Nod Factor induces an atypical receptor kinase, RINRK1, which specifically regulates infection thread formation in Lotus japonicus.
Abstract
During the legume-rhizobium symbiotic interaction, rhizobial invasion of legumes is primarily mediated by a plant-made tubular invagination called an infection thread (IT). Here, we identify a gene in Lotus japonicus encoding a Leu-rich repeat receptor-like kinase (LRR-RLK), RINRK1 (Rhizobial Infection Receptor-like Kinase1), that is induced by Nod factors (NFs) and is involved in IT formation but not nodule organogenesis. A paralog, RINRK2, plays a relatively minor role in infection. RINRK1 is required for full induction of early infection genes, including Nodule Inception (NIN), encoding an essential nodulation transcription factor. RINRK1 displayed an infection-specific expression pattern, and NIN bound to the RINRK1 promoter, inducing its expression. RINRK1 was found to be an atypical kinase localized to the plasma membrane and did not require kinase activity for rhizobial infection. We propose RINRK1 is an infection-specific RLK, which may specifically coordinate output from NF signaling or perceive an unknown signal required for rhizobial infection.
During the initiation of symbiotic nitrogen-fixation in legumes, signaling receptor complexes perceive nodulation factors (NFs) synthesized by rhizobia, which induce and coordinate rhizobial infection and nodule organogenesis (Oldroyd and Downie, 2008). NFs are lipochitin-oligosaccharides (LCOs), which, in different rhizobia, carry different acyl chains (C16-20) with variable degrees of saturation together with different substitutions on the oligosaccharide backbone (Lerouge et al., 1990; Spaink et al., 1991; Cullimore et al., 2001; Bek et al., 2010). These substitutions are determinants of the host-specific interactions between a given rhizobial strain and its host legume (Lerouge et al., 1990; Spaink et al., 1991).
In Lotus japonicus, NFs made by Mesorhizobium loti are perceived by a receptor complex containing NFR1 and NFR5 (Nod factor receptors 1 and 5; Madsen et al., 2003; Radutoiu et al., 2003). NFR1 and NFR5 encode LysM receptor kinases that are located in the plasma membrane, interact with each other, and directly bind to NFs using high-affinity binding sites in their ectodomains (Madsen et al., 2011; Broghammer et al., 2012). Knockout mutations in either NFR1 or NFR5 eliminate almost all NF-inducible responses, including induction of root hair deformation, perinuclear calcium oscillations (calcium spiking), calcium influx, and induction of genes required for the development and infection of nodules (Madsen et al., 2003; Radutoiu et al., 2003; Miwa et al., 2006; Høgslund et al., 2009 ), fitting with their function as signaling receptors. In Medicago truncatula, NFP (Nod factor perception) corresponds to NFR5, and mutations in this gene also cause complete loss of NF-inducible responses (Arrighi et al., 2006). However, although LYK3 corresponds to NFR1, lyk3 mutations block formation of infection threads but do not impact cortical cell divisions associated with nodule morphogenesis, suggesting that LYK3 might act as an entry NF receptor (Catoira et al., 2001; Smit et al., 2007). Although M. loti mutants imply a similar differentiation in NF perception in L. japonicus (Rodpothong et al., 2009), an equivalent NF receptor controlling entry has not been identified.
Coexpression of the L. japonicus NFR1 and NFR5 genes in roots of M. truncatula enables the formation of some uninfected nodules in response to M. loti (Radutoiu et al., 2007). Because M. loti normally does not induce nodule formation on M. truncatula, this indicates NFR1 and NFR5 play a role in host-specific nodulation and additional signaling is required for host recognition during the infection process. Symbiosis receptor kinase (SYMRK) is a plasma membrane-located receptor-like kinase (RLK) involved in symbiotic signaling in L. japonicus (Stracke et al., 2002), and can interact with NFR5, implying a heteromeric receptor complex that mediates NF signaling (Antolín-Llovera et al., 2014). Overexpression of NFR1, NFR5, or SYMRK can activate spontaneous nodule formation in the absence of rhizobia (Ried et al., 2014). In addition, some lipid raft proteins, such as flotilins (Haney and Long, 2010), and symbiosis-specific remorins (SYMREM) interact with symbiotic RLKs to help mediate rhizobial infection (Lefebvre et al., 2010; Tóth et al., 2012; Liang et al., 2018).
Rhizobial infection in most legumes is mediated by a tubular invagination called an infection thread. This often initiates in root-hair cells, in which rhizobia attach to the tip, inducing deformation of the tip region that leads to entrapment of the rhizobia in so-called ‘infection pockets’ formed by the root hair tip curling back on itself around the attached bacteria (van Brussel et al., 1992; Gage, 2004). Cell wall and membrane remodeling accompanied by cytoskeletal rearrangements are initiated at this early stage and lead to the inward growth of the infection thread toward the base of root hairs (van Spronsen et al., 1994; Brewin, 2004; Murray, 2011; Fournier et al., 2015). ITs then crosses the walls of the root hair and adjacent cortical cell and continues growing down through layers of cortical cells, eventually extending into those dividing cortical cells that correspond to the developing nodule primordium. Several proteins required for infection thread formation have been identified including a nodulation-specific pectate lyase (NPL; Xie et al., 2012); several components of the WAVE/SCAR-APR2/3 complex (NAP, PIR, SCARN, and ARPC1; Yokota et al., 2009; Miyahara et al., 2010; Hossain et al., 2012; Qiu et al., 2015) required for actin nucleation; a putative ubiquitin- E3 ligase LIN/CERBERUS; a coil-coil domain protein RPG; and Vapyrin, which contains a VAMP-associated protein (VAP)/ major sperm protein (MSP) domain and several ankyrin-repeat domains (Arrighi et al., 2008; Kiss et al., 2009; Yano et al., 2009; Murray et al., 2011). The phenotypes caused by mutations in these various genes show they are required for infection, but their precise biological/biochemical functions are not known. In addition, mutations causing other infection defects have been identified in L. japonicus (lot1, crinkle, sym8, sym104, sym105, and alb1), but the genes affected have not yet been described (Imaizumi-Anraku et al., 1997; Sandal et al., 2006; Murray, 2011; Oldroyd et al., 2011). Rhizobial secreted exopolysaccharides (EPSs) are important for infection thread initiation and formation, and EPS has been suggested to function as a signal in the chronic intracellular infection of plant cells of nitrogen-fixing root nodules (Gibson et al., 2008; Kelly et al., 2013). Recently, a LysM receptor kinase EPR3 (Exopolysaccharide receptor 3) was reported to be an EPS receptor in L. japonicus (Kawaharada et al., 2015, 2017b).
The transcription factors NIN (Nodule inception), ERN1 (ERF required for nodulation 1), NSP1 and NSP2 (Nodulation signaling pathway 1 and 2) also play crucial roles in rhizobial infection (Heckmann et al., 2006; Bek et al., 2010; Madsen et al., 2010; Cerri et al., 2017; Kawaharada et al., 2017a). NIN was the first essential nodulation gene identified in L. japonicus (Schauser et al., 1999), and it coordinates the regulation of rhizobial infection and nodule organogenesis (Vernié et al., 2015). The DNA-binding motif for NIN has been identified (Soyano et al., 2013, 2014), and NIN can directly bind to and activate expression of several infection-specific genes, such as NPL and SCARN, as well as genes required for nodule organogenesis, such as CRE1 and NF-YA1 (Xie et al., 2012; Soyano et al., 2013; Qiu et al., 2015; Vernié et al., 2015).
In this work, we identified a gene necessary for infection thread development encoding a Leu-rich-repeat (LRR) type RLK that we have named RINRK1 (Rhizobial infection receptor-like kinase 1). RINRK1 showed a specific expression pattern related to rhizobial infection. Because RINRK1 is required for full NIN induction and NIN is required for RINRK1 induction, there may be positive feedback involving RINRK1 and NIN, possibly resulting in amplification of NF signaling associated with rhizobial infection.
