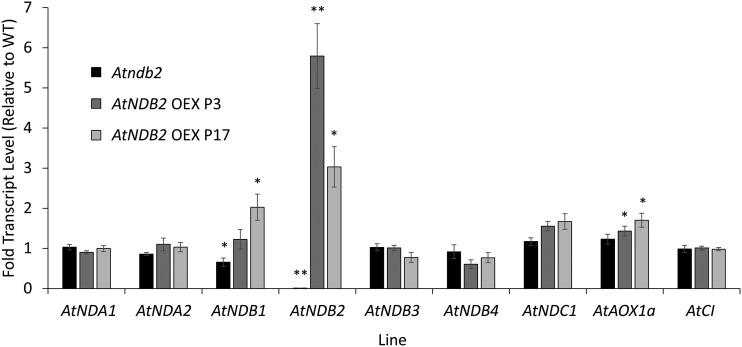
Molecular characterization of the Atndb2 T-DNA insertion line. Plants were grown for 3 weeks on agar plates, then a portion of shoot tissue was frozen for RNA extraction and the remaining shoot tissue was used for mitochondrial isolation. A, Example of a western blot for AtNDB2 (63 kD) and porin (31 kD) using 8 μg of purified mitochondrial protein. B, AtNDB2 transcript and protein levels in the Atndb2 T-DNA lines compared with the wild-type (WT) background. Transcript or protein levels were first normalized to a reference gene or protein (see “Materials and Methods”), then the mean value for the wild type was set to 1. C, Absolute transcript levels of all external-facing NDs (AtNDB1–AtNDB4) as well as AtAOX1A and a subunit of complex I (AtCI) in the wild type and the Atndb2 T-DNA line. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 4 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test).
AtNDB2 Overexpression Lines
AtNDB2 was overexpressed in both wild-type Columbia-0 plants and in a previously generated AtAOX1a overexpression line, denoted XX1 (Umbach et al., 2005). Overexpression of AtNDB2 in AtAOX1a overexpression plants was performed, as both genes are often coexpressed in response to stress.
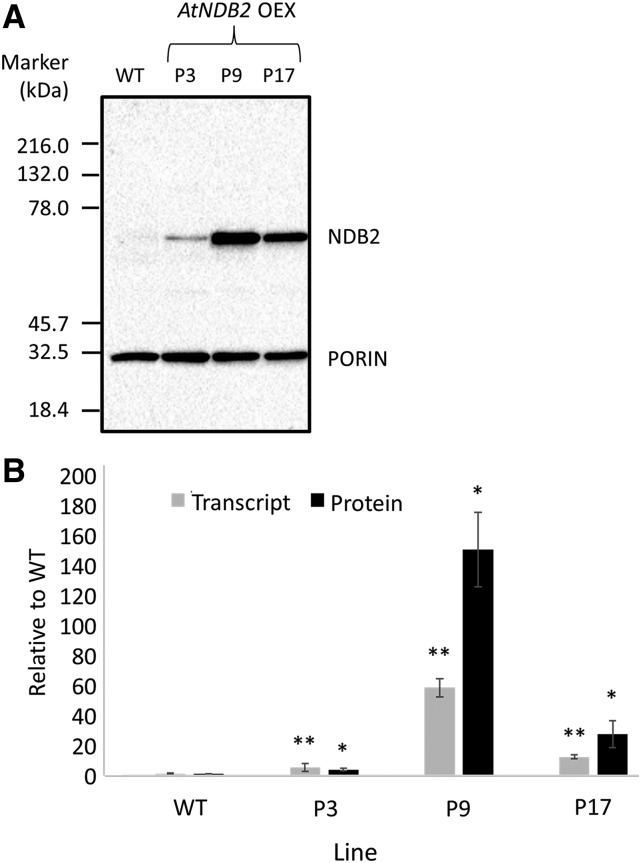
Three single-copy homozygous transgenic overexpression lines were produced in the wild-type background. AtNDB2 transcript abundance varied considerably in the different lines, with a range in leaves of 3- to 60-fold wild-type levels, and protein levels in isolated mitochondria varied accordingly (3- to 150-fold; Fig. 2).
Figure 2.
Molecular characterization of AtNDB2 overexpression lines. Plants were grown for 3 weeks on agar plates, then a portion of shoot tissue was frozen for RNA extraction and the remaining shoot tissue was used for mitochondrial isolation. A, Example of a western blot for AtNDB2 (63 kD) and porin (31 kD) using 8 μg of purified mitochondrial protein. B, AtNDB2 transcript and protein levels in the AtNDB2 overexpression lines (P3, P9, and P17) compared with the wild-type background (WT). Transcript or protein levels were first normalized to a reference gene or protein (see “Materials and Methods”), then the mean value for the wild type was set to 1. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 3 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test).
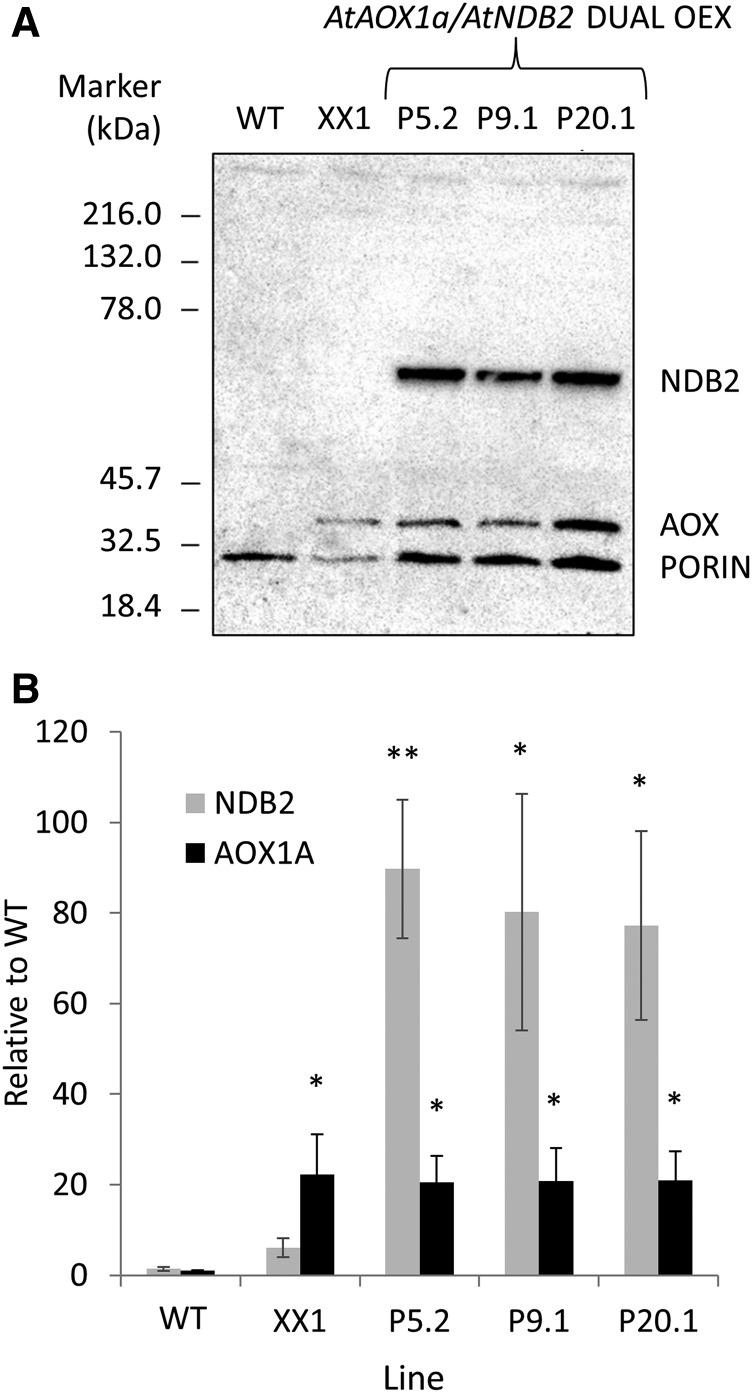
Mitochondria from three AtNDB2 and AtAOX1a dual overexpression lines (P5.2, P9.1, and P20.1) had approximately 80-fold increases in AtNDB2 protein (Fig. 3). Levels of AtAOX1a protein remained unchanged from that in the XX1 background, approximately 20-fold higher than that in wild-type plants (Fig. 3).
Figure 3.
Molecular characterization of dual AtAOX1A and AtNDB2 overexpression lines. Plants were grown for 3 weeks on agar and then used for mitochondrial isolation. A, Example of a western blot for AtNDB2 (63 kD), AtAOX (36 kD), and porin (31 kD) using 8 μg of purified mitochondrial protein. B, AtNDB2 and AtAOX protein levels in the dual overexpression lines (P5.2, P9.1, and P20.1) compared with the wild type (WT) and the single AOX1A overexpression background (XX1). Protein levels were first normalized to porin, and the mean value for the wild type was set to 1. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 3 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test).
