Abstract
The methylation of histone H3 at lysine 9 (H3K9me), performed by the methyltransferase Clr4/SUV39H, is a key event in heterochromatin assembly. In fission yeast, Clr4, together with the ubiquitin E3 ligase Cul4, forms the Clr4 methyltransferase complex (CLRC), whose physiological targets and biological role are currently unclear. Here, we show that CLRC‐dependent H3 ubiquitylation regulates Clr4's methyltransferase activity. Affinity‐purified CLRC ubiquitylates histone H3, and mass spectrometric and mutation analyses reveal that H3 lysine 14 (H3K14) is the preferred target of the complex. Chromatin immunoprecipitation analysis shows that H3K14 ubiquitylation (H3K14ub) is closely associated with H3K9me‐enriched chromatin. Notably, the CLRC‐mediated H3 ubiquitylation promotes H3K9me by Clr4, suggesting that H3 ubiquitylation is intimately linked to the establishment and/or maintenance of H3K9me. These findings demonstrate a cross‐talk mechanism between histone ubiquitylation and methylation that is involved in heterochromatin assembly.
Keywords: epigenetic gene silencing, fission yeast, heterochromatin, histone methylation, histone ubiquitylation
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics
Introduction
In eukaryotic cells, the formation of higher‐order chromatin structure, known as heterochromatin, plays an important role in diverse chromosomal processes. Heterochromatin assembly is intimately associated with changes in post‐translational histone‐tail modifications 1, 2, 3. Histone H3 lysine 9 methylation (H3K9me), a hallmark of heterochromatin structure, is catalyzed by SUV39H‐family histone methyltransferases (HMTases) and functions as a binding site for recruiting heterochromatin protein 1 (HP1) family proteins 4, 5, 6. Heterochromatic structures are epigenetically inherited through cell division in a metastable manner 7, 8.
In the fission yeast Schizosaccharomyces pombe (S. pombe), heterochromatin plays critical roles in the assembly of functional chromosomal domains such as centromeres, telomeres, and the mating‐type loci 3, 9. Clr4, a homolog of mammalian SUV39H, is the sole H3K9 methyltransferase expressed in S. pombe and plays a central role in heterochromatin assembly 4, 6, 10. H3K9me creates binding sites for the chromodomain (CD) proteins Swi6, Chp1, and Chp2, and these proteins further recruit a variety of chromatin proteins to form repressive higher‐order chromatin 11, 12, 13, 14. Clr4 also possesses a CD that can bind H3K9me, and it has been suggested that the ability of Clr4 and other HMTases to “write” and “read” H3K9me facilitates heterochromatin spreading and the maintenance of H3K9me during cell division 15.
In fission yeast, the assembly and maintenance of pericentromeric heterochromatin is directly linked to the RNA interference (RNAi) pathway 3, 16, 17, 18. Pericentromeric repeats are transcribed by RNA polymerase II, and the nascent RNAs are converted to small interfering RNAs (siRNAs) through the actions of the RNA‐dependent RNA polymerase complex (RDRC) and Dicer (Dcr1) ribonuclease. The siRNAs are then loaded onto Argonaute (Ago1), the catalytic component of the RNA‐induced transcriptional silencing (RITS) complex, which targets pericentromeric repeats through base‐pairing interaction between the siRNAs and nascent transcripts. In addition to Ago1, the RITS complex contains the GW motif protein Tas3 and the chromodomain protein Chp1, and the association of the RITS complex with chromatin is facilitated by the Chp1‐CD's H3K9me‐binding and nucleic acid‐binding activities 19. The deletion of RNAi pathway components leads to a loss of silencing and reduced H3K9me at the pericentromeric regions, indicating that the RNAi pathway and recruitment of the siRNA‐bound RITS complex to chromatin are coupled with the targeting of Clr4. While RNAi also targets the silent mating‐type loci and telomeres, alternative pathways act redundantly with the RNAi pathway to recruit the Clr4 HMTase activity 20, 21.
Clr4 forms a multi‐protein complex called the Clr4 methyltransferase complex (CLRC). The CLRC consists of the cullin scaffold protein Cul4, the β‐propeller protein Rik1, the WD‐40 protein Raf1 (Dos1/Cmc1/Clr8), the replication foci targeting sequence (RFTS)‐like domain‐containing protein Raf2 (Dos2/Cmc2/Clr7), and the RING‐box protein Rbx1 (Pip1) 22, 23, 24, 25, 26. All of these CLRC components except Rbx1 have been shown to be required for heterochromatic silencing. Cul4, Rik1, and Raf1 in fission yeast show a strong structural resemblance to the conserved CUL4‐DDB1‐DDB2 E3 ubiquitin ligase (CRL4DDB1) 27, 28. While Raf2 has no analogous component in the CRL4DDB1 complex, it interacts with Cul4, Rik1, and Raf1, and thus is proposed to be a hub for the core components in the CLRC 28, 29. Deleting genes encoding CLRC components results in the loss of both H3K9 methylation and siRNA 24, 30, indicating that a physical interaction between CLRC and RITS couples the siRNA production with H3K9 methylation. Stc1, a tandem zinc finger domain‐containing protein, was not identified among the CLRC components, but was shown to mediate the interaction between the CLRC and RITS, presumably through interactions with Raf2 and Ago1 31, 32. On the other hand, studies using cul4 mutant cells suggested that Rik1 is loaded onto heterochromatic repeats in an RNAi‐dependent manner and targets other CLRC components to heterochromatic loci 15, 25. Intriguingly, analyses of cells expressing mutant histone H3 (H3K9R) or CLRC components demonstrated that the CLRC complex promotes siRNA production, independently of H3K9 methylation 27, 29, 33, although the details of the mechanism remain unknown.
As predicted from its structural similarity to CRL4DDB1, the CLRC exhibits ubiquitin ligase activity in vitro 22. In addition, heterochromatin defects in Cul4 mutants cannot be rescued by expressing a Cul4 protein lacking Nedd8 modification, which is essential for its ubiquitin ligase activity 25, 34. These data show that the CLRC is an active ubiquitin ligase whose ligase activity is required for heterochromatin formation. Structural and functional studies also suggested that Raf1 plays a critical role in target recognition by the CLRC 27, 28. However, whether the CLRC acts as an E3 ubiquitin ligase in vivo and how ubiquitylation modulates Clr4's HMTase activity remain unclear.
Here, we demonstrated that affinity‐purified CLRC preferentially ubiquitylates lysine 14 on histone H3 (H3K14) in vitro, and the H3K14 ubiquitylation is tightly associated with H3K9me‐enriched heterochromatin in vivo. Importantly, the K14‐ubiquitylated H3 promotes H3K9's methylation by Clr4. This study demonstrates a cross‐talk mechanism between histone methylation and ubiquitylation for heterochromatin assembly.
Results
Histone H3 is ubiquitylated in vitro by the CLRC
To identify the physiological substrate(s) ubiquitylated by CLRC, we first affinity‐purified the CLRC and characterized it. For this purpose, an S. pombe strain expressing C‐terminally TAP‐tagged Rik1 (Rik1‐TAP) was constructed, and Rik1‐TAP and its associated proteins were affinity‐purified. Analysis of the purified fraction revealed protein bands that were specific to the Rik1‐TAP preparation but were absent from a control preparation using an untagged strain (mock) (Fig 1A). Mass spectrometry analysis of the purified fraction identified a number of specific proteins, including Cul4, Nedd8, Raf2, Raf1, and Clr4, in agreement with previous studies (Fig 1A) 22, 24, 25, 27. In addition, a limited number of histone H2B or H4 peptides were detected in the purified fraction, as previously observed 22, 24. Rik1 is reported to form a protein complex with Mms19 and Cdc20 35, but these two proteins were not identified in our Rik1‐TAP preparations.
Figure 1. Histone H3 is ubiquitylated in vitro by the CLRC.
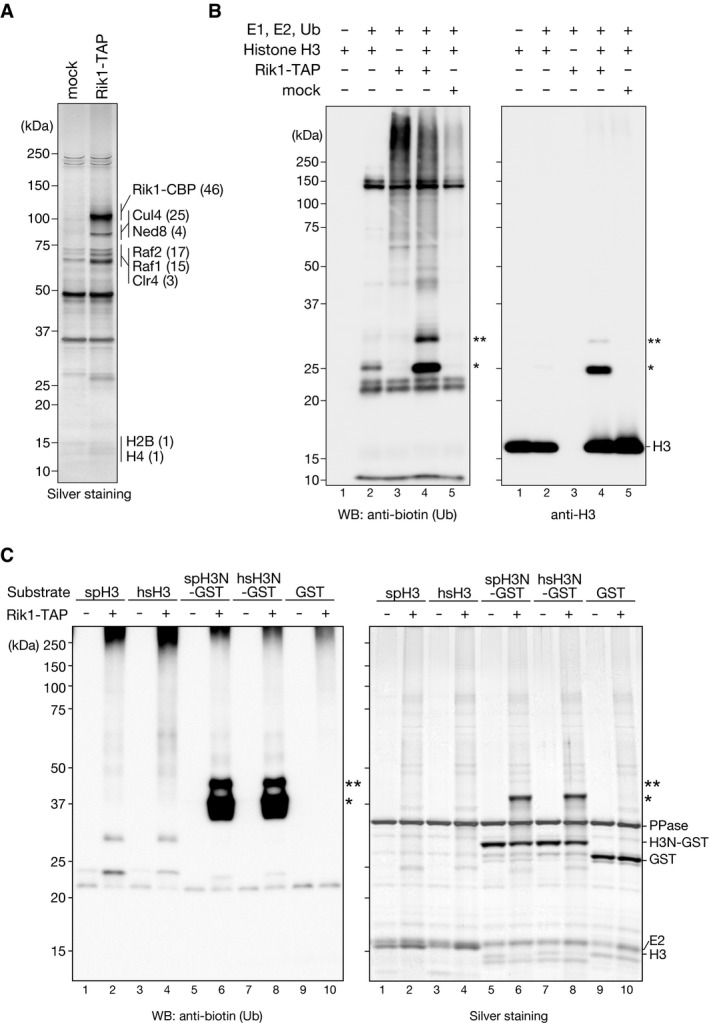
- Purified TAP‐tagged Rik1‐containing complexes analyzed by SDS–PAGE and silver staining. mock: a mock purification from an untagged strain. Proteins identified by LC‐MS/MS are indicated at right, with the number of identified unique peptides in parentheses.
- In vitro ubiquitylation assays using biotinylated ubiquitin, purified CLRC, and recombinant S. pombe histone H3 as the substrate. Proteins were analyzed by Western blotting (WB) using the indicated antibodies. Asterisks indicate ubiquitylated histone H3 species.
- Ubiquitylation assays using recombinant histone H3, the N‐terminal tail of histone H3 (residues 1‐36) fused with GST (H3N‐GST), or GST alone as the substrate. Both fission yeast (sp) and human (hs) full‐length H3 proteins and H3N‐GST fusion proteins were examined. The proteins were analyzed by SDS–PAGE followed by either silver staining or Western blotting with an anti‐biotin antibody. Asterisks indicate ubiquitylated histone H3 species.