RESULTS
Identification of a LRR-RLK Required for Rhizobial Infection
The infection thread-defective mutant itd4 (SL3055-2) was isolated from an ethyl methanesulfonate (EMS) mutagenized pool of L. japonicus Gifu B-129 as having a 10-fold reduction in the number of infection threads based on histochemical staining of lacZ-marked M. loti R7A (Lombardo et al., 2006). The mutation causing the infection defect was previously mapped to the short arm of linkage group I, using a mapping population generated by crossing itd4 with L. japonicus Miyakojima (MG20; Lombardo et al., 2006). Another L. japonicus infection-defective mutant, alb1, was described previously (Imaizumi-Anraku et al., 1997, 2000), and rough-mapping indicated ALB1 was also located on Chromosome I in the same region that precluded precise mapping in crosses with L. japonicus MG20. The alb1 mutation was reported not to be allelic with itd4, even though they mapped to a similar region, and had similar phenotypes (Lombardo et al., 2006). To check if this was correct, we conducted new crosses between the itd4 and alb1 mutants. From these crosses, 15 individual F1 plants (from 2 independent crosses) were obtained, all of which produced only white nodules 3 weeks after inoculation. Based on identification of the gene, these F1 plants were validated by sequencing PCR products, confirming that itd4 and alb1 are indeed allelic.
This region has a translocation between the distal part of the short arm of chromosome I of Gifu B-129 and the long arm of chromosome II of MG20. This causes strong suppression of recombination in this region precluding precise mapping (Hayashi et al., 2001). Therefore, we made a new mapping population by crossing itd4 (pollen donor) with L. burttii, which is an alternative parent to MG20 for mapping genes at this region (Kawaguchi et al., 2005; Sandal et al., 2012). From about 4000 F2 plants, 944 mutant progeny were obtained, suggesting inheritance of a recessive mutation; genotyping of these plants allowed us to map the mutation between the markers BM1741 and TM1842. Within this interval we identified no recombination with TM1840 (Supplemental Fig. S1A). Sequencing of candidate genes within this region identified one gene (Lj1g3v0415090.1) encoding a putative LRR-RLK, which contained a mutation causing a premature stop in itd4.
To confirm this identified mutation caused the infection defects, we cloned the full-length gene from Gifu B-129 including a 1.9-kb putative promoter region and 1.2 kb downstream of the translation stop. We transformed wild-type or itd4 mutant roots with empty vector (EV) or the cloned gene using Agrobacterium rhizogenes–mediated hairy root transformation. The transgenic roots were inoculated with M. loti and 18 d after inoculation, wild-type plants produced mature pink nodules (Table 1; Supplemental Fig. S2A). The roots of the itd4 mutant transformed with the EV retained the infection defect and did not produce pink mature nodules at this time point (Table 1;Supplemental Fig. S2A). Meanwhile, when the itd4 mutant was transformed with RINRK1, nearly all of transformed root systems had mature pink nodules 18 d after inoculation with M. loti R7A (Table 1; Supplemental Fig. S2A). At this time point, normal infection threads were formed on these complemented transgenic roots, and the pink nodules were fully infected (Supplemental Fig. S2, B–D). This complementation assay demonstrated that we had identified the gene causing the infection defect in itd4.
Table 1. Complementation of rinrk1-1 nodulation phenotype by hairy root transformation.
| Transformation Construct and Line | Nodulation Ratioa | Nodules (sd)b |
|---|---|---|
| pKGWR | ||
| Wild type | 21/21 | 4.75 (0.3) |
| rinrk1-1 | 0/18 | 0 |
| pRINRK1:RINRK1 | ||
| Wild type | 11/11 | 4.7 (0.6) |
| rinrk1-1 | 22/24 | 4.4 (1.7) |
| pRINRK1:RINRK1 (G364A) | ||
| Wild type | 15/15 | 4.6 (0.4) |
| rinrk1-1 | 6/10 | 5.0 (1.0) |
| pRINRK1:RINRK1 (K383E) | ||
| Wild type | 20/20 | 4.9 (0.4) |
| rinrk1-1 | 6/6 | 3.0 (0.9) |
Ratios indicate numbers of successfully transformed plants that formed nodules versus all plants of the indicated line that were successfully transformed with the indicated construct.
Mean nodule number per successfully transformed plant.
The predicted coding sequence consists of 1878 nucleotides in two exons (Supplemental Fig. S1B) encoding a 626-amino acid protein. The deduced sequence revealed a predicted signal peptide (SP) and three LRR motifs at the N-terminal region, a single transmembrane domain, and a cytoplasmic Ser/Thr protein kinase domain at the C terminus (Fig. 1A). The itd4 mutant carries a point mutation (G153 to A) converting W51 to a premature stop codon between the regions encoding the predicted signal peptide and the first LRR domain (Fig. 1A; Supplemental Fig. S1B). Although the mutation was previously called itd4 (Lombardo et al., 2006), for clarity, we renamed the gene RINRK1 and the mutant allele itd4 was renamed rinrk1-1.
Figure 1.
RINRKs encode LRR receptor-like kinases. A, RINRK1 and RINRK2 protein structure. RINRKs have an N-terminal signal peptide (SP), three LRR domains, a transmembrane (TM) domain, and a C-terminal RLK domain. The sites of the RINRK1-1 and RINRK2-1 mutations are shown. B to F, Confocal microscopy of root hairs of wild type (B), rinrk1-1 (C), rinrk2-1 (D), and rinrk1 rinrk2 (E and F) 7 d after inoculation with M. loti R7A expressing GFP. Bar = 20 μm. G, Mean number of infection events [infection foci (IF), infection thread (IT)]. H, mean number of nodules formed on wild-type, rinrk1-1, rinrk2-1, and rinrk1 rinrk2 mutants. The infection events were analyzed 7 and 14 d postinoculation (dpi), and the nodule numbers were scored 2, 3, and 5 wpi (weeks postinoculation; ± se, n > 15). *P < 0.05, **P < 0.01 between wild type and mutant at the same time point.
Identification of RINRK2, a homolog of RINRK1 with a Minor Role in Rhizobial Infection
Some infected nodules were found on SL3055-2 (rinrk1-1) after prolonged periods of infection. To determine if this delayed infection could be due to genetic redundancy, we used BLAST searches and identified proteins belonging to a legume-specific clade of homologs, including representatives from M. truncatula, common bean (Phaseolus vulgaris), and soybean (Glycine max; Supplemental Fig. S3). A L. japonicus paralog of RINRK1 (Lj4g3v1535150.1) was identified from this phylogenetic analysis, and we named it RINRK2. All of these legume RINRK homologs had a similar gene structure containing two exons and one intron at equivalent locations (https://phytozome.jgi.doe.gov/pz/portal.html). The predicted RINRK1 proteins shared about 69%–72% identity and about 80%–3% similarity from different legume species.
A mutant line (30018122) was identified, which carries a LORE1 retrotransposon insertion in the first exon of RINRK2 (Supplemental Fig. S1B); therefore, this mutant was designated as rinrk2-1. The rinrk2-1 mutant produced similar infection threads as wild type (Fig. 1D). We crossed rinrk1-1 and rinrk2-1 mutants and identified a double mutant (rinrk1 rinrk2). Infection events in the rinrk1-1, rinrk2-1 and rinrk1 rinrk2 mutants were analyzed by confocal microscopy using M. loti constitutively expressing GFP. This confirmed most infection events in rinrk1-1 were blocked, resulting in the accumulation of many enlarged infection foci (Fig. 1C) compared with the normal elongated infection threads in wild type (Fig. 1B). In the double mutant, almost no infection threads were observed and almost all infections appeared to be blocked at the stage of the infection foci (Fig. 1E). In the rinrk1-1 and rinrk1 rinrk2 mutants, occasional infection threads appeared to degrade and release bacteria into root hairs of the mutant (Fig. 1F). Infection events were quantified 7 and 14 d postinoculationwith M. loti R7A expressing cloned lacZ. This confirmed rinrk1-1 had fewer ITs and more infection foci (IF) than wild type and that rinrk2-1 was similar to wild type. The rinrk1 rinrk2 double mutant had even more IFs and fewer ITs than rinrk1-1 (Fig. 1G). Two weeks after inoculation with M. loti, wild-type and rinrk2-1 plants produced mature pink nodules, whereas the rinrk1-1 and rinrk1 rinrk2 mutants formed a few small white nodules. After 3 weeks a few pale-pink nodules appeared on the rinrk1-1 mutant, but this was not seen with the rinrk1 rinrk2 double mutant. Five weeks after inoculation, 10 out of 21 rinrk1-1 plants produced about two pale-pink nodules. In contrast, only two of 16 plants of the rinrk1 rinrk2 produced one pale-pink nodule (Fig. 1H; Supplemental Fig. S4, A–H). Examination of histochemically stained nodule sections revealed the pale-pink nodules were partially infected (Supplemental Fig. S4, I–N). The infections in the rinrk1 rinrk2 double mutant appeared to occur via crack entry (Supplemental Fig. S4N). These observations suggest RINRK2 has some functional redundancy with RINRK1 for rhizobial infection.
