Abstract
Background:
Antigen-specific T cell immune responses play a pivotal role in resolving acute and chronic hepatitis C virus (HCV) infections. Currently, no prophylactic or therapeutic vaccines against HCV are available. We previously demonstrated the preclinical potency of therapeutic HCV vaccines based on recombinant Semliki Forest virus (SFV) replicon particles. However, clinical trials do not always meet the high expectations of preclinical studies, thus, optimization of vaccine strategies is crucial. In efforts to further increase the frequency of HCV-specific immune responses in the candidate SFV-based vaccines, the authors assessed whether inclusion of three strong, so-called universal helper T cell epitopes, and an endoplasmic reticulum localization, and retention signal (collectively termed sigHELP-KDEL cassette) could enhance HCV-specific immune responses.
Methods:
We included the sigHELP-KDEL cassette in two of the candidate SFV-based HCV vaccines, targeting NS3/4A and NS5A/B proteins. We characterized the new constructs in vitro for the expression and stability of the transgene-encoded proteins. Their immune efficacy with respect to HCV-specific immune responses in vivo was compared with the parental SFV vaccine expressing the corresponding HCV antigen. Further characterization of the functionality of the HCV-specific CD8+ T cells was assessed by surface and intracellular cytokine staining and flow cytometry analysis.
Results:
Moderate, but significantly, enhanced frequencies of antigen-specific immune responses were achieved upon lower/suboptimal dosage immunization. In optimal dosage immunization, the inclusion of the cassette did not further increase the frequencies of HCV-specific CD8+ T cells when compared with the parental vaccines and the frequencies of effector and memory populations were identical.
Conclusion:
We hypothesize that the additional effect of the sigHELP-KDEL cassette in SFV-based vaccines depends on the immunogenicity, nature, and stability of the target antigen expressed by the vaccine.
Keywords: cancer vaccine, helper epitopes, hepatitis C virus, immunotherapy, Semliki Forest virus
Introduction
Hepatitis C virus (HCV) infection is a major cause of liver disease and because approximately 2 million people are newly infected annually,1 it represents a significant global health problem. Upon infection, HCV can persist in the majority of immune-competent hosts, leading to a state of chronic hepatic inflammation. Ultimately this chronic inflammation can progress to severe liver diseases, including hepatic cirrhosis and hepatocellular carcinoma.1–3 Recently, the standard-of-care treatment options for HCV improved with the approval of novel HCV-specific direct-acting antiviral drugs (DAAs), offering interferon-free regimens with very high curative rates.4 However, despite their therapeutic potency, DAAs are associated with a number of side effects including drug–drug interactions, development of resistance-associated variants,4,5 and importantly they do not induce protective immunity in patients.6–8 Therefore, it remains a compelling argument for the development of not only prophylactic but also therapeutic candidate vaccines against HCV in order to lower the chronicity rate and the disease burden.
A strong, broad, and persistent antigen-specific T cell response plays a pivotal role during acute and chronic HCV infection and is required for spontaneous viral clearance.9 Preclinical studies in mice and chimpanzees have used different vaccination strategies against HCV, based on, for example, HCV peptides, recombinant yeast or viral vectors, autologous dendritic cells loaded with lipopeptides, or plasmid DNA.10,11 Aiming to induce HCV-specific immunity, the authors developed therapeutic vaccines based on recombinant Semliki Forest virus (SFV) replicon particles. The authors demonstrated that SFV-based immunization targeting HCV nonstructural proteins (nsPs), which are highly immunogenic12 and genetically conserved,13,14 results in the induction of polyfunctional effector and memory CD8+ T cells and delays tumor growth of HCV nsPs-expressing tumors in vivo.15
However, as the outcomes of clinical trials do not always meet the high expectations based on the preclinical studies, the optimization of vaccine strategies is required. A strategy to improve vaccine potency, while not increasing or inducing toxicity, is the conjugation of antigens to nontoxic carrier proteins.16 The mechanism of enhancement depends on the biological function of the carrier protein involved. That occurs mainly through the provision of CD4+ T cell help, which is essential for the induction and maintenance of memory CD8+ T cell responses17–19 and/or by affecting antigen expression and stability,20 therefore, modulating antigen uptake and presentation by the antigen-presenting cells (APCs). Oosterhuis and colleagues previously demonstrated that the expression of a series of pan-helper T (Th) cell epitopes in a DNA-based vaccine against human papillomavirus (HPV) stimulated both Th cells and cytotoxic T lymphocytes (CTLs).21 In addition, inclusion of endoplasmic reticulum (ER) targeting signals within these DNA vaccines increased antigen stability and the combination of both modifications strongly enhanced the immunogenicity of their DNA vaccine.22 The authors previously demonstrated that the inclusion of Th and ER targeting signals (collectively termed sigHELP-KDEL cassette) also significantly enhanced the immunogenicity of an SFV vaccine expressing HPV early proteins, resulting in higher frequencies of functional HPV-specific CD8+ T cells and greater antitumor activity at a very low vaccine dosage.23
In the present study, the authors aim to assess the effect of sigHELP-KDEL inclusion in the efficacy of SFV-based HCV vaccines. Therefore, the authors combined this antigen design for two of the candidate SFV-based HCV vaccines, targeting NS3/4A and NS5A/B proteins. The new constructs were characterized in vitro and their immune efficacy with respect to HCV-specific immune responses in vivo was compared with the parental SFV vaccine expressing the corresponding HCV antigen.
Materials and methods
Cell lines
Baby hamster kidney cells (BHK-21, ATCC #CCL-10), were cultured in RPMI1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Lonza, Basel, Switzerland), 100 U/ml penicillin, and 100 µg/ml streptomycin (Life technologies). Hepa1-6VenusNS5A/B,15 Hepa1-6VenusnsPs,15 EL4VenusNS5A/B15, and EL4 cells were cultured in DMEM with GLUTAMAX (Life Techno-logies) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. All cell lines were cultured at 37°C with 5%CO2.