Respiratory Activities of Mitochondria from Plants with Altered AtNDB2 and AtAOX1a Protein Levels
Plants with Altered NDB2 Content
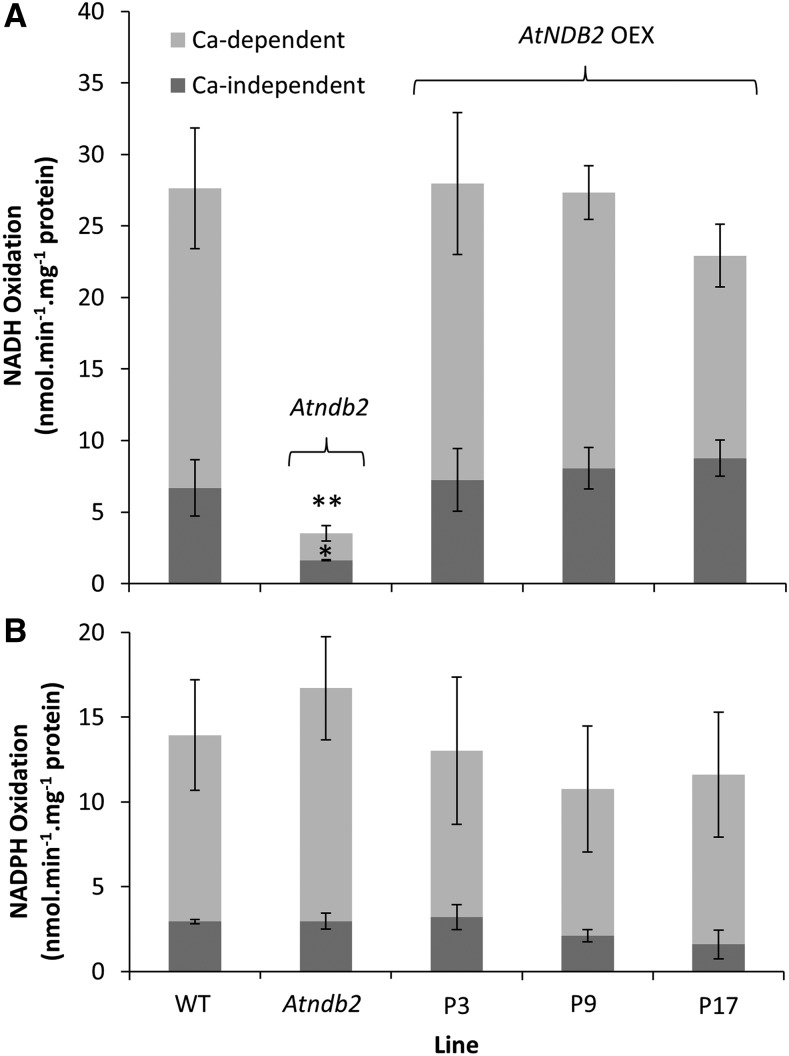
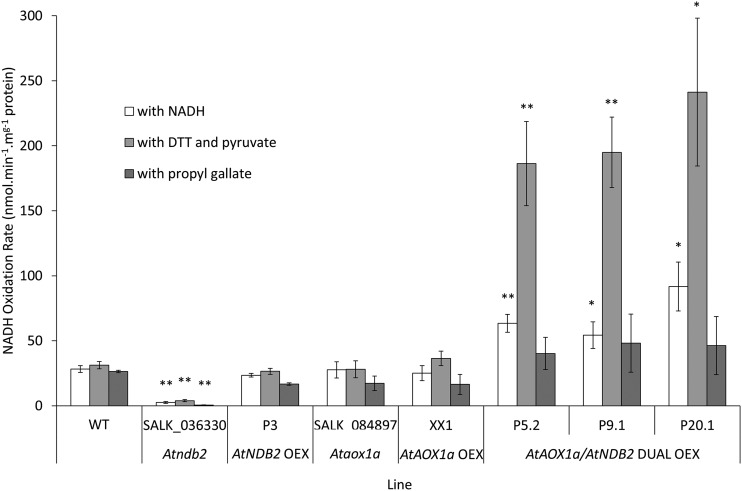
Mitochondria were purified from wild-type plants, the Atndb2 T-DNA line (SALK_036330), and the three AtNDB2 overexpression lines (P3, P9, and P17) grown under normal conditions, and NAD(P)H oxidation rates were measured. External NADH oxidation rates in mitochondria from wild-type plants were modest (20–30 nmol NADH min−1 mg−1 protein) and stimulated by Ca2+ (Fig. 4). In mitochondria from the T-DNA line, which had undetectable AtNDB2 protein, external NADH oxidation rates were less than 3 nmol min−1 mg−1 protein, whether Ca2+ was present or not. NADPH oxidation rates were unaffected by the mutation (Fig. 4B). This suggests that AtNDB2 is responsible for the majority of external NADH oxidation in Arabidopsis mitochondria. However, mitochondria from the three lines with increased levels of AtNDB2 protein displayed rates of external NADH oxidation similar to those from the wild type (Fig. 4A; Table 1). Again, rates of NADPH oxidation were unchanged. The lack of increase in NADH oxidation rates in lines with higher AtNDB2 protein content was puzzling. We tested whether downstream electron transport capacity was limiting by measuring oxygen consumption with NADH as a substrate and then adding a second substrate, succinate; this stimulated the rate of oxygen consumption substantially (Table 1), showing that the electron transport capacity of the cytochrome path (AOX activity was almost zero) was not limiting NADH oxidation rates per se in the mitochondria overexpressing AtNDB2. This suggests that the excess AtNDB2 protein was unable to engage the cytochrome chain in these mitochondria.
Figure 4.
Effects of AtNDB2 disruption and overexpression on NADH and NADPH oxidation rates by purified mitochondria. Plants of the wild type (WT), Atndb2 T-DNA (SALK_036330), and AtNDB2 overexpression lines (P3, P9, and P17) were grown for 3 weeks on agar and then used for mitochondrial isolation. Assays were carried out with purified mitochondria in a cuvette, with oxygen as the electron acceptor and either NADH (A) or NADPH (B) as the electron donor. Ca-independent activity was measured in the presence of 0.25 mm EGTA. Ca-dependent activity was calculated as the rate in the presence of 2 mm CaCl2 minus the Ca-independent rate. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 3–4 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test against the wild type).
Table 1. External NADH oxidation rates in AtNDB2 overexpression lines are not limited by flux through downstream mitochondrial eletron transport chain (mETC) components.
Plants were grown for 21 d on agar and then used for mitochondrial isolation. Purified mitochondria were assayed using an oxygen electrode (Oxygen Consumption). All assays were conducted in 1 mL of reaction medium (see “Materials and Methods”) supplemented with ADP (1 mm) and CaCl2 (2 mm). Where indicated, succinate (10 mm) and/or NADH (0.2 mm) were used as substrates. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 3 ± se).
| Genotype | Oxygen Consumption | ||
|---|---|---|---|
| Succinate + NADH | NADH Only | Succinate Only | |
| nmol min−1 mg−1 protein | |||
| Wild type | 49.5 ± 9.6 | 34.7 ± 8.4 | 35.6 ± 1.5 |
| NDB2 overexpression line P3 | 51.1 ± 6.2 | 39.4 ± 8.2 | 34.3 ± 4.9 |
| NDB2 overexpression line P17 | 63.3 ± 9.1 | 48.3 ± 6.8 | 33.3 ± 5.4 |
Whereas AtNDB2 transcript abundance was altered substantially in these plants, only minor changes were observed for the other components of the AP (Fig. 5). It is particularly interesting that no dramatic increases in transcript abundance of other AtNDB genes were observed in the T-DNA insertion line, suggesting that these components cannot compensate for the loss of AtNDB2 and external NADH oxidation. Small but significant increases in AtNDB1 and AtNDC1 transcript abundance were observed in the plants overexpressing AtNDB2, but transcripts of AtAOX1a and a subunit of complex I (AtCI) were unaltered in all lines (Fig. 5).
Figure 5.
Effects of AtNDB2 disruption and overexpression on transcript levels of type II dehydrogenase genes and AtAOX1a and AtCI. Plants were grown hydroponically in a growth cabinet for 6 weeks under control conditions (22°C, 80–120 μmol m−2 s−1, 16/8-h day/night periods). Each replicate consisted of a single, whole rosette. Transcripts of all NDs (AtNDA1 and AtNDA2, AtNDB1–AtNDB4, and AtNDC1) were normalized to two reference genes, Ubiquitin and Pdf2; therefore, values represent relative units (n = 6 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test). WT, Wild type.
Lines Overexpressing Both AtNDB2 and AtAOX1A
In our hands, plants of AtAOX1a overexpression line XX1 (Umbach et al., 2005), grown on plates or hydroponically, had between 25- and 50-fold higher AtAOX1a transcript levels than the wild type (mean fold change over a number of experiments was 45 ± 10), with mitochondria accumulating approximately 20-fold higher protein levels (Fig. 3) and at least a 10-fold higher AOX capacity (31 compared with 0.1–3 nmol oxygen min−1 mg−1 protein from the wild type) when measured with a combination of NADH and succinate as substrates and activated by pyruvate. These mitochondria, therefore, have substantially increased capacity for electron transport to oxygen. With the incorporation of the AtNDB2 overexpression construct into the XX1 background line, AtNDB2 protein in mitochondria increased 10- to 20-fold in the three lines tested (5.2, 9.1, and 20.1). The amount of AOX protein, however, did not change (Fig. 3).