RINRK1 Is Required for Normal Expression of Early Nodulin Genes
Several genes (e.g. NIN, ENDO40-1, NPL) are induced during rhizobial infection and nodule organogenesis in infected root hairs and nearby cells (Schauser et al., 1999; Grønlund et al., 2005; Xie et al., 2012). Therefore, we assayed (by reverse transcription-quantitative PCR [RT-qPCR]) the effect of the rinrk mutation on the expression pattern of some of these genes. NF induction of NIN was no different in wild type and rinrk2-1 showing a continuous increase in expression over 24 h. However, in the rinrk1-1 and rinrk1 rinrk2 mutants, NIN expression reached a maximum 6 h after NF addition but then did not increase in the next 12–24 h as seen in wild type (Fig. 2A), revealing a second phase of induction that is absent from the rinrk1 mutant and the rinrk1 rinrk2 double mutant. NPL was strongly induced in wild type and in the rinrk2 mutant, but this strong induction was absent from the rinrk1 and rinrk1 rinrk2 mutants (Fig. 2B). There was no significant difference between rinrk1 and rinrk1 rinrk2 for NIN and NPL induction (Fig. 2, A and B). Following inoculation with M. loti R7A, normal induction of NIN, RbohB, ENOD40-1, and N6 required RINRK1 (Fig. 2, C–F).
Figure 2.
A role for RINRKs in the induction of early nodulation genes by Nod-factor or M. loti. A and B, RT-qPCR analysis of NIN (A) and NPL (B) induction in wild-type (WT) and rinrk mutants after addition of 10 nm NFs. C to F, RT-qPCR analysis of NIN (C), ENOD40-1(D), RbohB (E), and N6 (F) induction in wild type and rinrk1-1 3, 7, and 14 d after inoculation of M. loti. Expression is relative to mock-treated samples and normalized to Ubiquitin levels. Means and ses (± se) were derived from three biological replicates, and asterisks indicate significant differences (*P < 0.05, **P < 0.01, Student’s t test) between mutant and wild type at the same time points. dpi, days postinoculation; hrs, Hours.
NFs added to L. japonicus roots induce nuclear-associated calcium spiking and a separate calcium influx at the root-hair tip (Miwa et al., 2006). Both responses are blocked in nfr1 and nfr5 mutants, whereas the calcium influx is retained in symrk mutants (Miwa et al., 2006). To assess if RINRK1 and RINRK2 were required for the NF-signaling, we assessed calcium responses in the rinrk1-1 and rinrk1 rinrk2 mutants. Both retained NF-induced calcium spiking and calcium influx (Supplemental Fig. S5), implying that RINRKs are not essential for these calcium responses.
RINRK1 and RINRK2 Are Induced by NF and Display an Infection-Specific Expression Pattern
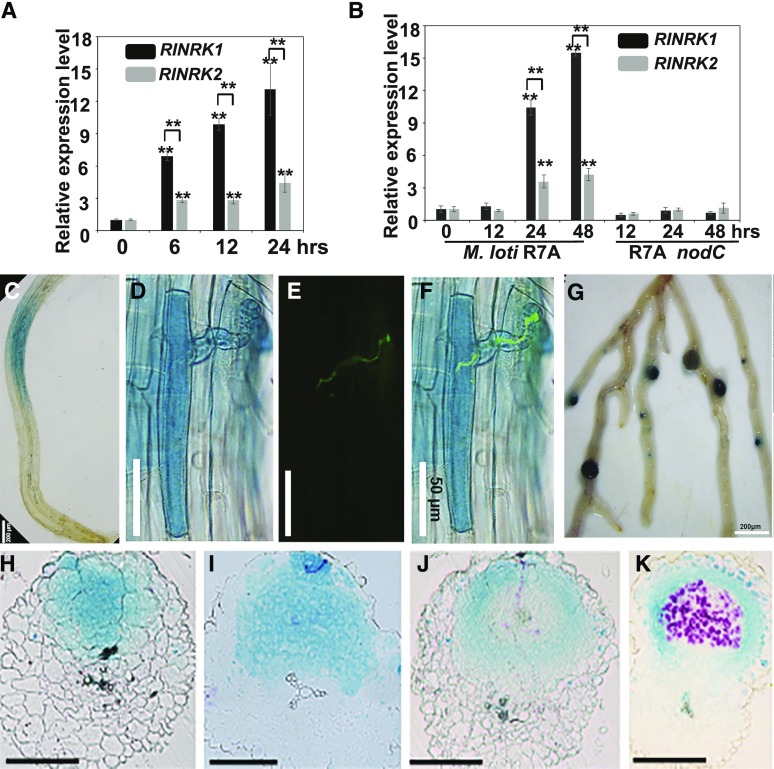
We used RT-qPCR to analyze expression of RINRK1 and RINRK2 in roots of wild-type L. japonicus at different time points after the addition of M. loti NF or following inoculation with M. loti R7A or the M. loti R7A nodC mutant that produces no NF. RINRK1 and RINRK2 expression increased 6 h after the addition of NFs from M. loti, and RINRK1 was more strongly induced than RINRK2 (Fig. 3A). M. loti induced RINRK1 and RINRK2 24 h after inoculation, and again, RINRK1 was more strongly induced than RINRK2. The M. loti R7A nodC mutant did not induce RINRK1 or RINRK2 expression (Fig. 3B).
Figure 3.
RINRKs are induced by NFs or M. loti and expression pattern of RINRK1. A and B, RINRK1 and RINRK2 expression in L. japonicus roots measured by RT-qPCR 6, 12, and 24 h after addition of 10 nm M. loti NFs (A) or 12, 24, and 48 h after inoculation with M. loti R7A or M. loti R7A nodC (B). Expression is relative to mock-treated samples and normalized to Ubiquitin. Means and SEs were derived from three biological replicates, and statistically significant differences are shown (**P < 0.01, Student’s t test). C to G, Histochemical localization of GUS activity in A. rhizogenes roots of L. japonicus wild type transformed with proRINRK1:GUS. Transformed hairy roots were analyzed 24 h after NF addition (C; n > 15), and 5 d (D–F) or 14 d (G) after inoculation of M. loti R7A carrying GFP or lacZ. The presence of infection threads was indicated by constitutively expressed GFP in M. loti R7A (E), and the images (D) and (E) were merged in (F). H to K, Sections of nodules indicating RINRK1 expression in whole cell layers of young uninfected nodules (H and I) and in epidermal and nodule parenchyma cells in mature nodules (J and K). The transgenic roots were costained with Magenta-Gal and X-Gluc to visualize M. loti (in purple) and proRINRK1:GUS expression (in blue), respectively, in nodules. Bars = 200 μm (C and G), 50 μm (D–F), and 150 μm (H–L).
Roots transformed with a RINRK1-promoter GUS fusion (proRINRK1:GUS) revealed RINRK1 was expressed in epidermal cells 24 h after NF addition; this expression occurred in the zone of the root that is normally most susceptible to rhizobial infection (Fig. 3C). Expression of proRINRK1:GUS was particularly strongly induced in infected root hairs, as observed 3 d after inoculation with a strain of M. loti R7A, that could be visualized due to its constitutive expression of GFP (Fig. 3, D–F). Expression of proRINRK1:GUS was also strongly induced in both young and mature nodules (Fig. 3G); sectioning and examination of these nodules by light microscopy revealed proRINRK1:GUS was expressed in all cell layers of young uninfected nodules (Fig. 3, H and I). However, in mature nodules, the GUS staining was observed in the nodule parenchyma cells but not in the fixation zone containing nitrogen-fixing bacteroids (Fig. 3, J and K).
