Abstract
Aging is a complex process emerging from integrated physiological networks. Recent work using principal component analysis (PCA) of multisystem biomarkers proposed a novel fundamental physiological process, “integrated albunemia,” which was consistent across human populations and more strongly associated with age and mortality risk than individual biomarkers. Here we tested for integrated albunemia and associations with age and mortality across six diverse nonhuman primate species and humans. PCA of 13 physiological biomarkers recovered in all species a primary axis of variation (PC1) resembling integrated albunemia, which increased with age in all but one species but was less predictive of mortality risk. Within species, PC1 scores were often reliably recovered with a minimal biomarker subset and usually stable between sexes. Even among species, correlations in PC1 structure were often strong, but the effect of phylogeny was inconclusive. Thus, integrated albunemia likely reflects an evolutionarily conserved process across primates and appears to be generally associated with aging but not necessarily with negative impacts on survival. Integrated albunemia is unlikely to be the only conserved emergent physiological process; our findings hence have implications both for the evolution of the aging process and of physiological networks more generally.
Keywords: Biomarkers, Physiological networks, Mortality risk, Nonhuman models, Principal component analysis
Aging is a complex process emerging from the interplay among multiple mechanisms and the breakdown in functioning of integrated physiological networks (1–5). This integrated nature underlies the hypothesized effects of several key phenomena associated with aging, such as physiological dysregulation (2,6–9), metabolic syndrome (10), and inflamm-aging (11,12). Understanding the broad causes and consequences of this process is an ongoing challenge, and a specific issue is to find appropriate measures that reflect the proposed integrative mechanisms (13,14). The complex-systems view of aging suggests that single biomarkers or single-system measures will incompletely reflect the aging process. Indeed, evidence shows that single biomarkers can be highly variable across populations, making them difficult to use as general indicators of aging (15). Yet most research on aging to date has examined single or few biomarkers of interest at a time. Recent work has increasingly sought multivariate approaches that can better capture the emergent patterns of physiological processes and their effects on aging, providing new insight into the aging process (9,14,16–18).
Recently, “integrated albunemia” was proposed as a novel physiological process implicated in aging in human populations (19). This phenomenon emerged as the primary axis of variation from principal component analysis (PCA) of 43 biomarkers and included markers of anemia, inflammation, and low albumin and calcium. Integrated albunemia consistently increased with age and predicted mortality and frailty across different populations and demographic groups. Notably, integrated albunemia was not predictable a priori from how single biomarkers or systems changed independently with age, it comprised biomarkers from multiple physiological systems, and it was more stable across populations than single biomarkers. Accordingly, it was proposed that integrated albunemia is an emergent physiological process, a stable epiphenomenon of physiological network structure, potentially implicating coordination of distant sub-networks (19). Other potential emergent physiological processes related to aging include inflamm-aging (11,20) and metabolic syndrome (10,21). A separate subsequent study using a different human dataset and different methods independently recovered a set of biomarkers aligned with integrated albunemia as predictors of mortality (22). Thus, integrated albunemia seems promising as an emergent physiological process associated with aging and risk of mortality, but further studies are needed to establish its wider implications.
In particular, we do not know if integrated albunemia is phylogenetically broad or specific to humans, nor how conserved physiological network characteristics are more generally. Further tests are needed to determine whether this is a promising direction for fundamental research in aging and if certain model organisms are useful. Nonhuman primates are important research models of aging and longevity and particularly useful for comparisons with humans due to similarities in fundamental biology (23–26). Research on nonhuman primates can provide important insights into broad patterns of primate aging and potential interventions (24). However, most studies on nonhuman primates are restricted to a few commonly studied species rather than cross-species comparisons, and few tackle ideas involving physiological integration (but see ref. (27)).
In the current study, we test for the presence of emergent physiological processes, in particular integrated albunemia, and associations with aging and mortality across primates. We further ask if cross-species similarity in emergent physiological processes is related to phylogenetic proximity. We apply a previously used PCA approach (19) to longitudinal biomarker datasets from several species of nonhuman primates and one human population. We hypothesize that integrated albunemia represents a fundamental process involved in aging across the primate lineage. If so, we should recover a similar primary axis of variation (markers of anemia and low albumin and calcium) associated with increased age and mortality risk across all or most species, and we would predict similarity of PCA axes to be stronger among more closely related species and diminish with phylogenetic distance. Alternatively, integrated albunemia might have more limited phylogenetic scope and only be present in species more closely related to humans. Finally, integrated albunemia might be a phenomenon specific to humans. More broadly, we examine if there are underlying physiological processes resulting from integrated regulatory networks that are evolutionarily conserved across species.
Method
Data Sets and Biomarker Selection
Our study included data on humans and six nonhuman primates: chimpanzees (Pan troglodytes), rhesus macaques (Macaca mulatta), pig-tailed macaques (Macaca nemestrina), common marmosets (Callithrix jacchus), Coquerel’s sifaka (Propithecus coquereli), and ring-tailed lemurs (Lemur catta) (Table 1). These six species span a range of taxonomic groupings and expected maximum lifespans. We acquired nonhuman primate biomarker data from the Internet Primate Aging Database (iPAD; https://www.ipad.primatedata.com/) (28), developed at the Wisconsin National Primate Center and now managed by CléMetric, LLC (Madison, WI). All observations came from nonexperimental captive animals considered healthy at the time of sampling, and research centers contributing to iPAD were approved and accredited by the relevant oversight bodies (USDA, AAALAC, EU Directives). Most nonhuman primate data came from a single research center; the exceptions were chimpanzees (three centers) and rhesus macaques (four centers). We acquired human biomarker data from a large-scale longitudinal population-based study, Invecchiare in Chianti (InCHIANTI), of older persons from the Chianti region of Tuscany, Italy (29). Use of this data was originally approved by ethics committees at the respective institutions responsible for data collection, and approval of secondary analysis came from the Comité d’éthique de la recherche sur l’humain du CHUS (project #14–059). We only considered adult (sexually mature) individuals in the study, and we determined adult age limits for each species from AnAge: the Animal Ageing and Longevity Database (http://genomics.senescence.info/species/) (30).
Table 1.
Primate Species Information
| Common Name | Scientific Name | Abbreviation | Species Group | IDs | % Female | Lifespan* | Ages-Female† | Ages-Male† |
|---|---|---|---|---|---|---|---|---|
| Humans | Homo sapiens | Human | Great Apes | 1304 | 55.6 | 122.5 | 21.3–100.9 | 23.4–97.2 |
| Chimpanzee | Pan troglodytes | Chimp | Great Apes | 400 | 56.3 | 59.4 | 9.6–57.9 | 9.6–43.6 |
| Rhesus Macaque | Macaca mulatta | Rhesus | Old World Monkeys | 178 | 48.3 | 40.0 | 5.6–33.4 | 5.6–31.6 |
| Pig-tailed Macaque | Macaca nemestrina | Pigtail | Old World Monkeys | 92 | 59.8 | 37.6 | 8.1–32.6 | 8.4–30.1 |
| Common Marmoset | Callithrix jacchus | Marmoset | New World Monkeys | 74 | 44.6 | 22.8 | 1.6–9.4 | 1.6–9.1 |
| Coquerel’s Sifaka‡ | Propithecus coquereli | Sifaka | Lemurs | 32 | 40.6 | 31.0 | 2.6–17.1 | 2.6–26.6 |
| Ring-tailed Lemur | Lemur catta | Ringtail | Lemurs | 50 | 44.0 | 37.3 | 2.6–24.1 | 3.1–22.6 |
Notes: IDs = number of individuals.