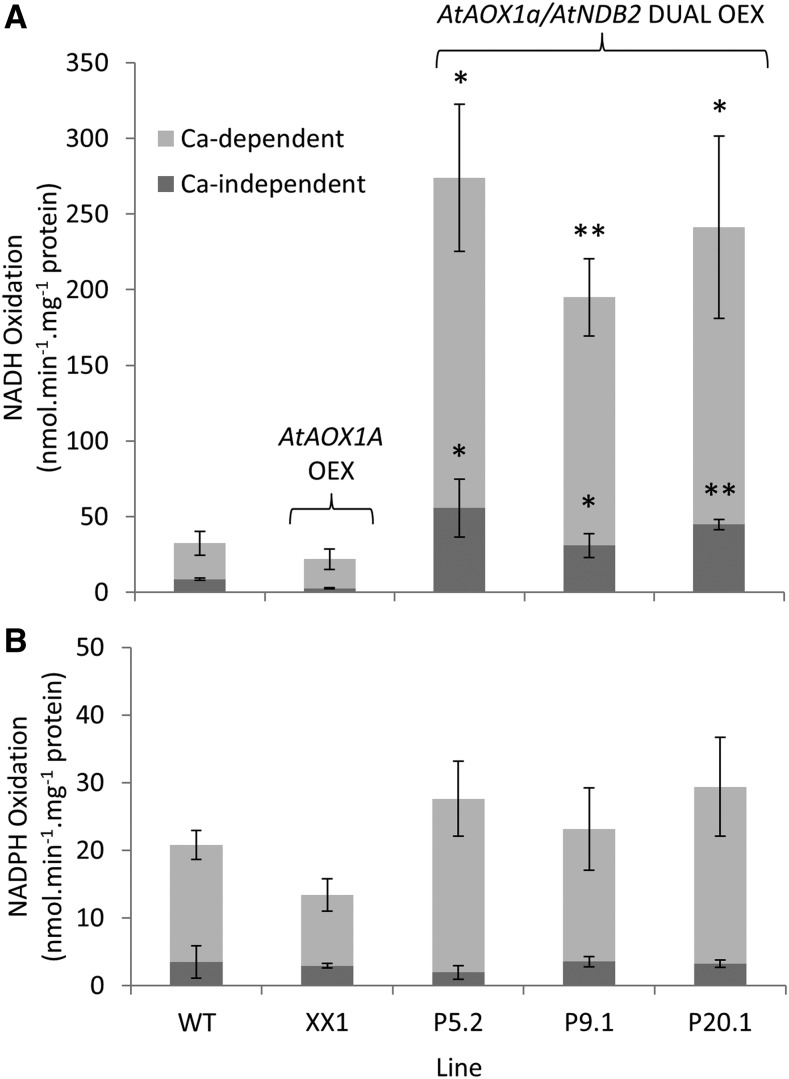
Each of the three lines overexpressing both AtNDB2 and AtAOX1a (dual overexpression lines) showed approximately double or triple the rate of NADH oxidation to oxygen, compared with mitochondria from wild-type and XX1 background plants, when assayed without AOX fully activated (Fig. 6). The addition of pyruvate to activate AOX increased NADH oxidation dramatically, with rates approaching 300 nmol NADH min−1 mg−1 (Fig. 6; Supplemental Fig. S2). Inhibition of AOX with propyl gallate prevented the activation and reduced NADH oxidation rates back to basal levels (Fig. 6; Supplemental Fig. S2). Both Ca2+-independent and Ca2+-dependent rates of NADH oxidation were increased in the dual overexpression lines, with calcium stimulating the rate approximately 5-fold (Fig. 7). External NADPH oxidation rates were not significantly different (Fig. 7).
Figure 6.
Effects of AOX activation on NADH oxidation rates by purified mitochondria with various AtNDB2 and AtAOX1A expression levels. Plants of the wild type (WT), Atndb2 T-DNA (SALK_036330), AtNDB2 overexpression line (P3), Ataox1a T-DNA (SALK_084897), and dual overexpression lines (P5.2, P9.1, and P20.1) were grown for 3 weeks on agar and then used for mitochondrial isolation. Assays were carried out in a cuvette with oxygen as the electron acceptor and NADH (0.2 mm) as the electron donor in the presence of ADP (1 mm) and CaCl2 (2 mm), with subsequent additions of dithiothreitol (DTT; 1 mm) and pyruvate (5 mm) to activate AOX and propyl gallate (0.25 mm) to inhibit AOX. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 3 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test).
Figure 7.
Biochemical characterization of dual overexpression lines. Plants of the wild type (WT), AtAOX1a overexpression line (XX1), and dual overexpression lines (P5.2, P9.1, and P20.1) were grown for 3 weeks on agar and then used for mitochondrial isolation. Spectrophotometric assays were carried out in a spectrophotometer with oxygen as the electron acceptor and either 0.2 mm NADH (A) or NADPH (B) as the electron donor. Pyruvate (5 mm) and DTT (1 mm) were present in all cases. Ca-independent activity was measured in the presence of EGTA (0.5 mm). Ca-dependent activity was calculated as the rate in the presence of CaCl2 (2 mm) minus the Ca-independent rate. Each replicate corresponds to a separate batch of plants used for mitochondrial isolations (n = 3–4 ± se). *, P < 0.05, **, P < 0.005 (unpaired, two-tailed Student’s t test against the wild type).
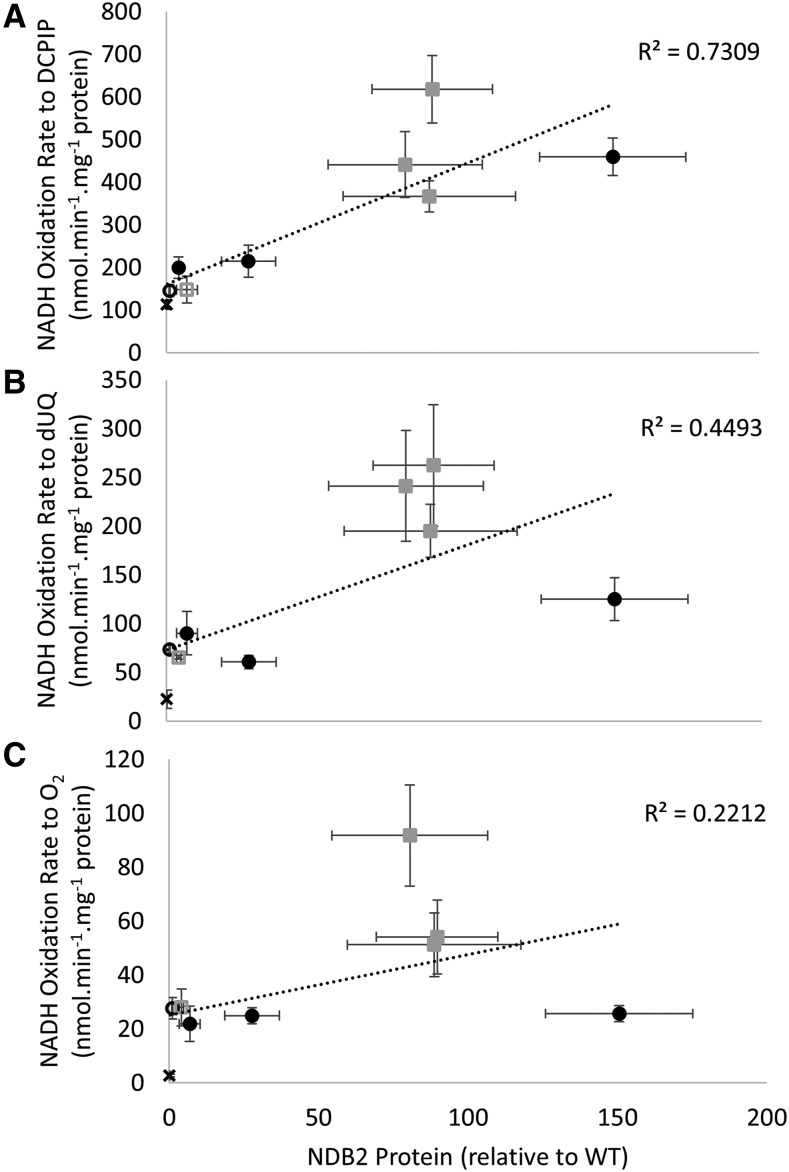
The results show that concomitantly increasing the capacity of AOX allows the overexpressed AtNDB2 protein to be active. To investigate this further, we compared NADH oxidation rates with different electron acceptors as a function of AtNDB2 protein abundance in isolated mitochondria (Fig. 8). 2,6-Dichlorophenolindophenol (DCPIP) accepts electrons directly from the flavoprotein moiety of dehydrogenases and can therefore provide a measure of the activity of the protein itself, without relying on its interaction with electron acceptors via the ubiquinone-binding site. Since DCPIP also interacts with the NADH dehydrogenase on the outer mitochondrial membrane, mitochondria from the T-DNA insertion line were included as a negative control. NADH-DCPIP oxidoreductase rates were linearly correlated with the amount of AtNDB2 protein, as measured by immunoblotting, over all the lines tested (Fig. 8A). However, when DCPIP was replaced with decylubiquinone or oxygen as electron acceptor, NADH oxidation rates no longer followed AtNDB2 protein abundance and were much faster in the dual overexpression lines than in the other lines (Fig. 8, B and C). These results suggest that in the single AtNDB2 overexpression lines, AtNDB2 activity was limited by its interaction with quinone, whether it was an artificially added quinone or the native UQ10. Concomitant overexpression of AtAOX1a apparently allows rapid turnover of reduced quinone and overcomes the limitation.
Figure 8.
Relationship between AtNDB2 protein content and NADH oxidation rates with different electron acceptors. Plants were grown for 3 weeks on agar and then used for mitochondrial isolation. Purified mitochondria were assayed in a quartz cuvette in a spectrophotometer. All assays were conducted in a final volume of 1 mL, with standard reaction medium (SRM) supplemented with NADH (0.2 mm), ADP (1 mm), CaCl2 (2 mm), and either DCPIP (0.1 mm; A), decylubiquinone (dUQ; 40 µm; B), or no artificial electron acceptor (C). Different markers represent different lines, including the wild type (WT; open circles), AtNDB2 overexpression lines (closed circles), AtAOX1a overexpression line XX1 (open squares), and AtAOX1a/AtNDB2 dual overexpression lines (closed squares). NADH oxidation rates for the Atndb2 T-DNA line (shown by X on the axis) are given to indicate background NADH oxidation rates that may represent remaining external NADH dehydrogenase activities, matrix-facing NADH dehydrogenase activities, or outer membrane NADH dehydrogenase activities, particularly for the DCPIP rates, which were measured with thawed mitochondria (a malate dehydrogenase latency assay for inner membrane integrity showed that these mitochondria were 70% intact). Data represent means of separate mitochondrial preparations used in enzyme assays or western blots (n = 3 ± se).
