Abstract
O-Vanillin and ethylenediamine were used for the synthesis of N,N′-bis(O-vanillinidene)ethylenediamine (O-VEDH2) Schiff base ligand, which was characterized by UV–visible and 1H NMR spectroscopy. The Schiff base ligand O-VEDH2 has been employed to synthesize novel [C18H22FeN2O6]NO3 (1) and C18H16CuN2O5 (2) complexes by the simple slow evaporation method. The single crystals were characterized by X-ray crystallography, as well as Fourier-transform infrared and UV–visible spectroscopy techniques. Complexes 1 and 2 crystallize in monoclinic and orthorhombic space groups with P121/n1 (14) and Pnma (62) point groups, respectively, and both show metal–organic frameworks like structures. Complexes 1 and 2 have optical band gaps of 4.1 and 2.9 eV, respectively, indicating their semiconducting properties. The dye degradation activity and H2O2 sensing of the complexes 1 and 2 were determined in different conditions. The photocatalytic test was performed in the presence of sunlight, methylene blue (MB), and complexes 1 and 2. An interesting result was obtained that complex 2 has degradation ability against MB and the rate constant (K) is 5.46 × 10–5 s–1, whereas complex 1 has H2O2 sensing properties. Both complexes are bound to Calf-thymus DNA by intercalation binding mode, and binding constants (Kb) of complexes 1 and 2 are 3.77 × 103 and 1.49 × 104 M–1, respectively.
Introduction
The Schiff base metal complexes are used extensively in the field of materials science, including catalyst, supramolecular chemistry, anticancer, antimicrobial, antioxidants, electrochemistry, photocatalysts, DNA binding, and energy materials.1−14 In general, the Schiff base can assemble the metal ions to develop metal–organic framework (MOF) compounds and shows different properties like dye adsorption, sensing, semiconductor, and biological properties.15−23
In recent years, researchers from around the world have been working to synthesize metal–organic framework complexes to promote the degradation or adsorption of dyes and to eliminate the dyes from chemically contaminated water.24−27 Because most of the synthetic dyes were used in a wide range of industries, such as paper, plastics, leather tanneries, food technology, and cosmetics,28,29 waste dyes are discharged into rivers and ponds, thereby contaminating the water. These dye contaminants can cause allergies, skin irritation, liver damage, kidney damage, and also central nervous system disorders in humans and animals.30 Currently, Yang et al. reported that three-dimensional (3D)-pillared layer copper(II) MOFs have been used for dual purposes, i.e., dye adsorption and separation.31
Nowadays, MOFs have often been used as different types of sensing materials, i.e., H2O2 sensing. Reactive oxygen species like H2O2 are nontoxic but produce free radicals such as superoxide anion (O2•–) and hydroxyl radical (•OH) at the time of decomposition, which plays an important role for the initiation of cancer, autoimmunity, neurodegenerative disorders, etc.32−34 Hydrogen peroxide is widely used in the food, cosmetic, pharmaceutical, paper, and chemical industries.35,36 Therefore, it is necessary to detect H2O2 for the healthy life of humans and animals. In the last few years, scientists have developed iron- and copper metal-based MOFs and reported variable sensing applications, i.e., fluorescent, luminescent, photocatalytic, and polar aliphatic volatile organic compounds.37−41
MOFs, as well as various Schiff base ligands, play an important role in bioapplications. The effective anticancer drugs have an active site primarily targeted to bind the DNA of the cancer cell, reducing the activity of the cancer cell and resulting in cell death.42−44 Anticancer drugs interact with cancer cell DNA through electrostatic, intercalative, or groove binding, which depends on the binding modes of anticancer drugs.25,45−47 The Schiff base ligands and the corresponding metal complexes have the active site that interacts with the DNA of the cancer cell.48,49 Recently, metal–organic framework systems have been used in biomedicine, as reported by Chen et al.50,51