Construction of SFV replicon vectors
Construction of pSFV-NS3/4A (12,839 bps) and pSFV-NS5A/B (13,700 bps) has been previously described.15 To generate the pSFV-sHELP-NS3/4A and pSFV-sHELP-NS5A/B constructs, a series of Th epitopes (HELP), ER localization signal (sig), HCV NS3/4A or NS5A/B antigens and ER retention signal (KDEL) were cloned into an SFV vector.15 The BssHII-sigHELP-NotI and the BssHII-sigHELP-XhoI fragments were amplified by PCR using the pVAX1-sigHELP-E7SHKDEL vector22 (kindly provided by K. Oosterhuis, J.B. Haanen and T.N. Schumacher, Netherlands Cancer Institute, Amsterdam, the Netherlands), as a template and ligated into the pSFVe vector,15 to generate pSFV-sHELP. Subsequently, pSFV-sHELP was linearized with NotI or XhoI restriction digestion and ligated to the NotI-NS3/4A-KDEL-NotI or the XhoI-NS5A/B-KDEL-XhoI insert fragments to generate pSFV-sHELP-NS3/4A (13,173 bps) and pSFV-sHELP-NS5A/B (14,217 bps) respectively. The inserts were amplified by PCR from the plasmid DNA containing the full-length cDNA of HCV H77 genotype 1a consensus sequence (H/FL) (kindly provided by Charles M. Rice, via Apath, LLC (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: p90HCVconsensuslongpU),24 and the four amino acid sequence KDEL was synthesized by PCR. PCR primers were synthesized by Eurogentec (Maastricht, the Netherlands). All restriction enzymes were purchased from Thermo Fisher Scientific (Landsmeer, the Netherlands). Correct DNA sequences were verified by Sanger sequencing analysis.
Production, purification and titer determination of recombinant SFV particles
The production, purification and titer determination of SFV particles were performed as previously described.25BHK-21 were co-electroporated with in vitro transcribed RNA encoding for the SFV replicase and the transgene (HCV antigens) simultaneously with a helper RNA encoding for the structural proteins of SFV, at a molar ratio 1:1. Transfected BHK-21 cells were cultured at 30°C, 5% CO2 for 48 h to produce SFV particles. The supernatant containing SFV particles was collected and purified on a discontinuous sucrose density gradient. Purified SFV particles were titrated on BHK-21 cells, using a polyclonal rabbit antireplicase (nsP3) antibody (kindly provided by Dr T. Ahola). Before use, the SFV particles were activated with α-chymotrypsin (Sigma, St Louis, USA) to cleave the mutated p62 spike protein. SFV particles were stored at –80°C.
Protein expression and stability by [35S] - methionine/cysteine pulse labeling
BHK-21 cells (5 × 105 cells per well in 6-well plates) were incubated with 5 × 106 infectious SFV replicon particles (SFV-NS3/4A, SFV-sHELP-NS3/4A, SFV-NS5A/B or SFV-sHELP-NS5A/B) or without virus. After 6 hours incubation, the supernatant was removed and the cells were washed with PBS. The cells were further cultured in L-methionine and L-cysteine-free DMEM for 30 min following the addition of [35S]-methionine/cysteine labeling mix (0.37 Mbq/well) (PerkinElmer, Groningen, the Netherlands). After 1 h of [35S]-methionine/cysteine labeling, the cells were washed with PBS and further cultured in a medium supplemented with 5 mM L-methionine and L-cysteine. After 1, 6, 18, or 24 h post [35S]-methionine/cysteine labeling, the cells were washed with cold PBS and lysed with TENT-SDS lysis buffer (1% SDS, 50 mM Tris-HCl, 5 mM EDTA, 150 mM NaCl, 0.5% Triton-X, pH 7,5) containing 0.2 mM phenyl-methane-sulphonyl-fluoride. The cell lysates were analyzed by 12% SDS-PAGE and autoradiography. Relative remaining expression of the transgene-encoded proteins was calculated with ImageJ analysis software.
Mice
Specific pathogen-free female inbred C57BL/6JOLaHsd mice (H-2b) were obtained from a commercial vendor (Harlan CPB, Zeist, the Netherlands). All mice were 8–10 weeks of age at the onset of the experiments. Housing and animal experiments were approved by the local Animal Experimentation Ethical Committee (the Institutional Animal Care and Use Committee of the University Medical Center Groningen, DEC number: 6405).
Prime-boost SFV immunizations
Mice were primed and boost immunized intramuscularly (i.m.) at a 2 weeks interval with a dose of 5 × 106 or 1 × 105 infectious SFV particles (SFV-NS3/4A, SFV-sHELP-NS3/4A, SFV-NS5A/B or SFV-sHELP-NS5A/B) in 50 µl (25 µl/thigh muscle) under anesthesia (isoflurane/O2). For negative controls, the same volume of PBS was injected.
Detection and phenotypic analysis of HCV NS3-specific CD8+ T cells
After removal of erythrocytes, peripheral blood cells and splenocytes were stained with HCV-NS3603-611-PE dextramers (Immudex, Copenhagen, Denmark) for 10 min at room temperature. T cell phenotypes were characterized with the following antibodies: anti-CD8-PE-Cy7 (clone 53–6.7), anti-CD44-PerCp-Cy5.5 (clone IM7), anti-CD62L-APC (clone Mel-14), and anti-CD127-eFluor450 (clone A7R34). All antibodies were obtained from eBioscience (Vienna, Austria). Surface staining was performed at 4°C for 20 min. Propidium Iodide (eBioscience) or DAPI (Thermo Fischer Scientific) were used as live/dead cell exclusion. Cells were washed twice in FACS buffer (PBS containing 0.5% bovine serum albumin) and samples were analyzed using an LSR-II cytometer (BD Biosciences). Data were analyzed using FlowJo analysis software (Tree Star).
Detection of IFN-γ and degranulation of HCV-specific CD8+ T cells
Splenocytes isolated from immunized mice were stimulated with 10 µg/ml of NS5A synthetic peptide (ILDSFDPL, H-2Db) in the presence of 1 µg/ml anti-CD28 (clone: PV-1, Bioceros, Utrecht, the Netherlands), anti-CD107a-efluor660 (clone: eBio1D4B), and anti-CD107b-efluor660 (clone: eBioABL-93) in round-bottom tubes cultured at an angle at 37°C with 5% CO2. Brefeldin A (1 mg/ml) was added to the cultures after 1 h and the cells were further incubated for 4 h. Next, the cells were harvested, washed and stained with LIVE/DEAD fixable violet dead cell stain kit (Life Technologies), followed by surface staining with antiCD8a- PE- Cy7 (clone: 53–6.7), at 4°C for 20 min. Cells were then fixed in 4% paraformaldehyde, permeabilized with Perm/Wash buffer (BD Biosciences) and stained intracellularly with anti-IFN-γ- PerCP- Cy5.5 (clone: XMG 1.2) at 4°C for 30 min. All antibodies were purchased from eBiosciences. FACs analysis was performed using an LSR-II flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Tree Star).
