Arabidopsis sterol 4α-methyl oxidase1 (SMO1) enzymes affect embryo development by balancing the appropriate activities of auxin and cytokinin.
Abstract
In the plant sterol biosynthetic pathway, sterol 4α-methyl oxidase1 (SMO1) and SMO2 enzymes are involved in the removal of the first and second methyl groups at the C-4 position, respectively. SMO2s have been found to be essential for embryonic and postembryonic development, but the roles of SMO1s remain unclear. Here, we found that the three Arabidopsis (Arabidopsis thaliana) SMO1 genes displayed different expression patterns. Single smo1 mutants and smo1-1 smo1-3 double mutants showed no obvious phenotype, but the smo1-1 smo1-2 double mutant was embryo lethal. The smo1-1 smo1-2 embryos exhibited severe defects, including no cotyledon or shoot apical meristem formation, abnormal division of suspensor cells, and twin embryos. These defects were associated with enhanced and ectopic expression of auxin biosynthesis and response reporters. Consistently, the expression pattern and polar localization of PIN FORMED1, PIN FORMED7, and AUXIN RESISTANT1 auxin transporters were dramatically altered in smo1-1 smo1-2 embryos. Moreover, cytokinin biosynthesis and response were reduced in smo1-1 smo1-2 embryos. Tissue culture experiments further demonstrated that homeostasis between auxin and cytokinin was altered in smo1-1 smo1-2 heterozygous mutants. This disturbed balance of auxin and cytokinin in smo1-1 smo1-2 embryos was accompanied by unrestricted expression of the quiescent center marker WUSCHEL-RELATED HOMEOBOX5. Accordingly, exogenous application of either auxin biosynthesis inhibitor or cytokinin partially rescued the embryo lethality of smo1-1 smo1-2. Sterol analyses revealed that 4,4-dimethylsterols dramatically accumulated in smo1-1 smo1-2 heterozygous mutants. Together, these data demonstrate that SMO1s function through maintaining correct sterol composition to balance auxin and cytokinin activities during embryogenesis.
Sterols are important components of eukaryotic membranes, where they are involved in the formation of microdomains, which are also called lipid rafts (Mongrand et al., 2004; Roche et al., 2008). The Arabidopsis (Arabidopsis thaliana) cyclopropylsterol isomerase1-1 mutant affects the polar localization of the auxin efflux carrier PIN-FORMED2 (PIN2) and the specific localization of the KNOLLE syntaxin to the cell plate by altering the membrane sterol composition (Men et al., 2008; Boutté et al., 2010). In animals, cholesterol acts as a signal to regulate embryo development (Porter et al., 1996). In plants, sterols have also been found to be essential for embryo development (Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Zhang et al., 2016). Plant sterols are precursors for the biosynthesis of brassinosteroids (BRs), which play important roles in plant growth and development (Bishop and Yokota, 2001). However, mutants of upstream sterol biosynthetic genes, such as fackel (fk), hydral (hyd1), cyclopropylsterol isomerase1-1, sterol methyltransferase1 (smt1)/cephalopod (cph), and cyp51A2, cannot be rescued by BR application (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Kim et al., 2005; Men et al., 2008). Plant sterols are required for the generation of reactive oxygen species, which play important roles in plant growth and cell death (Posé et al., 2009; Kim et al., 2010). Plant sterols, such as sitosterol and stigmasterol, when applied exogenously, can affect the expression of genes that are essential for cell expansion and division (He et al., 2003). The accumulation of a plant sterol biosynthetic intermediate (SBI), 4-carboxy-4-methyl-24-methylenecycloartanol, which is the product of the first C-4α-methyl oxidation reaction, leads to polar auxin transport (PAT)–related phenotypes (Mialoundama et al., 2013).
The sterol biosynthetic pathway differs substantially among fungi, animals, and plants. In fungi and animals, the sterol precursor 2,3-oxidosqualene is cyclized into lanosterol, whereas in plants it is cyclized into cycloartenol (Benveniste, 1986). Conversion of lanosterol/cycloartenol into functional sterols involves the removal of the two methyl groups at the C-4 position (Benveniste, 1986; Rahier, 2011). The reaction is performed by three enzymes, including a sterol C-4α-methyl oxidase (SMO), a C-4α-carboxysterol-C-3-dehydrogenase/C-4-decarboxylase, and a sterone ketoreductase, which are tethered into a complex by a scaffold protein Ergosterol biosynthetic protein28 (ERG28; Mo et al., 2002; Rahier, 2011). In fungi and animals, the two methyl groups are removed successively and catalyzed by the same set of enzymes, whereas removal of the two methyl groups in plants is interrupted by several other enzymatic steps involving two distinct SMO enzymes (SMO1 and SMO2; Darnet and Rahier, 2004; Rahier, 2011). Arabidopsis has three SMO1 genes and two SMO2 genes, and only the SMO2 genes can complement the yeast erg25 mutant (Darnet et al., 2001; Darnet and Rahier, 2004). Virus-induced gene silencing of SMOs in tobacco (Nicotiana benthamiana) has demonstrated that 4,4-dimethylsterols accumulate when SMO1s are silenced, whereas 4α-methylsterols accumulate when SMO2s are silenced, indicating that SMO1s and SMO2s function in the removal of the first and second C-4-methyl group, respectively (Darnet and Rahier, 2004; Rahier, 2011). Genetic analysis revealed that the smo2-1 smo2-2 double mutant is embryo lethal, and auxin is involved in SMO2 gene-regulated embryonic development in Arabidopsis (Zhang et al., 2016). Recent reports have indicated that ACYL-CoA-BINDING PROTEIN1 modulates sterol synthesis by interacting with SMO1-1 and SMO1-2 proteins (Lung et al., 2017, 2018). However, the roles of SMO1s in plant growth and development remain unclear.
Numerous studies have reported that mutants deficient in auxin biosynthesis, transport, and response are defective in embryogenesis (Hamann et al., 1999; Weijers et al., 2005; Cheng et al., 2007; Möller and Weijers, 2009; Robert et al., 2013). TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1)/TRYPTOPHAN AMINOTRANSFERASE RELATED and YUCCA (YUC) auxin biosynthesis genes are expressed in the integuments of the ovule to provide auxin for early embryogenesis (Robert et al., 2018). PAT is mediated by the asymmetric plasma membrane (PM) localization of the PIN auxin efflux and AUXIN RESISTANT1 (AUX1)/LIKE-AUX1 (LAX) auxin influx carriers (Wisniewska et al., 2006; Robert et al., 2015). At least four PIN proteins (PIN1, PIN3, PIN4, and PIN7) are expressed during embryogenesis (Friml et al., 2003). Prior to the globular stage, PIN1 is expressed in the proembryo without apparent polarity, whereas PIN7 is polarly localized to the apical PM of suspensor cells (Friml et al., 2003). At the globular stage, PIN7 localization is shifted to the basal PM of suspensor cells, likely mediating auxin transport into the suspensor (Friml et al., 2003). Simultaneously, PIN1 is localized to the basal PM of provascular cells and transports auxin into the hypophysis (Friml et al., 2003). At the transition stage, PIN1 is localized toward sites where cotyledons would be initiated, resulting in auxin maxima in the cotyledon primordium (Friml et al., 2003). The AUX1, LAX1, and LAX2 genes are also expressed during embryogenesis. At the 32-cell embryo stage, AUX1 and LAX2 are expressed in the inner cells of the embryo proper; at later stages, they are expressed in the provascular cells. Additionally, LAX2 is also expressed in the hypophysis and the uppermost suspensor cells (Robert et al., 2015). LAX1 expression during embryo development is similar to PIN1 (Robert et al., 2015). Consistently, loss of function of AUX/LAX genes results in defective embryogenesis (Robert et al., 2015). Therefore, localized auxin biosynthesis and its directional transport by PIN and AUX/LAX proteins lead to auxin accumulation in dynamic patterns during embryogenesis.
In addition to auxin, cytokinin is also required for cell differentiation and specification during embryogenesis (Müller and Sheen, 2008; Möller and Weijers, 2009). The ATP/ADP isopentenyltransferase (IPT) and LONELY GUY (LOG) gene family members encode enzymes involved in cytokinin biosynthesis. The cytokinin receptor family in Arabidopsis is composed of four His kinases (AHKs). At the early globular embryo stage, the cytokinin signaling marker two-component-output-sensor (TCS):GFP is expressed in the hypophysis; after the first division of the hypophysis, the TCS:GFP signal is maintained in the apical daughter cell but disappears in the basal daughter cell (Müller and Sheen, 2008). Homeostasis between auxin and cytokinin is required during embryonic development. Excessive auxin accumulation and reduced cytokinin levels lead to embryonic development defects (Zhang et al., 2017). Members of the WUSCHEL-RELATED HOMEOBOX (WOX) gene family play important roles in balancing auxin and cytokinin pathways (Zhang et al., 2017). WOX2 is expressed in the apical lineage of the embryo, whereas WOX8/WOX9 expression is restricted to the basal lineage (Haecker et al., 2004; Breuninger et al., 2008). WOX2 together with its redundant genes, WOX1, WOX3, and WOX5, are required for embryo pattern formation by regulating the homeostasis between auxin and the cytokinin pathways (Zhang et al., 2017).
In this study, we investigated the roles of the three Arabidopsis SMO1 genes (SMO1-1, SMO1-2, and SMO1-3) in embryo development. Single smo1 mutants and smo1-1 smo1-3 double mutants showed no obvious phenotype, but the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ heterozygous double mutants exhibited severe defects in embryogenesis. In addition, they showed a lumpy root phenotype, and the smo1-1 smo1-2/+ seedling root length was shorter than that of wild-type. We then demonstrated that the developmental defects of smo1-1 smo1-2 embryos were associated with dysregulated auxin biosynthesis, transport, and response. We further demonstrated that cytokinin activity was altered in the smo1-1 smo1-2 embryos.
RESULTS
The Three SMO1 Genes Show Different Expression Patterns, and Their Encoded Proteins Localize to the Endoplasmic Reticulum
In Arabidopsis, there are three predicted SMO1 proteins, which share more than 70% amino acid sequence identity (Supplemental Fig. S1A). The three SMO1-encoding genes are all located on the fourth chromosome, and the genetic distance between SMO1-2 and SMO1-3 is very close (Supplemental Fig. S1B). The predicted topology of these proteins also exhibits high similarity (Supplemental Fig. S1, C–E). To analyze their expression patterns, we generated constructs with promoter fragments of each of the SMO1 genes driving the GUS reporter gene and introduced these constructs into wild-type Arabidopsis. Strong GUS staining was observed in most tissues throughout the ProSMO1-1:GUS transgenic plants (Supplemental Fig. S2, A–D). In leaves, the GUS signal was strong in vascular tissues and stomata (Supplemental Fig. S2, A and C). In roots, ProSMO1-1:GUS expression was detected strongly in the stele (Supplemental Fig. S2B), and the GUS signal was strong at lateral root formation sites (Supplemental Fig. S2D). In flowers, ProSMO1-1:GUS was expressed in anthers and pistil (Supplemental Fig. S3, A–D). By contrast, the expression pattern of SMO1-2 was different from that of SMO1-1. In leaves, ProSMO1-2:GUS expression was detected in the vascular tissues, but not in stomata (Supplemental Fig. S2, E and G). In roots, ProSMO1-2:GUS expression was detected in the root tip, with the strongest expression in the root stem cell niche and columella cells (Supplemental Fig. S2, F and H). In flowers, SMO1-2 was highly expressed in the style, petals, and anther filaments (Supplemental Fig. S3, F–I). SMO1-3 showed the weakest expression among these SMO1 genes. In vegetative organs, ProSMO1-3:GUS expression was only detected in the vascular tissues of leaves and roots (Supplemental Fig. S2, K and L). In flowers, the ProSMO1-3:GUS signal was detected in petal vascular tissues, anther filaments, and the style (Supplemental Fig. S3, K–N).
