Isolation of open chromatin identifies MILDEW RESISTANCE LOCUS O3 and two novel DNA boxes as likely regulators of defense priming and systemic acquired disease resistance in Arabidopsis.
Abstract
Upon local infection, plants activate a systemic immune response called systemic acquired resistance (SAR). During SAR, systemic leaves become primed for the superinduction of defense genes upon reinfection. We used formaldehyde-assisted isolation of regulatory DNA elements coupled to next-generation sequencing to identify SAR regulators. Our bioinformatic analysis produced 10,129 priming-associated open chromatin sites in the 5′ region of 3,025 genes in the systemic leaves of Arabidopsis (Arabidopsis thaliana) plants locally infected with Pseudomonas syringae pv. maculicola. Whole transcriptome shotgun sequencing analysis of the systemic leaves after challenge enabled the identification of genes with priming-linked open chromatin before (contained in the formaldehyde-assisted isolation of regulatory DNA elements sequencing dataset) and enhanced expression after (included in the whole transcriptome shotgun sequencing dataset) the systemic challenge. Among them, Arabidopsis MILDEW RESISTANCE LOCUS O3 (MLO3) was identified as a previously unidentified positive regulator of SAR. Further in silico analysis disclosed two yet unknown cis-regulatory DNA elements in the 5′ region of genes. The P-box was mainly associated with priming-responsive genes, whereas the C-box was mostly linked to challenge. We found that the P- or W-box, the latter recruiting WRKY transcription factors, or combinations of these boxes, characterize the 5′ region of most primed genes. Therefore, this study provides a genome-wide record of genes with open and accessible chromatin during SAR and identifies MLO3 and two previously unidentified DNA boxes as likely regulators of this immune response.
Living organisms are continuously exposed to pathogens. To minimize the chance of successive infections, organisms have evolved the capacity to memorize previous attack and mount a more robust defense response upon reinfection (Dempsey et al., 2003; Spoel and Dong, 2012; Netea et al., 2016). For example, after localized foliar infection by a pathogen, plants develop a broad-spectrum systemic immune response called systemic acquired resistance (SAR; Spoel and Dong, 2012). During SAR, uninfected systemic leaves are primed for the superinduction of defense responses upon rechallenge (Kohler et al., 2002; Beckers et al., 2009; Spoel and Dong, 2012; Conrath et al., 2015). In Arabidopsis (Arabidopsis thaliana), defense priming involves an elevated level of microbial pattern receptor kinases (Tateda et al., 2014), accumulation of dormant signaling enzymes (e.g. mitogen-activated protein kinases; Beckers et al., 2009), and covalent modification to chromatin in the 5′ regulatory regions of defense-related genes, such as WRKY6, WRKY29, WRKY53, and PR1 (Jaskiewicz et al., 2011; Luna et al., 2012). Priming marker genes WRKY6, WRKY29, and WRKY53 belong to a family of >70 loci encoding transcription factors with a regulatory role in plant immunity (Wang et al., 2006; Jaskiewicz et al., 2011; Tsuda and Somssich, 2015), whereas PR1 encodes a sterol-binding protein with antimicrobial activity (Gamir et al., 2017).
The priming-associated modification of chromatin in the 5′ leader region of WRKY6, WRKY29, WRKY53, and PR1 comprises the hypomethylation of DNA (Luna et al., 2012; Furci et al., 2019) and the methylation and acetylation of specific Lys residues in the amino terminus of histones H3 and H4 (Jaskiewicz et al., 2011; Luna et al., 2012). Histone Lys methylation usually serves as a docking site for transcriptional regulatory proteins containing plant homeodomains (Peña et al., 2006). In contrast, histone Lys acetylation directly relaxes the interaction of nucleosome neighbors, loosens the ionic DNA-histone interaction, and provides docking sites for regulatory proteins containing bromodomains, such as transcription factor II D and the switch/Suc nonfermentable chromatin remodeling complex (Eberharter and Becker, 2002; Kanno et al., 2004). Regardless of their specific role in transcriptional regulation, the interaction of chromatin regulatory proteins with DNA leads to localized nucleosome eviction (Henikoff, 2008). Thus, nucleosome-free DNA or “open chromatin” marks regulatory activity (Giresi and Lieb, 2009) and can be used as a molecular tag to identify regulatory DNA and genes with an important role in defense priming and SAR.
Formaldehyde-assisted isolation of regulatory DNA elements (FAIRE) is a powerful technique for the isolation of nucleosome-depleted, regulatory DNA from various eukaryotes, including Arabidopsis (Giresi and Lieb, 2009; Gaulton et al., 2010; Schillheim et al., 2018). In FAIRE, chromatin is crosslinked with formaldehyde in vivo, extracted, sheared by sonication, and phenol-chloroform extracted. In contrast to nucleosome-rich chromatin, the DNA in nucleosome-depleted regulatory regions is less efficiently crosslinked to protein. Thus, during phenol-chloroform extraction, free regulatory DNA will accumulate in the aqueous phase, whereas crosslinked DNA-protein complexes will enrich in the interface. The DNA in the aqueous phase is then purified and quantitatively amplified with gene-specific primers (FAIRE-quantitative PCR [qPCR]), hybridized to a tiling DNA microarray (FAIRE-chip), or subjected to next-generation sequencing (FAIRE-seq; Giresi and Lieb, 2009).
To identify regulators of defense priming and SAR, we used FAIRE-seq to profile the chromatin of primed (uninfected) systemic leaves of Arabidopsis plants with local Pseudomonas syringae pv. maculicola (Psm) infection. Psm is a hemibiotroph pathogen that causes bacterial leaf spot on cruciferous plants, including Arabidopsis (Dong et al., 1991). Localized foliar infection of Arabidopsis leaves with Psm elicits the systemic accumulation of the defense signals N-hydroxypipecolic acid and salicylic acid (SA) that promote the establishment of defense priming and SAR (Bernsdorff et al., 2016; Hartmann et al., 2018). Our chromatin profiling discovered 10,129 priming-associated sites of open chromatin in the promoter and promoter-proximal region of 3,025 genes, revealed previously unknown candidates for priming-linked cis-regulatory DNA elements in the 5′ regulatory regions of those genes, and disclosed the Arabidopsis MILDEW RESISTANCE LOCUS O3 (MLO3) as a positive regulator of SAR in this plant.
RESULTS AND DISCUSSION
Open Chromatin Formation Accompanies Gene Priming and SAR
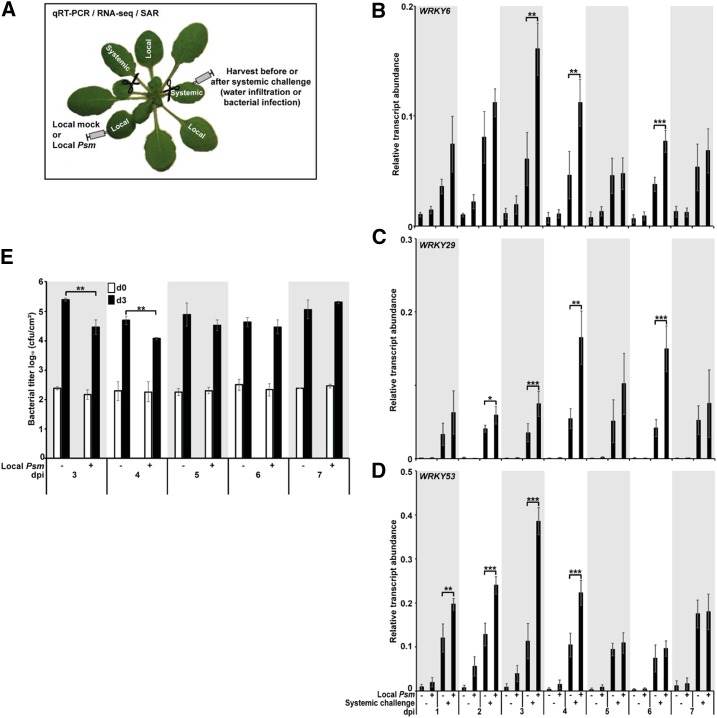
As a first step toward the global isolation of regulatory DNA associated with defense priming and SAR, we first determined a suitable time to perform the analysis. To do so, we used a syringe without a needle to infiltrate three leaves of a batch of 4- to 5-week-old Arabidopsis plants with ∼3 × 108 colony-forming units (cfu)/mL Psm in MgCl2 (this treatment is subsequently referred to in this work as “Local Psm”; for the experimental setup, see Fig. 1A). Three leaves of another batch of plants were infiltrated with MgCl2 in the absence of bacteria (this treatment is subsequently referred to in this work as “Local mock”; Fig. 1A). Seven days after treatment, at the same time of day, we infiltrated two systemic leaves of half of the Local mock and half of the Local Psm plants with water (this treatment is subsequently referred to in this work as “systemic challenge”; Fig. 1A). Infiltration of water into leaves provides a physical stress that is sufficient to activate defense genes (Kohler et al., 2002; Beckers et al., 2009; Jaskiewicz et al., 2011). At 2.5 h after the systemic challenge we harvested the infiltrated systemic leaves of the Local mock and Local Psm plants and analyzed them for expression of priming marker genes WRKY6, WRKY29, and WRKY53 by reverse transcription qPCR (RT-qPCR; Fig. 1, B–D). The result of the experiment showed that in Local Psm plants the systemic challenge-activated expression of WRKY6, WRKY29, and WRKY53 was significantly enhanced at days 3 and 4 day after the local Psm infection (Fig. 1, B–D). The primed activation of WRKY6 and WRKY53 expression was strongest at day 3 after local Psm infection and declined afterward (Fig. 1, B and D), with another significant increase in WRKY6 expression at day 6 after local Psm inoculation. For WRKY29, the enhanced expression was most pronounced at days 4 and 6 after local Psm infection (Fig. 1C). Together, these findings pointed to 3–4 d as a suitable time for isolating priming/SAR-regulatory DNA from systemic leaves of Local Psm plants.
Figure 1.
