SUMMARY
Activating mutations in Kras are nearly ubiquitous in human pancreatic cancer and initiate precancerous pancreatic intraepithelial neoplasia (PanINs) when induced in mouse acinar cells. PanINs normally take months to form, but are accelerated by deletion of acinar cell differentiation factors such as Ptf1a, suggesting that loss of cell identity is rate-limiting for pancreatic tumor initiation. Using a genetic mouse model that allows for independent control of oncogenic Kras and Ptf1a expression, we demonstrate that sustained Ptf1a is sufficient to prevent Kras-driven tumorigenesis, even in the presence of tumor-promoting inflammation. Furthermore, reintroducing Ptf1a into established PanINs reverts them to quiescent acinar cells in vivo. Similarly, Ptf1a re-expression in human pancreatic cancer cells inhibits their growth and colony-forming ability. Our results suggest that reactivation of an endogenous differentiation program can prevent and reverse oncogene-driven transformation in cells harboring tumor-driving mutations, introducing a potential paradigm for solid tumor prevention and treatment.
eTOC BLURB
Pancreatic cancer arises from mutations in exocrine acinar cells. Krah et al. demonstrate that preserving acinar cell identity protects these cells from oncogenic Kras-induced tumorigenesis. Furthermore, re-expressing the acinar differentiation gene Ptf1a in existing precancerous lesions in vivo, or human pancreatic cancer cells in vitro, induces quiescence and redifferentiation.
INTRODUCTION
Although pancreatic ductal adenocarcinoma (PDAC) is named for its duct-like characteristics, we and others have shown that its signature driver mutation, oncogenic Kras, induces tumorigenesis when introduced to exocrine acinar rather than duct cells (De La O et al., 2008; Guerra et al., 2007; Habbe et al., 2008; Ji et al., 2009; Kopp et al., 2012). Importantly, tumor initiation from Kras-mutant acinar cells involves a trans- or de-differentiation process, in which acinar-specific genes are downregulated and duct markers upregulated, that we refer to as “reprogramming” (De La O et al., 2008). Acinar cell-specific gene expression is normally driven by the transcription factor Ptf1a, which itself is downregulated during reprogramming to the PDAC precursor lesion pancreatic intraepithelial neoplasia (PanIN) (De La O et al., 2008; Krah et al., 2015). Together with several cooperating transcription factors, Ptf1a is essential for maintaining mature acinar identity and restraining Kras-mediated tumorigenesis (Hoang et al., 2016; Krah et al., 2015; Shi et al., 2009; von Figura et al., 2014). How loss of Ptf1a promotes tumor development is not yet known, but could be mediated by changes in the microenvironment: acute deletion of Ptf1a induces pro-inflammatory genes (Krah et al., 2015), and inflammation is itself known to promote PDAC (Krah and Murtaugh, 2016). Alternatively, Ptf1a target genes may act cell-autonomously to suppress the effects of oncogenic Kras; in this scenario, inflammation may promote PDAC via downregulation of Ptf1a (Molero et al., 2007). To directly test our hypothesis that enforcing acinar cell differentiation inhibits PDAC, we established experimental systems in which Ptf1a expression can be switched on or off in the presence of oncogenic Kras and inflammatory injury. Our results indicate that Ptf1a expression is sufficient to prevent and reverse tumor initiation in the pancreas, as well as to induce quiescence and differentiation of cells from invasive human PDAC.
RESULTS
A mouse model to independently control KrasG12D and Ptf1a expression
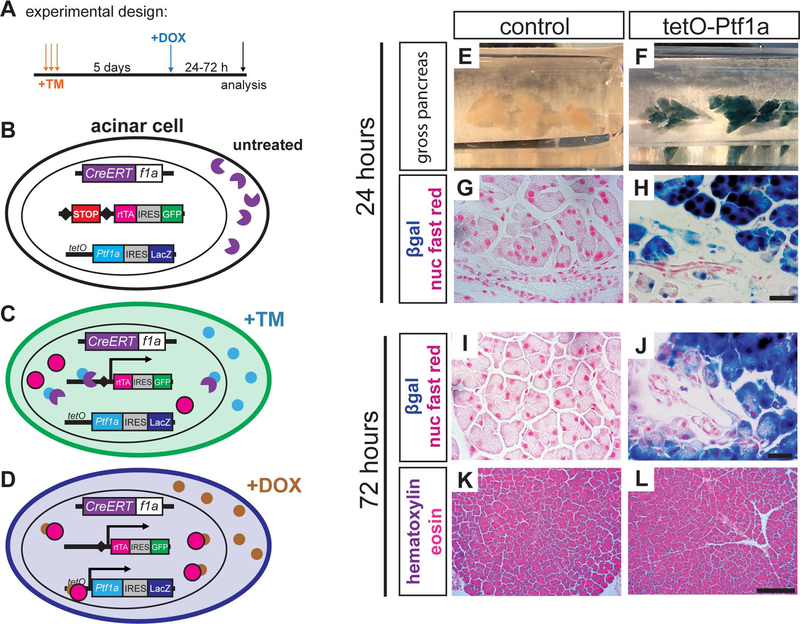
We established a mouse model permitting independent regulation of KrasG12D and Ptf1a: by tamoxifen (TM)-dependent excision of floxed STOP cassettes, Ptf1aCreERT induces expression of both KrasG12D and rtTA, the reverse tetracycline transactivator protein (Belteki et al., 2005) (Figure 1A–C). rtTA subsequently activates a tetO-Ptf1a transgene in a doxycycline (DOX)-inducible manner (Figure. 1D) (Willet et al., 2014). Expression of rtTA and tetO-Ptf1a can be monitored by their linked co-expression cassettes, IRES-GFP and IRES-LacZ, respectively. Staining for β-galactosidase (βgal) activity demonstrated acinar-specific activation of tetO-Ptf1a within 24 hours of DOX administration (Figure 1E–H). Importantly, tetO-Ptf1a expression alone had no detectable effect on pancreas histology (Figure 1K–L). These results indicate that both rtTA and transgenic Ptf1a can be rapidly induced in acinar cells following TM and DOX treatment, respectively.
Figure 1. A mouse model to sustain Ptf1a expression in acinar cells.
(A) Experimental schematic to induce sustained Ptf1a expression using alleles described below. (B) Prior to tamoxifen (TM) administration, CreERT fusion protein (purple) is expressed from the endogenous Ptf1a locus, but is sequestered to the cytoplasm. (C) When TM is administered, Cremediated recombination drives constitutive production of a reverse tetracycline transactivator protein (rtTA, magenta). Cells recombining this locus also permanently express GFP (green) via a downstream IRES-GFP element. (D) Upon administration of DOX, rtTA binds tetO and drives expression of Ptf1a; cells activating tetO-Ptf1a also express LacZ/β-galactosidase (blue). (E-F) Wholemount β-galactosidase (βgal)-stained pancreata of indicated genotypes, 24 hr after DOX administration. (G-H) Histology of above pancreata, highlighting βgal specific to acinar cells (100x, scale bare = 25 μm). (I-J) 72 hours following DOX administration, βgal remains restricted to acinar cells (100x, scale bare = 25 μm), with no histological changes caused by tetO-Ptf1a expression (K-L, 20x, scale bare = 200 μm).
Sustained Ptf1a expression prevents KrasG12D-mediated pancreatic oncogenesis
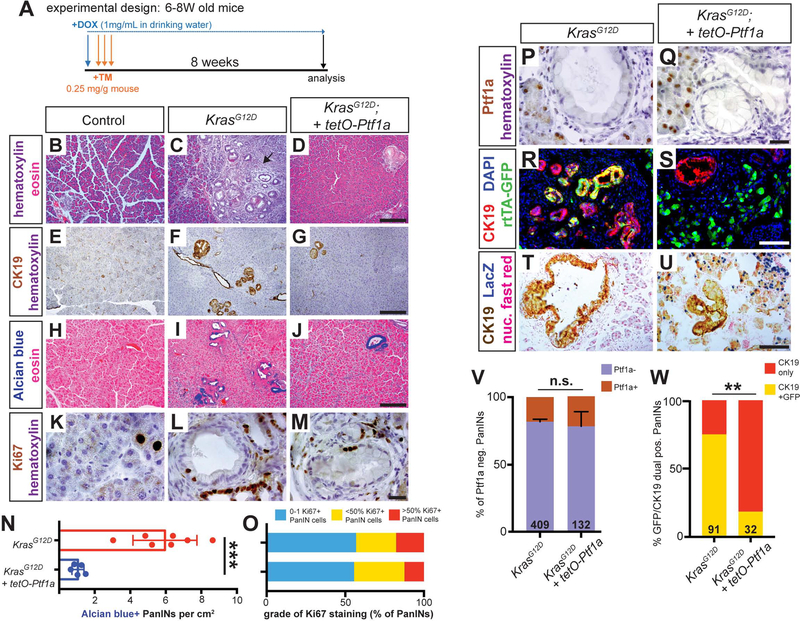
To test whether sustained Ptf1a blocks PDAC initiation, we subjected control, KrasG12D and KrasG12D + tetO-Ptf1a mice (Figure S1) to high-dose TM and an 8-week (8W) chase of continuous DOX (Figure 2A). All mice harbored Ptf1aCreERT and R26LSL-rtTA, and exhibit uniform Cre recombination between groups (Figure S2). After 8W, KrasG12D pancreata exhibited large areas of acinar cell loss and precancerous PanIN formation, which were greatly reduced in KrasG12D + tetO-Ptf1a mice (Figure 2B–J, N; Figure S3). These results suggest that preventing Ptf1a downregulation is sufficient to prevent acinar cell transformation.