In this study, we have designed and synthesized N,N′-bis(O-vanillinidene)ethylenediamine (O-VEDH2) Schiff base ligand and employed it for the synthesis of new [C18H22FeN2O6]NO3 (1) and C18H16CuN2O5 (2) complexes. The main aim of our research work is to synthesize transition-metal complexes using synthesized Schiff base ligand and study their multifunctional applications, i.e., dye degradation, H2O2 sensing, and DNA binding activity. The Schiff base ligand is characterized by UV–visible and 1H NMR spectroscopies. From optical measurements, complexes 1 and 2 have energy band gaps of 4.1 and 2.9 eV, respectively, and show semiconductor property. Complex 1 can actively sense H2O2, whereas complex 2 has no such sensing ability. On the other hand, complex 2 has photocatalytic activity; it degrades methylene blue (MB) and the rate constant (K) is 5.46 × 10–5 s–1, but complex 1 has no photocatalytic activity. Both the complexes bind to Calf-thymus DNA (CT-DNA) through intercalation binding mode, and their binding constants (Kb) were found to be 3.77 × 103 and 1.49 × 104 M–1, respectively.
Results and Discussion
Structural Description
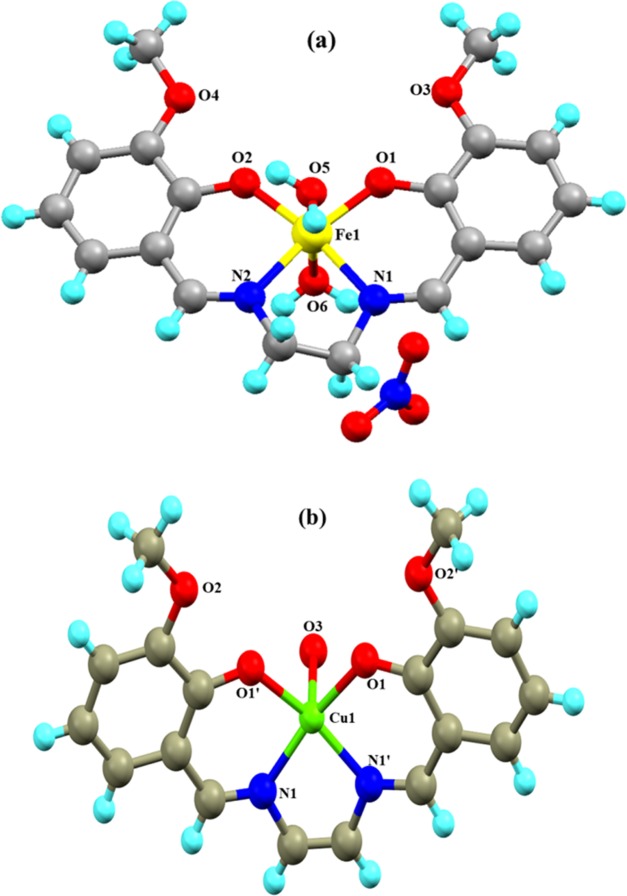
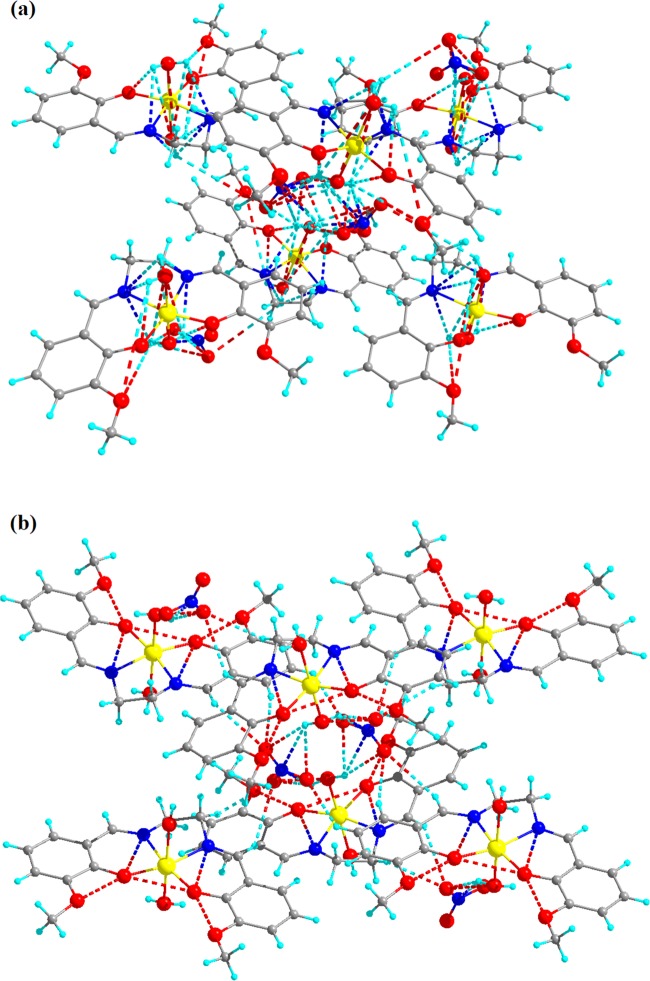
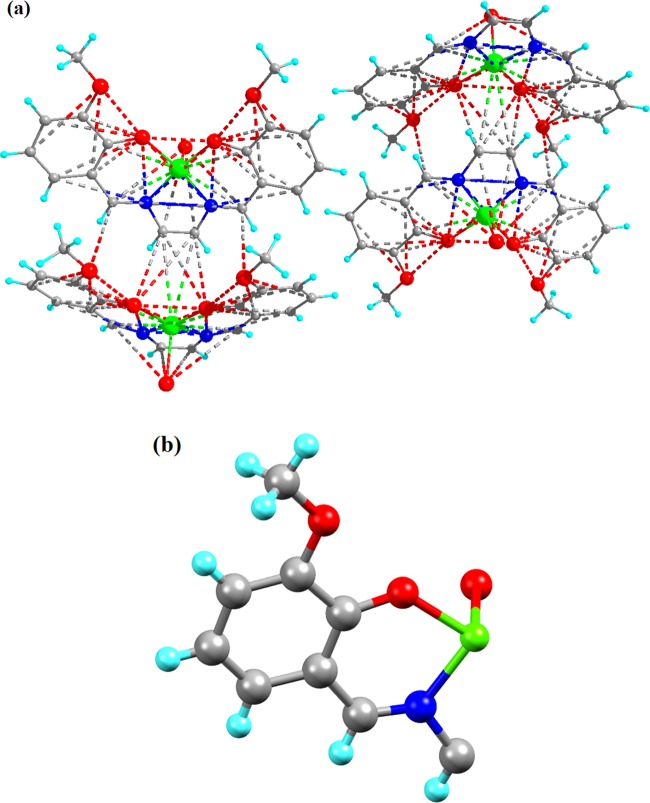
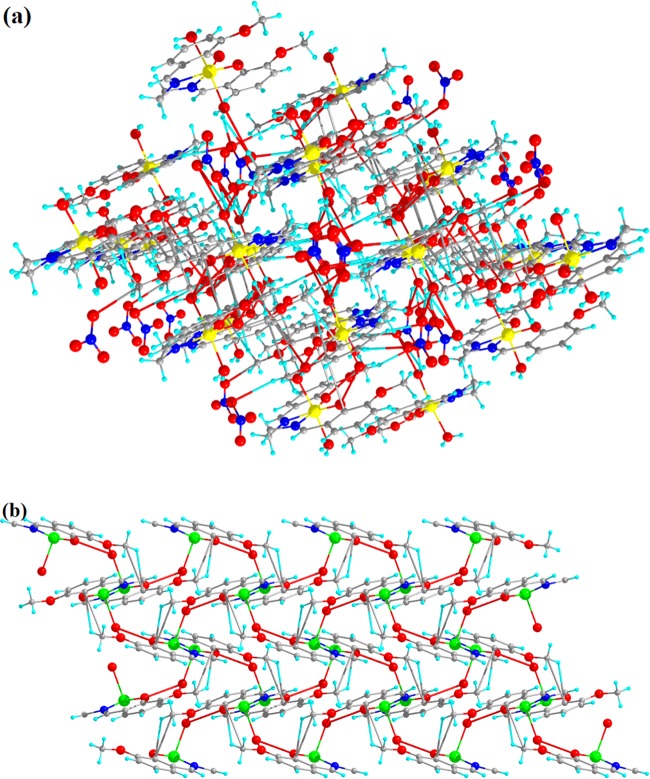
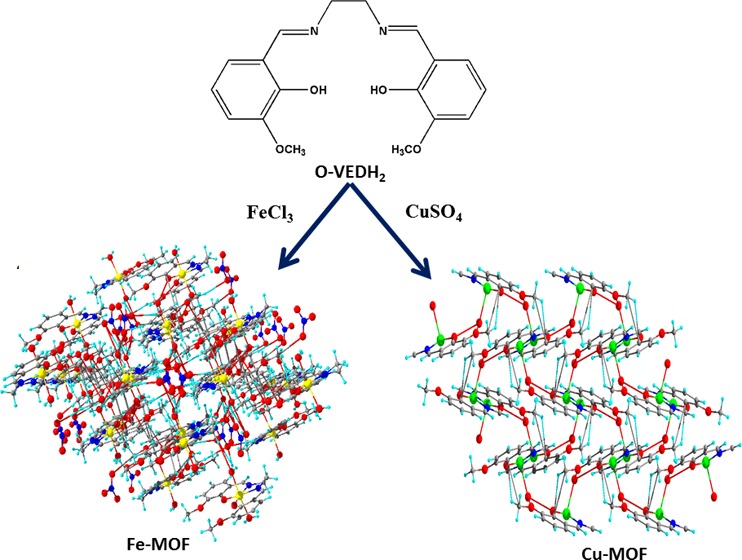
Complexes 1 and 2 crystallize in monoclinic and orthorhombic space groups with P121/n1 and Pnma, respectively, as shown in Figure 1. The phase data of the complexes are shown in Table 1, and the selected interatomic distances and angles are listed in Table 2. Both complexes have butterfly-like structures. Complex 1 was coordinated by two nitrogen atoms, N(1) and N(2), and two hydroxyl oxygen atoms, O(1) and O(2), of O-VEDH2 moiety and two oxygen atoms, O(5) and O(6), of two water molecules and form an octahedral geometry. Two water molecules are present in the axial position of complex 1. The oxidation state of the metal ion is stabilized by nitrate ion (NO3–) (surrounding the complex) and the corresponding protonated oxygen atom in complex 1, whereas complex 2 was coordinated by two nitrogen atoms, N(1) and N(1)a, and hydroxyl oxygen atoms, O(1) and O(1)a, from one O-VEDH2 moiety and one oxygen (O3) atom and developed square pyramidal structure. The oxidation states of the complexes 1 and 2 are found to be 3+ and 2+, which was also confirmed by the bond valence sum (BVS) calculation shown in Table 3.52 The bond lengths between the metal-oxygen and nitrogen of the Schiff base ligand of both complexes are attached through their normal modes of coordination, i.e., for complex 1, Fe1–O1, Fe1–O2, Fe1–O5, Fe1–O6, Fe1–N1, and Fe1–N2 are found to be 1.882(16), 1.866(21), 2.316(13), 2.088(26), 2.094(26), and 2.075(27) Å, respectively, and for complex 2, Cu1–N1/N1′, Cu1–O1/O1′, and Cu1–O3 are found to be 1.942(45), 1.932(28), and 2.330(28) Å, respectively. The intra- and intermolecular H-bonds of complex 1 H5A–O9, H6A–O4, H6A–O9, O1–H6B, H5B–O4, O5–H6B, O5–H6A, O6–H5A, O2–H6A, H6A–N3, H6A–O7, N1–H5A, N2–H6A, N1–H6B, N2–H5A, N2–H5B, O2–H5B, O7–H6B, H6B–N3, H6B–O9, O8–H6B, N3–H6B, N3–H6A, and H6A–O8 are found to be 4.669(48), 4.986(29), 4.382(48), 3.006(17), 4.572(21), 4.554(15), 4.558(13), 4.537(23), 3.045(19), 3.398(29), 3.185(54), 2.793(28), 2.738(23), 2.667(21), 2.771(26), 3.028(30), 2.732(14), 2.851(57), 2.655(29), 3.692(45), 4.753(35), 3.686(26), 2.764(26), and 3.927(39) Å, respectively, as shown in Figure 2a, whereas complex 2 has no intra- and intermolecular H bonding. Figures 2b and 3a represent van der Waals interaction between complexes 1 and 2, and their corresponding van der Waals bond lengths are shown in Table 4, which stabilized the complexes. Complex 2 has an asymmetric unit, as shown in Figure 3b, whereas complex 1 has no asymmetric unit. The most interesting feature was obtained that both the complexes have built metal–organic frameworks (MOFs) fabricated through Schiff base ligands linked to the entire two-dimensional net and a 3D layer framework for 1 and 2, as shown in Figure 4a,b, respectively.