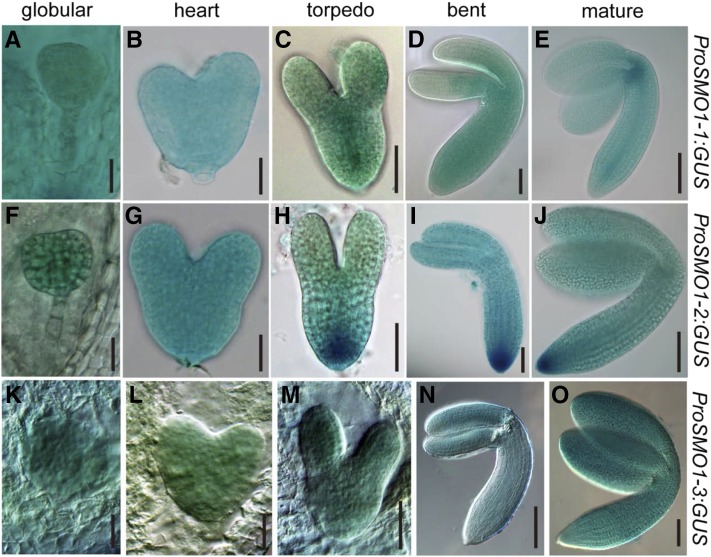
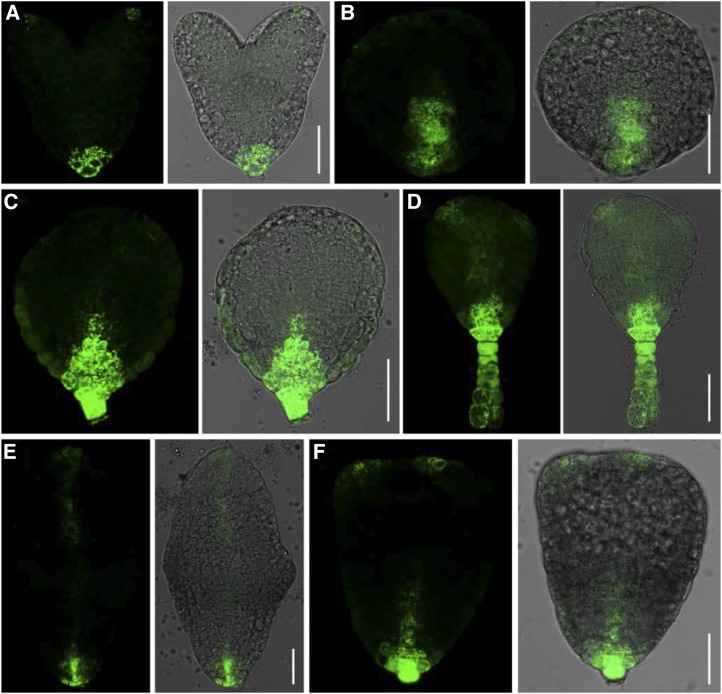
In siliques, the overall GUS staining patterns differed among the three SMO1 genes. SMO1-1 was highly expressed in the seedpods (Supplemental Fig. S3E), and SMO1-2 and SMO1-3 were expressed in the funiculi (Supplemental Fig. S3, J and O). SMO1 gene expression was detectable throughout embryo development from the globular stage to the mature stage, and SMO1-2 expression was the strongest among them (Fig. 1). At the globular embryo stage, SMO1-1 expression was detected in both the embryo and endosperm (Fig. 1A), but SMO1-2 and SMO1-3 were not detected in the endosperm (Fig. 1, F and K). At the heart embryo stage, the expression patterns of the three SMO1 genes were similar, but SMO1-3 expression was the weakest (Fig. 1, B, G, and L). From the torpedo to the mature embryo stages, SMO1-1 was strongly expressed in the provascular cells of the developing hypocotyl and in the shoot apical meristem (SAM; Fig. 1, C–E), SMO1-2 was strongly detected in the embryonic root meristem (Fig. 1, H–J), and SMO1-3 was expressed evenly in the embryo (Fig. 1, M–O). We also analyzed the expression patterns of the SMO1 genes in Arabidopsis embryos using the transcriptome database generated by the Raju Datla laboratory (Xiang et al., 2011). The expression patterns of these SMO1 genes in embryos were similar to the ProSMO1:GUS expression patterns we obtained (Supplemental Fig. S4). These results hint that the transcription of these SMO1 genes is coordinately regulated during embryogenesis and postembryonic development.
Figure 1.
Dynamic expression of SMO1 genes during embryo development. A to E, GUS staining of ProSMO1-1:GUS embryos at different developmental stages. F to J, Embryos from ProSMO1-2:GUS transgenic plant. K to O, Embryos from ProSMO1-3:GUS transgenic plant. Bars = 25 μm (A, B, F, G, K, and L), 50 μm (C, D, H, I, and M), and 100 μm (E, J, N, and O).
To study the subcellular localization of SMO1 proteins, we generated transgenic lines expressing a fusion protein between the SMO1 coding sequences and the EHANCED GFP (EGFP) gene, and the construct was driven by the 35S promoter. We observed T2 progeny from different transgenic lines of each construct and found a similar reticulate pattern of GFP signals in these lines. To verify that SMO1-EGFP localizes to the endoplasmic reticulum (ER), we conducted immunolocalization using the roots from Pro-35S:SMO1-EGFP transgenic plants. Immunoglobulin-binding protein (BiP; an ER-intrinsic chaperone) was used as an ER maker (Men et al., 2008). As shown in Supplemental Fig. S5, the SMO1-EGFP signal (indicated in green) overlapped with the immunoprocessed BiP signal (indicated in red; Supplemental Fig. S5). These results suggest that SMO1 proteins are localized to the ER.
The smo1-1 smo1-2 Double Mutant Is Embryo Lethal, and smo1-1/+ smo1-2 and smo1-1 smo1-2/+ Mutants Show a Lumpy Root Phenotype
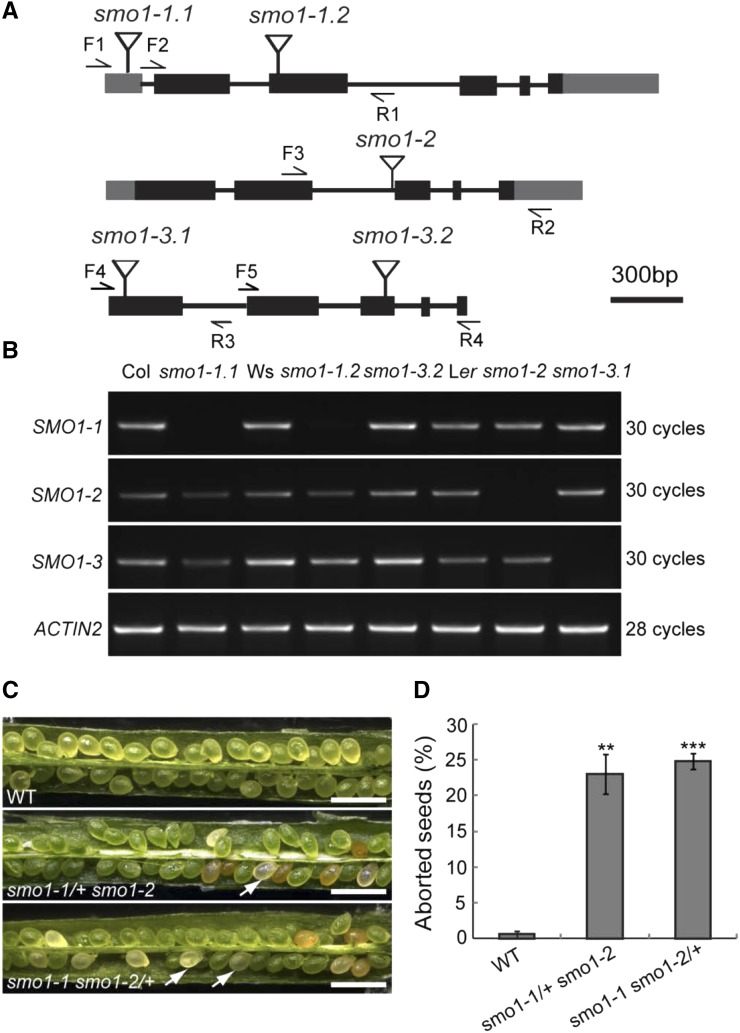
To study the biological functions of the SMO1 genes, we obtained transfer DNA (T-DNA) insertion mutants for SMO1-1 (SALK_021399, smo1-1.1; and FLAG_286D02, smo1-1.2), SMO1-2 (CSHL_GT13595, smo1-2), and SMO1-3 (CSHL_ET12310, smo1-3.1; and FLAG_425A03, smo1-3.2; Fig. 2A). The T-DNA insertions of these mutants were confirmed by PCR. Semiquantitative reverse-transcription (RT)-PCR analyses showed that all of the mutants were null mutant except for smo1-3.2 (Fig. 2B). We observed no abnormal phenotype for any of the single mutants. This result implies that these SMO1 genes are functionally redundant. Therefore, we crossed smo1-1, smo1-2, and smo1-3 plants to obtain double mutants, but due to genetic linkage between SMO1-2 and SMO1-3 genes, we did not cross smo1-2 with smo1-3. We obtained homozygous smo1-1 smo1-3 and heterozygous smo1-1/+ smo1-2 and smo1-1 smo1-2/+ progeny, but no homozygous smo1-1 smo1-2 progeny was obtained. These results implied that smo1-1 smo1-2 might be embryo lethal. To confirm this hypothesis, we observed the seed settings in siliques from the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ plants. In siliques from wild-type plants, 99.5% (n = 1,144) of the seeds developed normally, while siliques from the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ plants contained approximately one-quarter aborted white seeds (23.2%, n = 1,027, in smo1-1/+ smo1-2; 24.7%, n = 1,001, in smo1-1 smo1-2/+; Fig. 2, C and D). These data suggest that the smo1-1 smo1-2 double mutant is embryo lethal. We also observed the siliques from the smo1-1 smo1-3 double mutant, which had significantly shorter siliques and, accordingly, fewer seeds than wild-type (Supplemental Fig. S6, A–C). However, the ratio of unpollinated ovules and aborted seeds in the smo1-1 smo1-3 siliques was not significantly different from that of wild-type (Supplemental Fig. S6, D and E).