Figure 2. Sustained Ptf1a expression prevents KrasG12D-driven PanIN formation.
(A) Mice of indicated genotypes were started on DOX (1 mg/ml in drinking water) 24 hours before TM administration (0.25 mg/g body weight, daily over 3 consecutive days). Mice were euthanized 8 weeks (8W) after the final TM treatment. (B-D) H&E staining of pancreata from mice of indicated genotypes 8W after TM administration (20x). Arrow in C indicates inflammatory stromal reaction in KrasG12D pancreas. Staining for CK19 (E-G) and Alcian blue (H-J) highlights reduced PanIN burden in KrasG12D; tetO-Ptf1a pancreata compared to KrasG12D alone (all images 20x, scale bar = 200 μm). (K-M) Ki67 staining in PanINs of indicated mouse genotypes (100x, scale bare = 25 μm). (N) Quantification of Alcian blue+ PanIN burden, indicating drastic reduction in lesions in KrasG12D; tetO-Ptf1a pancreata over KrasG12D alone (P<0.001, n=5–7 mice per group). (O) Quantification of PanINs graded for proliferation based on Ki67 staining; no significant difference between genotypes (n.s.). (P-Q) Ptf1a staining is absent in PanINs of both genotypes, compared to adjacent acinar cells (100x, scale bare = 25 μm). (R-S) Immunofluorescence for the duct marker CK19 (red), GFP (R26rtTA. green), and DAPI (blue) (20x, scale bar is 100 μm). (T-U) Staining for CK19 (brown), βgal (tetO-Ptf1a, blue) and nuclear fast red (40x, scale bar = 100 μm). (V) Proportion of PanINs that are completely Ptf1a negative vs. those that harbor one or more Ptf1a-positive cell(s) (n.s., n=5 mice per genotype). (W) Proportion of CK19+/GFP+ dual-positive PanINs (Fisher’s exact test, p<0.01). Numbers on bar graphs indicate total lesions scored, across 3–5 mice per group. ** p<0.01, ***p<0.001.
As rare PanINs were still generated in KrasG12D + tetO-Ptf1a pancreata, we compared these lesions to PanINs induced by KrasG12D alone. Similar levels of proliferation were observed between genotypes (Figure 2K–M, O), and all PanINs were overwhelmingly Ptf1a-negative (Figure 2P–Q, V), suggesting that these rare PanINs had the same phenotype as those formed in mice expressing only KrasG12D. We therefore postulated that PanINs in KrasG12D + tetO-Ptf1a mice were “escapers,” recombining KrasG12D to initiate tumorigenesis, but not R26LSL-rtTA, which is required for sustained Ptf1a expression. To test this, we determined the frequency of CK19+ PanINs containing GFP+ cells. Most KrasG12D PanINs co-expressed CK19 and GFP, indicating derivation from acinar cells recombining both the KrasG12D and R26LSL-rtTA loci (Figure 2R, W). In contrast, almost all KrasG12D + tetO-Ptf1a PanINs were broadly GFP-negative (Figure 2S, W), indicating that they failed to recombine R26LSL-rtTA and therefore also did not express tetO-Ptf1a. Thus, the residual PanINs form in KrasG12D + tetO-Ptf1a pancreata due to incomplete Cre recombination. βgal staining further confirmed that KrasG12D + tetO-Ptf1a PanINs contained only very rare cells expressing tetO-Ptf1a (Figure 2T–U). Taken together, these results indicate that Ptf1a expression is sufficient to prevent PDAC initiation.
Pancreatitis is insufficient to overcome Ptf1a-mediated tumor suppression
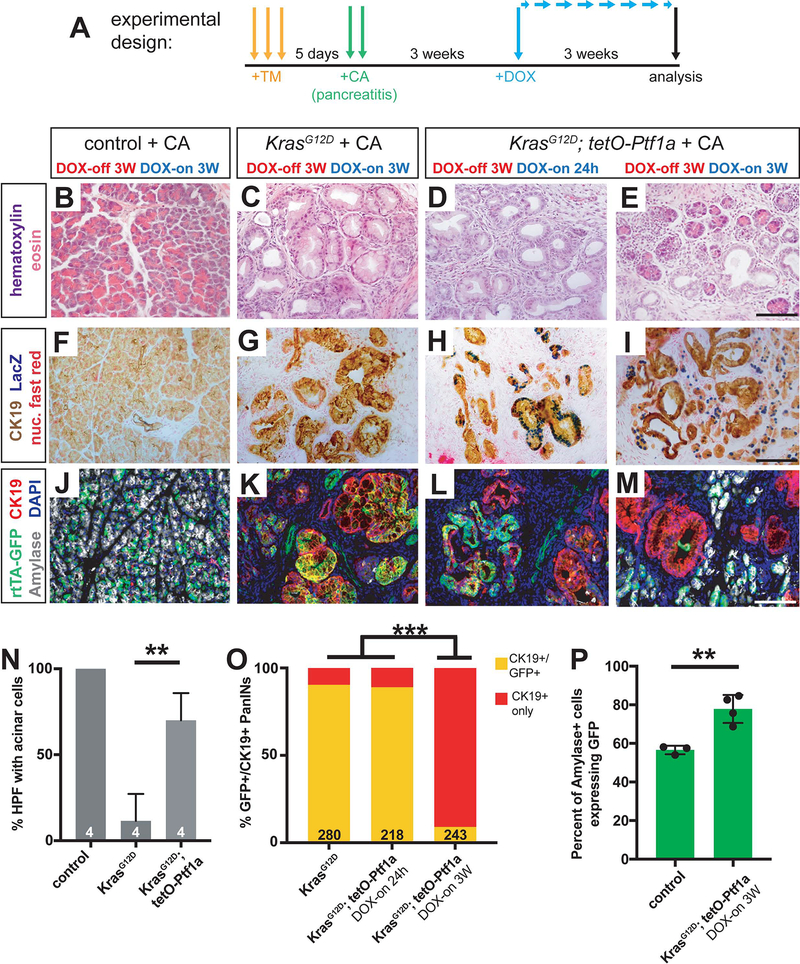
As noted above, deletion of Ptf1a enhances pancreatic inflammation (Krah et al., 2015), which could mediate its pro-tumorigenic consequences. To determine if inflammation could bypass Ptf1a tumor suppression, we subjected KrasG12D + tetO-Ptf1a mice to caerulein-induced pancreatitis, a model of increased PDAC risk (Fig 3A) (Guerra et al., 2007; Lowenfels et al., 1993). Pancreata of control mice were fully recovered at 3 weeks following induction of pancreatitis, while robust PanIN formation was seen in KrasG12D pancreata, as previously reported (Figure 3B–C) (Guerra et al., 2007; Kopp et al., 2012). In contrast, KrasG12D + tetO-Ptf1a pancreata exhibited reduced PanIN formation and lacked the widespread stromal expansion characteristic of caerulein-treated KrasG12D mice (Figure 3D–J, Q; Figure S4). As previously, residual PanINs in these mice arose from rtTA/Ptf1a-negative escaper cells (Figure 3K–P, R). Thus, Ptf1a is both necessary and sufficient to block PDAC initiation, even in the presence of tumor-promoting inflammation (Krah et al., 2015). These results also indicate that downregulation of Ptf1a is a necessary cell-autonomous event for PanIN initiation, as only cells incapable of sustaining Ptf1a expression (i.e. non-rtTA-expressing, GFP-negative cells) give rise to PanINs.
Figure 3. Maintenance of acinar identity inhibits inflammation-induced PanIN formation.
(A) Mice of indicated genotypes were started on DOX (1 mg/ml in drinking water) 24 hours before TM administration (0.25 mg/g body weight, daily over 3 consecutive days). Five days following the final TM dose, mice were administered six hourly injections of 0.1 μg/g caerulein on two consecutive days. Mice were euthanized for pancreatic histology at three weeks following the final caerulein injection. (B-D) H&E staining of pancreata from mice of indicated genotypes 3 weeks after caerulein-induced pancreatitis (20x, scale bar = 200 μm). (E-G) IHC for CK19 (20x, scale bar = 200 μm) and (H-J) Alcian blue staining (10x, scale bar = 500 μm), highlighting PanIN formation. (K-M) IHC for Ptf1a (100x, scale bar = 25 μm), showing lack of nuclear Ptf1a in normal ducts of control mice as well as in PanINs, compared to adjacent acinar cells. (N-P) Immunofluorescence for the duct marker CK19 (red), GFP (R26rtTA. green) and DAPI (blue), highlighting CK19+/GFP-negative escaper PanIN cells in KrasG12D; tetO-Ptf1a pancreata (20x, scale bar is 100 μm). (Q) Quantification of the genotype-dependent PanIN burden, indicating significant reduction in KrasG12D; tetO-Ptf1a pancreata (unpaired t-test, P<0.01). (R) Proportion of CK19+/GFP+ dual-positive PanINs (Fisher’s exact test, P<0.001).