Effect of local Psm infection on systemic expression of WRKY6, WRKY29, and WRKY53 and SAR. A, Experimental setup for analyzing gene expression (RT-qPCR and RNA-seq) and SAR. B to D, Expression of WRKY6, WRKY29, and WRKY53 in challenged systemic leaves of Local mock (− Local Psm) and Local Psm (+) plants at 2.5 h after the systemic challenge and at various days after the initial treatment (Local mock or Local Psm). dpi, days post initial treatment. Error bars represent the SD (n = 6); ***P ≤ 0.001; **P ≤ 0.01. E, Bacterial titer in systemic leaves of Local mock (− Local Psm) and Local Psm (+) plants at 0 h (d0) and 72 h (d3) after inoculation and at various days after the initial treatment (Local mock or Local Psm). cfu, colony-forming units. Error bars represent the SD (n = 3); **P ≤ 0.01.
To confirm or refute this conclusion, we investigated whether SAR to Psm was expressed from 3 to 7 d in Local Psm plants. At each of these days we infiltrated two systemic leaves of Local mock and Local Psm plants with Psm (∼3 × 108 cfu/mL; Fig. 1A). Immediately after the systemic inoculation and after 72 h, we determined the titer of Psm in the inoculated systemic leaves. As shown in Figure 1E, at days 3 and 4 after the initial treatment (infiltration of MgCl2 or Psm in MgCl2), the bacterial titer was significantly lower in inoculated systemic leaves of Local Psm plants as compared to Local mock plants. The bacterial titer in Local Psm plants was also lower at the days 5 and 6 post systemic infection, but this difference was not significant (Fig. 1E). At 7 d after inoculation, a reduction of the bacterial titer in systemic leaves of Local Psm plants was not detected anymore (Fig. 1E). These findings confirmed 3–4 d as a suitable time for isolating priming/SAR-regulatory DNA from systemic leaves of Local Psm plants.
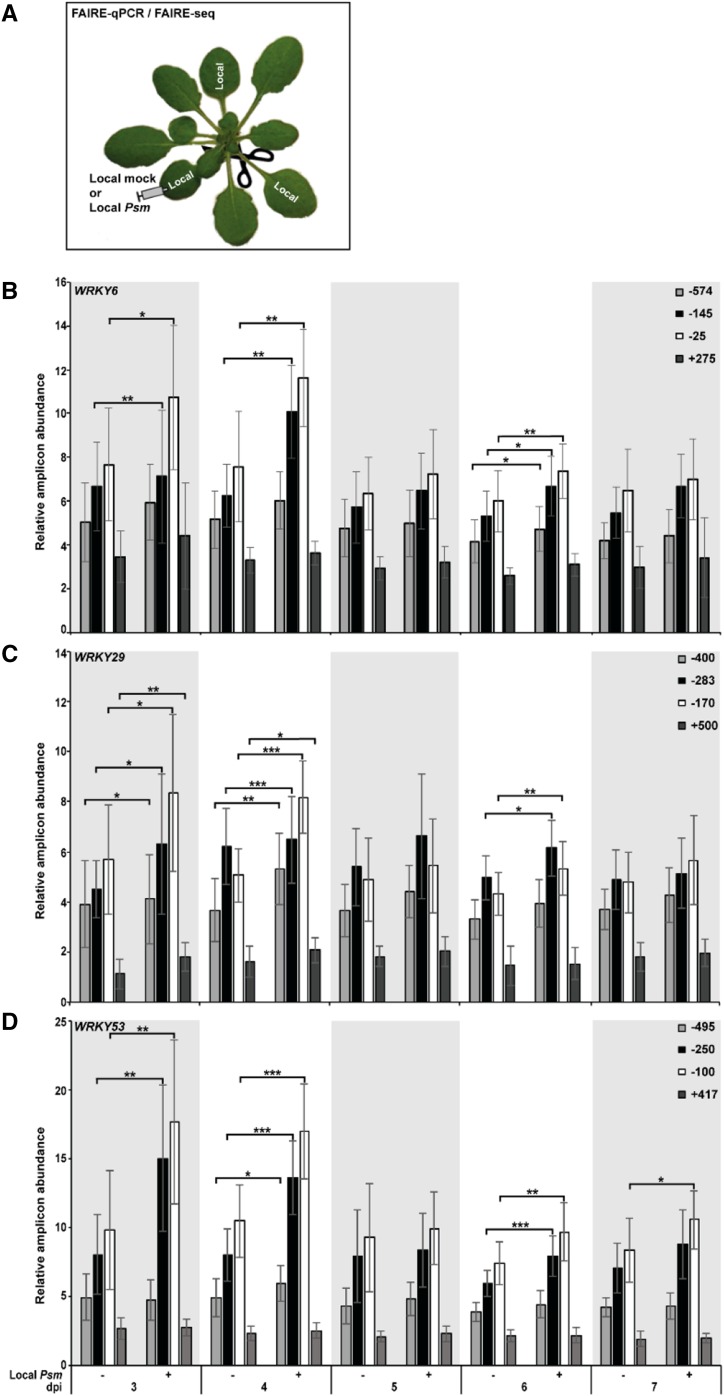
Next, we tested whether systemic defense priming and SAR to Psm is associated with the formation of open chromatin in the promoter/promoter-proximal region of priming-responsive genes. To do so, we harvested all systemic leaves from Local mock and Local Psm plants at various days after initial treatment (infiltration of MgCl2 or Psm in MgCl2; Fig. 2A). Harvested leaves were younger than the initially treated leaves (Fig. 2A). We subjected the systemic leaves to a FAIRE-qPCR protocol that we developed for mature Arabidopsis leaves (see "Materials and Methods"). Figure 2, B–D, shows that local Psm infection causes chromatin to open in the 5′ regulatory region of priming marker genes WRKY6, WRKY29, and WRKY53 in systemic leaves. Chromatin unpacking in the 5′ regulatory region of WRKY6, WRKY29, and WRKY53 was significantly higher in systemic leaves of Local Psm plants 3 and 4 d after the initial treatment, as was observed with WRKY6, WRKY29, and WRKY53 expression after challenge and SAR to Psm (Fig. 1, B–E). The presence of open chromatin declined afterward with another significant opening at day 6 after the local Psm infection (Fig. 2, B–D). The observed formation of open chromatin in the promoter/promoter-proximal region and near the transcriptional start site (TSS) of the priming marker genes WRKY6, WRKY29, and WRKY53 is consistent with a recent genome-wide study of the nucleosome coverage in SA-treated Arabidopsis plants (Singh et al., 2015). The authors demonstrated depletion of nucleosomes in the 5′ regulatory region and near the TSS of SA-induced genes and enhanced nucleosome density in the 5′ leader sequence and close to the TSS of SA-repressed genes. By contrast, the 5′ regulatory sequence of silent and constitutively expressed genes was not subjected to major changes in nucleosome occupancy (Singh et al., 2015).
Figure 2.
Local Psm infection in systemic leaves opens chromatin in the 5′ regulatory region of WRKY6, WRKY29, and WRKY53. A, Experimental setup for FAIRE-qPCR and FAIRE-seq. For the analyses, we harvested whole rosettes of leaves that were younger than the youngest Local mock or Local Psm leaf. B to D, Detection of open chromatin in the promoter and promoter-proximal region of WRKY6, WRKY29, and WRKY53 in unchallenged systemic leaves of Local mock (− Local Psm) and Local Psm (+) plants at various days after the initial treatment (Local mock or Local Psm). dpi, days post initial treatment. Numbers in graphs denote the midpoint of DNA amplicons relative to the TSS; error bars represent the SD (n = 5); ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
The results in Figures 1 and 2 disclosed that in systemic leaves of Local Psm plants, priming for enhanced WRKY6, WRKY29, and WRKY53 expression, SAR to Psm, and open chromatin formation were strong at day 3 and, to a seemingly lesser degree, day 4 after the local Psm infection. Therefore, we decided to perform the genome-wide isolation of regulatory DNA associated with defense priming and SAR in systemic leaves of Local Psm plants at day 3 after the initial Psm inoculation.
