Abstract
Network science has become increasingly important in life science over the last decade. The proposed Octave and MATLAB-compatible NOCAD toolbox provides a set of methods which enables the structural controllability and observability analysis of dynamical systems. In this paper, the functionality of the toolbox is presented, and the implemented functions demonstrated.
Keywords: Dynamical systems, Complex networks, Controllability and observability analysis, Robustness, MATLAB toolbox
Introduction
In the life sciences, the determination of driver nodes in networks that play a significant role in the emergence or treatment of diseases is an intensively researched field 1. In large-scale human liver metabolic networks (HLMN), the driver metabolites have essential functions, and the role of transport reactions and extracellular metabolites in terms of controlling HLMN has revealed the importance of the environment of human liver metabolism with regard to the health of the liver 2.
In terms of controlling the human signalling network, the role of different proteins was also systematically analysed with the toolset of network controlability in 3 to highlight the role of cancer-associated genes. Target control with objective-guided optimisation (TCO) was introduced to control a set of variables (or targets) of interest while the quantity of drivers and constrained nodes were minimised and maximised, respectively. This method is capable of determining the leading phenotype transitions in biological networks that can be identified as drug targets 4. Using statistical analysis, a subset of critical control nonprotein-coding RNAs (ncRNAs) enriched by human disease can also be determined 5. In intra-cellular networks, to understand the information flow, a natural control system was utilised and the robustness of such a control was analysed 6. The importance of determining the proper driver nodes in biological networks, or more generally in any dynamical system, is unequivocal, and the amount of research concerning network science has increased rapidly. A detailed study about the control principles in biological networks has already been published 7. The network science-based analysis of dynamical systems has spread rapidly as it provides simple and efficient tools to analyse the structural controllability of any linear or linearised system 1.
Although considerable research has utilised the method 8, a flexible software tool which may be used to support the research in this field has yet to be designed. Parallel research has resulted in a collection of applications, toolboxes, plug-ins and scripts that analyse and determine several structural properties of genes, protein-protein interaction or even social or urban networks. Most of these applications only analyse the structural properties of static networks and just a handful of them utilise these structural properties to draw conclusions concerning the dynamics of the system investigated. As our toolbox belongs to the second group, in the following section, the available applications and programs of this group are elaborated.
A brief summary of the available tools with expanded functionalities is given in Table 1. Applications or software packages implemented in Python and capable of analysing the controllability and observability of dynamical systems are: graph-control 9 and WDNfinder 10. The advantage of Python-based development lies in its widespread use and the countless methods and packages implemented in this language, including the tools developed for network analysis 11. Although in Python the focus is on developing a broad software package for complex system analysis, this has yet to be fulfilled and all of the available solutions have limitations. The graph-control toolbox only analyses the impact of network topology on the number of inputs and implements the fast matching algorithm 12. Even though WDNFinder only determines the minimum driver node set (MDS) and classifies nodes based on MDS, it is incapable of facilitating extended analysis.
Table 1. Toolboxes that implement some functions for dynamical analysis of complex systems based on their structural analysis.
| Software | Language | Applied on | GUI | Ref. | Last updated |
|---|---|---|---|---|---|
| netctrl | C++ | General networks | No | 16 | January 8, 2015 |
| CONTEST | MATLAB | General networks | No | 17 | February, 2009 |
| CytoCtrlAnalyser | Java | Biomolecular networks | Yes | 13 | May 25, 2017 |
| graph-control | Python | General networks | No | 9 | December 16, 2015 |
| WDNfinder | Python | Biological networks | No | 10 | June 24, 2018 |
| enaR | R | Ecological networks | No | 15 | May 18, 2018 |
Additionally, the CytoCtrlAnalyser 13 plug-in for Cytoscape 14 has been developed, which was implemented in Java and offers graphical interfaces for users as well. It evaluates control centrality, control capacity and classifies nodes for biomolecular networks. Furthermore, the Ecological Network Analysis with R software package (enaR) provides some dynamical analysis functions and can generate models to analyse ecological networks in the R environment 15. As can be seen, both software packages deal with special kinds of networks. The netctrl program can determine the driver nodes and switchboard dynamics model for any complex network 16. CONTEST is a MATLAB toolbox which can analyse the dynamics of complex systems, but these dynamics do not cover the structural controllability and observability properties 17 of the analysed system. Although the presented software packages ensure the design of a controllable and observable system, they do not provide the opportunity to analyse the designed system exhaustively. These functions are helpful in terms of supporting the work of experts, but are insufficient for the sophisticated analysis of systems.