Reactivation of Ptf1a eliminates cells from PanINs in vivo
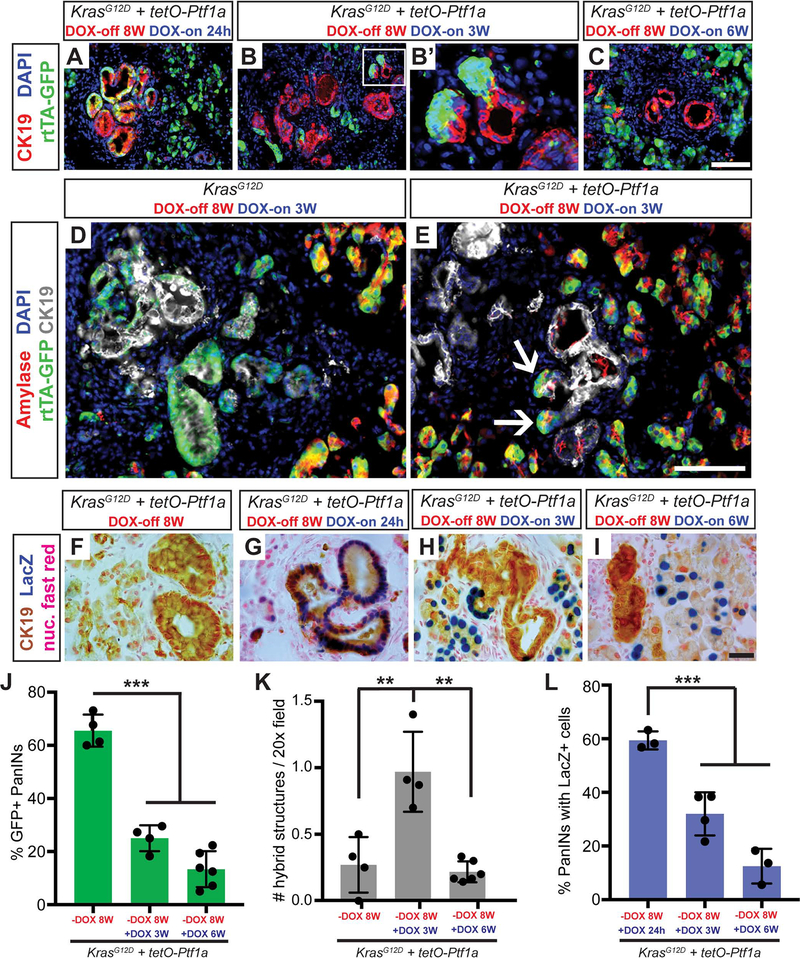
The therapeutic potential of these findings would be greatest if Ptf1a activation could prevent progression of already-established PDAC precursors and revert transformed cells to differentiation and quiescence. To begin to address this, we allowed PanINs to form in DOX-untreated KrasG12D and KrasG12D + tetO-Ptf1a mice for 8W (DOX-off), followed by DOX-induction of Ptf1a for 3W (DOX-on). Whereas large PanINs were present in KrasG12D pancreata, lesions of KrasG12D + tetO-Ptf1a mice appeared smaller and contained misplaced Ptf1a+ cells (Figure S5A–F). Inflammatory cells normally associated with PanINs, including F4/80+ macrophages and CD3+ T-cells (Clark et al., 2007), were found near residual CK19+ lesions but largely excluded from re-differentiated amylase+ acinar cells (Figure S5G–I). To specifically follow the fate of Ptf1a-reactivating cells within established lesions, we analyzed GFP (rtTA) and βgal (tetO-Ptf1a) production at multiple timepoints before and after DOX (Figure 4). Most PanINs that formed at 8W DOX-off, as well as those analyzed after 24 hrs DOX induction, contained abundant GFP+ cells (Figure 4A, J), i.e., they expressed rtTA. By contrast, 3W and 6W DOX-on treatment induced a progressive elimination of GFP+ cells from PanINs (Figure 4B–C, J). At 3W DOX-on, we observed a striking formation of hybrid structures in which clustered GFP+ cells appeared tightly connected to CK19+ lesions (Figure 4B’, K). In contrast to KrasG12D pancreata where amylase+ cells were excluded from PanINs, the emerging GFP+ cells in KrasG12D + tetO-Ptf1a pancreata were amylase+, suggesting re-differentiation into acini (Figure 4D–E, white arrows). These results were corroborated by the LacZ (tetO-Ptf1a) reporter: while DOX-off mice had no βgal+ pancreatic cells, 24 hr DOX-on induced widespread βgal+ cells in PanINs, which were then absent from lesions at 3–6W DOX-on (Figure 4F–I, L), suggesting that cells capable of expressing Ptf1a reverted to a CK19-negative phenotype. Together, these data suggest that Ptf1a re-expression leads to loss of the PanIN phenotype and potential re-differentiation to acinar cells.
Figure 4. Re-activation of Ptf1a in PanINs reverses premalignant cell phenotypes.
(A-C) Immunofluorescence for CK19 (red), GFP (R26rtTA, green), and DAPI (blue) in KrasG12D + tetO-Ptf1a pancreata following indicated treatments (20x, scale bar is 100 μm). (B’) Enlarged image highlighting GFP+ acinar cells emerging from CK19+ lesions. (D-E) Immunofluorescence for amylase (red), GFP (R26rtTA, green), CK19 (white), and DAPI (blue) in KrasG12D and KrasG12D + tetO-Ptf1a pancreata following indicated treatment (20x, scale bar is 100 μm). (E) Arrows indicate Amylase/GFP-dual positive cells emerging from CK19+ lesions. (F-I) Staining for CK19 (brown), βgal (tetO-Ptf1a, blue) and nuclear fast red in pancreata of indicated treatment groups (100x, scale bar is 25 μm). (J-L) Quantification of proportion of GFP+ PanINs (J), number of PanIN/acinar hybrid structures per field (K), and proportion of βgal+ PanINs (L) for mice of indicated treatment groups. ** p<0.01, *** p<0.001, as determined by ANOVA.
Re-expression of Ptf1a in PanINs leads to the emergence of normal acinar cells in vivo
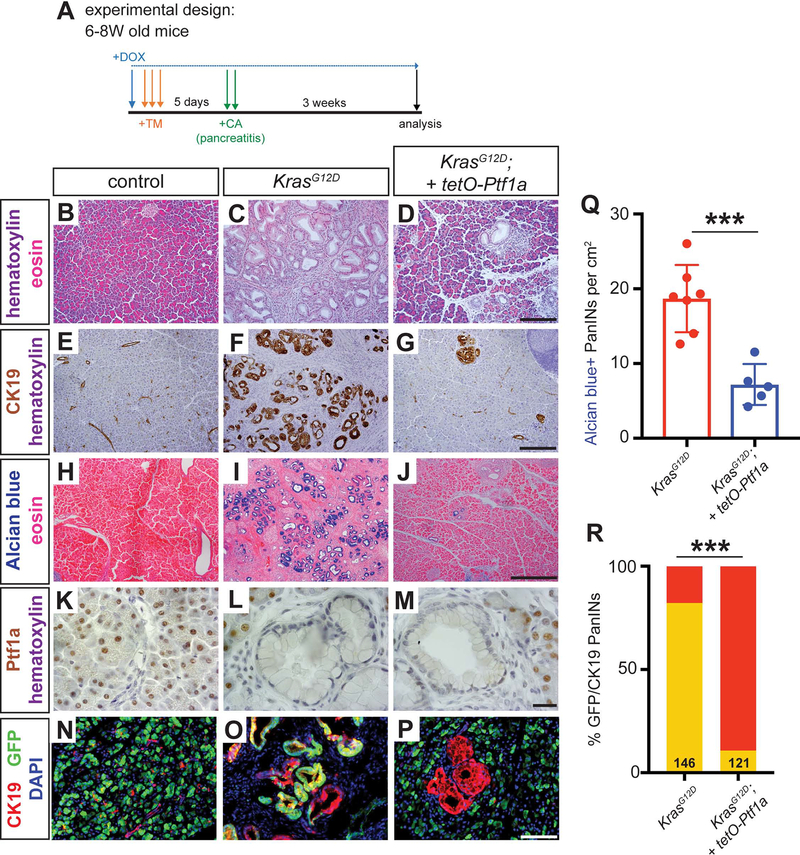
Although we observed amylase+ cells apparently emerging from PanINs in the above experiments (Figure 4B’, K), the majority of normal GFP+ acinar cells in these mice are likely to be derived from pre-existing GFP+ acini that never underwent KrasG12D-mediated transformation. To determine whether GFP+ PanINs directly re-differentiate into acinar cells, restoring normal exocrine histology, we used the Kras/caerulein experimental model in which nearly all acinar cells are destroyed or converted to PanINs prior to Ptf1a re-expression. Control, KrasG12D and KrasG12D + tetO-Ptf1a mice were administered high dose TM followed 5 days later by caerulein administration to induce pancreatitis (Figure 5A). During this time, and for the three weeks that followed, mice were kept in the DOX-off state, allowing for KrasG12D activity in the absence of transgenic Ptf1a. Following this 3W chase, mice were administered DOX for an additional 3W, to activate transgenic tetO-Ptf1a (Figure 5A).