Given that histones have been identified in several independent Rik1‐TAP purifications 22, 24 and that histone ubiquitylation has a direct effect on other histone‐modifying enzymes 36, 37, we chose histones as candidate CLRC substrates and examined whether CLRC ubiquitylates them. We performed an in vitro ubiquitylation assay using recombinant E1, E2 (UbcH5b), biotinylated ubiquitin, and histones. Incubation of the Rik1‐TAP preparation with these components in the absence of histone substrate resulted in a ladder of biotinylated ubiquitin (Fig 1B, lane 3, and Fig EV1A–C). The ladder probably represented the self‐ligation of polyubiquitin in the absence of substrate 38, 39, and its appearance suggested that the purified CLRC possessed ubiquitin ligase activity. When histone H3 was added as a substrate, several additional bands appeared, whose molecular weights corresponded to histone H3 modified with mono (~8.6 kDa)‐ or di‐ubiquitin (~17 kDa) (Fig 1B, lane 4, indicated by asterisks). This activity appeared to be specific for the Rik1‐TAP preparation, since we did not detect it in the control assay using a mock preparation obtained from the Rik1‐untagged strain (Fig 1B, lane 5, mock). Although a weak band (~25 kDa) was also detected in the control assay without E3 (Fig 1B, lane 2), it migrated slightly more slowly than the H3 species modified with mono‐ubiquitin, and appeared to be a background product. When histone H2A, H2B, or H4 was used in the assay, a very weak activity was detected for H2A (Fig EV1A, indicated by asterisks), and no obvious change was seen for H2B or H4 (Fig EV1B and C). Although H2A might also serve as a substrate of CLRC, the purified CLRC exhibited a stronger activity for H3, and thus, in this study, we focused on H3's ubiquitylation and its roles in heterochromatin assembly.
Figure EV1. In vitro ubiquitylation assay using affinity‐purified CLRC.
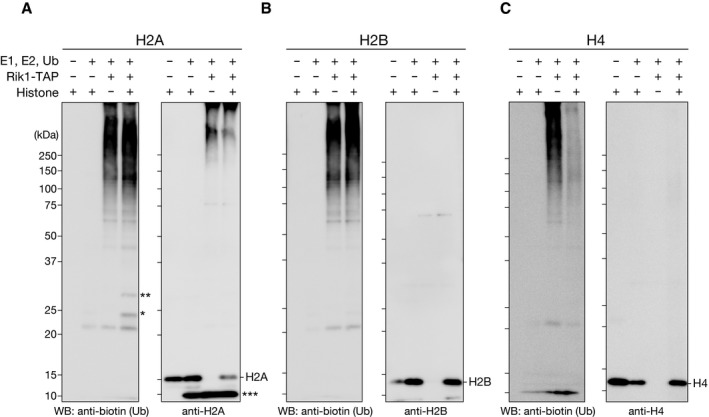
-
A–CIn vitro ubiquitylation assays, in which purified CLRC was incubated with biotinylated ubiquitin and recombinant histone H2A (A), H2B (B), or H4 (C). The proteins were resolved by SDS–PAGE and analyzed by Western blotting using the indicated antibodies. The single and double asterisks indicate mono‐ and di‐ubiquitylated histone species, respectively, and the triple asterisk indicates an unrelated cross‐reactive band for the anti‐H2A antibody.
Characterization of CLRC's H3 ubiquitylation activity
Cul4 functions as scaffold to form E3 ubiquitin ligase and is modified by the ubiquitin‐like protein Nedd8, which induces the E3 ubiquitin ligase activity 34. To determine whether Cul4's activity is involved in the CLRC's H3 ubiquitylation, we first created a cul4 mutant allele in which Lys680 (the site of Nedd8 conjugation) was mutated to arginine (cul4 K680R) as previously reported 25. However, the cul4 K680R mutant showed a severe growth defect (Fig EV2B), which made it difficult to assess Cul4's involvement in CLRC's H3 ubiquitylation. Instead, we isolated a milder cul4 mutant allele (cul4‐1), which did not exhibit a noticeable growth defect (Fig EV2B), but resulted in loss of the silencing of ade6 + or ura4 + marker genes inserted at the centromeric repeats (otr1R::ade6 +) and the mating‐type locus (kint2::ura4 +), respectively (Fig EV2A and B). In this cul4 mutant, its original 3′‐untranslated region was replaced with TEF terminator and a hygromycin‐resistant gene without altering cul4 coding region, which presumably led to a change in cul4 expression or mRNA stability. This mutation changed the profile of the Rik1‐TAP preparation with a clear reduction in the co‐purified Cul4 (Fig EV2C) and markedly reduced the H3 ubiquitylation activity (Fig EV2D). This result suggested that Cul4 plays a critical role in CLRC's H3 ubiquitylation activity.
Figure EV2. Mutation of cul4 affects CLRC's ubiquitylation activity.
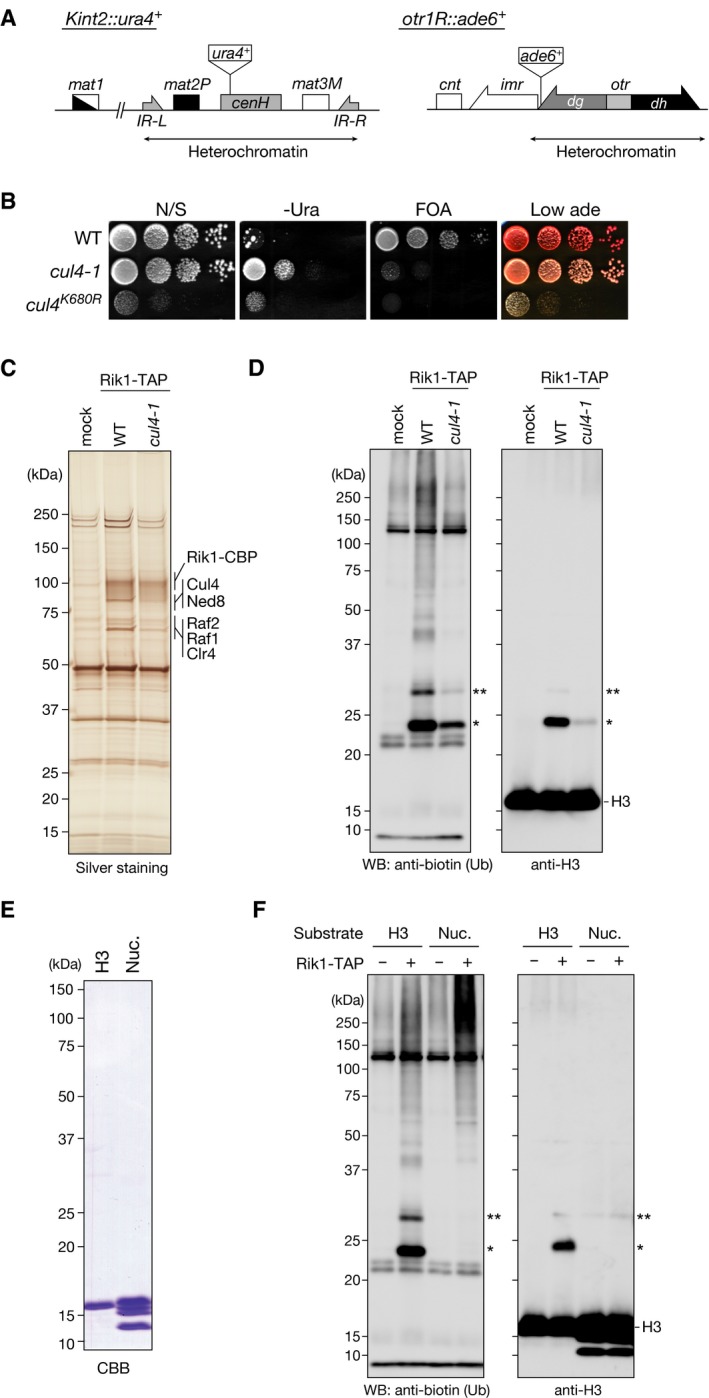
- Diagram of the mating‐type loci and part of centromere 1 in S. pombe. The positions of the silencing reporter genes (Kint2::ura4 + and otr1R::ade6 +) are shown.
- Heterochromatic silencing assays of wild‐type and cul4 mutant strains. Silencing at the mating‐type Kint2::ura4 + and otr1R::ade6 + was evaluated. Ten‐fold serial dilutions of the indicated strains were spotted onto non‐selective medium (N/S), medium lacking uracil (‐Ura), medium containing 5‐FOA (5‐FOA), and medium containing low adenine (Low ade).
- Purified TAP‐tagged Rik1‐containing complexes analyzed by SDS–PAGE and silver staining. mock: a mock purification from an untagged strain. Proteins identified by LC‐MS/MS are indicated at right.
- In vitro ubiquitylation assays using biotinylated ubiquitin, purified CLRC, and recombinant S. pombe histone H3 as the substrate. Proteins were analyzed by Western blotting using the indicated antibodies. The single and double asterisks indicate mono‐ and di‐ubiquitylated histone H3 species, respectively.
- Recombinant histone H3 protein and core histones in the reconstituted nucleosomes (Nuc.) were analyzed by SDS–PAGE and visualized by CBB staining.
- In vitro ubiquitylation assays using biotinylated ubiquitin, purified CLRC, and recombinant S. pombe histone H3 or reconstituted nucleosomes (Nuc.) as the substrate. Proteins were analyzed by Western blotting using the indicated antibodies. The single and double asterisks indicate mono‐ and di‐ubiquitylated histone H3 species, respectively.
To characterize the CLRC's ubiquitylation activity further, we prepared a fusion protein of the N‐terminal tail of H3 (residues A1‐K36) and GST (H3N‐GST). Since the human H3 histone‐tail sequence is slightly different from that of S. pombe, we prepared both human and S. pombe H3N‐GST fusion proteins, and examined them in the ubiquitylation assay. While both the human and S. pombe full‐length H3 histones were ubiquitylated by CLRC (Fig 1C, lanes 2 and 4), the H3N‐GSTs were more efficiently ubiquitylated than the full‐length histones (Fig 1C, lanes 6 and 8). These results suggested that the CLRC preferentially ubiquitylates the N‐terminal tail of H3 and that the C‐terminal region of H3 may have an inhibitory effect on the CLRC's activity for the H3 N‐terminal tail. We also noticed that CLRC did not efficiently ubiquitylate H3 in reconstituted nucleosomes (Fig EV2E and F), suggesting that nucleosomal DNA may also have an inhibitory effect on the CLRC's activity and additional factor(s) may help to promote it in vivo.
CLRC preferentially ubiquitylates histone H3 lysine 14
We next sought to identify the ubiquitylated lysine residues of the H3 N‐terminal tail. For this investigation, we performed mass spectrometry (LC‐MS/MS) analysis of the mono‐ and di‐ubiquitylated H3N‐GST species recovered from the CLRC‐mediated in vitro ubiquitylation assay (Figs 2A and EV3A). Because trypsin cleaves the C‐terminal Arg‐Gly‐Gly of ubiquitin, ubiquitylated lysine residues could be identified by the presence of a characteristic diGly remnant (Fig 2B) 40. In this analysis, four residues, K14, K18, K23, and K27, were identified as candidate H3 ubiquitylation sites (Fig EV3B). The number of identified peptides containing each of the candidate ubiquitylation sites suggested that K14 and K18 are the predominant H3 ubiquitylation sites.