*Confirmed maximum records from AnAge (http://genomics.senescence.info/species/), in years.
†Age ranges represented in the current data, in years.
‡Lifespan not available so estimated from records for congener Verraux’s sifaka (Propithecus verreauxi).
Biomarker availability varied among species, and we chose species to include based on data availability for multivariate analyses (number of markers, sample size, longitudinal samples, mortality data, etc.). In particular, we sought to optimize a balance between maximizing overall sample size and availability of similar biomarkers across species. We started with the 43 biomarkers used in the previous study in humans (19) and checked for availability and number of individuals with complete observations of biomarker subsets. A challenge in the cross-species comparisons is that inflammatory markers (C-reactive protein and interleukin-6) were stable components of human integrated albunemia (19), but were generally absent from primate data. We were thus constrained to estimate integrated albunemia without this inflammatory component. For each species, we removed observations that were biologically improbable based on previously published values for each species or taxonomic group or visible outliers outside of recorded ranges. We chose biomarkers for analysis based on availability in sufficient sample sizes across species, while avoiding redundancy. For example, we excluded hematocrit, which was always correlated with hemoglobin at r > .9, and we used neutrophil/lymphocyte ratio instead of neutrophil counts because neutrophils and lymphocytes together make up the great majority of white blood cells. The final 13 biomarkers span a range of physiological systems (Table 2). For more details on biomarkers availability and choice, see Additional Methods Details in Supplementary Material.
Table 2.
Physiological Biomarkers Used in Principal Component Analysis
| Biomarker (units) | System/Function | Abbreviation |
|---|---|---|
| Hemoglobin (g/dL)* | Blood | hb |
| Red blood cells (106/mm3)* | Blood | rbc |
| Mean corpuscular hemoglobin (pg) | Blood | mch |
| Mean corpuscular hemoglobin concentration (g/dL) | Blood | mchc |
| White blood cells (mm3) | Immune response | wbc |
| Lymphocytes (103/mm3) | Immune response | lympho_ct |
| Neutrophil/Lymphocyte ratio | Immune response | n/l |
| Calcium (mg/dL)* | Electrolytes | ca |
| Potassium (mmol/L) | Electrolytes | k |
| Albumin (g/dL)* | Proteins, liver, kidney | alb |
| Alkaline phosphatase (IU/L) | Proteins, liver, kidney | alkp |
| Creatinine (mg/dL) | Proteins, liver, kidney | creat |
| Total protein (g/dL)* | Proteins, liver, kidney | tot_prot |
Note: *Used in the five-biomarker subset analysis.
Analysis
Principal Components Analysis
Preliminary analyses indicated that individuals differed consistently in biomarker and principal component values in all species, so we sought to avoid repeated observations biasing our analysis towards better-sampled individuals. Therefore, we generated 1,000 samples of the complete dataset, each with one randomly sampled observation per adult (sexually mature) animal, ran PCA on each sample, and used the average loadings from these samples for subsequent analyses (eg, we looked for associations with age and mortality with PC1 characterized by these averaged loadings).
We first performed PCA on all biomarkers for each species separately, using observations on all individuals for each species. Before PCA, we log- or square-root-transformed all variables to the best approximation of normality and standardized all variables to mean 0 and standard deviation of 1. We focused interpretation and comparison on the first three principal components (PCs), which usually explained ~50% or more of the variance (Supplementary Tables S1 and S2). Note, however, that for PCA in complex biological systems, even low-variance axes often contain important biological information (31).
We next ran a PCA on all biomarkers with all species combined. To account for extreme differences in species sample sizes, we took the number of observations from the species with the fewest (Coquerel’s sifaka, 32 individuals), randomly sampled 32 observations for each of the other species, and ran the PCA on this combined-species subsample (this sampling was done after first sampling one observation per individual). We tested for average species differences in PC values with multilevel models of each PC with fixed effect of species and random effect of ID (to account for uneven sampling and repeated observations), followed by Tukey’s honest significant differences (HSD) post hoc tests to examine pairwise species differences in estimated marginal means for PC1.
Associations With Age and Mortality
The primary axis of variation was hypothesized to be integrated albunemia, so we focused on PC1, calculated from PCA for each species separately, in statistical models of associations with age and mortality risk. We modeled the fixed effects of age, sex, and their two-way interaction on PC1 using linear hierarchical models. When the interaction was not significant, we interpreted the effect of age on PC1 for both sexes together from a model with only the fixed effects of age and sex. The continuous variable “age” was centered to 0 on the sample mean and scaled to 1 SD (within species) to account for different lifespans across species. We included a random intercept for individual to account for repeated observations. For species that came from more than one population (chimpanzees and rhesus macaques), we further included a random effect of population. To get a general estimate of the effect of age on PC1 across species, we also ran linear hierarchical models with all species combined, with an additional random intercept for species. Because of large differences in species lifespans, we first calculated a standardized age index within species by dividing age in years by the maximum age for that species and used this index in the model. It is important to note that we did not account for phylogeny in this analysis, and a robust comparison across species would also require more sophisticated standardization of age. Hence, we present this analysis for illustrative purposes only and not for biological interpretation.
For species with data on mortality (humans, chimpanzees, rhesus macaques, pig-tailed macaques, common marmosets, and Coquerel’s sifaka), we also examined the association of PCs with mortality risk. We ran Cox proportional hazards (PH) models with the fixed effects of PC1, sex, and their two-way interaction. When the interaction was not significant, we interpreted the effect of PC1 on mortality risk for both sexes together from a model with only the fixed effects of PC1 and sex. For species that came from more than one population (chimpanzees and rhesus macaques), we again tested for a random effect of population, using Cox PH models with a random effect. We again ran a combined-species model with a random intercept for species and age standardized as described above, for illustrative purposes only, with the same caveat against over-interpretation.
Consistency of Principal Components
We tested for consistency of PCA results within and among species by examining the degree of correlation between different PC scores calculated for the same individuals using different loadings, obtained from PCA of a subset of biomarkers or of different populations (19,20,32). In other words, we can ask if populations A and B have similar PC structure by performing a PCA on each, calculating the PC scores for each individual obtained from either multiplying their biomarker values by loadings from population A or B, and correlating the PC scores obtained from the two loadings. If the scores are highly correlated (eg, r > .9), we interpret this to mean that we are essentially recovering the same loadings, resulting in similar PC scores. Lower correlations would suggest the PC structures differ, to varying degrees.