The contribution of this paper is to provide a novel toolbox, NOCAD 18, for the comprehensive analysis of linear or linearised dynamical systems based on the approach of network science. In the following section, the implemented functions and measurements are presented through examples of their application.
Methods
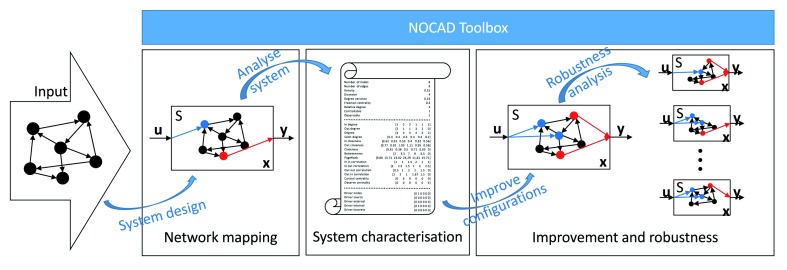
With the help of the presented Ocatave- and MATLAB-compatible toolbox, experts can create, analyse and improve any type of dynamical systems. As the structure of the dynamical systems is generally represented by their adjacency matrix and linear dynamical systems can be described by the state-space model that contains the state-transition, input, output and feedthrough matrices, the Octave/MATLAB programming language is a perfect environment to handle these matrices and provide comprehensive functionalities based on them. With the use of NOCAD 18, experts and researchers can effectively determine the input and output matrices of state-space models, calculate system-specific qualitative measurements (e.g. diameter, relative degree, control centrality and robustness of the system, etc.) and improve the system to satisfy the relative degree-based requirements. The workflow of the toolbox can be seen in Figure 1.
Figure 1. Workflow of the utilisation of the NOCAD toolbox.
The network mapping module provides two methods to create a dynamical system based on the topology of the state variables. The system characterisation module generates more than 49 measures to analyse, classify and characterise the developed system. The improvement and robustness module offers five algorithms to improve the system with additional inputs (observers) as well as outputs (controllers), and can analyse the robustness of the designed system.
The functions of the toolbox can be performed step-by-step given its modular structure. Each module has a specific task and one function from each module calls the others. A system can be analysed by calling the main functions from the modules. The advantage of this structure is its modularity as each module can be expanded easily and further modules also implemented in a simple way. A list of their functions and dependencies on each other is presented in the manual.
Implementation
According to the aforementioned approach, the implemented functions of the toolbox were divided into three modules as follows: (1) network mapping module, (2) system characterisation module and (3) improvements and robustness module.
The network mapping module creates a dynamical system from a given network structure, i.e. the necessary matrices of the state-space model are generated for the topology in such a way, that the created system is structurally controllable and structurally observable. The determination of the input and output matrices can be achieved by the path finding and signal sharing methods 19, which modify the result of the maximum matching algorithm.
The system characterisation module performs the calculation of 49 numerical measures to qualify the dynamical system based on its structure. The implemented measures, on the one hand, are well-known static measures (e.g. the number of nodes and edges, closeness and betweenness centralities), and, on the other hand, measures that characterise the dynamics of the system (e.g. structural controllability, observability, control centrality and relative degree). This module can also be used for the purpose of simple network analysis.
The improvement and robustness module integrates two main functions. On the one hand, it enables the input and output configurations of the system to be extended in such a way that the relative degree of the modified system does not exceed the initially defined threshold. For this purpose, this module implements five methods, namely the set covering-based grassroot and retrofit methods 20, the centrality measures-based method 20, the modified Clustering Large Applications based on Simulated Annealing algorithm (mCLASA), and the Geodesic Distance-based Fuzzy c-Medoid Clustering with Simulated Annealing algorithm (GDFCMSA) 20, 21. On the other hand, this module allows users to examine the robustness of the extended configurations by removing nodes from the network representation and by checking the structural controllability and structural observability of the damaged system.