Figure 2. Histone H3 lysine 14 is ubiquitylated in vitro by CLRC.
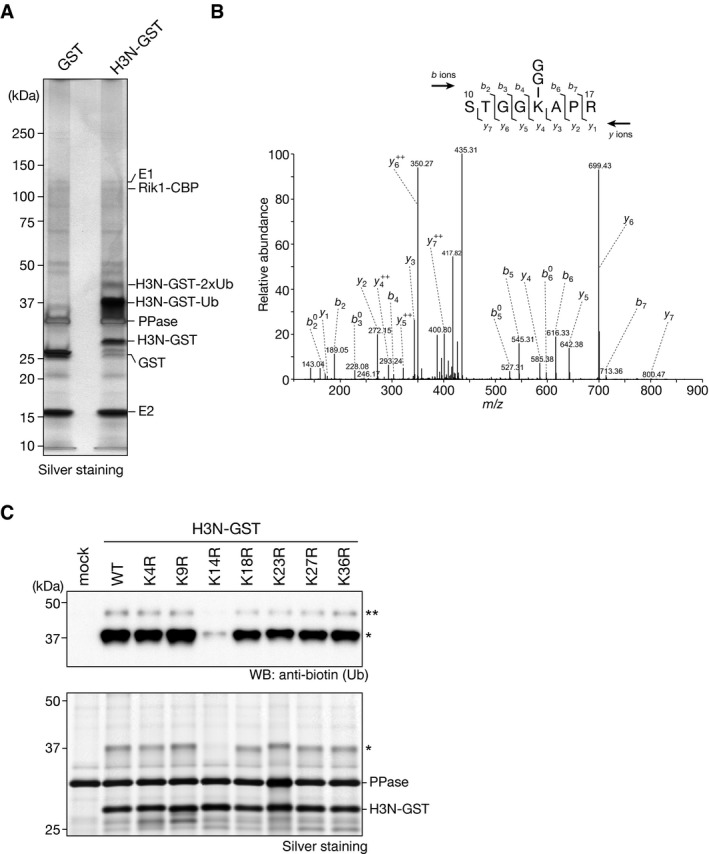
- Ubiquitylation assay using H3N‐GST. The components included in the assay and the ubiquitylated H3N‐GST species are indicated at right.
- MS/MS spectrum of the histone H3 peptide corresponding to residues 10–17. The observed y and b ions and fragment map are shown.
- Ubiquitylation assay using biotinylated ubiquitin and recombinant wild‐type H3N‐GST (WT) and arginine‐substituted H3N‐GST mutants as substrates. Proteins were analyzed by Western blotting and silver staining. mock: reaction without substrate. Asterisks indicate ubiquitylated H3N‐GST proteins.
Figure EV3. Analysis of the histone H3 ubiquitylation states.
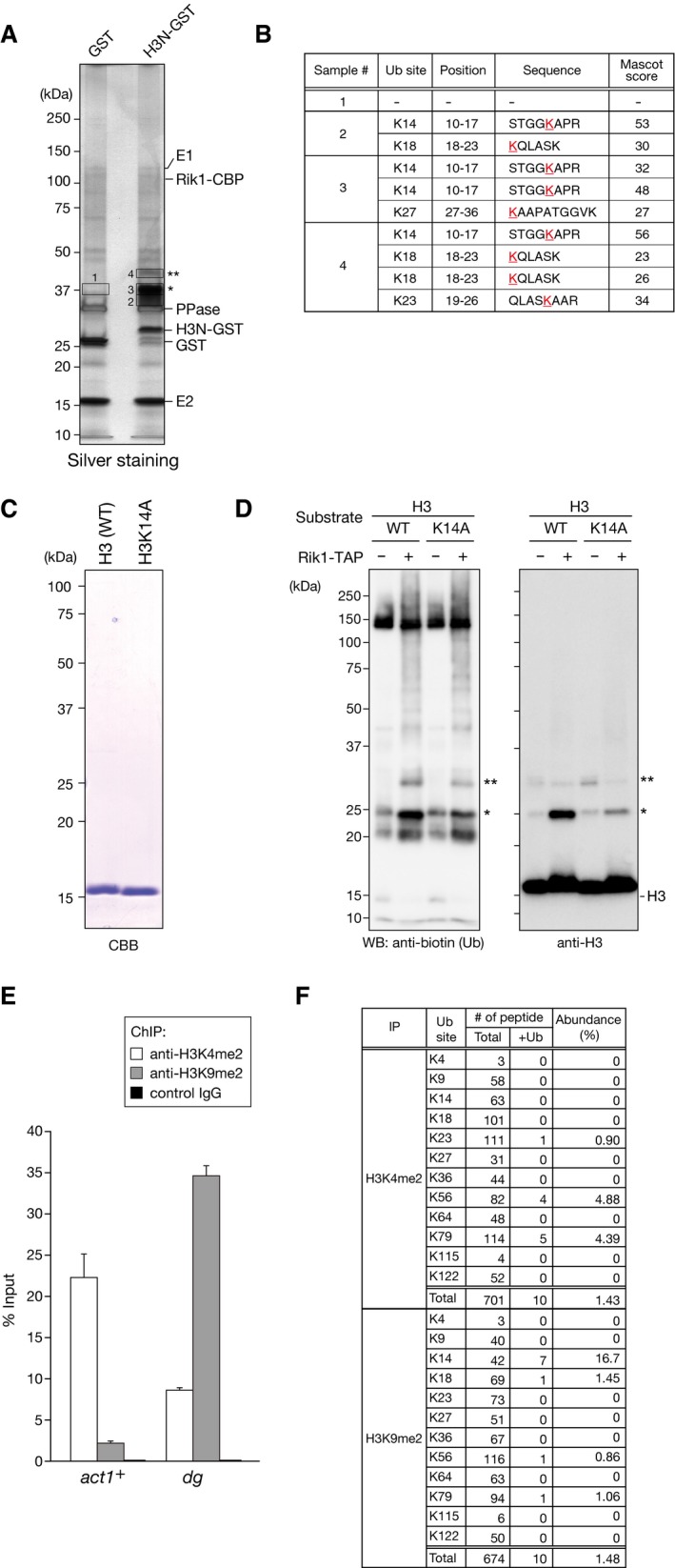
- Ubiquitylation assay using recombinant H3N‐GST. The proteins were analyzed by SDS–PAGE and silver staining. The components included in the assay are indicated at right, and the ubiquitylated H3N‐GST species are indicated by asterisks. The proteins subjected to LC‐MS/MS analysis are indicated by boxes.
- Summary of the ubiquitylated peptides identified by LC‐MS/MS.
- Recombinant S. pombe histone H3 (wild type; WT) and mutant histone H3 with K14A substitution (H3K14A) used in (D) were analyzed by SDS–PAGE and visualized by CBB staining.
- In vitro ubiquitylation assays using biotinylated ubiquitin, purified CLRC, and recombinant wild‐type (WT) or mutant (K14A) histone H3 as the substrate. Proteins were analyzed by Western blotting using the indicated antibodies. The single and double asterisks indicate mono‐ and di‐ubiquitylated histone H3 species, respectively.
- Chromatin precipitated with anti‐H3K4me2 or anti‐H3K9me2 antibodies was subjected to conventional ChIP analysis, and the levels of act1 + and pericentromeric dg loci relative to the input were determined. Error bars indicate standard errors from three technical replicates.
- Abundance of ubiquitylated lysine residues in chromatin‐associated histone H3 fractions. Since short peptides cleaved at unmodified lysine residues could not be efficiently detected in the LC‐MS/MS analysis, these data do not accurately reflect the abundance including unmodified lysines.
To confirm the results obtained by LC‐MS/MS analysis, we prepared H3N‐GST fusion proteins containing amino acid substitutions for each of the candidate lysine residues (K4R, K9R, K14R, K18R, K23R, K27R, and K36R) and assessed their effect on H3's ubiquitylation. In agreement with the LC‐MS/MS analysis (Fig EV3B), the K14R substitution resulted in a marked reduction in the CLRC‐mediated H3 ubiquitylation (Fig 2C, indicated by asterisks). Although the K18R, K23R, or K27R substitution appeared to slightly reduce the H3 ubiquitylation, the effect was much weaker than that of the K14R substitution. The other lysine substitutions (K4R, K9R, and K36R) had little or no impact on the CLRC‐mediated H3 ubiquitylation (Fig 2C). We also confirmed that K14A substitution also markedly reduced the CLRC‐mediated ubiquitylation of full‐length histone H3 (Fig EV3C and D). These results suggested that CLRC possesses an E3 ubiquitin ligase activity that preferentially targets histone H3K14, and that K14‐mono‐ubiquitylated H3 is a major product of CLRC's activity. At this point, we could not distinguish whether the di‐ubiquitylated H3 species contained two mono‐ubiquitins at different lysine residues or one di‐ubiquitin at a single lysine residue. In either case, our findings indicated that H3K14 ubiquitylation (H3K14ub) may play a role in promoting additional ubiquitylation.
Heterochromatin is enriched in H3K14ub in vivo
Since our in vitro assays suggested that the CLRC preferentially ubiquitylates histone H3K14, we next sought to assess the presence of H3K14ub in vivo. We speculated that H3K14ub would be concentrated at heterochromatic regions where the CLRC is predominantly localized 15. To examine this possibility, we carried out chromatin immunoprecipitation (ChIP) assays using antibodies against heterochromatic H3K9me2 or euchromatic H3K4me2 41, and performed LC‐MS/MS analyses to identify the histone modifications associated with these regions (Figs 3 and EV3E and F, and Appendix Table S1, Appendix Figs S1–S8). We confirmed that H3K9me2 was enriched in centromeric dg repeats, whereas H3K4me2 was preferentially enriched in the actively transcribed act1 + gene (Fig EV3E). Subsequent LC‐MS/MS analysis revealed that K14‐ubiquitylated H3 peptides were detected exclusively in the H3K9me2‐associated chromatin (Figs 3B–D and EV3F). It was also notable that nearly all of the tryptic K14‐ubiquitylated H3 peptides contained H3K9me2 or H3K9me3 (Fig 3C). These results demonstrated that H3K14ub is concentrated in H3K9me‐associated heterochromatin, where CLRC is predominantly localized, in vivo. Our LC/MS/MS analysis also revealed that K56‐ or K79‐ubiquitylated H3 peptides were preferentially concentrated in the H3K4me2‐associated chromatin (Figs 3C and D, and EV3F), although their biological functions and potential involvement in transcription are unknown.
Figure 3. H3K14 ubiquitylation is associated with H3K9me‐enriched chromatin.