Using the full dataset (including repeated observations), we compared correlations between PC scores from all 13 biomarkers and alternate scores obtained from a minimal subset of five targeted biomarkers (within species), between scores obtained from males or females only (within species), and between scores based on loadings from a species itself and each other species (among species). The five-biomarker subset was chosen to encapsulate key components of integrated albunemia with as few biomarkers as possible, and it consisted of hemoglobin, red blood cells, calcium, albumin, and total protein. Because we have repeated observations on individuals, representing pseudo-replication, we did not use Pearson’s correlation coefficient, which assumes independence of observations. Instead, we calculated the standardized slope coefficient from linear mixed models with a random intercept for each individual, where both x and y (the two different PC scores from different loadings) were centered to 0 and standardized to 1 SD. This coefficient is equivalent to a correlation coefficient but permits control for repeated sampling via a random effect.
We also tested if phylogeny affected PC1 structure among species by correlating the matrix of phylogenetic distances between species (calculated as time since divergence, taken from a large recent phylogeny of living primates (33,34)) with two possible measures of PC1 similarity. The first measure of similarity was the absolute difference in average species PC1 values from the combined-species PCA. The second measure of similarity was the correlation between PC1 scores for a given species calculated from loadings from a PCA of itself or PCA of each other species. We calculated Spearman’s rank correlation coefficient (ρ) among the cell values of the phylogenetic distance matrix and the two PC1 similarity matrices, excluding the diagonals. The matrices of phylogenetic distances and of absolute differences in average species PC1 values were symmetric, so significance of this correlation was only based on the upper triangle.
We performed all analysis in the R statistical environment v3.4.3 (35). We ran PCA with package “FactoMineR” (36), multilevel models with the package “lme4” (37), Cox PH models with the packages “survival” (38) and “coxme” (39), estimated marginal means with “emmeans” (40), and correlation plots with “corrplot” (41).
Results
PCA Results
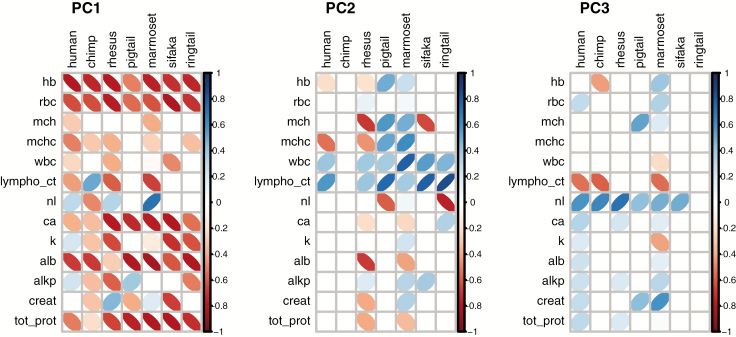
PCA consistently resulted in a main axis of variation characterized by biomarkers from multiple physiological systems. When PCA was performed on each species separately, PC1, PC2, and PC3 on average explained 26.2%, 16.4%, and 12.8% of variation, respectively (55.5% together); however, there was notable variation among species (Supplementary Table S1). The most important biomarkers in PC1 were generally ones associated with integrated albunemia. In all species, PC1 was significantly correlated with anemia markers (low hemoglobin and low red blood cell counts) and with low levels of calcium, albumin, and total protein (Figure 1, Supplementary Table S1). The strength and direction of correlation of other biomarkers with PC1 were more variable across species. Loadings for PC2, PC3, and all following PCs were more variable across species, and these should be interpreted with caution given the variation present in PC1 (ie, remaining variation to be explained by subsequent PCs depends on the variation explained by PC1, since subsequent axes must be orthogonal). PC2 largely seemed to reflect immune function and was significantly correlated with higher numbers of lymphocytes in all species, with higher white blood cells counts in all species except chimpanzees, and with lower N/L ratio in all species except common marmosets. Note that, in chimpanzees, the average loading value for lymphocyte counts (0.327, Supplementary Table S1) was significant, but the loading for this biomarker was relatively unstable between samples (Figure 1). MCH and MCHC were also usually implicated in PC2, but the direction of correlation differed among species. PC3 was characterized across species by high N/L ratio and low lymphocyte counts (Supplementary Table S1), but these loadings were sometimes unstable across samples. Some biomarkers did not load consistently on any PC across species but were important in some PCs in some species. For instance, creatinine, alkaline phosphatase, and potassium had highly variable loading strengths and directions across species.
Figure 1.
Loadings of each biomarker on PC1, PC2, and PC3, from PCA of each species separately. Darker narrower ellipses indicate stronger correlations. Positive correlations tilt to the right and are in blue, while negative correlations tilt to the left and are in red. Loadings represent averaged values from 1,000 PCAs on 1,000 random samples of one observation per ID. For easier visualization of biomarker importance, only loadings that consistently differed from 0 were included—loadings that were not > 2 SD away from 0 were set to 0. Standard deviations were calculated from the distribution of loading values obtained from the 1000 PCA runs, for each biomarker for each species. Note that, in PC2 and PC3, blank columns indicate that variable loadings were unstable between subsamples. See Supplementary Table S1 for all loading values.
From the PCA of all species combined (Supplementary Table S2, Supplementary Figure S1), the first three principal components (PCs) together explained 53.7% of the variation; PC1 explained 23.0%, PC2 16.7%, and PC3 14.0%. PC1 again largely resembled integrated albunemia and was characterized by low hemoglobin, low red blood cells, high MCH, low calcium, low potassium, low albumin, low alkaline phosphatase, high creatinine, and low total protein. Interestingly, PC2 was not strongly characterized by any biomarkers, while PC3 was most strongly associated with high numbers of lymphocytes and white blood cells and low N/L ratio, thus reflecting immune function and resembling PC2 in the separate-species PCAs. Despite apparent similarities in PC1 structure, species differed strongly in average values of the first three PCs (PC1: χ2 = 2060.5, p < .001; PC2: χ2 = 5776.4, p < .001; PC3: χ2 = 1222.1, p < .001) (Figure 2). Tukey’s HSD tests suggested that PC1 was more similar between certain species pairs (Supplementary Table S3 and Supplementary Figure S1). All species pairs differed significantly in average PC1 value with p < .001 except human-chimp (p = .048), rhesus-pigtail (p = .509), marmoset-sifaka (p = .994), marmoset-ringtail (p = .924), and sifaka-ringtail (p > .999).