51Cr release assay (BULK CTL assay)
Hepa1-6VenusNS5A/B and Hepa1-6VenusnsPs (stimulator cells) were cultured in the presence of 50 U/ml recombinant IFN-γ (Peprotech, London, UK) for 48 h. The stimulator cells were then irradiated (100 Gy) and co-cultured with effector cells (splenocytes isolated from naïve immunized mice) at a ratio of 1:25 in a T25 flask at 37°C with 5% CO2. 4 U/ml of recombinant human IL-2 (Peprotech, London, UK) was added on day 3 and 5 of the stimulator-effector cell co-cultures. After 7 days of culture, the effector cells (splenocytes) were harvested and co-cultured with 51Chromium (51Cr)-labeled target cells (EL4-pulsed with GAVQNEVTL or ILDSFDPL, or EL4VenusNS5A/B). Target cells were cultured in the presence of 50 U/ml recombinant murine IFN-γ 48 h prior to co-culture with the effector cells. Target cells were labeled with 51Cr (100 µCi/2 × 106 cells) (PerkinElmer, Groningen, the Netherlands) in the presence or absence of synthetic peptides (10 µg/ml) for 1 h at 37°C. Co-culture of effector and target cells was performed in 96 well plates at four different E:T ratios in triplicates at 37°C with 5%CO2 for 4 h. Subsequently, supernatants were harvested and 51Cr release from the cells was analyzed with a RiaStar manual gamma counter (Packard, Meriden, CT). The percentage of cytotoxicity was calculated according to the formula: % specific release = [(experimental release – spontaneous release)/(maximum release – spontaneous release)] count per minute (c.p.m).
Proliferation assay carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling
Splenocytes isolated from immunized mice were labeled with 5 µM CFSE ( Life Technologies) in PBS at 37°C for 10 min in the dark. Labeled cells were washed twice with PBS and cultured in medium only or with irradiated (100 Gy) Hepa1-6VenusnsPs cells at a ratio of 25:1 for 4 days at 37°C, 5% CO2. After 4 days of incubation, the cells were harvested and stained with anti-CD8-PE-Cy7 (clone 53–6.7, e Biosciences) at 4°C for 20 min. Cells were then washed twice in FACS buffer (PBS containing 0.5% bovine serum albumin) and analyzed with LSR-II flow cytometer (BD Biosciences). Data were analyzed using FlowJo analysis software (Tree Star).
Statistical analysis
Data were analyzed with GraphPad Prism 6.0 software. Differences between two groups were determined with the Mann–Whitney U test. p values of <0.05 were considered as statistically significant.
Results
Construction and characterization of SFV vectors encoding for HCV antigens and universal T helper epitopes
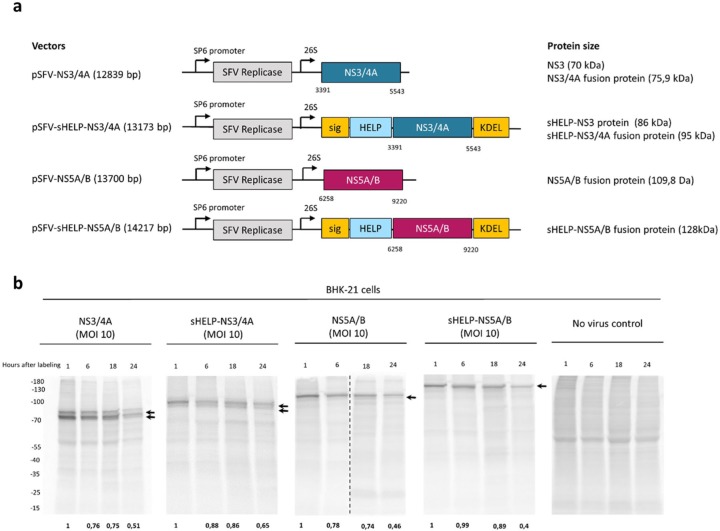
The authors previously reported that the inclusion of external CD4 Th epitopes and ER localization/retention signals in SFV replicon vectors targeting HPV could further enhance vaccine immunogenicity.23 Aiming to assess whether that modification may further augment the immunogenicity of SFV-based vaccines, the authors modified two of the previously developed SFV replicon-based HCV vaccines. The original vaccines express the nonstructural proteins NS3/4A or NS5A/B and induce potent T cell responses in vivo.15 A series of external Th epitopes that promiscuously bind to multiple MHC class II alleles were inserted, that is, the TTFC P30 pan DP epitope, the PADRE pan DR epitope and an HIV NEF pan DQ epitope, collectively termed HELP.22 In addition, a human growth hormone signaling peptide (sig)26 and a KDEL sequence27,28 were inserted to enable localization and retention of the antigen within the ER. Thus, the plasmid SFV DNA vectors pSFV-sHELP-NS3/4A (13,173 bp) and pSFV-sHELP-NS5A/B (14,217 bp) were constructed (Figure 1a) and the corresponding SFV replicon particles (SFV-sHELP-NS3/4A and SFV-sHELP-NS5A/B) were produced. Before testing the efficacy of these SFV replicon vectors in vivo, the expression and stability of the transgene-encoded proteins were characterized in vitro (Figure 1b). Infection of BHK-21 cells with SFV vectors encoding for sHELP-NS3/4A or NS3/4A resulted in expression of the sHELP-NS3/4A fusion protein (95 kDa) and NS3/4A protein (75.9 kDa) respectively, while two proteins of a smaller size, sHELP-NS3 (86 kDa) and NS3 (70 kDa) are generated upon cleavage of NS3 and NS4A by the HCV NS3 protease. In addition, BHK-21 cells infected with SFV vectors encoding for sHELP-NS5A/B or NS5A/B produced sHELP-NS5A/B (128 kDa) and NS5A/B (109.8 kDa) fusion proteins. Transgene-encoded HCV proteins were detectable already at 8 h post-SFV infection and up to 24 h, as indicated by [35S]-methionine/cysteine labeling (Figure 1b). Protein expression was also determined by western blotting stained with anti-NS3 and anti-NS5A antibodies (data not shown). Insertion of sigHELP-KDEL resulted in marginally enhanced stability of the HCV proteins. Of note, all the transgene-encoded HCV proteins are relatively stable up to 24 h, thus the sigHELP-KDEL modification did not have a pronounced effect in increasing the stability of these proteins, but it rather hindered the cleavage of the NS3/4A fusion protein by the virus protease, as shown by the total remaining transgene-encoded protein expression compared to time point 1 h post labeling (Figure 1b). The sigHELP-KDEL cassette also did not increase the amount of the transgene-encoded proteins and it resulted in similar expression levels of proteins when compared with cells infected with the parental SFV-based HCV replicons (Figure 1b).
