Abstract
Rab and Arl guanine nucleotide‐binding (G) proteins regulate trafficking pathways essential for the formation, function and composition of primary cilia, which are sensory devices associated with Sonic hedgehog (Shh) signalling and ciliopathies. Here, using mammalian cells and zebrafish, we uncover ciliary functions for Rab35, a multitasking G protein with endocytic recycling, actin remodelling and cytokinesis roles. Rab35 loss via siRNAs, morpholinos or knockout reduces cilium length in mammalian cells and the zebrafish left‐right organiser (Kupffer's vesicle) and causes motile cilia‐associated left‐right asymmetry defects. Consistent with these observations, GFP‐Rab35 localises to cilia, as do GEF (DENND1B) and GAP (TBC1D10A) Rab35 regulators, which also regulate ciliary length and Rab35 ciliary localisation. Mammalian Rab35 also controls the ciliary membrane levels of Shh signalling regulators, promoting ciliary targeting of Smoothened, limiting ciliary accumulation of Arl13b and the inositol polyphosphate 5‐phosphatase (INPP5E). Rab35 additionally regulates ciliary PI(4,5)P2 levels and interacts with Arl13b. Together, our findings demonstrate roles for Rab35 in regulating cilium length, function and membrane composition and implicate Rab35 in pathways controlling the ciliary levels of Shh signal regulators.
Keywords: Arl13b, cilia, left‐right asymmetry, Rab35, Smoothened
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Development & Differentiation; Membrane & Intracellular Transport
Introduction
Primary cilia are microtubule‐based organelles that protrude from the surface of most vertebrate cell types. Operating as antenna‐like structures, primary cilia detect and transmit chemical, light and mechanical signals from the extracellular environment to the intracellular space 1. Primary cilia are also critical for embryonic and postnatal development, serving key roles in important cell–cell communication signalling pathways (e.g. Sonic hedgehog, Wnt, PDGFα) 2. For example, in the limb bud and developing nervous system, bone and neural tube patterning relies on the trafficking of Sonic hedgehog (Shh) signalling proteins into and out of cilia 3. At the left‐right organiser (LRO), or the equivalent Kupffer's vesicle in zebrafish, the correct ratio of immotile and motile cilia 4 direct left‐right patterning of the body plane via mechanisms that involve directional fluid flow, mechano‐ or chemo‐sensation and planar cell polarity (PCP) signalling 5, 6, 7. Not surprisingly, defects in primary and motile cilia cause a wide range of mono‐ or multi‐symptomatic “ciliopathy” disorders such as primary ciliary dyskinesia, situs inversus, polycystic kidney disease, Joubert syndrome (JBTS), Meckel–Gruber syndrome (MKS) and Bardet–Biedl syndrome (BBS), which, collectively, affect most body tissues 8, 9.
Regulation of cilium structure, function and molecular composition is heavily dependent on various intracellular transport pathways. Chief amongst these is intraflagellar transport (IFT), which operates bidirectionally along the ciliary microtubules and is driven by kinesin‐2 and IFT dynein motors, together with IFT‐A, IFT‐B and BBSome cargo adaptor complexes 10, 11, 12, 13. Cilium structure and organisation are also reliant on cytosolic and membrane diffusion barriers at the ciliary base, as well as on secretory, exocytic (e.g. ciliary ectosome release), endocytic and recycling pathways that control ciliary membrane homeostasis 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24. Together, these transport and barrier‐associated processes dynamically control the molecular composition of the ciliary membrane and cytosol and, therefore, the appropriate sensory and signalling output of the organelle.
Intracellular membrane trafficking is highly regulated by members of the Rab and Arf [including Arl (Arf‐like)] families of guanine nucleotide‐binding proteins (G proteins) 25, 26, 27, 28. Acting as molecular switches, alternating between inactive GDP‐ and active (effector binding) GTP‐bound states, G proteins regulate vesicular membrane traffic steps to ensure correct cargo transport via integrated sorting, delivery, uptake and recycling pathways. G protein activity is tightly regulated, with guanine nucleotide exchange factors (GEFs) promoting GTP binding, and GTPase‐activating proteins (GAPs) enhancing the G protein's intrinsic GTPase activity which, when present, hydrolyses GTP to GDP 29. Currently, at least nine of the 66 Rabs that comprise this family of GTPases (Rab 5/8/10/11/17/23/28/29/34), as well as 3 Rab‐like proteins (Rabl2/4/5), are linked to cilium formation and/or function, and the control of ciliary membrane protein levels 30, 31, 32, 33, 34. For example, vertebrate and mammalian Rab11‐Rabin8‐Rab8 cascades are involved in early steps of cilium formation, and rhodopsin transport to the photoreceptor cell cilium (outer segment) 35, 36, 37. Furthermore, a number of cilium‐localised Rab, Rab‐like and Arl proteins are associated with IFT and the BBSome, and the trafficking of Hedgehog signalling intermediates out of cilia 32, 38, 39, 40. Amongst Arf/Arl proteins, ciliary membrane‐restricted Arl13b is linked to ciliogenesis, Shh signal regulation, IFT regulation, and the localisation and distribution of multiple ciliary proteins such as the inositol 1,4,5‐trisphosphate (InsP3) 5‐phosphatase, INPP5E, in part via its role as a GEF of Arl3 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52. Consistent with their important cilia‐related functions, a number of Arls and Rabs such as ARL13B, ARL3, ARL6, RAB23 and RAB28 are mutated in ciliopathy disorders 53, 54, 55, 56, 57.
With the aim of investigating new Rab proteins that regulate cilium formation and/or function, we focused on Rab35, a plasma membrane and endosomal protein with roles in cargo recycling, cytokinesis, actin cytoskeleton regulation, and autophagy, amongst others 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68. Initial clues towards a Rab35 ciliary role include its presence in a photoreceptor outer segment proteome, and high‐throughput siRNA or CRISPR screens implicating Rab35 as either a positive or negative regulator of ciliogenesis, or a regulator of Hedgehog signalling in NIH3T3 mouse fibroblasts 33, 69, 70, 71, 72. Here, using mammalian cell culture and zebrafish models, we show that Rab35 localises to the ciliary membrane and regulates primary and LRO cilium length. We also reveal that Rab35 controls the ciliary levels of Shh signal regulators, Smoothened, Arl13b, and INPP5E, as well as the INPP5E target, PI(4,5)P2. Furthermore, we identify the Rab35 GEF and GAP, DENND1B and TBC1D10A, respectively, as the regulators of Rab35 in the ciliary context. Together, our data uncover a novel conserved role for Rab35 in controlling cilium length and the ciliary levels of mammalian Shh signalling regulators.
Results
Rab35 localises to the ciliary membrane
Rab35 has been found in several proteomes of mammalian primary cilia 70, 71, 73. To more directly investigate a possible ciliary localisation of Rab35, we employed two well‐established mammalian cell models, namely mouse renal epithelial (IMCD3) and human retinal pigment epithelial (hTERT‐RPE1) cells, where ciliogenesis can be induced by serum withdrawal 74, 75. First, we analysed mouse IMCD3 and human hTERT‐RPE1 cells transiently expressing GFP‐tagged Rab35 and found that it localises along the ciliary axoneme in ~60% of transfected ciliated cells (Fig 1A and B). We also observed GFP‐Rab35 at the plasma membrane and in vesicular structures, consistent with previously described localisations in other cell types and its known endosomal functions 58, 60, 61, 62, 63, 64, 66. To further analyse the ciliary association of Rab35, we established a stable hTERT‐RPE1 cell line expressing GFP‐RAB35 and performed super‐resolution microscopy to localise RAB35 in relation to markers of the ciliary membrane (ARL13B) and axoneme (IFT88 and acetylated tubulin; Fig 1C). The GFP‐RAB35 signal coincides with that of ARL13B, with the radial extent of both signals being wider than that of the axonemal markers, indicating association of RAB35 with the ciliary membrane. Notably, in the proximal‐most part of the cilium, the radial extent of the GFP‐RAB35 signal is wider than that of ARL13B (Figs 1C and D, and EV1A and B), and in ~20% of cells, GFP‐RAB35 is also more concentrated in this region (Fig EV1C). This proximal staining is reminiscent of the ciliary pocket localisation for the membrane remodelling Eps15 homology domain (EHD) 1 protein 76 and super‐resolution imaging of GFP‐RAB35 expressing hTERT‐RPE1 cells stained for endogenous EHD1 revealed co‐localisation of both proteins in the proximal region of the cilium; importantly, these EHD1/GFP‐RAB35 co‐localising signals appeared broader than the radial extent of the ARL13B ciliary membrane signal, indicative of their association with the ciliary pocket membrane (Fig 1E). Taken together, these data suggest that GFP‐RAB35 localises to the ciliary pocket in addition to the ciliary membrane.
Figure 1. Rab35 localises to the ciliary membrane.
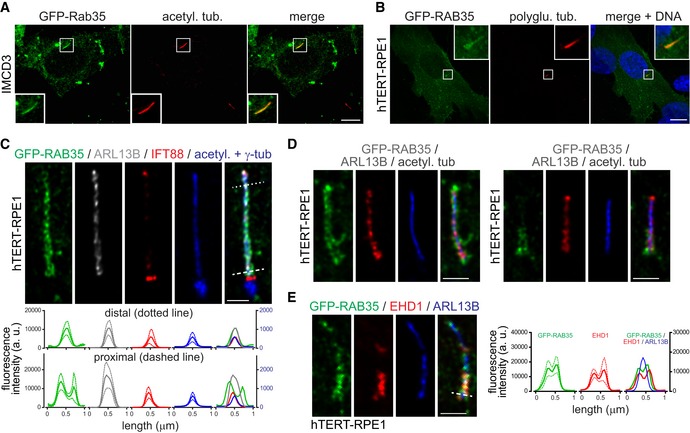
- Localisation of transiently expressed GFP‐Rab35 in IMCD3 cells after 48 h serum starvation and staining for acetylated tubulin (acetyl. tub.) and DNA. Insets show higher magnification images of the cilia region. Scale bars, 10 μm.
- Localisation of transiently expressed GFP‐RAB35 in hTERT‐RPE1 cells after 24 h serum starvation and staining for polyglutamylated tubulin (polyglu. tub.) and DNA. Insets show higher magnification images of the cilia region. Scale bars, 10 μm.
- Super‐resolution (FV‐OSR) imaging of hTERT‐RPE1 stably expressing GFP‐RAB35 after 24 h serum starvation and staining for GFP, ARL13B (cilia membrane), IFT88 (axoneme), acetylated tubulin (acetyl. tub.; axoneme), and γ‐tubulin (γ‐tub; centrosome). Top panels show representative images, and bottom panel shows line profile plots of fluorescence intensity (arbitrary units; a. u.) in the distal (dotted line) and proximal (dashed line) cilia regions. Data are mean ± SEM (n = 5 cilia), with the solid line in the line profile plots indicating mean and the dotted lines indicating SEM values. Scale bars, 1 μm.
- Representative FV‐OSR images of 24 h serum‐starved hTERT‐RPE1 stably expressing GFP‐RAB35 and stained for GFP, ARL13B and acetylated tubulin. Right panels show a cilium with proximal enrichment of GFP‐RAB35 and left panels a cilium with even GFP‐RAB35 localisation along the full length. Scale bars, 1 μm.
- Representative FV‐OSR images of 24 h serum‐starved hTERT‐RPE1 stably expressing GFP‐RAB35 and stained for GFP, EHD1 and ARL13B. Line profile plots of fluorescence intensity in the proximal cilia region (dashed line) are shown to the right. Data are mean ± SEM (n = 6 cilia), with the solid line in the line profile plots indicating mean and the dotted lines indicating SEM values. Scale bars, 1 μm.
Figure EV1. Rab35 localises to the ciliary membrane.

-
A, BSuper‐resolution (FV‐OSR) imaging of hTERT‐RPE1 stably expressing GFP‐RAB35 after 24 h serum starvation and staining for GFP and acetylated tubulin (acetyl. tub.) (A) or GFP, ARL13B and polyglutamylated tubulin (polyglu. tub.). Scale bars, 1 μm.
-
CRepresentative images of hTERT‐RPE1 cells stably expressing GFP‐RAB35 after 24 h serum starvation and staining for GFP, ARL13B, polyglutamylated tubulin (polyglu. tub.) and DNA. Higher magnification images of the cilia region shown to the bottom. Scale bars, 10 μm. Graph to the right shows quantification of cilia with GFP‐RAB35 localisation to the proximal region of the cilium marked with polyglutamylated tubulin staining (ARL13B > GFP‐RAB35 = polyglu. tub.) and cilia with GFP‐RAB35 localisation along the full length of the cilium marked with ARL13B staining (ARL13B = GFP‐RAB35 > polyglu. tub.). Note that in ˜15% of cilia all three markers stained the same cilia region (ARL13B = GFP‐RAB35 = polyglu. tub.). Data are mean ± SEM of three independent experiments (n ≥ 30 cilia per experimental condition).
Rab35 regulates cilium length
The ciliary localisation of Rab35 suggests that this G protein may regulate cilium formation, structure or function. Indeed, there are recent suggestions of a role for Rab35 in Hedgehog signalling regulation 33, 72; also, Rab35 was identified in an siRNA screen as a positive ciliogenesis modulator 69, although additional high‐throughput siRNA screening studies indicate that Rab35 is either a negative regulator of cilium formation 71, or plays no role at all in this process 77. To shed light on these seemingly contradictory results and test for a potential role of Rab35 in ciliogenesis, we employed siRNA‐mediated depletion of RAB35 in hTERT‐RPE1 cells and analysed cilia using acetylated tubulin staining. Two independent siRNAs targeting human RAB35 were used, both of which reduce RAB35 expression by > 90% (Fig 2A, and Appendix Fig S1A). Although RAB35 depletion does not affect the number of ciliated hTERT‐RPE1 cells observed after serum withdrawal (Fig 2B), the cilia are significantly shorter, with a median cilia length of ~2.1 μm in RAB35‐depleted cells, compared to ~2.8 μm in non‐depleted cells (Fig 2C and D). Expression of an siRNA‐resistant GFP‐RAB35 construct rescues the cilia length defect, thus ruling out the possibility that this phenotype is due to off‐target effects (Fig 2C and D). Similar results were observed in IMCD3 cells using a pool of siRNAs targeting mouse Rab35, where cilia length is reduced by ~20% (Figs 2E–H, and EV2B and C, and Appendix Fig S1B). To further validate these results and to address the possibility that residual Rab35 due to incomplete siRNA‐mediated depletion may still function in ciliogenesis, we generated Rab35 knockout (KO) NIH3T3 cell lines using the CRISPR/Cas9 system 78. We obtained two independent Rab35 KO clones (KO#1 and KO#2) using two different sgRNAs targeting exon 3 and confirmed Rab35 protein loss by immunoblot analysis and genomic locus disruption by sequencing (Fig 2I and Appendix Fig S1C). Analysis of cilia using acetylated tubulin staining revealed no differences in the number of ciliated cells between wild‐type (WT) and Rab35 KO NIH3T3 cells; however, like Rab35‐depleted cells, the KO cells display a shorter cilium, with a median length of ~1.8 μm compared to ~2.4 μm in the WT control (Fig 2J–L). Collectively, these findings suggest that Rab35 is not involved in initial cilium formation and demonstrate that Rab35 regulates the length of primary cilia.
Figure 2. Rab35 regulates cilium length.