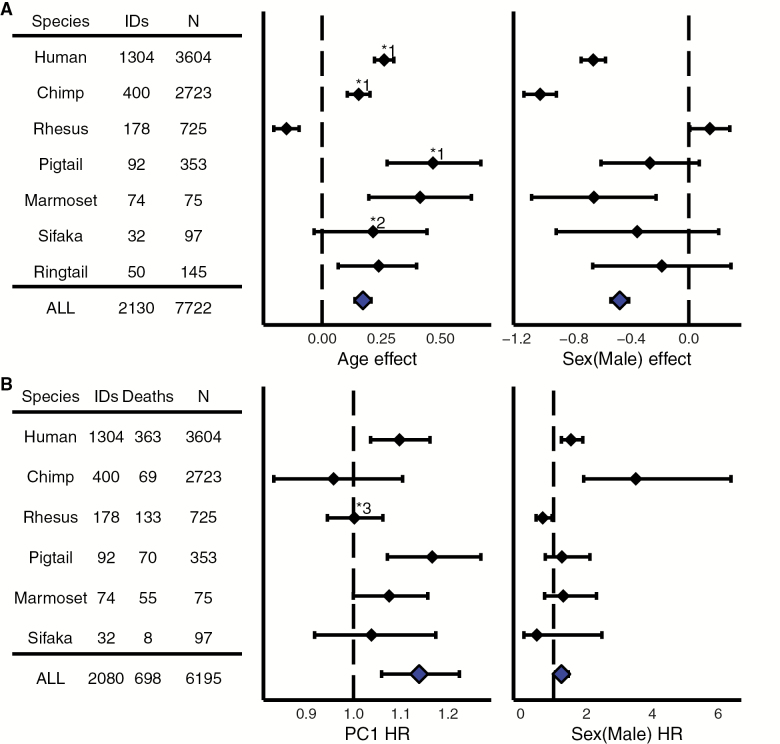
Figure 2.
Estimated effects of (A) age and sex on PC1 from linear mixed models and (B) PC1 and sex on mortality risk from Cox proportional hazards models. IDs = number of unique individuals; N = number of total observations; Deaths = confirmed mortality events; HR = hazard ratio per unit PC1. Solid diamonds indicate the estimated coefficient, and error bars are 95% confidence intervals. The dotted line references (A) estimated effect = 0 or (B) hazard ratio = 1. Both age and PC1 were centered to 0 and standardized to standard deviation = 1, where appropriate, to facilitate comparisons among species. Note that the combined-species (ALL) model results (at bottom in blue) are mainly presented for illustrative purposes, as the appropriate standardization for age and PC1 across species is unknown. Average sex differences in PC1 values are not shown, but indicated with an asterisk (*) where significant and discussed in the main text. *1 = stronger positive effect in males; *2 = positive effect in females only; *3 = higher PC1 scores predict lower risk in males only.
Associations With Age and Mortality
Linear models showed a significant increase in PC1 with age in both sexes in five of the seven species studied—humans, chimpanzees, pig-tailed macaques, common marmosets, and ring-tailed lemurs—and in females only in Coquerel’s sifaka (Figure 2A, Supplementary Figure S2). In contrast, there was a significant decrease in PC1 with age in rhesus macaques. In humans, chimpanzees, and common marmosets, males had lower values of PC1 than females at the mean age. In humans, there was additionally a positive quadratic effect of age (not shown), which indicated a steeper (faster) increase in PC1 at older ages. There were no significant (p > .05) quadratic effects of age in any other species. More detailed model results, with and without age x sex interactions, are presented in Supplementary Table S4. The combined-species model suggested that, across all species, PC1 generally increases with age and that males had lower PC1 values than females.
Higher values of PC1 were associated with increased risk of mortality in humans and pig-tailed macaques, whereas lower values of PC1 were associated with increased risk in male rhesus macaques only (Figure 2B). In humans and chimpanzees, males had higher average mortality risk than females, while in rhesus macaques, males had lower mortality risk. There was no significant effect of PC1 or sex on mortality risk in Coquerel’s sifakas or common marmosets (although there was a trend for PC1 to increase risk in marmosets, p = .051). More detailed model results, with and without age × sex interactions, are presented in Supplementary Table S5. The combined-species model suggested that, across all species, higher PC1 scores were associated with higher mortality risk and that males had higher mortality risk.
Consistency of Principal Components
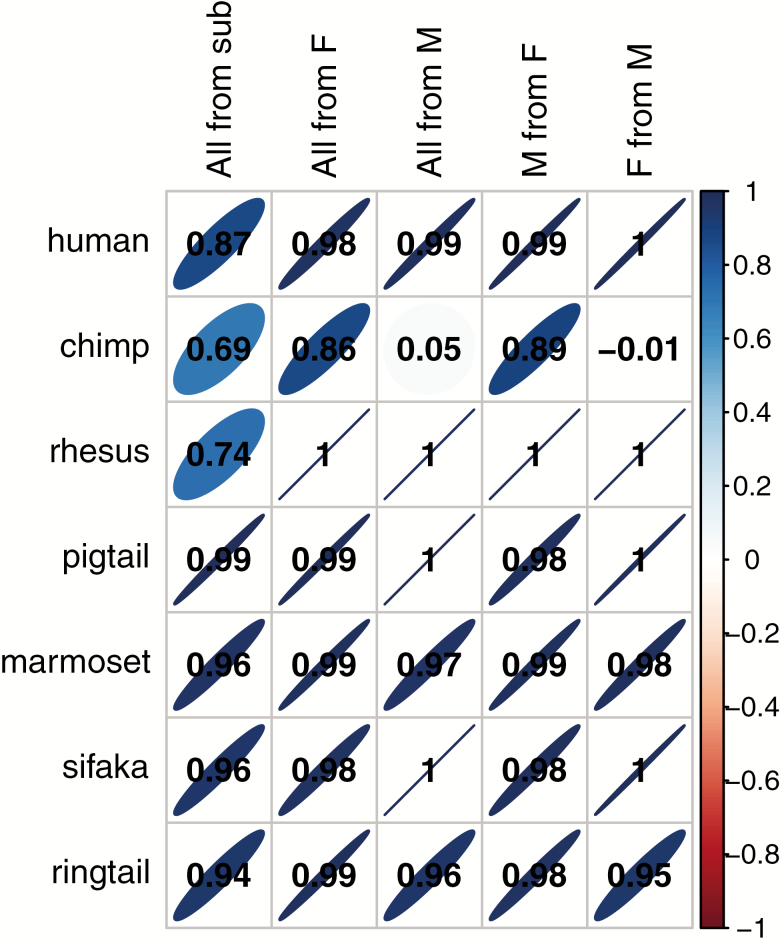
PC1 scores were usually strongly replicated using a simple five-biomarker subset (r > .69 in all species, Figure 3). However, r < .9 in chimpanzees, rhesus macaques, and humans suggested that biomarker composition was more important in these species. PC1 was also very stable between the sexes (r > .95) in all species except chimpanzees, where loadings from males only appeared to be a very poor reference for female or whole population scores (Figure 3). Conversely, loadings from female chimpanzees were robust references for male (r = .89) or whole population (r = .86) scores.
Figure 3.
Correlations between PC1 scores calculated from different possible loadings within species. Column names indicate the comparison groups. “All from sub” compares scores for all individuals from loadings from PCA of all biomarkers or the five-biomarker subsample. “All from F” compares scores for all individuals from loadings from PCA of all individuals or females only. “All from M” compares scores for all individuals from loadings from PCA of all individuals or males only. “M from F” compares scores for males from loadings from PCA of males or females only. “F from M” compares scores for females from loadings from PCA of females or males only. Darker narrower ellipses indicate stronger correlations, which indicate more consistent PC1 scores across different biomarker compositions and subpopulations. Positive correlations tilt to the right and are in blue, while negative correlations tilt to the left and are in red. Correlation coefficients are given in the cells and were calculated as standardized slope coefficients from linear mixed models accounting for repeated observations on individuals.