A Global Map of Open Chromatin during Defense Priming and SAR
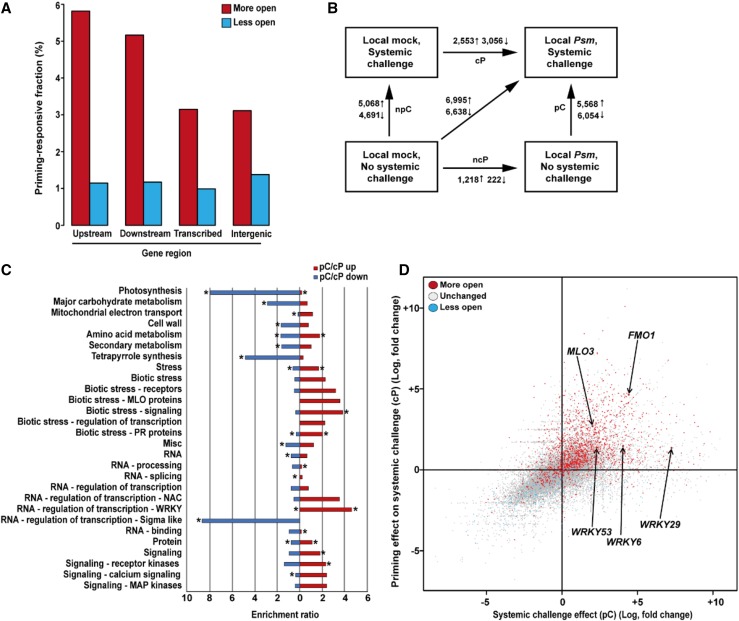
Using the experimental setup as described for FAIRE-qPCR (Fig. 2A), we isolated nucleosome-free, nuclear DNA from systemic leaves of an aliquot of Local mock and Local Psm plants 72 h after the initial treatment using our newly developed FAIRE-seq protocol for mature Arabidopsis leaves (see “Materials and Methods“). Isolated DNA elements were sequenced using next-generation sequencing (see “Materials and Methods“). Briefly, isolated FAIRE-DNA was subjected to two rounds of 50 bp single-end sequencing using an Illumina HiSeq 2000 platform. They produced ∼95 million reads totaling 4.76 billion bases in the first round, and 248 million reads totaling 12.66 billion bases in the second round of sequencing. FAIRE-DNA of a later repeat experiment was sequenced using 125 bp paired ends and an Illumina HiSeq 2500 platform to allow for the explicit detection of DNA fragment size. This sequencing run produced almost 548 million reads totaling 68.5 billion bases. In our bioinformatic analysis (see “Materials and Methods“), we cross-compared the data from untreated systemic leaves of the Local Psm plants to those of untreated systemic leaves of Local mock plants. Using a genome-wide sliding window approach (see “Materials and Methods“), we detected 42,146 DNA sites that were specifically more open and 13,814 DNA sites that were specifically less open in the chromatin of untreated systemic leaves of Local Psm plants when compared to systemic leaves of Local mock plants. Considering only loci centered within a region of 500 bp upstream of the TSS, 7,739 of the more open sites were aligned to 3,833 mRNA transcripts from 3,025 genes, producing a set of 10,129 transcript-specific putative regulatory relationships. This is accounting for cases in which a transcript-encoding sequence had multiple open loci and/or in which a FAIRE-positive DNA site was present in the 5′ regulatory region of multiple genes. Similarly, 1,528 of the less open loci were aligned to 957 transcripts from 785 genes, producing an additional set of 1,891 transcript-specific, putative regulatory relationships. DNA sites near genes (e.g. outside of, but within 500 bp of, the transcribed region) showed a higher proportion of FAIRE signals (5.8% for upstream and 5.2% for downstream) than transcribed (3.2%) and intergenic (3.1%) regions (Fig. 3A; Supplemental Table S1). Thus, according to our analysis, priming of systemic leaves of Local Psm plants seems to involve increased accessibility to regulatory chromatin of 3,025 genes and reduced accessibility to regulatory regions of 785 genes. The reason for the reduced chromatin accessibility in gene regulatory regions during defense priming is unknown. It may reflect the reported plant growth-to-defense transition (Pajerowska-Mukhtar et al., 2012) by shutting off genes with a role in Arabidopsis growth and development. And in fact, genes associated with the functional MapMan bins (Thimm et al., 2004; Usadel et al., 2009; Bolger et al., 2018) “photosynthesis,” “tetrapyrrole biosynthesis,” “carbohydrate metabolism,” and “sigma-like transcription factors” made up the biggest part of genes with reduced expression in challenged systemic leaves of Local Psm plants (Fig. 3C). Sigma-like transcription factors are nucleus-encoded regulators of plastid-encoded RNA polymerases with a role in chloroplast biogenesis (Börner et al., 2015).
Figure 3.
Identification of regulators of defense priming. A, In systemic leaves of Local Psm plants, priming is associated with formation of more open (red columns) and less open (blue columns) chromatin sites in different regions of the Arabidopsis genome. The diagram gives the proportion numbers given in Supplemental Table S1. The numerator is the number of regions with a given classification (upstream, downstream, transcribed, or intergenic), which are more open or less open. The denominator is the number of regions with the same classification in the entire genome. B, Changes in gene expression upon the indicated treatments. ↑, number of induced genes; ↓ number of repressed genes. C, Shown are top-level MapMan bins that were significantly enriched (red) or depleted (blue) in the pC/cP comparison and selected leaf bins that showed at least 2-fold enrichment in the categories “stress", “RNA”, and “signaling". *P ≤ 0.05. D, Relationship between chromatin state (more open, unchanged, or less open) and transcriptional response of genes. The chromatin state (determined by FAIRE-seq) in unchallenged systemic leaves of Local Psm plants was compared to that of unchallenged systemic leaves of Local mock plants. Chromatin state is indicated by color. The transcriptional response of genes (determined by RNA-seq) in challenged systemic leaves of Local Psm plants at 2.5 h after the systemic challenge was compared to that of challenged systemic leaves of Local mock plants (on the y axis) and to that of unchallenged systemic leaves of Local Psm plants (on the x axis). We used the response of all 24,447 genes, which we detected in any RNA-seq experiment, to compose this chart. MLO3, mildew resistance locus O3; FMO1, flavin monooxygenase 1; pC, effect of systemic challenge on priming; cP, effect of priming on systemic challenge.
A Global Map of Gene Expression during Defense Priming and SAR
Using the experimental setup in Figure 1A, we subjected systemic leaves of the Local mock and Local Psm plants to systemic challenge (water infiltration into leaves). After 2.5 h, we harvested systemic leaves of the four groups of plants (Local mock and Local Psm, both with and without systemic challenge; Fig. 3B) and subjected them to whole transcriptome shotgun sequencing (RNA-seq) analysis (see "Materials and Methods"). Briefly, we prepared a complementary DNA (cDNA) library from total RNA that we had extracted from systemic leaves of each of the four plant groups. The cDNA was sequenced using an Illumina HiSeq 2000 platform using 50 bp single-end reads, which produced ∼64.9 million reads totaling 32.48 billion bases. We aligned the acquired RNA-seq data to the Arabidopsis genome (Supplemental Dataset S1), assigned the genes to top-level MapMan bins (Supplemental Dataset S2), and did a transcript-based assessment of the FAIRE-seq data (for details, see "Materials and Methods"; Fig. 3D). Genes in the MapMan bins “stress,” “biotic stress signaling,” “WRKY transcription,” and “receptor kinases” were significantly induced in challenged systemic leaves of Local Psm plants (Fig. 3C), indicating a successful priming experiment. Further in-depth analysis of the data revealed complex patterns of open chromatin and altered gene expression, both of which depended on the physiological state of the plant (Fig. 3, B and D; Supplemental Fig. S1; Supplemental Dataset S1). Our data analysis also disclosed that most of the loci with open chromatin in the 5′ regulatory region (as detected by comparative FAIRE-seq analysis) and enhanced expression upon systemic challenge (as determined by comparative RNA-seq analysis) belonged to the “micro RNA,” “stress,” and “signaling” groups of Arabidopsis genes (Supplemental Fig. S2; Supplemental Dataset S2). Together, by cross-comparison of the FAIRE-seq data from untreated systemic leaves of Local Psm plants with the RNA-seq data from infiltrated systemic leaves of Local Psm plants, we found a coincidence of the presence of open chromatin near the TSSs of genes with their enhanced expression after systemic challenge (Fig. 3D; Supplemental Dataset S1).
Among the 25 genes with the highest FAIRE-seq signals before and RNA-seq signals after systemic challenge were those encoding Cys-rich receptor-like protein kinases 36 and 38, a putative Ser/Thr receptor kinase, Glu receptors 2.5 and 2.8, and a gene encoding a disease resistance protein in the toll-interleukin-receptor nucleotide binding site-Leu-rich repeat family (Supplemental Dataset S1). While the receptor kinases could contribute to the enhanced capacity of primed cells to perceive and respond to microbial patterns (Tateda et al., 2014), the Glu receptors probably play their reported role in systemic plant defense signaling (Toyota et al., 2018). The toll-interleukin-receptor nucleotide binding site-Leu-rich repeat family protein most likely has a role in gene-for-gene disease resistance that often causes intense defense priming and a very robust SAR response. Regardless of their role, these genes seem to be reliable, previously unknown marker genes for defense priming and SAR in Arabidopsis.
Other, but less striking, genes with open chromatin in the 5′ regulatory region and enhanced transcription upon systemic challenge were the known priming marker genes WRKY6 (AT1G62300), WRKY29 (AT4G23550), and WRKY53 (AT4G23810; Fig. 3D; Supplemental Dataset S1; Jaskiewicz et al., 2011). Because chromatin in the 5′ regulatory region of WRKY6, WRKY29, and WRKY53 did not open, or did not open much, in systemic leaves of Local mock plants (Supplemental Fig. S3), the observed formation of open chromatin in the 5′ leader region of a gene is an unlikely response to the physical stress during the inoculation procedure. It rather seems to be a consequence of the local Psm infection.
The expression level and accessibility of chromatin in the coding region or 5′ leader sequence did not significantly change for the housekeeping gene ACTIN2 and the plant defensive gene PDF1.2, respectively (Supplemental Fig. S4). PDF1.2 is epigenetically silenced by Psm-induced SA signaling (Luna et al., 2012) and has a role in the immune response of Arabidopsis to necrotrophic pathogens (Penninckx et al., 1996). Consistent with the constitutive expression of ACTIN2, FAIRE peaks did not differ much in the 5′ upstream and 3′ downstream region of ACTIN2 in untreated systemic leaves of Local mock and Local Psm plants (Supplemental Fig. S5A). In addition, FAIRE peaks were generally low in the 5′ regulatory region of PDF1.2 in untreated systemic leaves of both Local mock and Local Psm plants (Supplemental Fig. S5B). Thus, we conclude that formation of open chromatin in systemic leaves of Local Psm plants and enhanced gene expression upon systemic challenge of those leaves is a specific response of genes that are associated with defense priming and SAR.
Two priming/SAR-related genes with open chromatin near the TSS before systemic challenge and primed expression after the challenge wereFLAVIN-CONTAINING MONOOXYGENASE 1 (FMO1; AT1G19250) and MLO3 (AT3G45290; Fig. 3D; Supplemental Dataset S1). The flavin monooxygenase that is encoded by FMO1 catalyzes the hydroxylation of pipecolic acid to N-hydroxypipecolic acid, which is essential for defense priming and SAR (Hartmann et al., 2018). Consistently, the Arabidopsis fmo1 mutant is SAR deficient (Mishina and Zeier, 2006).