Figure 1.
Expression and stability of HCV NS3/4A and NS5A/B proteins in vitro. (a) Schematic representation of the plasmid SFV DNA vectors and the corresponding antigens (NS3/4A or NS5A/B). The numbers indicate the nucleotide position in the plasmid DNA containing the full genome of HCV 1a (H/FL). SP6 promoter; 26S, subgenomic 26S promoter. (b) BHK-21 cells were incubated with SFV replicon particles encoding for NS3/4A, sHELP-NS3/4A, NS5A/B, sHELP-NS5A/B or without SFV at an MOI of 10. Cells were pulsed with [35S]-methionine/cysteine for 1 h after incubation with the SFV particles for 6 h. Cell lysates were collected at 1, 6, 18, and 24 h post [35S]-methionine/cysteine pulse-chase labeling. Radioactively labeled proteins were revealed by autoradiography after 12% SDS-PAGE. Vertical dotted lines separate different gels that run at the same time. Arrows indicate the expression of the corresponding transgene-encoded proteins. Numbers at the bottom of the gels show remaining expression of the total transgene-encoded proteins when compared with their expression at time point 1 h after labeling. Data represent results from two independent experiments.
Frequencies and phenotypes of HCV-specific CD8+ T cells induced by SFV immunizations
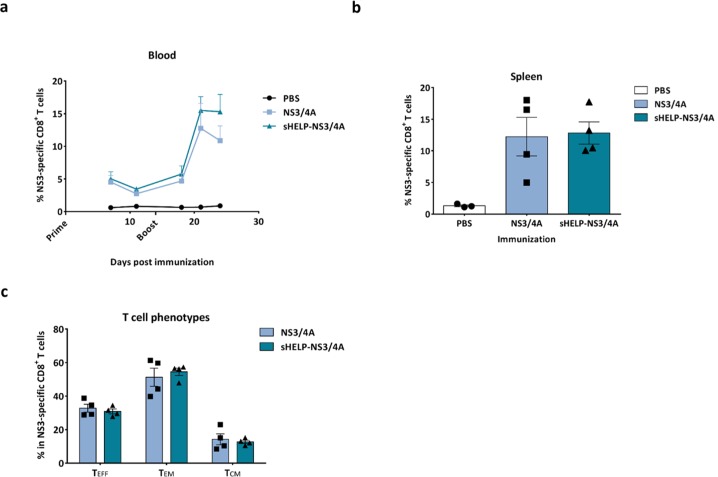
The authors then determined the immunogenicity of the SFV-based HCV vaccines with or without the sigHELP-KDEL modification in vivo. Naïve mice were primed and boosted intramuscularly at a 2 week interval with an optimal dose of 5 × 106 infectious SFV particles encoding for HCV antigens and sigHELP-KDEL fusion protein. The immune efficacy of the modified vaccines was compared to the parental SFV-based vaccines without the insertion of the Th epitopes series. The authors focused the initial analysis on T cell responses against an immunodominant epitope within the NS3 protein (NS3603-611), which is known as a strong binder for MHC class I, H-2Db molecule, and is detectable using different immunization approaches.15,29 Thus, the authors evaluated the frequencies, the kinetics, and the phenotype of NS3-specific CD8+ T cells upon immunization with SFV-NS3/4A or SFV-sHELP-NS3/4A replicon particles (Figure 2). The frequency of NS3-specific CD8+ T cells in peripheral blood of immunized mice on days 7, 11, 18, 21, and 24 after SFV immunizations were measured (Figure 2a). Surprisingly, the percentage of NS3-specific cells in the total CD8+ blood cells of mice immunized with SFV-sHELP-NS3/4A at all time points assessed was similar to the frequency observed after immunization with SFV-NS3/4A. All mice were sacrificed 10 days after the last immunization (day 24), and the frequency of NS3-specific CD8+ splenocytes was determined (Figure 2b). Consistent with the findings in blood, both vaccines induced equal numbers of NS3-specific CD8+ T cells. Aiming to further investigate whether the sigHELP-KDEL cassette may have altered the frequencies of the different CD8+ subsets, the authors further characterized the phenotypes of the vaccination-induced NS3-specific CD8+ T cells in the spleen. Notably, equal frequencies of effector (TEFF), effector memory (TEM) and central memory (TCM) NS3-specific CD8+ T cells were induced (Figure 2c).
Figure 2.
Frequencies and phenotypes of NS3-specific CD8+ T cells induced by SFV immunizations. Naïve mice were primed and boosted immunized intramuscularly at a 2 week intervals with 5 × 106 infectious particles of SFV-NS3/4A or SFV-sHELP-NS3/4A. (a) Kinetics of the percentage of NS3-specific cells in the total CD8+ blood cells. (b) Mice were sacrificed on day 24 after SFV immunizations and the percentage of NS3-specific cells in the total CD8+ splenocytes was determined. (c) NS3-specific CD8+ T cell subsets induced upon SFV immunization. Surface markers for the different subsets: effector T cells (TEFF) CD44+CD62L–CD127–, effector memory T cells (TEM) CD44+CD62L–CD127+, central memory T cells (TCM) CD44+CD62L+CD127+. The percentages of each T cell subset in the total NS3-specific CD44+CD8+ splenocytes are shown. Data represent mean + or ± SEM (n = 3–4).
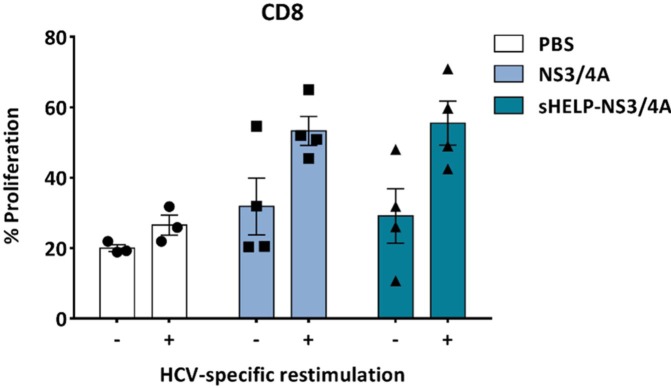
In addition, the proliferative capacity of the HCV-specific T cells upon antigen restimulation was assessed by a CFSE dilution assay. Splenocytes isolated from control mice or mice immunized twice with SFV-NS3/4A or SFV-sHELP-NS3/4A replicon particles were labeled with CFSE and cultured in the presence or absence of Hepa1-6VenusnsPs,15 a murine hepatoma cell line expressing all nonstructural proteins of HCV, for 4 days at a 25:1 ratio. Antigenic restimulation promoted the proliferation of CD8+ T cells from SFV immunized mice (Figure 3), but not CD4+ T cells (data not shown), when compared with control PBS injected mice. The effect was irrespective of the addition of the sigHELP-KDEL cassette in the SFV vaccines.