NIN Directly Binds to the RINRK1 Promoter and Induces RINRK1 Expression
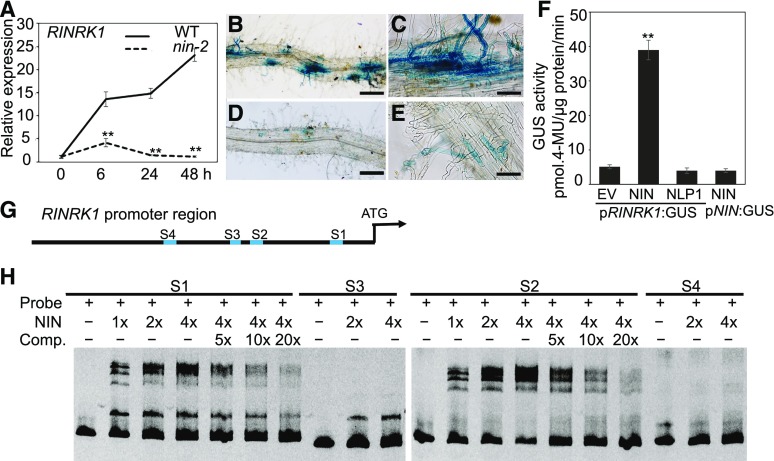
Since the transcription factor NIN is associated with the regulation of several genes during rhizobial infection (Soyano et al., 2014; Liu et al., 2019), we tested whether RINRK1 was regulated by NIN. First, our RT-qPCR analysis revealed NF induction of RINRK1 was strongly reduced in the nin-2 mutant background (Fig. 4A). We further used proRINRK1:GUS to assess NIN-dependent RINRK1 induction. At 3 d after inoculation, strong proRINRK1:GUS induction was seen in root hairs and epidermal cells of transformed roots of wild type (Fig. 4, B and C), but very low levels of GUS expression were observed in transformed roots of the nin-2 mutant (Fig. 4, D and E). To test directly for effects of NIN on RINRK expression, we coexpressed 35S:GFP-NIN (NIN) with proRINRK1:GUS in Nicotiana benthamiana leaf cells. As negative controls, we coexpressed with proRINRK1:GUS either the EV lacking NIN (EV) or a construct expressing the NIN-like gene NLP1, which induces expression of other promoters (Lin et al., 2018). Measurements of GUS activity indicated NIN but not NLP1 can activate proRINRK1:GUS expression in this system (Fig. 4F). As another negative control, we coexpressed NIN and proNIN:GUS and confirmed there was no induction of GUS activity in N. benthamiana leaves (Fig. 4F).
Figure 4.
NIN induces RINRK1 expression. A, RT-qPCR analysis of RINRK1 expression in L. japonicus wild-type (WT) or nin-2 roots 0, 6, 24, and 48 h after addition of 10 nm NFs. The bars show ses and significant differences (** P < 0.01) between mutant and wild type at the same time points. B to E, Assays of proRINRK1:GUS expression. proRINRK1:GUS was introduced into roots of L. japonicus wild type (B and C, n = 25) or the nin-2 mutant (D and E, n = 23) by A. rhizogenes–mediated hairy root transformation. After transgenic roots had grown, they were inoculated with M. loti R7A, and 3 d later the GUS activity in the roots was visualized by staining with X-gluc. Bar = 500 μm (B and D) or 100 μm (C and E). F, Transactivation assay of proRINRK1 expression by NIN in N. benthamiana. Leaves were coinfiltrated with strains of A. tumefaciens carrying pRINRK1:GUS and either the EV proUB:GFP (EV) or 35S:GFP-NIN (NIN) or 35S:GFP-NLP1 (NLP1) encoding the NIN-like protein NLP1 (Lin et al., 2018). GUS activity was determined quantitatively in leaf discs from three biological replicates (± se). The asterisks indicate a statistically significant difference (**P < 0.01, Student’s t test.) G, Diagram of the RINRK1 promoter region showing the putative NBS S1, S2, S3, and S4. H, Assays of NIN binding to the promoter of RINRK1. DNA fragments (1 nM) from parts of the RINRK1 promoter carrying the S1, S2, S3, or S4 regions were amplified by PCR, fluorescence labeled, and incubated with the indicated concentrations of the NIN protein for 20 min at 30°C. The protein-DNA complexes were separated by electrophoresis on native 6% polyacrylamide gels, and the fluorescent-labeled DNA was detected by fluorimetry. For S1 and S2, 5-, 10-, and 20-folds excess of unlabeled DNA fragments were added as competitors (Comp.) for binding.
Since the NF-induced expression of RINRK1 requires NIN, it seemed likely RINRK1 may be regulated by NIN. Genome wide NIN binding sites were identified previously using chromatin immunoprecipitation (Soyano et al., 2014), and that work identified a NIN-binding site upstream of RINRK1 (referred to as chr1.CM0166.760.r2.a in that work). In the promoter region of RINRK1 we identified four sequences (Fig. 4G; Supplemental Fig. S6) similar to a consensus NIN-binding site (NBS; Soyano et al., 2014). These four putative NBS sequences are −320 to −346 (S1), −709 to −734 (S2), −800 to −826 (S3), and −1296 to −1322 (S4) nucleotides upstream of the predicted translation start site (Fig. 4G). Electrophoresis mobility shift assays revealed specific retardation of RINRK1 promoter fragments containing S1 or S2 when incubated with the carboxyl-terminal half of NIN, which contains the predicted DNA-binding domain (Fig. 4H). Addition of unlabeled competitor DNA confirmed NIN specifically bound to these DNA fragments of the promoter region (Fig. 4H). No retardation was seen with promoter fragments containing S3 or S4 (Fig. 4H). Based on all these results, we conclude NIN can bind to the RINRK1 promoter to induce RINRK1 expression.
RINRK1 Is an Atypical Pseudokinase and Does Not Requires Its Kinase Activity for Rhizobial Infection
To determine the subcellular localization of the RINRK1 protein, we expressed a RINRK1-GFP fusion driven by its native promoter, but we were unable to detect any florescence signals, either in N. benthamiana leaves or L. japonicus roots. When RINRK1-GFP expression was driven by the constitutive cauliflower mosaic virus 35S promoter in transgenic L. japonicus roots, a GFP signal was observed in the plasma membranes of root, root hair cells and L. japonicus root protoplasts (Supplemental Fig. S7).
Comparison of the kinase domain of RINRK1 and RINRK2 with other RLKs revealed both have sequences and structural conservation typical of protein kinases but lack several conserved amino acid residues essential for kinase activity (Hanks et al., 1988); the Gly-rich loop (I), VALK domain (II), the HRD domain (VIb), and DFG (VII) motifs are conserved (Castells and Casacuberta, 2007; Supplemental Fig. S8). However, RINRK1 and RINRK2 and the nearest homologs of RINRK1 in M. truncatula (Medtr3g078250.1) and in Arabidopsis (Arabidopsis thaliana; At2g26730) possess an Asn (N) in the HRD domain where active protein kinases usually contain Asp (D; e.g. in NFR1, SYMRK, and AtBAK1). Inactive RLKs, such as STRUBBELIG, BAK1-Interacting Receptor-like Kinase 2 (BIR2), and Receptor Dead Kinase 1 (RDK1), also have Asn at this position (Halter et al., 2014; Kumar et al., 2017). Moreover, RINRK1 and RINRK2 have EYG residues instead of the DFG motif of subdomain VII, which is involved in cation binding and orientation of the ATP gamma phosphate for phosphate transfer (Hanks et al., 1988; Fig. 5A). These differences with active kinases suggest RINRK1 and RINRK2 lack kinase activity.