A Novel Positive Regulator of Defense Priming and SAR
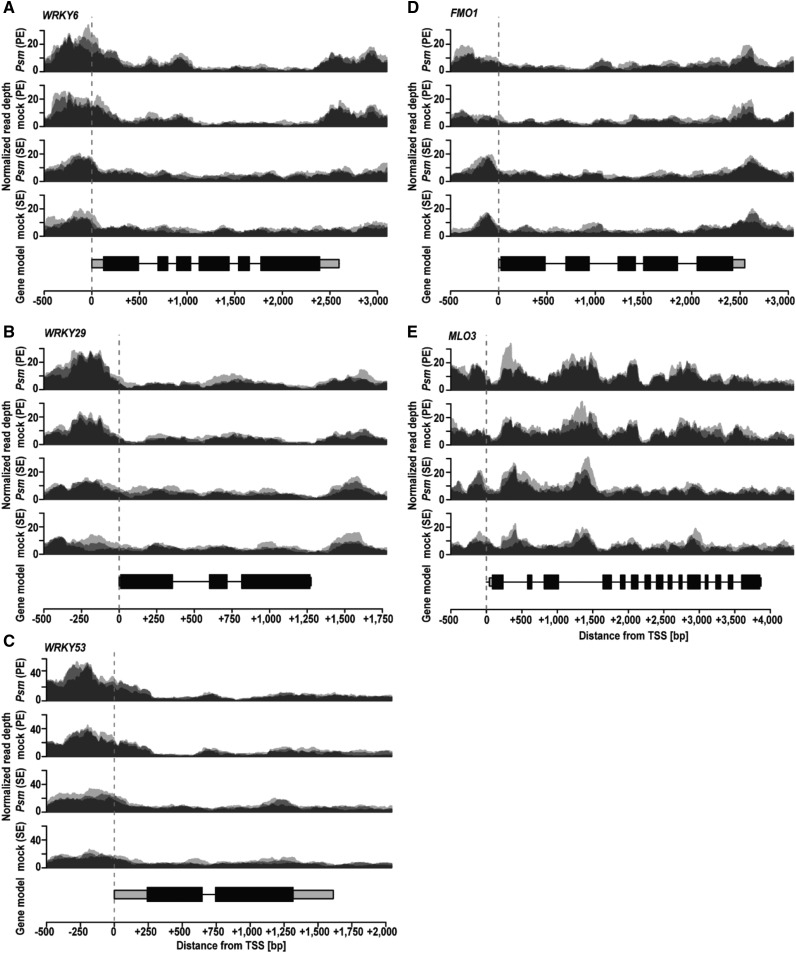
MLO3 belongs to a small family of genes (15 in Arabidopsis; Acevedo-Garcia et al., 2014) in which some members have a presumed suppressing role in plant immunity (Büschges et al., 1997; Acevedo-Garcia et al., 2014). In contrast to WRKY6, WRKY29, and WRKY5, and FMO1, MLO3 has not been associated with defense priming and SAR to date. A critical role in these phenomena was recently assigned to MLO2 (AT1G11310; Gruner et al., 2018), which also has more open chromatin near the TSS during Psm-induced priming and which shows enhanced expression upon systemic challenge (Supplemental Dataset S1). Because of the presumed suppressing role of most MLO genes in plant immunity (Büschges et al., 1997; Acevedo-Garcia et al., 2014), the presence of MLO3 in our FAIRE-seq/RNA-seq dataset (Supplemental Dataset S1) was surprising. Further analysis revealed that FAIRE peaks were generally high in the 5′ leader sequence and in some introns and low in coding regions of WRKY6, WRKY29, and WRKY53, FMO1, and MLO3 (Fig. 4; Supplemental Table S2). The expression level and accessibility of chromatin in the coding region or 5′ leader sequence did not significantly change for the housekeeping gene ACTIN2 or the plant defensive gene PDF1.2, respectively (Supplemental Fig. S4). PDF1.2 is epigenetically silenced by Psm-induced SA signaling (Luna et al., 2012) and has a role in the immune response of Arabidopsis to necrotrophic pathogens (Penninckx et al., 1996). Consistent with the constitutive expression of ACTIN2, FAIRE peaks did not differ much in the 5′ upstream and 3′ downstream regions of ACTIN2 in untreated systemic leaves of Local mock and Local Psm plants (Supplemental Fig. S5A). In addition, FAIRE peaks were generally low in the 5′ regulatory region of PDF1.2 in untreated systemic leaves of both Local mock and Local Psm plants (Supplemental Fig. S5B). Thus, we conclude that formation of open chromatin in systemic leaves of Local Psm plants and enhanced gene expression upon systemic challenge of those leaves is a specific response of genes that are associated with defense priming and SAR.
Figure 4.
Number of FAIRE reads in defense-related genes WRKY6 (A), WRKY29 (B), WRKY53 (C), FMO1 (D), and MLO3 (E) in unchallenged systemic leaves of Local mock and Local Psm plants. The variation in the height of the shading region indicates the sequence coverage levels in three independent biological replicates. Read depth was normalized based on total coverage assigned to the nuclear genome. The broken vertical line indicates the TSS. The numbers on the y axis give the distance from the TSS (in bp). −, upstream of the TSS; +, downstream of the TSS. The experiment was performed with six replicates of each sample (three for single-end and another three for paired-end sequencing). SE, single-end reads; PE, paired-end reads; Psm, Local Psm plants; mock, Local mock plants.
Locus-Specific Validation of Open Chromatin, Enhanced Expression, and Role in SAR
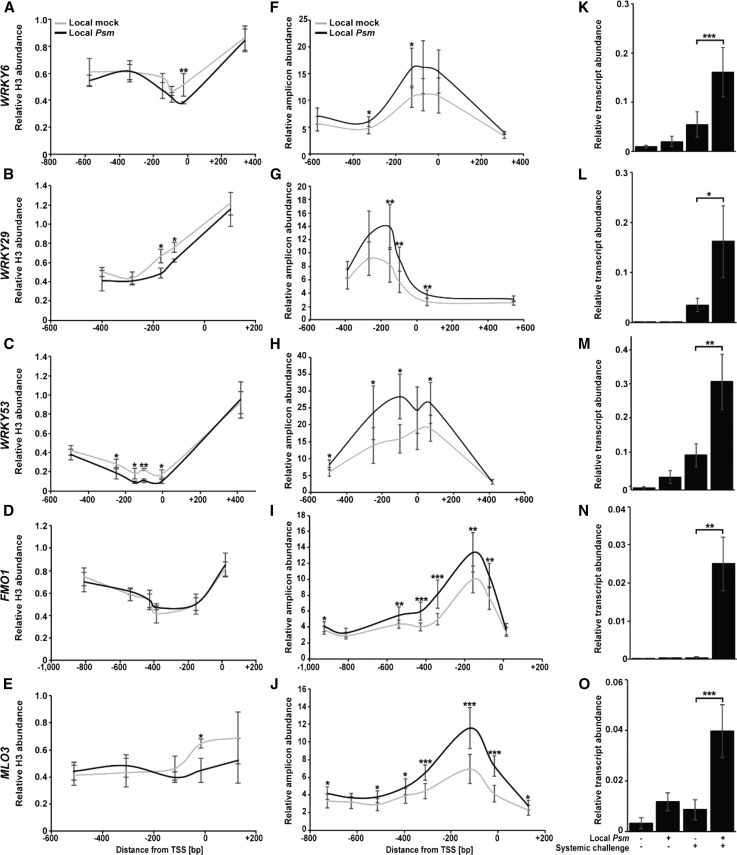
We validated priming-associated histone H3 reduction (Fig. 5, A–E) and open chromatin formation (Fig. 5, F–J) in the 5′ leader sequence of WRKY6, WRKY29, WRKY53, FMO1, and MLO3 by chromatin immunoprecipitation (ChIP; Jaskiewicz et al., 2011) and FAIRE-qPCR (Schillheim et al., 2018), respectively. We could not validate the reduction of histone H3 in the 5′ regulatory region of FMO1, which did not differ significantly between systemic leaves of Local mock and Local Psm plants (Fig. 5D). This indicates lower resolution of the ChIP technique when compared to FAIRE-qPCR (Fig. 5, A–J).
Figure 5.
Reduction of histone H3, formation of open chromatin, and enhanced responsiveness of defense genes in systemic leaves of Local Psm plants. A to E, Reduction of H3 in the 5′ leader sequence and near the TSS of defense genes in untreated systemic leaves of Local mock and Local Psm plants. Error bars represent the SD (n = 6). F to J, Formation of more open chromatin in the same region and in similarly treated plants. Error bars represent the SD (n ≥ 3). K to O, Gene expression upon systemic challenge (+) of Local mock (− Local Psm) and Local Psm (+ Local Psm) plants. Error bars represent the SD (n ≥ 4). ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
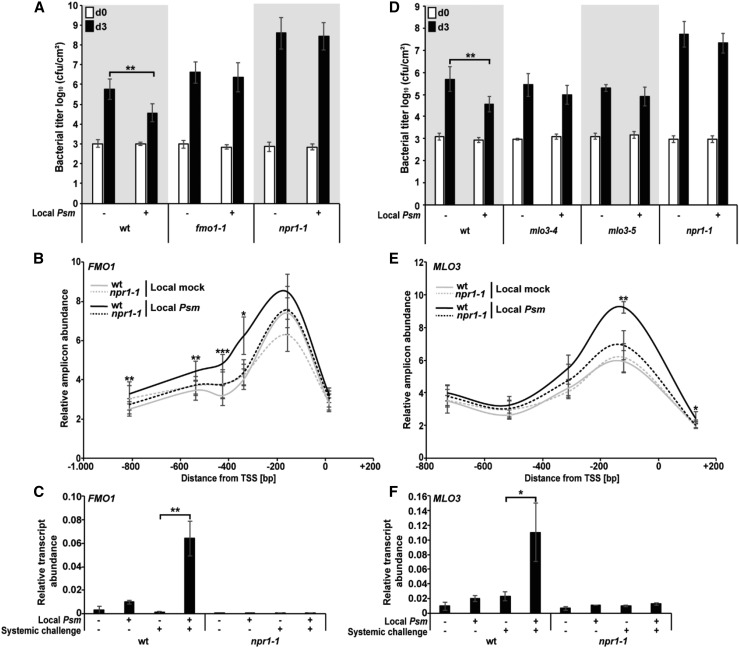
We also confirmed stronger expression of MLO3, FMO1, WRKY6, WRKY29, and WRKY53 upon systemic challenge of Local Psm plants (Fig. 5, K–O). Consistent with a presumed contribution of open chromatin formation and primed defense gene expression to SAR, this systemic immune response was absent in the Arabidopsis fmo1, nonexpressor of pathogenesis-related 1 (npr1), and mlo3 mutants (Fig. 6, A and D). fmo1 and npr1 were SAR defective in all the experiments, while the mlo3 mutant was SAR defective in seven of the nine experiments. These results support a positive role of MLO3 in the SAR response of Arabidopsis (Fig. 6D). However, a recent study reported the presence of SAR in mlo3 plants with local Psm infection (Kusch et al., 2019). The reason for the contradictory result is currently unclear. It may result from a different experimental setup. However, the finding that upon local Psm infection, systemic SA levels rise in the wild type and the npr1 mutant, but not in mlo3 (Supplemental Fig. S6), supports a critical role of MLO3 in SA-mediated defense responses in Arabidopsis.