The implemented methods are introduced in detail in the cited articles and the manual of the NOCAD toolbox.
Operation
In order to use the NOCAD toolbox 18, installation of Octave or MATLAB is required. Then the directories of the toolbox must be copied into the working directory, or the directories of the toolbox must be added to the paths. The functions were implemented in Octave 5.1.0 and MATLAB R2016a on a Windows 64-bit system. On other operating systems, or with other Octave or MATLAB versions, proper operation is not guaranteed. Our toolbox is independent of other MathWorks toolboxes, it uses only the octave-networks-toolbox 22 and the greedy set covering implementation 23.
Use cases
In this section, the main functionalities of the NOCAD toolbox 18 presented through examples of use cases. Although many biological networks are available from public databases, due to their complex nature, they are unsuitable for such a simple illustration. Therefore, the services of the NOCAD toolbox are presented on simple artificial networks.
The first step in each workflow is to create a state-space model from the adjacency matrix that presents the structural description of the system. This can be achieved by the use of path finding and signal sharing methods implemented in the first module. Both methods are modified versions of the maximum matching algorithm. An example of the application of the path finding method for the creation of a state-space model from the adjacency matrix ( A) is shown in Figure 2. In this figure, B denotes the resulting input matrix, C the output matrix, while D stands for representing the direct feedthrough.
Figure 2. An example network with determined input (blue) and output (red) configurations according to the path finding method.
The network represents the A state transition matrix. B denotes the input matrix in which the places of the nonzero elements are determined by the controller node allocation algorithm. Similarly, the C output matrix is defined with the observability analysis of the network of the state variables. The D matrix of the direct feedthrough contains only zeros.
As the configuration above is not complex enough to demonstrate the functions of the second module, a more complex configuration of the input and output nodes is used. The sample input and output configurations can be seen in Figure 3, where the input and the output nodes are denoted by blue and red, respectively.
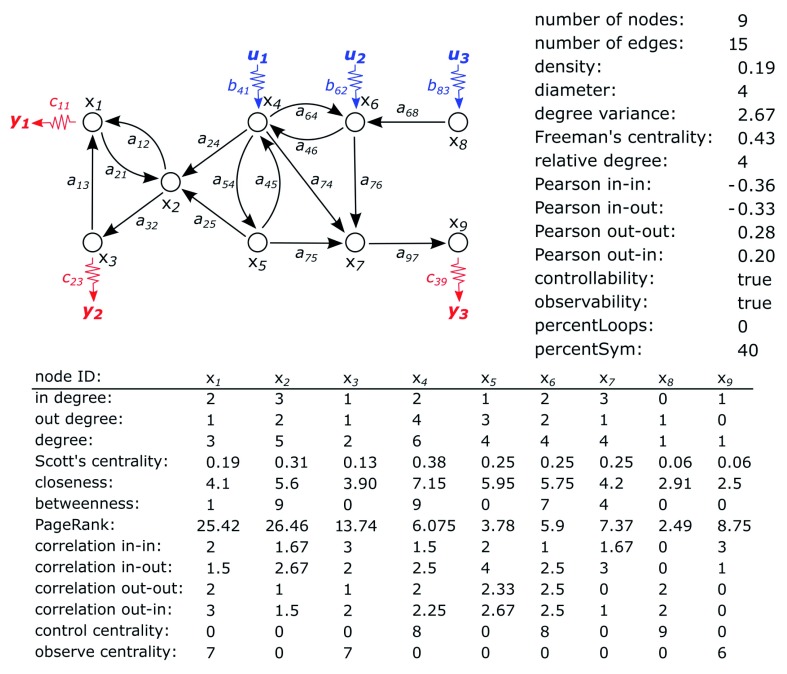
Figure 3. The complex configuration of the input and output nodes used for the demonstration of the system characterisation module of the NOCAD toolbox.