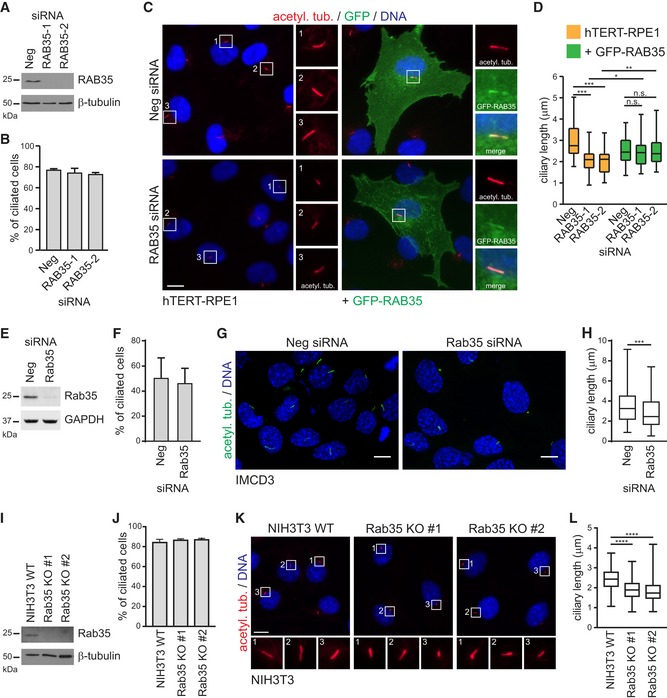
-
AImmunoblot analysis of RAB35 depletion in hTERT‐RPE1 cells transfected with two independent siRNAs targeting RAB35 (RAB35‐1, RAB35‐2) or a non‐targeting siRNA control (Neg). 24 h after transfection, cells were serum‐starved for further 48 h. β‐tubulin served as a loading control.
-
BQuantification of ciliation of hTERT‐RPE1 cells treated as (A) and using acetylated tubulin staining as a cilia marker. Data are mean ± SEM of four independent experiments (n ≥ 50 cilia per experimental condition).
-
C, DhTERT‐RPE1 cells and hTERT‐RPE1 cells stably expressing siRNA‐resistant GFP‐RAB35 were treated as in (A) and stained for GFP, acetylated tubulin (acetyl. tub.) and DNA. Representative images in (C) of cells treated with Neg or RAB35 siRNA. Regions within white boxes shown at higher magnifications to the right. Scale bars, 10 μm. Cilia length quantifications in (D) are shown as box‐and‐whisker plots. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment of three is shown (n ≥ 50 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, n.s.: non‐significant; P‐values: hTERT‐RPE1‐Neg vs. hTERT‐RPE1‐RAB35‐1 P < 0.0001, hTERT‐RPE1‐Neg vs. hTERT‐RPE1‐RAB35‐1 P < 0.0001, hTERT‐RPE1‐RAB35‐1 vs. GFP‐RAB35‐RAB35‐1 P = 0.0249, hTERT‐RPE1‐RAB35‐2 vs. GFP‐RAB35‐RAB35‐2 P = 0.0056).
-
EImmunoblot analysis of Rab35 depletion in IMCD3 cells transfected with pool of siRNAs targeting mouse Rab35 or a non‐targeting siRNA control (Neg). 24 h after transfection, cells were serum‐starved for further 48 h. GAPDH served as a loading control.
-
FQuantification of ciliation of IMCD3 cells treated as in (E) and using acetylated tubulin staining as a cilia marker. Data are mean ± SEM of three independent experiments (n ≥ 50 cilia per experimental condition).
-
G, HImmunofluorescence of IMCD3 representative images treated as in (E), cells are stained for acetylated tubulin (acetyl. tub.) and DNA. Cilia length quantification in (H) is shown as box‐and‐whisker plots and is the result of three independent experiments (n ≥ 50 cilia per experimental condition). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. Statistical significance according to unpaired t‐test with Mann–Whitney test (***P = 0.0002).
-
IImmunoblot analysis of NIH3T3 wild‐type (WT) and Rab35 knockout (KO) cell lines. β‐tubulin served as a loading control.
-
J–LNIH3T3 WT and Rab35 KO cell lines were serum‐starved for 24 h and stained for acetylated tubulin (acetyl. tub.) and DNA. Quantification of ciliation in (J). Data are mean ± SEM of three independent experiments (n ≥ 100 cilia per experimental condition). Representative images in (K). Regions within white boxes shown at higher magnifications at the bottom. Scale bars, 10 μm. Cilia length quantifications in (L) are shown as box‐and‐whisker plots. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment of three is shown (n ≥ 100 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (****P < 0.0001).
Source data are available online for this figure.
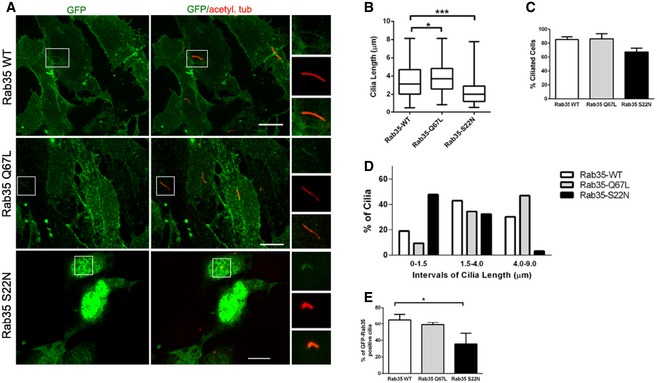
Figure EV2. Ciliary length is regulated by Rab35 nucleotide‐bound state.
-
ARepresentative images of IMCD3 cells transiently expressing wild‐type (WT), GDP‐bound (S22N) or GTP‐bound (Q67L) GFP‐tagged Rab35. 12 h after transfection, cells were serum‐starved for 48 h and stained for GFP and acetylated tubulin (acetyl. tub.) Higher magnification images of the cilia region shown in smaller panels. Scale bars, 10 μm.
-
B–EQuantification of ciliary length in μm (B), percentage of ciliation (C) and GFP‐Rab35 ciliary localisation (E) in IMCD3 cells transiently expressing the indicated GFP‐Rab35 constructs. Cilia length quantification in (B) is shown as box‐and‐whisker plots. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. (D) Histogram of cilia length distribution in which three categories of cilia length were considered: [0–1.5 μm length]; [1.5–4 μm], and [4–9 μm]; one representative experiment out of three is shown; n ≥ 50 cilia per experimental condition. Data in (C, E) are mean ± SEM of three independent experiments; n ≥ 100 cilia per experimental condition. Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, ***P < 0.001; Rab35‐WT vs. Rab35‐SN P < 0.0001, Rab35‐WT vs. Rab35‐QL P = 0.0105).
Rab35 ciliary localisation and function depend on its nucleotide‐bound state
Rab GTPases are regulated by their nucleotide‐bound state. To assess the requirements of GDP and GTP binding for the ciliary localisation of Rab35, we overexpressed GFP‐tagged dominant‐negative (GDP‐bound) or constitutively active (GTP‐bound) mutants in serum‐starved hTERT‐RPE1 cells 58. Both WT and GTP‐bound RAB35‐Q67L localise to cilia in ~60% of transfected ciliated cells. In contrast, ciliary signals for GDP‐bound RAB35‐S22N are much less frequent (~13%; Fig 3A and B). We also assessed whether overexpression of the Rab35 mutants affects hTERT‐RPE1 cilium formation and/or structure. We found that RAB35‐S22N overexpression exerts a dominant‐negative effect on ciliogenesis, resulting in a severe reduction in ciliation (down to 25% of cells; Fig 3C), with those cilia that form being significantly shorter (~20% reduction compared to GFP control; Fig 3D). Conversely, cells overexpressing RAB35‐Q67L display an increase in cilium length (Fig 3A and D). We obtained similar results in IMCD3 cells, where ~48% of cells overexpressing GFP‐tagged Rab35‐S22N exhibit cilia shorter than 1.5 μm (compared with 10 and 20% for cells expressing GFP‐tagged Rab35‐Q67L or Rab35‐WT, respectively), and 33% not forming a cilium (compared with 14% for cells expressing Rab35 WT; Fig EV2A–D). Moreover, like hTERT‐RPE1 cells, ciliary localisations were detected more frequently (60% of cilia) for GFP‐tagged Rab35‐WT and Rab35‐Q67L compared to Rab35‐S22N (~40% of cilia; Fig EV2E). We conclude from these data that GTP binding to Rab35 is required for its ciliary localisation and function in cilium length regulation.
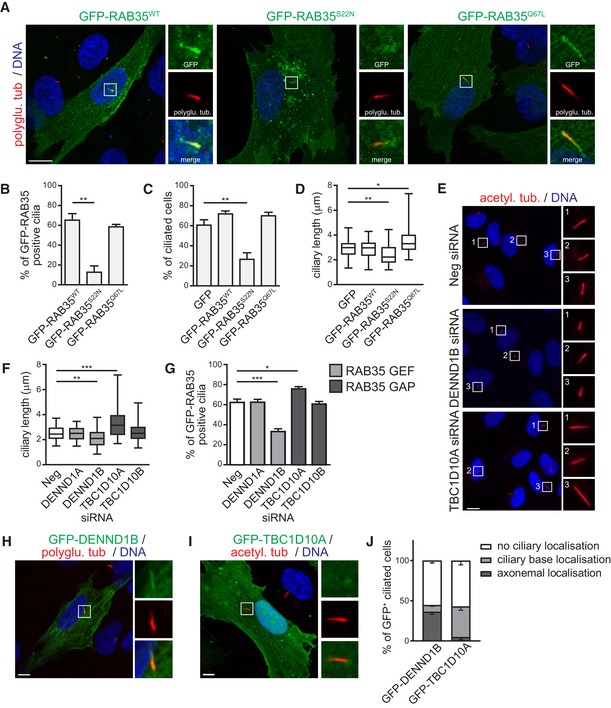
Figure 3. Rab35 ciliary localisation and function depends on its nucleotide‐bound state.
-
ARepresentative images of hTERT‐RPE1 cells transiently expressing wild‐type (WT), GDP‐bound (S22N) or GTP‐bound (Q67L) GFP‐tagged RAB35. Cells were serum‐starved for 24 h and stained for polyglutamylated tubulin (polyglu. tub.) and DNA. Higher magnification images of the cilia region shown in smaller panels. Scale bars, 10 μm.
-
B–DQuantification of GFP‐RAB35 ciliary localisation (B), ciliation (C), and ciliary length in hTERT‐RPE1 cells transiently expressing indicated GFP‐RAB35 constructs or GFP. Data in (B, C) are mean ± SEM of three independent experiments. Statistical significance according to ANOVA followed by Bonferroni post hoc test (**P < 0.01; P‐values: (B) P = 0.0015, (C) P = 0.0062). Cilia length quantification in (D) is shown as box‐and‐whisker plots. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n ≥ 50 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, **P < 0.01; P‐values: GFP vs. GFP‐RAB35‐S22N P = 0.0015, GFP vs. GFP‐RAB35‐Q67L P = 0.0158).
-
E, FAnalysis of cilia length in hTERT‐RPE1 cells depleted of GEF (DENND1A, DENND1B) and GAP (TBC1D10A, TBC1D10B) regulators of RAB35. Cells transfected with indicated siRNAs were serum‐starved for 48 h and stained for acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (E). Regions within white boxes shown at higher magnifications to the right. Scale bars, 10 μm. Cilia length quantification in (F) is shown as box‐and‐whisker plots. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n ≥ 50 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (**P < 0.01, ***P < 0.001; P‐values: Neg vs. DENND1B P = 0.0033, Neg vs. TBC1D10A P = 0.0008).
-
GQuantification of ciliary localisation of GFP‐RAB35 in hTERT‐RPE1 cells stably expressing GFP‐RAB35 treated as in (E). Data are mean ± SEM of five independent experiments. Statistical significance according to ANOVA followed by Bonferroni post hoc test (*P < 0.05, ***P < 0.001; P‐values: Neg vs. DENND1B P < 0.0001, Neg vs. TBC1D10A P = 0.0322).
-
H–JLocalisation of transiently expressed GFP‐DENND1B (H) or GFP‐TBC1D10A (I) in hTERT‐RPE1 cells after 24 h serum starvation and staining with indicated antibodies. Higher magnification images of the cilia region are shown in smaller panels. Scale bars, 5 μm. Quantification of localisation to ciliary base or along axoneme in (J). Data are mean ± SEM of three independent experiments (n ≥ 30 cilia per experimental condition).
Rab35 GEF DENND1B and Rab35 GAP TBC1D10A regulate cilium length and Rab35 ciliary localisation
The activity of Rab GTPases is regulated by specific guanine nucleotide exchange factors (GEFs) and GTPase‐activating proteins (GAPs) that control their nucleotide‐bound state. Several such Rab35 regulators have been identified, including GEFs of the DENND1 family and GAPs of the TBC1D10/EPI64 family 59, 64, 67, 79. To test whether Rab35 regulators affect cilium length or Rab35 ciliary targeting, we used siRNAs to deplete their expression in hTERT‐RPE1 cells. Quantitative real‐time PCR analysis confirmed efficient depletion of DENND1A, DENND1B, TBC1D10A and TBC1D10B (Appendix Fig S2A). For DENND1C and TBC1D10C, we did not detect expression using qPCR in hTERT‐RPE1 cells and, therefore, they were excluded from subsequent experiments. First, we analysed ciliary length and found that DENND1B depletion leads to significantly shorter cilia (~20% reduction), which phenocopies what we observed for RAB35 depletion or overexpression of GDP‐bound RAB35‐S22N (Fig 3E and F). Conversely, TBC1D10A depletion leads to longer cilia, similar to the overexpression of GTP‐bound RAB35‐Q67L (Fig 3E and F). These ciliary length phenotypes were not observed when DENND1A or TBC1D10B was depleted (Fig 3F and Appendix Fig S2B). Given that Rab35 ciliary localisation is GTP‐dependent and that GEFs are known to recruit their cognate Rab GTPases to target membranes 80, we analysed the effect of GEF and GAP depletion on Rab35 localisation. We found that the number of GFP‐RAB35‐positive cilia is considerably reduced in DENND1B‐depleted cells, while TBC1D10A depletion increases the number of GFP‐RAB35 positive cilia (Fig 3G). Finally, we investigated the localisation of the Rab35 GEF and GAP regulators in ciliated hTERT‐RPE1 cells using transiently expressed GFP‐tagged constructs. Consistent with their roles in cilium length control and Rab35 ciliary localisation, pools of both DENND1B and TBC1D10A were found at the cilium and/or ciliary base in ~45% of transfected cells, whereas no ciliary localisation was observed for DENND1A or TBC1D10B (Fig 3H and I, and Appendix Fig S2C and D). Interestingly, DENND1B localises preferential along the axoneme, while TBC1D10A is found more often at the ciliary base (Fig 3J). Together, these findings identify ciliary roles and localisations for the DENND1B GEF and TBCD10A GAP regulators, suggesting that they control Rab35 GTP status in cilia and copperfasten our conclusion that Rab35 serves cilia‐related functions.
Rab35 ciliary function is conserved in zebrafish
We investigated whether Rab35 has a conserved ciliary localisation and function in an animal model, namely zebrafish. We focused our studies on the left‐right organiser (LRO) or Kupffer's vesicle (KV), a fluid‐filled vesicular organ transiently present in the early embryonic life of the fish that is essential to establish internal body laterality 6. First, we injected mRNA encoding mCherry‐tagged mRab35 at the one‐cell stage and analysed its localisation in KV cells. At 8 somite stage [13 h post‐fertilisation (hpf)], we observed a signal, albeit faint, corresponding to mCherry‐Rab35 along KV monocilia (Fig 4A). Thus, Rab35 also localises to the cilium in zebrafish KV cells.
Figure 4. Zebrafish Rab35 regulates KV cilia length and left‐right organ patterning.