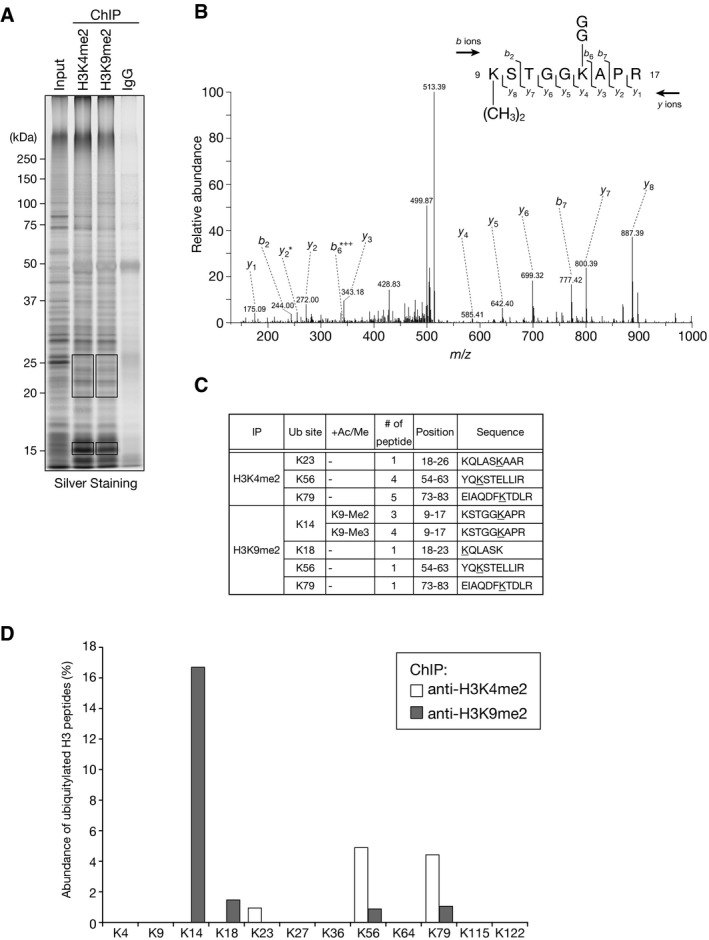
- Chromatin precipitated with anti‐H3K4me2 or anti‐H3K9me2 antibodies was analyzed by SDS–PAGE and silver staining. The input sample and mock precipitation using total mouse IgG are also shown. The gel slices indicated by boxes were excised and subjected to LC‐MS/MS analysis.
- MS/MS spectrum of the histone H3 peptide corresponding to residues 9–17. The observed y and b ions and fragment map are shown.
- Summary of the ubiquitylated peptides identified by LC‐MS/MS.
- Relative abundance of ubiquitylated lysine residues in chromatin‐associated histone H3 fractions. Chromatin precipitated with anti‐H3K4me2 or anti‐H3K9me2 antibodies was subjected to LC‐MS/MS analysis, and the abundance of ubiquitylated lysine residues in each fraction is shown (see also Fig EV3F). Since short peptides cleaved at unmodified lysine residues could not be efficiently detected in the LC‐MS/MS analysis, these data do not accurately reflect the abundance including unmodified lysines.
H3K14 is critical for heterochromatin assembly in vivo
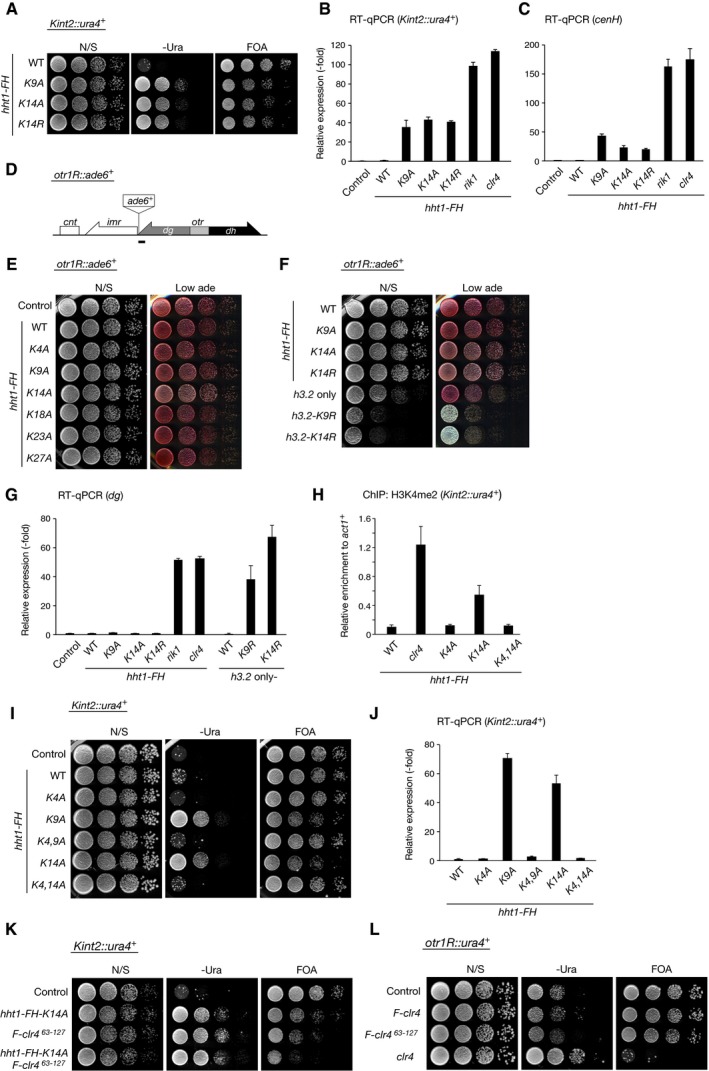
We next asked whether H3K14 is essential for heterochromatin assembly in fission yeast. Schizosaccharomyces pombe encodes three copies of genes that encode histone H3 (H3.1, H3.2, and H3.3). We introduced an alanine substitution mutation at individual lysine residues within the H3.1 N‐terminal tail and assessed the effect on the silencing of kint2::ura4 + (Fig 4A). Introduction of the K9A mutation clearly abolished the kint2::ura4 + silencing (Fig 4B), and ChIP analysis showed that the level of H3K9me2 at heterochromatic regions was reduced to half or less that of wild type (Fig 4C). Notably, the K14A or K14R mutation also led to defective silencing and decreased H3K9me2 levels, comparable to that of the K9A mutation (Figs 4B and C, and EV4A). The status of silencing defect caused by amino acid substitution for K9 or K14 was also confirmed by quantitative reverse‐transcriptase PCR (RT–qPCR) (Fig EV4B and C). Alanine substitution of the other lysine residues in the H3.1 N‐terminal tail had no effect on gene silencing (Fig 4B). While these effects were presumably due to a dominant‐negative effect of the mutations in H3.1, ChIP analysis revealed that the levels of Rik1 associated with the mating‐type cenH or centromeric dg locus were higher than that of wild‐type cells (Fig 4D). This may imply that amino acid substitution for K9 or K14 might affect CLRC's enzymatic activity and/or inhibit its turnover at heterochromatic regions. We also noticed that these amino acid substitutions had only a limited effect on the centromeric otr1R::ade6 + silencing and dg expression (Fig EV4D–G). As previously reported 42, clear silencing defect of otr1R::ade6 + and elevated dg expression were observed for cells harboring single H3 gene (hht2) containing K9 or K14 amino acid substitution (Fig EV4F and G). The different effects on two heterochromatic regions might be attributable to the difference in pathways to promote H3K9 methylation at these regions.
Figure 4. H3K14 is critical for heterochromatic silencing.
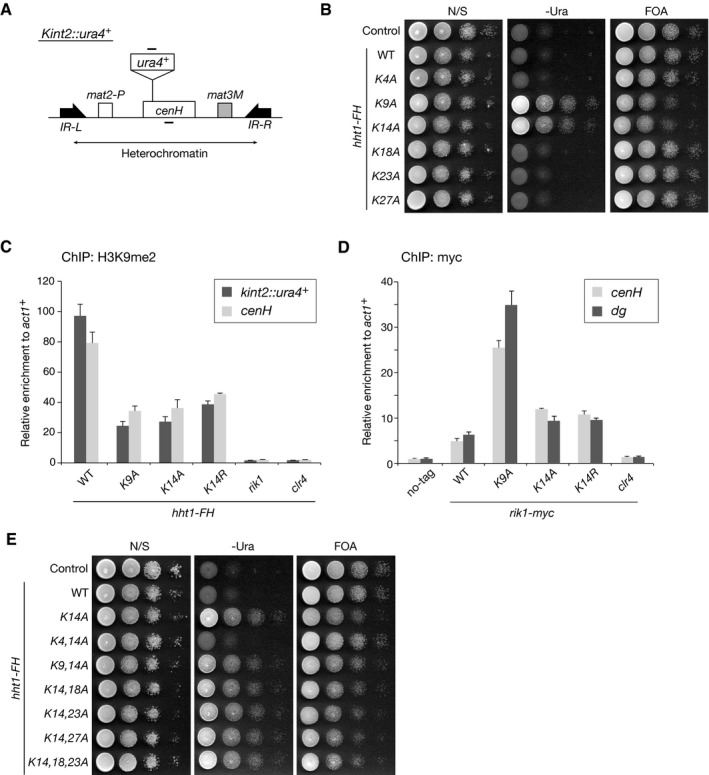
- Diagram of the mating‐type loci in S. pombe. The position of the silencing reporter gene (Kint2::ura4 +) is shown. The target regions in the ChIP analysis are indicated by black bars above (Kint2::ura4 +) and below (cenH) in the diagram.
- Heterochromatic silencing assays of strains expressing C‐terminally FLAG‐ and His‐tagged, wild‐type or mutant H3 (Hht1‐FH). Silencing at the mating‐type Kint2::ura4 + was evaluated. Ten‐fold serial dilutions of the indicated strains were spotted onto non‐selective medium (N/S), medium lacking uracil (‐Ura), and medium containing 5‐FOA (5‐FOA).
- ChIP analysis of H3K9me2 levels associated with the mating‐type loci, relative to the control act1 + locus. Error bars indicate standard errors from three technical replicates.
- ChIP analysis of Rik1‐myc levels associated with the mating‐type cenH and centromeric dg loci, relative to the control act1 + locus. Error bars indicate standard errors from three technical replicates.
- Heterochromatic silencing assays comparing the effects of H3 lysine substitution mutants. Silencing at the mating‐type kint2::ura4 + was evaluated as in (B).
Figure EV4. H3K14 is critical for heterochromatin assembly in vivo .
-
AHeterochromatic silencing assays of wild‐type and mutant Hht1‐FH strains. Silencing at the mating‐type Kint2::ura4 + was evaluated.
-
B, CThe levels of Kint2::ura4 + and the mating‐type cenH transcripts were quantified by RT–qPCR and calculated as fold change to the wild‐type expression. Error bars indicate standard errors from three technical replicates.
-
DDiagram of the right half region of centromere 1 in S. pombe. The positions of the silencing reporter genes (otr1R::ade6 +) are shown. The target regions in the RT–qPCR are indicated by black bars below in the diagram.
-
E, FHeterochromatic silencing assays of wild‐type and mutant Hht1‐FH strains. Silencing at the centromeric otr1R::ura4 + was evaluated.
-
GThe levels of centromeric dg transcripts were quantified by RT–qPCR and calculated as fold change to the wild‐type expression. Error bars indicate standard errors from three technical replicates.
-
HChIP analysis of H3K4me2 levels associated with the mating‐type loci (Kint2::ura4 +), relative to the control act1 + locus. Error bars represent the range of the standard deviations from three independent experiments.