The system presented in Figure 3 consists of 9 state variables and 15 directed connections between them. Quality measures calculated by the System characterisation module of the NOCAD toolbox can be seen in Figure 4, Figure 5, and Figure 6.
Figure 4. The example network with system and node centrality measures.
Figure 5. Measures of node clustering and the representation of the topology of the network.
Figure 6. Calculated edge centrality measures for the given topology.
In Figure 4, measures qualifying the whole system with one value are presented. The density shows that the number of edges is almost a fifth of the possible maximum, and the diameter of the system (i.e. the longest shortest path in the network that presents its structure) is 4. The degree variance is 2.67, while the Freeman’s centrality is 0.43. The relative degree of the system is also 4. The Pearson coefficient shows that the in-in and in-out correlations are assortative in nature, while out-out and out-in correlations are likely to be disassortative. The system is controllable and observable. As no loop is present in the network, the percentage of loops relative to edges is 0%. As there are 6 edges that have symmetric edge pairs and the number of connections is 15, the percentage of the symmetric edge pairs relative to the edges is 40%.
Node centrality measures assigned to the state variables of the system are also presented in Figure 4. One of the most important values is the highest degree of the nodes, which belongs to state variable x 4. As Scott’s centrality is a normalised degree, the most important node is once again x 4. The closeness of node x i is calculated as the ratio of the number of nodes reachable from x i to the sum of their distances from x i. The higher value indicates the more central position of the node, and, once again, node x 4 is the most central element. The betweenness centrality shows how many shortest paths intercept the given node. If a node has a high value, then it is a critical node in the structure. The highest value belongs to nodes x 2 and x 4. The PageRank assigns a percentage value for each node, based on their centrality roles if Markov-chains are modelled. The measure referred to as correlation shows the proportion of the number of edges of neighbours’ and the number of neighbours. This information is useful when determining the assortativity of the system. The control centrality and observe centrality measures determine how many state variables can be influenced or observed by the nodes.
In Figure 5, the first vectors (referred to as driver and sensor nodes) show the driver and sensor nodes as logical vectors. The following four vectors classify these nodes as source, external, internal and inaccessible driver and sensor nodes. These types of nodes are introduced in 24 in detail. In the next section of the figure, the controlling and observing matrices are presented. Generally, these matrices are sparse matrices, as only the columns of drivers and sensors contain nonzero values. In Figure 5, we converted them into row vectors for their appropriate visualisation. The values show the number of derivations necessary to influence or observe a state variable in the system. Next, the similarity of the driver and sensor nodes is presented. The similarity of driver nodes x 4 and x 6 is 0.81. In this case, the reason why it is less than 1 is that although they control the same set of nodes, the numbers of derivations that influence them are different. In terms of sensor similarity, sensor nodes x 2 and x 3 observe the same set of nodes and they do this almost simultaneously, so their similarity is 0.91. R and R are the simple reachability matrices. They show which nodes can be controlled or observed by a given node. In R , the i th column shows which nodes can control node i. From the other viewpoint, elements in row i highlight those nodes which can be controlled by node i. In this example, node x 8 can influence every node, but it does not guarantee structural controllability. The R matrix can be interpreted analogously with regard to observability.
Finally, measures of edge centrality are seen in Figure 6. The betweenness has the same meaning as in the case of nodes, that is, it yields the number of shortest paths that intercept the edge 25. From this perspective, the most critical edge is the edge a 46 with a value of 10. The endpoint similarity shows how similar the influenced and observed sets of the state variables with regard to the endpoints of edges are. This metric has a high value if the edge is part of a cycle or creates a bridge in the network. As no bridges are present in this network, only cycles can be recognised by this measure. The edge similarity shows how similar the roles of edges are, and it allows redundancies, to be located. In the topology presented, nodes x 1, x 2 and x 3, or nodes x 4, x 5, x 6 and x 7 also create parts of the network that possess redundancy.