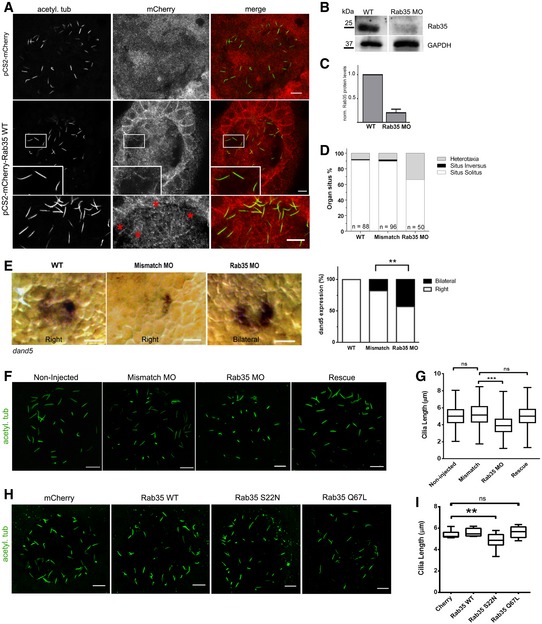
-
AImmunostaining of representative images of KV embryos at 8 somite stage in which mRNA encoding for mCherry or mCherry‐Rab35 WT was injected. Staining of acetylated tubulin was used as cilia marker. Inset shows a higher magnification image of the cilia region, displaying co‐localisation of mCherry‐Rab35 and acetylated tubulin. Scale bar, 10 μm.
-
B, CImmunoblot analysis with anti‐Rab35 and anti‐GAPDH antibodies of 40 μg of total lysates of zebrafish embryos injected with Rab35 morpholino (MO) at 8 somite stage, compared with non‐injected embryos. The plot in (C) shows Rab35 protein level in non‐injected and Rab35 morphants, normalised to GAPDH protein level. Data are mean ± SEM of three independent experiments (n > 50 embryos per condition).
-
DOrgan situs was determined by observing the heart position at 30 h post‐fertilisation (hpf), and the liver position at 53 hpf, by in situ hybridisation with a probe for foxa3, in controls (mismatch MO and non‐injected [WT]) and Rab35 MO. Situs solitus means left heart and liver; situs inversus means right heart and liver; and heterotaxia means any other possible combination of the heart and liver position.
-
EWhole‐mount in situ hybridisation with the dand5 probe at 8 somite stage (13 hpf) in wild‐type non‐injected embryos and mismatch morphants, both presenting right‐sided distribution of dand5, and Rab35 morphants presenting bilateral localisation of the dand5 probe. Scale bar, 50 μm. Graph shows quantification of embryos (%) with bilateral or right‐sided dand5 localisation from both conditions. Statistical significance according to Fisher Exact test (**P = 0.0002).
-
F–IImmunostaining of representative images of KV embryos at 8 somite stage in which: (F) Mismatch or Rab35 MO was injected, as well as a co‐injection of Rab35 MO with mRNA encoding mCherry‐Rab35 WT [Rescue]; (H) mRNA encoding mCherry or mCherry‐tagged Rab35 WT, Rab35‐Q67L or Rab35‐S22N were injected. Acetylated tubulin is shown in green as cilia marker. Quantification of KV cilia length in (F) and (H) is shown, respectively, in (G) and (I) as box‐and‐whisker plots of three independent experiments. N ≥ 8 embryos and n > 300 cilia per experimental condition. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (**P < 0.01, ***P < 0.001, ns. not significant; Mismatch vs. Rab35 MO, P < 0.001; Cherry vs. Rab35‐SN, P = 0.0063). Scale bars, 10 μm.
Source data are available online for this figure.
Next, we depleted rab35 using a translation blocker antisense morpholino and compared the phenotypes with those obtained with a mismatch morpholino and non‐injected embryos. Zebrafish has one Rab35‐encoding gene, Rab35b, that leads to a transcript of 201 amino acids, which is highly homologous to the mouse and human orthologs. Rab35b was successfully silenced by MO injection (Fig 4B and C) with a dose determined by titration to cause less than 30% mortality (Appendix Fig S3A). Importantly, Rab35 morphants display aberrant left‐right (LR) patterning of internal organs (Fig 4D). Normal organ patterning is referred to as situs solitus, while defects in laterality can take many forms 81. A full reversal in the placement of all organs is termed situs inversus, while heterotaxia describes uncoupled defects in some of the organs. At 30 hpf, we observed that in 34% of Rab35 morphants, the heart is misplaced at the centre or the right side of the body, compared with 1 and 10% of non‐injected and mismatch MO control embryos, respectively. We also scored liver positioning and found that 34% of Rab35 morphants display misplacement of this organ, compared with 1 and 8% of controls (Figs 4D and EV3A and B). Also, morphological analysis at 48 hpf revealed that Rab35 morphants possess pericardial oedema, which is a typical feature of ciliary impairment (Fig EV3C–E) 82, 83, 84. To investigate LR phenotypes at an earlier time point, we analysed whether the asymmetric expression of dand5, one of the earliest transcriptional responses to proper KV function, was affected in Rab35 morphants and found that 45% of embryos exhibit bilateral expression (Fig 4E). Thus, Rab35 function is required for the establishment of LR asymmetry in early zebrafish development.
Figure EV3. Rab35 morphants present abnormal left‐right patterning and pericardial oedema.
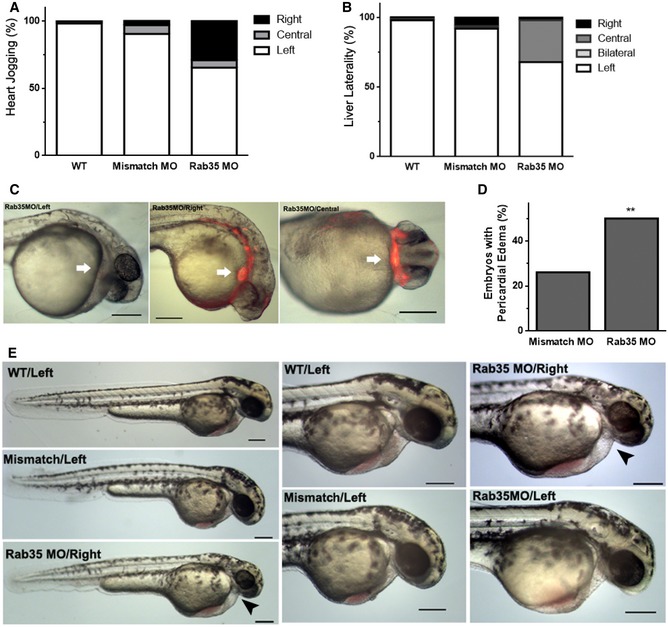
-
A, BEffects of Rab35 morpholino (MO) on the heart jogging and liver laterality of zebrafish embryos treated with 140 μM of MO, compared with wild‐type non‐injected embryos (WT) and mismatch MO, scored at 30 hpf and 53 hpf, respectively. Values are expressed as percentages (n = 3).
-
CLateral or ventral view of Rab35 morphant larvae, with right‐, central‐ or left‐sided heart, at 30 hpf where heart position (red fluorescence; white arrow) is depicted as well as the absence of pericardial oedema. Scale bars, 200 μm.
-
DMismatch MO control or Rab35 MO at 140 μM was injected in zebrafish embryos and pericardial oedema phenotype was quantified at 48 hpf. Statistical significance according to Fisher's exact test (**P = 0.0004; N > 20 embryos).
-
ELateral view of non‐injected and Rab35 and mismatch MO morphant larvae at 48 hpf, with indication of heart positioning (left or right). Higher magnifications of lateral view are shown where the heart and pericardial oedema (when present; black arrowhead) can be appreciated. Scale bars, 200 μm.
Since KV cilia produce directional fluid flow responsible for the establishment of LR asymmetry of dand5 7, we analysed the length and number of KV cilia in Rab35 morphants. We found that cilia are abnormally short when Rab35 is depleted, compared with mismatch MO‐injected or non‐injected embryo controls (Fig 4F and G). Importantly, this phenotype is rescued by co‐injection of Rab35 mRNA (Fig 4F and G). Consistent with the results in mammalian cell lines, we did not observe a significant change in the number of cilia per KV in Rab35 morphants (Appendix Fig S3E).
To assess whether the regulatory role of Rab35 in KV cilia length depends on its GTPase activity, we overexpressed mCherry‐tagged dominant‐negative (GDP‐bound Rab35‐S22N) or constitutively active (GTP‐bound Rab35‐Q67L) Rab35 by injecting one‐cell stage embryos with the corresponding mRNA. We observed that KV cilia length is significantly reduced in embryos overexpressing Rab35‐S22N, when compared with the overexpression of Rab35‐WT or mCherry alone (Fig 4H and I). Together, these findings show that Rab35 regulates cilia length in zebrafish and is required for the asymmetric expression of dand5 and subsequent left‐right organ patterning during embryo development.
Rab35 regulates Arl13b levels at the ciliary membrane
Having established a conserved function for Rab35 in mammalian and zebrafish cilium length control, we next wondered if Rab35 regulates the localisation of specific proteins at the ciliary membrane. First, we analysed the localisation of endogenous ARL13B in RAB35‐depleted hTERT‐RPE1 cells and found that the ciliary levels of ARL13B are elevated 1.5‐ to twofold upon RAB35 depletion (Fig 5A and B). The increase in ARL13B ciliary levels was observed with two different ARL13B antibodies (Figs 5A and B, and EV4A and B) and was rescued in the stable hTERT‐RPE1 cell line expressing an siRNA‐resistant form of RAB35 (Fig 5A and B). Given the effect of Rab35 depletion on both ciliary length and ARL13B, we wondered whether these two phenotypes are correlated; however, closer analysis at a single‐cell level revealed no correlation between ciliary ARL13B intensity and ciliary length (Fig EV5A), suggesting that Rab35 may function in more than one cilia‐related pathway. Interestingly, immunoblot analysis of whole‐cell lysates showed increased total ARL13B protein levels in RAB35‐depleted cells (Fig 5C), suggesting that RAB35 may regulate ARL13B protein stability and/or degradation. Similar results were also observed with the Rab35 KO NIH3T3 cell lines, where ciliary Arl13b levels are elevated 1.5‐fold compared to wild‐type NIH3T3 cells (Fig 5D–F).
Figure 5. Rab35 regulates Arl13b levels at the ciliary membrane.
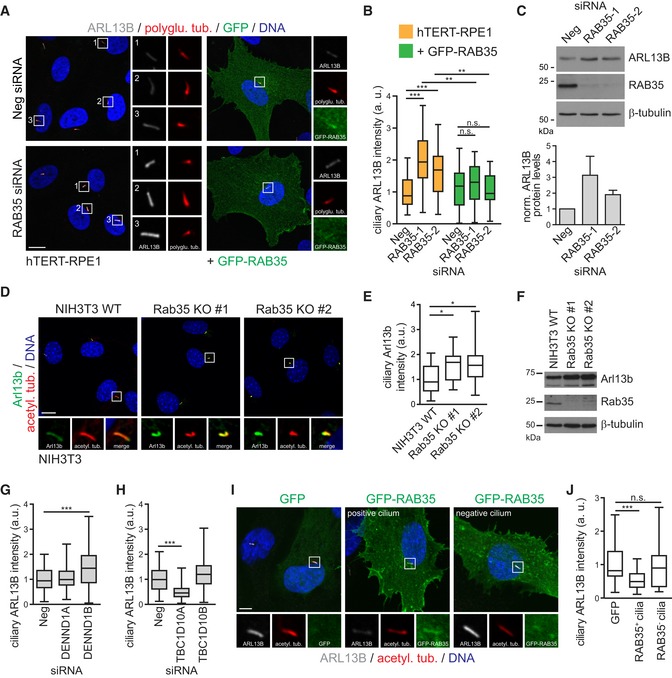
-
A, BhTERT‐RPE1 cells and hTERT‐RPE1 cells stably expressing siRNA‐resistant GFP‐RAB35 transfected with non‐targeting siRNA control (Neg) or Rab35 siRNA, were serum‐starved for 48 h and stained for ARL13B, GFP, polyglutamylated tubulin (polyglu. tub.) and DNA. Representative images are shown in (A). Regions within white boxes shown at higher magnifications to the right. Scale bars, 10 μm. (B) Box‐and‐whisker plots show quantification of ciliary ARL13B intensity in arbitrary units (a. u.). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 25 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (**P < 0.01, ***P < 0.001, n.s.: non‐significant; P‐values: hTERT‐RPE1‐Neg vs. hTERT‐RPE1‐RAB35‐1 P < 0.0001, hTERT‐RPE1‐Neg vs. hTERT‐RPE1‐RAB35‐1 P = 0.0001, hTERT‐RPE1‐RAB35‐1 vs. GFP‐RAB35‐RAB35‐1 P = 0.0014, hTERT‐RPE1‐RAB35‐2 vs. GFP‐RAB35‐RAB35‐2 P = 0.0081).
-
CImmunoblot analysis of total ARL13B protein levels in control and RAB35‐depleted cells. β‐tubulin served as a loading control. The graph shows ARL13B protein levels normalised (norm.) to β‐tubulin. Data are mean ± SEM of four independent experiments.
-
D, ENIH3T3 wild‐type (WT) and Rab35 knockout (KO) cell lines were serum‐starved for 24 h and stained for Arl13b, acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (D). Higher magnifications of regions within white boxes shown at the bottom. Scale bars, 10 μm. (E) Box‐and‐whisker plots show quantification of ciliary Arl13b intensity in arbitrary units (a. u.). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 25 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05; P‐values: NIH3T3 WT vs. Rab35 KO#1 P = 0.0118, NIH3T3 WT vs. Rab35 KO#2 P = 0.0111).
-
FImmunoblot analysis of total ARL13B protein levels in serum‐starved NIH3T3 WT and Rab35 KO cell lines. β‐tubulin served as a loading control.
-
G, HQuantification of ciliary ARL13B intensity in hTERT‐RPE1 cells depleted of GEF (G) and GAP (H) regulators of RAB35. Cells transfected with indicated siRNAs were serum‐starved for 48 h and stained for ARL13B, acetylated tubulin and DNA. Representative images are shown in (Fig EV4C and D). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 30 cilia per experimental condition). Statistical significance according ANOVA followed by Bonferroni post hoc test (***P < 0.001; P‐values: (G) P = 0.0005, (H) P < 0.0001).
-
I, JCiliary ARL13B intensity quantification of hTERT‐RPE1 cells transiently overexpressing GFP or GFP‐RAB35. Cells were serum‐starved for 24 h and stained for ARL13B, acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (I). Higher magnification images of the cilia region are shown at the bottom. Scale bars, 5 μm. Cells with (RAB35+ cilia) and without (RAB35− cilia) GFP‐RAB35 ciliary localisation are compared in (J). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 25 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (***P = 0.0004).
Source data are available online for this figure.
Figure EV4. RAB35, DENND1B and TBC1D10A regulate ciliary ARL13B levels in hTERT‐RPE1 cells.
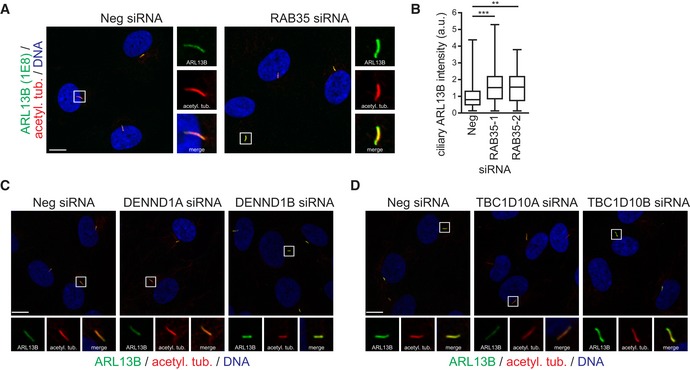
-
A, BhTERT‐RPE1 cells transfected with non‐targeting siRNA control (Neg) or RAB35 siRNAs were serum‐starved for 48 h and stained for ARL13B (rat monoclonal antibody 1E8), acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (A). Regions within white boxes shown at higher magnifications to the right. Scale bars, 10 μm. (B) Box‐and‐whisker plots show quantification of ciliary ARL13B intensity in arbitrary units (a. u.). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 50 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (**P < 0.01, ***P < 0.001; P‐values: Neg vs. RAB35‐1 P < 0.0017, Neg vs. RAB35‐2 P = 0.0003).