Figure 1.
(a) Complex 1 and (b) complex 2.
Table 1. Phase Data of Complexes 1 and 2a,b,c,d.
| parameter | complex 1 | complex 2 |
|---|---|---|
| formula | [C18H22FeN2O6]NO3 | C18H16CuN2O5 |
| formula weight (g/mol) | 480.2 g/mol | 403.8 g/mol |
| temperature (K) | 273 K | 99 K |
| wavelength | 0.71073 Å | 0.71073 Å |
| crystal system | monoclinic | orthorhombic |
| space group | P121/n1 (14) | Pnma (62) |
| unit cell dimensions a, b, c (Å) | a = 11.8135(2), b = 13.3278(1), c = 13.8168(2) | a = 9.212(2), b = 24.643(5), c = 7.363(2) |
| α, β, γ (°) | α = 90, β = 111.607(2), γ = 90 | α = 90, β = 90, γ = 90 |
| V (Å3) | 2022.56(6) | 1671.5(7) |
| Z | 4 | 1 |
| density (g/cm3) | 1.577 | 0.411 |
| radiation type | Mo Kα | Mo Kα |
| μ (mm–1) | 6.504 | 0.335 |
| absorption correction | multiscan absorption correction | multiscan absorption correction |
| Tmin, Tmax | 0.295, 1.00 | 0.571, 0.733 |
| Rint | 0.029 | 0.0878 |
| θ (max) (Å–3) | 71.360 | 24.996 |
| refinement R[F2> 2σ(F2)], wR(F2), S | 0.0502(3642), 0.1362(3879), 1.043 | 0.0568(1411), 0.1556(1496), 1.074 |
| goodness of fit on F2 | 1.041 | 1.052 |
| no of reflections | 3879 | 1496 |
| no of parameters | 284 | 122 |
| data completeness | 0.986 | 0.995 |
R1 = ∑(||Fo| – |Fc||)/∑|Fo|.
wR2 = [∑[w(Fo2 – Fc2)2]/∑w[(Fo2)2]]1/2.
S = [∑[w(Fo2 – Fc2)2]/(n – p)]1/2.
w = 1/[∑2(Fo2) + (m*p)2 + n*p], p = [max(Fo2,0) + 2*Fc2]/3, m and n are constants.
Table 2. Selected Bond Distances (Å) and Bond Angles (°) for Complexes 1 and 2a.
| complex 1 | complex 2 | ||
|---|---|---|---|
| atoms 1,2 | d 1,2 (Å) | atoms 1,2 | d 1,2 (Å) |
| Fe1–O1 | 1.882(16) | Cu1–N1/N1′ | 1.942(45) |
| Fe1–O2 | 1.866(21) | Cu1–O1/O1′ | 1.932(28) |
| Fe1–O5 | 2.316(13) | Cu1–O3 | 2.330(28) |
| Fe1–O6 | 2.088(26) | ||
| Fe1–N1 | 2.094(26) | ||
| Fe1–N2 | 2.075(27) | ||
| atoms 1,2,3 | angle 1,2,3 (°) | atoms 1,2,3 | angle 1,2,3 (°) |
|---|---|---|---|
| O1–Fe1–O5 | 89.57(8) | O1–Cu1–O1′ | 90.48(16) |
| O1–Fe1–O2 | 101.45(8) | O1′–Cu1–O3 | 98.47(11) |
| O1–Fe1–O6 | 93.26(9) | O1–Cu1–O3 | 98.42(11) |
| O1–Fe1–N1 | 88.99(8) | O1–Cu1–N1 | 92.30(15) |
| O1–Fe1–N2 | 168.06(9) | O1–Cu1–N1′ | 166.99(18) |
| O2–Fe1–O5 | 92.04(8) | O1′–Cu1–N1′ | 92.30(15) |
| O2–Fe1–O6 | 91.91(9) | O1′–Cu1–N1 | 167.01(18) |
| O2–Fe1–N1 | 169.56(9) | N1–Cu1–O3 | 93.67(17) |
| O2–Fe1–N2 | 89.63(9) | N1′–Cu1–O3 | 93.73(17) |
| O6–Fe1–O5 | 174.60(9) | N1′–Cu1–N1 | 82.3(3) |
| O6–Fe1–N1 | 87.28(10) | ||
| N1–Fe1–O5 | 88.17(8) | ||
| N2–Fe1–O5 | 85.57(9) | ||
| N2–Fe1–O6 | 90.77(10) | ||
| N2–Fe1–N1 | 79.98(9) |
Symmetry codes: primed atoms are related by symmetry −x + 1, y, −z.
Table 3. Bond Valence Sumsb for Complexes 1 and 2a.
| complex 1 |
complex 2 |
|||
|---|---|---|---|---|
| atom | Fe(II) | Fe(III) | Cu(II) | Cu(I) |
| Fe1 | 3.29 | 3.14 | ||
| Cu1 | 2.01 | 1.76 | ||
The oxidation state of a particular atom is the nearest integer to the underlined value.
The underlined value is the closest to the charge for which it was calculated.
Figure 2.
(a) Intra- and intermolecular H-bonding and (b) van der Waals interaction of complex 1.
Figure 3.
(a) van der Waals interaction and (b) asymmetric unit of complex 2.
Table 4. van der Waals Interaction of Complexes 1 and 2.
| complex 1 |
complex 2 |
||
|---|---|---|---|
| atoms 1,2 | d 1,2 (Å) | atoms 1,2 | d 1,2 (Å) |
| N3–H6B | 2.655(29) | O3–C007 | 3.981(62) |
| O8–H6B | 1.919(39) | O3–C00E | 3.657(117) |
| O8–H16A | 2.647(29) | H00E–O2 | 3.960(36) |
| O6–O8 | 2.729(48) | H007–O2 | 3.395(38) |
| O8–H14 | 2.957(43) | H007–O1 | 3.721(31) |
| H7–O9 | 2.929(44) | C007–O2 | 3.664(70) |
| O9–H5 | 2.5834(36) | C00E–O1 | 3.482(125) |
| O9–H6A | 2.266(48) | H00G–N1 | 3.477(46) |
| N3–6A | 2.764(26) | H00G–O1 | 3.160(31) |
| O7–H6A | 2.560(46) | N1–H00E | 3.858(50) |
| O7–H6B | 2.851(57) | N1–N1 | 2.558(70) |
| H4–O7 | 2.538(36) | N1–N00C | 3.645(70) |
| N1–C006 | 2.930(62) | ||
| O1–N1 | 2.794(48) | ||
| O1–O1 | 2.740(35) | ||
| O1–O2 | 2.589(39) | ||
| O2–H00D | 2.649(26) | ||
| O2–C00D | 3.692(67) | ||
| C009–O2 | 3.617(49) | ||
| O1–C00B | 3.614(56) | ||
| O1–C00C | 3.694(59) | ||
| O1–C009 | 2.423(54) | ||
| O1–C007 | 2.952(56) | ||
| O1–H007 | 3.895(30) | ||
Figure 4.
MOF at (010) b axis in central projection for (a) complex 1 and (b) complex 2.
Optical Properties
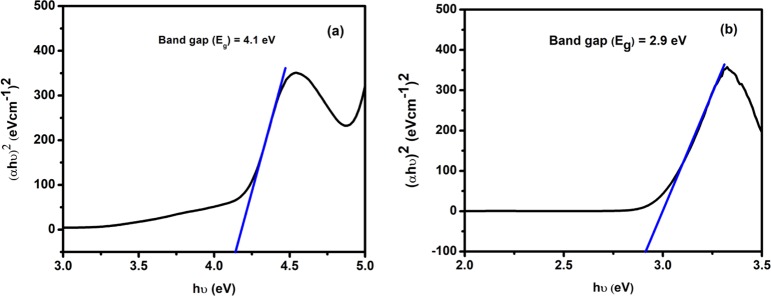
To explain the conductivity of the isolated complexes, the optical band gap energy of the complexes was calculated by the Tauc formula according to eq 1
| 1 |
where α denotes the absorption coefficient, which is calculated as α = (2.303 × absorption)/t, t = cubet thickness (1 cm); A is a constant; Eg indicates the band gap energy; exponent n depends on the type of transition; and h denotes Planck’s constant.