Figure 3.
The proliferation of HCV-specific T cells upon in vitro restimulation. Splenocytes isolated from SFV immunized mice were labeled with CFSE and cultured in the presence (+) or absence (–) of irradiated Hepa1-6VenusnsPs cells at a 25:1 ratio for 4 days. Subsequently, the percentage of proliferating cells was analyzed by flow cytometry after staining with anti-CD8a. The bars represent the percentage of proliferating daughter CFSE+CD8+ cells (sum of generation 1–7) within the total CFSE+CD8+ cells (sum of generations 0–7). Data represent mean ± SEM (n = 3–4).
The effect of the sigHELP-KDEL cassette on low dosage SFV immunization and responses to less immunodominant HCV antigens
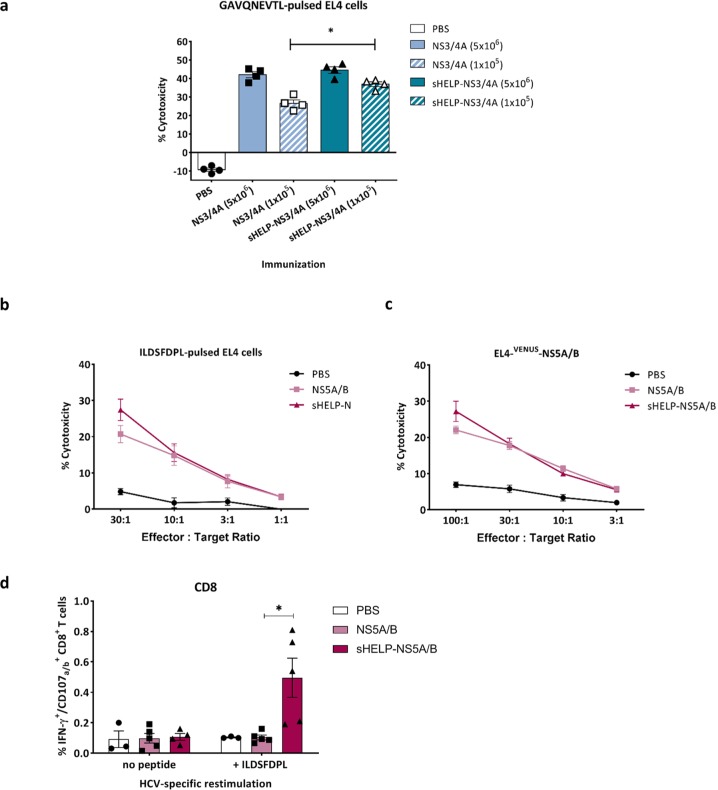
Immunization with an optimal dose of 5 × 106 particles of SFV-NS3/4A induces a very potent T cell response, as demonstrated by the high frequency of 13% NS3-specific CD8+ T cells in the spleen (at day 7 post-immunization (Figure 2a). Therefore, the effect of sigHELP-KDEL cassette inclusion might not be detectable, since the NS3-specific T cell responses may have already reached an optimal level that cannot be further upregulated. To further assess the contribution of sigHELP-KDEL in HCV vaccines, the authors studied its effect, using a suboptimal immunization dose, that is, 105 infectious particles (Figure 4a). HCV-specific CTL activity was measured based on the cytotoxic activity of restimulated NS3-specific CTLs in a 51Cr-release CTL assay. As expected, the optimal dose of SFV immunizations with 5 × 106 infectious particles induced higher HCV-specific cytotoxicity as compared to suboptimal SFV immunization. At the low, suboptimal dose, inclusion of sigHELP-KDEL cassette induced a slight, but significantly higher cytotoxic activity against GAVQNETL-pulsed EL4 cells, when compared with the parental vaccine (Figure 4a), while no effect of the sigHELP-KDEL inclusion was observed upon optimal dose immunization.
Figure 4.
Induction of HCV-specific CTLs upon SFV immunizations. Mice were primed and boosted intramuscularly with (a) 5 × 106 or 105 SFV-NS3/4A or SFV-sHELP-NS3/4A infectious particles, (b) to (d) 5 × 106 SFV-NS5A/B or SFV-sHELP-NS5A/B infectious particles, or PBS at a 2 week interval. Mice were sacrificed 10 days after the last immunization, and splenocytes were isolated for an in vitro cytotoxicity assay (a) to (c) and intracellular cytokine staining using flow cytometry (d). (a) Isolated splenocytes were cultured with Hepa1-6VenusnsPs cells at a ratio of 25:1. After a 7 day culture, splenocytes were co-cultured for 4 h with 51Cr labeled target EL4 cells pulsed with GAVQNEVTL peptide at 25:1 ratio. (b) and (c) Isolated splenocytes were cultured with Hepa1-6VenusNS5A/B cells at a ratio of 25:1. After a 7 day culture, splenocytes were co-cultured for 4 h with 51Cr labeled target EL4 cells pulsed with ILDSFDPL peptide or EL4-VENUS-NS5A/B at the indicated E:T ratios. (d) Splenocytes were stimulated with ILDSFDPL for 4 h and subjected to surface and intracellular cytokine staining. The frequencies of IFN–γ+CD107a/b+ within the CD8+ T cells are shown. Data represent mean ± SEM (n = 3–5). *p < 0.05.
Further, the authors aimed to assess whether the inclusion of sigHELP-KDEL enhances immune responses against other HCV antigens, including NS5A/B proteins that are generally known to contain less immunodominant epitopes, unlike the HCV NS3 protein. Mice were prime boosted with 5 × 106 infectious particles of SFV-NS5A/B or SFV-sHELP-NS5A/B. The cytotoxic activity of effector T cells, induced upon immunization, was measured after restimulation with Hepa1-6VenusNS5A/B cells, a murine hepatoma cell line that expresses the whole NS5A/B fusion protein of HCV.15 Both SFV vaccines induced HCV-specific CTLs that killed target cells expressing either the ILDSFDPL epitope of NS5A15 (Figure 4b) or the whole NS5A/B fusion protein (Figure 4c). The cytotoxic potential after immunization with SFV-sHELPNS5A/B was slightly higher, yet not significantly different. HCV-specific CTLs induced against the NS5A/B protein of HCV, most likely target the ILDSFDPL epitope30 as indicated by the equal level of cytotoxicity induced against the two different target cell lines (Figure 4b and c). Of note, immunization with SFV-sHELPNS5A/B increased significantly the frequencies of polyfunctional CD8+ T cell subsets that produce IFN-γ and CD107a/b upon HCV-specific stimulation with ILDSFDPL peptide, when compared with the parental vaccine (Figure 4d).