Figure 5. Ptf1a induces acinar cell re-differentiation from PanIN cells after caerulein-induced pancreatitis.
(A) Mice of indicated genotypes were administered TM (0.25 mg/g body weight, daily over 3 consecutive days), and five days later administered six hourly injections of 0.1 μg/g caerulein for two consecutive days. Three weeks after the last caerulein injection, mice were administered DOX (1 mg/ml in drinking water) for either 24 hours or 3 weeks before pancreatic histology was examined. (B-E) H&E staining of pancreata from mice of indicated genotypes and treatment regimens (40x, scale bar is 100 μm). (F-I) Staining for CK19 (brown), βgal (tetO-Ptf1a, blue) and nuclear fast red in pancreata of indicated treatment groups (40x, scale bar is 100 μm). (J-M) Immunofluorescence for CK19 (red), GFP (R26rtTA, green), Amylase (white), and DAPI (blue) in Control, KrasG12D and KrasG12D + tetO-Ptf1a pancreata following indicated treatment (20x, scale bar is 100 μm). (N) Quantification of percentage of high-powered fields (HPF, 40x) containing acinar cells with normal morphology following the treatment protocol in (A) (unpaired t-test, P<0.001). (O) Percentage of CK19+ PanINs co-expressing GFP in KrasG12D and KrasG12D + tetO-Ptf1a pancreata following the indicated treatments (Fisher’s exact test, P<0.001). The number on the bar graph indicate the number of lesions counted between 3–4 mice per group. (P) Quantification of the percentage of amylase+ cells that co-express GFP in Control and KrasG12D + tetO-Ptf1a pancreata following the treatment protocol outlined in (A) (unpaired t-test, P<0.01, n=3–4 per group).
Compared to control pancreata, which completely recovered from pancreatitis, those of KrasG12D mice comprised almost exclusively PanINs, as previously described (Figure 5B–C, Figure 6A–B). In contrast, KrasG12D + tetO-Ptf1a pancreata exhibited normal-appearing acinar cells emerging from PanINs after undergoing the same treatment protocol (Figure 5E, Figure 6C). Importantly, although a single day of DOX treatment (3W post-caerulein treatment, 1 day DOX-on) was sufficient to induce the tetO-Ptf1a transgene within PanINs, as indicated by LacZ expression (Figure 5F–H), neither residual nor induced acinar cells were detected at this initial time, similar to our previous observation in KrasG12D-caerulein mice (Figure 5D, Figure 3). In the subsequent 3W DOX-on period, normal-appearing acinar cells re-differentiated throughout the pancreas of KrasG12D + tetO-Ptf1a mice specifically (Figure 5E, N), and could be seen as LacZ-positive cells abutting residual PanINs (Figure 5I).
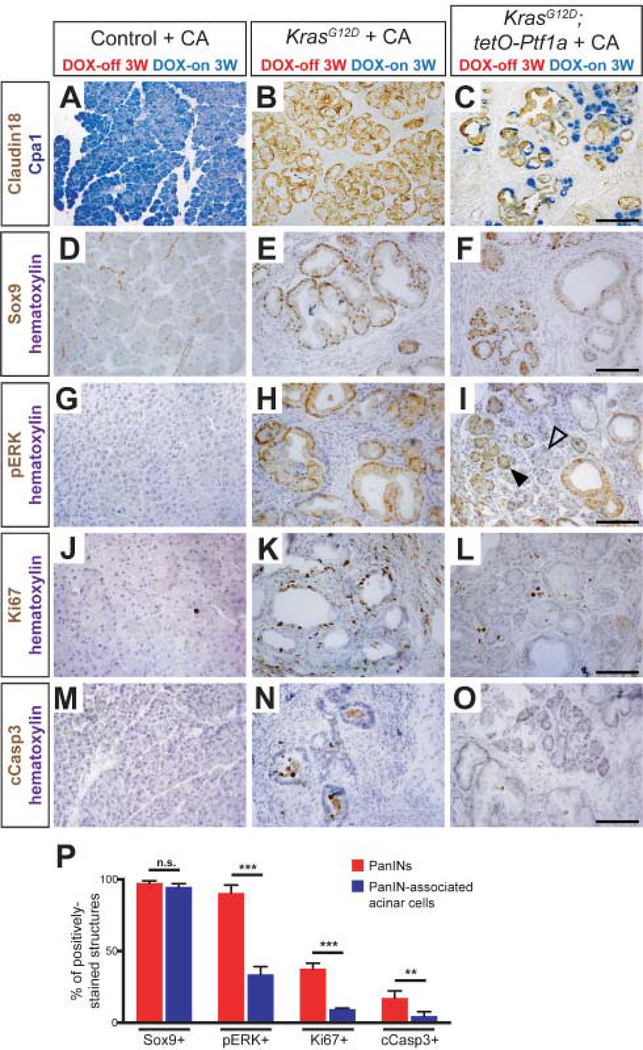
Figure 6. Characterization of PanIN-associated acinar cells.
(A-C) Dual immunohistochemistry for the acinar cell marker Cpa1 and the PanIN-specific marker Claudin18 in mice of indicated genotypes. Images are 20x magnification (scale bar is 200 μm). Immunohistochemistry for Sox9 (D-F), phospho-ERK (G-I), Ki67 (J-L), and cleaved Caspase3 (M-O) in mice of indicated genotypes. Closed and open arrowheads in I indicate pERK-positive and -negative acini, respectively. All images are 40x magnification (scale bars are 100 μm). (P) Quantification of the percent of PanINs (red bar) and PanIN-associated acinar cells (blue bar) that express Sox9, p-ERK, Ki67, and cleaved Caspase3. All comparisons were made by an unpaired t-test (n.s. = not significant, **P<0.01, *** P<0.001).
To follow the fate of re-differentiating acinar cells more quantitatively, we performed immunofluorescence for GFP (R26rtTA), CK19, and amylase in the same genotypes/treatment groups. As expected, control pancreata had abundant GFP+/amylase+ cells together with GFP-negative/CK19+ ducts intercalating through the acinar epithelium (Figure 5J). In contrast, nearly all GFP+ cells in KrasG12D pancreata, as well as KrasG12D + tetO-Ptf1a after 1 day DOX treatment, were CK19+/amylase-negative, i.e. duct-like (Figure 5K, L). After three weeks DOX-on, however, the situation was reversed: almost all GFP+ cells were now CK19-negative/amylase+, consistent with a PanIN-to-acinar fate switch (Figure 5M, O).
This was corroborated by comparing the fraction of amylase+ acinar cells expressing GFP in control vs. KrasG12D + tetO-Ptf1a at the end of 3-weeks DOX treatment. If acinar cells in KrasG12D + tetO-Ptf1a mice arose from a residual, Kras-wild-type survivor population, their GFP-labeling frequency should be lower than that of controls: assuming that rtTA transgene activation correlates with that of KrasLSL-G12D, cells that failed to activate KrasG12D should be enriched for cells that failed to undergo any Cre recombination events, including that of rtTA. Thus rtTA-negative acinar cells should predominate among “recovered” acini due to their relative lack of KrasG12D expression (i.e. lack of Cre activity). Instead, we found a significant increase in the GFP+ fraction of KrasG12D + tetO-Ptf1a acinar cells, relative to control (Figure 5P), suggesting that the new acini preferentially arose from PanIN cells that re-expressed transgenic Ptf1a. Taken together, these data indicate that PanIN cells re-expressing Ptf1a are able to re-differentiate into acinar cells, despite harboring oncogenic KrasG12D.
Partial normalization of gene expression and signaling status upon PanIN-to-acinar re-differentiation
Re-differentiating acinar cells in KrasG12D + tetO-Ptf1a pancreata, expressing the digestive enzyme (and Ptf1a target gene) Carboxypeptidase-A1 (Cpa1), arise in direct apposition to residual Claudin-18+ PanIN cells (Figure 6A–C). To characterize the properties of newly-formed acinar clusters, we compared them to the well-formed PanINs found in KrasG12D pancreata at the same 6W post-caerulein timepoint (Figure 6P). As previously reported, Sox9 was expressed in duct cells of controls and PanINs of KrasG12D pancreata (Figure 6D–E). Unexpectedly, nearly all PanIN-associated acinar cells in KrasG12D + tetO-Ptf1a mice maintained strong Sox9 expression (Figure 6F). Nuclear staining for phosphorylated ERK1/2 (pERK), a key effector of RAS signaling, was seen in the majority of KrasG12D PanINs, but only within a minority of KrasG12D + tetO-Ptf1a re-differentiated acinar cells (Figure 6H, I). Interestingly, a subset of morphologically normal acinar cells did remain pERK+ (Figure 6I), suggesting that shutdown of this pathway may not be essential for acinar cell re-differentiation.
Acinar cells re-differentiating from KrasG12D + tetO-Ptf1a PanINs also returned to a state of cellular quiescence: while PanINs in KrasG12D pancreata were heterogeneously positive for the proliferation marker Ki67, most PanIN-associated acini were Ki67-negative, similar to control acinar cells (Figure 6J–L). Apoptotic cells, staining for cleaved Caspase-3 (cCasp3), were occasionally observed in the lumens of PanINs, potentially marking cells undergoing anoikis, but were effectively absent from both control acini and re-differentiating KrasG12D + tetO-Ptf1a acinar structures (Figure 6M–O).