Equation 1 was used to plot (αhυ)2 vs hυ (Figure 5) from which band gap energy (Eg) of the complexes was calculated by extrapolating the linear portion of the curve to (αhυ)2 = 0. The band gap energy of complexes 1 and 2 were 4.1 and 2.9 eV, respectively. In the solar cell, optoelectronic, and electronic applications, this result was significant because the small band gap energy expanded the electronic progress between highest occupied molecular orbital and lowest unoccupied molecular orbital energy levels and increased the electrical conductivity of the complexes.53−56 The band gap value suggests that complexes are semiconductor materials and complex 2 has shown photocatalytic activity due to the lower band gap energy value (2.9 eV).
Figure 5.
Band gap energies of (a) complex 1 and (b) complex 2.
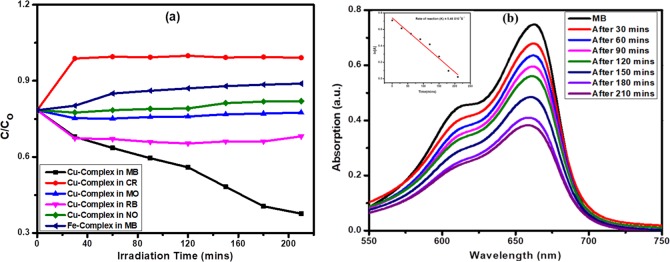
Photocatalytic Activity
We continued our experiment
to investigate the photocatalytic activity of the complexes against
different types of organic dyes because the band gap of the complexes
suggests that complexes are semiconductor materials. The photocatalytic
activity of the complexes was investigated against MB, Congo red (CR),
methyl orange (MO), rhodamine B (RB), and naphthalol orange (NO) in
the presence of sunlight (Figure 6a). Among the two complexes, complex 2 has photocatalytic activity against MB (Figure 6b) due to lower band gap energy. The degradation
rate of MB by complex 2 in 210 min was 50%. Photocatalytic
degradation of MB by complex 2 follows the pseudo-first-order
kinetics, and rate constant was determined by the following relation: 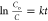 , where C0 and C denote the MB dye concentration
at times t = 0 and t = t, respectively. The inset in Figure 6b shows a good linear plot of
, where C0 and C denote the MB dye concentration
at times t = 0 and t = t, respectively. The inset in Figure 6b shows a good linear plot of 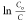 vs t, in which the rate constant
was 5.46
× 10–5 s–1.
vs t, in which the rate constant
was 5.46
× 10–5 s–1.
Figure 6.
(a) Kinetics of the dye degradation and (b) MB degradation by complex 2 and rate of reaction.
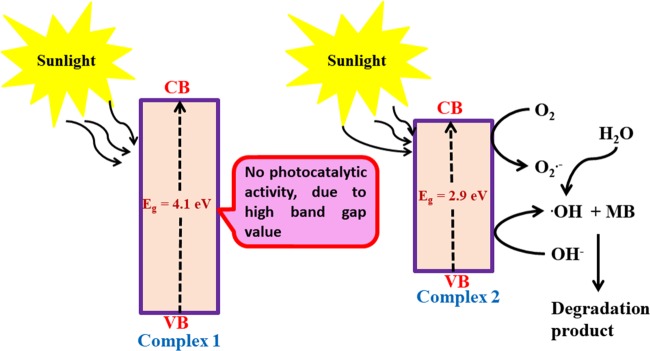
Complex 1 has no photocatalytic activity due to higher band gap energy value, as a result of which the electron cannot make an effective transition from valence band to conduction band in the presence of sunlight. The proposed mechanistic pathway for the degradation of MB in the presence of complex 2 is shown in Figure 7.
Figure 7.
Possible mechanism and reason for dye degradation by complex 2 in the presence of sunlight.
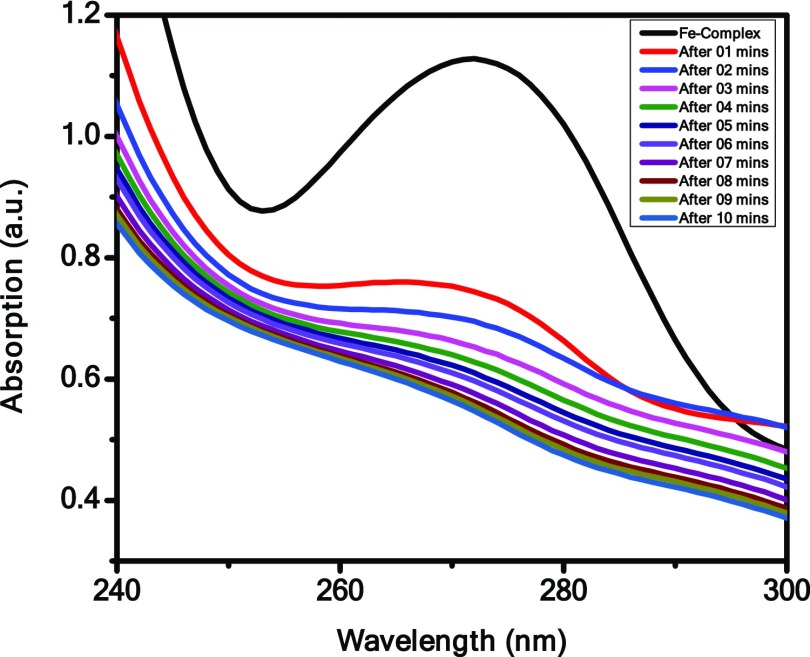
H2O2 Sensing by Complexes
To detect the sensing capacity of the complexes in the presence of H2O2 by adding 1 mL of 20 mM H2O2 to 3 mL solution of the complexes, the electronic spectrum was recorded at regular intervals. Figure 8 shows that absorbance of complex 1 decreases with increasing time in the presence of H2O2. After 10 min, we have observed that the characteristic peak of complex 1 at 274 nm disappears and the color intensity of complex 1 turns from brown to colorless. It indicates that complex 1 has H2O2 decomposing ability, whereas complex 2 has no sensing ability. This function of complex 1 can be used as the measure to rapidly detect H2O2 in unknown samples. The addition of H2O2 to the Fe(III) complex results in the formation of free radicals, which initiated the degradation of iron complex, and Fe(III) gets reduced to Fe(II) and decreases the absorbance of complex 1.
Figure 8.
H2O2 sensing by Complex 1.
The possible reaction mechanism of H2O2 decomposition by complex 1 is given below.
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
In the possible proposed reaction mechanism, eq 5 was an intermediate of the decomposition of H2O2 by complex 1.57
DNA Binding
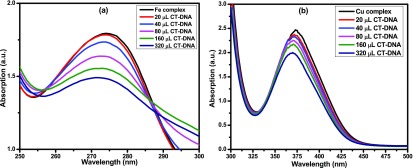
Before adding CT-DNA to complexes, the stabilities of complexes 1 and 2 in Tris–HCl buffer solution were measured by a UV–visible spectrophotometer at room temperature for 1 h (each 15 min interval measured by UV) and the absorption peak was not changed. Figure 9 indicates that the UV–vis absorption spectrum of complexes 1 and 2 have absorption peak at 274 and 374 nm, respectively, which can be assigned d–d transition. To demonstrate the binding characteristics of the complexes with CT-DNA, we used electronic absorption spectroscopy through changes in absorbance and shift in wavelength. The wavelengths of complexes 1 and 2 exhibit blue shift (for complex 1, the wavelength shifts from 274 to 274, 273, 272, 271, and 271 nm with increasing concentration of CT-DNA of 20, 40, 80, 160, and 320 μL, respectively, and similarly in complex 2, also wavelength shifts from 374 to 371, 371, 370, 370, and 369 nm with increasing concentration of CT-DNA of 20, 40, 80, 160, and 320 μL, respectively), when the concentration of CT-DNA was increased.
Figure 9.
Absorption spectra of complexes in the absence and presence of CT-DNA: (a) complex 1 and (b) complex 2.