Clearly, overexpressing both AtNDB2 and AtAOX1a provides for very rapid electron transport rates via a complete alternative path in these mitochondria. What happens in vivo will obviously depend on posttranslational control of AOX via pyruvate and redox poise and provision of extramitochondrial NADH, but the dual overexpression lines possess the potential for dramatically enhanced respiration rates via external NADH dehydrogenase and through AOX.
Growth and Stress Response of Plants with Altered AtNDB2 and AtAOX1a
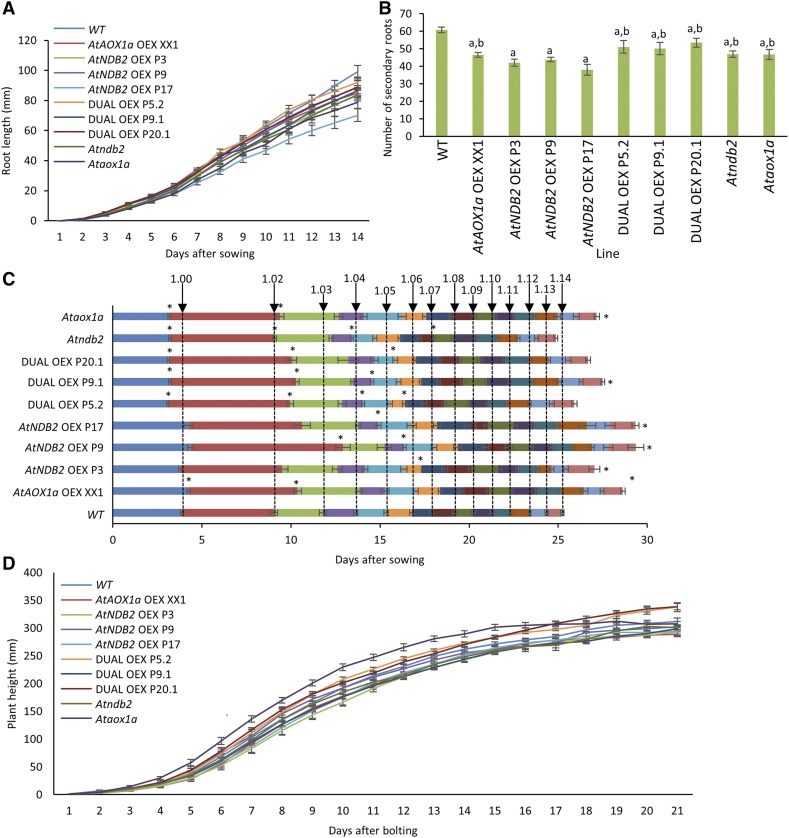
Atndb2 T-DNA and AtNDB2 overexpression plants showed no obvious phenotypic deviation from the wild type when grown under controlled, nonstressed conditions (Supplemental Fig. S3), nor did the XX1 AtAOX1a overexpression line or the three dual overexpression lines (Supplemental Fig. S4). However, a more detailed analysis using an adaptation of the Boyes et al. (2001) method revealed subtle differences in development between the lines. Plants were maintained on either an agar- or soil-based medium, and subsequent destructive and nondestructive measurements were made over 14 or 48 d, respectively. Root measurements performed in agar demonstrated a significant decrease in root length in the AtAOX1a overexpression line XX1 and the AtNDB2 overexpression lines P3 and P17 as well as in both Ataox1a and Atndb2 T-DNA lines (Fig. 9A). Interestingly, this phenotype was not seen in any of the three dual overexpression lines. All transgenic lines showed decreases in the number of secondary roots compared with the wild type, but this was more pronounced in the AtNDB2 single overexpression lines (Fig. 9B). When plants were grown in soil, germination of the dual overexpression and T-DNA lines occurred significantly earlier than that of the wild type, whereas in the single AtAOX1a overexpression line (XX1), germination was delayed (Fig. 9C). The emergence of the first rosette leaves was significantly delayed in all lines except the AtNDB2 overexpression lines P3 and P17, which nonetheless showed a trend toward delayed emergence. Emergence of the fourth rosette leaf was delayed in two AtNDB2 overexpression lines and two AtAOX1a/AtNDB2 dual overexpression lines. Although there were small differences in rosette development for the Atndb2 T-DNA and two dual overexpression lines (P5.2 and P20.1), these were not apparent by the end of vegetative development There were no significant differences in floral development between lines, although differences were seen in inflorescence height among the Ataox1a T-DNA line, two of the dual overexpression lines (P9.1 and P20.1), and the single AtAOX1a overexpression line (Fig. 9D). The Ataox1a T-DNA bolted earlier and reached its maximum height before all other lines, whereas the single AtAOX1a overexpression line was delayed and had a smaller stem length. Two dual overexpression lines (P5.2 and P20.1) followed similar patterns of increased stem height between days 16 and 21.
Figure 9.
Dual overexpression of AtAOX1A and AtNDB2 reversed early growth delays observed with individual AtAOX1A or AtNDB2 overexpression. Measurements were performed for 14 d using plants grown on agar (A and B) or for 44 d using plants grown in soil (C and D). All plants were grown in a growth room at 22°C with 14-h daylength and photosynthetically active radiation (PAR) of 100 to 150 μmol m−2 s−1, using Sylvania Luxline Plus T5 fluoro lights. A, Roots were measured every day after complete emergence of the cotyledon (n = 16–19 ± se). Values found to be statistically significant (P < 0.05; unpaired, two-tailed Student’s t test against the wild type [WT]) were as follows: day 3, P3, P17, and 084; day 4, P3, P9, and P17; day 5, P17 and 084; day 6, P17; day 7, P17 and 084; day 8, P17; day 9, P17, 330, and 084; day 10, P17, 330, and 084; day 11, P17, 330, and 084; day 12, P17; day 13, P17 and 330; day 14 XX1, P3, P17, 330, and 084. B, Secondary roots were counted at 14 d using a light microscope (n = 16–19 ± se). Bars not sharing a common letter are statistically significantly different from each other (P < 0.001; unpaired, two-tailed Student’s t test). C, Growth stages were recorded based on the milestones outlined by Boyes et al. (2001); n = 7–16 ± se). D, Plant height was measured after bolting and measurements continued until plant height had plateaued (n = 7–23 ± se). Values found to be statistically significant (P < 0.01; unpaired, two-tailed Student’s t test against the wild type) were as follows: day 4, 084; day 5, 084; day 6, 084; day 7, 084; day 8, 20.1 and 084; day 9, 20.1 and 084; day 10, 084; day 11, XX1, P3, 9.1, and 084; day 12, 9.1 and 084; day 13, XX1, P3, 9.1, and 084; day 14, P3, 9.1, and 084; day 15, 5.2, 9.1, and 084; day 16, 5.2, 20.1, and 084; day 17, 20.1 and 084; day 18, 20.1; day 19, XX1, P3, and 20.1; day 20, 20.1; day 21, 20.1.
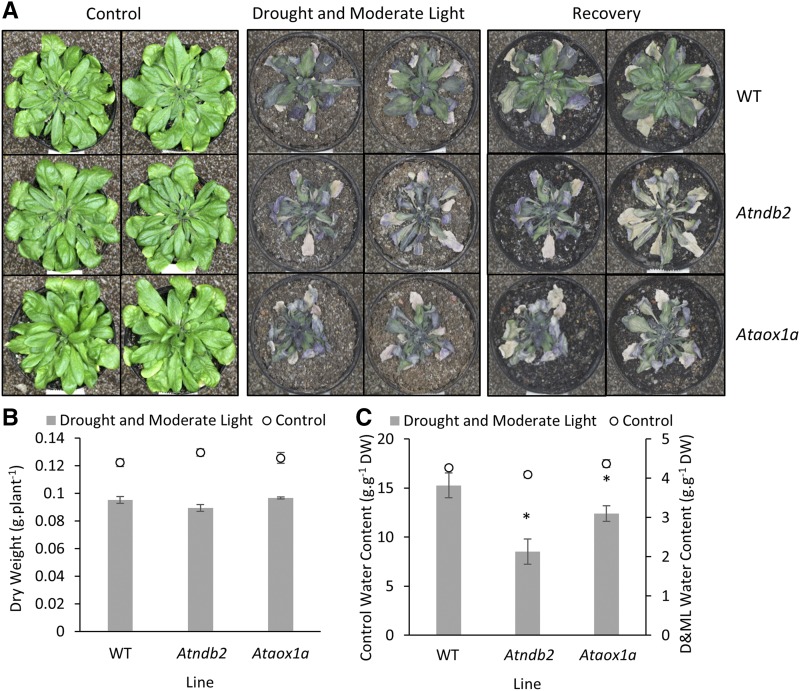
The effect of alterations in AtNDB2 and AtAOX1a on the response of plants to environmental stress was examined by subjecting the different lines to a combination of drought and increased light intensity (Figs. 10 and 11). An Ataox1a T-DNA insertion line was used as an additional control, as it was previously shown to be more sensitive to these combined stress treatments (Giraud et al., 2008; Ng et al., 2013). Wild-type, Atndb2, and Ataox1a lines were subjected to 12 d without watering and with increased light levels (300 μmol m−2 s−1) for the final 6 d, followed by rewatering and a return to normal conditions. Under these conditions, the wild-type plants substantially recovered from the stress upon rewatering (Fig. 10). However, the two T-DNA lines were more sensitive to the stress conditions and were not able to recover after being returned to control conditions (Fig. 10). The T-DNA lines similarly showed reduced water content relative to the wild-type plants at the end of the stress treatment (i.e. before recovery).
Figure 10.