For the demonstration of the last module, configurations provided by the first module are used again ( Figure 2). Results provided by this module can be seen in Figure 7. In this case, five methods were applied to the system to extend the configuration as follows: the required relative degree was set at 2, while the alpha parameter of the cost function was set at 0.5 21. Results show that all the methods determine the same set of driver nodes for the system, that is, they are sufficient to influence state variables x 4 and x 8. The resultant cost is 1.5556, the relative degree is 2 which satisfies the requirements, and the mean of the relative degrees is 1.1111. In this configuration, six different nodes can be identified which can be damaged separately and the system remains controllable. This is expressed by the value of robustness (66.6%). The most important nodes in terms of controllability are x 2, x 4 and x 7. In the case of observability, methods yield different solutions with the exception of the centrality measures-based and mCLASA algorithms which provide the best configuration in this case. Although the cost as well as the maximum and mean of the relative degree were identical in the case of retrofit set covering-based and GDFCMSA methods as well, the robustness analysis of these configurations exhibits a higher degree of vulnerability.
Figure 7. Improvement and robustness analysis of the system.
Conclusions
In this article the Octave- and MATLAB-compatible NOCAD toolbox 18 was proposed to support the network-based controllability and observability analysis of dynamical systems. The toolbox offers two methods to design a structurally controllable and observable system based on the state-transition matrix. The designed system can be analysed by 49 qualitative measures both from structural and dynamical points of view. The toolbox serves five methods to improve the designed system by adding new inputs and outputs to it, thus, its relative degree can be decreased. Then the robustness of the individual designs can also be evaluated. The modular structure of the toolbox supports the facile improvement of the modules by adding new functions and the toolbox can be extended by new modules as well. Even though the modules are built on each other, most of their functions can also be used independently from each other.
Although our goal in this paper is to draw the attention of researchers of life sciences to the services provided by the NOCAD toolbox, it can be utilised in practice in various fields of sciences as well, for example, it enables social networks to be controlled in the economy, transaction networks to be analysed in finance or dynamical systems to be designed in engineering.
Data availability
All data underlying the results are available as part of the article and no additional source data are required.
Software availability
Source code available from: https://github.com/abonyilab/NOCAD.
Archived source code at time of publication: https://doi.org/10.5281/zenodo.2656674 18
License: GNU General Public License v3.0
Author contributions
Dániel Leitold reviewed the literature on network science, developed the algorithms, implemented the Octave and MATLAB functions, designed as well as performed the experiments, and wrote the related sections. Ágnes Vathy-Fogarassy participated in the formalisation of the methodology. János Abonyi developed the algorithms, implemented the Octave and MATLAB functions and proofread the paper.
Funding Statement
This research was supported by the National Research, Development and Innovation Office NKFIH, through the project OTKA-116674 (Process mining and deep learning in the natural sciences and process development) and the EFOP-3.6.1- 16-2016- 00015 Smart Specialization Strategy (S3) Comprehensive Institutional Development Program. Dániel Leitold was supported by the ÚNKP-18-3 New National Excellence Program of the Ministry of Human Capacities.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved with reservations]
References
- 1. Liu YY, Slotine JJ, Barabási AL: Controllability of complex networks. Nature. 2011;473(7346):167–73. 10.1038/nature10011 [DOI] [PubMed] [Google Scholar]