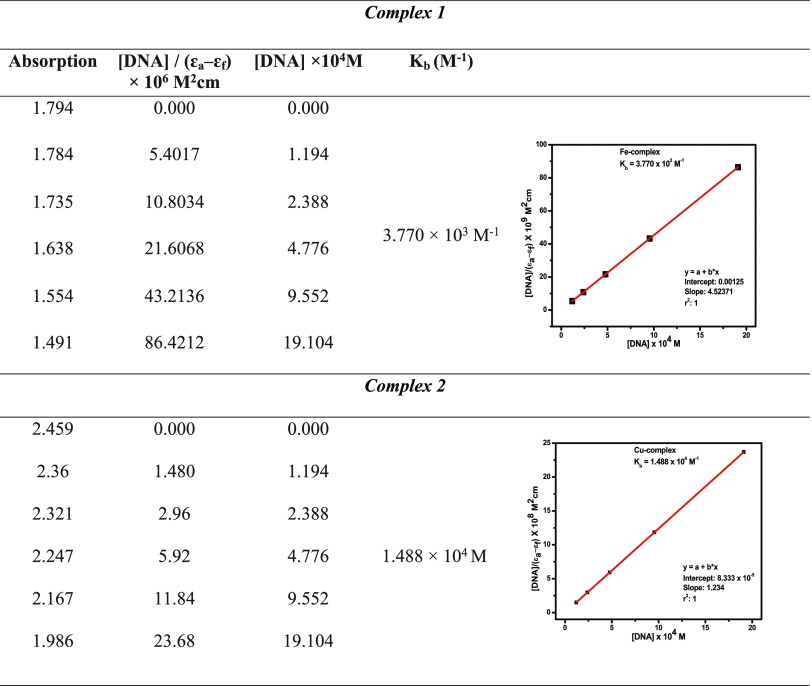
The intercalability of a metal complex depended on the conjugation, planarity, ligand donor atoms, and the formation of the geometry of the complex.25 The absorption spectra of the complex binding to DNA through intercalation reveal that blue and red shifts occur due to strong staking interaction between ligand of complexes and base pairs of DNA. The binding constant Kb of the complexes was determined by eq 8(58)
| 8 |
where εa, εf, and εb denote extinction coefficients of the complex, CT-DNA, and bound complex, respectively. The binding constants of complexes 1 and 2 were 3.770 × 103 and 1.488 × 104 M–1, respectively, as shown in Table 5. Therefore, binding constant values lie within the range of a characteristic of the CT-DNA binding by complexes through intercalative mode.59,60
Table 5. Absorption Titration Data of Complexes 1 and 2.
Conclusions
The Fe(III) and Cu(II) complexes of the designed Schiff base ligand N,N′-bis(O-vanillinidene)ethylenediamine (O-VEDH2) were successfully synthesized and characterized by single X-ray crystallography and Fourier-transform infrared (FT-IR) spectroscopy. Several applications of the metal complexes were investigated, such as MOFs, conductivity, photocatalytic activity, H2O2 sensing, and DNA binding activity. Complexes 1 and 2 have MOF structures and semiconductor properties, and their optical band gap energy values were 4.1 and 2.9 eV, respectively. Complex 1 has sensing ability to decompose H2O2, but it has no photocatalytic activity, whereas complex 2 has photocatalytic activity against MB degradation but no sensing ability to decompose H2O2. Both the complexes were bound to CT-DNA via intercalative mode of interaction, and binding constants were 3.770 × 103 and 1.488 × 104 M–1, respectively.
Experimental Section
Materials and Methods
All chemical reagents used were commercially available and used without further purification. The solvents were used as received.
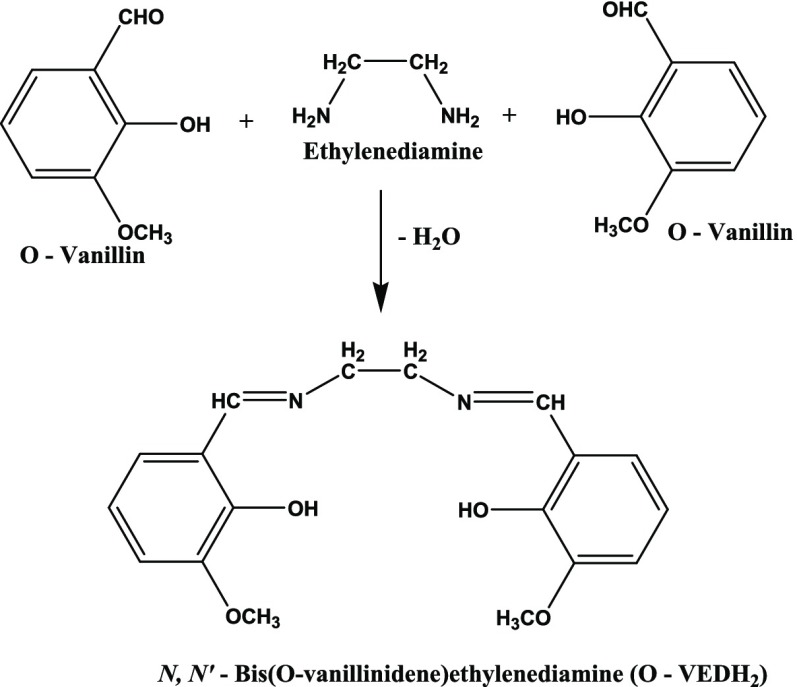
Synthesis of Schiff Base Ligand
A methanolic solution of O-Vanillin (0.608 g) was mixed with the methanolic solution of ethylenediamine (0.133 g) under constant stirring for 15 min at room temperature, resulting in the formation of a yellow precipitate. The precipitate was filtered, washed with methanol, and finally dried in vacuum. The yield of Schiff base ligand is 80% (0.486 g). 1H NMR (400 MHz, CDCl3) δ: 8.34 (s, 1H), 6.92 (d, J = 4 Hz, 1H), 6.87 (m, 1H), 6.82 (d, J = 8.3 Hz, 1H), 4.73 (s, J = 7.1 Hz, 1H), 3.97 (t, J = 12.1 Hz, 2H), and 3.50 (s, 3H), as shown in Figure S1 (Supporting Information). Solubility: acetonitrile, dimethyl sulfoxide, dimethylformamide (DMF). Scheme 1 presents the synthesis of Schiff base ligand N,N′-bis(O-vanillinidene)ethylenediamine (O-VEDH2).
Scheme 1. Preparation of Schiff Base Ligand N,N′-Bis(O-vanillinidene)ethylenediamine (O-VEDH2).
Synthesis of [C18H22FeN2O6]NO3 (1)
FeCl3 (0.487g, 3 mmol) was dissolved in 10 mL of acetonitrile, followed by dropwise addition of O-VEDH2 (0.305 g, 1 mmol) (separately soluble in MeCN solution) and stirred for another 2 h until a clear solution is obtained. Finally, the resulting solution was filtered and one part of the brown filtrate was kept for slow evaporation and another part is layered in diethyl ether. Within a week, high-quality black single crystals resulted under both conditions in high yield, which were washed with Et2O (two to three times) and dried under vacuum. Yield: 35% (0.17 g). Anal. calcd for complex 1 C18H22FeN3O9: C, 45.01; H, 4.62; N, 8.75%. Found: C, 45.11; H, 4.82; N, 8.56%. Selected IR data in cm–1: 633 (s), 739 (s), 1080 (m), 1218 (m), 1384 (b), 1442 (s), 1550 (s), 1618 (b) [s = strong, m = medium, b = broad] (for a detailed discussion of IR spectrum of 1, see Figure S2a).
Synthesis of C18H16CuN2O5 (2)
A similar method was followed for the synthesis of complex 2. Copper sulfate (0.250 g, 1 mmol) was dissolved in 10 mL of methanol followed by dropwise addition of acetonitrile solution (0.61 g, 2 mmol) of O-VEDH2 with the other conditions remaining constant. Within a week, high-quality green single crystals appeared, which were washed with Et2O (two to three times) and dried under vacuum. Yield: 30% (0.075 g). Anal. calcd for complex 2 C18H16CuN2O5: C, 53.54; H, 3.99; N, 6.94%. Found: C, 53.23; H, 3.85; N, 6.85%. Selected IR data in cm–1: 499 (b), 601 (s), 772 (s), 1080 (m), 1218 (s), 1394 (m), 1437 (s), 1544 (s), 1636 (b) [s = strong, m = medium, b = broad] (for a detailed discussion of IR spectrum of 2, see Figure S2b).
Physical Measurements
1H NMR spectroscopy was performed on a JEOL-400 MHz spectrometer. Infrared spectra were recorded in the solid-state (KBr pellets) using a PerkinElmer FT-IR spectrometer in the 400–4000 cm–1 range. Bruker SMART APEX performed elemental analysis (C, H, N) and X-ray crystallography. UV–visible spectrophotometer (Shimadzu UV-1800) was used to analyze the electronic spectrum of complexes.