Figure 2.
Expression and phenotypic analyses of T-DNA insertion mutants of SMO1 genes. A, Structures of SMO1 genes with the T-DNA insertion sites. Gray boxes indicate 5′ and 3′ untranslated region, dark boxes indicate coding regions, lines indicate introns, and flags indicate T-DNA insertion site. F, Forward primer; R, reverse primer. B, Transcript levels of SMO1-1, SMO1-2, and SMO1-3 genes in wild-type and T-DNA insertion mutants. ACTIN2 gene was used as an internal control. Col, ecotype Columbia of Arabidopsis; Ler, ecotype Landsberg erecta of Arabidopsis; Ws, ecotype Wassilewskija of Arabidopsis. C, Eight-DPA siliques of wild-type (WT), smo1-1/+ smo1-2, and smo1-1 smo1-2/+ mutants. Arrows indicate aborted seeds. Bars = 1 mm. D, Percentage of aborted seeds in dissected siliques of wild-type, smo1-1/+ smo1-2, and smo1-1 smo1-2/+ mutants. Values are means ± sd of three independent experiments (n = 10 siliques from 5 plants for each experiment). Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; **P < 0.01; ***P < 0.001).
During vegetative growth, smo1-1 smo1-3 plants were slightly shorter than wild-type (Supplemental Fig. S6F), but their rosette size did not differ (Supplemental Fig. S6G). The smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants showed no phenotype in terms of plant height and rosette size, but they displayed a root phenotype (Supplemental Fig. S7, A–E). The smo1-1 smo1-2/+ seedling roots were clearly short (Supplemental Fig. S7, A and C), and approximately 52.6% (n = 114) of the smo1-1 smo1-2/+ seedlings showed a lumpy root phenotype (Supplemental Fig. S7, A and B). Although the smo1-1/+ smo1-2 seedling roots were not short, a small proportion of them (13.2%, n = 53) also displayed a lumpy root phenotype. This root phenotype was similar to those of sterol biosynthetic mutants, such as hyd1, hyd2/fk, smt1/cph, and cotyledon vascular pattern1 smt3 (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002, 2004; Souter et al., 2002; Willemsen et al., 2003; Carland et al., 2010). Similar to these mutants, the root phenotypes of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants could not be rescued by BR (Supplemental Fig. S7F). These results suggest that the phenotypes of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants are caused by sterol deficiency.
Patterning Defects of smo1-1 smo1-2 Embryos
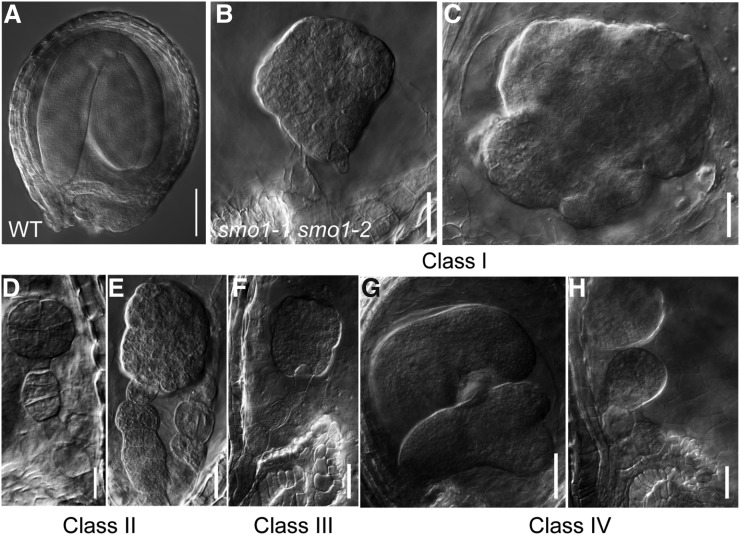
To further determine the defects of the smo1-1 smo1-2 embryos, we observed the embryo development of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants. Through observations of 8-DPA siliques of wild-type, smo1-1/+ smo1-2, and smo1-1 smo1-2 mutants, we found that almost all the embryos had developed into the mature stage in wild-type seeds (100%, n = 230; Fig. 3A); however, embryos of the white seeds from smo1-1/+ smo1-2 and smo1-1 smo1-2/+ siliques were aborted and showed an abnormal morphology (Fig. 3, B–H). These abnormal embryos can be categorized into the following four classes; class I, these embryos had a ball-shaped embryo proper with no obvious cotyledonary primordia and no SAM formation (68.2%, n = 606, in smo1-1/+ smo1-2; 61.7%, n = 433, in smo1-1 smo1-2/+; Fig. 3, B and C); class II, embryos grouped into this class showed defects in both the embryo proper and suspensor, and there were longitudinal divisions in the suspensor cells, which resulted in an enlarged suspensor with a multicell-layer girth (20.3%, n = 606, in smo1-1/+ smo1-2; 23.1%, n = 433, in smo1-1 smo1-2/+; Fig. 3, D and E); class III, these embryos were also globular-like but were smaller than those of class I, suggesting that they were arrested at earlier stages (7.8%, n = 606, in smo1-1/+ smo1-2; 7.4%, n = 433, in smo1-1 smo1-2/+; Fig. 3F); and classIV, two embryos existed in one seed, which could develop to the globular or late heart stage (3.8%, n = 606, in smo1-1/+ smo1-2; 7.9%, n = 433, in smo1-1 smo1-2/+; Fig. 3, G and H). Together, these results showed that loss of SMO1-1 and SMO1-2 functions affects embryo patterning and suspensor cell identity maintenance. To verify whether these embryo phenotypes are caused by the lack of SMO1-1 and SMO1-2 function, SMO1 genomic fragment-GFP translational fusions (ProSMO1-1:SMO1-1-EGFP, ProSMO1-2:SMO1-2-EGFP) were introduced into the smo1-1 smo1-2/+ and smo1-1/+ smo1-2 mutants, respectively. The defects in embryo development were rescued by these constructs (Supplemental Fig. S8).
Figure 3.
Phenotypes of smo1-1 smo1-2 embryos. Nomarski images of whole-mount cleared seeds from mature siliques of wild-type (WT; A) and smo1-1 smo1-2 heterozygous double mutants (B to H). Shown are representative images of class I to class IV smo1-1 smo1-2 embryos. Bars = 50 μm (A and G) and 20 μm (B–F, and H).
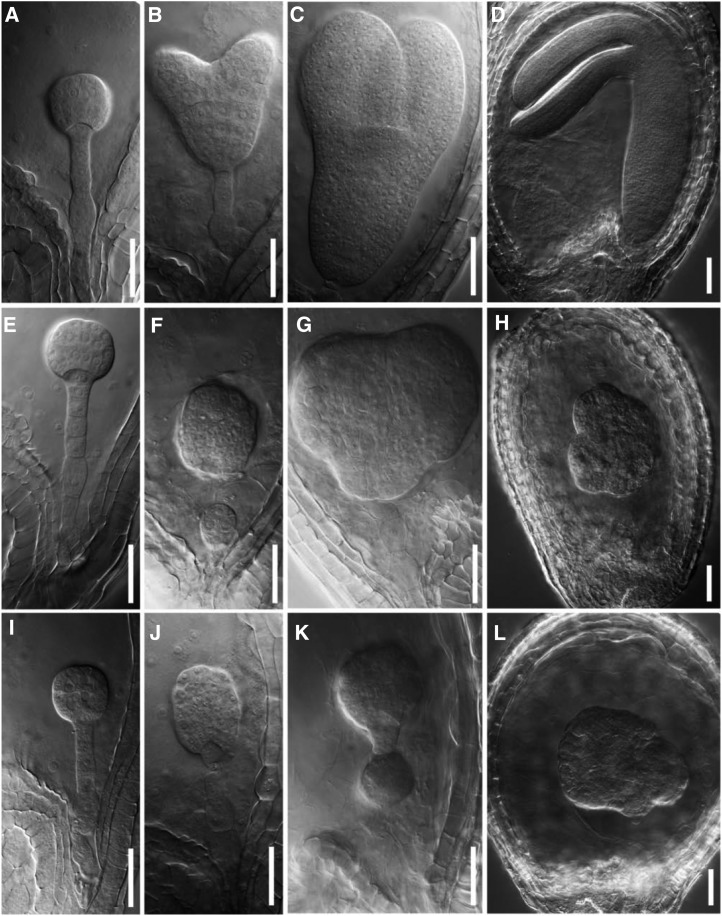
To explore the developmental stage at which smo1-1 smo1-2 embryo was arrested, we examined cleared seeds from siliques at different developmental stages of the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants. Consistent with the above observations, normal embryo development was observed in wild-type (Fig. 4, A–D), while embryos from the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ siliques showed a normal morphology until the late globular stage (Fig. 4, E and I). When most of the embryos developed into the heart stage in wild-type (Fig. 4B), abnormal embryos were found in the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ siliques (Fig. 4, F and J), which developed more slowly than those of wild-type and displayed aberrant suspensor cell division (Fig. 4F) or an abnormal embryo proper (Fig. 4J). From the heart stage onward, the cotyledon primordium was visible in the wild-type (Fig. 4, B–D), but the abnormal embryos from smo1-1/+ smo1-2 and smo1-1 smo1-2/+ siliques could not establish the correct bilaterally symmetrical structure (Fig. 4, G and K) and resulted in no cotyledon formation (Fig. 4, H and L). Therefore, after the late globular embryo stage, embryo development of the smo1-1 smo1-2 double mutants was arrested, and they failed to transit to the heart embryo stage. These developmental defects of smo1-1 smo1-2 embryos were similar to but more severe than those of the smo2-1 smo2-2, fk, hyd1, and smt1/cph mutants (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Zhang et al., 2016). It had been reported that SMO2 genes are essential for endosperm development (Zhang et al., 2016); therefore, we observed endosperm development in the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ seeds. From the 1-cell embryo to the globular embryo stage, the proliferation of endosperm nuclei was not obviously different between wild-type and putative smo1-1 smo1-2 double mutants (Supplemental Fig. S9). Together, these results demonstrate that the SMO1-1 and SMO1-2 genes play important roles during embryo development but are not essential for endosperm development.
Figure 4.
Wild-type and putative smo1-1 smo1-2 embryos at different developmental stages. Nomarski images of whole-mount cleared embryos at different developmental stages of wild type (A–D), smo1-1/+ smo1-2 (E–H), and smo1-1 smo1-2/+ (I–L). Note that smo1-1 smo1-2 embryo was abnormal from the heart stage onward (F–H and J–L). Bars = 50 μm.