Figure 6.
Attenuated SAR in fmo1, npr1, and mlo3 and absence of open chromatin and primed FMO1 and MLO3 expression in npr1. A and D, SAR bioassay in wild type (wt) and the fmo1, npr1, and mlo3 mutants. Error bars represent the SD (n = 5). B and E, Reduced promoter opening in systemic leaves of the npr1 mutant upon local mock or local Psm infection. Error bars represent the SD (n = 3). C and F, Absence of primed FMO1 and MLO3 expression in systemic leaves of locally Psm-inoculated npr1 plants. Error bars represent the SD (n = 4). Cfu, colony-forming units; d0, bacterial titer immediately after Psm inoculation (day 0); d3, bacterial titer at 3 d post Psm inoculation; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
The positive role of MLO3 (Fig. 6D) and MLO2 (Gruner et al., 2018) in SAR is particularly interesting. In Arabidopsis, loss-of-function mutation of MLO2 or MLO2, MLO6, and MLO12 results in partial or full immunity to powdery mildew disease (Büschges et al., 1997; Acevedo-Garcia et al., 2014), thus raising the question of the biological meaning of MLO evolution. For MLO2 and MLO3, the answer to this question may be their positive roles in SAR.
In Arabidopsis, loss of function of MLO4 (At1G11000) or MLO11 (At5G53760) causes aberrant root thigmomorphogenesis (Chen et al., 2009; Bidzinski et al., 2014), which is the growth and developmental response of plants to mechanical stimulation. What is more, loss of MLO7 (At2G17430) function leads to pollen tube overgrowth in this plant (Kessler et al., 2010). Consistently, in systemic leaves of Local Psm plants, FAIRE signals were absent for MLO4 and MLO11 and were low for MLO7 (Supplemental Dataset S1). Thus, the discoveries that MLO3 (Figs. 3D, 5O, and 6F) and MLO2 (Gruner et al., 2018) are priming-responsive defense genes and positive regulators of SAR (Fig. 6D; Gruner et al., 2018) are consistent with the different roles of MLO family members in plant biology.
NPR1 is an essential key regulator of defense priming and SAR (Cao et al., 1997; Kohler et al., 2002) and a likely determinant of nucleosome density in the promoter and promoter-proximal region of NPR1-dependent target genes of SA (Singh et al., 2015). Interestingly, the opening of chromatin in the 5′ regulatory regions of FMO1, MLO3, WRKY6, WRKY29, and WRKY53 was impaired in systemic leaves of the Psm-infected SAR-negative npr1 mutant (Fig. 6, B and E; Supplemental Fig. S7). This further suggests a critical epigenetic role of NPR1 in the regulation of those priming-associated genes (Jaskiewicz et al., 2011). Consistently, the expression of FMO1, MLO3, WRKY6, WRKY29, and WRKY53 was not significantly enhanced upon systemic challenge of the npr1 mutant when this genotype had been inoculated before with Psm (Fig. 6, C and F; Supplemental Fig. S7, D–F).
In Silico Identification of Consensus DNA Boxes
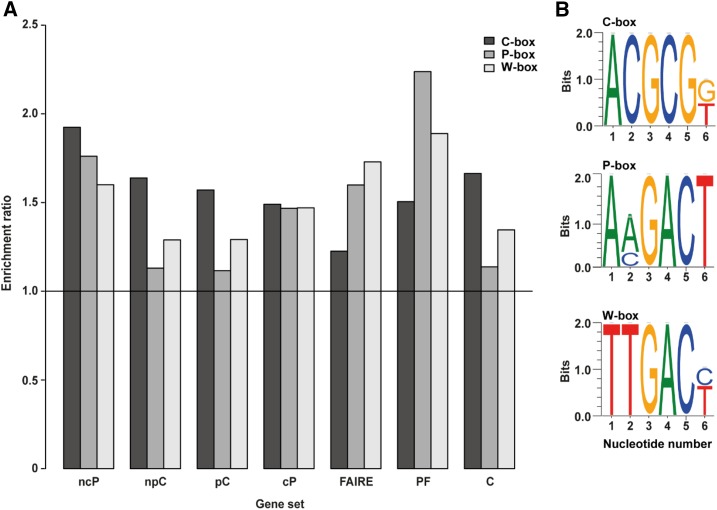
To investigate the governing basis of gene priming, we searched in silico for consensus DNA motifs in the 5′ regulatory genome region of genes that were induced in the various gene sets: ncP (induced during priming in the absence of challenge), npC (induced by systemic challenge in the absence of priming), pC (induced by systemic challenge in primed condition), and cP (induced because of priming in systemic challenge condition; Fig. 3B; Supplemental Dataset S1). In addition to the ncP, npC, pC, and cP data, we included the FAIRE-seq gene set, the priming-associated gene set, PF (ncP-up, cP-up, and FAIRE-up), and the challenge-related gene set, C (npC-up and pC-up) in the analysis (Fig. 7A). The PF set contains mainly those genes with open chromatin in systemic leaves of Local Psm plants (as deduced from the FAIRE-seq dataset) and enhanced expression upon systemic challenge (as deduced from the RNA-seq dataset). The challenge-related gene set, C, contained the genes with expression upon challenge or enhanced expression upon systemic challenge (as deduced from the RNA-seq dataset), independent of priming. We found that three distinct DNA sequence motifs are enriched to different extents in the 5′ regulatory region of genes in these datasets (Fig. 7). One of the sequence motifs is the known W-box (Fig. 7B), a regulatory DNA element that recruits WRKY transcription factors to target genes (Tsuda and Somssich, 2015), including NPR1 (AT1G64280; Yu et al., 2001). W-box was most enriched in the PF and FAIRE sets of genes (Fig. 7A). This finding is consistent with the enrichment of W-box in nucleosome-depleted regions in the 5′ regulatory region of SA-induced Arabidopsis genes (Singh et al., 2015).
Figure 7.
Consensus DNA sequence motifs. A, Enrichment ratio of DNA boxes in the 5′ regulatory region of genes that respond in various immunological conditions (Fig. 3B). FAIRE, increase in FAIRE coverage with priming; PF, genes with open chromatin and enhanced expression upon systemic challenge (ncP-up, cP-up, and FAIRE-up); C, genes with (enhanced) expression upon (systemic) challenge (npC-up and pC-up). The PF dataset contains 439 and the C dataset 4,332 genes. B, C-, P-, and W-box consensus DNA sequences.
The two other consensus sequences, which are novel, seem to be more specific. One of them, which we call P-box (Fig. 7B), was particularly enriched in the 5′ regulatory region of the PF (ncP-up, cP-up, and FAIRE-up) set of priming-associated genes and least present in the npC, pC, and C gene sets (Fig. 7A). The second, which we named C-box (Fig. 7B), was most enriched in the promoter of the priming-specific ncP gene set (Fig. 7A). C-box was also enriched in the npC and C sets of genes and, consistently, least present in the FAIRE-seq gene set (Fig. 7A). Comparative analysis of the RNA-seq and FAIRE-seq datasets suggested that the effect of the P-box on transcription, as reflected in the RNA-seq dataset, might be a consequence of the chromatin opening, as reflected in the FAIRE-seq dataset. The C-box is consistently enriched in all challenge/systemic challenge (RNA-seq) gene sets, but rarely present in the priming (FAIRE-seq) dataset (Fig. 7A). This suggests that P-box governs priming, whereas C-box influences transcription.
Occurrence of C-, P-, and W-Boxes
To complement our bioinformatic analysis, we modeled the change in transcript level and FAIRE peaks for each individual Arabidopsis gene using the copy numbers of C-, P- and W-boxes as explanatory variables (Supplemental Fig. S8). In this analysis, too, the W-box was associated with both gene priming (as seen in the FAIRE dataset) and enhanced transcription (as seen in all RNA-seq datasets, that is, ncP, cP, npC, and pC), whereas C- and P-boxes showed more specialization. Presence of the C-box was associated with a major change in gene expression after systemic challenge of Local mock or Local Psm plants (npC and pC datasets), whereas P-box was particularly associated with priming and enhanced expression after systemic challenge (ncP and cP datasets). Nonetheless, considerable complexity was observed, with presence of the C-box and multiple copies of the P-box being substantially enhanced in all sets of genes that respond in the physiological conditions assayed in this work.
Next, we further investigated the relationship between the presence of the above-mentioned DNA boxes in the 5′ leader regions of genes and (enhanced) transcription upon challenge. To do so, we selected eight representative priming/SAR-associated genes (that is, genes with open chromatin in systemic leaves of Local Psm plants, enhanced expression upon systemic challenge, and a known or presumed role in SAR) for motif analysis in their 5′ regulatory region. The promoter of these genes contained up to two P- or W-boxes or mixes of these boxes (Table 1), further suggesting that these discrete DNA-sequence motifs or combinations of these motifs, in the 5′ leader sequence could determine whether expression of a given gene will be unprimed or primed. Notably, DNA-binding protein HsfB1, which is required for defense priming and SAR (Pajerowska-Mukhtar et al., 2012; Pick et al., 2012) and which specifically binds to cis-element translocon 1 (TL1; GAAGAAGAA; Pajerowska-Mukhtar et al., 2012), has a P-box in its 5′ regulatory region (Table 1). Similarly, the promoter of the Arabidopsis ascorbate peroxidase (APX) 2 gene, which has two P-boxes in its 5′ regulatory region, confers transcriptional memory to a heterologous luciferase reporter gene (Liu et al., 2018). Moreover, priming marker genes WRKY6, WRKY29, and WRKY53, whose protein products bind to W-box in the promoter of target genes (Tsuda and Somssich, 2015), have two copies of the P-box, two copies of the W-box, and two copies of the P- and W-box, respectively, in their 5′ regulatory regions (Table 1). Furthermore, the promoter of MLO3, which has been identified as a positive regulator of SAR in this work (Fig. 6D), harbors both a P- and a W-box (Table 1). Together, these findings point to P- and W-boxes as putative cis-regulatory DNA motifs with a role in defense priming. Consistently, the Arabidopsis NPR1 gene has three copies of the W-box in its 5′ regulatory region that are recognized by SA-induced WRKY transcription factors (Yu et al., 2001). Mutations in these W-boxes abolished their recognition by WRKY transcription factors, rendered the promoter unable to activate a downstream β-glucuronidase reporter gene, and compromised the ability of NPR1 to complement the npr1 mutant for SA-induced defense gene expression and SAR (Yu et al., 2001). The detected enrichment of W-box in the 5′ leader sequence of genes in the priming (PF) and systemic challenge (C) datasets illustrates the power of our study to identify putative regulatory DNA motifs in the regulatory region of genes with a role in priming and SAR.