Figure 5.
RINRK1 is an atypical kinase and is not a substrate of NFR1 or SYMRK. A, Sequence comparison of RINRK1 and RINRK2 with other legume symbiosis RLKs and RLKs described in other plants. The sequences of Gly-rich loop (I) and subdomains II, VIb, and VII are marked. B, In vitro kinase assay with recombinant expressed kinase domains of MBP-tagged RINRK1(RINRK1-CD) and purified His-tagged NFR1, NFR5, and SYMRK (NFR1-CD, NFR5-CD, and SYMRK-CD). The tagged proteins were expressed in E. coli and purified. Samples were separated by SDS-PAGE followed by phosphor-imaging analysis. C, Complementation tests of rinrk1-1 with mutant forms of RINRK1 lacking residues normally required for kinase activity. Nodules are shown on hairy roots of the rinrk1-1 mutant transformed with RINRK1, RINRK1G364A, or RINRK1K383E. The top show bright field images, and the bottom are epifluorescence microscopy images showing DsRed expression in the same transgenic roots. Bars = 5 mm.
We tested the in vitro kinase activity of RINRK1 compared with NFR1, NFR5, and SYMRK (NFR1-CD, NFR5-CD, or SYMRK-CD). The NFR1 and SYMRK kinase domains showed strong auto-phosphorylation, whereas no auto-phosphorylation was observed for the RINRK1 and NFR5 kinase domains (Fig. 5B; Supplemental Fig. S9). Cloned RINRK1 carrying mutations of residues predicted to be essential for kinase activity (RINRK1G364A or RINRK1K383E) could still rescue the infection defects of the rinrk1-1 mutant (Fig. 5C; Table1), supporting the idea that kinase activity is not essential for RINRK1 function. We tested whether RINRK1 is a substrate of NFR1 or SYMRK by coincubating RINRK1 with NFR1 or SYMRK in the presence of γ32P-labeled ATP. It appears not to be a substrate, because we found no phosphorylated band corresponding to RINRK1; under the same conditions, NFR5 was phosphorylated by NFR1 (Fig. 5B; Supplemental Fig. S9).
RINRK1 Is Not Required for Spontaneous Nodule Formation Induced by Overexpression of SYMRK
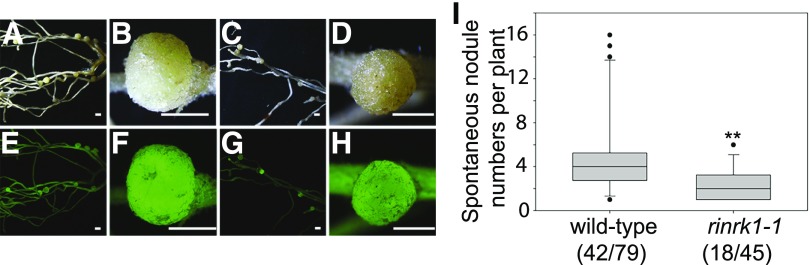
Overexpression of NFR1, NFR5, or SYMRK in L. japonicus leads to spontaneous nodule formation, and this has been used to show SYMRK functions downstream of NFR1 and NFR5 (Ried et al., 2014). Overexpression of RINRK1 (proUB:RINRK1) did not induce spontaneous nodules (143 transgenic root systems). We also tested if proUB:SYMRK could induce spontaneous nodule formation in the rinrk1-1 mutant. Transformation of hairy roots with proUB-SYMRK induced spontaneous nodules in both L. japonicus wild type and the rinrk1-1 mutant (Fig. 6, A–H). Spontaneous nodules formed on the transformed roots of the rinrk1-1 mutant. There were fewer nodules than seen with wild-type roots transformed with proUB-SYMRK (Fig. 6I); a similar reduction in SYMRK-induced spontaneous nodules was observed in nfr1 and nfr5 mutants (Ried et al., 2014). We conclude RINRK1 is not required for the induction of nodule organogenesis.
Figure 6.
RINRK1 is not required for nodule organogenesis. A to H, Spontaneous nodule formation on hairy roots of L. japonicus wild-type (A, B, E, and F) and the rinrk1-1 mutant (C, D, G, and H) transformed with proUB:SYMRK. The top show bright field images, and the bottom are epifluorescence microscopy images showing GFP expression in the same transgenic roots. Scale bars = 5 mm (A, C, E, and G) and 1 mm (B, D, F, and H). I, The numbers of spontaneous nodules formed in wild type and rinrk1-1 following hairy root transformation by A. rhizogenes carrying proUB:SYMRK. Plot represents the nodule numbers per nodulated plants in absence of M. loti 42 d post transformation (dpt). Number of nodulated plants per total plants is specified under each line label. Black dots represent the individual data points outside of the plotting area. (**statistical differences P < 0.01, Student’s t test).
DISCUSSION
NF recognition is essential for all aspects of symbiotic nitrogen fixation in most legumes, coordinating the processes in the root cortex necessary for nodule organogenesis and in the root epidermis where rhizobial infection is initiated (Oldroyd and Downie, 2008). Evidence from both plant and bacterial genetic studies suggests at least two stringencies of NF perception with less stringent ‘early’ recognition initiating nodule organogenesis and more stringent ‘late’ recognition being required for bacterial infection (Ardourel et al., 1994; Morieri et al., 2013). Such differential perception of NF suggests multiple receptor complexes that can differentially perceive NF and activate the very different processes required for organogenesis and infection. Here we identify two new RLKs (RINRK1 and RINRK2) that are induced by NF and can promote rhizobial infection but not nodule organogenesis. Mutation of rinrk1 and rinrk1 rinrk2 revealed what appeared to be two phases of NF induction of the NIN gene: an initial early phase of induction (up to 6 h) that was retained in the rinrk1 and rinrk1 rinrk2 mutants, and a second phase of induction (12–24 h) that was lost in these mutants. This is entirely consistent with RINRK1 and RINRK2 functioning in infection thread formation. However, RINRK1 and RINRK2 were not required for NF induced calcium oscillations, suggesting these RINRKs are not required for NF activation of the signaling pathway leading to nodule development. This does not exclude the possibility RINRK1 could interact with known NF receptors (e.g. NFR1 or NFR5) to generate a specific output that relates NF perception to rhizobial infection.
The RINRKs are required for full induction of NIN and other early nodulation genes such as ENOD40-1, N6 and RbohB. The interpretation of patterns of induction of these genes in M. loti infection-defective mutants may be made a little more complicated by the observation that expression of these genes is associated with infection events; therefore, reducing the number of infection events in the rinrk mutants could decrease overall expression levels of infection-related genes. However, another infection-thread–deficient mutant scarn, which has similar infection phenotypes as rinrk1 rinrk2, showed normal M. loti induction of NIN, NPL, and ENOD40-1 (Qiu et al., 2015). This observation suggests the dynamics of these gene expression patterns in rinrk1 and rinrk1 rinrk2 is not directly linked to a reduction in infection but rather is related to their biological function.