Defective smo1-1 smo1-2 Embryos Are Associated with an Impaired Auxin Response, Increased Auxin Biosynthesis, and Defective PAT
Because SMO2s affect embryogenesis via auxin-related mechanisms (Zhang et al., 2016), we wondered whether the auxin pathway is affected in smo1-1 smo1-2 embryos. Therefore, we crossed DR5rev:GFP (an auxin responsive reporter) into the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants and examined the GFP signals in the embryos. In the heart stage wild-type embryos, GFP signals were observed in the hypophysis and cotyledon primordial tips (Fig. 5A). In contrast, strong and abnormal GFP signals were observed in segregated smo1-1 smo1-2 embryos (Fig. 5, B–F). In the apical region of the smo1-1 smo1-2 embryos, DR5rev:GFP signals were either completely absent (Fig. 5, B and C) or accumulated in one point (Fig. 5E), or were present throughout the apical epidermis (Fig. 5F). In the basal region of the smo1-1 smo1-2 embryos, DR5rev:GFP signals were enhanced in the hypophysis (Fig. 5, C, D, and F) and expanded into the lower suspensor cells (Fig. 5D). These results indicate that auxin activity and distribution are altered in smo1-1 smo1-2 embryos.
Figure 5.
Expression patterns of DR5rev:GFP in wild-type and putative smo1-1 smo1-2 embryos. A, DR5rev:GFP distribution in wild-type embryos at the heart stage. B to F, DR5rev:GFP distribution in putative smo1-1 smo1-2 embryos. Note that DR5rev:GFP signal was absent at the apical part of the ball-shaped smo1-1 smo1-2 embryos (B and C), exhibited ectopic expression in suspensor cells (D), accumulated in one point at the apical part of the embryo (E), or was evenly distributed at the apical outer layer of the embryo proper (F). Bars = 50 μm.
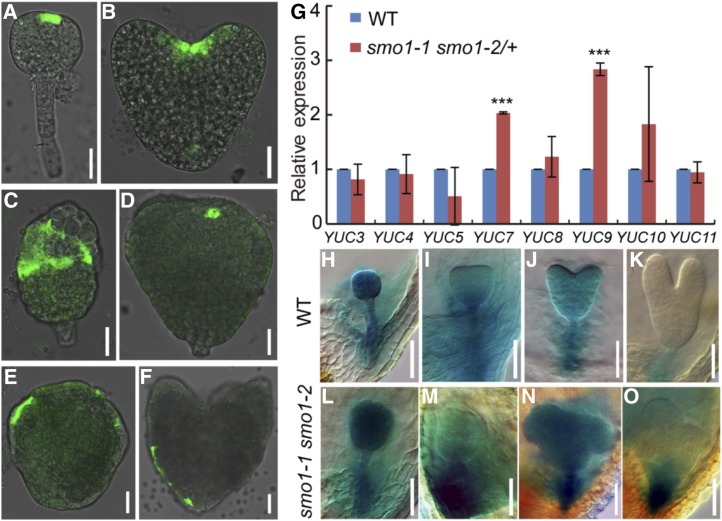
TAA1/TRYPTOPHAN AMINOTRANSFERASE RELATED and YUC genes are key auxin biosynthesis genes and essential for embryogenesis (Cheng et al., 2006, 2007; Stepanova et al., 2008; Robert et al., 2018). To detect whether the expression pattern of TAA1 was affected in smo1-1 smo1-2 embryos, we introduced ProTAA1:GFP-TAA1 (GFP-TAA1) into the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants. In wild-type embryos, at the globular stage, the GFP signal was detected in the apical protoderm layer (Fig. 6A); at the heart stage, the GFP signal was detected in the SAM (Fig. 6B). Compared to wild-type, the smo1-1 smo1-2 embryos exhibited abnormal GFP-TAA1 expression (Fig. 6, C–F). In these embryos, GFP-TAA1 was expressed in most of the protoderm cells of the upper half (Fig. 6, C and E), only expressed in one or two cells of the apical layer (Fig. 6D), or even detected in some protoderm cells of the lower half (Fig. 6F). We then analyzed the expression of YUC genes in 1- to 3-DPA smo1-1 smo1-2/+ seeds by RT-qPCR. In comparison to wild-type, the expression of YUC7 and YUC9 genes was significantly increased in the smo1-1 smo1-2/+ seeds (Fig. 6G). Subsequently, we crossed ProYUC9:GUS transgenic plants with smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants and detected the expression of the GUS reporter gene in different embryo developmental stages. In wild-type, GUS was expressed throughout the embryo at the globular stage (Fig. 6H); then, its expression became weaker at the transition and heart stage (Fig. 6, I and J), whereas at the torpedo stage, its expression was only detectable in the suspensor (Fig. 6K). In contrast, the expression of ProYUC9:GUS was increased in the smo1-1 smo1-2 embryos, and the GUS signal was either strongly detected throughout the embryos (Fig. 6, L and N) or strongly detected in the suspensor (Fig. 6, M and O). Together, these results indicate that in the smo1-1 smo1-2 embryos, expression of auxin biosynthesis genes is enhanced, and their expression domains are also altered.
Figure 6.
Expression analyses of TAA1 and YUC genes in wild-type (WT) and putative smo1-1 smo1-2 embryos. A and B, Expression patterns of ProTAA1:GFP-TAA1 in wild-type embryos at globular (A) and heart (B) stages. C to F, Expression patterns of ProTAA1:GFP-TAA1 in putative smo1-1 smo1-2 embryos. G, Relative transcript levels of YUC genes in early developing seeds dissected from 1- to 3-DPA siliques. The expression levels were normalized to that of TAP42 INTERACTING PROTEIN OF 41 KDA (TIP41). Values are means ± sd of three independent experiments. For each experiment, approximately 0.05 g of seeds dissected from 1- to 3-DPA siliques of wild-type and smo1-1 smo1-2/+ mutant, respectively, was used to extract total RNA, 1 μg of total RNA was reverse transcribed into cDNA, 1 μL of each cDNA sample was mixed with 7.5 µL of SYBR Green Real-Time PCR Master Mix (DBI Bioscience) and then analyzed by reverse-transcription quantitative PCR (RT-qPCR). Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; ***P < 0.001). H to K, Expression patterns of ProYUC9:GUS in wild-type embryos at globular (H), triangular (I), heart (J), and torpedo (K) stages. L to O, Expression patterns of ProYUC9:GUS in putative smo1-1 smo1-2 embryos. Bars = 25 μm (A–F) and 50 μm (H–O).
To explore whether an auxin biosynthesis inhibitor could rescue the embryonic defects in smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants, we treated pistils of opening flowers from smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants with 500 µm l-kynurenine (Kyn) and quantified the aborted seeds at 10 d posttreatment. There was no difference between the wild-type before and after treatment. However, the percentage of aborted seeds was dramatically reduced in smo1-1/+ smo1-2 (after treatment: 15.2%, n = 594; before treatment: 23.3%, n = 505) and smo1-1 smo1-2/+ (after treatment: 17.7%, n = 607; before treatment: 23.2%, n = 589) siliques after Kyn treatment (Supplemental Fig. S10). These results suggest that exogenous application of auxin biosynthesis inhibitor can partially rescue the seed abortion phenotype of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ siliques.
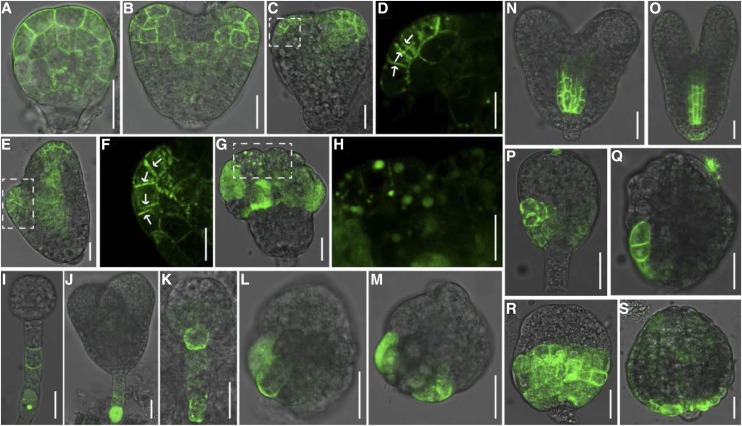
In addition to auxin biosynthesis, polar localization of PIN auxin efflux carriers, especially PIN1 and PIN7, is essential for the establishment and maintenance of correct auxin gradients during early embryo development (Friml et al., 2003). To explore whether PIN1 and PIN7 localization was altered in the smo1-1 smo1-2 embryos, we introduced PIN1-GFP and PIN7-GFP into the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants. In wild-type embryos, at the globular stage, PIN1-GFP was localized in the PM of proembryo cells without obvious polarity (Fig. 7A); at the heart stage, PIN1-GFP was detected in the basal PM of the provascular cells, whereas in the epidermis layer, PIN1-GFP was polarly localized in PMs toward the sites where cotyledons would be initiated (Fig. 7B). In contrast, PIN1-GFP localization was disrupted in the putative smo1-1 smo1-2 embryos (Fig. 7, C–H). PIN1-GFP was found at two opposite neighbor PMs in putative smo1-1 smo1-2 embryos (Fig. 7, D and F). Moreover, it was found within intracellular aggregates in the smo1-1 smo1-2 embryo cells (Fig. 7, G and H). At the globular and heart embryo stages, PIN7-GFP was localized in the basal PM of suspensor cells in wild-type embryos (Fig. 7, I and J). However, in putative smo1-1 smo1-2 embryos, PIN7-GFP was detected in the hypophysis (Fig. 7K) or in cells in the basal part of the embryo proper (Fig. 7, L and M). These results demonstrate that polar localization of PIN auxin efflux carriers is altered in the smo1-1 smo1-2 embryos.
Figure 7.
Localization of auxin efflux and influx proteins in wild-type and putative smo1-1 smo1-2 embryos. A and B, PIN1-GFP localization in wild-type embryos at globular (A) and heart (B) stages. C to H, PIN1-GFP localization in putative smo1-1 smo1-2 embryos. D, F, and H, Higher-magnification images of the boxed regions in C, E, and G. Arrows indicate disorganized PIN1 localization in the plasma membranes. I and J, PIN7-GFP localization in wild-type embryos at globular (I) and heart (J) stages. K to M, PIN7-GFP localization in putative smo1-1 smo1-2 embryos. N and O, AUX1-yellow fluorescent protein (YFP) localization in wild-type embryos at heart (N) and torpedo (O) stages. P to S, AUX1-YFP localization in putative smo1-1 smo1-2 embryos. Bars = 25 μm (A–C, E, G, and I to S), 5 μm (F), and 15 μm (D and H).