Table 1. Occurrence of DNA boxes in the 5′ regulatory region of selected Arabidopsis genes associated with the biotic and abiotic stress responses.
AGI, Arabidopsis Genome Initiative; NIMIN1, noninducible immunity 1-interacting 1 gene; PR1, pathogenesis-related 1 gene; HSFB1, heat shock transcription factor HsfB1 gene.
| AGI code | Name | C-Box | P-Box | W-Box |
|---|---|---|---|---|
| AT1G62300 | WRKY6 | 0 | 2 | 0 |
| AT4G23550 | WRKY29 | 0 | 0 | 2 |
| AT4G23810 | WRKY53 | 0 | 2 | 2 |
| AT1G19250 | FMO1 | 0 | 0 | 1 |
| AT3G45290 | MLO3 | 0 | 1 | 1 |
| AT1G02450 | NIMIN1 | 1 | 2 | 1 |
| AT1G14610 | PR1 | 0 | 1 | 0 |
| AT4G36990 | HSFB1 | 0 | 1 | 0 |
Our genome-wide in silico analysis disclosed a correlation between the copy number of P-box in the 5′ regulatory region of a gene and priming (Supplemental Fig. S8). However, whether the presence of P-box, either alone or in combination with W-box, indeed confers gene priming, how it does so, and whether other DNA sequences in gene-regulatory regions may influence its activity, remains to be seen. Regulation by additional, so far unknown, DNA sequence features is likely, because FMO1, which in the primed state has open chromatin in the 5′ regulatory region (Figs. 3D, 4D, and 5I), enhanced expression upon systemic challenge (Fig. 5N), and an important role in defense priming and SAR (Mishina and Zeier, 2006; Hartmann et al., 2018), contains a W-box, but no P-box, in its 5′ leader sequence (Table 1). Intriguingly, when we performed a genome-wide analysis for the presence of C-, P-, and W-boxes in the 5′ promoter region of genes in 62 species of diverse plant families, we found that the three DNA-boxes were mainly present in the Brassicaceae family of plants (Supplemental Fig. S9). They were less abundant in other families of eudicot or in monocotyledonous plants and seem to be absent from mosses and algae (Supplemental Fig. S9). This suggests either specific evolution of the C-, P-, and W-boxes in the Brassicaceae or loss of these DNA motifs in essentially all species except those in the Brassicaceae family.
CONCLUSIONS
We used FAIRE-seq to provide a genome-wide map of regulatory DNA elements in defense priming and SAR in Arabidopsis. We identified 10,129 priming/SAR-associated sites of open chromatin in the 5′ leader region of 3,025 genes. Many of them included at least one P- or C-box, which we identified in silico as putative regulatory DNA sequences of priming and SAR. P-box was mainly, although not exclusively, associated with priming, whereas C-box was preferably, but not solely, linked to (systemic) challenge. The presence of P- or W-box, or combinations of these boxes, seems to be associated with gene priming. A scan of our list of genes with priming-specific open chromatin in the 5′ upstream and 3′ downstream regulatory regions and enhanced expression upon challenge disclosed MLO3 as a novel positive regulator of defense priming and SAR. We expect that our datasets will enable the identification of additional novel loci important to priming and systemic immunity and help to further elucidate the biology of these processes.
MATERIALS AND METHODS
Growth of Arabidopsis Plants
Arabidopsis (Arabidopsis thaliana) wild-type accession Columbia-0 and the npr1-1 (At1g64280), fmo1-1 (SALK_026163), mlo3-4 (SALK_027770), and mlo3-5 (SALK_116848) mutants (all in Columbia-0 genetic background) were grown on soil in short-day conditions (8 h light, 100 µmol m−2 s−1) at 20°C.
Cultivation of Bacteria
Psm strain ES4326 was grown on King‘s B medium (20 g/L tryptone, 10 mL/L glycerol, 1.5 g/L K2HPO4, and 1.5 g/L MgSO4; King et al., 1954) supplemented with 100 μg/mL streptomycin and 10 g/L agar. After incubation for 2 d at 28°C, four to five colonies were transferred to a 250-mL flask containing 50 mL King’s B medium supplemented with 100 μg/mL streptomycin. The flask was incubated overnight at 28°C at 220 rpm on a rotary shaker. The next day, the bacterial culture was transferred to a 50-mL plastic tube and centrifuged at 1,800g and 16°C for 8 min. The supernatant was discarded and the pellet suspended in 50 mL of 10 mm MgCl2. After centrifugation at 1,800g and 16°C for 8 min, the pellet was suspended in 50 mL of 10 mm MgCl2. One mL of bacterial suspension was diluted with 10 mm MgCl2 to an OD600 of 0.0002, resulting in ∼3 × 108 colony-forming units (cfu)/mL.
Plant Treatment
Using a syringe without a needle, three leaves of 4- to 5-week-old Arabidopsis plants were infiltrated with 10 mm MgCl2 (Local mock) or ∼3 × 108 cfu/mL Psm in 10 mm MgCl2 (Local Psm; for the experimental setups, see Figs. 1A and 2A). For open chromatin analysis, we harvested the whole rosette of leaves that were younger than the youngest Local mock or Local Psm leaf at the indicated times and subjected them to FAIRE-seq or FAIRE-qPCR as described below. For gene expression analysis, two systemic leaves per plant and treatment were left untreated (no systemic challenge) or challenged by infiltration of tap water at the indicated times after the initial treatment (systemic challenge). Systemic leaves were harvested at 2.5 h after the systemic challenge and subjected to RNA-seq or RT-qPCR analysis as described below.
SAR Assay
Using a syringe without a needle, three leaves of 4- to 5-week-old Arabidopsis plants were infiltrated with ∼3 × 108 cfu/mL Psm in 10 mm MgCl2 or with 10 mm MgCl2 only (control). After 72 h, two distal leaves of each plant were infiltrated with Psm (∼3 × 108 cfu/mL) in 10 mm MgCl2 (systemic challenge). After another 72 h, leaf discs (0.5 cm diameter) were punched out of inoculated systemic leaves and homogenized in 10 mm MgCl2. A serial dilution of the homogenate was plated on King’s B agar (see “Cultivation of Bacteria”) and developing colonies counted after incubation for 48 h at 28°C. For determining the duration of SAR (Fig. 1E), the challenge infection of distal leaves was done 3–7 d after the initial inoculation, because it takes 1–2 d for Psm-induced SAR to develop (Gruner et al., 2018). Bacterial titer in systemic leaves was determined by plating serial dilutions as described above (see “Cultivation of Bacteria”).
ChIP
ChIP was performed as described (Jaskiewicz et al., 2011). For immunoprecipitation of histone H3, 1 µg of a histone H3-specific antibody (ab1791, Abcam) was used per sample. Relative DNA enrichment was quantified by qPCR using gene-specific primers (Supplemental Dataset S3), related to input, and normalized to the level of the coding sequence of the ACTIN2 gene.
Analysis of Gene-Specific mRNA Transcript Abundance by RT-qPCR
RNA was isolated from frozen leaves using the TRIZOL method (Chomczynski, 1993). Two µL of RNA were subjected to DNase (Thermo Fisher Scientific) digestion followed by cDNA synthesis using RevertAid reverse transcriptase (Thermo Fisher Scientific). mRNA transcript abundance was determined by RT-qPCR on an ABI Prism 2300 sequence detector system (Applied Biosystems) in a 96-well format or on a Light Cycler 480 (Roche) with a 384-well format using gene-specific primers (Supplemental Dataset S3) and SYBR Green fluorescence (Platinum SYBR Green qPCR Mix, Thermo Fisher Scientific). Data were normalized to the mRNA transcript level of ACTIN2.
RNA-Seq Analysis
Arabidopsis plants (4 to 5 weeks old) were subjected to Local mock or Local Psm treatment (see “Plant Treatment” and Fig. 1A). After 72 h, distal leaves were left untreated or were challenged by infiltration of tap water. After 2.5 h, the remote leaves were collected and analyzed for transcript abundance. In total, we processed 12 samples (four treatments [Local mock and Local Psm, each with and without systemic challenge], with three replicates; each sequenced replicate represented nine leaves [three leaves per plant of three plants]). RNA was extracted as described (see “Analysis of Gene-Specific mRNA Transcript Abundance”) followed by concentration of the RNA using the RNA Clean & Concentrator Kit (Zymo Research). The concentration step included an on-column DNase digestion according to the manufacturer’s instructions (Zymo Research). cDNA library preparation and sequencing were performed at the Genomics Core Facility of the European Molecular Biology Laboratory using the standard Illumina sequencing library kit (TruSeq RNA v1 kit) according to the manufacturer’s instructions (Illumina). Samples were sequenced on an Illumina HiSeq 2000 platform using 50 bp single-end reads, resulting in ∼64.9 million reads totaling 32.48 billion bases. For the read counts and alignment statistics of the next-generation sequencing datasets, see Supplemental Dataset S4.