RINRKs lack several conserved motifs or amino acid residues that are important for kinase activity, and our in vitro assays suggest RINRK1 lacks auto-phosphorylation. We showed mutant forms of RINRK1, in which the conserved Lys in kinase domain II is altered to Glu RINRK1K383E or the conserved Gly at 364 is converted to Ala RINRK1G364A, still can rescue the rinrk1-1 nodulation defect. Mutating Lys 383 to Glu (RINRK1K383E) should impair the binding of the nucleotide before phosphoryl transfer. The mutation RINRK1G364A is predicted to affect the P loop, which is involved in orienting the ATP through interactions with its phosphates. Mutating the equivalent Lys to Glu (K350E) in NFR1 blocked complementation of the nfr1-3 mutant; in contrast, the pseudokinase NFR5 carrying the equivalent mutation (NFR5K339E) retained the ability to complement nfr5-2 mutant (Madsen et al., 2010). Our in vitro and in planta data therefore indicate the intracellular region of RINRK1, despite its predicted kinase-like structure, does not have kinase activity. Although we did not test if RINRK2 has kinase activity, it is clear based on its sequence that, like RINRK1, RINRK2 lacks residues critical for kinase activity. Its nearest homolog in Arabidopsis, At2g26730, has also been suggested to be a kinase-dead protein, and it has been estimated the Arabidopsis genome contains 10%–20% atypical or kinase-dead RLKs (Castells and Casacuberta, 2007). These proteins were proposed to have diverged to become signal transduction proteins despite their lack of kinase activity (Castells and Casacuberta, 2007). Typical examples are BIR2, which acts in BAK1-mediated plant immunity (Halter et al., 2014); STRUBBELIG (SUB), which acts in leaf and floral organ development (Chevalier et al., 2005); POLLEN-SPECIFIC RECEPTOR-LIKE KINASE 5 (PRK5), which acts in PAMP associated plant immunity (Wrzaczek et al., 2015); and RDK1, which acts in in abiotic stress signaling (Kumar et al., 2017). The mechanistic basis of signaling via atypical RLKs is not understood, but all these proteins are essential for signal transduction and are proposed to function via protein-protein interactions. For example, BIR2 interacts with BAK1 and RDK1 interacts with ABI1 in Arabidopsis (Halter et al., 2014; Kumar et al., 2017). Identification and characterization of proteins that interact with RINRK1 and RINRK2 will be needed to help us understand how these signaling kinase-like proteins promote formation and propagation of infection threads.
NIN is the key regulator that coordinates rhizobial infection and nodule organogenesis (Vernié et al., 2015). NIN is required for the induction of many of the genes known to be associated with rhizobial infection in root hairs of M. truncatula (Liu et al., 2019). Several genes with NIN binding sites were identified previously (Soyano et al., 2014), and we confirmed NIN binds upstream of RINRK1. Since NIN is required for normal RINRK1 expression in L. japonicus and can promote RINRK1 expression in N. benthamiana, we can conclude NIN directly regulates RINRK1 expression. However, RINRK is required for full induction of NIN by NF. Since RINRK induction requires NIN and full NIN induction requires RINRK, we propose there may be a positive feedback loop involving RINRK and NIN; such positive feedback could enhance NF responses, and this could facilitate rhizobial infection.
As a NF-induced LRR-type pseudokinase, RINRK1 most probably acts to produce a signaling output in response to NF to promote the initiation and maintenance of infection threads along which rhizobia can grow. Thus RINRK1 may form part of a branch of the nodulation signaling pathway that promotes infection rather than nodule organogenesis. This could facilitate the spatial and temporal coordination of infection-related signaling. We do not yet know how this branch could be activated. One possibility is that RINRK1 could interact with component(s) of the NF recognition complex and function along with an active kinase to phosphorylate target proteins. Another possibility is that RINRK1 may be involved in perceiving an unknown rhizobial signal that activates formation of infection threads once the nodulation signaling pathway has been activated by NF. The potential positive feedback loop, resulting from NIN induction of RINRK1 expression and RINRK1 promoting NIN expression, could facilitate the commitment required by the plant to promote a structure that enables root infection by rhizobia.
MATERIAL AND METHODS
Plant Materials and Strains
Lotus japonicus Gifu B-129 (Handberg and Stougaard, 1992) and Lotus burttii (Kawaguchi et al., 2005; Fukai et al., 2012) were used for this study. The rinrk1-1 (itd4/SL3055-2) EMS-induced mutant was a derivative of Gifu B-129 (Lombardo et al., 2006). rinrk2-1 was obtained from a pool of L. japonicus mutants with LORE1 transposon insertions (Urbański et al., 2012). rinrk1-1 and rinrk2-1 were crossed, then F1 plants were generated, and the rinrk1 rinrk2 double mutant was identified among the F2 plants by genotyping and sequencing. L. japonicus seeds were prepared and inoculated with Mesorhizobium loti R7A as described previously (Qiu et al., 2015). Plasmids were introduced by transformation into Escherichia coli DH10B or DH5α for cloning, into E.coli Rosetta (DE3) for protein expression, into Agrobacterium rhizogenes AR1193 (Stougaard et al., 1987) for expression in L. japonicus roots and into A. tumefaciens EHA105 for expression in Nicotiana benthamiana.
Map-Based Cloning
Rinrk1-1 (SL3055-2) was crossed with L. burttii, and nodulation phenotype was scored in F2 progeny from self-pollinated F1 plants. Genomic DNA was extracted (Perry and Parniske, 2005) using simple sequence repeats markers described previously (Kawaguchi et al., 2005).
Nodulation and Infection Thread Analysis
L. japonicus seedlings were grown in 1:1 mix of vermiculite and perlite and after 1 week were inoculated with M. loti R7A carrying pMP2444 (GFP). Nodules were counted 3 to 6 weeks after inoculation. Infection events were visualized by laser scanning confocal microscopy (Olympus FV1000) using GFP-marked M. loti R7A, and counterstained with propidium iodide. For nodule sectioning, nodules were fixed in 2.5% (v/v) glutaraldehyde and embedded in Technovit 7100 (Kulzer GmbH) resin according to the manufacturer’s instructions, and 5–10 mm transverse sections were prepared with a microtome (Leica RM2265). Then sections were stained with 0.5% (w/v) toluidine blue O in 0.5% (w/v) sodium tetraborate buffer before pictures were taken using a Nikon Eclipse Ni light microscope.
Expression Analysis
Nodulation marker gene expression samples were generated from whole roots of about 10 seedlings of wild-type or rinrk mutants that had been grown for 7 d on Fahraeus N-free agar plates and then treated with 10 nm NF purified from M. loti R7A or inoculated with M. loti R7A, respectively. All plants were grown under a 16-h-light /8-h-dark cycle. Total RNA was extracted from the roots using the TRIpure Isolation Reagent (Aidlab) based on the manufacturer’s protocol and then quantified using a Nano-drop 2000 (Thermo). RNA was reverse transcribed by TransScript one-step genomic DNA (gDNA) Removal and cDNA synthesis SuperMix (Trans Gen Biotech). Quantitative expression was analyzed by real-time PCR using TOYOBO SYBR Green Realtime PCR Master Mix (TOYOBO) and a step-one Plus PCR system (ABI). Subsequently, relative expression was normalized to the reference gene ubiquitin (Lj5g3v2060710.1). ses and statistical significance based on three biological replicates were calculated using the 2−ΔΔCt method.
Complementation and Subcellular Localization of RINRK1 Protein
RINRK1 genomic DNA (5.8 kb) including the 1.9-kb native promoter and 1.2-kb terminator was amplified from Gifu leaves using primer RINRK1-gDNA(MfeI)-F and RINRK1-gDNA(MfeI)-R (Supplemental Table S1). The PCR products were cloned into pBluescript SK to generate RINRK1 pBluescript SK, which was digested with MfeI, and RINRK1 was cloned in pKGWR to form RINRK1 gDNA pKGWR. The point mutations of RINRK1K364E and RINRK1G383A were generated with a Site-Directed Mutagenesis Kit (YEASWA Biotechnology) following the manufacturer’s instructions. The constructs were confirmed by DNA sequencing. All the constructs in destination vectors were introduced into A. rhizogenes AR1193 by electroporation and then introduced into roots of the rinrk1-1 mutant by hair root transformation on one-half strength B5 medium. The transgenic plants were transferred into pots with perlite:vermiculite (1:1 mix) and after 5–7 d inoculated with M. loti R7A/lacZ.
For subcellular localization assays, the RINRK1 genomic DNA (without stop codon) was amplified from Gifu B-129 by RINRK1-attB-F and RINRK1-ns-attB-R, and the PCR products were cloned into pDONR207 by BP reaction to generate RINRK1 gDNA pDONR207 and then recombined into the destination vector pK7FWG2 (Karimi et al., 2002) by LR reaction to generate RINRK1 pK7FWG2. This construct was transformed into A. rhizogenes AR1193 by electroporation and then introduced into wild-type roots by hairy-root transformation. For transient expression in L. japonicus root protoplasts, the RINRK1 genomic DNA was amplified from Gifu B-129 by RINRK1-GFP-F and RINRK1-GFP-R, and the PCR products were digested with SpeI and then inserted into pA7-GFP using ClonExpress II One Step Cloning Kit (Vazyme) to generate pA7-RINRK1-GFP. The sequence was confirmed by DNA sequencing, the constructs were transiently expressed in L. japonicus root protoplasts using DNA-PEG-calcium transfection, and images were taken by laser scanning confocal microscopy (Leica TCS SP8; Jia et al., 2018).