-
C, DRepresentative images of hTERT‐RPE1 cells transfected with indicated siRNAs. Cells were serum‐starved for 48 h and stained for ARL13B, acetylated tubulin (acetyl. tub.) and DNA. Regions within white boxes shown at higher magnifications in smaller panels. Scale bars, 10 μm.
Figure EV5. Correlation analysis of ciliary length and ciliary intensity levels of ARL13B and INPP5E.
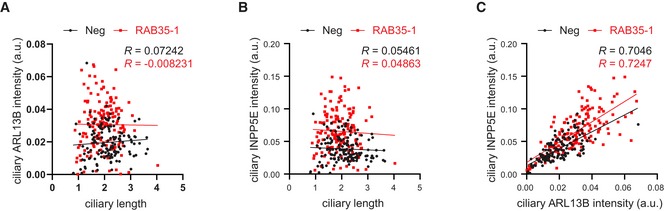
-
A–ChTERT‐RPE1 cells transfected with non‐targeting siRNA control (Neg) or RAB35 siRNA were serum‐starved for 48 h and stained for ARL13B, INPP5E, acetylated tubulin and DNA. Ciliary length was measured using acetylated tubulin staining as cilia marker and mean ciliary intensities of ARL13B and INPP5E were determined. Scatterplots correlating the ciliary ARL13B levels (A) or INPP5E (B) with the ciliary length, or ciliary INPP5E levels with the ARL13B levels (C). Data points in scatterplots represent single cells (Neg: n = 139, Rab35‐1: n = 151). Lines correspond to linear regressions of the data sets, with Pearson correlation coefficient R values indicated.
Next, using siRNAs targeting the cilia‐related RAB35 GEFs and GAPs discussed above, we assessed the requirement of GTP binding for RAB35‐mediated regulation of ARL13B localisation. We found that depletion of the GEF DENND1B leads to an increase in ciliary ARL13B levels, which phenocopies what we observed for RAB35 depletion (Figs 5G and EV4C). Conversely, depletion of the GAP TBC1D10A has the opposite effect, resulting in a significant reduction of ARL13B in cilia (Figs 5H and EV4D). We also found that ARL13B localisation is unaffected in cells depleted for either DENND1A or TBC1D10B. Thus, RAB35 control of ARL13B levels at the ciliary membrane depends on GTP binding and is regulated by the same GAP and GEF regulators that control RAB35 ciliary association (see Fig 3 above). We also tested whether the RAB35‐mediated effect on ciliary ARL13B levels requires the ciliary localisation of RAB35. By analysing hTERT‐RPE1 cells overexpressing GFP‐RAB35 and comparing ARL13B ciliary levels in GFP‐positive and negative cilia, we observed a significant decrease in ciliary ARL13B levels in cells with GFP‐RAB35 ciliary localisation (Fig 5I and J). Collectively, these findings suggest that Rab35 negatively regulates Arl13b levels at the ciliary membrane.
Rab35 interacts with Arl13b
Given that Rab35 and Arl13b co‐localise at the ciliary membrane (Fig 1C) and that cilia‐localised Rab35 regulates Arl13b ciliary levels (Fig 5I and J), we assessed their potential interaction via co‐immunoprecipitation (co‐IP) assays in HEK293T cells. We found that ARL13B‐FLAG specifically co‐precipitates with GFP‐RAB35 but not GFP alone, and that this interaction is greatly enhanced in cells where ciliogenesis is induced by serum withdrawal (Fig 6A). Reciprocal co‐IP of mCherry‐RAB35 with ARL13B‐GFP confirmed the interaction (Fig 6C). Next, we investigated the domains or motifs of Arl13b required for Rab35 interaction. Arl13b consists of an N‐terminal guanine nucleotide‐binding (G) domain and an unusually long C‐terminal tail with coiled coil (CC) and proline‐rich (PR) domains (Fig 6B) 85. All these domains were previously shown to be important for protein–protein interactions; for example, IFT‐B proteins interact with Arl13b via its PR domain, while INPP5E binds to the G‐domain 86. Arl13b also contains a C‐terminal RVEP motif for ciliary targeting and N‐terminal palmitoylation for ciliary membrane anchoring 45, 86, 87, 88. By co‐expressing various GFP‐tagged ARL13B mutants and truncations with mCherry‐RAB35 in HEK293T cells, we found that ARL13B membrane association is crucial for RAB35 interaction because ARL13B lacking the palmitoylation motif (ARL13B‐C8S, C9S) fails to co‐IP with RAB35, as do ARL13B truncations lacking the N terminus (Fig 6C). On the other hand, RAB35 interaction with ARL13B does not require the C‐terminal PR domain or the RVEP motif, whereas ARL13B truncations containing only the G‐ and/or CC‐ domain show reduced levels of RAB35 co‐IP. Together, these findings show that several ARL13B sequence motifs are essential for the interaction with RAB35, namely the N‐terminal palmitoylation motif, as well as a binding region in the C‐terminal tail mapped to amino acids 245–356. Finally, we investigated whether the nucleotide‐bound state of RAB35 affects the interaction with ARL13B. For this, we immunoprecipitated GFP‐RAB35 with anti‐GFP beads and then exchanged this GTPase with either GTPγS, a non‐hydrolysable GTP analogue, or GDP. After incubation with lysate from HEK293T cells expressing ARL13B‐FLAG, we found that GFP‐RAB35 loaded with GTPγS pulled down significantly more ARL13B‐FLAG than GDP‐loaded GFP‐RAB35 (Fig 6D). Since ARL13B is also a GTPase and its binding to GTP could also influence the interaction, we performed a similar pull‐down experiment and found that the interaction of GTPγS‐loaded ARL13B‐GFP with mCherry‐RAB35 is greatly increased compared to GDP‐loaded ARL13B‐GFP (Fig 6E). Thus, a stronger RAB35‐ARL13B interaction is observed when either protein is GTP‐bound. Taken together, these findings demonstrate an interaction between Rab35 and Arl13b, which further supports a role for Rab35 in regulating Arl13b at the ciliary membrane.
Figure 6. Rab35 interacts with Arl13b.
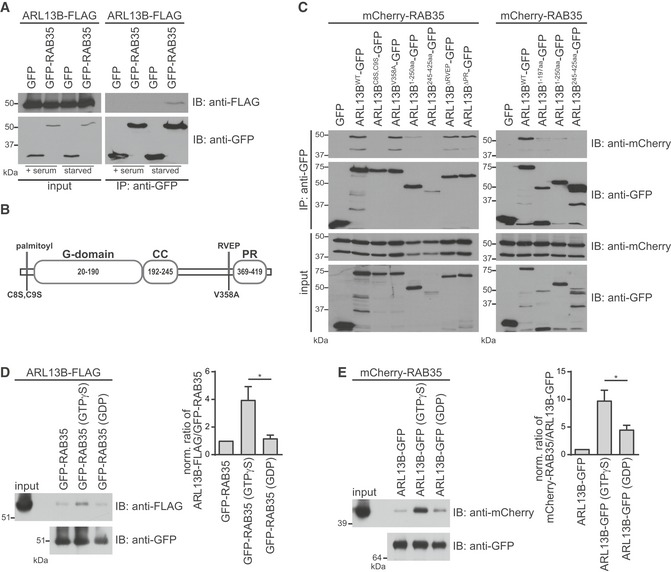
- HEK293T cells were transiently co‐transfected with ARL13B‐FLAG and GFP or GFP‐RAB35. Cells were cultured either in full growth medium (+serum) or serum‐starved for the final 8 h before harvesting. Immunoprecipitations (IP) were performed using anti‐GFP beads. Cell lysates (2.5% of input) and immunoprecipitates were loaded on the same gel, subjected to immunoblot (IB) analysis with indicated antibodies and detected with the same exposure.
- Schematic representation of the human ARL13B structure depicting the guanine nucleotide‐binding (G), coiled coil (CC) and proline‐rich (PR) domains as well as the point mutations affecting N‐terminal palmitoylation or C‐terminal ciliary‐targeting RVEP motif.
- HEK293T cells were transiently co‐transfected with mCherry‐RAB35 together with GFP or indicated ARL13B‐GFP wild‐type, mutant or truncation constructs. Cells were serum‐starved for the final 8 h before harvesting. IPs were performed using anti‐GFP beads and interacting proteins detected by immunoblotting (IB).
- HEK293T cells were transiently transfected with ARL13B‐FLAG and serum‐starved for the final 8 h before harvesting. HEK293T cell lysates expressing ARL13B‐FLAG were subjected to pull‐down with GFP‐RAB35 bound to anti‐GFP beads and preloaded with either no nucleotide, GTPγS or GDP. Bound proteins and cell lysates (1% of input) were analysed by immunoblotting. The graph shows the ratio of precipitated ARL13B and RAB35, normalised to the no nucleotide control. Data are mean ± SEM of three independent experiments. Statistical significance according ANOVA followed by Tukey post hoc test (*P = 0.0333).
- HEK293T cells were transiently transfected with mCherry‐RAB35 and serum‐starved for the final 8 h before harvesting. HEK293T cell lysates expressing ARL13B‐FLAG were subjected to pull‐down with ARL13B‐GFP bound to anti‐GFP beads and preloaded with either no nucleotide, GTPγS or GDP. Bound proteins and cell lysates (1% of input) were analysed by immunoblotting. The graph shows the ratio of precipitated ARL13B and RAB35, normalised to the no nucleotide control. Data are mean ± SEM of three independent experiments. Statistical significance according ANOVA followed by Tukey post hoc test (*P = 0.0466).
Source data are available online for this figure.
Rab35 regulates INPP5E ciliary localisation and PI(4,5)P2 ciliary levels in hTERT‐RPE1 cells
Given Rab35 function in regulating Arl13b ciliary levels, we asked whether Rab35 also affects the localisation of other ciliary proteins. Arl13b was previously shown to mediate the ciliary targeting and distribution of various ciliary proteins 42, 47, 89 such as farnesylated INPP5E, whose ciliary targeting involves the GDP dissociation inhibitor (GDI)‐like solubilising factor PDE6D and GTP‐bound Arl3 that acts as a cargo‐release factor 89, 90. Arl13b acts as a GEF for Arl3, activating Arl3 specifically in the cilium due to its own restricted ciliary localisation, and ensures the specific release of INPP5E from PDE6D in the ciliary compartment 51. We analysed the endogenous localisation of INPP5E in RAB35‐depleted hTERT‐RPE1 cells and found that INPP5E is significantly increased at the cilium, resembling the phenotype observed for ARL13B localisation in these cells (Fig 7A and B). Interestingly, closer analysis at a single‐cell level revealed a clear correlation between ciliary INPP5E and ARL13B levels in RAB35‐depleted cells, although no correlation was found for ciliary INPP5E intensity and cilium length (Fig EV5B and C). To determine whether the increased INPP5E ciliary levels in RAB35‐depleted cells depend on ARL13B, we examined INPP5E localisation in cells depleted of RAB35 and ARL13B. We found that ARL13B loss reverses the RAB35 depletion phenotype, resulting in decreased INPP5E ciliary levels, similar to that of cells depleted for ARL13B alone (Fig 7C) 89. Similar results were also observed with the Rab35 KO NIH3T3 cell lines, where ciliary Inpp5e levels are elevated 1.5‐fold compared to wild‐type NIH3T3 cells (Fig 7D and E). These data show that Rab35 regulates INPP5E ciliary localisation in an Arl13b‐dependent manner and indicate that the increased ciliary Arl13b levels observed in Rab35‐depleted cells result in increased Arl13b activity towards its ciliary targets.
Figure 7. Rab35 depletion disrupts the ciliary levels of INPP5E and PIP(4,5)P2 .
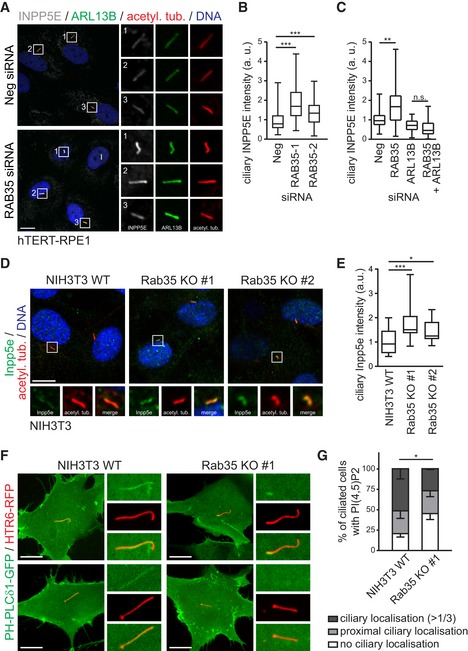
-
A, BhTERT‐RPE1 cells transfected with non‐targeting siRNA control (Neg) or RAB35 siRNA, were serum‐starved for 48 h and stained for INPP5E, ARL13B, acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (A). Regions within white boxes shown at higher magnifications to the right. Scale bars, 10 μm. (B) Box‐and‐whisker plots show quantification of ciliary INPP5E intensity in arbitrary units (a. u.). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 50 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (***P < 0.001; P‐values: Neg vs. RAB35‐1 P < 0.0001, Neg vs. RAB35‐2 P = 0.0002).
-
CFor co‐depletion experiments, hTERT‐RPE1 cells were first transfected with Neg or RAB35 siRNA. After 24 h, a second transfection with Neg or ARL13B siRNA was performed and cells were serum‐starved for 48 h and stained as in (A). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 40 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (**P = 0.0017).
-
D, ENIH3T3 wild‐type (WT) and Rab35 knockout (KO) cell lines were serum‐starved for 24 h and stained for Inpp5e, acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (D). Higher magnifications of regions within white boxes shown at the bottom. Scale bars, 10 μm. (E) Box‐and‐whisker plots show quantification of ciliary Inpp5e intensity in arbitrary units (a. u.). Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n > 25 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, ***P < 0.001; NIH3T3 WT vs. Rab35 KO#1 P < 0.0001, NIH3T3 WT vs. Rab35 KO#2 P = 0.0345).
-
F, GLocalisation of the PI(4,5)P2 sensor PH‐PLCδ1‐GFP to cilia in live NIH3T3 WT and Rab35 KO cells. Cilia are marked with HTR6‐RFP. Representative images are shown in (F). Higher magnification images of the cilia region shown to the right. Scale bars, 10 μm. Quantification of localisation in (G). Data are mean ± SEM of three independent experiments (n > 100 cilia per experimental condition). Statistical significance according to two‐way ANOVA (*P = 0.0242).
The phosphoinositide 5‐phosphatase INPP5E catalyses the conversion of phosphatidylinositol (4,5)‐bisphosphate (PI(4,5)P2) to phosphatidylinositol 4‐phosphate (PI4P). Due to its ciliary localisation, INPP5E establishes a distinct localisation pattern for phosphoinositides along the ciliary axoneme with PI(4,5)P2 being more enriched in the proximal region of the cilium 91, 92. Given the effect of Rab35 loss on ciliary INPP5E levels, we examined the PI(4,5)P2 localisation in the Rab35 KO NIH3T3 cells using the PI(4,5)P2‐specific probe PH‐PLCδ1‐GFP 93. In wild‐type NIH3T3 cells, the majority of cilia are positive for PI(4,5)P2 with ~28% of cilia displaying signal only at the most‐proximal region and ~52% of cilia with staining extending beyond the first third of the axoneme. Importantly, in the Rab35 KO cell line the number of PI(4,5)P2‐positive cilia is significantly reduced with 45% of cilia displaying no ciliary PI(4,5)P2 localisation (Fig 7F and G). These results are consistent with increased INPP5E in cilia observed after Rab35 depletion and loss.