Effects of drought and moderate-light treatment on plants containing AtNDB2 or AtAOX1a T-DNA insertions. Plants of the wild type (WT), Atndb2 T-DNA, and Ataox1a T-DNA lines were grown in a controlled-temperature growth cabinet at 22°C with 16-h daylength and PAR of 80 to 120 μmol m−2 s−1 using custom-made panels of red and blue light-emitting diode (LED) light modules (Phoenix Biosystems). The position of each pot was rotated daily. After 36 d, water was withheld, with continued pot rotation. After 6 d, plants were transferred to moderate light (300 μmol m−2 s−1), with drought continued in the same manner for a further 6 d, at which point plants were either harvested or rewatered and returned to control growth conditions to check recovery. A, Photographs of representative plants grown under control conditions (left), at the end of the drought and moderate-light treatment (middle), and after 14 d of recovery (right). B and C, Dry weights (DW; B) and water contents (C) of plants harvested at the end of the drought and moderate-light treatment (n = 4 ± se). *, P < 0.05 (unpaired, two-tailed Student’s t tests compared with the wild type).
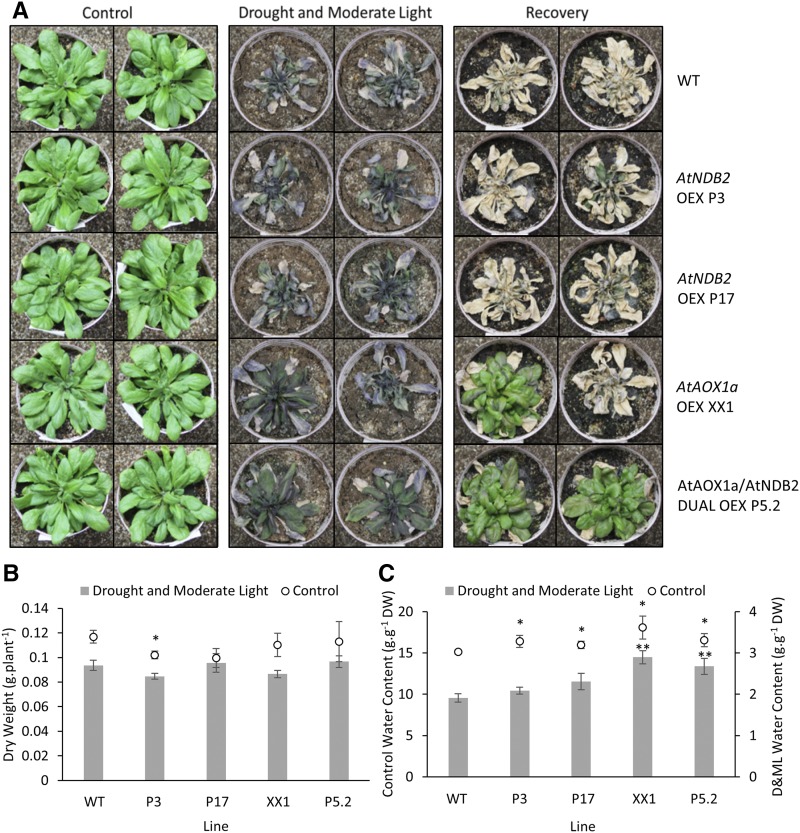
Figure 11.
Effects of extended drought and moderate-light treatment on plants overexpressing AtNDB2 and/or AtAOX1a. Plants of the wild type (WT), AtNDB2 overexpression lines (P3 and P17), AtAOX1a overexpression line (XX1), and dual overexpression line (P5.2) were grown in a controlled-temperature growth cabinet at 22°C with 16-h daylength and PAR of 80 to 120 μmol m−2 s−1 using custom-made panels of red and blue LED light modules (Phoenix Biosystems). The position of each pot was rotated daily. After 32 d, water was withheld, with continued pot rotation. After 6 d, plants were transferred to moderate light (300 μmol m−2 s−1), with drought continued in the same manner for a further 7 d (i.e. a slightly prolonged stress, compared with Fig. 10), at which point plants were either harvested or rewatered and returned to control growth conditions to check recovery. A, Photographs of representative plants grown under control conditions (left), at the end of the drought and moderate-light treatment (middle), and after 14 d of recovery (right). B and C, Dry weights (DW; B) and water contents (C) of plants harvested at the end of the drought and moderate-light treatment (n = 4 ± se). *, P < 0.05 (unpaired, two-tailed Student’s t tests compared with the wild type).
A more severe treatment was given to the overexpression lines: two single AtNDB2 overexpression lines, the AtAOX1a overexpression line, and a dual overexpression line were subjected to a longer drought period and 7 d of increased light levels. This treatment was severe enough to prevent the wild-type plants and the single AtNDB2 overexpression lines from recovering when rewatered (Fig. 11). However, the AtAOX1a and dual overexpression lines showed greater recovery, presumably because of the higher water content of their leaves measured at the end of the stress period. A repeat of the above experiment was performed including all three dual overexpression lines. All tested dual overexpression lines had significantly higher rates of recovery after a moderate light and drought stress compared with the wild type (Supplemental Fig. S5; Supplemental Table S1).
DISCUSSION
AtNDB2 Is the Major External-Facing NADH Dehydrogenase of Arabidopsis Mitochondria
The results presented here clearly show that AtNDB2 is the main external-facing NADH dehydrogenase in Arabidopsis mitochondria. Mitochondria from the Atndb2 T-DNA insertion line had dramatically decreased rates of exogenous NADH oxidation, whereas overexpressing AtNDB2 increased NADH oxidation, especially when coexpressed with AtAOX1a. This is consistent with AtNDB2 being the most highly expressed of the four external NDs at the transcript level (Fig. 1; Elhafez et al., 2006). Both Ca2+-dependent and Ca2+-independent NADH activities decreased in the T-DNA line, which is consistent with data for the enzyme when it was expressed heterologously in E. coli (Geisler et al., 2007) and correlative data in Arabidopsis (Smith et al., 2011). The small amount of NADH oxidation activity that remained in the Atndb2 T-DNA plants was probably attributable to other external NDs, most likely AtNDB4, which is also capable of calcium-independent NADH oxidation (Geisler et al., 2007). Based on our results, under normal growth conditions, AtNDB4 only accounts for a small proportion of external NADH oxidation, at least in leaf mitochondria; it may be expressed more highly in other tissues. In this context, it is interesting that when expression of AtNDB4 was reduced by RNA interference, there was substantial up-regulation of AtNDB2 transcript and protein (Smith et al., 2011). It is intriguing that altering the expression of AtNDB4, responsible for such a small fraction of total external NADH oxidation, according to our results, leads to a response in the much more highly expressed AtNDB2 protein. Smith et al. (2011) suggested that AtNDB2 expression induction was a general stress response to the knockdown of AtNDB4 rather than a compensatory mechanism, since AtAOX1a was also up-regulated. However, it is possible that the two isoforms have different roles or are expressed in different tissues; this requires more detailed investigations of the localization and regulation of the different NDB proteins.
AtNDB2 and AtAOX1a Appear To Be Intimately Linked in Arabidopsis
Overexpression of AtNDB2 alone resulted in increased amounts of AtNDB2 protein in mitochondria, but much of it was not engaged in NADH oxidation through to oxygen. Only when AtAOX1a protein amount and activity were also enhanced did we observe large increases in external NADH oxidation, suggesting that AtAOX1a can either protect AtNDB2 from oxidative damage or that rapid reoxidation by AtAOX1a of the reduced quinone arising from AtNDB2 occurs. Although coordinated increases in the expression of AtAOX1a and AtNDB2 transcripts have been shown under various stress and other conditions (Clifton et al., 2005; Smith et al., 2009, 2011), this is the first clear demonstration of a functional association between AtAOX1a and AtNDB2.
In transgenic Nicotiana sylvestris lines overexpressing an external NADPH dehydrogenase from potato (Solanum tuberosum), the plants demonstrated increased AOX protein and activity (Liu et al., 2008), possibly to prevent overreduction of the ubiquinone pool and potential damage to mitochondrial proteins as a consequence of a higher rate of electron flux into the mETC. Occasionally we have similarly observed increased AtAOX1a levels in some of the Arabidopsis single AtNDB2 overexpression lines. These observations suggest that subtle differences exist between species in their response to changes in mitochondrial electron transport activity and may point to differences between the different NDB proteins and their interactions with ubiquinone.
A simple explanation for our results is that AtNDB2 activity was restricted in mitochondria from the single AtNDB2 overexpression line by the activity of mETC components downstream of the dehydrogenase and the ubiquinone pool, yet the addition of succinate as a second substrate stimulated oxygen uptake in these mitochondria (Table 1). It is possible that AtNDB2 is especially sensitive to ubiquinone redox poise and requires a strongly oxidized ubiquinone pool for it to be active, a condition guaranteed by increased AOX activity in mitochondria from the dual overexpression lines. The differing electron acceptor experiments support this interpretation (Fig. 8). Alternatively, the results may point to a special relationship between AtNDB2 and AtAOX1a, and it is noteworthy that in many circumstances expression of these two genes is coordinately regulated (Clifton et al., 2005; Supplemental Fig. S6). There is also a hint from recent studies of mitochondrial protein complexes in Arabidopsis that AtNDB and AtAOX proteins may be physically connected to form alternative respirasomes (Senkler et al., 2017) to streamline the bypass of proton-pumping and ROS-generating complexes of the mETC. Whereas AtNDB2 and AtAOX1a have not been found specifically associated together in wild-type Arabidopsis, it is possible that overexpression of the two proteins together may result in such a physical association. Even if this is the case, it is clear from the competition between external NADH and succinate (Table 1; Supplemental Table S2) that AtNDB2 and AtAOX1a interact via a ubiquinone pool that is at least partially accessible to other respiratory proteins such as complex II. It should also be noted that while mitochondria from wild-type Arabidopsis grown under nonstressed conditions express very little AOX protein and activity, as do those from Ataox1a T-DNA knockout plants, both readily oxidize external NADH. Likewise, mitochondria overexpressing AtAOX1a had normal levels of external NADH oxidation. It appears that the link between the two proteins is important only when they are both up-regulated, as occurs under many, although not all (Svensson et al., 2002), stress conditions, and it is only in rare cases that AtNDB2 expression is enhanced without concurrent enhancement of AtAOX1a expression (Supplemental Fig. S6). Acclimation to cold is one such circumstance, and in this case AtUCP1 was up-regulated (Armstrong et al., 2008), which would also modulate the redox state of the ubiquinone pool. The link between NDs and AOX is also evident from a bioinformatics study looking for type II NADH dehydrogenases in the kingdom Animalia, where, although ND genes were only found in primitive metazoa, animal species containing the ND gene also had an AOX gene (Matus-Ortega et al., 2011).