In addition to restoring acinar cell differentiation and quiescence, Ptf1a induction produced changes in the PanIN microenvironment: re-differentiating acinar cells in KrasG12D + tetO-Ptf1a pancreata were associated with reduced density of F4/80+ macrophages and αSMA+ fibroblasts compared to PanINs (Figure S6A–H). Consistent with a reduction in fibrogenic cells, re-differentiating acini were found in areas with diminished Sirius Red staining (Figure S6I–L). Thus, Ptf1a re-expression fundamentally normalizes many aspects of the PanIN-reprogrammed acinar cell, including digestive enzyme production and quiescence, as well as restoring a non-inflamed microenvironment.
Ptf1a expression reduces clonogenicity and proliferation of human PDAC cell lines
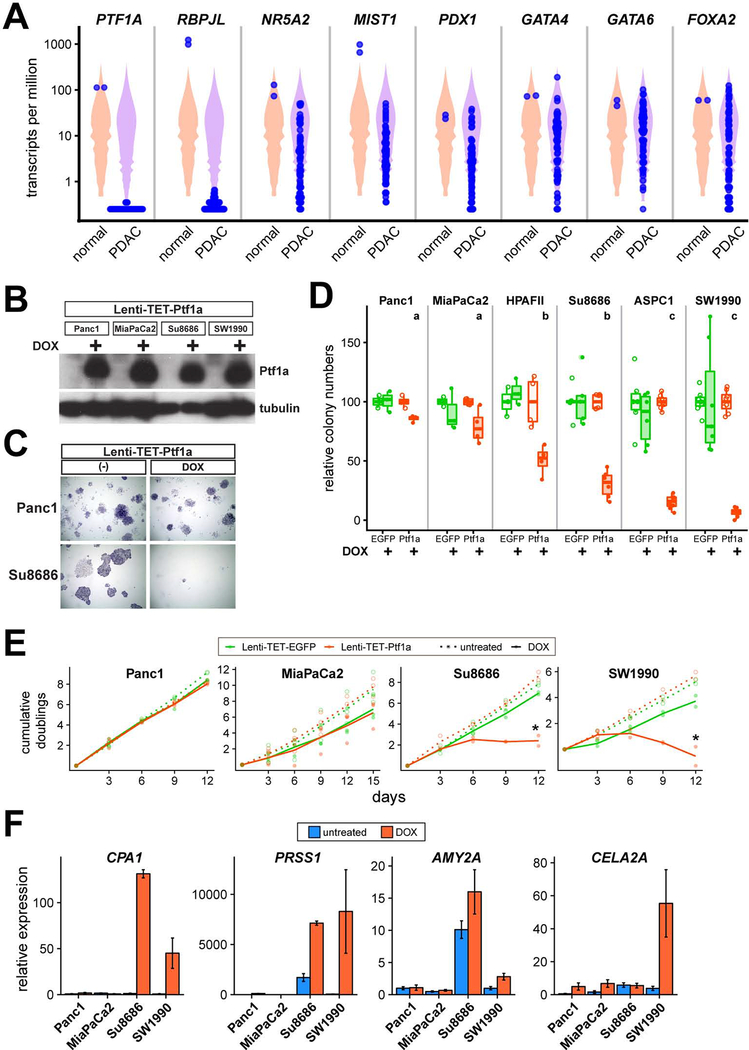
Our results suggest that a hypothetical therapy to maintain or restore Ptf1a expression could effectively prevent progression of human PDAC, or even reverse it. To identify human pancreatic cancer cell lines in which PTF1A has been silenced, we analyzed RNA-seq data from two large surveys of human cancer cell lines (n=41 PDAC cell line samples, 26 analyzed in duplicate), together with normal pancreas RNA-seq data from human anatomical surveys (n=2) (Barretina et al., 2012; Klijn et al., 2015; Mele et al., 2015; Uhlen et al., 2015). Remarkably, PTF1A is not detectably expressed in any of the PDAC lines, nor is its acinar-specific cofactor RBPJL (Figure 7A). Other acinar-specific or acinar-enriched TFs implicated as regulating target genes with Ptf1a, including MIST1/BHLHA15, NR5A2, PDX1, GATA4 and GATA6, exhibited much more variable expression across PDAC lines, in some cases expressed at levels comparable to normal pancreas and completely silenced in only a minority of lines (Figure 7A). Thus, whereas some of these TFs may be “repurposed” in pathological conditions to functions other than regulating acinar gene expression (Cobo et al., 2018), PDAC lines appear to be intolerant of even low level Ptf1a expression. Alternatively, these lines may have arisen from a ductal cell of origin, in which PTF1A is physiologically silent, and have coincidentally activated acinar TFs for unrelated reasons. To explore the therapeutic potential of Ptf1a reactivation, we sought to determine if human PDAC cells are indifferent or sensitive to ectopically expressed Ptf1a.
Figure 7. Ptf1a inhibits growth of human PDAC cells.
(A) Expression of PTF1A and other acinar regulators in normal human pancreas (n=2) and PDAC cell lines (n=41), derived from public RNA-seq datasets. Expression plotted in transcripts per million, with individual samples plotted in blue over violin plots representing all expressed genes in each sample. (B) Western blot of Ptf1a and tubulin (control) expression in Lenti-TET-Ptf1a derivatives of Panc1, MiaPaCa2, Su8686 and SW1990 cells, treated −/+ DOX (1 μg/ml) for 48 hrs. (C) Representative images from clonogenic growth assays of Panc1 and Su8686 Lenti-TET-Ptf1a cells, grown 2 weeks −/+ DOX and stained with crystal violet. (D) Quantification of relative clone numbers in indicated PDAC cell lines, transduced with Lenti-TET-EGFP (green) or Lenti-TET-Ptf1a (orange), grown 10–14 days −/+ DOX (open vs. closed circles). Distinct lowercase letters indicate p<0.05 from ANOVA followed by Tukey’s post-hoc test of DOX-treated Lenti-TET-Ptf1a cells. (E) Cumulative population doublings of indicated PDAC cells, transduced with EGFP or Ptf1a (green vs orange). Cells were counted and replated at constant densities every 3 days, and treated from day 1 onwards −/+ DOX (open vs. closed circles for individual experiments, dotted vs. solid lines representing means). * P<0.05, DOX-treated Ptf1a vs EGFP cells. (F) Expression of indicated genes in PDAC cell lines transduced with Lenti-TET-Ptf1a, grown 48 hrs −/+ DOX (blue vs. orange), and analyzed by RT-qPCR (ΔΔCt method, normalized to PPIA expression within each sample and untreated Panc1 cells across samples).
We cloned mouse Ptf1a into a “polyswitch” lentiviral vector that permits selection of infected cells as well as DOX-inducible transgene expression (Giry-Laterriere et al., 2011), together with EGFP as a negative control. After infection and polyclonal selection of a panel of commonly-used human PDAC lines (all validated by STR genotyping), we confirmed tightly inducible Ptf1a protein expression in Lenti-TET-Ptf1a cells (Figure 7B), as well as EGFP in control Lenti-TET-EGFP cells (data not shown). As an initial test of sensitivity to Ptf1a expression, we performed clonal growth assays in which cells were plated at low density in the absence of DOX, switched the next day to medium lacking or containing DOX, and grown until easily-visible colonies appeared in control cultures (Lenti-TET-EGFP, no DOX). We observed a remarkably varied response, with some lines such as Panc1 and MiaPaCa2 exhibiting no deficit in colony formation upon Ptf1a induction, while others such as Su8686 and SW1990 were profoundly inhibited (Figure 7C–D). Using the former lines as exemplars of “Ptf1a-insensitive” PDAC cells, and the latter as representative Ptf1a-sensitive lines, we performed bulk growth assays to determine if Ptf1a overexpression is sufficient to prevent tumor cell proliferation under permissive (non-clonal) conditions. Consistent with our clonogenicity results, Panc1 and MiaPaCa2 cells were unaffected by Ptf1a expression, while Su8686 and SW1990 cells underwent complete growth arrest upon Ptf1a induction (Figure 7E). Taken together, these results indicate that at least a subset of human PDAC remains profoundly sensitive to Ptf1a expression, potentially explaining the dramatic silencing of this gene across PDAC lines.
To begin to understand the mechanism underlying the differential response to Ptf1a, we compared the ability of Ptf1a to activate known acinar-specific target genes (Hoang et al., 2016), between insensitive and sensitive cell lines. We found little or no induction of these targets in either Panc1 or MiaPaCa2 cells, while a more complex pattern applied in the Ptf1a-sensitive Su8686 and SW1990 cells: CPA1 and PRSS1 were both strongly activated by Ptf1a, while AMY2A was weakly activated (while exhibiting high basal expression in Su8686 specifically) and CELA2A was induced only in SW1990 (Figure 7F). Thus, the differential Ptf1a-sensitivity of human PDAC cell growth is mirrored by differential transcriptional competence at Ptf1a target genes.