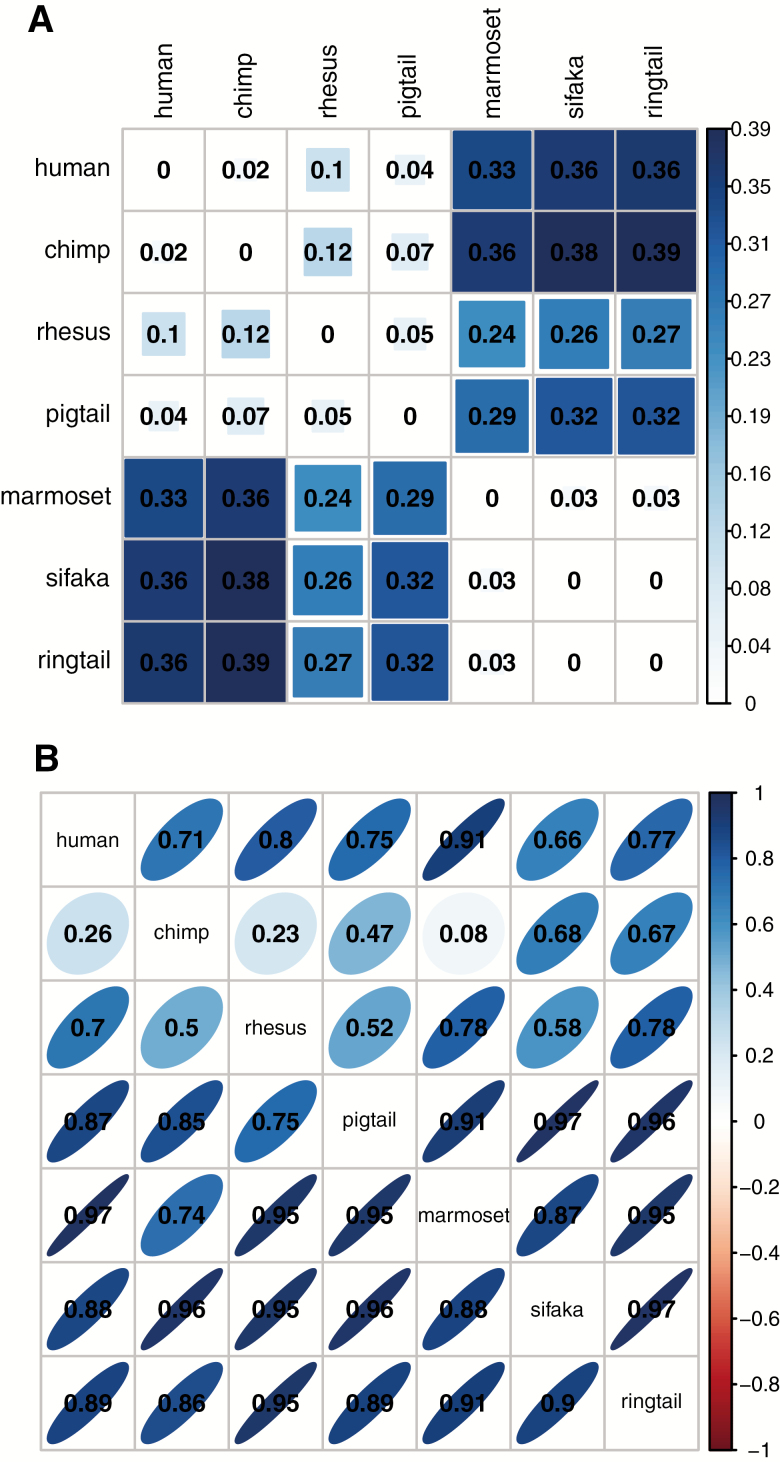
Phylogeny had variable effects depending on the method used to calculate similarity in PC1. Phylogenetic distance was significantly positively correlated with the absolute differences in species average PC1 values (Spearman’s ρ = 0.552, p = .009; Figure 4A), that is, more closely related species had more similar PC1 values when PCA was performed on all species together. However, PC1 scores calculated from loadings of different species in separate-species PCA were correlated to variable degrees in a less structured way (.08 < r < .97; Figure 4B). Chimpanzee scores were conspicuously poorly predicted by other species’ loadings (.08 < r < .68; all other species: .50 < r < .97). Somewhat surprisingly, human PC1 scores were best predicted by common marmoset loadings (r = .91), out of the nonhuman primates. More closely related species did not have significantly more similar PC1 loadings; in fact, after correcting for repeated observations, there was a significant association in the opposite direction, with more distantly related species being more highly correlated (ρ = −0.434, p = .004). However, this result disappears if chimpanzees (who had conspicuously low correlations overall) were omitted, and the pattern should be considered with caution, especially given the low number of species. Additionally, we attempted to link PC1 scores to species lifespan, but sample size was insufficient given the phylogenetic relationships (more closely related species have more similar lifespans), and results are not presented.
Figure 4.
Matrices of: (A) the absolute difference in average species PC1 score from combined-species PCA; and (B) the correlations between PC1 scores calculated from average loadings of each species itself and from average loadings of other species, from separate-species PCA. In (A), darker larger squares indicate larger differences, and the largest difference in the data is set as the maximum. In (B), darker narrower ellipses indicate stronger correlations, which indicate more consistent PC1 scores between species. Positive correlations tilt to the right and are in blue, while negative correlations tilt to the left and are in red. Rows correspond to the species analyzed; columns correspond to the species used to calibrate the loadings. Correlations were calculated as standardized slope coefficients from linear mixed models accounting for repeated observations on individuals.
Discussion
In this study, we found a strikingly consistent primary axis of variation across diverse primate species, resembling one previously described in humans as integrated albunemia (19). This axis generally increased with increasing age, as in humans, but was not universally linked to mortality risk. These results suggest that integrated albunemia indeed reflects an evolutionarily conserved emergent physiological phenomenon and could be a candidate measure of aging, but one with different health consequences in different species. Nonetheless, the conserved aspects of this process are remarkable, given the diversity of species, and encourage further research into (a) integrated albunemia as an evolutionarily broad phenomenon and (b) the use of nonhuman primates as models of emergent physiological processes.
When we applied principal components analysis to a standardized set of clinical biomarkers in seven taxonomically diverse primate species, we always found a physiological axis that resembled the proposed phenomenon of integrated albunemia previously documented in humans (19). Similar biomarkers (anemia markers, low albumin, low calcium) emerged as being important in all species, whether PCA was performed on each species separately or on all species together. Hence, a process comprising integrated albunemia is present in all species but possibly integrated with other aspects of physiology (eg, metabolism and immune function) differently among species. Alternatively, biomarkers that were associated with PC1 in some species but not others might reflect anomalies of sample composition. For example, while all individuals in the dataset were considered healthy, if there were many older individuals who trended towards kidney issues, this might explain an unusual association of creatinine with the integrated albunemia axis (eg, in rhesus macaques) even in the absence of a direct physiological link with the process. In order to rule out such possibilities, it would be necessary to validate PC1 in multiple subpopulations of each species and ensure more consistent age representation across all species.