Crystallographic Data Collection and Refinement
Single precious stone X-beam information from complexes 1 and 2 was collected at 293 and 99 K on the Bruker SMART APEX CCD diffractometer using SMART/SANTI programming.61 Intensity data were collected with Mo Kα graphite monochromatized radiation (0.71073 Å) at 100 K. The structures were understood by a coordinate strategy using the SHELXS-201362 program, which was incorporated in WinGX.63,64 Exact corrections to the absorption were linked to SADABS.65 All nonhydrogen particles were refined with an anisotropic displacement coefficient. The hydrogen carbon-bonded particles were incorporated in geometric positions, and the given warm parameters are one to two times those of the molecule to which they were attached. An initial search for reciprocal space exposed that complexes 1 and 2 crystallize in monoclinic and orthorhombic space groups with P121/n1 and Pnma, respectively. A and B alerts are observed in the checkCIF file of the complexes due to the presence of high disorder in the structure and solvent molecules.
The results of the precious crystal data and structure-refined information of complexes 1 and 2 are available in Table 1.
Dye Degradation
Rhodamine B (RB), Congo Red (CR), methyl orange (MO), naphthalol orange (NO), and methylene blue (MB) were separately dissolved in water. The complex (10 mg) was soaked in 10 mL of dye solution and kept under sunlight. Furthermore, we examined the complex’s specific photocatalytic activity, and readings with UV spectroscopy were taken regularly.
Hydrogen Peroxide-Sensing Capacity of the Complexes
The detection of H2O2 by both complexes is investigated by the protocol developed by Bera et al. (2013).66 Initially, complexes 1 and 2 were dissolved in CH3CN and DMF, respectively. A suitable dilute solution of the complexes (3 mL) was noted for the initial of UV–visible spectrum. In the complex solution, 1 mL of 20 mM H2O2 was added. The readings were taken at regular intervals with UV–visible spectroscopy.
DNA Binding Study
Calf-thymus (CT) DNA was dissolved in Tris–HCl buffer (pH = 7.25) solution, and its purity was checked from the absorbance ratio A260/A280, which is in the range 1.8–1.9. Metal complexes 1 and 2 were dissolved, and absorption titration experiments were performed using increased concentration of CT-DNA with the constant amount with the metal complex.
Acknowledgments
This work was fully supported by Madhya Pradesh Council of Science & Technology, Govt. of India, Madhya Pradesh (File No. A/R&D/RP-2/ Phy & Engg./2017-18/271); Indira Gandhi National Tribal University, Amarkantak, Madhya Pradesh; and University of Gour Banga, Malda, India. The authors acknowledge IIT Indore and IACS Kolkata, Madhya Pradesh, India, for measurement of single X-ray crystallography. They also acknowledge GGU (Central University), Chhattisgarh, India, for FT-IR measurement. The authors thank Dr Hari Singh Gour University (Central University), Madhya Pradesh, India, for 1H NMR measurement.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b02268.
1H NMR of Schiff base ligand (Figure S1); FT-IR of (a) complex 1 and (b) complex 2 (Figure S2); MOF at (100) “a” axis in central projection of complex 2 (Figure S3); selected geometric information of the complex 1 (Table S1); selected geometric information of the complex 2 (Table S2) (PDF)
Designed and synthesized of new N,N’-bis(O-vanillinidene)ethylenediamine (O-VEDH2) Schiff base ligand; synthesis of novel [C18H22FeN2O6]NO3 (1) and C18H16CuN2O5 (2) complex employing by O-VEDH2
Accession Codes
CCDC 1937318 and 1937320 contain the supplementary crystallographic data for complexes 1 and 2, respectively. These data are available free of charge at http://www.ccdc.cam.ac.uk/conts/retrieving.html or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, U.K.; Fax: (+44) 1223-336-033; E-mail:deposit@ccdc.cam.ac.uk. Supporting Information (SI) available: Figures S1–S3 and Tables S1 and S2.
Author Contributions
The Schiff base ligand and metal complexes were synthesized by M.K.G. and Dr S.P. The applications of the complexes were investigated by M.K.G. The manuscript was designed and written by M.K.G. and Dr T.K.G. All of the authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Vieira A. P.; Wegermann C. A.; Ferreira A. M. D. C. Comparative studies of Schiff base-copper (II) and zinc (II) complexes regarding their DNA binding ability and cytotoxicity against sarcoma cells. New J. Chem. 2018, 42, 13169–13179. 10.1039/C7NJ04799A. [DOI] [Google Scholar]
- a Zhang J.; Xu L.; Wong W. Y. Energy materials based on metal Schiff base complexes. Coord. Chem. Rev. 2018, 355, 180–198. 10.1016/j.ccr.2017.08.007. [DOI] [Google Scholar]; b Rana S.; Biswas J. P.; Sen A.; Clémancey M.; Blondin G.; Latour J. M.; Rajaraman G.; Maiti D. Selective C–H halogenation over hydroxylation by non-heme iron (iv)-oxo. Chem. Sci. 2018, 9, 7843–7858. 10.1039/C8SC02053A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek M. T.; Zabiszak M.; Nowak M.; Jastrzab R. Lanthanides: Schiff base complexes, applications in cancer diagnosis, therapy, and antibacterial activity. Coord. Chem. Rev. 2018, 370, 42–54. 10.1016/j.ccr.2018.05.012. [DOI] [Google Scholar]
- Silva P. P.; Guerra W.; Silveira J. N.; Ferreira A. M. D. C.; Bortolotto T.; Fischer F. L.; Terenzi H.; Neves A.; Pereira-Maia E. C. Two new ternary complexes of copper (II) with tetracycline or doxycycline and 1, 10-phenanthroline and their potential as antitumoral: cytotoxicity and DNA cleavage. Inorg. Chem. 2011, 50, 6414–6424. 10.1021/ic101791r. [DOI] [PubMed] [Google Scholar]
- Al Zoubi W.; Ko Y. G. Organometallic complexes of Schiff bases: Recent progress in oxidation catalysis. J. Organomet. Chem. 2016, 822, 173–188. 10.1016/j.jorganchem.2016.08.023. [DOI] [Google Scholar]
- Matsunaga S.; Shibasaki M. Multimetallic schiff base complexes as cooperative asymmetric catalysts. Synthesis 2013, 45, 421–437. 10.1055/s-0032-1316846. [DOI] [Google Scholar]
- Silva P. P.; Guerra W.; dos Santos G. C.; Fernandes N. G.; Silveira J. N.; da Costa Ferreira A. M.; Bortolotto T.; Terenzi H.; Bortoluzzi A. J.; Neves A.; Pereira-Maia E. C. Correlation between DNA interactions and cytotoxic activity of four new ternary compounds of copper (II) with N-donor heterocyclic ligands. J. Inorg. Biochem. 2014, 132, 67–76. 10.1016/j.jinorgbio.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Cozzi P. G. Metal–Salen Schiff base complexes in catalysis: practical aspects. Chem. Soc. Rev. 2004, 33, 410–421. 10.1039/B307853C. [DOI] [PubMed] [Google Scholar]
- Gupta K. C.; Sutar A. K. Catalytic activities of Schiff base transition metal complexes. Coord. Chem. Rev. 2008, 252, 1420–1450. 10.1016/j.ccr.2007.09.005. [DOI] [Google Scholar]