- 2. Liu X, Pan L: Detection of driver metabolites in the human liver metabolic network using structural controllability analysis. BMC Syst Biol. 2014;8(1):51. 10.1186/1752-0509-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu X, Pan L: Identifying driver nodes in the human signaling network using structural controllability analysis. IEEE/ACM Trans Comput Biol Bioinform. 2015;12(2):467–72. 10.1109/TCBB.2014.2360396 [DOI] [PubMed] [Google Scholar]
- 4. Guo WF, Zhang SW, Shi QQ, et al. : A novel algorithm for finding optimal driver nodes to target control complex networks and its applications for drug targets identification. BMC Genomics. 2018;19(Suppl 1):924. 10.1186/s12864-017-4332-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nacher JC, Akutsu T: Controllability methods for identifying associations between critical control ncrnas and human diseases. Methods Mol Biol.In Computational Biology of Non-Coding RNA.2019;1912:289–300. 10.1007/978-1-4939-8982-9_11 [DOI] [PubMed] [Google Scholar]
- 6. Ravindran V, Nacher JC, Akutsu T, et al. : Network controllability analysis of intracellular signalling reveals viruses are actively controlling molecular systems. Sci Rep. 2019;9(1): 2066. 10.1038/s41598-018-38224-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li M, Gao H, Wang J, et al. : Control principles for complex biological networks. Brief Bioinform. 2018. 10.1093/bib/bby088 [DOI] [PubMed] [Google Scholar]
- 8. Liu YY, Barabási AL: Control principles of complex systems. Rev Mod Phys. 2016;88(3): 035006. 10.1103/RevModPhys.88.035006 [DOI] [Google Scholar]
- 9. Chaturvedi V: Controllability of networks. Reference Source [Google Scholar]
- 10. Chu Y, Wang Z, Wang R, et al. : Wdnfinder: A method for minimum driver node set detection and analysis in directed and weighted biological network. J Bioinform Comput Biol. 2017;15(5):1750021. 10.1142/S0219720017500214 [DOI] [PubMed] [Google Scholar]
- 11. Zinoviev D: Recognize-Construct-Visualize-Analyze-Interpret. Pragmatic Bookshelf.2018. Reference Source [Google Scholar]
- 12. Faradonbeh MKS, Tewari A, Michailidis G: Optimality of fast-matching algorithms for random networks with applications to structural controllability. IEEE Trans Control Netw Syst. 2017;4(4):770–780. 10.1109/TCNS.2016.2553366 [DOI] [Google Scholar]
- 13. Wu L, Li M, Wang J, et al. : Cytoctrlanalyser: a cytoscape app for biomolecular network controllability analysis. Bioinformatics. 2018;34(8):1428–1430. 10.1093/bioinformatics/btx764 [DOI] [PubMed] [Google Scholar]
- 14. Shannon P, Markiel A, Ozier O, et al. : Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borrett SR, Lau MK: enaR: an R package for ecosystem network analysis. Methods Ecol Evol. 2014;5(11):1206–1213. 10.1111/2041-210X.12282 [DOI] [Google Scholar]
- 16. Nepusz T, Vicsek T: Controlling edge dynamics in complex networks. Nat Phys. 2012;8(7):568–573. 10.1038/nphys2327 [DOI] [Google Scholar]
- 17. Taylor A, Higham DJ: Contest: A controllable test matrix toolbox for matlab. ACM Trans Math Softw. 2009;35(4):26 10.1145/1462173.1462175 [DOI] [Google Scholar]
- 18. Abonyi J: abonyilab/nocad v2.0.2019. 10.5281/zenodo.2656674 [DOI] [Google Scholar]
- 19. Leitold D, Vathy-Fogarassy Á, Abonyi J: Controllability and observability in complex networks–the effect of connection types. Sci Rep. 2017;7(1): 151. 10.1038/s41598-017-00160-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leitold D, Vathy-Fogarassy Á, Abonyi J: Evaluation of the complexity, controllability and observability of heat exchanger networks based on structural analysis of network representations. Energies. 2019;12(3):513 10.3390/en12030513 [DOI] [Google Scholar]
- 21. Leitold D, Vathy-Fogarassy A, Abonyi J: Network distance-based simulated annealing and fuzzy clustering for sensor placement ensuring observability and minimal relative degree. Sensors (Basel). 2018;18(9): pii: E3096. 10.3390/s18093096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bounova G: Octave networks toolbox.2015. Reference Source [Google Scholar]
- 23. Gori F, Folino G, Jetten MS, et al. : MTR: taxonomic annotation of short metagenomic reads using clustering at multiple taxonomic ranks. Bioinformatics. 2011;27(2):196–203. 10.1093/bioinformatics/btq649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruths J, Ruths D: Control profiles of complex networks. Science. 2014;343(6177):1373–1376. 10.1126/science.1242063 [DOI] [PubMed] [Google Scholar]
- 25. Freeman LC: A set of measures of centrality based on betweenness. Sociometry. 1977;40(1):35–41. 10.2307/3033543 [DOI] [Google Scholar]