Thus, despite some variation, we refer to this primary axis as “integrated albunemia” from here on for simplicity, although we cannot exactly replicate the study in humans, which also included elevated inflammatory markers and others that were unavailable for our study. In humans, a systemic inflammatory response generally results in an increase in circulating neutrophils and decrease in circulating lymphocytes, leading to high N/L associated with a number of acute health issues and potential prognostic value for some conditions (42–44). Thus, in this study, we included the neutrophil/lymphocyte ratio (N/L) as a potential marker of inflammation, and indeed it has the predicted positive association with PC1 in humans. However, the interpretation of N/L as an inflammatory marker is indirect, and its value in this role in other species is inconsistent, with two nonhuman primates showing positive association with PC1, two showing a negative association, and two showing no association. The current study also lacked some of the markers that could reflect metabolic syndrome (lipids and blood sugar), which emerged as the second axis of variation in the original article on human integrated albunemia (19). Nevertheless, we used one of the same human datasets and recovered a similar PC1 for humans and other species as emerged from a much larger set of biomarkers. Accordingly, while the addition of other biomarkers would likely change the exact composition of PC1, we believe our general conclusions are likely to be robust to the differences in biomarkers included in the analysis. A remarkable aspect of our result is the consistent emergence of multi-system biomarkers in PC1, implying consistent connections between physiological systems in different species, rather than the emergence of similar clusters of system-specific biomarkers, as one might predict a priori (19).
Integrated albunemia increased with age in all species except rhesus macaques (although only in females in Coquerel’s sifakas), suggesting a process that is generally associated with aging across primates. Our ability to detect overall significant effects could be limited by sample size in Coquerel’s sifakas (with only 32 individuals), which had estimated positive effects of age on PC1 in both sexes of similar magnitude to humans and chimpanzees, but whose larger confidence intervals overlap with 0. In general, it is important to note that the species with fewer individuals and that only come from single research centers represent much less diverse populations than species represented by more individuals and multiple research populations. Chimpanzees and common marmosets were also similar to humans in that males had lower values of PC1 than females. Integrated albunemia was less broadly predictive of survival. It was only significantly associated with increased risk of mortality in humans and pig-tailed macaques, although it was close to significance in common marmosets. Rhesus macaque males showed the opposite pattern: higher integrated albunemia scores were actually associated with lower mortality risk. Males had higher mortality risk in humans and chimpanzees, while males had lower risk in rhesus macaques. Rhesus macaques thus stood out as showing opposing patterns from the general trends in several ways and perhaps merit further study; in particular, it is surprising that a mammal would broadly show males at lower risk of mortality than females. We explored some possible factors that might affect this pattern, such as differences among research populations and individual biomarker associations with age but did not find an obvious explanation. Interestingly, correlating each of the biomarkers individually with age highlighted a high degree of variation among species in how each biomarker tends to change with age (Supplementary Table S6) and implies that the association between integrated albunemia and age is more stable than most individual biomarkers, as found across human populations (19). Only one biomarker (albumin) changed in the same direction in all species, and one biomarker (calcium) differed only for rhesus macaques. While calcium is important in PC1, it would seem surprising if one biomarker could drive the strongly divergent pattern in this species. Indeed, rerunning analyses without calcium resulted in the same divergent pattern for rhesus macaques. Our results were also robust to including body mass as covariate in linear models, to account for potential differences due to body size. In this case, PC1 still increased with age in the same species; the only slight difference was that the pattern became nonsignificant in ring-tailed lemurs, but the effect size remained similar (in fact, slightly larger). Ultimately, despite species variation and some sample size limitations, our overall results strongly suggest that integrated albunemia is a conserved process that changes with increasing age; the generality of the negative health consequences documented in humans is unclear.
Our results in humans generally align well with other human studies using different techniques (19,22,45), which have found that anemia markers, albumin, and calcium were prominently associated with human aging. It is worth noting that other studies that examined a number of physiological biomarkers came to variable conclusions about the most important markers of aging (46–48). However, these studies employed very different methods typically including non-physiological variables and focusing on single or few biomarker values rather than the relationships among biomarkers. The fact that we replicate previous findings in humans to a large degree indicates that we are capturing the same process, despite using fewer and a different subset of biomarkers (notably, missing inflammatory markers entirely), and again highlights a notable consistency.
The stability of integrated albunemia within species was generally high between sexes or using a very minimal biomarker subset, suggesting the phenomenon is generally quite robust. In particular, the ability to use fewer biomarkers or limited subsets of the population to get accurate measures has important practical implications. The notable exception to the general robustness was in chimpanzees, where loadings from males were extremely poor references and the subset of biomarkers was only moderately correlated. The reasons for this conspicuous difference in one species are not immediately evident, but a possible factor is that, despite reasonable numbers of observations on males and females overall, there were very few observations of older male chimpanzees (indeed all observations on very old animals were from females). Hence, this poor correlation could largely mean that younger male chimpanzees are not representative of females or the larger population, especially if we are trying to measure a process predicted to be involved in aging.
Perhaps more surprising than overall high correlations within species, integrated albunemia was sometimes strongly correlated even across different species. Notably, for humans, scores from loadings of common marmosets replicated scores from human loadings with high accuracy (r = .91). Somewhat unexpectedly, the more closely related chimpanzees were not the best correlated with human biomarker structure, even though chimpanzees showed very similar patterns of age and mortality to humans. Again, chimpanzees were conspicuously poorly correlated with other species in general, and, for the reasons mentioned above, could be affected by demographic representation. Given their similarity in integrated albunemia scores and in associations with age and mortality, common marmosets might prove to be particularly useful research models for emergent physiological processes in humans. This adds to previous recommendations to better develop common marmosets as models for human aging (49,50). Notably, in this study, common marmosets appear to be much better models for emergent patterns of human physiology than rhesus macaques, which are more commonly used as research models.
The effect of phylogeny on integrated albunemia was inconclusive. When all species were combined into a single PCA, more closely related species had more similar PC1 values, as predicted. However, when PCA was performed on each species separately and scores from different species’ loading correlated, phylogenetic proximity was actually negative correlated with similarity in integrated albunemia loadings. As noted earlier, this result disappears if chimpanzees are omitted, and there is no a priori explanation for this result, so it should be considered with caution. While these results are inconclusive about the effect of phylogeny, we had too few species to test for this effect rigorously. Further work including more species and analyzing structure at multiple taxonomic levels will help determine if evolutionary relationships indeed influence the similarity in the structure and health consequences of integrated albunemia in different species. Our results from the combined-species PCA suggest some potential hypotheses about emergent physiological processes at higher taxonomic levels (Supplementary Figure S1). For example, there appears to be a major distinction between PC1 values for species in the Parvorder Catarrhini (the taxonomic group including Old World Monkeys and Apes (34)) and other species, perhaps suggesting that important derived physiological characteristics appeared at this stage. More broadly, our study illustrates the benefits of linking research in aging to other disciplines, such as evolutionary biology.