Simultaneous Increase in AtNDB2 and AtAOX1a Expression Does Not Affect Plant Growth under Normal Conditions
Data obtained in this study represent a detailed analysis of the impact that overexpressing either AtAOX1a and/or AtNDB2 has on plant growth under control conditions. Interestingly, some minor negative phenotypic effects were observed with single overexpression of either gene, which were not evident when both genes were overexpressed together (Fig. 9). A few studies have previously assessed the impact of overexpressing AOX on growth, and no obvious phenotypic effects were reported under control conditions (Fiorani et al., 2005; Umbach et al., 2005; Murakami and Toriyama, 2008). Most of the analyses focused on effects under stress (Bartoli et al., 2006; Pasqualini et al., 2007; Li et al., 2013; Liu et al., 2014; Dahal and Vanlerberghe, 2017). Smith et al. (2009) observed a slight growth penalty in relative growth rate of leaves at 4 to 5 weeks of age in AtAOX1a overexpression lines under control conditions but no effect on root growth. Similarly, a slight growth penalty (15%–17% in inflorescence stem length) was also evident in some lines of transgenic cassava (Manihot esculenta) overexpressing AtAOX1a (Zidenga et al., 2012). However, in transgenic Arabidopsis plants where AtNDB4 expression had been reduced (Smith et al., 2011), both AtAOX1a and AtNDB2 expression were increased and no growth penalty was evident in shoots or roots under control growth conditions. These observations are mostly consistent with the results of the detailed phenotype analysis in the dual overexpression lines in this study (Fig. 9).
Mitochondria from the dual overexpression plants were capable of extremely high rates of uncoupled electron transport via AtNDB2 and AtAOX1A, and this has the potential to influence the efficiency of respiration in vivo. However, AOX in these mitochondria was still controlled through activation by 2-oxoacids, such as pyruvate. As the activation of AOX was required for rapid external NADH oxidation in the dual overexpression lines, regulating AOX activity also effectively controls NDB2 activity, although the concentration of cytosolic NADH, and possibly Ca2+, will also affect NDB2 activity in vivo. In this way, the metabolic state of the cell may prevent enhanced activity of the AP when it is likely to be detrimental to the plant, and this may be why the overexpression lines show only modest growth and developmental changes under normal growth conditions.
The Atndb2 T-DNA knockout plants, with a greatly reduced capacity for extramitochondrial NADH oxidation, also showed only small differences in growth and development when grown in the absence of environmental stress. Yet knockdown of AtNDB1 produced plants with dramatically less external NADPH oxidation capacity and led to a decrease of almost 20% in rosette dry weight (Wallström et al., 2014a). Interestingly, an approximately 80% loss of external NADPH oxidation had a negative impact on growth, whereas a 90% loss of external NADH oxidation did not, suggesting that the former may be more important during normal growth and development. External NADH oxidation may be more important during stress conditions, considering the higher stress responsiveness of the AtNDB2 gene relative to AtNDB1. In both Atndb1 (Wallström et al., 2014a) and Atndb2 T-DNA lines (our results), a small residual (10%–20%) capacity for oxidation of external NAD(P)H was retained, possibly due to the activity of other dehydrogenases. Complete loss of activity would presumably result in a much more severe phenotype.
Altered AtNDB2 and AtAOX1a Expression Influences Stress Responses of Arabidopsis
Data sets from Genevestigator were screened to identify which stresses elicit a large and significant up-regulation of transcript levels of AtNDB2 and each of the other external type II ND genes in Arabidopsis (Supplemental Fig. S6). Most of the conditions that up-regulated AtNDB2 also up-regulated AtAOX1a, reconfirming the coexpression relationship between these two genes. Cold, drought, high-light, and salt treatments all led to increased AtNDB2 expression above that of AtAOX1a in at least some instances, whereas Suc and Glc treatments, as well as elevated CO2 and circadian cycles that returned to light after a period of darkness, all seem to induce AtNDB2 transcript. This suggests that AtNDB2 may be particularly important when carbon and/or reductant supply exceeds cellular demand, perhaps because of its ability to dissipate excess reducing equivalents. On this basis, we chose a combination of drought and increased light exposure to investigate the effect of altered AtNDB2 and AtAOX1a on stress responses.
Whereas the reduction in external NADH oxidation rates observed in the Atndb2 T-DNA plants did not impact plant growth under normal controlled conditions, it did lower the tolerance of the plant to the combination of drought and moderately high-light stress (Fig. 10). These conditions are likely to cause photoinhibition, which necessitates the dissipation of excess reducing power in the chloroplast for the plant to survive (Raghavendra and Padmasree, 2003; Yoshida et al., 2007, 2010). One way of achieving this is via the operation of a malate/oxaloacetate cycle across the chloroplast envelope, which delivers NAD(P)H to the cytosol. Oxidation of cytosolic NADH via AtNDB2 may be crucial for this cycle to be maintained.
Overexpression of AtNDB2 and AtAOX1a together allowed the plants to survive more severe drought and high-light stress, although no more so than overexpression of AtAOX1a alone (Fig. 11). Overexpression of AtNDB2 alone had little effect, but this is not surprising since the mitochondria of these plants have only marginally elevated capacity for external NADH oxidation. In this context, it should be noted that whereas wild-type and single AtNDB2 overexpression plants essentially lack AOX activity, the AtAOX1a overexpression plants do possess normal capacity for external NADH oxidation, suggesting that both activities are needed to provide resistance to the stress imposed. This would explain why, in contrast to the wild type, neither the Atndb2 nor the Ataox1a T-DNA line was able to recover from the milder stress treatment (Fig. 10). The plants overexpressing AtAOX1a showed higher relative water content at the beginning and end of the stress treatment, which may suggest an increased capacity for the synthesis of compatible solutes in these plants. In other studies, it has been suggested that AP during high-light stress allows an increase in tricarboxylic acid cycle flux toward amino acid metabolism (Florez-Sarasa et al., 2016). A metabolic analysis of the AtAOX1a overexpression plants during drought and light stress is worthy of further investigation.
CONCLUSION
We have shown that AtNDB2 is the major external NADH dehydrogenase in Arabidopsis and that, together with AtAOX1a, it plays a critical role in the ability of the plant to cope with photoinhibitory stress conditions. Furthermore, our results indicate that both AP components need to be present simultaneously for the stress response, but not necessarily for normal growth, consistent with the well-documented coordinated expression of the two proteins under environmental stress in Arabidopsis. It has been proposed that AOX expression could be a useful indicator of an improved stress response in crop breeding (Arnholdt-Schmitt et al., 2006). Our results suggest that the expression of both AOX and NDB genes needs to be considered.
MATERIALS AND METHODS
Plant Growth Conditions
For all plate-grown Arabidopsis (Arabidopsis thaliana) plants, seeds were sterilized and then stratified at 4°C in the dark for 3 d. Seeds were plated on one-half-strength Murashige and Skoog medium (Sigma-Aldrich) containing 2% (w/v) Suc and 0.8% (w/v) agar at a density of approximately one plant per cm2.
For hydroponically grown plants, the method of Conn et al. (2013) was used in a growth room at 22°C with 14-h daylength and PAR of 100 to 150 μmol m−2 s−1, using Sylvania Luxline Plus T5 fluoro lights, or in a glasshouse during early spring, with temperatures varying between 20°C and 24°C during the night and day, respectively, with 11-h daylength and PAR of 100 to 150 μmol m−2 s−1.
For soil-grown plants, a cocopeat soil mix (PIRSA-SARDI) was supplemented either with slow-release fertilizer with trace elements (Osmocote) or with a hydroponic growth solution (Conn et al., 2013) and used to fill approximately 6-cm-diameter pots, each harboring a single Arabidopsis plant.
T-DNA Insertion Lines
The SALK_036330 T-DNA insertion line, which contained a T-DNA insertion in an intron of AtNDB2, was obtained from the Arabidopsis Biological Resource Center. PCR screening was used to identify lines homozygous for the insertion using primers designed to target the T-DNA insert within the AtNDB2 gene or to bridge the T-DNA insertion site. Results from a PCR screen are shown in Supplemental Figure S7, clearly distinguishing homozygotes from heterozygotes and null segregants. The precise location of the T-DNA insertion was determined by sequencing the PCR products obtained using LBb1.3 and RB primers for this line (SIGnAL Web site, http://signal.salk.edu/tdnaprimers.2.html).
An Ataox1a T-DNA insertion line (SALK_084897), characterized in previous studies (Giraud et al., 2008; Strodtkötter et al., 2009), was also used in some analyses.
Generation of Transgenic Lines
A full-length transcript of AtNDB2 was amplified (Supplemental Table S3) from cDNA of Arabidopsis ecotype Columbia-0 and cloned into either the Gateway binary overexpression vector, pK7WG2, for the single AtNDB2 overexpression lines (Karimi et al., 2002) or pEarlygate (Earley et al., 2006) to transform plants already harboring an AtAOX1a overexpression cassette (XX1 line from Umbach et al., 2005), using the floral dip method (Clough and Bent, 1998). Positive transformants were selected either on kanamycin (for single overexpression lines; Sigma-Aldrich) or glufosinolate (for dual overexpression lines; Sigma-Aldrich) and checked by PCR for the presence of the AtNDB2 overexpression cassette. T2 seeds were screened for copy number based on segregation ratios and χ2 analysis (Supplemental Fig. S8). Single-copy transgenic lines were identified and self-fertilized until homozygous lines were obtained. Homozygous T3 and T4 seeds (P3, P9, and P17 single overexpression lines; P5.2, P9.1, and P20.1 dual overexpression lines) were used in subsequent experiments.