Discussion
Preclinical studies suggest that CD8+ CTLs are the key effector cells required for HCV clearance.31,32 Taking into account the importance of CD4+ T cell help with respect to CTL differentiation, in this study, the authors aimed to apply the sigHELP-KDEL22,23,33 modification to two of the HCV candidate vaccines and assess whether inclusion of sigHELP-KDEL cassette can enhance the HCV-specific T cell responses.
The authors generated SFV-based vaccines encoding for NS3/4A and NS5A/B antigens of HCV with or without the addition of the fusion protein, sigHELP-KDEL and compared the immune efficacy of these different SFV-based vaccines in vivo. Inclusion of universal helper T cell epitopes and the re-localization of antigens did not increase the frequencies of HCV-specific CD8+ T cells when compared with the parental vaccines, while the frequencies of effector and memory populations were equal upon immunization at the optimal dose (5 × 106 infectious particles) for both HCV antigens. However, upon lower dosage immunization (105 infectious particles) the authors observed a minor yet significant increase in the cytotoxic activity of the NS3-specific CD8+ T cells induced upon immunization with the SFV vaccine encoding for sHELP-NS3/4A (Figure 4a). This finding, in combination with the high frequency of NS3-specific CD8+ T cells (13% on day 7, Figure 2a) upon optimal dose SFV immunization, confirms that the NS3-specific T cell proliferative response may have reached such a high level that insertion of the sigHELP-KDEL cassette cannot further augment the antigen-specific responses. It is probable that in mouse models the contribution of sigHELP-KDEL in potent cancer vaccines, such as SFV, encoding immunodominant epitopes, can only be demonstrated in low suboptimal dose immunizations regimes.
One of the main factors for the chronification of the HCV infection is the loss of antiviral effector functions of virus-specific CD8+ T cells, known as T cell exhaustion. A number of studies assessed the role of extrinsic factors, including the reduced CD4+ T cell help in virus-specific CD8+ T cell exhaustion.34–36 Thus, for rational design of HCV immunotherapies, sufficient CD4+ T cell help is crucial and antigen modifications such as the sigHELP-KDEL cassette that provides external CD4+ T cell help22 might have a beneficial effect for clinical translation. This pan-HELP cassette contains three ‘universal’ CD4+ T cell helper peptides that are known to promiscuously bind multiple MHC class II alleles across the population, thus in the clinical setting these peptides are likely to be beneficial mainly through the use of pre-existing immunity.37–39 All, or part, of these epitopes have been used in preclinical studies as a strategy to enhance vaccine immune responses mainly as assessed by humoral immunity augmentation40 but a limited number of studies have shown the potential of the strategy to enhance CTL responses.21–23
The authors previously demonstrated that the efficacy of SFV-based HPV vaccines is influenced by the stability of the transgene expressed23 and that inclusion of sigHELP-KDEL further stabilized these HPV proteins. The authors observed that insertion of sigHELP-KDEL into the HCV vaccines resulted in minor enhanced stability of the NS3/4A or NS5A/B proteins. However, as these proteins are by nature already rather stable antigens anchored to ER membranes,41 this small effect on stability could be another or an additional, explanation on why immunization with sigHELP-KDEL targeting HCV nsPs is as efficacious as SFV immunization without sigHELP-KDEL.
The role of CD4+ T cell help with respect to memory CTL differentiation is crucial. However, in several infection models, the primary CTL response appears to be CD4+ T helper cell-independent, since it is generally considered that CD4+ T cell help is only required when pattern recognition receptors or other inflammatory signals are lacking.42,43 Consistent with this, in weakly adjuvanted vaccination strategies, such as DNA vaccination, the external CD4+ T cell help is required to qualitatively improve the CTL responses.21,22,33 In cancer setting CTLs primed with adequate CD4+ T cells help acquire intrinsic properties for overcoming potential resistance hurdles.44,45 SFV-based vaccines represent a rather self-adjuvanted vaccination strategy, where external CD4+ T cell help might not be as necessary. In addition, clinical studies suggest that the universality of helper epitopes such as PADRE are highly dependent on the dose, regimen, route of immunization, as well as the nature of the targeted antigen.46,47 Nevertheless, further analysis of the activation of CD4+ T cells is required to demonstrate if immunization with SFV vaccines including the sigHELP-KDEL cassette actually results in a stronger CD4+ T cell response as a consequence of the inclusion of external Th epitopes.
The minimal enhancing effect of sigHELP-DEL in the SFV-NS3/4A vaccine is probably due to the presence of the immunodominant NS3 epitope (NS3603-611), which is a strong binder for MHC class I H-2Db molecule. This epitope was detected not only in these studies15 but also in other immunization approaches.29 The strong T cell response against this epitope could be partially explained due to the presence of Th epitopes located at the 5′ end of the CTL epitope, as predicted in the Immune Epitope Database and analysis resource (IEDB) (Supplementary Table 1). Several immunodominant tumor and viral antigens contain Th and CTL epitopes in close proximity. This colocalization of Th and CTL epitopes appears to be positively correlated to the dominancy of the specific CTL epitope.48,49 Potentially, such strong responses cannot be further enhanced. This is also different in the HPV vaccine. In this case, T cell responses against the E7 protein are mainly against the E749-57 CTL epitope, which according to prediction algorithms is not in close proximity to Th epitopes and also has a much lower binding affinity to MHC class I molecules when compared with NS3603-611.
In summary, in this preclinical study, optimal dose SFV immunization targeting HCV antigens NS3/4A or NS5A/B with or without sigHELP-KDEL induced equal levels of HCV-specific CD8+ T cells with functional effector and memory phenotypes, suggesting that the inclusion of sigHELP-KDEL in the candidate HCV vaccines does not further augment HCV-specific T cell responses in this MHC setting. This result contrasts with the authors previous finding showing that the insertion of sigHELP-KDEL in SFV vaccines targeting HPV can enhance CTL responses.23 The difference may be attributed to the differences in antigenic nature of the antigens expressed, influencing epitope binding affinity and immunodominance, and possibly the presence of internal Th epitopes as well as the stability of the protein. Understanding the mechanism(s) responsible for the enhancing effect of sigHELP-KDEL will be useful for future development of therapeutic vaccines. Clinically, it is to be expected that the HLA phenotype of the patient will probably dictate if sigHELP-KDEL modifications of SFV vaccines will further augment vaccine efficacy.