Rab35 depletion impairs SAG‐induced ciliary translocation of Smoothened
Having established Arl13b as a target of Rab35, we wondered whether Rab35 serves Arl13b‐related roles in ciliary signalling pathways. Specifically, we focussed on the Shh pathway because of the conserved role of Arl13b in regulating this pathway 41, 42, 94. Shh signalling relies on proper trafficking of signalling molecules into and out of the cilium in response to pathway activation. Under basal conditions, the Shh receptor Patched (PTCH) localises to cilia and prevents the ciliary localisation of the critical pathway activator, Smoothened (SMO). Binding of Shh ligand to PTCH induces its ciliary exit, which in turn leads to the ciliary accumulation and activation of SMO, and subsequently the processing of downstream cilium‐localised Gli transcription factors 95. To determine the requirement of RAB35 for SMO ciliary translocation, we treated RAB35‐depleted hTERT‐RPE1 cells with the SMO agonist SAG, which binds to SMO and activates the pathway independently of PTCH 96. In both control and RAB35‐depleted cells, SMO localisation to cilia is rarely observed under basal conditions (DMSO vehicle control). After SAG treatment, whilst SMO is present in ~75% of cilia in control‐depleted cells, only 40% of cilia are SMO‐positive in RAB35‐depleted cells (Fig 8A and B) and those depleted cilia that retain SMO localisation display significantly decreased SMO ciliary levels (see data on Smo+ cilia in Fig 8C). Thus, Rab35 loss interferes with SAG‐induced ciliary accumulation of SMO.
Figure 8. Rab35 depletion disrupts the ciliary localisation of Smoothened.
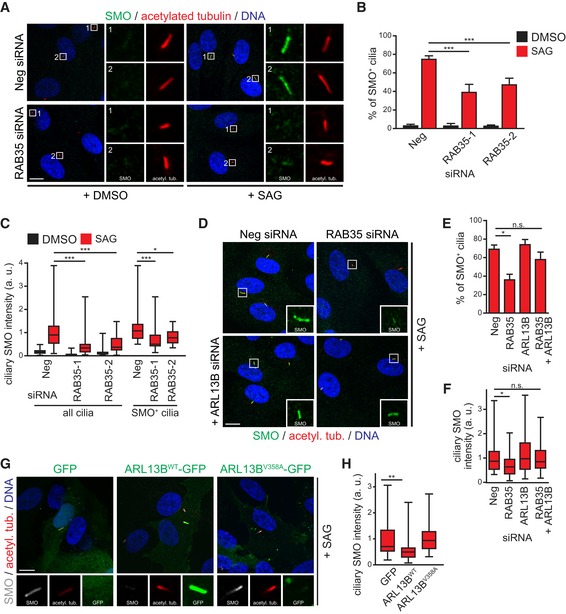
-
A–ChTERT‐RPE1 cells transfected with indicated siRNAs were serum‐starved for 48 h and treated with SMO agonist (SAG) or vehicle control (DMSO) for the last 24 h. Cells were stained for SMO, acetylated tubulin and DNA. Representative images are shown in (A). Regions within white boxes shown at higher magnifications to the right. Scale bars, 10 μm. Graph in (B) shows the percentages of SMO‐positive (SMO+) cilia. Data are mean ± SEM of five independent experiments. Statistical significance according to ANOVA followed by Bonferroni post hoc test (***P < 0.001; P‐values: Neg + SAG vs. RAB35‐1 + SAG P < 0.0001, Neg + SAG vs. RAB35‐2 + SAG P = 0.0033) (C) box‐and‐whisker plots show quantification of the average SMO intensity in the ciliary area marked by acetylated tubulin staining. All cilia: cilia identified with acetylated tubulin staining. SMO+ cilia: cilia with discernible SMO localisation. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n ≥ 30 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test [*P < 0.05, ***P < 0.001; P‐values: Neg + SAG vs. RAB35‐1 + SAG (all cilia) P < 0.0001, Neg + SAG vs. RAB35‐2 + SAG (all cilia) P = 0.0024, Neg + SAG vs. RAB35‐1 + SAG (SMO+ cilia) P < 0.0007, Neg + SAG vs. RAB35‐2 + SAG (SMO+ cilia) P = 0.0205].
-
D–FFor co‐depletion experiments, hTERT‐RPE1 cells were first transfected with Neg or RAB35 siRNA. After 24 h, a second transfection with Neg or ARL13B siRNA was performed and cells were treated as in (A). Representative images are shown in (D). Regions within white boxes shown at higher magnifications as insets. Scale bars, 10 μm. Graph in (E) shows the percentages of SMO‐positive cilia. Data are mean ± SEM of five independent experiments. Statistical significance according to ANOVA followed by Bonferroni post hoc test (*P = 0.0151, n.s.; non‐significant). Box‐and‐whisker plots in (F) show quantification of ciliary SMO intensity. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n ≥ 30 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (*P = 0.0142, n.s.; non‐significant).
-
G, HCiliary SMO intensity quantification in hTERT‐RPE1 cells transiently expressing GFP, wild‐type ARL13B‐GFP (WT) or the ciliary‐targeting defective mutant ARL13B‐V358A. Cells were serum‐starved and treated with SAG for 24 h and stained for SMO, acetylated tubulin (acetyl. tub.) and DNA. Representative images are shown in (G). Higher magnification images of the cilia region are shown at the bottom. Scale bars, 10 μm. Box‐and‐whisker plots in (H) show quantification of ciliary SMO intensity. Horizontal lines show 25, 50 and 75th percentiles; whiskers extend to minimum and maximum values. One representative experiment out of three is shown (n ≥ 25 cilia per experimental condition). Statistical significance according to Kruskal–Wallis followed by Dunn's post hoc test (**P = 0.0046).
Previous studies have shown that Arl13b loss leads to aberrant SMO accumulation in cilia, suggesting that Arl13b limits the ciliary accumulation of SMO, possibly by promoting its ciliary export 42, 86. We wondered, therefore, whether elevated ciliary ARL13B levels in RAB35‐depleted cells are required for the SMO localisation defect in these cells. To answer this question, we analysed SAG‐treated hTERT‐RPE1 cells co‐depleted of RAB35 and ARL13B and observed near wild‐type levels of ciliary SMO (Fig 8D–F). To test whether the ARL13B‐mediated effect on SMO requires its ciliary localisation, we employed a GFP‐tagged ciliary‐targeting defective mutant of ARL13B (V358A) 87 and found that SAG‐induced SMO ciliary translocation is greatly reduced in hTERT‐RPE1 cells expressing ARL13B‐WT but not ARL13B‐V358A or GFP (Fig 8G and H). Thus, the reduced levels of ciliary SMO observed in SAG‐induced RAB35‐depleted cells depend on elevated levels of ARL13B in the cilium.
Discussion
In this study, we identify Rab35 as a new regulator of cilium structure and function. We show evolutionarily conserved localisation of Rab35 at the ciliary membrane and demonstrate a role for Rab35 in restricting the ciliary accumulation of Arl13b and INPP5E and promoting the ciliary accumulation of SMO as well as PI(4,5)P2. In addition, we demonstrate that loss of Rab35 causes left‐right asymmetric patterning defects in zebrafish. Thus, a ciliary function is now added to the multiple cellular roles of Rab35 in diverse processes such as endocytic recycling, actin remodelling, autophagy and cytokinesis.
A number of our observations support a role for Rab35 as a positive regulator of cilium length. First, cilia are abnormally short in Rab35‐depleted, knockout and dominant‐negative Rab35‐S22N‐expressing mammalian cells, in Rab35‐depleted zebrafish KV cells, as well as in Rab35‐S22N‐expressing KV embryos. Second, cilia are abnormally long in mammalian cells expressing constitutively active Rab35‐Q67L. Third, depletion of a Rab35 GAP (TBC10D1A) elongates cilia, whereas depletion of a Rab35 GEF (DENND1B) truncates cilia. Thus, Rab35 joins a number of other Rabs, namely Rab8, Rab11 and Rab23 (reviewed in 32) required for normal cilium structure. In the context of cilium length regulation, it is interesting that the truncated RAB35‐deficient cilia exhibit increased ARL13B levels, given reports of abnormally long cilia in cells overexpressing Arl13b 44, 97. However, it should be recognised that the level of overexpression in those studies likely reflects a much higher ciliary concentration of Arl13b compared to the 1.5‐ to twofold increase that was observed for endogenous Arl13b in Rab35‐disrupted cells. It has also been shown that cilia lacking Arl13b can be abnormally long 98, thereby highlighting the lack of a correlation between cilium length and Arl13b levels. Additionally, Rab35 disruption may impact more than one cilia‐related pathway, and thus, the cilium length defect in cells lacking Rab35 may not arise directly from the elevated ciliary Arl13b levels. Indeed, this notion is supported by our observation of no correlation between the ciliary length and ARL13B level phenotypes in Rab35‐disrupted cells (Fig EV5A). It is also noteworthy that Rab35 knockout and/or depleted mammalian or zebrafish KV cells display normal cilia number. Our finding that Rab35 positively regulates cilium length but is not required for ciliogenesis agrees with data from a genome‐wide siRNA screen in mouse IMCD3 cells 77, although two additional high‐throughput depletion studies in RPE1 cells report, conflictingly, that Rab35 is a positive or negative regulator of ciliary incidence 69, 71.
Consistent with a function for Rab35 at the cilium, we identify ciliary roles for specific Rab35 GEF and GAP proteins, DENND1B and TBC1D10A, respectively. We found that DENND1B‐ and TBC1D10A‐depleted cells display opposing cilium length defects (truncated/elongated), thereby phenocopying what is observed in WT cells overexpressing GDP or GTP‐bound Rab35. Also, loss of DENND1B or TBC1D10A results in opposing effects on ARL13B ciliary levels (reduced/increased), consistent with the ARL13B phenotype in RAB35‐depleted cells (discussed further below). Notably, ciliary defects were not found in DENND1A‐ or TBC1D10B‐depleted cells, and DENND1C and TBC1D10C are not expressed in our ciliated cell model. Thus, only a subset of DENND1 and TBC1D10 subfamily members of Rab35 regulators have ciliary roles, at least in those cells we investigated. Multiple DENN domain‐containing (DENND1A/B/C, Folliculin) GEFs and TBC domain‐containing (TBC1D10A/10B/10C/13/24) GAPs control Rab35 activation, most likely because of its multitasking roles and requirement for regulation at distinct intracellular sites 59, 60. Of the Rab35 GEFs and GAPs, we identify in the ciliary context, DENND1B regulates a fast endocytic recycling pathway and endocytic uptake of cell surface T‐cell receptors, and TBC1D10A impairs exosome secretion 99, 100, 101, 102. It is important to note that some Rab35 regulators such as TBC1D10A target other Rabs (e.g. Rab27a, Rab8a) 103, 104 and, therefore, it remains possible that part of the ciliary phenotype in Rab35 GAP/GEF‐depleted cells could result from dysregulation of Rabs other than Rab35. Indeed, overexpression of the dominant‐negative mutant of Rab35 (Rab35 S22N) leads not only to a reduction in cilia length but also in the percentage of ciliated cells. This could be caused by the sequestration of GEFs and/or effectors that are common to other Rabs.
By analysing embryonic KV cell cilia, we found that the role of Rab35 in controlling cilium length is conserved in zebrafish. We found that Rab35 faintly localises to KV cell cilia, supporting the ciliary localisations we find in mammalian cells. The low levels of Rab35 in KV cell cilia suggest that Rab35 may transiently localise to cilia, a notion supported by our results in mammalian cells where Rab35 ciliary localisation is observed in only ~60% of Rab35‐expressing cells. Importantly, the reduction in cilia length upon Rab35 depletion correlates with abnormal patterning of the internal organs. These results suggest that KV fluid flow is likely perturbed as a consequence of shortened cilia in Rab35‐depleted embryos, as evidenced by abnormal bilateral expression of dand5. Both theoretical and experimental studies have demonstrated that short KV cilia cause KV flow defects leading to the disruption of the whole left‐right cascade of gene expression, namely by changing dand5 and nodal asymmetric expression patterns 7, 105, 106, 107.
We also demonstrate roles for Rab35 in regulating the ciliary membrane levels of Sonic hedgehog signal regulators, Arl13b, SMO and INPP5E. Specifically, RAB35 prevents overaccumulation of ARL13B and INPP5E in cilia, whilst promoting the signal‐induced accumulation of SMO to cilia (summarised in Fig 9B). Our observations are consistent with the role of Arl13b in promoting the ciliary release of lipidated INPP5E from its transport carrier 89 and provide an explanation for why more ciliary ARL13B in RAB35‐depleted cells correlates with abnormally high levels of ciliary INPP5E. In addition, reduced ciliary SMO levels in RAB35‐depleted cells, where ciliary ARL13B levels are elevated, correlates with the opposite phenotype (i.e. increased SMO accumulation) in cells with a loss‐of‐function mutation in Arl13b 42. However, in cancer cells, Arl13b promotes the ciliary localisation of SMO, although this difference may be explained by the transformed state of these cells 50, 108, 109. One scenario to explain our observations is that Rab35 principally targets Arl13b, possibly as a direct effector or a more distant upstream regulator. Thus, the SMO and INPP5E localisation defects in RAB35‐depleted cells would derive from misregulation of the proposed roles of Arl13b in INPP5E ciliary targeting, as well as retrograde IFT and the ciliary removal of Shh signalling molecules 86, 89. A role for Rab35 in Arl13b regulation also agrees with their interaction in co‐immunoprecipitation assays. Furthermore, Rab35 regulation of mammalian Arl13b ciliary levels is consistent with the left‐right defects observed in zebrafish Rab35 (this study) and Arl13b morphants, as well as in zebrafish overexpressing Arl13b 44, 97, 110.
Figure 9. Model of Rab35 function at the cilium.
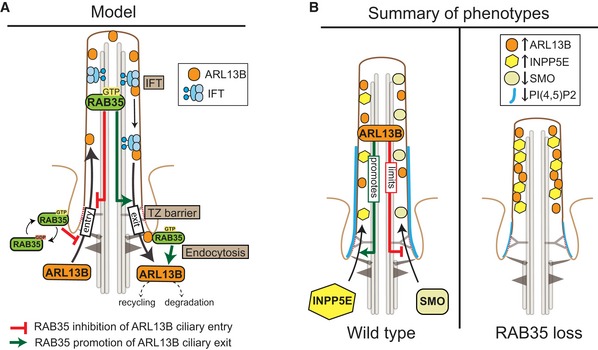
- Model of how activated (GTP‐bound) Rab35 at the ciliary and/or periciliary (pocket) membrane limits Arl13b ciliary levels. In one scenario, Rab35 inhibits Arl13b entry into cilia, possibly through an IFT pathway. In a second scenario, Rab35 promotes Arl13b exit/retrieval from ciliary (and periciliary) membranes via: (i) interaction with retrograde IFT, (ii) modulation of the transition zone membrane diffusion barriers that prevent Arl13b from exiting the cilum, and/or (iii) endocytic processes at or near the ciliary pocket that target Arl13b for degradation or recycling to non‐ciliary plasma membrane destinations.
- Ciliary membrane and ciliary length phenotypes following Rab35 disruption. In wild‐type cells, Arl13b, INPP5E and Smoothened (SMO; under conditions of Shh signal activation) all localise to the ciliary membrane. Arl13b promotes the ciliary localisation of INPP5E 89, whilst limiting (or altering) the ciliary distribution of SMO 42. PI(4,5)P2 (blue line) is enriched within the proximal portion of the ciliary membrane. In Rab35‐depleted cells, ciliary Arl13b levels are elevated, causing a concomitant increase and decrease in the ciliary levels of INPP5E and SMO, respectively. Rab35 disruption also leads to a reduction in the length of the proximal ciliary PI(4,5)P2 signal and a reduction in the frequency of PI(4,5)P2‐positive cilia (thin blue line). Misregulation of ciliary membrane protein composition in Rab35‐disrupted cells leads to a decrease in cilium length.