-
IHeterochromatic silencing assays of wild‐type and mutant Hht1‐FH strains. Silencing at the mating‐type Kint2::ura4 + was evaluated.
-
JThe levels of Kint2::ura4 + transcripts were quantified by RT–qPCR and calculated as fold change to the wild‐type expression. Error bars indicate standard errors from three technical replicates.
-
KHeterochromatic silencing assays comparing the effects of H3K14 substitution and Clr4 mutation (∆63–127). Silencing at the mating‐type Kint2::ura4 + was evaluated.
-
LHeterochromatic silencing assays of wild‐type and clr4 mutant strains. Silencing at the centromeric otr1R::ura4 + was evaluated.
To further examine the relationship between K9 and K14, we introduced combinatorial amino acid substitutions in the H3.1 N‐terminal tail (Fig 4E). No additive effect was observed for cells expressing mutant H3.1 with both the K9A and K14A substitutions (Fig 4E, K9, 14A), suggesting that the silencing defect caused by K9A or K14A substitution was brought about through the same pathway. Although the K4A mutation showed a suppressing effect on K14A (Fig 4E, K4, 14A), a similar effect was seen for K9A (Fig EV4I and J). Thus, this effect was probably due to the reduction in H3K4me2, which was associated with active transcription of the inserted ura4 + marker gene (Fig EV4H). Taken together, these results are consistent with previous findings 42, 43 and confirmed the importance of H3K14 in H3K9me generation and heterochromatin assembly.
H3K14ub promotes H3K9me
Based on the above results, we next examined the possible connection between H3K14ub and H3K9me. To test whether H3K14ub modulates H3K9me, we performed in vitro histone methyltransferase (HMTase) assays using recombinant Clr4 and ubiquitylated H3N‐GST as a substrate (Fig 5A). H3N‐GST was first subjected to an in vitro ubiquitylation assay using affinity‐purified CLRC (as in Fig 1C). After the reaction, the resultant H3N‐GST was purified and subjected to an in vitro HMTase assay, in which the incorporation of a radioactively labeled methyl group transferred from 3H‐SAM was detected by autoradiography. In the control experiments without CLRC‐mediated ubiquitylation, Clr4 methylated the wild‐type (WT) H3N‐GST (Fig 5B, right panel, lane 2). Strikingly, even though only 10–15% of the H3N‐GST was ubiquitylated by the CLRC under our assay conditions and the rest remained unmodified (Fig 5B, left panel, lanes 3 and 4), Clr4 exclusively methylated the ubiquitylated H3N‐GST (Fig 5B, right panel, lane 4). In addition, the methylation of the ubiquitylated H3N‐GST was much stronger than that of the control H3N‐GST (Fig 5B, right panel, lanes 2 and 4), suggesting that CLRC‐mediated ubiquitylation of H3 increases Clr4's HMTase activity for the H3 as a substrate.
Figure 5. H3K14 ubiquitylation promotes H3K9 methylation.
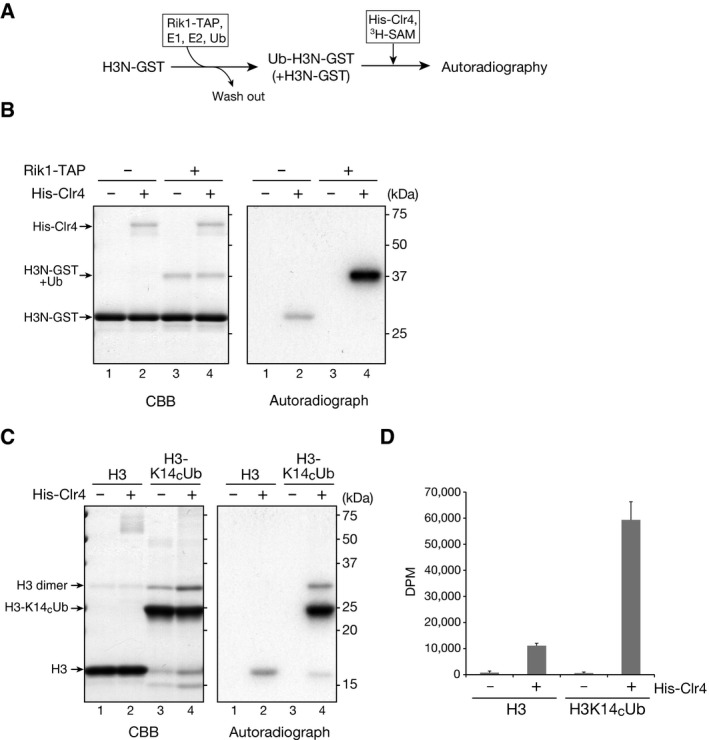
-
AExperimental scheme.
-
BIn vitro HMTase assays. Recombinant H3N‐GST pre‐ubiquitylated by the CLRC was purified and used in the HMTase assay with 6×His‐tagged recombinant Clr4 (His‐Clr4). Proteins were analyzed by SDS–PAGE and Coomassie Brilliant Blue (CBB) staining (left) and autoradiography (right).
-
C, DIn vitro HMTase assay using disulfide‐linked ubiquitylated histone H3. H3K14CUb and control histone H3 (H3C110A) were used as substrates in the HMTase assay and analyzed as in (B). The 3H‐methyl group incorporation was measured using a liquid scintillation counter (D). Error bars indicate the standard deviations calculated from three independent experiments.
To confirm the effect of H3K14ub on H3K9me, we prepared K14‐ubiquitylated histone H3 (H3K14CUb) by disulfide cross‐linking (see Materials and Methods) and used it as a substrate in the HMTase assay. Consistent with the results in Fig 5B, H3K14CUb was more efficiently methylated by Clr4 than was unmodified H3 (Fig 5C, right panel), and the incorporation of 3H‐labeled methyl groups was approximately six times that of the control H3 (Fig 5D). These results were consistent with those using CLRC‐treated H3N‐GST (Fig 5B). We also examined whether H3K9me modulates H3K14ub by using histone H3 containing a K9me3 analog (H3Kc9me3) as a substrate (see Materials and Methods). In the in vitro ubiquitylation assay, CLRC's ubiquitin ligase activity for histone H3 was not noticeably changed (Fig EV5, indicated by asterisks), suggesting that H3K9me does not affect K3K14ub. Taken together, these results indicated that H3K14ub promotes H3K9me and that this effect is unidirectional.
Figure EV5. Effect of H3K9me on CLRC‐mediated ubiquitylation.
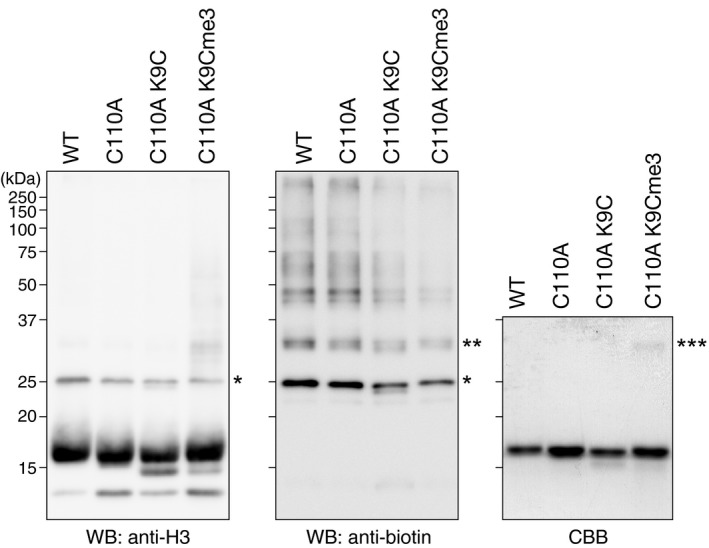
Ubiquitylation assays using biotinylated ubiquitin and recombinant wild‐type and mutant human H3.2, and the K9me3 analog of human H3.2 (C110A K9Cme3). The proteins were resolved by SDS–PAGE and analyzed by Western blotting (left and center). The recombinant protein substrates were also analyzed by Coomassie Brilliant Blue (CBB) staining (right). The single and double asterisks indicate mono‐ and di‐ubiquitylated H3.2, and the triple asterisks indicate cross‐linked dimers of human H3.2 C110A K9C.
Clr4 N‐terminal regions recognize ubiquitylated H3
These in vitro HMTase assays indicated that ubiquitylated histone H3 is recognized by Clr4. Clr4 possesses an evolutionarily conserved N‐terminal chromodomain (CD) and C‐terminal SET domain, and less conserved SET‐associated regions (Fig 6A) 44, 45. To investigate how Clr4 recognizes ubiquitylated histone H3, we produced a series of N‐terminally truncated Clr4 mutants as GST fusion proteins (Fig 6A and B) and examined their ability to recognize ubiquitylated H3 (H3K14CUb) in the HMTase assay using control histone H3 and H3K14CUb as substrates. Although GST‐fused full‐length (FL) Clr4 showed a specificity for H3K14CUb (Fig 6C, right panel, lanes 3 and 4), the CD‐deleted Clr4 mutant (Clr4_∆1) lost this specificity and methylated both control H3 and H3K14CUb with comparable efficiency (Fig 6C, right panel, lanes 5 and 6). Further N‐terminally deleted Clr4 mutants (Clr4_∆2 and Clr4_∆3) appeared to show the opposite preference; the level of methylated control H3 was apparently higher than that of H3K14CUb (Fig 6C, right panel, lanes 7–10). The deletion of the N‐terminal SET‐associated region (Clr4_∆4) abolished Clr4's HMTase activity (Fig 6C, right panel, lanes 11 and 12), suggesting that it has a critical role in this activity.
Figure 6. Clr4 N‐terminal regions are involved in recognizing ubiquitylated H3 and heterochromatic silencing.