Supplemental Material
Supplemental material, Supplementary_Table_1_1 for Alphavirus-based hepatitis C virus therapeutic vaccines: can universal helper epitopes enhance HCV-specific cytotoxic T lymphocyte responses? by Georgia Koutsoumpli, Peng Peng IP, Ilona Schepel, Baukje Nynke Hoogeboom, Annemarie Boerma and Toos Daemen in Therapeutic Advances in Vaccines and Immunotherapy
Acknowledgments
The author’s thanks to Charles M. Rice (The Rockefeller University) for providing the H/FL plasmid. The authors thank Koen Ooosterhuis, John B. Haanen, Ton N. Schumacher (Netherlands Cancer Institute, Amsterdam, the Netherlands) for providing the pVAX1-sigHELP-E7SHKDEL vector.
Footnotes
Author’s Note: Toos Daemen is a cofounder of ViciniVax, a spinoff company from the UMCG developing therapeutic cancer vaccines.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project was funded by the Graduate School of Medical Sciences (GSMS), University Medical Center Groningen (UMCG), Groningen, the Netherlands.
Conflict of interest statement: The other authors declare no conflict of interest.
ORCID iD: Georgia Koutsoumpli  https://orcid.org/0000-0003-2615-0413
https://orcid.org/0000-0003-2615-0413
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Georgia Koutsoumpli, Department of Medical Microbiology, Tumor Virology and Cancer Immunotherapy, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Peng Peng Ip, Department of Medical Microbiology, Tumor Virology and Cancer Immunotherapy, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Ilona Schepel, Department of Medical Microbiology, Tumor Virology and Cancer Immunotherapy, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Baukje Nynke Hoogeboom, Department of Medical Microbiology, Tumor Virology and Cancer Immunotherapy, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Annemarie Boerma, Department of Medical Microbiology, Tumor Virology and Cancer Immunotherapy, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Toos Daemen, Department of Medical Microbiology, Tumor Virology and Cancer Immunotherapy, University of Groningen, University Medical Center Groningen, Antonius Deusinglaan 1, Groningen, 9713 AV, the Netherlands.
References
- 1. World Health Organization. Global hepatitis report 2017. Geneva: World Health Organization, http://apps.who.int/iris/bitstream/handle/10665/255017/WHO-HIV-2017.06-eng.pdf?sequence=1 (2017, accessed 19 June 2018). [Google Scholar]
- 2. Tu T, Bühler S, Bartenschlager R. Chronic viral hepatitis and its association with liver cancer. Biol Chem 2017; 398: 817–837. [DOI] [PubMed] [Google Scholar]
- 3. Bartenschlager R, Baumert TF, Bukh J, et al. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res 2018; 248: 53–62. [DOI] [PubMed] [Google Scholar]
- 4. Spengler U. Direct antiviral agents (DAAs) - a new age in the treatment of hepatitis C virus infection. Pharmacol Ther 2018; 183: 118–126. [DOI] [PubMed] [Google Scholar]
- 5. de Leuw P, Stephan C. Protease inhibitor therapy for hepatitis C virus-infection. Expert Opin Pharmacother 2018; 19: 577–587. [DOI] [PubMed] [Google Scholar]
- 6. Grebely J, Prins M, Hellard M, et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect Dis 2012; 12: 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trembling PM, Tanwar S, Rosenberg WM, et al. Treatment decisions and contemporary versus pending treatments for hepatitis C. Nat Rev Gastroenterol Hepatol 2013; 10: 713–728. [DOI] [PubMed] [Google Scholar]
- 8. Sarrazin C, Isakov V, Svarovskaia ES, et al. Late relapse versus hepatitis C virus reinfection in patients with sustained virologic response after sofosbuvir-based therapies. Clin Infect Dis 2017; 64: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holz L, Rehermann B. T cell responses in hepatitis C virus infection: historical overview and goals for future research. Antiviral Res 2015; 114: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ip PP, Nijman HW, Wilschut J, et al. Therapeutic vaccination against chronic hepatitis C virus infection. Antiviral Res 2012; 96: 36–50. [DOI] [PubMed] [Google Scholar]
- 11. Guo X, Zhong JY, Li JW. Hepatitis C virus infection and vaccine development. J Clin Exp Hepatol 2018; 8: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Missale G, Bertoni R, Lamonaca V, et al. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest 1996; 98: 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 2005; 42: 962–973. [DOI] [PubMed] [Google Scholar]
- 14. Powdrill MH, Bernatchez JA, Götte M. Inhibitors of the hepatitis C virus RNA-dependent RNA polymerase NS5B. Viruses 2010; 2: 2169–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ip PP, Boerma A, Regts J, et al. Alphavirus-based vaccines encoding nonstructural proteins of hepatitis C virus induce robust and protective T-cell responses. Mol Ther 2014; 22: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin K, Roosinovich E, Ma B, et al. Therapeutic HPV DNA vaccines. Immunol Res 2010; 47: 86–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu A, Zeng Q, Kang TH, et al. Innovative DNA vaccine for human papillomavirus (HPV)-associated head and neck cancer. Gene Ther 2011; 18: 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arashkia A, Roohvand F, Memarnejadian A, et al. Construction of HCV-polytope vaccine candidates harbouring immune-enhancer sequences and primary evaluation of their immunogenicity in BALB/c mice. Virus Genes 2010; 40: 44–52. [DOI] [PubMed] [Google Scholar]
- 19. Pishraft-Sabet L, Kosinska AD, Rafati S, et al. Enhancement of HCV polytope DNA vaccine efficacy by fusion to an N-terminal fragment of heat shock protein gp96. Arch Virol 2015; 160: 141–152. [DOI] [PubMed] [Google Scholar]
- 20. Huang CY, Chen CA, Lee CN, et al. DNA vaccine encoding heat shock protein 60 co-linked to HPV16 E6 and E7 tumor antigens generates more potent immunotherapeutic effects than respective E6 or E7 tumor antigens. Gynecol Oncol 2007; 107: 404–412. [DOI] [PubMed] [Google Scholar]