As to how Rab35 might limit the ciliary membrane levels of Arl13b, one possibility is that Rab35, operating in the cilium, interacts with ciliary transport pathways that establish and maintain the organelle's composition (Fig 9A). For example, Rab35 may facilitate an IFT function that either promotes the ciliary removal, or limits the ciliary entry, of Arl13b. In support of this notion, Arl13b undergoes IFT, interacts with IFT‐B, and requires IFT for its ciliary targeting in Caenorhabditis elegans 86, 88, 111. Alternatively, Rab35 could regulate membrane diffusion barriers at the transition zone that keep Arl13b in the cilium 88, 112, 113. Another possibility is that ciliary Rab35 controls Arl13b localisation and cilium structure by regulating ectosomes that bud from ciliary membranes to control their composition and length 20, 21, 22, 23, 24, 114; indeed, some ciliary ectosomes are Arl13b‐positive 108. However, this model is mitigated by our finding that Rab35‐disrupted cells possess reduced ciliary levels of PI(4,5)P2, which is a positive regulator of ciliary ectosome formation 20.
A second non‐mutually exclusive possibility is that Rab35 functions at the ciliary pocket and/or nearby endosomal compartments to regulate Arl13b uptake from the periciliary membrane and subsequent sorting into recycling or degradation pathways (Fig 9A). Indeed, the ciliary pocket is a site of endocytosis, and a role for Rab35 in promoting Arl13b uptake and subsequent degradation is consistent with our observation of increased total Arl13b levels in Rab35‐depleted cells, as well as known roles for Rab35 in endocytic events such as endosomal sorting of proteins for recycling or degradation 17, 115, 116, 117, 118, 119. An uptake function for Rab35 could act on Arl13b molecules that are either en route to the cilium or exiting from the cilium as part of a natural cycling process of Arl13b transport between ciliary and non‐ciliary compartments. Consistent with its established roles at the endosome 58, 60, 100, 120, Rab35 could also limit ciliary Arl13b levels by facilitating its recycling to non‐ciliary plasma membrane destinations. Notably, Arl13b is known to co‐localise with recycling endosomal markers and regulate endocytic recycling traffic 121.
In summary, we have uncovered a new and conserved role for Rab35 at the cilium of mammalian and zebrafish cells. We show that Rab35 is required for normal cilium length and the ciliary membrane levels of vertebrate Sonic hedgehog signalling regulators SMO, Arl13b and INPP5E, as well as organ patterning. Given the strong association of Sonic hedgehog signalling defects in ciliary disease, our findings also suggest possible roles for Rab35 in ciliopathy mechanisms.
Materials and Methods
Cell culture and transfection
All cell lines were grown at 37°C under 5% CO2. Human hTERT‐immortalised retinal pigmented epithelial cells, hTERT‐RPE1, were cultured in DMEM/F12 medium supplemented with 10% foetal bovine serum (FBS), 2 mM L‐glutamine and 0.348% sodium bicarbonate. Human embryonic kidney cells, HEK293T, were cultured in DMEM with 10% FBS. Murine inner medullary collecting duct cells, IMCD3, were cultured in DMEM/F12 medium supplemented with 10% FBS (Sigma), 100 U/ml penicillin G and 100 μg/ml streptomycin (Gibco). NIH3T3 cells were grown in DMEM supplemented with 10% calf serum, and ciliogenesis in these cells was induced by serum starvation with medium containing 1% calf serum for 24 h. Cilia formation in hTERT‐RPE1, IMCD3 or HEK293T cells was induced by serum withdrawal for 24–48 h, 48 h or 8 h, respectively. Smoothened agonist SAG (Santa Cruz) was added at 200 nM for 24 h in starvation media to induce Sonic hedgehog pathway activation.
HEK293T cells were transiently transfected with plasmid DNA using the calcium phosphate precipitation method: Briefly, ~1.5 × 106 cells were plated in a 10‐cm dish 16–20 h before transfection. For each dish, 1–3 μg plasmid DNA was diluted in 500 μl 0.25 M CaCl2, mixed with 500 μl 2× HEBS (280 mM NaCl, 1.5 mM Na2HPO4, 50 mM HEPES, 10 mM KCl, 12 mM dextrose, pH 7.10) and added to the cells. After 8 h, media was exchanged and cells were harvested 2 days after transfection for cell lysis and co‐immunoprecipitation assays. For transfection of hTERT‐RPE1 cells, 150,000 cells/well were seeded in 12‐well plates. After 16 h, cells were transiently transfected with 0.5 μg plasmid DNA using 1.5 μl TransIT‐LT1 (Mirus Bio), serum‐starved 6 h after transfection and fixed after 24–48 h for immunofluorescence. Stable hTERT‐RPE1 cells were generated by transfecting 1 μg plasmid using 2 μl Lipofectamine 3000 and 2 μl P3000 reagent (Invitrogen) and subsequent selection with 500 μg/ml G418 (Invitrogen). For transfection of IMCD3 cells, 300,000 cells/well were seeded in 24‐well plates. After 16 h, cells were transiently transfected with 1 μg plasmid DNA using 1 μl of Lipofectamine 2000 (Invitrogen), serum‐starved 12 h after transfection and fixed after 48 h for immunofluorescence. For live imaging, 800,000 NIH3T3 cells were seeded in a 35‐mm Ibidi glass plate. They were transiently transfected 16 h after with 2 μg of DNA mix containing equal parts of HTR6‐RFP and PI(4,5)P2 sensor PH‐PLCδ1‐GFP with 4 μl of TurboFect (Thermo Fisher) in starvation media. After 24 h, cells were imaged.
RNA interference and qPCR
For depletion experiments, hTERT‐RPE1 cells were seeded in 12‐well plates (150,000 cells/well) and immediately transfected with Silencer Select siRNAs at a final concentration of 20 nM (Ambion) using 1.5 μl Lipofectamine 2000 (Invitrogen). For RAB35/ARL13B co‐depletion experiments, hTERT‐RPE1 cells were first transfected with either Neg or RAB35 siRNA (2 wells for each siRNA). After 24 h, cells were serum‐starved and transfected with either Neg or ARL13B siRNA. Regarding IMCD3 depletion experiments, cells were seeded in 24‐well plates (300,000 cells/well) and immediately transfected with 1.25 μl of Dharmafect 1 (Dharmacon) with 80 nM of siGENOME siRNAs pools (Dharmacon). Efficiencies of mRNA depletion were measured by quantitative real‐time PCR (qPCR). For this, total RNA was purified with a NucleoSpin RNA Minikit (Macherey‐Nagel) or RNeasy kit (Qiagen) following the manufacturer's instructions. cDNA was synthesised with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) or SuperScript II (Life Technologies) using 200–500 ng of RNA per reaction. qPCR analysis was performed using SYBR green detection in an Applied Biosystems 7500 system or a LightCycler 96 system (Roche Life Sciences). GAPDH mRNA levels were used for normalisation and results were obtained using the ΔΔC t method 122. Primers and siRNAs used in this study are listed in Appendix Tables S3 and S4, respectively.
Generation of Rab35 knockout cell lines using the CRISPR/Cas9 system
The CRISPR/Cas9 system was used to generate NIH3T3 Rab35 knockout cell lines as previously described 78. The sgRNA sequences targeting mouse Rab35 (sgRNA1: 5′‐TCGGACTGTGGAGATCAACG‐3′; sgRNA2: 5′‐CTACATCACCACAATCG GAG‐3′) were chosen based on their On‐target and Off‐target‐scores 123, 124 using the webtool Benchling (https://benchling.com) and inserted into the vector pSpCas9(BB)‐2A‐Puro (PX459) V2.0 (a gift from Feng Zhang, Addgene plasmid # 62988) 78, which also contains expression cassettes for Cas9 and a puromycin selectable marker. NIH3T3 cells (50,000 cells/well) were transfected in 12‐well plates with 1 μg plasmid DNA using 2 μl Lipofectamine 2000. 24 h after transfection, cells were treated with puromycin for 48 h to remove untransfected cells. Subsequently, single‐cell clones were isolated, expanded and screened by immunoblot for loss of Rab35 protein expression.
Plasmids and antibodies
Plasmids and antibodies used in this study are listed in Appendix Tables S1 and S2, respectively. Mammalian expression constructs comprising a GFP‐, mCherry‐ or FLAG‐tag were generated by PCR cloning of full‐length cDNAs into pEGFP‐C1/N1 (Clontech), pENTR‐D‐TOPO‐C1 (Invitrogen), pmCherry‐C1/N1 (Clontech), pCMV‐3Tag‐1 (Agilent Technologies) and pCS2 (+) 105. Point mutations for the siRNA‐resistant construct and the GDP‐ or GTP‐bound mutants were inserted by site‐directed mutagenesis (Agilent Technologies) using primers indicated in Appendix Table S3. All constructs were confirmed by DNA sequencing.
Zebrafish culture and injections of morpholino oligonucleotides and/or mRNA
WT zebrafish line [AB background and Tg(β‐actin2:loxP‐DsRed‐loxP‐GFP; s928Tg)] were maintained and staged as described in Tavares et al 4. Embryos were kept at 32°C in the dark and in E3 embryo medium. Morpholinos blocking translation of Rab35 or Mismatch (Appendix Table S5) were used as previously described 4 at 140–280 μM, by injecting one‐cell stage embryos. These were left to develop at 32°C or 25°C until the desired stage. While preparing this manuscript, a paralogue for Rab35 was identified, Rab35a or Rab35‐like, which encodes a shorter protein (195 amino acid) with a divergent C‐terminal region (not found in mouse, human or Drosophila) and lacking key basic residues characteristic of Rab35. We ensured that the sequence of the used morpholino does not match any sequence in the Rab35‐like gene. Mouse Rab35 coding sequence was cloned into a pCS2 (+) vector using the primers indicated in Appendix Table S3 and a mCherry tag was added at the N terminus of Rab35. Linearised plasmid DNA was used to produce in vitro mRNA with the mMESSAGE mMACHINE T7 transcription kit (Invitrogen), which was then purified with the RNA Clean and Concentrator‐5 kit (Zymo). At one‐cell stage, embryos were injected with 200 pg of mRNA and left to develop at 32°C or 25°C until the desired stage. To analyse KV cilia, embryos were fixed at 8 somite stage [13 h post‐fertilisation (hpf)] with 4% PFA at 4°C overnight and processed for immunofluorescence. To evaluate the left‐right patterning of internal organs, embryos were observed from their ventral side at 30 hpf and their heart positioning was evaluated using a stereoscopic zoom microscope (SMZ745, Nikon Corporation, Japan). At 53 hpf, whole‐mount in situ hybridisation was performed as described previously 105 for foxa3 probe and embryos were scored for liver patterning. To evaluate KV function, whole‐mount in situ hybridisation was performed with the dand5 probe at 8 somite stage embryos using the same protocol, and embryos were scored for dand5 expression laterality using a stereoscopic zoom microscope (SMZ745, Nikon Corporation, Japan).
Immunofluorescence staining
Mammalian cell lines were grown on glass coverslips, fixed with 3% PFA in PBS for 15 min and permeabilised for 5 min in either 0.2% Triton X‐100/PBS or ice‐cold methanol. Blocking was performed for 30 min with 3% BSA (hTERT‐RPE1, NIH3T3) or 1% BSA (IMCD3) in 0.1% Triton X‐100/PBS, or with 3% BSA in PBS (for staining with rat anti‐Arl13b antibody). Antibodies (Appendix Table S2) were diluted in blocking solution. Cells were incubated with primary antibodies at room temperature for 2 h. Samples were washed three times with PBS and incubated with appropriate secondary antibodies conjugated to fluorescent dyes for 1 h at room temperature. DAPI (4′,6‐diamidino‐2‐phenylindole, Sigma‐Aldrich) was included with secondary antibodies for DNA staining. Samples were washed three times with PBS, and coverslips were mounted on glass slides in Mowiol (Sigma‐Aldrich).
Zebrafish embryos were fixed with 4% PFA at 8 somite stage (13 hpf) and stored overnight at 4°C. They were then washed 5 times for 5 min with PBS with 1% Triton X‐100 and dechorionated using sharp forceps. Permeabilisation was performed using Proteinase K (10 mg/ml) for 1 min, re‐fixed for 20 min with 4% PFA and washed in cold acetone for 7 min at −20°C. Blocking was performed for 2 h with 1% BSA/1% DMSO/1% Triton X‐100 in PBS supplemented with 1.5% FBS. Antibodies against acetylated α‐tubulin (mouse) and mCherry (rabbit) were diluted at 1:300 and incubated overnight at 4°C with rotation. On the second day, embryos were washed in blocking solution for 4 h at room temperature with rotation. Then, embryos were incubated overnight with the appropriate secondary antibodies conjugated to Alexa Fluor dyes (1:500) in equal parts of blocking and DAPI solution at 4°C with rotation. On the third day, embryos were washed in blocking solution 2 times for 10 min, washed 30 min in PBS with 1% Triton X‐100, fixed for 5 min in 4% PFA, re‐washed in PBS with 1% Triton X‐100 and mounted between a coverslip and glass slide in PBS.
Microscopy
Widefield images were acquired as z‐stacks at 0.4 μm intervals using a Zeiss AxioImager M1 microscope equipped with a Zeiss Plan Neofluar 40×/NA1.3 oil‐immersion objective and a QImaging Retiga R6 CCD camera or Zeiss CellObserver equipped with an Apochromat 63×/NA1.4 oil‐immersion objective and a CoolSNAP HQ2 camera. Confocal images of hTERT‐RPE1 and NIH3T3 cells were acquired on an Olympus Fluoview FV1000 microscope with a 60×/1.35 NA UPlanSApo oil‐immersion objective. For IMCD3 cells, confocal Z‐series stacks were acquired on a Leica SP5 confocal, mounted on a DM6000 inverted microscope, using a 63× 1.3NA oil‐immersion objective and a Zeiss LSM 710 confocal microscope equipped with a Plan‐Apochromat 63/1.40 Oil Ph3 lens. Live imaging of NIH3T3 cells to detect Pi(4,5)P2 was performed using a spinning disc confocal microscope (Nikon TI) with full temperature atmosphere control (37°C + Humidity + CO2) equipped with a Photometrics 512 EMCCD camera and a Nikon 100× Oil Apo TIRF NA1.4. For whole‐mount embryos, confocal Z‐series stacks were made on the same microscopes with a 40× 0.85NA objective. Super‐resolution imaging was performed using an Olympus FV3000RS Advanced Laser Scanning microscope equipped with a PLAPON60XOSC2 NA 1.40 oil‐immersion objective and a FluoView Olympus Super Resolution (FV‐OSR) software module. FV‐OSR images were captured as z‐stacks at 220 nm intervals and with a pixel size of 43 nm. A noise reduction filter was applied to the images in the FV3000 acquisition software, and images were deconvolved using Olympus cellSens software.