DISCUSSION
Pancreatic cancer evolves through premalignant stages lasting a decade or more, providing a window for prevention and reversion of this deadly disease with the development of improved tools for detection and management (Yachida et al., 2010). Several recent mouse modeling studies have shown the importance of acinar cell differentiation factors, including Ptf1a, in restraining PDAC initiation (Flandez et al., 2014; Krah et al., 2015; Roy et al., 2016; Shi et al., 2009; von Figura et al., 2014). Consistent with these findings, human genome-wide association studies have identified PDAC risk-modifying polymorphisms in NR5A2 and PDX1, transcriptional partners and regulators of Ptf1a (Petersen et al., 2010; Wolpin et al., 2014). Ptf1a itself is downregulated in human PanINs, and required in mice to maintain homeostasis, restrain pancreatic inflammation, and suppress RAS-related gene signatures (Hoang et al., 2016; Krah et al., 2015). Together, these studies suggest that maintaining acinar cell differentiation could be an attractive therapeutic approach to limit pancreatic tumor initiation and progression.
Such an approach is supported by the findings reported here, that sustained Ptf1a expression can prevent and reverse the early stages of pancreatic tumorigenesis in vivo, as well as induce growth arrest in human PDAC cells in vitro. Our results indicate that downregulation of Ptf1a is essential for PanIN formation and maintenance, as only Ptf1a-negative “escaper” cells contribute to PanINs in KrasG12D + tetO-Ptf1a pancreata, in two different experimental schemas of PDAC initiation (Figures 2 and 3). Additionally, transgenic reintroduction of Ptf1a into established lesions reverts their phenotype to amylase+/CPA1+ acinar cells, establishing that differentiation can override KrasG12D-mediated transformation (Figures 4–6). This re-differentiation phenotype is similar to that observed following silencing of KrasG12D expression or pharmacological inhibition of MEK1/2 in PanINs (Collins et al., 2012; Collins et al., 2014), and fits with a model in which Ptf1a antagonizes KRAS signaling activity by inhibiting expression of RAS-dependency genes, while also promoting tissue reorganization, maintenance, and homeostasis (Hoang et al., 2016; Krah et al., 2015). Importantly, while KRAS activation is an effectively irreversible genetic alteration in PDAC, downregulation of human PTF1A occurs at the transcriptional level, and is in principle reversible. Indeed, our cell culture findings indicate that many human PDAC cell lines are highly susceptible to Ptf1a-induced growth arrest, potentially explaining the universal silencing of PTF1A expression observed in these cells (Figure 7). Strategies aimed at restoring endogenous PTF1A should complement efforts to develop inhibitors of KRAS itself and its downstream effectors.
Our results, and those of other investigators, also suggest that antagonism between Ptf1a and KRAS occurs at multiple levels. At early stages of tumor initiation, KRAS signals through MEK/MAPK to drive acinar-to-PanIN reprogramming, and MEK inhibition induces PanIN-to-acinar re-differentiation (Collins et al., 2014). Presumably this re-differentiation process proceeds through Ptf1a activity, although this has not yet been tested. In full blown PDAC, however, MEK inhibition alone has only modest anti-cancer activity, and gene expression profiling does not indicate that it induces acinar re-differentiation (Fedele et al., 2018; Gysin et al., 2012). Thus, the silencing of human PTF1A in PDAC cell lines is likely independent of ongoing KRAS/MEK/MAPK signaling, suggesting a transition between active and passive inhibition of PTF1A. Understanding these inhibitory mechanisms may point to a strategy for reactivating PTF1A in full-blown cancer. It is notable as well that some PDAC cell lines appear to be highly resistant to the anti-proliferative effects of Ptf1a, through mechanisms that also require further investigation.
While this work was in progress, an independent study demonstrated that overexpression of Ptf1a in Panc1 cells could induce acinar-specific gene expression as well as inhibit clonogenic growth (Jakubison et al., 2018), whereas we find Panc1 to be Ptf1a-resistant. The precise reason for this discrepancy is not known, but some differences in methodologies exist. For example, the approach of Jakubison et al. (2018) may have achieved higher level expression of Ptf1a, overcoming an inhibitory threshold not matched by our expression system. In addition, whereas we introduced Ptf1a via polyclonal lentiviral transduction, Jakubison et al. (2018) employed stable plasmid transfection and clonal selection. It is possible that subclones of Ptf1a-sensitive cells exist within the Panc1 bulk population; indeed, we observe a small but reproducible decrease in clonogenicity in Ptf1a-induced Panc1 cells (Figure 7D). It will be important to determine whether this and other cell lines sporadically switch between sensitive and insensitive states, as manipulating such a switch could increase the translational utility of Ptf1a re-activation.
Taken together, our results highlight the potency of Ptf1a as an inhibitor of pancreatic tumorigenesis, sufficient not only to prevent but also to reverse tumor initiation. Chemoprevention strategies are under active investigation in PDAC (Miller et al., 2016), and our in vivo misexpression studies suggest that maintained expression of Ptf1a will be pivotal to their success. While a route to therapeutic Ptf1a re-activation is not obvious, our human cell line studies underscore the potential utility of such an approach. Future cell line experiments will also focus on identifying key Ptf1a target genes and the mechanisms by which they inhibit growth of pancreatic cancer cells. Looking more broadly, we hypothesize that factors like Ptf1a act broadly to suppress cancer in any organ where tumor initiation involves loss of differentiation (Krah and Murtaugh, 2016; Roy and Hebrok, 2015).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, L. Charles Murtaugh (murtaugh@genetics.utah.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice:
Experimental mouse alleles used in this study include the following: Ptf1aCreERT (Ptf1atm2(CreER1)CVW) (Krah et al., 2015), KrasLSL-G12D (Krastm4tyj) (Hingorani et al., 2003), Rosa26rtTA-IRES-EGFP (Belteki et al., 2005), and tetO-Ptf1a (Willet et al., 2014). Shorthand references to compound mouse models are as follows:
control: Ptf1aCreERT/+; R26RrtTA/rtTA
KrasG12D: Ptf1aCreERT/+; KrasLSL-G12D/+; R26RrtTA/rtTA
KrasG12D + tetO-Ptf1a: Ptf1aCreERT/+; KrasLSL-G12D/+; tetO-Ptf1a (hemizygous); R26RrtTA/rtTA
Primers for genotyping are provided in Table S1. To induce Cre-mediated recombination, tamoxifen (Cayman Chemical, Ann Arbor, MI) in corn oil was administered via oral gavage at 0.25 mg/g body weight on three consecutive days. To induce tetO-Ptf1a expression, all experimental mice were administered 1 mg/mL of Doxycycline (Research Products International, Mt. Prospect, IL) with 1% D-sucrose (Fisher Scientific) in their drinking water. Water bottles containing Doxycycline were protected from light and refreshed two times per week. All mice utilized in this study were between 6 and 12 weeks of age at the beginning of experiments. No mice with health status reports, as determined by an in-house veterinarian, were utilized in any analysis. For each experiment and experimental group, we utilized approximately identical number of male and female mice. Previous studies have not identified a gender-dependent PanIN burden in any mouse model of PDAC initiation or progression. Furthermore, human PDAC affects men and women with similar incidence, leading to ~7% of cancer related deaths in both genders (Siegel et al., 2018).
All experiments involving mice were performed according to institutional IACUC and NIH guidelines.
Cell lines:
The following human cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA), directly or indirectly through colleagues: 293T, Panc1, MiaPaCa2, Su8686, SW1990, HPAFII, ASPC1. All lines were authenticated by STR genotyping at the University of Utah DNA Sequencing Core, and were confirmed free of mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza, Walkersville, MD).
METHOD DETAILS
Tissue processing and histology:
Pancreata were dissected into ice cold PBS, separated into multiple parts and processed for frozen and paraffin sections, as previously described (De La O et al., 2008; Krah et al., 2015). For paraffin sectioning, tissues were fixed in zinc-buffered formalin (Z-fix; Anatech, Battle Creek, MI) at room temperature overnight, followed by processing (dehydration in ethanol washes) into Paraplast-Plus (Leica Biosystems, Buffalo Grove, IL). Frozen specimens were fixed for 1–2 hr in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in 1x PBS on ice, followed by processing into Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, CA). Paraffin and frozen sections were 6–8-microns with ≥100 μm spacing between individual sections, all placed on a single slide.