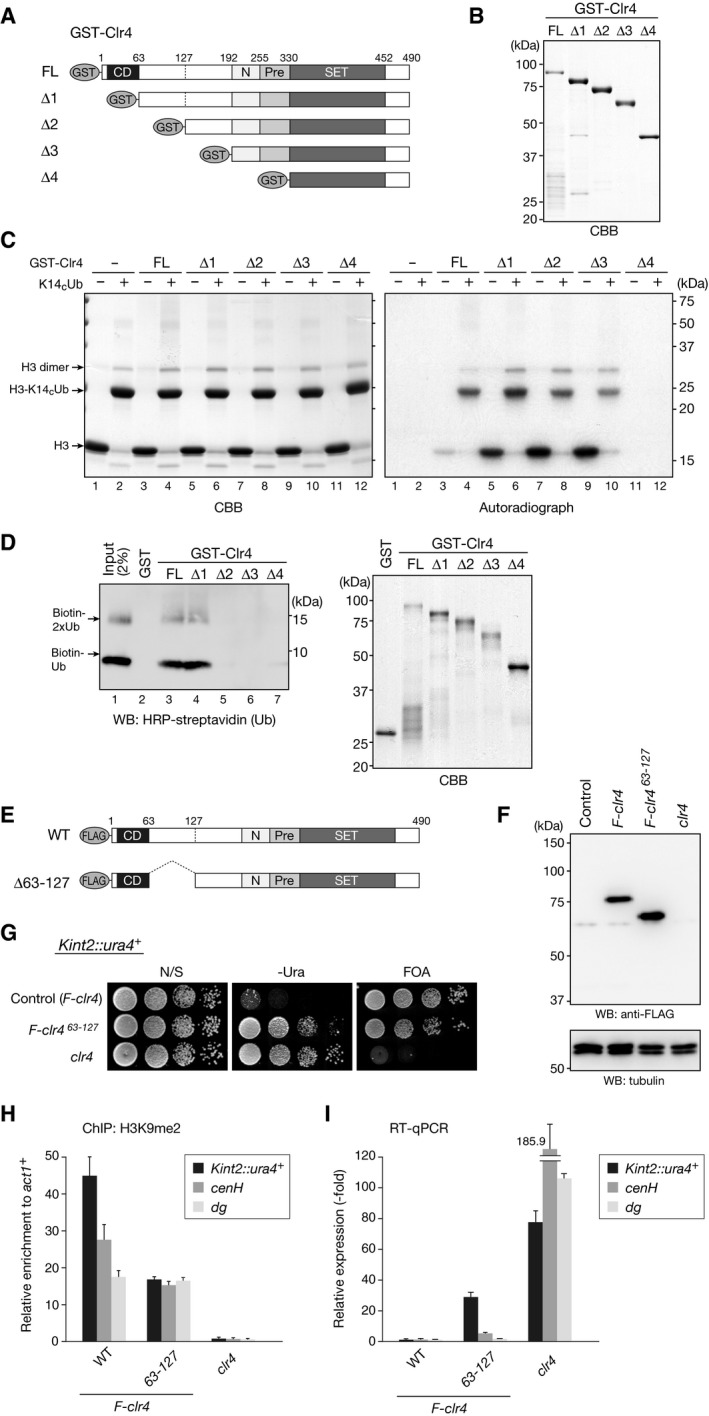
- Schematic of recombinant Clr4 proteins used in the in vitro HMTase assay: GST‐fused full‐length Clr4 (FL) and truncated Clr4 proteins (∆1‐∆4).
- Recombinant proteins used in (C) and (D) were resolved by 10% SDS–PAGE and visualized by CBB staining.
- In vitro HMTase assay using disulfide‐linked ubiquitylated histone H3. H3K14CUb and control histone H3 (H3C110A) were used as substrates in the HMTase assay and analyzed as in Fig 5C.
- In vitro pull‐down assays of GST‐Clr4 proteins with biotinylated ubiquitin. Bound ubiquitin was analyzed by Western blotting with HRP‐conjugated streptavidin (left). Precipitated GST proteins were analyzed by SDS–PAGE and CBB staining (right).
- Schematic of FLAG‐tagged, wild‐type, and mutant (∆63–127) Clr4 proteins.
- Whole cell lysates prepared from control cells, cells expressing FLAG‐tagged wild‐type Clr4 (F‐clr4) or mutant Clr4 (F‐clr4 ∆63–127), or clr4∆ cells were subjected to immunoblotting using the indicated antibodies.
- Heterochromatic silencing assays of wild‐type and clr4 mutant strains. Silencing at the mating‐type Kint2::ura4 + was evaluated.
- ChIP analysis of H3K9me2 levels associated with the silencing marker gene inserted in the mating‐type loci (Kint2::ura4 +) and the mating‐type cenH and centromeric dg loci, relative to the control act1 + locus. Error bars indicate standard errors from three technical replicates.
- The levels of Kint2::ura4 +, mating‐type cenH, and centromeric dg transcripts were quantified by RT–qPCR and calculated as fold change to the wild‐type expression. Error bars indicate standard errors from three technical replicates.
To confirm this result, we tested whether Clr4 binds directly to ubiquitin. We carried out GST pull‐down assays using GST‐fused full‐length (FL)‐deleted and N‐terminally deleted (∆1‐∆4) Clr4 proteins (Fig 6A and B) and biotinylated ubiquitin. GST was used as a control. Subsequent Western blots showed that the FL‐ and CD‐deleted ∆1 proteins could bind to ubiquitin, whereas further deleted mutants (∆2‐∆4) failed to bind ubiquitin (Fig 6D). These results suggested that the CD‐adjacent N‐terminal region can bind ubiquitin and contributes to the recognition of ubiquitylated H3. To further investigate the potential role of the CD‐adjacent region for Clr4 function, we created fission yeast strains expressing FLAG‐tagged wild‐type or mutant (∆63–127) Clr4 from its original promoter (Fig 6E and F). Notably, the deletion led to defective silencing for Kint2::ura4 + and reduced H3K9me2 levels at Kint2::ura4 + and cenH loci, although this effect was milder than that of clr4∆ cells (Fig 6G–I). These results suggested that the CD‐adjacent N‐terminal region plays an important role in Clr4's function. Interestingly, the effect of ∆63–127 deletion on Kint2::ura4 + was comparable to that of cells expressing hht1‐K14A (Fig EV4K), and this deletion had a negligible impact on centromeric silencing (otr1R::ura4 +) and dg expression (Figs 6H and I, and EV4L), implying that Clr4's enzymatic activity may be differentially regulated at each heterochromatin region.
Discussion
Here, we demonstrated that a novel modification regulates H3K9me. Our study revealed that the CLRC preferentially ubiquitylates histone H3K14 and, more importantly, that H3K14ub promotes H3K9me generation for heterochromatin assembly.
H3K14 was previously shown to play a critical role in heterochromatin assembly, and this residue is also the acetylation site associated with transcriptional activation. Either mutations in clr3, which encodes an H3K14‐specific deacetylase, or the H3K14A substitution, which mimics the acetylated lysine, cause reduced H3K9me and defects in heterochromatic silencing 6, 42, 43, 46, 47. These findings suggest that the deacetylation of H3K14 by Clr3 is required for H3K9's methylation by Clr4. In contrast, Clr4 efficiently methylates a K14‐acetylated H3 peptide 6, and the H3K14R substitution, which mimics unacetylated lysine, also leads to a severe silencing defect 42. These results indicated that the unacetylated state of H3K14 is not sufficient for efficient H3K9me. These apparently conflicting findings suggested that there is a gap in our understanding of how H3K14 is involved in heterochromatin assembly. Here, we demonstrated that H3K14 is modified by ubiquitylation, which promotes H3K9's methylation by Clr4. Since H3K9me did not noticeably affect the H3K14ub by CLRC (Fig EV5), H3K14ub is likely to be a prerequisite modification for H3K9me.
The chromodomain (CD) is a highly conserved protein module that is critical for targeting proteins to their action sites in chromatin. CD's best‐known role is its binding to methylated histone tails. As demonstrated for the CDs in HP1 family proteins, Clr4‐CD also binds K9‐methylated histone H3 15, and this activity is critical for CLRC's heterochromatic association and for stable inheritance of the H3K9me mark 15, 48. Since Clr4 and its homologs contain both CD (H3K9me reader) and SET (H3K9me writer) domains, a direct read–write mechanism has been suggested for heterochromatin spreading and epigenetic inheritance 15, 48. According to this model, Clr4 recruited by preexisting H3K9me provides new K9me on H3 in the same or neighboring nucleosomes. However, in the present study we demonstrated that Clr4's HMTase activity was enhanced by the presence of H3K14ub (Fig 5). These results suggested that the simple read–write mechanism could be revised to include the involvement of ubiquitylation. Since we also demonstrated that Clr4‐CD and its neighboring regions contribute to the recognition of ubiquitylated H3 (Fig 6A–C), it is possible that Clr4‐CD has multiple functions that modulate its HMTase activity for heterochromatin spreading and epigenetic inheritance 19. In this study, we also demonstrated that Clr4 binds directly to ubiquitin via its N‐terminal region (Fig 6D) and that the CD‐adjacent region is critical for heterochromatic silencing (Fig 6E–I). Considering that N‐terminally deleted Clr4 mutants showed a preference for unmodified H3, it is also possible that CD or a CD‐adjacent region plays an inhibitory role for the SET domain's HMTase activity for unmodified H3 and that ubiquitylated H3 somehow suppresses this activity. This notion is consistent with a very recent study on SUV39H1 49. Another recent study demonstrated that Clr4's catalytic activity is inhibited by an internal loop and that the automethylation of a specific lysine in the loop promotes a conformational switch that enhances Clr4's H3K9me activity 50. Thus, the coordination of auto‐regulation mechanisms may help to suppress Clr4's promiscuous activity and provide an additional system ensuring the read–write mechanism. While we clearly showed the importance of H3K14Ub and Clr4's CD‐adjacent region for Clr4's function, the effect was limited to the mating‐type region, and only a negligible effect was seen for centromeric heterochromatin (Fig EV4E and L). It is possible that there are additional CLRC's targets for ubiquitylation, which may redundantly promote Clr4's activity. Alternatively, Clr4's activity may also be regulated by mechanisms other than ubiquitylation 19, 51.
An important issue requiring further investigation is the dynamics of H3K14ub. Since H3K9me is coupled with the deposition of newly synthesized histones after DNA synthesis, H3K14's ubiquitylation and de‐ubiquitylation may be cell‐cycle‐dependent, as well. Although 20 de‐ubiquitylating enzymes are expressed in S. pombe, a specific de‐ubiquitylating enzyme(s) that targets heterochromatin have not been identified yet. Multiple enzymes may act cooperatively in this process. Another issue requiring clarification is the mechanism by which the CLRC is recruited to specific regions of chromatin. Although Raf1 and Raf2 have been implicated in substrate recognition 27, 28, 29, the identity of their target(s) remains unclear. Raf2 contains a domain that shows weak similarity to the RFTS of DNMT1 (a methyltransferase involved in maintaining DNA methylation) 29, and DNMT1's RFTS is known to play a role in recognizing ubiquitylated H3 52, 53. Raf2's RFTS‐like domain may also have a role in targeting ubiquitylated H3. Alternatively, it is possible that Raf2's RFTS has a distinct role in CLRC function, unrelated to H3 ubiquitylation 29.
In Neurospora crassa, the H3K9 methyltransferase Dim‐5 forms a complex with Cul4 and DDB1 called the DCDC, which is required for Dim‐5‐mediated H3K9me 54. In human cells, CUL4 is associated with SUV39H1 55, and depleting CUL4 or DDB1 impairs H3K9me 56. In addition, CUL4‐DDB1 has been shown to have ubiquitin ligase activity for H3 57, and proteomic surveys using human cells revealed that H3K14 is ubiquitylated in vivo 40. It is therefore likely that H3 ubiquitylation mediated by CUL4‐DDB1 plays an important role in the methylation of H3K9 in other systems. The ubiquitin ligase activity of CUL4‐DDB1 promotes H3 poly‐ubiquitylation 58, which appears to be distinct from the CLRC‐mediated ubiquitylation we observed. In our in vitro ubiquitylation assay, the major products detected were mono‐ and di‐ubiquitylated species, and other multiply ubiquitylated species were not clearly detected (Fig 1C). However, it is possible that the combination of appropriate DCAF(s) (DDB1 and CUL4‐associated factors) modulates CUL4‐DDB1's activity and provides substrate specificity 59.