RINRK1 Promoter:GUS Analysis
A 1.9-kb region upstream of the RINRK1 translation start codon was amplified from Gifu leaf DNA using primers pRINRK1-attB-F and pRINRK1-attB-R. The PCR products were cloned into pDONR207 and then recombined into the destination vector pKGWFS7 to form proRINRK1:GUS. The construct in pDONR207 was confirmed by DNA sequencing. proRINRK1:GUS was introduced into A. rhizogenes AR1193 by electroporation and then transformed into roots of wild type by hairy root transformation on 1/2 B5 medium. The transformed chimeric plants were transplanted into vermiculite/perlite pots and after 5–7 d inoculated with M. loti R7A containing lacZ or GFP. The roots were harvested at different time points for GUS staining, and some samples were then stained with Magental-gal (Sangon Biotech) for visualization of lacZ-tagged M. loti (Lotus japonicus handbook, 2005). The double stained nodules were sectioned, and images were taken with a Nikon Eclipse Ni light microscope.
Electrophoresis Mobility Shift Assays
The RINRK1 promoter regions: S1(−283 to −461 bp), S2 (−618 to −771 bp), S3(−763 to −924 bp) and S4(−1223 to −1378 bp) were amplified by primers shown in Supplemental Table S1 using proRINRK1:GUS as a template. The PCR products were fluorescently labeled at the 5′ ends with Cy5 (Yingjun Corp.) and purified by gel extraction (OMEGA Bio-TEK), and then fluorescence-labeled DNA was detected using a Biophotometer Plus (Eppendorf). NIN carrying a C-terminal His tag was purified as described previously (Xie et al., 2012). Fluorescence-labeled DNA (1 nm) was incubated with the purified NIN protein in 20 μL of binding buffer (20 mm Tris, pH 7.5; 5% (w/v) glycerol; 10 mm MgCl2; 0.25 mm dithiothreitol; 0.8 μg bovine serum albumin; and 1 μg salmon sperm DNA). After incubation at 30°C for 20 min, the products were electrophoresed at 4°C on a 6% native polyacrylamide gel in Tris-borate/EDTA buffer for 2 h at 100 V. Fluorescence in the gel was detected with a Starion FLA-9000 (FujiFilm).
Transient Activation of proRINRK1:GUS Expression in N. benthamiana Leaves by NIN
LjNIN pK7WGF2 (Qiu et al., 2015) or MtNLP1 (Lin et al., 2018) and proRINRK:GUS, or LjLIN and proNIN:GUS (Qiu et al., 2015) were introduced into A. tumefaciens strain EHA105 and then coinfiltrated into N. benthamiana leaves as described previously (Qiu et al., 2015). Samples from at least 5 leaves were collected 48 h after inoculation, frozen in liquid nitrogen, and used for protein extraction. GUS activity was measured with 4-methylumbelliferyl-β-d-glucuronide as substrate (Sigma-Aldrich) using Thermo scientific Varioskan flash. Mean values and standard deviations were determined from three biological replicates.
In Vitro Kinase Assays
RINRK1-CD pDONR207 was recombined into pMGWA by LR reaction to generate RINRK1-CD-MBP. NFR1, NFR5, and SYMRK cytoplasmic domains were tagged with 6xHis in pHGWA (Busso et al., 2005). These constructs were introduced into E. coli BL21-Codon Plus (DE3)-RIL (Stratagene). Protein expression was induced during exponential growth using 0.2 mm isopropylthio-β-galactoside for 9 h at 20°C. The protein was purified using MBP or His beads (Smart) according to the manufacturer’s instructions. For auto-phosphorylation assays, 1 μg purified RINRK1-MBP protein was incubated in 20 μL kinase buffer (50 mm Tris-HCl, pH 7.4; 10 mm MgCl2;1 mm dithiothreitol; 200 µm ATP; 10 µCi [γ – 32P]ATP) at 30°C for 1 h. Then the proteins were separated by SDS-PAGE, and phosphorylated proteins were analyzed by autoradiography and/or phosphor-imaging by the Starion FLA-9000 (FujiFlim).
All constructs in entry vectors were sequenced, and primers used in this paper were available in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers MN109982 for RINRK1 and MN109983 for RINRK2 or from the L. japonicus genome version 3.0 (http://www.kazusa.or.jp/lotus/) for RINRK1 Lj1g3v0415090.1 and RINRK2 Lj4g3v1535150.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Positional cloning of RINRK1 and gene structure of RINRK1 and RINRK2.
Supplemental Figure S2. RINRK1 can rescue rinrk1-1 infection deficiency.
Supplemental Figure S3. Phylogenic analysis of RINRK proteins.
Supplemental Figure S4. Nodule phenotype of wild type and rinrk mutants.
Supplemental Figure S5. Analysis of calcium oscillation in wild type and rinrk mutants.
Supplemental Figure S6. Alignment of putative NIN-binding sites (NBS) in the RINRK1 promoter region.
Supplemental Figure S7. Assay of RINRK1 subcellular localization using transgenic expression of RINRK1-GFP in L. japonicus roots and root protoplasts.
Supplemental Figure S8. Alignment of the sequence of RINRK and related kinases with typical and atypical kinase domains.
Supplemental Figure S9. A Coomassie staining of the corresponding SDS-PAGE gel.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Jeremy Murray (Institute of Plant Physiology and Ecology, China) and Jie Zhang (Institute of Microbiology, China) for constructive suggestions on the work and helpful comments on the manuscript. We thank Tracey Welham, Anne B. Heckman, Fabien Lombardo, and Trevor Wang (Joint Intelligence Committee, United Kingdom) as well as Martin Parniske (University of Munich, Germany) for the mutant and for help with the preliminary mutant screen.
Footnotes
This work was funded by National Key R&D Program of China (2016YFA0500500), Chinese Academy of Sciences (CAS) (the Strategic Priority Research Program: XDB27040208 and IPP 153D31KYSB20160074), and National Natural Science Foundation of China (NSFC) (31470344) and received support from Biotechnology and Biological Sciences Research Council (BBSRC) (Award E017045/1) and from the John Innes Foundation (JIF) (to J.A.D.).
Articles can be viewed without a subscription.