PIN proteins recycle between the PM and endosomal compartments, and their polar localization depends on directed vesicle trafficking (Geldner et al., 2001, 2003). The abnormal localization of PIN1-GFP in smo1-1 smo1-2 embryos (Fig. 7, C–H) suggested that SMO1 may be involved in the vesicle trafficking. To explore this possibility, we treated the wild-type and smo1-1 smo1-2/+ seedlings with Brefeldin A (BFA) and examined the formation of BFA bodies. BFA is used as a vesicle trafficking inhibitor, which blocks PIN1 cycling (Geldner et al., 2001). After 100 µm BFA treatment for 40 min, although PIN1-GFP-containing BFA bodies was accumulated in both wild-type and smo1-1 smo1-2/+ seedling root cells, the smo1-1 smo1-2/+ mutant showed somewhat fewer BFA bodies than the wild-type (Supplemental Fig. S11, A, B and E). The effects of BFA treatment on vesicle trafficking were reversible. After washout of BFA for 90 min, BFA bodies in both wild-type and smo1-1 smo1-2/+ root cells were markedly decreased (Supplemental Fig. S11, C and D). However, the smo1-1 smo1-2/+ root cells (26.4%, n = 11) retained significantly more BFA bodies than the wild-type (16.9%, n = 7; Supplemental Fig. S11E). These results demonstrate that SMO1 deficiency affects PIN1 protein cycling between the PM and endosomes.
AUX1/LAX proteins, in concert with PIN proteins, maintain balanced auxin transport and are required for establishing a correct embryo pattern (Robert et al., 2015). To explore whether AUX1 localization was altered in the smo1-1 smo1-2 embryos, we introduced AUX1-YFP into the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants. In wild-type heart and torpedo stage embryos, AUX1-YFP was strongly detected in the PM of provascular cells and weakly detected in the hypophysis and the several epidermal cells around the hypophysis (Fig. 7, N and O). By contrast, in putative smo1-1 smo1-2 embryos, AUX1-YFP was strongly detected in the PM of either several or all of the epidermal cells in the lower half of the embryo proper, and no AUX1-YFP signal was observed in the inner cells (Fig. 7, P–S).
Together, these results demonstrate that enhanced auxin biosynthesis and disrupted PAT in smo1-1 smo1-2 embryos result in abnormal auxin activity, and exogenous application of an auxin biosynthesis inhibitor can partially rescue its embryo lethality.
Cytokinin Biosynthesis and Response Are Altered in smo1-1 smo1-2 Embryos
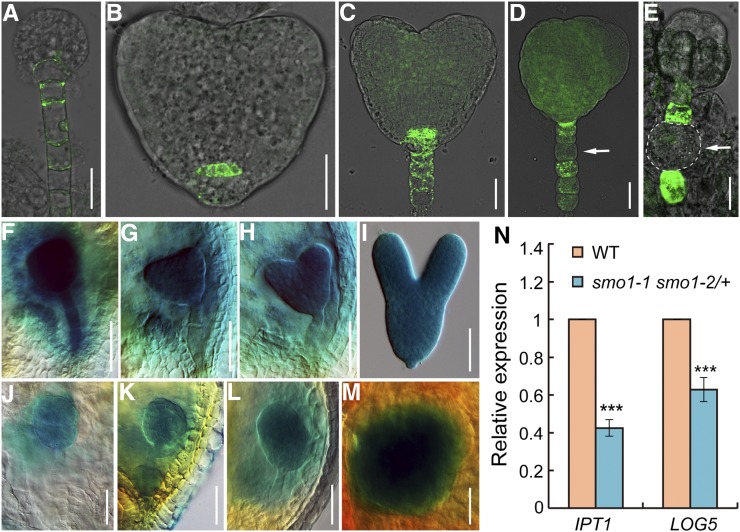
Auxin and cytokinin interact in many aspects of plant growth and development. Similar to auxin, cytokinin is also a key regulator during plant embryogenesis. To investigate whether defective embryo development of the smo1-1 smo1-2 mutants was caused by cytokinin deficiency, we crossed the TCS:GFP (a cytokinin reporter) marker line (Müller and Sheen, 2008) with smo1-1/+ smo1-2 and smo1-1 smo1-2/+ plants and examined the GFP signals in embryos. In wild-type embryos, at the globular stage, GFP signal was detected in the hypophyseal and suspensor cells (Fig. 8A). At the heart stage, the GFP signal was observed in the quiescent center cells and suspensor cells (Fig. 8, B and C). However, in the smo1-1 smo1-2 embryos, TCS:GFP signals were only detected in the suspensor cells, and sometimes the GFP signal was not evenly distributed in all suspensor cells (Fig. 8, D and E).
Figure 8.
Expression analyses of cytokinin-related genes. A to C, TCS:GFP distribution in wild-type (WT) embryos at globular (A) and heart (B and C) stages. D and E, TCS:GFP distribution in putative smo1-1 smo1-2 embryos. Arrows indicate the suspensor cells without GFP signal. F to I, Expression patterns of ProAHK4:GUS in wild-type embryos at globular (F), triangular (G), heart (H), and torpedo (I) stages. J to M, Expression patterns of ProAHK4:GUS in putative smo1-1 smo1-2 embryos. N, Relative transcript levels of cytokinin biosynthesis genes IPT1 and LOG5 in early developing seeds dissected from 1- to 3-DPA siliques. The expression levels were normalized to that of TIP41. Values are means ± sd of three independent experiments. For each experiment, approximately 0.05 g of seeds dissected from 1- to 3-DPA siliques of wild-type and smo1-1 smo1-2/+ mutant, respectively, was used to extract total RNA, and 1 μg of total RNA was use for RT-qPCR analysis. Significant differences were analyzed by Student’s t test (one-tailed, two-sample equal variance; ***P < 0.001). Bars = 25 μm (A–E) and 50 μm (F–M).
We further examined the expression patterns of a relevant cytokinin receptor gene, AHK4, because among the four cytokinin receptors (AHKs), only the AHK4 gene is detected during embryo development (Müller and Sheen, 2008). We crossed ProAHK4:GUS transgenic plants (Higuchi et al., 2004) with smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants and detected the expression of the GUS reporter gene in different embryonic developmental stages. In wild-type, GUS was strongly expressed throughout the embryo at the globular stage (Fig. 8F). From the transition to the heart stage, although the expression of AHK4 was still strong in the embryo proper, its expression in the suspensor was reduced (compare Fig. 8, G and H with Fig. 8F). At the torpedo stage, AHK4 expression was slightly decreased compared with the early stages (Fig. 8I). In contrast, ProAHK4:GUS expression was obviously decreased in the smo1-1 smo1-2 embryos (Fig. 8, J–M). Moreover, sometimes the GUS signal was not evenly detected in the embryo proper; it was either stronger in the apical part of the embryo proper that gave rise to one cotyledon primordium (Fig. 8L) or only observed in the inner region (Fig. 8M). These data suggest that cytokinin activity is decreased in smo1-1 smo1-2 embryos.
Furthermore, we wondered whether cytokinin biosynthesis was affected in smo1-1 smo1-2 mutants. IPT and LOG are key cytokinin biosynthesis genes and are essential for maintenance of the apical meristems. Therefore, we examined the expression of the IPT1 and LOG5 genes in 1- to 3-DPA seeds by RT-qPCR. In comparison to wild-type, expression of the IPT1 and LOG5 genes was significantly decreased in smo1-1 smo1-2/+ seeds (Fig. 8N).
To explore whether cytokinin could rescue the embryo defects of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants, we treated their pistils with 500 µm 6-benzylamino purine (6-BA) and quantified the aborted seeds 10 d posttreatment. There was no difference between the wild-type before and after treatment. However, the percentage of aborted seeds was dramatically reduced in smo1-1/+ smo1-2 (after treatment: 18.5%, n = 467; before treatment: 25.6%, n = 420) and smo1-1 smo1-2/+ (after treatment: 17.4%, n = 320; before treatment: 24.9%, n = 433) after 6-BA treatment (Supplemental Fig. S12). These results suggest that exogenous cytokinin application can partially rescue the seed abortion phenotype of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants.
Together, these data demonstrate that mutation of SMO1-1 and SMO1-2 genes also impairs cytokinin biosynthesis and response during embryo development.
The Auxin and Cytokinin Balance Is Perturbed in smo1-1/+ smo1-2 and smo1-1 smo1-2/+ Mutants
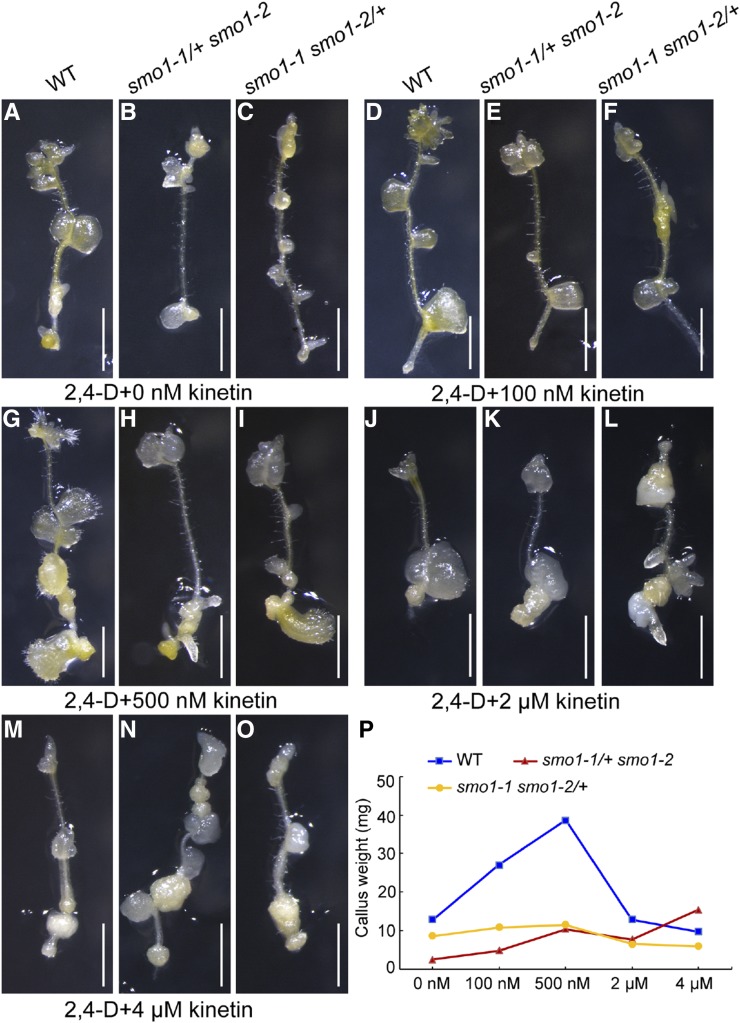
The appropriate ratio of auxin and cytokinin is important for embryo development (Zhang et al., 2017). The smo1-1 smo1-2 embryos displayed enhanced auxin biosynthesis and response and decreased cytokinin biosynthesis and response, suggesting that their balance between auxin and cytokinin was altered. To test this, we performed tissue culture of root segments from smo1-1/+ smo1-2 and smo1-1 smo1-2/+ seedlings. When cultured in Murashige and Skoog (MS) medium supplemented with 0 nm, 100 nm, 500 nm, 2 µm, and 4 µm kinetin, in the presence of 450 nm 2,4-dichlorophenoxy acetic acid (2,4-d), we observed that at lower concentrations of kinetin (0 to 500 nm), the wild-type root fragments developed calli more rapidly than the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants (Fig. 9, A–I and P). In contrast, at higher kinetin concentrations (2 and 4 µm), the performance of the mutants was comparable to the wild-type (Fig. 9, J–P). Consistently, when cultured in MS medium supplemented with 0 nm, 100 nm, 500 nm, 2 µm, and 4 µm 2,4-d, in the presence of 450 nm 6-BA, the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ root fragments developed calli more rapidly than wild-type at lower concentrations of 2,4-d (500 nm and 2 µm; Supplemental Fig. S13). These data further demonstrate that smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants have higher endogenous auxin activity and lower cytokinin activity than wild-type, and the appropriate ratio of auxin versus cytokinin activity is altered in these mutants.