Cross‐talk between histone ubiquitylation and methylation is well known to occur for H2B mono‐ubiquitylation and H3K4 methylation 36. It was also recently reported that histone H2A's mono‐ubiquitylation promotes H3K27's methylation 37 and that H3's ubiquitylation plays a role in the maintenance of DNA methylation 52, 53. Although it has been suggested that ubiquitin functions as an adaptor to recruit other modifying enzymes, the underlying molecular mechanisms of this action are not fully understood. Further analysis of the functional interactions between H3K14ub and H3K9me will provide additional insight into the cross‐talk between histone ubiquitylation and methylation and the role it plays in chromatin assembly.
Materials and Methods
Strains and plasmids
The S. pombe strains used in this study are described in Appendix Table S2. The media and genetic methods used in the study were described previously 60. The cul4‐1 strain was generated by inserting the hygromycin‐resistant gene into the 3′ untranslated region of the cul4 gene. For the deletion or epitope tagging of target genes, the PCR‐based module method was used 61. To produce H3N‐GST fusion proteins in E. coli, the portion of the histone H3 coding sequence corresponding to amino acids 1–36 was amplified by PCR and cloned into the pCold IV vector (Takara), followed by downstream insertion of the GST coding sequence. Site‐directed mutagenesis was used to introduce each of the amino acid substitutions.
Antibodies
The following antibodies were used in this study: anti‐biotin (7075; Cell Signaling Technology), anti‐histone H2A (ab13923; Abcam), anti‐histone H2B (63‐125; BioAcademia), anti‐histone H3 (rat, 1G1; kindly provided by H. Kimura 62), anti‐histone H4 (mouse, 9C5; kindly provided by H. Kimura), anti‐H3K9me2 (mouse; kindly provided by T. Urano [63]: MABI0307, MAB Institute, Inc.), anti‐H3K4me2 (MABI0303; MAB Institute, Inc.), horseradish peroxidase (HRP)‐conjugated anti‐FLAG M2 (A8592; Sigma), anti‐tubulin (T5168; Sigma), HRP‐conjugated streptavidin (21130; Thermo), HRP‐conjugated anti‐mouse IgG (112–035‐072; Jackson ImmunoResearch), HRP‐conjugated anti‐rabbit IgG (A6667; Sigma), and HRP‐conjugated anti‐rat IgG (NA935; GE Healthcare). Anti‐GST rabbit polyclonal antibodies were prepared and affinity‐purified using recombinant GST protein.
Tandem affinity purification
Rik1‐TAP purification was performed as described previously 64, with some modifications. Exponentially growing cells (6.4 × 1011 cells) were harvested by centrifugation, washed once in phosphate‐buffered saline (PBS), and transferred to 50‐ml tubes (approximately 8 g per tube, 16 tubes). The pelleted cells were frozen with liquid nitrogen and stored at −80°C before further purification. All subsequent steps were performed at 4°C. The frozen cells in each tube were resuspended in 10 ml of TAP lysis buffer (50 mM HEPES [pH 7.5], 300 mM potassium acetate, 20 mM β‐glycerophosphate, 5 mM magnesium acetate, 1 mM EGTA, 1 mM EDTA, 0.1% Nonidet P‐40 [NP‐40], 1 mM dithiothreitol [DDT], 1 mM phenylmethylsulfonyl fluoride [PMSF]), supplemented with protease inhibitor cocktail (Complete; Roche), and lysed by glass‐bead breakage in a Multi Beads Shocker (Yasui Kikai). The crude lysate in each tube was diluted with 15 ml of TAP lysis buffer and incubated with gentle agitation on a disk rotator for 30 min. The lysates were then centrifuged at 12,000 ×g for 20 min, and the cleared lysates (approximately 20 mg/ml protein) were pooled and divided into 12 aliquots. Each aliquot was further purified as previously described 64. For LC‐MS/MS analysis, the eluted proteins were precipitated by adding 2 volumes of ethanol and resolved by SDS–PAGE. For the ubiquitylation assay, the eluted proteins were dialyzed against Ubc buffer (50 mM HEPES [pH 7.3], 150 mM NaCl, 10% glycerol, 1 mM DTT) and concentrated using Amicon Ultra (Merck Millipore). The eluate derived from 6.4 × 1011 cells was concentrated to approximately 150 μl.
Recombinant protein production
The cDNAs of S. pombe histones (H2A [hta1 +], H2B [htb1 +], H3 [hht1 +], and H4 [hhf1 +]) and human H3 were PCR‐amplified and cloned into the pET‐3a vector (Merck Millipore). Recombinant histones were expressed in BL21 (DE3) E. coli and purified as described previously 65. To generate the H3K9me3 analog, human histone H3.2 containing K9C and C110A substitutions was expressed and purified as for the other histones. The purified H3 K9C/C110A protein was subjected to alkylation as previously described 66. Recombinant GST‐fused and 6×His‐tagged proteins were expressed in BL21 (DE3) E. coli and purified, respectively, by Glutathione Sepharose 4B (GE Healthcare) and TALON Metal Affinity Resin (Clontech), according to the manufacturers’ instructions. The eluted proteins were further purified by anion exchange chromatography (SOURCE 15Q; GE Healthcare).
In vitro ubiquitylation assays
In vitro ubiquitylation assays were performed using a Ubiquitinylation Kit (Enzo Life Sciences) according to the manufacturer's instructions with some modifications. The E1 (12.5 or 25 nM) and E2 (175 or 350 nM recombinant human UbcH5b) enzymes were mixed with 2.5–10 μl of the concentrated Rik1‐TAP purified fraction and 0.5 or 1 μM substrate in 25 μl of reaction buffer, and the mixtures were incubated at 30°C for 2 h. The reactions were terminated by adding equal volumes of 2× SDS sample buffer. The proteins were resolved by SDS–PAGE and analyzed by Western blotting. Can Get Signal (Toyobo) was used to enhance the Western blotting signals.
Nucleosome reconstitution
Nucleosomes used in the in vitro ubiquitylation assays were prepared as described previously 67.
Nano‐liquid chromatography‐tandem mass spectrometry (LC‐MS/MS)
The proteins in each gel slice were subjected to reduction with 10 mM dithiothreitol (DDT), at 56°C for 1 h, alkylation with 55 mM iodoacetamide at room temperature for 45 min in the dark, and digestion with 10 μg/ml modified trypsin (Promega) or 20 μg/ml ArgC (Promega) at 37°C for 16 h. The resulting peptides were extracted with 1% trifluoroacetic acid and 50% acetonitrile, dried under a vacuum, and dissolved in 2% acetonitrile and 0.1% formic acid. The peptides were then fractionated by C18 reverse‐phase chromatography (ADVANCE UHPLC; AMR Inc.) and applied directly into a hybrid linear ion trap mass spectrometer (LTQ Orbitrap Velos Pro; Thermo Fisher Scientific) with Advanced Captive Spray SOURCE (AMR Inc.). The mass spectrometer was programmed to carry out 11 successive scans, with the first consisting of a full MS scan from 400–2,000 m/z by FT‐ICR at a resolution of 60,000, and the second to eleventh consisting of data‐dependent scans of the top ten abundant ions obtained in the first scan, by ion trap. Automatic MS/MS spectra were obtained from the highest peak in each scan by setting the relative collision energy to 35% and the exclusion time to 20 s for molecules in the same m/z value range. The molecular masses of the resulting peptides were searched against the non‐redundant NCBI database using the MASCOT program. MS/MS spectra were also analyzed using MaxQuant software (version 1.5.2.8). Searches were performed using the default parameters with the following modifications: (i) The minimum peptide length was 5 amino acids, (ii) the minimum score for modified peptides was 10, and (iii) variable modification was allowed.
Immunoprecipitation of H3K4me2‐ or H3K9me2‐associated chromatin from S. pombe
Exponentially growing cells (1 × 1010 cells) were treated with 5 mM N‐ethylmaleimide (Nacalai Tesque) for 30 min, and then fixed with 1% formaldehyde for 30 min at 25°C. After quenching the fixation by adding 150 mM glycine, the cells were harvested and washed once with PBS and once with PBS containing 25% glycerol. The pelleted cells were frozen with liquid nitrogen and stored at −80°C until use. The pelleted cells were resuspended in 10 ml of IP buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X‐100, and 0.1% Na‐deoxycholate) supplemented with 1 mM PMSF and protease inhibitor cocktail (Complete; Roche), and lysed by glass‐bead breakage in a Multi Beads Shocker (Yasui Kikai). The cell extract was diluted to 80 ml with IP buffer containing 1 mM PMSF and protease inhibitor cocktail, and sonicated with a Bioruptor (CosmoBio) for 240 s set at level H. After sonication, the cell extract was centrifuged at 12,000 ×g for 10 min, and the supernatants were used for immunoprecipitation. Anti‐mouse IgG antibody‐conjugated magnetic beads (DYNAL, 500 μl) were preincubated with 10 μg of primary antibody, anti‐H3K4me2 (MABI0303; MABI) or anti‐H3K9me2 (MABI0307; MABI), for 1 h at 4°C, followed by incubation with the cell extracts for 2 h at 4°C. Total mouse IgG (Millipore) was used as a control. After immunoprecipitation, the beads were washed once with IP buffer, and twice each with wash buffer A (50 mM HEPES [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% Triton X‐100, and 0.1% Na‐deoxycholate), wash buffer B (10 mM Tris [pH 8.0], 250 mM LiCl, 0.5% NP‐40, and 0.5% Na‐deoxycholate), and TES buffer (10 mM Tris [pH 7.5], 1 mM EDTA, and 250 mM NaCl). The bound proteins were eluted with 500 μl of TES buffer containing 1% SDS, precipitated by adding 2 volumes of ethanol, resolved by SDS–PAGE, and subjected to Western blotting or LC‐MS/MS analysis.
Silencing assays
Silencing assays were conducted using unsaturated cultures grown in YEA medium. Serial 10‐fold dilutions were spotted on plates with minimal non‐selective medium containing L‐glutamic acid as a nitrogen source instead of ammonium chloride (N/S), minimal medium lacking uracil (‐Ura), or minimal medium containing 5‐fluoroorotic acid (5‐FOA). The plates were then incubated at 30°C for 2–4 days.
Chromatin immunoprecipitation (ChIP) analysis
ChIP was performed as described previously 63. The primers used in the ChIP analyses are described in Appendix Table S3.
Real‐time quantitative reverse transcription PCR (RT–qPCR)
RT–qPCR analyses were performed as described previously 19. The primers used in the RT–qPCR analyses are described in Appendix Table S3.
In vitro HMTase assay
HMTase assays were performed as described previously 68 with some modifications. The substrate (6.2 μM) was mixed with 0.5 μM recombinant His‐Clr4 or GST‐Clr4 in 25 μl of reaction buffer (50 mM Tris–HCl [pH 8.0], 10% [v/v] glycerol, 1 mM DTT, 1 mM PMSF, and 0.5 μCi of S‐[Methyl‐3H]‐Adenosyl‐L‐methionine [PerkinElmer]) and incubated at 30°C for 1 h. The reactions were terminated by adding 4× SDS sample buffer, and the proteins were resolved by SDS–PAGE and visualized by autoradiography. To quantify the level of methylated histones, the reactions were quenched with cold 5% (v/v) trichloroacetic acid and subjected to filter‐binding assays followed by scintillation counting.