Image analysis
Fluorescence intensity profile plots were created using the Twin Slicer tool of the Huygens Professional software (Scientific Volume Imaging B.V) by drawing lines across cilia in the proximal or distal region and exporting the values into Excel and GraphPad Prism. Cilia length measurements in hTERT‐RPE1 cells were performed in Fiji/ImageJ (NIH) 125 using acetylated alpha tubulin as a cilia marker, and ≥ 50 cilia were analysed for each experimental condition. Cilia length measurements in IMCD3 and Kupffer's vesicle (KV) cells were also performed in Fiji/ImageJ (NIH) 125 in 3D, using the Segmentation plugin and the Simple Neurite Tracer tool 126, and acetylated alpha tubulin as a cilia marker. Representative maximum projections of confocal images of KV and IMCD3 cells stained with anti‐acetylated alpha tubulin are shown. For IMCD3 cells, > 50 cilia were analysed for each experimental condition, whereas for Zebrafish embryos, 8–12 embryos were analysed per condition and per experiment, which corresponds to > 250 cilia per condition and experiment. Staining intensity of ciliary membrane proteins was quantified with Cellprofiler 2.1.1 software using an automated image analysis pipeline for the identification of the ciliary area in each cell 127. Single‐plane confocal images of cells stained for an axonemal marker (acetylated or polyglutamylated tubulin) and the cilia membrane protein of interest (ARL13B, INPP5E or SMO) were used. First, background subtraction was performed using a rolling ball algorithm in the RunImageJ module. To identify cilia, the area stained by the axonemal marker was segmented using the IdentifyPrimaryObjects module with the RobustBackground thresholding method. Average pixel intensity within these regions was measured for the protein of interest using the MeasureObjectIntensity module, and ≥ 25 cilia were analysed for each experimental condition. Intensity values were normalised to the mean of the control sample, which was either cells treated with non‐targeting siRNA for depletion experiments or cells expressing GFP alone for overexpression experiments. For quantification of SMO ciliary intensity levels and SMO+ cilia, cells were stained for acetylated tubulin and SMO. Acetylated tubulin staining was used to identify the ciliary area (acetyl. tub+ cilia) of all cilia as described above and the average pixel intensity of SMO staining was measured within these regions. To identify cilia that displayed a discernible enrichment of SMO (SMO+ cilia), images stained for SMO were thresholded using the IdentifyPrimaryObjects module with the RobustBackground thresholding method. This resulted in the segmentation of ciliary area stained by SMO above the threshold, which were then correlated with the acetyl. tub‐positive cilia.
Cell lysis, immunoprecipitation and immunoblotting
IMCD3 cells were lysed in cold lysis buffer [50 mM Tris–HCl pH 7.4, 1% IGEPAL, 150 mM NaCl, 1 mM EDTA, 1 mM ethylene glycol tetraacetic acid (EGTA), 2 mM MgCl2, 1 mM dithiothreitol (DTT)] in the presence of EDTA‐free protease and phosphatase inhibitors (Roche) for 20 min on ice, followed by centrifugation at 12,000 × g for 20 min at 4°C. Protein concentration in total cell lysates was determined using the DC protein assay kit (Bio‐Rad). Zebrafish embryos at 8 somite stage were dechorionated using sharp forceps, then disrupted in media with 0.1% EDTA to remove yolk and dissociate cells. Next, they were lysed in cold extraction buffer (0.5 M Tris–HCl pH 6.8, 3% SDS, 1% glycerol, 1 mM DTT) supplemented with complete EDTA‐free protease inhibitor cocktail (Roche) with the help of a sonicator. NIH3T3 and hTERT‐RPE1 cells were lysed on ice for 20 min in lysis buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1% NP‐40, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 2 mM Na3VO4) supplemented with complete EDTA‐free protease inhibitor cocktail (Sigma‐Aldrich) and lysates were cleared by centrifugation. For immunoprecipitation, one 10‐cm dish of transfected HEK293T cells per immunoprecipitation reaction were lysed at 4°C for 20 min in co‐IP buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1% NP‐40, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 2 mM Na3VO4) supplemented with complete EDTA‐free protease inhibitor cocktail (Sigma‐Aldrich) and lysates were cleared by centrifugation. Protein concentrations were determined with the Bradford assay (Sigma‐Aldrich). Equal protein amounts (~0.5–1 μg in 800 μl) were subjected to immunoprecipitation with 5 μl GFP‐Trap beads (Chromotek) at 4°C for 4 h. Beads were washed three times with extraction buffer, and the bound proteins were eluted in Laemmli sample buffer. For pull‐down experiments, HEK293T cells expressing either GFP‐RAB35 or ARL13B‐GFP were lysed at 4°C for 20 min in 1.6 ml buffer (50 mM HEPES pH 7.2, 150 mM NaCl, 1 mM MgCl2, 1% Triton X‐100) supplemented with complete EDTA‐free protease inhibitor cocktail (Sigma‐Aldrich). Lysates were cleared by centrifugation, divided equally into three tubes (500 μl each), and subjected to immunoprecipitation with 5 μl GFP‐Trap beads (Chromotek) at 4°C for 2 h. Beads were washed once with nucleotide exchange buffer (50 mM HEPES ph 7.2, 150 mM NaCl, 1 mM MgCl2, 1% Triton X‐100, 2.5 mM EDTA), resuspended in 50 μl nucleotide exchange buffer and incubated for 10 min on ice. A sample without nucleotide addition was used as negative control, and to other samples, either GTPγS or GDP (Sigma‐Aldrich) was added at a final concentration of 1 mM and incubated for 20 min on ice. To lock the nucleotide‐bound state, MgCl2 was added to a final concentration of 20 mM and incubate for 10 min on ice. Beads were washed once in IP buffer (50 mM HEPES pH 7.2, 150 mM NaCl, 3 mM MgCl2, 1% Triton X‐100), and 500 μl of cell lysates, which was prepared from HEK293T cells expressing either ARL13B‐FLAG or mCherry‐RAB35 in IP buffer, was added and incubated at 4°C for 2 h. Beads were washed three times with IP buffer, and bound proteins were eluted in Laemmli sample buffer. Protein samples were separated by SDS–PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% milk/0.1% Tween‐20/PBS and probed with primary antibodies and HRP‐conjugated secondary antibodies (Appendix Table S2), followed by detection with ECL (Thermo Fisher). Band intensities on immunoblots were quantified using Fiji/ImageJ (NIH) 125. For the analysis of ARL13B total protein levels, the ratios of ARL13B and β‐tubulin (loading control) levels were calculated and normalised to the control treated with non‐targeting siRNA to allow the comparison of independent experiments.
Statistical data
Graphs were drawn, and statistical analysis was performed using Prism (GraphPad). Data are presented as mean ± standard error of the mean (SEM) or box‐and‐whisker plots with horizontal lines showing 25, 50 and 75th percentiles and whiskers extending to minimum and maximum values. For data in which more than two datasets were compared, ANOVA followed by Bonferroni post hoc test or Kruskal–Wallis test followed by Dunn's post hoc test were used. For data comparing two datasets, unpaired t‐test with Welch's correction was used. For zebrafish data, Fisher exact test was performed. Differences were considered significant if P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
Animal welfare statement
The procedures performed in zebrafish were approved by the Portuguese Veterinary General Administration (DGAV—Direção Geral de Alimentação e Veterinária) and guaranteed the welfare of the animals.
Author contributions
SK, CS, DCB and OEB conceived the project and designed the experiments. SK conducted the hTERT‐RPE1 cell experiments assessing Rab35 localisation, ciliogenesis in Rab35/DENND1B/TBC1D10A‐disrupted cells, and the ciliary membrane levels of Arl13b/INPP5E/SMO, as well as the co‐immunoprecipitation experiments in HEK293T cells. SK generated the Rab35 knockout NIH3T3 cell lines and conducted the experiments assessing ciliogenesis and ciliary membrane levels of Arl13b/INPP5E. CS conducted the IMCD3 cell experiments that assessed Rab35 localisation and ciliogenesis in Rab35‐disrupted cells and Rab35 overexpressing cells, with the contribution of RN, and the experiments assessing PI(4,5)P2 levels in Rab35 knockout cells. CS, SP, BT and RJ designed and performed the zebrafish experiments with SSL supervision. JSR generated constructs used on experiments with zebrafish and mouse cell lines. JCS contributed reagents and advice on image analysis. JSA contributed reagents and advice on experimental design. AE contributed with reagents, advice on experimental design and shared preliminary data. SK and CS compiled the figures. SK, CS, DCB and OEB wrote the manuscript. All authors reviewed and provided input to the final text.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
The authors are grateful to K. Kontani (University of Tokyo), G. Pereira (University of Heidelberg), A. Smolenski (University College Dublin), J. Reiter (University of California), C. Westlake (NIH) and E. van Wijk (Radboud University) for reagents, and the UCD Conway Institute imaging facility and the Danish Molecular Biomedical Imaging Center (DaMBIC, University of Southern Denmark) for imaging support. We would like to thank the technical support of Instituto Gulbenkian de Ciência Advanced Imaging Facility (AIF‐UIC), supported by funding from Fundação para a Ciência e a Tecnologia (FCT, IP; PPBI‐POCI‐01‐0145‐FEDER‐022122), co‐financed by Lisboa Regional Operational Programme (Lisboa 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER) and Fundação para a Ciência e a Tecnologia (FCT; Portugal), as well as Telmo Pereira and Petra Pintado from CEDOC's imaging and zebrafish facilities, respectively. This work was funded by a Science Foundation Ireland Principal investigator (11/PI/1037) award to OEB, by the FCT Investigator Program (IF/00501/2014/CP1252/CT0001) to DCB, by a grant from the Danish Council for Independent Research (#8021‐00425B) to JSA, by a postdoctoral fellowship from FCT (SFRH/BPD/32323/2006) to CS and by iNOVA4Health—UID/Multi/04462/2013, a program financially supported by FCT/Ministério da Educação e Ciência, through national funds and co‐funded by FEDER under the PT2020 Partnership Agreement.
EMBO Reports (2019) 20: e47625
Contributor Information
Stefanie Kuhns, Email: stefaniek@bmb.sdu.dk.
Cecília Seixas, Email: cecilia.seixas@nms.unl.pt.
Duarte C Barral, Email: duarte.barral@nms.unl.pt.
Oliver E Blacque, Email: oliver.blacque@ucd.ie.
References
- 1. Satir P, Pedersen LB, Christensen ST (2010) The primary cilium at a glance. J Cell Sci 123: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goetz SC, Anderson KV (2010) The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11: 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bangs F, Anderson KV (2016) Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol 9: a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tavares B, Jacinto R, Sampaio P, Pestana S, Pinto A, Vaz A, Roxo‐Rosa M, Gardner R, Lopes T, Schilling B et al (2017) Notch/Her12 signalling modulates, motile/immotile cilia ratio downstream of Foxj1a in zebrafish left‐right organizer. Elife 6: e25165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shinohara K, Hamada H (2017) Cilia in left‐right symmetry breaking. Cold Spring Harb Perspect Biol 9: a028282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Essner JJ (2005) Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left‐right development of the brain, heart and gut. Development 132: 1247–1260 [DOI] [PubMed] [Google Scholar]
- 7. Sampaio P, Ferreira RR, Guerrero A, Pintado P, Tavares B, Amaro J, Smith AA, Montenegro‐Johnson T, Smith DJ, Lopes SS (2014) Left‐right organizer flow dynamics: how much cilia activity reliably yields laterality? Dev Cell 29: 716–728 [DOI] [PubMed] [Google Scholar]
- 8. Waters AM, Beales PL (2011) Ciliopathies: an expanding disease spectrum. Pediatr Nephrol 26: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rubbo B, Lucas JS (2017) Clinical care for primary ciliary dyskinesia: current challenges and future directions. Eur Respir Rev 26: 170023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wingfield JL, Lechtreck K‐F, Lorentzen E (2018) Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem 62: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nachury MV (2018) The molecular machines that traffic signaling receptors into and out of cilia. Curr Opin Cell Biol 51: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jensen VL, Leroux MR (2017) Gates for soluble and membrane proteins, and two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr Opin Cell Biol 47: 83–91 [DOI] [PubMed] [Google Scholar]
- 13. Ishikawa H, Marshall WF (2017) Intraflagellar transport and ciliary dynamics. Cold Spring Harb Perspect Biol 9: a021998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reiter JF, Blacque OE, Leroux MR (2012) The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13: 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia‐Gonzalo FR, Reiter JF (2017) Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol 9: a028134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukhopadhyay S, Badgandi HB, Hwang S‐H, Somatilaka B, Shimada IS, Pal K (2017) Trafficking to the primary cilium membrane. Mol Biol Cell 28: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pedersen LB, Mogensen JB, Christensen ST (2016) Endocytic control of cellular signaling at the primary cilium. Trends Biochem Sci 41: 784–797 [DOI] [PubMed] [Google Scholar]
- 18. Monis WJ, Faundez V, Pazour GJ (2017) BLOC‐1 is required for selective membrane protein trafficking from endosomes to primary cilia. J Cell Biol 216: 2131–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scheidel N, Kennedy J, Blacque OE (2018) Endosome maturation factors Rabenosyn‐5/VPS45 and caveolin‐1 regulate ciliary membrane and polycystin‐2 homeostasis. EMBO J 37: e98248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Setou M, Rohatgi R, Reiter JF, Ikegami K et al (2017) Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168: 264–279 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nager AR, Goldstein JS, Herranz‐Pérez V, Portran D, Ye F, Garcia‐Verdugo JM, Nachury MV (2017) An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168: 252–263 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood CR, Huang K, Diener DR, Rosenbaum JL (2013) The cilium secretes bioactive ectosomes. Curr Biol 23: 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, Barr MM (2014) C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol 24: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao M, Ning J, Hernandez‐Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ (2015) Uni‐directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife 4: 183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Donaldson JG, Jackson CL (2011) ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 12: 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- 27. Zhen Y, Stenmark H (2015) Cellular functions of Rab GTPases at a glance. J Cell Sci 128: 3171–3176 [DOI] [PubMed] [Google Scholar]
- 28. Pfeffer SR (2017) Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell 28: 712–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cherfils J, Zeghouf M (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309 [DOI] [PubMed] [Google Scholar]
- 30. Yoshimura S‐I, Egerer J, Fuchs E, Haas AK, Barr FA (2007) Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol 178: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsiao Y‐C, Tuz K, Ferland RJ (2012) Trafficking in and to the primary cilium. Cilia 1: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blacque OE, Scheidel N, Kuhns S (2018) Rab GTPases in cilium formation and function. Small GTPases 9: 76–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pusapati GV, Kong JH, Patel BB, Krishnan A, Sagner A, Kinnebrew M, Briscoe J, Aravind L, Rohatgi R (2018) CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev Cell 44: 271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu S, Liu Y, Meng Q, Wang B (2018) Rab34 small GTPase is required for Hedgehog signaling and an early step of ciliary vesicle formation in mouse. J Cell Sci 131: jcs213710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J, Deretic D (2014) Molecular complexes that direct rhodopsin transport to primary cilia. Prog Retin Eye Res 38: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Knödler A, Feng S, Zhang J, Zhang X, Das A, Peränen J, Guo W (2010) Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA 107: 6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC et al (2011) Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex‐dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA 108: 2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jensen VL, Carter S, Sanders AAWM, Li C, Kennedy J, Timbers TA, Cai J, Scheidel N, Kennedy BN, Morin RD et al (2016) Whole‐organism developmental expression profiling identifies RAB‐28 as a novel ciliary GTPase associated with the BBSome and intraflagellar transport. PLoS Genet 12: e1006469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW et al (2014) IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell 31: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liew GM, Ye F, Nager AR, Murphy JP, Lee JS, Aguiar M, Breslow DK, Gygi SP, Nachury MV (2014) The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell 31: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caspary T, Larkins CE, Anderson KV (2007) The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 12: 767–778 [DOI] [PubMed] [Google Scholar]