Figure 9.
Callus growth of wild-type (WT), smo1-1/+ smo1-2, and smo1-1 smo1-2/+ root segments. A to O, Root segments were cultured on MS medium containing 450 nm 2,4-d plus different concentrations of kinetin. After 21 d in culture, the induced calli were photographed. Bars = 1 mm. (P) Callus weight at 30 d postculture. n = 14.
WOX5 Is Ectopically Expressed in smo1-1 smo1-2 Embryos
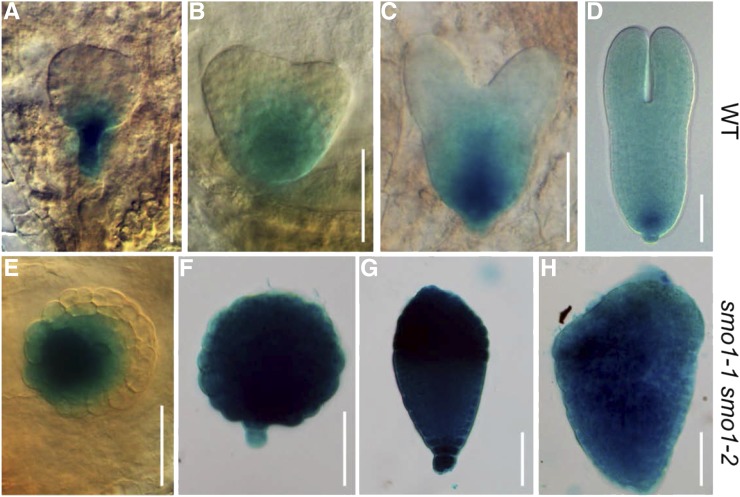
Auxin and cytokinin interact to regulate the expression of key transcription factors, including WOX5, during initiation of the root apical meristem (RAM) in early embryogenesis (Müller and Sheen, 2008; Zhang et al., 2017). Therefore, we explored the expression of WOX5 in smo1-1 smo1-2 embryos using ProWOX5:GUS transgenic plants (Sarkar et al., 2007). In wild-type embryos, the expression of WOX5 was restricted to the lens-shaped cell that gave rise to the quiescent center from the globular to torpedo stage (Fig. 10, A–D), whereas in smo1-1 smo1-2 embryos, WOX5 expression was broader and often expanded throughout the embryo proper (Fig. 10, E–H). These results suggest that restricted expression of WOX5 in the RAM cannot be established in the smo1-1 smo1-2 embryo.
Figure 10.
Expression patterns of ProWOX5:GUS in wild-type (WT) and putative smo1-1 smo1-2 embryos. A to D, Expression patterns of ProWOX5:GUS in wild-type embryos at globular (A), triangular (B), heart (C), and torpedo (D) stages. E to H, Expression patterns of ProWOX5:GUS in putative smo1-1 smo1-2 embryos. Bars = 50 μm.
smo1-1/+ smo1-2 and smo1-1 smo1-2/+ Mutants Accumulate 4,4-Dimethylsterols
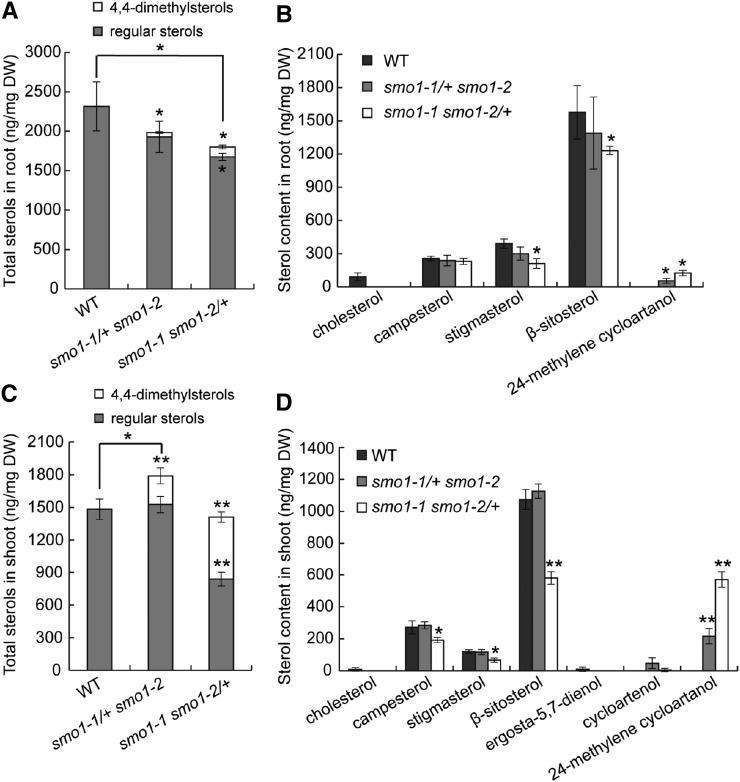
To examine whether sterol profiles were affected in the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants, we performed sterol analyses of seedling shoots and roots. Total sterol content was decreased in both smo1-1/+ smo1-2 and smo1-1 smo1-2/+ roots, with the latter showing significant differences (Fig. 11A). Sterol composition was also altered in the roots of these mutants, which accumulated large amounts of 24-methylene cycloartanol (a 4,4-dimethylsterol, which was not detected in the wild-type), accompanied by reduced levels of regular sterols, including cholesterol, stigmasterol, and β-sitosterol, especially in the smo1-1 smo1-2/+ roots (Fig. 11B). In smo1-1 smo1-2/+ mutant shoots, total sterol content was comparable to wild-type, but approximately 41% of the sterols were 4,4-dimethylsterols (Fig. 11C), and regular sterols, including campesterol, stigmasterol, and β-sitosterol, were markedly reduced (Fig. 11D). In contrast, in the smo1-1/+ smo1-2 mutant shoots, total sterol content was increased as a consequence of the accumulation of 4,4-dimethylsterols (≈15% of total sterols; Fig. 11, C and D). Moreover, two types of 4,4-dimethylsterols, cycloartenol and 24-methylene cycloartanol, accumulated in these mutant shoots (Fig. 11D). This finding suggests that in addition to 24-methylene cycloartanol, cycloartenol may also be a substrate of SMO1 enzymes.
Figure 11.
Sterol content of wild-type (WT), smo1-1/+ smo1-2, and smo1-1 smo1-2/+ seedlings. A and C, Total sterol (including regular sterols and 4,4-dimethylsterol precursors) content in roots (A) and shoots (C). B and D, Composition analyses of sterols found in roots (B) and shoots (D). Values are means ± sd of four experiments (A and B) and five experiments (C and D). For each experiment, samples were prepared from shoots or roots of 7-d-old wild-type, smo1-1/+ smo1-2, and smo1-1 smo1-2/+ seedlings. Significant differences were analyzed by two-tailed Wilcoxon rank-sum test (nonparametric due to n < 20; *P < 0.05; **P < 0.01). DW, dry weight.
DISCUSSION
The Three SMO1 Genes Play Redundant as Well as Specific Roles in Embryonic and Postembryonic Development
In this study, we found that all three SMO1 genes were expressed at all stages during embryo development, but only SMO1-1 was clearly detected in endosperm (Fig. 1). Moreover, SMO1-1 and SMO1-2 clearly showed complementary expression in both embryo and seedling root (Fig. 1; Supplemental Fig. S2). In general, the expression of SMO1-3 was weaker than SMO1-1 and SMO1-2. These findings suggest that the three SMO1 genes play redundant as well as specific roles in embryo and root development, and SMO1-3 may play less important roles than SMO1-1 and SMO1-2. Indeed, single smo1 mutants showed no obvious phenotype, and smo1-1 smo1-3 double mutants displayed very mild phenotypes in silique length, pollination rate, and plant height, but the smo1-1 smo1-2 double mutant was embryo lethal. We also found that the smo1-1 smo1-2/+ seedling showed a more severe root phenotype than the smo1-1/+ smo1-2 seedling (Supplemental Fig. S7, A–C). This phenotype was correlated with the altered sterol composition in these mutants, as the smo1-1 smo1-2/+ mutant showed a greater accumulation of precursor and reduction of regular sterols (Fig. 11). These findings suggest that SMO1-1 plays more important roles than SMO1-2 in root development.
SMO1s Play Similar Yet More Important Roles than SMO2s in Embryogenesis
Mutants of sterol biosynthetic pathway genes upstream of SMO2, such as smt1, fk, and hyd1, exhibit severe embryonic defects, but these embryos can germinate into seedlings (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002). By contrast, smo2-1 smo2-2 embryos are lethal (Zhang et al., 2016). Similar to smo2-1 smo2-2 mutants, smo1-1 smo1-2 was also embryo lethal, but its embryonic phenotypes were more severe and diverse (Fig. 3). Interestingly, two types of novel embryonic phenotypes were observed in the smo1-1 smo1-2 embryos: abnormal division of suspensor cells (class II) and twin embryos (class IV). We noticed that the class II smo1-1 smo1-2 embryos were very similar to the previously reported cph/smt1 hyd1 and cph/smt1 fk double mutants (Schrick et al., 2002). These findings suggest that SMO1s and SMO2s are essential for embryogenesis, and SMO1s may play more important roles than SMO2s in this process. Although SMO1s and SMO2s are both 4α-methyl oxidases, they have different substrate specificity. The results of our sterol analyses in this study and previous studies (Zhang et al., 2016) clearly revealed that 4,4-dimethylsterols accumulated in SMO1-deficient mutants, whereas 4α-methylsterols accumulated in SMO2-deficient mutants. Additionally, total sterol content was significantly reduced in smo1-1 smo1-2/+ mutant roots and increased in smo1-1/+ smo1-2 mutant shoots, but the levels in the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants roots and shoots were comparable to those in wild-type. These differences in sterols may account for the embryo phenotypic differences between these mutants. Indeed, a previous report has shown that the levels of the major plant sterols, such as sitosterol and stigmasterol, vary in different tissues; the highest levels of sitosterol are found in rapidly dividing and differentiating tissues, especially in the young embryo, whereas the highest levels of stigmasterol are found in fully differentiated leaves (Schrick et al., 2011). Sterol analysis of smo1-1 smo1-2 and smo2-1 smo2-2 embryos will provide additional information.