Analysis of RNA-Seq Data
The 12 RNA-seq samples (see “RNA-Seq Analysis”) were preprocessed using Trimmomatic (v0.32; Bolger et al., 2014) with the recommended settings for single-end data, but replacing the sliding window trimmer with the maximum information trimmer (MAXINFO:40:0.4). The resulting files were aligned using the Burrows-Wheeler Alignment tool (v0.5.9-r16; Li and Durbin, 2009) against the representative nuclear gene models of the Arabidopsis genome (TAIR10; Berardini et al., 2015). Resulting counts were extracted using Samtools (v0.0.18; Li et al., 2009) and a custom script. The count table was analyzed using EdgeR (Robinson et al., 2010), comparing adjacent conditions, as well as the control versus combined-treatment condition, to determine significantly activated or repressed genes at a false discovery rate threshold of 0.05.
FAIRE-Seq Analysis
Arabidopsis wild-type (accession Columbia-0) plants, 4 to 5 weeks old, were subjected to Local mock or Local Psm treatments (see “Plant Treatment” and Fig. 2A). At 72 h post-treatment, the whole rosettes of remote leaves of 20 or more plants per treatment were collected in a 120-mL plastic tube. The tube was filled to 80 mL with crosslinking buffer (10 mm HEPES, pH7.8, 400 mm Suc, 5 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride (PMSF), and 3% [v/v] formaldehyde) and subjected to successive vacuum infiltration for 1, 1.5, and 1 min. Excessive formaldehyde was quenched by addition of freshly prepared Gly to a final concentration of 125 mm followed by vacuum infiltration for 1, 1.5, and 1 min. Leaves were transferred to a beaker and thoroughly washed with tap water. After drying between paper towels on the bench, leaves were quick frozen in liquid nitrogen. Frozen leaves were ground to a fine powder using a mortar and pestle. The powder was split into 10-mL aliquots in 50-mL plastic tubes, and 35 mL nuclei extraction buffer (10 mm HEPES, pH 7.8, 10 mm KCl, 10 mm MgCl2, 250 mm Suc, 5 mm β-mercaptoethanol, and 0.2 mm PMSF) was added to each tube, followed by incubation at 4°C for 20 min on a rotary plate. The mixture was subsequently filtered through four and eight layers of miracloth. After centrifugation for 15 min at 4°C and 2,880g, the supernatant was discarded and the pellet was suspended in 500–700 µL nuclei extraction buffer. Adequate samples were merged in a fresh 2-mL test tube. After centrifugation for 10 min at 4°C and 12,000g, the supernatant was discarded. The pellet was suspended in 900 µL of nuclei extraction buffer and centrifuged for 10 min at 4°C and 1,500g. This washing step was repeated three times. The pellet was then suspended in 700 µL Suc buffer (1.7 m Suc, 10 mm HEPES, pH 7.8, 0.15% [v/v] Triton X-100, 2 mm MgCl2, 5 mm β-mercaptoethanol, and 0.1 mm PMSF). One mL Suc buffer was transferred to a fresh 2-mL test tube and overlaid with the suspended pellet. After centrifugation for 1 h at 4°C and 16,000g, the supernatant was discarded and the pellet washed and suspended in washing buffer (10 mm HEPES, pH7.8, 10 mm KCl, 10 mm MgCl2, 250 mm Suc, 1% [v/v] Triton X-100, 5 mm β-mercaptoethanol, and 0.2 mm PMSF). The sample was transferred to a fresh 1.5-mL microfuge tube and centrifuged and the pellet was washed three times for 10 min at 4°C and 1,500g. The supernatant was removed and the pellet dissolved in 850 µL of extraction buffer (100 mm Tris-HCl, pH8.0, 100 mm NaCl, and 50 mm EDTA, pH 8.0). The suspension was transferred to a fresh 1.5-mL test tube and sonicated 10 times using high-voltage pulses (30 s on, 30 s off). Cell debris was removed by centrifugation for 5 min at room temperature and 16,100g. Then, 700 µL of the supernatant was transferred to a fresh 1.5-mL tube and 80 µL of the supernatant was mixed with 540 µL of extraction buffer and incubated at 65°C overnight to reverse formaldehyde crosslinks (input control). The remaining supernatant was kept frozen at −20°C overnight. The next day, both samples were centrifuged at room temperature for 5 min at 16,100g. Then, 550 µl µL of the supernatant was supplemented with the same volume of phenol-chloroform, mixed, and centrifuged for 15 min at 4°C and 20,800g, and 350 µL of the aqueous phase was transferred to a fresh tube. After addition of one volume chloroform, samples were mixed and centrifuged as before. DNA in 200 µL of the supernatant was precipitated by addition of two volumes ice-cold 96% (v/v) ethanol and incubation at −20°C for 30 min. DNA was collected by centrifugation for 20 min at 4°C and 20,800g. The pellet was washed with 70% (v/v) ethanol and dried on the bench for 1 h. The DNA pellet was suspended in 200 µL of deionized water and incubated at 70°C for 15 min. For sequencing, samples were successively treated with RNase (10 mg/mL; Qiagen) for 45 min at 25°C and with proteinase (20 mg/mL; Life Technologies) for 60 min at 55°C. After overnight incubation at 65°C, DNA was purified with a DNA & Concentrator Kit (Zymo Research).
The first group of six FAIRE samples (three replicates of systemic leaves each of 20 or more Local mock and Local Psm plants; experiment #1) was sequenced at the Genomics Core Facility of the EMBL. Sequencing was done in two rounds, using 50 bp single-end reads on an Illumina HiSeq 2000 platform, from a single library preparation using NEBNext ChIP-Seq Library Prep Master Mix Set (New England Biolabs) for each sample. The initial sequencing run produced ∼95 million reads totaling 4.76 billion bases. It is referred to as dataset FAIRE-1A. The subsequent run produced 248 million reads totaling 12.66 billion bases. It is referred to as dataset FAIRE-1B.
The DNA libraries of the second group of six FAIRE samples (three replicates of systemic leaves each of 26 or more Local mock and Local Psm plants; experiment #2) were prepared with the TruSeq ChIP Sample Preparation kit and sequenced at FASTERIS SA. Sequencing was performed using 125 bp paired-end sequencing on an Illumina HiSeq 2500 to enable explicit detection of the DNA fragment size. This sequencing run produced ∼548 million reads, totaling 68.5 billion bases. It has been referred to as dataset FAIRE-2. For the read counts and alignment statistics of the next-generation sequencing datasets, see Supplemental Dataset S4.
Analysis of FAIRE-Seq Data
The FAIRE-1A dataset was preprocessed using Trimmomatic as described above (see “Analysis of RNA-Seq Data”), and then aligned to the Arabidopsis genome (TAIR10; Berardini et al., 2015) using the Burrows-Wheeler Alignment tool (v0.5.9-r16; Li and Durbin, 2009). The resulting alignments were in silico extended to 200 bp, and coverage was calculated for the promoter region of each protein-coding gene, covering from 250 bp upstream to the TSS, using custom scripts. This coverage was then normalized by dividing by the total coverage genome wide, to allow for a differing overall dataset size of each sample.
When this process was repeated for the FAIRE-1B dataset, it became apparent that the resulting coverage patterns differed substantially from the earlier FAIRE-1A dataset for these samples. Since the prepared libraries had been stored for several months between the sequencing runs, it was suspected that there might have been some degradation, which reduced the relative content of longer fragments. To minimize artifacts from this issue, the in-silico extension of the FAIRE-1B dataset was reduced to 100 bp, which resulted in a coverage pattern that was more similar to that of the FAIRE-1A dataset. The data of corresponding samples in the FAIRE-1A and FAIRE-1B datasets were then combined.
The FAIRE-2 dataset was processed in a similar manner to the FAIRE-1A/1B datasets, with the following changes: Paired-end settings were used for preprocessing and alignment, and the in silico read extension was done by extending each read as far as its corresponding mate, resulting in a more accurate estimate of each fragment.
Transcript-Based Assessment of FAIRE-Seq Data
Based on results of pilot FAIRE-qPCR analyses, the region from 400 bp upstream to 200 bp downstream of the TSS was determined to be most indicative of defense priming. Therefore, the total normalized coverage of this region was determined for each mRNA transcript in each FAIRE sample. Inspection of these coverage values indicated that while their distribution was not perfectly Gaussian, fitting them to a Gaussian distribution gave a residual sum of squares value of 0.9965. This value, and an additional manual inspection of both the distribution histogram and quantile-quantile plot against a normal distribution indicated that a parametric test could safely be used. To determine whether the treatment had a significant effect, the samples were grouped according to treatment versus control and FAIRE-seq experiment #1 versus FAIRE-seq experiment no. 2. They were then analyzed using two-way ANOVA, modeling the observed coverage using treatment versus control and experiment as explanatory variables. Those transcripts that had an F-statistic for the control treatment versus treatment factor with P < 0.05 were considered significant, with the sign of the coefficient indicating the direction of change (more open versus less open). A gene model was considered significant if any transcript isoform was significant. To enable comparison of the FAIRE-seq and RNA-seq data, only those genes with detected expression in at least one RNA sample were retained.
Whole-Genome Assessment of FAIRE-Seq Data
To determine the positional relationship between FAIRE-seq signals and the coding regions of the genome, a genome-wide assessment of the FAIRE-seq data was also performed. A sliding window approach was used, with 600-bp window size, with the window moving by 100 bp for each assessment. Each window was classified, based on its center position, as within a transcript, within 500 bp upstream of a transcript, or within 500 bp downstream of a transcript. Windows that fell within the transcribed, upstream, or downstream regions of multiple transcripts were assigned accordingly to each transcript. Regions not falling into any of these three categories were considered intergenic. The significance of a region was assessed as before, using two-way ANOVA and an F-statistic cutoff of P < 0.05.