Immunohistochemistry (IHC) and immunofluorescence followed established protocols (De La O et al., 2008; Krah et al., 2015) and included high temperature antigen retrieval (Vector Unmasking Solution; Vector Laboratories, Burlingame, CA), prior to staining all paraffin sections. Primary antibodies are listed in the Key Resource Table and were diluted in blocking solution consisting of 5% donkey serum (Jackson Immunoresearch, West Grove, PA), 0.3% Triton X-100 and 0.2% sodium azide in PBS. Secondary antibodies, raised in donkey (Jackson Immunoresearch) were diluted 1:250 in blocking solution. Vectastain reagents and diaminobenzidine (DAB) substrate (Vector Laboratories) were used for all IHC experiments (see Key Resource Table). Immunofluorescence sections were counterstained with DAPI and mounted in Fluoromount-G (Southern Biotech, Birmingham, AL), and photographed on an Olympus IX71 microscope, using MicroSuite software (Olympus America, Waltham, MA). Images were processed in Adobe Photoshop, with exposure times and adjustments identical between genotypes and treatment groups.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep polyclonal anti-Amylase | BioGenesis | 0480–0104 |
| Goat polyclonal anti-Cpa1 | R&D Systems | AF2765 |
| Rabbit monoclonal anti-Cleaved-caspase3 | Cell Signaling | 9664S |
| Rabbit monoclonal anti-Claudin-18 | Invitrogen | 700178 |
| Rat monoclonal anti-Cytokeratin-19 | Developmental Studies Hybridoma Bank (DSHB) | TROMA-III |
| Rabbit monoclonal anti-Cytokeratin-19 | Abcam | AB133496 |
| Chicken polyclonal anti-GFP | Aves Labs | GFP-1010 |
| Rabbit polyclonal anti-GFP | Abcam | Ab290 |
| Mouse monoclonal anti-Ki67 | BD Biosciences | 550609 |
| Rabbit polyclonal anti-Ptf1a | Chris Wright, Vanderbilt University | N/A |
| Rabbit polyclonal anti-phospho-ERK1/2 (T202/Y204) | Cell Signaling | 9101 |
| Rabbit polyclonal anti-Sox9 | Millipore | AB5535 |
| Mouse monoclonal anti-β-tubulin | DSHB | 12G10 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Cayman Chemical | 13258 |
| Doxycycline | Research Products International |
D43020–25.0 Lot 22362 |
| Zinc formalin fixative | Anatech | 174 |
| Paraformaldehyde 16% Solution | Electron Microscopy Sciences | 15710 |
| Caerulein | Bachem | 4030451 |
| Deposited Data | ||
| Normal human pancreas RNA-seq data (TPM), GTEX | GTEx Portal | https://storage.googleapis.com/gtex_analysis_v7/rna_seq_data/GTEx_Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_median_tpm.gct.gz |
| Normal human pancreas RNA-seq data (TPM), Human Protein Atlas | EMBL-EBI Expression Atlas | https://www.ebi.ac.uk/gxa/experiments/E-MTAB-2836/Downloads?ref=aebrowse |
| Human cancer cell line RNA-seq data (TPM), Genentech | EMBL-EBI Expression Atlas | https://www.ebi.ac.uk/gxa/experiments/E-MTAB-2706/Downloads?ref=aebrowse |
| Human cancer cell line RNA-seq data (TPM), Cancer Cell Line Encyclopedia | EMBL-EBI Expression Atlas | https://www.ebi.ac.uk/gxa/experiments/E-MTAB-2770/Downloads?ref=aebrowse |
| Experimental Models: Cell Lines | ||
| HEK 293T | ATCC | CRL-3216 |
| ASPC1 | ATCC | CRL-1682 |
| HPAFII | ATCC | CRL-1997 |
| Panc1 | ATCC | CRL-1469 |
| MiaPaCa2 | ATCC | CRL-1420 |
| Su8686 | ATCC | CRL-1837 |
| SW1990 | ATCC | CRL-2172 |
| Experimental Models: Mouse Strains | ||
| Mouse: Ptf1aCreERTM (Ptf1atm2(CreER1)CVW) | Jackson Laboratory | 019378 |
| Mouse: KrasLSL-G12D (Krastm4tyj) | Jackson Laboratory | 008179 |
| Mouse: Rosa26rtTA-EGFP (B6.Cg-Gt(ROSA)26Sortm1(rtTA,EGFP)NagyI) |
Jackson Laboratory | 005670 |
| Mouse: tetO-Ptf1a | Raymond MacDonald, UT-Southwestern | N/A |
| Oligonucleotides | ||
| see Table S2 | ||
| Recombinant DNA | ||
| pENTR221-mPtf1a | this paper | N/A |
| pME-EGFP | (Kwan et al., 2007) | N/A |
| pCLX-pTF-R1-DEST-R2-EBR65 | Addgene | # 45952 |
| pCLX-pTF-mPtf1a | this paper | N/A |
| pCLX-pTF-EGFP | this paper | N/A |
| psPAX2 | Addgene | # 12259 |
| pCAG-VSVG | Addgene | # 35616 |
| Other | ||
| Citrate-based Antigen Unmasking Solution | Vector Laboratories Inc | H-3300 |
For Alcian blue staining, paraffin sections were pre-incubated 15 min in 3% acetic acid, stained 10 min in 1% Alcian blue in 3% acetic acid), and washed extensively in 3% acetic acid and dH2O. Following staining, all slides were washed in 0.5% acetic acid, dehydrated and equilibrated into xylene, and mounted with Permount (Thermo-Fisher Scientific, Waltham, MA).
Caerulein treatment:
Acute pancreatitis was induced by i.p. injection of caerulein (Bachem, Torrance, CA), 0.1 μg/g in filter sterilized saline, six times (every 60 minutes) daily over two consecutive days, as previously described (Keefe et al., 2012; Kopp et al., 2012; Krah et al., 2015). Controls were injected with an equal volume of sterile saline. Pancreata from all caerulein-treated mice were harvested three weeks following final injection and processed as described above.
Quantification of tissue staining:
PanIN scoring:
To measure the number of PanINs per pancreas, the entire surface area of each Alcian blue/eosin-stained section was photographed at 4x original magnification, followed by photo-merging in Adobe Photoshop. The surface area was measured using ImageJ software (NIH). Alcian blue+ PanINs were counted manually under the microscope and marked on composite images in Adobe Photoshop. PanIN burden was calculated as total number of Alcian blue+ lesions per cm2. As previously described, metaplastic lesions that did not stain with Alcian blue were not counted (Krah et al., 2015). To avoid double-counting tortuous lesions that could occupy multiple regions in 3-D space, no more than one lesion was scored within an anatomically distinct pancreatic lobule (De La O et al., 2008; Krah et al., 2015).
Quantification of immunofluorescence images:
To quantify the R26rtTA recombination frequency, we imaged 10–12 randomly selected 20x fields per specimen (across multiple sections). Using ImageJ (NIH), cell co-expressing GFP with the acinar differentiation marker, Amylase, were detected by additive image overlay with DAPI and anti-GFP, and counted using the Analyze Particles function as described previously (Keefe et al., 2012; Krah et al., 2015). To ensure counting accuracy, random images were manually spot-checked, using Adobe Photoshop. All calculations were performed in Microsoft Excel and results graphed as individuals with error bars representing the standard deviation. The p-values were determined by two-tailed, unpaired t-test in Graphpad Prism 7.
To quantify the number of GFP-positive PanINs, 10 randomly selected fields were imaged per mouse. Each PanIN was manually scored according to the number of GFP+/CK19+ cells present. If more than two cells (or regions) co-expressed CK19 and GFP, the PanIN was considered GFP-positive.
Quantification of histological images:
To quantify cell proliferation in PanINs, each lesion was scored according to its number of Ki67+ nuclei. Each lesion was categorized as low-(0–1 Ki67+ nuclei), mid- (<50% Ki67+ nuclei), or high-grade (>50% Ki67+ nuclei). At least 10 fields with PanINs were counted per animal (n=3 mice per genotype). To quantify the number of Ptf1a-negative vs. positive PanINs, as many lesions as possible were imaged from Ptf1a-stained KrasG12D and KrasG12D + tetO-Ptf1a pancreata (n=5 mice per genotype). If two or more cells contained Ptf1a+ nuclei, the PanIN was considered Ptf1a-positive. The number of βgal+ PanINs was determined by counting βgal+ cells per lesion; if the lesion contained 2 or more βgal (tetO-Ptf1a)-positive cells, the lesion was considered positive. To score reappearance of acinar cells after tetO-Ptf1a activation in PanIN-bearing mice, at least 15 high-power fields (40x objective) were scored per sample after anti-amylase immunohistochemistry, and scored positive if they contained at least one cluster of ≥ 4 amylase+ acinar cells.
RNA-seq analysis:
Datasets representing transcripts-per-million (TPM) count data from normal human organs (Mele et al., 2015; Uhlen et al., 2015) and cancer cell lines (Barretina et al., 2012; Klijn et al., 2015) were obtained from public databases, as listed in Key Resource Table. Data were filtered to include only normal pancreas or PDAC samples, and processed and analyzed using R/Bioconductor (full code available in Data S1). In brief, genes were filtered for minimal expression in either normal pancreas or PDAC, counts were quantile-normalized across samples, normal and PDAC samples were separately merged, and global and gene-specific expression was analyzed. Sample-specific counts for all genes are provided in Table S2.
Tissue culture methods:
Lentivirus production:
The Gateway cloning system (Thermo-Fisher Scientific) was used to generate a mouse Ptf1a entry vector (pENTR221-mPtf1a), via PCR and BP cloning (primer sequences in Key Resources Table). An EGFP entry vector (pME-EGFP) was the gift of Dr. Kristen Kwan (Kwan et al., 2007). These were transferred to the pCLX-pTF-R1-DEST-R2-EBR65 destination vector (a gift of Dr. Patrick Salmon, Addgene plasmid # 45952) to create DOX-inducible lentivectors. Lentivirus particles were generated by CaPO4-mediated transfection of 293T cells, with psPAX2 and pCAG-VSVG packaging plasmids (gifts of Dr. Didier Trono, Addgene plasmid # 12259, and Dr. Patrick Salmon, Addgene # 35616, respectively).