In addition, smo2-1 smo2-2/+ plants display dwarf phenotypes similar to the downstream mutants of the sterol biosynthetic pathway (Zhang et al., 2016), but the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ plants are normal in plant height and rosette size. As there are three SMO1 genes, we speculate that further knockout of the SMO1-3 gene in the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants might generate a dwarf phenotype.
SMO1s Maintain a Proper Activity Ratio of Cytokinin/Auxin during Embryogenesis
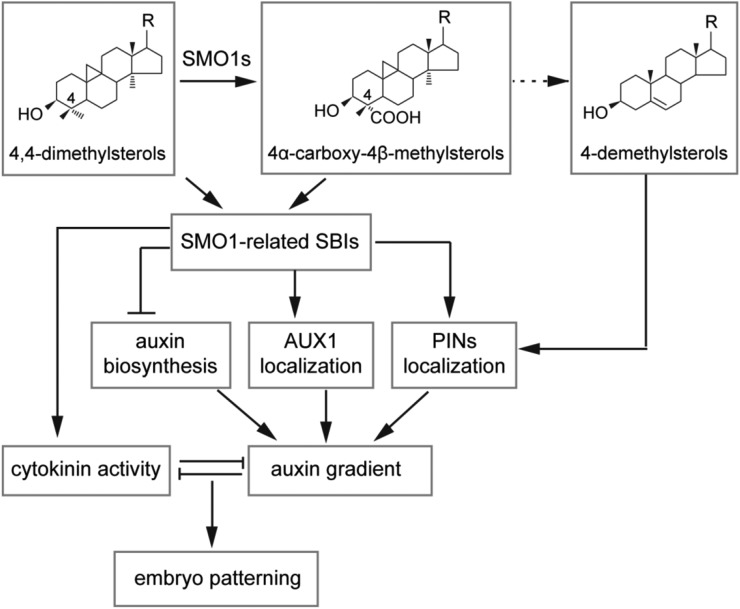
Auxin and cytokinin interact to regulate many plant growth and developmental processes, such as the initiation of SAM and RAM, branching, and embryogenesis. It has been reported that auxin signaling directly activates the transcription of the cytokinin response regulator genes ARABIDOPSIS RESPONSE REGULATOR7 (ARR7) and ARR15 to reduce the cytokinin response during early embryogenesis (Müller and Sheen, 2008). The Aux/ indole-3-acetic acid protein SHORT HYPOCOTYL2 acts as a central switch in controlling the balance of auxin and cytokinin signaling (Moubayidin et al., 2010). Classical tissue cultural experiments have demonstrated that a high cytokinin/auxin ratio is essential for shoot formation (Skoog and Miller, 1957). The balance between the cytokinin and auxin pathway is also essential for the formation of SAM during embryogenesis (Su et al., 2015; Zhang et al., 2017). We found that the overall phenotype of the smo1-1 smo1-2 embryos featured an absence of cotyledon and SAM (Fig. 3). Further investigations showed that in the smo1-1 smo1-2 embryos, cytokinin biosynthesis and response were downregulated, whereas auxin biosynthesis and response were upregulated. Tissue culture experiments also demonstrated a low cytokinin/auxin ratio in these mutants. Consistently, exogenous application of either auxin biosynthesis inhibitor or cytokinin partially rescued the smo1-1 smo1-2 embryonic lethality (Supplemental Figs. S10 and S12). Although it is unclear whether the cytokinin reduction was a direct result of sterol deficiency or enhanced auxin biosynthesis and response, we can conclude that SMO1s function through maintaining the correct contents of SMO1-related SBIs and regular sterols to maintain the appropriate activity ratio between cytokinin and auxin during embryo development (Fig. 12).
Figure 12.
A working model for SMO1s in embryonic development. SMO1 enzymes catalyze the conversion of 4,4-dimethylsterols into 4α-carboxy-4β-methylsterols, which are further converted into end-product 4-demethylsterols, including β-sitosterol, campesterol, stigmasterol, and cholesterol. Correct composition of these sterols is required for polar localization of PIN proteins. SMO1-related SBIs may inhibit auxin biosynthesis, affect the polar localization of AUX1 and PIN proteins, and affect cytokinin activity, thereby maintaining an appropriate ratio between auxin and cytokinin activities, which is required for embryo development. R, Side chain of the sterol.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0, Landsberg erecta, and Wassileskija-4 were used as wild-type controls in this study. The T-DNA insertion lines smo1-1.1 (SALK_021399), smo1-1.2 (FLAG_286D02), smo1-2 (CSHL_GT13595), smo1-3.1 (CSHL_ET12310), and smo1-3.2 (FLAG_425A03) were obtained from the Nottingham Arabidopsis Stock Center and the Versailles Arabidopsis Stock Center. DR5rev:GFP (Friml et al., 2003), PIN1-GFP (Benková et al., 2003), PIN7-GFP (Friml et al., 2003), AUX1-YFP (Swarup et al., 2004), ProTAA1:GFP-TAA1 (Stepanova et al., 2008), ProYUC9:GUS (Hentrich et al., 2013), TCS:GFP (Müller and Sheen, 2008), ProAHK4:GUS (Higuchi et al., 2004), and ProWOX5:GUS (Sarkar et al., 2007) were crossed into smo1-1/+ smo1-2 and smo1-1 smo1-2/+ plants.
Seeds were sterilized for 5 min in 70% (v/v) ethanol and for 10 min in 1% (v/v) NaClO and were washed five times with sterile water. After incubation for 3-4 d at 4°C, the seeds were sowed on MS medium supplemented with 1% (w/v) Suc and solidified by 0.8% (w/v) agar. Plates were transferred to 22°C for germination in a growth chamber. Seedlings were transferred to soil ∼7 d later and grown in the culture room under a 16-h light/8-h dark cycle at 22°C.
Vector Construction and Plant Transformation
ProSMO1-1:GUS was generated by insertion of a 1,954-bp genomic fragment containing the SMO1-1 promoter sequence and 5′ untranslated region into pGreenII0229-GUS (Men et al., 2008) at the EcoRI and BamHI sites. A 1,755-bp genomic fragment containing the SMO1-2 promoter sequence and 5′ untranslated region was inserted at the XhoI and BamHI sites, and a 1,471-bp genomic fragment containing the SMO1-3 promoter sequence was inserted at the XhoI and BamHI sites.
To obtain Pro-35S:SMO1-1-EGFP, Pro-35S:SMO1-2-EGFP, and Pro-35S:SMO1-3-EGFP constructs, the EGFP coding sequences were inserted into the pBA002 vector (provided by Nam-Hai Chua) between the SpeI and SacI sites. Then, an 894-bp SMO1-1 coding region, 897-bp SMO1-2 coding region, and 873-bp SMO1-3 coding region, respectively, were cloned into the pBA002-EGFP construct. For ProSMO1-1:SMO1-1-GFP and ProSMO1-2:SMO1-2-GFP, the SMO1-1 promoter region (1,470-bp upstream sequence of the start codon) and SMO1-2 promoter region (1,755-bp upstream sequence of the start codon) were amplified and cloned into the pGreenII0229-EGFP vector, respectively. Then, the SMO1-1 and SMO1-2 coding regions were cloned into the above-obtained ProSMO1-1:EGFP and ProSMO1-2:EGFP. The sequences of the primers are listed in Supplemental Table S1.
The above constructs were introduced into Agrobacterium tumefaciens strain C58C1 (pMP90/pJIC Sa-Rep) and transformed into ecotype Columbia-0 plants using the floral dip method (Clough and Bent, 1998). Transgenic plants were screened by spraying with 0.02% (v/v) Basta (Sangon Biotech) and PCR amplification.
RT-qPCR
For analyses of SMO1-1, SMO1-2, and SMO1-3 transcripts in wild-type and smo1 mutants, total RNA was extracted from 7-d-old seedlings using the Plant Total RNA Purification Kit (TIANGEN). Next, 1 μg of total RNA was reverse transcribed into cDNA using EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech). The ACTIN2 gene was used as an internal control. The primers are listed in Supplemental Table S1.
RT-qPCR experiments were performed in triplicate using SYBR Green Real-Time PCR Master Mix (DBI Bioscience). Three biological replicates of 1- to 3-DPA seeds from wild-type and smo1-1 smo1-2/+ were isolated, and total RNA samples were extracted as described in the Plant Total RNA Purification Kit (TIANGEN). One microgram of total RNA was reverse transcribed into cDNA using EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech). One microliter of each cDNA sample was mixed with 7.5 µL of SYBR Green Real-Time PCR Master Mix and then analyzed on a fluorescent quantitative PCR machine (Eppendorf). The TIP41 gene was used as an internal control. We used the cycle threshold (Ct) 2-∆∆Ct method to calculate the relative expression level. The RT-qPCR primers are listed in Supplemental Table S1.
GUS Histochemical Staining
To assay the GUS activity, embryos at different developmental stages were dissected from siliques using a Leica M165FC dissection microscope. The samples were incubated overnight at 37°C in the staining solution (1.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-GlcA; 50 mm sodium phosphate buffer, pH 7.0; 0.1% [v/v] Triton X-100; 0.5 mm potassium ferricyanide; and 0.5 mm potassium ferrocyanide). After incubation, the samples were cleared in a clearing solution of 8:3:1 (w:v:v) chloral hydrate:distilled water:glycerol. Then the samples were photographed using an Olympus BX63 microscope under differential interference contrast.
Histology and Fluorescence Detection in Embryos
Seeds and ovules were cleared using a clearing solution of 8:3:1 (w:v:v) chloral hydrate:distilled water:glycerol as described by Men et al. (2008). Samples were observed under an Olympus BX63 microscope using differential interference contrast. Wild-type and mutant embryos containing the GFP signals (DR5rev:GFP, PIN1-GFP, PIN7-GFP, AUX1-YFP, ProTAA1:GFP-TAA1, and TCS-GFP) were dissected from the seeds at different developmental stages and mounted in 8% (v/v) Glc solution. The GFP signals were observed using a confocal laser-scanning microscope (Leica TCS SP5).
Immunocytochemistry
Immunolocalization was conducted using the roots from Pro-35S:SMO1-EGFP transgenic plants as previously described by Men et al. (2008), with 8–10 roots from the different transgenic lines. A mouse monoclonal anti-BiP (used as an ER maker) antibody (Enzo Life Sciences) was diluted 1,000 times. Donkey anti-mouse Tetramethylrhodamine was used as the secondary antibody (Jackson ImmunoResearch) and diluted 1:200.