- Matsunaga S.; Shibasaki M. Recent advances in cooperative bimetallic asymmetric catalysis: dinuclear Schiff base complexes. Chem. Commun. 2014, 50, 1044–1057. 10.1039/C3CC47587E. [DOI] [PubMed] [Google Scholar]
- Yang J.; Shi R.; Zhou P.; Qiu Q.; Li H. Asymmetric Schiff bases derived from diaminomaleonitrile and their metal complexes. J. Mol. Struct. 2016, 1106, 242–258. 10.1016/j.molstruc.2015.10.092. [DOI] [Google Scholar]
- Kostova I.; Saso L. Advances in research of Schiff-base metal complexes as potent antioxidants. Curr. Med. Chem. 2013, 20, 4609–4632. 10.2174/09298673113209990149. [DOI] [PubMed] [Google Scholar]
- Erxleben A. Transition metal salen complexes in bioinorganic and medicinal chemistry. Inorg. Chim. Acta. 2018, 472, 40–57. 10.1016/j.ica.2017.06.060. [DOI] [Google Scholar]
- Lee S. J.; Lee S. W. Cd (II), Ni (II), and Co (II) complexes based on a pyridyl–amine Schiff-base ligand:[M (L) 2 (NO3)]·(NO3)(M= Cd, Ni, Co), cis-[CoL2Cl2]·(C6H6), and [Co (L) 3]·(ClO4) 2 (CH3CN) 2 (H2O)(L = N-(2-pyridylmethylene) benzene-1, 4-diamine,(2-py) CHNC6H4NH2). Polyhedron 2019, 159, 259–264. 10.1016/j.poly.2018.12.003. [DOI] [Google Scholar]
- Liu Y. H.; Li A.; Shao J.; Xie C. Z.; Song X. Q.; Bao W. G.; Xu J. Y. Four Cu (II) complexes based on antitumor chelators: Synthesis, structure, DNA binding/damage, HSA interaction and enhanced cytotoxicity. Dalton Trans. 2016, 45, 8036–8049. 10.1039/C6DT00451B. [DOI] [PubMed] [Google Scholar]
- Mollick S.; Mandal T. N.; Jana A.; Fajal S.; Desai A. V.; Ghosh S. K. Ultrastable Luminescent Hybrid Bromide Perovskite@MOF Nanocomposites for the Degradation of Organic Pollutants in Water. ACS Appl. Nano Mater. 2019, 2214 10.1021/acsanm.8b02214. [DOI] [Google Scholar]
- Cheng R.; Ou S.; Xiang B.; Li Y.; Liao Q. Equilibrium and molecular mechanism of anionic dyes adsorption onto copper (II) complex of dithiocarbamate-modified starch. Langmuir 2009, 26, 752–758. 10.1021/la9039489. [DOI] [PubMed] [Google Scholar]
- Xiao J. D.; Jiang H. L. Metal–Organic Frameworks for Photocatalysis and Photothermal Catalysis. Acc. Chem. Res. 2018, 521 10.1021/acs.accounts.8b00521. [DOI] [PubMed] [Google Scholar]
- Naskar K.; Dey A.; Maity S.; Bhunia M. K.; Ray P. P.; Sinha C. Novel Porous Polycatenated Iodo-Cadmium Coordination Polymer for Iodine Sorption and Electrical Conductivity Measurement. Cryst. Growth Des. 2019, 1806 10.1021/acs.cgd.8b01806. [DOI] [Google Scholar]
- Du M.; Li L.; Li M.; Si R. Adsorption mechanism on metal organic frameworks of Cu-BTC, Fe-BTC and ZIF-8 for CO2 capture investigated by X-ray absorption fine structure. RSC Adv. 2016, 6, 62705–62716. 10.1039/C6RA07582G. [DOI] [Google Scholar]
- Yang J.; Zhao F.; Zeng B. One-step synthesis of a copper-based metal–organic framework–graphene nanocomposite with enhanced electrocatalytic activity. RSC Adv. 2015, 5, 22060–22065. 10.1039/C4RA16950F. [DOI] [Google Scholar]
- Rajalakshmi S.; Kiran M. S.; Nair B. U. DNA condensation by copper (II) complexes and their anti-proliferative effect on cancerous and normal fibroblast cells. Eur. J. Med. Chem. 2014, 80, 393–406. 10.1016/j.ejmech.2014.04.064. [DOI] [PubMed] [Google Scholar]
- Torres J. F.; Macias M. A.; Franco-Ulloa S.; Miscione G. P.; Cobo J.; Hurtado J. Cu (II) and Zn (II) Complexes with Dinitrobenzoates and Pyrazolyl Ligands: Structural and Thermal Stability Influence of NH Moiety. Cryst. Growth Des. 2019, 246 10.1021/acs.cgd.9b00246. [DOI] [Google Scholar]
- Wu M. Z.; Shi J. Y.; Chen P. Y.; Tian L.; Chen J. Two 3D Cobalt (II) Metal–Organic Frameworks with Micropores for Selective Dye Adsorption. Inorg. Chem. 2019, 58, 3130–3136. 10.1021/acs.inorgchem.8b03194. [DOI] [PubMed] [Google Scholar]
- Tanaka S.; Sato H.; Ishida Y.; Deng Y.; Haraguchi T.; Akitsu T.; Sugiyama M.; Hara M.; Moon D. Photo-Control of Adsorption of Dye Metal Complexes Incorporating Chiral Schiff Base Ligands Containing Azo-Groups on TiO2. J. Korean Chem. Soc. 2018, 62, 328–332. [Google Scholar]
- Wen T.; Zhang D. X.; Zhang J. Two-dimensional copper (I) coordination polymer materials as photocatalysts for the degradation of organic dyes. Inorg. Chem. 2013, 52, 12–14. 10.1021/ic302273h. [DOI] [PubMed] [Google Scholar]
- Beheshti A.; Fard E. S. M.; Kubicki M.; Mayer P.; Abrahams C. T.; Razatofighi S. E. Design, synthesis and characterization of copper-based coordination compounds with bidentate (N, N and N, O) ligands: reversible uptake of iodine, dye adsorption and assessment of their antibacterial properties. CrystEngComm 2019, 21, 251–262. 10.1039/C8CE01348A. [DOI] [Google Scholar]
- Yue C.-Y.; Lei X.-W.; Han Y.-F.; Lu X.-X.; Tian Y.-W.; Xu J.; Liu X.-F.; Xu X. Transition-Metal-Complex Cationic Dyes Photosensitive to Two Types of 2D Layered Silver Bromides with Visible-Light-Driven Photocatalytic Properties. Inorg. Chem. 2016, 55, 12193–12203. 10.1021/acs.inorgchem.6b01770. [DOI] [PubMed] [Google Scholar]
- Pathak S.; Ghosh M. K.; Ghorai T. K. Luminescence, Dye Degradation and DNA Binding Properties of a Dinuclear Nona-Coordinated Y (III) Complex. ChemistrySelect 2018, 3, 13501–13506. 10.1002/slct.201801826. [DOI] [Google Scholar]
- Umamaheswari C.; Lakshmanan A.; Nagarajan N. S. Green synthesis, characterization and catalytic degradation studies of gold nanoparticles against congo red and methyl orange. J. Photochem. Photobiol., B 2018, 178, 33–39. 10.1016/j.jphotobiol.2017.10.017. [DOI] [PubMed] [Google Scholar]
- Yang L.; Li X.; Sun C. Y.; Wu H.; Wang C. G.; Su Z. M. A stable pillared-layer Cu (ii) metal–organic framework with magnetic properties for dye adsorption and separation. New J. Chem. 2017, 41, 3661–3666. 10.1039/C7NJ00389G. [DOI] [Google Scholar]
- Winterbourn C. C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D.; Dasgupta P.; Roy D. S.; Palchoudhuri S.; Chatterjee I.; Ali S.; Dastidar S. G. A Sensitive In vitro Spectrophotometric Hydrogen Peroxide Scavenging Assay using 1, 10-Phenanthroline. Free Rad. Antiox. 2016, 6, 124–132. 10.5530/fra.2016.1.15. [DOI] [Google Scholar]
- Dröge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- Liu R.; Wei Y.; Zheng J.; Zhang H.; Sheng Q. A hydrogen peroxide sensor based on silver nanoparticles biosynthesized by Bacillus subtilis. Chin. J. Chem. 2013, 31, 1519–1525. 10.1002/cjoc.201300487. [DOI] [Google Scholar]
- Tredwin C. J.; Naik S.; Lewis N. J.; Scully C. B. E. C. Hydrogen peroxide tooth-whitening (bleaching) products: review of adverse effects and safety issues. Br. Dent. J. 2006, 200, 371–376. 10.1038/sj.bdj.4813423. [DOI] [PubMed] [Google Scholar]
- Zeng L.; Miller E. W.; Pralle A.; Isacoff E. Y.; Chang C. J. A selective turn-on fluorescent sensor for imaging copper in living cells. J. Am. Chem. Soc. 2006, 128, 10–11. 10.1021/ja055064u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig W. P.; Mukherjee S.; Rudd N. D.; Desai A. V.; Li J.; Ghosh S. K. Metal–organic frameworks: functional luminescent and photonic materials for sensing applications. Chem. Soc. Rev. 2017, 46, 3242–3285. 10.1039/C6CS00930A. [DOI] [PubMed] [Google Scholar]