Gene Expression Analysis
RNA was extracted from 100 mg of frozen, powdered rosette leaves (MacRae, 2007; Shavrukov et al., 2013). Traces of genomic DNA were removed with RQ1 RNase-free DNase (Promega). Reverse transcription of total RNA (1 µg) was performed with the iScript cDNA Synthesis Kit (Bio-Rad). Reverse transcription quantitative PCR was carried out essentially as described by Soole and Smith (2015), using the KAPA SYBR-fast qPCR Universal ReadyMix system (Geneworks). Biological replicates were made up of pooled rosettes of plate-grown samples (three replicates, each from separate batches of plates) and single rosettes from hydroponically grown samples (six replicates). Reverse transcription quantitative PCR assays were run in duplicate for each cDNA sample. Transcripts were normalized to two reference genes, ubiquitin (At2g17190) and protein phosphatase2A (At1g13320.1).
Mitochondrial Isolation
Plants were grown for 3 weeks on agar plates in a controlled-temperature growth cabinet at 22°C with 16-h daylength and PAR of 80 to 120 μmol m−2 s−1 using custom-made panels of red and blue LED light modules (Phoenix Biosystems). Mitochondrial isolation and purification were carried out with 15 g of shoot tissue using a protocol described by Dry and Wiskich (1987), with modifications to isolation and wash media as well as discontinuous Percoll density gradient concentrations as per Juszczuk et al. (2007). Density gradients were centrifuged at 30,000g for 40 min in a swinging-bucket rotor (SW32Ti; Beckman Coulter). The final wash and resuspension of mitochondrial protein were carried out in wash media lacking bovine serum albumin.
Enzyme Assays
Unless otherwise specified, freshly prepared mitochondria were assayed in SRM (0.45 m mannitol, 50 mm N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid [TES], 50 mm KCl, 10 mm KH2PO4, and 2 mm MgCl2, pH 7.2).
The integrity of fresh and frozen mitochondrial preparations was estimated using cytochrome c oxidase and NAD-malate dehydrogenase latency assays (Rasmusson and Møller, 1990).
NADH and NADPH oxidation was monitored using a modernized Aminco DW-2 spectrophotometer (Olis) with split beam at 340 nm. Assays contained 50 µg of mitochondrial protein in 1 mL of SRM supplemented with 0.2 mm NAD(P)H, 1 mm ADP, and either 0.5 mm EGTA (for Ca2+-independent rates) or 2 mm CaCl2 (for maximum rates with Ca2+). These measurements were conducted either with oxygen as the electron acceptor or supplemented with 40 µm decylubiquinone as an artificial electron acceptor, similar to that described by Geisler et al. (2007). In a similar assay, 0.1 mm DCPIP was used as an alternative electron acceptor and reduction was monitored at 600 nm using a Halo DB-20 double-beam spectrophotometer (Dynamica). DCPIP assays also contained 1 mm KCN and 0.25 mm propyl gallate, and the background rate in the absence of mitochondria was subtracted from the rate in their presence.
Oxygen consumption was monitored polarographically using Oxygraph Plus chambers coupled to O2View software (Hansatech Oxygraph). Assays contained 100 µg of mitochondrial protein in 1 mL of reaction medium supplemented with various substrates as detailed in individual figure and table legends. For NADH oxidation rates, sequential additions included 1 mm NADH, 1 mm ADP, and 2 mm CaCl2. For succinate oxidation rates, sequential additions included 10 mm succinate, 0.1 mm ATP, and 1 mm ADP. To activate AOX, additions of 1 mm DTT and 5 mm pyruvate were made. COX was inhibited by 0.5 mm KCN, and AOX was inhibited by 0.12 mm propyl gallate.
Immunoblotting
Mitochondrial samples were boiled at 95°C for 2 min in SDS-PAGE loading buffer (62.5 mm Tris, 30% [v/v] glycerol, 5% [v/v] β-mercaptoethanol, 2% [w/v] SDS, and 0.002% [w/v] Bromophenol Blue), centrifuged for a few seconds, then supernatants were loaded onto 10% (w/v) SDS-PAGE gels. Once resolved, the proteins were transferred to a 0.45-µm nitrocellulose membrane (Trans-Blot Transfer Medium; Bio-Rad) before probing with the AOA antibody raised against Sauromatum guttatum AOX in mouse that reacts with all AOX proteins tested to date (Elthon and McIntosh, 1987), the NDB2 antibody raised against Arabidopsis NDB2 in rabbit (Oryctolagus cuniculus; Carrie et al., 2008), or the PORIN antibody raised in mouse (Mus musculus; Armstrong et al., 2008). Protein band densities were analyzed using ImageLab 4.0 (Bio-Rad), and the porin protein band in each sample was used as an internal normalizing control for mitochondrial protein content.
Phenotyping Methods
For fresh weight (FW) measurements, plants were harvested and weighed immediately. For hydroponically grown plants, where root measurements were possible, the roots were rinsed in deionized water before blotting dry and weighing. For dry weight (DW) measurements, plant tissues were dried at 80°C until all water was removed, then cooled to room temperature and weighed. Water content was calculated as (FW − DW)/DW (Munns, 2010).
A detailed growth analysis of various overexpression lines was conducted as part of this study, according to Boyes et al. (2001). For root analyses, plants were grown on plates in a growth room at 22°C with 14-h daylength and PAR of 100 to 125 μmol m−2 s−1, using Sylvania Luxline Plus T5 fluoro lights. Seeds were arranged in a single row, approximately 1 cm apart, and plates were oriented vertically. Plates were rearranged within the cabinet daily to minimize positional effects. Root lengths were measured daily from 8 to 14 d. Secondary root measurements were made on day 14 using a light microscope. For growth milestones, plants were grown in individual pots of soil and subirrigated as necessary. Rosette leaf emergence was recorded once leaf diameter reached at least 1 mm. Stem measurements began as soon as the floral stem could be seen and recorded every alternate day.
Drought and Moderate-Light Experimental Conditions
Plants were grown in a controlled-temperature growth cabinet at 22°C with 16-h daylength and PAR of 80 to 120 μmol m−2 s−1 using custom-made panels of red and blue LED light modules (Phoenix Biosystems). Pots of soil were sealed at the bottom to prevent soil loss or water leakage, and each contained the same volume of soil and water and a single Arabidopsis plant. Plants were rearranged within the cabinet daily to minimize positional effects. After 36 d, water was withheld, except to top up pots to the same weight. This ensured that all pots contained the same amount of water despite becoming drier every day. After 6 d, plants were transferred to moderate light (300 μmol m−2 s−1) with continued water limitation. For experiments with T-DNA lines, the combined drought and moderate-light treatment continued for 6 d, after which time the wild-type plants could still recover. For experiments with overexpression lines, the combined treatment continued for 7 d, after which time the wild-type plants could not recover. For recovery, plants were returned to normal light conditions and rewatered to control levels.
Statistical Methods
Statistical comparison of data groups was conducted using two-tailed unpaired Student’s t tests or by one-way or two-way ANOVA (with posthoc Tukey’s tests) using GraphPad Prism. Information specific to each data set, including replication and statistical analyses, is provided in the figure legends.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At3g22370 (AtAOX1a), At1g07180 (AtNDA1), At2g29990 (AtNDA2), At4g28220 (AtNDB1), At4g05020 (AtNDB2), At4g21490 (AtNDB3), At2g20800 (AtNDB4), At5g08740 (AtNDC1), At5g08530 (AtCI), At4g05320 (AtUBQ), and At1g13320 (AtPDF2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence information for the AtNDB2 T-DNA insertion site.
Supplemental Figure S2. Examples of AOX activation and inhibition effects on NADH oxidation rates in purified mitochondria.
Supplemental Figure S3. Manipulation of AtNDB2 expression had little effect on plant growth under normal conditions.
Supplemental Figure S4. Dual overexpression of AtNDB2 and AtAOX1a had little effect on plant growth under control conditions.
Supplemental Figure S5. Effects of extended drought and moderate-light treatment on plants overexpressing AtNDB2 and AtAOX1a.
Supplemental Figure S6. Up-regulation of AtNDB2 transcript level relative to other AP genes in abiotic stress experiments.
Supplemental Figure S7. Example of a genomic screen for T-DNA insert zygosity.
Supplemental Figure S8. Screening transgenic Arabidopsis lines for AtNDB2 overexpression constructs.
Supplemental Table S1. Survival rates of dual Aox1a/Ndb2 overexpression plants watered for recovery at the end of the drought and moderate-light treatment.
Supplemental Table S2. External NADH oxidation competes with succinate oxidation in dual overexpression lines.
Supplemental Table S3. Primer sets used for AtNDB2 full-length gene amplification and transcript level analysis.
Acknowledgments
We thank Jim Siedow for providing seed of the AtAOX1a-overexpressing line and Harvey Millar for providing the AOA (AOX) and porin antibodies.
Footnotes
This work was supported by the Australian Research Council (DP140103090 and IH140100013) and an Australian Postgraduate Award to C.D.W.
Articles can be viewed without a subscription.