FAIRE-qPCR
FAIRE-qPCR data are from three biological replicates. Each replicate consisted of two leaf samples that were untreated, systemic leaves of Local Psm or Local mock plants. Three aliquots were used per leaf sample and processed simultaneously. For each aliquot, two spatula tips of frozen, ground leaf material (∼80 mg) were suspended in 850 µL of DNA extraction buffer (100 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 50 mm EDTA, pH 8.0). Samples were shaken at 25°C for 3 min, followed by sonication as described (see “FAIRE-Seq Analysis”). After centrifugation for 5 min at 16,100g and at room temperature, supernatants of the three aliquots of a same sample were pooled in a 50-mL plastic tube. Three 700-µL aliquots of pooled supernatant were transferred to a fresh 1.5-mL tube, and 80 µL of the supernatant was mixed with 540 µL of extraction buffer and incubated at 65°C overnight to reverse formaldehyde crosslinks (input control). The remaining supernatant was kept frozen at −20°C overnight. DNA was extracted as described for FAIRE-seq. Relative abundance of DNA fragments was determined by qPCR using site-specific primers (see Supplemental Material) relative to input and normalized to the level of a coding sequence of ACTIN2.
Promoter Motif Analysis
Promoter motif analysis was performed on the significantly activated genes from the four RNA-seq comparisons, plus the genes showing significant opening in the FAIRE-seq analysis, to determine whether these sets were enriched for specific DNA sequence motifs. To increase the reliability, two additional sets, focusing on the shared priming-responsive genes (intersection between the two “priming effect” RNA-seq activated sets and the FAIRE “opening” sets) and the shared challenge-responsive genes (intersection between the two “challenge effect” RNA-seq activated sets) were created. The complete set of genes that showed expression in any RNA-seq dataset was used as background.
For each gene, the region from 500 bp upstream of the TSS until the TSS was extracted and the canonical 6-mer and 8-mer content was determined. Each gene set was then analyzed to determine the degree of enrichment for each 6-mer for that set versus the background gene set. In both the priming-responsive and challenge-responsive sets, multiple 6-mers corresponding to the well-known W-box (WRKY-binding motif) were found within the top 10 most enriched motifs. It was, however, possible to find an additional hypothetical motif for each 6-mer list, which could explain the top four entries. These motifs, referred to as the P-box and C-box, were then checked to determine whether their presence in a promoter sequence could be associated with a stronger response in the individual RNA and FAIRE comparisons. For the W-box and P-box tests, the number of copies in each promoter region was considered as a four-way factor, corresponding to 0 copies, one copy, two copies, and three or more copies. Since the C-box motif is relatively rare, it was tested using a two-way factor, corresponding to 0 copies and 1 or more copies. A Wilcoxon rank-sum test was used to determine whether the distribution of estimated change in expression (for RNA-seq) or coverage (for FAIRE) differed between genes containing no copies of the motif and those genes containing either any number of copies (for challenge) or specifically one, two, or three or more copies (for priming/WKRY).
MapMan Over-Representation Analysis
The sets of genes induced or repressed in each differential expression comparison (ncP, npC, cP, and pC) and those assessed as more open in the FAIRE-seq analysis (FAIRE) were tested for over-representation within top-level MapMan bins (Thimm et al., 2004; Usadel et al., 2009; Bolger et al., 2018) using Fisher’s exact test (Supplemental Dataset S2). The resulting P-values were corrected for multiple testing using the so-called Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995).
Analysis of C-, P-, and W-Box Presence in the 5′ Leader Region of Genes in Different Plant Species
The genomes and GFF3 structural annotations of 62 unrestricted plant genomes were downloaded from Phytozome v12.1.5 (http://www.phytozome.net). Reciprocal best BLAST, with an e-value cutoff of 1e-20, was used to determine the most likely 1:1 putative ortholog between Arabidopsis and genes in each other plant species. Each putatively orthologous gene pair was assessed to determine whether the absence or presence of the promoter motif of interest (P-, C-, or W-box, or all their possible 6-mers) in a 500-bp window upstream of the TSS was conserved between the species pairs. The total conservation of each promoter motif per species pair was then calculated using Fisher's exact test.
Statistical Analysis
All experiments were repeated at least three times. Normal distribution was assumed for all statistical analyses. The unpaired Student´s t test (two-sided) was applied using Sigma Stat (Systat Software) to determine whether the observed differences were statistically significant. If the unpaired Student’s t test was inappropriate, the Wilcoxon rank-sum test was used to confirm the statistical analysis. Changes were considered statistically significant when P ≤ 0.05.
Accession Numbers
The Arabidopsis Genome Initiative accession numbers for the genes and gene products mentioned in this article are as follows: At1G62300 (WRKY6), At4G23550 (WRKY29), At4G23810 (WRKY53), At1G19250 (FMO1), At1G11310 (MLO2), At3G45290 (MLO3), At1G11000 (MLO4), At1G61560 (MLO6), At2G17430 (MLO7), At5G53760 (MLO11), At2G39200 (MLO12), At1G64280 (NPR1), At5G44420 (PDF1.2), At3G09640 (APX2), and At3G18780 (ACTIN2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Number of genes in the four RNA-seq datasets and the FAIRE dataset.
Supplemental Figure S2. Assignment to top-level MapMan bins of genes in the four RNA-seq datasets and the FAIRE dataset.
Supplemental Figure S3. Local mock inoculation does not enhance, or only slightly enhances, chromatin opening or WRKY6, WRKY29, WRKY53 responsiveness in systemic leaves.
Supplemental Figure S4. In systemic leaves of Local mock or Local Psm plants, the expression level and chromatin state in the coding region or 5′ leader sequence did not differ for ACTIN2 or PDF1.2, respectively.
Supplemental Figure S5. Number of FAIRE reads in unchallenged systemic leaves of Local mock and Local Psm plants.
Supplemental Figure S6. SA level in wild type, npr1, and mlo3 plants.
Supplemental Figure S7. Formation of open chromatin in the 5′ regulatory region and priming for enhanced expression of WRKY6, WRKY29, WRKY53 are absent in npr1.
Supplemental Figure S8. Relationship between occurrences of DNA boxes in the 5′ regulatory region of gene and the transcriptional and FAIRE response to treatment.
Supplemental Figure S9. Presence or absence of P-, C-, and W-Boxes in putatively orthologous genes of 62 plant species compared to Arabidopsis.
Supplemental Table S1. Sites of more open and less open chromatin in systemic leaves of Local Psm plants as categorized by genomic region.
Supplemental Table S2. FAIRE signals in the coding region of primed (WRKY6, WRKY29, and WRKY53) and an unprimed (PDF1.2) defense gene.
Supplemental Dataset S1. FAIRE-seq and RNA-seq datasets.
Supplemental Dataset S2. Assignment of Arabidopsis genes responding in different immunological conditions to top-level MapMan bins.
Supplemental Dataset S3. Primers used in this study.
Supplemental Dataset S4. Read counts and alignment statistics of the next-generation sequencing datasets.
Acknowledgments
We thank X. Dong for making us aware of FAIRE, and we appreciate valuable discussions with, and help and support from, F. Leissing, G.J.M. Beckers, M.R. Jaskiewicz, and C.J. Langenbach. R. Panstruga is thanked for providing Arabidopsis seeds.
Footnotes
This work was supported by a German Research Foundation grant (CO 186/13-1) and by the Excellence Initiative of the German federal and state governments.
Articles can be viewed without a subscription.
References
- Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol 204: 273–281 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300 [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Döring A-C, Gruner K, Schuck S, Bräutigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidzinski P, Noir S, Shahi S, Reinstädler A, Gratkowska DM, Panstruga R (2014) Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant Cell Environ 37: 2738–2753 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger ME, Arsova B, Usadel B (2018) Plant genome and transcriptome annotations: From misconceptions to simple solutions. Brief Bioinform 19: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim Biophys Acta 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. (1997) The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chen Z, Noir S, Kwaaitaal M, Hartmann HA, Wu MJ, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM (2009) Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 21: 1972–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 532–534, 536–537 [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53: 97–119 [DOI] [PubMed] [Google Scholar]
- Dempsey PW, Vaidya SA, Cheng G (2003) The art of war: Innate and adaptive immune responses. Cell Mol Life Sci 60: 2604–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB (2002) Histone acetylation: A switch between repressive and permissive chromatin. EMBO Rep 3: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furci L, Jain R, Stassen J, Berkowitz O, Whelan J, Roquis D, Baillet V, Colot V, Johannes F, Ton J (2019) Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. eLife 8: e40655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamir J, Darwiche R, Van’t Hof P, Choudhary V, Stumpe M, Schneiter R, Mauch F (2017) The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J 89: 502–509 [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. (2010) A map of open chromatin in human pancreatic islets. Nat Genet 42: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD (2009) Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods 48: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Zeier T, Aretz C, Zeier J (2018) A critical role for Arabidopsis MILDEW RESISTANCE LOCUS O2 in systemic acquired resistance. Plant J 94: 1064–1082 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T, et al. (2018) Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173: 456–469.e16 [DOI] [PubMed] [Google Scholar]
- Henikoff S. (2008) Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9: 15–26 [DOI] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhänsel C (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K (2004) Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell 13: 33–43 [DOI] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Kohler A, Schwindling S, Conrath U (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch S, Thiery S, Reinstädler A, Gruner K, Zienkiewicz K, Feussner I, Panstruga R (2019) Arabidopsis mlo3 mutant plants exhibit spontaneous callose deposition and signs of early leaf senescence. Plant Mol Biol [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-C, Lämke J, Lin SY, Hung MJ, Liu KM, Charng YY, Bäurle I (2018) Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J 95: 401–413 [DOI] [PubMed] [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J (2012) Next-generation systemic acquired resistance. Plant Physiol 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, van der Meer JWM (2016) Adaptation and memory in innate immunity. Semin Immunol 28: 317–318 [DOI] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, Dong X (2012) The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick T, Jaskiewicz M, Peterhänsel C, Conrath U (2012) Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol 159: 52–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillheim B, Jansen I, Baum S, Beesley A, Bolm C, Conrath U (2018) Sulforaphane modifies histone H3, unpacks chromatin, and primes defense. Plant Physiol 176: 2395–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Bag SK, Bhardwaj A, Ranjan A, Mantri S, Nigam D, Sharma YK, Sawant SV (2015) Global nucleosome positioning regulates salicylic acid mediated transcription in Arabidopsis thaliana. BMC Plant Biol 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Tateda C, Zhang Z, Shrestha J, Jelenska J, Chinchilla D, Greenberg JT (2014) Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26: 4171–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE (2015) Transcriptional networks in plant immunity. New Phytol 206: 932–947 [DOI] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ 32: 1211–1229 [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]