While we did not examine further dimensions of variation in detail, other PCA axes appeared to be less stable than integrated albunemia. In PCA of each species separately, results for the second and third axes were much more variable in that loadings were less consistent across samples, and there were rarely strong correlations and more commonly weak or no correlations within species. Among species, PC2 and PC3 were relatively poorly correlated and highly variable across species, often even showing negative correlations (as PC loadings were standardized so that specific biomarkers showed the same sign across all species; Supplementary Table S1).
In conclusion, revisiting our original hypothesis, integrated albunemia appears to be a fundamental emergent physiological process, present in all primates studied here, and it has some association with age in most species but was not clearly associated with survival outside of humans. While significant variation exists in the characterization and consequences of integrated albunemia in different species, this process clearly has a broader evolutionary role across primates and is not restricted to humans or closely related species. Our current results complement another recent study using parts of the same primate database to demonstrate conserved patterns of physiological dysregulation across primates, with strong implications for aging and mortality (27). Together, these studies provide evidence for evolutionarily broad patterns of aging and for nonhuman primates as valuable models of emergent processes and aging. A particularly promising model system might be common marmosets, which matched human results closely and whose relatively small size and short lifespans make them more practical research models. Further study into other species might reveal even broader conservation of this process and provide insight into basic mechanisms of animal and human aging.
Funding
This work was supported by the Canadian Institutes of Health Research (grants #s 145585 and 153011, to A.A.C.), the Natural Sciences and Engineering Research Council of Canada (grant # 402079-2011, to A.A.C.), the National Institutes of Health/National Institute on Aging (contract # HHSN271201300026C, to J.W.K.), the National Institutes of Health (grant # P51 RR000167/OD011106, to J.W.K.), and the Intramural Research Program of the National Institute on Aging, NIH, Baltimore, MD, United States. A.A.C. is also supported by a Canadian Institutes of Health Research New Investigator Salary Award and is a member of the Centre de recherche du CHUS and Centre de recherche sur le vieillissement, funded by Fonds de recherche du Québec – Santé. The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health, and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336). The Follow-up 1 (2001–2003) was funded by the U.S. National Institute on Aging (Contracts: N.1-AG-1-1 and N.1-AG-1-2111); the Follow-up 2 and 3 studies (2004–2010) were financed by the U.S. National Institute on Aging (Contract: N01-AG-5-0002).
Conflict of Interest
J.W.K. is a consultant to CléMetric, LLC, a health data analytics and management company. The authors declare no other conflict of interest.
Supplementary Material
Acknowledgments
We are grateful to the sites that contributed to iPAD and to Ms.m Wendy Newton (WNPRC) for gathering and entering much of the data for iPAD. T.W.W., E.R., and A.A.C. conceptualized and developed the study, with help from V.L. and J.W.K. J.W.K. and L.F. manage the data used. T.W.W. and E.R. analyzed the data with help from V.L. T.W.W. wrote the manuscript, and all authors contributed to revisions. We also thank two anonymous reviewers for their helpful feedback on an earlier version of the manuscript.
References
- 1. Cohen AA, Martin LB, Wingfield JC, McWilliams SR, Dunne JA. Physiological regulatory networks: ecological roles and evolutionary constraints. Trends Ecol Evol. 2012;27:428–435. doi: 10.1016/j.tree.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 2. Fried LP, Xue QL, Cappola AR, et al. Nonlinear multisystem physiological dysregulation associated with frailty in older women: implications for etiology and treatment. J Gerontol A Biol Sci Med Sci. 2009;64:1049–1057. doi: 10.1093/gerona/glp076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Han J-DJ, Hou L, Sun N, Xu C, McDermott J, Wang D. The system capacity view of aging and longevity. Quant Biol. 2017;5(3):251–259. doi: 10.1007/s40484-017-0115-4 [DOI] [Google Scholar]
- 4. Kriete A. Robustness and aging–a systems-level perspective. Biosystems. 2013;112:37–48. doi: 10.1016/j.biosystems.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 5. West GB, Bergman A. Toward a systems biology framework for understanding aging and health span. J Gerontol A Biol Sci Med Sci. 2009;64:205–208. doi: 10.1093/gerona/gln066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen AA, Milot E, Yong J, et al. A novel statistical approach shows evidence for multi-system physiological dysregulation during aging. Mech Ageing Dev. 2013;134:110–117. doi: 10.1016/j.mad.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cornman JC, Glei DA, Goldman N, Weinstein M. Physiological dysregulation, frailty, and risk of mortality among older adults. Res Aging. 2017;39:911–933. doi: 10.1177/0164027516630794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crimmins EM, Johnston M, Hayward M, Seeman T. Age differences in allostatic load: an index of physiological dysregulation. Exp Gerontol. 2003;38:731–734. doi:10.1016/S0531-5565(03)00099-8 [DOI] [PubMed] [Google Scholar]
- 9. Arbeev KG, Ukraintseva SV, Bagley O, et al. “Physiological Dysregulation” as a promising measure of robustness and resilience in studies of aging and a new indicator of preclinical disease. J Gerontol A Biol Sci Med Sci. 2019;74:462–468. doi: 10.1093/gerona/gly136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grundy SM, Cleeman JI, Daniels SR, et al. ; American Heart Association; National Heart, Lung, and Blood Institute Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404 [DOI] [PubMed] [Google Scholar]
- 11. Franceschi C, Bonafè M, Valensin S, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi:10.1111/j.1749-6632.2000.tb06651.x [DOI] [PubMed] [Google Scholar]
- 12. Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol Ser A. 2014;69(Suppl_1):S4–S9. doi: 10.1093/gerona/glu057 [DOI] [PubMed] [Google Scholar]
- 13. Seplaki CL, Goldman N, Glei D, Weinstein M. A comparative analysis of measurement approaches for physiological dysregulation in an older population. Exp Gerontol. 2005;40:438–449. doi: 10.1016/j.exger.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 14. Belsky DW, Moffitt TE, Cohen AA, et al. Eleven telomere, epigenetic clock, and biomarker-composite quantifications of biological aging: do they measure the same thing? Am J Epidemiol. 2018;187(6):1220–1230. doi: 10.1093/aje/kwx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen AA, Legault V, Fuellen G, Fülöp T, Fried LP, Ferrucci L. The risks of biomarker-based epidemiology: associations of circulating calcium levels with age, mortality, and frailty vary substantially across populations. Exp Gerontol. 2018;107:11–17. doi: 10.1016/j.exger.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gruenewald TL, Seeman TE, Ryff CD, Karlamangla AS, Singer BH. Combinations of biomarkers predictive of later life mortality. Proc Natl Acad Sci U S A. 2006;103:14158–14163. doi: 10.1073/pnas.0606215103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura E, Miyao K. A method for identifying biomarkers of aging and constructing an index of biological age in humans. J Gerontol A Biol Sci Med Sci. 2007;62:1096–1105. doi:10.1093/gerona/62.10.1096 [DOI] [PubMed] [Google Scholar]