Synthesis of K14C‐ubiquitylated histone H3
Disulfide‐linked ubiquitylated histone H3 was synthesized as described previously 69. Human recombinant histone H3.2 proteins containing K14C and C110A substitutions were expressed in BL21 (DE3) E. coli and purified as described previously 67. Purified H3.2 proteins were reacted with 2,2′‐dithiobis(5‐nitropyridine) (DTNP) and dialyzed against sterile water. The ubiquitin–cysteamine fusion protein was produced as described previously 66 with some modifications. The DNA fragment encoding human ubiquitin was inserted into the pTXB1 vector, and the C‐terminally Intein‐CBD (chitin‐binding domain)‐fused ubiquitin produced in BL21 (DE3) E. coli was purified with a chitin column (New England Biolabs). Ubiquitin was cleaved from the Intein‐CBD portion by incubating with cysteamine‐dihydrochloride (Sigma‐Aldrich). The eluted ubiquitin with a C‐terminal aminoethanethiol linker was further purified by gel filtration chromatography (HiLoad 26/60 Superdex 75 pg; GE Healthcare), dialyzed against sterile water, and lyophilized. The purified DTNP‐reacted histone H3 and ubiquitin–cysteamine were allowed to react at pH 6.9 to form the disulfide‐linked ubiquitylated histone H3 (H3K14CUb). H3K14CUb was purified with a MonoS column (GE Healthcare) and used as a substrate in the in vitro histone methyltransferase (HMTase) assay.
GST pull‐down assays
GST‐fused, full‐length, and truncated Clr4 proteins were incubated with biotinylated ubiquitin (BML‐UW8705; Enzo Life Sciences) in binding buffer (25 mM Tris–HCl [pH 8.0], 100 mM NaCl, 10% glycerol, 0.2% NP‐40) at 25°C for 2 h. GST fusion proteins and associated proteins were pulled down by adding 5 μl of Glutathione Magnetic Beads (88821; Thermo Scientific). After beads were washed with washing buffer (125 mM Tris–HCl [pH 8.0], 150 mM NaCl, 0.01% Tween 20), bound proteins were eluted by boiling with 2× SDS sample buffer. The eluted proteins were resolved by 15% SDS–PAGE, and the pulled‐down biotinylated ubiquitin was detected by Western blotting using HRP‐conjugated streptavidin (21130; Thermo Scientific).
Author contributions
EO and JN designed the research. EO, RN, YY, MT, GN, and AS performed the experiments. SM, HK, and HT contributed to generate histone‐related reagents. KE gave valuable advice and helped with supervision of EO. EO and JN wrote the manuscript, and all authors commented and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank H. Kimura, T. Urano, and H. Kato for the antibodies and R. Allshire for strains. We also thank E. Suzuki, T. Kawaguchi, A. Hachisuka, and R. Nakamura for technical help, and Y. Nishiyama, A. Hayashi, K. Hiragami‐Hamada, K. Kataoka, and K. Tanaka for critical reading of the manuscript. The authors have no financial interests related to this work. This research was supported by KAKENHI (JP23114005, JP26291072, JP17H03713, and JP18H05532 to J.N., and JP18H05534 to H.K.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to J.N.), the Uehara Memorial Foundation (to J.N.), the Takeda Science Foundation (to J.N.), and the Swedish Cancer Society and the Swedish Research Council (to K.E).
EMBO Reports (2019) 20: e48111
References
- 1. Richards EJ, Elgin SC (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500 [DOI] [PubMed] [Google Scholar]
- 2. Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- 3. Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- 4. Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- 5. Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- 6. Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- 7. Nakayama J, Klar AJ, Grewal SI (2000) A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101: 307–317 [DOI] [PubMed] [Google Scholar]
- 8. Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI (2002) Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- 9. Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- 10. Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet 19: 192–195 [DOI] [PubMed] [Google Scholar]
- 11. Sadaie M, Iida T, Urano T, Nakayama J (2004) A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J (2008) Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol 28: 6973–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI (2009) Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA 106: 8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang K, Mosch K, Fischle W, Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- 16. Moazed D (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goto DB, Nakayama J (2012) RNA and epigenetic silencing: insight from fission yeast. Dev Growth Differ 54: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama J (2012) Intrinsic nucleic acid‐binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell 47: 228–241 [DOI] [PubMed] [Google Scholar]
- 20. Jia S, Noma K, Grewal SI (2004) RNAi‐independent heterochromatin nucleation by the stress‐activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- 21. Kanoh J, Sadaie M, Urano T, Ishikawa F (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819 [DOI] [PubMed] [Google Scholar]
- 22. Horn PJ, Bastie JN, Peterson CL (2005) A Rik1‐associated, cullin‐dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19: 1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ (2005) Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol 15: 1448–1457 [DOI] [PubMed] [Google Scholar]
- 24. Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D (2005) A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3‐K9 methyltransferase and is required for RNAi‐mediated heterochromatin formation. RNA Biol 2: 106–111 [DOI] [PubMed] [Google Scholar]
- 25. Jia S, Kobayashi R, Grewal SI (2005) Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7: 1007–1013 [DOI] [PubMed] [Google Scholar]
- 26. Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein‐Hansen J, Bonaduce MJ, Klar AJ (2005) The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe . Genetics 171: 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buscaino A, White SA, Houston DR, Lejeune E, Simmer F, de Lima Alves F, Diyora PT, Urano T, Bayne EH, Rappsilber J et al (2012) Raf1 Is a DCAF for the Rik1 DDB1‐like protein and has separable roles in siRNA generation and chromatin modification. PLoS Genet 8: e1002499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuscu C, Zaratiegui M, Kim HS, Wah DA, Martienssen RA, Schalch T, Joshua‐Tor L (2014) CRL4‐like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing. Proc Natl Acad Sci USA 111: 1795–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White SA, Buscaino A, Sanchez‐Pulido L, Ponting CP, Nowicki MW, Allshire RC (2014) The RFTS domain of Raf2 is required for Cul4 interaction and heterochromatin integrity in fission yeast. PLoS ONE 9: e104161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- 31. Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez‐Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J et al (2010) Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He C, Pillai SS, Taglini F, Li F, Ruan K, Zhang J, Wu J, Shi Y, Bayne EH (2013) Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification. Proc Natl Acad Sci USA 110: E1879–E1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gerace EL, Halic M, Moazed D (2010) The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell 39: 360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997 [DOI] [PubMed] [Google Scholar]
- 35. Li F, Martienssen R, Cande WZ (2011) Coordination of DNA replication and histone modification by the Rik1‐Dos2 complex. Nature 475: 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chandrasekharan MB, Huang F, Sun ZW (2010) Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans‐histone H3 methylation. Epigenetics 5: 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J (2014) Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21: 569–571 [DOI] [PubMed] [Google Scholar]
- 38. Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM (1995) Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem 270: 30408–30414 [DOI] [PubMed] [Google Scholar]
- 39. Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ (1999) Recruitment of a ROC1‐CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol Cell 3: 527–533 [DOI] [PubMed] [Google Scholar]
- 40. Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ et al (2011) Systematic and quantitative assessment of the ubiquitin‐modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin‐ and RNAi‐mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- 42. Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC (2003) Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol 13: 1748–1757 [DOI] [PubMed] [Google Scholar]
- 43. Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF (2013) Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. EMBO J 32: 2321–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB et al (1999) Functional mammalian homologues of the Drosophila PEV‐modifier Su(var)3‐9 encode centromere‐associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD et al (2000) Regulation of chromatin structure by site‐specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- 46. Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol 22: 2170–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K (2005) Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J 24: 2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ragunathan K, Jih G, Moazed D (2015) Epigenetics. Epigenetic inheritance uncoupled from sequence‐specific recruitment. Science 348: 1258699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muller MM, Fierz B, Bittova L, Liszczak G, Muir TW (2016) A two‐state activation mechanism controls the histone methyltransferase Suv39h1. Nat Chem Biol 12: 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M, Gygi SP, Moazed D (2018) Automethylation‐induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature 560: 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Akoury E, Ma G, Demolin S, Bronner C, Zocco M, Cirilo A, Ivic N, Halic M (2019) Disordered region of H3K9 methyltransferase Clr4 binds the nucleosome and contributes to its activity. Nucleic Acids Res 47: 6726–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F et al (2013) Uhrf1‐dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502: 249–253 [DOI] [PubMed] [Google Scholar]
- 53. Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y et al (2017) Structure of the Dnmt1 reader module complexed with a unique two‐mono‐ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol Cell 68: 350–360 e7 [DOI] [PubMed] [Google Scholar]
- 54. Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Knip M, Sack R, Selker EU (2010) DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet 6: e1001196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang Y, Liu R, Qiu R, Zheng Y, Huang W, Hu H, Ji Q, He H, Shang Y, Gong Y et al (2015) CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation‐based epigenetic silencing. Oncogene 34: 104–118 [DOI] [PubMed] [Google Scholar]
- 56. Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H (2006) CUL4‐DDB1 ubiquitin ligase interacts with multiple WD40‐repeat proteins and regulates histone methylation. Nat Cell Biol 8: 1277–1283 [DOI] [PubMed] [Google Scholar]
- 57. Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument‐Bromage H, Tempst P, Xiong Y, Zhang Y (2006) Histone H3 and H4 ubiquitylation by the CUL4‐DDB‐ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 22: 383–394 [DOI] [PubMed] [Google Scholar]
- 58. Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z (2013) A Cul4 E3 ubiquitin ligase regulates histone hand‐off during nucleosome assembly. Cell 155: 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N (2006) Molecular architecture and assembly of the DDB1‐CUL4A ubiquitin ligase machinery. Nature 443: 590–593 [DOI] [PubMed] [Google Scholar]
- 60. Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- 61. Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR‐based gene targeting in Schizosaccharomyces pombe . Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- 62. Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, Kimura H, Obuse C (2010) Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol 12: 719–727 [DOI] [PubMed] [Google Scholar]
- 63. Oya E, Kato H, Chikashige Y, Tsutsumi C, Hiraoka Y, Murakami Y (2013) Mediator directs co‐transcriptional heterochromatin assembly by RNA interference‐dependent and ‐independent pathways. PLoS Genet 9: e1003677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kitano E, Hayashi A, Kanai D, Shinmyozu K, Nakayama J (2011) Roles of fission yeast Grc3 protein in ribosomal RNA processing and heterochromatic gene silencing. J Biol Chem 286: 15391–15402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- 66. Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM (2007) The site‐specific installation of methyl‐lysine analogs into recombinant histones. Cell 128: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi‐Takanaka Y, Kimura H, Kurumizaka H (2010) Structural basis of instability of the nucleosome containing a testis‐specific histone variant, human H3T. Proc Natl Acad Sci USA 107: 10454–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sadaie M, Shinmyozu K, Nakayama J (2008) A conserved SET domain methyltransferase, Set11, modifies ribosomal protein Rpl12 in fission yeast. J Biol Chem 283: 7185–7195 [DOI] [PubMed] [Google Scholar]
- 69. Chatterjee C, McGinty RK, Fierz B, Muir TW (2010) Disulfide‐directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol 6: 267–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