Chemical Application
To test the effect of 24-epibrassinolide (BL) on root growth, wild-type and mutant seeds were germinated on plates containing MS medium supplemented with different concentrations (0 pm, 10 pm, 20 pm, 50 pm, 0.1 nm, 1 nm, and 10 nm) of BL for 7 d. Approximately 30 seedlings were used for the measurement. BL was dissolved in 95% (v/v) ethanol. A mock treatment was performed using an equal volume of 95% (v/v) ethanol. To examine the effect of the exogenous auxin biosynthesis inhibitor Kyn and cytokinin on the aborted seeds from the smo1-1/+ smo1-2 and smo1-1 smo1-2/+ siliques, 10 opening flowers from five plants were dipped in a 500 μm Kyn or 6-BA solution for 30 s, and these flowers were continuously treated for 3 d. Ten days after treatment, the aborted seeds in these siliques were scored. Kyn and 6-BA were dissolved in 0.1 m HCl and 1 m NaOH, respectively. A mock treatment was performed using an equal volume of 0.1 m HCl and 1 m NaOH. BL, Kyn, and 6-BA were purchased from Sigma-Aldrich.
BFA Treatment
Five-day-old seedlings were incubated in half-strength MS liquid medium containing 100 µm BFA (Selleck Chemicals) for 40 min and then observed. For BFA washout analyses, 5-d-old seedlings were first treated with 100 µm BFA for 40 min and then washed with half-strength MS liquid medium for 90 min. After treatment, the seedlings were photographed using a confocal laser-scanning microscope (Leica TCS SP5). For quantitative analyses of BFA bodies, stele cells of 7–11 roots were scored.
Tissue Culture
All media for tissue culture were supplemented with 1 mg/L biotin and different concentrations of auxin and cytokinin. The medium contained 4.4 g/L MS salts, including vitamins (Duchefa Biochemie), 1% (w/v) Suc, 0.05% (w/v) 2-(N-morpholino) ethane-sulfonic acid (pH 5.8 adjusted with KOH), and 0.8% (w/v) plant agar (Duchefa Biochemie). Two kinds of media were used, one containing 450 nm 2,4-d and different concentrations of kinetin (0 nm, 100 nm, 500 nm, 2 µm, and 4 µm), and the other containing 450 nm 6-BA and different concentrations of 2,4-d (0 nm, 100 nm, 500 nm, 2 µm, and 4 µm). Biotin and the plant hormone stocks were added to autoclaved medium at a 1:1,000 dilution. For auxin and cytokinin treatment assay, approximately 1-cm-long root segments were excised from 7-d-old seedlings and then cultured on the above-mentioned medium.
Sterol Measurements
Sterol analyses from 7-d-old seedling shoots and roots were performed as previously described by Zhang et al. (2016).
Statistical Analyses
Three independent repetitions were performed for all experiments. Statistical analyses were performed using one-tailed Student’s t test. All values are presented as means ± sd. Significant differences are noted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or Arabidopsis Genome Initiative database under accession numbers SMO1-1 (AT4G12110), SMO1-2 (AT4G22756), and SMO1-3 (AT4G22755). T-DNA insertion lines used for mutant analyses were as follows: smo1-1.1 (SALK_021399), smo1-1.2 (FLAG_286D02), smo1-2 (CSHL_GT13595), smo1-3.1 (CSHL_ET12310), and smo1-3.2 (FLAG_425A03).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Bioinformatics analyses of SMO1 gene family.
Supplemental Figure S2. Tissue specific expression of SMO1 genes in seedling.
Supplemental Figure S3. Expression patterns of SMO1 genes in inflorescence and siliques.
Supplemental Figure S4. Relative expression levels of SMO1 genes in embryos at different developmental stages.
Supplemental Figure S5. ER localization of SMO1 proteins.
Supplemental Figure S6. Phenotypic analyses of smo1-1 smo1-3 double mutant.
Supplemental Figure S7. Phenotypic analyses of smo1-1/+ smo1-2 and smo1-1 smo1-2/+ mutants.
Supplemental Figure S8. smo1-1 smo1-2 complementation experiments.
Supplemental Figure S9. Endosperm development of wild-type and putative smo1-1 smo1-2 seeds.
Supplemental Figure S10. Exogenous application of an auxin biosynthesis inhibitor partially rescues the embryo lethal phenotype of smo1-1 smo1-2.
Supplemental Figure S11. SMO1 deficiency affects PIN1-GFP cycling in root cells.
Supplemental Figure S12. Exogenous application of cytokinin partially rescues the embryo lethal phenotype of smo1-1 smo1-2.
Supplemental Figure S13. Callus growth of wild-type, smo1-1/+ smo1-2, and smo1-1 smo1-2/+ root segments.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Nottingham Arabidopsis Stock Center and Versailles Arabidopsis Stock Center for the T-DNA insertions. We thank John Innes Centre and Nam-Hai Chua for vectors. We thank Yuling Jiao for the ProAHK4:GUS seeds, Thomas Laux for the ProWOX5:GUS seeds, Stephan Pollmann for the ProYUC9:GUS, and Jiří Friml for DR5rev:GFP and PIN1-GFP seeds. Lipidomic analyses were performed on Bordeaux Metabolome Facility-MetaboHUB (ANR-11-INBS-0010).
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31570247, 31870230, and 91417308 to S.M.).
Articles can be viewed without a subscription.
Note added in proof
Previously, Lung et al. (2017, 2018) obtained similar expression data in guard cells of sepals and anthers of ProSMO1-1:GUS and vascular tissues of ProSMO1-2:GUS transgenic plants, although we were using different constructs which were under the control of both 5′ and 3′ regulating sequences of these genes. Besides, the subcellular localization results of SMO1-1-EGFP and SMO1-2-EGFP on ER obtained by immunolocalization using an anti-BiP antibody (an ER-intrinsic chaperone used as an ER maker) were consistent with the results got by Lung et al. (2017, 2018) using ER-Tracker Red. The functionality of these fusion proteins has been confirmed by complementing the smo1-1 smo1-2 embryo lethal phenotype in this paper.
References
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Benveniste P. (1986) Sterol biosynthesis. Annu Rev Plant Physiol 37: 275–308 [Google Scholar]
- Bishop GJ, Yokota T (2001) Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol 42: 114–120 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Frescatada-Rosa M, Men S, Chow CM, Ebine K, Gustavsson A, Johansson L, Ueda T, Moore I, Jürgens G, et al. (2010) Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J 29: 546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Carland F, Fujioka S, Nelson T (2010) The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Darnet S, Rahier A (2004) Plant sterol biosynthesis: Identification of two distinct families of sterol 4α-methyl oxidases. Biochem J 378: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnet S, Bard M, Rahier A (2001) Functional identification of sterol-4alpha-methyl oxidase cDNAs from Arabidopsis thaliana by complementation of a yeast erg25 mutant lacking sterol-4alpha-methyl oxidation. FEBS Lett 508: 39–43 [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- He JX, Fujioka S, Li TC, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang JC (2003) Sterols regulate development and gene expression in Arabidopsis. Plant Physiol 131: 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S (2013) The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74: 626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J (2000) A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann KA, Feyereisen R (2005) Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol 138: 2033–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Lee H, Oh CJ, Lee HY, Eum HL, Kim HS, Hong YP, Lee Y, Choe S, An CS, et al. (2010) Postembryonic seedling lethality in the sterol-deficient Arabidopsis cyp51A2 mutant is partially mediated by the composite action of ethylene and reactive oxygen species. Plant Physiol 152: 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung SC, Liao P, Yeung EC, Hsiao AS, Xue Y, Chye ML (2017) Acyl-CoA-binding protein ACBP1 modulates sterol synthesis during embryogenesis. Plant Physiol 174: 1420–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung SC, Liao P, Yeung EC, Hsiao AS, Xue Y, Chye ML (2018) Arabidopsis ACYL-COA-BINDING PROTEIN1 interacts with STEROL C4-METHYL OXIDASE1-2 to modulate gene expression of homeodomain-leucine zipper IV transcription factors. New Phytol 218: 183–200 [DOI] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Mialoundama AS, Jadid N, Brunel J, Di Pascoli T, Heintz D, Erhardt M, Mutterer J, Bergdoll M, Ayoub D, Van Dorsselaer A, et al. (2013) Arabidopsis ERG28 tethers the sterol C4-demethylation complex to prevent accumulation of a biosynthetic intermediate that interferes with polar auxin transport. Plant Cell 25: 4879–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo C, Valachovic M, Randall SK, Nickels JT, Bard M (2002) Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc Natl Acad Sci USA 99: 9739–9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA (1996) Cholesterol modification of hedgehog signaling proteins in animal development. Science 274: 255–259 [DOI] [PubMed] [Google Scholar]
- Posé D, Castanedo I, Borsani O, Nieto B, Rosado A, Taconnat L, Ferrer A, Dolan L, Valpuesta V, Botella MA (2009) Identification of the Arabidopsis dry2/sqe1-5 mutant reveals a central role for sterols in drought tolerance and regulation of reactive oxygen species. Plant J 59: 63–76 [DOI] [PubMed] [Google Scholar]
- Rahier A. (2011) Dissecting the sterol C-4 demethylation process in higher plants. From structures and genes to catalytic mechanism. Steroids 76: 340–352 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J (2013) Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J (2015) Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142: 702–711 [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, Wójcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, et al. (2018) Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche Y, Gerbeau-Pissot P, Buhot B, Thomas D, Bonneau L, Gresti J, Mongrand S, Perrier-Cornet JM, Simon-Plas F (2008) Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J 22: 3980–3991 [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, Schmidt J, Jürgens G (2002) Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J 31: 61–73 [DOI] [PubMed] [Google Scholar]
- Schrick K, Fujioka S, Takatsuto S, Stierhof YD, Stransky H, Yoshida S, Jürgens G (2004) A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J 38: 227–243 [DOI] [PubMed] [Google Scholar]
- Schrick K, Cordova C, Li G, Murray L, Fujioka S (2011) A dynamic role for sterols in embryogenesis of Pisum sativum. Phytochemistry 72: 465–475 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–130 [PubMed] [Google Scholar]
- Souter M, Topping J, Pullen M, Friml J, Palme K, Hackett R, Grierson D, Lindsey K (2002) hydra Mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Su YH, Liu YB, Bai B, Zhang XS (2015) Establishment of embryonic shoot-root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front Plant Sci 5: 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al. (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R (2005) Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17: 2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E, Cao Y, Babic V, Cloutier M, Keller W, Wang E, et al. (2011) Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol 156: 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun S, Nie X, Boutté Y, Grison M, Li P, Kuang S, Men S (2016) Sterol methyl oxidases affect embryo development via auxin-associated mechanisms. Plant Physiol 171: 468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tucker E, Hermann M, Laux T (2017) A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev Cell 40: 264–277.e4 [DOI] [PubMed] [Google Scholar]