References
- Armstrong AF, Badger MR, Day DA, Barthet MM, Smith PMC, Millar AH, Whelan J, Atkin OK (2008) Dynamic changes in the mitochondrial electron transport chain underpinning cold acclimation of leaf respiration. Plant Cell Environ 31: 1156–1169 [DOI] [PubMed] [Google Scholar]
- Arnholdt-Schmitt B, Costa JH, de Melo DF (2006) AOX: A functional marker for efficient cell reprogramming under stress? Trends Plant Sci 11: 281–287 [DOI] [PubMed] [Google Scholar]
- Bartoli CG, Yu J, Gómez F, Fernández L, McIntosh L, Foyer CH (2006) Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J Exp Bot 57: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al. (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, et al. (2013) Protocol: Optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal K, Vanlerberghe GC (2017) Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol 213: 560–571 [DOI] [PubMed] [Google Scholar]
- Djajanegara I, Holtzapffel R, Finnegan PM, Hoefnagel MHN, Berthold DA, Wiskich JT, Day DA (1999) A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett 454: 220–224 [DOI] [PubMed] [Google Scholar]
- Djajanegara I, Finnegan PM, Mathieu C, McCabe T, Whelan J, Day DA (2002) Regulation of alternative oxidase gene expression in soybean. Plant Mol Biol 50: 735–742 [DOI] [PubMed] [Google Scholar]
- Dry IB, Wiskich JT (1987) 2-Oxoglutarate dehydrogenase and pyruvate dehydrogenase activities in plant mitochondria: Interaction via a common coenzyme A pool. Arch Biochem Biophys 257: 92–99 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Elhafez D, Murcha MW, Clifton R, Soole KL, Day DA, Whelan J (2006) Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: Intraorganelle location and expression. Plant Cell Physiol 47: 43–54 [DOI] [PubMed] [Google Scholar]
- Elthon TE, McIntosh L (1987) Identification of the alternative terminal oxidase of higher plant mitochondria. Proc Natl Acad Sci USA 84: 8399–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A, Latimer S, Schmollinger S, Block A, Dussault PH, Vermaas WFJ, Merchant SS, Basset GJ (2015) A dedicated type II NADPH dehydrogenase performs the penultimate step in the biosynthesis of vitamin K1 in Synechocystis and Arabidopsis. Plant Cell 27: 1730–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: A study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez-Sarasa I, Ribas-Carbo M, Del-Saz NF, Schwahn K, Nikoloski Z, Fernie AR, Flexas J (2016) Unravelling the in vivo regulation and metabolic role of the alternative oxidase pathway in C3 species under photoinhibitory conditions. New Phytol 212: 66–79 [DOI] [PubMed] [Google Scholar]
- Geisler DA, Broselid C, Hederstedt L, Rasmusson AG (2007) Ca2+-binding and Ca2+-independent respiratory NADH and NADPH dehydrogenases of Arabidopsis thaliana. J Biol Chem 282: 28455–28464 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LHM, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GR, Maxwell DP, Villarimo AR, McIntosh L (2004) Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep 23: 497–503 [DOI] [PubMed] [Google Scholar]
- Ho LHM, Giraud E, Uggalla V, Lister R, Clifton R, Glen A, Thirkettle-Watts D, Van Aken O, Whelan J (2008) Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol 147: 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczuk IM, Flexas J, Szal B, Dabrowska Z, Ribas-Carbo M, Rychter AM (2007) Effect of mitochondrial genome rearrangement on respiratory activity, photosynthesis, photorespiration and energy status of MSC16 cucumber (Cucumis sativus) mutant. Physiol Plant 131: 527–541 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Li CR, Liang DD, Xu RF, Li H, Zhang YP, Qin RY, Li L, Wei PC, Yang JB (2013) Overexpression of an alternative oxidase gene, OsAOX1a, improves cold tolerance in Oryza sativa L. Genet Mol Res 12: 5424–5432 [DOI] [PubMed] [Google Scholar]
- Liu J, Li Z, Wang Y, Xing D (2014) Overexpression of ALTERNATIVE OXIDASE1a alleviates mitochondria-dependent programmed cell death induced by aluminium phytotoxicity in Arabidopsis. J Exp Bot 65: 4465–4478 [DOI] [PubMed] [Google Scholar]
- Liu YJ, Norberg FEB, Szilágyi A, De Paepe R, Akerlund HE, Rasmusson AG (2008) The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP+ ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol 49: 251–263 [DOI] [PubMed] [Google Scholar]
- MacRae E. (2007) Extraction of plant RNA. In Hilario E, Mackay J, eds, Protocols for Nucleic Acid Analysis by Nonradioactive Probes. Humana Press, Totowa, NJ, pp 15–24 [Google Scholar]
- Matus-Ortega MG, Salmerón-Santiago KG, Flores-Herrera O, Guerra-Sánchez G, Martínez F, Rendón JL, Pardo JP (2011) The alternative NADH dehydrogenase is present in mitochondria of some animal taxa. Comp Biochem Physiol Part D Genomics Proteomics 6: 256–263 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA (2011) Organization and regulation of mitochondrial respiration in plants. In Merchant SS, Briggs WR, Ort D, eds, Annual Review of Plant Biology, Vol 62 Annual Reviews, Palo Alto, CA, pp 79–104 [DOI] [PubMed] [Google Scholar]
- Munns R. (2010) Plant water content and relative water content. PrometheusWiki: http://www.publish.csiro.au/prometheuswiki (April 27, 2017). [Google Scholar]
- Murakami Y, Toriyama K (2008) Enhanced high temperature tolerance in transgenic rice seedlings with elevated levels of alternative oxidase, OsAOX1a. Plant Biotechnol 25: 361–364 [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini S, Paolocci F, Borgogni A, Morettini R, Ederli L (2007) The overexpression of an alternative oxidase gene triggers ozone sensitivity in tobacco plants. Plant Cell Environ 30: 1545–1556 [DOI] [PubMed] [Google Scholar]
- Polidoros AN, Mylona PV, Pasentsis K, Scandalios JG, Tsaftaris AS (2005) The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress. Redox Rep 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Møller IM (1990) NADP-utilizing enzymes in the matrix of plant mitochondria. Plant Physiol 94: 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Møller IM (2011) Mitochondrial electron transport and plant stress. In Kempken F, ed, Plant Mitochondria, Advances in Plant BiologyVol 1 Springer, New York, pp 357–381 [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Selinski J, Hartmann A, Höfler S, Deckers-Hebestreit G, Scheibe R (2016) Refined method to study the posttranslational regulation of alternative oxidases from Arabidopsis thaliana in vitro. Physiol Plant 157: 264–279 [DOI] [PubMed] [Google Scholar]
- Selinski J, Hartmann A, Kordes A, Deckers-Hebestreit G, Whelan J, Scheibe R (2017) Analysis of posttranslational activation of alternative oxidase isoforms. Plant Physiol 174: 2113–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun HP (2017) The mitochondrial complexome of Arabidopsis thaliana. Plant J 89: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Shavrukov Y, Bovill J, Afzal I, Hayes JE, Roy SJ, Tester M, Collins NC (2013) HVP10 encoding V-PPase is a prime candidate for the barley HvNax3 sodium exclusion gene: Evidence from fine mapping and expression analysis. Planta 237: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Smith CA, Melino VJ, Sweetman C, Soole KL (2009) Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol Plant 137: 459–472 [DOI] [PubMed] [Google Scholar]
- Smith C, Barthet M, Melino V, Smith P, Day D, Soole K (2011) Alterations in the mitochondrial alternative NAD(P)H dehydrogenase NDB4 lead to changes in mitochondrial electron transport chain composition, plant growth and response to oxidative stress. Plant Cell Physiol 52: 1222–1237 [DOI] [PubMed] [Google Scholar]
- Soole KL, Smith CA (2015) Analysis of type II NAD(P)H dehydrogenases. In Whelan J and Murcha MW, eds, Plant Mitochondria. Humana Press, New York, pp 151–164 [Google Scholar]
- Strodtkötter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, et al. (2009) Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297 [DOI] [PubMed] [Google Scholar]
- Svensson AS, Johansson FI, Møller IM, Rasmusson AG (2002) Cold stress decreases the capacity for respiratory NADH oxidation in potato leaves. FEBS Lett 517: 79–82 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Gonzàlez-Meler MA, Sweet CR, Siedow JN (2002) Activation of the plant mitochondrial alternative oxidase: Insights from site-directed mutagenesis. Biochim Biophys Acta 1554: 118–128 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC. (2013) Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan V, Soole K (2010) Effect of short- and long-term phosphate stress on the non-phosphorylating pathway of mitochondrial electron transport in Arabidopsis thaliana. Funct Plant Biol 37: 455–466 [Google Scholar]
- Wagner AM, Krab K, Wagner MJ, Moore AL (2008) Regulation of thermogenesis in flowering Araceae: The role of the alternative oxidase. Biochim Biophys Acta 1777: 993–1000 [DOI] [PubMed] [Google Scholar]
- Wallström SV, Florez-Sarasa I, Araújo WL, Aidemark M, Fernández-Fernández M, Fernie AR, Ribas-Carbó M, Rasmusson AG (2014a) Suppression of the external mitochondrial NADPH dehydrogenase, NDB1, in Arabidopsis thaliana affects central metabolism and vegetative growth. Mol Plant 7: 356–368 [DOI] [PubMed] [Google Scholar]
- Wallström SV, Florez-Sarasa I, Araújo WL, Escobar MA, Geisler DA, Aidemark M, Lager I, Fernie AR, Ribas-Carbó M, Rasmusson AG (2014b) Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in Arabidopsis thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport. Plant Cell Physiol 55: 881–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K (2007) Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol 48: 606–614 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Shibata M, Terashima I, Noguchi K (2010) Simultaneous determination of in vivo plastoquinone and ubiquinone redox states by HPLC-based analysis. Plant Cell Physiol 51: 836–841 [DOI] [PubMed] [Google Scholar]
- Zidenga T, Leyva-Guerrero E, Moon H, Siritunga D, Sayre R (2012) Extending cassava root shelf life via reduction of reactive oxygen species production. Plant Physiol 159: 1396–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]