- Yu Y.; Ma J. P.; Zhao C. W.; Yang J.; Zhang X. M.; Liu Q. K.; Dong Y. B. Copper (I) metal–organic framework: visual sensor for detecting small polar aliphatic volatile organic compounds. Inorg. Chem. 2015, 54, 11590–11592. 10.1021/acs.inorgchem.5b02150. [DOI] [PubMed] [Google Scholar]
- Wang D.; Wang M.; Li Z. Fe-based metal–organic frameworks for highly selective photocatalytic benzene hydroxylation to phenol. ACS Catal. 2015, 5, 6852–6857. 10.1021/acscatal.5b01949. [DOI] [Google Scholar]
- Kreno L. E.; Leong K.; Farha O. K.; Allendorf M.; Van Duyne R. P.; Hupp J. T. Metal–organic framework materials as chemical sensors. Chem. Rev. 2011, 112, 1105–1125. 10.1021/cr200324t. [DOI] [PubMed] [Google Scholar]
- Hemmert C.; Pitié M.; Renz M.; Gornitzka H.; Soulet S.; Meunier B. Preparation, characterization and crystal structures of manganese (II), iron (III) and copper (II) complexes of the bis [di-1, 1-(2-pyridyl) ethyl] amine (BDPEA) ligand; evaluation of their DNA cleavage activities. J. Biol. Inorg. Chem. 2001, 6, 14–22. 10.1007/s007750000177. [DOI] [PubMed] [Google Scholar]
- Wang J.; Liu X.; Sun Y.; Yan L.; He S. Synthesis, crystal structures, thermal properties, and DNA-binding studies of transition metal complexes with imidazole ligands. J. Coord. Chem. 2011, 64, 1554–1565. 10.1080/00958972.2011.575937. [DOI] [Google Scholar]
- Zuber G.; Quada J. C.; Hecht S. M. Sequence selective cleavage of a DNA octanucleotide by chlorinated bithiazoles and bleomycins. J. Am. Chem. Soc. 1998, 120, 9368–9369. 10.1021/ja981937r. [DOI] [Google Scholar]
- Pages B. J.; Ang D. L.; Wright E. P.; Aldrich-Wright J. R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. 10.1039/C4DT02700K. [DOI] [PubMed] [Google Scholar]
- Pathak S.; Ghosh M. K.; Mandal M.; Mandal V.; Bhattacharyya A.; Ghorai T. K. Synthesis of a new acetate bridged Cu (ii) building block generated 1D polymer and studies on structural, magnetic, antibacterial and anticancer properties. New J. Chem. 2019, 43, 2019–2029. 10.1039/C8NJ04937H. [DOI] [Google Scholar]
- Santini C.; Pellei M.; Gandin V.; Porchia M.; Tisato F.; Marzano C. Advances in copper complexes as anticancer agents. Chem. Rev. 2013, 114, 815–862. 10.1021/cr400135x. [DOI] [PubMed] [Google Scholar]
- Viqueira J.; Durán M. L.; García-Vázquez J. A.; Castro J.; Platas-Iglesias C.; Esteban-Gómez D.; Alzuet-Piña G.; Moldes A.; Nascimento O. R. Modulating the DNA cleavage ability of copper (ii) Schiff bases through ternary complex formation. New J. Chem. 2018, 42, 15170–15183. 10.1039/C8NJ03292K. [DOI] [Google Scholar]
- García-Ramos J. C.; Vértiz-Serrano G.; Macías-Rosales L.; Galindo-Murillo R.; Toledano-Magaña Y.; Bernal J. P.; Cortés-Guzmán F.; Ruiz-Azuara L. Isomeric Effect on the Pharmacokinetic Behavior of Anticancer CuII Mixed Chelate Complexes: Experimental and Theoretical Approach. Eur. J. Inorg. Chem. 2017, 2017, 1728–1736. 10.1002/ejic.201601199. [DOI] [Google Scholar]
- Chen W.; Wu C. Synthesis, functionalization, and applications of metal–organic frameworks in biomedicine. Dalton Trans. 2018, 47, 2114–2133. 10.1039/C7DT04116K. [DOI] [PubMed] [Google Scholar]
- Rojas S.; Devic T.; Horcajada P. Metal organic frameworks based on bioactive components. J. Mater. Chem. B 2017, 5, 2560–2573. 10.1039/C6TB03217F. [DOI] [PubMed] [Google Scholar]
- Brown I. D.; Altermatt D. Bond-valence parameters obtained from a systematic analysis of the inorganic crystal structure database. Acta Crystallogr., Sect. B: Struct. Sci. 1985, 41, 244–247. 10.1107/S0108768185002063. [DOI] [Google Scholar]
- Ahmed A. H.; Hassan A. M.; Gumaa H. A.; Mohamed B. H.; Eraky A. M.; Omran A. A. Copper (II)-oxaloyldihydrazone complexes: Physico-chemical studies: Energy band gap and inhibition evaluation of free oxaloyldihydrazones toward the corrosion of copper metal in acidic medium. Arab. J. Chem. 2016, 15 10.1016/j.arabjc.2016.05.015. [DOI] [Google Scholar]
- Turan N.; Gündüz B.; Körkoca H.; Adigüzel R.; Çolak N.; Buldurun K. Study of structure and spectral characteristics of the Zinc (II) and Copper (II) complexes with 5, 5-Dimethyl-2-(2-(3-nitrophenyl) hydrazono) cyclohexane-1, 3-dione and their effects on optical properties and the developing of the energy band gap and investigation of antibacterial activity. J. Mex. Chem. Soc. 2014, 58, 65–75. [Google Scholar]
- Sengupta S. K.; Pandey O. P.; Srivastava B. K.; Sharma V. K. Synthesis, structural and biochemical aspects of titanocene and zirconocene chelates of acetylferrocenyl thiosemicarbazones. Transition Met. Chem. 1998, 23, 349–353. 10.1023/A:1006986131435. [DOI] [Google Scholar]
- Fu M. L.; Guo G. C.; Liu X.; Liu B.; Cai L. Z.; Huang J. S. Syntheses, structures and properties of three selenoarsenates templated by transition metal complexes. Inorg. Chem. Commun. 2005, 8, 18–21. 10.1016/j.inoche.2004.10.021. [DOI] [Google Scholar]
- Chi G. T.; Nagy Z. K.; Huddersman K. D. Kinetic modelling of the Fenton-like oxidation of maleic acid using a heterogeneous modified polyacrylonitrile (PAN) catalyst. Prog. React. Kinet. Mech. 2011, 36, 189–214. 10.3184/146867811X13021847366179. [DOI] [Google Scholar]
- Wolfe A.; Shimer G. H. Jr; Meehan T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. 10.1021/bi00394a013. [DOI] [PubMed] [Google Scholar]
- Martins D. A.; Gouvea L. R.; Batista D. D. G. J.; Da Silva P. B.; Louro S. R.; Maria de Nazaré C. S.; Teixeira L. R. Copper (II)–fluoroquinolone complexes with anti-Trypanosoma cruzi activity and DNA binding ability. BioMetals 2012, 25, 951–960. 10.1007/s10534-012-9565-3. [DOI] [PubMed] [Google Scholar]
- Patel M. N.; Gandhi D. S.; Parmar P. A. Synthesis, biological aspects and SOD mimic activity of square pyramidal copper (II) complexes with the 3rd generation quinolone drug sparfloxacin and phenanthroline derivatives. Inorg. Chem. Commun. 2011, 14, 128–132. 10.1016/j.inoche.2010.10.003. [DOI] [Google Scholar]
- Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008, 64, 112. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M.SHELXL2013; University of Göttingen: Germany, 2013.
- Farrugia L. J. WinGX and ORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854. 10.1107/S0021889812029111. [DOI] [Google Scholar]
- Farrugia L. J.WinGX, version 3; Department of Chemistry, University of Glasgow: Glasgow, Scotland, 2013.
- Sheldrick G. M.SADABS; University of Göttingen: Göttingen, Germany, 1999.
- Bera R. K.; Raj C. R. A facile photochemical route for the synthesis of triangular Ag nanoplates and colorimetric sensing of H2O2. J. Photochem. Photobiol., A 2013, 270, 1–6. 10.1016/j.jphotochem.2013.07.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.