- 18. Brown PJ, Wall MM, Chen C, et al. Biological age, not chronological age, is associated with late-life depression. J Gerontol A Biol Sci Med Sci. 2018;73:1370–1376. doi: 10.1093/gerona/glx162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cohen AA, Milot E, Li Q, et al. Detection of a novel, integrative aging process suggests complex physiological integration. PLoS One. 2015;10(3):e0116489. doi: 10.1371/journal.pone.0116489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morrisette-Thomas V, Cohen AA, Fülöp T, et al. Inflamm-aging does not simply reflect increases in pro-inflammatory markers. Mech Ageing Dev. 2014;139:49–57. doi: 10.1016/j.mad.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dusseault-Belanger F, Cohen AA, Hivert MF, Courteau J, Vanasse A. Validating metabolic syndrome through principal component analysis in a medically diverse, realistic cohort. Metab Syndr Relat Disord. 2013;11:21–28. doi: 10.1089/met.2012.0094 [DOI] [PubMed] [Google Scholar]
- 22. Moeller M, Pink C, Endlich N, et al. Mortality is associated with inflammation, anemia, specific diseases and treatments, and molecular markers. PLoS One. 2017;12:e0175909. doi: 10.1371/journal.pone.0175909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colman RJ, Kemnitz JW. Aging experiments using nonhuman primates. In: Yu BP, ed. Methods in Aging Research. Boca Raton, FL: CRC Press; 1998:249–267. [Google Scholar]
- 24. Didier ES, MacLean AG, Mohan M, Didier PJ, Lackner AA, Kuroda MJ. Contributions of nonhuman primates to research on aging. Vet Pathol. 2016;53:277–290. doi: 10.1177/0300985815622974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lane MA. Nonhuman primate models in biogerontology. Exp Gerontol. 2000;35:533–541. doi: 10.1016/S0531-5565(00)00102-9 [DOI] [PubMed] [Google Scholar]
- 26. Verdier JM, Acquatella I, Lautier C, et al. Lessons from the analysis of nonhuman primates for understanding human aging and neurodegenerative diseases. Front Neurosci. 2015;9:64. doi: 10.3389/fnins.2015.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dansereau G, Wey TW, Legault V, et al. Conservation of physiological dysregulation signatures of aging across primates. Aging Cell. 2019;18:e12925. doi: 10.1111/acel.12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smucny DA, Allison DB, Ingram DK, et al. ; Primate Aging Database Working Group Changes in blood chemistry and hematology variables during aging in captive rhesus macaques (Macaca mulatta). J Med Primatol. 2001;30:161–173. doi: 10.1111/j.1600-0684.2001.tb00005.x [DOI] [PubMed] [Google Scholar]
- 29. Ferrucci L, Bandinelli S, Benvenuti E, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x [DOI] [PubMed] [Google Scholar]
- 30. Tacutu R, Thornton D, Johnson E, et al. Human Ageing Genomic Resources: new and updated databases. Nucleic Acids Res. 2017;46(D1):D1083–D1090. doi: 10.1093/nar/gkx1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leung DL, Fried LP, Ferrucci L, Cohen AA. Measuring loss of homeostasis in aging. In: Morales AJ, Gershenson C, Braha D, Minai AA, Bar-Yam Y, eds. Unifying Themes in Complex Systems IX. Vol 9. Springer Proceedings in Complexity. Cambridge, MA: Springer International Publishing; 2018:XXIII, 506. doi: 10.1007/978-3-319-96661-8 [DOI] [Google Scholar]
- 32. Cohen AA, Dhingra N, Jotkar RM, Rodriguez PS, Sharma VP, Jha P. The Summary Index of Malaria Surveillance (SIMS): a stable index of malaria within India. Popul Health Metr. 2010;8:1. doi: 10.1186/1478-7954-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- 34. Perelman P, Johnson WE, Roos C, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. http://www.R-project.org. [Google Scholar]
- 36. Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25(1):1–18. doi: 10.18637/jss.v025.i01 [DOI] [Google Scholar]
- 37. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. doi: 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 38. Therneau T. A Package for Survival Analysis in S. 2015. https://CRAN.R-project.org/package=survival. Accessed December 13, 2017. [Google Scholar]
- 39. Therneau TM. Coxme: Mixed Effects Cox Models. 2015. http://CRAN.R-project.org/package=coxme. Accessed May 29, 2017. [Google Scholar]
- 40. Lenth R. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. 2018. https://CRAN.R-project.org/package=emmeans. Accessed November 28, 2018. [Google Scholar]
- 41. Wei T, Simko V.. R Package “Corrplot”: Visualization of a Correlation Matrix.; 2017. https://github.com/taiyun/corrplot. Accessed April 12, 2018. [Google Scholar]
- 42. Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607 [DOI] [PubMed] [Google Scholar]
- 43. Guthrie GJ, Charles KA, Roxburgh CS, Horgan PG, McMillan DC, Clarke SJ. The systemic inflammation-based neutrophil-lymphocyte ratio: experience in patients with cancer. Crit Rev Oncol Hematol. 2013;88:218–230. doi: 10.1016/j.critrevonc.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 44. Zahorec R. Ratio of neutrophil to lymphocyte counts–rapid and simple parameter of systemic inflammation and stress in critically ill. Bratisl Lek Listy. 2001;102:5–14. [PubMed] [Google Scholar]
- 45. Mamoshina P, Kochetov K, Putin E, et al. Population specific biomarkers of human aging: a big data study using South Korean, Canadian, and Eastern European Patient Populations. J Gerontol A Biol Sci Med Sci. 2018;73:1482–1490. doi: 10.1093/gerona/gly005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haring R, Feng YS, Moock J, et al. Self-perceived quality of life predicts mortality risk better than a multi-biomarker panel, but the combination of both does best. BMC Med Res Methodol. 2011;11:103. doi: 10.1186/1471-2288-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martin-Ruiz C, Jagger C, Kingston A, et al. Assessment of a large panel of candidate biomarkers of ageing in the Newcastle 85+ study. Mech Ageing Dev. 2011;132:496–502. doi: 10.1016/j.mad.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 48. Walter S, Mackenbach J, Vokó Z, et al. Genetic, physiological, and lifestyle predictors of mortality in the general population. Am J Public Health. 2012;102:e3–10. doi: 10.2105/AJPH.2011.300596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The marmoset as a model of aging and age-related diseases. ILAR J. 2011;52:54–65. doi: 10.1093/ilar.52.1.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee HJ, Gonzalez O, Dick EJ, et al. Marmoset as a model to study kidney changes associated with aging. J Gerontol A Biol Sci Med Sci. 2019;74:315–324. doi: 10.1093/gerona/gly237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.