References
- Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24: 422–427 [DOI] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Amor BB, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Neibel F, Journet E-P, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Godfroy O, de Billy F, Saurat O, Jauneau A, Gough C (2008) The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc Natl Acad Sci USA 105: 9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek AS, Sauer J, Thygesen MB, Duus JO, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S (2010) Improved characterization of nod factors and genetically based variation in LysM Receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol Plant Microbe Interact 23: 58–66 [DOI] [PubMed] [Google Scholar]
- Brewin NJ. (2004) Plant cell wall remodelling in the rhizobium–legume symbiosis. Crit Rev Plant Sci 23: 293–316 [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso D, Delagoutte-Busso B, Moras D (2005) Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem 343: 313–321 [DOI] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM (2007) Signalling through kinase-defective domains: The prevalence of atypical receptor-like kinases in plants. J Exp Bot 58: 3503–3511 [DOI] [PubMed] [Google Scholar]
- Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D, Dénarié J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Cerri MR, Wang Q, Stolz P, Folgmann J, Frances L, Katzer K, Li X, Heckmann AB, Wang TL, Downie JA, et al. (2017) The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol 215: 323–337 [DOI] [PubMed] [Google Scholar]
- Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K (2005) STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci USA 102: 9074–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore JV, Ranjeva R, Bono JJ (2001) Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci 6: 24–30 [DOI] [PubMed] [Google Scholar]
- Fournier J, Teillet A, Chabaud M, Ivanov S, Genre A, Limpens E, de Carvalho-Niebel F, Barker DG (2015) Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol 167: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai E, Soyano T, Umehara Y, Nakayama S, Hirakawa H, Tabata S, Sato S, Hayashi M (2012) Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J 69: 720–730 [DOI] [PubMed] [Google Scholar]
- Gage DJ. (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC (2008) Molecular determinants of a symbiotic chronic infection. Annu Rev Genet 42: 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønlund M, Roussis A, Flemetakis E, Quaedvlieg NE, Schlaman HR, Umehara Y, Katinakis P, Stougaard J, Spaink HP (2005) Analysis of promoter activity of the early nodulin Enod40 in Lotus japonicus. Mol Plant Microbe Interact 18: 414–427 [DOI] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24: 134–143 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496 [Google Scholar]
- Haney CH, Long SR (2010) Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc Natl Acad Sci USA 107: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Miyahara A, Sato S, Kato T, Yoshikawa M, Taketa M, Hayashi M, Pedrosa A, Onda R, Imaizumi-Anraku H, et al. (2001) Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res 8: 301–310 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. Plos One 4: e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Liao J, James EK, Sato S, Tabata S, Jurkiewicz A, Madsen LH, Stougaard J, Ross L, Szczyglowski K (2012) Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol 160: 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Kawaguchi M, Koiwa H, Akao S, Syono K (1997) Two ineffective-nodulating mutants of Lotus japonicus—different phenotypes caused by the blockage of endocytotic bacterial release and nodule maturation. Plant Cell Physiol 38: 871–881 [Google Scholar]
- Imaizumi-Anraku H, Kouchi H, Syono K, Akao S, Kawaguchi M (2000) Analysis of ENOD40 expression in alb1, a symbiotic mutant of Lotus japonicus that forms empty nodules with incompletely developed nodule vascular bundles. Mol Gen Genet 264: 402–410 [DOI] [PubMed] [Google Scholar]
- Jia N, Zhu Y, Xie F (2018) An efficient protocol for model legume root protoplast isolation and transformation. Front Plant Sci 9: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Pedrosa-Harand A, Yano K, Hayashi M, Murooka Y, Saito K, Nagata T, Namai K, Nishida H, Shibata D, et al. (2005) Lotus burttii takes a position of the third corner in the lotus molecular genetics triangle. DNA Res 12: 69–77 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, Vinther M, Andersen SU, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, James EK, Kelly S, Sandal N, Stougaard J (2017a) The ethylene responsive factor required for nodulation 1 (ERN1) transcription tactor is required for infection-thread formation in Lotus japonicus. Mol Plant Microbe Interact 30: 194–204 [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Nielsen MW, Kelly S, James EK, Andersen KR, Rasmussen SR, Füchtbauer W, Madsen LH, Heckmann AB, Radutoiu S, Stougaard J (2017b) Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun 8: 14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW (2013) Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol Plant Microbe Interact 26: 319–329 [DOI] [PubMed] [Google Scholar]
- Kiss E, Oláh B, Kaló P, Morales M, Heckmann AB, Borbola A, Lózsa A, Kontár K, Middleton P, Downie JA, Oldroyd GE, Endre G (2009) LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol 151: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Kumar R, Baek D, Hyun TK, Chung WS, Yun DJ, Kim JY (2017) An Arabidopsis thaliana RECEPTOR DEAD KINASE1 (RDK1) gene functions as a positive regulator in plant responses to ABA. Mol Plant 10: 223–243 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, et al. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Liang P, Stratil TF, Popp C, Marín M, Folgmann J, Mysore KS, Wen J, Ott T (2018) Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc Natl Acad Sci USA 115: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Li X, Luo Z, Mysore KS, Wen J, Xie F (2018) NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nat Plants 4: 942–952 [DOI] [PubMed] [Google Scholar]
- Liu CW, Breakspear A, Guan D, Cerri MR, Jackson K, Jiang S, Robson F, Radhakrishnan GV, Roy S, Bone C, et al. (2019) NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol 179: 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo F, Heckmann AB, Miwa H, Perry JA, Yano K, Hayashi M, Parniske M, Wang TL, Downie JA (2006) Identification of symbiotically defective mutants of Lotus japonicus affected in infection thread growth. Mol Plant Microbe Interact 19: 1444–1450 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J, Parniske M (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65: 404–417 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA (2006) Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Miyahara A, Richens J, Starker C, Morieri G, Smith L, Long S, Downie JA, Oldroyd GE (2010) Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol Plant Microbe Interact 23: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Morieri G, Martinez EA, Jarynowski A, Driguez H, Morris R, Oldroyd GE, Downie JA (2013) Host-specific Nod-factors associated with Medicago truncatula nodule infection differentially induce calcium influx and calcium spiking in root hairs. New Phytol 200: 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: Rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Murray JD, Muni RRD, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, et al. (2011) Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Perry J, Parniske M (2005) 96-Well DNA isolation method. In Stougaard J, Udvardi M, Parniske M, Spaink H, Saalbach G, Webb J, Chiurazzi M, Márquez AJ, eds, Lotus japonicus Handbook. Srpinger Netherlands, Heidelberg, Germany, pp 129–131 [Google Scholar]
- Qiu L, Lin JS, Xu J, Sato S, Parniske M, Wang TL, Downie JA, Xie F (2015) SCARN a novel class of SCAR protein that is required for root-hair infection during legume nodulation. PLoS Genet 11: e1005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried MK, Antolín-Llovera M, Parniske M (2014) Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodpothong P, Sullivan JT, Songsrirote K, Sumpton D, Cheung KW, Thomas-Oates J, Radutoiu S, Stougaard J, Ronson CW (2009) Nodulation gene mutants of Mesorhizobium loti R7A-nodZ and nolL mutants have host-specific phenotypes on Lotus spp. Mol Plant Microbe Interact 22: 1546–1554 [DOI] [PubMed] [Google Scholar]
- Sandal N, Petersen TR, Murray J, Umehara Y, Karas B, Yano K, Kumagai H, Yoshikawa M, Saito K, Hayashi M, et al. (2006) Genetics of symbiosis in Lotus japonicus: Recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol Plant Microbe Interact 19: 80–91 [DOI] [PubMed] [Google Scholar]
- Sandal N, Jin H, Rodriguez-Navarro DN, Temprano F, Cvitanich C, Brachmann A, Sato S, Kawaguchi M, Tabata S, Parniske M, et al. (2012) A set of Lotus japonicus Gifu x Lotus burttii recombinant inbred lines facilitates map-based cloning and QTL mapping. DNA Res 19: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard S (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M (2013) Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M (2014) Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci USA 111: 14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP, Sheeley DM, van Brussel AA, Glushka J, York WS, Tak T, Geiger O, Kennedy EP, Reinhold VN, Lugtenberg BJ (1991) A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 354: 125–130 [DOI] [PubMed] [Google Scholar]
- Stougaard J, Abildsten D, Marcker KA (1987) The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet 207: 251–255 [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Tóth K, Stratil TF, Madsen EB, Ye J, Popp C, Antolín-Llovera M, Grossmann C, Jensen ON, Schüssler A, Parniske M, Ott T (2012) Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PLoS One 7: e30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbański DF, Małolepszy A, Stougaard J, Andersen SU (2012) Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J 69: 731–741 [DOI] [PubMed] [Google Scholar]
- van Brussel AA, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJ, Kijne JW (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- van Spronsen PC, Bakhuizen R, van Brussel AA, Kijne JW (1994) Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process. Eur J Cell Biol 64: 88–94 [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho-Niebel F, Oldroyd GE (2015) The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Vainonen JP, Stael S, Tsiatsiani L, Help-Rinta-Rahko H, Gauthier A, Kaufholdt D, Bollhöner B, Lamminmäki A, Staes A, et al. (2015) GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J 34: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA 109: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Shibata S, Chen WL, Sato S, Kaneko T, Jurkiewicz A, Sandal N, Banba M, Imaizumi-Anraku H, Kojima T, et al. (2009) CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis. Plant J 60: 168–180 [DOI] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, Jurkiewicz A, Rueda P, Radutoiu S, Held M, Hossain MS, Szczyglowski K, Morieri G, et al. (2009) Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]