- 21. Oosterhuis K, Ohlschläger P, van den Berg JH, et al. Preclinical development of highly effective and safe DNA vaccines directed against HPV 16 E6 and E7. Int J cancer 2011; 129: 397–406. [DOI] [PubMed] [Google Scholar]
- 22. Oosterhuis K, Aleyd E, Vrijland K, et al. Rational design of DNA vaccines for the induction of human papillomavirus Type 16 E6- and E7-specific cytotoxic T-cell responses. Hum Gene Ther 2012; 23: 1301–1312. [DOI] [PubMed] [Google Scholar]
- 23. Ip PP, Boerma A, Walczak M, et al. Antigen design enhances the immunogenicity of Semliki Forest virus-based therapeutic human papillomavirus vaccines. Gene Ther 2015; 22: 560–567. [DOI] [PubMed] [Google Scholar]
- 24. Kolykhalov AA, Agapov EV, Blight KJ, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 1997; 277: 570–574. [DOI] [PubMed] [Google Scholar]
- 25. Daemen T, Regts J, Holtrop M, et al. Immunization strategy against cervical cancer involving an alphavirus vector expressing high levels of a stable fusion protein of human papillomavirus 16 E6 and E7. Gene Ther 2002; 9: 85–94. [DOI] [PubMed] [Google Scholar]
- 26. Martial JA, Hallewell RA, Baxter JD, et al. Human growth hormone: complementary DNA cloning and expression in bacteria. Science 1979; 205: 602–607. [DOI] [PubMed] [Google Scholar]
- 27. Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell 1987; 48: 899–907. [DOI] [PubMed] [Google Scholar]
- 28. Rivera VM, Wang X, Wardwell S, et al. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science 2000; 287: 826–830. [DOI] [PubMed] [Google Scholar]
- 29. Ahlén G, Nyström J, Pult I, et al. In vivo clearance of hepatitis C virus nonstructural 3/4A–expressing hepatocytes by DNA vaccine–primed cytotoxic T lymphocytes. J Infect Dis 2005; 192: 2112–2116. [DOI] [PubMed] [Google Scholar]
- 30. Holmstrom F, Pasetto A, Nahr V, et al. A synthetic codon-optimized hepatitis C virus nonstructural 5A DNA vaccine primes polyfunctional CD8+ T cell responses in wild-type and NS5A-transgenic mice. J Immunol 2013; 190: 1113–1124. [DOI] [PubMed] [Google Scholar]
- 31. Shoukry NH, Grakoui A, Houghton M, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med 2003; 197: 1645–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nitschke K, Luxenburger H, Kiraithe MM, et al. CD8+ T-cell responses in hepatitis B and C: the (HLA-) A, B, and C of hepatitis B and C. Dig Dis 2016; 34: 396–409. [DOI] [PubMed] [Google Scholar]
- 33. Ahrends T, Spanjaard A, Pilzecker B, et al. CD4+ T cell help confers a cytotoxic T cell effector program including coinhibitory receptor downregulation and increased tissue invasiveness. Immunity 2017; 47: 848–861.e5. [DOI] [PubMed] [Google Scholar]
- 34. Cabrera R, Tu Z, Xu Y, et al. An immunomodulatory role for CD4+CD25+ regulatory T lymphocytes in hepatitis C virus infection. Hepatology 2004; 40: 1062–1071. [DOI] [PubMed] [Google Scholar]
- 35. Boettler T, Spangenberg HC, Neumann-Haefelin C, et al. T cells with a CD4+CD25+ regulatory phenotype suppress in vitro proliferation of virus-specific CD8+ T cells during chronic hepatitis C virus infection. J Virol 2005; 79: 7860–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wieland D, Hofmann M, Thimme R. Overcoming CD8+ T-cell exhaustion in viral hepatitis: lessons from the mouse model and clinical perspectives. Dig Dis 2017; 35: 334–338. [DOI] [PubMed] [Google Scholar]
- 37. Panina-Bordignon P, Tan A, Termijtelen A, et al. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol 1989; 19: 2237–2242. [DOI] [PubMed] [Google Scholar]
- 38. Raju R, Navaneetham D, Okita D, et al. Epitopes for human CD4+ cells on diphtheria toxin: structural features of sequence segments forming epitopes recognized by most subjects. Eur J Immunol 1995; 25: 3207–3214. [DOI] [PubMed] [Google Scholar]
- 39. Diethelm-Okita BM, Okita DK, Banaszak L, et al. Universal epitopes for human CD4+cells on tetanus and diphtheria toxins. J Infect Dis 2000; 181: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 40. Fraser CC, Altreuter DH, Ilyinskii P, et al. Generation of a universal CD4 memory T cell recall peptide effective in humans, mice and non-human primates. Vaccine 2014; 32: 2896–2903. [DOI] [PubMed] [Google Scholar]
- 41. Popescu CI, Rouillé Y, Dubuisson J. Hepatitis C virus assembly imaging. Viruses 2011; 3: 2238–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bevan MJ. Helping the CD8+ T-cell response. Nat Rev Immunol 2004; 4: 595–602. [DOI] [PubMed] [Google Scholar]
- 43. Castellino F, Germain RN. Cooperation between CD4+ and CD8+ T cells: when, where, and how. Annu Rev Immunol 2006; 24: 519–540. [DOI] [PubMed] [Google Scholar]
- 44. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013; 39: 1–10. [DOI] [PubMed] [Google Scholar]
- 45. Borst J, Ahrends T, Bąbała N, et al. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018; 18: 635–647. [DOI] [PubMed] [Google Scholar]
- 46. Wierecky J, Müller MR, Wirths S, et al. Immunologic and clinical responses after vaccinations with peptide-pulsed dendritic cells in metastatic renal cancer patients. Cancer Res 2006; 66: 5910–5918. [DOI] [PubMed] [Google Scholar]
- 47. Chu CS, Boyer J, Schullery DS, et al. Phase I/II randomized trial of dendritic cell vaccination with or without cyclophosphamide for consolidation therapy of advanced ovarian cancer in first or second remission. Cancer Immunol Immunother 2012; 61: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paul S, Piontkivska H. Frequent associations between CTL and T-Helper epitopes in HIV-1 genomes and implications for multi-epitope vaccine designs. BMC Microbiol 2010; 10: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun X, Fujiwara M, Shi Y, et al. Superimposed epitopes restricted by the same HLA molecule drive distinct HIV-specific CD8+ T cell repertoires. J Immunol 2014; 193: 77–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Table_1_1 for Alphavirus-based hepatitis C virus therapeutic vaccines: can universal helper epitopes enhance HCV-specific cytotoxic T lymphocyte responses? by Georgia Koutsoumpli, Peng Peng IP, Ilona Schepel, Baukje Nynke Hoogeboom, Annemarie Boerma and Toos Daemen in Therapeutic Advances in Vaccines and Immunotherapy