- 42. Larkins CE, Aviles GDG, East MP, Kahn RA, Caspary T (2011) Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 22: 4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K (2016) Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci 130: 563–576 [DOI] [PubMed] [Google Scholar]
- 44. Lu H, Toh MT, Narasimhan V, Thamilselvam SK, Choksi SP, Roy S (2015) A function for the Joubert syndrome protein Arl13b in ciliary membrane extension and ciliary length regulation. Dev Biol 397: 225–236 [DOI] [PubMed] [Google Scholar]
- 45. Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley‐Fisher C, Cottell D, Katada T, Kontani K, Blacque OE (2010) Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans . J Cell Biol 188: 953–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y, Wei Q, Zhang Y, Ling K, Hu J (2010) The small GTPases ARL‐13 and ARL‐3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol 189: 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y, Zhang Q, Wei Q, Zhang Y, Ling K, Hu J (2013) SUMOylation of the small GTPase ARL‐13 promotes ciliary targeting of sensory receptors. J Cell Biol 200: 357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He K, Ma X, Xu T, Li Y, Hodge A, Zhang Q, Torline J, Huang Y, Zhao J, Ling K et al (2018) Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules. Nat Commun 9: 3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seixas C, Choi SY, Polgar N, Umberger NL, East MP, Zuo X, Moreiras H, Ghossoub R, Benmerah A, Kahn RA et al (2016) Arl13b and the exocyst interact synergistically in ciliogenesis. Mol Biol Cell 27: 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bay SN, Long AB, Caspary T (2018) Disruption of the ciliary GTPase Arl13b suppresses Sonic hedgehog overactivation and inhibits medulloblastoma formation. Proc Natl Acad Sci USA 115: 1570–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gotthardt K, Lokaj M, Koerner C, Falk N, Gießl A, Wittinghofer A (2015) A G‐protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. Elife 4: e11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Q, Li Y, Zhang Y, Torres VE, Harris PC, Ling K, Hu J (2016) GTP‐binding of ARL‐3 is activated by ARL‐13 as a GEF and stabilized by UNC‐119. Sci Rep 6: 24534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attié‐Bitach T, Holden KR, Dobyns WB et al (2008) Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet 83: 170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alkanderi S, Molinari E, Shaheen R, Elmaghloob Y, Stephen LA, Sammut V, Ramsbottom SA, Srivastava S, Cairns G, Edwards N et al (2018) ARL3 mutations cause Joubert syndrome by disrupting ciliary protein composition. Am J Hum Genet 103: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May‐Simera H, Compton DS et al (2004) Mutations in a member of the Ras superfamily of small GTP‐binding proteins causes Bardet‐Biedl syndrome. Nat Genet 36: 989–993 [DOI] [PubMed] [Google Scholar]
- 56. Roosing S, Rohrschneider K, Beryozkin A, Sharon D, Weisschuh N, Staller J, Kohl S, Zelinger L, Peters TA, Neveling K et al (2013) Mutations in RAB28, encoding a farnesylated small GTPase, are associated with autosomal‐recessive cone‐rod dystrophy. Am J Hum Genet 93: 110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jenkins D, Seelow D, Jehee FS, Perlyn CA, Alonso LG, Bueno DF, Donnai D, Josifova D, Mathijssen IMJ, Morton JEV et al (2007) RAB23 mutations in Carpenter syndrome imply an unexpected role for hedgehog signaling in cranial‐suture development and obesity. Am J Hum Genet 80: 1162–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kouranti I, Sachse M, Arouche N, Goud B, Echard A (2006) Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol 16: 1719–1725 [DOI] [PubMed] [Google Scholar]
- 59. Chaineau M, Ioannou MS, McPherson PS (2013) Rab35: GEFs, GAPs and effectors. Traffic 14: 1109–1117 [DOI] [PubMed] [Google Scholar]
- 60. Klinkert K, Echard A (2016) Rab35 GTPase: a central regulator of phosphoinositides and F‐actin in endocytic recycling and beyond. Traffic 17: 1063–1077 [DOI] [PubMed] [Google Scholar]
- 61. Kobayashi H, Fukuda M (2012) Rab35 regulates Arf6 activity through centaurin‐β2 (ACAP2) during neurite outgrowth. J Cell Sci 125: 2235–2243 [DOI] [PubMed] [Google Scholar]
- 62. Kobayashi H, Etoh K, Ohbayashi N, Fukuda M (2014) Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL‐L1 during neurite outgrowth. Biol Open 3: 803–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Allaire PD, Seyed Sadr M, Chaineau M, Seyed Sadr E, Konefal S, Fotouhi M, Maret D, Ritter B, Del Maestro RF, McPherson PS (2013) Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J Cell Sci 126: 722–731 [DOI] [PubMed] [Google Scholar]
- 64. Allaire PD, Marat AL, Dall'Armi C, Di Paolo G, McPherson PS, Ritter B (2010) The Connecdenn DENN domain: a GEF for Rab35 mediating cargo‐specific exit from early endosomes. Mol Cell 37: 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wheeler DB, Zoncu R, Root DE, Sabatini DM, Sawyers CL (2015) Identification of an oncogenic RAB protein. Science 350: 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cauvin C, Rosendale M, Gupta‐Rossi N, Rocancourt M, Larraufie P, Salomon R, Perrais D, Echard A (2016) Rab35 GTPase triggers switch‐like recruitment of the Lowe syndrome lipid phosphatase OCRL on newborn endosomes. Curr Biol 26: 120–128 [DOI] [PubMed] [Google Scholar]
- 67. Chesneau L, Dambournet D, Machicoane M, Kouranti I, Fukuda M, Goud B, Echard A (2012) An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr Biol 22: 147–153 [DOI] [PubMed] [Google Scholar]
- 68. Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocancourt M, El Marjou A, Formstecher E, Salomon R, Goud B, Echard A (2011) Rab35 GTPase and OCRL phosphatase remodel lipids and F‐actin for successful cytokinesis. Nat Cell Biol 13: 981–988 [DOI] [PubMed] [Google Scholar]
- 69. Kim J, Lee JE, Heynen‐Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza‐Blanc P, Gleeson JG (2010) Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 464: 1048–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu Q, Tan G, Levenkova N, Li T, Pugh EN Jr, Rux JJ, Speicher DW, Pierce EA (2007) The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6: 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT et al (2015) A dynamic protein interaction landscape of the human centrosome‐cilium interface. Cell 163: 1484–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Breslow DK, Hoogendoorn S, Kopp AR, Morgens DW, Vu BK, Kennedy MC, Han K, Li A, Hess GT, Bassik MC et al (2018) A CRISPR‐based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet 50: 460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mayer U, Küller A, Daiber PC, Neudorf I, Warnken U, Schnölzer M, Frings S, Möhrlen F (2009) The proteome of rat olfactory sensory cilia. Proteomics 9: 322–334 [DOI] [PubMed] [Google Scholar]
- 74. Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, Peters DJM, Doxsey S (2004) Pericentrin forms a complex with intraflagellar transport proteins and polycystin‐2 and is required for primary cilia assembly. J Cell Biol 166: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mai W, Chen D, Ding T, Kim I, Park S, Cho S‐Y, Chu JSF, Liang D, Wang N, Wu D et al (2005) Inhibition of Pkhd1 impairs tubulomorphogenesis of cultured IMCD cells. Mol Biol Cell 16: 4398–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang Y‐S et al (2015) Early steps in primary cilium assembly require EHD1/EHD3‐dependent ciliary vesicle formation. Nat Cell Biol 17: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wheway G, Schmidts M, Mans DA, Szymanska K, Nguyen T‐MT, Racher H, Phelps IG, Toedt G, Kennedy J, Wunderlich KA et al (2015) An siRNA‐based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol 17: 1074–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ran FA, Ann Ran F, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fuchs E, Haas AK, Spooner RA, Yoshimura S‐I, Lord JM, Barr FA (2007) Specific Rab GTPase‐activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J Cell Biol 177: 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Blümer J, Rey J, Dehmelt L, Mazel T, Wu Y‐W, Bastiaens P, Goody RS, Itzen A (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol 200: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hackett B (2002) Formation and malformation of the vertebrate left‐right axis. Curr Mol Med 2: 39–66 [DOI] [PubMed] [Google Scholar]
- 82. Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, Ikegami K et al (2012) CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet 44: 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fogelgren B, Lin S‐Y, Zuo X, Jaffe KM, Park KM, Reichert RJ, Bell PD, Burdine RD, Lipschutz JH (2011) The exocyst protein Sec10 interacts with Polycystin‐2 and knockdown causes PKD‐phenotypes. PLoS Genet 7: e1001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kramer‐Zucker AG (2005) Cilia‐driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132: 1907–1921 [DOI] [PubMed] [Google Scholar]
- 85. Hori Y, Kobayashi T, Kikko Y, Kontani K, Katada T (2008) Domain architecture of the atypical Arf‐family GTPase Arl13b involved in cilia formation. Biochem Biophys Res Commun 373: 119–124 [DOI] [PubMed] [Google Scholar]
- 86. Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K (2017) Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci 130: 563–576 [DOI] [PubMed] [Google Scholar]
- 87. Higginbotham H, Eom T‐Y, Mariani LE, Bachleda A, Hirt J, Gukassyan V, Cusack CL, Lai C, Caspary T, Anton ES (2012) Arl13b in primary cilia regulates the migration and placement of interneurons in the developing cerebral cortex. Dev Cell 23: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cevik S, Sanders AAWM, Van Wijk E, Boldt K, Clarke L, van Reeuwijk J, Hori Y, Horn N, Hetterschijt L, Wdowicz A et al (2013) Active transport and diffusion barriers restrict Joubert Syndrome‐associated ARL13B/ARL‐13 to an Inv‐like ciliary membrane subdomain. PLoS Genet 9: e1003977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S (2012) ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA 109: 19691–19696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fansa EK, Kösling SK, Zent E, Wittinghofer A, Ismail S (2016) PDE6δ‐mediated sorting of INPP5E into the cilium is determined by cargo‐carrier affinity. Nat Commun 7: 11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Garcia‐Gonzalo FR, Phua SC, Roberson EC, Garcia G III, Abedin M, Schurmans S, Inoue T, Reiter JF (2015) Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev Cell 34: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chávez M, Ena S, Van Sande J, de Kerchove d'Exaerde A, Schurmans S, Schiffmann SN (2015) Modulation of ciliary phosphoinositide content regulates trafficking and sonic Hedgehog signaling output. Dev Cell 34: 338–350 [DOI] [PubMed] [Google Scholar]
- 93. Stauffer TP, Ahn S, Meyer T (1998) Receptor‐induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol 8: 343–346 [DOI] [PubMed] [Google Scholar]
- 94. Li Y, Tian X, Ma M, Jerman S, Kong S, Somlo S, Sun Z (2016) Deletion of ADP ribosylation factor‐like GTPase 13B leads to kidney cysts. J Am Soc Nephrol 27: 3628–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mukhopadhyay S, Rohatgi R (2014) G‐protein‐coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol 33: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen JK, Taipale J, Young KE, Maiti T, Beachy PA (2002) Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA 99: 14071–14076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pintado P, Sampaio P, Tavares B, Montenegro‐Johnson TD, Smith DJ, Lopes SS (2017) Dynamics of cilia length in left‐right development. R Soc Open Sci 4: 161102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wiegering A, Dildrop R, Kalfhues L, Spychala A, Kuschel S, Lier JM, Zobel T, Dahmen S, Leu T, Struchtrup A et al (2018) Cell type‐specific regulation of ciliary transition zone assembly in vertebrates. EMBO J 37: e97791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Marat AL, McPherson PS (2010) The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J Biol Chem 285: 10627–10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shah M, Baterina OY Jr, Taupin V, Farquhar MG (2013) ARH directs megalin to the endocytic recycling compartment to regulate its proteolysis and gene expression. J Cell Biol 202: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hsu C, Morohashi Y, Yoshimura S‐I, Manrique‐Hoyos N, Jung S, Lauterbach MA, Bakhti M, Grønborg M, Möbius W, Rhee J et al (2010) Regulation of exosome secretion by Rab35 and its GTPase‐activating proteins TBC1D10A‐C. J Cell Biol 189: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yang C‐W, Hojer CD, Zhou M, Wu X, Wuster A, Lee WP, Yaspan BL, Chan AC (2016) Regulation of T cell receptor signaling by DENND1B in TH2 cells and allergic disease. Cell 164: 141–155 [DOI] [PubMed] [Google Scholar]
- 103. Hokanson DE, Bretscher AP (2012) EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol Biol Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Itoh T, Fukuda M (2006) Identification of EPI64 as a GTPase‐activating protein specific for Rab27A. J Biol Chem 281: 31823–31831 [DOI] [PubMed] [Google Scholar]
- 105. Lopes SS, Lourenço R, Pacheco L, Moreno N, Kreiling J, Saúde L (2010) Notch signalling regulates left‐right asymmetry through ciliary length control. Development 137: 3625–3632 [DOI] [PubMed] [Google Scholar]
- 106. Smith DJ, Montenegro‐Johnson TD, Lopes SS (2014) Organized chaos in Kupffer's vesicle: how a heterogeneous structure achieves consistent left‐right patterning. Bioarchitecture 4: 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Montenegro‐Johnson TD, Baker DI, Smith DJ, Lopes SS (2016) Three‐dimensional flow in Kupffer's Vesicle. J Math Biol 73: 705–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hoang‐Minh LB, Dutra‐Clarke M, Breunig JJ, Sarkisian MR (2018) Glioma cell proliferation is enhanced in the presence of tumor‐derived cilia vesicles. Cilia 7: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shao J, Xu L, Chen L, Lu Q, Xie X, Shi W, Xiong H, Shi C, Huang X, Mei J et al (2017) Arl13b promotes gastric tumorigenesis by regulating Smo trafficking and activation of the Hedgehog signaling pathway. Cancer Res 77: 4000–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Duldulao NA, Lee S, Sun Z (2009) Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development 136: 4033–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Williams CL, McIntyre JC, Norris SR, Jenkins PM, Zhang L, Pei Q, Verhey K, Martens JR (2014) Direct evidence for BBSome‐associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun 5: 5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Garcia‐Gonzalo FR, Corbit KC, Sirerol‐Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ et al (2011) A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet 43: 776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS (2011) A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol 14: 61–72 [DOI] [PubMed] [Google Scholar]
- 114. Carter SP, Blacque OE (2019) Membrane retrieval, recycling and release pathways that organise and sculpt the ciliary membrane. Curr Opin Cell Biol 59: 133–139 [DOI] [PubMed] [Google Scholar]
- 115. Haley R, Wang Y, Zhou Z (2018) The small GTPase RAB‐35 defines a third pathway that is required for the recognition and degradation of apoptotic cells. PLoS Genet 14: e1007558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Egami Y, Fukuda M, Araki N (2011) Rab35 regulates phagosome formation through recruitment of ACAP2 in macrophages during FcγR‐mediated phagocytosis. J Cell Sci 124: 3557–3567 [DOI] [PubMed] [Google Scholar]
- 117. Vaz‐Silva J, Gomes P, Jin Q, Zhu M, Zhuravleva V, Quintremil S, Meira T, Silva J, Dioli C, Soares‐Cunha C et al (2018) Endolysosomal degradation of Tau and its role in glucocorticoid‐driven hippocampal malfunction. EMBO J 37: e99084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Molla‐Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A et al (2010) The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci 123: 1785–1795 [DOI] [PubMed] [Google Scholar]
- 119. Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MPR, Pedersen LB, Benmerah A, Andersen CY, Larsen LA et al (2013) TGF‐β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep 3: 1806–1814 [DOI] [PubMed] [Google Scholar]
- 120. Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD (2008) Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME‐4, a coated‐pit protein. EMBO J 27: 1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Barral DC, Garg S, Casalou C, Watts GFM, Sandoval JL, Ramalho JS, Hsu VW, Brenner MB (2012) Arl13b regulates endocytic recycling traffic. Proc Natl Acad Sci USA 109: 21354–21359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 123. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R et al (2016) Optimized sgRNA design to maximize activity and minimize off‐target effects of CRISPR‐Cas9. Nat Biotechnol 34: 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O et al (2013) DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat Biotechnol 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Longair MH, Baker DA, Armstrong JD (2011) Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27: 2453–2454 [DOI] [PubMed] [Google Scholar]
- 127. Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J et al (2006) Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6