PDAC cell line culture:
Cell lines were grown in RPMI-1640 medium (Corning Life Sciences, Corning, NY) containing 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA) and 1x Penicillin-Streptomycin-Glutamine supplement (Corning). For lentiviral transduction, target cell lines were incubated overnight with viral particles at ≤ 0.1 multiplicity of infection (m.o.i.), in the presence of 8 μg/ml polybrene. After 48 hours, infected cells were selected as a polyclonal population with Blasticidin (5–10 μg/ml; Invivogen, San Diego, CA). Transgene expression was induced by culture in 1 μg/ml doxycycline (DOX).
Clonogenicity assays:
Cells were plated at 500–2000 cells/well of 6-well plate, in the presence or absence of DOX, and grown for 8–15 days with media changed every 3 days. Plating density and growth duration were kept constant between experimental groups of each cell line. For visualization, cells were fixed briefly in 70% EtOH, stained for 30 min in 0.5% (w/v) crystal violet (Millipore-Sigma) in 50% EtOH, and air-dried. Colonies comprising clusters of ≥10 cells were counted on a dissected stereo microscope.
Bulk growth assays:
Cells were plated at an initial density of 1.5–1.7×104 cells/cm2 in 35 mm dishes (identical plating density between experimental groups), with DOX treatment beginning 1 day after plating, and were split, counted and re-plated at the same density every 3 days.
RT-PCR:
RNA was harvested from growing cells using the RNeasy Mini kit (QIAGEN, Germantown, MD), and converted to cDNA with MLV reverse transcriptase and random primers (both from Thermo-Fisher). Quantitative PCR (qPCR) reactions were set up with PerfeCta SYBR Green SuperMix (Quanta Biosciences, Beverly, MA), and run on a QuantStudio 12K Flex instrument (Thermo-Fisher) in the University of Utah Genomics Core Facility. Primer sequences, obtained through PrimerBank (Wang et al., 2012), are provided in Table S1. Relative expression was determined using the ΔΔCt method (Schmittgen and Livak, 2008), using the cyclophilin A gene (PPIA) as a reference control.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics:
Statistical analyses were performed in Graphpad Prism 7 and R. A two-tailed t-test was used to calculate p-values for PanIN burden and percentage of Ptf1a-negative PanINs (Figure 2). A Fisher exact test was used to calculate p-values for nominal data, such as relative frequencies of GFP/CK19 PanINs (Figure 2 and Figure 3). Where multiple groups were compared against one another, p-values were determined by ANOVA and post-hoc t-test. All error bars, regardless of the statistical test used, indicate the standard deviation from the mean. Statistical significance was defined as a P-value of <0.05 for the indicated analysis, as determined by Graphpad Prism 7 or R.
DATA AND CODE AVAILABILITY
Data and code used for analysis of normal pancreas and PDAC cell line gene expression (Figure 7A) are provided in Data S1.
Supplementary Material
HIGHLIGHTS.
Maintained expression of differentiation factor Ptf1a blocks pancreatic tumorigenesis
Tumor-promoting inflammation cannot overcome Ptf1a-mediated tumor suppression
Ptf1a expression re-differentiates pancreatic cancer precursors to exocrine cells
Ptf1a expression inhibits growth and clonogenicity of human pancreatic cancer cells
ACKNOWLEDGMENTS
We are grateful to members of our laboratories as well as Howard Crawford, Gabrielle Kardon and Matthew Firpo for helpful input. This work was supported by the National Institutes of Health, through the following grants: F30-CA192819 (N.M.K.), R01-DK061220 and R01-CA194941 (L.C.M. and R.J.M.); U01-DK089570 (C.V.E.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, and Nagy A (2005). Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res 33, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, and Vonderheide RH (2007). Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res 67, 9518–9527. [DOI] [PubMed] [Google Scholar]
- Cobo I, Martinelli P, Flandez M, Bakiri L, Zhang M, Carrillo-de-Santa-Pau E, Jia J, Sanchez-Arevalo Lobo VJ, Megias D, Felipe I, et al. (2018). Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature 554, 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, and Pasca di Magliano M (2012). Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 122, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Yan W, Sebolt-Leopold JS, and Pasca di Magliano M (2014). MAPK signaling is required for dedifferentiation of acinar cells and development of pancreatic intraepithelial neoplasia in mice. Gastroenterology 146, 822–834 e827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La O JP, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, and Murtaugh LC (2008). Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A 105, 18907–18912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, Araki K, Ozerdem U, Simeone DM, Miller G, et al. (2018). SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discov 8, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandez M, Cendrowski J, Canamero M, Salas A, del Pozo N, Schoonjans K, and Real FX (2014). Nr5a2 heterozygosity sensitises to, and cooperates with, inflammation in KRas(G12V)-driven pancreatic tumourigenesis. Gut 63, 647–655. [DOI] [PubMed] [Google Scholar]
- Giry-Laterriere M, Cherpin O, Kim YS, Jensen J, and Salmon P (2011). Polyswitch lentivectors: “all-in-one” lentiviral vectors for drug-inducible gene expression, live selection, and recombination cloning. Hum Gene Ther 22, 1255–1267. [DOI] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, and Barbacid M (2007). Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302. [DOI] [PubMed] [Google Scholar]
- Gysin S, Paquette J, and McMahon M (2012). Analysis of mRNA profiles after MEK1/2 inhibition in human pancreatic cancer cell lines reveals pathways involved in drug sensitivity. Mol Cancer Res 10, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, et al. (2008). Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A 105, 18913–18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. (2003). Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450. [DOI] [PubMed] [Google Scholar]
- Hoang CQ, Hale MA, Azevedo-Pouly AC, Elsasser HP, Deering TG, Willet SG, Pan FC, Magnuson MA, Wright CV, Swift GH, et al. (2016). Transcriptional Maintenance of Pancreatic Acinar Identity, Differentiation, and Homeostasis by PTF1A. Mol Cell Biol 36, 3033–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubison BL, Schweickert PG, Moser SE, Yang Y, Gao H, Scully K, Itkin-Ansari P, Liu Y, and Konieczny SF (2018). Induced PTF1a expression in pancreatic ductal adenocarcinoma cells activates acinar gene networks, reduces tumorigenic properties, and sensitizes cells to gemcitabine treatment. Mol Oncol 12, 1104–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B, Tsou L, Wang H, Gaiser S, Chang DZ, Daniluk J, Bi Y, Grote T, Longnecker DS, and Logsdon CD (2009). Ras activity levels control the development of pancreatic diseases. Gastroenterology 137, 1072–1082, 1082 e1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, Degenhardt J, Mayba O, Gnad F, Liu J, et al. (2015). A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 33, 306–312. [DOI] [PubMed] [Google Scholar]
- Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris J.P.t., Pan FC, Akiyama H, Wright CV, Jensen K, et al. (2012). Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah NM, De La OJ, Swift GH, Hoang CQ, Willet SG, Chen Pan F, Cash GM, Bronner MP, Wright CV, MacDonald RJ, et al. (2015). The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah NM, and Murtaugh LC (2016). Differentiation and Inflammation: ‘Best Enemies’ in Gastrointestinal Carcinogenesis. Trends Cancer 2, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andren-Sandberg A, and Domellof L (1993). Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med 328, 1433–1437. [DOI] [PubMed] [Google Scholar]
- Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. (2015). Human genomics. The human transcriptome across tissues and individuals. Science 348, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MS, Allen P, Brentnall TA, Goggins M, Hruban RH, Petersen GM, Rao CV, Whitcomb DC, Brand RE, Chari ST, et al. (2016). Pancreatic Cancer Chemoprevention Translational Workshop: Meeting Report. Pancreas 45, 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero X, Adell T, Skoudy A, Padilla MA, Gomez JA, Chalaux E, Malagelada JR, and Real FX (2007). Pancreas transcription factor 1alpha expression is regulated in pancreatitis. Eur J Clin Invest 37, 791–801. [DOI] [PubMed] [Google Scholar]
- Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA, Bueno-de-Mesquita HB, Gallinger S, Gross M, et al. (2010). A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet 42, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N, Takeuchi KK, Ruggeri JM, Bailey P, Chang D, Li J, Leonhardt L, Puri S, Hoffman MT, Gao S, et al. (2016). PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes Dev 30, 2669–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shi G, Zhu L, Sun Y, Bettencourt R, Damsz B, Hruban RH, and Konieczny SF (2009). Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 136, 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2018). Cancer statistics, 2018. CA Cancer J Clin 68, 7–30. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- von Figura G, Morris J.P.t., Wright CV, and Hebrok M (2014). Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut 63, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, and Seed B (2012). PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 40, D1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet SG, Hale MA, Grapin-Botton A, Magnuson MA, MacDonald RJ, and Wright CV (2014). Dominant and context-specific control of endodermal organ allocation by Ptf1a. Development 141, 4385–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, Beane-Freeman L, Bracci PM, Buring J, et al. (2014). Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 46, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. (2010). Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code used for analysis of normal pancreas and PDAC cell line gene expression (Figure 7A) are provided in Data S1.